Abstract
Tick-borne rickettsioses are world-spreading infectious zoonoses. Ticks serve as reservoirs and vectors for Rickettsia and play a key role in transmission of rickettsioses. Most of the Chinese rickettsiosis patients are reported from Northeastern China but the distribution of tick and tick-borne Rickettsia species in Northeastern China remain poorly studied. In this study, a total of 1,286 ticks were captured from the seven counties of Harbin, an area in Northeastern China, and the tick-borne Rickettsia species were identified by PCR and sequencing of rrs, gltA, groEL, ompA and 17-kDa antigen-encoding genes. Of the 5 identified tick species, Haemaphysalis longicornis and Ixodes persulcatus were the predominant tick species in the livestock and vegetation, respectively. Rickettsia raoultii and “Candidatus Rickettsia tarasevichiae” were the two detectable Rickettsia species in the ticks with a 28.8% positive rate but no rickettsiae were found in ticks of Haemaphysalis concinna. R. raoultii detected in 37.6% of the Dermacentor nuttalli, Dermacentor silvarum and H. longicornis ticks while “Ca. R. tarasevichiae” was only present in 22.8% of the I. persulcatus ticks. In particular, the positive rate of both R. raoultii and “Ca. R. tarasevichiae” in ticks from the livestock (40.7%) was significantly higher than that from the vegetation (19.5%). The results indicate that the tick and tick-borne Rickettsia species are diverse in different regions of Harbin due to geographic difference and the ticks from livestock may play a more important role in transmission of rickettsioses to human.
Author summary
Rickettsiosis is a tick-borne infectious disease of global importance. The disease has been prevailing in Northeastern China but the distribution of tick and tick-borne Rickettsia species from different areas of Northeastern China remain poorly studied. We collected a total of 1,286 ticks in the seven counties with different geographic environments of Harbin, an area of Northeastern China, and all the ticks were classified as Dermacentor nuttalli, Dermacentor silvarum, Haemaphysalis concinna, Haemaphysalis longicornis or Ixodes persulcatus. A total of 28.8% of the ticks tested positive for either Rickettsia raoultii or “Candidatus Rickettsia tarasevichiae”, in which 37.6% of the D. nuttalli, D. silvarum and H. longicornis ticks were positive for R. raoultii while 22.8% of the I. persulcatus ticks were positive for “Ca. R. tarasevichiae”. The positive rate of both R. raoultii and “Ca. R. tarasevichiae” in ticks from the livestock (40.7%) was significantly higher than that from the vegetation (19.5%). All the data indicate that ticks in the Harbin area have a high infection rate with Rickettsia species and domestic animals may have a tick-livestock rickettsial circulation that may play an important role in transmission of rickettsioses.
Introduction
Rickettsiae are a large group of Gram-negative obligate intracellular prokaryotic microbes that can cause rickettsioses in human and many animals [1]. These microbes are widely distributed throughout the world, and maintained and transmitted by arthropods such as ticks, fleas, mites and lice [2]. Information about many Rickettsia species is available in GenBank, in which approximate 20 species are well characterized as human pathogens [1–3]. Particularly in the recent years, novel Rickettsia species that cause human diseases have been continuously reported, such as R. monacensis in Europe and South Korea [4, 5], and R. sibirica subsp. sibirica, R. raoultii, R. subsp. XY99 and “Candidatus R. tarasevichiae” in China [6–9]. At present, the genus Rickettsia is classified into four groups: the spotted fever group (SFG) that include R. conorii, R. rickettsii and R. japonica, the typhus group (TG) that include R. typhi and R. prowazekii, the ancestral group (AG) with species such as R. bellii and R. canadensis) and transitional group (TRG) that contains R. felis and R. akari [2,3]. Fever, headache, nausea, anorexia, rash and occasional eschar at the tick biting sites are common clinical manifestations of rickettsioses caused by most rickettsiae [10, 11]. Therefore, it is difficult to distinguish diagnosis of rickettsiosis caused by different Rickettsia species based on the clinical signs and symptoms of rickettsiosis patients. More specific and accurate laboratory diagnostic methods, for example, PCR and sequencing and genetic analysis, have been widely employed to diagnose human rickettsiosis in clinic [9].
Until now, the four Rickettsia species, R. heilongjiangensis, R. monacensis, R. raoultii, and R. sibirica, have been identified by cultivation methods, while the seven Rickettsia species, R. aeschlimannii, R. conorii, R. felis, R. massiliae, R. slovaca, “Ca. R. tarasevichiae” and Ca. R. jingxinensis, have also been confirmed by genetic molecular methods over the past 30 years in mainland of China [12–18]. In addition, according to phylogenic analysis of target gene loci, several potential novel Rickettsia species, such as Ca. R. hebeiii, Ca. R. tibetani, Ca. R. gannanii and R. subsp. XY99, have been reported in different areas of China according to the phylogenetic analysis of target gene loci [8, 19–21]. Among the rickettsiae, R. heilongjiangensis, Ca. R. hebeiii, Ca. R. tibetani, Ca. R. gannanii, R. subsp. XY99 and Ca. R. jingxinensis were first identified in ticks from mainland of China. More importantly, R. heilongjiangensis, R. raoultii, R. sibirica, R. subsp. XY99, and “Ca. R. tarasevichiae” have been confirmed as the causative agents of human rickettsioses in mainland of China [22].
Ticks act as the most important arthropod vectors in the world for transmission of microbial pathogens to humans [23]. Previous studies revealed the extensive diversity of rickettsiae in different tick species and geographic areas [1–3]. Since 1982, many different species of Rickettsia have been identified as pathogens in rickettsiosis patients in mainland of China, especially in the areas of Northeastern China [22]. In addition, in the recent years, many more rickettsiosis patients have been reported in China due to the application of more sensitive and precise laboratory diagnostic methods and most of these patients were from Northeastern China [17, 22].
The Harbin area is located in the southwest of Heilongjiang province, which is the most northeast province of China. In this area, rickettsiosis cases have been frequently reported in the recent years [6, 9, 24, 25]. However, until now, no information about the circulation of Rickettsia in ticks of this area has been available. Therefore, in the present study, we investigated the circulation of ticks and tick-borne Rickettsia in natural environments from different regions of Harbin area and the risk of rickettsial infections in the local populations were also estimated.
Materials and methods
Ethics statement
The collection of ticks from the body surface of cattle, goats and horses in this study was verbally approved by the animal owners and performed in strict accordance with the National Guidelines for Experimental Animal Welfare of China (2006–398).
