In grapevines, a microRNA-encoded peptide specifically increases the expression of the miRNA that encodes it and enhances the number of adventitious roots of tissue culture plantlets.
Abstract
One of the biggest challenges in clonal propagation of grapevine (Vitis vinifera) is difficulty of rooting. Adventitious root initiation and development are the critical steps in the cutting and layering process of grapevine, but the molecular mechanism of these processes remains unclear. Previous reports have found that microRNA (miRNA)-encoded peptides (miPEPs) can regulate plant root development by increasing the transcription of their corresponding primary miRNA. Here, we report the role of a miPEP in increasing adventitious root formation in grapevine. In this study, we performed a global analysis of miPEPs in grapevine and characterized the function of vvi-miPEP171d1, a functional, small peptide encoded by primary-miR171d. There were three small open reading frames in the 500-bp upstream sequence of pre-miR171d. One of them encoded a small peptide, vvi-miPEP171d1, which could increase the transcription of vvi-MIR171d. Exogenous application of vvi-miPEP171d1 to grape tissue culture plantlets promoted adventitious root development by activating the expression of vvi-MIR171d. Interestingly, neither exogenous application of the vvi-miPEP171d1 peptide nor overexpression of the vvi-miPEP171d1 coding sequence resulted in phenotypic changes in Arabidopsis (Arabidopsis thaliana). Similarly, application of synthetic ath-miPEP171c, the small peptide encoded by the Arabidopsis ortholog of vvi-MIR171d, inhibited the growth of primary roots and induced the early initiation of lateral and adventitious roots in Arabidopsis, while it had no effect on grape root development. Our findings reveal that miPEP171d1 regulates root development by promoting vvi-MIR171d expression in a species-specific manner, further enriching the theoretical research into miPEPs.
A type of small proteins with two to 100 amino acids are peptides (Tavormina et al., 2015). Although most reported plant peptides are derived from nonfunctional precursors, different studies have pointed out the existence of peptides originating from function proteins (Schmelz et al., 2006; Pearce et al., 2010; Chen et al., 2014). Peptides play diverse roles in plant growth, development, and environmental response by interfering with signaling cascades or as important messengers in cell-to-cell communication (Murphy et al., 2012; Qu et al., 2015). In addition, plant peptides have been recently used as a new kind of fertilizer in fruit trees such as litchi (Litchi chinensis) and mango (Mangifera indica; Zhou et al., 2009; Li et al., 2010).
Similar to protein-coding genes, microRNAs (miRNAs) are initially transcribed by RNA polymerase II with a multisubunit complex to form a primary miRNA (pri-miRNA; Lee et al., 2004; Kim et al., 2011) and then subjected to RNA splicing (Bielewicz et al., 2013) and modification with a 5′-7mGTP-cap and a 3′-polyadenylated tail (Xie et al., 2005). Another feature similar to coding genes is that the promoter of miRNAs contains TATA box cis-elements, which are recognized for the assembly of the RNA polymerase II preinitiation complex (Xie et al., 2005; Megtam et al., 2006). The transcription of MIRNA genes with postprocessing modifications leads to the production of pri-miRNAs containing a few hundred bases that harbor a typical stem loop structure. The structured region of the transcript surrounding the miRNA sequence is recognized and processed by Dicer-like1 in plants (Tang et al., 2003; Kurihara and Watanabe, 2004). Subsequently, the miRNA:miRNA* duplex that comprises the mature miRNA and a fragment sequence from the opposing arm with similar size (miRNA*) is transported into the cytoplasm (Bartel, 2004). Then, the miRNA* is degraded, and the miRNA negatively regulates gene expression based on the complementarity between the miRNA and the target sequence by two possible mechanisms: transcript cleavage or translational inhibition (Brodersen et al., 2008; Reis et al., 2015).
It is generally thought that the sequences upstream and downstream of the stem-loop region are useless and rapidly degraded after the excision of the pre-miRNA. However, an increasing number of reports indicate that there is a type of peptide translated from short open reading frames (sORFs) in the 5′ leader sequence of an mRNA, pri-miRNA, or other transcripts (Hanada et al., 2013; von Arnim et al., 2014; Lauressergues et al., 2015). Although this type of peptide maturation process has not been investigated in detail, its effect on plant morphogenesis or regulatory functions has been proven (Hanada et al., 2013; von Arnim et al., 2014; Lauressergues et al., 2015). The peptides produced from pri-miRNA are called miRNA-encoded peptides (miPEPs; Lauressergues et al., 2015). These miPEPs can promote the transcription of their corresponding pri-miRNAs rather than enhance miRNA stability, thereby increasing mature miRNA accumulation (Lauressergues et al., 2015; Couzigou et al., 2017).
As the underground organs of plants, the function of roots is to fix terrestrial plants in soil and absorb water and nutrients from soil. Therefore, roots are essential for plant survival and growth. With the rapid development of modern biotechnology, plant tissue culture and cutting techniques have made great progress. Meanwhile, many problems have arisen, such as difficulty in rooting for some species. As an important fruit-bearing crop, grapevine (Vitis vinifera) varieties are widely planted around the world, and the main propagation method is vegetative propagation, including techniques such as cutting and layering. However, the root formation ability of different grapevine varieties differs, and the state of root development also varies, which are extremely unfavorable for the development and resistance of grapevine. The root systems produced by grapevine cuttings and layering consist mainly of adventitious roots and lateral roots. Therefore, the study of the molecular mechanisms of adventitious root formation and development has important theoretical and practical value for the development of the grape industry.
Adventitious root formation is a complex process in which roots emerge from stems, leaves, or hypocotyls. Adventitious roots may develop spontaneously or upon environmental stress or hormonal treatment and are crucial for clonal propagation (da Costa et al., 2018). With the continuous improvement of high-throughput sequencing technology and the deepening of miRNA research, functional studies of miRNAs in root formation and development have been carried out in various plants, such as maize (Zea mays), tomato (Solanum lycopersicum), and rice (Oryza sativa; Ma et al., 2013; Kong et al., 2014; Lakhotia et al., 2014). Among these miRNAs, miR171, a conserved miRNA family, regulates scarecrow-like (SCLs) genes belonging to the GRAS (GAI [gibberellin insensitive], RGA [repressor gibberellin], and SCR [scarecrow]) family (Bolle, 2004) and has been reported to play an important role in root formation and growth (Li et al., 2019). GRAS family members participate in a variety of developmental processes, including determining shoot and root cell fate (Carlsbecker et al., 2010) and controlling meristem maintenance (Stuurman et al., 2002; Schulze at al., 2010; Wang et al., 2010; Engstrom et al., 2011; Curaba et al., 2013; Huang et al., 2017; Jiang et al., 2018). Four HAM (hair meristem) copies belonging to the GRAS family exist in Arabidopsis (Arabidopsis thaliana), AtHAM1, AtHAM2, and AtHAM3 mRNAs are targeted by miR171, and Atham1, Atham2, and Atham3 mutants exhibit a substantial reduction in elongation of the primary root (Engstrom et al., 2011). In apple (Malus domestica) rootstock, miR171/HAM may interact with WOX (WUSCHEL-related homeobox) to participate in cell fate determination during adventitious root formation (Li et al., 2019). In Medicago and Arabidopsis, it has been shown that the different miR171 members participate in root stem cell niche maintenance by restricting the translation of HAM mRNA (Wang et al., 2010; Lauressergues et al., 2015).
miPEPs increase the expression of their corresponding mature miRNA by stimulating the transcription of pri-miRNA (Lauressergues et al., 2015). Synthetic miPEPs have been proven to modify root development. For example, miPEP165a, encoded by pri-miR165a of Arabidopsis, could inhibit the growth of lateral roots and promote the growth of primary roots, and in Medicago truncatula, miPEP171b could reduce lateral root formation and modify root development and the symbiotic relationship between mycorrhizae and roots (Lauressergues et al., 2015; Couzigou et al., 2017). Although few reports about miPEPs are available, the discovery of miPEPs reveals a new layer of gene regulation and may present a key advantage in agronomy. More experiments are needed to verify the existence of the miPEPs in other species.
In this study, by detecting the temporal and spatial expression of four vvi-MIR171genes during adventitious root development of grapevine cultivar Muscat Hamburg cuttings, we found that vvi-MIR171d had a different expression pattern from the other vvi-MIR171 genes. To study the function of vvi-MIR171d, we first analyzed the 500-bp sequence upstream of premiR171d and found three sORFs. Through transient expression and promoter activity assays driven by different lengths upstream of premiR171d, we found that the peptide encoded by the first sORF could increase the expression of vvi-MIR171d, and we named it vvi-miPEP171d1. External application of synthetic vvi-miPEP171d1 showed that it could increase adventitious root number in grape tissue culture plantlets but had no effect on Arabidopsis. Our results indicated that vvi-miPEP171d1 had a role in the formation of adventitious roots in grapevine. Overall, we demonstrated the presence of a functional miPEP in grapevine, providing a theoretical basis for studying miPEPs in grapevine.
RESULTS
Identification of miPEPs in Grapevine
Mature miRNAs are formed from the stem loop regions of long primary transcripts of MIRNA genes as templates by RNA polymerase II, so the production of mature miRNAs is dependent on the presence of pri-miRNAs (Jonesrhoades et al., 2006). Recent studies have found that there are sORFs in plant pri-miRNA sequences that can encode a class of regulatory short peptides (miPEPs; Lauressergues et al., 2015). To explore whether there are short peptides encoded by pri-miRNAs in grapevine, first we need to know the characteristic of reading frames in pri-miRNA sequences, especially upstream of pre-miRNA. The positions of the 159 grape miRNAs on the chromosomes reported in the miRBase database were used to BLAST against the grape genome to obtain a certain length of vvi-MIRNA sequence. Due to the different lengths of pri-miRNAs, to reflect the reading frame characteristics of all pri-miRNAs as accurately as possible, the 500-bp sequence upstream of pre-miRNAs was used as the prediction range. Twenty vvi-MIRNA genes were randomly selected, forward primers were designed within 500- to 600-bp sequences upstream of the corresponding precursor, and reverse primers were designed at the 3′ end of the precursor (Supplemental Fig. S1A). The results showed that 16 vvi-MIRNAs could be identified from the complementary DNA (cDNA) synthesized from RNA extracted from multiple tissues (Supplemental Fig. S1B). Thus, we analyzed the possible transcription initiation sites in the 500 bp upstream of the pre-miRNAs using DNAMAN6.0. Multiple transcription initiation sites were identified in the pri-miRNAs. Excluding the reading frames with more than 100 or less than two amino acids, the number of transcription initiation sites in the pri-miRNAs ranged from one to 13 (Fig. 1; Supplemental Table S1).
