Highlights
-
•
Comprehensive evaluation of 2 SARS-CoV-2 antibody assays with excellent agreement.
-
•
Rigorously validated antibody tests are reliable to detect antibody kinetic change.
-
•
Three types of seroconversion were observed in hospitalized COVID-19 patients.
-
•
Convalescent sera show a wide range of antibody levels.
-
•
Current antibody testing is not useful in early screening for COVID-19.
Keywords: Coronavirus disease 19 (COVID-19), Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), Serology, Immunoassay
Abbreviations: COVID-19, corona virus disease-2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; ED, emergency department; EUA, Emergency Use Authorization; CEFA, cyclic enhanced fluorescence assay; MIA, microsphere immunoassay; RT-PCR, real-time reverse transcription polymerase chain reaction; SARS-CoV, severe acute respiratory syndrome coronavirus; MERS-CoV, Middle East respiratory syndrome coronavirus; ICU, intensive care unit. IV: Index value
Abstract
Background
In the ongoing COVID-19 pandemic, there is an urgent need for comprehensive performance evaluation and clinical utility assessment of serological assays to understand the immune response to SARS-CoV-2.
Methods
IgM/IgG and total antibodies against SARS-CoV-2 were measured by a cyclic enhanced fluorescence assay (CEFA) and a microsphere immunoassay (MIA), respectively. Independent performance evaluation included imprecision, reproducibility, specificity and cross-reactivity (CEFA n = 320, MIA n = 364). Clinical utility was evaluated by both methods in 87 patients at initial emergency department visit, 28 during subsequent hospitalizations (106 serial samples), and 145 convalescent patients. Totally 916 patients and 994 samples were evaluated.
Results
Agreement of CEFA and MIA was 90.4%-94.5% (Kappa: 0.81–0.89) in 302 samples. CEFA and MIA detected SARS-CoV-2 antibodies in 26.2% and 26.3%, respectively, of ED patients. Detection rates increased over time reaching 100% after 21 days post-symptom onset. Longitudinal antibody kinetic changes by CEFA and MIA measurements correlated well and exhibited three types of seroconversion. Convalescent sera showed a wide range of antibody levels.
Conclusion
Rigorously validated CEFA and MIA assays are reliable for detecting antibodies to SARS-CoV-2 and show promising clinical utility when evaluating immune response in hospitalized and convalescent patients, but are not useful for early screening at patient’s initial ED visit.
1. Introduction
The ongoing global pandemic of Coronavirus Disease-2019 (COVID-19) has rapidly spread with globally over 3.7 million confirmed cases and over 259,000 total deaths as of May 5, 2020 [1]. Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV-2), the cause of COVID-19, is highly contagious and can result in significant mortality among susceptible individuals with comorbidities. Acute symptoms and signs of SARS-CoV-2 infection are highly nonspecific and include fever, cough, fatigue, myalgia, and dyspnea with some patients progressing to pneumonia [2], [3], [4]. However some other individuals are asymptomatic carriers [5], [6], [7]. These characteristics of the disease create an urgent need to develop serological tests to identify asymptomatic silent infections, evaluate patient immune response, better predict disease progression and improve our understanding of the epidemiology, including transmission patterns, of SARS-CoV-2. Serological testing could also play an important role for de-isolation procedures [8] and implementation of convalescent plasma therapy for ill COVID-19 patients [9], [10].
Throughout the COVID pandemic, a wide variety of serological tests have entered the global market, including, but not limited to, colloidal gold immunochromatographic assays, magnetic chemiluminescent immunoassays, enzyme-linked immunosorbent assays (ELISA), and rapid test cassettes and dipsticks [4], [11], [12]. Due to the growing public health emergency and in an effort to facilitate rapid expansion of testing capacity, the United States Food and Drug Administration (FDA) issued a policy on mid-March 2020 [13] and a revised policy in early May [14], allowing for the development of COVID-19 diagnostic testing in the clinical health care and commercial settings through the Emergency Use Authorization (EUA) program. Over 100 manufacturers have notified the FDA that they are offering or plan to offer serological tests in the United States, but as yet only 12 assays have received EUA clearance [15]. Furthermore, there has been a lack of rigorous validation and performance evaluation of the available serological assays in COVID-19 negative and positive populations as well as a lack of thorough comparison between different serological testing platforms. Such data are urgently needed to evaluate the clinical utility and also the limitations of serological tests, as there has been significant controversy over the diagnostic and prognostic value of antibody testing. In addition, the potential role of serological antibody testing in epidemiological studies and in the accurate identification of convalescent plasma donors for COVID-19 patients is not known.
As a collaborative effort between Weill Cornell Medicine (WCM) and Wadsworth Center at the New York State Department of Health (NYS DOH), this study aimed to perform rigorous evaluation of two semi-quantitative SARC-CoV-2 serological tests [cyclic enhanced fluorescence assay (CEFA) and microsphere immunoassay (MIA, FDA EUA approved)] and characterize antibody responses in emergency department (ED), hospitalized and convalescent patients during the COVID-19 outbreak in New York City, the current epicenter in the US of the COVID-19 pandemic.
2. Materials and methods
2.1. Sources of specimen and data acquisition
This study was approved by the Institutional Review Board (#20-03021671) of Weill Cornell Medicine (site 1). The testing at Wadsworth Center at the New York State Department of Health (NYS DOH) (site 2) is waived for public health purposes.
