Abstract
Metabolic engagement is intrinsic to immune cell function. Prostaglandin E2 (PGE2) has been shown to modulate macrophage activation, yet how PGE2 might affect metabolism is unclear. Here we show that PGE2 caused mitochondrial membrane potential (Δψm) to dissipate in interleukin-4 activated macrophages (M(IL-4)). Effects on Δψm were a consequence of PGE2-initiated transcriptional regulation of genes in the malate-aspartate shuttle (MAS), particularly GOT1. Reduced Δψm caused alterations in the expression of 126 voltage regulated genes (VRGs) including Resistin like molecule-α (RELMα), a key marker of M(IL-4), and genes that regulate cell cycle. The transcription factor ETS variant 1 (ETV1) played a role in the regulation of 38% of the VRGs. These results reveal ETV1 as a Δψm-sensitive transcription factor, and Δψm as a mediator of mitochondrial-directed nuclear gene expression.
Keywords: Mitochondrial membrane potential, ETV1, GOT1, Malate-aspartate shuttle, Macrophages, PGE2, IL-4, cAMP, Alternative activation, Immunometabolism
INTRODUCTION
Changes in metabolism are intrinsic to, and required for, immune cell activation (Buck et al., 2016; O’Neill and Pearce, 2016). When macrophages are exposed to lipopolysaccharide (LPS) and interferon-γ (IFN-γ) or interleukin-4 (IL-4), they undergo major and largely non-overlapping changes in gene expression and exhibit distinct metabolic profiles dominated by aerobic glycolysis (Warburg metabolism) and augmented oxidative phosphorylation (OXPHOS), respectively. In both cases, inhibition of the dominant metabolic pathway inhibits the activation response (Buck et al., 2016; O’Neill and Pearce, 2016).
Consistent with their broad roles in tissue biology during homeostatic conditions, infection, cancer, and traumatic damage, macrophages express a range of receptors that allow them to respond to exogenous signals and assume distinct functions (Amit et al., 2015). Differing gene expression profiles under this spectrum of stimulatory conditions have been defined (Xue et al., 2014), but little is known about whether metabolism is fine-tuned to support and enable these various activation states. In this context we have been interested in alternatively activated macrophages, a term originally used to describe macrophages activated via the IL-4 receptor (M2 or M(IL-4) macrophages), but since evolved to include cells stimulated through other pathways (Ginhoux et al., 2015). For example, a variety of reports indicate that PGE2 can both independently and in concert with IL-4 promote alternative activation (Luan et al., 2015; Na et al., 2015). PGE2 is implicated in cancer development (Zelenay et al., 2017) and it also suppresses the production of inflammatory cytokines in LPS-activated macrophages (Tang et al., 2017). Moreover, PGE2 plays positive roles in the maintenance of gastric mucosal integrity (Miyoshi et al., 2017), immunity to Mycobacterium tuberculosis (Mayer-Barber et al., 2014) and in the prevention of fibrosis (Harada et al., 2017). Consequently, we were interested in whether PGE2 would accentuate or modify the metabolic profile of M(IL-4) as it relates to gene expression in these cells.
Increased OXPHOS, a hallmark of M(IL-4) cells, is intimately linked to mitochondrial membrane potential (Δψm). In mitochondria, potential across the inner mitochondrial membrane is established as the electron transport chain (ETC) exports protons from the matrix into the intermembrane space, providing the motive force for ATP synthesis. The activity of the tricarboxylic acid (TCA) cycle, the availability of nicotinamide adenine dinucleotide (NADH) and flavin adenine dinucleotide (FADH2) to fuel the ETC, and the expression of uncoupling proteins, provide physiologic mechanisms for regulating Δψm. Recent work has identified additional roles for Δψm, beyond ATP synthesis, in mitochondrial ROS and cellular proliferation (Martínez-Reyes et al., 2016), acetylation of mitochondrial matrix proteins (Yang et al., 2016), plus stemness and longevity of CD8 T cells (Sukumar et al., 2016). Thus Δψm is linked to different cell fates.
Here we found that macrophages costimulated with IL-4 plus PGE2 exhibited altered gene expression accompanied by changes in metabolism, which included a marked decrease in Δψm. Diminished Δψm was induced by changes in the malate-aspartate shuttle (MAS), a mitochondrial pathway that regulates redox balance. We discovereda Δψm-sensitive gene program regulated largely by the ETV1 transcription factor(TF). ETV1 controlled “voltage regulated genes” (VRGs) included Retnla, a key markof alternative activation, and genes involved in cell cycle, which may explainobserved PGE2-induced reductions in cellular proliferation. Thus, by exploring an unappreciated facet of PGE2 biology, we have identified a Δψm-dependentmechanism of mitochondria-nucleus communication in which nuclear geneexpression is regulated by Δψm-dependent control of ETV1.
RESULTS
PGE2 modifies macrophage alternative activation
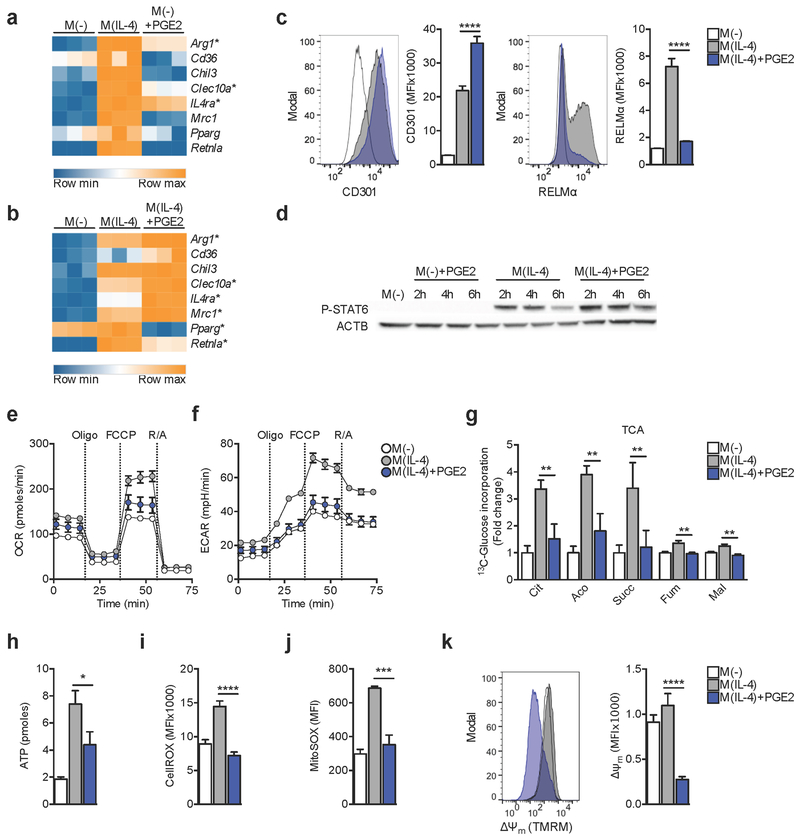
In light of existing evidence indicating that PGE2 can both independently and in concert with IL-4 promote alternative activation (Luan et al., 2015; Na et al., 2015), we set out to study the transcriptome of macrophages exposed to this lipid mediator. In mouse bone marrow derived macrophages (BMMs) PGE2 induced the expression of some genes associated with alternative (M(IL-4)) activation, including Arg1, IL4ra and Clec10a (CD301) (Figure 1A). However, other genes encoding marks of M(IL-4), such as Retnla (RELMα), Cd36, Chil3, Mrc1, and Pparg, were not induced. Indeed, most genes regulated by PGE2 were not related to M(IL-4) activation (Figure S1A–B). Nevertheless, PGE2 accentuated IL-4 induced expression of Arg1, IL4ra, Clec10a, CD36 and Mrc1 (Figure 1B) when both stimuli where used simultaneously (costimulation), although it suppressed the expression of Retnla and Pparg (Figure 1B). Examination of CD301 and RELMα expression by flow cytometry confirmed that IL-4 induced markers can be accentuated or suppressed by PGE2 (Figure 1C). PGE2 did not alter IL-4 induced phosphorylation of STAT6 (Figure 1D) (Gordon and Martinez, 2010). Thus PGE2 has marked STAT6-independent effects on M(IL-4) activation.
Figure 1. PGE2 modifies IL-4 induced macrophage activation and metabolism.
(A-B) RNA-seq analysis of gene expression in bone marrow macrophages (BMMs) stimulated for 6 h with: IL-4 (M(IL-4)) or PGE2 (M(−)+PGE2)(A) or M(IL-4) or M(IL-4)+PGE2 (B), compared to no stimulus controls (M(−)) (A,B).
(C) Expression of CD301 and RELMα proteins, as measured by flow cytometry, in BMMs stimulated for 24 h with indicated treatments. MFI = mean fluorescence intensity.
(D) Western blot of phosphorylated STAT6 (P-STAT6) at indicated times in BMMs treated as indicated. ACTB, actin loading control.
(E-F) Extracellular flux analysis (EFA) of oxygen consumption rates (OCR) (E) or extracellular acidification rates (ECAR) (F) at 24 hr post stimulation. For EFA, BMMs were sequentially treated with oligomycin (Oligo), FCCP and rotenone plus antimycin A (R/A) as indicated.
(G) 13C-Glucose LC-MS trace into TCA cycle metabolites from cells stimulated for 24 h with indicated treatments.
(H-K) ATP concentration (H), cytoplasmic ROS (I), mitochondrial ROS (J), and Δψm as measured by TMRM incorporation (K), in BMMs stimulated as indicated for 24 h.
(A-B) Significant (adjusted p value < 0.1) > 2 fold up or down regulated genes based on 3 biological replicates denoted by *. (C) Mean ± SEM from 5 biological replicates (p***<0.0005, p****<0.0001). (G) Mean ± SEM from 3 biological replicates, normalized to M(−) (p**<0.005). (H-K) Mean ± SEM from 5 biological replicates (p*<0.05, p***<0.0005, p****<0.0001). Data are representative of 2 (D) or 3 (C, E, F, H-K) independent experiments. See also Figure S1.
Metabolic reprogramming intrinsically regulates macrophage function (O’Neill and Pearce, 2016). Therefore we asked whether PGE2-induced changes in M(IL-4) reflected alterations in metabolism. Elevated baseline oxygen consumption rates (OCR), maximal respiration, and glycolytic reserve are marks of M(IL-4) (Huang et al., 2014), and each was diminished by PGE2 (Figure 1E,F; S1C–E). Consistent with this, PGE2 caused decreased glucose 13C incorporation into TCA cycle intermediates (Figure 1G); this was not a reflection of reduced glucose uptake (Figure S1F), nor of altered activity of Phosphofructokinase 1 (PFK1) (Rider et al., 2004) (Figure S1G), and could not readily be explained by changes in the expression of genes involved in glycolysis, the TCA cycle, or OXPHOS (Figure S1H). These metabolic changes were associated with reduced cellular ATP (Figure 1H), and diminished cytoplasmic and mitochondrial reactive oxygen species (ROS) (Figure 1I, J), overall indicating that PGE2 has a dampening effect on mitochondrial metabolism. The development of Δψm is critical for mitochondrial function, and we reasoned that this parameter might also be affected. Using tetramethylrhodamine, methyl ester (TMRM), we found that Δψm of macrophages treated with IL-4 plus PGE2 was lower than that of M(IL-4) cells (Figure 1K).
PGE2 modulates mitochondrial membrane potential.
