Abstract
Background
To use competing analyses to estimate the prognostic value of KRAS mutation status in colorectal cancer (CRC) patients and to build nomogram for CRC patients who had KRAS testing.
Method
The cohort was selected from the Surveillance, Epidemiology, and End Results database. Cumulative incidence function model and multivariate Fine-Gray regression for proportional hazards modeling of the subdistribution hazard (SH) model were used to estimate the prognosis. An SH model based nomogram was built after a variable selection process. The validation of the nomogram was conducted by discrimination and calibration with 1,000 bootstraps.
Results
We included 8,983 CRC patients who had KRAS testing. SH model found that KRAS mutant patients had worse CSS than KRAS wild type patients in overall cohort (HR = 1.10 (95% CI [1.04–1.17]), p < 0.05), and in subgroups that comprised stage III CRC (HR = 1.28 (95% CI [1.09–1.49]), p < 0.05) and stage IV CRC (HR = 1.14 (95% CI [1.06–1.23]), p < 0.05), left side colon cancer (HR = 1.28 (95% CI [1.15–1.42]), p < 0.05) and rectal cancer (HR = 1.23 (95% CI [1.07–1.43]), p < 0.05). We built the SH model based nomogram, which showed good accuracy by internal validation of discrimination and calibration. Calibration curves represented good agreement between the nomogram predicted CRC caused death and actual observed CRC caused death. The time dependent area under the curve of receiver operating characteristic curves (AUC) was over 0.75 for the nomogram.
Conclusion
This is the first population based competing risk study on the association between KRAS mutation status and the CRC prognosis. The mutation of KRAS indicated a poor prognosis of CRC patients. The current competing risk nomogram would help physicians to predict cancer specific death of CRC patients who had KRAS testing.
Keywords: KRAS, Colorectal cancer, Nomograms, Prognosis, SEER, Competing risk
Introduction
Colorectal cancer (CRC) is the second and third most common cancer of women and men worldwide, respectively (Bray et al., 2018). The amount of deaths due to CRC ranked the second among all cancer types in 2018 (Bray et al., 2018). CRC is a heterogeneous disease with various genetic events (Inamura, 2018; Punt, Koopman & Vermeulen, 2017). Target therapy such as anti-epidermal growth factor receptor (EGFR) therapy has been developed for metastatic CRC (Chan et al., 2017).
KRAS is an effector molecule that makes the signal transduction from ligand-bound EGFR to the nucleus (Liu, Wang & Li, 2019). KRAS has intrinsic GTPase activity and it binds to GTP to active downstream pathway, such as RAS/RAF/MAPK and PI3K/AKT pathways, to promote cell proliferation. Normally, the GTPase activating proteins would enhance the GTPase activity of KRAS and transform the status of GTP-bound KRAS into a status of GDP-bound KRAS, terminating the downstream signaling. However, some types of KRAS mutation could impair the GAP binding to KRAS and lead to a continuous GTP-bound KRAS status to promote the proliferation related pathways and cancer development (Cox & Der, 2010). The mutation of KRAS would also impair the efficacy of EGFR-targeted therapy (Liu, Wang & Li, 2019). KRAS mutation is found in about 33–45% of CRC (Tan & Du, 2012). Hence, the KRAS testing is recommended for CRC patients who would receive anti-EGFR therapy. The anti-EGFR therapy is limited to KRAS wild type (WT) CRC patients but not KRAS mutant (MT) patients (Markman et al., 2010).
Despite the KRAS mutation status as a biomarker for the anti-EGFR therapy of CRC patients, whether it is an independent prognostic factor in CRC was controversial. In metastatic CRC, there were studies showed that KRAS MT patients had worse progression-free survival (PFS) (Modest et al., 2016; Souglakos et al., 2009) or overall survival (OS) (Modest et al., 2016) than KRAS WT patients, while other study found there was no association between KRAS mutation status and PFS or OS of CRC patients (Kim et al., 2016). Among stage II and III CRC, there were studies found KRAS mutation would worsen the OS (Richman et al., 2009) or disease-free survival (DFS) (Deng et al., 2015; Lee et al., 2015) of patients while other study found KRAS mutation was not associated with the OS or recurrence-free survival (RFS) of CRC patients (Roth et al., 2010). In stage III colon cancer, a study found KRAS mutation status was not associated with the OS or RFS or DFS of patients (Ogino et al., 2009), while more recently studies found the KRAS mutation would worsen the DFS (Sperlich et al., 2018) or survival after recurrence (SAR) (Taieb et al., 2019) of patients. To be noted, most of these studies included with limited amount of CRC patients who had KRAS testing.
The Surveillance, Epidemiology, and End Results (SEER) database of the National Cancer Institute is a national collaboration program of United States, covering 34.6% of the national population. It collects the incidence, survival and treatment data of cancer patients. There was a SEER based study (Charlton et al., 2017) on the association between KRAS mutation status and the OS of patients with left or right side CRC. However, despited that CRC is an aggressive disease, the median age at diagnosis for colon cancer patients is 68 years in men and 72 years in women, respectively; for rectal cancer patients it is 63 years in both men and women (Society, 2017). In this case, competing risk events might be involved, as the elders might die from diseases other than CRC such as cardiovascular disease (Zhang, 2017). Competing risk models such as the cumulative incidence function (CIF) model and Fine-Gray regression for proportional hazards modeling of the subdistribution hazard (SH) model (Austin, Lee & Fine, 2016) should be used for the prognostic analyses of population based studies of CRC.
A nomogram is a useful method to predict the probability of patients’ clinical outcomes (Balachandran et al., 2015). It has compared favorably to traditional TNM staging systems in the prognostic prediction in a series of cancers (Bobdey et al., 2018; He et al., 2018). To our knowledge, there is currently no nomogram constructed for predicting the outcomes of CRC patients who had KRAS testing.
Here we performed a SEER based study to evaluate the association between KRAS mutation status and the cancer specific survival (CSS) of CRC patients by using competing risk analyses. We also drew an SH model based nomogram for the cancer specific death prediction of CRC patients who had KRAS testing.
