Abstract
Circular RNA (circRNA) abnormal expression and regulation are involved in the occurrence and development of a variety of tumors. However, the role of circRNAs still remains unknown in gastrointestinal stromal tumors (GISTs). In the present study, the differential circRNA expression profile of GISTs was screened by human circRNAs chip and verified by qRT-PCR. The circRNA–miRNA–mRNA regulatory network was constructed using the cytoHubba plugin based on the Cytoscape software. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed to explore circRNA functions. Six significantly differential circRNAs were also verified in 20 pairs of GISTs and adjacent tissues by qRT-PCR. The result showed that a total of 543 differentially expressed circRNAs were identified in GISTs, of which 242 were up-regulated and 301 were down-regulated. Additionally, the circRNA–miRNA–mRNA network contained six circRNAs, 30 miRNAs, and 308 mRNAs, and the targeted mRNAs were associated with “regulation of biological process,” “intracellular organelle,” “protein binding,” and enriched in Wnt signaling pathway. Furthermore, qRT-PCR demonstrated that hsa_circRNA_061346, hsa_circRNA_103114, and hsa_circRNA_103870 were significantly up-regulated in GISTs (n = 20), and hsa_ circRNA_405324, hsa_circRNA_406821, and hsa_circRNA_000361 were dramatically down-regulated in GISTs (n = 20). In addition, all of these circRNAs were shown to have high diagnostic values, and most of them were significantly associated with tumor size, mitotic figure, and malignant degrees in GISTs (P < 0.05). Therefore, we concluded that circRNAs were abnormally expressed in GISTs, and the circRNA–miRNA–mRNA regulatory network plays an important role in the occurrence and development of GISTs. Also, the identified six candidate circRNAs might be critical circRNAs and may present as potential diagnostic biomarkers for GISTs.
Keywords: gastrointestinal stromal tumors, circular RNA, miRNA, mRNA, biomarker
Introduction
Gastrointestinal stromal tumors are rarely one of the gastrointestinal carcinomas that originate from mesenchymal tissue. GISTs are characterized by expression of CD117 receptor in cells and have variable biological phenotypes ranging from benign to highly malignant (Gautam, 2020). As one of the most common non-epithelial neoplasms, they are mainly located in the stomach (55.6%) and small intestine (31.8%) (Tao et al., 2020). Radical surgery is the preferred treatment, and molecular target therapy, such as imatinib, can improve the survival of advanced patients with c-kit and/or PDGFRα mutations (Gupta and Rateria, 2020). However, a few effective tumor biomarkers are used for GIST diagnosis and prediction (Etherington and DeMatteo, 2019).
Circular RNA is a novel class of endogenous non-coding RNA characterized with 3′- and 5′-ends covalently linked in a closed-loop structure (Mahmoudi et al., 2020), which makes circRNAs resistant to exonucleases and more stable than traditional linear RNA, such as lncRNA and miRNA (Ding et al., 2020). Accordingly, the circRNAs can be divided into four types according to the source (Meng et al., 2017): exonic circRNAs (ecircRNA), intronic circRNA (ciRNA), exonic–intronic circRNA (EIciRNA), and intergenic circRNAs. Among them, 80% of circRNAs are ecircRNA. circRNAs may act as microRNA (miRNA/miR) sponges by competitively binding to miRNA response elements to influence downstream target gene expression, as well as affecting gene function at a post-translational level, and the same circRNA can regulate multiple miRNAs. Also, the same miRNAs can regulate multiple mRNA genes, thereby forming a large circRNA–miRNA–mRNA competitive network to affect the development of tumors (Xu S. et al., 2018). Also, some circRNAs can exert their activities via interaction with some proteins. Even then, some ecircRNAs may participate in the assembly and protein ribosomes translation. It was reported that circRNA is involved in various biological processes, including signal transduction and transcription, cell cycle regulation, RNA-binding protein, responses to stress, protein metabolism, cellular immunity, and cell structure (Jiang et al., 2018). Recent studies (Lu et al., 2018; Chaichian et al., 2020) have also demonstrated that circRNA abnormal expression and regulation are involved in the occurrence and development of a variety of tumors. Therefore, circRNAs are of great importance as a biomarker for cancer diagnosis, cancer prediction, and treatment feedback, and may even serve as targets for cancer treatment.
In this study, we first analyzed the circRNA differential expression profile in GISTs using circRNA chip and identified six potential key circRNAs by qRT-PCR. Also, the circRNA–miRNA–mRNA network was constructed, and the GO and KEGG pathway were performed via bioinformatics analysis. Our study provides a novel insight into the molecular mechanisms of GISTs from the circRNA–miRNA–mRNA network view, and these circRNAs gave new direction for diagnosis and treatment of GISTs.
Materials and Methods
Patients and Samples
Twenty pairs of GISTs and adjacent tissues were collected from The Second Xiangya Hospital of Central South University. All pathological specimens were experienced pathologists confirmed, did not accept the pre-operative radiotherapy, chemotherapy, and imatinib targeted therapy. The clinicopathological features are shown in Table 1. All tissues were collected during surgical operation and instantly stored in liquid nitrogen. The present project was permitted by the ethics committee of The Second Xiangya Hospital of Central South University, and informed consents were obtained from all the participants.
TABLE 1.