Figure 1.
Positions of predicted miPEPs on the Vitis vinifera chromosomes. Chr1 to Chr19 represent chromosome number from chromosome 1 to chromosome 19. The numbers in parentheses represent the number of predicted miPEPs encoded by the small reading frame in the 500 bp upstream of the pre-miRNA at this position, and the numbers left side of chromosomes represent the starting positions of the miRNAs on that chromosome.
If there were miPEPs in grapevine, it could be speculated that a part of pri-miRNAs in grape cells would be used to form mature miRNA, and the other part would be transported to the cytoplasm to participate in the translation process. To validate this hypothesis, the selected 20 vvi-MIRNAs were first confirmed by PCR and sequencing (Supplemental Fig. S1C), and then reverse transcription quantitative PCR (RT-qPCR)was performed to analyze their abundance using primers designed to target the pri-miRNA and pre-miRNA sequences, respectively (Supplemental Fig. S2A). The results showed that the relative content of almost all the pri-miRNAs in the cells was lower than that of the corresponding pre-miRNAs (Supplemental Fig. S2B). This might have resulted from the fact that pri-miRNA takes a relatively long time to form pre-miRNA by shearing and modification before mature miRNA takes shape. Therefore, the different abundance levels of the two existing sequences after vvi-MIRNA transcription indicated that pri-miRNAs and pre-miRNAs might perform different functions in grapevine.
Analysis of the miR171 Family in Grapevine
The conserved and ancient miRNA family (Zhang et al., 2006), miR171, targeted SCL (SCR-like) family members was first reported in Arabidopsis (Llave and Carrington, 2002) and then reported in other species (Huang et al., 2017; Li et al., 2017), indicating complex functions for this miRNA family. Here, the vvi-miR171 family members identified in the miRBase database were mapped on grape chromosomes: vvi-miR171b and vvi-miR171c were distributed on chromosome 12, vvi-miR171h and vvi-miR171i were located on chromosome 17, and five additional members were distributed on five different chromosomes. In addition, vvi-miR171j could not be found on any chromosome (Fig. 1). The phylogenetic analysis of the MIR171 gene family from 17 species showed six branches, and the vvi-MIR171 family members were scattered on four branches. Vvi-MIR171i had a closer evolutionary relationship to some MIR171 family members from rapeseed (Brassica napus), and vvi-MIR171b was closer to a MIR171 family member from black cottonwood (Populus trichocarpa; Supplemental Fig. S3), indicating that the evolutionary distance in the MIR171 gene family was not related to the genetic relationships between the species itself; they had their special evolutionary process.
Expression Patterns of MIR171 Family Members and Their Targets in Grapevine
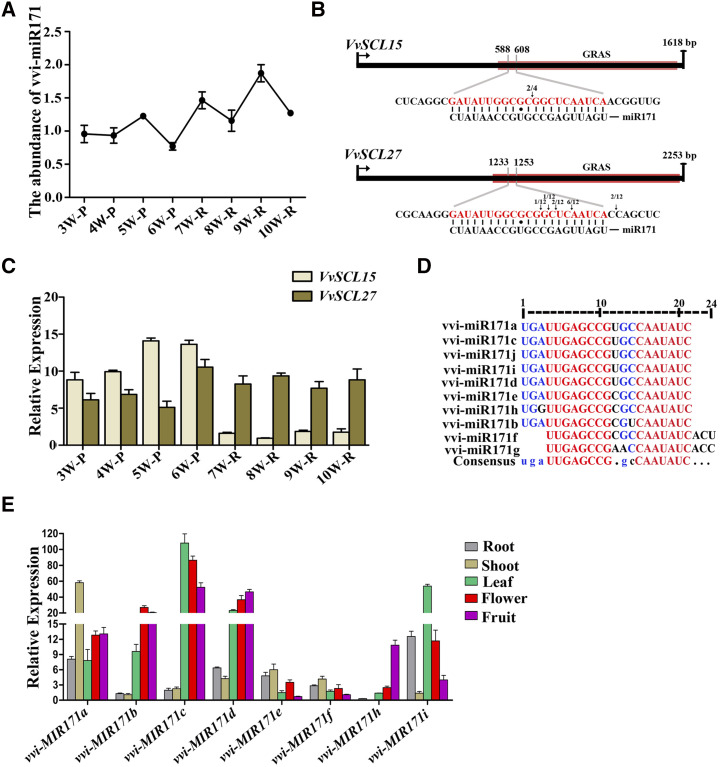
To study the effects of vvi-miR171 on grape roots, we first analyzed the changes of vvi-miR171 abundance during the process of adventitious root formation by stem-loop RT-qPCR. The results showed that the abundance of vvi-miR171 in adventitious roots was higher than that in phloem, and the vvi-miR171 abundance level in phloem was reduced before the appearance of adventitious roots (Fig. 2A). Individual miRNAs cannot regulate root development but rather must be associated with their target genes. By rapid amplification of 5′ complementary DNA ends (5′ RACE), we confirmed that the target genes of vvi-miR171 were VvSCL15 and VvSCL27, and VvSCL15 was cleaved at a single site, while VvSCL27 was cleaved at multiple sites (Fig. 2B). During the formation of adventitious roots, the expression level of VvSCL15 in the phloem before adventitious root appearance was high and decreased in adventitious roots, while the expression level of VvSCL27 in adventitious roots was higher than that in phloem before adventitious root appearance (Fig. 2C). To date, 10 vvi-miR171 family members have been identified in grapevine. According to mature miRNA sequence alignment, the miR171 family members have the same length (21 nucleotides), and the sequences of vvi-miR171a, vvi-miR171c, vvi-miR171d, vvi-miR171i, and vvi-miR171j were the same, vvi-miR171b, vvi-miR171e, and vvi-miR171h had single-base differences at different sites, while vvi-miR171g and vvi-miR171f lacked three bases at the 5′ end and had three additional bases at the 3′ end compared to the other members (Fig. 2D). These differences might be caused by the identification of different cleavage sites during the formation of the mature miRNAs.
Figure 2.
Expression patterns of vvi-miR171 and target genes during adventitious root formation and growth of cv Muscat Hamburg cuttings and the expression pattern of the vvi-MIR171 family in different tissues. A, Relative abundance of vvi-miR171 during adventitious root formation and growth of cuttings by stem-loop RT-qPCR. B, Cleavage sites of vvi-miR171 at complementary sequences of VvSCL15 and VvSCL27 mRNA as determined by 5′ RACE. The numbers of 5′ RACE clones corresponding to each site are indicated by arrows. The red boxes represent the GRAS domains of VvSCL15 and VvSCL27. Short black bars represent base matches, and black circles represent base mismatches. C, Expression patterns of VvSCL15 and VvSCL27 during adventitious root formation and growth of cuttings. The 3W to 10W indicate the time that the samples were collected beginning in the third week (14th d) after cuttings were prepared, and once a week until the 10th week. P, Phloem; R, root. D, Multiple sequence alignment of the vvi-miR171 family members. E, Expression pattern of vvi-MIR171 family in different tissues. Data are plotted as means ± sd, and error bars show the sd among three biological replicates (n = 3).
By analyzing the sequences of vvi-MIR171 gene family, we found that vvi-MIR171g is located in the exon of nodulation signaling pathway2 gene (Supplemental Fig. S4), and vvi-MIR171j cannot be found on a defined chromosome (Fig. 1), so we did not study them further. We analyzed the expression patterns of the remaining vvi-MIR171 family members. The results showed that each member was expressed in many tissues, such as roots, stems, leaves, flowers, and fruits, and different members showed different expression patterns in the same tissue (Fig. 2E). These results indicated that the miR171 family members of grapevine were highly conservative, with only a few base mutations during the evolutionary process, but they might play different regulatory roles due to their temporal and spatial expression differences.
The Role of miR171 in Grape Root Development
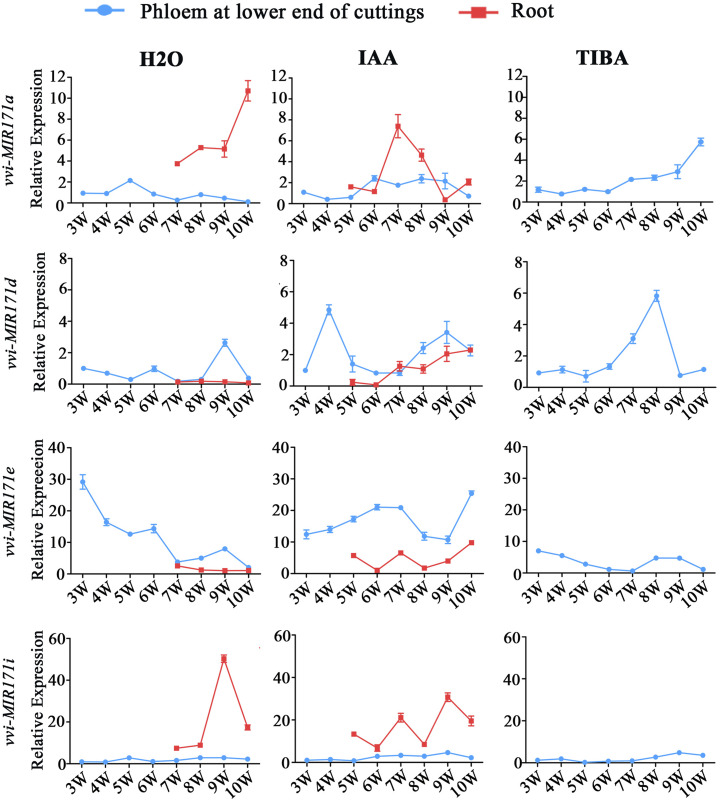
Propagation by cuttings is an important method of grape reproduction, and the formation of adventitious roots is crucial for this process. According to the expression patterns of miR171 family members in different tissues, the expression of vvi-MIR171a, vvi-MIR171d, vvi-MIR171e, and vvi-MIR171i mRNAs were relatively high in roots, so we speculated that they might play an important role in adventitious root formation and development. To verify this speculation, we examined the spatiotemporal expression patterns of the four vvi-MIR171s during the rooting process of cv Muscat Hamburg cuttings (Supplemental Fig. S5). The results showed that the expression levels of vvi-MIR171a and vvi-MIR171i in the phloem changed little; when the adventitious roots formed, their expression levels in adventitious roots increased. For indole acetic acid (IAA)-treated cuttings, the expression level of vvi-MIR171a decreased in the late growth stage of adventitious roots; this effect might be caused by the growth cessation and maturation of the adventitious roots. The expression of vvi-MIR171d and vvi-MIR171e in adventitious roots was lower than that in phloem, and there was a peak in the expression level of vvi-MIR171d before the appearance of adventitious roots (Fig. 3). Moreover, in grapevine 'Red Globe' cuttings (Supplemental Fig. S6), the spatiotemporal expressions of the four vvi-MIR171 genes were similar to those in Muscat Hamburg cuttings (Supplemental Fig. S7). From this result, we speculated that vvi-MIR171a and vvi-MIR171i might play a role in the growth of adventitious roots, while vvi-MIR171d could affect the formation of adventitious roots.
