Based on diel and circadian leaf transcriptomes in barley wild-type and clock mutant lines we predicted a structure of the barley circadian oscillator and interactions of its individual components with day/night cues.
Abstract
The circadian clock is a complex transcriptional network that regulates gene expression in anticipation of the day/night cycle and controls agronomic traits in plants. However, in crops, how the internal clock and day/night cues affect the transcriptome remains poorly understood. We analyzed the diel and circadian leaf transcriptomes in the barley (Hordeum vulgare) cultivar ‘Bowman’ and derived introgression lines harboring mutations in EARLY FLOWERING3 (ELF3), LUX ARRHYTHMO1 (LUX1), and EARLY MATURITY7 (EAM7). The elf3 and lux1 mutants exhibited abolished circadian transcriptome oscillations under constant conditions, whereas eam7 maintained oscillations of ≈30% of the circadian transcriptome. However, day/night cues fully restored transcript oscillations in all three mutants and thus compensated for a disrupted oscillator in the arrhythmic barley clock mutants elf3 and lux1. Nevertheless, elf3, but not lux1, affected the phase of the diel oscillating transcriptome and thus the integration of external cues into the clock. Using dynamical modeling, we predicted a structure of the barley circadian oscillator and interactions of its individual components with day/night cues. Our findings provide a valuable resource for exploring the function and output targets of the circadian clock and for further investigations into the diel and circadian control of the barley transcriptome.
The circadian clock is a time-keeping mechanism that reflects the day/night cycle through an endogenous transcriptional rhythm to anticipate dawn and dusk (McClung, 2006). This clock synchronizes internal rhythms with external light and temperature cycles (Harmer, 2009; Greenham and McClung, 2015). The prevalence of circadian rhythms in all domains of life suggests that circadian clocks provide an adaptive advantage for organisms (Edgar et al., 2012). The Arabidopsis (Arabidopsis thaliana) oscillator contains an interconnected regulatory network of transcriptional repressors and activators (Hsu et al., 2013; Fogelmark and Troein, 2014). These components are expressed sequentially to regulate output genes through regulatory elements present in target promoters (Harmer et al., 2000; Covington et al., 2008; Michael et al., 2008b).
In the Arabidopsis circadian oscillator, two morning-expressed MYB transcription factors, CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), inhibit the expression of TIMING OF CAB EXPRESSION1/PSEUDO-RESPONSE REGULATOR1 (TOC1/PRR1) that in turn represses the transcription of CCA1 and LHY in the night (Alabadí et al., 2001; Gendron et al., 2012; Huang et al., 2012). During the day, sequentially expressed PRR9, PRR7, and PRR5 repress the transcription of CCA1 and LHY (Nakamichi et al., 2010, 2012; Liu et al., 2016). CCA1 and LHY in turn promote the expression of PRR9 and PRR7 (Farré et al., 2005). However, this effect might be indirect because transient induction analysis demonstrated that CCA1 can directly repress the expression of both genes PRR9 and PRR7 (Kamioka et al., 2016). Similarly, REVEILLE8 (RVE8), a close homolog of CCA1/LHY directly activates transcription of PRR5 and TOC1, and likely other evening-phased genes (Farinas and Mas, 2011; Rawat et al., 2011; Nakamichi et al., 2012; Hsu et al., 2013). The downregulation of CCA1 and LHY by PRR proteins allows the induction of EARLY FLOWERING3 (ELF3), ELF4, and LUX ARRHYTHMO (LUX), which encode the Evening Complex (EC) of proteins (Hazen et al., 2005, Kikis et al., 2005). The EC acts at dusk as a transcriptional repressor of PRR9 expression (Helfer et al., 2011; Nusinow et al., 2011; Herrero et al., 2012). Furthermore, the evening-expressed GIGANTEA (GI) protein was modeled as a negative regulator of the EC, which in turn inhibits TOC1 expression (Huang et al., 2012; Pokhilko et al., 2012).
The complex network of transcriptional regulators at the core of the clock underscores the role of transcriptional regulation as a central regulatory mechanism for circadian oscillation. Consequently, the Arabidopsis clock is a master regulator of transcription and controls approximately 30% to 40% of the global gene expression in a time-of-day–specific cycling pattern where transcription of functionally related genes often peaks in clusters (Harmer et al., 2000; Covington et al., 2008; Michael et al., 2008a; Staiger et al., 2013). Expression of such functional clusters often precedes or coincides with the underlying physiological event (Covington et al., 2008; Michael et al., 2008a), suggesting that circadian control anticipates diel regulation to improve physiological performance (Greenham and McClung, 2015).
In Arabidopsis, the circadian system controls many agronomically important processes, such as metabolism, growth, photosynthesis, and flowering time (Greenham and McClung, 2015). Consequently, it has been suggested that the circadian clock is key to improving adaptation and performance of crop plants (Hsu and Harmer, 2014; Bendix et al., 2015). Putative circadian oscillator genes have been identified in the monocot crop barley (Hordeum vulgare) based on their homology with the Arabidopsis clock genes (Campoli et al., 2012; Calixto et al., 2015). Although the circadian oscillator genes diversified via duplication independently between the monocot and eudicot clades, their structure and expression patterns remained highly similar (Campoli et al., 2012; Hsu and Harmer, 2014; Bendix et al., 2015). For example, in monocots, the morning-expressed MYB-like transcription factor LHY is the only ortholog of the Arabidopsis paralogs CCA1 and LHY (Takata et al., 2009; Campoli et al., 2012). HvLHY overexpression in Arabidopsis causes arrhythmia, suggesting circadian functionality (Kusakina et al., 2015). The PRRs duplicated independently from three ancient PRR genes after the divergence of monocots and eudicots such that the orthologous relationship within the PRR3/7 and PRR5/9 clades of Arabidopsis and monocot plants cannot be immediately resolved (Takata et al., 2010). Partial complementation of Arabidopsis prr7-11 by HvPRR37 suggests that the barley gene might retain some functionality of the Arabidopsis ortholog (Kusakina et al., 2015). However, PRR37 orthologs in monocots—PPD1 in barley and wheat (Triticum aestivum; Turner et al., 2005; Beales et al., 2007), and SbPRR37 in sorghum (Sorghum bicolor; Murphy et al., 2011)—are major determinants of photoperiod sensitivity and flowering time. In Arabidopsis, PRR proteins stabilize the CONSTANS protein, a floral promoter, and repress CYCLING DOF FACTOR genes, thereby promoting flowering under long days (Nakamichi et al., 2007; Hayama et al., 2017). However, natural variation in PRR genes in Arabidopsis do not have any notable effect on flowering time (Ehrenreich et al., 2009). Furthermore, the genes underlying the two early maturity mutants, early maturity8 (eam8) and eam10, have been identified as barley homologs of the Arabidopsis clock genes ELF3 and LUX1, respectively (Faure et al., 2012; Zakhrabekova et al., 2012; Campoli et al., 2013). Mutations in both genes cause photoperiod insensitivity and early flowering under long- and short-day conditions in barley (Faure et al., 2012; Zakhrabekova et al., 2012; Campoli et al., 2013). In addition, several ELF4-like homologs that exist in barley, including HvELF4-like 4, can complement an Arabidopsis Atelf4 null mutant (Hicks et al.. 2001; Kolmos et al., 2009). Whereas a number of putative clock components in barley have been identified, there is little information on the contribution of the clock versus day/night cues on the global transcriptome in barley.
We generated diel and circadian RNA-sequencing (RNA-seq) datasets of four barley genotypes—the spring barley ‘Bowman’ and three derived introgression lines with mutations in HvELF3 (BW290), HvLUX1 (BW284), and EAM7 (BW287; Faure et al., 2012; Campoli et al., 2013). The candidate gene for EAM7 has not yet been identified, but loss of EAM7 function accelerates flowering by abolishing sensitivity to the photoperiod (Gallagher et al., 1991). We used the RNA-seq time-course data to analyze the effects of barley clock genes on diel and circadian transcriptome oscillations, including changes in phase and period under constant conditions and light and dark cycles. Dynamical modeling allowed us to predict a molecular structure of the barley circadian oscillator and to uncover how circadian oscillator components interact with day/night cues to regulate the global transcriptome in barley.
RESULTS
Diel and Circadian Oscillations of the Barley Transcriptome
We analyzed the diel and circadian global leaf transcriptome of the barley 'Bowman' and the derived introgression lines carrying mutations in HvELF3 (BW290), HvLUX1 (BW284), and HvEAM7 (BW287). Plants were grown under cycles of 12-h light and 12-h night (LD) and the second leaf of replicate plants was harvested every 4 h over 24 h. Additional samples were taken in at 2-h intervals at dusk in all genotypes and additionally at dawn in cv Bowman (Supplemental Fig. S1). Thereafter, plants were transferred to constant light and temperature conditions (LL) and leaf samples were taken every 4 h for 36 h starting from the first subjective night. The sampling strategy was optimized for modeling the circadian oscillator using systems identification, an approach we have used previously to model the circadian Arabidopsis (Dalchau et al., 2010; Herrero et al., 2012; Mombaerts et al., 2019). Systems identification models the network based on the dynamical information in time series data sets. The dynamical information is greatest when the system is perturbed by environmental or genetic stimulation. We therefore optimized our RNA-seq strategy to capture the data with a time series that included light-dark transitions to perturb the oscillator and sampled several genotypes with alterations in putative oscillator genes. Whereas it is typical to observe circadian dynamics in prolonged constant conditions to distinguish free-running from entrained behaviors, circadian dynamics occur in both LD and LL and the LD conditions have greater perturbation to inform the modeling. We sampled for 36 h in LL because barley circadian transcript oscillations rapidly dampen upon transfer to constant conditions, which results in a high signal-to-noise ratio of the rhythms that would have compromised the systems identification (Campoli et al., 2012; Hughes et al., 2017). Our strategy of sampling before the onset of dampening of the signal reduces the false-negative discovery rate by increasing the signal-to-noise. This is at the expense of possibly increasing the false-negative rate by having a shorter time series in constant light (Hughes et al., 2017). Consequently, by taking a conservative approach to network identification, we can be more confident of the connections we have identified.
Individual libraries were single-end–sequenced on a HiSeq 2500 System (Illumina) with 10 million reads per library and reads were mapped against a custom reference sequence consisting of 68,739 transcripts (Digel et al., 2015). The nomenclature of the gene models used in this study (Digel et al., 2015) was cross referenced with the identifiers of the HORVU gene models annotated on the barley pseudochromosomes (Mascher et al., 2017). Raw read counts normalized to counts per million (CPM) were used for the downstream rhythmic analysis and modeling. We determined the oscillating patterns of gene expression, including period as the duration of one complete oscillation, and phase as the time point of transcript peak expression (Yang and Su, 2010; Wu et al., 2016). To increase the analytical power for the rhythmic analysis, the 24-h diel dataset, but not the 36-h LL data, was duplicated to imitate 48 h of sampling data.