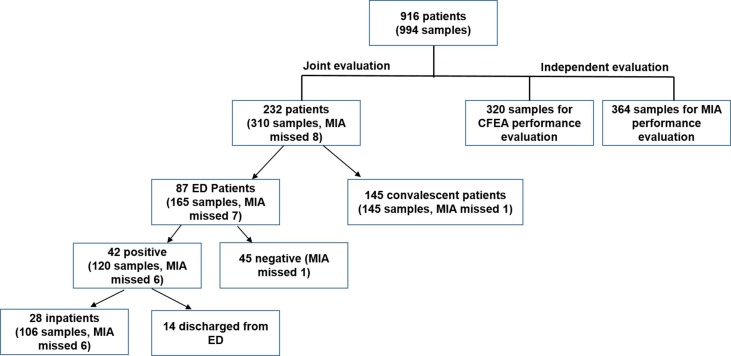
Different cohorts of patient serum samples were included in this study for evaluating analytical and clinical performance of the two assays. A chart of patients and samples used in this study is shown in Fig. 1 .
Fig. 1.
Chart of numbers of patients and samples used in the study.
2.2. Samples for testing assay specificity (independent cohorts)
Serum specimens (n = 320), collected from the pre-COVID 19 ED patients in July 2019, were tested to validate the specificity of the CEFA assay. Serum from 256 pre-COVID-19 healthy blood donors collected before 2019 were used to validate the specificity of the MIA assay.
2.3. Samples for testing assay cross-reactivity
Thirty sera from patients treated for recent non-COVID-19 respiratory infections (including 8 Coronavirus NL63, 2 Coronavirus 229E, and 2 Coronavirus OC43) and 78 serum samples from patients with antibodies to known microbial agents or with autoantigens were tested to validate the cross-reactivity of the MIA assay. Additional serum samples from patients who tested positive for one of the respiratory viruses in the Respiratory Pathogen PCR Panel (RPP) (including 2 Coronavirus NL63, 2 Coronavirus 229E, and 1 Coronavirus OC43) but negative for SARS-CoV-2 RT-PCR were analyzed by both CEFA (N = 16) and MIA (N = 15) methods (Supplemental table 1).
2.4. ED and hospitalized patients
A total 165 remnant serum samples from 87 patients presenting to site 1 ED from March 6 to April 4, 2020, displaying signs and symptoms suspicious for COVID-19, were evaluated. Among them, 42 patients were SARS-CoV-2 infected and 45 were uninfected patients, as determined by RT-PCR (Altona Diagnostics USA, Inc., Plain City, OH) at site 1. Twenty-eight COVID-19 positive patients had more than one remnant sample available for serial antibody testing. Patient demographics, medical history and care notes from the ED visit during which SARS-CoV-2 RT-PCR testing was performed and pertinent subsequent hospital admissions, noting admission to an intensive care unit (ICU) with or without intubation, were queried within the electronic medical record.
2.5. Convalescent patients
The convalescent serum samples were from 145 patients who tested positive by SARS-CoV-2 RT-PCR or had a COVID-19-like illness but had not undergone SARS-CoV-2 RT-PCR testing. Based on patients’ self-description, they had been symptom free for at least 14 days.
2.6. SARS-CoV-2 IgM and IgG cyclic enhanced fluorescence assay (CEFA)
The IgM and IgG antibodies against SARS-CoV-2 in serum were measured using the Pylon COVID-19 IgM and IgG assays on the Pylon 3D analyzer ([16], ET HealthCare, Palo Alto, CA). The antigen for the CEFA method is the S-Receptor Binding Domain (RBD) and recombinant Nucleocapsid Protein (NP). IgM and IgG were tested on different unitized test strips containing wells with pre-dispensed reagents. [Additional methodology described in supplemental material]. The assay is calibrated every month or when a new lot of unitized test strips and reagents are received. High and low QC materials provided by the manufacturer and positive and negative patient samples are run on a daily basis when patient samples are tested.
2.7. NYS DOH SARS-CoV microsphere immunoassay (MIA) assay
This assay has received US FDA EUA clearance. Specimens were assessed for the presence of total antibody using the SARS-CoV-2 MIA. Antigen for the MIA method is the recombinant SARS-CoV-2 NP. Analysis was performed using a Luminex 100 Analyzer (Luminex Corporation, Austin, TX. [Additional methodology described in supplemental material]. Positive and negative patient samples are run on a daily basis when patient samples are tested.
2.8. Cut off values
The cut off values of both methods were determined by mean of non-COVID-19 samples plus 6 Standard Deviation (SD). The index value (IV) was determined by the instrument readout of the test sample divided by instrument readout at cut off. An indeterminate IV was mean (of IgG for CEFA and total antibodies for MIA, respectively) plus 3SD divided by the instrument readout at cut off. Samples with an IV ≥ 1 and 1.78 were designated as positive for CEFA and MIA, respectively.
2.9. Imprecision and reproducibility
For the CEFA method, high and low levels of IgM and IgG QC, respectively, were run on 20 days. Mean, SD, CV were calculated. Six patient samples with negative, indeterminate, and positive (low, medium and high levels) IgM and IgG were run on five consecutive days. For the MIA method, mean, SD, CV were calculated for positive and negative QC which were run on 30 days.