Figure 3.
Abundance of vvi-MIR171a, vvi-MIR171d, vvi-MIR171e, and vvi-MIR171i mRNAs during adventitious root formation and growth of cv Muscat Hamburg cuttings treated with IAA, 2,3,5-triiodobenzoic acid (TIBA), and control group. The 3W to 10W indicate the time that the samples were collected beginning in the third week (14th d) after cuttings were prepared, and once a week until the 10th week. Data are plotted as means ± sd, and error bars show the sd among three biological replicates (n = 3).
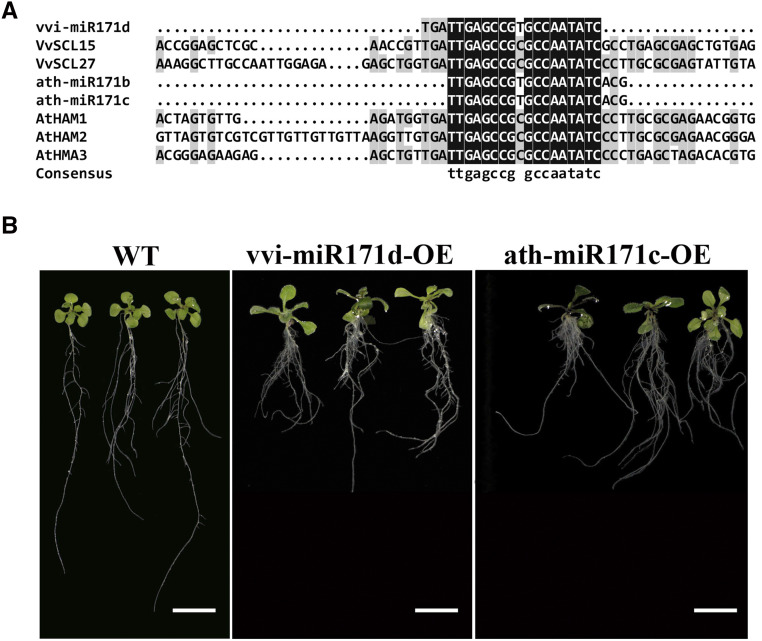
Notably, ath-MIR171b, ath-MIR171c, and vvi-MIR171d have the same mature miRNA sequence (Fig. 4A). Previous studies have shown that the number of branches were reduced, and the growth of primary roots was inhibited in ath-miR171c/b-overexpression Arabidopsis lines compared with the wild type (Wang et al., 2010). To explore the role of vvi-MIR171d in root development, we overexpressed vvi-miR171d and ath-miR171c in Arabidopsis. Compared with the wild type, the transgenic lines exhibited a phenotype of shorter primary roots, adventitious roots appeared earlier, and the density of lateral roots increased at the morphogical lower end of the primary root (Fig. 4B).
Figure 4.
Multiple sequence alignment of vvi-miR171d, ath-miR171b, ath-miR171c, and their target genes and phenotypes of transgenic Arabidopsis. A, Multiple sequence alignment of vvi-miR171d, ath-miR171b, ath-miR171c, and their corresponding target genes VvSCL15, VvSCL27, AtHAM1, AtHAM2, and AtHAM3. B, Representative photographs of transgenic Arabidopsis cultured vertically on MS medium for 15 d. Scale bars = 1 cm. WT, Wild type.
miPEPs have been shown to modify plant phenotype by the external application of synthetic peptides (Lauressergues et al., 2015). We cultured Arabidopsis on Murashige and Skoog (MS) medium containing 0.2 μm ath-miPEP171c, which was predicted by Lauressergues et al. (2015). Compared with the control group, the primary roots of plants treated with ath-miPEP171c were inhibited significantly, and the number of lateral roots and adventitious roots increased significantly in the early stage of root development (Supplemental Fig. S8), and in the later development of roots, the difference was not significant. These results suggested that ath-miPEP171c was active and could modify the root development in Arabidopsis.
Exploration of vvi-miPEP171d in Grapevine
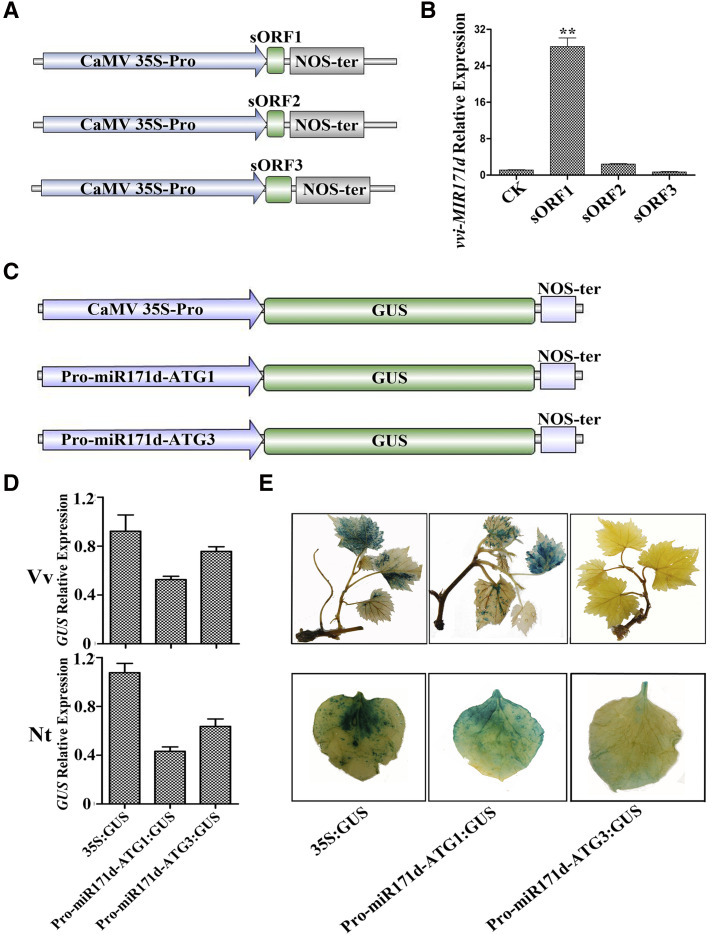
Based on the above results, we speculated that vvi-MIR171d might play a different role from the other three vvi-MIR171 genes in the formation of adventitious roots. Is there a miPEP in grape that specifically regulates the expression of vvi-MIR171d? First, we designed primers at different positions in the sequence upstream of pre-miR171d and combined them with a reverse primer that targeted pre-miR171d for PCR amplification (Supplemental Fig. S9A). The gel electrophoresis results showed that the 500-bp sequence upstream of pre-miR171d was part of the transcript of the vvi-MIR171d gene (Supplemental Fig. S9B). Then, the level of pri-miR171d in the cytoplasm and nucleus were determined separately by RT-qPCR, showing that most of pri-miR171d was present in the nucleus, but pri-miR171d was also present in the cytoplasm (Supplemental Fig. S9C). Through the previous analysis, we knew that there were three reading frames upstream of premiR171d: the first reading frame encoded seven amino acids and was named as sORF1 (vvi-miPEP171d1); the second reading frame encoded 17 amino acids and was named as sORF2 (vvi-miPEP171d2); and the third reading frame encoded six amino acids, and was named as sORF3 (vvi-miPEP171d3; Supplemental Fig. S9D). Since miPEPs can alter the transcription levels of their corresponding pri-miRNAs (Lauressergues et al., 2015) to determine which reading frame was functional, separate overexpression vectors for each of these three reading frames were constructed with the Cauliflower mosaic virus (CaMV) 35S promoter (Fig. 5A). The three reading frames were transiently overexpressed in grape tissue culture plantlets with the empty vector as the control, and then the quantity of pri-miR171d was detected. The RT-qPCR results showed that sORF1 increased the expression level of vvi-MIR171d, while sORF2 and sORF3 had no effect on the expression of vvi-MIR171d (Fig. 5B). In addition, the abundance of vvi-miR171 decreased after sORF1 treatment compared with that of the control, while the abundance did not change in the sORF2 and sORF3 treatment groups (Supplemental Fig. S10A). This result suggested that the first reading frame was active.
Figure 5.
Exploration of vvi-miPEP171d in grapevine. A, Schematic diagrams of OE-sORF1, OE-sORF2, and OE-sORF3 constructs with the CaMV 35S promoter. B, Expression of vvi-MIR171d in grape tissue culture plantlets after transient expression of sORF1, sORF2, or sORF3. Asterisks indicate significance at **P < 0.01, according to the two-tailed Student's t test. Data are plotted as means ± sd, and error bars show the sd among three biological replicates (n = 3). C Schematic diagrams of promoter-MIR171d extending to ATG1 or ATG3 constructs. D Expression of GUS in grape tissue plantlets and Nicotiana benthamiana leaves. Data are plotted as means ± sd, and error bars show the sd among three biological replicates (n = 3). E Activity analysis of GUS in grape tissue culture plantlets and N. benthamiana leaves.