The clinicopathological features of GISTs patients.
| Variables | Cases (n) |
| Total | 20 |
| Age (years) | |
| ≥60 | 12 |
| <60 | 8 |
| Gender | |
| Male | 15 |
| Female | 5 |
| Tumor size (cm) | |
| ≤5 | 9 |
| >5 | 11 |
| Mitotic figure (HPF) | |
| ≤5/50 | 13 |
| >5/50 | 7 |
| Malignant degrees | |
| Low/Moderate risk | 12 |
| High risk | 8 |
CircRNA Chip Detection
Total RNAs were extracted by RNeasy Mini Kit (Qiagen, Hilden, Germany). Total RNA from each sample was quantified using the NanoDrop ND-1000. The sample preparation and microarray hybridization were performed based on the Arraystar’s standard protocols. Total RNAs were digested with Rnase R to remove linear RNAs and enrich CircRNAs. Then, the enriched CircRNAs were amplified and transcribed into fluorescent cRNA (Arraystar Super RNA Labeling Kit; Arraystar). The labeled cRNAs were hybridized onto the Arraystar Human circRNA Array V2 (8 × 15 K; Arraystar). Agilent Feature Extraction software (version 11.0.1.1) was used to analyze acquired array images. Quantile normalization and subsequent data processing were performed using the R software limma package. Differentially expressed circRNAs with statistical significance between two groups were identified through volcano plot filtering. Differentially expressed circRNAs between two samples were identified through fold change filtering. Hierarchical clustering was performed to show the distinguishable circRNA expression pattern among samples.
Quantitative Real-Time PCR
Total RNAs were extracted using RNeasy Mini kit (Qiagen, Hilden, Germany). Then, RNA was reversed into complementary DNA (cDNA) by SuperScript III Reverse Transcriptase (Invitrogen). qRT-PCR was performed with 95.0°C for 3 min, and 39 circles of 95.0°C for 10 s and 60°C for 30 s using SYBR Green PCR Master Mix system. The relative expression levels were calculated using the 22−ΔΔCt method. RNA levels were normalized to GAPDH expression. The forward (F) and reverse (R) primer sequences for qRT-PCR were designed and synthesized by Shanghai Kangcheng Co., Ltd. (Chinese).
CircRNA–miRNA–mRNA Interaction Prediction
The fundamental structure of circRNAs was predicted using Cancer-Specific circRNA (CSCD1). circRNA–miRNA interactions were predicted using TargetScan and miRanda databases, and miRNA target gene was predicted using TargetScan, miRanda v5, and miBase prediction databases. Candidate miRNAs and mRNAs should be overlapped in at least two databases. Arraystar’s miRNA target prediction software site: miRanda v52, TargetScan3, and mibase4. The circRNA–miRNA–mRNA competitive network (cirCeNET) was visualized by Cytoscape software (version 3.6.15).
Gene Ontology (GO) and KEGG Pathway Analysis
Gene ontology and KEGG pathways analysis was used to determine the function of candidate mRNAs in circRNA–miRNA–mRNA competitive network. DAVID6 was used to predict the enriched functional categories and enriched signaling pathways. GO term, including BP, CC, MF, and KEGG pathway with P < 0.05 and FDR < 0.05 were considered as statistically significant.
Statistical Analysis
All data were analyzed by SPSS17.0 statistics software. Paired t-test was employed for the comparison of two groups. Chi-square test was used to investigate the relationship between circRNA expression and clinicopathologic features of GISTs patients. P < 0.05 was considered as statistically significant.
Results
Differential CicrRNA Expression Profiles Were Established Successfully
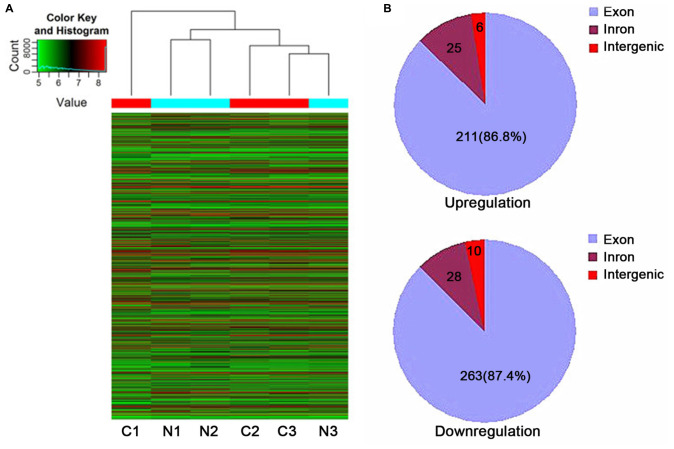
The box plot showed similar distributions of tissues. In the volcano plots, differentially expressed circRNAs were categorized using fold change and P values. The scatter plots demonstrated the variation of differentially expressed circRNAs. Hierarchical cluster analysis showed differentially expressed circRNAs in GISTs with fold change >1.5 and P < 0.05 (Figure 1A). After normalization and data analysis, compared with adjacent tissues, a total of 543 differentially expressed circRNAs were identified, including 242 up-regulated circRNAs and 301 down-regulated circRNAs, of which, exonic circRNAs accounted for 86.8% in up-regulated circRNAs and 87.4% in down-regulated circRNAs (Figure 1B). The top 20 significantly up- and down-regulated circRNAs are listed in Tables 2 and 3.
FIGURE 1.
Characterization of circRNA expression in GISTs. (A) Hierarchical cluster map. (B) The origin of upregulated and downregulated circRNAs. N, adjacent normal tissues; C, GIST tissues.
TABLE 2.