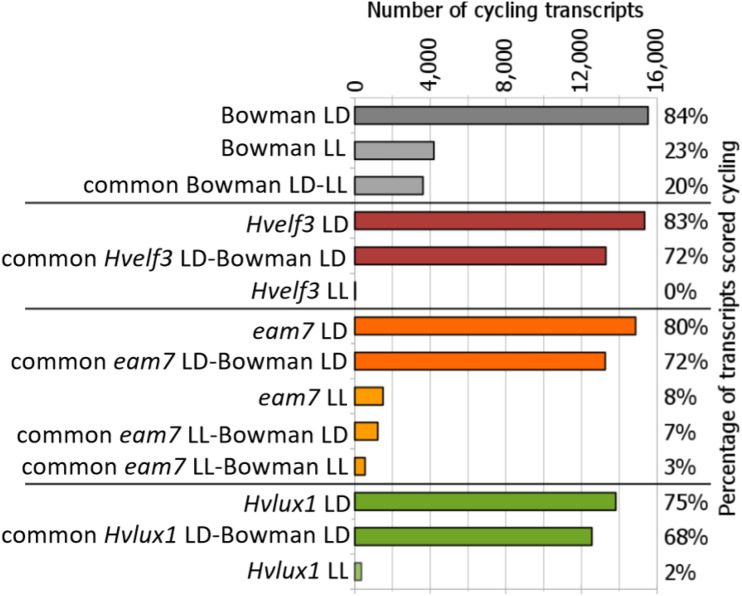
We identified 18,500 transcripts with expression levels >5 CPM in at least two libraries. Among 18,500 transcripts expressed across all the investigated lines, 84% were scored as rhythmic under LD in cv Bowman (Fig. 1; Supplemental Dataset S1). The duplicated LD data sets may have resulted in elevated false-positive rates of diel-regulated genes estimated per genotype. However, we found that out of all expressed genes ∼70% were scored as rhythmic in cv Bowman and at least one other mutant genotype, which represent independent biological data (Fig. 1).
Figure 1.
Fraction of transcripts with oscillating transcription pattern in cv Bowman and cv Bowman-derived introgression lines with mutations in Hvelf3, eam7, and Hvlux1 under LD and LL. Fractions refer to a total of 18,500 transcripts expressed in all genotypes.
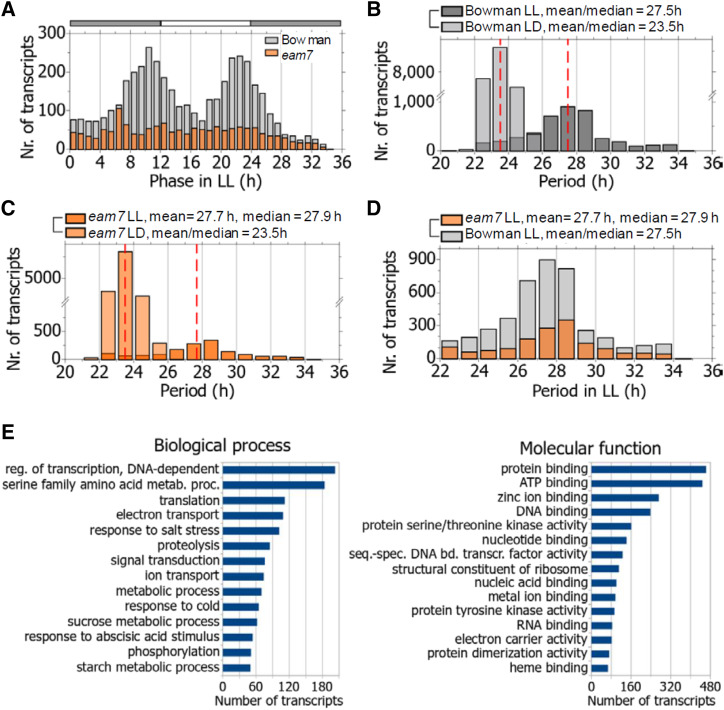
Under LL, ∼23% of the 18,500 transcripts were rhythmic, which is a feature of clock-regulated genes (Fig. 1; Supplemental Dataset S1). The gene ontology (GO) analyses revealed that, in cv Bowman under LL, the circadian-controlled transcripts were primarily related to the processes of regulation of DNA-dependent transcription, translation, electron transport, signal transduction, responses to salt stress and cold, and metabolic processes, including amino acid, Suc and starch metabolism (Fig. 2E; Supplemental Dataset S1). The molecular functions of the circadian-controlled transcripts in cv Bowman in LL were primarily represented by protein-, zinc ion-, and ATP binding, DNA- and nucleotide binding, and sequence-specific DNA-binding transcription factor activity GO terms (Fig. 2E).
Figure 2.
Distribution of the period and the phase of the oscillating transcriptome in constant light and their involvement in biological processes and molecular functions. A, Phase distribution in LL of cv Bowman and eam7. Gray-orange bars indicate the subjective night (gray) and subjective day (orange) in LL. B and C, Period distribution of the oscillating transcriptome under LL in comparison with LD in (B) cv Bowman and (C) eam7. D, Comparison of the period distribution in LL between cv Bowman and eam7. E, Top-15 categories of the GO terms for biological processes and molecular function of the transcripts oscillating in cv Bowman under LL.
We found that the majority of the transcripts expressed rhythmically under LL were also rhythmic under LD (20% of all the transcripts, 87% of LL transcripts). This demonstrated that approximately one-quarter of the cv Bowman transcriptome is modulated by the circadian clock, whereas the largest proportion of the rhythmic transcripts in LD required day/night cues for their rhythmic expression.
The large impact of external transitions on transcriptome oscillations independent of the clock was further supported by analysis of Hvelf3 plants. In Hvelf3, no transcript rhythms were detected under LL, demonstrating that HvELF3 function is required for self-sustained transcriptome oscillations in barley (Fig. 1; Supplemental Dataset S1). Environmental cues under LD restored oscillatory dynamics in the Hvelf3 loss-of-function line with 83% of the global transcriptome being rhythmic in the Hvelf3 plants (Fig. 1). The number and the identity of oscillating transcripts were similar between Hvelf3 and cv Bowman plants under diel cycles (Fig. 1). In Hvlux1 plants, only 2% of the expressed transcripts were rhythmic under LL suggesting that, like HvELF3, HvLUX1 is required for free-running oscillations under LL (Fig. 1). Once again, LD cycles were sufficient to restore transcriptional rhythms in the Hvlux1 mutant, i.e. 75% of the transcriptome oscillated in Hvlux1 plants under LD (Fig. 1). Mutation of the EAM7 locus in BW287 reduced the pervasiveness of circadian transcriptional oscillations, but did not completely abolish them because 8% of the expressed transcripts cycled under LL in eam7, which was approximately one-third of the number of the oscillating transcripts in cv Bowman (Fig. 1; Supplemental Dataset S1). Under LD, 80% of the global transcriptome was rhythmic in eam7 and 72% of the rhythmic transcripts were common between eam7 and the cv Bowman plants (Fig. 1).
Our data demonstrate that cycles of light and temperature and the circadian oscillator drive rhythmic expression in barley. HvELF3, HvLUX1, and EAM7 contribute to free-running oscillations under constant conditions whereas environmental rhythms are sufficient to drive rhythmic expression in the absence of a free-running oscillator.
EAM7 Is a Modulator of a Bimodal Phase Distribution under LL and Shortens the Free-Running Period
To investigate temporal expression patterns of the circadian-regulated transcripts under free-running conditions, we estimated the phase and the period of every circadian-regulated transcript in cv Bowman and eam7, the two genotypes that sustained free-running circadian rhythms. In cv Bowman, the distribution of the circadian transcriptome expression phase followed a bimodal pattern with the highest number of transcripts peaking shortly before the transitions to subjective days and nights (Fig. 2A; Supplemental Dataset S1). By contrast, in eam7, this phase pattern of the cumulative circadian transcriptome was not evident (Fig. 2A). These findings indicate that EAM7 is required to modulate the characteristic bimodal pattern of the circadian transcriptome expression in barley. The period estimates of the oscillating transcripts under LL ranged between 22 h and 34 h in cv Bowman and eam7 and followed a bell-shaped distribution with mean periods of 27.5 h and 27.7 h in cv Bowman and eam7, respectively (Fig. 2, B–D). The transcript periods were not statistically different between cv Bowman and eam7. In both cv Bowman and eam7, the sd of the period distribution was higher under LL (6 h) than under LD (2.5 h; Fig. 2, B and C). This could arise from either the uncoupled nature of cellular oscillations in free-running conditions or as a consequence from the period estimation as the signal amplitude was lower in LL than in LD.
Regulation of the Transcriptome-Wide Phase in Day/Night Cycles
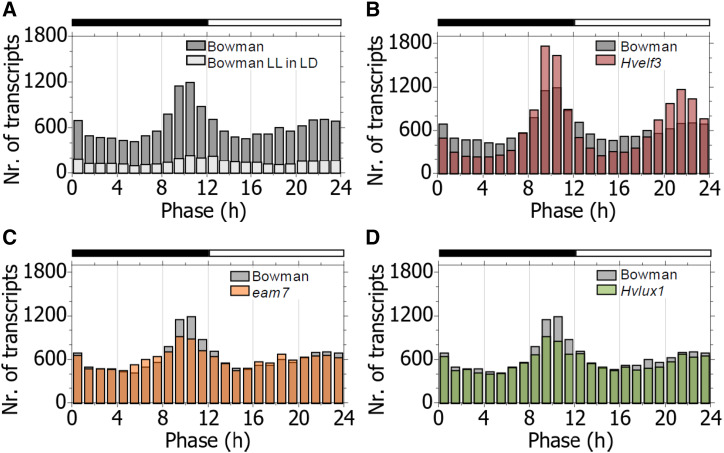
Next, we investigated the transcriptome oscillations under the diel LD conditions. In all genotypes, including those that were arrhythmic in LL, the mean of the period distribution was consistent with the enforced 24-h diel cycle and ranged between 23.5 h and 23.6 h (Supplemental Fig. S2). The phase was bimodally distributed over the day/night cycle in cv Bowman so that for the highest number of transcripts, the peak of expression occurred before dawn and dusk, and the number of transcripts with their peak expression during the night and day was the lowest (Fig. 3A). This pattern was similar to the phase distribution under LL (Fig. 2A). The transcripts that oscillated in both LL and LD were also bimodally distributed under the diel cycles, although the bimodal pattern of these genes was less pronounced under LD (Fig. 3A). This suggested that the bimodal distribution of transcriptome-wide gene expression is, at least partly, under the control of the circadian clock.
Figure 3.
Distribution of the phase of the oscillating transcriptome in night/day cycles. A, Phase distribution in cv Bowman in LD cycles for the global oscillating transcriptome and those transcripts detected oscillating in both night/day cycles and constant light (cv Bowman LL in LD). B to D, Phase distribution in diel cycles in Hvelf3 mutant (B), eam7 mutant (C), and Hvlux1 mutant (D) in comparison with cv Bowman.