2.10. SARS-CoV-2 RT-PCR and respiratory Pathogen PCR panel (RPP)
SARS-CoV-2 RT-PCR assay was performed using the RealStar SARS-CoV-2 RT-PCR kit 1.0 (Altona Diagnostics USA, Inc., Plain City, OH), which qualitatively detects SARS-CoV-2 RNA extracted from nasopharyngeal swab specimens. Respiratory Pathogen 2 PCR (RPP) was performed using the FilmArray Respiratory Pathogen 2 Panel (BioFire Diagnostics, LLC, Salt Lake City, UT) for the simultaneous qualitative detection and identification of multiple respiratory viruses and bacteria. [Additional information provided in supplemental material].
2.11. Statistical analysis
Data are presented as mean ± SD or median with interquartile range (IQR) for continuous variables, and proportion for categorical variables. Fisher’s exact test; t-test (equal variances) or Wilcoxon Rank-sum test were used for hypothesis testing. Ninety-five percent (95%) confidence interval (95%) of rates were calculated based on exact binomial distribution. The differences of antibody levels between groups were estimated and compared by Generalized Estimating Equations (GEE) model with log10-transformed raw fluorescence reading. p-values < 0.05 were considered significant. The degree of agreement between test methods was quantified by Kappa statistics. The dilution study was evaluated by linear regression. Analyses were performed in statistical software SAS Version 9.4 (SAS Institute, Cary, NC) or GraphPad Prism Version 8.4.1 (GraphPad Software, La Jolla, CA).
3. Results
3.1. ED and hospitalized patient demographics
The demographic and clinical characteristics of 42 COVID-19 RT-PCR positive patients and 45 negative patients are summarized in Table 1 . The demographics did not vary significantly between the COVID-19 positive and negative patients in terms of age, race/ethnicity or major comorbidities. Of those patients admitted to the hospital, none of the COVID-19 negative patients required ICU care or intubation. In comparison, 54.8% and 57.1% of the COVID-19 positive patients required ICU care or intubation, respectively.
Table 1.
ED and hospitalized patient Demographics, Negative COVID-19 versus Positive COVID-19 by RT-PCR.
| Demographic | Total (n = 87) | Negative (n = 45) | Positive (n = 42) | p-value |
|---|---|---|---|---|
| Mean Age, in years (SD) | 54.7 (18.56) | 53.0 (20.7) | 56.5 (16.0) | 0.3922 |
| Female/Male, n (%) | 30/57 (34.5/65.5) | 21/24 (46.7/53.3) | 9/33 (21.4/78.6) | 0.0232* |
| Comorbidities, any below, n (%) | 47 (55.3) | 23 (51.1) | 24 (64.9) | 0.2640 |
| Hypertension | 41 (47.1) | 20 (44.4) | 21 (50.0) | 0.6700 |
| Diabetes | 15 (17.2) | 5 (11.1) | 10 (23.8) | 0.1579 |
| Hyperlipidemia | 27 (31.0) | 11 (24.4) | 16 (38.1) | 0.1352 |
| CHF | 6 (6.9) | 4 (8.9) | 2 (4.8) | 0.6774 |
| CAD | 11 (22) | 4 (23.5) | 7 (21.2) | 1.0000 |
| Asthma | 11 (12.6) | 7 (15.6) | 4 (9.5) | 0.5237 |
| COPD | 7 (8.0) | 5 (11.1) | 2 (4.8) | 0.6774 |
| Race/Ethnicity, n (%) | ||||
| Caucasian/White | 43 (49.4) | 24 (53.3) | 19 (45.2) | 0.3842 |
| African- American/Black | 9 (11.0) | 4 (8.9) | 5 (13.5) | 0.7247 |
| Asian | 5 (6.1) | 4 (8.9) | 1 (2.7) | 0.6863 |
| Unknown/Declined | 15 (17.2) | 7 (15.6) | 78 (19.0) | 0.5707 |
| Other | 10 (11.5) | 3 (6.7) | 7 (16.7) | 0.2871 |
| Hispanic | 5 (5.7) | 3 (6.7) | 2 (4.8) | 0.6230 |
| Mean Emergency severity index (SD) | 2.9 (0.39) | 2.9 (0.29) | 2.8 (0.45) | 0.1635 |
| Fever at ED visit, n (%) | 39 (44.8) | 17 (37.8) | 22 (52.4) | 0.2375 |
| Mean Lymphocyte in ED, x103/mL (SD) | 2.62 (13.3) | 1.39 (0.75) | 3.93 (19.6) | 0.3766 |
| Mean Neutrophil in ED, x103/mL (SD) | 5.50 (4.24) | 5.21 (3.89) | 5.81 (4.62) | 0.5139 |
| Discharged Home from ED, n (%) | 21 (24.1) | 17 (37.8) | 4 (9.5) | 0.0025* |
| Mean, SpO2 % (SD) | 95.0 (14) | 97.0 (2) | 90.0 (19) | 0.0191* |
| Given O2 in ED, n (%) | 32 (36.8) | 7 (15.6) | 25 (59.5) | 0.0001* |
| ICU Admission, n (%) | 23 (26.4) | 0 (0.0) | 23 (54.8) | <0.0001* |
| Intubated, n (%) | 24 (27.6) | 0 (0.0) | 24 (57.1) | <0.0001* |
3.2. Performance of the CEFA and MIA methods
For the CEFA method, 20-day precision of IgM high QC (IV = 13.8), IgM low QC (IV = 0.3), IgG high QC (IV = 134.0), and IgG low QC (IV = 0.7) yielded CVs of 13.3%, 13.6%, 5.6% and 19.8%, respectively. In addition, antibody detectability remained the same for six patients on five consecutive days. Analysis from 320 pre-COVID-19 samples revealed specificity of 99.4% and 98.8% for IgM and IgG, respectively. Furthermore, one patient who (among the 16 patients positive for one of the respiratory viruses in the RPP panel) was human metapneumovirus positive, but negative for SARS-CoV-2 RNA, was found to have SARS-CoV-2 IgM and IgG antibodies (Supplemental table 1).