Top 20 significantly up-regulated circRNAs.
| circRNA | P-value | FDR | FC (abs) | Regulation | source | chrom | strand | type | GeneSymbol |
| hsa_circRNA_061346 | 0.049017552 | 0.541205763 | 6.9877315 | up | circBase | chr21 | − | exonic | APP |
| hsa_circRNA_103114 | 0.027813714 | 0.541205763 | 6.8848564 | up | circBase | chr21 | − | exonic | APP |
| hsa_circRNA_103223 | 0.040961154 | 0.541205763 | 6.5339969 | up | circBase | chr22 | − | exonic | DDX17 |
| hsa_circRNA_100582 | 0.02164493 | 0.541205763 | 4.5762691 | up | circBase | chr10 | + | exonic | ZEB1 |
| hsa_circRNA_001481 | 0.047719242 | 0.541205763 | 4.5058956 | up | circBase | chr5 | − | sense overlapping | EMB |
| hsa_circRNA_103128 | 0.03858124 | 0.541205763 | 4.1057473 | up | circBase | chr21 | + | exonic | DYRK1A |
| hsa_circRNA_103870 | 0.039001555 | 0.541205763 | 4.0573338 | up | circBase | chr5 | + | exonic | SMA4 |
| hsa_circRNA_404643 | 0.041732555 | 0.541205763 | 4.0082859 | up | 25070500 | chr1 | − | exonic | PIK3C2B |
| hsa_circRNA_004182 | 0.039090826 | 0.541205763 | 3.428553 | up | circBase | chr2 | + | intronic | CRIM1 |
| hsa_circRNA_103977 | 0.03760255 | 0.541205763 | 3.426974 | up | circBase | chr5 | + | exonic | ARHGAP26 |
| hsa_circRNA_104076 | 0.047784401 | 0.541205763 | 3.2920026 | up | circBase | chr6 | − | exonic | KIF13A |
| hsa_circRNA_101504 | 0.043980697 | 0.541205763 | 3.215775 | up | circBase | chr15 | + | exonic | PDIA3 |
| hsa_circRNA_102510 | 0.029244589 | 0.541205763 | 3.1147657 | up | circBase | chr19 | + | exonic | LSM14A |
| hsa_circRNA_002164 | 0.048545781 | 0.541205763 | 3.082576 | up | circBase | chr18 | − | exonic | SS18 |
| hsa_circRNA_103224 | 0.006590126 | 0.541205763 | 3.0780956 | up | circBase | chr22 | − | exonic | DDX17 |
| hsa_circRNA_100915 | 0.030601002 | 0.541205763 | 3.0777302 | up | circBase | chr11 | − | exonic | PICALM |
| hsa_circRNA_403471 | 0.003668259 | 0.541205763 | 3.060178 | up | 25242744 | chr5 | + | exonic | ARHGAP26 |
| hsa_circRNA_102248 | 0.047545686 | 0.541205763 | 3.056733 | up | circBase | chr17 | + | exonic | TBCD |
| hsa_circRNA_404446 | 0.005776341 | 0.541205763 | 3.0361211 | up | 25070500 | chr1 | − | exonic | CAPZB |
| hsa_circRNA_102378 | 0.045317666 | 0.541205763 | 2.9997955 | up | circBase | chr18 | + | exonic | ZNF532 |
TABLE 3.
Top 20 significantly down-regulated circRNAs.
| circRNA | P-value | FDR | FC (abs) | Regulation | source | chrom | strand | type | GeneSymbol |
| hsa_circRNA_405324 | 0.013412911 | 0.541205763 | 4.3231649 | down | 25070500 | chr15 | + | sense overlapping | STARD9 |
| hsa_circRNA_405443 | 0.028911656 | 0.541205763 | 3.3381633 | down | 25070500 | chr16 | + | intronic | NDE1 |
| hsa_circRNA_406309 | 0.01797211 | 0.541205763 | 3.1653952 | down | 25070500 | chr3 | + | intronic | CMSS1 |
| hsa_circRNA_100075 | 0.029545308 | 0.541205763 | 3.1382274 | down | circBase | chr1 | − | exonic | EMC1 |
| hsa_circRNA_406821 | 0.020949363 | 0.541205763 | 3.0380172 | down | 25070500 | chr6 | + | exonic | ARMC2 |
| hsa_circRNA_000361 | 0.027240076 | 0.541205763 | 3.0118108 | down | circBase | chr3 | − | antisense | PLCL2 |
| hsa_circRNA_082335 | 0.035559522 | 0.541205763 | 3.008493 | down | circBase | chr7 | + | exonic | KLHDC10 |
| hsa_circRNA_102207 | 0.013654637 | 0.541205763 | 3.0013356 | down | circBase | chr17 | + | exonic | AFMID |
| hsa_circRNA_405825 | 0.015220854 | 0.541205763 | 2.522762 | down | 25070500 | chr2 | + | exonic | KLF11 |
| hsa_circRNA_074660 | 0.017052344 | 0.541205763 | 2.4763773 | down | circBase | chr5 | − | exonic | ATOX1 |
| hsa_circRNA_104924 | 0.01941688 | 0.541205763 | 2.4549169 | down | circBase | chr9 | + | exonic | MVB12B |
| hsa_circRNA_056037 | 0.027008503 | 0.541205763 | 2.4478589 | down | circBase | chr2 | − | exonic | BUB1 |
| hsa_circRNA_406780 | 0.028354789 | 0.541205763 | 2.3707217 | down | 25070500 | chr6 | − | sense overlapping | DNPH1 |
| hsa_circRNA_024371 | 0.025742814 | 0.541205763 | 2.36132 | down | circBase | chr11 | + | exonic | PAFAH1B2 |
| hsa_circRNA_061284 | 0.026417014 | 0.541205763 | 2.30245 | down | circBase | chr21 | + | exonic | USP25 |
| hsa_circRNA_100456 | 0.009103973 | 0.541205763 | 2.2394652 | down | circBase | chr1 | + | exonic | KCNK2 |
| hsa_circRNA_083919 | 0.037676327 | 0.541205763 | 2.213904 | down | circBase | chr8 | + | exonic | UNC5D |
| hsa_circRNA_406295 | 0.017374416 | 0.541205763 | 2.2027287 | down | 25070500 | chr3 | + | sense overlapping | SUCLG2-AS1 |
| hsa_circRNA_035426 | 0.040284453 | 0.541205763 | 2.2016635 | down | circBase | chr15 | + | exonic | TCF12 |
| hsa_circRNA_405296 | 0.023543643 | 0.541205763 | 2.1972163 | down | 25070500 | chr15 | + | sense overlapping | TUBGCP5 |
qRT-PCR Validation
The top eight most upregulated circRNAs with fold change >4 and P < 0.05 and the top eight most downregulated circRNAs with fold change >3 and P < 0.05 are shown in the cluster heat map (Figure 2A). qRT-PCR assay was used to assess the accuracy of circRNAs chip data. After filtering circRNAs with low raw intensity, six candidate circRNAs, including three up-regulated circRNAs (hsa_ circRNA_061346, hsa_circRNA_103114, hsa_circRNA_103870) and three down-regulated circRNAs (hsa_circRNA_405324, hsa_circRNA_406821, hsa_circRNA_ 000361) were selected for qRT-PCR analysis. The results showed that qRT-PCR results were consistent with the circRNAs chip data (Figure 2B), indicating the reliability of circRNAs chip data.