The analysis of the clock mutants, however, suggested that the bimodal phase distribution under LD is controlled by both the circadian clock and day/night cues. In Hvelf3, the phase was bimodally distributed under diel cycles similar to cv Bowman; however, the quantitative characteristics of the phase distribution differed. Specifically, in Hvelf3, the phase distribution showed higher peaks at dawn and dusk and deeper troughs during the night or the day than in cv Bowman (Fig. 3B). A large number of the transcripts that peaked around the night-to-day and day-to-night transition in Hvelf3 (Fig. 3B) peaked during the day or the night in cv Bowman. This demonstrated that HvELF3 modulates timing of peak expression of multiple transcripts in day/night cycles. This effect was apparently completely or partially independent of the oscillator defect that causes arrhythmia in the Hvelf3 plants under LL, because the phase distribution in Hvlux1 mutants under LD was similar to that in cv Bowman (Fig. 3, C and D), even though self-sustained circadian oscillations were also absent in this genotype under LL conditions (Fig. 1). This was also evident from the transcriptome-wide comparison of the phase between the barley clock mutants with cv Bowman under LD (Supplemental Fig. S3). Here, the phase distributions strongly correlated between Hvlux1 and cv Bowman (Pearson correlation ρ = 0.97, R2 = 0.94), whereas the phase distributions in Hvelf3 and cv Bowman were correlated to a lower degree (Pearson correlation ρ = 0.93, R2 = 0.86), even though both mutant genotypes harbor an arrested oscillator under LL conditions (Fig. 1).
Day/night cycles had strong effects on the phase distribution of the transcriptome as demonstrated by the analysis of the eam7 transcriptome. Whereas the phase distribution was not bimodal in eam7 under LL (Fig. 2A), under LD, the phase distribution was bimodal, similar to that in cv Bowman (Fig. 3C). Consistently, the phase distributions under LD were highly correlated between eam7 and cv Bowman (Pearson correlation ρ = 0.96, R2 = 0.92, Supplemental Fig. S3). Consequently, external cues under LD controlled the phase of the global transcriptome in eam7 to peak at the night/day transitions despite the circadian defects observed in eam7 under LL. Together, these results demonstrate that the bimodal distribution of the phase in diel cycles is controlled by both day/night cues and the clock component HvELF3. The genetic defects and their underlying circadian phenotypes in Hvlux1 and eam7 have limited effects on the phase of the global oscillating transcriptome in diel cycles despite their strong transcriptional phenotypes under LL.
Dynamical Models Predict Components and Regulatory Interactions of the Barley Clock
We then sought to infer the regulatory relationships between components of the barley circadian clock. To this end, we modeled a transcriptional network based on the RNA-seq time series data. Our data suggested that HvELF3 and HvLUX1 are integral components of the barley oscillator as they were necessary to sustain transcriptome oscillations under LL (Fig. 1). Therefore, we hypothesized that modeling a transcriptional network around HvELF3 and HvLUX1 could identify the regulatory relationships that shape the circadian clock in barley. We followed an approach that searches the dynamic dependencies of HvELF3 and HvLUX1 expression on other transcripts. We used linear time invariant (LTI) models for interpreting expression data without relying on a priori knowledge of the transcriptional network (Dalchau et al., 2010; Herrero et al., 2012; Mombaerts et al., 2019; Supplemental Information). LTI models require transcriptional data sets that display robust changes in expression over time under free-running conditions. Therefore, only the expression datasets from cv Bowman and eam7 could be used in modeling, because their transcriptomes oscillated under LL conditions. In both cv Bowman and eam7, the transcripts encoding HvELF3 had a very low signal-to-noise ratio due to low rhythmicity under LL and could not be used for modeling. We therefore rooted the network around HvLUX1, which displayed robust oscillatory dynamics (Supplemental Fig. S4).
To reduce the identification of erroneous interactions, we filtered all circadian transcripts for those that were homologous to Arabidopsis genes representing transcription factors that were labeled circadian, and thus show circadian expression but are not necessarily components of the core clock (www.geneontology.org). Indeed, whereas our modeling methodology is computationally inexpensive, the uncertainty about the structure of the network is increasing exponentially with the number of genes considered. Additionally, we filtered the resulting 131 transcripts (Supplemental Dataset S2) for those that exhibited unambiguous dynamics and a high signal-to-noise ratio of expression in both cv Bowman and eam7. This filter was applied because of the transitional nature of constant light data, which typically shows a large decrease of amplitude after a few hours in barley (Campoli et al., 2012), and the dependency of noise on gene expression levels. Hvlux1 and Hvelf3 datasets were not considered in the following network analysis because these mutations led to the arrhythmic transcriptomes. This resulted in 42 transcripts in cv Bowman and 41 in eam7, of which 35 transcripts were in common between cv Bowman and eam7 and used for modeling (Supplemental Dataset S3). For the 35 shared gene transcripts, we predicted all possible pairwise (or single source-target) dynamic dependencies based on the transcript abundance over time for both the cv Bowman and the eam7 datasets. For each of these 35 pairwise comparisons, we fitted a model that captures the expression pattern of HvLUX1 in each of the two genotypes.
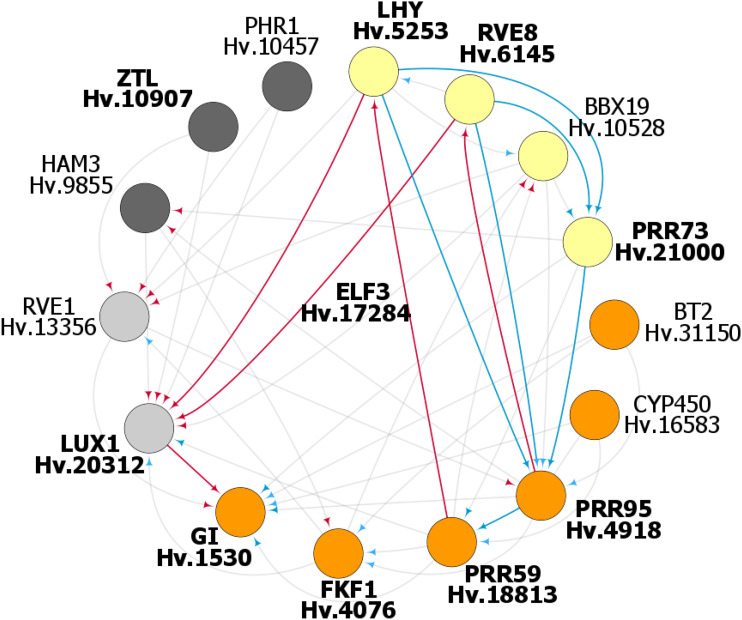
We then investigated the consistency between the models obtained for cv Bowman and eam7 using the v-gap metric (Supplemental Fig. S5; Supplemental Dataset S4). This approach estimates differences between models and allowed us to identify regulatory interactions that were maintained or abolished in the eam7 mutant (Mombaerts et al., 2019). Following this approach, we identified 20 transcripts and 79 regulatory links in cv Bowman, of which 15 transcripts and 49 regulatory links could be cross validated in eam7 (Fig. 4; Supplemental Figs. S5 and S6; Supplemental Dataset S4). Five transcripts could not be confirmed in eam7 because they were arrhythmic in eam7, or the regulatory link could not be modeled with high confidence (i.e. model fitness), or the models for cv Bowman and eam7 displayed a large difference (i.e. v-gap). Among the five genes that could not be confirmed in eam7 transcripts, those with homology to RVE8, RVE6, and RVE4 (Hv.12868, HORVU7Hr1G001830.3), and RVE1 (Hv.25709, HORVU6Hr1G066450.5), were the most prominent, with eight and seven links to putative clock genes in cv Bowman, respectively. Both transcripts displayed rhythmic oscillations under LL in cv Bowman, but were arrhythmic in eam7. Among the 15 cross-validated candidates, nine transcripts were encoded by barley genes homologous to known Arabidopsis core oscillator genes. In addition to HvLUX1 as a core of the model (Hv.20312, HORVU3Hr1G114970), the predicted components of barley circadian clock were barley homologs of LHY (Hv.5253, HORVU7Hr1G070870; Mizoguchi et al., 2002), RVE8 (Hv.6145, HORVU6Hr1G066000; Hsu et al., 2013), PRR95 (Hv.4918, HORVU5Hr1G081620; Farré et al., 2005), PRR59 (Hv.18813, HORVU4Hr1G021000; Nakamichi et al., 2010), FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 (Hv.4076, HORVU7Hr1G099010; Baudry et al., 2010), GI (Hv.1530, HORVU3Hr1G021140; Dalchau et al., 2011), and ZEITLUPE (ZTL; Hv.10907, HORVU6Hr1G022330; Más et al., 2003). Such a result supports that putative circadian-clock genes are themselves strongly driven by circadian processes.
Figure 4.
The putative circadian network of the barley oscillator as predicted from time series expression data. Genetic evidence but no model prediction allowed placing HvELF3 as a core clock component. The figure displays the inferred components and interactions that constitute the barley circadian transcriptional network (also see Supplemental Information). Circadian clock components are represented by circles and sorted in clockwise direction for the time point of peak expression starting with HvLHY at dawn (yellow, morning; orange, evening; gray, night). The regulatory interactions are represented by directional arrows, where activation is marked in blue and inhibition in red. The components printed in bold and the links highlighted in color are consistent with key components and key regulatory principles present in circadian clock models from Arabidopsis.
On the resulting conjunction network, we noted that HvPRR95 appeared as a hub with eight connections, whereas HvLUX1, as the origin of the graph, had 11 connections. This indicated a significant role of HvPRR95 in the regulation of the core circadian genes. Therefore, we repeated the search for regulators of HvPRR95, computed their interactions in both datasets, and tested for their consistency. Consequently, four genes were added to the final network, namely REVEILLE-like7 (HvRVE7, Hv.13356, HORVU2Hr1G104580), Cytochrome P450 (Hv.16583), HvPRR73 (HORVU4Hr1G057550; Farré et al., 2005), and BTB/POZ and TAZ domain-containing protein2 (HvBT2, Hv.31150, HORVU3Hr1G092090; Fig. 4; Supplemental Dataset S4). Our modeling did not place barley homologs of other known Arabidopsis clock genes TOC1, ELF4, HvELF3, and PRR37 in the barley clock model. In the case of HvTOC1 and barley homologs of ELF4, the inferred regulatory interactions to other putative clock components were weak and did not pass the cutoff filter. The weak transcript oscillations of HvELF3 precluded its modeling as part of the barley clock. However, we placed HvELF3 in the core clock model based on the genetic evidence that HvELF3 is required for clock function in barley (arrhythmic phenotype of Hvelf3 mutant). Furthermore, PRR37, the photoperiod response gene Ppd-H1, did not display rhythmic oscillations under LL and was therefore not placed into the circadian oscillator. Based on the timing of the peak expression starting with HvLHY expression at subjective dawn, we arranged the predicted components into a model of the barley circadian clockwork (Fig. 4).