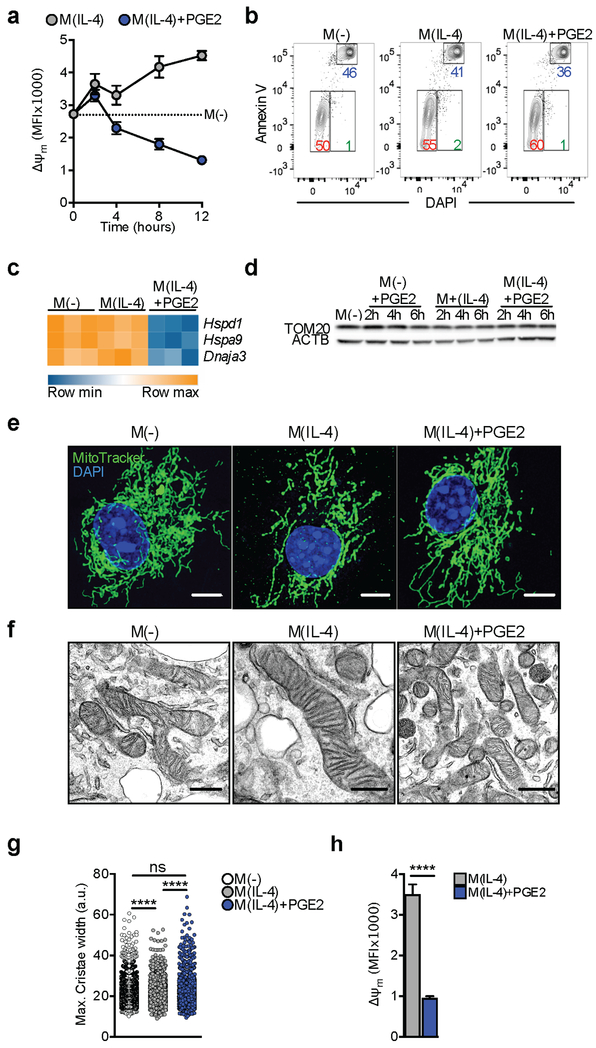
We found that effects of PGE2 on Δψm developed between 2 h and 4 h post stimulation, and intensified thereafter (Figure 2A; S2A–B). Diminished Δψm can presage cell death (Gottlieb and Bernstein, 2016), but M(IL-4) treated with PGE2 survived normally in culture (Figure 2B; S2C). Moreover, the mitochondrial unfolded protein response, an indicator of mitochondrial damage (Gottlieb and Bernstein, 2016), was not induced by PGE2 since expression of Hspd1, Hspa9 and Dnaja3 was diminished rather than increased (Figure 2C). Reduction in Δψm was not a reflection of reduction in mitochondrial mass, as measured by TOM20 amount or MitoTracker staining (Figure 2D, S2D). Decreased Δψm can lead to mitochondrial fission (Willems et al., 2015), but mitochondria remained elongated in M(IL-4) plus PGE2 (Figure 2E), and we found no evidence of proteolytic cleavage of the long isoform of OPA1 (Figure S2E), which is required for fission (Willems et al., 2015). Furthermore, ultrastructural examination revealed no pathological changes in M(IL-4) treated with PGE2, but did show that cristae were narrower in M(IL-4) than in M(−), and that this difference was lost once the M(IL-4) were costimulated with PGE2 (Figure 2F–G), a finding that is consistent with the observed differences in maximal respiration in these cells (Figure 1E) and our understanding of the relationship between maximal respiration and cristae morphology (Buck et al., 2016). Thus, PGE2 reduced Δψm independently of effects on viability or major alterations in mitochondrial content or morphology.
Figure 2. PGE2-driven loss of mitochondrial membrane potential is not associated with mitochondrial damage.
(A) Analysis of Δψm via incorporation of TMRM over time into BMMs stimulated as indicated. Mean ± SEM from 3 biological replicates.
(B) Representative flow cytometry plots with percentages of viable (red), apoptotic (green) and necrotic (blue) BMMs, based on Annexin V and DAPI staining of cells stimulated for 24 h as indicated.
(C) mRNA of mitoUPR genes after 6 h of indicated stimulation.
(D) Western blot for TOM20 in BMMs before and after indicated stimulations. ACTB, actin loading control.
(E) Representative confocal images of BMMs stained with MitoTracker Deep Red, and DAPI after 24 h of stimulation as indicated. Bar = 5 μm.
(F, G) Representative electron micrographs (F, bar = 500 nm) and cristae width (G) measured from electron micrographs, of BMMs treated for 24 h as indicated. Each symbol in G represents an individual measurement.
(H) Analysis of Δψm (as in A) in peritoneal macrophages after 24 h. Bars are mean ± SEM from 3 biological replicates (p****<0.0001).
Data are representative of 3 (A-B, E, F, H) or 2 (D) independent experiments. (G-H) p****<0.0001. See also Figure S2.
We further examined the effects of PGE2 on Δψm. We found that a PGE2-induced reduction in Δψm was also apparent in peritoneal macrophages (pMacs) stimulated with IL-4 (Figure 2H). Moreover, the ability of PGE2 to affect macrophage Δψm was not dependent on IL-4-induced events, since Δψm in M(−) was also reduced by PGE2 (Figure S2F). Nor was it restricted to murine macrophages, as the same was true for human peripheral blood monocytes cultured with PGE2 (Figure S2G). However, PGE2 had no effect on Δψm when used to costimulate M(LPS+IFNγ) (Figure S2H).
Overall, our data indicate that PGE2 diminishes Δψm in respiring cells, and that while this is associated with reduced OXPHOS, it is not an indicator of mitochondrial damage.
Changes in Δψm result from altered mitochondrial shuttle activity
We wanted to understand the basis of the effect of PGE2 on Δψm. We reasoned that cAMP, which is induced by PGE2 (Rodriguez et al., 2013), could play a role in diminished Δψm. We found that either 8-Br-cAMP, a membrane permeable form of cAMP, or the cAMP-inducer forskolin, recapitulated the effects of PGE2 in lowering Δψm and cellular ATP concentration while suppressing RELMα and increasing CD301 expression in M(IL-4) (Figure S3A–B). On the other hand, PKA inhibition increased Δψm (Figure S3C). Thus PGE2-induced changes in Δψm are likely mediated by cAMP.
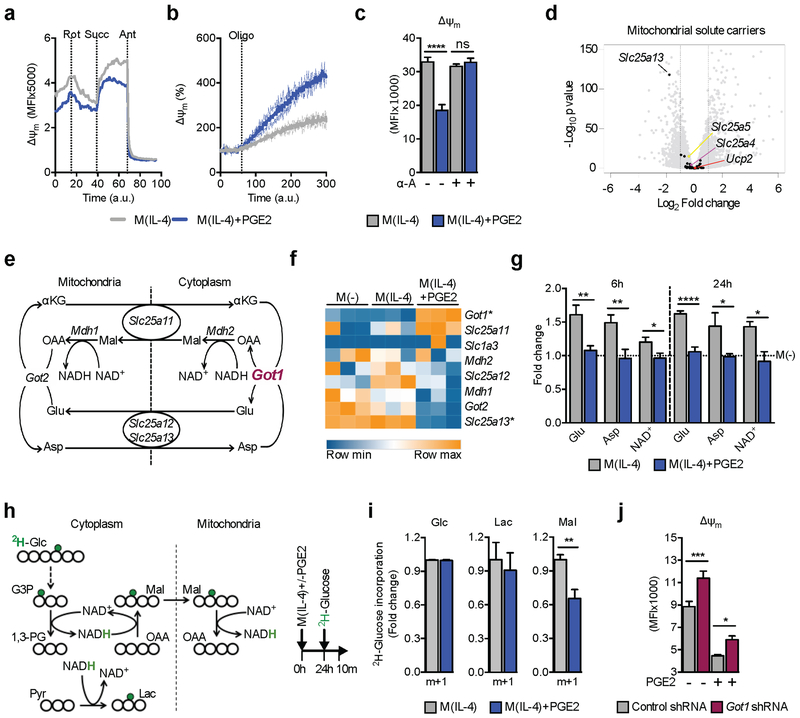
Δψm is established by the transport of protons from the mitochondrial matrix into the inter-membrane space by ETC complexes (C) I, III and IV. However, we detected no overall decrease in expression of ETC complexes following PGE2 treatment (Figure S3D). We next determined whether dysfunction at one of the complexes is responsible for PGE2-induced changes in Δψm. In permeabilized macrophages, we found that CI inhibition with rotenone decreased Δψm, while subsequently providing succinate for CII restored Δψm comparably in both M(IL-4) and M(IL-4) plus PGE2 (Figure 3A). Moreover, CIII inhibition with antimycin abrogated Δψm (Figure 3A) in both types of activated macrophages. These data indicate that PGE2 does not affect the activity of CI, CII or CIII, and imply that CI and CIII are equally capable of generating Δψm in M(IL-4) with or without PGE2. Next we asked whether inhibiting CV, which should increase Δψm by preventing the dissipation of the proton gradient to generate ATP, would allow an equivalent increase in Δψm in the presence or absence of PGE2. We found that Δψm increased more rapidly in M(IL-4) plus PGE2 than in M(IL-4) without PGE2 (Figure 3B; S3E). The rapid increase in Δψm as a result of CV inhibition in PGE2 treated cells could be consistent with heightened ATP demand, leading to decreased ATP in these cells (Figure 1H). We addressed this by pharmacologically increasing ATP in the cell (Figure S3F), assuming that if Δψm was depleted by heightened ATP demand, an increase in ATP would remove this constraint. However, increasing ATP had no effect on Δψm (Figure S3G). We do not have an explanation for the increased rate of Δψm recovery following CV inhibition in M(IL-4) plus PGE2 vs. M(IL-4). Nevertheless, the data show clearly that M(IL-4) treated with PGE2 have a functional ETC capable of restoring Δψm to basal amounts. Thus, PGE2 is inducing changes to the mitochondrial milieu that actively limits Δψm, without negatively affecting individual complex activity.
Figure 3. PGE2 regulates mitochondrial membrane potential through changes in the malate-aspartate shuttle (MAS).
(A-B) Real time contribution of ETC complexes to Δψm in BMMs stimulatd as indicated. Changes in Δψm after sequential treatment with rotenone (Rot), succinate (Succ) and antimycin (Ant) (A), or in response to oligomycin (B).
(C) Δψm in BMMs stimulated and treated with α-amanitin (α-A) as indicated for 24 h. Mean ± SEM from 6 biological replicates.
(D) Mitochondrial solute carrier gene expression in M(IL-4)+PGE2 stimulated for 6 h compared to M(IL-4).
(E) Schematic of the MAS.
(F) MAS gene expression in BMMs stimulated for 6 h as indicated. Statistically significant (adjusted p value < 0.1) up or down regulated (> 2 fold) genes from 3 biological replicates denoted by *.
(G) LC-MS quantitation of MAS metabolites in BMMs stimulated as indicated for 6 h or 24 h, normalized to M(−) (dotted line). Mean ± SEM from 3 biological replicates.
(H-I) Schematic (H) and results of using GC-MS to trace deuterium (green, H) in BMMs cultured in singly deuterated glucose (2H-Glc) into malate and lactate (I), plus depiction of stimulation strategy.
(J) Δψm in BMMs transduced with a control or Got1 targeting shRNA after 24 h stimulation with IL-4 +/− PGE2 as indicated. Means ± SEM from 6–7 biological replicates.
Data in A-C, G, I and J are representative of 3 independent experiments. (C, G, I-J) p*<0.05, p**<0.005, p***<0.0005, p****<0.0001. See also Figure S3.