FIGURE 2.
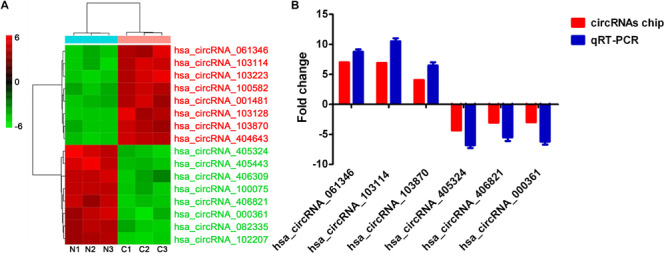
qRT-PCR validation. (A) The cluster heat map of the top eight most up-regulated and down-regulated circRNAs. (B) Comparison of Arraystar human circRNAs chip data and qRT-PCR results. circRNAs expression levels were normalized to GAPDH. N, adjacent normal tissues; C, GIST tissues.
CircRNA–miRNA–mRNA Network Construction
The fundamental structure modes of the six candidate circRNAs predicted by CSCD are shown in Figure 3. To estimate the function of six candidate circRNAs, circRNA–miRNA interactions were constructed with TargetScan and miRanda databases. The top five targeted miRNAs of six candidate circRNAs are exhibited in Figure 4, and the detailed potential circRNA–miRNA interaction sites of targeted miRNAs with the highest context score percentile are shown in Figure 5. Then, the circRNA–miRNA–mRNA competitive network (cirCeNET) was visualized by Cytoscape software (version 3.6.1) based on circRNA–miRNA interactions and miRNA–mRNA interactions (Figure 6). This network contained six circRNAs, 30 miRNAs, and 308 mRNAs, which provided a comprehensive perspective into the links between circRNA, miRNA, and mRNAs in GISTs.
FIGURE 3.
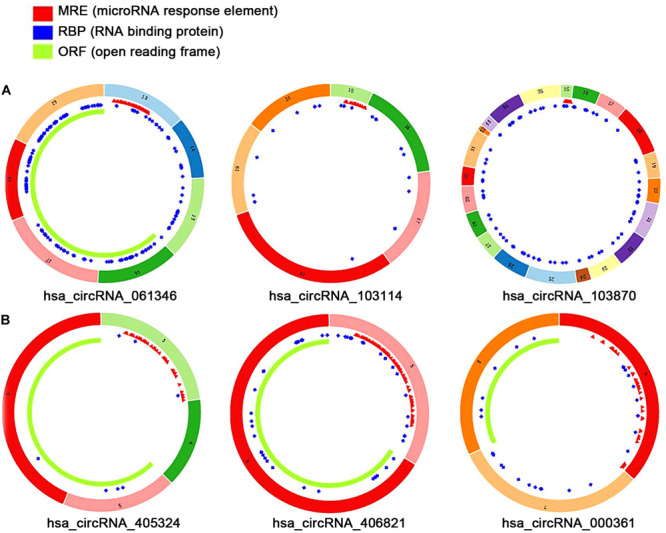
The fundamental structure modes of the six candidate circRNAs predicted by CSCD. (A) Up-regulated circRNAs: hsa_circRNA_061346, hsa_circRNA_103114, hsa_circRNA_103870. (B) Down-regulated circRNAs: hsa_circRNA_405324, hsa_ circRNA _406821, hsa_circRNA_000361.
FIGURE 4.
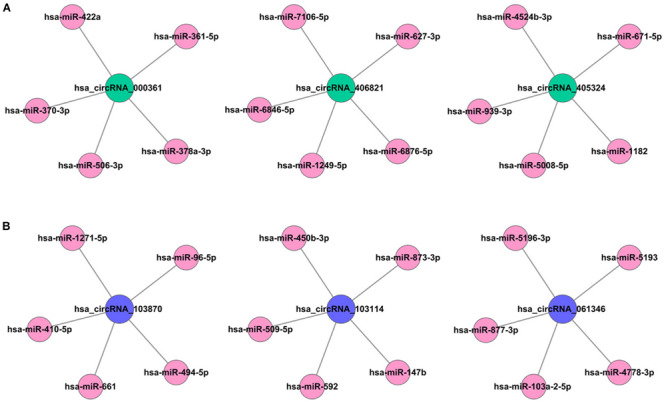
circRNA–miRNA regulatory network. (A) The top five targeted miRNAs of up-regulated circRNAs. (B) The top five targeted miRNAs of down-regulated circRNAs.