To further validate our results, the region from the vvi-MIR171d promoter to the ATG start site of either sORF1 (PromiR171d-ATG1) or sORF3 (PromiR171d-ATG3) was fused with the GUS gene, and the CaMV 35S promoter was used to drive the GUS gene as the control (Fig. 5C). A transient transformation assay was performed in grape tissue culture plantlets and Nicotiana benthamiana leaves to observe GUS activity. The results showed that PromiR171d-ATG1 could initiate transcription of the GUS gene in grape tissue culture plantlets and N. benthamiana leaves, and the activity of the GUS protein could be detected, while the PromiR171d-ATG3 could also initiate the transcription of the GUS gene, but the activity of the GUS protein was almost undetectable (Fig. 5, D and E). The abundance of vvi-premiR171d in the PromiR171d-ATG3 treatment also increased significantly (Supplemental Fig. S10B). These results further demonstrated that the short peptide encoded by the first reading frame was active. Since the sequence from the promoter to the ATG start site of sORF3 could initiate GUS transcription, but the sequence from the promoter to the ATG start site of sORF3 (including the stop codon of sORF1) could terminate the translation of the GUS protein prematurely, thus the activity of the GUS protein could not be detected. We named the short peptide encoded by sORF1 as vvi-miPEP171d1. Furthermore, the abundance of vvi-miR171 showed a slight decrease compared with the control group, which was consistent with the result of vvi-miPEP171d1 transient overexpression (Supplemental Fig. S10C).
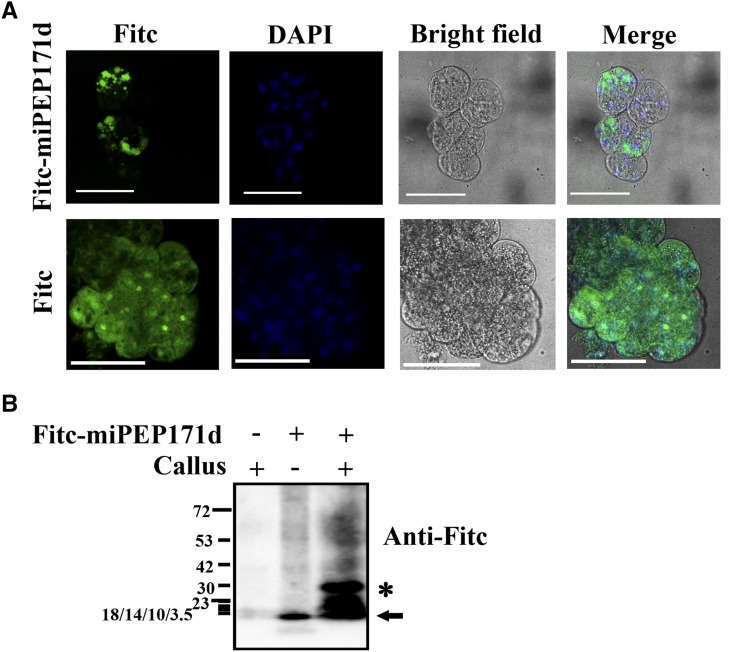
To explore whether synthetic vvi-miPEP171d1 can enter cells after the external application, grape embryogenic calli were treated with FITC (fluorescein isothiocyanate)-miPEP171d1. We found that the fluorescence signal was aggregated in the cytoplasm and partially in the nucleus, while the fluorescence signal in the cells treated with FITC was evenly distributed throughout the cells (Fig. 6A). The crude protein was extracted from the treated embryogenic calli and detected with FITC antibody. FITC-miPEP171d1 could be detected (Fig. 6B), which further demonstrated that vvi-miPEP171d1 could be absorbed by grape embryogenic callus cells.
Figure 6.
Subcellular localization of vvi-miPEP171d1. A, The same cell was photographed for DAPI and GFP fluorescence and bright-field microscopy, and then the images were merged. Scale bars = 25 μm. B, Western blot analysis of Fitc-miPEP171d1 entering cells. The black arrow indicates the position of vvi-miPEP171d1, and asterisk indicates nonspecific binding. “+” represents addition and “−” represents no addition. Short horizontal lines on the left represent bands of proteins with different molecular weights in the marker, and the number next to the band represents the molecular weight of the protein. The concentration of the SDS-PAGE gel was 15%, the electrophoresis time was about 60 min (when the loading and small molecular weight protein band reached to the middle length of SDS-PAGE gel), and the 3.5 to 23 kDa protein marker bands overlapped together.
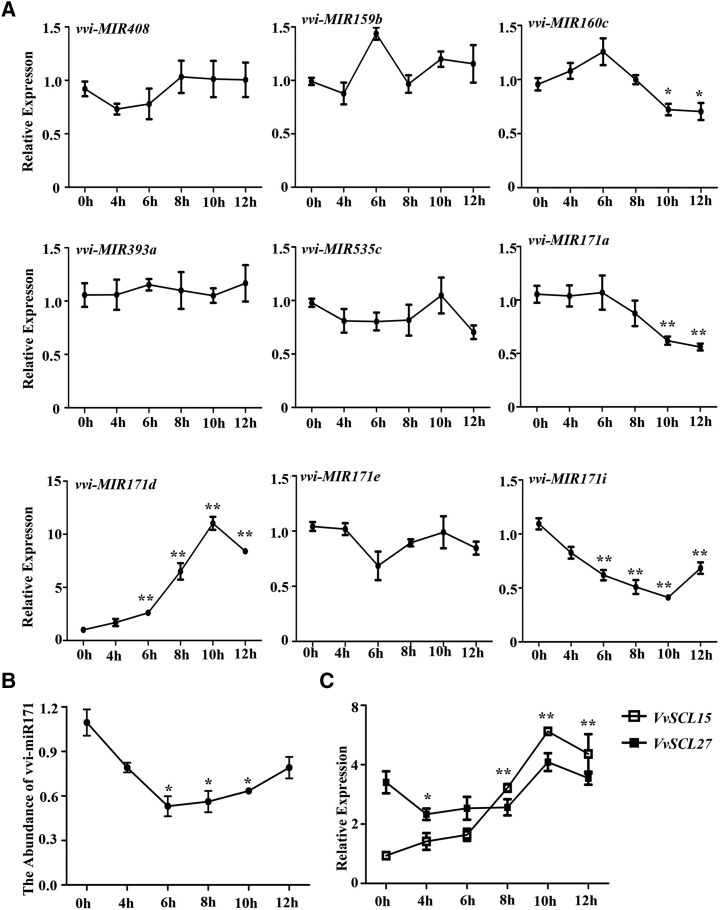
Based on the above results, we treated grape tissue culture plantlets with synthetic vvi-miPEP171d1 to explore its effect on the expression of vvi-MIR171 family members and other vvi-MIRNA genes. The results of RT-qPCR showed that the expression of vvi-MIR171d increased, and the expression level increased with treatment duration. In addition, the expression levels of vvi-MIR171a and vvi-MIR171i were reduced and that of vvi-MIR171i was reduced more significantly, while the expression of vvi-MIR171e was barely changed (Fig. 7A). We also detected the expression levels of some other vvi-MIRNA genes and found that the expression of vvi-MIR160c was reduced at later stages, while that of the other vvi-MIRNAs was hardly affected (Fig. 7A). Under vvi-miPEP171d1 treatment, the abundance of pri-miR171d and pre-miR171d increased with the treatment duration, and the abundance of pre-miR171d was always higher than that of pri-miR171d (Supplemental Fig. S10D). These results suggested that vvi-miPEP171d1 could induce vvi-MIR171d expression specifically and increased the abundance of vvi-premiR171d.
Figure 7.
Effects of vvi-miPEP171d1 on the expression of some vvi-MIRNA genes, vvi-miR171, and VvSCL15 and VvSCL27. A, Accumulation of vvi-MIR171a, vvi-MIR171d, vvi-MIR171e, vvi-MIR171i, vvi-MIR408, vvi-MIR159b, vvi-MIR160c, vvi-MIR393a, and vvi-MIR535 mRNAs in grape tissue culture plantlet roots treated with 0.2 μm vvi-miPEP171d1. B, Abundance of vvi-miR171 in grape tissue culture plantlet roots treated with 0.2 μm vvi-miPEP171d1. C, Expression of VvSCL15 and VvSCL27 in grape tissue culture plantlet roots treated with 0.2 μm vvi-miPEP171d1. Data are plotted as means ± sd, and error bars represent sd among three biological replicates (n = 3). Asterisk indicate significance at *P < 0.05 and **P < 0.01, respectively, according to the two-tailed Student's t test.
Then, we scored the abundance of vvi-miR171 and the expression levels of the target genes VvSCL15 and VvSCL27. The abundance of vvi-miR171 decreased at the beginning of the vvi-miPEP171d1 treatment and then increased with the treatment duration (Fig. 7B). The expression level of VvSCL27 decreased, then increased to a peak, and then finally decreased. The expression of VvSCL15 increased and decreased after reaching a peak (Fig. 7C). To further explore the different regulatory effect of vvi-miR171a, vvi-miR171d, vvi-miR171e, and vvi-miR171i on VvSCL15 and VvSCL27, we coexpressed VvSCL15 or VvSCL27 in N. benthamiana leaves with each of the four vvi-MIR171s. The results showed that the efficiency of vvi-miR171a, vvi-miR171d, vvi-miR171e, or vvi-miR171i guiding the cleavage of VvSCL15 and VvSCL27 mRNAs was different: VvSCL15 mRNA was mainly regulated by vvi-miR171a and vvi-mR171i, and VvSCL27 mRNA was mainly regulated by vvi-miR171d and vvi-miR171e (Supplemental Fig. S10E). Thus, we could conclude that vvi-miPEP171d1 induced the expression of vvi-MIR171d, which decreased the abundance of VvSCL27 mRNA; meanwhile, the expression of vvi-MIR171i decreased, resulting in relatively high abundance of mRNA of its main target gene VvSCL15 (Fig. 7, B and C).