Methods
Cohort information
The SEER based cohort was selected using SEER*Stat 8.3.5 software (SEER ID: daid). The access to Collaborative Stage Site-Specific Factor 9 (KRAS mutation status) was granted by the National Cancer Institute (NCI). We included patients who met the inclusion criteria as the follows: (1) it should be a CRC patient who had KRAS testing; (2) it should include sufficient clinicopathological information of the variables in current study (Table 1). As the information of KRAS testing was collected since 2010, we only included patients who were diagnosed equal to or after 2010. Finally, as shown in Fig. S1, to find an adequate follow-up time, the patients diagnosed between 2010 and 2012 were included. For tumor location, left side means the tumors in splenic flexure, descending colon, sigmoid and rectosigmoid junction, and right side means the tumors in cecum, ascending colon, hepatic flexure and transverse. We defined the median follow-up as the median observed survival time. The last follow-up time was December 31, 2015.
Table 1. The characteristic of each included variables in KRAS MT and KRAS WT patients.
| Characteristics | KRAS MT | KRAS WT | p Value | ||
|---|---|---|---|---|---|
| No. of patients | % | No. of patients | % | ||
| Age | 0.045 | ||||
| <29 | 38 | 1.05 | 70 | 1.30 | |
| 30–39 | 145 | 4.01 | 246 | 4.58 | |
| 40–49 | 503 | 13.91 | 738 | 13.75 | |
| 50–59 | 820 | 22.68 | 1,261 | 23.50 | |
| 60–69 | 992 | 27.43 | 1,435 | 26.74 | |
| 70–79 | 749 | 20.71 | 995 | 18.54 | |
| >=80 | 369 | 10.20 | 622 | 11.59 | |
| Sex | 0.023 | ||||
| Female | 1,697 | 46.93 | 2,387 | 44.48 | |
| Male | 1,919 | 53.07 | 2,980 | 55.52 | |
| Race | <0.001 | ||||
| White | 2,757 | 76.24 | 4,264 | 79.45 | |
| African Americans | 518 | 14.33 | 598 | 11.14 | |
| Others | 328 | 9.07 | 490 | 9.13 | |
| Unknown | 13 | 0.36 | 15 | 0.28 | |
| Location | <0.001 | ||||
| Left | 1,208 | 33.41 | 2,268 | 42.26 | |
| NOS | 97 | 2.68 | 160 | 2.98 | |
| Rectum | 665 | 18.39 | 992 | 18.48 | |
| right | 1,646 | 45.52 | 1,947 | 36.28 | |
| Tumor size | 0.086 | ||||
| <=2 cm | 399 | 11.03 | 647 | 12.06 | |
| >6 | 674 | 18.64 | 997 | 18.58 | |
| 2–4 | 855 | 23.64 | 1,363 | 25.40 | |
| 4–6 | 1,054 | 29.15 | 1,476 | 27.50 | |
| N | 634 | 17.53 | 884 | 16.47 | |
| Surgery | 0.005 | ||||
| No | 816 | 22.57 | 1,079 | 20.10 | |
| Yes | 2,800 | 77.43 | 4,288 | 79.90 | |
| Stage | <0.001 | ||||
| 0/(Tis) | 13 | 0.36 | 12 | 0.22 | |
| I | 202 | 5.59 | 384 | 7.15 | |
| II | 461 | 12.75 | 817 | 15.22 | |
| III | 887 | 24.53 | 1,494 | 27.84 | |
| IV | 2,020 | 55.86 | 2,615 | 48.72 | |
| Unknown | 33 | 0.91 | 45 | 0.84 | |
| Grade | <0.001 | ||||
| Low grade (I & II) | 2,502 | 69.19 | 3,490 | 65.03 | |
| High grade (III & IV) | 712 | 19.69 | 1,370 | 25.53 | |
| NOS | 402 | 11.12 | 507 | 9.45 | |
| Regional nodes positive | 0.025 | ||||
| >=10 | 1,196 | 33.08 | 1,726 | 32.16 | |
| 0 | 903 | 24.97 | 1,485 | 27.67 | |
| 1–3 | 880 | 24.34 | 1,212 | 22.58 | |
| 4–9 | 637 | 17.62 | 944 | 17.59 | |
| Radiotherapy | 0.383 | ||||
| No | 3,058 | 84.57 | 4,576 | 85.26 | |
| Yes | 535 | 14.80 | 772 | 14.38 | |
| Unknown | 23 | 0.64 | 19 | 0.35 | |
| Chemotherapy | 0.002 | ||||
| No | 924 | 25.55 | 1,534 | 28.58 | |
| Yes | 2,692 | 74.45 | 3,833 | 71.42 | |
| Marital status | 0.237 | ||||
| Married | 1,988 | 54.98 | 2,958 | 55.11 | |
| Unmarried | 1,464 | 40.49 | 2,204 | 41.07 | |
| Unknown | 164 | 4.54 | 205 | 3.82 | |
Note:
KRAS MT, KRAS mutant; KRAS WT, KRAS wild type; the widowed or single (never married or having a domestic partner) or divorced or separated patients was defined as unmarried; Tis, Tumor in situ; p value referred to the difference between MT and WT KRAS patients; the significant p values were bolded.
Statistical analyses
The chi-square test was applied for the comparisons of difference variables between KRAS WT and KRAS MT CRC patients. The cumulative incidences of death (CID) was estimated for cancer related deaths and non-cancer related deaths. Multivariate SH model, which involved all variables, was used to assess the CSS of CRC patients. SH model based nomogram was constructed to predict the 1-year, 2-year and 3-year CSS of CRC patients who had KRAS testing. To be noted, many prediction factors in one model might cause over-fitting. Hence, we used the variable selection to improve the interpretation and the accuracy of prediction of the competing nomogram (Ha et al., 2014). Penalized variable selection was performed by using methods of least absolute shrinkage and selection operator (LASSO), measure–correlate–predict (MCP) and smoothly clipped absolute deviation (SCAD) to select variables for SH model based nomogram. This nomogram was internally validated by discrimination and calibration with 1,000 times bootstraps (Balachandran et al., 2015). The calibration curves and the area under the curve of receiver operating characteristic curve (AUC) were used for discrimination and calibration, respectively.
The statistical analyses of current study were performed by a series of packages in R version 3.5.1. The detailed using of those packages could be found in our previous published study (Dai et al., 2020). We considered a p-value less than 0.05 as statistically significant.