FIGURE 5.
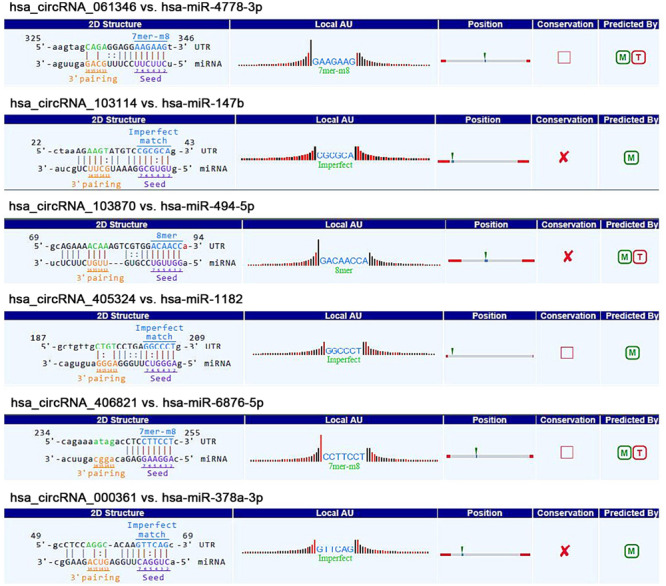
The detailed potential circRNA–miRNA interaction sites of targeted miRNAs with highest context score percentile based on TargetScan and miRanda data.
FIGURE 6.
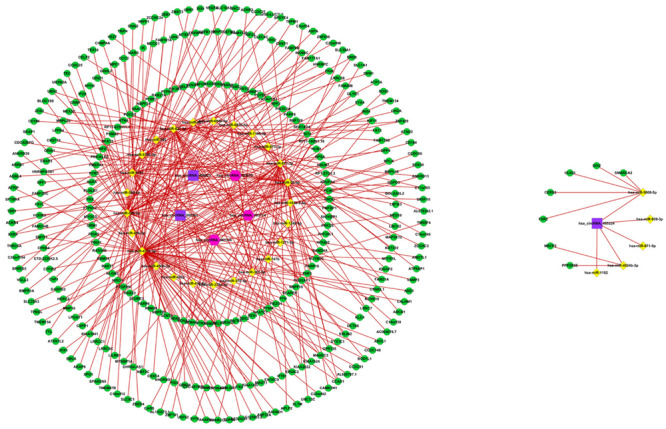
circRNA–miRNA–mRNA regulatory network. Up-regulated circRNAs, down-regulated circRNAs, miRNA, and mRNA are presented as square, hexagon, diamond, and circle, respectively.
GO and KEGG Pathway Analysis
The GO analysis demonstrated that the term with the highest enrichment score was regulation of biological process (GO:0050789) for biological process terms (BP), intracellular organelle (GO:0043229) for cellular component terms (CC), and protein binding (GO:0005515) for molecular function terms (MF), respectively. The top 10 enrichment scores are shown in Figures 7A–C. The KEGG pathway with the highest enrichment score was the Wnt signaling pathway. The top 10 enriched KEGG pathways are shown in Figure 7D.
FIGURE 7.
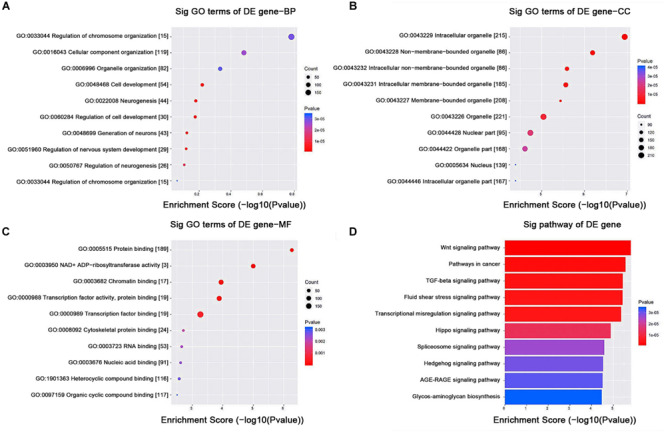
GO and KEGG pathway analysis. (A–C) GO annotation of targeted mRNAs with the top 10 enrichment scores for biological process, cellular component, and molecular function, respectively. (D) The top 10 enriched KEGG pathways. GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
qRT-PCR Validation
Six significantly differential circRNAs were also verified in 20 pairs of GISTs and adjacent tissues by qRT-PCR. The results showed that circRNA_061346, circRNA_103114, and circRNA_103870 were significantly up-regulated in GIST tissues (Figure 8) (P < 0.05), and circRNA_405324, circRNA_ 406821, and circRNA_000361 were dramatically down-regulated in GIST tissues (Figure 9) P < 0.05), compared with corresponding adjacent tissues.
FIGURE 8.
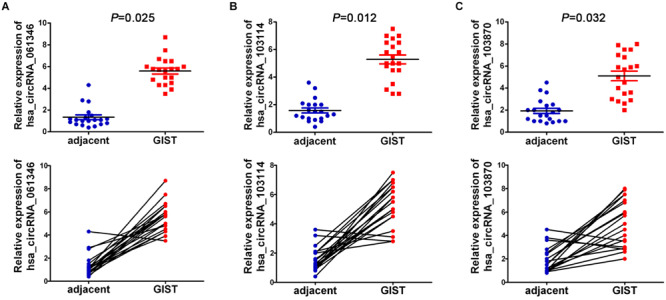
The relative expression of up-regulated circRNAs of 20 GIST tissues were detected by qRT-PCR, (A) hsa_circRNA_061346; (B) hsa_circRNA_103114; (C) hsa_ circRNA_103870. circRNA expression levels were normalized to GAPDH.
