Abstract
Fibroblast growth factors (FGFs) 9 and 10 are essential during the pseudoglandular stage of lung development. Mesothelium-produced FGF9 is principally responsible for mesenchymal growth, whereas epithelium-produced FGF9 and mesenchyme-produced FGF10 guide lung epithelial development, and loss of either of these ligands affects epithelial branching. Because FGF9 and FGF10 activate distinct FGF receptors (FGFRs), we hypothesized that they would control distinct developmental processes. Here, we found that FGF9 signaled through epithelial FGFR3 to directly promote distal epithelial fate specification and inhibit epithelial differentiation. By contrast, FGF10 signaled through epithelial FGFR2b to promote epithelial proliferation and differentiation. Furthermore, FGF9-FGFR3 signaling functionally opposed FGF10-FGFR2b signaling, and FGFR3 preferentially used downstream phosphoinositide 3-kinase (PI3K) pathways, whereas FGFR2b relied on downstream mitogen-activated protein kinase (MAPK) pathways. These data demonstrate that, within lung epithelial cells, different FGFRs function independently; they bind receptor-specific ligands and direct unique developmental functions through activation of distinct downstream signaling pathways.
Introduction
Mouse lung development begins with the emergence of two buds from the foregut at embryonic day 9.5 (E9.5). Between E9.5 and E16.5, the pseudoglandular stage of development forms the primary airway branching pattern of the lung. This is followed by the canalicular (E16.5–17.5), saccular (E18.5 to postnatal day 4 (P4)), and finally alveolar (P4-P21) stages that progressively form a functional adult lung (1–3). The lung bud contains three primary cell types: epithelium, mesenchyme, and mesothelium. The innermost layer, the epithelium, gives rise to the branched network of ducts that will conduct air to the alveoli, the distal gas-exchange units of the lung. Mesenchyme surrounding the epithelial ducts will form the vascular and stromal components of the lung. The mesothelium is the single layer of cells that envelopes all lung lobes. The mesothelium, mesenchyme, and epithelium all interact through complex signaling networks to orchestrate lung development (4, 5).
Fibroblast Growth Factor (FGF) signaling pathways function in all stages of lung development. There are 18 signaling FGF ligands that activate four transmembrane tyrosine kinase receptors (FGFRs). Fgfrs are expressed in both mesenchyme and epithelium, and these cell types preferentially produce alternative splice variants of FGFRs, which modifies their ligand-binding specificities and thus the responsiveness of these cell types to different FGF ligands (6). During the early pseudoglandular stage (E9.5-E14.5), FGF10 is produced in the mesenchyme and FGF9 is produced in the mesothelium and epithelium (5, 7).
FGF10 primarily signals to the b splice variant of FGFR2 (FGFR2b) in the lung epithelium (8, 9). In the absence of FGF10 or FGFR2b, the initial lung buds form but fail to grow or branch (10–14). However, when FGF10-FGFR2b signaling is reduced or conditionally disrupted after initial bud formation, branching is impaired and, in some studies, epithelial cell death is increased (15–20). Downstream of FGF10-FGFR2b, the extracellular signal-regulated kinase (ERK) mitogen-activated protein kinase (MAPK) signaling cascade directs epithelial proliferation and bud outgrowth (21, 22).
Fgf9 is expressed in lung epithelium during the first half of the pseudoglandular stage of development, from E9.5-E12.5, and in the mesothelium throughout most of the pseudoglandular stage, E9.5-E14.5 (23–26). Mice lacking FGF9 (Fgf9−/−) die at birth and their lungs are small with reduced mesenchyme and airway branching (27, 28). Investigation of the differential roles of epithelial- and mesenchymal-derived FGF9 signaling showed that mesothelial-derived FGF9 mainly functions to direct mesenchymal proliferation. Mechanistic studies identified FGFR1 and FGFR2 as the mesenchymal receptors that respond to FGF9, phosphorylated ERK as a downstream signal, and mesenchymal Wnt-β-Catenin activation as a feedforward signal that is essential to maintain mesenchymal Fgfr1 and Fgfr2 expression (5, 26, 29, 30).
By contrast, inactivation of epithelial FGF9 primarily affects epithelial budding (26), and overexpression of FGF9 leads to epithelial dilation (28, 31, 32). Initial models showed that FGF9 overexpression in pseudoglandular-stage epithelium induces Fgf7 and Fgf10 in adjacent mesenchyme, and it was hypothesized that this increase in FGF7 and FGF10 causes the observed epithelial dilation (28, 32). However, evidence from several studies indicate that the effects of FGF9 on pseudoglandular-stage epithelium may result from FGF9 signaling directly to the epithelium. For example, isolated lung epithelium from E12.5 embryos directly responds to FGF9 (32), and lung explants lacking mesenchymal FGFR1 and FGFR2 fail to show a mesenchymal response to FGF9 (growth), but still show epithelial dilation in response to FGF9, consistent with FGF9 signaling directly to epithelium (26, 29). A study from del Moral et al. reported that epithelial explants lacking FGFR2 fail to respond to FGF9 and concluded that FGF9 induces distal epithelial proliferation directly through activation of epithelial FGFR2b (32). However, this is inconsistent with receptor specificity studies (8) and with in vivo overexpression of FGF9 in the lung epithelium, which results in mesenchymal hyperplasia and epithelial dilation without affecting epithelial proliferation (31).
The epithelial FGFR that responds to FGF9 during pseudoglandular stage lung development is thus not known. In adult lung, overexpression of FGF9 in lung epithelium results in adenocarcinoma without affecting lung stroma (mesenchyme), and the induction of adenocarcinoma by FGF9 is exclusively dependent on FGFR3 (33, 34). Collectively, these data show that FGF9 signals through FGFR1 and FGFR2 in pseudoglandular-stage mesenchyme and FGFR3 in adult epithelium. Consistent with this, in vitro receptor specificity studies show that FGF9 can efficiently activate mesenchymal FGFR1c and FGFR2c and epithelial FGFR3b splice variants (8, 35, 36). We therefore hypothesized that FGF9 signaling to FGFR3 may also function during embryonic lung development.
Previous studies convincingly show that the original notions of strict and independent epithelial-to-mesenchymal and mesenchymal-to-epithelial FGF signaling do not account for all of the FGF signals in pseudoglandular-stage lung development (26, 29, 32). However, why several non-redundant FGFs are required for lung epithelial development and how signaling by these different FGFs is coordinated in lung epithelial cells, is not known. Here, we examined how FGF9 and FGF10 signaling contribute unique functions during pseudoglandular-stage lung development. We identified a partially antagonistic relationship between FGF10-FGFR2b and FGF9-FGFR3 signaling in which FGF9 directly promoted distal epithelial fate specification and inhibited epithelial differentiation through FGFR3, whereas FGF10 promoted epithelial proliferation and differentiation through FGFR2. We further showed that these two ligand-receptor pairs relied on different downstream signaling pathways.
Results
FGF9 signals to lung epithelium in the absence of functional mesenchymal FGF receptors
To determine whether FGF9 can directly signal to the lung epithelium (the tissue in which it is produced) in whole-lung explants, we tested explants with genetically impaired mesenchymal FGFR1 and FGFR2 signaling for their responsiveness to FGF9. We used the Dermo1Cre (Twist2Cre) allele to target lung mesenchyme (28, 37) and mated these mice with mice carrying floxed alleles of β-Catenin (Ctnnb1) or Fgfr1 and Fgfr2 (37–39). Lungs with the genotype Dermo1Cre/+; β-Cateninf/f (Dermo1-βCat-KO), Dermo1Cre/+; Fgfr1f/f; Fgfr2f/f (Dermo1-FGFR1/2-KO), and Fgf9LacZ/LacZ (Fgf9−/−) (40) were placed in explant culture on embryonic day 11.5 (E11.5) for 48 h. Due to loss of a mesenchymal FGFR–Wnt–β-Catenin feed-forward signaling loop, all of these genotypes lack mesenchymal FGFR1 and FGFR2 signaling (26, 29). Wild-type explants treated with FGF9 showed mesenchymal expansion, epithelial dilation, and reduced numbers of buds (Fig. 1, A and B), consistent with previous observations (26, 28, 29, 32). Additionally, in vivo overexpression of FGF9 from E10.5 to E12.5 induces mesenchymal but not epithelial proliferation (31). By contrast, Dermo1-βCat-KO, Dermo1-FGFR1/2-KO, and Fgf9−/− explants all showed an absence of mesenchymal growth in response to FGF9 but still showed marked epithelial dilation and reduced epithelial branching (Fig. 1, A, C to E). These data showed that lung epithelium could respond to FGF9 independently of mesenchymal FGF signaling in whole-organ explants.
Fig. 1. FGF9 signals to the lung epithelium in the absence of functional mesenchymal FGF receptors.
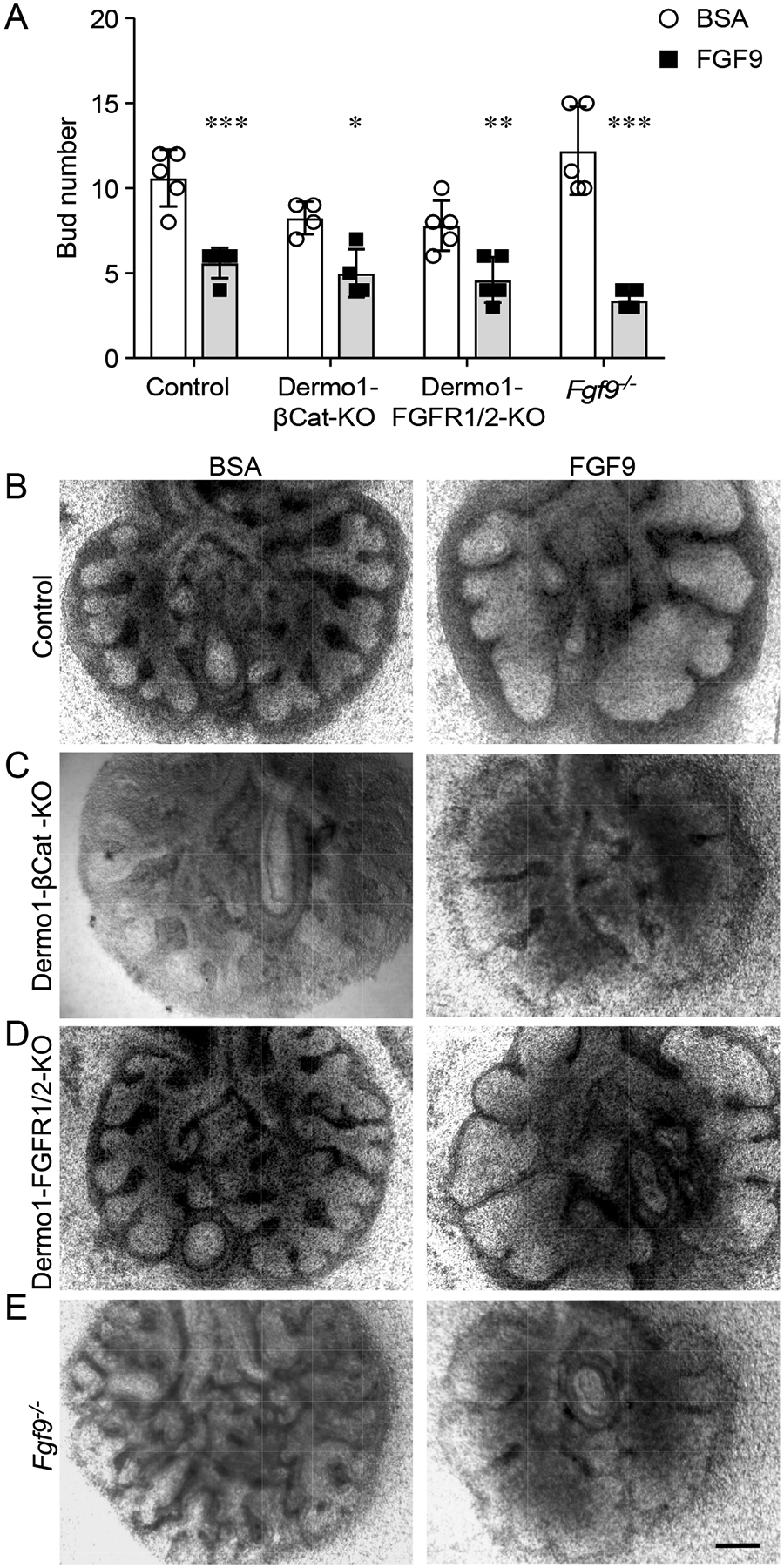
(A) Quantification of bud number in response to BSA or FGF9 in lung explants from E11.5 embryos of the indicated genotypes and wild-type controls cultured for 48 h. (B–E) Explants from wild-type control (B), Dermo1-βCat-KO (C), Dermo1-FGFR1/2-KO (D), and Fgf9−/− (E) mice cultured in media with BSA or FGF9. 2-Way ANOVA, Sidak’s multiple comparisons test, n= 4–5 explants per genotype and condition. *p< 0.01, ** p< 0.02, *** p< 0.0001). Scale bar, 200 μm.