Effects of vvi-miPEP171d1 on Grapevine Growth and Development
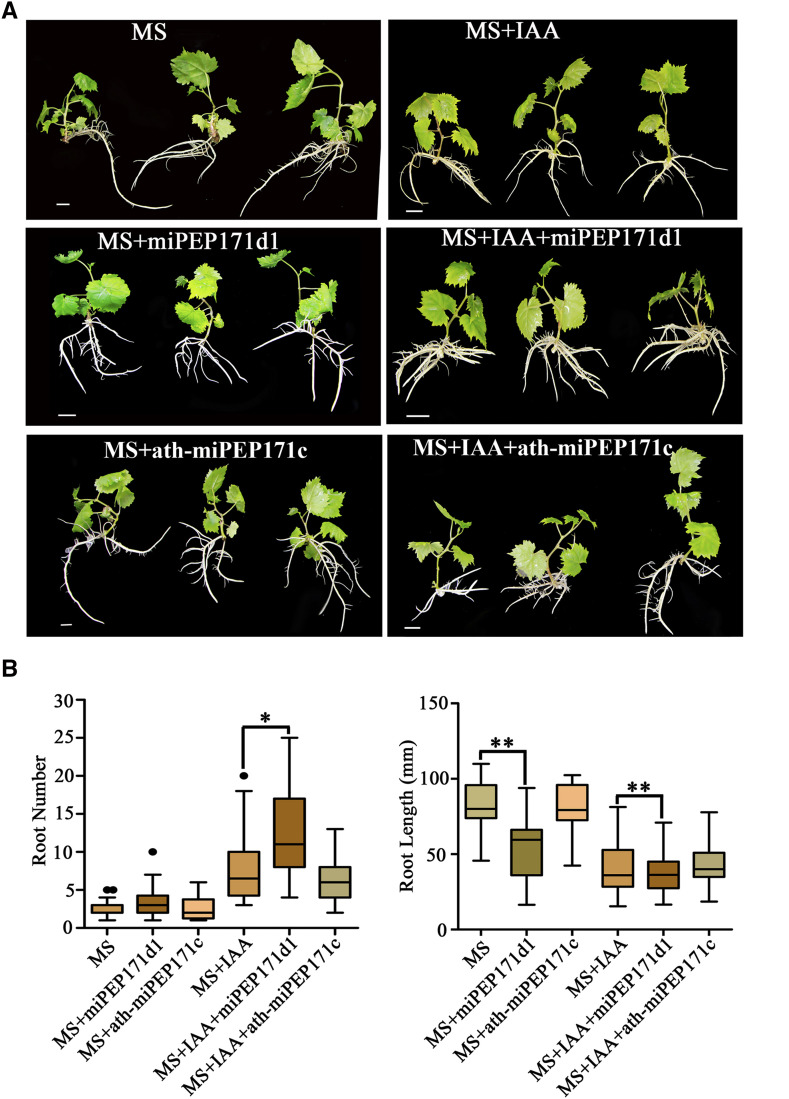
To identify the function of vvi-miPEP171d1 in grape adventitious root formation and development, we cultured grape plantlets on 1 mg/l IAA MS medium containing 0.2 μm synthetic vvi-miPEP171d1 or ath-miPEP171c and IAA-free MS medium containing 0.2 μm synthetic vvi-miPEP171d1 or ath-miPEP171c. The results showed that, for the group without IAA, the adventitious root number increased under vvi-miPEP171d1 treatment, and the length of the adventitious roots became significantly shorter, while the plantlets treated with ath-miPEP171c showed no phenotypic changes compared with the control group (Fig. 8). For the IAA-added group, the number of adventitious roots increased and root length decreased significantly under vvi-miPEP171d1 treatment, and the plantlets treated with ath-miPEP171c also showed no phenotypic changes compared with the control group (Fig. 8). The abundance of vvi-miR171 was slightly reduced in the vvi-miPEP171d1-treated group, while that in the ath-miPEP171c-treated group did not differ (Supplemental Fig. S11A). In the first two rooting stages, the expression of vvi-miR171 in the vvi-miPEP171d1 treatment group was lower than that in the control group, but in the later stage, they were not distinguishable. The expression of VvSCL15 and VvSCL27 was higher than that in the control group in the first two stages and then approached that in the control group in the third stage (Supplemental Fig. S11, B and C). These results suggested that exogenous application of synthetic vvi-miPEP171d1 could increase the formation of adventitious roots, while ath-miPEP171c had no effect on the formation and development of grape adventitious roots.
Figure 8.
Phenotypes of grape tissue culture plantlets cultured with 0.2 μm vvi-miPEP171d1 or ath-miPEP171c. A, Representative photographs of grape tissue culture plantlets cultured on MS medium containing 1 mg/L IAA and 0.2 μm vvi-miPEP171d1 or ath-miPEP171c or MS medium containing 0.2 μm vvi-miPEP171d1 or ath-miPEP171c. Scale bars = 1 cm. B, Adventitious root number and root length of 25-d-old grape tissue culture plantlets cultured on MS medium containing 1 mg/L IAA and 0.2 μm vvi-miPEP171d1 or ath-miPEP171c or MS medium containing 0.2 μm vvi-miPEP171d1 or ath-miPEP171c, and MS or MS containing 1 mg/L IAA supplemented with sterile water as control. Black dots represent outliers. Asterisk indicate significance at *P < 0.05 and **P < 0.01, respectively, according to the two-tailed Student's t test. Data are plotted as means ± sd, and error bars show the sd.
To explore whether vvi-miPEP171d1 is universal and can function in other species, Arabidopsis plants were cultured on MS medium with or without 0.2 μm synthetic vvi-miPEP171d1. The results showed that the treated plants had little change compared to the control. Furthermore, after overexpression of vvi-miPEP171d1 in Arabidopsis, the vvi-miPEP171d1-OE lines did not differ from the wild type in height, branch (Supplemental Fig. S12A), root length, or lateral root number (Supplemental Fig. S12, B and C). These results indicated that vvi-miPEP171d1 could only act in grapevine and had no effect in Arabidopsis, which indicated the functional specificity of this miPEP in different species.
DISCUSSION
Peptides perform multiple functions in plant growth and development, and most of them are derived from nonfunctional precursors or functional proteins (Schmelz et al., 2006; Pearce et al., 2010; Matsubayashi, 2011). Recently, a novel type of peptide termed miPEPs has been reported. These miPEPs are encoded by pri-miRNA transcripts, and this type of peptide has a regulatory function (Lauressergues et al., 2015). Unlike those of peptides derived from nonfunctional precursor proteins or functional proteins, the transcripts encoding this novel type of peptide should contain one or more sORFs (Dinger et al., 2008). In this study, we analyzed the open reading frames in the 500-bp sequences upstream of grape miRNA precursors that have been registered in the miRBase database, and we found that at least one open reading frames exist in each pri-miRNA (Fig. 1; Supplemental Table S1). Therefore, we speculate that there are peptides encoded by pri-miRNAs in grapevine, and these peptides may regulate grapevine growth and development.
Among the miRNAs that are involved in the development of plant roots (Couzigou et al., 2017), miR171 is a conserved miRNA within different plant species that has been associated with cell fate determination and root organ development (Wang et al., 2010; Engstrom et al., 2011). In apple, mdm-miR171a exhibited a low expression level before the appearance of adventitious roots (Li et al., 2019). To date, research on the effects of miR171 on grape root development is still lacking. In this study, the abundance of vvi-miR171 and its targets VvSCL15 and VvSCL27 mRNAs were detected during the root formation and development of grape cuttings, and we found that the abundance of vvi-miR171 was suppressed, and those of VvSCL15 and VvSCL27 mRNAs were increased in the phloem at the lower end of the cuttings before the appearance of adventitious roots (Fig. 1, A and C). Many studies have found that the expression of SCL is elevated in the initial stage of root development. For example, the SCR) reporter line END199 shows GUS activity in the central cells of the outer layer during lateral root formation (Malamy and Benfey, 1997; Konishi and Sugiyama, 2006). CsSCL1 and PrSCL1 are highly expressed in rooting-competent cuttings of Pinus radiata and Castanea sativa. (Sánchez et al., 2007), and CsSCL1 mRNA is specifically located in the cambial zone and derivative cells during the formation of adventitious roots, while in rooting-incompetent shoots, it is diffuse and evenly distributed through the phloem and parenchyma (Vielba et al., 2011). In this study, consistent with previous reports, we demonstrated that vvi-miR171 affected the formation of adventitious roots by regulating VvSCL15 and VvSCL27 in grapevine.
Pri-miRNAs can encode peptides. Overexpression of miPEP171b and miPEP165a in M. truncatula and Arabidopsis, respectively, increased the abundance of their respective endogenous miRNAs by stimulating the transcription of the pri-miRNA, which downregulated the expression of its corresponding target genes and affected root growth and development (Lauressergues et al., 2015). Exogenous application of synthetic miPEP172c could stimulate miR172c expression and increase nodule number in soybean (Glycine max; Couzigou et al., 2016). There are 50 miPEPs with lengths of three to 59 amino acids sharing no common signature, which suggests that each miPEP is specific for its corresponding miRNAs (Lauressergues et al., 2015). Through sequence analysis, we found three sORFs in the 500-bp sequence upstream of premiR171d (Supplemental Fig. S9D). The transient overexpression experiment and GUS activity assay demonstrated that the first sORF could specifically enhance the expression of vvi-MIR171d (Fig. 5, B and D), and synthetic vvi-miPEP171d1 treatment increased adventitious root number of the grape tissue culture plantlets (Fig. 8; Supplemental Fig. S11B). These results indicated the presence of miPEP in grapevine and confirmed our speculation that vvi-miR171d could play an important role in the formation of adventitious roots.
