Pseudo Response Regulator proteins and the Evening Complex transmit daylength information to regulate photoperiodic hypocotyl growth by directly repressing transcription of key growth regulators.
Abstract
The circadian clock measures and conveys daylength information to control rhythmic hypocotyl growth in photoperiodic conditions to achieve optimal fitness, but it operates through largely unknown mechanisms. Here, we show that Pseudo Response Regulators (PRRs) coordinate with the Evening Complex (EC), a transcriptional repressor complex within the clock core oscillator, to specifically regulate photoperiodic hypocotyl growth in Arabidopsis (Arabidopsis thaliana). Intriguingly, a distinct daylength could shift the expression phase and extend the expression duration of PRRs. Multiple lines of evidence have further demonstrated that PRRs directly bind the promoters of PHYTOCHROME-INTERACTING FACTOR4 (PIF4) and PIF5 to repress their expression, hence PRRs act as transcriptional repressors of the positive growth regulators PIF4 and PIF5. Importantly, mutation or truncation of the TIMING OF CAB EXPRESSION1 (TOC1) DNA binding domain, without compromising its physical interaction with PIFs, still caused long hypocotyl growth under short days, highlighting the essential role of the PRR-PIF transcriptional module in photoperiodic hypocotyl growth. Finally, genetic analyses have demonstrated that PIF4 and PIF5 are epistatic to PRRs in the regulation of photoperiodic hypocotyl growth. Collectively, we propose that, upon perceiving daylength information, PRRs cooperate with EC to directly repress PIF4 and PIF5 transcription together with their posttranslational regulation of PIF activities, thus forming a complex regulatory network to mediate circadian clock-regulated photoperiodic growth.
Seedlings of terrestrial flowering plants display diel rhythmic growth upon responding to recurring natural stimuli immediately after protruding from the soil. The photoperiod, i.e. the daylength, is the most prominent environmental factor that shapes plant architecture and determines growth phase transition. Photoperiod information, which reflects seasonal changes, can be processed by circadian clock-dependent mechanisms to shape the gene expression pattern, with an acrophase at a specific time of the day, and thus modulate a wide range of plant growth and developmental processes, including flowering time (Yanovsky and Kay, 2002; Valverde et al., 2004; Sawa et al., 2007; Sawa and Kay, 2011; Andrés and Coupland, 2012; Lee et al., 2017). In particular, the seedling hypocotyl displays robust growth rhythms under certain photoperiodic conditions. The length of the hypocotyl is reversely associated with daylength, which has long been considered a coordinative mechanism between the circadian clock and daily photoreception (Nozue et al., 2007; Niwa et al., 2009; Nomoto et al., 2012). Nevertheless, the regulatory network underlying this coordinative mechanism is largely unknown.
Phytochrome-interacting factors (PIFs), a group of basic helix-loop-helix transcription factors (Huq and Quail, 2002), can profile the hypocotyl photoperiodic growth dynamics and are regarded as converging regulators to explain the coincidence between external environmental cues and the circadian clock (Millar, 2016; Quint et al., 2016). Under photoperiodic conditions, the protein abundance and activity of PIFs, especially PIF4 and PIF5, are concurrently regulated by light signaling and the circadian clock via a combination of transcriptional and posttranscriptional mechanisms (Fujimori et al., 2004; Shen et al., 2007; Nusinow et al., 2011; Nakamichi et al., 2012; Nieto et al., 2015; Soy et al., 2016; Zhu et al., 2016; Martin et al., 2018). Light signals modulate PIF protein abundance by triggering physical interaction between PIFs and phytochromes and subsequent degradation of PIFs (Al-Sady et al., 2006; Shen et al., 2007), while the circadian clock mainly shapes the circadian transcriptional waves of PIF4 and PIF5 (Nusinow et al., 2011; Nakamichi et al., 2012; Nieto et al., 2015; Soy et al., 2016; Zhu et al., 2016; Martin et al., 2018). Thus, the diurnal regulation of PIF4 and PIF5 transcription plays a critical role in photoperiodic hypocotyl cell elongation. The circadian clock Evening Complex (EC), which is composed of EARLY FLOWERING4 (ELF4), ELF3, and LUX ARRHYTHMO (LUX), inhibits PIF4 and PIF5 expression in the early evening and the first part of night, thus directly allowing the circadian clock to diurnally regulate hypocotyl growth (Nusinow et al., 2011). As the transcriptional peak phase of PIF5 is ahead of PIF4 for about 2 to 4 h, when EC proteins have not yet highly accumulated, it raises a possibility that other clock components are also involved in the progressive repression of PIF4 and PIF5. Hence, the intricate regulation of PIF4 and PIF5 transcription remains to be fully unraveled (Nusinow et al., 2011; Nakamichi et al., 2012; Liu et al., 2013, 2016; Zhu et al., 2016; Martin et al., 2018).
The Arabidopsis (Arabidopsis thaliana) Pseudo Response Regulator (PRR) gene family is composed of five members (PRR9, PRR7, PRR5, PRR3, and TIMING OF CAB EXPRESSION 1 [TOC1]), each of which peaks at a specific time of day in a consecutive manner from dawn to dusk (Matsushika et al., 2000; Nakamichi et al., 2010). PRR proteins were proposed to regulate photoperiodic hypocotyl elongation mainly via two pathways. One is the transcriptional regulation of PIF4 and PIF5 by PRR5 and PRR7 (Liu et al., 2013; Nakamichi et al., 2012), and the other is the transcriptional activation activities of PIFs, which are tightly regulated by the circadian clock via physical interaction between PIFs and PRRs (Soy et al., 2016; Zhu et al., 2016; Martin et al., 2018). Currently, the underlying mechanisms of the long-hypocotyl phenotype of prr mutants in short-day (SD) conditions or in response to temperature are thought to be mainly due to their posttranscriptional regulation of PIFs via physical interaction and their antagonistic regulation of PIFs by binding to a set of cotargets including PHYTOCROME INTERACTING FACTOR 3-LIKE1 (PIL1), YUCCA8 (YUC8), and CYCLING DOF FACTOR5 (CDF5; Martin et al., 2018; Soy et al., 2016; Zhu et al., 2016). In addition, TOC1 can physically interact with ELF3 (Huang et al., 2016), the bridging protein of the EC, but it is still unclear whether ELF3 and TOC1 work in the same pathway or independently to regulate photoperiodic hypocotyl growth. Moreover, how PRRs respond to distinct daylength information at the transcriptional and posttranscriptional level and subsequently transmit photoperiod information to control hypocotyl cell elongation is still largely unknown.
Here, we show that PRRs and the EC act additively in regulating photoperiodic hypocotyl growth in Arabidopsis, and daylength information can alter the expression phase and duration of PRRs. We further unveiled PIF4 and PIF5 as direct transcriptional targets of PRRs, and their transcriptional patterns were accordingly altered by daylength information via PRRs. Importantly, by using the TOC1 DNA binding domain mutation or truncation alleles, we show that the PRR-PIF transcription module is essential for regulating hypocotyl growth in photoperiodic conditions. Together with the posttranslational regulation of PIF abundance and activity by PRRs and the EC, we thus propose a complex regulatory network that mediates circadian clock-regulated photoperiodic hypocotyl growth by a combination of transcriptional and posttranscriptional mechanisms.