Collection and identification of ticks
Adult ticks were captured from the ear, neck, armpit, chest, abdomen and crissum of cattle, goats and horses using tweezers (1–10 ticks per animal), and collected from the different types of vegetative covers by flagging with a white cotton flag (60 cm × 1 m) along its linear transection in the seven counties from the Harbin area of Northeastern China during April to May of 2019 (Fig 1) [26]. The tick species were firstly identified according to their morphology as previously described [27, 28]. Each of the ticks was soaked in 70% ethanol for disinfection and then washed three times with autoclaved double distilled water (ddH2O) for homogenization. The total DNAs in each of the homogenized samples were extracted by using an Insect DNA Extraction Kit (D0926, Omega, USA) according to the manufacturer’s protocol and then dissolved in 80 μL TE-buffer in the kit. Using several dilutions of each of the total DNAs as templates, PCR was performed to further identify the ticks using universal primers (Table 1) targeting the 16S ribosomal RNA (rrs) genes from different tick species with a High-Fidelity PCR Kit (TaKaRa, China), in which a proof-reading Pfu DNA polymerase was used [29]. In the PCR, a recombinant pUC19 plasmid containing the entire rrs gene segment of D. nuttalli, provided by our laboratory, wild-type pUC19 plasmid and TE-buffer were used as the positive, negative and blank controls, respectively. To prevent cross-contamination, DNA extraction, PCR mixture preparation, amplification and agarose gel electrophoresis were performed in separate rooms, and autoclaved pipettes and filter-containing tips were used. The PCR products were sequenced by Sangon Biotech Co. in China.
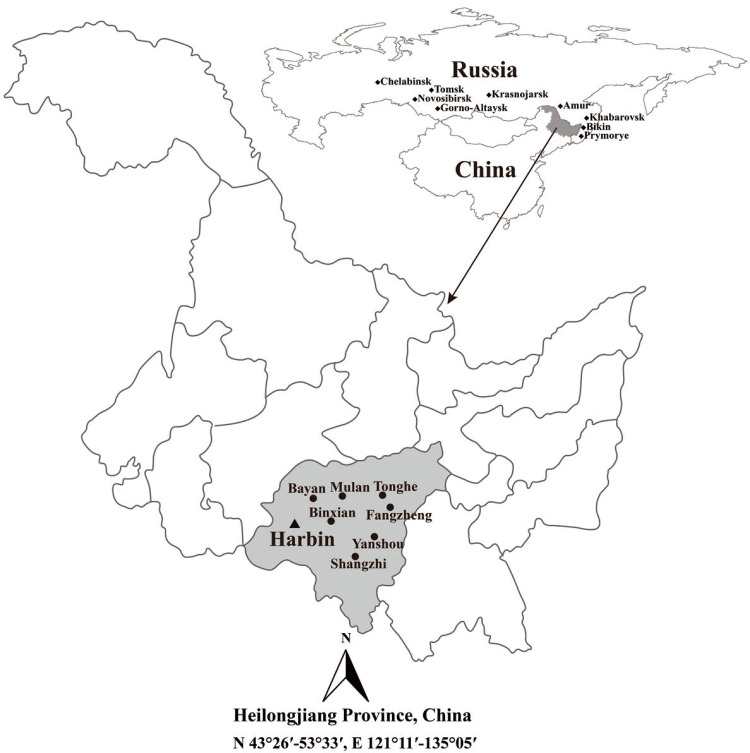
Fig 1. Location of the seven counties for tick sampling in the Harbin area of Heilongjiang province in Northeastern China.
This map was plotted by combination of Surfer software version-4 (Golden Software, USA) and Photoshop CS 8.0.1 (Adobe Systems, USA). The black dots indicate the sampling regions in this study. The black diamonds indicate the endemic regions of rickettsioses in Russia.
Table 1. Primers used in this study.
| Gene | Primer | Sequence (5’→3’) | Amplicon (bp) | Reference |
|---|---|---|---|---|
| Tick rrs | 16SF | F: GTATTTTGACTATACAAAGGTATTG | 300 | 29 |
| 16SR | R: TATTACGCTGTTATCCCTAGAGTATT | |||
| gltA | Ric-CS409d | F: CCTATGGCTATTATGCTTGC | 720 | 30 |
| Ric-CS535d | F: GCAATGTCTTATAAATATTC | |||
| Ric-CS1258n | R: ATTGCAAAAAGTACAGTGAACA | |||
| Ric-CS2d | F: ATGACCAATGAAAATAATAAT | 1,200 | 9 | |
| Ric-CSEndr | R: CTTATACTCTCTATGTACA | |||
| 17-kDa | Rr17k.1p | F: TTTACAAAATTCTAAAAACCAT | 450 | 31 |
| Rr17k.90p | F: GCTCTTGCAACTTCTATGTT | |||
| Rr17k.539n | R: TCAATTCACAACTTGCCATT | |||
| Rickettsial rrs | Ric-16SF | F: GAACGAACGCTATCGGTATGC | 1,390 | This study |
| Ric-16SR1 | R: AATTTTACCGTGGTTGGCTGC | |||
| Ric-16SR2 | R: TGCCTCTTGCGTTAGCTCAC | |||
| groEL | Ric-ESL-F1 | F: GGTAAATGGGCAGGYACCGAA | 1,580 | This study |
| Ric-ESL-R1 | R: GAAGCAACRGAAGCAGCATCTTG | |||
| Ric-ESL-F2 | F: ATCGTTATGAAAGAAAGCGAYG | |||
| Ric-ESL-R2 | R: AGWGCAGTACGCACTACTTTAGC | |||
| ompA | Rr190k.71p | F: TGGCGAATATTTCTCCAAAA | 530 | 31 |
| Rr190k.602n | R: AGTGCAGCATTCGCTCCCCCT | |||
| Rr190k.720n | R: TGCATTTGTATTACCTATTGT |
F: Forward primer. R: Reverse primer. Y = C or T. W = A or T
Detection of rickettsiae in ticks
The total DNAs in the homogenized samples of the ticks were extracted as above. Using the DNAs as templates, rickettsial DNAs were detected by nested-PCRs with the primers targeting a 720-bp citrate synthase encoding gene (gltA) and a 450-bp 17 kilodalton antigen encoding gene (17-kDa) segment as previously described [30, 31]. Subsequently, both the 720-bp gltA and 17-kDa gene segment positive total DNA samples were selected for identification of Rickettsia species by PCR with the primers (Table 1) targeting the nearly entire rrs gene (1,390 bp), gltA gene (1,200 bp) and 60-kDa heat shock protein encoding gene (groEL, 1,580 bp), and the partial segment of outer membrane protein A encoding gene (ompA, 530 bp) [9, 31]. In particular, the reported primers targeting rickettsial rrs gene by PCR and groEL genes in semi-nested PCR produced smaller products (813 and 217 bp) [32, 33]. To increase the sensitivity, specificity and efficiency of rickettsial identification, the primers targeting rickettsial rrs and groEL gene by semi-nested PCR with larger products were designed after analysis of the rickettsial rrs and groEL gene sequences in GenBank. The PCR products were examined by agarose gel using an Image Analyzer (Bio-Rad, USA) after electrophoresis. The amplified DNA fragments in the gels with expected sizes were extracted using a Gel Extraction Kit (Qiagen, USA) and then cloned into pMD19-T plasmid (TaKaRa) according to the manufacturers’ protocols for sequencing to identify rickettsial species. For PCR detection, the High-Fidelity PCR Kit and prevention of cross-contamination were the same as above. The DNAs from Rickettsiae japonica and Haemaphysalis concinna, provided by our laboratory, and TE-buffer were used as the positive, negative and blank controls in the PCR, respectively.