The expression pattern of vvi-MIR171d was quite different from those of the other three members during adventitious root formation in cuttings, especially in the stage before adventitious root emergence (Figs . 2, A and C, and 3). Thus, we speculated that vvi-MIR171d might play a special role in the formation of adventitious roots. It has been reported that miRNA family members can generate different miPEPs (Lauressergues et al., 2015); thus, treatment with different synthetic miPEPs can enable distinct functional analysis of each member of a miRNA family. In this study, after treatment with synthetic vvi-miPEP171d1, we found that only the expression of vvi-MIR171d was induced and that of vvi-MIR171e and other vvi-MIRNAs was scarcely changed; the expression of vvi-MIR171a and vvi-MIR171i was decreased significantly, and that of both target mRNAs showed different changes, especially VvSCL15 mRNA (Fig. 7; Supplemental Fig. S11), consistent with their expression patterns during the rooting process in cuttings: the expression of vvi-MIR171d was induced, while that of vvi-MIR171a and vvi-MIR171i was reduced, and VvSCL15 was increased during the adventitious root initiation stage (Figs. 2C and 3; Supplemental Fig. S7). Based on previous reports about the role of SCL in the formation of adventitious roots, we speculated that the increased expression of VvSCL15 might be the reason for the increased adventitious root number after miPEP171d1 treatment.
The increased content of pri-miR171d does not mean that the content of miR171 is increased. There are 10 members of the miR171 family in grapevine, and more experiments are needed to explore the relationship among them. The miR171 mature sequences of these 10 members are highly conserved, and the conservation determines the preference of the target gene and the way it acts on the target gene. 5′ RACE experiments showed that the target genes of vvi-miR171 family members were VvSCL15 and VvSCL27 (Fig. 2B). The different expression patterns of eight vvi-MIR171 family members in many tissues suggested that they played different regulatory roles in grape development and growth (Fig. 2E). There are different bases among the 10 mature sequences of vvi-miR171 (produced by 10 premiR171), indicating their different physiological functions as small molecular regulators. The efficiency of different miR171 members guiding the cleavage of diverse target genes was detected by the coinjection assay on N. benthamiana leaves, and from the result we could see that the efficiencies varied (Supplemental Fig. S10E). Compared with the mature sequence, precursors of different miR171 family members are less conserved in sequence, and divergent in the evolutionary tree (Supplemental Fig. S3). The specificity of the precursors determines the specific hairpin structure, and precursors of miRNA have different hairpin structures in different species, which results in functional differences (Van Wynsberghe et al., 2011). In Arabidopsis, the activation of miR171 was thought to be due to direct binding to the promoter region by its own target gene AtSCL protein, thereby forming a homeostatic feedback loop (Xue et al., 2014). The efficiency of vvi-miR171d guiding the cleavage of the two target mRNAs was different, and the two target genes might be involved in regulating the expression of other vvi-MIR171 members, leading to differences in their expression patterns from that of vvi-MIR171d, thus forming a complex regulatory network. It is interesting and important to do further studies. In addition, the expression patterns of 8 vvi-MIR171 family members in many tissues were different (Fig. 2E), and different vvi-MIR171 genes had different expression levels during adventitious root development (Fig. 3; Supplemental Fig. S7), indicating that the dosage effects and the spatiotemporal expression specificity of vvi-miR171 family members might form a complex regulatory network to maintain and regulate VvSCL mRNA abundance (Li and Mao, 2007), and further research could be performed to demonstrate its exact features.
Through genetic studies, Atham1, Atham2,and Atham3 triple mutant plants were found to display a reduced primary root phenotype, similar to that of 35Spro-MIR171c plants (Wang et al., 2010; Engstrom et al., 2011), and tomato plants overexpressing SlGRAS24 showed short primary roots and few lateral roots (Huang et al., 2017), which shows that the expression of a MIRNA or their target genes beyond a certain range will affect root development. Plants balance gene expression levels through complex regulatory networks. Compared with homologous or heterologous miRNA overexpression, treatment with miPEPs could be used to analyze gene or miRNA function and their regulatory mechanisms, and this method could more accurately reflect the endogenous regulation in plants (Fig. 8).
The grape tissue culture plantlets treated with ath-miPEP171c showed no difference from the control (Fig. 8), and Arabidopsis seedlings cultured with vvi-miPEP171d1 showed no change in root phenotype. Furthermore, heterologous overexpression of vvi-miPEP171d1 in Arabidopsis did not affect the roots or aerial part compared with the wild type (Supplemental Fig. S12). These results again suggest that each miPEP is specific for its corresponding miRNA (Lauressergues et al., 2015) and that miPEPs are species specific. According to the results of the promoter-reporter and FITC-miPEP171d1 infecting embryogenic calli penetration experiments, vvi-miPEP171d1 might regulate its own promoter activity, just as Waterhouse and Hellens (2015) assumed. However, how miPEPs enter the nucleus and increase the activity of their own promoter needs further investigation.
In conclusion, we demonstrated that the species specificity of the miPEP vvi-miPEP171d1 could affect the growth and development of roots by regulating the expression of its corresponding vvi-MIR171d in grapevine. While the ath-miPEP171c could regulate root development in Arabidopsis, but not grapevine. These results enrich the theoretical understanding of miRNA regulation networks and provide new ideas for root development studies in perennial fruit trees.
MATERIALS AND METHODS
Plant Material
Micropropagated ‘Thompson Seedless’ grape (Vitis vinifera) plantlets were grown in tissue culture at Shanghai Jiao Tong University, Shanghai, China. The tissue culture plantlets were maintained under 16 h light and 8 h dark at 23°C and subcultured every 30 d on MS medium containing 1.0 mg/L IAA, 0.7% (w/v) agar, and 3% (w/v) Suc. The pH of the medium was adjusted to 5.8 before sterilization.
For external application of vvi-miPEP171d1, tissue culture plantlets subcultured for ∼25 d under the above conditions were subjected to the following treatments: about 5-cm tips were cut and subcultured on solid MS medium containing 0.2 μm vvi-miPEP171d1 or 0.2 μm ath-miPEP171c; the solid MS medium either contained 1 mg/l IAA or was hormone-free. The control groups were placed in the same media without the peptides. Forty uniform grape tissue culture plantlets were selected for each treatment, and three biological replicates were performed. After being subcultured 25 d, the number of adventitious roots of each treatment was counted. In addition, a total of 200 uniform tissue culture cuttings were selected and sampled at three time points: 3 (S0), 6 (S1), and 9 d (S2). The tissue culture cuttings grown on the MS medium with 1.0 mg/L IAA for 25 d were immersed in 0.2 μm vvi-miPEP171d1 solution for 30 min and then removed to place in an empty container. Samples were collected at regular intervals, frozen in liquid nitrogen, and stored in a −80°C ultralow temperature freezer for further use.
The Arabidopsis (Arabidopsis thaliana) Columbia accession (Col) was used as the wild type. Arabidopsis seeds were sterilized and sown on MS medium containing 0.2 μm vvi-miPEP171d1 or ath-miPEP171c, and sterile water was added to the medium of the control group. The seedlings were transferred to the corresponding medium after germination, and 10 Arabidopsis seedlings were placed in each dish. The culture dish was placed vertically, and the growth of the seedlings was observed regularly and photographed.
The grape cuttings were 1-year-old hardwood branches of 'Muscat Hamburg' and 'Red Globe'. This work was carried out in the artificial climate room of the experimental base of the School of Agriculture and Biology, Shanghai Jiao Tong University, on November 25, 2017, and the temperature of the artificial climate room was controlled at 25°C. The work was repeated on March 20, 2018. At pruning in winter, the 1-year-old healthily growing hardwood branches were selected, marked, and then stored under river sand in a pit prepared in advance. One day before the cuttings were started, the prepared branches were removed from the pit and cut into pieces with two buds. The morphologically upper end of the cuttings near the node was cut flat, and the morphologically lower end was cut diagonally at a distance of 2 to 3 cm from the bud. Then 120 trimmed cuttings per treatment were dipped into different solutions: (1) a solution containing 100 mg/L IAA; (2) a solution containing 80 mg/L 2,3,5-triiodobenzoic acid; and (3) water as the control group. After soaking for 12 h, the cuttings were taken from the container, the upper end was sealed with wax, and the cuttings were inserted into the river sand at a 45-degree angle at a depth of ∼5 cm. After insertion, the river sand was pressed firmly and watered thoroughly. The river sand was kept moist throughout the experiment. After 14 d of cutting incubation, we began to collect samples once a week. The morphologically lower end of each sampled cutting was washed and photographed. The number of roots of each cutting was counted, and the adventitious roots and ∼5 cm of the phloem at the morphologically lower end of the cuttings were collected separately, frozen with liquid nitrogen, and stored in a −80°C ultra-low temperature freezer for further use.
Synthetic Peptide Assay
Peptides were synthesized by Shanghai RuiMian Biological Technology. The purity of the synthetic peptides is ≥95% (w/w).
Peptide Sequence
The peptide sequences are as follows: vvi-miPEP171d1—MGYGTTP, FITC-KMGYGTTP (1.22 kDa); ath-miPEP171c—MLSLSHFHIC.
RNA Extraction and Synthesis of cDNA
Nuclear and cytoplasmic fractions were isolated according to Xu and Copeland (2012), and then the RNA and total RNA samples were extracted using a modified hexadecyl trimethyl ammonium bromide method (Li et al., 2009) and treated with DNase I (TaKaRa) to remove DNA contamination. Total genomic DNA was extracted from young leaves using a modified hexadecyl trimethyl ammonium bromide method and treated with RNase A to remove RNA. The concentration of RNA was estimated using a Nanodrop Spectrophotometer (Thermo Fischer Scientific). RNA was used as template to synthesize the first-strand cDNA using the SuperScript reverse transcriptase (TaKaRa) according to the manufacturer's instructions. For vvi-miR171, a stem-loop primer was designed for synthesizing the first-strand cDNA. Total RNA from all three biological replicates was independently used for RT-qPCR.
Phylogenetic Analyses of the miR171 Family
We downloaded the precursor sequences of the miR171 family of V. vinifera (vvi), Oryza sativa (osa), Medicago truncatula (mtr), Brassica napus (bna), Triticum aestivum (tae), Populus trichocarpa (ptr), Zea mays (zma), Sorghum bicolor (sbi), Selaginella moellendorffii (smo), Glycine max (gma), Solanu mlycopersicum (sly), Citrus sinensis (csi), Nicotiana tabacum (nta), Prunus persica (ppe), Malus domestica (mdm), and Fragaria vesca (fve) from miRBase database (http://www.mirbase.org/). Multiple sequence alignments of the collected miR171 precursor sequences were performed, and a phylogenetic tree was constructed using MEGA 6.0 software and the neighbor-joining method. A bootstrap test was performed with 1000 replicates. The confidence values are shown on the branches.