FGF9 signals to lung epithelium through FGFR3
To further investigate whether FGF9 could directly target lung epithelium, E11.5 wild-type distal lung epithelial bud tips were isolated and cultured in Matrigel in the presence or absence of FGF9. After 48 h of culture, isolated epithelial explants failed to expand in the absence of FGF9 and formed an enlarged cyst-like structure in the presence of FGF9 (Fig. 2, A and B). Consistent with previous studies (32), these data show that functional FGF receptor(s) that can respond to FGF9 must be present on the surface of distal lung epithelium. Based on previous studies showing that FGF9 signals through FGFR3 in adult lung epithelium to induce adenocarcinoma and specificity studies that identify the FGF9 family as uniquely able to activate the b splice variant of FGFR3 (8, 33–36), we hypothesized that FGFR3 mediated the response of embryonic lung epithelium to FGF9. Immunostaining of E12.5 and E13.5 lung sections showed the presence of both FGFR2 and FGFR3 on the basal surface of the distal developing airway epithelium and in the mesenchyme (fig. S1). This similar epithelial expression pattern suggested that epithelial cells produce both receptors. At E13.5, both receptors were detected in the epithelium, and FGFR2 was also present throughout mesenchyme, whereas FGFR3 was also present in a subset of lung mesenchymal cells (fig. S1). The failure of epithelial explants derived from Fgfr3−/− mice to respond to FGF9 (Fig. 2, A and B), and the observation that FGFR3 is produced by embryonic lung epithelium, is consistent with a requirement for FGFR3 for the epithelial response to FGF9.
Fig. 2. FGF9 signals to the lung epithelium through FGFR3.
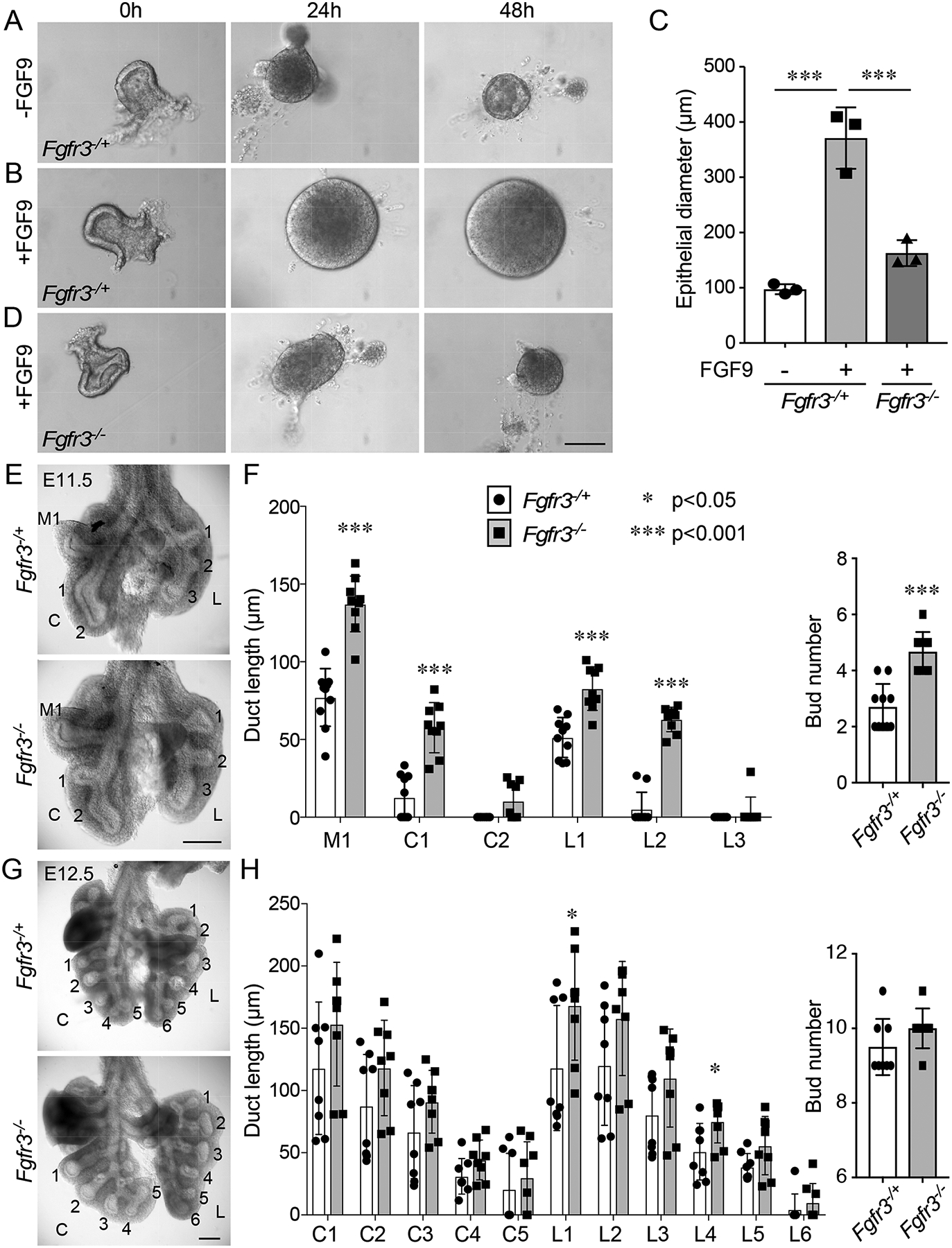
(A) Isolated E11.5 distal lung epithelium from Fgfr3−/+ and Fgfr3−/− embryos cultured in Matrigel in the presence or absence of FGF9 for the indicated periods of time. (B) Quantification of lung epithelial diameter in tissue from (A) at 48 h. ANOVA with Tukey’s multiple comparison test. n = 3 epithelial explants per genotype. (C) Anterior views of gross dissections of Fgfr3−/+ and Fgfr3−/− lungs at E11.5. (D) Quantification of duct length and bud number in Fgfr3−/+ and Fgfr3−/− lungs at E11.5. Duct lengths and number of buds were counted in middle (M), caudal (C), and left (L) lobe regions. (E) Anterior views of gross dissections of Fgfr3−/+ and Fgfr3−/− lungs at E12.5. (F) Quantification of duct length and bud number in Fgfr3−/+ and Fgfr3−/− lungs at E12.5. The caudal lobe and left lobe duct lengths and bud numbers were counted. Bud length was analyzed using the Wilcoxon rank sum test, and bud number was analyzed using the Student’s t-test. n = 9–10 lungs per genotype at E11.5 and 8 lungs per genotype at E12.5. * p< 0.05; *** p< 0.001. Scale bars, 100 μm (A), 200 μm (C and E).
To determine if FGFR3 was required in vivo during early pseudoglandular-stage development, lungs were isolated from E11.5 and E12.5 heterozygous (Fgfr3−/+) and knockout (Fgfr3−/−) embryos. Lungs from Fgfr3−/− embryos appeared larger than those from Fgfr3−/+ embryos at both time points. At E11.5, quantification of bud number and length revealed increased numbers of buds and increased bud length in Fgfr3−/− compared to Fgfr3−/+ lungs (Fig. 2, C and D). At E12.5, there was a trend towards increased bud length that was only significant in the left lobe (L1 and L4) ducts (Fig. 2, E and F). Bud number was not significantly different at E12.5. Consistent with this phenotype, Fgfr3−/+ E11.5 explants (cultured for 48 h) showed a greater decrease in bud number in response to treatment with FGF9 compared to Fgfr3−/− explants treated with FGF9 (fig. S2, A to C). We interpret this as failure of FGF9 to suppress branching in the absence of FGFR3. In the absence of added FGF9, bud number was similar between Fgfr3−/+ and Fgfr3−/− explants. These data show that FGFR3 affects lung epithelial development during early embryonic stages but that this phenotype is masked or compensated at later stages.
FGF9 directs distal epithelial specification and differentiation but not proliferation or viability
We previously showed that lung epithelial proliferation was not significantly affected in Fgf9−/− embryos at E10.5-E13.5 (27) or in embryos in which FGF9 overexpression was induced from E10.5-E12.5 (31). To confirm that FGF9 signaling does not affect lung epithelial proliferation at the pseudoglandular stage and to extend the analysis to include any effects on cell death, we examined the presence of phosphorylated histone H3 (PHH3) and terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) in the lungs of control (Fgf9−/+) and Fgf9−/− (Fgf9LacZ allele) embryos at E10.5, E11.5, and E12.5 (fig. S3, A and B). Quantification of labeled nuclei per total nuclei in epithelium showed no significant change in cell death (Table 1A) or proliferation (Table 1B) in control and Fgf9−/− lungs at these time points.
Table 1.
Cell death and cell proliferation in lung epithelium.
| A | TUNEL+ cells/1000 epithelial cells | ||
|---|---|---|---|
| E10.5 | E11.5 | E12.5 | |
| Control | 10.6# ±6.7 | 0.0 ±0 | 0.6 ±0.5 |
| Fgf9−/− | 1.7 ±2.9 | 0.8 ±1.4 | 0.9 ±0.7 |
| p | 0.1 | 0.4 | 0.7 |
| B | PHH3+ cells/1000 epithelial cells | ||
| E10.5 | E11.5 | E12.5 | |
| control | 65.8 ±29.0 | 44.5 ±32.7 | 52.6 ±23.2 |
| Fgf9−/− | 81.1 ±26.0 | 73.6 ±8.9 | 31.2 ±12.0 |
| p | 0.5 | 0.2 | 0.2 |
Mean ± SD. Three control and three Fgf9−/− lungs were sectioned and all epithelial cells in all sections from each lung were counted (294–1411 epithelial cells/lung).
Because the loss of Fgf9 did not affect epithelial proliferation or cell death, we next examined epithelial specification and differentiation along the proximal-distal axis of lung branches. SOX9 marks embryonic lung epithelial progenitors that undergo branching morphogenesis. SOX9 progenitors give rise to SOX2-producing cells that will become the future conducting and alveolar airway epithelium (41, 42). SOX2 therefore marks the non-branching proximal lung epithelium, whereas SOX9 is restricted to the distal branching epithelium (43, 44).
At E13.5, the proximal SOX2 domain was greatly expanded and the SOX9 domain was restricted to the most distal regions in Fgf9−/− lungs compared to heterozygous control lungs (Fig. 3, A and B). To test the effects of FGF9 overexpression on proximal-distal specification, Spc-rtTA; Tre-Fgf9-Ires-Gfp (SPC-FGF9-OE) mice and Spc-rtTA control mice were induced with doxycycline from E10.5-E14.5. In contrast to the loss-of-function experiment, when FGF9 was overexpressed, all airway epithelia produced SOX9, whereas SOX2 was absent from all but the very proximal tracheal epithelium (Fig. 3C). Furthermore, overexpression of FGF9 or FGF10 (E10.5-E12.5) did not affect the production of NKX2–1, a marker of lung epithelial identity (fig. S4A). We also evaluated the distribution of TRP63 and KRT5, markers of basal cells within the lung epithelium, in FGF9- and FGF10-overexpressing lungs. TRP63 was only present in proximal airway epithelium (fig. S4B), and KRT5 was not detected in the lung at this stage, but it was present in the esophagus (fig. S4C). Together, these data suggest that FGF9 may suppress the conversion of SOX9-producing progenitors to non-branching, SOX2-producing respiratory epithelium during the pseudoglandular stages of lung development without affecting epithelial identity.
Fig. 3. FGF9 directs distal epithelial specification and differentiation in th e pseudoglandular-stage lung.