Analysis of sequence homology of the genes from ticks and rickettsiae
The obtained nucleotide sequences from the target gene segments of ticks and rickettsiae were edited and assembled using the SeqMan program (DNASTAR, Madison, WI) and aligned using the Clustal W method in the Lasergene program [34], and then compared with the corresponding sequences in GenBank using BLAST software. The following sequences were used for comparison: MN448327-MN448342 for the rrs genes of ticks (300 bp) while MN450395-MN450401 for the gltA genes (1,200 bp), MN446743-MN446749 for the rrs genes (1,390 bp), MN450402-MN450408 for the groEL genes (1,580 bp), and MN450409-MN450415 for the ompA genes (530 bp) of rickettsiae.
Genetic and phylogenic analysis of the ticks and rickettsiae
The best-fit nucleotide substitution models for phylogenetic analysis based on the target genes from the ticks and rickettsiae were determined using jModel Test [35]. Phylogenic trees were constructed using the Maximum likelihood (ML) method in the PhyML v3.0 software [36]. The boot strap support values calculated from 1000 replicates were used to test the reliability of branches in the trees and values over 70% were considered as significant difference for presentation. All phylogenic trees were mid-point rooted for purpose of clarity.
Statistical data analysis
Statistical analysis of the obtained data was performed using the Statistical Package for Social Sciences Version 21.0 software (SPSS, Chicago, IL, USA). The Chi-square test or Fisher’s exact test was used for calculating the P values to determine differences of the positive rates in the ticks and rickettsiae. Statistical significance was defined as P<0.05.
Results
Species and distribution of the collected ticks
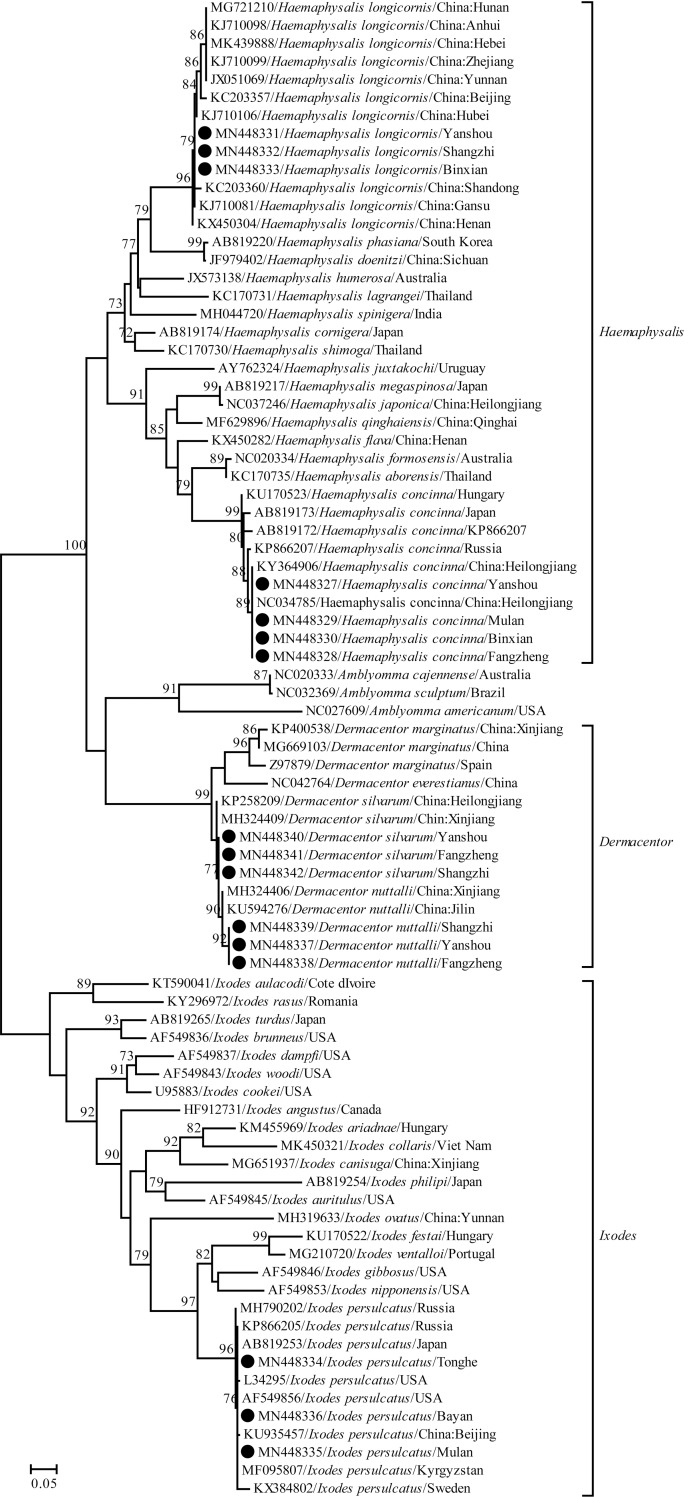
A total of 1,286 adult ticks were collected from the seven sampling regions of the Harbin area and all the ticks were classified as one of five different species belonging to three different genera of ticks according to their morphological characteristics and rrs gene sequencing data, namely Dermacentor nuttalli (9.6%, 123/1286), Dermacentor silvarum (12.8%, 165/1286), Haemaphysalis concinna (8.2%, 106/1286), Haemaphysalis longicornis (30.6%, 393/1286) and Ixodes persulcatus (38.8%, 499/1286) (Fig 1 and Table 2). H. longicornis was the predominant tick species (63.8%, 393/616) in Yanshou (65.3%, P = 112.05–166.33, P < 0.05), Shangzhi (50.8%, P = 13.25 and 53.10, P < 0.05) and Binxian (76.7%, P = 71.88, P < 0.05) county which have similar geographic environments and similar vegetative covers while I. persulcatus was the predominant (88.6%, 294/332) or unique tick species in Tonghe (87.6%, P = 181.88, P < 0.05) and Mulan (87.6%, P = 213.16, P < 0.05) regions or in Bayan region, which also have the same climates and similar geographic environments (Table 2). Among the ticks, 563 were captured from different domestic animals, while 723 were collected from different types of vegetative covers (Table 3). The same five tick species were captured from cattle and goats while three of the five tick species were found on horses, with the absence of D. nuttalli and H. concinna (Table 3). H. longicornis (51.2%, 288/563) was the predominant tick species on cattles (54.8%, P = 64.58–145.87, P < 0.05), goats (47.0%, P = 40.83–75.16, P < 0.05) and horses (50.0%, P = 9.23 and 18.36, P < 0.05), while I. persulcatus (62.1%, 381/614) was the predominant tick species in forest shrub (67.6%, P = 213.37 and 234.02, P < 0.05) and hilly grass/shrub (50.7%, P = 20.84–80.35, P < 0.05), but it could not be detected in farm grassland (Table 3). The identification of ticks based on phylogenic analysis with the rrs gene segment (300 bp) from the five tick species and sequences from GenBank is shown in Fig 2.
