Abstract
Purpose
The aim of this study was to compare a recently established whole brain MR spectroscopic imaging (wbMRSI) technique using spin-echo planar spectroscopic imaging (EPSI) acquisition and the Metabolic Imaging and Data Analysis System (MIDAS) software package with single voxel spectroscopy (SVS) technique and LCModel analysis for determination of relative metabolite concentrations in aging human brain.
Methods
A total of 59 healthy subjects aged 20–70 years (n ≥ 5 per age decade for each gender) underwent a wbEPSI scan and 3 SVS scans of a 4ml voxel volume located in the right basal ganglia, occipital grey matter and parietal white matter. Concentration ratios to total creatine (tCr) for N-acetylaspartate (NAA/tCr), total choline (tCho/tCr), glutamine (Gln/tCr), glutamate (Glu/tCr) and myoinositol (mI/tCr) were obtained both from EPSI and SVS acquisitions with either LCModel or MIDAS. In addition, an aqueous phantom containing known metabolite concentrations was also measured.
Results
Metabolite concentrations obtained with wbMRSI and SVS were comparable and consistent with those reported previously. Decreases of NAA/tCr and increases of line width with age were found with both techniques, while the results obtained from EPSI acquisition revealed generally narrower line widths and smaller Cramer-Rao lower bounds than those from SVS data.
Conclusion
The wbMRSI could be used to estimate metabolites in vivo and in vitro with the same reliability as using SVS, with the main advantage being the ability to determine metabolite concentrations in multiple brain structure simultaneously in vivo. It is expected to be widely used in clinical diagnostics and neuroscience.
Keywords: Spin-echo planar spectroscopic imaging, Metabolic Imaging and Data Analysis System, Aging
Introduction
Magnetic resonance spectroscopy (MRS) is often used as a complementary measurement to magnetic resonance imaging (MRI) for clinical studies to obtain information about the metabolic status of brain tissue. Cerebral metabolites measured by MRS, such as N-acetylaspartate (NAA), total choline (tCho), total creatine (tCr), composite (Glx) of glutamine (Gln) and glutamate (Glu), and myoinositol (MI) provide information about metabolic alterations in the brain associated with physiological or pathological processes [1–3]. The most commonly used spectroscopic technique is single voxel spectroscopy (SVS) that measures metabolites in a user-defined voxel with a typical size of 4–8ml [4]. Advantages of SVS include the relative ease of implementation, ability to achieve good field homogeneity and the availability of different software for quantification of the metabolite concentrations [5, 6]. The LCModel is the most popular software used to analyze SVS data [6]. A clear disadvantage of SVS is the limited spatial coverage, which restricts its application. An alternative acquisition method is whole brain MR spectroscopic imaging (wbMRSI) that on average enables evaluation of brain metabolites over approximately 70% of the brain volume [7, 8], thereby enabling sampling of multiple brain regions simultaneously with sufficient spatial resolution in a clinical acceptable scan time [9–11]. Good reproducibility of both SVS and wbMRSI has been reported in a study on 10 volunteers by Zhang et al. [12]. A further detailed comparison of wbMRSI and SVS, as well as the methods used for evaluation of brain metabolites in human has not yet been reported, which would provide basic reference information for choosing the appropriate MRS technique in future studies. Therefore, in this study, the data acquisition methods of wbMRSI and SVS as well as the spectral analysis methods using MIDAS [13, 14] and the LCModel [6] were compared, based on the measurements of brain metabolite contents in healthy aging humans as well as in an aqueous phantom.
Methods
Subjects
Healthy volunteers who had no history of brain trauma, neurological disorders or other systemic diseases according to a self report were recruited from the local community. Efforts were made to obtain a population sample with an even age distribution for this prospective cross-sectional study. To exclude potential cognitive or depressive impairments each volunteer underwent two screening tests prior to the MR examination, the Beck Depression Inventory (BDI-II) [15] and the DemTect screening test [16]. Subjects with abnormal results of screening tests (n = 1), incomplete MR examinations (n = 3) or morphological alterations of the brain (n = 2) were excluded. Finally, 59 subjects aged between 20 and 70 years (29 males and 30 females, mean age 44 ± 14 years, n ≥ 5 per age decade for each gender) with a body mass index (BMI) less than 30 were included. This study was approved by the local institution review board and conducted according to the principles in the Declaration of Helsinki. Written consent was obtained from each subject before the examinations. A 2.7l aqueous phantom (model 2152220, General Electric Medical, Milwaukee, WI, USA) was also studied, which contained 12.5mmol/l NAA, 3mmol/l Cho, 10mmol/l Cr, 12.5mmol/l Glu, 7.5mmol/l MI, and 5mmol/l lactate that simulate metabolite concentrations in the human brain.