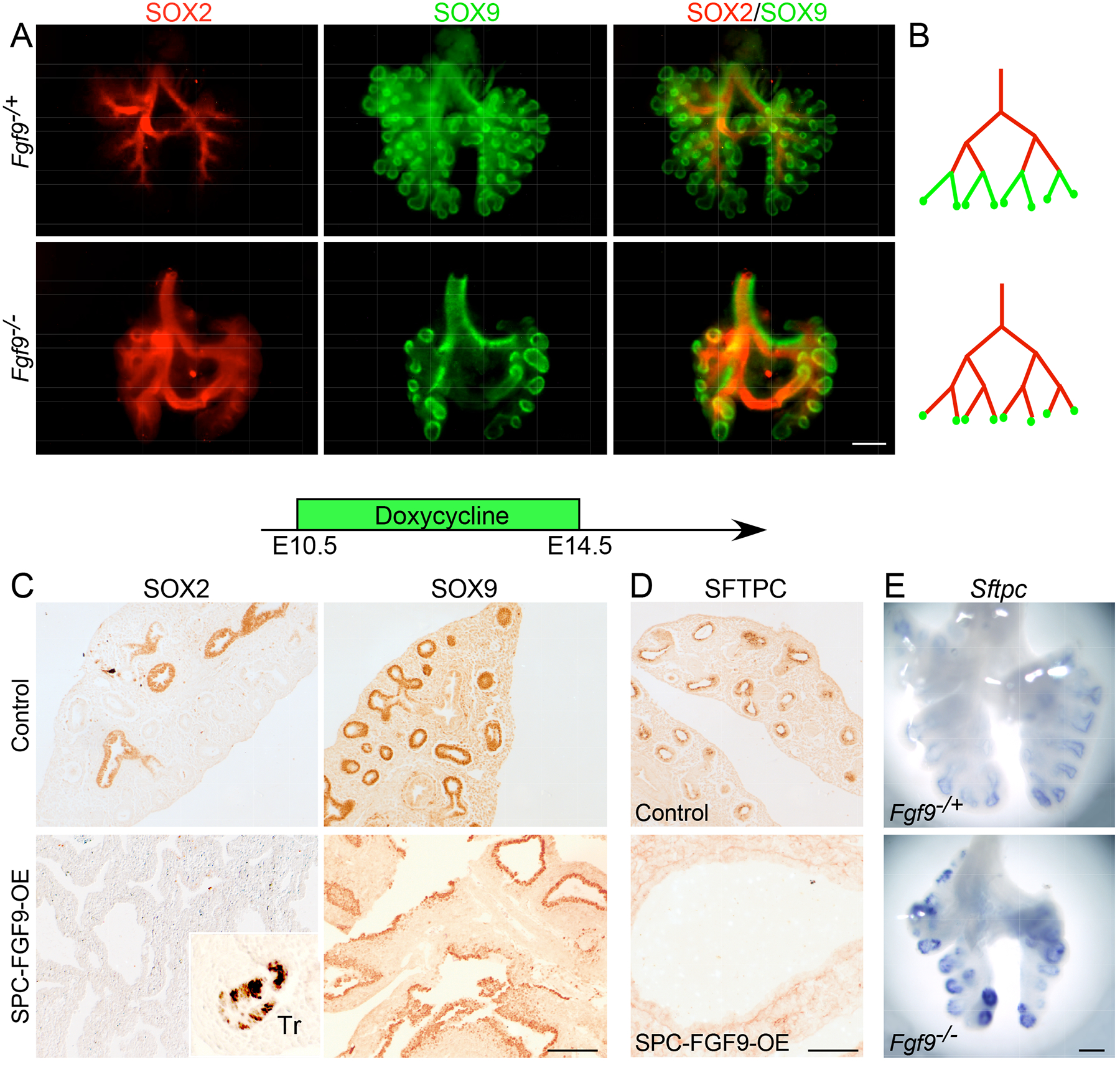
(A) Immunofluorescence imaging showing SOX2 and SOX9 distribution in whole E13.5 Fgf9−/+ and Fgf9−/− lungs. (B) Diagram of the distribution patterns shown in (A). (C, D) Immunostaining for SOX2 and SOX9 (C) and SFTPC (D) in control (Sftpc-rtTA) and SPC-FGF9-OE lung sections that were induced with doxycycline from E10.5–14.5 and harvested at E14.5. (E) Comparison of Sftpc expression (in situ hybridization) in Fgf9−/+ and Fgf9−/− lungs at E12.5. All images are representative of at least 3 embryos. Scale bars, 200 μm (A and E) and 100 μm (C and D).
We next examined the effects of FGF9 on epithelial differentiation by examining expression of Sftpc, an early marker of epithelial differentiation (31). Consistent with FGF9 functioning to suppress epithelial differentiation in distal regions of buds, inducing overexpression of FGF9 from E10.5 to E14.5 inhibited all SFTPC production (Fig. 3D). By contrast, at E12.5, distal epithelium showed increased expression of Sftpc in Fgf9−/− lungs compared to Fgf9−/+ control lungs (Fig. 3E). Markers of late-stage distal epithelial cell differentiation, HOPX and LPCAT1, were not detected at this stage and were not affected by overexpression of FGF9 (fig. S4, D and E).
Because FGFR3 is likely the relevant receptor for FGF9 in vivo, we also examined the effects of loss of FGFR3 on proximal-distal epithelial specification. Similar to Fgf9−/− lungs, Fgfr3−/− lungs showed a distal expansion of the SOX2 distribution pattern (fig. S5A). However, unlike the Fgf9−/− lungs, the SOX9 distribution pattern in Fgfr3−/− lungs remained similar to the control, resulting in an increased region of epithelium that contained both SOX2 and SOX9 (fig. S5B arrow). Similar to the Fgf9−/− lungs, distal epithelium showed increased production of SFTPC in Fgfr3−/− lungs (fig. S5C). These data support a model in which FGF9 signals to FGFR3, but also suggests that FGF9 may transmit additional signals, either through another epithelial FGFR or, indirectly, through mesenchymal FGFR1 and FGFR2, which should still be active in the Fgfr3−/− lungs.
To determine whether FGF9 could induce SOX9 in canalicular-stage bronchial epithelium, SPC-FGF9-OE and control mice were induced with doxycycline from E16.5-E18.5. In control mice, SOX2 was distributed in all conducting airways, and SOX9 was located in only the most distal epithelial cells, whereas in SPC-FGF9-OE mice, SOX2 was only present in the most proximal airway epithelium, and the SOX9 distribution pattern was expanded to most of the airway epithelium (Fig. 4, A to C). Similar to earlier time points, NKX2–1 was present in all lung epithelial cells of FGF9-overexpressing lungs, confirming that lung epithelial identity was retained (Fig. 4D). The basal cell markers, TRP63 and KRT5, were only expressed in the proximal epithelium and were not affected by overexpression of FGF9 (Fig. 4E and fig. S6A). Production of LPCAT1, a marker of distal progenitors and alveolar type 2 (AT2) cells (45), was observed in the distal epithelium in controls and throughout the lung epithelium in FGF9-overexpressing mice (fig. S6B). Production of HOPX, an AT1 cell marker (46), was observed in the distal epithelium of control lungs and was sparsely distributed throughout the epithelium of FGF9-overexpressing lungs (fig. S6C). Collectively, these data suggest that at a late embryonic time point, FGF9 can drive specified proximal bronchial epithelium to a less differentiated, distal-like state with expanded domains of SOX9 and LPCAT1 production and restricted domains of HOPX production.
Fig. 4. FGF9 can reverse epithelial specification without affecting epithelial identity.
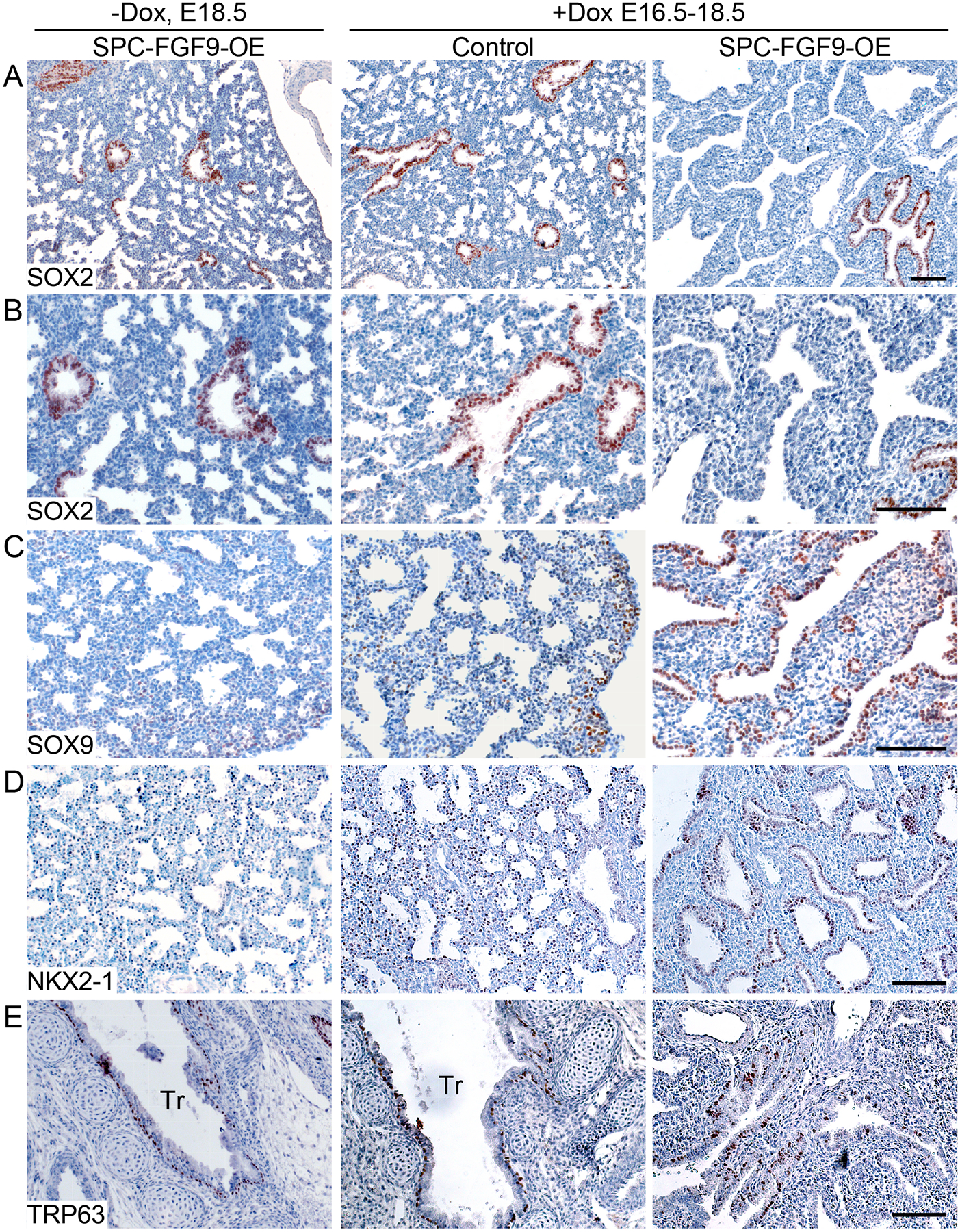
SPC-FGF9-OE mice and Sftpc-rtTA control mice were induced with doxycycline (Dox) from E16.5–18.5, and lung sections were immunostained for SOX2 (A, B), SOX9 (C), NKX2–1 (D), KRT5 (E), and TRP63 (F). Additional controls included SPC-FGF9-OE mice that were not induced with doxycycline (left). All images are representative of at least 3 embryos. Scale bars, 100μm.
FGF9 and FGF10 have distinct effects on pseudoglandular-stage lung epithelium
To directly compare the effects of overexpressing FGF9 and FGF10 on pseudoglandular-stage lung development, we induced Spc-rtTA; Tre-Fgf10 (SPC-FGF10-OE) and SPC-FGF9-OE mice with doxycycline from E10.5 to E12.5. In both models, the overall size of the lungs was larger compared to Spc-rtTA controls (Fig. 5A). Both FGF9- and FGF10-induced lungs showed mesenchyme expansion and distal airway dilation, although this was more prominent in the lungs overexpressing FGF9. However, when proximal-distal specification was examined by immunostaining for SOX2 and SOX9, SPC-FGF9-OE lungs clearly showed expansion of the SOX9-producing epithelium and loss of the SOX2-producing epithelium (Fig. 3C and 5B), whereas SPC-FGF10-OE lungs showed relatively normal patterns of SOX2 and SOX9 distribution, which were comparable to control lungs (Fig. 5B).
Fig. 5. FGF9 and FGF10 have distinct effects on pseudoglandular stage lung epithelium.