Table 2. Ticks collected from different sampling regions.
| Region | Coordinate | Geography and climate | Tick (n) | Species / number |
|---|---|---|---|---|
| Yanshou | N 45°31’ | Cold temperate continental monsoon climate, abundant rainfall, low mountain-hilly terrain, broadleaf forest and shrub grassland. | 239 | D. nuttalli / 21 |
| E 128°44’ | D. silvarum / 42 | |||
| H. concinna / 20 | ||||
| H. longicornis / 156* | ||||
| Fangzheng | N 45°50’ | The geography and climate are the same as in Yanshou. | 133 | D. nuttalli / 36 |
| E 128°49’ | D. silvarum / 70 | |||
| H. concinna / 27 | ||||
| Shangzhi | N 45°12’ | Broadleaf-conifer forest. The others are the same as in Yanshou. | 201 | D. nuttalli / 66 |
| E 128°01’ | D. silvarum / 33 | |||
| H. longicornis / 102* | ||||
| Binxian | N 45°44’ | The geography and climate are the same as in Yanshou. | 176 | H. concinna / 41 |
| E 127°25’ | H. longicornis / 135* | |||
| Tonghe | N 45°58’ | Mountain terrain. The others are the same as in Shangzhi. | 161 | D. silvarum / 20 |
| E 128°45’ | I. persulcatus / 141* | |||
| Mulan | N 45°56’ | Mid-temperate continental monsoon climate. The others are the same as in Tonghe. | 171 | H. concinna / 18 |
| E 128°01’ | I. persulcatus / 153* | |||
| Bayan | N 46°04’ | Plain and low mountain terrain and shrub grassland and conifer forest. The others are the same as in Mulan. | 205 | I. persulcatus / 205 |
| E 127°23’ |
*H. longicornis and I. persulcatus were the predominant tick species in the local regions (P<0.05).
Table 3. Ticks collected from different sources.
| Source | Animal/Geomorphy | Tick (n) | Species/number |
|---|---|---|---|
| Livestock | Cattle | 261 | D. nuttalli / 16 |
| D. silvarum / 21 | |||
| H. concinna / 27 | |||
| H. longicornis / 143* | |||
| I. persulcatus / 54 | |||
| Goat | 202 | D. nuttalli / 25 | |
| D. silvarum / 30 | |||
| H. concinna / 17 | |||
| H. longicornis / 95* | |||
| I. persulcatus / 35 | |||
| Horse | 100 | D. silvarum / 21 | |
| H. longicornis / 50* | |||
| I. persulcatus / 29 | |||
| Vegetation | Forest shrub | 411 | D. silvarum / 71 |
| H. concinna / 62 | |||
| I. persulcatus / 278# | |||
| Hilly grass/shrub | 203 | D. nuttalli / 20 | |
| D. silvarum / 22 | |||
| H. longicornis / 58 | |||
| I. persulcatus / 103# | |||
| Farm grass | 109 | D. nuttalli / 62 | |
| H. longicornis / 47 |
* or #the two tick species were significantly more than the other tick species from the livestock and shrub/grasslands, respectively (P<0.05).
Fig 2. Identification of the ticks based on phylogenic analysis with the rrs gene.
The black dots indicate the representative sequences of rrs gene segments (300 bp) from the ticks in the different sampling regions.
Rickettsiae in the collected ticks
Nested-PCR showed that 43.9% (54/123) of the D. nuttalli ticks, 46.7% (77/165) of the D. silvarum ticks, 31.8% (125/393) of the H. longicornis, in the ticks and 22.8% (114/499) in the I. persulcatus ticks, but not the H. concinna ticks, were positive for both the Rickettsia-specific 720-bp gltA and 450-bp 17-kDa gene segments and the total positive rate of all ticks was 28.8% (370/1286) (Table 4) PCR and sequence analysis of rickettsial rrs, gltA (1200 bp), groEL and ompA genes showed that only two species of Rickettsia, R. raoultii in the D. nuttalli, D. silvarum and H. longicornis (69.2%, 256/370) ticks and “Ca. R. tarasevichiae” in the I. persulcatus ticks (30.8%, 114/370), were identified from the 370 rickettsial gltA (720 bp) and 17-kDa gene positive samples (Table 4). However, the two Rickettsia species were not found in the same sample of the Rickettsia-positive ticks. DNA of R. raoultii was detected in the ticks from all the different sources but its positive rate in ticks from the domestic animals (32.0%, 180/563) was significantly higher than that from the vegetative covers (10.5%, 76/723) (P = 91.42, P<0.05). In addition, R. raoultii was the unique Rickettsia species detected in ticks from the Yanshou, Fangzheng, Shangzhi and Binxian regions and the “Ca. R. tarasevichiae” was only found in the ticks from Mulan and Bayan regions, probably due to their noticeable geographic differences. The total positive rate of both R. raoultii and “Ca. R. tarasevichiae” in ticks from the domestic animals (40.7%, 229/563) was significantly higher than that from the vegetative covers (19.5%, 141/723) (P = 69.24, P<0.05).
Table 4. Rickettsia species in ticks from different sampling regions.
| Region | Source | Tick species / n | Rickettsia species / infection rate (n / %) | ||
|---|---|---|---|---|---|
| R. raoultii | “Ca. R. tarasevichiae” | ||||
| Yanshou | Livestock | Cattle | D. nuttalli / 4 | 3 / 75.0 | 0 |
| D. silvarum / 9 | 5 / 55.6 | 0 | |||
| H. longicornis / 59 | 35 / 59.3 | 0 | |||
| Goat | D. silvarum / 15 | 11 / 73.3 | 0 | ||
| H. concinna / 17 | 0 | 0 | |||
| H. longicornis / 41 | 20 / 48.8 | 0 | |||
| Vegetation | Forest shrub | D. silvarum / 18 | 4 / 22.2 | 0 | |
| H. concinna / 3 | 0 | 0 | |||
| Hilly grass/shrub | H. concinna / 3 | 0 | 0 | ||