In addition to the barley homologs of known Arabidopsis oscillator genes, our analysis suggested several previously uncharacterized components of barley circadian clock. These included the B-Box Zinc Finger Protein19 (HvBBX19, Hv.10528, HORVU5Hr1G081190) and HvRVE7 (Hv.13356, HORVU2Hr1G104580; Fig. 4). In our model, both HvBBX19 and HvRVE7 regulate HvPRR95 and are regulated by HvLHY (Fig. 4). The modeling predicted that HvRVE7 represses HvPRR95, and HvBBX19 activates HvPRR73 and HvPRR95. Other predicted components of the barley circadian clock were a homolog of HAIRY MERISTEM3 (HAM3; Hv.9855, HORVU6Hr1G063650) of BT2, CYTOCHROME 450 (Hv.16583, HORVU2Hr1G025160), and PHOSPHATE STARVATION RESPONSE1 (Hv.10457, HORVU4Hr1G051080). However, all of these players were predicted to regulate clock components, but were not themselves regulated by the clock genes (Fig. 4). To summarize, our analysis was able to predict components of the barley clock, which are close homologs of the Arabidopsis clock components (Campoli et al., 2012; Calixto et al., 2015) and additionally identified HvBBX19, HvRVE7, and HvHAM3 as putative components of the barley clock.
Relationship between Internal and External Cues to Regulate the Global Transcriptome in Barley
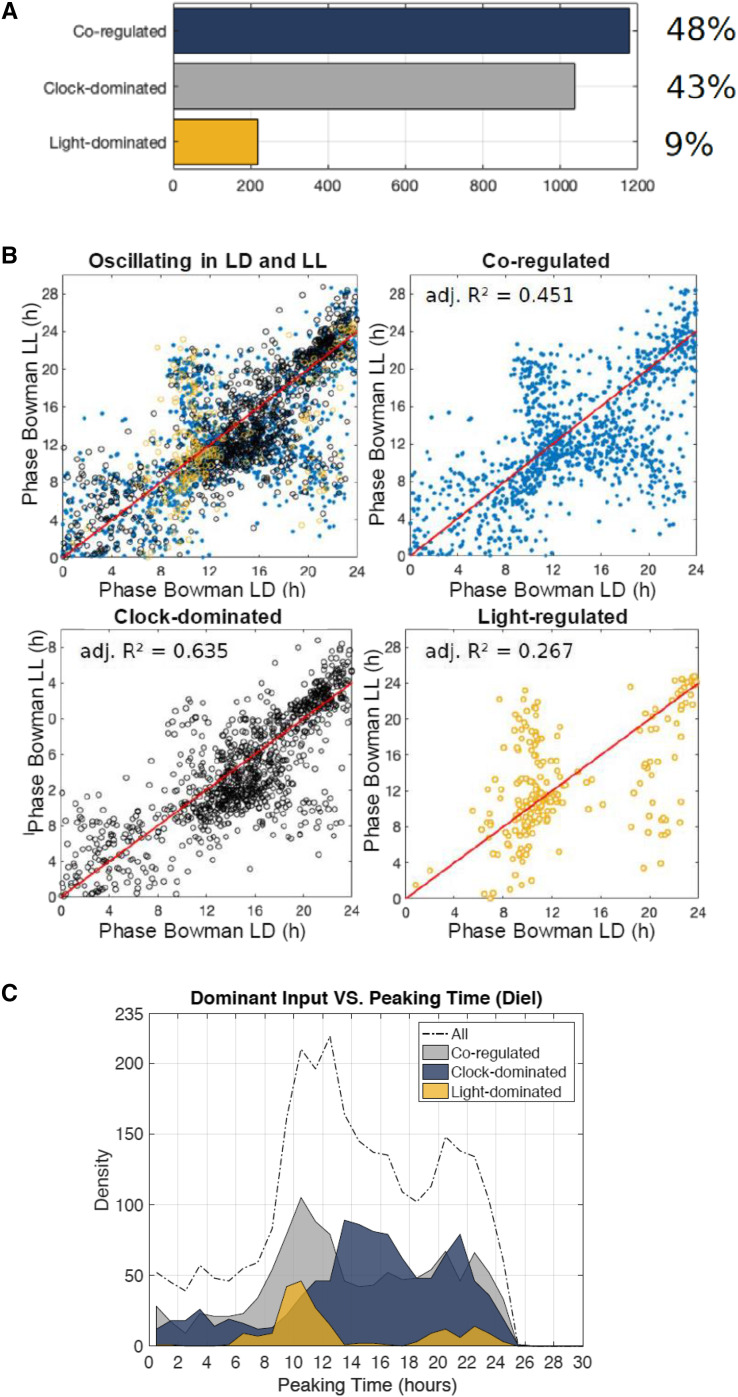
To quantify the relationship between the circadian oscillator and light signaling in regulating the rhythmicity of barley transcripts, LTI models that integrate both inputs explicitly were computed for each transcript. As a morning clock gene, the expression pattern of HvLHY accounted for the contribution of the circadian oscillator, whereas the light/dark cycle was integrated as a rectangular input (1 = light ON, 0 = light OFF; Supplemental Fig. S7). The expression pattern of the output transcript was approximated by finding the combination of inputs that fit the data best. Then, the contribution of each input was formally compared using a Bode analysis (Dalchau et al., 2010). The analysis estimated that 43% of the transcripts that oscillate in both day/night cycles and constant light were predominantly controlled by the circadian clock in light/dark cycles and that 48% were co-regulated by the circadian clock and light/dark cues (Fig. 5A; Supplemental Dataset S1). Only 9% of the transcripts were primarily controlled by light/dark cues (Fig. 5A; Supplemental Dataset S1). This is consistent with the expected under-representation of light/dark-controlled transcripts in a set of genes that oscillate in the absence of environmental cues.
Figure 5.
Relationship between external and internal cues to regulate the phase of the barley transcriptome. A, Fractions of transcripts identified as clock-dominated, co-dominated by the clock and light, and light-dominated by the Bode analysis. B, Phase relationships among diel cycles (LD) and constant light (LL) for all transcripts oscillating in LD and LL, those co-regulated by the circadian clock, those dominated by the circadian clock, and those that are dominated by light. C, Phase distribution of co-regulated, clock-dominated, and light-dominated transcripts in LD.
We next investigated the phase relationship between driven and free-running conditions for transcripts predicted to be under clock control, light control, and co-regulated by light and the clock by the Bode analysis (Fig. 5, B and C; Supplemental Dataset S1). The clock-dominated transcripts revealed the highest correlation (R2 = 0.64; Fig. 5B) and the light-dominated transcripts the lowest correlation (R2 = 0.27) of the phase between day/night cycles and constant light (Fig. 5B). The correlation of the phase of co-regulated transcripts under LD and LL was intermediate (R2 = 0.45; Fig. 5B). This suggests that transcripts dominated by the circadian clock maintained a similar expression phase under changing light conditions, whereas the phase of transcripts dominated by light cues reflected the changes in light. These findings suggest that the Bode analysis predicted the main regulatory principles that determine the phase of oscillating transcription in day/night cycles. Namely, it suggests that ∼40% of the transcripts with clock-maintained oscillations reveal a phase dominated by the circadian clock in diel cycles. For the remaining 60% of the transcripts with clock-maintained oscillations, the peak of their expression is under the control of light signaling pathways or co-regulated by light signaling and clock. This finding highlights the importance of light signaling pathways to regulate the phase of oscillating transcription even for the transcripts, the rhythmicity of which is maintained by the circadian clock.
DISCUSSION
The circadian clock was estimated to control a large proportion (∼25%) of the barley transcriptome under constant conditions, which is similar to estimates for the proportion of circadian transcripts in Arabidopsis, rice (Oryza sativa), and poplar (Populus trichocarpa; Covington et al., 2008; Michael et al., 2008b; Filichkin et al., 2011; Gehan et al., 2015). Despite the strong control of the clock on transcript oscillations under LL, day/night cues had a major influence on shaping expression patterns of circadian transcripts under diel conditions. First, the expression phase under LL conditions was generally not a strong predictor of the transcript phase under LD conditions. The expression phase of transcripts was therefore a plastic trait where LD conditions delayed or advanced expression phase as compared to LL depending on the transcript. Second, the Bode analysis demonstrated that the majority of circadian transcripts were regulated by light/temperature or a combination of the clock and light/temperature cues under LD conditions. It is well known that the circadian clock is dynamically plastic and constantly entrained by metabolic and environmental cues for synchronization with the cycles of the environment (Webb et al., 2019).
Here, however, we demonstrate that day/night cues entrain the clock and can also largely compensate for the lack of a functioning oscillator. The Hvelf3 and Hvlux1 mutants, with no cycling transcriptome under LL conditions, were characterized by transcriptome oscillations under LD comparable to wild-type cv Bowman. In this context, it is interesting to note that Hvelf3 and Hvlux1 mutants, which have a disrupted circadian clock, have been used to breed for barley cultivars adapted to northern European environments with strong daily and seasonal changes in light and temperatures (Faure et al., 2012; Campoli et al., 2013; Pankin et al., 2014). Neither of these two arrhythmic mutants has been reported to display any obvious impairment in photosynthesis and growth under conditions of pronounced photo- and thermocycles, in contrast to the corresponding Arabidopsis mutants (Faure et al., 2012; Campoli et al., 2013; Habte et al., 2014). Similarly, Izawa et al. (2011) reported that an osgi mutant in the field is not affected in photosynthesis and yield. Only under atypical growing conditions with late transplanting dates in the field was fertility significantly reduced in osgi plants, indicating a loss of seasonal adaptability. Our data suggest that diel cycles can compensate for circadian defects in the barley clock mutants by increasing the number of oscillating transcripts compared to free-running conditions and strongly influencing the time point of transcript peak expression. These findings suggest that the circadian oscillator has limited control over expression dynamics of circadian transcripts under conditions of pronounced diel oscillations in barley.
Whereas the number of cycling transcripts was not different among the Hvelf3 and Hvlux1 mutants and cv Bowman, we observed quantitative variation in the phase distribution under diel conditions between the three genotypes. HvELF3 altered the timing of transcript oscillations in day/night cycles by suppressing expression at the light and dark interfaces. This effect was apparently completely or partially independent of the oscillator defect that causes arrhythmia in the Hvelf3 plants under LL. Loss of HvELF3, but not of HvLUX1, altered the expression phase under diel cycles, although both mutants had a disrupted circadian clock. Therefore, HvELF3 modifies the light- and temperature-controlled diel transcriptome oscillations in barley. The role of HvELF3 in mediating light and temperature cues is supported by the loss of photoperiod sensitivity in the Hvelf3 mutant (Faure et al., 2012) and resembles the role of ELF3 in antagonizing light input to the clock during the night in Arabidopsis (McWatters et al., 2000; Covington et al., 2001; Thines and Harmon, 2010). However, our experimental setup with light and temperature covarying between day and night did not allow the detection of light- or temperature-specific effects on the diel transcriptome. It has already been shown that photic versus thermal entrainment results in a different behavior of the Arabidopsis circadian clock (Boikoglou et al., 2011). Future studies on the barley circadian clock should therefore test for the effects of clock genes on the circadian and diel transcriptomes under thermal versus photic entrainment.