For the MIA method, 30-day precision of positive and negative QC was determined to be 7.7% and 14.9%, respectively. Within-assay reproducibility was assessed over a range of values and the CV ranged from 2.4 to 6% for the positive samples and 9.6% for the negative sample. Specificity, determined from 256 pre-COVID-19 healthy donors, was 99.2%. The specificity was 100% based on evaluation of 30 specimens from patients treated for recent non-COVID-19 respiratory infections, 78 serum specimens with antibodies to known microbial agents or autoantigens, and 15 serum samples from patients positive for one of the respiratory viruses in the RPP panel.
A total of 302 samples from ED, subsequently hospitalized and convalescent patients were compared between the two methods. The overall agreement was 94.5% if indeterminate results were excluded (Kappa = 0.89 [95%CI: 0.84–0.94]), and 90.4% (Kappa = 0.81 [95%CI: 0.74–0.87]), if indeterminate results were counted as negative, as shown in Table 2 .
Table 2.
Agreement between two immunoassay methods based on total antibody detection.
| Total # of samples with both Pylon and ELISA results:302 samples | ||||
|---|---|---|---|---|
| Site 2 Luminex MIA | ||||
| Site 1 Pylon CEFA |
Positive | Negative | Indeterminate | |
| Positive | 139 | 10 | 11 | |
| Negative | 5 | 119 | 5 | |
| Indeterminate | 3 | 9 | 1 | |
| Agreement between Pylon CEFA and Luminex MIA: 94.5% (excluding indeterminate results) (Kappa = 0.89, 95% CI: 0.84–0.94); 90.4% (indeterminate counted as negative) (Kappa = 0.81, 95% CI: 0.74–0.87). Kappa between 0.81 and 1.00: Almost perfect agreement. | ||||
3.3. Clinical utility assessment of the antibody testing
-
1.
Antibody testing in patients during their ED visit
CEFA and MIA had detection rates of 26.2% (95%CI: 15.3–41.1%) and 26.3% (95%CI: 15.0–42%), respectively (p > 0.05), when patients first presented to the ED and tested COVID-19 positive by RT-PCR (number of days since any symptom onset: mean = 6.5, 95%CI: 5.18–7.73).
-
2.
Longitudinal change of IgM and IgG in the hospitalized patients
We investigated 42 ED patients (120 samples by CEFA and 114 samples by MIA), in which 28 patients (106 samples by CEFA and 100 samples by MIA) were subsequently admitted. The detection rates of IgM, IgG, or both at different times after symptom onset are shown in Table 3 . Only 25% (CEFA, 95% CI: 4.4–59.1%) and 14.3% (MIA, 95% CI: 0.7–51.3%) of the samples in early phase (0–3 days after symptom onset) were antibody positive, however the detection rate increased to 69.0% (CEFA, 95% CI:53.8–80.9) and 65.0% (MIA, 95% CI: 49.5–77.9%) during days 8–14, 93.3% (95% CI:70.2–99.7%) during days 15–20, and finally reached 100% (CEFA 95% CI: 84.5–100.0%, MIA 95% CI: 83.2–100.0%) in samples that were obtained more than 21 days after symptom onset. The sensitivities of the two methods were not significantly different during any time frame.
Table 3.
Sensitivities of CEFA and MIA to detect IgM, IgG and total antibodies.
| Methods | Days after any symptom onset |
||||||
|---|---|---|---|---|---|---|---|
| 0–3 | 4–7 | 8–14 | 15–20 | 21–32 | Over 32 | ||
| Pylon CEFA | Number of samples* | 8 | 33 | 42 | 15 | 21 | 1 |
| IgM positive | 1 | 5 | 23 | 13 | 19 | 0 | |
| IgG positive | 2 | 6 | 28 | 13 | 21 | 1 | |
| IgG and IgM positive | 1 | 4 | 22 | 12 | 19 | 0 | |
| Total positive | 2 | 7 | 29 | 14 | 21 | 1 | |
|
Sensitivity % (95% CI) |
25.0 (4.4–59.1) |
21.2 (10.7–37.8) |
69.0 (53.8–80.9) |
93.3 (70.2–99.7) |
100.0 (84.5–100.0) |
100.0 (5.1–100.0) |
|
| Luminex MIA | Number of samples* | 7 | 32 | 40 | 15 | 19 | 1 |
| Total positive | 1 | 8 | 26 | 14 | 19 | 1 | |
|
Sensitivity % (95% CI) |
14.3 (0.7–51.3) |
25.0 (13.3–42.1) |
65.0 (49.5–77.9) |
93.3 (70.2–99.7) |
100.0 (83.2–100.0) |
100.0 (5.1–100.0) |
|
95%Cl was calculated by the hybrid Wilson/Brown method. Sensitivities were not significant different between the two methods at any time frame.