Results
Cohort information
As shown in Table 1, there were totally 8,983 CRC patients (3,616 KRAS MT patients and 5,367 KRAS WT patients) included in current study. Significant differences were found between KRAS MT and KRAS WT patients among variables of age, race, location, surgery, tumor stage, grade, positive regional nodes amount, and chemotherapy experience (p < 0.05). In detail, compared with KRAS WT patients, the KRAS MT patients had more African American race (14.33% vs. 11.14%), more occurrence in right side of the colon (45.52% vs. 36.28%), less surgery performance (77.43% vs. 79.90%), more metastatic site (55.86% vs. 48.72%), lower grade (grade III & IV: 19.69% vs. 25.53%), and more chemotherapy experience (74.45% vs. 71.42%). The median follow-time were 30 months and 36 months for KRAS MT and KRAS WT, respectively. In KRAS MT patients, the death rate caused by cancer and other reasons were 49.89% and 13.69%, respectively. In KRAS WT patients, the death rate caused by cancer and other reasons were 42.59% and 14.83%, respectively.
KRAS MT patients had worse outcomes than KRAS WT patients
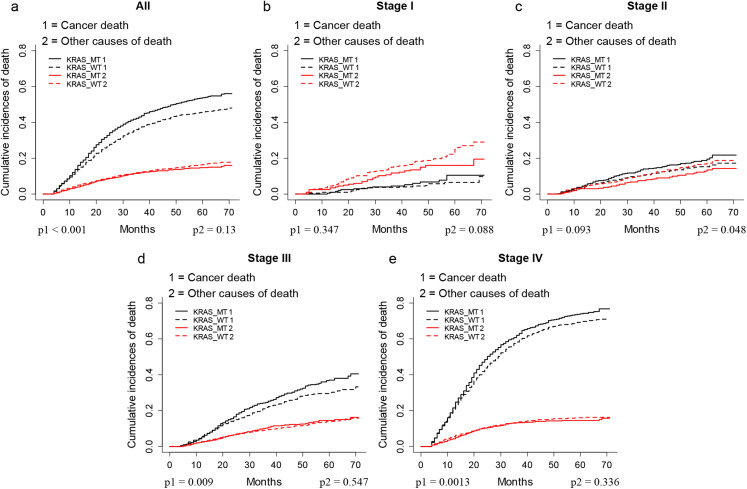
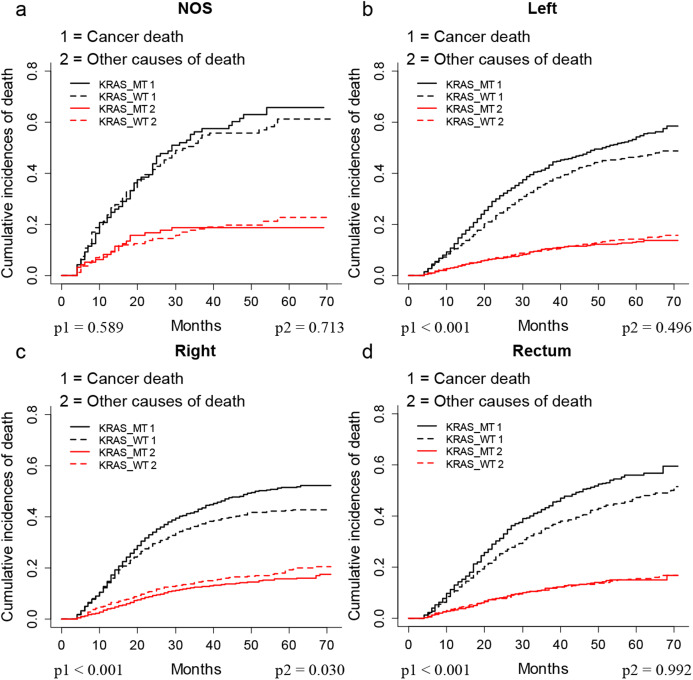
The CIF plots showed that the KRAS MT patients had a worse CSS than KRAS WT patients (p < 0.001, Fig. 1A). We further performed subgroup analysis of KRAS mutation status among different AJCC 7th stages and tumor locations. The CIF plots found that KRAS mutation had no association with the CSS of stage I (p = 0.347, Fig. 1B) and stage II (p = 0.093, Fig. 1C) CRC patients while it contributed to worse CSS in stage III (p = 0.009, Fig. 1D) and stage IV (p = 0.0013, Fig. 1E) CRC patients. In addition, the CIF plots showed that KRAS mutation was a hazard factor for the CSS of patients with cancers in the location of left colon, right colon and rectum (p < 0.001, Fig. 2).
Figure 1. CSS of CRC patients with different stages according to KRAS status by CIF plot.
CIF plots of KRAS status and the prognosis of CRC in overall population (A) and stage I–IV CRC patients (B–E).
Figure 2. CSS of CRC patients with differed location according to KRAS status by CIF plot.
CIF plots of KRAS status and the prognosis of CRC in locations of unknown (A), left colon (B), right colon (C) and rectum (D) NOS, not otherwise specified.
As shown in Table 2, the multivariate SH model showed that KRAS MT patients had worse CSS (Hazard ratio (HR) = 1.10, 95% CI (95% confidence index) = 1.04–1.17, p < 0.05) than KRAS WT patients. Further subgroup analysis found the KRAS mutation was an independent risk factor for the CSS of stage III (HR = 1.28 (95% CI [1.09–1.49]), p < 0.05) and stage IV (HR = 1.14 (95% CI [1.06–1.23]), p < 0.05) CRC patients. Moreover, we found KRAS shorten the CSS in patients with cancers occurred at left colon (HR = 1.28 (95% CI [1.15–1.42]), p < 0.05) and rectum (HR = 1.23 (95% CI [1.07–1.43]), p < 0.05) but not right colon (HR = 1.07 (95% CI [0.97–1.19]), p > 0.05).