RESULTS
PRRs Act Additively with the EC to Regulate Photoperiodic Hypocotyl Growth
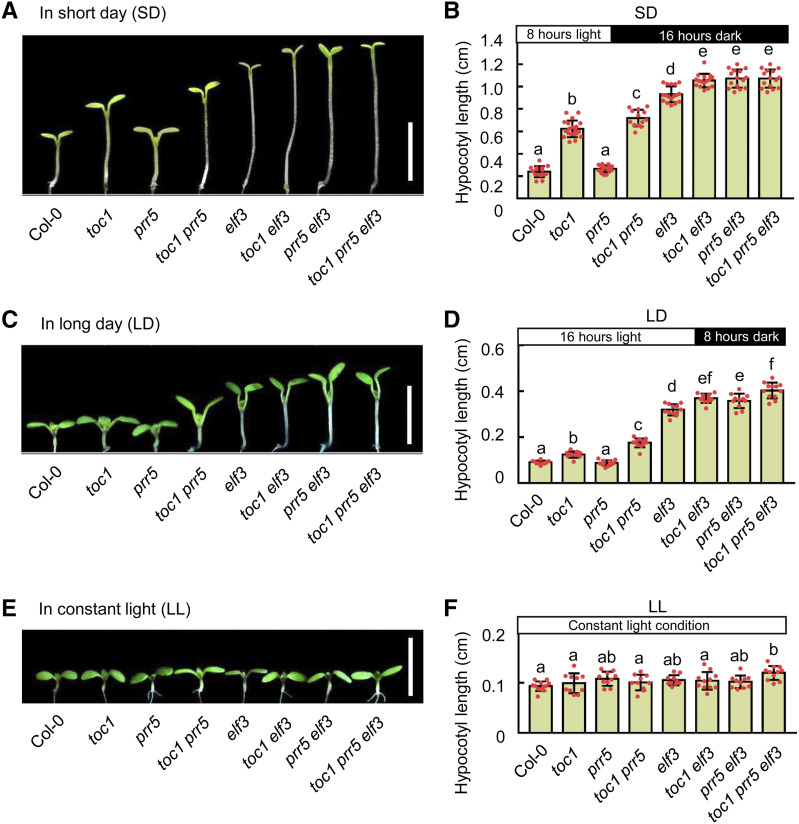
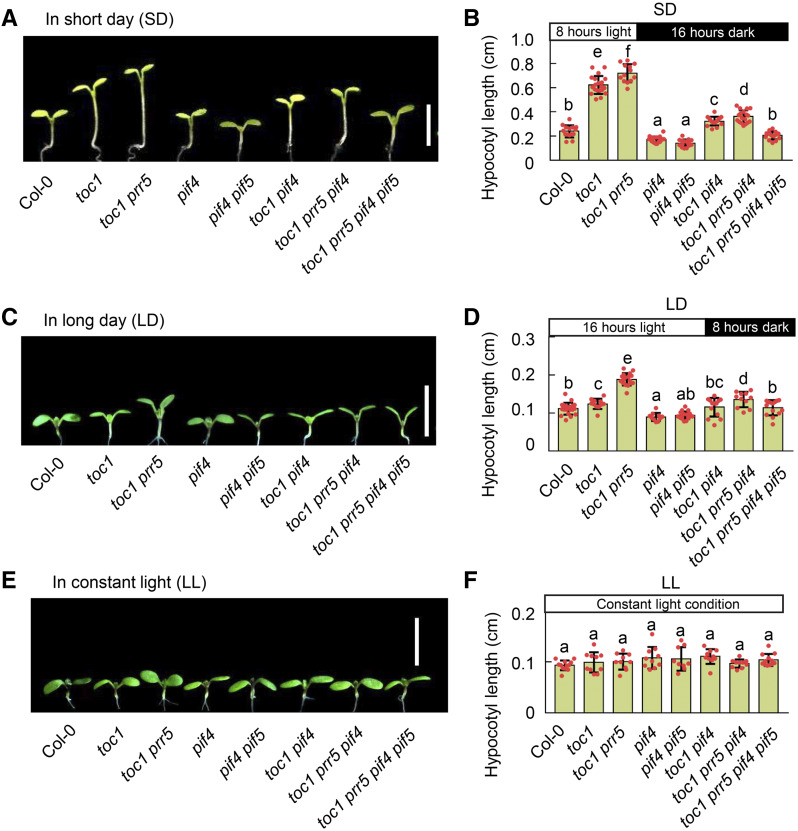
Both PRRs and the EC are involved in hypocotyl growth regulation (Sato et al., 2002; Kaczorowski and Quail, 2003; Yamamoto et al., 2003; Nusinow et al., 2011; Nieto et al., 2015; Soy et al., 2016; Zhu et al., 2016; Martin et al., 2018; Li et al., 2019). TOC1 can also physically interact with the EC component ELF3 (Huang et al., 2016). Nevertheless, the relationship between PRRs and the EC in regulating hypocotyl growth, especially under photoperiodic conditions, is unclear. To systematically address this question, we generated higher-order Arabidopsis mutants between PRRs and EC components. After growth for 5 d at different conditions, we measured the hypocotyl length and found that toc1, prr5, toc1 prr5, and elf3 mutants displayed dramatically longer hypocotyl phenotypes under both SD (8 h light/16 h dark) and long-day (LD; 16 h light/8 h dark) conditions relative to Col-0, but not under constant-light (LL) conditions (Fig. 1). Strikingly, the hypocotyls of toc1 elf3 and prr5 elf3 double mutants were significantly longer than those of the single mutants, suggesting that they act additively to regulate hypocotyl growth only under photoperiod conditions. Notably, the hypocotyl lengths of the toc1 prr5 elf3 triple mutant were modestly but significantly longer than those of the toc1 prr5 and elf3 mutants under both LD and SD conditions (Fig. 1, A–D), further supporting the notion that PRRs and the EC additively regulate hypocotyl growth. Since ELF3 has been shown to interact with PIF4 to regulate hypocotyl growth independent of the EC (Nieto et al., 2015), we further examined the genetic relationship between PRRs and the EC by using LUX, a DNA binding protein in the EC (Hazen et al., 2005; Nusinow et al., 2011). Consistently, the toc1 prr5 lux triple mutant displayed significantly longer hypocotyls than either the toc1 prr5 or lux mutants in both SD and LD conditions (Supplemental Fig. S1), which further confirmed that PRRs and the EC additively regulate photoperiodic hypocotyl growth. In addition, the transcript phases of PRR9 and PRR7 displayed an inverse pattern to that of the EC, but the hypocotyls of the prr7 prr9 double mutant were significantly longer than that of Col-0 (Nakamichi et al., 2005), specifically under photoperiodic conditions, though not in constant light (Supplemental Fig. S2). Altogether, multiple lines of genetic evidence clearly demonstrated that PRRs act additively with the EC to regulate hypocotyl growth under photoperiodic conditions.
Figure 1.
TOC1 and PRR5 coordinate with EC to regulate photoperiodic hypocotyl growth. A, C, and E, Hypocotyl phenotypes of Col-0, toc1, elf3, toc1 elf3, prr5, prr5 elf3, toc1 prr5, and toc1 prr5 elf3 seedlings grown under SD conditions (A), LD conditions (C), and continuous white light (E) for 5 d after germination. Seedling images in A, C, and E were digitally abstracted and multiple images were made into a composite for comparison. Scale bars = 5 mm. B, D, and F, Quantitative analyses of the hypocotyl length of seedlings shown in A, C, and E, respectively. Lowercase letters indicate statistically significant differences among averages as determined by Tukey’s HSD mean-separation test (P < 0.05). Data are the means ± sd of >15 seedlings.
Daylength Information Alters the Expression Patterns of PRRs and the EC
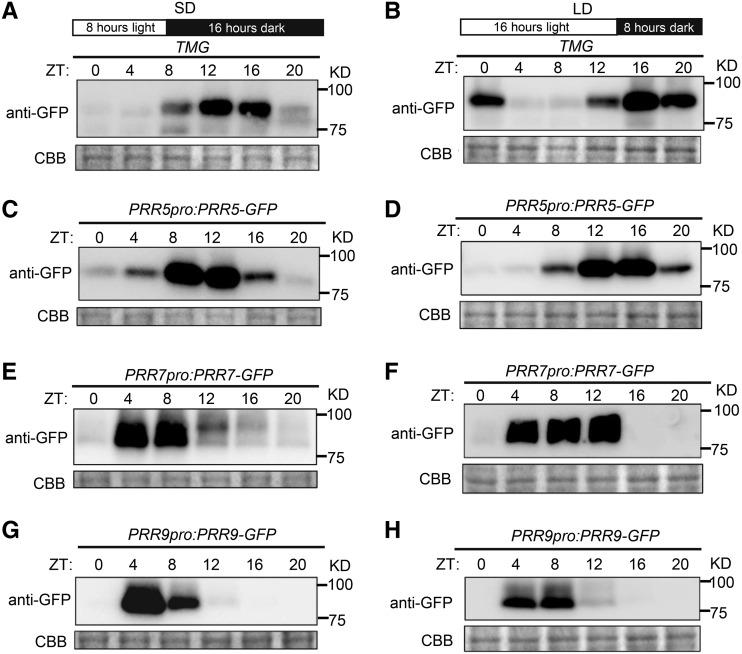
In general, the hypocotyl length decreases with increasing daylength. However, the ratio of hypocotyl length in SD to that in LD conditions was significantly increased in the toc1 mutant compared to that in prr5, elf3, or Col-0 plants (Fig. 1, A–D). This prompted us to compare the expression patterns of TOC1 and other PRR family members under SD and LD conditions. Previously, it has been shown that the transcript and protein abundance of each PRR gene peak sequentially from dawn to dusk in the order PRR9, PRR7, PRR5, PRR3, and TOC1 (Matsushika et al., 2000; Fujiwara et al., 2008; Martin et al., 2018). However, whether the distinct daylength information could change their mRNA or protein patterns remains unclear. By using a time-course reverse-transcription quantitative PCR (RT-qPCR) assay and the publicly accessible database (http://diurnal.mocklerlab.org), we found that the expression pattern of TOC1 was overall shifted by about 4 h in SD versus LD conditions, while the PRR5 mRNA expression pattern was not significantly altered by the daylength difference (Supplemental Figs. S3 and S4). Interestingly, when we compared the protein expression patterns of TOC1 and PRR5 between SD and LD conditions using previously generated TMG (the TOC1 Mini Gene driven by its native promoter) and PRR5pro:PRR5-GFP transgenic lines (Más et al., 2003; Fujiwara et al., 2008), we found that the duration and peak times of TOC1 and PRR5 proteins are highly variable under these two distinct conditions. This might have been caused by posttranscriptional regulation, given that the PRR5 mRNA pattern did not display a phase shift. Moreover, both PRR5 and TOC1 proteins were barely detectable at zeitgeber time (ZT) 20 in SD conditions but were still present at appreciable levels in LD conditions (Fig. 2, A–D). Remarkably, the high TOC1 protein level could even extend to ZT0 in the night under LD conditions (Fig. 2, A and B). In addition, the protein abundance of two other PRR family members, PRR9 and PRR7, started to rise from ZT4, and persisted over the daytime in both LD and SD conditions. The PRR7 protein was maintained at a higher level with increasing daylength (Fig. 2, E–H). Interestingly, among EC components, transcripts of LUX and ELF4 displayed a shifted pattern similar to that of TOC1 in SD conditions, while ELF3 only showed an increased expression level, without pattern shifting in SD conditions (Supplemental Fig. S4, E–G). Thus, it appeared that the daylength information could either shift the expression phase or extend the expression period of PRRs and EC at both the transcriptional and posttranscriptional levels, which might contribute to the daylength-dependent photoperiodic hypocotyl growth.
Figure 2.
PRR protein expression patterns in differential photoperiod conditions. A to H, Immunoblots showing TOC1, PRR5, PRR7, and PRR9 protein abundances in seedlings of TMG, PRR5pro:PRR5-GFP, PRR7pro:PRR7-GFP, and PRR9pro:PRR9-GFP, respectively, grown in SD or LD conditions for 10 d. Coomassie Brilliant Blue (CBB) staining indicates the protein loading amount. Data are representative of three biological replicates with similar results.