FIGURE 9.
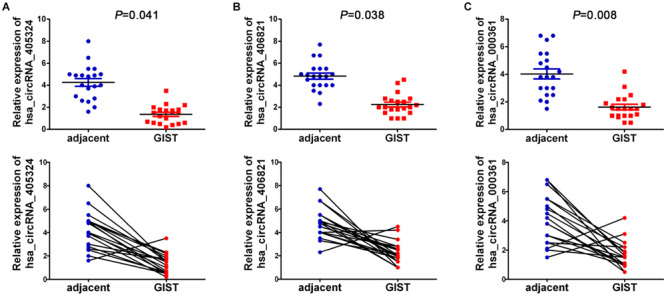
The relative expression of down-regulated circRNAs of 20 GIST tissues was detected by qRT-PCR, (A) hsa_circRNA_405324; (B) hsa_circRNA _406821; (C) hsa_circRNA_000361. circRNA expression levels were normalized to GAPDH.
Diagnosis Values of CircRNA
In order to determine the diagnostic value of six candidate circRNAs in GISTs, the ROC curve was employed. Statistical analysis demonstrated that all six candidate circRNAs had high diagnostic efficiency with AUC = 0.9925, AUC = 0.9824, AUC = 0.9231, AUC = 0.9300, AUC = 0.9463, AUC = 0.9138 for circRNA_061346, circRNA_103114, circRNA_103870 and circRNA_405324, circRNA_406821, circRNA_000361, respectively (Figure 10) (P < 0.05).
FIGURE 10.
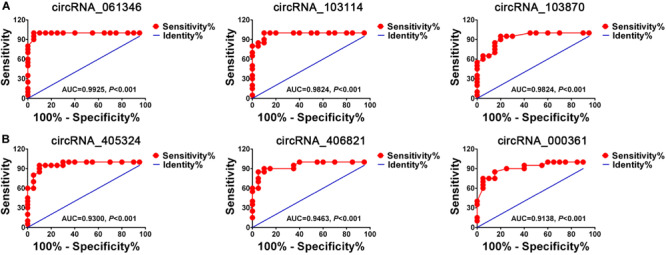
The diagnostic value of six candidate circRNAs in GISTs. (A) ROC curve for three up-regulated circRNAs. (B) ROC curve for three down- regulated circRNAs.
Correlation of CircRNA Expressions With Clinical Pathologic Features
In order to investigate the correlation of six candidate circRNA expressions with clinical–pathologic features, the median circRNA expression was used to divide the 20 pairs of GISTs tissues into higher and lower circRNA expression groups. Chi-square assay was employed for statistical analysis. The results suggested that circRNA_061346 and circRNA_103114 expressions were positively associated with tumor size, mitotic figure, malignant degrees, and circRNA_103870 expression was positively associated with tumor size, mitotic figure, but without relation to malignant degrees (Figure 11). On the contrary, circRNA_405324 expression was negatively associated with tumor size, mitotic figure, malignant degrees, and circRNA_406821 was negatively correlated with mitotic figure, malignant degrees, but not with tumor size; nevertheless, circRNA_000361 expression was only negatively related with mitotic figure (Figure 11B). However, there was no correlation with age, gender, and tumor location.
FIGURE 11.
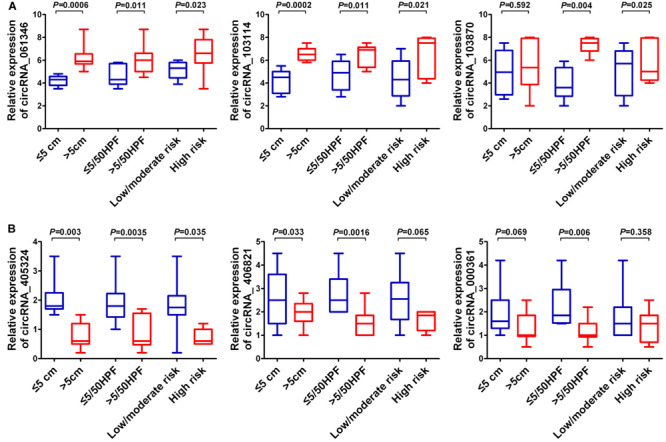
Correlation of six candidate circRNAs with clinical pathologic features. (A) Box plots for three up-regulated circRNAs. (B) Box plots for three down-regulated circRNAs. Tumor size: ≤5 cm, >5 cm; mitotic figure: ≤5/50 HPF, <5/50 HPF; malignant degrees: low risk, moderate risk, high risk.
Discussion
Gastrointestinal stromal tumors are a rare malignant tumor that occurs principally in the stomach, small intestine, and colon–rectum (Flavahan et al., 2019). K-ras gene mutation might be correlated with the mechanism of development and infiltration of GISTs, but the pathogenesis of GISTs is inadequately understood (Lasota et al., 2019). A growing body of research (Wang et al., 2018; Cai et al., 2019; Zaghlool et al., 2020) demonstrates that the development of cancer is often accompanied by abnormal expression of circRNA. Also, circRNA is characterized by inherent stability, highly conservative, and universality. Therefore, circRNA is of great importance as a biomarker for cancer screening, cancer diagnosis, cancer prediction, feedback of treatment, and prognosis. Additionally, circRNA’s abnormal expression and the circRNA–miRNA–mRNA regulatory network regulation have been increasingly demonstrated in a variety of tumors, such as circRNA_CAMK2A–miR-615-5p–fibronectin 1 network in lung adenocarcinoma metastasis (Du et al., 2019), circRNA_0006948–miR-490-3p–HMGA2 network in esophageal squamous cell carcinoma (Pan et al., 2019), circRNA_ACAP2–miR-29a/b-3p–COL5A1 network in breast cancer (Zhao et al., 2019), circRNA_51217–miRNA-646–TGFβ1/p-Smad2/3 network in prostate cancer (Xu et al., 2019), etc. At present, there are few reports about the circRNA–miRNA–mRNA regulatory network in GISTs.