PCR Analysis
To analyze whether the 500-bp sequence upstream of each pre-miRNA was part of the pri-miRNA transcript, 20 pairs of primers were designed with forward primers beyond 500 bp upstream of the pre-miRNA and reverse primers at the 3′ end of the pre-miRNA. For vvi-MIR171d, the primers used for amplification were a reverse primer at the 3′ end of premiR171d and forward primers at different sites beyond 350 bp upstream of premiR171d (Supplemental Fig. S9A; Supplemental Table S2). DNA and cDNA were used as templates to carry out PCR assays. PCR amplification was performed as follows: preincubation at 94°C for 5 min followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 54°C for 30 s, and extension at 72°C for 1 min. The final extension was carried out at 72°C for 10 min, and the reaction was held at 4°C. The VvGAPDH gene was used as a control, and the total RNA was determined to be clean of DNA by observing the different sizes of bands in the two PCRs (Supplemental Figs. S1, B and C, and S9B).
Plasmid Construction
DNA fragments of the vvi-MIR171d promoter, the precursor of vvi-miR171a, vvi-miR171d, vvi-miR171e, and vvi-miR171i, the predicted coding sequences in vvi-MIR171d, the partial coding frame sequences of VvSCL15 and VvSCL27 containing vvi-miR171 target sequence, and the precursor of ath-miR71c of Arabidopsis were amplified from grape genomic DNA or cDNA or Arabidopsis cDNA. Primer sequences used for amplification are listed in Supplemental Table S2. PCR-amplified fragments of vvi-premiR171a, vvi-premiR171d, vvi-premiR171e, vvi-premiR171i, three reading frames predicted in vvi-MIR171d (sORF1, sORF2, and sORF3), the partial coding frame of VvSCL15 and VvSCL27, and Arabidopsis premiR171c (ath-premiR171c) were cloned into pRI 101-AN, all under the CaMV 35S promoter. The vvi-MIR171d promoter sequences were fused with GUS in vector pCambia1305. After confirmation by sequencing, all vectors were then transformed into Agrobacterium tumefaciens strain GV3101 and used for further transient expression or plant transformation experiments.
Transient Expression and Plant Transformation
Leaves of 3- or 4-week-old Nicotiana benthamiana plants or grape tissue culture plantlets were infiltrated with Agrobacterium strain GV3101 harboring different constructs for transient expression. The Agrobacterium overnight culture was centrifuged, and the supernatant was removed. The concentration of Agrobacterium was adjusted to an absorbance value of 1.0 at a wavelength of 600 nm (OD600 = 1.0) using suspension buffer (10 mm MgCl2, 10 mm MES [pH 5.6], and 0.1 mm acetosyringone). Infiltration was performed after the resuspended Agrobacterium was placed at room temperature for 2 to 4 h. After 24 h of incubation in the dark, the N. benthamiana plants or grape tissue culture plantlets were transferred to light and cultured for 2 d. The infiltrated tissues were collected for RNA analysis or GUS analysis.
The floral dipping method was used to create transgenic plants (Bechtold et al., 1993). Transgenic seeds were screened on MS plates containing 25 mg/L kanamycin. Approximately 10 independent T1 lines for each gene were generated. Homozygous T4 seeds of four representative lines for each construct were routinely used for phenotypic analysis.
Histochemical Staining for GUS Activity
The histochemical GUS assay was performed as follows: plant tissues were placed into GUS reaction buffer (0.5 mg/mL X-GlcA, 1% [v/v] methyl alcohol, 0.2 m phosphate buffer, pH 7.0). After vacuum infiltration for 10 min, the plant tissues were incubated in GUS reaction buffer overnight at 37°C. Stained tissues were cleared in 70% (v/v) ethanol to remove chlorophyll and photographed. At least three independent trials were performed.
Determination of Cleavage Sites Using 5′ RACE
The cDNA was obtained by reverse transcription of total RNA extracted from leaves and roots and specific primers of the target genes. The cDNA was then purified using the 5′ RACE kit (Gibco BRL Biotechnology) according to the manufacturer’s instructions. Nested PCR analyses were performed using purified cDNA as templates and the primers listed in Supplemental Table S2. The amplified products were purified on 1% (w/v) agar gels and cloned to the pMD19-Tsimple vector and then transformed into competent Escherichia coli strain DH5α. We selected 15 positive clones from each predicted target gene for confirmation by sequencing analysis.
Western Blot
Total protein extracts were obtained as previously described (Rudd et al., 1996), and 50 mg were loaded and separated by SDS-PAGE. Primary antibodies were used at 1:1000 (v/v) dilution and horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G was used as secondary antibody at 1:3000 (v/v) dilution. The western blot analysis was performed as described by Lauressergues et al. (2015).
Expression Analysis
For RT-qPCR, total RNA was treated with DNase I (TaKaRa), and 1 µg was reverse transcribed to obtain first-strand cDNA according to the manufacturer’s instruction (TaKaRa). The cDNA was diluted 10 times with nuclease-free water, and 1 µL was used as template for qPCR or stem-loop qPCR performed using TB Green II mix (TaKaRa) in a CFX Connect Bio-Rad instrument. The relative abundance of the mRNA was calculated by the 2−ΔΔCT method and normalized by VvGADPH, U6 (U6 belongs to small nuclear RNA and is transcribed by RNA polymerase III, and the sequence is rich in U) of grapevine (Luo et al., 2018) and Actin of N. benthamiana as reference (Livak and Schmittgen, 2001). The primers used are listed in Supplemental Table S2.
Vvi-miPEP171d1 Uptake Assay in Grape Embryogenic Callus
Vvi-miPEP171d1 labeled with FITC at the N terminus was synthesized from Shanghai RuiMian Biological Technology (≥95% purity). The grape embryogenic calli were incubated with 0.1 mg/L FITC-miPEP171d1 or FITC in sterilized water at 23°C for 10 h. After treatment, the calli were washed five times with sterile water by gently shaking for 2 min. The nuclei were stained with 4′,6-diamidino-2-phenylindole. The calli were imaged using a Leica laser confocal scanning microscope (LAS AF Lite). Fluorescence was visualized with excitation at 488 nm.
Statistical Analysis
All data in this study were obtained from three independent experiments. Data were plotted as means ± sd, and error bars were sd. The data were analyzed with the two-tailed Student’s t test using GraphPad Prism 5.01 software.
Accession Numbers
Accession numbers for this study are as follows: vvi-miR171a (MIMAT0005691), vvi-miR171d (MIMAT0005694), vvi-miR171e (MIMAT0005695), vvi-miR171i (MIMAT0005698), VvSCL15 (LOC100251313), VvSCL27 (LOC100267664), ath-miR171c (MIMAT0000921), AtHAM1 (NM_130079), AtHAM2 (NM_115927), At HAM3 (NM_116232).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. PCR analysis of vvi-MIRNA gene transcripts.
Supplemental Figure S2. Relative abundance of pre-miRNA and pri-miRNA.
Supplemental Figure S3. Phylogenetic analysis of miR171 family members in 17 species.
Supplemental Figure S4. Schematic diagram of vvi-miR171g overlapping with the exon of nodulation signaling pathway2 gene.
Supplemental Figure S5. Morphological features of the rooting process of cv Muscat Hamburg cuttings.
Supplemental Figure S6. Morphological features of the rooting process of cv Red Globe cuttings.
Supplemental Figure S7. The expression patterns of vvi-MIR171a, vvi-MIR171d, vvi-MIR171e, and vvi-MIR171i during adventitious root formation and growth of Red Globe cuttings treated with IAA and 2,3,5-triiodobenzoic acid and the control group.
Supplemental Figure S8. Phenotype of Arabidopsis seedlings cultured with 0.2 μm ath-miPEP171c.
Supplemental Figure S9. Analysis of the vvi-MIR171d transcript.
Supplemental Figure S10. Analysis of the relative abundance of miR171, pri-miR171d, and premiR171d and efficiency ofvvi-miR171a, vvi-miR171d, vvi-miR171e, and vvi-miR171i guiding the cleavage of VvSCL15 and VvSCL27 mRNAs.
Supplemental Figure S11. The effects of vvi-miPEP171d1 on adventitious root formation and growth of grape tissue culture plantlets.
Supplemental Figure S12. The phenotype of transgenic Arabidopsis plants.
Supplemental Table S1. Peptide sequences encoded by the short putative open reading frames in 500 bp upstream of vvi-pre-miRNAs.
Supplemental Table S2. Primers used in the study.
Footnotes
This work was supported by the National Key Research and Development Program of China (grant no. 2018YFD1000300), the National Natural Science Foundation of China (grant no. 31972383 and 31701888), and the Special Funds of Modern Industrial Technology System for Agriculture (grant no. CARS–29–zp–7).
Articles can be viewed without a subscription.