PIF4 and PIF5 Are Potential Common Transcriptional Targets of PRRs and EC
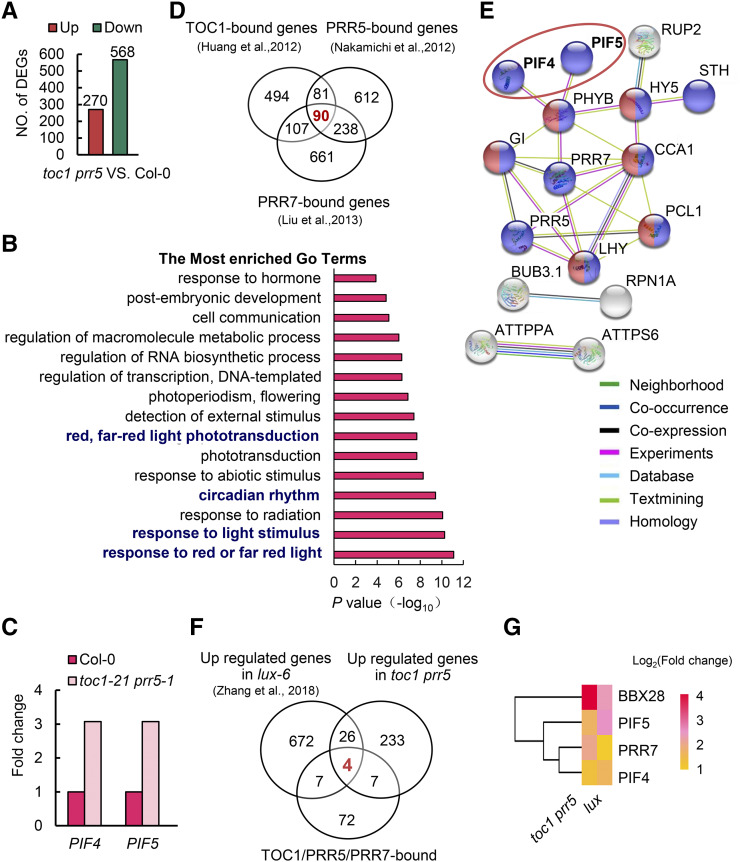
To further elucidate the underlying mechanisms of PRR coordination with the EC to regulate photoperiodic hypocotyl growth, we identified their direct transcriptional targets, as both are transcription regulators (Gendron et al., 2012; Huang et al., 2012; Nakamichi et al., 2012). RNA-sequencing (RNA-seq) with 10-d-old seedlings of toc1 prr5 grown under 12 h light/12 h dark conditions was conducted with tissues harvested at ZT15, the exact same time point used for TOC1 chromatin immunoprecipitation sequencing (ChIP-seq; Huang et al., 2012) and close to the time point for PRR5 ChIP-seq (Nakamichi et al., 2012). In total, we identified 838 differentially expressed genes (DEGs) in the toc1 prr5 double mutant using 2-fold cutoff (false discovery rate <0.05) compared to Col-0 (Fig. 3A; Supplemental Dataset 1). The randomly selected four upregulated genes and four downregulated genes validated by RT-qPCR displayed expression patterns similar to that in the RNA-seq data (Supplemental Fig. S5). Notably, CIRCADIAN CLOCK ASSOCIATED1 (CCA1), LATE ELONGATED HYPOCOTYL (LHY), GIGANTEA (GI), and some other core circadian clock genes were among the 270 upregulated genes, consistent with the fact that they are direct targets of TOC1 within the interlocked circadian clock oscillator (Huang et al., 2012). Functional assignment of the DEGs by gene ontology (GO) enrichment analysis further revealed that the DEGs were mainly involved in response to red or far-red light, response to light stimulus, circadian rhythms, and red/far-red light phototransduction (Fig. 3B), implicating a dual role for TOC1 and PRR5 in regulating the circadian clock and light signaling. Among the DEGs, we found that transcript levels of PIF4 and PIF5 were significantly increased in the toc1 prr5 mutant (Fig. 3C). Previous ChIP-seq analysis identified 772 TOC1-bound genes (Huang et al., 2012), 1,021 PRR5-bound genes (Nakamichi et al., 2012), and 1,096 PRR7-bound genes (Liu et al., 2013). As the PRRs play redundant roles in regulating photoperiodic hypocotyl growth, we thus compared the ChIP-seq data of PRR7, PRR5, and TOC1, and obtained 90 commonly bound genes (Fig. 3D; Supplemental Fig. S6). The interaction network analysis using the STRING database (http://string-db.org/) showed that the 90 common genes could form a major cluster, including known circadian clock genes, such as CCA1, LHY, and GI, and genes involved in photomorphogenesis, including PIF4, PIL6/PIF5, and PHYTOCHROME B (PHYB; Fig. 3E). The potential direct target genes of PRRs were further revealed by comparing our RNA-seq data with the PRR7/PRR5/TOC1 common target genes. Strikingly, PIF4 and PIF5 were found among the 11 genes (P < 3.5 × 10−9, hypergeometric test; Supplemental Fig. S6) overlapped between the upregulated genes in the toc1 prr5 mutant and the 90 common target genes, indicating that PIF4 and PIF5 were potential direct target genes of TOC1 and PRR5. Furthermore, when we compared these 11 overlapping genes with the upregulated genes in the lux-6 mutant, PIF4 and PIF5 were again among the only four common cotargets (Fig. 3, F and G). Hence, PIF4 and PIF5 became promising target genes of the EC and PRRs in mediating their regulation of photoperiodic hypocotyl growth.
Figure 3.
PIF4 and PIF5 are potential direct transcriptional targets of TOC1 and PRR5. A, DEGs between the toc1 prr5 mutant and wild-type Col-0 in RNA-seq. The samples were harvested at ZT15 from 10-d-old seedlings grown in 12-h-light/12-h-dark photocycles. B, GO analysis of the overlapping genes between upregulated DEGs in the toc1 prr5 mutant and the genes bound by TOC1. C, Expression profiles of PIF4 and PIF5 in the toc1 prr5 mutant. Data from RNA-seq. D, Venn diagram showing the number of common genes bound by TOC1, PRR5, and PRR7. E, Protein interaction network analysis of the 90 genes co-bound by TOC1, PRR5, and PRR7 in D using the STRING database (http://string-db.org/), showing a major cluster including PIF4, PIF5, and other known circadian core components. Colored nodes represent query proteins and the first shell of interactors, white nodes the second shell of interactors, empty nodes the proteins of unknown 3D structure, and solid nodes proteins for which some 3D structure is known or predicted. Edges represent protein-protein associations taken from curated databases (light blue) or determined by experiment (magenta), gene neighborhood (green), gene co-occurrence (dark blue), text mining (light green), coexpression (black), or protein homology (light purple). F, Venn diagram showing the number of overlapping genes among the TOC1, PRR5, and PRR7 cobound genes, upregulated DEGs in the toc1 prr5 mutant, and upregulated DEGs in the lux-6 mutant. G, Heat map showing four common cotargets in upregulated DEGs in toc1 prr5 and lux-6 mutants. The scale represents the log2 (fold change).
PRRs Directly Bind PIF4 and PIF5 Promoters to Repress Their Transcription
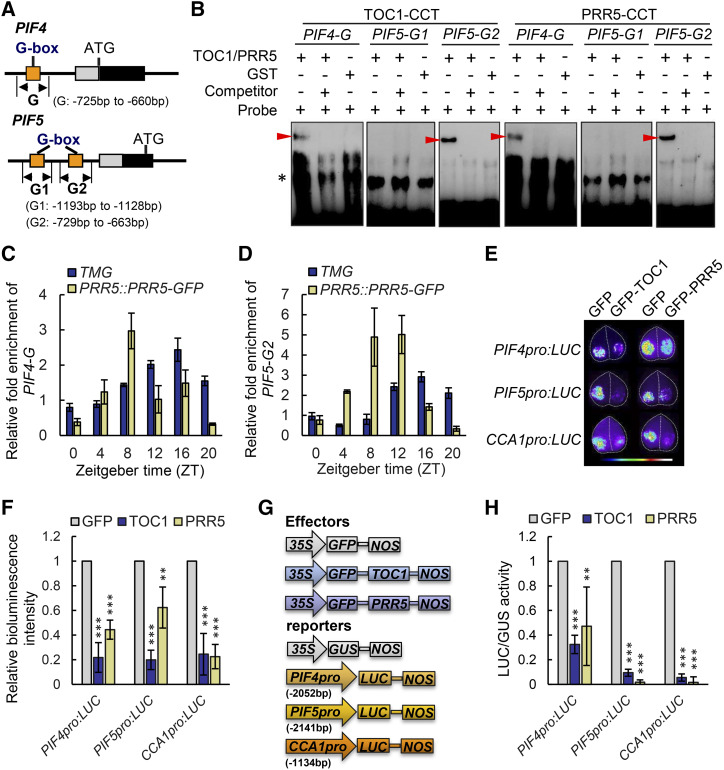
As PIF4 and PIF5 are two potential common transcriptional targets of PRRs and EC, we determined whether PRRs could directly repress PIF4 and PIF5 transcription. Promoter analysis suggested that one potential TOC1 and PRR5 binding element, PIF4-G (G-box, CACGTG; Gendron et al., 2012), was found at −707 bp upstream of the PIF4 start codon, and two G-boxes (CACGTG), PIF5-G1 and PIF5-G2, are found at −1,151 and −718 bp, respectively, upstream of the PIF5 start codon (Fig. 4A). We then conducted electrophoretic mobility shift assays (EMSAs) with the purified glutathione S-transferase (GST)-tagged CCT (CONSTANS CONSTANS-like TOC1) domain of TOC1 and PRR5, which is the DNA-binding domain of PRRs (Gendron et al., 2012). Both GST-TOC1-CCT and GST-PRR5-CCT could more efficiently bind the PIF4-G and PIF5-G2 regions compared to GST alone (Fig. 4B), and they could bind the CCA1 promoter (as a positive control; Supplemental Fig. S7A), but not the PIF5-G1 region. Importantly, the binding could be abolished by the nonlabeled competitive probe, suggesting that TOC1 and PRR5 could specifically bind the promoters of PIF4 and PIF5 (Fig. 4B; Supplemental Fig. S7A). Results of ChIP-qPCR analysis further confirmed that the amplicons containing the PIF4 promoter G-box and PIF5 promoter G2 regions were significantly enriched in TMG lines ranging from ZT12 to ZT20 and in PRR5:PRR5-GFP from ZT8 to ZT16 (Fig. 4, C and D), in line with the TMG and PRR5 protein expression window. Similar binding enrichment was observed for the amplicons for the CCA1 promoter, but not the negative control ASCORBATE PEROXIDASE3 (APX3; (Supplemental Fig. S7, B and C). These results are consistent with previous ChIP-seq studies (Huang et al., 2012; Nakamichi et al., 2012). Taken together, TOC1 and PRR5 could directly bind PIF4 and PIF5 promoters in vitro and in vivo.