The timing of PGE2 effects on Δψm (Figure 2A) was consistent with induced transcriptional changes rather than direct biochemical events being causative. To explore this, we measured Δψm in M(IL-4) with or without PGE2 when transcription was inhibited by α-amanitin (α-A) (Figure S3H), and found that PGE2 had no effect on Δψm (Figure 3C). We next searched for gene expression changes that could account for reduced Δψm, focusing on mitochondrial solute carriers as candidates that modulate Δψm. We found no effect of PGE2 on the expression of Slc25a5, Slc25a4, or Ucp2, which encode proteins capable of dissipating Δψm (Figure 3D) (Woyda-Ploszczyca and Jarmuszkiewicz, 2017). However, we noted a reduction in expression of Slc25a13 in M(IL-4) treated with PGE2 (Figure 3D). This gene encodes a component of the MAS, a system that facilitates redox balance between mitochondria and cytoplasm (Figure 3E). Since early reports linked MAS to Δψm (Kauppinen et al., 1987), we reasoned that MAS function might be altered after PGE2 treatment. Indeed, PGE2 caused changes in expression of many of the MAS genes (Figure 3F), and reductions in overall cellular amounts of the MAS metabolites Glutamate (Glu), Aspartate (Asp) and NAD+ at early and late times post stimulation (Figure 3G). We next evaluated NADH mitochondrial shuttling by tracing the transfer of a deuterium from glucose (Glc), through glyceraldehyde 3 phosphate (G3P) and NADH, into malate (Mal) (Figure 3H) (Lewis et al., 2014). Our data show that M(IL-4) with or without PGE2 took up similar amounts of 2H-Glc and comparably reduce pyruvate to lactate (Figure 3I). However, NADH incorporation into Mal was diminished in response to PGE2 (Figure 3I). Our data indicate that MAS activity is changed by PGE2, suggesting that this shuttle could regulate Δψm following PGE2 treatment. If altered MAS function underlied the effects of PGE2, then changes in MAS gene expression should occur prior to the loss of Δψm. Indeed, expression of Got1 and Slc25a13 were altered by PGE2 shortly after treatment (Figure S3I). We then hypothesized that the effects of PGE2 should be reversed by loss of function of GOT1, which is increased in response to PGE2 (Figure 3F, S3I). Consistent with this, we found that Got1-shRNA mediated suppression of GOT1 expression (Figure S3J) reversed the inhibitory effects of PGE2 on Δψm (Figure 3J), and also increased Δψm in M(IL-4) that had not been treated with PGE2, suggesting a broader regulatory role for the MAS in M(IL-4) biology.
Together, our data indicate that reduced Δψm downstream of PGE2 reflects changes in the expression of genes encoding mitochondrial redox shuttles with associated changes in shuttle activity, and point to the MAS as being instrumental in the regulation of Δψm under these conditions.
Δψm controls the expression of a set of nuclear genes
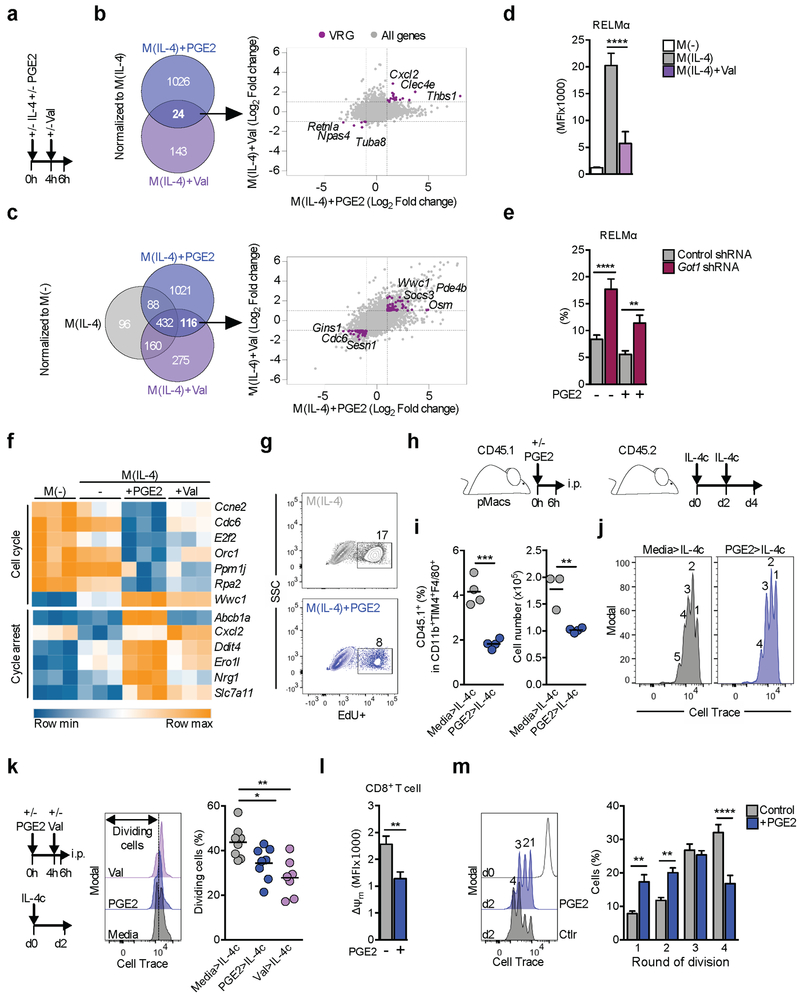
Mitochondria are signaling hubs (Chandel, 2015), and we questioned whether PGE2-mediated changes in IL-4-induced gene expression were functionally related to diminished Δψm. To separate the effects of changes in Δψm from transcriptional regulation induced by PGE2 signaling we used low dose valinomycin (Val), a K+ ionophore, which collapses Δψm, without affecting plasma membrane permeability or viability (Figure S4A). We added Val to macrophages stimulated 4 h earlier with IL-4 in order to initiate loss of Δψm at approximately the time it occurs following PGE2 addition (Figure 2A) and compared changes in gene expression in these cells 2 h later with those induced at 6 h post IL-4 and PGE2 costimulation (Figure 4A). We reasoned that genes regulated in the same way by PGE2 and Val, both in comparison to M(IL-4) (Figure 4B), and in comparison to resting macrophages but excluding the effects of IL-4 (Figure 4C), were likely to be responding to changes in Δψm. In this manner, we identified 126 voltage regulated genes (VRGs), that had a significant >2 fold change in expression, and coincided in M(IL-4) treated with PGE2 or Val (purple dots in Figure 4B – 24 genes and Figure 4C – 116 genes; together 126 unique genes). Retnla was amongst these, and its expression was downregulated by PGE2 and Val (Figure 4B). Flow cytometry confirmed that RELMα expression was suppressed by Val (Figure 4D), and by FCCP (Figure S4B), confirming RELMα as a VRG. Although induced RELMα production in M(IL-4) was typically low after viral transduction, we found that reduced GOT1 expression, which partially restored Δψm (Figure 3J), resulted in increased RELMα expression in PGE2-treated M(IL-4) (Figure 4E). Moreover, RELMα was increased in M(IL-4) following suppression of GOT1 expression (Figure 4E). This paralleled effects of GOT1 suppression on Δψm in M(IL-4) (Figure 3J), providing support for a link between the MAS, Δψm and RELMα expression.
Figure 4. Mitochondrial membrane potential controls the expression of a subset of genes associated with proliferation in PGE2 treated cells.
(A) Experimental protocol for B-D and F.
(B-C) Venn diagrams show significant (adjusted p value < 0.1) up or down regulated (> 2 fold) genes in M(IL-4)+PGE2 or M(IL-4)+Val cells compared to M(IL-4) (B), or to M(−) (C). Genes regulated by both PGE2 and Val were assumed to be Δψm regulated genes (“voltage regulated genes”, VRGs, purple dots).
(D) RELMα, measured by flow cytometry in BMMs treated as in (A).
(E) RELMα expression in BMMs transduced with a control or Got1 targeting shRNA, stimulated with IL-4 +/− PGE2. Means ± SEM from 5–7 biological replicates.
(F) Heatmap of VRGs matching “Cell Cycle” or “Cycle arrest” expression signatures, based on Ingenuity Pathway Analysis of RNA-seq data from 3 biological replicates from BMMs treated as in (A).
(G) Representative flow cytometry plots of EdU incorporation into BMMs after 24 h of treatment treated as indicated.
(H) Schematic of pMac labeling and transfer protocol.
(I) Transferred pMacs were recovered on day 4 after recipients received 2 injections of IL-4c. Percentages and numbers of CD45.1+ cells within live CD11b+F4/80+TIM4+ pMacs were calculated. Symbols are biological replicates.
(J) Representative Cell Trace dilution plots from data in (I).
(K) Schematic of pMac labeling and transfer protocol, with representative CellTrace dilution plots and percentages of cells that diluted Cell Trace under different conditions. Symbols are biological replicates.
(L) Δψm in CD8+ T cells activated with CD3 and CD28 +/− PGE2 24 h previously, as measured using TMRM and flow cytometry. MFI = mean fluorescence intensity.
(M) Proliferation of cells from (L), measured at 48 h post activation. Representative plots (left) and compiled data from all replicates (right). Mean r SEM from 5–6 biological replicates.
Data are representative of 3 (D, E, G, I-K) or 2 (L-M) independent experiments. (D-E, I, K, M) p*<0.05, p**<0.005, p***<0.0005, p****<0.0001. See also Figure S4.
Ingenuity Pathway Analysis revealed that aspects of cell cycle biology were enriched in the VRG set (Figure S4C). Specifically, multiple VRGs relate to cell cycle, and a signature induced by tunicamycin, which can lead to cell cycle arrest (Figure 4F). This prompted us to explore the effects of PGE2 on macrophage proliferation. Macrophages proliferate following IL-4 stimulation (Jenkins et al., 2011) and we found that in vitro, EdU incorporation in M(IL-4) was reduced by approximately 40% upon PGE2 costimulation (Figure 4G; S4D). We examined the effect of PGE2 on proliferation in vivo using a system in which pMacs were collected, labeled with Cell Trace, treated with PGE2 in vitro for 6 h (by which point Δψm was diminished) and then transferred by intraperitoneal (i.p.) injection into congenically marked mice (Figure 4H). Recipients were then injected with IL-4 complex (IL-4c) i.p. and 4 days later pMacs, including those that were transferred (Figure S4E), were recovered. As expected, the number of host CD45.2+ pMacs between the groups was comparable (Figure S4F), however, the number and percentage of transferred CD45.1+ pMacs were reduced by PGE2 pre-treatment (Figure 4I). Moreover, pMacs treated with PGE2 underwent fewer divisions in response to IL-4c (Figure 4J). We hypothesized that reduced Δψm would restrict the initial expansion of pMacs. Consequently, we examined proliferation on day 2, and included a group treated with Val to directly implicate a loss of Δψm (Figure 4K). Both PGE2 and Val pre-treatment reduced the percentage of transferred pMacs found to be dividing after IL-4c (Figure 4K), demonstrating that loss of Δψm negatively impacts macrophage proliferation in vivo. Our data support previous contentions that Δψm regulates cell cycle (Martínez-Reyes et al., 2016) by showing that a set of cell cycle related genes are regulated by PGE2-induced changes in Δψm. Consistent with this, reductions in Δψm driven by PGE2 or Val result in diminished cellular proliferation.