Table 2. Subgroup analysis of KRAS mutation status and the prognosis of CRC patients.
| Group | Patients amount | KRAS MT vs. KRAS WT |
|---|---|---|
| Multivariate SH model | ||
| HR (95% CI) | ||
| All | 8,983 | 1.10 [1.04–1.17] |
| Stage I | 586 | 1.30 [0.62–2.70] |
| Stage II | 1,278 | 1.27 [0.95–1.69] |
| Stage III | 2,381 | 1.28 [1.09–1.49] |
| Stage IV | 4,635 | 1.14 [1.06–1.23] |
| Unknown location | 257 | 1.01 [0.69–1.49] |
| Left | 3,476 | 1.28 [1.15–1.42] |
| Right | 3,593 | 1.07 [0.97–1.19] |
| Rectum | 1,657 | 1.23 [1.07–1.43] |
Note:
KRAS MT, KRAS mutant; KRAS WT, KRAS wild type; HR, Hazard ratios; 95% CI, 95% confidence intervals; the significant results were bolded.
Multivariate SH analyses of each variable for the CSS of KRAS MT and KRAS WT CRC patients
As shown in Table 3, the multivariate SH model identified the absence of surgery, higher tumor stage and grade, and unmarried status as risk factors for both KRAS MT and KRAS WT CRC patients (HR > 1, p < 0.05). We observed there was no significant association between sex and the prognosis in neither KRAS MT nor KRAS WT CRC patients (p > 0.05).
Table 3. Multivariate SH analyses of each variables in KRAS MT and WT patients.
| Characteristics | SH model | |
|---|---|---|
| KRAS MT | KRAS WT | |
| HR (95% CI) | HR (95% CI) | |
| Age | ||
| <29 | Reference | Reference |
| 30–39 | 0.58 [0.36–0.94] | 1.19 [0.85–1.66] |
| 40–49 | 0.71 [0.46–1.11] | 1.05 [0.77–1.44] |
| 50–59 | 0.72 [0.46–1.13] | 1.11 [0.82–1.51] |
| 60–69 | 0.59 [0.38–0.92] | 1.03 [0.76–1.40] |
| 70–79 | 0.57 [0.37–0.90] | 0.99 [0.72–1.36] |
| >=80 | 0.58 [0.36–0.94] | 0.93 [0.66–1.31] |
| Sex | ||
| Female | Reference | Reference |
| Male | 1.07 [0.98–1.18] | 0.97 [0.89–1.06] |
| Race | ||
| White | Reference | Reference |
| African Americans | 1.16 [1.02–1.33] | 1.03 [0.90–1.18] |
| Others | 1.02 [0.87–1.20] | 1.10 [0.96–1.27] |
| Unknown | 0.35 [0.05–2.57] | 0.99 [0.29–3.37] |
| Location | ||
| Left | Reference | Reference |
| NOS | 0.97 [0.71–1.31] | 1.15 [0.90–1.48] |
| Rectum | 0.87 [0.75–1.01] | 0.95 [0.83–1.08] |
| Right | 1.05 [0.94–1.17] | 1.22 [1.10–1.35] |
| Tumor size | ||
| <=2 cm | Reference | Reference |
| 2–4 cm | 0.95 [0.79–1.15] | 1.00 [0.85–1.19] |
| 4–6 cm | 1.00 [0.83–1.20] | 1.10 [0.94–1.29] |
| >6 cm | 1.06 [0.87–1.28] | 1.25 [1.05–1.48] |
| N | 1.04 [0.86–1.27] | 1.01 [0.85–1.21] |
| Surgery | ||
| No | Reference | Reference |
| Yes | 0.77 [0.65–0.91] | 0.86 [0.75–0.995] |
| Stage | ||
| 0/(Tis) | Reference | Reference |
| I | 1.55 [0.22–10.88] | 1.07 [0.14–8.48] |
| II | 4.62 [0.69–30.73] | 2.96 [0.39–22.73] |
| III | 6.96 [1.06–45.70] | 4.08 [0.54–30.82] |
| IV | 18.90 [2.88–123.93] | 11.31 [1.50–85.33] |
| Unknown | 5.52 [0.78–39.14] | 5.01 [0.64–39.46] |
| Grade | ||
| Low grade (I & II) | Reference | Reference |
| High grade (III & IV) | 1.35 [1.20–1.52] | 1.38 [1.25–1.53] |
| NOS | 0.95 [0.81–1.10] | 1.07 [0.93–1.23] |
| Regional nodes positive | ||
| >=10 | Reference | Reference |
| 0 | 0.48 [0.39–0.59] | 0.37 [0.31–0.46] |
| 1–3 | 0.61 [0.52–0.73] | 0.50 [0.43–0.57] |
| 4–9 | 0.80 [0.68–0.95] | 0.70 [0.61–0.80] |
| Radiotherapy | ||
| No | ||
| Yes | 1.07 [0.92–1.26] | 1.00 [0.87–1.15] |
| Unknown | 1.04 [0.56–1.92] | 0.45 [0.19–1.09] |
| Chemotherapy | ||
| No | Reference | Reference |
| Yes | 0.77 [0.66–0.90] | 0.99 [0.86–1.13] |
| Marital status | ||
| Married | Reference | Reference |
| Unmarried | 1.19 [1.08–1.32] | 1.11 [1.01–1.21] |
| Unknown | 0.88 [0.70–1.11] | 1.12 [0.91–1.37] |
Note:
KRAS MT, KRAS mutant; KRAS WT, KRAS wild type; HR, Hazard ratios; 95% CI, 95% confidence intervals; the widowed or single (never married or having a domestic partner) or divorced or separated patients was defined as unmarried; Tis, Tumor in situ; significant results were bolded.
Prognostic discrepancies were found in other variables between KRAS MT and WT CRC patients. The older age was a protective factor for KRAS MT patients (HR < 1, p < 0.05) but was not associated with the prognosis of KRAS WT patients (p > 0.05). We found that the race of African American was a risk factor for KRAS MT patients but not for KRAS WT patients. The right side colon cancer was observed to have worse CSS than left side colon cancer in KRAS WT patients (HR > 1, p < 0.05) but not in KRAS MT patients (p > 0.05). Moreover, we found that the chemotherapy was only a protective factor for KRAS MT patients (HR < 1, p < 0.05) but not for KRAS WT patients (p > 0.05).