| H.longicornis / 38 | 3 / 7.9 | 0 | |||
| Farm grassland | D. nuttalli / 17 | 6 / 35.3 | 0 | ||
| H.longicornis / 18 | 3 / 16.7 | 0 | |||
| Total | 239 | 90 / 37.7 | 0 | ||
| Fangzheng | Livestock | Cattle | D.silvarum / 12 | 6 / 50.0 | 0 |
| H.concinna / 27 | 0 | 0 | |||
| Goat | D.silvarum / 15 | 8 / 53.3 | 0 | ||
| Horse | D.silvarum / 21 | 12 / 57.1 | 0 | ||
| Vegetation | Forest shrub | D.silvarum / 22 | 14 / 63.6 | 0 | |
| Farm grassland | D. nuttalli / 36 | 10 / 27.8 | 0 | ||
| Total | 133 | 50 / 37.6 | 0 | ||
| Shangzhi | Livestock | Goat | D. nuttalli / 25 | 16 / 64.0 | 0 |
| H.longicornis / 54 | 10 / 18.5 | 0 | |||
| Cattle | D. nuttalli / 12 | 7 / 58.3 | 0 | ||
| H.longicornis / 28 | 5 / 17.9 | 0 | |||
| Vegetation | Forest shrub | D. silvarum / 31 | 11 / 35.5 | 0 | |
| Hilly grass/shrub | D. nuttalli / 20 | 9 / 45.0 | 0 | ||
| D. silvarum / 2 | 0 | 0 | |||
| H.longicornis / 20 | 2 / 10.0 | 0 | |||
| Farm grassland | D. nuttalli / 9 | 3 / 33.3 | 0 | ||
| Total | 201 | 63 / 31.3 | 0 | ||
| Binxian | Livestock | Cattle | H.longicornis / 56 | 23 / 41.1 | 0 |
| Horse | H.longicornis / 50 | 19 / 38.0 | 0 | ||
| Vegetation | Forest shrub | H.concinna / 41 | 0 | 0 | |
| Farm grassland | H.longicornis / 29 | 5 / 17.2 | 0 | ||
| Total | 176 | 47 / 26.7 | 0 | ||
| Tonghe | Livestock | Cattle | I.persulcatus / 40 | 0 | 12 / 30.0 |
| Vegetation | Forest shrub | I. persulcatus / 28 | 0 | 4 / 14.3 | |
| Hilly grass/shrub | D.silvarum / 20 | 6 / 30.0 | 0 | ||
| I. persulcatus / 73 | 0 | 12 / 16.4 | |||
| Total | 161 | 6 / 3.7 | 28 / 17.4 | ||
| Mulan | Livestock | Goat | I.persulcatus / 17 | 0 | 5 / 29.4 |
| Horse | I.persulcatus / 29 | 0 | 16 / 55.2 | ||
| Vegetation | Forest shrub | H.concinna / 18 | 0 | 0 | |
| I.persulcatus / 77 | 0 | 13 / 16.9 | |||
| Hilly grass/shrub | I.persulcatus / 30 | 0 | 5 / 16.7 | ||
| Total | 171 | 0 | 39 / 22.8 | ||
| Bayan | Livestock | Cattle | I. persulcatus / 14 | 0 | 8 / 57.1 |
| Goat | I. persulcatus / 18 | 0 | 8 / 44.4 | ||
| Vegetation | Forest shrub | I. persulcatus / 173 | 0 | 31 / 17.9 | |
| Total | 205 | 0 | 47 / 22.9 | ||
Genetic and phylogenic analysis of the identified rickettsiae
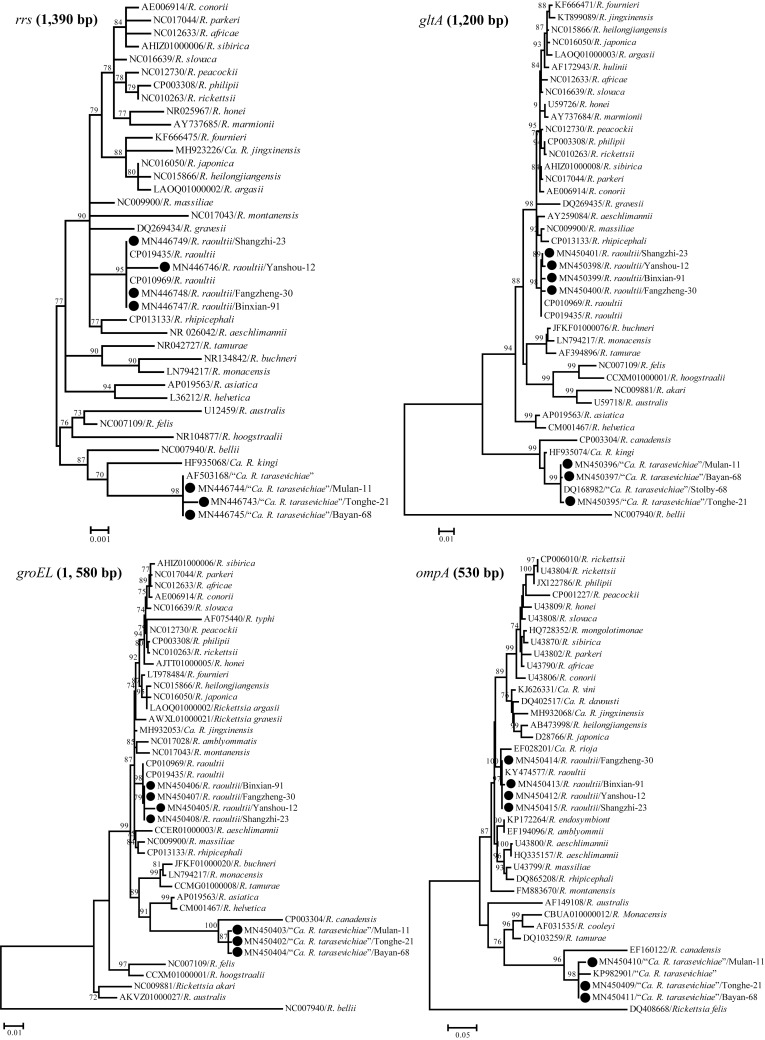
Sequencing data revealed that the rrs, gltA (1,200 bp), groEL and ompA gene segments from the 256 strains of R. raoultii and 114 strains of “Ca. R. tarasevichiae” identified in this study presented 99.6%-100% nucleotide sequence identities. The nucleotide sequence identities of the rrs, gltA (1,200 bp), groEL and ompA gene segments from the 256 R. raoultii strains displayed 99.6%-100%, 99.4%-100%, 99.2%-100% and 99.2%-100% nucleotide sequence identity, respectively, compared with the corresponding genes in the whole genome of R. raoultii strain IM16 (GenBank: CP019435.1), an isolate from a patient in Northern China. For the rrs, gltA (1,200 bp) and ompA gene segments from the 114 “Ca. R. tarasevichiae” strains, the nucleotide sequence identities were 99.9%-100%, 99.8%-100% and 98.8%-100%, respectively, compared with the rrs and gltA genes of a “Ca. R. tarasevichiae” strain (GenBank: AF503168.1 and AF503167.2) [37], and the ompA gene of “Ca. R. tarasevichiae” strain M-R217 (GenBank: KU361217.1), respectively. Since no groEL gene sequences of “Ca. R. tarasevichiae” could be found in GenBank and a previous study reported that the rrs and gltA genes of “Ca. R. tarasevichiae” had the highest nucleotide sequence identities (98.0% and 96.0%) with those of Rickettsia canadensis among different Rickettsia species [37], the nucleotide sequence identities of groEL gene segments from the 114 “Ca. R. tarasevichiae” strains were compared with the groEL gene of R. canadensis strain McKiel (GenBank: CP000409.1) and the sequence identities were 96.8%-97.0%. The phylogenetic tree based on comparison of the four rickettsial genes with those from GenBank is shown in Fig 3.
Fig 3. Phylogenic tree based on the rrs, gltA, groEL and ompA genes of rickettsiae.
The genetic identity among different Rickettsia species was inferred by maximum-likelihood method implemented in PhyML v3.0, and rooted by midpoint method. The black dots indicate the nucleotide sequences of rrs (1,390 bp), gltA (1,200 bp), groEL (1,580 bp) and ompA (530 bp) gene segments from R. raoultii and “Ca. R. tarasevichiae” in the different sampling regions.
Discussion
Ticks act as the main reservoir hosts of many microbial pathogens as well as the major transmission vector of the pathogens to both human and animals in tropical and subtropical areas [23, 38]. Rickettsia is a large group of heterogeneous obligate intracellular microbes and ticks serve as the major host and vector for most Rickettsia species [1–3]. Since many Rickettsia species can cause different types of human and animal rickettsioses and the geographic distribution of tick species and tick-borne Rickettsia species are considerably various, investigation of tick-borne rickettsiae in different areas is significant for prevention and control of rickettsioses in different areas.