Multiple cell types express PGE2 receptors and we were interested in whether the effects of PGE2 on macrophage Δψm, respiration and proliferation would also be apparent in other immune cells. We examined effects of PGE2 on naïve CD8+ T cells stimulated with anti-CD3 plus anti-CD28, as stimulation increases OXPHOS (Geltink et al., 2017) and drives extensive proliferation. We found that PGE2 caused a reduction in Δψm (Figure 4L), basal OCR, maximal respiration (Figure S4G), and proliferation (Figure 4M), providing mechanistic insight into PGE2 mediated CD8+ T cell inhibition (Basingab et al., 2016).
ETV-1 is a Δψm-sensitive transcription factor
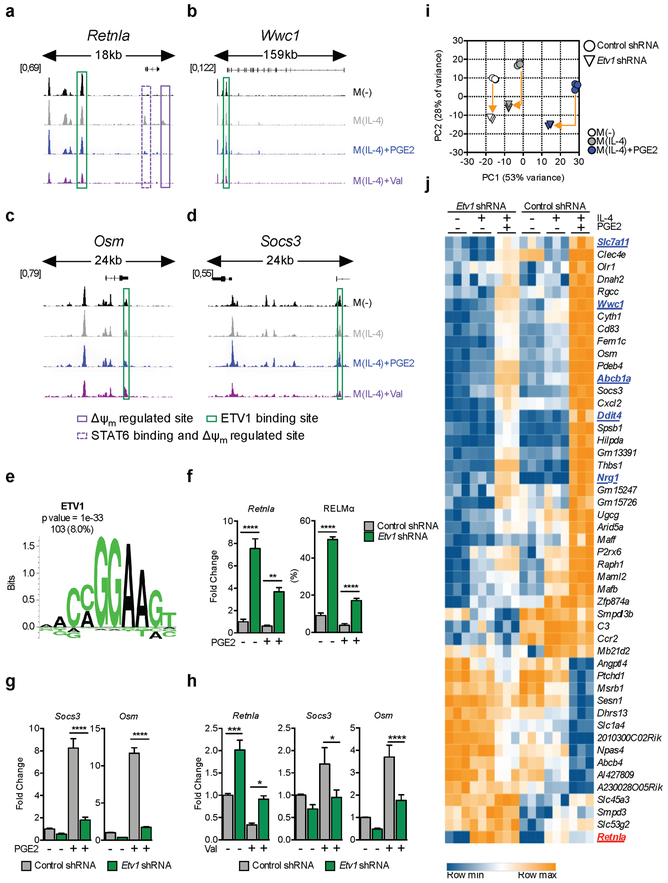
Mitochondrial metabolism can alter chromatin accessibility through its effects on the acetylation of histones. Consequently, we asked whether PGE2 and Val altered the accessibility of regulatory regions in the loci of VRGs. Using ATAC-seq we looked for shared changes in chromatin accessibility between cells treated with IL-4 plus PGE2 or plus Val. In this manner, we found 627 statistically supported regions across the genome which exhibited >2 fold variation in accessibility in response to diminished Δψm (Figure S5A,B). Of these sites, 93% were less accessible upon treatment with PGE2 or Val (Figure S5B), supporting the view that suppressed mitochondrial metabolism negatively impacts chromatin accessibility. Among them, we observed at least 2 distinct sites in the Retnla locus that were closed in resting macrophages but open in M(IL-4), and which were made less accessible by PGE2 or Val (Figure 5A, highlighted in purple). A consensus STAT6-binding site was found in one of these peaks (Figure 5A, dotted purple line), providing a mechanistic basis for why this region of chromatin regulates RELMα expression. Thus, Δψm-induced reduction in chromatin accessibility could account for PGE2 and Val induced changes in expression of Retnla. Nevertheless, of the VRGs, only Retnla and 5 additional loci (Fignl1, Ero1l, Zfp521, Gins1 and Tuba8 – Figure S5C) were regulated in this way, while other VRGs were not – for example Wwc1, Osm and Socs3 (Figure 5B–D).
Figure 5. ETV1 is a mitochondrial membrane potential regulated transcription factor.
(A-D) Regions of accessible chromatin, measured by ATAC-seq, in VRGs Retnla (A), Wwc1 (B), Osm (C) and Socs3 (D), in BMMs treated as indicated for 6 h. Δψm-regulated peaks +/− STAT6 binding sites (solid and dotted purple lines respectively), or with predicted ETV1 binding sites (green), are highlighted.
(E) Enriched ETV1 motif in accessible chromatin regions up to 20kb upstream from transcription start sites of VRGs. Enrichment score (p value) and number plus percentage of matching regulatory regions (out of 1286 detected) are indicated.
(F-J) BMMs transduced with a control or Etv1 shRNA were stimulated with IL-4 +/− PGE2 or +/− Val (F-J). At 6 h RNA was extracted and tested by qRT-PCR for indicated genes (F-H), or by RNAseq (I-J). RELMα was assessed by flow cytometry after 24 h (F). RNAseq data from 3 biological replicates are presented as a principal component analysis (PCA), (I) and heat map (J) of 48 VRGs regulated (adjusted p value < 0.1, >2 fold change) in M(IL-4)+PGE2 by Etv1-shRNA. In (I), arrows provide a measure for the magnitude of variation for each treatment, and in (J), genes associated with cell cycle are in blue and Retnla is in red.
(F-H) Mean ± SEM from 3–8 biological replicates, p*<0.05, p**<0.005, p***<0.0005, p****<0.0001. See also Figure S5.
Given the fact that for the majority of VRGs, expression and chromatin accessibility were not correlated, we hypothesized that the mechanism linking Δψm to transcriptional regulation could be through the gain or loss of activation of specific TFs. To address this possibility, we identified TF binding sites within accessible chromatin in VRG promoter regions (defined as ≤ 20 Kb upstream from transcription start sites), reasoning that shared TF binding signatures could provide evidence for shared regulatory mechanisms. This footprint analysis identified enriched binding motifs for ETV1 (Figure 5E), FLI1 and KLF5 (Figure S5D). ETV1 was the most enriched of these, with 103 potential binding sites in 84% of the identified VRGs, including Retnla, Wwc1, Osm and Socs3 (Figure 5A–D – highlighted in green). Other TFs typically associated with PGE2 stimulation, such as CREB or Wnt signaling (Rodriguez et al., 2013), were not enriched in these promoter sites.
Based on these findings, we reasoned that ETV1 might play a role in regulating expression of VRGs, and therefore examined effects of loss or gain of function of Etv1 using lentivirally delivered Etv1-specific shRNA (Figure S5E) or an shRNA-resistant V5 peptide-tagged ETV1 mutant (ETV1V5WT ) respectively, in M(IL-4) with and without PGE2. Compared to macrophages transduced with control shRNA, Etv1-shRNA reversed the effects of PGE2 on RELMα expression (Figure 5F). Moreover, Etv1-shRNA promoted RELMα expression beyond the amounts seen in cells stimulated with IL-4 alone (Figure 5F), marking ETV1 as a negative regulator of RELMα that controls Retnla expression following Δψm reduction. Conversely, overexpressed ETV1 (Figure S5F), which was exclusively nuclear-localized under all stimulation conditions (Figure S5G), reverted the effects of Etv1-shRNA treatment and profoundly suppressed RELMα expression. We selected two additional VRGs in which the ETV1 motif was enriched, Socs3 and Osm, and found that effects of PGE2 on their expression was also reversed by Etv1-shRNA (Figure 5G). Likewise, gene expression changes on Retnla, Socs3 and Osm downstream of dissipation of Δψm with Val were reversed by Etv1-shRNA (Figure 5H). We then explored the global effects of Etv1-shRNA on gene expression via RNA-seq. Suppression of ETV1 expression had an effect in M(−) and M(IL-4) (Figure 5I), but had the biggest impact on M(IL-4) cells treated with PGE2 (Figure 5I). Indeed, effects of PGE2 on the expression of 48 out of 126 VRGs, including Retnla (in red) as well as 5 genes earlier associated with cell cycle (underlined in blue), were reversed by suppression of Etv1 (Figure 5J). Consequently, we explored the role of ETV1 on macrophage proliferation and found that Etv1-shRNA negatively impacted incorporation of EdU+ regardless of the presence or absence of PGE2 (Figure S5H), suggesting that this protein has a role in cell cycle beyond the phenotype associated with Δψm.
We sought an explanation for how ETV1 might respond to Δψm. Nuclear localization of ETV1 under all circumstances intimated that regulated movement into the nucleus is not responsible for Δψm-driven changes in ETV1 activity. Based on the known propensity of Ets family TFs to interact with other proteins (Verger and Duterque-Coquillaud, 2002), we explored the possibility that ETV1 activation was regulated by association with a Δψm–sensitive partner that transfers information from mitochondria to nucleus. We expressed ETV1V5WT in murine embryonic fibroblasts (MEFs; Figure S5I), and used anti-V5 Abs to immunoprecipitate ETV1 and any interacting proteins. As a negative control, we used identical conditions that included competing concentrations of V5 epitope peptide. Tryptic peptides from these immunoprecipitates were identified by mass spectrometry and compared to negative controls (FDR 1%; Figure S5J). Ten proteins were identified as interacting with ETV1, defined as having greater than 4-fold enrichment and with 2 or more detected peptides when compared to the negative control (Figure S5J). In this analysis, ETV1 was the most highly enriched protein, while no ETV1 peptides were detected in the negative controls (Figure S5J). Amongst the interacting proteins was single-stranded DNA-binding protein (SSBP1), which is a mitochondrial protein linked directly to Δψm, which can dissociate from mitochondria (Jiang et al., 2016; Tan et al., 2015). We postulate that SSBP1 acting in concert with ETV1, could serve to mediate Δψm-dependent effects on transcription.
Taken together, our data show that PGE2 causes a reduction in Δψm, that in turn leads to the change in expression of 126 VRGs including RELMα. We have shown that ETV1 plays a role in the regulation of 38% of these VRGs, revealing ETV1 as a TF that, possibly through interaction with other proteins, detects changes in Δψm, and Δψm as a mediator of mitochondrial-directed nuclear gene expression.
DISCUSSION
The electrochemical gradient across the inner mitochondrial membrane is the force that drives mitochondrial ATP synthesis. This process is fueled by NADH and FADH2, which are made by the TCA cycle, and by mitochondrial shuttle systems which balance redox between mitochondria and cytoplasm. As we sought to understand the reported role of PGE2 on macrophage alternative activation, we uncovered the ability of PGE2 to alter immune cell function by regulating Δψm. Our data indicate that loss of Δψm following PGE2 treatment results from changes in MAS function, and leads to the regulation of expression of 126 genes (the VRGs). For a minority of these genes, including Retnla, expression was associated with a Δψm-driven loss of chromatin accessibility. However, accessibility of binding sites for ETV1 was the dominant common feature underpinning VRG regulation, suggesting that ETV1 is a Δψm-sensitive TF. Our work reveals a previously unappreciated ability of PGE2 to regulate Δψm and a Δψm-based mechanism through which mitochondria are able to regulate nuclear gene expression.