Nomogram construction and validation
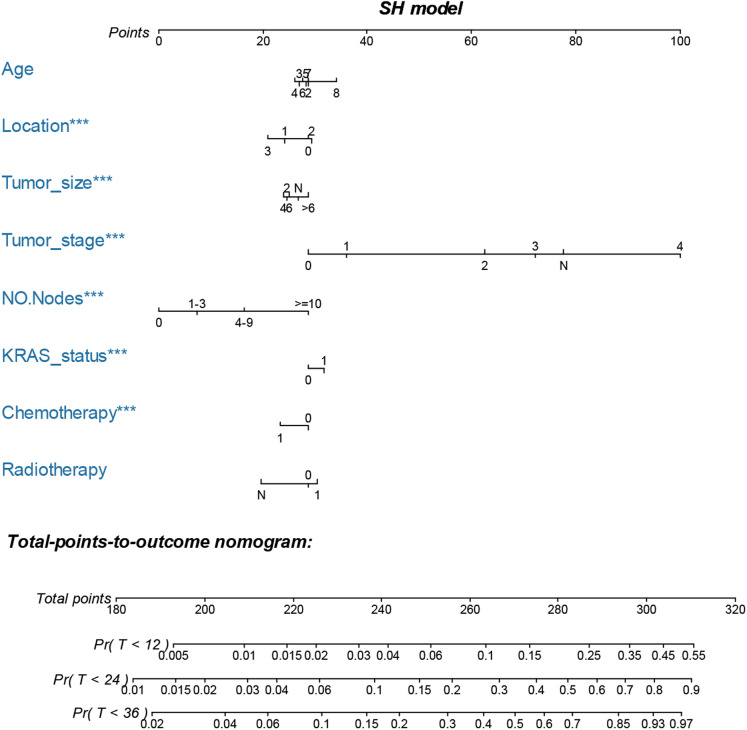
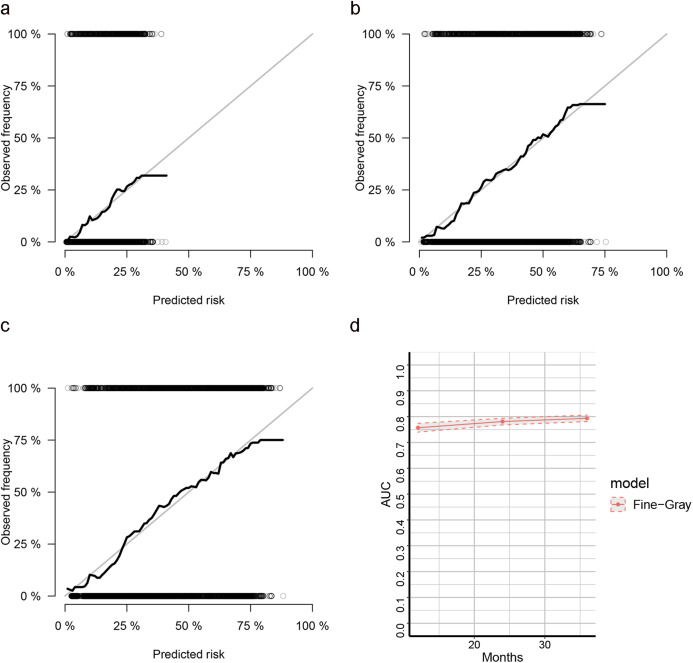
The LASSO, SCAD and MCP analyses all selected age, location, tumor size and stage, regional positive nodes amount, KRAS mutation status, chemotherapy experience and radiotherapy experience as the key prognostic variables of our nomogram (Table 4). These variables were then used to construct the multivariate SH model based nomogram to predict the 1-year, 2-year and 3-year CRC specific death (Fig. 3). Internal validation showed good calibration (Figs. 4A–4C, there were good agreements between the nomogram predicted CRC death and actual observed CRC death) and discrimination (AUC > 0.75, Fig. 4D) of current nomogram.
Table 4. Variable selection: estimated coefficients (SEs) for the current SH model.
| Characteristics | LASSO | SCAD | MCP |
|---|---|---|---|
| Age | −0.022 | −0.025 | −0.028 |
| Sex | 0.000 | 0.000 | 0.000 |
| Race | 0.000 | 0.000 | 0.000 |
| Location | −0.032 | −0.041 | −0.044 |
| Surgery | −0.069 | 0.000 | 0.000 |
| Tumor size | 0.017 | 0.027 | 0.025 |
| Tumor stage | 0.226 | 0.233 | 0.233 |
| Grade | 0.000 | 0.000 | 0.000 |
| Regional nodes positive | 0.097 | 0.105 | 0.105 |
| KRAS status | 0.13 | 0.182 | 0.183 |
| Chemotherapy | 0.314 | 0.384 | 0.383 |
| Radiotherapy | −0.061 | −0.095 | −0.095 |
| Marital status | 0.000 | 0.000 | 0.000 |
Note:
LASSO, least absolute shrinkage and selection operator; SCAD, smoothly clipped absolute deviation (SCAD); MCP, measure–correlate-predict (MCP).
Figure 3. Nomogram for predicting 1-year, 2-year and 3-year CSS of CRC patients who had KRAS testing.
The nomogram is used by summing the points identified on the top scale for each independent variable and drawing a vertical line from the total points scale to the 1-year, 2-year and 3-year CSS to obtain the probability of survival. The total points projected to the bottom scale indicate the % probability of the 3-year survival. Age: 2, 20–29 years, 3, 30–39 years, 4, 40–49 years, 5, 50–59 years, 6, 60–69 years and 7, 70–79 years; Race: 1, Caucasian, 2, African American, 3, Other race and N, Unknown race; Tumor size: 2, “0–2 cm”, 4, “2–4 cm”, 6, “4–6 cm”, >6 = “>6 cm”, N, Unknown size; Tumor stage, 0, 0 stage (Tumor in situ), 1, I stage, 2, II stage, 3, III stage, 4, IV stage and N, Unknown stage; No. Nodes, the number of positive regional lymph nodes; KRAS status: 0, Wild type and 1, Mutation; Chemotherapy, 0, none/unknown and 1, yes; Radiotherapy, 0, none/unknown or refused, 1, beam radiation or combination of beam with implants or isotopes or radiation with method or source not specified or radioactive implants or radioisotopes and N, Recommended, unknown if administered.
Figure 4. Calibration curves for cox-based and SH based nomograms.
(A–C) The calibration plots for predicting 1-year, 2-year and 3-year CSS of CRC patients; (D) the AUC plots for SH-based nomogram.