Figure 4.
TOC1 and PRR5 directly bind the PIF4 and PIF5 promoters to repress their transcription. A, Schematic diagram of the promoter regions of PIF4 and PIF5. Orange boxes represent the putative G-box elements. G, G1, and G2 represent the respective DNA fragments used for generating EMSA probes and ChIP-qPCR detection. B, EMSA with the CCT domain of TOC1 and PRR5 incubated with a probe designed for the PIF4-G, PIF5-G1, and PIF5-G2 regions of the PIF4 and PIF5 genes as shown in A, and the 100-fold unlabeled competitor. GST alone was used as a negative control. Arrowheads mark the shifted bands. C and D, Time-course ChIP-qPCR assay showing that TOC1 and PRR5 bind to the PIF4-G (C) and PIF5-G2 (D) regions diurnally, which was well associated with their respective protein abundances. Data are the means ± sd. E, Transient transcriptional expression analysis showing that PIF4 and PIF5 were repressed by TOC1 and PRR5 in epidermal cells of N. benthamiana leaves. CCA1pro:LUC was used as a positive control. Data are representative of three biological replicates with similar results. Leaf images were digitally abstracted and multiple images were made into a composite for comparison. F, Quantification of bioluminescence intensity as shown in E. Data are the means ± sd. Asterisks denote statistically significant difference among means: *P < 0.05, **P < 0.01, and ***P < 0.001, determined by Student’s t test. G and H, Transient transcriptional expression assay in Arabidopsis protoplasts showing a schematic diagram of the effector and reporter vectors (G) and respective quantification of relative LUC/GUS activity (H). The relative LUC/GUS activity in protoplasts cotransformed with GFP and reporter vector was defined as one. CCA1pro:LUC was used as a positive control, while 35S:GUS was used as an internal control. Data are the means ± sd. Asterisks in H denote statistically significant differences among means: *P < 0.05, **P < 0.01, and ***P < 0.001 by Student’s t test.
Whether TOC1 and PRR5 could directly repress PIF4 and PIF5 transcription was determined by monitoring the bioluminescence signals of PIF4pro:LUC and PIF5pro:LUC using well-established transient expression systems in the leaves of Nicotiana benthamiana and in Arabidopsis protoplast. Results of the transient expression analyses clearly indicated that the transcriptional activities of PIF4 and PIF5 could be repressed by PRRs (Fig. 4, E–H; Supplemental Fig. S8). Collectively, our results supported the notion that PIF4 and PIF5 are direct transcriptional targets of PRRs.
PRRs Cooperate with the EC in Timing Photoperiodic Transcription of PIF4 and PIF5
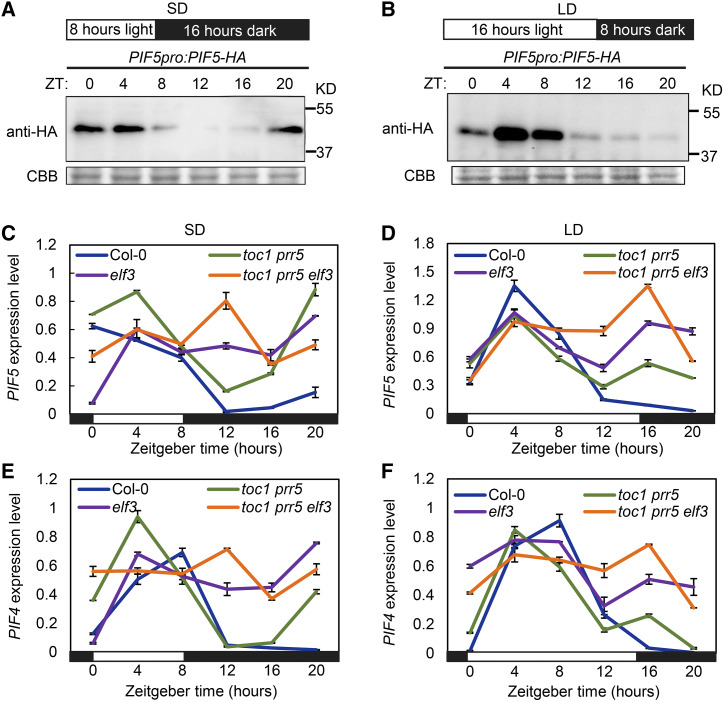
As PIF4 and PIF5 are the common transcriptional targets of PRRs and the EC, and daylength could alter the expression patterns of PRRs and the EC, we questioned whether PRR proteins could coordinate with the EC in conveying daylength information to control photoperiodic hypocotyl growth through the timing of PIF4 and PIF5 transcription. To test this, PIF5pro:PIF5-HA transgenic plants were generated to investigate the temporal protein pattern of PIF5 under SD and LD conditions. Intriguingly, the PIF5 protein abundance was inversely associated with TOC1 and PRR5 protein abundance (Fig. 2, A–D) under both SD (Fig. 5A) and LD (Fig. 5B) conditions, consistent with the idea that TOC1 and PRR5 directly repressed PIF5 transcription. Similarly, PIF4 protein has been observed to accumulate during the light period and decrease in the dark period from ZT12 to ZT20, then increase before dawn under SD conditions, but not under a 12 h light/12 h dark photoperiod. As PIF4 and PIF5 protein accumulation was well associated with their transcription, PIF4 and PIF5 transcript levels were examined in the toc1 prr5 double mutant and toc1 prr5 elf3 triple mutant. Results of RT-qPCR indicated that PIF4 and PIF5 transcript levels were similar to that of Col-0 at the subjective daytime in both toc1 prr5 and toc1 prr5 elf3 mutants, but modestly increased at the subjective early night and accrued more significantly at late night, especially at ZT20 in both photoperiodic conditions (Fig. 5, C–F). As the EC represses PIF4 and PIF5 transcription from dusk to early night, PIF4 and PIF5 transcript levels displayed a modest but consistent increase in the toc1 prr5 elf3 triple mutant compared to those in toc1 prr5 or elf3 mutants, especially under LD conditions (Fig. 5, C–F). Similarly, the transcript levels of PIF4 and PIF5 were also significantly elevated in prr7 prr9 and prr5 prr7 prr9 mutants under both SD and LD conditions (Supplemental Fig. S9). Together, our results support a notion that PRRs, in concert with the EC, repress the transcription of PIF4 and PIF5, hence to shape their transcriptional patterns in mediating circadian clock-regulated photoperiodic hypocotyl growth.
Figure 5.
TOC1 and PRR5 coordinate with the EC to transmit daylength information for shaping PIF4 and PIF5 transcription. A and B, Immunodetection of PIF5 protein levels in PIF5pro:PIF5-HA transgenic seedlings using extracts from seedlings grown in SD (A) and LD (B) conditions for 10 d. Coomassie Brilliant Blue (CBB) staining indicates the protein loading amount. Data are representative of three biological replicates with similar results. C to F, RT-qPCR analysis showing PIF5 (C and D) and PIF4 (E and F) transcript levels in Col-0, toc1 prr5, elf3, and toc1 prr5 elf3 seedlings grown for 10 d in SD (C and E) or LD (D and F) conditions. Data are the means ± sd. White and black rectangles below the graphs represent day and night, respectively.
Direct Transcriptional Inhibition of PIF4 and PIF5 by TOC1 Is Required for Its Regulation of Photoperiodic Hypocotyl Growth
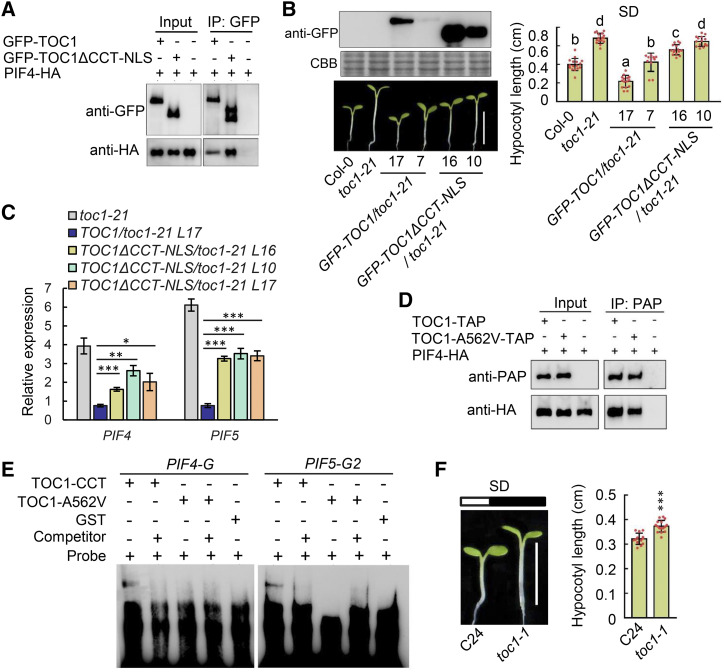
As the physical interaction of PRRs with PIFs antagonizes PIF function under a diurnal cycle (Soy et al., 2016; Zhu et al., 2016; Martin et al., 2018), a truncated TOC1 without the CCT DNA-binding domain (Gendron et al., 2012) was used to test whether PRR-mediated PIF4/5 repression was required in photoperiodic hypocotyl growth. Similar to the full-length TOC1, GFP-TOC1ΔCCT-NLS was predominantly localized in nuclear speckles both in the epidermal cells of infiltrated N. benthamiana leaves and in the hypocotyl cells of stable transgenic Arabidopsis plants (Supplemental Fig. S10). Importantly, the truncated TOC1 protein without its DNA binding domain could still physically interact with PIF4 and PIF5, with an affinity similar to that observed for full-length TOC1 (Fig. 6A; Supplemental Fig. S11A), as the CCT domain was dispensable in mediating TOC1-PIF interactions in yeast (Zhu et al., 2016). However, transcriptional repression of PIF4 and PIF5 by the truncated TOC1 protein without its CCT domain was severely compromised compared to repression by the full-length TOC1 (Supplemental Fig. S12). Notably, overexpression of full-length TOC1, but not TOC1ΔCCT, could fully rescue the long-hypocotyl phenotype of the toc1-21 mutant grown in SD conditions, even when the TOC1 ectopic expression levels were comparable to or lower than the endogenous TOC1 (Fig. 6B). Consistently, the transcript levels of PIF4 and PIF5 were significantly repressed by overexpression of full-length TOC1 but not of TOC1ΔCCT (Fig. 6C). Compared to that in toc1-21 mutants, the moderately shortened hypocotyl phenotypes in the TOC1ΔCCT transgenic lines was likely due to TOC1ΔCCT-PIF interaction and sequestration of PIF function (Soy et al., 2016; Zhu et al., 2016; Martin et al., 2018).
