Abstract
The COVID-19 pandemic is a serious and global public health concern. It is now well known that COVID-19 cases may result in mild symptoms leading to patient recovery. However, severity of infection, fatality rates, and treatment responses across different countries, age groups, and demographic groups suggest that the nature of infection is diverse, and a timely investigation of the same is needed for evolving sound treatment and preventive strategies. This paper reports an the analysis of age distribution patterns in six groups of Indian COVID-19 patient populations based on their likely geographical origin of infection viz. the United Kingdom, North America, the European Union, the Middle East, and Asian countries. It was observed that patient groups stratified in this way had a distinct age profile and that some of these groups e.g. patient groups from Asia, the European Union, and the United Kingdom formed a different cluster than those from North America, the Middle East, and other regions. Patient age profiles of a population were found to be highly predictive of the group they belong to, and there are indications of their distinct recovery and fatality rates across gender. Altogether this study provides a scalable framework to estimate the source of infection in a new population of COVID-19 patients with unknown origin. It is also concluded that greater public availability of age and other demographic profile details of patients may be helpful in gaining robust insights into COVID-19 infection origins. Datasets and scripts used in this work are shared at http://covid.sciwhylab.org.
Keywords: COVID-19, Age profiles, Pandemic analysis, Origin of infection, Fatality
1. Introduction
The COVID-19 pandemic is a major challenge and has already caused close to 5.5 million infections and more than 350,000 deaths worldwide [1]. In order to understand and combat such a large-scale outbreak, the treatment and containment strategies must also consider the diversity of infection, and potential differences such as the manner in which a population responds to the many variants that might exist. A number of studies on the genetic diversity of these viruses have been performed [[2], [3], [4]]. The biomedical community does not yet know much about the diversity of COVID-19 variants and the likely impact on clinical outcomes, but the transmission dynamics and mutation rates of viral genomes are of critical significance [5,6]. For example at the time of writing this report the fatality rates in across the countries range widely from as low as < 4% to as high as >15% [7]. Although the exact reasons of these dramatically different mortality rates and possible differences in affected age groups are not very well understood, it is widely recognized that the collection and presentation of accurate clinical data as well as elaborate epidemiological studies are needed in order to understand and control both current and future outbreaks such as COVID-19 [8,9]. In many demographic studies, patient age has been recognized as one of the key factors predictive of clinical outcome, including fatality rate and severity of symptoms [8,10,11]. However, the way in which a given viral variant might actually impact different patient age groups has not been well investigated. A detailed impact analysis of this kind requires sequencing of thousands of samples across various ethnicities and age groups, which is yet to be attempted. In this work, an alternative approach of labeling viral variants and their relationship to the age profile of patients is presented. Using the travel history and potential origin of infection for Indian COVID-19 positive cases, six groups of patients have been identified from different geographical locations from where the infection was potentially carried. Patient age profiles of these groups originating from North America, the United Kingdom (UK), the European Union (EU), Asia, the Middle East, and other regions reveal that patients from each region have clearly distinct age distributions and that these groups tend to cluster to form two super groups. There are also indications that the recovery and fatality ratios of patients may be related to the patient groups defined here. Finally, an attempt was made to quantify how well a new population group can be assigned to one of these six categories based on a simple age statistic of these populations. Results from randomly sampled populations from these data are highly promising in terms of these predictabilities. Thus, this study establishes two clusters of three patient groups each from COVID-19 infected cases, and proposes a framework to assign patient groups based on age distribution in a patient cohort. It is expected that further data on group wise clinical responses, coupled with automatic annotations of patient groups proposed here, can help in improving therapeutic strategies towards the COVID-19 pandemic.