Discussion
The KRAS testing for metastatic CRC patients was recommended by the National Comprehensive Cancer Network (NCCN). The rate of KRAS testing for metastatic or non-metastatic CRC patients was increased in recent years according to SEER database (Fig. S2). However, the association between KRAS mutation status and the prognosis of CRC patients remains unclear. A SEER based study (Charlton et al., 2017) found that there was no association between KRAS mutation status and the OS of CRC. This might be a result of the limited follow up time, as they included the 2010–2012 data meanwhile had a last follow-up time of December 2013. Compared with this study, we included with 2010–2012 data while the last follow-up time was December 2015. The median survival time of our cohort was 33 months and the overall death rate of current study was 62.1%, indicating that our follow up time was relative sufficient. Furthermore, CRC patients were often diagnosed at an old age, therefore, competing risk analysis was more appropriate in the SEER based study. Our competing risk model found KRAS MT would shorten the CSS in CRC patients. Further subgroup analysis found that KRAS MT patients had worse CSS than KRAS WT patients among stage III or stage IV CRC patients or patients with left side colon cancer or rectal cancer. Moreover, the current study firstly built a competing nomogram for CRC patients who had KRAS testing.
Age was observed as a risk factor for the OS of CRC patients (Charlton et al., 2017; Van Eeghen et al., 2015). However, CRC patients are usually elders who might have high potential risk of deaths from other diseases. Our competing risk model found the older age was not associated with worse CSS of CRC patients. Moreover, older KRAS MT patients might have better CSS than young patients.
Left colon cancer was found to be more sensitive to anti-EGFR targeted therapy than right colon cancer (Venook et al., 2017). The right side colon cancer was found to have more BRAF mutation than left side colon cancer, which might cause the resistant to anti-EGFR therapy (Van Brummelen et al., 2017) and worsen the prognosis (Salem et al., 2017). Hence, for left-sided colon cancer, KRAS WT CRC patients are more likely to be benefit from anti-EGFR targeted therapy and have better outcomes than KRAS MT patients. Indeed, we found KRAS mutation was an independent risk factor for left side colon cancer but not right side colon cancer. Moreover, in KRAS WT patients, we found right colon cancer had worse CSS than left side colon cancer meanwhile in KRAS MT patients, there was no significant prognostic difference between right and left side colon cancers.
We built an SH model-based nomogram to predict the probability of cancer specific death after a variable selection. Our nomogram was well validated. The predictors of current nomogram were easy to be obtained in clinical use. The increasing concern about competing risk had promoted researchers to develop competing risk nomograms for a groups of cancers (Brockman et al., 2015; Kattan, Heller & Brennan, 2003; Kutikov et al., 2010; Shen, Sakamoto & Yang, 2016; Yang, Shen & Sakamoto, 2013).
There were certain limitations in our study. First, prognostic differences were found between KRAS codon 12 and codon 13 mutations (Imamura et al., 2012). However, the detailed KRAS mutation pattern was not registered in SEER. The detailed anti-EGFR therapy and chemotherapy strategy were also missed. Second, other genetic variables, such as BRAF mutation and microsatellite instability (MSI), were also frequently occurred in CRC and associated with the prognosis of CRC (Jung, Kim & Kim, 2016; Sanz-Garcia et al., 2017). These data were also not available in SEER. Third, selection bias might exist in current study as we only included patients with complete information of included variables.
Conclusion
This is the first population based competing risk study on the association between KRAS mutation status and the CRC prognosis. We found that KRAS mutation would worsen the CSS for patients with stage III and stage IV CRC, and for patients with cancers in the locations of left side colon and rectum. We constructed an SH based nomogram with good discrimination and calibration which might help the clinicians to predict the 1-year, 2-year and 3-year cancer specific death of CRC patients who had KRAS testing.
Supplemental Information
The current study included with 2010–2012 group (arrow), which exhibited similar median survival time as 2010 group.
The meanings of the code are available at the SEER website (https://seer.cancer.gov: "SEER RESEARCH DATA RECORD DESCRIPTION").
Funding Statement
This grant was supported by the National Natural Science Foundation of China (81372178; 81502386; 81772944; 81572715) and the High level health innovative talents program in Zhejiang and Natural Science Foundation of Zhejiang (LZ17H60003). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare that they have no competing interests.
Author Contributions
Dongjun Dai conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Yanmei Wang performed the experiments, analyzed the data, prepared figures and/or tables, and approved the final draft.
Liyuan Zhu performed the experiments, analyzed the data, prepared figures and/or tables, and approved the final draft.
Hongchuan Jin conceived and designed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Xian Wang conceived and designed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Data Availability
The following information was supplied regarding data availability:
The raw data from the SEER database is available in the Supplemental File.