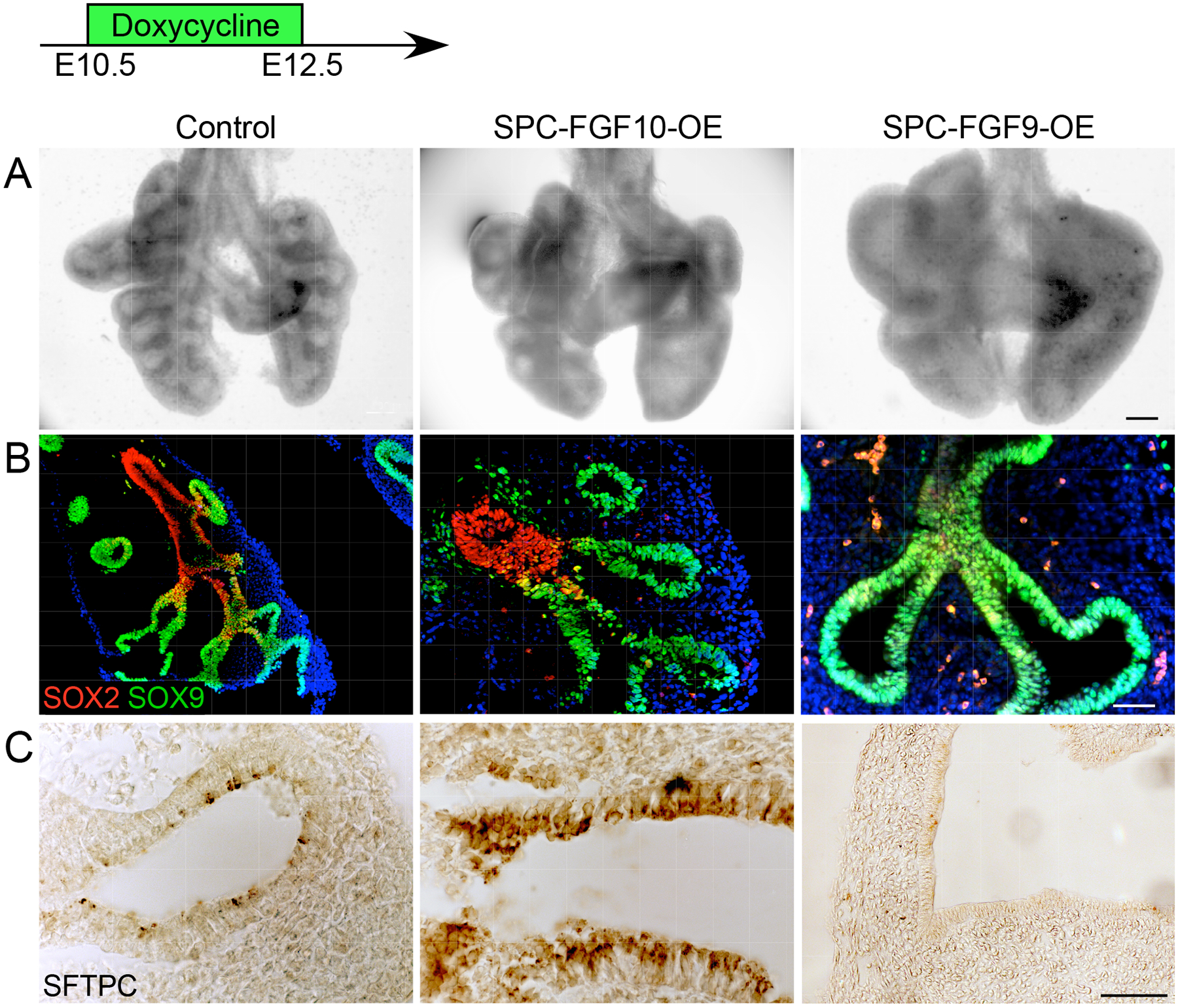
(A) Anterior views of whole E12.5 lungs from mice expressing control (Sftpc-rtTA), FGF10 overexpression (SPC-FGF10-OE), or FGF9 overexpression (SPC-FGF9-OE) transgenes. All embryos were induced with doxycycline from E10.5–12.5. (B) Immunofluorescence showing SOX2 and SOX9 distribution. (C) Immunostaining showing SFTPC distribution. All images are representative of at least 3 embryos. Scale bars, 200 μm (A), 100 μm (B), 50 μm (C).
We next examined the effects of FGF9 and FGF10 overexpression on epithelial differentiation by immunostaining for SFTPC. Control lungs showed detectable SFTPC production at E12.5 (Fig. 5C). SPC-FGF9-OE lungs, induced with doxycycline from E10.5 to E12.5, showed an absence of SFTPC production. However, SPC-FGF10-OE lungs showed normal or possibly increased SFTPC (Fig. 5C) and LPCAT1 (fig. S4E) production. To complement the FGF10 activation studies, we also looked at lungs in which the FGF10 receptor, FGFR2, was inactivated in the lung epithelium. Mice carrying the ShhCreER; ROSAtdTomato (Ai9) alleles were injected with tamoxifen at E10.5, and embryos were harvested at E13.5 (Fig. 6A). tdTomato-positive cells were primarily located in the distal epithelium and in epithelial tips (Fig. 6B). We then generated a ShhCreER; Fgfr2f/f (SHH-FGFR2-CKO) mouse line in which the FGF10 receptor, FGFR2, could be conditionally inactivated in this distal lung epithelial domain after the initiation of branching morphogenesis. Female mice carrying SHH-FGFR2-CKO embryos were injected with tamoxifen at E10.5, and the lungs were collected at E13.5. Compared to ShhCreER control lungs, SHH-FGFR2-CKO lungs were larger, with more mesenchyme and elongated ducts (Fig. 6C). Consistent with the FGF10 overexpression data, when FGFR2 was inactivated there were relatively small effects on the distribution pattern of SOX2 (Fig. 6D). The extent of SOX9-producing epithelium, however, was somewhat expanded (Fig. 6E). Epithelial differentiation, assessed by immunostaining, showed notably reduced distribution of SFTPC in SHH-FGFR2-CKO compared to control lungs (Fig. 6F).
Fig. 6. Conditional epithelial inactivation of Fgfr2 shows similar phenotypes to Fgf9 overexpression.
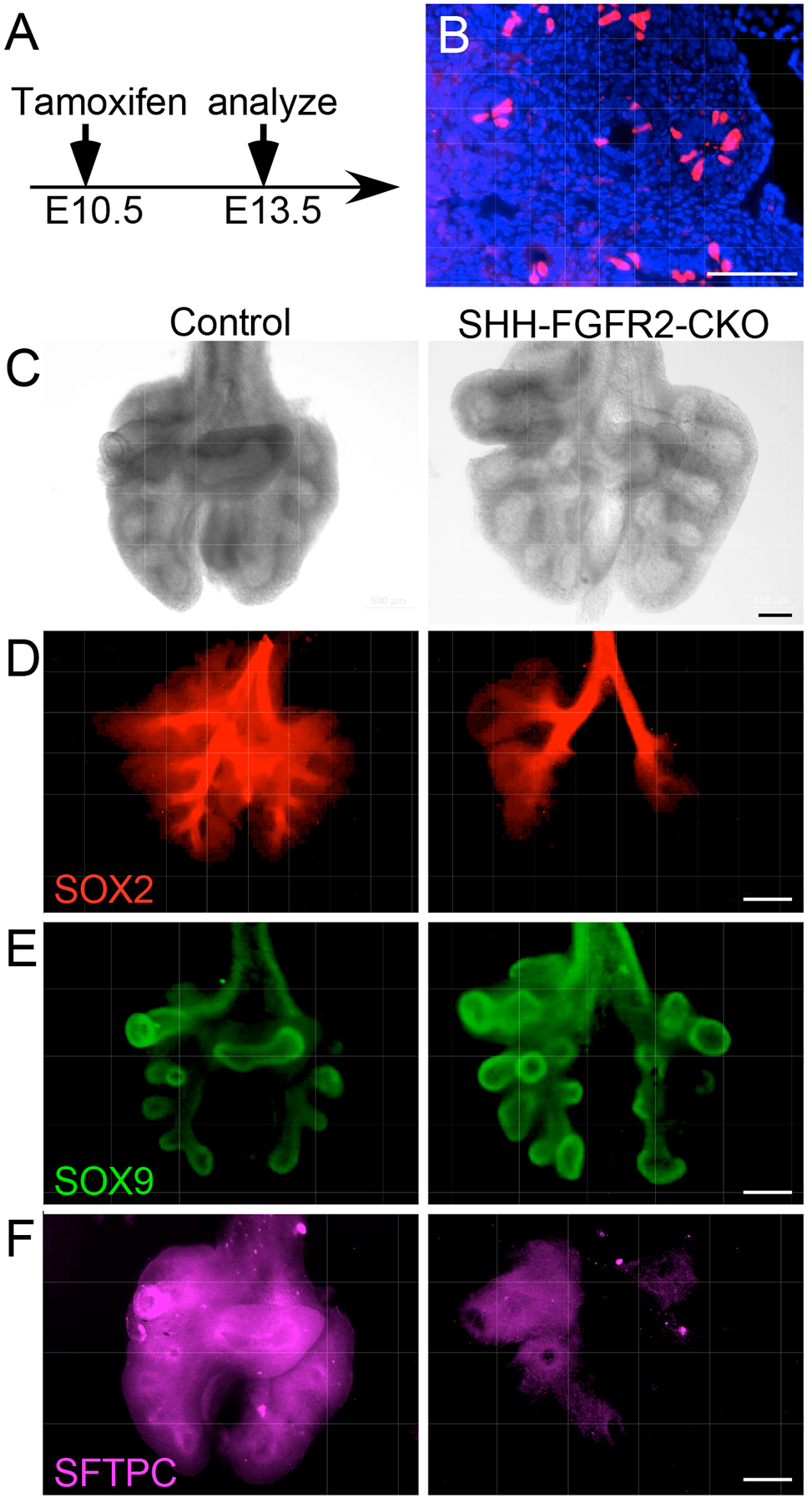
(A) Experimental plan. All embryos were induced with Tamoxifen (intraperitoneal injection of dam) at E10.5 and harvested at E13.5. (B) Frozen section of a lung from ShhCreER; ROSAtdTomato embryo showing lineage-labeled cells in the epithelial tips. (C–F) Anterior views of E13.5 whole control lungs [ShhCreER (C, D) or ShhCreER; Fgfr2f/+ (E, F)] and lungs lacking epithelial FGFR2 (SHH-FGFR2-CKO). Brightfield images (C), and immunostaining for SOX2 (D), SOX9 (E,) and SFTPC (F) are shown. All images are representative of at least 3 embryos. Scale bars, 100 μm (B), 50 μm (C), 200 μm (D to F).
These data indicate that FGF9 has much greater effects on epithelial specification than does FGF10, with FGF9 functioning to maintain a SOX9-producing distal epithelium, whereas FGF10 directs duct outgrowth and branching. Furthermore, the epithelial differentiation phenotype associated with inactivation of epithelial FGFR2 (reduced production of SFTPC) resembles that of FGF9 activation (reduced production of SFTPC). Together, these data suggest that FGF9-FGFR3 signaling primarily directs specification, and FGF10-FGFR2b signaling primarily directs proliferation, but not specification, in the pseudoglandular-stage lung epithelium.
FGF10-FGFR2b and FGF9-FGFR3 activate different signaling pathways
FGFRs activate four primary downstream signaling pathways – RAS-MAPK, phosphoinositide 3-kinase (PI3K), phospholipase C γ (PLCγ) and signal transducer and activator of transcription (STAT) – to mediate cell proliferation, migration, differentiation, and survival (6, 47, 48). However, most signaling studies have focused on FGFR1 and to some extent FGFR2, and very few studies have addressed differences in downstream signaling between the four FGFRs (47). Because activation of FGFR2 and FGFR3 in embryonic lung epithelium have distinct outcomes, we hypothesized that these FGFRs may preferentially use different downstream signaling pathways. To address this question, we examined the response of lung explants to FGF9 and FGF10 in the presence of inhibitors of the PI3K, MAPK, and STAT3 signaling pathways.
Wild-type lung explants were placed in culture on embryonic day 11.5 (E11.5) for 48 h. Over this period, the explants increased in size, and epithelial ducts branched and extended distally (Fig. 1A and 7A). As shown in several studies and here, treatment of explants with FGF9 resulted in mesenchymal growth, reduced branching, and epithelial dilation (Fig. 1A, 7A) (26, 28, 29, 32, 49, 50), whereas treatment with FGF10 resulted in increased branching (Fig. 7A) (10, 49–52). In the presence of the mitogen-activated protein kinase kinase (MEK) inhibitor, U0126, which blocks FGFR downstream signaling mediated by the MAPK ERK, explants showed reduced branching and epithelial dilation (Fig. 7B), a phenotype that resembles that of wild-type explants treated with FGF9 (Fig. 7A). By contrast, treatment of explants with a PI3K inhibitor, Ly2294002, resulted in increased branching (Fig. 7C), a phenotype that resembles that of wild-type explants treated with FGF10 (Fig. 7A). Treatment with U0126 had little effect on the response to FGF9, which still induced epithelial dilation and suppressed branching, but strongly inhibited FGF10-induced branching (Fig. 7B). Ly2294002 suppressed the epithelial dilation induced by FGF9 and had very little effect on branching induced by FGF10 (Fig. 7C). Of note, FGF9 also strongly suppressed the increased branching induced by Ly2294002 (Fig. 7C), suggesting more complexity in the downstream signaling pathways activated by FGF9 or indirect effects of FGF9 signaling through mesenchyme. Treating wild-type explants with both MEK and PI3K inhibitors suppressed both epithelial branching and dilation (Fig. 7D) and caused epithelial cell death (Fig. 7, E and F). STAT3 is produced in the developing lung (22, 53) and can be activated by FGFRs (54). In the presence of the STAT3 inhibitor, Stattic (55), control explants showed modestly increased branching, but cotreatment with Stattic and FGF10 did not increase branching compared to treatment with FGF10 alone (fig. S7, A and B). STAT3 inhibition also had little effect on the epithelial dilation and reduction in branching induced by FGF9 (fig. S7, A and C).
Fig. 7. Preferential use of distinct cellular signaling pathways by FGF9 and FGF10.