In Northeastern China, at least eighteen species belonging to seven genera of ticks have been reported due to the profuse and manifold vegetative covers serving as habitats for ticks [39]. Among the ticks, I. persulcatus, H. longicornis and D. silvarum are the most predominant tick species in the area [17, 39]. In the present study, five tick species, D. nuttalli, D. silvarum, H. concinna, H. longicornis and I. persulcatus, could be found in the seven counties/regions of the Harbin area, but the number of tick species from these sampling regions presented a notable difference. For example, I. persulcatus was the unique tick species found in the Bayan region. H. longicornis was the predominant tick species in Yanshou, Shangzhi and Binxian regions (63.8%) while I. persulcatus was the predominant tick species in Tonghe and Mulan regions (88.6%), probably due to their distinct differences in climates, geographic environments and vegetative covers. On the other hand, H. longicornis was significantly more associated with the domestic animals (51.2%, 288/563) than with the different types of vegetative covers (33.7%, 105/312) (P<0.05), and it could not even be found from the forest shrub. In contrast, I. persulcatus was significantly less associated with the domestic animals (21.0%, 118/563) than with the forest shrub and hilly grass/shrub (62.1%, 381/614) (P<0.05). Previous reports showed that H. longicornis prefers to parasitize on artiodactyl/perissodactyl mammalian herbivores, such as cattle, goats and horses, while adult I. persulcatus parasites on multiple wild and domestic mammalian animals and several kinds of birds [40, 41]. During the free-living stage, H. longicornis likes to conceal in ground grasses, while I. persulcatus is encountered in forests of taiga in the mountains. The distribution of I. persulcatus and H. longicornis found in this study was corresponding with previous reports. These data indicate that H. longicornis and I. persulcatus are the predominant tick species in the Harbin area, while the different geographic environments, vegetative covers, climates and sampling sources can account for the diversity in distribution of different tick species.
Although a total of 1286 ticks belonging to five tick species were collected in this study, only two Rickettsia species (R. raoultii and “Ca. R. tarasevichiae”) could be found. Previous studies reported that H. concinna is an important vector of rickettsiae such as R. raoultii, “Ca. R. tarasevichiae”, R. heilongjangensis and R. hulinii [17, 42, 43]. However, no rickettsiae were detected in any of the H. concinna ticks collected in this study. R. raoultii was firstly detected in D. nuttalli and R. pumilio ticks in 1999 in the former Soviet Union [44]. Subsequently, R. raoultii was found in 12 species belonging to 6 genera of ticks in Europe, North Africa and Asia and the tick species belonging to the genus Dermacentor have been confirmed as the major reservoir and vector of this rickettsial species [38, 45–51]. R. raoultii is widely distributed in Northeastern China and the Far-East/Siberia areas of Russia and ticks belonging to Dermacentor species were confirmed as the common natural hosts [49–54]. In this study, R. raoultii was detected in D. nuttalli, D. silvarum, and H. longicornis ticks in Harbin, an area of Northeastern China. “Ca. R. tarasevichiae” is an emerging tick-borne Rickettsia species initially found in I. persulcatus ticks in Russia [37]. In this study, “Ca. R. tarasevichiae” was also solely found in I. persulcatus ticks. The tick and tick-borne Rickettsia species found in this study were similar to those reported from Russia, probably due to the adjacency of Northeastern China and the Far-East/Siberia areas of Russia, with similar natural environments and habitats for ticks. Several rrs and groEL gene segments of R. raoultii strains and rrs gene segments of “Ca. R. tarasevichiae” strains were different from the others in tree branch lengths of the phylogenetic tree, probably due to single nucleotide polymorphisms (SNPs) in the genes from different strains [55, 56]. All the data indicate that the different geographic environments act as the major influenting factor for distribution of tick and tick-borne Rickettsia species.
Both R. raoultii and “Ca. R. tarasevichiae” have been confirmed as causative agents of human rickettsioses. Most of rickettsiosis patients have a common pathological change of blood vessel endothelial injury at early stage during infection [57]. R. raoultii can cause human disease called tick-borne lymphadenopathy (TIBOLA) with the clinical features necrotic erythema, eschar and cervical adenopathies [45, 58]. The initial TIBOLA Chinese cases were reported in Northeastern China in 2014 [24]. “Ca. R. tarasevichiae” is a member of rickettsiae in the spotted fever group and clinical signs and symptoms of patients infected by this pathogen are fever, headache, nausea, eschar and lymphadenopathy [9]. The “Ca. R. tarasevichiae”-infected Chinese patients were also initially found in Northeastern China in 2013 [59]. In the past years, nearly all of the emerging or re-emerging tick-borne human rickettsioses have been found in Northeastern China including Heilongjiang province [6, 9, 24, 25]. In this study, approximately 30% of the collected ticks from the Harbin area, which is located in the southwest of Heilongjiang province, were found to carry either R. raoultii or “Ca. R. tarasevichiae”. In particular, the total positive rate of both R. raoultii and Ca. R. tarasevichiae in ticks from the domestic animals (40.7%) was significantly higher than that from the different types of vegetative covers (19.5%). Except for the preference of host and habitat, co-feeding of domestic animals is also a risk factor that increases tick-borne rickettsial infections among the animals. These data indicate that the circulation of rickettsial infections in the domestic animals in the Harbin area of Northeastern China is an important subject for investigation and may play an important role in prevention and control of transmission of tick-borne rickettsioses in local populations.
Flagging is a typical method for collection of ticks from vegetation, but it is unable to capture all the given groups of ticks in the sampling sites due to many influening factors, such as different types of vegetation, behavior and habitat characteristics of different tick species, and climate [60]. The capture of ticks by tweezers from the livestock is also influenced by the preferred infestation positions on the different animals and developmental stages of the ticks. However, the large-sample of 1286 ticks collected in this study should still reflect the general distribution and predominant species of ticks in the different geographic regions of the Harbin area. Taken together, this study revealed the predominant tick species (H. longicornis and I. persulcatus) and tick-borne Rickettsia species (R. raoultii and “Ca. R. tarasevichiae”) in the Harbin area of Northeastern China, as well as the more important role of domestic animals in transmission of rickettsioses, as reflected by the higher positive rates of Rickettsia-infected ticks.