References
- Austin, Lee & Fine (2016).Austin PC, Lee DS, Fine JP. Introduction to the analysis of survival data in the presence of competing risks. Circulation. 2016;133(6):601–609. doi: 10.1161/CIRCULATIONAHA.115.017719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran et al. (2015).Balachandran VP, Gonen M, Smith JJ, DeMatteo RP. Nomograms in oncology: more than meets the eye. Lancet Oncology. 2015;16(4):e173–e180. doi: 10.1016/S1470-2045(14)71116-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobdey et al. (2018).Bobdey S, Mair M, Nair S, Nair D, Balasubramaniam G, Chaturvedi P. A nomogram based prognostic score that is superior to conventional TNM staging in predicting outcome of surgically treated T4 buccal mucosa cancer: time to think beyond TNM. Oral Oncology. 2018;81:10–15. doi: 10.1016/j.oraloncology.2018.04.002. [DOI] [PubMed] [Google Scholar]
- Bray et al. (2018).Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- Brockman et al. (2015).Brockman JA, Alanee S, Vickers AJ, Scardino PT, Wood DP, Kibel AS, Lin DW, Bianco FJ, Jr, Rabah DM, Klein EA, Ciezki JP, Gao T, Kattan MW, Stephenson AJ. Nomogram predicting prostate cancer-specific mortality for men with biochemical recurrence after radical prostatectomy. European Urology. 2015;67(6):1160–1167. doi: 10.1016/j.eururo.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan et al. (2017).Chan DLH, Segelov E, Wong RS, Smith A, Herbertson RA, Li BT, Tebbutt N, Price T, Pavlakis N. Epidermal growth factor receptor (EGFR) inhibitors for metastatic colorectal cancer. Cochrane Database of Systematic Reviews. 2017;6:CD007047. doi: 10.1002/14651858.CD007047.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton et al. (2017).Charlton ME, Kahl AR, Greenbaum AA, Karlitz JJ, Lin C, Lynch CF, Chen VW. KRAS testing, tumor location, and survival in patients with stage IV colorectal cancer: SEER 2010–2013. Journal of the National Comprehensive Cancer Network. 2017;15(12):1484–1493. doi: 10.6004/jnccn.2017.7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox & Der (2010).Cox AD, Der CJ. Ras history: the saga continues. Small GTPases. 2010;1(1):2–27. doi: 10.4161/sgtp.1.1.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai et al. (2020).Dai D, Shi R, Wang Z, Zhong Y, Shin VY, Jin H, Wang X. Competing risk analyses of medullary carcinoma of breast in comparison to infiltrating ductal carcinoma. Scientific Reports. 2020;10(1):560. doi: 10.1038/s41598-019-57168-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng et al. (2015).Deng Y, Wang L, Tan S, Kim GP, Dou R, Chen D, Cai Y, Fu X, Zhu J, Wang J. KRAS as a predictor of poor prognosis and benefit from postoperative FOLFOX chemotherapy in patients with stage II and III colorectal cancer. Molecular Oncology. 2015;9(7):1341–1347. doi: 10.1016/j.molonc.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha et al. (2014).Ha ID, Lee M, Oh S, Jeong JH, Sylvester R, Lee Y. Variable selection in subdistribution hazard frailty models with competing risks data. Statistics in Medicine. 2014;33(26):4590–4604. doi: 10.1002/sim.6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He et al. (2018).He C, Zhang Y, Cai Z, Duan F, Lin X, Li S. Nomogram to predict cancer-specific survival in patients with pancreatic acinar cell carcinoma: a competing risk analysis. Journal of Cancer. 2018;9(22):4117–4127. doi: 10.7150/jca.26936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura et al. (2012).Imamura Y, Morikawa T, Liao X, Lochhead P, Kuchiba A, Yamauchi M, Qian ZR, Nishihara R, Meyerhardt JA, Haigis KM, Fuchs CS, Ogino S. Specific mutations in KRAS codons 12 and 13, and patient prognosis in 1075 BRAF wild-type colorectal cancers. Clinical Cancer Research. 2012;18(17):4753–4763. doi: 10.1158/1078-0432.CCR-11-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamura (2018).Inamura K. Colorectal cancers: an update on their molecular pathology. Cancers. 2018;10(1):26. doi: 10.3390/cancers10010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, Kim & Kim (2016).Jung SH, Kim SH, Kim JH. Prognostic impact of microsatellite instability in colorectal cancer presenting with mucinous, signet-ring, and poorly differentiated cells. Annals of Coloproctology. 2016;32(2):58–65. doi: 10.3393/ac.2016.32.2.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattan, Heller & Brennan (2003).Kattan MW, Heller G, Brennan MF. A competing-risks nomogram for sarcoma-specific death following local recurrence. Statistics in Medicine. 2003;22(22):3515–3525. doi: 10.1002/sim.1574. [DOI] [PubMed] [Google Scholar]
- Kim et al. (2016).Kim HS, Heo JS, Lee J, Lee JY, Lee MY, Lim SH, Lee WY, Kim SH, Park YA, Cho YB, Yun SH, Kim ST, Park JO, Lim HY, Choi YS, Kwon WI, Kim HC, Park YS. The impact of KRAS mutations on prognosis in surgically resected colorectal cancer patients with liver and lung metastases: a retrospective analysis. BMC Cancer. 2016;16(1):120. doi: 10.1186/s12885-016-2141-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutikov et al. (2010).Kutikov A, Egleston BL, Wong YN, Uzzo RG. Evaluating overall survival and competing risks of death in patients with localized renal cell carcinoma using a comprehensive nomogram. Journal of Clinical Oncology. 2010;28(2):311–317. doi: 10.1200/JCO.2009.22.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee et al. (2015).Lee D-W, Kim KJ, Han SW, Lee HJ, Rhee YY, Bae JM, Cho N-Y, Lee K-H, Kim T-Y, Oh D-Y, Im S-A, Bang Y-J, Jeong S-Y, Park K-J, Park J-G, Kang GH. KRAS mutation is associated with worse prognosis in stage III or high-risk stage II colon cancer patients treated with adjuvant FOLFOX. Annals of Surgical Oncology. 2015;22(1):187–194. doi: 10.1245/s10434-014-3826-z. [DOI] [PubMed] [Google Scholar]
- Liu, Wang & Li (2019).Liu P, Wang Y, Li X. Targeting the untargetable KRAS in cancer therapy. Acta Pharmaceutica Sinica B. 2019;9(5):871–879. doi: 10.1016/j.apsb.2019.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markman et al. (2010).Markman B, Javier Ramos F, Capdevila J, Tabernero J. EGFR and KRAS in colorectal cancer. Advances in Clinical Chemistry. 2010;51:71–119. doi: 10.1016/s0065-2423(10)51004-7. [DOI] [PubMed] [Google Scholar]