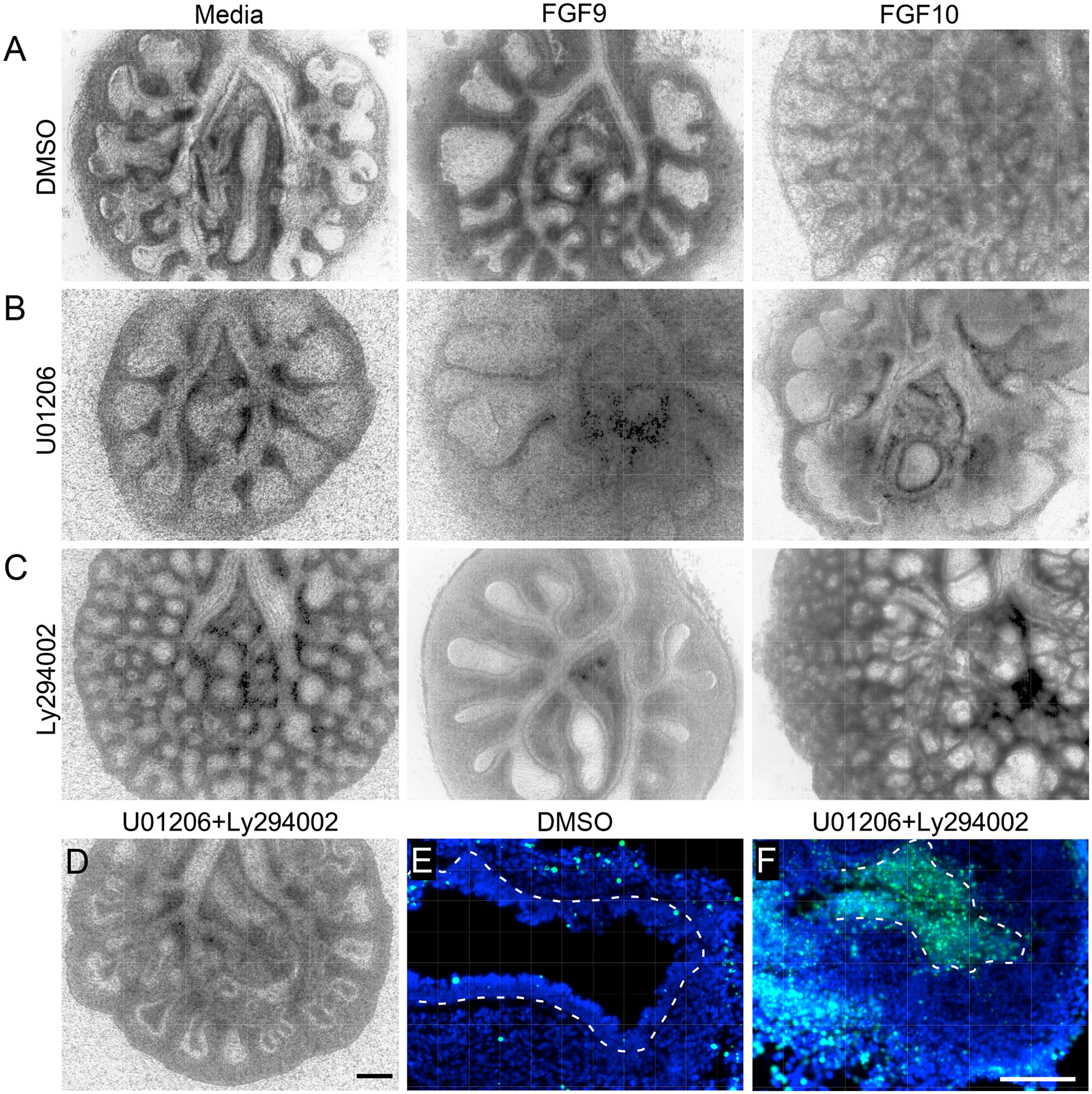
(A–C) E11.5 wild-type lung explants cultured for 48 h in media alone, with FGF9, or with FGF10 as indicated, in the presence of DMSO (A), the MEK inhibitor U01206 (B), or the PI3K inhibitor Ly294002, (C). (D–F) E11.5 wild-type lung explants cultured for 48 h and treated with U01206 and Ly294002 (D and F) or DMSO (E). Brightfield image of whole lung explant (D) and TUNEL stained sections (E and F) are shown. All images are representative of at least 3 embryos. Scale bars, 200 μm (A to D), 100 μm (E and F).
To further examine downstream signaling, epithelial explants from Dermo1-FGFR1/2-KO mice were treated with FGF9 or FGF10 (Fig. 8A), sectioned, and then immunostained for phosphorylated (active) forms of ERK (dpERK) and AKT (pAKT) (Fig. 8, B and C). Treatment with FGF9 had little effect on dpERK immunostaining but robustly induced pAKT. By contrast, treatment with FGF10 robustly induced dpERK but had little effect on pAKT. These data support a model in which different FGFRs activate different downstream signaling pathways in lung epithelial cells to induce distinct developmental responses.
Fig. 8. Activation of AKT and ERK signaling by FGF9 and FGF10 in lung explants lacking mesenchymal FGFR1 and FGFR2.
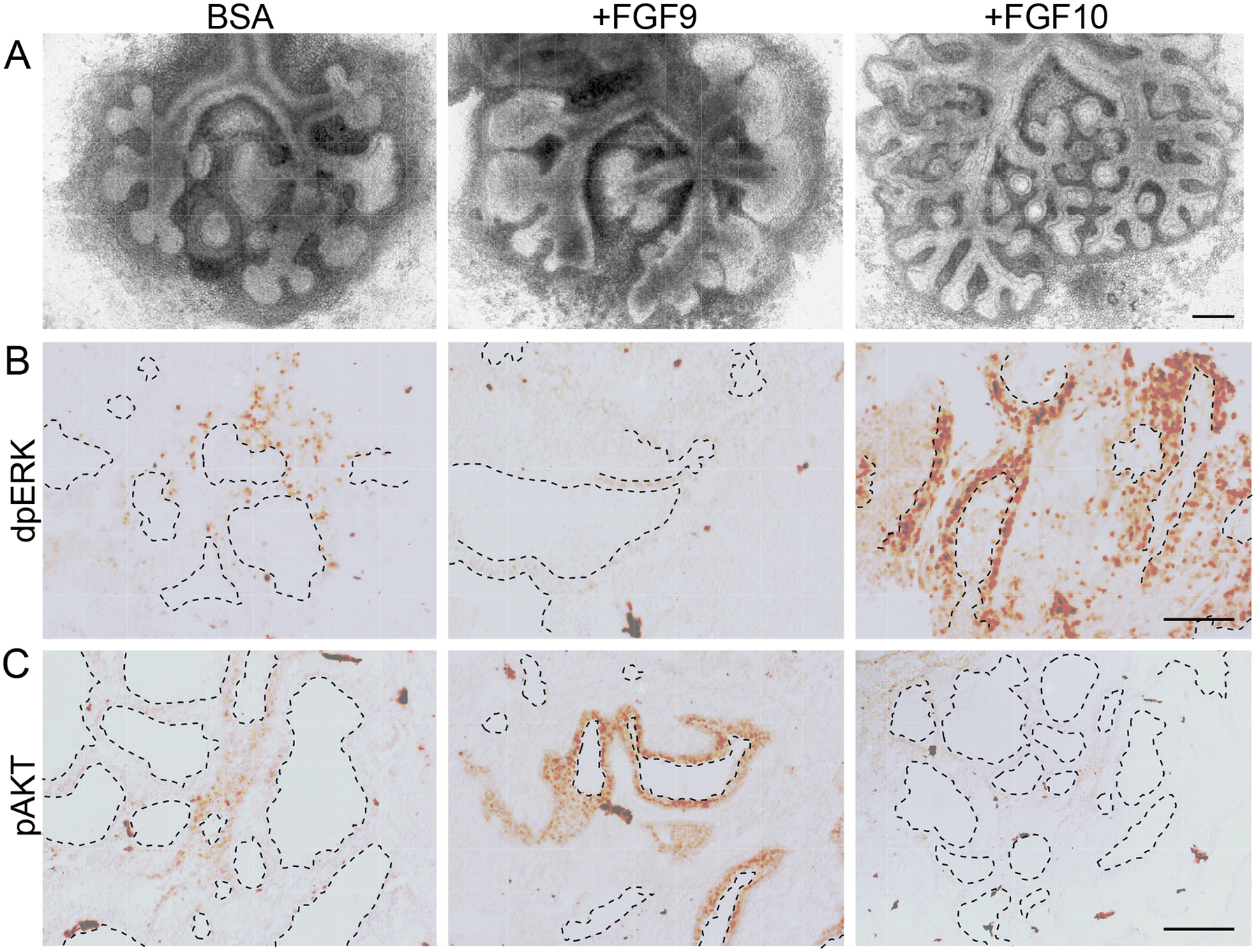
(A) Images of E11.5 whole lung explants from Dermo1-FGFR1/2-KO mice cultured for 48 h and treated with BSA, FGF9, or FGF10. (B, C) Sections of lung explants treated as in (A) immunostained for doubly phosphorylated (active) ERK (dpERK, B) and phosphorylated (active) AKT (pAKT, C). All images are representative of at least 3 embryos. Dashed lines outline ductal lumens. Scale bars, 200 μm (A); 100 μm (B and C).
Discussion
Development of the lung airways involves epithelial growth, branching, and differentiation, all of which must be coordinated to make a functional lung. Many of the signals that direct lung epithelial development arise from the mesenchyme; however, several studies have identified factors that act as autocrine or juxtacrine signals within the lung epithelium (5). For example, Bone morphogenetic protein 4 (Bmp4) is expressed in pseudoglandular-stage epithelial tips and directs epithelial proliferation and proximal-distal patterning (56–58). Wnt7b is also expressed in lung epithelium, where it directs canonical Wnt signaling and expression of Bmp4 and Fgfr2 (59, 60). Here, we focused on FGF9 and FGF10, factors that are produced in developing airway epithelium and mesenchyme, respectively, that signal to epithelium to direct proximal-distal specification, differentiation, branching, and outgrowth.
FGF9 signals through FGFR3 in early pseudoglandular-stage lung epithelium
Although it has been established that FGF9 signals to the lung epithelium during the pseudoglandular stage, the receptor that responds to FGF9 has not been rigorously defined. Initial studies suggested that FGFR2b is responsible for the FGF9 response because Fgfr2−/− lung explants failed to respond to FGF9 (32). However, FGF9 does not bind to FGFR2b in vitro, and FGFR2b is an established in vivo receptor for FGF10, which has effects on lung epithelium (proliferation and outgrowth) that are distinct from those of FGF9 (specification and differentiation). The data showing that FGF9 induces epithelial proliferation in lung explants from Fgf10−/− embryos (32) can be explained by indirect effects through the induction by FGF9 of Fgf7 expression in adjacent mesenchyme and FGF7-mediated activation of epithelial FGFR2b. Consistent with this model, del Moral et al. (32) showed that FGF9 strongly induces Fgf7 expression (287%) in primary lung mesenchyme; White et al. (28) showed that overexpression of FGF9 in lung epithelium in vivo induces both Fgf7 and Fgf10 in adjacent mesenchyme; Lebeche et al. (61) showed that both Fgf7 and Fgf10 are expressed in primary lung bud mesenchyme and are important for growth of the lung bud; and Bellusci et al. and Park et al. showed that FGF7 is a stronger inducer of epithelial proliferation than is FGF10 (10, 62). Additional explanations for why Fgfr2−/− lung explants failed to respond to FGF9 include the possibility that: 1) the Fgfr2b−/− tissue used was compromised due to lack of earlier FGF10-FGFR2b signaling; 2) FGFR3 is required for FGF9 to signal through FGFR2b by, for example, forming heterodimers; or 3) FGFR2b signaling is required for the production of FGFR3 in lung epithelium.
Here, using isolated epithelium and whole-lung explants deficient in mesenchymal FGFR signaling, we confirmed that FGF9 signals directly to pseudoglandular-stage lung epithelium through FGFR3. We showed that epithelial explants lacking FGFR3 failed to respond to FGF9, that Fgfr3−/− lungs had increased duct length and bud number at E11.5, and finally, that the ability of FGF9 to suppress branching was reduced in Fgfr3−/− explants. At E12.5, there remained a trend towards increased duct length, and Fgfr3−/− lungs appeared larger than heterozygous controls. However, Fgfr3−/− mice are viable and, as adults, do not have identified defects in lung development, although FGFR3 together with FGFR4 is important for postnatal alveologenesis (63–65). It is therefore likely that the Fgfr3−/− embryonic phenotype is compensated for or masked by other factors later in development. Furthermore, in the developing lung, epithelial expression of Fgf9 is only detected at E10.5-E12.5 and is restricted to the mesothelium at later times (23–26). This suggests that epithelial FGF9 is only needed during the early pseudoglandular stage to direct epithelial development and that mesothelial FGF9 continues to function throughout the pseudoglandular stage to direct mesenchymal development.
FGF9 directly promotes distal epithelial fate specification and inhibits epithelial differentiation
Here, we show that inactivation of FGF9 does not affect epithelial proliferation or death during pseudoglandular-stage development. We therefore hypothesized that FGF9 may direct some aspect of epithelial specification and/or differentiation, and that the relative distribution of SOX2 and SOX9, markers of proximal and distal lung epithelium, respectively, would be affected by the amount of FGF9 signaling in the epithelium. In support of this hypothesis, Fgf9−/− and FGF9 overexpression mouse models clearly showed that FGF9 directed the proximal-distal distribution of SOX2- and SOX9-expressing epithelial cells without affecting proliferation or cell death. Specifically, FGF9 overexpression expanded the population of SOX9-producing epithelial progenitors, and prevented these cells from producing SFTPC, a marker of differentiation. By contrast, FGF10 overexpression or FGFR2 inactivation in epithelium had little effect on the distribution of SOX2- and SOX9-expressing epithelial cells, although FGFR2 signaling was required to maintain SFTPC production. In contrast to our results with inducible Fgfr2 inactivation in distal lung epithelium beginning at E10.5, Abler et al. (15) reported that inactivation of Fgfr2 with Sftpc-Cre results in smaller lungs, increased epithelial death, reduced Sox9, and expansion of the Sox2 expression domain. However, unlike the limited and delayed gene targeting with ShhCreER and a single tamoxifen injection at E10.5, inactivation of Fgfr2 with Sftpc-Cre efficiently targets epithelium beginning at E9.75 (15, 66). It is thus likely that inactivation of Fgfr2 at an earlier developmental time leads to epithelial apoptosis and secondary effects on proximal-distal patterning, one of which could include the loss of Fgf9-expressing cells in the distal lung epithelium.