Total 42 SARS2-CoV RT-PCR positive patients, 28 of them had serial samples. Total 120 samples for CEFA and 114 samples for MIA.
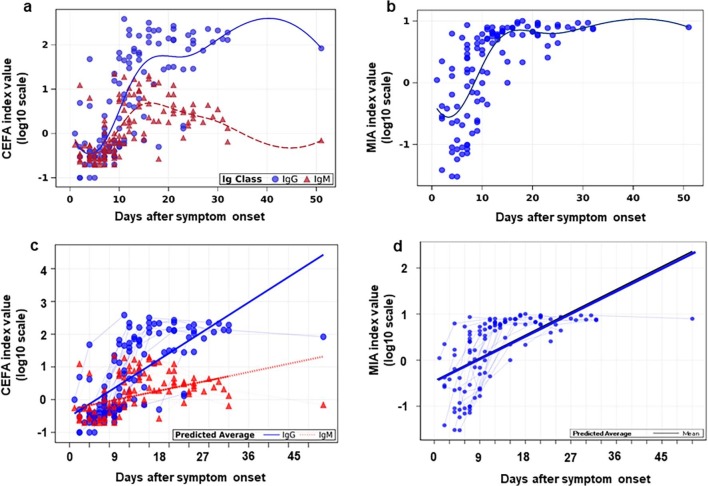
The plot of index values (log10 scale) and a simulated trending curve of IgM/IgG and total antibody are shown in Fig. 2 a and b, respectively. Overall, IgM (CEFA, Fig. 2a) reached peak levels between days 10 and 20 after symptom onset and declined afterwards. IgG (CEFA, Fig. 2a) and total antibody (MIA, Fig. 2b) reached a peak in a similar time frame as IgM but then plateaued. The comparison of the slope utilizing the GEE method revealed the rate of change of IgM IV (log10 scale) is significantly slower than that of IgG (p < 0.0001, Fig. 2c). The changing rate of total antibody is shown in Fig. 2d.
Fig. 2.
Antibody index values (log10 scale) of samples from 42 SARS2-CoV RT-PCR positive patients measured by CEFA and MIA methods over time. Each antibody level measured by CEFA (a) and total antibody level measured by MIA (b) was plotted over days after any symptom onset. Longitudinal samples from each individual patient measured by CEFA were connected by the solid (IgG, solid cirles) and dotted (IgM, solid triangles) lines measured by CEFA (c). Longitudinal samples from each individual patient measured by MIA were connected by the solid (total antibodies) line (d). Average line predicted by GEE method.
Longitudinal changes of normalized index value (log10 scale) measured by the CEFA assay (IgG and IgM) and the MIA method (total antibodies) were plotted in the 28 inpatients, including 18 patients on ventilators (Fig. 3 a) and 10 patients not on ventilators (Fig. 3b). Overall the changing trend of antibody levels correlated well between the CEFA and MIA measurements. All patients on ventilators and 5 patients not on ventilators had a significant increase in antibody levels during their disease course. Among them, the IgG and total antibody levels in 10 patients reached to a plateau around day 15–20 post-symptom onset. IgM levels in 5 patients started declining 16 – 21 days post-symptom onset. Three types of seroconversion were observed: synchronous seroconversion of IgG and IgM (17 patients), IgM seroconversion earlier than that of IgG (1 patient), and IgG seroconversion earlier than that of IgM (5 patients).
Fig. 3.
Antibody kinetic changes of SARS2-CoV RT-PCR positive patients who were on ventilators (a) and not on ventilators (b). Dashed black line is the normalized index values of positive cutoff. —□—CEFA IgG; —ο— CEFA IgM; —Δ—MIA total antibodies.
Five patients, discharged less than 14 days after admission, did not have detectable antibody levels. Thus, it is unclear if these patients had sufficient time to develop an antibody response.
Further analysis suggests that changes in IgM, IgG and total antibody levels over time are not significantly different between the ventilator and non-ventilator groups in our dataset (p = 0.30 for IgM, p = 0.84 for IgG, p = 0.1 for total antibody).
3.4. Antibody levels in convalescent patients
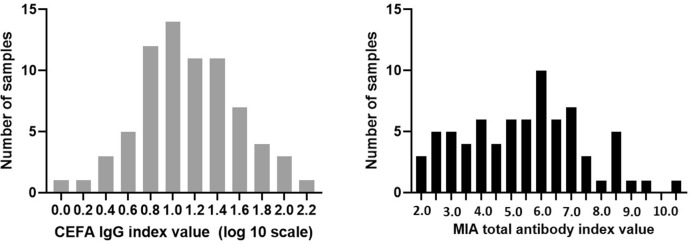
A total of 144 samples from convalescent patients who tested positive by SARS-CoV-2 RT-PCR or had COVID-19 exposure or symptoms were tested by both CEFA and MIA methods, which showed large variations of antibody levels in convalescent sera. The distribution of IgG index values and total antibody index values among 75 positive samples by both methods are shown in Fig. 4 a and b, respectively.