2. Methods
2.1. Data sets
This study is based on a crowd-sourced data on COVID-19 patients in India compiled at www.covid19india.org [12]. JSON:API provided patient details including age, gender, recovery status, and possible source of infection. The resource is basically a compilation of daily updates provided by State (provincial) Governments from throughout India, and it attempts to present crucial information in a unified format, most importantly: (a) patient identification number assigned by the data administrators (b) state-wise patient identification number (c) age of the patient (d) gender of the patient (e) travel history or any relevant detail of the patient that may reveal the source of infection (f) link to the potential patient(s) who could have been the source of this patient's infection and (g) current status of the patient e.g. recovered or deceased. Several other aspects of data available in the sheet are not relevant for the current study. The first of these JSON:API files (raw_data1.json) contains data about 17,305 patients, which is the primary source of data used in this study. This data is frozen on April 19, 2020 and is likely to remain unchanged. Origin of infection is available only for the first few thousand cases, in the early stages of pandemic. Therefore, the current study has utilized information from the first 5000 patients, of which many do not have age information, leaving behind fewer patients' data, as detailed in the results. For parsing JSON:API files, the R package called rjson, available from the CRAN repository, was installed and utilized.
2.2. Classification and labeling of likely COVID-19 variants
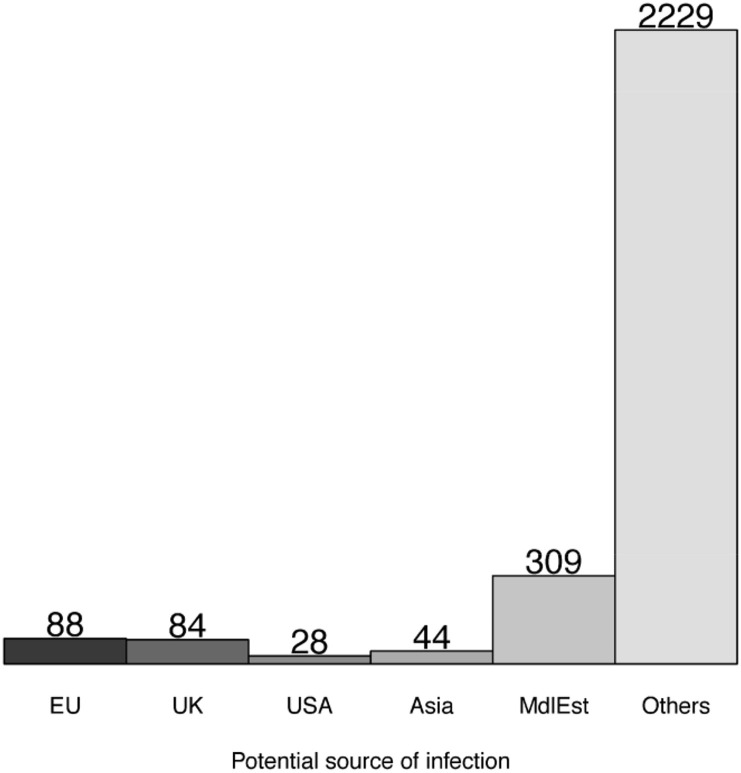
At least in the Stage 1 of the pandemic, every patient can be traced back to the source of infection; for example travel history to a specific country with high infection rate. In the subsequent stages of pandemic, the number of cases with unknown source of infection grow, and the data becomes less useful for creating predictive models of infection origin. The current study will develop patterns from all the patient groups that could be traced with some confidence to their likely source of infection, so that the insights can be developed by the application of these findings to new patient groups for a better clinical response. In this study, the original travel history to a country has been taken as the source of infection. Patients with no travel history but those whose likely contact with a person with a travel history, has been established and were labeled by the original source in order to expand the annotated data. In this way, all known sources of infection directly or traceable to a travel history have been used for labeling the data. Five specific groups of travel history have been defined by pooling them to broader geographical locations. Thus, all the patients with travel history to a member of European Union have been labeled as the EU group. Patients with travel history to the USA and Canada, or those likely infected by them, were pooled into the North America (N_Am) group and those from an Asian country including China, Singapore, and Malaysia were likewise labeled as the Asia group. Similarly, Middle Eastern (MdlEst) cases were those with travel history to Iran or other Middle Eastern countries such as the United Arab Emirates (UAE) and Saudi Arabia. Such patients were excluded from the Asian group. Wherever the source of infection could not be ascertained or provided, the patients were placed in the “Other” group. Thus, five patient groups from a specific travel history or their infected cases along with a sixth group with untraceable source were identified in this study viz. “N_Am”, “UK”, “EU”, “Asia”, “MdlEst” and “Other” groups. The final number of patients in this study within each category with clear age annotation data is as shown in Fig. 1 .