Consistent with FGF9 promoting the development of distal (SOX9-producing) epithelium and suppressing differentiation, when Sox9 is inactivated in the epithelium using ShhCre, Sftpb expression increases in the distal epithelium as early as E11 and there are no effects on epithelial proliferation at E13 (67). Of note, del Moral et al. (32) did not observe a change in SFTPC production in E12.5 explants that were cultured for 48 h in the presence of FGF9; however, because Sftpc expression initiates as early as E10, this explant stage may be too late to see a suppressive effect of FGF9 on Sftpc expression. In a more recent paper, Jones et al. (20) showed that inhibiting FGF10 signaling at E12.5 for 9 h by overexpressing a soluble FGFR2b ligand trap leads to reduced expression of Sftpc, like what we observed with conditional inactivation of FGFR2 in the epithelium from E10.5 to E13.5. Together these data support a model in which FGF9 suppresses distal epithelial differentiation.
An antagonistic relationship between FGF10-FGFR2 and FGF9-FGFR3 signaling in the early pseudoglandular-stage lung
Previous work comparing FGFR signaling in vitro suggested that differences between receptors are most likely due to differences in the strength of the tyrosine kinase domain rather than to qualitative differences in downstream signaling pathways (54, 68). Our comparison between mouse lines that affect FGF9–FGFR3 signaling and those that affect FGF10–FGFR2 signaling revealed distinctly different and, in some cases, opposite phenotypes, suggesting that these ligand-receptor pairs activate distinct signaling pathways in vivo (Fig. 9). To investigate potential differences in FGF9 and FGF10 signaling on three of the major FGFR downstream signaling pathways (RAS-MAPK, PI3K, and STAT), we tested the response of lung explants to pathway inhibitors and treatment with FGF9 or FGF10. This analysis showed that inhibition of RAS-MAPK signaling blocked FGF10-induced branching and resulted in phenotypes that resembled those due to FGF9 activation, whereas inhibition of PI3K blocked the epithelial dilation induced by FGF9 and, in the absence of FGF9, resulted in a phenotype that resembled that of FGF10 activation (increased branching). However, in the presence of the PI3K inhibitor LY294002, FGF9 still potently suppressed branching, suggesting that at the concentrations used, FGF9 effectively competed with this inhibitor or signaled through additional pathways, such as the STAT3 signaling pathways. However, inhibition of STAT3 did not affect FGF9-induced epithelial dilation or suppression of branching.
Fig. 9. Model for FGF9 and FGF10 signaling during pseudoglandular-stage lung development.
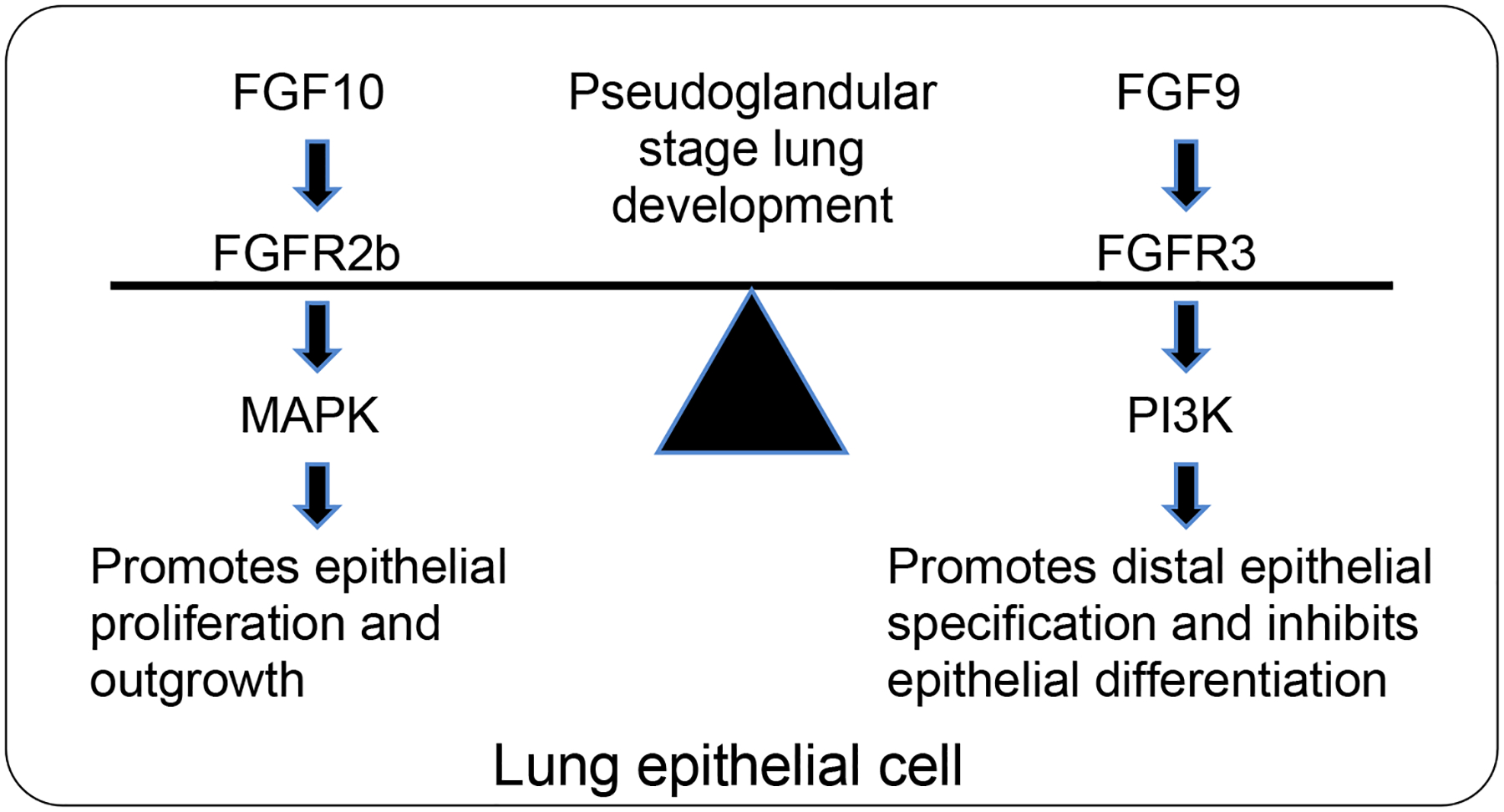
FGF10 signaling through FGFR2b preferentially activates signaling through the MAPK ERK and is in balance with FGF9 signaling through FGFR3, which preferentially activates PI3K.
The experiments with FGF9 and inhibitors of downstream signaling were complicated by the fact that FGF9 also signals to lung mesenchyme, which could activate indirect signals that affect epithelial development. However, the effects of FGF9 on epithelium were seen in both wild-type lung explants and in explants lacking mesenchymal FGFR signaling. Together, these observations suggest a model in which, in the epithelium, FGFR2 and FGFR3 are activated by FGF10 and FGF9, respectively, and preferentially activate distinct downstream signaling pathways. Finally, the effects of FGF9-FGFR3 signaling and FGF10-FGFR2b signaling appear to functionally oppose each other with respect to epithelial branching, proximal-distal specification, and differentiation.
Materials and Methods
Mice
All mouse strains, Fgf9LacZ, Fgfr1f/f, Fgfr2f/f, Fgfr3−/−, β-Cateninf/f, Dermo1Cre/+, ShhCreER/+, Spc-rtTA, Tre-Fgf9-Ires-Gfp, Tre-Fgf10, have previously been described (28, 37–40, 69–73). The following mouse lines can be obtained from the Jackson lab repository: β-Cateninf/f (Stock No: 004152), ShhCreER/+ (Stock No: 005623), Spc-rtTA (Stock No: 006225), Tre-Fgf10 (Stock No: 025671), Fgf9f/f (Stock No: 030675). We refer to the Fgf9LacZ allele as Fgf9-. Mice were of mixed sexes. Fgfr3−/−, Spc-rtTA, Tre-Fgf9-Ires-Gfp, and Tre-Fgf10 mice are on an FVB/NJ inbred background. Fgf9LacZ (Fgf9-), ShhCreER/+; Fgfr2f/f, Dermo1Cre/+; Fgfr1f/f; Fgfr2f/f mice are maintained on a mixed C57BL/6J x 129X1/SvJ genetic background. Wild-type lung explants are from mixed genetic backgrounds derived from C57BL/6J, 129X1/SvJ, and FVB/NJ mice. For tamoxifen administration, pregnant female mice were injected i.p. with 100 μg tamoxifen (#T5648, Millipore-Sigma) dissolved in corn oil (#C8267, Millipore-Sigma). For doxycycline administration, mice were given doxycycline chow (#S3888, 200 mg/kg doxycycline, Bio-Serv).
Mice were group housed with littermates, in breeding pairs, or in a breeding harem (2 females to 1 male), with food and water provided ad libitum. Mice were housed in a specific pathogen-free facility and handled in accordance with standard use protocols, animal welfare regulations, and the NIH Guide for the Care and Use of Laboratory Animals. All studies performed were in accordance with the Institutional Animal Care and Use Committee at Washington University in St. Louis (protocol #20160113, 20190110).
Antibodies
The following antibodies were used for immunostaining: anti-phospho-histone H3 (PHH3, 1:500, #H9908, Millipore-Sigma), anti-SRY-box-containing gene 9 (SOX9, 1:2000, #AB5535, Millipore Sigma), anti-SRY-box-containing gene 2 (SOX2, 1:250, #sc-17320, Santa Cruz Biotechnology), anti-proSP-C (SFTPC, 1:500, #AB3786, Millipore-Sigma), anti-phospho-AKT (pAKT, 1:150, #9271, Cell Signaling Technology), anti-phospho-p44/42 MAPK (dpERK, 1:200, #9101s, Cell Signaling Technology), anti-Keratin 5 (KRT5, 1:500, #905501, Biolegend Inc.), anti-LPCAT1 (1:200, #16112–1-AP, Proteintech), anti-NKX2–1 (1:200, #M3575, Agilent DAKO), anti-HOP (HOPX, 1:200, #sc-398703, Santa Cruz), anti-p63 (TRP63, 1:200, #CM163A, Biocare Medical).
Section and whole-mount immunostaining
Lung tissue was dissected and fixed following published protocols (43, 67, 74). Dissected embryo torsos were opened along the sternum and fixed in phosphate-buffered saline (PBS, pH 7.4) with 0.5% paraformaldehyde (PFA, #15714-S, Electron Microscopy Sciences) for 2 h for E12.5 and younger embryos and 3 h for E14.5 embryos on a rocker at room temperature. Embryos were then washed with PBS and lungs were dissected, photograph and used for cryo-section or whole-mount immunostaining. Images of whole lungs were captured on a stereo microscope (Olympus SZX12-ILLD100, Olympus).
Lung explant cultures
Lung explant cultures were performed as described (28). E11.5 embryonic lungs were dissected and cultured on Transwell filters (#3462, Corning Life Sciences) for 48 h in DMEM, 2 μg/ml heparin, at 37°C, 5% CO2. Tissues were treated with mouse or human FGF9 (#100–23, #100–23, PeproTech Inc.) or human FGF10 (#100–26, PeproTech Inc.) at 2.5 ng/ml, or with inhibitors. MEK inhibitor (U01206, #9903, Cell Signaling), 25 μM; PI3K inhibitor (Ly294002, #9901 Cell Signaling), 25 μM; STAT3 inhibitor (Stattic, #S7947, Millipore-Sigma), 10 μM. Lung explants were photographed on a stereo microscope (Olympus SZX12-ILLD100) or inverted microscope (Leica, DM IL LED). Duct length was measured using ImageJ software. Data shown is representative of at least three independent experiments.
Lung epithelial cultures
Lung epithelial cultures were performed as described (32). E11.5 mouse lungs were isolated and treated with undiluted dispase (#354235, Corning Life Science) for 30 min at 4°C. The distal epithelium was separated from the mesenchyme using fine tungsten needles and transferred into Matrigel™ (#356231, Corning Life Science) with culture medium (DMEM, 10% FBS, penicillin-streptomycin, L-glutamine) in a 4-well Nunc plate (#176740, ThermoFisher). Polymerization of Matrigel™ was initiated at 37°C for 30 min. After polymerization, epithelial explants were treated with either medium or medium containing recombinant FGF9 (2.5 ng/ml) and were cultured for 48 h at 37 °C in a 5% CO2 incubator. Explants were photographed on a stereo microscope (Olympus SZX12-ILLD100) or inverted microscope (Leica, DM IL LED). Epithelial diameter was measured using ImageJ software.