It is interesting that only HvELF3 but not HvLUX1 had strong effects on the diel transcriptome, because out of the Arabidopsis core components of the EC ELF3–ELF4–LUX, only LUX has been identified as a transcription factor with direct DNA binding activity (Helfer et al., 2011). The different effects observed in Hvelf3 and Hvlux1 regarding the diel transcriptome may also be caused by the different nature of the underlying mutations, Whereas the Hvelf3 mutant line carries a premature stop codon leading to a truncated HvELF3 protein, the Hvlux1 mutant is characterized by a single amino acid exchange in the Myb-domain, which is important for the binding to cognate DNA sequences and regulation of their target genes (Faure et al., 2012; Campoli et al., 2013). In addition, a paralogue of HvLUX1 termed HvLUX2 may have partly redundant function and might have compensated for the mutation in HvLUX1 (Pankin et al., 2014). On the other hand, chromatin immunoprecipitation experiments demonstrated that ELF3 had many more significant binding sites than LUX, suggesting that ELF3 also binds independently of LUX (Ezer et al., 2017). Therefore, HvELF3 and HvLUX1 might have independent targets in the barley genome.
Based on RNA time series data, we modeled a possible barley clock as a basis for understanding its effects on physiology, metabolism, and agronomic performance. It is important to emphasize that the resulting interactions between the individual components of the clock represent one of the possible solutions of the barley circadian clock circuit, which may serve as a null model in future studies aimed to experimentally resolve composition and regulation of this clock. We used simple dynamical models to capture gene regulatory dynamics without making a priori assumptions on the structure of the network. These dynamical models have been successfully used in the past to describe circadian processes of Arabidopsis under conditions that are similar to those of our dataset (Dalchau et al., 2010; Herrero et al., 2012; Trejo Banos et al., 2015; Mombaerts et al., 2016, 2019). It is also important to stress that our approach could only model genes with circadian expression oscillations, whereas it is well known that post-transcriptional regulation and the rate of protein degradation and activity is an essential constituent of the clock mechanism in Arabidopsis (Kim et al., 2003; Más et al., 2003). For example, the expression of the important circadian oscillator component ZTL is not rhythmic (Somers et al., 2000), but ZTL protein is post-translationally regulated by light and its function is modified by GI at specific times of the day (Kim et al., 2007).
Our modeling strategy used HvLUX1 to reveal the circadian circuitry, which therefore appeared as a major hub in the barley clock. Nevertheless, this predicted central role of HvLUX1 is consistent with the loss of self-sustained rhythms in the Hvlux1 mutant. Unlike HvELF3 and HvELF4, HvLUX1 encodes a protein with a known DNA-binding domain, suggesting that the transcriptional regulation of the EC converges on HvLUX1 (Nusinow et al., 2011). Our model predicted that HvLUX1 represses HvGI and is itself repressed by HvLHY, consistent with the suggested repression of HvGI by the EC and CCA1/LHY repressing the EC in Arabidopsis (Hsu et al., 2013; Fogelmark and Troein, 2014).
Further, the regulatory predictions suggested that HvLHY and HvRVE8 are activators of HvPRR73 and HvPRR95 in the morning and, at the same time, repress HvLUX1. The morning activation of the HvPRRs through HvLHY and HvRVE8, together with the repression of HvLHY and HvREV8 through the HvPRRs later in the day, are also key regulatory relationships within the Arabidopsis clock (Hsu et al., 2013; Fogelmark and Troein, 2014). This suggests that the regulatory links among HvLHY, HvRVE8, and the HvPRRs are conserved between barley and Arabidopsis, despite the independent evolutionary history of LHY-like and PRR-like genes in the barley and Arabidopsis clades (Takata et al., 2009, 2010; Campoli et al., 2012).
Our model suggested that HvPRR73, the first PRR expressed in barley in the morning, activates HvPRR95, which, in turn, activates HvPRR59, such that HvPRR73, HvPRR95, and HvPRR59 are expressed in a sequential cascade. This resembles predictions by Pokhilko et al. (2012), who described the PRRs as a series of activators in the Arabidopsis clock, whereas other models have predicted that direct interactions between the PRRs are negative and directed from the later PRRs in the sequence to the earlier ones (Huang et al., 2012; Carré and Veflingstad 2013; Fogelmark and Troein, 2014). However, the sequential regulation of the PRRs during the day appears to be a common feature of the circadian clock in both barley and Arabidopsis, whereas the sequence of expression of PRR genes is altered between Arabidopsis and barley. In Arabidopsis, the sequence of PRR expression starts with PRR9, followed by PRR7, PRR5, and PRR3, and ends with PRR1 (Matsushika et al., 2000; Hsu et al., 2013). However, in our data the sequential PRR expression wave started with PRR73 and ended with PRR59 (PRR1 was not scored rhythmic in our data). Interestingly, PRR37, corresponding to the major photoperiod response gene PPD1 in wheat and barley, showed no circadian oscillations and is therefore probably not part of the circadian clock in barley. This is consistent with the finding that mutations in PPD1 do not affect the circadian clock in barley and wheat (Campoli et al., 2012; Shaw et al., 2012).
Our modeling placed several members of the RVE family, including barley homologs of the principal clock activators RVE8, RVE6, and RVE4, into the barley clock (Kuno et al., 2003; Zhang et al., 2007; Rawat et al., 2009). The mutation in eam7 had a limited effect on the central oscillator, but several RVE homologs lost rhythmicity or were strongly downregulated and this correlated with a lengthening of the period. The suggested that EAM7 is a component of the slave oscillator that only regulates a subset of clock-controlled transcripts. RVE-like genes have been implicated in such slave oscillators (Kuno et al., 2003) and rve mutants are characterized by a period lengthening (Hsu et al., 2013); however, none of the expressed RVE genes carried mutations that would alter the protein sequence.
BBX19, RVE7, and HAM3 were identified as three new candidate oscillator components in barley. Whereas the three proteins have already been proposed to have connections to the Arabidopsis oscillator, they have not been modeled as an integral part of the circadian clock but rather as clock outputs in Arabidopsis (Kuno et al., 2003; Wang et al., 2014). BBX19 acts as a gatekeeper of EC formation by mediating degradation of ELF3 and is part of a regulatory loop with CCA1 and/or LHY (Wang et al., 2015; Tripathi et al., 2017), supporting the link between BBX19 and LHY in our model. The model also predicted that barley homologs of BT2, CYTOCHROME 450, and PHOSPHATE STARVATION RESPONSE1 are part of the core circadian oscillator in barley. However, these proteins were only predicted to regulate other clock components and were not regulated themselves by the clock. They therefore displayed a low connectivity within the circadian network, consistent with their known functions outside the central clock (Bari et al., 2006; Ren et al., 2007; Bak et al., 2011). Therefore, these components might provide input into the circadian network but are probably not components of the barley oscillator. However, the function of the predicted barley clock components, their role in the barley circadian clock, and interactions generated by the network modeling need to be experimentally verified using natural and induced mutants and transgenic lines.
CONCLUSION
Our comparison of diel and circadian transcriptomes in the different barley clock mutants revealed that fluctuations of light and temperature have a major effect on the diel oscillating transcriptome and can compensate for circadian defects in arrhythmic barley clock mutants. HvELF3, but not HvLUX1, controlled the expression phase of a large number of transcripts under diel conditions and this effect was independent from the oscillator arrest under LL. Dynamical modeling suggested novel putative clock genes and connections between clock genes as a basis for experimental explorations into the nature and functions of the barley circadian clock. Finally, our findings and the dataset provide a valuable resource for mining for the output targets of the barley clock components HvELF3, HvLUX1, and EAM7 and to understand the role of the diel cues and clock in controlling the barley transcriptome and plant performance.
MATERIALS AND METHODS
Genetic Material and Growth Conditions
Four spring barley (Hordeum vulgare) genotypes were used in this study, the wild-type spring barley ‘Bowman’ (wild type) and three derived introgression lines with mutations in HvELF3 (BW290), HvLUX1 (BW284), and EAM7 (BW287; Faure et al., 2012; Campoli et al., 2013). BW290 carries an introgression of the eam8 allele (eam8.k) that is characterized by a bp mutation leading to a premature stop codon in HvELF3, which is orthologous to ELF3 in Arabidopsis (Arabidopsis thaliana; Faure et al., 2012; Hicks et al., 2001). BW284 carries the eam10 locus characterized by a single nonsynonymous nucleotide polymorphism in the conserved Myb-domain of the barley LUX/ARRHYTHMO homolog (Campoli et al., 2013). The candidate gene for EAM7 has not yet been identified, but eam7 is characterized by accelerated flowering and reduced sensitivity to the photoperiod (Stracke and Börner, 1998). The lines were sown in soil (Einheitserde) in a 96-well format. Seeds were maintained at 4°C for 3 d, followed by germination in 12-h light/12-h dark cycle at 20°C with a photon flux density 300 μmol m−2 s−1 during the day and 18°C during the night, and grown for three weeks.
Sampling, Extraction of Total RNA, and Sequencing
After three weeks of growth, the second expanded leaf after the cotyledon leaf was harvested from two plants, resulting in two biological replicates per genotype and time point every 4 h for 24 h starting with lights off, except for dusk and dawn, when extra samples were taken for cv Bowman every 2 h (Supplemental Fig. S1). After completion of a 12-h dark/12-h light diel cycle, the growth chamber was switched to constant light and 20°C and sampling continued for an additional 36 h starting from the first subjective dusk. Total RNA was extracted from ground tissue using a hybrid protocol of TRIZOL (Invitrogen) and purification columns from a RNeasy RNA extraction kit (Qiagen). Extracted total RNA was DNAse-treated (Ambion). The concentration and integrity of the extracted RNA was determined on a BioAnalyzer (Agilent) before library preparation. The library preparation was carried out following the TruSeq protocol and single-end sequenced on a HiSeq 2500 System (Illumina) with 10 million reads per library.
Mapping of Reads and Rhythmicity Analysis
The quality of the sequencing data were verified using the software FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The reads were mapped against a custom barley reference transcriptome (Digel et al., 2015) and raw read counts were obtained using the software implementing the full pipeline for RNA-seq analysis RobiNA (v1.2.3) with default settings (Lohse et al., 2012). Raw read counts were normalized to CPM with the R/Bioconductor package edgeR (http://bioinf.wehi.edu.au/edgeR/) and used for the downstream rhythmic analysis and modeling. To cross reference a nomenclature of the gene models used in this study (Digel et al., 2015) with the identifiers of the HORVU gene models annotated on the barley pseudochromosomes (Mascher et al., 2017), we used reciprocal BLASTn v2.9.0+ (e value cutoff 10e-05). The reciprocal best BLAST (https://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/) hit pairs were extracted as matching gene model identifiers.