Figure 6.
Direct transcriptional inhibition of PIF4 and PIF5 by TOC1 is required for its regulation of photoperiodic hypocotyl growth. A, Physical interactions between TOC1, TOC1ΔCCT (1–532 amino acids)-NLS, and PIF4 in vivo were detected by coimmunoprecipitation after transient coexpression in N. benthamiana. B, Hypocotyl phenotypes of toc1-21, GFP-TOC1/toc1-21, and GFP-TOC1ΔCCT-NLS/toc1-21 transgenic seedlings grown under SD conditions for 5 d after germination. Seedling images were digitally abstracted and multiple images were made into a composite for comparison. The protein levels of GFP-TOC1 and GFP-TOC1ΔCCT-NLS in these transgenic seedlings were also detected by immunoblot (top left). Representative seedlings were photographed (bottom left), and the hypocotyl lengths of the seedlings shown were quantified (right). Scale bar = 5 mm. Data are the means ± sd of >20 seedlings. Lowercase letters indicate statistically significant differences among averages by Tukey’s HSD mean-separation test (P < 0.05). C, RT-qPCR analysis of PIF4 and PIF5 expression in toc1-21, GFP-TOC1 toc1-21, and GFP-TOC1ΔCCT-NLS toc1-21 transgenic seedlings grown for 10 d in SD conditions at ZT12. Data are the means ± sd. Asterisks denote statistically significant differences among means: *P < 0.05, **P < 0.01, and ***P < 0.001 by Student’s t test. D, Physical interaction between TOC1-A562V and PIF4 was detected by coimmunoprecipitation after they were transiently coexpressed in leaves of N. benthamiana. The immunoprecipitates with human IgG beads were analyzed by immunoblot with anti-prostatic acid phosphatase or anti-HA antibody, as indicated. E, EMSA with CCT and CCT-A562V of TOC1 and GST incubated with a probe designed to the PIF4-G and PIF5-G2 regions, and 100-fold unlabeled competitor. Arrowheads mark the shifted bands. F, Hypocotyl phenotypes of wild-type (C24 ecotype) and toc1-1 seedlings grown for 5 d in SD conditions. Representative seedlings were photographed (left) and measured (right). Data are the means ± sd of >20 seedlings. Asterisks denote statistically significant differences among means: ***P < 0.001 by Student’s t test.
A missense allele of toc1-1 caused by an A562V mutation in the TOC1 DNA binding domain (Strayer et al., 2000) was further employed to distinguish the direct transcriptional role of TOC1 on PIF4 and PIF5 from its posttranslational regulation of PIFs via sequestration. Similar to TOC1ΔCCT, the TOC1 A562V protein could still physically interact with PIF4 and PIF5 like the wild-type TOC1 (Fig. 6D; Supplemental Fig. S11B). However, the TOC1 A562V had much reduced ability to bind PIF4 and PIF5 promoters in the EMSA (Fig. 6E), similar to the results of a previous report on the binding of the CCA1 promoter by TOC1 A562V (Gendron et al., 2012). That the toc1-1 mutant still displayed long-hypocotyl phenotypes (Dowson-Day and Millar, 1999) under SD conditions (Fig. 6F) further supported the idea that the TOC1-PIF transcriptional module played a pivotal role in regulating photoperiodic hypocotyl growth.
PIF4 and PIF5 Are Epistatic to PRRs in Regulating Photoperiodic Hypocotyl Growth
As PIF4 and PIF5 are direct PRR transcriptional targets, and PRRs physically interact with PIFs to sequester their activity (Martin et al., 2018; Soy et al., 2016; Zhu et al., 2016), we proposed that PIF4 and PIF5 act as major downstream factors to mediate circadian clock-regulated photoperiodic hypocotyl growth. Thus, we determined whether PIF4 and PIF5 were required for PRR-mediated circadian clock regulation of hypocotyl elongation by generating a variety of higher-order mutants. In agreement with a previous report (Soy et al., 2016), the long-hypocotyl phenotypes in toc1 and toc1 prr5 mutants could be partially reverted by a single introgression of pif4 under either LD or SD conditions. Moreover, the long-hypocotyl phenotype in the toc1 prr5 mutant could be completely rescued to the wild-type (Col-0) level by an introgression of pif4 pif5 mutations under either LD or SD conditions (Fig. 7, A–D), indicating a redundancy of PIF4 and PIF5 in mediating photoperiodic hypocotyl growth. The hypocotyl length in various mutants, including toc1, toc1 pif4, pif4, toc1 prr5, pif4 pif5, toc1 prr5 pif4, and toc1 prr5 pif4 pif5, were indistinguishable from that of Col-0 under continuous light conditions (Fig. 7, E and F), further reinforcing the notion that the repression of PIF4 and PIF5 by PRRs at both the transcriptional and posttranscriptional levels is required to concurrently regulate photoperiodic hypocotyl growth by the circadian clock. Given a previous report showing that mutations of PIF4 and PIF5 inhibit the long hypocotyls of prr mutants (Soy et al., 2016; Martin et al., 2018) under SD conditions, our evidence further demonstrates that PIF4 and PIF5 function downstream of PRRs to mediate photoperiodic hypocotyl growth.
Figure 7.
PIF4 and PIF5 are epistatic to TOC1 and PRR5 for photoperiodic hypocotyl growth. A, C, and E, Hypocotyl phenotypes of Col-0, toc1, pif4, toc1 pif4, toc1 prr5, toc1 prr5 pif4, pif4 pif5, and toc1 prr5 pif4 pif5 seedlings (5 d after germination) grown under SD conditions (A), LD conditions (C), or continuous white light (E). Representative seedlings were photographed. Seedling images were digitally abstracted and multiple images made into a composite for comparison. Scale bars = 5 mm. B, D, and F, Hypocotyl lengths of the seedlings shown in A, C, and E, respectively, were measured and quantified. Lowercase letters indicate statistically significant differences among means as determined by Tukey’s HSD mean-separationi test (P < 0.05). Data are the means ± sd of >15 seedlings.
DISCUSSION
By sensing photoperiod, the plant circadian clock regulates a plethora of daily rhythmic physiological events (Yanovsky and Kay, 2002; Valverde et al., 2004; Sanchez and Kay, 2016). The hypocotyl displays a robust rhythmic elongation pattern under photoperiodic conditions by a coincidental mechanism between the circadian clock and external light signals (Nozue et al., 2007; Niwa et al., 2009; Nomoto et al., 2012). Nevertheless, how the circadian clock coordinates with the external photoperiod to facilitate optimal hypocotyl growth remains largely unknown. PIF4 and PIF5 have been characterized as potential targets of PRR5 and PRR7 (Liu et al., 2013; Nakamichi et al., 2012). However, the mechanisms involved in temporal transcriptional regulation of PIF4 and PIF5 by PRR proteins, especially under distinct photoperiodic cycles, are still largely unclear. In this study, we found that PRRs genetically act additively with the EC to regulate photoperiodic hypocotyl growth. We further demonstrated that PRRs directly bound the promoters of PIF4 and PIF5 to repress their transcription, and the altered temporal patterns of PRRs by daylength information could subsequently change PIF4 and PIF5 mRNA expression patterns, thus mediating photoperiodic hypocotyl growth (Fig. 8). By using specific TOC1 alleles, our results unequivocally showed that the transcriptional regulation of PIF4 and PIF5 is critical for PRR-regulated photoperiodic hypocotyl growth. In addition to posttranslational regulation of PIF abundance and activities by PRRs and ELF3 (Martin et al., 2018; Nieto et al., 2015; Soy et al., 2016; Zhu et al., 2016), here we show that PRRs cooperate with the EC to control PIF4 and PIF5 temporal transcription patterns, which mediates the crosstalk between the circadian clock and light signaling to achieve optimal hypocotyl growth and fitness under photoperiodic conditions.
Figure 8.
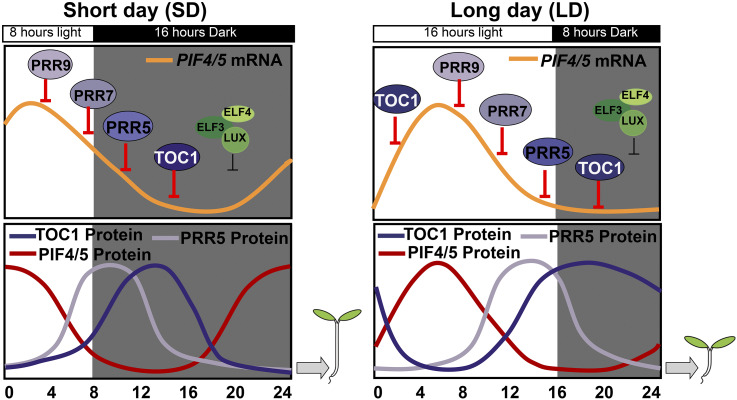
A proposed working model for PRRs-PIF4/5 transcriptional module-mediated photoperiodic hypocotyl growth. PRRs, as core circadian clock components, can directly and sequentially bind the promoters of PIF4 and PIF5 to repress their transcription in an independent manner with the Evening Complex. Diurnal rhythms of PIF4/5 protein abundance are determined by the coordination of light signaling-mediated protein stability and circadian clock-regulated transcriptional expression. Hence, TOC1 and other PRRs represent a primary molecular node between the circadian clock and photoperiod to control photoperiodic hypocotyl growth.