Fig. 4.
Distribution of positive antibody levels in convalescent serum measured by CEFA for IgG (log10 scale, 4a) and MIA for total antibody (4b).
4. Discussion
This study sheds light on the performance and potential clinical utility of two SARS-CoV-2 antibody testing methodologies during the COVID-19 outbreak in New York City. It also serves as an important example of a collaborative effort between an academic clinical laboratory and state government agency.
Overall, both CEFA and MIA assays demonstrated excellent analytical performance in their independent validations as well as concordance in ED, inpatient and convalescent samples. While the result reporting differs between the two assays, where one detects IgM and IgG separately and the other targets total antibody, we found that the detection rate at any given time period post-symptom onset did not differ significantly, with both assays demonstrating sensitivities ≤ 25% when tested < 7 days after symptoms began, reaching 100% > 21 days after symptom onset. These results are consistent with previous studies [17] evaluating recombinant immunofluorescence or chemiluminescent immunoassays [18], [19]. In contrast, RT-PCR testing demonstrates its highest level of sensitivity during the first week of symptoms, and then gradually declines in the next few weeks [20], [21]. We demonstrate that serological testing has limited diagnostic value in screening symptomatic patients during their first ED visit. As reporting of symptom onset in ED patients is subject to recall bias, a variation in time to antibody detection was observed, with most RT-PCR SARS-CoV-2 positive patients demonstrating detectable antibody levels 3–7 days post hospital admission. Therefore, if there is high clinical suspicion of COVID-19 in patients negative by RT-PCR, repeated or serial monitoring of serum antibodies may be helpful in confirming disease status and prudent to achieve optimal sensitivity in combination with RT-PCR.
Both methods demonstrated high specificity which is critical for evaluating hospitalized patients and understanding of the epidemiology. Neither CEFA nor MIA showed the cross-reactivity in the samples from other coronavirus infected patients. The limitation was the relatively small sample size due to the limited access, and future larger studies are warranted to further examine the assay cross-reactivity. In addition, while the CEFA and MIA assays exhibited overall excellent agreement with Kappa > 0.81, a small proportion of samples disagreed. A possible explanation for the disagreement was the difference in the underlying principle of the assays. The antigens for the CEFA method is the S protein RBD and NP targeting IgM and IgG in separate assays. The antigen for the MIA method is the NP targeting total antibodies. Currently, it is unknown which assay is more correlated with protected role of antibody, which warrant future studies.
The kinetic change of antibody levels was consistent between the two methodologies. All mechanically ventilated patients displayed an increase in IgM and IgG levels during the course of their hospitalization. Three types of seroconversion were observed. Interesting, 74% patients in our study showed synchronous seroconversion, 5 showed IgG prior to IgM, whereas only 1 out of 23 patients had IgM seroconversion earlier than IgG. These patterns of seroconversion are similar to a previous report showing that the majority of patients (73.1%) had either synchronous seroconversion or IgG seroconversion earlier than IgM [22]. However, due to the nature of this study, serum specimens of most patients could not be collected on a daily basis, at the later stages of illness or post-recovery. Therefore, a complete dynamic pattern of antibody levels could not be studied and thus, the full time course of the antibody response remains unknown in these patients. A more thorough investigation is needed to monitor the kinetic change of antibody levels over an extended period of time, including convalescence.
Prior studies reported most individuals infected with SARS-CoV-1 mounted an antibody response [23], [24], [25] whereas MERS-CoV infected patients with mild or asymptomatic infections exhibited varied immune responses, which at times were undetectable by antibody assays [26], [27]. While there is growing data on the antibody response to SARS-CoV-2 infection [18], [28], [29], the level and duration of the anti-SARS-CoV-2 antibodies in asymptomatic and mildly symptomatic COVID-19 patients is still uncertain. In the present dataset, all 18 patients in the mechanically ventilated group seroconverted. This is in contrast to 5 (CEFA) and 4 (MIA) out of 10 patients in the non-ventilated group who had undetectable IgM and IgG levels during their hospital stay. Four patients were discharged < 10 days post-symptom onset and 1 patient had undetectable antibodies when discharged on day 13 after symptom onset. Thus, this retrospective study cannot provide additional data on these patients. However, the perceived suboptimal antibody responses is likely due to short hospitalizations resulting in the lack of specimen for analysis during the intermediate and late stages of the disease. More specimens from mildly ill patients will need to be collected for longer time periods in order to better understand patterns of SARS-CoV-2 immune response. This information is essential to discussions regarding reentry into the workforce by asymptomatic individuals post-infection.