For the analysis of the day/night data, the sequence of samples was inverted to start with the night followed by the day samples (12, 14, 16, 20, 0, 2, 4, and 8 h), and this data set was therefore termed “LD.” The oscillating patterns of gene expression and period (as a duration of one complete cycle) and phase (as the location of time of the peak of the curve) of the curves were determined using the ARSER algorithm in the R package, MetaCycle (Yang and Su, 2010; Wu et al., 2016). To increase the analytical power for the rhythmic analysis, the 24-h diel dataset, but not the 36-h LL data, was duplicated to imitate 48 h of sampling data. The settings for the ARSER algorithm were adjusted for each genotype individually so that the period was normally distributed with two symmetric tails and the number of transcripts passing the cutoff (Benjamini–Hochberg corrected false discovery rate of 0.1; Benjamini and Hochberg, 1995) was maximal with the given range of period estimation being minimal. For cv Bowman and BW287 the settings are shown in Figure 2, B and C, and comprised mean = 28, upper/lower limit = 22/34 for the constant light data. In diel cycles, the mean and upper/lower limits were set to 24 and 21/27, respectively. Significant differences between the period lengths of transcripts in cv Bowman and eam7 were tested with a one-factorial ANOVA (P < 0.05).
Modeling the Barley Circadian Clock
To identify putative barley clock genes, we blasted all expressed barley transcripts against the Arabidopsis transcriptome (geneontology.org). For each barley transcript, the best Arabidopsis BLAST hit was retained, and we then selected only those barley transcripts for which the Arabidopsis homolog carried the GO annotation “circadian” and “transcription” (Supplemental Dataset S1). The resulting 138 barley transcripts were then further filtered for those that exhibited unambiguous dynamics and a high signal-to-noise ratio of expression. This filtering step was necessary to ensure that we did not identify dynamics out of noise. Hence, genes for which the amplitude of oscillation was <20 CPM in the last 24 h were removed. The choice of such a filter is motivated by both the transitional nature of constant light data, which typically shows a large decrease of amplitude after few hours in barley, and the dependency of noise on gene expression levels. Furthermore, genes that were constantly up-/downregulated without exhibiting further significant dynamics were also discarded. This was performed by detrending the final 24 h of constant light data before applying the same filtering criterion. After filtering, out of a total of 138, 49 and 48 genes passed the filtering criterions in the wild-type and BW287 datasets, respectively. BW284 (Hvlux1) and BW290 (Hvelf3) datasets were not considered in the following network analysis because these mutations led to the arrhythmic transcriptomes. Finally, seven genes (Hv.21080, Hv.22191, Hv.23289, Hv.32914, Hv.33010, Hv.6793, and MLOC_7084.3) were manually discarded from both subset lists of candidates as they were not DNA-binding transcription factors but rather enzymes in a metabolic process, leaving 42 in cv Bowman and 41 in BW287, of which 35 transcripts were in common between the cv Bowman and BW287 final datasets used for modeling. The HvELF3 transcript did not pass the filtering and, therefore, could not be used to infer dynamical interactions.
To model the barley circadian clock based on the time-course gene transcription data, we adopted an approach based on LTI models. LTI models do not rely on prior knowledge of the transcriptional network to provide accurate predictions and have been shown to provide reliable predictions of the dynamical processes involved in the Arabidopsis circadian network (Dalchau, 2012; Herrero et al., 2012; Mombaerts et al., 2016, 2019). To provide a comprehensive evaluation of the LTI model, the performance of the modeling strategy was evaluated and compared under conditions that replicated those of the experiments using widely used benchmark models (Supplemental Information).
We used first-order models to represent the system dynamics between two genes at a time using an LTI model with the following equation:
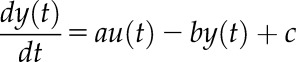 |
where  corresponds to the degradation rate of y and
corresponds to the degradation rate of y and 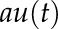 represents the influence of another transcription factor through the synthesis rate of y. The model, therefore, evaluates whether the rate of change of a particular gene y depends on another gene u. Estimating a model means finding (a), (b), and (c) that produce a vector y(t) as close as possible to the real data. The estimation of parameters was performed using the function “pem” implemented in the software MATLAB (Mathworks; https://www.mathworks.com/) that minimizes the prediction error of the data. LL data were used for the estimation, as they represent the autonomous behavior of the oscillator.
represents the influence of another transcription factor through the synthesis rate of y. The model, therefore, evaluates whether the rate of change of a particular gene y depends on another gene u. Estimating a model means finding (a), (b), and (c) that produce a vector y(t) as close as possible to the real data. The estimation of parameters was performed using the function “pem” implemented in the software MATLAB (Mathworks; https://www.mathworks.com/) that minimizes the prediction error of the data. LL data were used for the estimation, as they represent the autonomous behavior of the oscillator.
The goodness-of-fit of the model, with the data, was calculated as follows:
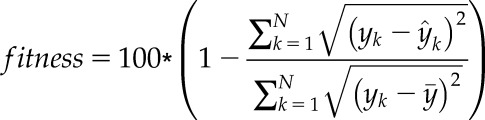 |
where 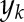 is the data (output),
is the data (output), 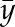 is the average value of the data, and
is the average value of the data, and 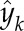 is the estimated output. The MATLAB function compare was used to compute the fitness of the model. Each potential link between two genes was validated if the associated model reproduced the dynamics involved with a sufficient degree of precision, which corresponds to a fitness threshold estimated at 60% (Supplemental Information).
is the estimated output. The MATLAB function compare was used to compute the fitness of the model. Each potential link between two genes was validated if the associated model reproduced the dynamics involved with a sufficient degree of precision, which corresponds to a fitness threshold estimated at 60% (Supplemental Information).
To investigate the potential regulators of HvLUX1, a collection of independent first-order LTI models was estimated separately between each of the transcripts and HvLUX1 in the cv Bowman background. In each case, the parameters were estimated so that they together provide the best possible fit to the HvLUX1 time-course data. This step takes the following form:
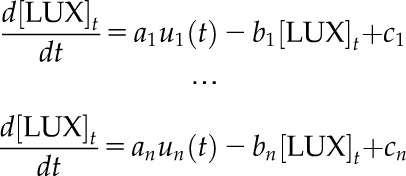 |
where n corresponds to the number of candidates (42 models in total). Each model was characterized by a fitness metric that ranges from 0% to 100%, representing its capability to describe the regulatory dynamics between genes. A gene, therefore, would be further considered as a regulator for HvLUX1 if the model is capable of reproducing the shape of HvLUX1 with a sufficient degree of precision. A fitness threshold, evaluated from in silico benchmarks systems (Supplemental Information), was used to validate the models. In this case, the fitness threshold was set to 60% to limit false-positive predictions of regulatory interactions while accounting for sufficient gene regulatory models to describe the system of interest. Finally, 20 models passed the validation step (Supplemental Dataset S5). The methodology is summarized in Supplemental Figure S4.
To further narrow down the predicted regulatory interactions, we estimated the consistency of the candidate models using the filtered eam7 (BW287) dataset. For this purpose, we evaluated first-order LTI models for each of the previously identified regulations and retained those with the goodness-of-fit > 60% in the eam7 (BW287) experimental condition (Supplemental Fig. S4). To keep links with the highest confidence only, the dynamical consistency of the LTI models based on these two independent datasets (cv Bowman and BW287) was evaluated using the nu (ν) gap metric (gapmetric, MATLAB; Vinnicombe, 1993; Mombaerts et al., 2019). We further considered models that had a v-gap <0.2 following Carignano et al. (2015). As a result, six regulatory interactions were filtered out (Hv.10528 to Hv.27754, Hv.1530 [GI] to Hv.19411, Hv.19411 to Hv.20312 [LUX], Hv.19759 [TOC1] to Hv.20312 [LUX], Hv.5253 [LHY] to Hv.27754, and Hv.9855 to Hv.18813 [PRR59]). HvPRR95 (Hv.4918) appeared as a hub with eight connections, so we repeated the search for regulators of HvPRR95 (Eq. 3), computed their interactions in both datasets, and checked their consistency.
The relative contribution of light-signaling and circadian-clock pathways in generating oscillating transcriptome was evaluated using a Bode analysis (bode function in MATLAB) with the threshold of 7 dB to discern between the two alternative regulatory inputs (Dalchau et al., 2010; Supplemental Fig. S5). Here, we use the magnitude response of the signal to assess the relative contribution of the inputs 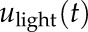 and
and 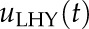 in each of the validated models, at a frequency of 24 h (or 0.262 rad h ^−1). Following Dalchau et al. (2010), if the magnitude of the response of the light input was 7-dB higher than the contribution of the clock (represented by HvLHY potentially delayed), the circadian-regulated gene (the output of the model) was considered driven mostly by light; in the opposite case, the transcript expression was considered as driven by the clock. If the magnitude difference was <7 dB, then the circadian-regulated gene was considered regulated by both inputs equally. The methodology is summarized in Supplemental Figure S7B.
in each of the validated models, at a frequency of 24 h (or 0.262 rad h ^−1). Following Dalchau et al. (2010), if the magnitude of the response of the light input was 7-dB higher than the contribution of the clock (represented by HvLHY potentially delayed), the circadian-regulated gene (the output of the model) was considered driven mostly by light; in the opposite case, the transcript expression was considered as driven by the clock. If the magnitude difference was <7 dB, then the circadian-regulated gene was considered regulated by both inputs equally. The methodology is summarized in Supplemental Figure S7B.
To this end, we used the 2,759 transcripts that were identified as oscillating in both diel and free-running conditions in the wild-type cv Bowman background to calculate another set of LTI models as described earlier. As a reference, we selected a formerly identified clock gene peaking in the morning, HvLHY (Hv.5253), with a range of delays integrated into the model to implicitly represent the clock input following Dalchau et al. (2010). The structure of such models is schematically represented in Supplemental Figure S7A. This way, the light input is incorporated on two levels: explicitly through the light input and implicitly through the clock pathway using the following equation:
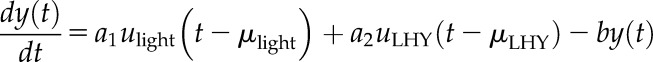 |
where 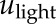 was assumed to be binary (1 = light; 0 = dark). We fixed the light delay
was assumed to be binary (1 = light; 0 = dark). We fixed the light delay 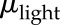 to 0 h to represent the effect of rapid light signaling on the transcripts, and computed delays ranging from 0 to 8 h, every 0.2 h, for HvLHY. The delay that provided the best fit to the data was selected independently for each transcript. Ultimately, models were validated if they succeeded in capturing the regulatory dynamics involved with a goodness-of-fit > 60%.
to 0 h to represent the effect of rapid light signaling on the transcripts, and computed delays ranging from 0 to 8 h, every 0.2 h, for HvLHY. The delay that provided the best fit to the data was selected independently for each transcript. Ultimately, models were validated if they succeeded in capturing the regulatory dynamics involved with a goodness-of-fit > 60%.