Sensing and transmitting daylength information have long been proposed as interplay between the circadian clock and the external photoperiod, with mainly unclear mechanisms. Hypocotyls display diel rhythmic growth patterns after emerging from the soil in natural photoperiodic conditions, but the underlying molecular mechanism remains unclear. Differential daylength information, i.e. LD versus SD, can drastically change the expression pattern and period of PRR transcripts and proteins, indicating that daylength information can be transmitted at least through PRRs and the EC via both transcriptional and posttranscriptional mechanisms. The altered expression pattern of PRRs, particularly for TOC1 and PRR5, subsequently causes altered expression of PIF4 and PIF5 transcripts and proteins, hence affecting daylength-dependent hypocotyl growth patterns (Fig. 5). That PRRs and the EC act additively in the regulation of PIF4 and PIF5 transcription could be explained by their differential binding sites within the PIF4 and PIF5 promoters, but it is not due to the physical interaction between TOC1 and ELF3 (Huang et al., 2016). Hence, the biological significance of TOC1 physically interacting with ELF3 awaits to be further explored. Intriguingly, daylength information does not alter either the transcript level or expression pattern of PRR5 (Supplemental Fig. S3B), but the overall expression pattern of PRR5 protein was shifted by ∼4 h earlier in SD conditions (Fig. 2, C and D), indicating that daylength information sensing and transmission also occurs at the posttranscriptional level for photoperiodic hypocotyl growth. A similar case has been observed for photoperiod-regulated flowering time in which the CONSTANS (CO) protein level is tightly controlled by a coincident mechanism between the circadian clock and photoperiod (Valverde et al., 2004; Song et al., 2012). It will be of great interest in future studies to decipher how daylength information affects the expression patterns of PRRs.
The expression of PIF4 and PIF5 oscillates with a peak after dawn, and then decreases gradually (Nusinow et al., 2011). The EC represses the expression of PIF4 and PIF5 at nighttime, but aside from the EC, how PIF4 and PIF5 are regulated by other circadian clock components at the transcriptional level is still not clear. Our present findings filled this knowledge gap, and we proposed that in LD conditions, the extended expression time frame and the shifted expression pattern together maximize PRR repression of PIF expression, thus inhibiting hypocotyl growth. Under SD conditions, PRR5 and TOC1 proteins do not accumulate before the subjective dawn range, from ZT20 to ZT24, which causes a high abundance of PIF4 and PIF5 to promote hypocotyl growth. Taken together, our findings revealed a key underlying mechanism by which the PRRs-PIF4/5 transcriptional module finely orchestrates circadian photoperiodic responsive hypocotyl growth in Arabidopsis.
Very recently, CCA1 and LHY, the two morning-phased circadian core components, were shown to recruit SHORT HYPOCOTYL UNDER BLUE1 (SHB1) to promote PIF4 transcription by directly binding to the PIF4 promoter (Sun et al., 2019). Our EMSA results (Figs. 3B and 5E; Supplemental Fig. S7) and previous evidence clearly demonstrated that PRRs can bind the G-box cis-elements of CCA1, PIF4, and PIF5 promoters to repress their transcription. Collectively, the transcription of PIF4 and PIF5 was intricately modulated by the circadian clock, among which CCA1 and LHY act as daytime transcriptional activators, while PRRs and the EC cooperatively act as transcription repressors to sequentially repress PIF4 and PIF5 transcription (Fig. 6C). Meanwhile, PRRs and ELF3 also inhibit the activity of PIFs at the posttranslational level by physically interacting with PIF proteins. Together, the complex regulatory network, integrating both transcriptional and posttranscriptional regulation of PRRs and EC on PIFs, collectively limits the function of PIFs from morning to early evening, to precisely time the higher growth rate in the late night. Intriguingly, GI, another key circadian clock protein, was recently reported to play a pivotal role in modulating light signaling through physical interaction with PIFs (Nohales et al., 2019). GI protein not only negatively regulates PIF protein stability, but also occupies PIF genomic target loci in the early evening (Nohales et al., 2019). Hence, it is conceivable that the circadian clock tightly coordinates photoperiodic hypocotyl growth by integrating multiple circadian mechanisms of PIF regulation at both the transcriptional and posttranscriptional levels. As PIF4 and PIF5 serve as a central cellular signaling hub by integrating phytohormones, light signaling, and circadian signals to control many downstream physiological processes, such as senescence (Song et al., 2014; Nohales et al., 2019), shade avoidance, and temperature signaling (Ma et al., 2016; Pedmale et al., 2016), it will be of great interest in the future to investigate whether the PRR-PIF4/5 transcriptional module plays other roles besides photoperiodic hypocotyl growth control.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Except where indicated, all of the Arabidopsis (Arabidopsis thaliana) plants used in this study were in the Col-0 background, including the wild type, toc1-21 (Ding et al., 2007), prr5-1 (Wang et al., 2010), prr5-1 prr7-11 (Yamashino et al., 2008), prr5-1 prr9-10 (Yamashino et al., 2008), prr7-11 prr9-10 (Yamashino et al., 2008), prr5-1 prr7-11 prr9-10 (Yamashino et al., 2008), elf3-1(Nusinow et al., 2011), lux-6 (Zhang et al., 2018), TMG (Más et al., 2003), PRR5pro:PRR5-GFP (Fujiwara et al., 2008), PRR7pro:PRR7-GFP (Fujiwara et al., 2008), PRR9pro:PRR9-GFP (Fujiwara et al., 2008), pif4-2 (Leivar et al., 2008), pif4-2 pif5-3 (CS68096). toc1-21 prr5-1, toc1-21 elf3-1, prr5-1 elf3-1, toc1-21 prr5-1 elf3-1, toc1-21 prr5-1 lux-6, toc1-21 pif4-2, toc1-21 prr5-1 pif4-2, and toc1-21 prr5-1 pif4-2 pif5-3, were generated by crossing. The sterilized Arabidopsis seeds were stratified at 4°C for 3 d, and then transferred to a 22°C growth chamber with light/dark cycles of 12 h/12 h, 16 h/8 h, or 8 h/16 h, as indicated.
Plasmids Construction
For the transient transcriptional repression assays in Nicotiana benthamiana, the amplicons of PIF4 and PIF5 promoters from ∼2,000 bp upstream of their start codons were amplified from Col-0 genomic DNA, then inserted into the promoter-free pLUC-N-1300 vector between the Pst I and Kpn I sites to generate the PIF4pro:LUC-N-1300 and PIF5pro:LUC-N-1300 constructs, respectively. To prepare the vectors of PIF4pro:LUC and PIF5pro:LUC for Arabidopsis protoplast transient expression analysis, the same sequences of PIF4 and PIF5 promoters were digested with BamH I and Bsu36 I and then cloned into the pLUC-999 vector.
Hypocotyl Length Measurements
Sterilized seeds were placed on Murashige and Skoog medium (PhytoTech, M524) for 3 d of incubation at 4°C, then incubated in specific light photoperiod conditions (12-h-light/12-h-dark, 16-h-light/8-h-dark, or 8-h-light/16-h-dark cycles; white light, 200 μmol m−2 s−1; Digital light meter, TES-1332A [TES Electrical and Electronic Corp.]) for an additional 5 d. Seedlings were photographed and hypocotyl lengths were measured using Image J software (http://rsb.info.nih.gov/ij).
Protein Detection Method for PRRs
Seedlings of TMG, PRR5pro:PRR5-GFP, PRR7pro:PRR7-GFP, and PRR9pro:PRR9-GFP transgenic lines were grown under SD or LD conditions (8 h light/16 h dark or 16 h light/8 h dark, respectively; light intensity, 200 μmol m−2 s−1; Digital light meter, TES-1332A) for 10 d, and samples were harvested at 4-h intervals during a 24-h cycle. Total proteins were extracted with immunoprecipitation buffer (50 mm Tris-C, [pH 7.5], 150 mm NaCl, 0.5% [v/v] Nonidet P-40, 1 mm EDTA, 1 mm dithiotihreitol [DTT], 1 mm phenylmethylsulfonyl fluoride [PMSF], 5 μg mL−1 leupeptin, 1 μg mL−1 aprotinin, 1 μg mL−1 pepstatin, 5 μg mL−1 antipain, 5 μg mL−1 chymostatin, 2 mm NaVO3, 2 mm NaF, 50 μm MG132, 50 μm MG115, and 50 μm ALLN [proteosome inhibitor V]). Supernatants were resolved using an 8% SDS-PAGE gel. The respective proteins were detected by western blotting using GFP antibody (Abcam; ab6556).
RNA-Seq Analysis
For the RNA-seq assays, plants were grown under 12-h-light/12-h-dark conditions at 22°C for 10 d and harvested at ZT15. RNA-sequencing and differential gene expression analyses were performed at Bionova. In brief, RNA quality was evaluated on a Bioanalyzer 2100 instrument (Agilent). Sequencing libraries were prepared following the protocol of the Directional RNA Library Prep Kit (E7760S, New England Biolabs). The 150-nucleotide (nt) paired-end high-throughput sequencing was performed on an Illumina Hiseq X TEN. Low-quality sequencing reads were removed. Clean reads were mapped to the Arabidopsis reference genome (TAIR10, www.arabidopsis.org) with Tophat2 (https://ccb.jhu.edu/software/tophat/index.shtml) software, and DEGs were identified using edgeR in the R package (http://www.bioconductor.org/packages/release/bioc/html/edgeR.html) with fold change >2 and false discovery rate <0.05 between the case group and control group samples. GO enrichment analysis was performed using TopGO in the R package (http://bioconductor.org/).