In the present study, we illuminate the molecular mechanisms of circRNAs in the occurrence and development of GISTs for the first time. We first performed circRNA chip analysis to assess differential cicrRNA expression profiles in GIST tissues and corresponding non-cancer tissues. A totally of 543 differentially expressed circRNAs were identified, of which 242 were significantly upregulated and 301 were significantly downregulated in GISTs tissues. Additionally, in order to fully elucidated the function of the circRNA-related ceRNA in GISTs, six candidate circRNAs including three up-regulated circRNAs (hsa_circRNA_061346, hsa_circRNA_103114, hsa_ circRNA_103870) and three down-regulated circRNAs (hsa_circRNA_405324, hsa_ circRNA_406821, hsa_circRNA_000361) were identified to be involved in the ceRNA network. The ceRNA network consists of six circRNAs, 30 miRNAs, and 308 mRNAs. In this network, previous studies (Xu Z.H. et al., 2018; Zhang et al., 2019) show that many miRNAs, such as miR-4778-3p, miR-147b, miR-1182, and miR-378a-3p, were involved in tumor cell growth, invasion, and metastasis. Also, many targeted genes, such as ZEB1, SOX5, AKAP1, CHP1, CNBP, VEGFR, and MAGT1, play a vital function in the cell shape, movement, invasion, adhesion, and polarity formation, so as to involve in many kinds of diseases such as malignant tumors, wound healing, and so on (Rinaldi et al., 2017; Caramel et al., 2018; Hu et al., 2018).
Furthermore, 20 GIST tissues and adjacent tissues were collected to verify the expression of identified six candidate circRNAs. qRT-PCR results showed that hsa_circRNA_061346, hsa_circRNA_103114, and hsa_circRNA_103870 were significantly up-regulated in GISTs, and hsa_ circRNA_405324, hsa_circRNA _406821, hsa_circRNA_ 000361 were dramatically down-regulated in GISTs. In addition, all of these circRNAs were shown to have high diagnostic values, and most of them were significantly associated with tumor size, mitotic figure, and malignant degrees in GISTs. The six candidate circRNAs might be critical circRNAs participating in the occurrence and development of GISTs and can serve as novel potential diagnostic biomarkers for GISTs patients. Through a large literature review, very limited data are available about these circRNAs’s functions and their deregulation in cancer.
However, there are several limitations to our study. First, only 20 patients were enrolled in our study, the sample size is relatively small, and the result showed an association, rather than a definite, causal relationship. Also, the relation analysis of clinical factors and circRNAs needs to be supported by large samples. Second, in our study, we only conducted a network based on identified six critical circRNAs, miRNA, and target mRNA, but a total of 543 circRNAs were identified in GISTs. Other circRNAs may contribute as well. Third, our paper starts with a general analysis of circRNAs in GISTs, but the mechanism is not discussed in detail. Therefore, in our future work, further studies with larger groups of patients, a network based on 543 circRNAs are needed to confirm these findings, and the concrete mechanism of circRNAs in GISTs also needs to be further explored.
Conclusion
In the present study, the differential circRNA expression profile of GISTs was established, and a total of 543 differentially expressed circRNAs were screened. In addition, the circRNA–miRNA–mRNA regulatory network was constructed. hsa_circRNA_ 061346, hsa_circRNA_103114, hsa_circRNA_103870 and hsa_circRNA_405324, hsa_ circRNA_406821, hsa_circRNA_000361 were identified as critical circRNAs in the occurrence and development of GISTs and may present as potential diagnostic biomarkers for GISTs. In brief, our study provides a new insight into the pathogenesis of GISTs from the circRNA–miRNA–mRNA regulatory network view.
Data Availability Statement
The datasets generated for this study can be found in NCBI GEO accession GSE147303.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of The Second Xiangya Hospital of Central South University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
LZ coordinated all aspects of the research. FZ and DC were responsible for the clinical sample collection. FZ participated in most molecular and cellular experiments and manuscript preparation. LS and ZZ were responsible for data analysis. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We highly appreciate the kind help and support from the Shanghai Kangcheng Co., Ltd. (Chinese). Personally, for my parents and wife and daughter, I really appreciate all your love and support up to my work. Thank you for all you did to us, really thanks, I love you all. Patient clinicopathological data used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- CircRNA
circular RNA
- DAVID
Database for Annotation Visualization and Integrated Discovery
- GISTs
gastrointestinal stromal tumors
- GO
gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MiRNAs
microRNAs
- MREs
microRNA response element
- ROC curve
receiver operating characteristic curve.
Funding. The project received support from the Hunan Provincial Science and Technology Department (2017SK1032, LZ), fundamental research funds for the Central Universities of Central South University (2019zzts356, FZ), Changsha Science and Technology Plan Project (kq1907080, YT), and Changsha Science and Technology Plan Project (kq1907078, FZ).