We assessed the accuracy of our LTI-based network reconstruction algorithm on the circadian model from Pokhilko et al. (2010) as detailed in the Supplemental Information. The performance of the modeling strategy was evaluated based on the Area Under the Curve of the Receiver Operating Characteristic and Area Under the Precision-Recall Curve criteria (Supplemental Information; Supplemental Fig. S8).
The scripts for the modeling of the barley clock are available under: https://github.com/Lmombaerts/CircadianBarley.
Accession Number
The ArrayExpress accession no. E-MTAB-8372 was deposited in the European Molecular Biology Laboratory data library (https://www.ebi.ac.uk/arrayexpress/). Accession numbers of genes are given in Supplemental Dataset S1.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Sampling scheme for the LD and LL samples in Bowman, BW284 (Hvlux1), BW287 (eam7), and BW290 (Hvelf3) grown under 12-h light/12-h dark cycles (20°C day/16°C night).
Supplemental Figure S2. Transcriptome-wide period estimates under diel day/night conditions.
Supplemental Figure S3. Phase correlation of transcripts of cv Bowman with the clock mutants Hvelf3, Hvlux1, and eam7 grown under LD conditions.
Supplemental Figure S4. Schematic of the strategy used in this article to investigate the potential dependencies of HvLUX1 through identification of dynamics.
Supplemental Figure S5. Predicted regulatory interactions between the regulators of HvLUX1 that have been identified in cv Bowman and further validated using the eam7 dataset.
Supplemental Figure S6. Expression profiles of predicted core clock components of the barley oscillator in free-running conditions of constant light.
Supplemental Figure S7. Methodology and examples of the Bode analysis.
Supplemental Figure S8. Performance of the modeling strategy compared to other clock models.
Supplemental Dataset S1. CPM for transcripts and information about rhythmicity, period, amplitude, and phase, as well as gene annotation.
Supplemental Dataset S2. Oscillating barley transcripts homologous to Arabidopsis gene models with the annotations “circadian” and “transcription.”
Supplemental Dataset S3. Transcripts with unambiguous dynamics and a high signal-to-noise ratio of expression in cv Bowman and BW287 (eam7).
Supplemental Dataset S4. Predicted 49 regulatory links between 15 genes in the circadian oscillator network in barley (Fig. 4).
Supplemental Dataset S5. LTI model parameters of the 20 models that passed the validation step (>60% fitness).
Supplemental Information. Benchmarking models.
Acknowledgments
We cordially thank Kerstin Luxa, Caren Dawidson, Thea Rütjes, and Andrea Lossow for excellent technical assistance.
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (DFG; German Research Foundation) under Germany´s Excellence Strategy (EXC–2048/1 Project ID 390686111), the German Federal Ministry for Education and Research (ERASysApp), and the Biotechnology and Biological Sciences Research Council (no. BB/L014130/1 to L.M. and A.A.R.W. and BB/M00113X/1 to J.M.G. and A.A.R.W.).
Articles can be viewed without a subscription.
References
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA(2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Bak S, Beisson F, Bishop G, Hamberger B, Höfer R, Paquette S, Werck-Reichhart D(2011) Cytochromes p450. Arabidopsis Book 9: e0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible W-R(2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudry A, Ito S, Song YH, Strait AA, Kiba T, Lu S, Henriques R, Pruneda-Paz JL, Chua NH, Tobin EM, et al. (2010) F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell 22: 606–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beales J, Turner A, Griffiths S, Snape JW, Laurie DA(2007) A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor Appl Genet 115: 721–733 [DOI] [PubMed] [Google Scholar]
- Bendix C, Marshall CM, Harmon FG(2015) Circadian clock genes universally control key agricultural traits. Mol Plant 8: 1135–1152 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y(1995) Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Boikoglou E, Ma Z, von Korff M, Davis AM, Nagy F, Davis SJ(2011) Environmental memory from a circadian oscillator: The Arabidopsis thaliana clock differentially integrates perception of photic vs. thermal entrainment. Genetics 189: 655–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calixto CPG, Waugh R, Brown JWS(2015) Evolutionary relationships among barley and Arabidopsis core circadian clock and clock-associated genes. J Mol Evol 80: 108–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campoli C, Pankin A, Drosse B, Casao CM, Davis SJ, von Korff M(2013) HvLUX1 is a candidate gene underlying the early maturity 10 locus in barley: Phylogeny, diversity, and interactions with the circadian clock and photoperiodic pathways. New Phytol 199: 1045–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campoli C, Shtaya M, Davis SJ, von Korff M(2012) Expression conservation within the circadian clock of a monocot: Natural variation at barley Ppd-H1 affects circadian expression of flowering time genes, but not clock orthologs. BMC Plant Biol 12: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carignano A, Webb AAR, Gonçalves J, Jin J (2015) Assessing the effect of unknown widespread perturbations in complex systems using the nu-gap. In Proceedings of the 54th IEEE Conference Decision and Control, Institute of Electrical and Electronics Engineers, Piscataway, NJ, pp 3193–3198 [Google Scholar]
- Carré I, Veflingstad SR(2013) Emerging design principles in the Arabidopsis circadian clock. Semin Cell Dev Biol 24: 393–398 [DOI] [PubMed] [Google Scholar]
- Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL(2008) Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol 9: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington MF, Panda S, Liu XL, Strayer CA, Wagner DR, Kay SA(2001) ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell 13: 1305–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalchau N, Baek SJ, Briggs HM, Robertson FC, Dodd AN, Gardner MJ, Stancombe MA, Haydon MJ, Stan G-B, Gonçalves JM, et al. (2011) The circadian oscillator gene GIGANTEA mediates a long-term response of the Arabidopsis thaliana circadian clock to sucrose. Proc Natl Acad Sci USA 108: 5104–5109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalchau N, Hubbard KE, Robertson FC, Hotta CT, Briggs HM, Stan G-B, Gonçalves JM, Webb AAR(2010) Correct biological timing in Arabidopsis requires multiple light-signaling pathways. Proc Natl Acad Sci USA 107: 13171–13176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalchau N.(2012) Understanding biological timing using mechanistic and black-box models. New Phytol 193: 852–858 [DOI] [PubMed] [Google Scholar]
- Digel B, Pankin A, von Korff M(2015) Global transcriptome profiling of developing leaf and shoot apices reveals distinct genetic and environmental control of floral transition and inflorescence development in barley. Plant Cell 27: 2318–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, Xu Y, Pan M, Valekunja UK, Feeney KA, et al. (2012) Peroxiredoxins are conserved markers of circadian rhythms. Nature 485: 459–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich IM, Hanzawa Y, Chou L, Roe JL, Kover PX, Purugganan MD(2009) Candidate gene association mapping of Arabidopsis flowering time. Genetics 183: 325–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezer D, Jung J-H, Lan H, Biswas S, Gregoire L, Box MS, Charoensawan V, Cortijo S, Lai X, Stöckle D, et al. (2017) The evening complex coordinates environmental and endogenous signals in Arabidopsis. Nat Plants 3: 17087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinas B, Mas P(2011) Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J 66: 318–329 [DOI] [PubMed] [Google Scholar]
- Farré EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA(2005) Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol 15: 47–54 [DOI] [PubMed] [Google Scholar]
- Faure S, Turner AS, Gruszka D, Christodoulou V, Davis SJ, von Korff M, Laurie DA(2012) Mutation at the circadian clock gene EARLY MATURITY 8 adapts domesticated barley (Hordeum vulgare) to short growing seasons. Proc Natl Acad Sci USA 109: 8328–8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filichkin SA, Breton G, Priest HD, Dharmawardhana P, Jaiswal P, Fox SE, Michael TP, Chory J, Kay SA, Mockler TC(2011) Global profiling of rice and poplar transcriptomes highlights key conserved circadian-controlled pathways and cis-regulatory modules. PLoS One 6: e16907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogelmark K, Troein C(2014) Rethinking transcriptional activation in the Arabidopsis circadian clock. PLOS Comput Biol 10: e1003705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher LW, Soliman KM, Vivar H(1991) Interactions among loci conferring photoperiod insensitivity for heading time in spring barley. Crop Sci 31: 256–261 [Google Scholar]
- Gehan MA, Greenham K, Mockler TC, McClung CR(2015) Transcriptional networks—crops, clocks, and abiotic stress. Curr Opin Plant Biol 24: 39–46 [DOI] [PubMed] [Google Scholar]
- Gendron JM, Pruneda-Paz JL, Doherty CJ, Gross AM, Kang SE, Kay SA(2012) Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc Natl Acad Sci USA 109: 3167–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenham K, McClung CR(2015) Integrating circadian dynamics with physiological processes in plants. Nat Rev Genet 16: 598–610 [DOI] [PubMed] [Google Scholar]
- Habte E, Müller LM, Shtaya M, Davis SJ, von Korff M(2014) Osmotic stress at the barley root affects expression of circadian clock genes in the shoot. Plant Cell Environ 37: 1321–1327 [DOI] [PubMed] [Google Scholar]
- Harmer SL.(2009) The circadian system in higher plants. Annu Rev Plant Biol 60: 357–377 [DOI] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang H-S, Han B, Zhu T, Wang X, Kreps JA, Kay SA(2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113 [DOI] [PubMed] [Google Scholar]
- Hayama R, Sarid-Krebs L, Richter R, Fernández V, Jang S, Coupland G(2017) PSEUDO RESPONSE REGULATORs stabilize CONSTANS protein to promote flowering in response to day length. EMBO J 36: 904–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen SP, Schultz TF, Pruneda-Paz JL, Borevitz JO, Ecker JR, Kay SA(2005) LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc Natl Acad Sci USA 102: 10387–10392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfer A, Nusinow DA, Chow BY, Gehrke AR, Bulyk ML, Kay SA(2011) LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr Biol 21: 126–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero E, Kolmos E, Bujdoso N, Yuan Y, Wang M, Berns MC, Uhlworm H, Coupland G, Saini R, Jaskolski M, et al. (2012) EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24: 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks KA, Albertson TM, Wagner DR(2001) EARLY FLOWERING3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis. Plant Cell 13: 1281–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PY, Harmer SL(2014) Wheels within wheels: The plant circadian system. Trends Plant Sci 19: 240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PY, Devisetty UK, Harmer SL(2013) Accurate timekeeping is controlled by a cycling activator in Arabidopsis. eLife 2: e00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Pérez-García P, Pokhilko A, Millar AJ, Antoshechkin I, Riechmann JL, Mas P(2012) Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336: 75–79 [DOI] [PubMed] [Google Scholar]