References
- Bartel DP.(2004) MicroRNAs genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G(1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci III 316: 1194–1199 [Google Scholar]
- Bielewicz D, Kalak M, Kalyna M, Windels D, Barta A, Vazquez F, Szweykowska-Kulinska Z, Jarmolowski A(2013) Introns of plant pri-miRNAs enhance miRNA biogenesis. EMBO Rep 14: 622–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolle C.(2004) The role of GRAS proteins in plant signal transduction and development. Planta 218: 683–692 [DOI] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O(2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190 [DOI] [PubMed] [Google Scholar]
- Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, Lindgren O, Moreno-Risueno MA, Vaten A, Thitamadee S, et al. (2010) Cell signaling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465: 316–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YL, Lee CY, Cheng KT, Chang WH, Huang RN, Nam HG, Chen YR(2014) Quantitative peptidomics study reveals that a wound-induced peptide from PR-1 regulates immune signaling in tomato. Plant Cell 26: 4135–4148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couzigou JM, Lauressergues D, André O, Gutjahr C, Guillotin B, Bécard G, Combier JP(2017) Positive gene regulation by a natural protective miRNA enables arbuscular mycorrhizal symbiosis. Cell Host Microbe 21: 106–112 [DOI] [PubMed] [Google Scholar]
- Couzigou JM, André O, Guillotin B, Alexandre M, Combier JP(2016) Use of microRNA-encoded peptide miPEP172c to stimulate nodulation in soybean. New Phytol 211: 379–381 [DOI] [PubMed] [Google Scholar]
- Curaba J, Talbot M, Li Z, Helliwell C(2013) Over-expression of microRNA171 affects phase transitions and floral meristem determinancy in barley. BMC Plant Biol 13: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa CT, Gaeta ML, de Araujo Mariath JE, Offringa R, Fett-Neto AG(2018) Comparative adventitious root development in pre-etiolated and flooded Arabidopsis hypocotyls exposed to different auxins. Plant Physiol Biochem 127: 161–168 [DOI] [PubMed] [Google Scholar]
- Dinger ME, Pang KC, Mercer TR, Mattick JS(2008) Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLOS Comput Biol 4: e1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrom EM, Andersen CM, Gumulak-Smith J, Hu J, Orlova E, Sozzani R, Bowman JL(2011) Arabidopsis homologs of the petunia hairy meristem gene are required for maintenance of shoot and root indeterminacy. Plant Physiol 155: 735–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Higuchi-Takeuchi M, Okamoto M, Yoshizumi T, Shimizu M, Nakaminami K, Nishi R, Ohashi C, Iida K, Tanaka M, Horii Y, Kawashima M, et al. (2013) Small open reading frames associated with morphogenesis are hidden in plant genomes. Proc Natl Acad Sci USA 110: 2395–2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Peng S, Xian Z, Lin D, Hu G, Yang L, Ren M, Li Z(2017) Overexpression of a tomato miR171 target gene SlGRAS24 impacts multiple agronomical traits via regulating gibberellin and auxin homeostasis. Plant Biotechnol J 15: 472–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Chen Q, Zhang Q, Zhang Y, Hao N, Ou C, Wang F, Li T(2018) Pyr-miR171f-targeted PyrSCL6 and PyrSCL22 genes regulate shoot growth by responding to IAA signaling in pear. Tree Genet Genomes 14: 20 [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B(2006) MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol 57: 19–53 [DOI] [PubMed] [Google Scholar]
- Kim YJ, Zheng B, Yu Y, Won SY, Mo B, Chen X(2011) The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J 30: 814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Zhang M, Xu X, Li X, Li C, Ding Z(2014) System analysis of microRNAs in the development and aluminium stress responses of the maize root system. Plant Biotechnol J 12: 1108–1121 [DOI] [PubMed] [Google Scholar]
- Konishi M, Sugiyama M(2006) A novel plant-specific family gene, ROOT PRIMORDIUM DEFECTIVE 1, is required for the maintenance of active cell proliferation. Plant Physiol 140: 591–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Watanabe Y(2004) Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci USA 101: 12753–12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakhotia N, Joshi G, Bhardwaj AR, Katiyar-Agarwal S, Agarwal M, Jagannath A, Goel S, Kumar A(2014) Identification and characterization of miRNAome in root, stem, leaf and tuber developmental stages of potato (Solanum tuberosum L.) by high-throughput sequencing. BMC Plant Biol 14: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauressergues D, Couzigou JM, Clemente HS, Martinez Y, Dunand C, Bécard G, Combier JP(2015) Primary transcripts of microRNAs encode regulatory peptides. Nature 520: 90–93 [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN(2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23: 4051–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Mao L(2007) Evolution of plant microRNA gene families. Cell Res 17: 212–218 [DOI] [PubMed] [Google Scholar]
- Li MF, Li XF, Han Z, Shu HR, Li TZ(2009) Molecular analysis of two Chinese pear (Pyrus bretschneideri Rehd.) spontaneous self-compatible mutants, Yan Zhuang and Jin Zhui. Plant Biol (Stuttg) 11: 774–783 [DOI] [PubMed] [Google Scholar]
- Li SG, Chen YY, Du ZJ, Zhang L, Luo HY, Wang JB(2010) Effects of peptide on flowering, fruit development, yield and fruit quality of Litchi chinensis Sonn. Redai Zuowu Xuebao 31: 567–571 [Google Scholar]
- Li HY, Zhang J, Yang Y, Jia NN, Wang CX, Sun HM(2017) miR171 and its target gene SCL6 contribute to embryogenic callus induction and torpedo-shaped embryo formation during somatic embryogenesis in two lily species. Plant Cell Tissue Organ Cult 130: 591–600 [Google Scholar]
- Li K, Liu Z, Xing L, Wei Y, Mao J, Meng Y, Bao L, Han M, Zhao C, Zhang D(2019) miRNAs associated with auxin signaling, stress response, and cellular activities mediate adventitious root formation in apple rootstocks. Plant Physiol Biochem 139: 66–81 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD(2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Llave C, Carrington JC(2002) Cleavage of scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297: 2053–2056 [DOI] [PubMed] [Google Scholar]
- Luo M, Gao Z, Li H, Li Q, Zhang C, Xu W, Song S, Ma C, Wang S(2018) Selection of reference genes for miRNA qRT-PCR under abiotic stress in grapevine. Sci Rep 8: 4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y.(2011) Post-translational modifications in secreted peptide hormones in plants. Plant Cell Physiol 52: 5–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Shao C, Wang H, Jin Y, Meng Y(2013) Construction of small RNA-mediated gene regulatory networks in the roots of rice (Oryza sativa). BMC Genomics 14: 510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN(1997) Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44 [DOI] [PubMed] [Google Scholar]
- Megtam M, Baev V, Rusinov V, Jensen ST, Kalantidis K, Hatzigeorgiou AG(2006) MicroRNA promoter element discovery in Arabidopsis. RNA 12: 1612–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E, Smith S, De Smet I(2012) Small signaling peptides in Arabidopsis development: How cells communicate over a short distance. Plant Cell 24: 3198–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce G, Yamaguchi Y, Barona G, Ryan CA(2010) A subtilisin-like protein from soybean contains an embedded, cryptic signal that activates defense-related genes. Proc Natl Acad Sci USA 107: 14921–14925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu LJ, Li L, Lan Z, Dresselhaus T(2015) Peptide signaling during the pollen tube journey and double fertilization. J Exp Bot 66: 5139–5150 [DOI] [PubMed] [Google Scholar]
- Reis RS, Hart-Smith G, Eamens AL, Wilkins MR, Waterhouse PM(2015) MicroRNA regulatory mechanisms play different roles in Arabidopsis. J Proteome Res 14: 4743–4751 [DOI] [PubMed] [Google Scholar]
- Rudd JJ, Franklin F, Lord JM, Franklin-Tong VE(1996) Increased phosphorylation of a 26-KD pollen protein is induced by the self-incompatibility response in Papaver rhoeas. Plant Cell 8: 713–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez C, Vielba JM, Ferro E, Covelo G, Solé A, Abarca D, de Mier BS, Díaz-Sala C(2007) Two SCARECROW-LIKE genes are induced in response to exogenous auxin in rooting-competent cuttings of distantly related forest species. Tree Physiol 27: 1459–1470 [DOI] [PubMed] [Google Scholar]
- Schmelz EA, Carroll MJ, LeClere S, Phipps SM, Meredith J, Chourey PS, Alborn HT, Teal PEA(2006) Fragments of ATP synthase mediate plant perception of insect attack. Proc Natl Acad Sci USA 103: 8894–8899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze S, Schäfer BN, Parizotto EA, Voinnet O, Theres K(2010) LOST MERISTEMS genes regulate cell differentiation of central zone descendants in Arabidopsis shoot meristems. Plant J 64: 668–678 [DOI] [PubMed] [Google Scholar]
- Stuurman J, Jäggi F, Kuhlemeier C(2002) Shoot meristem maintenance is controlled by a GRAS-gene mediated signal from differentiating cells. Genes Dev 16: 2213–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Reinhart BJ, Bartel DP, Zamore PD(2003) A biochemical framework for RNA silencing in plants. Genes Dev 17: 49–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavormina P, De Coninck B, Nikonorova N, De Smet I, Cammue BPA(2015) The plant peptidome: An expanding repertoire of structural features and biological function. Plant Cell 27: 2095–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wynsberghe PM, Chan SP, Slack FJ, Pasquinelli AE(2011) Analysis of microRNA expression and function. Methods Cell Biol 106: 219–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim AG, Jia Q, Vaughn JN(2014) Regulation of plant translation by upstream open reading frames. Plant Sci 214: 1–12 [DOI] [PubMed] [Google Scholar]
- Vielba JM, Díaz-Sala C, Ferro E, Rico S, Lamprecht M, Abarca D, Ballester A, Sánchez C(2011) CsSCL1 is differentially regulated upon maturation in chestnut microshoots and is specifically expressed in rooting-competent cells. Tree Physiol 31: 1152–1160 [DOI] [PubMed] [Google Scholar]
- Waterhouse PM, Hellens RP(2015) Plant biology: Coding in non-coding RNAs. Nature 520: 41–42 [DOI] [PubMed] [Google Scholar]
- Wang L, Mai YX, Zhang Y-C, Qian L, Yang H-Q(2010) MicroRNA171c-targeted SCL6-II, SCL6-III, and SCL6-IV genes regulate shoot branching in Arabidopsis. Mol Plant 3: 794–806 [DOI] [PubMed] [Google Scholar]
- Xie Z, Allen E, Fahlgren N, Calamar A, Givan SA, Carrington JC(2005) Expression of Arabidopsis MIRNA genes. Plant Physiol 138: 2145–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Copeland C(2012) Nuclear Extraction from Arabidopsis thaliana. Bio Protoc 2: e306 [Google Scholar]
- Xue XY, Zhao B, Chao LM, Chen DY, Cui WR, Mao YB, Wang LJ, Chen XY(2014) Interaction between two timing microRNAs controls trichome distribution in Arabidopsis. PLoS Genet 10: e1004266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Pan X, Cannon CH, Cobb GP, Anderson TA(2006) Conservation and divergence of plant microRNA genes. Plant J 46: 243–259 [DOI] [PubMed] [Google Scholar]
- Zhou ZX, Du ZJ, Chen YY, Zhao JJ, Wang WL(2009) Effects of polypeptide treatment on the calcium nutrition absorption of mango fruit. Guangdong Agricultural Sciences 2: 21–22 [Google Scholar]