- Modest et al. (2016).Modest DP, Ricard I, Heinemann V, Hegewisch-Becker S, Schmiegel W, Porschen R, Stintzing S, Graeven U, Arnold D, Von Weikersthal LF, Giessen-Jung C, Stahler A, Schmoll HJ, Jung A, Kirchner T, Tannapfel A, Reinacher-Schick A. Outcome according to KRAS-, NRAS- and BRAF-mutation as well as KRAS mutation variants: pooled analysis of five randomized trials in metastatic colorectal cancer by the AIO colorectal cancer study group. Annals of Oncology. 2016;27(9):1746–1753. doi: 10.1093/annonc/mdw261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino et al. (2009).Ogino S, Meyerhardt JA, Irahara N, Niedzwiecki D, Hollis D, Saltz LB, Mayer RJ, Schaefer P, Whittom R, Hantel A, Benson AB, Goldberg RM, Bertagnolli MM, Fuchs CS, for the Cancer and Leukemia Group B, North Central Cancer Treatment Group, Canadian Cancer Society Research Institute, Southwest Oncology Group KRAS mutation in stage III colon cancer and clinical outcome following intergroup trial CALGB 89803. Clinical Cancer Research. 2009;15(23):7322–7329. doi: 10.1158/1078-0432.CCR-09-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punt, Koopman & Vermeulen (2017).Punt CJ, Koopman M, Vermeulen L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nature Reviews Clinical Oncology. 2017;14(4):235–246. doi: 10.1038/nrclinonc.2016.171. [DOI] [PubMed] [Google Scholar]
- Richman et al. (2009).Richman SD, Seymour MT, Chambers P, Elliott F, Daly CL, Meade AM, Taylor G, Barrett JH, Quirke P. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. Journal of Clinical Oncology. 2009;27(35):5931–5937. doi: 10.1200/JCO.2009.22.4295. [DOI] [PubMed] [Google Scholar]
- Roth et al. (2010).Roth AD, Tejpar S, Delorenzi M, Yan P, Fiocca R, Klingbiel D, Dietrich D, Biesmans B, Bodoky G, Barone C, Aranda E, Nordlinger B, Cisar L, Labianca R, Cunningham D, Van Cutsem E, Bosman F. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. Journal of Clinical Oncology. 2010;28(3):466–474. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- Salem et al. (2017).Salem ME, Weinberg BA, Xiu J, El-Deiry WS, Hwang JJ, Gatalica Z, Philip PA, Shields AF, Lenz HJ, Marshall JL. Comparative molecular analyses of left-sided colon, right-sided colon, and rectal cancers. Oncotarget. 2017;8(49):86356–86368. doi: 10.18632/oncotarget.21169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Garcia et al. (2017).Sanz-Garcia E, Argiles G, Elez E, Tabernero J. BRAF mutant colorectal cancer: prognosis, treatment, and new perspectives. Annals of Oncology. 2017;28(11):2648–2657. doi: 10.1093/annonc/mdx401. [DOI] [PubMed] [Google Scholar]
- Shen, Sakamoto & Yang (2016).Shen W, Sakamoto N, Yang L. Melanoma-specific mortality and competing mortality in patients with non-metastatic malignant melanoma: a population-based analysis. BMC Cancer. 2016;16(1):413. doi: 10.1186/s12885-016-2438-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Society (2017).Society AC. Atlanta: American Cancer Society; 2017. Colorectal cancer facts & figures 2017–2019. [Google Scholar]
- Souglakos et al. (2009).Souglakos J, Philips J, Wang R, Marwah S, Silver M, Tzardi M, Silver J, Ogino S, Hooshmand S, Kwak E, Freed E, Meyerhardt JA, Saridaki Z, Georgoulias V, Finkelstein D, Fuchs CS, Kulke MH, Shivdasani RA. Prognostic and predictive value of common mutations for treatment response and survival in patients with metastatic colorectal cancer. British Journal of Cancer. 2009;101(3):465–472. doi: 10.1038/sj.bjc.6605164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperlich et al. (2018).Sperlich A, Balmert A, Doll D, Bauer S, Franke F, Keller G, Wilhelm D, Mur A, Respondek M, Friess H, Nitsche U, Janssen KP. Genetic and immunological biomarkers predict metastatic disease recurrence in stage III colon cancer. BMC Cancer. 2018;18(1):998. doi: 10.1186/s12885-018-4940-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taieb et al. (2019).Taieb J, Shi Q, Pederson L, Alberts S, Wolmark N, Van Cutsem E, De Gramont A, Kerr R, Grothey A, Lonardi S, Yoshino T, Yothers G, Sinicrope FA, Zaanan A, Andre T. Prognosis of microsatellite instability and/or mismatch repair deficiency stage III colon cancer patients after disease recurrence following adjuvant treatment: results of an ACCENT pooled analysis of seven studies. Annals of Oncology. 2019;30(9):1466–1471. doi: 10.1093/annonc/mdz208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan & Du (2012).Tan C, Du X. KRAS mutation testing in metastatic colorectal cancer. World Journal of Gastroenterology. 2012;18(42):5171–5180. doi: 10.3748/wjg.v18.i42.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Brummelen et al. (2017).Van Brummelen EMJ, De Boer A, Beijnen JH, Schellens JHM. BRAF mutations as predictive biomarker for response to anti-EGFR monoclonal antibodies. Oncologist. 2017;22(7):864–872. doi: 10.1634/theoncologist.2017-0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eeghen et al. (2015).Van Eeghen EE, Bakker SD, Van Bochove A, Loffeld RJ. Impact of age and comorbidity on survival in colorectal cancer. Journal of Gastrointestinal Oncology. 2015;6:605–612. doi: 10.3978/j.issn.2078-6891.2015.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venook et al. (2017).Venook AP, Niedzwiecki D, Lenz H-J, Innocenti F, Fruth B, Meyerhardt JA, Schrag D, Greene C, O’Neil BH, Atkins JN, Berry S, Polite BN, O’Reilly EM, Goldberg RM, Hochster HS, Schilsky RL, Bertagnolli MM, El-Khoueiry AB, Watson P, Benson AB, III, Mulkerin DL, Mayer RJ, Blanke C. Effect of first-line chemotherapy combined with cetuximab or bevacizumab on overall survival in patients with KRAS wild-type advanced or metastatic colorectal cancer: a randomized clinical trial. JAMA. 2017;317(23):2392–2401. doi: 10.1001/jama.2017.7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Shen & Sakamoto (2013).Yang L, Shen W, Sakamoto N. Population-based study evaluating and predicting the probability of death resulting from thyroid cancer and other causes among patients with thyroid cancer. Journal of Clinical Oncology. 2013;31(4):468–474. doi: 10.1200/JCO.2012.42.4457. [DOI] [PubMed] [Google Scholar]
- Zhang (2017).Zhang Z. Survival analysis in the presence of competing risks. Annals of Translational Medicine. 2017;5(3):47. doi: 10.21037/atm.2016.08.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The current study included with 2010–2012 group (arrow), which exhibited similar median survival time as 2010 group.
The meanings of the code are available at the SEER website (https://seer.cancer.gov: "SEER RESEARCH DATA RECORD DESCRIPTION").
Data Availability Statement
The following information was supplied regarding data availability:
The raw data from the SEER database is available in the Supplemental File.