Fig. 1.
Number of patients included in this study for each of the six groups of potential infection.
2.3. Age distribution characterization of patient groups
In many but not all of the cases, age of the patient has been provided in the JSON:API source file of covid19india.org in the attribute type “agebracket” and the same has been used for the purpose of this study. Within each of the six patient groups, defined above, the patient age from this attribute has been used as a raw value for conducting statistical tests of significant difference using Student's t-test in the R programing language [13]. Further, the raw age values were converted to their relative frequency histograms by computing the percentage of patients from within that group that fell within the 10-year class interval of eight age groups from 0 to 80 years range. These relative frequency histograms are used as group representative features for clustering these groups and making predictions of patient group label from a randomly sampled population (see below).
2.4. Clustering of patient groups
In order to estimate the similarity between the six patient groups defined above, a hierarchical clustering using the “Heatmap” function of R language was used. Two clustering approaches were used to evaluate if they both produce similar outcomes. In the first approach the bin-wise population densities defined from their histograms were used to represent each patient group and an average linkage clustering was performed. Next, the all-against-all pair-wise Pearson correlations between patient groups were calculated from the bin-wise population density and then the clustering over the rows of this pairwise matrix was carried out. Some of the patient groups are expected to be more similar than others and such patient group clusters are explored from these two clustering patterns.
2.5. Recovery/fatality analysis
Recovery rate within each group of patients has been defined from within the closed number of cases and includes fewer instances than the overall number of patients because many cases were still not closed or were not labeled as such in the database. Thus, the fatality rate is defined as the number of deceased patients within the patient group relative to the total number of closed cases in that category. An additional parameter of recovery index was defined to estimate survival probability within each patient group as follows:
| Recovery Index (R) = log2 (Nr/(1 + Nd )) |
where Nr and Nd are the numbers of patients recovered and deceased, respectively.
2.6. Bootstrapping samples and predictability of patient group from a patient population
This work aims to establish a framework in which a new group of patients such as a community or village can be assigned to one of the six patient groups defined above, so that treatment strategies may be optimized for that group. However, since we only have one population in each group in the current data,we cannot develop a model with pre-estimated confidence of its predictability. To overcome this problem, an approximate approach of creating multiple samples of populations were developed wherein fifty instances of patients were created from the already labeled patient groups to create one population. Subsequently, fifty training and fifty test sets of population from each of the six groups were created and represented by the normalized age histograms of the sampled populations.
For each of the six patient groups, positive class data corresponded to fifty training and fifty test examples. An equal number of negative population instances were taken from the remaining groups. A multiple linear regression model was trained over a set of these corresponding fifty positive plus fifty negative training examples and tested on another set of one hundred (fifty plus fifty) example populations. Performance levels of the training and the test data give us the approximate confidence levels at which, the group labels can be estimated for new patient populations. For the six patient groups, separate prediction models were created to assess how well a given population from that patient group could be labeled in contrast to other populations.
The predictive model in all cases for this work is a Multiple Linear regression (MLR) model, implemented using the lm function of the stat package in R [14]. Essentially, an MLR fits a linear equation with histogram bin frequencies as the independent variable, and the class label of origin of infection as the dependent variable, whereas the regression coefficients are determined during training so as to provide best classification of the training data based on known class labels of a population. These regression coefficients are preserved and the model's performance is tested on new sampled populations using these trained parameters.