RT-qPCR for Gene Expression Analysis
Seedlings were grown under specific light photoperiod conditions (12 h light/12 h dark, 16 h light/8 h dark, or 8 h light/16 h dark; light intensity, 200 μmol m−2 s−1) for 10 d, and samples were harvested at 4-h intervals during a 24-h period. Total RNA was extracted using TRIzol Reagent (Life Technologies) as described by the manual. One microgram of RNA was used for reverse transcription with the PrimeScript RT Reagent Kit with gDNA Eraser (Takara). qPCR was performed using SYBR Green Real-Time PCR Master Mix (Toyobo) according to the manufacturer’s instructions on a QuantStudio 3 instrument (Applied Biosystems). The following PCR program was used: 95°C for 2 min, followed by 40 cycles at 95°C for 15 s, 55°C for 15 s, and 72°C for 15 s, followed by a melting-curve analysis. Gene expression was normalized by the geometric mean of ACTIN2 and TUB4 expression as previously described (Li et al., 2019). Experiments were repeated with at least two biological and two technical replicates. Data represent the means ± sd of two technical replicates. Primers used for qPCR are listed in Supplemental Table S1.
Transient Transcriptional Repression Activity Assay in N. benthamiana
Agrobacterium tumefaciens AGL carrying various fusion expression vectors (effectors GFP-TOC1, GFP-PRR5, GFP-PRR7, GFP-PRR9, or GFP; reporters PIF4pro: LUC-1300, PIF5pro: LUC-1300, and CCA1pro: LUC-1300) were cultured overnight. Each reporter vector paired with the GFP-TOC1, GFP-PRR5, GFP-PRR7, GFP-PRR9, or GFP effector vector was then cotransformed into N. benthamiana leaves using a syringe infiltration method. The luciferase signal was detected using a CCD camera (LN/1300-EB/1, Princeton Instruments) 2 d after infiltration. The bioluminescence intensity of LUC signals was quantified by MetaMorph Microscopy Automation and Image Analysis Software (Molecular Devices).
Arabidopsis Protoplast Transient Expression Analysis
Protoplasts were isolated from rosette leaves of 4-week-old Arabidopsis plants (Col-0). For transient expression assays, 200 μL of protoplast was transferred to a 2-mL microfuge tube containing 5 μg of effector plasmid, 3 μg of reporter plasmid, and 2 μg of 35S:GUS plasmid, which was used as an internal control. The effector, reporter, and GUS were cotransformed into protoplasts at a ratio of 5:3:2, and the LUC/GUS ratio was presented as normalized gene expression. PIF4pro:LUC-1300, PIF5pro:LUC-1300, and CCA1pro:LUC-1300 were used as reporters, and 35S:GFP-TOC1, 35S:GFP-PRR5, 35S:GFP-PRR7, 35S:GFP-PRR9, and 35S:GFP were used as effectors. The protoplasts were incubated for 16–24 h at 22°C. Luminescence measurements were acquired with a luciferase assay system (E1500, Promega) on a GloMax 20/20 luminometer (Promega). The GUS activity was detected with 4-methylumbelliferone glucuronide substrate (Alfa) on a GloMax 20/20 luminometer.
ChIP Assays
ChIP assays were performed using TMG and PRR5pro:PRR5-GFP transgenic lines grown under 22°C in a growth chamber with 12 h light/12 h dark cycles for 2 weeks, and seedlings were harvested at 4-h intervals during a 24-h period (ZT0, ZT4, ZT8, ZT12, ZT16, and ZT20) as noted. ChIP experiments were performed as described (Huang et al., 2012). GFP antibody (ab11120, Invitrogen) was used for immunoprecipitation. The immunoprecipitates were analyzed by qPCR. Data are presented as means ± sd from n = 3 biological replicates. Primers used in this assay are shown in Supplemental Table S1.
Purified GST-Tagged CCT Domain of TOC1 and PRR5 Proteins
GST-TOC1 or PRR5-CCT plasmids were transformed into Escherichia coli BL21 strain, induced with 1 mm isopropylthio-β-galactoside, and cultured overnight at 16°C. The cells were collected by centrifuging at 10,000 rpm for 10 min, then the cells were resuspended in 10 mL of extraction buffer (50 mm Tris-Cl, pH 8.0, 250 mm NaCl, 5 mm EDTA, 1 mm PMSF, 5 μg mL−1 leupeptin, 1 μg mL−1 aprotinin, and 1 μg mL−1 pepstatin). Lysozyme was added, and the reaction was incubated on ice for 30 min, after which 100 μL of 1 m DTT and 1 mL of 10% (w/v) sarkosyl were added and thoroughly mixed. The lysate was sonicated until it became transparent, and 2.3 mL of Triton-X-100 was added and mixed for 5 min. After centrifuging at 10,000 rpm for 10 min, the supernatant was incubated with 500 μL of GST-resin at 4°C for 3 h. The beads were washed with wash buffer (50 mm Tris-Cl [pH 8.0], 150 mm NaCl, 1 mm EDTA, 3 mm DTT, 1 mm PMSF, and 0.5% [v/v] Triton X-100) five times. The GST-resin was eluted with a reduced glutathione solution to obtain a GST-TOC1 or PRR5-CCT protein solution.
EMSA
The Lightshift Chemiluminescent EMSA kit (Thermo Scientific) was used for all assays, with 5 μL GST-TOC1-CCT, GST-PRR5-CCT, or GST protein and 0.5 μL of each biotin-labeled probe. Protein and probe were incubated in 1× Lightshift binding buffer [0.05 μg µL−1 poly(dI-dC), 2.5% (v/v) glycerol, 0.05% (v/v) Nonidet P-40, 50 mm KCl, and 5 mm MgCl2] for 1 h at 4°C on 6% gels. Gel running, transfer, and imaging were done according to the Lightshift kit directions, as previously described (Gendron et al., 2012).
Co-immunoprecipitation Assay
Agrobacteria containing 35S:TOC1-GFP or TOC1 CCT domain deletions, 35S:PRR5-GFP or PRR5 CCT domain deletions, and CsVMV:PIF4-HA or CsVMV:PIF5-HA were coinfiltrated into 4-week-old N. benthamiana leaves. The infiltrated leaves were ground to a fine powder in liquid nitrogen after infiltration for 3 d. Total protein was extracted with ice-cold immunoprecipitation buffer (50 mm Tris-Cl [pH 7.5], 150 mm NaCl, 0.5% [v/v] Nonidet P-40, 1 mm EDTA, 1 mm DTT, 1 mm PMSF, 5 μg mL−1 leupeptin, 1 μg mL−1 aprotinin, 1 μg mL−1 pepstatin, 5 μg mL−1 antipain, 5 μg mL−1 chymostatin, 2 mm NaVO3, 2 mm NaF, 50 μm MG132, 50 μm MG115, and 50 μm ALLN). The cleared supernatant was incubated with Protein A beads (15918-014, Invitrogen) with captured anti-GFP (ab11120, Invitrogen) antibody at 4°C for 2 h. The immune complex was released from the resin by 6× SDS loading buffer. Supernatants were resolved using an 8% SDS-PAGE gel. GFP-tagged TOC1 and PRR5 and hemagglutinin (HA)-tagged PIF4 and PIF5 were detected by western blotting using GFP antibody (ab6556, Abcam) and HA antibody (3F10, Roche), respectively.
Statistical Analysis
Differences between means were statistically analyzed by one-way ANOVA using Tukey’s honestly significant difference (HSD) mean-separation test (IBM SPSS Statistics Software) or Student’s t test (Microsoft Excel), as indicated in the figure legends. Statistically significant differences were defined as those with P < 0.05. Significance levels are indicated as *P < 0.05, **P < 0.01, and ***P < 0.001.
Accession Numbers
The Arabidopsis Genome Initiative numbers for the genes mentioned in this article are as follows: AT5G61380 (TOC1), AT5G24470 (PRR5), AT5G02810 (PRR7), AT2G46790 (PRR9), AT2G43010 (PIF4), AT3G59060 (PIF5), AT4G28720 (YUC8), AT3G15540 (IAA19), AT4G16780 (ATHB2), AT2G25930 (ELF3), AT2G40080 (ELF4), AT3G46640 (LUX). RNA-seq data reported in this study have been deposited in the Gene Expression Omnibus database under accession number GSE99290.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. TOC1 and PRR5 regulate photoperiodic hypocotyl growth independent of LUX.
Supplemental Figure S2. The hypocotyl phenotypes of prr57, prr59, prr79, and prr579 mutants in different photoperiod conditions.
Supplemental Figure S3. Time-course expression pattern of TOC1/PRR5 in SD or LD conditions.
Supplemental Figure S4. Time-course expression pattern of PRRs and EC components in SD or LD conditions.
Supplemental Figure S5. Validation of RNA-seq results by RT-qPCR.
Supplemental Figure S6. PIF4 and PIF5 were found among the 11 overlapping genes between upregulated genes in the toc1 prr5 mutant and genes cobound by TOC1, PRR5, and PRR7.
Supplemental Figure S7. TOC1 and PRR5 bind the CCA1 promoter but not the APX3 promoter.
Supplemental Figure S8. PRR7 and PRR9 directly repress PIF4 and PIF5 transcription.
Supplemental Figure S9. The transcriptional pattern of PIF4 and PIF5 in prr mutants under different photoperiod conditions.
Supplemental Figure S10. Subcellular localization of GFP-TOC1 and GFP-TOC1ΔCCT-NLS proteins.
Supplemental Figure S11. Physical interactions between TOC1, TOC1ΔCCT, TOC1-A562V, and PIF5.
Supplemental Figure S12. Transcriptional inhibition of PIF4 and PIF5 by TOC1ΔCCT was significantly attenuated.
Supplemental Table S1. Primers used in this study.
Supplemental Dataset S1. DEGs in the toc1 prr5 double mutant identified by RNA-seq.
Acknowledgments
We are grateful to Jyan Chyun Jang and David E. Somers (Ohio State University) for their constructive and critical comments on this manuscript, and we thank Jingquan Li (Key Laboratory of Plant Molecular Physiology and Plant Science Facility of the Institute of Botany, Chinese Academy of Sciences) for excellent technical assistance with confocal microscopy.
Footnotes
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (CAS; grant no. XDB27030206), the National Natural Sciences Foundation of China (grant nos. 31670290 and 31570292 to L.W.), and the Youth Innovation Promotion Association CAS (grant no. 2017110 to Y.Z.).