It has been suggested that worse outcomes and increased disease severity with COVID-19 may correspond with increased SARS-CoV-2 IgG levels or a higher titer of SARS-CoV-2 antibodies, when compared to those with less severe disease [20], [30], [31]. Although the current study found that the positivity rate of antibodies increased with time after symptom onset for both methods, there was no statistical difference in antibody levels based on ventilation status. Assay variation may explain the lack of correlation between antibody levels and severity of disease, as our methods currently are validated only for qualitative SARS-CoV-2 antibody analysis and its semi-quantitative use may not give the analytical resolution needed for such studies. Bias may also exist in that patients with milder symptoms did not present to the hospital and thus, there is no comparison between this missing cohort and the cohort of hospitalized patients which displayed more severe symptoms. Functional assays, such as neutralizing antibody assays, have been proposed to evaluate the efficacy of vaccinations and due to the complicated interplay between virus and the immunologic host response [32], neutralizing antibody assays [30] may be better suited for such correlation studies. Furthermore, this study did not extensively follow longitudinally patient antibody levels and consequently, the study was likely underpowered in its ability to see this secondary outcome measure. Additional independent large-cohort studies would be needed to further study these previously described findings.
Both the FDA and CDC stress the importance of serological testing for the detection of prior infection in asymptomatic individuals and those presenting late in illness as well as identifying individuals who have mounted an immune response to SARS-CoV-2 [33], [34]. Our data suggest that there is a wide range of antibody levels in convalescent serum. Of note, the distribution of antibody positivity in convalescent sera examined in this study cannot be used to determine antibody prevalence and levels in the general population, as these were random samples from patients recovered from COVID-19 and COVID-19-like illnesses. Cases have been reported that convalescent plasma therapy improved the clinical outcomes by neutralizing viremia in severe COVID-19 cases [10], [35]. Further studies are needed to investigate whether IgG or total antibody levels correlate with neutralizing antibody levels, thereby identifying potential therapeutic serum donors.
In summary, we performed a thorough analytical validation and clinical evaluation of two semi-quantitative SARS-CoV-2 immunoassays. Our results demonstrated the clinical utilities of serological testing in evaluating inpatients and convalescent patients. The sensitivity of the current immunoassays are not suitable for early-stage screening. Future studies are needed to investigate the immune response in asymptomatic and mildly ill patient population, and to understand the correlation between total antibody and neutralizing antibody levels.
CRediT authorship contribution statement
He S. Yang: Conceptualization, Methodology, Validation, Investigation, Writing - review & editing, Data curation, Visualization. Sabrina E. Racine-Brzostek: Conceptualization, Methodology, Investigation, Writing - review & editing, Data curation. William T. Lee: Validation, Formal analysis, Data curation. Danielle Hunt: Validation, Formal analysis, Data curation. Jim Yee: Investigation, Software. Zhengming Chen: Formal analysis. Jeffrey Kubiak: Investigation. Miguel Cantu: Investigation. Layla Hatem: Investigation. Elaine Zhong: Investigation. Danielle D'Ambrosio: Investigation. Amy Chadburn: Writing - review & editing. Lars Westblade: Investigation. Marshall Glesby: Writing - review & editing. Massimo Loda: Supervision, Writing - review & editing. Melissa M. Cushing: Supervision, Writing - review & editing. Zhen Zhao: Conceptualization, Methodology, Validation, Investigation, Writing - review & editing, Data curation, Visualization, Supervision.
Declaration of Competing Interest
ZZ received seed instruments and sponsored travel from ET Healthcare.
The manufacturers did not review the article and had no input on data analysis prior to the manuscript submission.
Acknowledgments
Acknowledgement:
The authors thank the contribution of the following scientists from Wadsworth Center, Diagnostic Immunology Laboratory New York State Department of Health: K. McDonough, K. Kulas, R. Bievenue, S. Bush, K. Carson, V. Demarest, A. Furuya, K. Howard, M. Marchewka, R. Stone. We thank Dr. Fred Apple from University of Minnesota and Dr Alan Wu from University of California San Francisco for their help with manuscript review.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cca.2020.06.004.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20(5):533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai X., Chen J., Hu J., Long Q., Deng H., Fan K. A Peptide-based Magnetic Chemiluminescence Enzyme Immunoassay for Serological Diagnosis of Corona Virus Disease 2019 (COVID-19) J. Infect. Dis. 2020;May 8:jiaa243. doi: 10.1093/infdis/jiaa243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu X., Yang R. COVID-19 transmission through asymptomatic carriers is a challenge to containment. Influenza Other Respir. Viruses. 2020 doi: 10.1111/irv.12743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang A., Tong Z.D., Wang H.L., Dai Y.X., Li K.F., Liu J.N. Detection of Novel Coronavirus by RT-PCR in Stool Specimen from Asymptomatic Child. China. Emerg. Infect. Dis. 2020;26(6):1337–1339. doi: 10.3201/eid2606.200301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu J., Ji P., Pang J., Zhong Z., Li H., He C. Clinical characteristics of 3,062 COVID-19 patients: a meta-analysis. J. Med. Virol. 2020;Apr 15 doi: 10.1002/jmv.25884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng M.P., Papenburg J., Desjardins M., Kanjilal S., Quach C., Libman M. Diagnostic Testing for Severe Acute Respiratory Syndrome-Related Coronavirus- 2: A Narrative Review. Ann. Intern. Med. 2020;Apr 13:M20–1301. doi: 10.7326/M20-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bloch E.M., Shoham S., Casadevall A., Sachais B.S., Shaz B., Winters J.L. Deployment of convalescent plasma for the prevention and treatment of COVID-19. J. Clin. Invest. 2020;Apr 7:138745. doi: 10.1172/JCI138745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duan K., Liu B., Li C., Zhang H., Yu T., Qu J. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc. Natl. Acad. Sci. U S A. 2020;117(17):9490–9496. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.J. Xiang, M. Yan, H. Li, T. Liu, C. Lin, S. Huang, et al. Evaluation of Enzyme-Linked Immunoassay and Colloidal Gold- Immunochromatographic Assay Kit for Detection of Novel Coronavirus (SARS-Cov-2) Causing an Outbreak of Pneumonia (COVID-19). medRxiv 2020:2020.02.27.20028787.