We found that changes in expression of MAS genes, and in particular Got1, were in part responsible for the PGE2-induced decrease in Δψm. ATAC-seq coupled with transcription factor enrichment analysis showed that MAS genes are likely to be regulated by AP-1 (data not shown), consistent with reports that PGE2 is able to promote gene expression by activating AP-1 in a cAMP and PKA mediated manner (Iwahashi et al., 2000). We found that consistent with altered MAS, amounts of NAD+, Glu and Asp were reduced in PGE2-treated M(IL-4). Glu has been implicated previously in M(IL-4) activation (Jha et al., 2015), at least in part through the production of αKG (Liu et al., 2017). Increased αKG enhances the expression of M(IL-4) genes by promoting histone-demethylation (Liu et al., 2017). Broadly consistent with this, our results show that the STAT6 binding site in the Retnla locus is made accessible by IL-4 treatment, a process that likely requires histone demethylation, while PGE2 is able to block this chromatin-remodeling event. Moreover, GOT1 has been found to affect DNA-methylation by decreasing the ratio of αKG to 2-hydroxyglutarate (2-HG) (Xu et al., 2017), a mechanism that could be impacting the Retnla locus, in addition to GOT1 effects on Δψm. Based on recent reports of the importance of Asp for cellular proliferation(Sullivan et al., 2015), it is conceivable that reductions in the amount of this amino acid in PGE2-treated cells could contribute to the observed reduction in proliferative potential. Given the strong regulatory role of the MAS in M(IL-4), the functional consequences of loss of function of other genes in the pathway warrants further study.
In the case of IL-4 stimulated macrophages, PGE2-mediated signaling has been reported to potentiate the STAT6-dependent gene signature via a CREB-dependent pathway (Luan et al., 2015). Our data concur with these findings, but differ in showing that Retnla expression is repressed by PGE2. We can recapitulate the effect of PGE2 on RELMα using 8-Br-cAMP and forskolin, thereby reinforcing the view that PGE2 and associated signaling events negatively affect RELMα expression. Moreover, our transcriptomic analysis revealed that it is an over-simplification to say that PGE2 enhances or inhibits alternative activation, but rather our data showed that PGE2 broadly modified the effects of IL-4. Functionally, RELMα has been shown to induce the expression of lysyl hydroxylase 2 in fibroblasts, which induces collagen crosslinking, a process that is important for tissue repair, but which in excess can contribute to fibrosis (Knipper et al., 2015). In this context, it is possible to speculate that the reported regulatory effects of PGE2 on fibrosis (Harada et al., 2017) are linked to its ability to suppress RELMα expression.
Our conclusion that PGE2-induced loss of Δψm affects nuclear gene expression in M(IL-4) is based on our use of Val as a distinct, parallel approach to dissipate Δψm. Val treatment emphasizes the role of Δψm and not other mechanisms of mitochondrial-nuclear communication. It causes an acute loss of Δψm in the absence of secondary effects resulting from complete mitochondrial DNA depletion (as seen in ρ0 cells - (Martínez-Reyes et al., 2016)) or plasma membrane depolarization (as seen with FCCP (Kenwood et al., 2014)). Unlike with PGE2, M(IL-4) cells treated with Val had higher amounts of ROS and increased amounts of NAD+ compared to M(IL-4) controls (data not shown), making it unlikely that changes in ROS or NAD+ abundance per se explain the overlapping effects of PGE2 and Val. However, the similar effect of PGE2 and Val provided a mechanistic link between Δψm and the distinct VRG expression signature. Our data show that regulatory regions around VRGs are enriched for binding sites for the Ets family TF ETV1. ETV1 is known to regulate neurogenesis (Bartel et al., 2000), repress the expression of cell cycle regulators (Lunardi et al., 2015), and is implicated in various cancers (Oh et al., 2012). To the best of our knowledge ETV1 has not been linked to immune cell function, yet our genetic loss and gain of function experiments show that it has marked effects on macrophage biology, particularly on the expression of RELMα. We have made initial steps to understand how ETV1 is regulated by Δψm. Consistent with its annotation in curated databases (UniProt ID: P41164), we found ETV1 to be inherently nuclear-localized. With the caveat that we used an over-expression vector, this intimates that regulated movement between the cytoplasm and nucleus is not responsible for Δψm-driven changes in ETV1 activity. Ets proteins are recognized to have extensive interactions with other TFs (Verger and Duterque-Coquillaud, 2002). Therefore we reasoned that an interacting partner could confer Δψm-sensitivity on ETV1. Using mass spectrometry we identified a set of ETV1-interacting partners, one of which was SSBP1. In previous studies SSBP1 was shown to translocate from mitochondria to the nucleus and alter nuclear gene transcription during mitochondrial stress (Tan et al., 2015). Thus it is conceivable that in the context of decreased Δψm, SSBP1 translocates from the mitochondria to the nucleus and alters ETV1 activity.
Our studies to address the role of PGE2 in alternative macrophage activation led us to the discovery that PGE2 is able to affect mitochondrial biology by lowering Δψm. We linked this PGE2-mediated effect to induced changes in the MAS. We found that changes in Δψm can in turn affect the expression of a set of genes transcriptionally regulated by ETV1. Our data indicate that in addition to more widely understood mitochondrial mechanisms that can affect nuclear gene expression, the transcriptional activity of ETV1 can be regulated by Δψm. Future work will attempt to further elucidate underlying mechanisms controlling this process.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Professor Edward James Pearce (pearceed@ie-freiburg.mpg.de).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse Models
C57BL/6J (RRID: IMSR_JAX:000664) and CD45.1 congenic (RRID: IMSR_JAX:002014) mouse strains were purchased from The Jackson Laboratory, maintained at the Max Planck Institute for Immunobiology and Epigenetics and cared for according to the Institutional Animal Use and Care Guidelines with approval by the animal care committee of the Regierungspraesidium Freiburg, Germany. All animals used for tissue harvest or experimental procedures were female and aged between 6–7 weeks at the start of the experiment. Animals were humanely sacrificed by carbon dioxide asphyxiation followed by cervical dislocation and bone marrow, peritoneal lavage or spleens were harvested. Mice were bred under specific pathogen free standards, and transferred to open top cages for experimental procedures.
Primary cell cultures
Mature BMMs were obtained at day 7 of culture of bone marrow cells in 20 ng/mL CSF-1 (Peprotech) in RPMI medium containing 10% fetal bovine serum, 4 mM L-glutamine, 100 U/mL penicillin and streptomycin (complete RPMI) (all Gibco). BMMs were stimulated in complete RPMI supplemented with CSF-1 for indicated times with a combination of 20 ng/mL IL-4 (Peprotech), 10 μM PGE2 (SIGMA), 100 μM 8-Br-cAMP (Peprotech), 20 μM Forskolin (Peprotech), 50 ng/mL IFN-γ (Peprotech), 20 ng/mL LPS (SIGMA), 100 μM Ap5a (SIGMA), 1.5 μM flurorcarbonyl cyanide phenylhydrazon (FCCP - SIGMA), 10 nM valinomycin (SIGMA), 10 μM EdU (Life Technologies), 10 μM KT5720 (TOCRIS), 10 μM H-89 (SIGMA) or 2 μM α-amanitin (SIGMA) under 5% CO2, atmospheric oxygen, at 37°C in a humidified incubator. In specific experiments, glucose-free complete RPMI was used, supplemented with 10 mM Glucose (SIGMA),10 mM fully labeled 13C-Glucose (Euroiso-top) or 10 mM singly deuterated 2H-Glucose (SIGMA).
Peritoneal macrophages were obtained by performing a peritoneal lavage on naïve animals with 2% fetal bovine serum (Gibco) in PBS. Cells were counted, plated in complete RPMI supplemented with CSF-1 and selected based on adherence. Stimulation was performed using the same conditions as with BMMs.
Naive CD8+ T cells were obtained from total splenocytes using the naïve CD8 T cell isolation kit (Stem Cell technologies) according to the manufacturers protocol. In specific experiments isolated T cells were stained with CFDA-SE Cell Tracer (Thermo Fisher Scientific) following the manufacturer’s protocol. T cells were then activated using plate bound 5 mg/mL anti-CD3 (clone: 17A2, BioLegend) plus soluble 0.5 mg/mL anti-CD28 (clone: 37.51, BioLegend) in complete RPMI supplemented with 100 U/mL hrIL-2 (Peprotech), 55 μM beta-mercaptoethanol, under 5% CO2, atmospheric oxygen, at 37°C in a humidified incubator.
Fresh buffy coats from healthy donors were collected from the Blood Donation Center of the University Hospital Freiburg, under approval by the ethical committee of the University of Freiburg. Researchers were blinded to the identity of the donors, and age or sex matching was not performed. Sample size is indicated in the figure legends.
Cell lines
HEK293T/17 cells and NIH3T3 MEFs (all ATCC) maintained in DMEM containing 10% fetal bovine serum, 4 mM L-glutamine, 100 U/mL penicillin and streptomycin (all Gibco) under 5% CO2, atmospheric oxygen, at 37°C in a humidified incubator.
METHOD DETAILS
RNA-seq
BMMs were stimulated and total RNA was extracted with the RNAqueous-Micro Total RNA Isolation Kit (Thermo Fisher Scientific) and quantified using Qubit 2.0 (Thermo Fisher Scientific) following the manufacturer’s instructions. Libraries were prepared using the TruSeq stranded mRNA kit (Illumina) and sequenced in a HISeq 3000 (Illumina) by the Deep-sequencing Facility at the Max-Planck-Institute for Immunobiology and Epigenetics. Sequenced libraries were processed using STAR (Dobin et al., 2013), for trimming and mapping, and featureCounts (Liao et al., 2014) to quantify mapped reads. Raw mapped reads were processed in R (Lucent Technologies) with DESeq2 (Love et al., 2014), to determine differentially expressed genes and generate normalized read counts to visualize as heatmaps using Morpheus (Broad Institute). Pathway enrichment analysis was performed using Ingenuity Pathway Analysis (Qiagen), which makes predictions using pair-wise comparisons between conditions, incorporating in the analysis the statistically significant changes in expression within each of those comparisons.
Flow cytometry
Used fluorochrome-conjugate monoclonal antibodies included: CD301 (Milteny Biotech, clone: REA687), CD11b (Biolegend, clone: M1/70), F4/80 (Biozol, clone: BM8), TIM4 (BioLegend, clone: F31–5G3), CD45.1 (BioLegend, clone: A20). Staining was performed in 1% fetal bovine serum in PBS for 30 min on ice, including anti-CD16/CD32 (Biozol); dead cells were excluded with the LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (Thermo scientific). For intracellular RELMα staining, cells were processed with the Transcription Factor Staining Buffer Set (Thermo Fisher Scientific) following the manufacturer protocol. RELMα primary antibody staining (Peprotech Cat# 500-P214) was detected using Alexa Fluor-421 anti-rabbit secondary antibody (Life technologies). Tagged ETV1 were labeled using a monoclonal anti-V5 antibody (Life Technologies, RRID: AB_2556564) and detected with an anti-Mouse IgG2a PE secondary (eBioscience). EdU staining was performed using the Click-iT® EdU Alexa Fluor 647 Flow Cytometry Assay Kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. Dead and Apoptotic cells were detected using Annexin V, Alexa Fluor 550 conjugate (Thermo Fisher Scientific) following the manufacturer’s protocol. Cells were analyzed using LSR Fortessa flow cytometers (BDBiosciences) and data were processed using FlowJo software (FlowJo).
Metabolic assays
Extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) were measured using the 96 well Extracellular Flux Analyzer (Seahorse Bioscience). 1×105 BMMs exposed to different treatments or 2×105 T cells were spun onto each well of seahorse X96 cell culture microplates (coated with poly-D-lysine for T cell assays) and preincubated at 37°C for a minimum of 45 min in the absence of CO2 in unbuffered RPMI with 25 mM glucose, 1 mM pyruvate and 2 mM L-glutamine with pH adjusted to 7.4. For long T cell assays (> 8 h) media was supplemented with 100 U/ml hrIL-2 (Peprotech). For activation of T cells, anti-CD3 + anti-CD28 coated beads were used at 1:1 ratio of beads:T cells. OCR and ECAR were measured under basal conditions, after activation, and after the addition of the following drugs: 1 μM oligomycin, 1.5 μM flurorcarbonyl cyanide phenylhydrazon (FCCP) and 100 nM rotenone + 1 μM antimycin A (all SIGMA) as indicated. Results were collected with Wave software version 2.4 (Agilent).