Cell death analysis
Cyosectioned slides (7 μm), prepared as described above, were assayed by DeadEnd™ Fluorometric TUNEL System (#G3250, Promega). Slides were mounted with DAPI containing Vectashield mounting medium (#H-1200, Vector Laboratories) for fluorescent detection. Three sections, 10 μm apart, were counted for three lungs at each time point.
In situ hybridization
The Sftpc (Spc) probe (75) was synthesized and labeled with a kit from Roche Applied Science. Whole mount in situ hybridization was performed as described (28, 29). Following color reaction and methanol dehydration lungs were photographed on an Olympus SZX12-ILLD100 stereo microscope.
Statistical analysis
For quantification of lung tissue and explant morphologies at least 4 individual samples were measured. Data was analyzed using the unpaired Student’ s t test, ANOVA, or Wilcoxon rank-sum test (see below). The data are reported as the mean ± SD and changes with p values less than 0.05 were considered to be statistically significant.
Duct length was compared in Fgfr3−/− animals and Fgfr3−/+ lungs at each lobe/duct location using a nonparametric Wilcoxon rank-sum test. This test was chosen because the data are underlying continuous but distinctly non-Gaussian. Specifically, in most lobe/duct locations the values are skewed to the left and some have a floor of a few 0 lengths. The data include repeated measures within animals (multiple lobe and duct locations). They are not well suited to multi-level or repeated measures modeling. E11.5 data has 7 and E12.5 has 11 simultaneous Wilcoxon tests.
Supplementary Material
Fig. S1. FGFR2 and FGFR3 are present in embryonic airway epithelium.
Fig. S2. The absence of FGFR3 partially rescues the suppression of branching induced by FGF9.
Fig. S3. Loss of FGF9 does not affect epithelial cell death or proliferation.
Fig. S4. Overexpression of FGF9 or FGF10 in early-stage lung development does not affect epithelial cell identity.
Fig. S5. FGFR3 directs distal epithelial specification and differentiation in pseudoglandular-stage lung.
Fig. S6. Overexpression of FGF9 in late-stage lung development does not affect epithelial cell identity.
Fig. S7. Inhibition of STAT3 has minimal effects on FGF9 and FGF10 signaling.
Acknowledgements:
We thank L. Li for technical help, K. Trinkaus and the Siteman Cancer Center Biostatistics Shared Resource (https://siteman.wustl.edu/research/shared-resources-cores/biostats-core/) for statistical consultation, J. Que for providing initial embryos with epithelial Fgfr2 inactivation. S. De Langhe for providing Tre-Fgf10 mice. A. Hagan and K.V. Woo for critically reading the manuscript.
Funding: This work was supported by grant HL111190 from the National Institutes of Health. Mouse lines were generated with assistance from the Washington University Mouse Genetics Core, the Digestive Disease Research Core Center (National Institutes of Health grant P30 DK052574), and the Washington University Musculoskeletal Research Center (National Institutes of Health grant P30 AR057235). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and Materials Availability: All data needed to evaluate the conclusions in this paper are present in the paper or in the Supplementary Materials. Mouse genetic models may require a material transfer agreement (MTA). All mice generated at Washington University in St. Louis are available with a standard Uniform Biological Material Transfer Agreement (UBMTA). Dermo1Cre/+, Fgfr2f/f, Fgfr3−/− and Tre-Fgf9-Ires-Gfp mouse lines are available from D. Ornitz under a MTA with Washington University in St. Louis. An MTA between Washington University in St. Louis and KOMP exists for Fgf9LacZ mice produced by the Ornitz lab with ES cells obtained from KOMP (https://www.komp.org/geneinfo.php?geneid=57953). Fgfr1f/f can be obtained from D. Ornitz with permission from J. Partanen at the University of Helsinki (juha.m.partanen@helsinki.fi).
References and Notes
- 1.Morrisey EE, Hogan BL, Preparing for the first breath: genetic and cellular mechanisms in lung development. Dev Cell 18, 8–23 (2010); published online EpubJan 19 (S1534–5807(09)00527–9 [pii]). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amy RW, Bowes D, Burri PH, Haines J, Thurlbeck WM, Postnatal growth of the mouse lung. J. Anat 124, 131–151. (1977). [PMC free article] [PubMed] [Google Scholar]
- 3.Schittny JC, Development of the lung. Cell Tissue Res. 367, 427–444 (2017); published online EpubMar ( 10.1007/s00441-016-2545-0). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hines EA, Sun X, Tissue crosstalk in lung development. J. Cell. Biochem 115, 1469–1477 (2014); published online EpubSep ( 10.1002/jcb.24811). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ornitz DM, Yin Y, Signaling networks regulating development of the lower respiratory tract. Cold Spring Harb Perspect Biol 4, 1–19 (2012); published online EpubMay ( 10.1101/cshperspect.a008318). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ornitz DM, Itoh N, The Fibroblast Growth Factor signaling pathway. Wiley Interdiscip Rev Dev Biol 4, 215–266 (2015); published online EpubMay-Jun ( 10.1002/wdev.176). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan T, Volckaert T, Chanda D, Thannickal VJ, De Langhe SP, Fgf10 Signaling in Lung Development, Homeostasis, Disease, and Repair After Injury. Front Genet 9, 418 (2018) 10.3389/fgene.2018.00418). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM, Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J. Biol. Chem 281, 15694–15700 (2006); published online EpubJun 9 ( 10.1074/jbc.M601252200). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cardoso WV, Itoh A, Nogawa H, Mason I, Brody JS, FGF-1 and FGF-7 induce distinct patterns of growth and differentiation in embryonic lung epithelium. Dev. Dyn 208, 398–405 (1997). [DOI] [PubMed] [Google Scholar]
- 10.Bellusci S, Grindley J, Emoto H, Itoh N, Hogan BL, Fibroblast growth factor 10 (FGF10) and branching morphogenesis in the embryonic mouse lung. Development 124, 4867–4878 (1997). [DOI] [PubMed] [Google Scholar]
- 11.De Moerlooze L, Spencer-Dene B, Revest J, Hajihosseini M, Rosewell I, Dickson C, An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development 127, 483–492 (2000).10631169 [Google Scholar]
- 12.Volckaert T, Campbell A, Dill E, Li C, Minoo P, De Langhe S, Localized Fgf10 expression is not required for lung branching morphogenesis but prevents differentiation of epithelial progenitors. Development 140, 3731–3742 (2013); published online EpubAug 7 ( 10.1242/dev.096560). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sekine K, Ohuchi H, Fujiwara M, Yamasaki M, Yoshizawa T, Sato T, Yagishita N, Matsui D, Koga Y, Itoh N, Kato S, Fgf10 is essential for limb and lung formation. Nat. Genet 21, 138–141 (1999); published online EpubJan ( 10.1038/5096). [DOI] [PubMed] [Google Scholar]
- 14.Wu J, Chu X, Chen C, Bellusci S, Role of Fibroblast Growth Factor 10 in Mesenchymal Cell Differentiation During Lung Development and Disease. Front Genet 9, 545 (2018) 10.3389/fgene.2018.00545). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abler LL, Mansour SL, Sun X, Conditional gene inactivation reveals roles for Fgf10 and Fgfr2 in establishing a normal pattern of epithelial branching in the mouse lung. Dev. Dyn 238, 1999–2013 (2009); published online EpubJul 17 ( 10.1002/dvdy.22032 [doi]). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramasamy SK, Mailleux AA, Gupte VV, Mata F, Sala FG, Veltmaat JM, Del Moral PM, De Langhe S, Parsa S, Kelly LK, Kelly R, Shia W, Keshet E, Minoo P, Warburton D, Bellusci S, Fgf10 dosage is critical for the amplification of epithelial cell progenitors and for the formation of multiple mesenchymal lineages during lung development. Dev Biol 307, 237–247 (2007); published online EpubJul 15 ( 10.1016/j.ydbio.2007.04.033 [doi]). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mailleux AA, Kelly R, Veltmaat JM, De Langhe SP, Zaffran S, Thiery JP, Bellusci S, Fgf10 expression identifies parabronchial smooth muscle cell progenitors and is required for their entry into the smooth muscle cell lineage. Development 132, 2157–2166 (2005); published online EpubMay ( [DOI] [PubMed] [Google Scholar]
- 18.Hokuto I, Perl AKT, Whitsett JA, Prenatal, but not postnatal, inhibition of fibroblast growth factor receptor signaling causes emphysema. J. Biol. Chem 278, 415–421 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Peters K, Werner S, Liao X, Wert S, Whitsett J, Williams L, Targeted expression of a dominant negative FGF receptor blocks branching morphogenesis and epithelial differentiation of the mouse lung. Embo J 13, 3296–3301 (1994); published online EpubJul 15 ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones MR, Dilai S, Lingampally A, Chao CM, Danopoulos S, Carraro G, Mukhametshina R, Wilhelm J, Baumgart-Vogt E, Al Alam D, Chen C, Minoo P, Zhang JS, Bellusci S, A Comprehensive Analysis of Fibroblast Growth Factor Receptor 2b Signaling on Epithelial Tip Progenitor Cells During Early Mouse Lung Branching Morphogenesis. Front Genet 9, 746 (2019) 10.3389/fgene.2018.00746). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boucherat O, Nadeau V, Berube-Simard FA, Charron J, Jeannotte L, Crucial requirement of ERK/MAPK signaling in respiratory tract development. Development 141, 3197–3211 (2014); published online EpubAug ( 10.1242/dev.110254). [DOI] [PubMed] [Google Scholar]
- 22.Carraro G, El-Hashash A, Guidolin D, Tiozzo C, Turcatel G, Young BM, De Langhe SP, Bellusci S, Shi W, Parnigotto PP, Warburton D, miR-17 family of microRNAs controls FGF10-mediated embryonic lung epithelial branching morphogenesis through MAPK14 and STAT3 regulation of E-Cadherin distribution. Dev Biol 333, 238–250 (2009); published online EpubSep 15 ( 10.1016/j.ydbio.2009.06.020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colvin JS, Feldman B, Nadeau JH, Goldfarb M, Ornitz DM, Genomic organization and embryonic expression of the mouse fibroblast growth factor 9 gene. Dev. Dyn 216, 72–88 (1999). [DOI] [PubMed] [Google Scholar]
- 24.Caprioli A, Villasenor A, Wylie LA, Braitsch C, Marty-Santos L, Barry D, Karner CM, Fu S, Meadows SM, Carroll TJ, Cleaver O, Wnt4 Is essential to normal mammalian lung development. Dev Biol, (2015); published online EpubAug 27 ( 10.1016/j.ydbio.2015.08.017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weaver M, Batts L, Hogan BL, Tissue interactions pattern the mesenchyme of the embryonic mouse lung. Dev Biol 258, 169–184 (2003); published online EpubJul 1 ( [DOI] [PubMed] [Google Scholar]
- 26.Yin Y, Wang F, Ornitz DM, Mesothelial- and epithelial-derived FGF9 have distinct functions in the regulation of lung development. Development 138, 3169–3177 (2011); published online EpubAug ( 10.1242/dev.065110). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colvin JS, White A, Pratt SJ, Ornitz DM, Lung hypoplasia and neonatal death in Fgf9-null mice identify this gene as an essential regulator of lung mesenchyme. Development 128, 2095–2106 (2001). [DOI] [PubMed] [Google Scholar]
- 28.White AC, Xu J, Yin Y, Smith C, Schmid G, Ornitz DM, FGF9 and SHH signaling coordinate lung growth and development through regulation of distinct mesenchymal domains. Development 133, 1507–1517 (2006); published online EpubApr ( 10.1242/dev.02313). [DOI] [PubMed] [Google Scholar]
- 29.Yin Y, White AC, Huh SH, Hilton MJ, Kanazawa H, Long F, Ornitz DM, An FGF-WNT gene regulatory network controls lung mesenchyme development. Dev Biol 319, 426–436 (2008); published online EpubJul 15 ( 10.1016/j.ydbio.2008.04.009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Langhe SP, Carraro G, Tefft D, Li C, Xu X, Chai Y, Minoo P, Hajihosseini MK, Drouin J, Kaartinen V, Bellusci S, Formation and differentiation of multiple mesenchymal lineages during lung development is regulated by beta-catenin signaling. PLoS One 3, e1516 (2008) 10.1371/journal.pone.0001516). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin Y, Castro AM, Hoekstra M, Yan TJ, Kanakamedala AC, Dehner LP, Hill DA, Ornitz DM, Fibroblast Growth Factor 9 Regulation by MicroRNAs Controls Lung Development and Links DICER1 Loss to the Pathogenesis of Pleuropulmonary Blastoma. PLoS Genet 11, e1005242 (2015); published online EpubMay ( 10.1371/journal.pgen.1005242). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.del Moral PM, De Langhe SP, Sala FG, Veltmaat JM, Tefft D, Wang K, Warburton D, Bellusci S, Differential role of FGF9 on epithelium and mesenchyme in mouse embryonic lung. Dev Biol 293, 77–89 (2006); published online EpubMay 1 ( 10.1016/j.ydbio.2006.01.020). [DOI] [PubMed] [Google Scholar]