- 12.Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C. Diagnosing COVID-19: The Disease and Tools for Detection. ACS Nano. 2020;14(4):3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 13.Food and Drug Administration, Policy for Diagnostic Tests for Coronavirus Disease-2019 during the Public Health Emergency, Rockville, MD, 2020.
- 14.Administration FaD, Policy for Coronavirus Disease-2019 Tests During the Public Health Emergency, 2020.
- 15.Administration FaD, Emergency Use Authorizations [Internet], FDA, 2020.
- 16.Fred S. Apple A.H.B.W., Sandoval Yader, Sexter Anne, Love Sara A., Myers Gary, Schulz Karen, Duh Show-Hong, Christenson Robert H. Sex-specific 99th percentile upper reference limits for high sensitivity cardiac troponin assays derived using a universal sample bank. Clin. Chem. 2020;66(3):434–444. doi: 10.1093/clinchem/hvz029. [DOI] [PubMed] [Google Scholar]
- 17.Guo L., Ren L., Yang S., Xiao M., Chang Yang F, Early Profiling. Humoral Response to Diagnose Novel Coronavirus Disease (COVID-19) Clin. Infect. Dis. 2020;Mar 21:ciaa310. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M.A. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;Apr 1 doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 19.Jin Y., Wang M., Zuo Z., Fan C., Ye F., Cai Z. Diagnostic value and dynamic variance of serum antibody in coronavirus disease 2019. Int. J. Infect. Dis. 2020;94:49–52. doi: 10.1016/j.ijid.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin. Infect. Dis. 2020;Mar 28:ciaa344. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sethuraman N., Jeremiah S.S., Ryo A. Interpreting Diagnostic Tests for SARS-CoV-2. JAMA. 2020;May 6 doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- 22.Long Q.X., Liu B.Z., Deng H.J., Wu G.C., Deng K., Chen Y.K. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;Apr 29 doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 23.Chan P.K., Ng K.C., Chan R.C., Lam R.K., Chow V.C., Hui M. Immunofluorescence assay for serologic diagnosis of SARS. Emerg. Infect. Dis. 2004;10:530–532. doi: 10.3201/eid1003.030493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang S.C., Wang J.T., Huang L.M., Chen Y.C., Fang C.T., Sheng W.H. Longitudinal analysis of Severe Acute Respiratory Syndrome (SARS) coronavirus-specific antibody in SARS patients. Clin. Diagn. Lab. Immunol. 2005;12:1455–1457. doi: 10.1128/CDLI.12.12.1455-1457.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 26.Okba N.M.A., Raj V.S., Widjaja I., GeurtsvanKessel C.H., de Bruin E., Chandler F.D. Sensitive and Specific Detection of Low-Level Antibody Responses in Mild Middle East Respiratory Syndrome Coronavirus Infections. Emerg. Infect. Dis. 2019;25:1868–1877. doi: 10.3201/eid2510.190051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Al Kahlout R.A., Nasrallah G.K., Farag E.A., Wang L., Lattwein E., Muller M.A. Comparative Serological Study for the Prevalence of Anti-MERS Coronavirus Antibodies in High- and Low-Risk Groups in Qatar. J. Immunol. Res. 2019;2019:1386740. doi: 10.1155/2019/1386740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haveri A., Smura T., Kuivanen S., Osterlund P., Hepojoki J., Ikonen N. Serological and molecular findings during SARS-CoV-2 infection: the first case study in Finland, January to February 2020. Euro. Surveill. 2020;25(11):2000266. doi: 10.2807/1560-7917.ES.2020.25.11.2000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.F. Wu, A. Wang, M. Liu, Q. Wang, J. Chen, S. Xia, et al., Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. medRxiv 2020:2020.03.30.20047365.
- 31.B. Zhang, X. Zhou, C. Zhu, F. Feng, Y. Qiu, J. Feng, et al., Immune phenotyping based on neutrophil-to-lymphocyte ratio and IgG predicts disease severity and outcome for patients with COVID-19. medRxiv 2020:2020.03.12.20035048. [DOI] [PMC free article] [PubMed]
- 32.Madore D.V., Meade B.D., Rubin F., Deal C., Lynn F., Meeting C. Utilization of serologic assays to support efficacy of vaccines in nonclinical and clinical trials: meeting at the crossroads. Vaccine. 2010;28:4539–4547. doi: 10.1016/j.vaccine.2010.04.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coronavirus (COVID-19) Update: Serological Tests. Available at: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-serological-tests. Accesed: Apr 7 2020.
- 34.Serology Testing for COVID-19. Available at: https://www.cdc.gov/coronavirus/2019-ncov/lab/serology-testing.html. Accesed: Apr 4 2020.
- 35.Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J. Treatment of 5 Critically Ill Patients With COVID-19 With Convalescent Plasma. JAMA. 2020;27(323(16)):1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.