For metabolic tracing and metabolite quantitation BMMs stimulated as described above were cultured in glucose-free media supplemented with 10 mM 13C-glucose (Euroiso-top) for 6 h or 24 h, or supplemented with 10 mM 2H-glucose (SIGMA) for 10 minutes. Cells were washed with ice cold 0.9% (w/v) NaCl buffer and metabolites were extracted twice with Methanol (80%) and analyzed by Liquid chromatography (Agilent 1290 infinity II UHPLC) coupled to tandem mass spectrometry (Agilent 6495B), or by Gas chromatography coupled to tandem mass spectrometry (Agilent 5977) by the Metabolomics Core, Max-Plank-Institute for Immunobiology and Epigenetics, Freiburg, Germany or with a Thermo Scientific Q Exactive mass spectrometer coupled to a UltiMate 3000 UPLC chromatography system at the University of California – Los Angeles Metabolomics Centre, Los Angeles, USA.
Glucose uptake, mitochondrial content, cellular ROS and mitochondrial ROS were measured by incubating with 100 nM 2NBDG for 2 min, 100 nM MitoTracker Green for 30 min, 5 μM CellROX deep red for 30 min, all in complete medium, 5 μM MitoSOX for 10 min in HBSS, all at 37°C (all Thermo Fisher Scientific), respectively. Cells were then cooled on ice and analyzed using LSR Fortessa flow cytometers (BDBiosciences) and data were processed using FlowJo software (FlowJo). For ATP measurements, cells were resuspended in water (1000 cells/μL), boiled for 10 min at 95°C and then lysates were analyzed with the ATP determination kit (Thermo Fisher Scientific) following the manufacturer’s instructions.
Mitochondrial membrane potential was measured by incorporation of Tetramethylrhodamine, Methyl Ester, Perchlorate (TMRM) (Thermo Fisher Scientific) into living cells. Cells were incubated for 30 min at 37°C with 50 nM TMRM with and without 15 μM FCCP. Cells were then washed and analyzed by flow cytometry in the presence of 2 μg/mL DAPI as a viability exclusion dye. For real time measurement of ETC complex activity, BMMs were harvested after 24 h stimulation and placed in a solution containing 220 mM mannitol, 70 mM sucrose, 10 mM KH2PO4, 5 mM MgCl2, 2 mM HEPES, 1 mM EGTA and 0.2% (w/v) bovine serum albumin (all SIGMA) with 50 nM TMRM and 1 nM Agilent Seahorse XF Plasma Membrane Permeabilizer (Agilent). For real time CV measurements cells were maintained in complete media and then treated with 1 μM oligomycin and later 15 μM FCCP. Cells were analyzed using LSR Fortessa flow cytometers (BDBiosciences) and data were processed using FlowJo software (FlowJo). For real time measurements, cells were maintained at 37°C using a custom built sample holder, and treated sequentially with 100 nM rotenone, 10 mM sodium succinate, 1 μM antimycin A (all Sigma) as indicated.
Confocal Microscopy
Macrophages were cultured for 6 h or 24 h as described above on glass cover slips, then stained with 100 nM MitoTracker Deep Red (Thermo Fisher Scientific) for 30 min at 37°C, then washed, briefly incubated with 2 μg/mL DAPI (Thermo Fisher Scientific) and then fixed with 4% formaldehyde (SIGMA). In selected experiments, cells were permeabilized and then stained in 0.05% Saponin (SIGMA) and 0.2% BSA with anti-V5 antibody detected with anti-mouse AF546 (all Life Technologies). Cover slips were mounted on slides using ProLong Diamond (Thermo Fisher Scientific) and left to settle at 4°C overnight. LSM 880 Airyscan microscope (Zeiss) was used to acquire confocal images. Confocal images were acquired with ZEN software (Zeiss), deconvolved with HuygensSoftware, and analyzed using Imaris imaging software.
Electron Microscopy
1×106 BMMs were fixed in 2.5% glutaraldehyde (SIGMA) in 100 mM sodium cocodylate (SIGMA) then washed in cocodylate buffer. Following this step, samples were processed and imaged at the Electron Microscopy Laboratory at the University of Padova. After dehydration samples were embedded in Eponate 12 resin (Ted Pella) and sections were cut. Images were acquired using a FEI Tecnai 12 Transmission electron microscope equipped with a TIETZ digital camera. Maximal cristae width was measured using ImageJ software and averaged over 50 independent images, acquisition of EM micrographs.
Western Blotting
Cells were washed with ice cold PBS and lysed in 1 × Cell Signaling lysis buffer (20 mM Tris-HCl, [pH 7.5], 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM beta-glycerophosphate, 1 mM Na3VO4, 1 mg/mL leupeptin – Cell Signaling Technologies) supplemented with 1 mM PMSF (SIGMA). Samples were frozen and thawed 3 times followed by centrifugation at 20,000 × g for 10 min at 4°C. Cleared protein lysates were denatured with LDS loading buffer supplemented with 50 mM dithiothreitol (SIGMA) for 10 min at 70°C and loaded on precast 4% to 12% bis-tris protein gels (Thermo Fisher Scientific). Proteins were transferred onto nitrocellulose membranes using the iBLOT 2 system (Thermo Fisher Scientific) following the manufacturer’s protocols. Membranes were blocked with 5% (w/v) BSA in TBS with 0.1% Tween-20 and incubated with the appropriate antibodies in 5% (w/v) BSA in TBS with 0.1% Tween-20 overnight at 4°C. Used antibodies included: OPA1 (Abcam, Cat# ab177941), TOM20 (CST, clone: D8T4N), P-STAT6 (Tyr641) (CST, Cat# 9361S), ACTB (CST, clone: 13E5) and the Total OXPHOS rodent WB Ab cocktail (Abcam, Cat# ab110413). All primary antibody incubations were followed by incubation with secondary HRP-conjugated antibody (Pierce) in 5% milk in TBS with 0.1% Tween-20 and visualized using SuperSignal West Pico or femto Chemiluminescent Substrate (Pierce) on Hyperfilm Mp (Millipore).
ATAC-seq
Libraries were prepared using the Nextera DNA library Prep Kit (Illumina) adapting a published protocol (Buenrostro et al., 2015). Briefly, 5×104 BMMs treated for 6 h as described were washed in PBS and then lysed in 10 mM Tris-HCl, pH 7.4,10 mM NaCl, 3 mM MgCl2 and 0.1% Igepal CA-630 (all SIGMA). Nuclei were then spun down and then resuspend in 25 μl TD (2x reaction buffer), 2.5 μL TDE1 (Nextera Tn5 Transposase) and 22.5 μL nuclease-free water, incubated for 30 min at 37°C. DNA was purified with the Qiagen MinElute PCR Purification Kit (Thermo Fisher Scientific). PCR amplification was performed with the NEBNext High-Fidelity 2x PCR Master Mix (New England Labs) using custom Nextera PCR Primers containing barcodes. Adaptors were removed with AMPure XP beads according to manufacturer’s protocol. Libraries were quantified with the Qubit and submitted for sequencing with a HISeq 3000 (Illumina) by the staff at the Deep-sequencing Facility at the Max-Planck-Institute for Immunobiology and Epigenetics.
Sequenced samples were trimmed with Trimmomatic (Bolger et al., 2014), mapped using Bowtie2 (Langmead and Salzberg, 2012) and replicate mapped files merged with SAM tools (Li et al., 2009). Coverage files were generated with deepTools (Ramírez et al., 2016). Open chromatin and differentially regulated chromatin was detected with MACS2 (Zhang et al., 2008) with a p value < 1×10−7 and a q value of less than 0.1 and a 2 fold enrichment threshold. Bed files were analyzed with Bedtools (Quinlan and Hall, 2010), and visualized alongside coverage files on IGV (Robinson et al., 2011). Footprint analysis was carried out using pyDNAse (Piper et al., 2013) and transcription factor enrichment was done with HOMER (Heinz et al., 2010). Motifs were visualized with Seq2Logo (Thomsen and Nielsen, 2012).
Differential gene expression analysis by qRT-PCR
Total RNA was extracted with the RNAqueous-Micro Total RNA Isolation Kit, quantified using a Qubit and cDNA synthesized with the High-Capacity RNA-to-cDNA Kit following the manufacturer’s instructions (all Thermo Fisher Scientific). qPCR was performed on the QuantStudio 3 (Thermo Fisher Scientific) using the DreamTaq Green PCR Master Mix (2X) (BIORAD) and the following probe sets (all Applied Biosystems): Hprt (Mm03024075_m1), Retnla (Mm01193966_m1), Osm (Mm01193966_m1), Socs3 (Mm00545913_s1), Etv1 (Mm00514804_m1), Slc25a13 (Mm00489442_m1) and Got1 (Mm01195792_g1).Results were analyzed by the 2−ΔΔCt method using Hprt as a house-keeping gene.
Lentiviral and retroviral production and cell transduction
HEK293T cells were transfected using Lipofectamine 3000 (Thermo Fisher Scientific) with lentiviral packaging vectors pCAG-eco and psPAX.2 plus empty pLKO.1 control with a Puromycin selection cassette (all Adgene) or a shRNA containing pLKO.1 targeting Got1 (GE Dharmacon Cat# RMM3981–201819865 – TRCN0000119792) or Etv1 (GE Dharmacon Cat# RMM3981–201787707 – TRCN0000075476). ETV1 sequence was obtained from NCBI (NM_007960.5) and modified to include only the CDS, preceded by an EcoRI site and a Kozak sequence, and followed by a linker (GGCTCCGGGTCAGGA), the V5 tag (GGAAAGCCAATTCCCAACCCACTTTTGGGATTGGACTCCACT), a stop codon (TAA) and a XhoI site. The original site of shRNA binding (AGGCCGGCGATGAACTATGACAAACTTAG) was heavily altered (AGACCAGCAATGAATTACGATAAGTTGTC) without any change in coded amino acids. Complete modified DNA was purchased as a gBlock from Integrated DNA technologies. Overexpression construct was expressed in a pMX-IRES-GFP vector (gift from Dr. Takashi Saito, Riken Center for Integrative Medical Science, Japan), which was packaged into a retrovirus with a pCL-eco packaging vector (Addgene). Virus was collected in the supernatant of the cells. Bone marrow cultures were transduced by centrifugation for 90 min (1300 × g) in the presence of polybrene (8 mg/ml) on day 2–3 of culture. 48 h selection of transduced cells was performed with 6 μg/mL puromycin (SIGMA). MEFs were transduced by incubating the cells with viral supernatant in the presence of polybrene (8 mg/ml) for 5h. 48 h selection of transduced cells was performed with 1 μg/mL puromycin (SIGMA) and subsequently GFP+ cells were sorted using a FACS Aria II.