- 33.Yin Y, Betsuyaku T, Garbow JR, Miao J, Govindan R, Ornitz DM, Rapid induction of lung adenocarcinoma by fibroblast growth factor 9 signaling through FGF receptor 3. Cancer Res. 73, 5730–5741 (2013); published online EpubSep 15 ( 10.1158/0008-5472.CAN-13-0495). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yin Y, Ren X, Smith C, Guo Q, Malabunga M, Guernah I, Zhang Y, Shen J, Sun H, Chehab N, Loizos N, Ludwig DL, Ornitz DM, Inhibition of fibroblast growth factor receptor 3-dependent lung adenocarcinoma with a human monoclonal antibody. Dis Model Mech 9, 563–571 (2016); published online EpubMay 01 ( 10.1242/dmm.024760). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, Gao G, Goldfarb M, Receptor specificity of the fibroblast growth factor family. J. Biol. Chem 271, 15292–15297 (1996); published online EpubJun 21 (8663044). [DOI] [PubMed] [Google Scholar]
- 36.Santos-Ocampo S, Colvin JS, Chellaiah AT, Ornitz DM, Expression and biological activity of mouse fibroblast growth factor-9. J. Biol. Chem 271, 1726–1731 (1996). [DOI] [PubMed] [Google Scholar]
- 37.Yu K, Xu J, Liu Z, Sosic D, Shao J, Olson EN, Towler DA, Ornitz DM, Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development 130, 3063–3074 (2003); published online EpubJul ( [DOI] [PubMed] [Google Scholar]
- 38.Trokovic R, Trokovic N, Hernesniemi S, Pirvola U, Vogt Weisenhorn DM, Rossant J, McMahon AP, Wurst W, Partanen J, FGFR1 is independently required in both developing mid- and hindbrain for sustained response to isthmic signals. Embo J 22, 1811–1823 (2003); published online EpubApr 15 ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, Sommer L, Boussadia O, Kemler R, Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development 128, 1253–1264 (2001); published online EpubApr ( [DOI] [PubMed] [Google Scholar]
- 40.Huh SH, Warchol ME, Ornitz DM, Cochlear progenitor number is controlled through mesenchymal FGF receptor signaling. Elife 4, 1–17 (2015); published online EpubApr 27 ( 10.7554/eLife.05921). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang J, Chen J, Developmental programs of lung epithelial progenitors: a balanced progenitor model. Wiley Interdiscip Rev Dev Biol 3, 331–347 (2014); published online EpubSep-Oct ( 10.1002/wdev.141). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perl AK, Kist R, Shan Z, Scherer G, Whitsett JA, Normal lung development and function after Sox9 inactivation in the respiratory epithelium. Genesis 41, 23–32 (2005); published online EpubJan ( 10.1002/gene.20093). [DOI] [PubMed] [Google Scholar]
- 43.Alanis DM, Chang DR, Akiyama H, Krasnow MA, Chen J, Two nested developmental waves demarcate a compartment boundary in the mouse lung. Nat Commun 5, 3923 (2014) 10.1038/ncomms4923). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gontan C, de Munck A, Vermeij M, Grosveld F, Tibboel D, Rottier R, Sox2 is important for two crucial processes in lung development: branching morphogenesis and epithelial cell differentiation. Dev Biol 317, 296–309 (2008); published online EpubMay 1 ( 10.1016/j.ydbio.2008.02.035). [DOI] [PubMed] [Google Scholar]
- 45.Laresgoiti U, Nikolic MZ, Rao C, Brady JL, Richardson RV, Batchen EJ, Chapman KE, Rawlins EL, Lung epithelial tip progenitors integrate glucocorticoid- and STAT3-mediated signals to control progeny fate. Development 143, 3686–3699 (2016); published online EpubOct 15 ( 10.1242/dev.134023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barkauskas CE, Cronce MJ, Rackley CR, Bowie EJ, Keene DR, Stripp BR, Randell SH, Noble PW, Hogan BL, Type 2 alveolar cells are stem cells in adult lung. J Clin Invest 123, 3025–3036 (2013); published online EpubJul 1 ( 10.1172/jci68782). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brewer JR, Mazot P, Soriano P, Genetic insights into the mechanisms of Fgf signaling. Genes Dev. 30, 751–771 (2016); published online EpubApr 1 ( 10.1101/gad.277137.115). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goetz R, Mohammadi M, Exploring mechanisms of FGF signalling through the lens of structural biology. Nat Rev Mol Cell Biol 14, 166–180 (2013); published online EpubMar ( 10.1038/nrm3528). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Danopoulos S, Thornton ME, Grubbs BH, Frey MR, Warburton D, Bellusci S, Al Alam D, Discordant roles for FGF ligands in lung branching morphogenesis between human and mouse. J Pathol 247, 254–265 (2019); published online EpubFeb ( 10.1002/path.5188). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.El Agha E, Kheirollahi V, Moiseenko A, Seeger W, Bellusci S, Ex vivo analysis of the contribution of FGF10+ cells to airway smooth muscle cell formation during early lung development. Dev Dyn, (2017); published online EpubApr 07 ( 10.1002/dvdy.24504). [DOI] [PubMed] [Google Scholar]
- 51.Li C, Hu L, Xiao J, Chen H, Li JT, Bellusci S, Delanghe S, Minoo P, Wnt5a regulates Shh and Fgf10 signaling during lung development. Dev Biol 287, 86–97 (2005); published online EpubNov 1 ( [DOI] [PubMed] [Google Scholar]
- 52.Izvolsky KI, Shoykhet D, Yang Y, Yu Q, Nugent MA, Cardoso WV, Heparan sulfate-FGF10 interactions during lung morphogenesis. Dev Biol 258, 185–200 (2003); published online EpubJun 1 ( [DOI] [PubMed] [Google Scholar]
- 53.Kameyama H, Kudoh S, Hatakeyama J, Matuo A, Ito T, Significance of Stat3 Signaling in Epithelial Cell Differentiation of Fetal Mouse Lungs. Acta histochemica et cytochemica 50, 1–9 (2017); published online EpubFeb 28 ( 10.1267/ahc.16032). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hart KC, Robertson SC, Kanemitsu MY, Meyer AN, Tynan JA, Donoghue DJ, Transformation and Stat activation by derivatives of FGFR1, FGFR3, and FGFR4. Oncogene 19, 3309–3320 (2000); published online EpubJul 6 ( 10.1038/sj.onc.1203650). [DOI] [PubMed] [Google Scholar]
- 55.Schust J, Sperl B, Hollis A, Mayer TU, Berg T, Stattic: a small-molecule inhibitor of STAT3 activation and dimerization. Chem. Biol 13, 1235–1242 (2006); published online EpubNov ( 10.1016/j.chembiol.2006.09.018). [DOI] [PubMed] [Google Scholar]
- 56.Weaver M, Yingling JM, Dunn NR, Bellusci S, Hogan BL, Bmp signaling regulates proximal-distal differentiation of endoderm in mouse lung development. Development 126, 4005–4015 (1999). [DOI] [PubMed] [Google Scholar]
- 57.Wang Y, Tian Y, Morley MP, Lu MM, Demayo FJ, Olson EN, Morrisey EE, Development and regeneration of Sox2+ endoderm progenitors are regulated by a Hdac1/2-Bmp4/Rb1 regulatory pathway. Dev Cell 24, 345–358 (2013); published online EpubFeb 25 ( 10.1016/j.devcel.2013.01.012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weaver M, Dunn NR, Hogan BL, Bmp4 and Fgf10 play opposing roles during lung bud morphogenesis. Development 127, 2695–2704 (2000). [DOI] [PubMed] [Google Scholar]
- 59.Rajagopal J, Carroll TJ, Guseh JS, Bores SA, Blank LJ, Anderson WJ, Yu J, Zhou Q, McMahon AP, Melton DA, Wnt7b stimulates embryonic lung growth by coordinately increasing the replication of epithelium and mesenchyme. Development 135, 1625–1634 (2008); published online EpubMay ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shu W, Guttentag S, Wang Z, Andl T, Ballard P, Lu MM, Piccolo S, Birchmeier W, Whitsett JA, Millar SE, Morrisey EE, Wnt/beta-catenin signaling acts upstream of N-myc, BMP4, and FGF signaling to regulate proximal-distal patterning in the lung. Dev Biol 283, 226–239 (2005); published online EpubJul 1 ( [DOI] [PubMed] [Google Scholar]
- 61.Lebeche D, Malpel S, Cardoso WV, Fibroblast growth factor interactions in the developing lung. Mech. Dev 86, 125–136 (1999). [DOI] [PubMed] [Google Scholar]
- 62.Park WY, Miranda B, Lebeche D, Hashimoto G, Cardoso WV, FGF-10 is a chemotactic factor for distal epithelial buds during lung development. Dev. Biol 201, 125–134 (1998). [DOI] [PubMed] [Google Scholar]
- 63.Li R, Herriges JC, Chen L, Mecham RP, Sun X, FGF receptors control alveolar elastogenesis. Development, (2017); published online EpubNov 09 ( 10.1242/dev.149443). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Srisuma S, Bhattacharya S, Simon DM, Solleti SK, Tyagi S, Starcher B, Mariani TJ, Fibroblast Growth Factor Receptors Control Epithelial-mesenchymal Interactions Necessary for Alveolar Elastogenesis. Am J Respir Crit Care Med, (2010); published online EpubJan 21 ( 10.1164/rccm.200904-0544OC [doi]). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weinstein M, Xu X, Ohyama K, Deng CX, FGFR-3 and FGFR-4 function cooperatively to direct alveogenesis in the murine lung. Development 125, 3615–3623 (1998). [DOI] [PubMed] [Google Scholar]
- 66.Okubo T, Hogan BL, Hyperactive Wnt signaling changes the developmental potential of embryonic lung endoderm. J Biol 3, 11 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chang DR, Martinez Alanis D, Miller RK, Ji H, Akiyama H, McCrea PD, Chen J, Lung epithelial branching program antagonizes alveolar differentiation. Proc Natl Acad Sci U S A 110, 18042–18051 (2013); published online EpubNov 5 ( 10.1073/pnas.1311760110). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Raffioni S, Thomas D, Foehr ED, Thompson LM, Bradshaw RA, Comparison of the intracellular signaling responses by three chimeric fibroblast growth factor receptors in PC12 cells. Proc. Natl. Acad. Sci. U. S. A 96, 7178–7183 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin Y, Liu G, Wang F, Generation of an Fgf9 conditional null allele. Genesis 44, 150–154 (2006); published online EpubMar ( 10.1002/gene.20194). [DOI] [PubMed] [Google Scholar]
- 70.Colvin JS, Bohne BA, Harding GW, McEwen DG, Ornitz DM, Skeletal overgrowth and deafness in mice lacking fibroblast growth factor receptor 3. Nat. Genet 12, 390–397 (1996); published online EpubApr ( 10.1038/ng0496-390). [DOI] [PubMed] [Google Scholar]
- 71.Harfe BD, Scherz PJ, Nissim S, Tian H, McMahon AP, Tabin CJ, Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell 118, 517–528 (2004); published online EpubAug 20 ( 10.1016/j.cell.2004.07.024). [DOI] [PubMed] [Google Scholar]
- 72.Clark JC, Tichelaar JW, Wert SE, Itoh N, Perl AK, Stahlman MT, Whitsett JA, FGF-10 disrupts lung morphogenesis and causes pulmonary adenomas in vivo. Am J Physiol Lung Cell Mol Physiol 280, L705–715. (2001). [DOI] [PubMed] [Google Scholar]
- 73.Tichelaar JW, Lu W, Whitsett JK, Conditional expression of fibroblast growth factor-7 in the developing and mature lung. J. Biol. Chem 275, 11858–11864 (2000). [DOI] [PubMed] [Google Scholar]
- 74.Ostrin EJ, Little DR, Gerner-Mauro KN, Sumner EA, Rios-Corzo R, Ambrosio E, Holt SE, Forcioli-Conti NR, Akiyama H, Hanash SM, Kimura S, Huang SXL, Chen J, Beta-Catenin maintains lung epithelial progenitors after lung specification. Development, (2018); published online EpubFeb 13 ( 10.1242/dev.160788). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu J, Tian J, Grumelli SM, Haley KJ, Shapiro SD, Stage-specific effects of cAMP signaling during distal lung epithelial development. J. Biol. Chem 281, 38894–38904 (2006); published online EpubDec 15 ( 10.1074/jbc.M609339200). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. FGFR2 and FGFR3 are present in embryonic airway epithelium.
Fig. S2. The absence of FGFR3 partially rescues the suppression of branching induced by FGF9.
Fig. S3. Loss of FGF9 does not affect epithelial cell death or proliferation.
Fig. S4. Overexpression of FGF9 or FGF10 in early-stage lung development does not affect epithelial cell identity.
Fig. S5. FGFR3 directs distal epithelial specification and differentiation in pseudoglandular-stage lung.
Fig. S6. Overexpression of FGF9 in late-stage lung development does not affect epithelial cell identity.
Fig. S7. Inhibition of STAT3 has minimal effects on FGF9 and FGF10 signaling.