Acknowledgments
We are grateful to Prof. Stijn van der Veen, a native English speaker working at our university, to improve the English of our manuscript.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by grants from Foshan University (KLPREAD201801-09 and KLPREAD201801-08), Education Bureau of Guangdong Province (2014KTSPT037 and 2018KQNCX277) and the Guangdong Provincial Science and Technology Plan (2012A020100001). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Parola P, Paddock CD, Socolovschi C, Labruna MB, Mediannikov O, Kernif T, et al. Update on tick-borne rickettsioses around the world: a geographic approach. Clin Microbiol Rev. 2013; 26: 657–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fournier PE, Raoult D. Current knowledge on phylogeny and taxonomy of Rickettsia spp. Ann N Y Acad Sci. 2009; 1166: 1–11. [DOI] [PubMed] [Google Scholar]
- 3.Merhej V, Angelakis E, Socolovschi C, Raoult D. Genotyping, evolution and epidemiological findings of Rickettsia species. Infect Genet Evol. 2014; 25: 122–137. [DOI] [PubMed] [Google Scholar]
- 4.Portillo A, Santibáñez S, García-Álvarez L, Palomar AM, Oteo JA. Rickettsioses in Europe. Microbes Infect. 2015; 17: 834–838. [DOI] [PubMed] [Google Scholar]
- 5.Kim YS, Choi YJ, Lee KM, Ahn KJ, Kim HC, Klein T, et al. First isolation of Rickettsia monacensis from a patient in South Korea. Microbiol Immunol. 2017; 61: 258–263. [DOI] [PubMed] [Google Scholar]
- 6.Jia N, Jiang JF, Huo QB, Jiang BG, Cao WC. Rickettsia sibirica subspecies sibirica BJ-90 as a cause of human disease. N Engl J Med. 2013; 369: 1176–1178. [DOI] [PubMed] [Google Scholar]
- 7.Li H, Zhang PH, Huang Y, Du J, Cui N, Yang ZD, et al. Isolation and identification of Rickettsia raoultii in human cases: a surveillance study in 3 medical centers in China. Clin Infect Dis. 2018; 66: 1109–1115. [DOI] [PubMed] [Google Scholar]
- 8.Li H, Cui XM, Cui N, Yang ZD, Hu JG, Fan YD, et al. Human infection with novel spotted fever group Rickettsia genotype, China, 2015. Emerg Infect Dis. 2016; 22: 2153–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jia N, Zheng YC, Jiang JF, Ma L, Cao WC. Human infection with Candidatus Rickettsia tarasevichiae. N Engl J Med. 2013; 369: 1178–1180. [DOI] [PubMed] [Google Scholar]
- 10.Fang R, Blanton LS, Walker DH. Rickettsiae as emerging infectious agents. Clin Lab Med. 2017; 37: 383–400. [DOI] [PubMed] [Google Scholar]
- 11.Santibáñez S, Portillo A, Santibáñez P, Palomar AM, Oteo JA. Usefulness of rickettsial PCR assays for the molecular diagnosis of human rickettsioses. Enferm Infect Microbiol Clin. 2013; 31: 283–288. [DOI] [PubMed] [Google Scholar]
- 12.Yu X, Jin Y, Fan M, Xu G, Liu Q, Raoult D. Genotypic and antigenic identification of two new strains of spotted fever group rickettsiae isolated from china. J Clin Microbiol. 1993; 31: 83–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W, Liu L, Jiang X, Guo X, Garnier M, Raoult D, et al. Molecular identification of spotted fever group rickettsiae in ticks collected in central China. Clin Microbiol Infect. 2009; 15: 279–280. [DOI] [PubMed] [Google Scholar]
- 14.Wei QQ, Guo LP, Wang AD, Mu LM, Zhang K, Chen CF, et al. The first detection of Rickettsia aeschlimannii and Rickettsia massiliae in Rhipicephalus turanicus ticks, in northwest China. Parasit Vectors. 2015; 8: 631–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian ZC, Liu GY, Shen H, Xie JR, Luo J, Tian MY. First report on the occurrence of Rickettsia slovaca and Rickettsia raoultii in Dermacentor silvarum in China. Parasit Vectors. 2015; 5: 19–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, Lu G, Kelly P, Zhang Z, Wei L, Yu D, et al. First report of Rickettsia felis in China. BMC Infect Dis. 2014; 14: 682–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu H, Li Q, Zhang X, Li Z, Wang Z, Song M, et al. Characterization of rickettsiae in ticks in Northeastern China. Parasit Vectors. 2016; 9: 498–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ge Y, Yin H, Rikihisa Y, Pan W, Yin H. Molecular detection of tick-borne rickettsiales in goats and sheep from Southeastern China. Vector Borne Zoonotic Dis. 2016; 16: 309–316. [DOI] [PubMed] [Google Scholar]
- 19.Zou Y, Wang Q, Fu Z, Liu P, Jin H, Yang H, et al. Detection of spotted fever group Rickettsia in Haemaphysalis longicornis from Hebei Province, China. J Parasitol. 2011; 97: 960–962. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Liu Z, Yang J, Chen Z, Liu J, Li Y, et al. Rickettsia raoultii-like bacteria in Dermacentor spp. ticks, Tibet, China. Emerg Infect Dis. 2012; 18: 1532–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J, Tian Z, Liu Z, Niu Q, Han R, Li Y, et al. Novel spotted fever group rickettsiae in Haemaphysalis qinghaiensis ticks from Gansu, Northwest China. Parasit Vectors. 2016; 9: 146–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang LQ, Liu K, Li XL, Liang S, Yang Y, Yao HW, et al. Emerging tick-borne infections in mainland China: an increasing public health threat. Lancet Infect Dis. 2015; 15: 1467–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dantas-Torres F, Chomel BB, Otranto D. Ticks and tick-borne diseases: a one health perspective. Trends Parasitol. 2012; 28: 437–446. [DOI] [PubMed] [Google Scholar]
- 24.Jia N, Zheng YC, Ma L, Huo QB, Ni XB, Jiang BG, et al. Human infections with Rickettsia raoultii, China. Emerg Infect Dis. 2014; 20: 866–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Fu XY, Jiang JF, et al. Severe illness caused by Rickettsia sibirica subspecies sibirica BJ-90 infection, China. Emerg Microbes Infect. 2017; 6: e107–e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ereqat S, Nasereddin A, Al-Jawabreh A, Azmi K, Harrus S, Mumcuoglu K, et al. Molecular detection and identification of spotted fever group rickettsiae in ticks collected from the West Bank, Palestinian territories. PLoS Negl Trop Dis. 2016; 10: e0004348–0004358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dantas-Torres F, Latrofa MS, Annoscia G, Giannelli A, Parisi A, Otranto D. Morphological and genetic diversity of Rhipicephalus sanguineus sensu lato from the new and old worlds. Parasit Vectors. 2013; 6: 213–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barker SC, Murrell A. Systematics and evolution of ticks with a list of valid genus and species names. Parasitology. 2004; 129: S15–S36. [DOI] [PubMed] [Google Scholar]
- 29.Cheng WY, Zhao GH, Jia YQ, Bian QQ, Du SZ, Fang YQ, et al. Characterization of Haemaphysalis flava (Acari: ixodidae) from Qingling subspecies of giant panda (Ailuropoda melanoleuca qinlingensis) in Qinling mountains (Central China) by morphology and molecular markers. PLoS One. 2013; 8: e69793–e69798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roux V, Rydkina E, Eremeeva M, Raoult D. Citrate synthase gene comparison, a new tool for phylogenetic analysis, and its application for the rickettsiae. Int J Syst Bacteriol. 1997; 47: 252–261. [DOI] [PubMed] [Google Scholar]