Cell transfer and IL-4c injection
Peritoneal lavage from CD45.1 congenic female mice were collected and pooled, then treated in vitro for 6 h in complete RPMI supplemented with 20 μM PGE2, 10 nM Valinomycin or no supplement. Cultured cells were stained with Cell Trace Violet (Life Technologies), following the manufacturer’s instructions, and then washed 3 times with PBS to remove excess dye and non-adherent cells. Adherent cells were recovered by scrapping, counted and then 5×105 cells/mouse were transferred via intra-peritoneal injection to littermates randomly assigned to experimental groups. PBS control or IL-4 complex (5 μg of recombinant IL-4 [Peprotech] + 25 μg of anti-IL-4 antibody [clone: 11B11, BioXCell] in PBS per mouse) was injected to mice on day 0 and day 2. Peritoneal lavage was harvested on day 2 or day 4.
Protein immunoprecipitation and Proteomics
MEFs transduced with a V5 tagged ETV1 protein were washed twice with ice cold PBS and lysed in 1 × Cell Signaling lysis buffer (20 mM Tris-HCl, [pH 7.5], 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM beta-glycerophosphate, 1 mM Na3VO4, 1 mg/mL leupeptin – Cell Signaling Technologies) supplemented with 1 mM PMSF (SIGMA). Samples were frozen and thawed 3 times followed by centrifugation at 20,000 × g for 10 min at 4°C. Immunoprecitipation (IP) of V5 tagged ETV1 was performed in triplicate. Cleared protein lysates were diluted to 0.2 × Cell Signaling lysis buffer in PBS supplemented with phosphatase and protease inhibitors (Cell Signaling Technologies) and incubated for 3 h at 4°C with anti-V5 antibody (clone D3H8Q – Cell Signaling Technologies). Negative IP controls, performed in duplicate, were carried out under identical conditions, but with the addition of 16 μg of purified V5 peptide (New England peptide, GKPIPNPLLGLDST). Samples were subsequently incubated with Protein G dynabeads (Invitrogen, 10003D) for 1.5 h, which were then repeatedly washed with 0.2 × Cell lysis buffer in PBS with protease and phosphatase inhibitors. Recovered beads were washed once in 750 μl 100 mM Tris-HCl, pH 7.5 and 500 μl 100 mM Tris-HCl, pH 7.5 (supplemented with 0.01% Rapigest (Waters)), respectively. Following addition of 500 μl 50 mM ammonium biocarbonate (supplemented with 0.01% Rapigest) beads were transferred to a 1.5 ml low-bind tube and finally resuspended in 50 μl urea digestion buffer (4 M urea, 2 mM DTT and 100 mM Tris pH8.0). Magnetic beads were digested (Thermomixer) with 400 ng endoproteinase Lys-C for 1.5 hours at 28°C in order to shave-off proteins associated with the V5 antibody. The Lys-C digested peptides were transferred to another 1.5 ml low-bind tube and pooled with a final wash of the beads (100 μl 50 mM ammonium bicarbonate, 10 min, 25°C, Thermomixer) followed by addition of iodoacetamide to a final concentration of 3 mM and incubation for 2 hr at 25°C. The sample was ultimately digested with trypsin (400 ng, Promega) for 13 hours at 25°C and digestion was stopped by the addition of TFA (ad 1%). Tryptic peptides were then cleaned-up/desalted and eluted using C18-STAGE tips as described previously (Rappsilber et al., 2007). General nanoLC-MS setup and DDA analysis was performed essentially as reported (Musa et al., 2017) with modifications detailed in the following. QExactive mass spectrometer and Easy nanoLC-1000 were employed for all experiments. Tryptic peptides were separated by a 2h linear gradient of 5–80% buffer B (80% acetonitrile, 0.1% formic acid) at constant flowrate of 300 nL/min. For MS data acquisition the “sensitive” method from Kelstrup et al. (Kelstrup et al., 2012) was adopted. MS raw files were analyzed by MaxQuant software (1.5.7.4) and peak lists were searched against the mouse Uniprot FASTA (October 2017) database (concatenated with a database containing common contaminants) by the Andromeda search engine embedded in MaxQuant (Cox and Mann, 2008; Cox et al., 2011). MS1-based label free quantification (LFQ) was done using maxLFQ algorithm (Cox et al., 2014). We require a minimum of two peptide ratios in order to consider a given protein as valid (protein and peptide ID FDR=0.01). Perseus platform (Tyanova et al., 2016) was used to perform data normalization, filtering and statistical testing. Student’s t-test was utilized to define differentially expressed proteins employing a 4-fold change as a cut-off at a 1% FDR. Negative IP control replicates and IP sample triplicates were measured twice on the mass spectrometer, and the resulting 4 or 6 datasets respectively used to calculate protein enrichment across the IP samples.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis was performed using prism 6 software (Graph pad) and results are represented as mean ± SEM, unless otherwise indicated. Comparisons for two groups were calculated using unpaired two-tailed Student’s t tests, comparisons of more than two groups were calculated using one-way ANOVA with Sidak’s correction for multiple comparison tests. Comparisons between materials from human biopsies, were analyzed with paired two-tailed Student’s t tests. We observed normal distribution and no difference in variance between groups in individual comparisons. Selection of sample size was based on extensive experience with metabolic assays.
DATA AND SOFTWARE AVAILABILITY
Next generation sequencing data can be accessed at Gene Ontology Omnibus under GSE119521.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD010997.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti mouse RELMα | Peprotech | Cat#500-P214, RRID:AB 1268491 |
| Anti mouse IL-4 (clone: 11B11) | BioXCell | Cat# BE0045-25MG, RRID:AB 1107707 |
| Anti mouse CD3 (clone: 17A2) | BioLegend | Cat# 100223, RRID: AB 1877072 |
| Anti mouse CD28 (clone: 37.51) | BioLegend | Cat# 102112, RRID: AB_312877 |
| Anti V5 | Life Technologies | Cat# R960-25 RRID: AB 2556564 |
| Anti V5 (clone: D3H8Q) | Cell Signaling Technologies | Cat#13202S |
| Total OXPHOS rodent WB Ab cocktail | Abeam | Cat#ab110413, RRID:AB 2629281 |
| Bacterial and Virus Strains | ||
| Etv1-hpRNA (pLKO.1 backbone) | GE Dharmacon | RMM3981-201787707–TRCN0000075476 |
| Got1-hpRNA (pLKO.1 backbone) | GE Dharmacon | RMM3981-201819865–TRCN0000119792 |
| Biological Samples | ||
| Healthy control blood | UniKlinik Freiburg | NA |
| Chemicals, Peptides, and Recombinant Proteins | ||
| CSF-1 | Peprotech | Cat# 315-02 |
| IL-4 | Peprotech | Cat# 214-14-100 |
| V5 peptide (GKPIPNPLLGLDST) | New England Peptide | NA |
| PGE2 | SIGMA | Cat# P0409 |
| Valinomycin | SIGMA | Cat# V3639 |
| Ap5a | SIGMA | CAS 4097-04-5 |
| Tetramethylrhodamine, Methyl Ester, Perchlorate (TMRM) | Thermo Fisher Scientific | Cat# T668 |
| Agilent Seahorse XF Plasma Membrane Permeabilizer | Agilent | Cat# 102504-100 |
| Oligomycin | SIGMA | CAS# 1404-19-9 |
| FCCP | SIGMA | CAS# 370-86-5 |
| Rotenone | SIGMA | CAS# 83-79-4 |
| Antimycin A | SIGMA | CAS# 1397-94-0 |
| Critical Commercial Assays | ||
| Seahorse extracellular flux analyzer Xfe 96 | Agilent | https://www.agilent.com/en/products/cell-analysis-(seahorse)/seahorseanalyzers/seahorse-xfe96-analyzer |
| Nextera DNA library Prep Kit | Illumina | Cat#FC-121-1030 |
| Deposited Data | ||
| RNA-Seq | This paper | GEO:GSE119509 |
| ATAC-Seq | This paper | GEO: GSE119463 |
| Proteomics | This paper | PRIDE: PXD010997 |
| Experimental Models: Organisms/Strains | ||
| C57BL/6J mice | Jackson Laboratories | RRID: IMSR_JAX:000664 |
| CD45.1 congenic mice | Jackson Laboratories | RRID: IMSR_JAX:002014 |
| Recombinant DNA | ||
| pLKO.1 (control backbone for transductions) | Addgene | Plasmid #8453 |
| pMX-IRES-GFP | Gift from Dr. Takashi Saito, Riken Center for Integrative Medical Science, Japan | Cat#RTV013 |
| Etv1 | NCBI | NM_007960.5 |
| Software and Algorithms | ||
| Wave software version 2.4 | Agilent | https://www.agilent.com/en-us/products/cell-analysis-(seahorse)/software-download-for-wave-desktop |
| FlowJO | FlowJO | https://www.flowjo.com/ |
| DESeq2 | Love, et. al. 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Imaris | Bitplane | http://www.bitplane.com/imaris/imaris |
| IGV | Broad Institute | http://software.broadinstitute.org/software/igv/ |
| Bedtools | Quinlan, et. al. 2010 | http://quinlanlab.org/tutorials/bedtools/bedtools.html |
| HOMER | Heinz, et. al. 2010 | http://homer.ucsd.edu/homer/index.html |
| pyDNAse | Piper, et. al. 2013 | http://pythonhosted.org/pyDNasel |
| Ingenuity Pathway Analysis | Qiagen | https://www.qiagenbioinformatics.com/ |
| MaxQuant software | Cox, et. al. 2008 | http://www.biochem.mpg.de/5111795/maxquant |
| maxLFQ algorithm | Cox, et. al. 2011 | http://www.biochem.mpg.de/5111795/maxquant |
| Andromeda | Cox, et. al. 2014 | http://www.biochem.mpg.de/5111795/maxquant |
| Perseus platform | Tyanova, et. al. 2016 | http://www.biochem.mpg.de/5111810/perseus |
| MACS2 | Zhang, et. al. 2008 | https://github.com/taoliu/MACS |
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Pearce laboratories and Drs. Angelika Rambold, Anne Rensing-Ehl, Devon Ryan, Senthilkumar Ramamoorthy and Barbara Hummel for invaluable advice and assistance, and the UCLA Metabolomics Centre, the Electron Microscopy Laboratory at the University of Padova, and the Metabolomics, Imaging and DeepSequencing facilities at the MPI-IE for technical support. We acknowledge the transcriptional analysis that Dr. Max Artyomov performed on the early stages of the study. This work was funded by the NIH (AI110481 to E.J.P.), and the Max Planck Society. M.M. is supported by the Swiss National Science Foundation, A.M.K., M.C. and F.B. by the Alexander von Humboldt Fellowship Foundation.
Footnotes
DECLARATION OF INTERESTS
E.J.P. and E.L.P. are founders of Rheos Medicines. E.L.P. is a member of the SAB for ImmunoMet Therapeutics.