2.7. Prediction performance estimates
Each trained model performance was estimated on independent data sets generated from the same populations using random sampling as described above. For each model, the positive class refers to sampled populations from one specific patient group and the negative class refers to all other sampled populations. Binary class labels assigned in this way are estimated using MLR model using training/testing protocols. The prediction performances are estimated first by taking the Pearson correlation between binary class labels and predicted analogue values produced by the trained MLR. In the same way, Area Under the Curve (AUC) of Receiver Operative Characteristic (ROC) was computed from the predicted analogue values for each pair of binary classes. Note that AUC of ROC is a standard measure to assess performances of predictive models, where binary class labels are predicted with a variable confidence level, allowing choices of sensitivity and specificity based on the thresholds applied on the predicted analog values. To assess the confidence in these models, the sampling, training and test steps were repeated 10 times, and the prediction performance scores are presented as a box plot for each of these iterations.
3. Results
3.1. Age distributions of patient groups
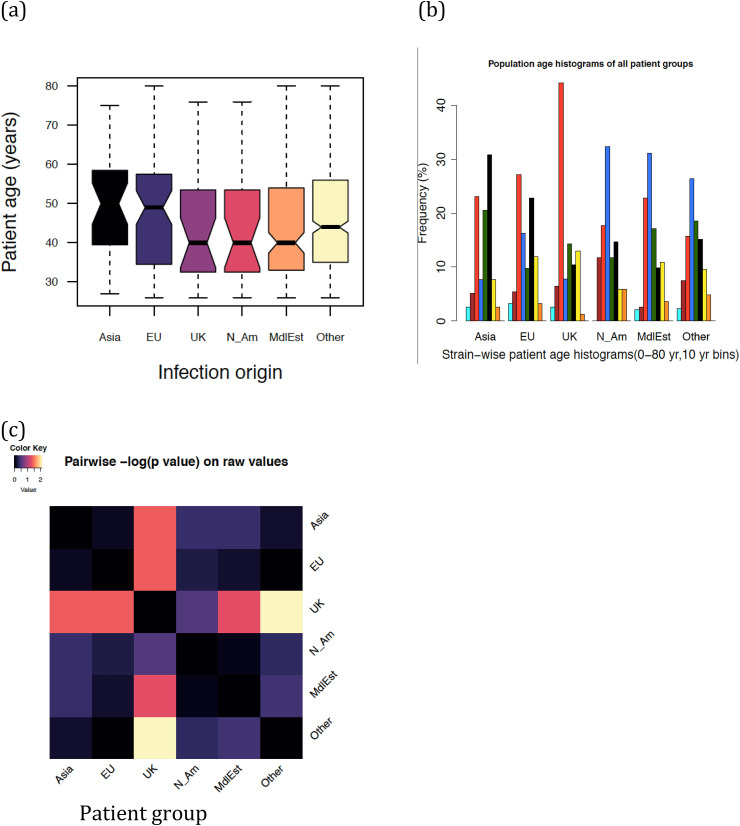
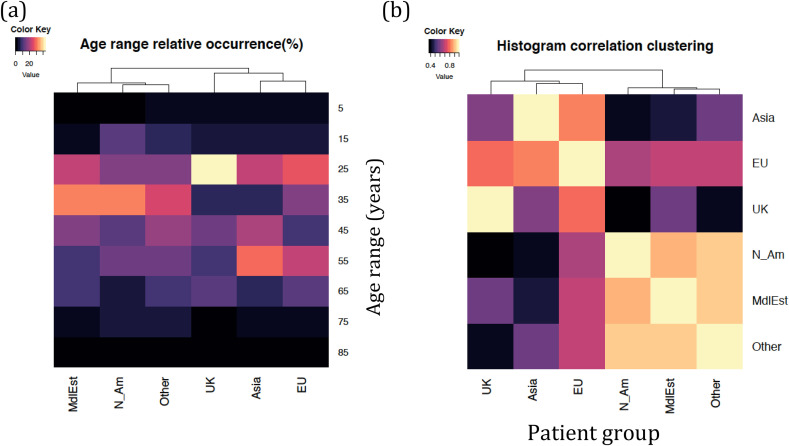
Fig. 2 (a–c) provide the first assessment of patient age distributions within each group of patients. Fig. 2(a) shows a clear difference between the overall distribution of patients from EU and Asia infections on the one hand and MidEst and N_Am on the other. Fig. 2(b) shows a more detailed distribution, which suggests that even though the average age values place UK closer to MidEst and N_Am, individual histogram bins suggest the case of UK to be more complex, and a rigorous clustering over age histograms suggest that UK clusters better with EU and Asia groups. Fig. 2(c) reiterates the same pattern, even as it utilizes the difference of means between the data (t-test) as a distance metric. In Fig. 3 , we explore a detailed pattern of clustering between age groups in two different ways. Fig. 3(a) shows how the patients from different infection origins cluster in terms of their age histograms. Clearly, the UK group, which looked similar to N_Am and MdlEst in terms of average patient age, tended to cluster closer to the EU and Asia group on account of a greater population density in the age range 30–40, which suggests more younger people being infected in each of these three groups. A second test of clustering, shown in Fig. 3(b) was performed by first computing histogram-to-histogram correlations between all six against six groups of patients, and then clustering them based on correlation values (see Methods). This method also produced clusters similar to those shown in Fig. 3(a), suggesting that the age group distribution patterns may be better clustered into UK/EU/Asia on the one hand and MdlEst/N_Am/Other on the other. Patient recovery data and its gender-wise variation also supports the existence of these two clusters (see below).