- Hughes ME, Abruzzi KC, Allada R, Anafi R, Arpat AB, Asher G, Baldi P, de Bekker C, Bell-Pedersen D, Blau J, et al. (2017) Guidelines for genome-scale analysis of biological rhythms. J Biol Rhythms 32: 380–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa T, Mihara M, Suzuki Y, Gupta M, Itoh H, Nagano AJ, Motoyama R, Sawada Y, Yano M, Hirai MY, et al. (2011) Os-GIGANTEA confers robust diurnal rhythms on the global transcriptome of rice in the field. Plant Cell 23: 1741–1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamioka M, Takao S, Suzuki T, Taki K, Higashiyama T, Kinoshita T, Nakamichi N(2016) Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis circadian clock. Plant Cell 28: 696–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikis EA, Khanna R, Quail PH(2005) ELF4 is a phytochrome-regulated component of a negative-feedback loop involving the central oscillator components CCA1 and LHY. Plant J 44: 300–313 [DOI] [PubMed] [Google Scholar]
- Kim W-Y, Geng R, Somers DE(2003) Circadian phase-specific degradation of the F-box protein ZTL is mediated by the proteasome. Proc Natl Acad Sci USA 100: 4933–4938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y, Han L, David K, Putterill J, Nam HG, Somers DE(2007) ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449: 356–360 [DOI] [PubMed] [Google Scholar]
- Kolmos E, Nowak M, Werner M, Fischer K, Schwarz G, Mathews S, Schoof H, Nagy F, Bujnicki JM, Davis SJ(2009) Integrating ELF4 into the circadian system through combined structural and functional studies. HFSP J 3: 350–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuno N, Møller SG, Shinomura T, Xu X, Chua NH, Furuya M(2003) The novel MYB protein EARLY-PHYTOCHROME-RESPONSIVE1 is a component of a slave circadian oscillator in Arabidopsis. Plant Cell 15: 2476–2488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusakina J, Rutterford Z, Cotter S, Martí MC, Laurie DA, Greenland AJ, Hall A, Webb AA(2015) Barley Hv CIRCADIAN CLOCK ASSOCIATED 1 and Hv PHOTOPERIOD H1 are circadian regulators that can affect circadian rhythms in Arabidopsis. PLoS One 10: e0127449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TL, Newton L, Liu M-J, Shiu S-H, Farré EM(2016) A G-box-like motif is necessary for transcriptional regulation by circadian pseudo-response regulators in Arabidopsis. Plant Physiol 170: 528–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Bolger AM, Nagel A, Fernie AR, Lunn JE, Stitt M, Usadel B(2012) RobiNA: A user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res 40(Web Server issue): W622–W627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más P, Kim W-Y, Somers DE, Kay SA(2003) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426: 567–570 [DOI] [PubMed] [Google Scholar]
- Mascher M, Gundlach H, Himmelbach A, Beier S, Twardziok SO, Wicker T, Radchuk V, Dockter C, Hedley PE, Russell J, et al. (2017) A chromosome conformation capture ordered sequence of the barley genome. Nature 544: 427–433 [DOI] [PubMed] [Google Scholar]
- Matsushika A, Makino S, Kojima M, Mizuno T(2000) Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: Insight into the plant circadian clock. Plant Cell Physiol 41: 1002–1012 [DOI] [PubMed] [Google Scholar]
- McClung CR.(2006) Plant circadian rhythms. Plant Cell 18: 792–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWatters HG, Bastow RM, Hall A, Millar AJ(2000) The ELF3 Zeitnehmer regulates light signalling to the circadian clock. Nature 408: 716–720 [DOI] [PubMed] [Google Scholar]
- Michael TP, Breton G, Hazen SP, Priest H, Mockler TC, Kay SA, Chory J(2008a) A morning-specific phytohormone gene expression program underlying rhythmic plant growth. PLoS Biol 6: e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Mockler TC, Breton G, McEntee C, Byer A, Trout JD, Hazen SP, Shen R, Priest HD, Sullivan CM, et al. (2008b) Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet 4: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song H-R, Carré IA, Coupland G(2002) LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell 2: 629–641 [DOI] [PubMed] [Google Scholar]
- Mombaerts L, Carignano A, Robertson FC, Hearn TJ, Junyang J, Hayden D, Rutterford Z, Hotta CT, Hubbard KE, Maria MRC, et al. (2019) Dynamical differential expression (DyDE) reveals the period control mechanisms of the Arabidopsis circadian oscillator. PLOS Comput Biol 15: e1006674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts L, Mauroy A, Goncalves J (2016) Optimising time-series experimental design for modelling of circadian rhythms: The value of transient data. In Findeisen R, Bullinger E, Balsa-Canto E, and Bernaerts K, eds, 6th International Federation of Automatic Control (IFAC) Conference on Foundations of Systems Biology in Engineering, Elsevier, Dordrecht, p. 4 [Google Scholar]
- Murphy RL, Klein RR, Morishige DT, Brady JA, Rooney WL, Miller FR, Dugas DV, Klein PE, Mullet JE(2011) Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proc Natl Acad Sci USA 108: 16469–16474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua N-H, Sakakibara H(2010) PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22: 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Kamioka M, Suzuki T, Yamashino T, Higashiyama T, Sakakibara H, Mizuno T(2012) Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc Natl Acad Sci USA 109: 17123–17128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kita M, Niinuma K, Ito S, Yamashino T, Mizoguchi T, Mizuno T(2007) Arabidopsis clock-associated pseudo-response regulators PRR9, PRR7 and PRR5 coordinately and positively regulate flowering time through the canonical CONSTANS-dependent photoperiodic pathway. Plant Cell Physiol 48: 822–832 [DOI] [PubMed] [Google Scholar]
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farré EM, Kay SA(2011) The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankin A, Campoli C, Dong X, Kilian B, Sharma R, Himmelbach A, Saini R, Davis SJ, Stein N, Schneeberger K, et al. (2014) Mapping-by-sequencing identifies HvPHYTOCHROME C as a candidate gene for the early maturity 5 locus modulating the circadian clock and photoperiodic flowering in barley. Genetics 198: 383–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhilko A, Hodge SK, Stratford K, Knox K, Edwards KD, Thomson AW, Mizuno T, Millar AJ(2010) Data assimilation constrains new connections and components in a complex, eukaryotic circadian clock model. Mol Syst Biol 6: 416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhilko A, Fernández AP, Edwards KD, Southern MM, Halliday KJ, Millar AJ(2012) The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol Syst Biol 8: 574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat R, Schwartz J, Jones MA, Sairanen I, Cheng Y, Andersson CR, Zhao Y, Ljung K, Harmer SL(2009) REVEILLE1, a Myb-like transcription factor, integrates the circadian clock and auxin pathways. Proc Natl Acad Sci USA 106: 16883–16888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat R, Takahashi N, Hsu PY, Jones MA, Schwartz J, Salemi MR, Phinney BS, Harmer SL(2011) REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet 7: e1001350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S, Mandadi KK, Boedeker AL, Rathore KS, McKnight TD(2007) Regulation of telomerase in Arabidopsis by BT2, an apparent target of TELOMERASE ACTIVATOR1. Plant Cell 19: 23–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw LM, Turner AS, Laurie DA(2012) The impact of photoperiod insensitive Ppd-1a mutations on the photoperiod pathway across the three genomes of hexaploid wheat (Triticum aestivum). Plant J 71: 71–84 [DOI] [PubMed] [Google Scholar]
- Somers DE, Schultz TF, Milnamow M, Kay SA(2000) ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101: 319–329 [DOI] [PubMed] [Google Scholar]
- Staiger D, Shin J, Johansson M, Davis SJ(2013) The circadian clock goes genomic. Genome Biol 14: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke S, Börner A(1998) Molecular mapping of the photoperiod response gene ea7 in barley. Theor Appl Genet 97: 797–800 [Google Scholar]
- Takata N, Saito S, Saito CT, Nanjo T, Shinohara K, Uemura M(2009) Molecular phylogeny and expression of poplar circadian clock genes, LHY1 and LHY2. New Phytol 181: 808–819 [DOI] [PubMed] [Google Scholar]
- Takata N, Saito S, Saito CT, Uemura M(2010) Phylogenetic footprint of the plant clock system in angiosperms: evolutionary processes of pseudo-response regulators. BMC Evol Biol 10: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B, Harmon FG(2010) Ambient temperature response establishes ELF3 as a required component of the core Arabidopsis circadian clock. Proc Natl Acad Sci USA 107: 3257–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trejo Banos D, Millar AJ, Sanguinetti G(2015) A Bayesian approach for structure learning in oscillating regulatory networks. Bioinformatics 31: 3617–3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi P, Carvallo M, Hamilton EE, Preuss S, Kay SA(2017) Arabidopsis B-BOX32 interacts with CONSTANS-LIKE3 to regulate flowering. Proc Natl Acad Sci USA 114: 172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A, Beales J, Faure S, Dunford RP, Laurie DA(2005) The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310: 1031–1034 [DOI] [PubMed] [Google Scholar]
- Vinnicombe G.(1993) Frequency domain uncertainty and the graph topology. IEEE Trans Automat Contr 38: 1371–1383 [Google Scholar]
- Wang C-Q, Guthrie C, Sarmast MK, Dehesh K(2014) BBX19 interacts with CONSTANS to repress FLOWERING LOCUS T transcription, defining a flowering time checkpoint in Arabidopsis. Plant Cell 26: 3589–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C-Q, Sarmast MK, Jiang J, Dehesh K(2015) The transcriptional regulator BBX19 promotes hypocotyl growth by facilitating COP1-mediated EARLY FLOWERING3 degradation in Arabidopsis. Plant Cell 27: 1128–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb AAR, Seki M, Satake A, Caldana C(2019) Continuous dynamic adjustment of the plant circadian oscillator. Nat Commun 10: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Anafi RC, Hughes ME, Kornacker K, Hogenesch JB(2016) MetaCycle: An integrated R package to evaluate periodicity in large scale data. Bioinformatics 32: 3351–3353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Su Z(2010) Analyzing circadian expression data by harmonic regression based on autoregressive spectral estimation. Bioinformatics 26: i168–i174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakhrabekova S, Gough SP, Braumann I, Müller AH, Lundqvist J, Ahmann K, Dockter C, Matyszczak I, Kurowska M, Druka A, et al. (2012) Induced mutations in circadian clock regulator Mat—a facilitated short-season adaptation and range extension in cultivated barley. Proc Natl Acad Sci USA 109: 4326–4331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chen Y, Wang ZY, Chen Z, Gu H, Qu LJ(2007) Constitutive expression of CIR1 (RVE2) affects several circadian-regulated processes and seed germination in Arabidopsis. Plant J 51: 512–525 [DOI] [PubMed] [Google Scholar]