REFERENCES
- Amit I, Winter DR, and Jung S (2015). The role of the local environment and epigenetics in shaping macrophage identity and their effect on tissue homeostasis. Nat Immunol 17, 18–25. [DOI] [PubMed] [Google Scholar]
- Bartel FO, Higuchi T, and Spyropoulos DD (2000). Mouse models in the study of the Ets family of transcription factors. Oncogene 19, 6443–6454. [DOI] [PubMed] [Google Scholar]
- Basingab FS, Ahmadi M, and Morgan DJ (2016). IFNγ-Dependent Interactions between ICAM-1 and LFA-1 Counteract Prostaglandin E2-Mediated Inhibition of Antitumor CTL Responses. Cancer Immunol Res 4, 400–411. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck MD, O’Sullivan D, Geltink RIK, Curtis JD, Chang C-H, Sanin DE, Qiu J, Kretz O, Braas D, van der Windt GJW, et al. (2016). Mitochondrial Dynamics Controls T Cell Fate through Metabolic Programming. Cell 166, 63–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, and Greenleaf WJ (2015). ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol 109, 21.29.1–.29.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandel NS (2015). Evolution of Mitochondria as Signaling Organelles. Cell Metabolism 22, 204–206. [DOI] [PubMed] [Google Scholar]
- Cox J, and Mann M (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, and Mann M (2014). Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteomics 13, 2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, and Mann M (2011). Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geltink RIK, O’Sullivan D, Corrado M, Bremser A, Buck MD, Buescher JM, Firat E, Zhu X, Niedermann G, Caputa G, et al. (2017). Mitochondrial Priming by CD28. Cell 171, 385–390.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Schultze JL, Murray PJ, Ochando J, and Biswas SK (2015). New insights into the multidimensional concept of macrophage ontogeny, activation and function. Nat Immunol 17, 34–40. [DOI] [PubMed] [Google Scholar]
- Gordon S, and Martinez FO (2010). Alternative Activation of Macrophages: Mechanism and Functions. Immunity 32, 593–604. [DOI] [PubMed] [Google Scholar]
- Gottlieb RA, and Bernstein D (2016). Mitochondrial remodeling: Rearranging, recycling, and reprogramming. Cell Calcium 60, 88–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada M, Takeda N, and Toko H (2017). Prostaglandin E2 Signaling as a Target of Anti-Cardiac Fibrosis in Heart Failure Treatment. International Heart Journal 58, 3–4. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SC-C, Everts B, Ivanova Y, O’Sullivan D, Nascimento M, Smith AM, Beatty W, Love-Gregory L, Lam WY, O’Neill CM, et al. (2014). Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat Immunol 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwahashi H, Takeshita A, and Hanazawa S (2000). Prostaglandin E2 stimulates AP-1-mediated CD14 expression in mouse macrophages via cyclic AMP-dependent protein kinase A. J.I 164, 5403–5408. [DOI] [PubMed] [Google Scholar]
- Jenkins SJ, Ruckerl D, Cook PC, Jones LH, Finkelman FD, van Rooijen N, MacDonald AS, and Allen JE (2011). Local Macrophage Proliferation, Rather than Recruitment from the Blood, Is a Signature of TH2 Inflammation. Science 332, 1284–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha AK, Huang SC-C, Sergushichev A, Lampropoulou V, Ivanova Y, Loginicheva E, Chmielewski K, Stewart KM, Ashall J, Everts B, et al. (2015). Network Integration of Parallel Metabolic and Transcriptional Data Reveals Metabolic Modules that Regulate Macrophage Polarization. Immunity 42, 419–430. [DOI] [PubMed] [Google Scholar]
- Jiang H-L, Sun H-F, Gao S-P, Li L-D, Huang S, Hu X, Liu S, Wu J, Shao Z-M, and Jin W (2016). SSBP1 Suppresses TGFβ-Driven Epithelial-to-Mesenchymal Transition and Metastasis in Triple-Negative Breast Cancer by Regulating Mitochondrial Retrograde Signaling. Cancer Research 76, 952–964. [DOI] [PubMed] [Google Scholar]
- Kauppinen RA, Sihra TS, and Nicholls DG (1987). Aminooxyacetic acid inhibits the malate-aspartate shuttle in isolated nerve terminals and prevents the mitochondria from utilizing glycolytic substrates. Biochim. Biophys. Acta 930, 173–178. [DOI] [PubMed] [Google Scholar]
- Kelstrup CD, Young C, Lavallee R, Nielsen ML, and Olsen JV (2012). Optimized fast and sensitive acquisition methods for shotgun proteomics on a quadrupole orbitrap mass spectrometer. J. Proteome Res. 11, 3487–3497. [DOI] [PubMed] [Google Scholar]
- Kenwood BM, Weaver JL, Bajwa A, Poon IK, Byrne FL, Murrow BA, Calderone JA, Huang L, Divakaruni AS, Tomsig JL, et al. (2014). Identification of a novel mitochondrial uncoupler that does not depolarize the plasma membrane. Mol Metab 3, 114–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipper JA, Willenborg S, Brinckmann J, Bloch W, Maaß T, Wagener R, Krieg T, Sutherland T, Munitz A, Rothenberg ME, et al. (2015). Interleukin-4 Receptor α Signaling in Myeloid Cells Controls Collagen Fibril Assembly in Skin Repair. Immunity 43, 803–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Meth 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis CA, Parker SJ, Fiske BP, McCloskey D, Gui DY, Green CR, Vokes NI, Feist AM, Vander Heiden MG, and Metallo CM (2014). Tracing Compartmentalized NADPH Metabolism in the Cytosol and Mitochondria of Mammalian Cells. Molecular Cell 55, 253–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Liu P-S, Wang H, Li X, Chao T, Teav T, Christen S, Di Conza G, Cheng W-C, Chou C-H, Vavakova M, et al. (2017). α-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat Immunol 18, 985–994. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan B, Yoon Y-S, Le Lay J, Kaestner KH, Hedrick S, and Montminy M (2015). CREB pathway links PGE2 signaling with macrophage polarization. Proceedings of the National Academy of Sciences 201519644–201519646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunardi A, Varmeh S, Chen M, Taulli R, Guarnerio J, Ala U, Seitzer N, Ishikawa T, Carver BS, Hobbs RM, et al. (2015). Suppression of CHK1 by ETS Family Members Promotes DNA Damage Response Bypass and Tumorigenesis. Cancer Discov 5, 550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Reyes I, Diebold LP, Kong H, Schieber M, Huang H, Hensley CT, Mehta MM, Wang T, Santos JH, Woychik R, et al. (2016). TCA Cycle and Mitochondrial Membrane Potential Are Necessary for Diverse Biological Functions. Molecular Cell 61, 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer-Barber KD, Andrade BB, Oland SD, Amaral EP, Barber DL, Gonzales J, Derrick SC, Shi R, Kumar NP, Wei W, et al. (2014). Host-directed therapy of tuberculosis based on interleukin-1 and type I interferon crosstalk. Nature 511, 99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H, VanDussen KL, Malvin NP, Ryu SH, Wang Y, Sonnek NM, Lai C-W, and Stappenbeck TS (2017). Prostaglandin E2 promotes intestinal repair through an adaptive cellular response of the epithelium. The EMBO Journal 36, 5–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musa YR, Boller S, Puchalska M, Grosschedl R, and Mittler G (2017). Comprehensive Proteomic Investigation of Ebf1Heterozygosity in Pro-B Lymphocytes Utilizing Data Independent Acquisition. J. Proteome Res. 17, 76–85. [DOI] [PubMed] [Google Scholar]
- Na YR, Jung D, Yoon BR, Lee WW, and Seok SH (2015). Endogenous prostaglandin E2 potentiates anti-inflammatory phenotype of macrophage through the CREB-C/EBP-β cascade. Eur. J. Immunol 45, 2661–2671. [DOI] [PubMed] [Google Scholar]
- O’Neill LAJ, and Pearce EJ (2016). Immunometabolism governs dendritic cell and macrophage function. J Exp Med 213, 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Shin S, and Janknecht R (2012). ETV1, 4 and 5: an oncogenic subfamily of ETS transcription factors. Biochim. Biophys. Acta 1826, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper J, Elze MC, Cauchy P, Cockerill PN, Bonifer C, and Ott S (2013). Wellington: a novel method for the accurate identification of digital genomic footprints from DNase-seq data. Nucleic Acids Res 41, e201–e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, and Manke T (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44, W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, and Ishihama Y (2007). Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2, 1896–1906. [DOI] [PubMed] [Google Scholar]
- Rider MH, Bertrand L, Vertommen D, Michels PA, Rousseau GG, and Hue L (2004). 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase: head-to-head with a bifunctional enzyme that controls glycolysis. Biochem. J 381, 561–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP (2011). Integrative genomics viewer. Nat. Biotechnol 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M, Domingo E, Municio C, Alvarez Y, Hugo E, Fernandez N, and Sanchez Crespo M (2013). Polarization of the Innate Immune Response by Prostaglandin E2: A Puzzle of Receptors and Signals. Molecular Pharmacology 85, 187–197. [DOI] [PubMed] [Google Scholar]
- Sukumar M, Liu J, Mehta GU, Patel SJ, Roychoudhuri R, Crompton JG, Klebanoff CA, Ji Y, Li P, Yu Z, et al. (2016). Mitochondrial Membrane Potential Identifies Cells with Enhanced Stemness for Cellular Therapy. Cell Metabolism 23, 63–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LB, Gui DY, Hosios AM, Bush LN, Freinkman E, and Vander Heiden MG (2015). Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell 162, 552–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K, Fujimoto M, Takii R, Takaki E, Hayashida N, and Nakai A (2015). Mitochondrial SSBP1 protects cells from proteotoxic stresses by potentiating stress-induced HSF1 transcriptional activity. Nature Communications 6, 6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T, Scambler TE, Smallie T, Cunliffe HE, Ross EA, Rosner DR, O’Neil JD, and Clark AR (2017). Macrophage responses to lipopolysaccharide are modulated by a feedback loop involving prostaglandin E2, dual specificity phosphatase 1 and tristetraprolin. Nature Publishing Group 7, 1547–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen MCF, and Nielsen M (2012). Seq2Logo: a method for construction and visualization of amino acid binding motifs and sequence profiles including sequence weighting, pseudo counts and two-sided representation of amino acid enrichment and depletion. Nucleic Acids Res 40, W281–W287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, and Cox J (2016). The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Meth 13, 731–740. [DOI] [PubMed] [Google Scholar]
- Verger A, and Duterque-Coquillaud M (2002). When Ets transcription factors meet their partners. Bioessays 24, 362–370. [DOI] [PubMed] [Google Scholar]
- Willems PHGM, Rossignol R, Dieteren CEJ, Murphy MP, and Koopman WJH (2015). Redox Homeostasis and Mitochondrial Dynamics. Cell Metabolism 1–12. [DOI] [PubMed] [Google Scholar]
- Woyda-Ploszczyca AM, and Jarmuszkiewicz W (2017). The conserved regulation of mitochondrial uncoupling proteins: From unicellular eukaryotes to mammals. Biochim. Biophys. Acta 1858, 21–33. [DOI] [PubMed] [Google Scholar]
- Xu T, Stewart KM, Wang X, Liu K, Xie M, Kyu Ryu J, Li K, Ma T, Wang H, Ni L, et al. (2017). Metabolic control of TH17 and induced Treg cell balance by an epigenetic mechanism. Nature 548, 228–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J, Schmidt SV, Sander J, Draffehn A, Krebs W, Quester I, De Nardo D, Gohel TD, Emde M, Schmidleithner L, et al. (2014). Transcriptome-Based Network Analysis Reveals a Spectrum Model of Human Macrophage Activation. Immunity 40, 274–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Nagasawa K, Münch C, Xu Y, Satterstrom K, Jeong S, Hayes SD, Jedrychowski MP, Vyas FS, Zaganjor E, et al. (2016). Mitochondrial Sirtuin Network Reveals Dynamic SIRT3-Dependent Deacetylation in Response to Membrane Depolarization. Cell 167, 985–1000.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelenay S, van der Veen AG, Böttcher JP, Snelgrove KJ, Rogers N, Acton SE, Chakravarty P, Girotti MR, Marais R, Quezada SA, et al. (2017). Cyclooxygenase-Dependent Tumor Growth through Evasion of Immunity. Cell 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Next generation sequencing data can be accessed at Gene Ontology Omnibus under GSE119521.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD010997.