Fig. 2.
Age distributions in each group of patients based on the potential origin of infection (a) boxplots showing three quartiles and data ranges of patient age in each group (b) relative frequency histograms in each patient group after pooling them into eight bins (c) distribution of -log(pvalues) between the ages for each pair of patient groups.
Fig. 3.
Detailed view and further clustering of patient groups in terms of their age brackets (a) patient groups further clustered based on the frequency histograms in each infection group (b) alternative clustering of patient groups represented by their histogram-histogram correlations with all other groups (correlation vector). See Methods for details.
3.2. Gender specific recovery and fatality rates
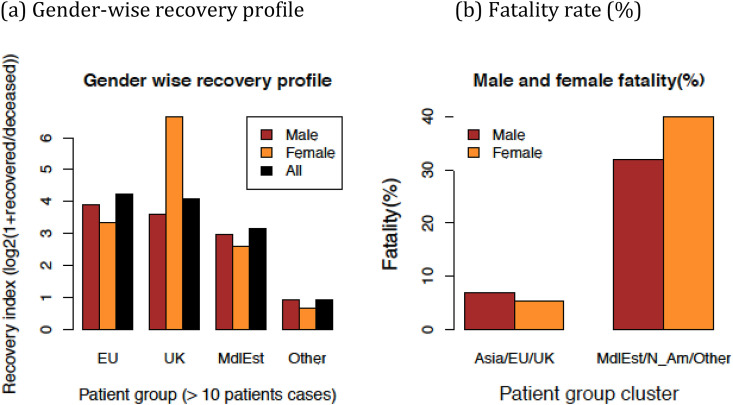
In order to investigate if the patient groups are only characterized by their age-wise population distribution or their recovery rates are also group specific; recovery and fatality rates were computed for each of the patient groups and then for the group clusters, as shown in Fig. 4 . It is interesting to observe that the patient recovery rates also indicate clustering patterns similar to those based on the age histograms (sufficient recovery data was available and shown only for the four of the six patient groups for individual group comparison). For example Fig. 4(a) shows that overall, male and female recovery rates were higher in the case of Asia/EU/UK groups compared to the "Others" group. The N_Am and Asia groups do not have sufficient data to compute these values since an insufficient number of patients had a clinical results (recovery or deceases) available in these groups. Merging the data from the patient group clusters, these differences in the fatality rate become evident. We observe not only higher fatality rates in the MdlEst/N_Am/Other group, but also subtle differences between male versus female fatality rates that are suggestive that the fatality rate is lower for females in this cluster compared with Asia/EU/UK cluster. However, given the small number of patient data available in each of these cases, the confidence in this statistic cannot be ascertained with a proper test, and a more reliable conclusion about this must wait for more data to emerge over time or it may be independently confirmed by the hospitals or health agencies with better access to patient age values.