- 31.Ishikura M, Ando S, Shinagawa Y, Matsuura K, Hasegawa S, Nakayama T, et al. Phylogenetic analysis of spotted fever group rickettsiae based on gltA, 17-kDa, and rOmpA genes amplified by nested PCR from ticks in Japan. Microbiol Immunol. 2003; 47: 823–832. [DOI] [PubMed] [Google Scholar]
- 32.Roux V, Raoult D. Phylogenetic analysis of the genus by 16S rDNA sequencing. Res Microbiol. 1995; 146: 385–396. [DOI] [PubMed] [Google Scholar]
- 33.Park HS, Lee JH, Jeong EJ, Kim JE, Hong SJ, Park TK, et al. Rapid and simple identification of Orientia tsutsugamushi from other group rickettsiae by duplex PCR assay using groEL Gene. Microbiol Immunol. 2005; 49: 545–549. [DOI] [PubMed] [Google Scholar]
- 34.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994; 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Posada D. jModel Test: phylogenetic model averaging. Mol Biol Evol. 2008; 25: 1253–1256. [DOI] [PubMed] [Google Scholar]
- 36.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010; 59: 307–321. [DOI] [PubMed] [Google Scholar]
- 37.Shpynov S, Fournier PE, Rudakov N, Raoult D. “Candidatus Rickettsia tarasevichiae” in Ixodes persulcatus ticks collected in Russia. Ann NY Acad Sci 2003; 990: 162–172. [DOI] [PubMed] [Google Scholar]
- 38.Parola P, Paddock CD, Raoult D. Tick-borne rickettsioses around the world: emerging diseases challenging old concepts. Clin Microbiol Rev. 2005; 18: 719–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Z, Yang X, Bu F, Yang X, Yang X, Liu J. Ticks (Acari ixodoidea: argasidae ixodidae) of China. Exp Appl Acarol. 2010; 51: 393–404. [DOI] [PubMed] [Google Scholar]
- 40.Lawrencea KE, Summersa SR, Heathb ACG, McFaddenc AMJ, Pulfordc DJ, Taitd AB, et al. Using a rule-based envelope model to predict the expansion of habitat suitability within New Zealand for the tick Haemaphysalis longicornis, with future projections based on two climate change scenarios. Vet Parasitol. 2017; 243: 226–234. [DOI] [PubMed] [Google Scholar]
- 41.Sirotkin MB, Korenberg EI. Influence of abiotic factors on different developmental stages of the taiga tick Ixodes persulcatus and the sheep tick Ixodes ricinus. Entomol Rev. 2018; 98: 496–513. [Google Scholar]
- 42.Cheng C, Fu W, Ju W, Yang L, Xu N, Wang YM, et al. Diversity of spotted fever group Rickettsia infection in hard ticks from Suifenhe, Chinese-Russian border. Ticks Tick Borne Dis. 2016; 7: 715–719. [DOI] [PubMed] [Google Scholar]
- 43.Zhang JZ, Fan MY, Wu YM, Fournier PE, Roux V, Raoult D. Genetic classification of Rickettsia heilongjiangii and Rickettsia hulinii, two Chinese spotted fever group rickettsiae. J Clin Microbiol. 2000; 38: 3498–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rydkina E, Roux V, Rudakov N, Gafarova M, Tarasevich I, Raoult D. New rickettsiae in ticks collected in territories of the former Soviet Union. Emerg Infect Dis. 1999; 5: 811–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mediannikov O, Matsumoto K, Samoylenko I, Drancourt M, Roux V, Rydkina E, et al. Rickettsia raoultii sp. nov., a spotted fever group rickettsia associated with Dermacentor ticks in Europe and Russia. Int J Syst Evol Microbiol. 2008; 58: 1635–1639. [DOI] [PubMed] [Google Scholar]
- 46.Han R, Yang J, Niu Q, Liu Z, Chen Z, Kan W, et al. Molecular prevalence of spotted fever group rickettsiae in ticks from Qinghai Province, northwestern China. Infect Genet Evol. 2018; 57: 1–7. [DOI] [PubMed] [Google Scholar]
- 47.Yin X, Guo S, Ding C, Cao M, Kawabata H, Sato K, et al. Spotted fever group rickettsiae in Inner Mongolia, China, 2015–2016. Emerg Infect Dis. 2018; 24: 2105–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song S, Chen C, Yang M, Zhao S, Wang B, Hornok S, et al. Diversity of Rickettsia species in border regions of northwestern China. Parasit Vectors. 2018; 11: 634–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wen J, Jiao D, Wang JH, Yao DH, Liu ZX, Zhao G, et al. Rickettsia raoultii, the predominant Rickettsia found in Dermacentor silvarum ticks in China-Russia border areas. Exp Appl Acarol. 2014; 63: 579–585. [DOI] [PubMed] [Google Scholar]
- 50.Samoylenko I, Shpynov S, Raoult D, Rudakov N, Fournier PE. Evaluation of Dermacentor species naturally infected with Rickettsia raoultii. Clin Microbiol Infect. 2009; 15: 305–306. [DOI] [PubMed] [Google Scholar]
- 51.Igolkina Y, Krasnova E, Rar V, Savelieva M, Epikhina T, Tikunov A, et al. Detection of causative agents of tick-borne rickettsioses in Western Siberia, Russia: identification of Rickettsia raoultii and Rickettsia sibirica DNA in clinical samples. Clin Microbiol Infect. 2018; 24: 199.e9–199.e12. [DOI] [PubMed] [Google Scholar]
- 52.Dong X, Chen XP, Liu N, Dumler SJ, Zhang YZ. Co-circulation of multiple species of rickettsiales bacteria in one single species of hard ticks in Shenyang, China. Ticks Tick Borne Dis. 2014; 5: 727–733. [DOI] [PubMed] [Google Scholar]
- 53.Wei F, Song M, Liu H, Wang B, Wang S, Wang Z, et al. Molecular detection and characterization of zoonotic and veterinary pathogens in ticks from Northeastern China. Front Microbiol. 2016; 7:1913–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo WP, Wang YH, Lu Q, Xu G, Luo Y, Ni X, et al. Molecular detection of spotted fever group rickettsiae in hard ticks, northern China. Transbound Emerg Dis. 2019; 66: 1587–1596. [DOI] [PubMed] [Google Scholar]
- 55.Janecek E, Streichan S, Strube C. SNP-based real-time pyrosequencing as a sensitive and specific tool for identification and differentiation of Rickettsia species in Ixodes ricinus ticks. BMC Infect Dis. 2012; 12: 261–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clark TR, Noriea NF, Bublitz DC, Ellison DW, Martens C, Lutter EI, et al. Comparative genome sequencing of Rickettsia rickettsii strains that differ in virulence. Infect Immun. 2015; 83: 1568–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walker DH, Ismail N. Emerging and re-emerging rickettsioses: endothelial cell infection and early disease events. Nat Rev Microbiol. 2008; 6: 375–386. [DOI] [PubMed] [Google Scholar]
- 58.Parola P, Rovery C, Rolain JM, Brouqui P, Davoust B, Raoult D. Rickettsia slovaca and R. raoultii in tick-borne rickettsioses. Emerg Infect Dis. 2009; 15: 1105–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu W, Li H, Lu QB, Cui N, Yang ZD, Hu JG, et al. Candidatus Rickettsia tarasevichiae infection in Eastern Central China: a case series. Ann Intern Med. 2016; 164: 641–648. [DOI] [PubMed] [Google Scholar]
- 60.Dantas-Torres F, Lia RP, Capelli G, Otranto D. Efficiency of flagging and dragging for tick collection. Exp Appl Acarol. 2013; 61:119–127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.