Articles can be viewed without a subscription.
References
- Al-Sady B, Ni W, Kircher S, Schäfer E, Quail PH(2006) Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell 23: 439–446 [DOI] [PubMed] [Google Scholar]
- Andrés F, Coupland G(2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13: 627–639 [DOI] [PubMed] [Google Scholar]
- Dowson-Day MJ, Millar AJ(1999) Circadian dysfunction causes aberrant hypocotyl elongation patterns in Arabidopsis. Plant J 17: 63–71 [DOI] [PubMed] [Google Scholar]
- Ding Z, Doyle MR, Amasino RM, Davis SJ(2007) A complex genetic interaction between Arabidopsis thaliana TOC1 and CCA1/LHY in driving the circadian clock and in output regulation. Genetics 176: 1501–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori T, Yamashino T, Kato T, Mizuno T(2004) Circadian-controlled basic/helix-loop-helix factor, PIL6, implicated in light-signal transduction in Arabidopsis thaliana. Plant Cell Physiol 45: 1078–1086 [DOI] [PubMed] [Google Scholar]
- Fujiwara S, Wang L, Han L, Suh SS, Salomé PA, McClung CR, Somers DE(2008) Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J Biol Chem 283: 23073–23083 [DOI] [PubMed] [Google Scholar]
- Gendron JM, Pruneda-Paz JL, Doherty CJ, Gross AM, Kang SE, Kay SA(2012) Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc Natl Acad Sci USA 109: 3167–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen SP, Schultz TF, Pruneda-Paz JL, Borevitz JO, Ecker JR, Kay SA(2005) LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc Natl Acad Sci USA 102: 10387–10392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Alvarez S, Bindbeutel R, Shen Z, Naldrett MJ, Evans BS, Briggs SP, Hicks LM, Kay SA, Nusinow DA(2016) Identification of evening complex associated proteins in Arabidopsis by affinity purification, and mass spectrometry. Mol Cell Proteomics 15: 201–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Pérez-García P, Pokhilko A, Millar AJ, Antoshechkin I, Riechmann JL, Mas P(2012) Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336: 75–79 [DOI] [PubMed] [Google Scholar]
- Huq E, Quail PH(2002) PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J 21: 2441–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczorowski KA, Quail PH(2003) Arabidopsis PSEUDO-RESPONSE REGULATOR7 is a signaling intermediate in phytochrome-regulated seedling deetiolation and phasing of the circadian clock. Plant Cell 15: 2654–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BD, Kim MR, Kang MY, Cha JY, Han SH, Nawkar GM, Sakuraba Y, Lee SY, Imaizumi T, McClung CR, et al. (2017) The F-box protein FKF1 inhibits dimerization of COP1 in the control of photoperiodic flowering. Nat Commun 8: 2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Al-Sady B, Carle C, Storer A, Alonso JM, Ecker JR, Quail PH(2008) The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20: 337–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Wang Y, Zhang Y, Tian W, Chong K, Jang JC, Wang L(2019) PRR5, 7 and 9 positively modulate TOR signaling-mediated root cell proliferation by repressing TANDEM ZINC FINGER 1 in Arabidopsis. Nucleic Acids Res 47: 5001–5015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Carlsson J, Takeuchi T, Newton L, Farré EM(2013) Direct regulation of abiotic responses by the Arabidopsis circadian clock component PRR7. Plant J 76: 101–114 [DOI] [PubMed] [Google Scholar]
- Liu TL, Newton L, Liu MJ, Shiu SH, Farré EM(2016) A G-box-like motif is necessary for transcriptional regulation by circadian pseudo-response regulators in Arabidopsis. Plant Physiol 170: 528–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Li X, Guo Y, Chu J, Fang S, Yan C, Noel JP, Liu H(2016) Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc Natl Acad Sci USA 113: 224–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Rovira A, Veciana N, Soy J, Toledo-Ortiz G, Gommers CMM, Boix M, Henriques R, Minguet EG, Alabadi D, et al. (2018) Circadian waves of transcriptional repression shape PIF-regulated photoperiod-responsive growth in Arabidopsis. Curr Biol 28: 311–318 [DOI] [PubMed] [Google Scholar]
- Más P, Kim WY, Somers DE, Kay SA(2003) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426: 567–570 [DOI] [PubMed] [Google Scholar]
- Matsushika A, Makino S, Kojima M, Mizuno T(2000) Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: Insight into the plant circadian clock. Plant Cell Physiol 41: 1002–1012 [DOI] [PubMed] [Google Scholar]
- Millar AJ.(2016) The intracellular dynamics of circadian clocks reach for the light of ecology and evolution. Annu Rev Plant Biol 67: 595–618 [DOI] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua NH, Sakakibara H(2010) PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22: 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Kamioka M, Suzuki T, Yamashino T, Higashiyama T, Sakakibara H, Mizuno T(2012) Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc Natl Acad Sci USA 109: 17123–17128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kita M, Ito S, Yamashino T, Mizuno T(2005) PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol 46: 686–698 [DOI] [PubMed] [Google Scholar]
- Nieto C, López-Salmerón V, Davière JM, Prat S(2015) ELF3-PIF4 interaction regulates plant growth independently of the Evening Complex. Curr Biol 25: 187–193 [DOI] [PubMed] [Google Scholar]
- Niwa Y, Yamashino T, Mizuno T(2009) The circadian clock regulates the photoperiodic response of hypocotyl elongation through a coincidence mechanism in Arabidopsis thaliana. Plant Cell Physiol 50: 838–854 [DOI] [PubMed] [Google Scholar]
- Nohales MA, Liu W, Duffy T, Nozue K, Sawa M, Pruneda-Paz JL, Maloof JN, Jacobsen SE, Kay SA(2019) Multi-level modulation of light signaling by GIGANTEA regulates both the output and pace of the circadian clock. Dev Cell 49: 840–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomoto Y, Kubozono S, Miyachi M, Yamashino T, Nakamichi N, Mizuno T(2012) A circadian clock- and PIF4-mediated double coincidence mechanism is implicated in the thermosensitive photoperiodic control of plant architectures in Arabidopsis thaliana. Plant Cell Physiol 53: 1965–1973 [DOI] [PubMed] [Google Scholar]
- Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN(2007) Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361 [DOI] [PubMed] [Google Scholar]
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farré EM, Kay SA(2011) The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedmale UV, Huang SC, Zander M, Cole BJ, Hetzel J, Ljung K, Reis PAB, Sridevi P, Nito K, Nery JR, et al. (2016) Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 164: 233–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M, Delker C, Franklin KA, Wigge PA, Halliday KJ, van Zanten M(2016) Molecular and genetic control of plant thermomorphogenesis. Nat Plants 2: 15190. [DOI] [PubMed] [Google Scholar]
- Sanchez SE, Kay SA(2016) The plant circadian clock: From a simple timekeeper to a complex developmental manager. Cold Spring Harb Perspect Biol 8: a027748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato E, Nakamichi N, Yamashino T, Mizuno T(2002) Aberrant expression of the Arabidopsis circadian-regulated APRR5 gene belonging to the APRR1/TOC1 quintet results in early flowering and hypersensitiveness to light in early photomorphogenesis. Plant Cell Physiol 43: 1374–1385 [DOI] [PubMed] [Google Scholar]
- Sawa M, Kay SA(2011) GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proc Natl Acad Sci USA 108: 11698–11703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M, Nusinow DA, Kay SA, Imaizumi T(2007) FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318: 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Khanna R, Carle CM, Quail PH(2007) Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol 145: 1043–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Yang C, Gao S, Zhang W, Li L, Kuai B(2014) Age-triggered and dark-induced leaf senescence require the bHLH transcription factors PIF3, 4, and 5. Mol Plant 7: 1776–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T(2012) FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336: 1045–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soy J, Leivar P, González-Schain N, Martín G, Diaz C, Sentandreu M, Al-Sady B, Quail PH, Monte E(2016) Molecular convergence of clock and photosensory pathways through PIF3-TOC1 interaction and co-occupancy of target promoters. Proc Natl Acad Sci USA 113: 4870–4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Más P, Panda S, Kreps JA, Kay SA(2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771 [DOI] [PubMed] [Google Scholar]
- Sun Q, Wang S, Xu G, Kang X, Zhang M, Ni M(2019) SHB1 and CCA1 interaction desensitizes light responses and enhances thermomorphogenesis. Nat Commun 10: 3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G(2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006 [DOI] [PubMed] [Google Scholar]
- Wang L, Fujiwara S, Somers DE(2010) PRR5 regulates phosphorylation, nuclear import and subnuclear localization of TOC1 in the Arabidopsis circadian clock. EMBO J 29: 1903–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Sato E, Shimizu T, Nakamich N, Sato S, Kato T, Tabata S, Nagatani A, Yamashino T, Mizuno T(2003) Comparative genetic studies on the APRR5 and APRR7 genes belonging to the APRR1/TOC1 quintet implicated in circadian rhythm, control of flowering time, and early photomorphogenesis. Plant Cell Physiol 44: 1119–1130 [DOI] [PubMed] [Google Scholar]
- Yamashino T, Ito S, Niwa Y, Kunihiro A, Nakamichi N, Mizuno T(2008) Involvement of Arabidopsis clock-associated pseudo-response regulators in diurnal oscillations of gene expression in the presence of environmental time cues. Plant Cell Physiol 49: 1839–1850 [DOI] [PubMed] [Google Scholar]
- Yanovsky MJ, Kay SA(2002) Molecular basis of seasonal time measurement in Arabidopsis. Nature 419: 308–312 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang Y, Wei H, Li N, Tian W, Chong K, Wang L(2018) Circadian Evening Complex represses jasmonate-induced leaf senescence in Arabidopsis. Mol Plant 11: 326–337 [DOI] [PubMed] [Google Scholar]
- Zhu JY, Oh E, Wang T, Wang ZY(2016) TOC1-PIF4 interaction mediates the circadian gating of thermoresponsive growth in Arabidopsis. Nat Commun 7: 13692. [DOI] [PMC free article] [PubMed] [Google Scholar]