Fig. 4.
Recovery and fatality rate distribution of individual patient group and their two major clusters formed by merging corresponding patient populations.
3.3. Predictability of patient group label from new patient communities
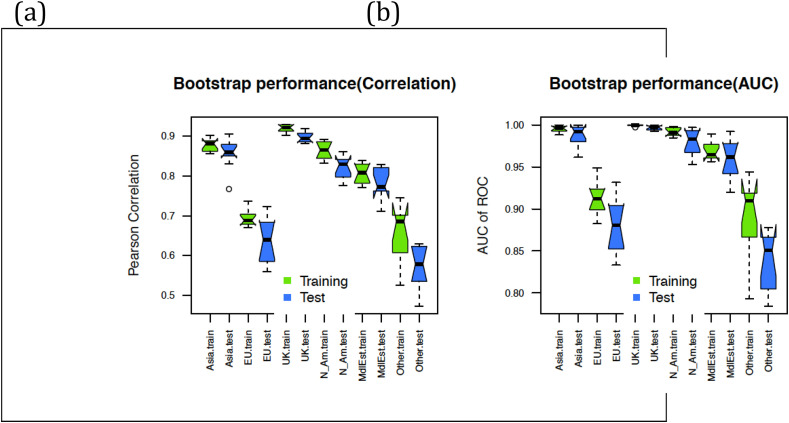
Next we assess whether the results obtained above can be used for the de novo assignment of infection origin group labels, whenever a new population age distribution becomes available from a community of patients. For example, can we predict if the patients from a village carry COVID-19 virus infection from Asia, the UK, or any of the six groups studied here? Random samples of populations were created to evaluate this, and the prediction models were created on random samples of populations. Subsequently, these trained models were tested on new random sets as explained in the Methods. Fig. 5 shows the results from such prediction models in terms of correlation between predicted class and group labels for each patient group. It is observed that the population group prediction in each of these categories is as high as 80–90% AUC. The worst case is that of the Middle East and Other groups which also show a similar Pearson correlation of 0.60 and above, with AUC always reaching more than 88% on the average for the 100 sample populations generated in each case (distribution for 100 iterations shown as a box plot). Even though the Bootstrapping type of cross-validation is prone to overestimate performance levels because of redundancy between the training and test examples, such high levels of predictability from a small number of feature values (8 probability density bins) is highly promising.
Fig. 5.
Predictability of infection type for a population from the age distribution profiles of patients randomly sampled from each category. (a) Prediction performance measured by correlation between predicted value and sample class. (b) Prediction performance measured by AUC of ROC for the same model as (a) (see Methods).
4. Discussion
There are many COVID-19 related compilations of data in the public domain emerging from agencies as varied as WHO and state governments. Infection origin studies like this could have been carried out on country-specific data, where all infections are more or less of the same origin. However, in such a scenario, the age profile differences observed between different conditions would be further confounded by additional factors such as: (i) the degree and target population being tested, (ii) ethnic variations from one place to the other, and (iii) general population age variations. Taking the population data from a single country and origin of infection over the patient groups located in the same country reduces the differences between testing strategies, background population demographics, genetic variations, and time points of infection in a better way. Some of the caveats still remain. The first of them pertains to the scarcity of data on age, gender, and recovery status. There are many patients in the database for which none or only some of this information are available. Better recording of these data and making them available in a timely manner will help in furthering these studies. From the analytic perspective, the traveling population age profile from the UK, USA, and EU may in itself be different from those traveling from the Middle East and Asian countries, for which the data was not available for this study. These issues may need further investigation in the future. However, despite these caveats, this study provides a framework to utilize rudimentary patient population profiles such as patient age distribution to segregate them in terms of source of infection, and potentially develop combative strategies informed by these differences.