References
- Cai H., Li Y., Niringiyumukiza J. D., Su P., Xiang W. (2019). Circular RNA involvement in aging: an emerging player with great potential. Mech. Age. Dev. 178 16–24. [DOI] [PubMed] [Google Scholar]
- Caramel J., Ligier M., Puisieux A. (2018). Pleiotropic roles for ZEB1 in cancer. Cancer Res. 78 30–35. [DOI] [PubMed] [Google Scholar]
- Chaichian S., Shafabakhsh R., Mirhashemi S. M., Moazzami B., Asemi Z. (2020). Circular RNAs: a novel biomarker for cervical cancer. J. Cell. Physiol. 235 718–724. [DOI] [PubMed] [Google Scholar]
- Ding H., Xu Q., Wang B., Lv Z., Yuan Y. (2020). MetaDE-based analysis of circRNA expression profiles involved in gastric cancer. Digest. Dis. Sci. 12 1–12. [DOI] [PubMed] [Google Scholar]
- Du J., Zhang G., Qiu H., Yu H., Yuan W. (2019). The novel circular RNA circ-CAMK2A enhances lung adenocarcinoma metastasis by regulating the miR-615-5p/fibronectin 1 pathway. Cell Mol. Biol. Lett. 24 72–78. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Etherington M. S., DeMatteo R. P. (2019). Tailored management of primary gastrointestinal stromal tumors. Cancer 10 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan W. A., Drier Y., Johnstone S. E., Hemming M. L., Tarjan D. R., Hegazi E., et al. (2019). Altered chromosomal topology drives oncogenic programs in SDH-deficient GISTs. Nature 575 229–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam A. (2020). Gastrointestinal stromal tumor of the stomach infiltrating the hilum of spleen: case report. Clin. Surg. J. 3 17–21. [Google Scholar]
- Gupta S. K., Rateria N. (2020). Gastrointestinal stromal tumors (GIST): an overview. Indian J. Surg. 24 1–7. [Google Scholar]
- Hu J., Tian J., Zhu S., Sun S., Yu J., Tian H., et al. (2018). Sox5 contributes to prostate cancer metastasis and is a master regulator of TGF-β-induced epithelial mesenchymal transition through controlling Twist1 expression. Br. J. Cancer 118 88–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Sun D., Hou J., Hou J. C., Ji Z. L. (2018). CircRNA: a novel type of biomarker for cancer. Breast Cancer 25 1–7. [DOI] [PubMed] [Google Scholar]
- Lasota J., Kowalik A., Felisiak-Golabek A., Ziêba S., Wang Z.-F. (2019). New mechanisms of mTOR pathway activation in KIT-mutant malignant GISTs. Appl. Immunohist. 27 54–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C. X., Sun X. M., Li N., Wang W. G., Kuang D. X., Tong P., et al. (2018). CircRNAs in the tree shrew (Tupaia belangeri) brain during postnatal development and aging. Aging 1 833–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi E., Kiltschewskij D., Fitzsimmons C., Cairns M. J. (2020). Depolarization-associated CircRNA regulate neural gene expression and in some cases may function as templates for translation. Cells 9 25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng S., Zhou H., Feng Z., Xu Z., Tang Y., Li P., et al. (2017). CircRNA: functions and properties of a novel potential biomarker for cancer. Mol. Cancer 16 94–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z., Lin J., Wu D., He X., Wang W., Hu X., et al. (2019). Hsa_circ_0006948 enhances cancer progression and epithelial-mesenchymal transition through the miR-490-3p/HMGA2 axis in esophageal squamous cell carcinoma. Aging 11 11937–11954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi L., Sepe M., Delle D. R., Conte K., Arcella A., Borzacchiello D., et al. (2017). Mitochondrial AKAP1 supports mTOR pathway and tumor growth. Cell Death Dis. 8 e2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao K., Zeng X., Liu W., Wang S., Gao J., Shuai X., et al. (2020). Primary gastrointestinal stromal tumor mimicking as gynecologic mass: characteristics, management, and prognosis. J. Surg. Res. 246 584–590. [DOI] [PubMed] [Google Scholar]
- Wang H., Xiao Y., Wu L., Ma D. (2018). Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-000911/miR-449a pathway in breast carcinogenesis. Intern. J. Oncol. 52 743–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Sun Y., You B., Huang C. P., Ye C., Chang C. (2019). Androgen receptor reverses the oncometabolite R-2-hydroxyglutarate-induced prostate cancer cell invasion via suppressing the circRNA-51217/miRNA-646/TGFβ1/p-Smad2/3 signaling. Cancer Lett. 472 151–164. [DOI] [PubMed] [Google Scholar]
- Xu S., Zhou L. Y., Ponnusamy M., Zhang L., Dong Y. H., Zhang Y. H., et al. (2018). A comprehensive review of circRNA: from purification and identification to disease marker potentia. PeerJ 6:e5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z. H., Yao T. Z., Liu W. (2018). miR-378a-3p sensitizes ovarian cancer cells to cisplatin through targeting MAPK1/GRB2. Biomed. Pharmacother. 107 1410–1417. [DOI] [PubMed] [Google Scholar]
- Zaghlool S. B., Kühnel B., Elhadad M. A., Kader S., Halama A., Thareja G., et al. (2020). Epigenetics meets proteomics in an epigenome-wide association study with circulating blood plasma protein traits. Nat. Commun. 11 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Li P., Hu J., Jhao J. P., Ma R., Li W., et al. (2019). Role and mechanism of miR-4778-3p and its targets NR2C2 and Med19 in cervical cancer radioresistance. Biochem. Biophys. Res. Commun. 508 210–216. [DOI] [PubMed] [Google Scholar]
- Zhao B., Song X., Guan H. (2019). CircACAP2 promotes breast cancer proliferation and metastasis by targeting miR-29a/b-3p-COL5A1 axis. Life Sci. 14:117179. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated for this study can be found in NCBI GEO accession GSE147303.