One natural question that emerges from this study and for any data-driven study is that of causality. Why should people with different travel history respond to a virus differently? If the population groups studied here are considered to be more or less uniform, because they are all taken from the same country, the differences are likely be due to the mutations in coronavirus in the country where patients were potentially infected. If this is indeed true, it would have huge implications for therapeutic and vaccination strategies, as the treatments developed for one strain and target population may not be effective on other mutated variants. However, it would be a bit too early to draw an exact conclusion about all the variations being attributed to the mutations in the virus. As stated in the caveats above, the socio-economic backgrounds of people traveling to different countries may be one big confounding factor, among others. In view of these exciting possibilities and potential sources and implications of variations, further studies on the age profiles of COVID-19 patients are required and continued.
5. Conclusions
The study establishes that affected patient groups stratified by their travel and contact history have distinct patterns of patient age profiles. Out of the six categories of such patients, two clusters of patient groups were identified which also had differential patterns of male versus female fatality ratio across age groups, although only subtle differences could be recorded due to the available data size. A framework to assign a new patient population to one of these predefined categories in a predictive manner was also introduced. Overall, if the data can also be linked to the detailed clinical history and genetic constructs of clinical variants, a powerful combative strategy against Coronavirus may be developed which incorporates these findings.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This study has been supported by the following grants to the corresponding author: SERB, Department of Science and Technology, India (Grant no: EMR/2017/005485) and , Department of biotechnology, India (Garnt no. BT/PR24208/BID/7/801/2017)
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.imu.2020.100364.
Ethical statement
The authors must make a clear statement that the laws which apply to them in their own country were followed.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.https://www.who.int/emergencies/diseases/novel-coronavirus-2019
- 2.Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect Genet Evol. 2020;81:104260. doi: 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang C., Liu Z., Chen Z., Huang X., Xu M., He T. The establishment of reference sequence for SARS-CoV-2 and variation analysis. J Med Virol. 2020 doi: 10.1002/jmv.25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li X., Wang W., Zhao X., Zai J., Zhao Q., Li Y., Chaillon A. Transmission dynamics and evolutionary history of 2019-nCoV. J Med Virol. 2020;92:501–511. doi: 10.1002/jmv.25701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu J., Liu J., Zhao X., Liu C., Wang W., Wang D. Clinical characteristics of imported cases of COVID-19 in Jiangsu Province: a multicenter descriptive study. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa199. ciaa199. 10.1093/cid/ciaa199. (Online ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.https://coronavirus.app/map?mode=infected
- 8.Bhopal R. COVID-19 worldwide: we need precise data by age group and sex urgently. BMJ. 2020;369:m1366. doi: 10.1136/bmj.m1366. [DOI] [PubMed] [Google Scholar]
- 9.Lipsitch M., Swerdlow D.L., Finelli L. Defining the epidemiology of COVID- 19 — studies needed. N Engl J Med. 2020;382:1194–1196. doi: 10.1056/NEJMp2002125. [DOI] [PubMed] [Google Scholar]
- 10.Lauc G., Sinclair D. Biomarkers of biological age as predictors of COVID-19 disease severity. Aging (Albany NY) 2020;12(8):6490–6491. doi: 10.18632/aging.103052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahase E. COVID-19: death rate is 0.66% and increases with age, study estimates. BMJ. 2020;369:m1327. doi: 10.1136/bmj.m1327. [DOI] [PubMed] [Google Scholar]
- 12.https://www.covid19india.org/
- 13.Team R.C. R Foundation for Statistical Computing; Vienna, Austria: 2013. R: A language and environment for statistical computing. URL. [Google Scholar]
- 14.Chambers J.M. Wadsworth & Brooks/Cole; Pacific Grove, California: 1992. Linear models. [Chapter 4] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.