MR Examination
All subjects underwent MR examinations at 3T (Magnetom Verio, Siemens Medical Solutions, Erlangen, Germany) with a 12-channel phased-array head coil. Considering the future clinical applications of the wbMRSI each MR examination was conducted to keep the scan time as short as possible. The scan protocol included a three-dimensional T1-weighted magnetization prepared rapid gradient echo (MPRAGE) acquisition (1mm isotropic resolution, acquisition time of 5min); a first version of spin-echo planar spectroscopic imaging (EPSI) sequence (TR/TE= 1550/17.6ms, 50× 50 voxels in-plane and 18 slices, over a field-of-view of 280× 280× 180mm3, flip angle 71° and acquisition time of 16min). This included acquisition of a second dataset without water suppression (flip angle of 10°) that was used for different processing functions and providing internal water as a reference for the normalization of the metabolite concentrations, as reported previously in detail [11]. In the same session, 3 SVS scans (stimulated echo acquisition mode [STEAM], TR/TE= 1550/20ms, 192 acquisitions, voxel volume 4ml, scan time 5min each) in 3 regions of interest (ROI), i.e. the right basal ganglia (BG), occipital grey matter (oGM) and parietal white matter (pWM), were carried out. Each SVS scan also included a second acquisition of spectral data of nonsuppressed water with 4 excitations. For SVS an automatic shimming was used. For wbMRSI a manual shimming until a line width less than 30Hz was used. The same protocol was applied to the aqueous phantom, except that only one SVS was carried out at a ROI located in the phantom center.
Data Processing
All SVS data without filtering or other pre-processing were analyzed by using LCModel (named as SVS-LCM method) to estimate metabolite concentrations, which analyzes each MR spectrum as a linear combination of model spectra of metabolite solutions in vitro and was provided by Dr. Provencher (http://s-provencher.com/lcmodel.shtml). The water-unsuppressed data were used for eddy current correction and for calculating a water reference signal that was used to estimate metabolite concentration by spectral analysis.
The EPSI data were processed using the MIDAS software package to obtain volumetric metabolite maps, which included data resampling, spatial reconstruction, B0 correction, spatial registration, brain and scalp mask formation for lipid k-space extrapolation [17], spectral fitting, and signal normalization [13, 14]. Spectral data were reconstructed into 64 × 64 × 32 voxels with a basic interpolated voxel volume of 0.107ml. Light spectral apodization of 2Hz was applied to the wbMRSI data. The final spectral data were corrected for phase and B0 variations using values determined from the spectral analysis. The processing also included calculation of the fractional tissue volume contributing to each MRSI voxel, by applying a tissue segmentation procedure [18, 19] to the T1-weighted MPRAGE data to map gray matter (GM), white matter (WM), and cerebrospinal fluid (CSF), followed by a resampling and convolution by the MRSI spatial response function to coincide with the MRSI voxel volume and location. The locations of the three ROIs were obtained by extracting the SVS voxel locations from the raw data headers from the SVS acquisitions, which were then mapped to the wbMRSI dataset in subject space using the map integrated spectrum (MINT) module of the MIDAS software [20]. To estimate brain metabolite concentrations at the three ROIs from the wbMRSI data, the individual basic voxel spectra within each of the SVS ROIs were integrated. Prior to integration, spectra of the basic voxels were excluded if they had a spectral line width larger than 12Hz. The resultant spectra of with and without water-suppression were then analyzed in two different ways: a) using the FITT program included in MIDAS, in which a default Gaussian line shape was used for spectral fitting (named as wbMRSI method) and b) using LCModel (named as hybrid method).
Concentrations of each metabolite were estimated by all three methods either as a ratio to water signal and presented in institutional units (i.u.) or as a ratio to tCr. Correction for CSF volume contribution was applied as Met’= Met /(1 − fcsf), where Met is the uncorrected metabolite value and fcsf is the fractional volume of CSF in each SVS ROI, which was also calculated by the MINT program. As quality criteria metabolite values with a spectral line width larger than 14Hz, or a Cramer-Rao lower bound (CRLB) >20% for NAA, tCho and tCr and a CRLB >30% for MI, Glu, and Gln, were not considered for further analysis.
Statistical Analysis
The two-sided t-tests with Bonferroni corrected significance level (α= 0.05 /7= 0.007) were performed to estimate gender differences of the 6 metabolites and the spectral line widths in subjects at each ROI. For metabolite concentrations, spectral line widths and CRLB of each ROI derived with SVS-LCM, wbMRSI, and hybrid methods, respectively, mean values, standard deviations or coefficient of variance (COV, derived by mean over standard deviation) were calculated by averaging the corresponding values over the subjects. Repeated measures analysis of variance (rA-NOVA) with Bonferroni post hoc test was used to compare the metabolite values of each ROI measured with the three methods, where the values determined as a ratio to internal water were not considered, because the internal water reference signals were different from each other depending on the method used. Linear regression analysis with uncorrected significance level (α = 0.05) was used to estimate age dependence of brain regional metabolite concentrations obtained both as a ratio to tCr and to internal water obtained with each method. Statistical analyses were performed with SPSS, Version 24 (IBM, Armonk, NY, USA) and with Origin (OriginLab, Northampton, MA, USA) for creating graphics.
Results
Phantom Measurements
The MR spectra and the position of selected ROI in T1-weighted image of the aqueous phantom are shown in Fig. 1. The spectrum of the SVS acquisition and the LCModel fit curve of the ROI are shown as empty squares (Fig. 1a). The integrated spectrum of wbMRSI data over the same ROI volume are shown as filled squares, which was mapped by extracting the SVS voxel locations from the raw data headers from the SVS acquisition (Fig. 1b) and the raw data of the same integrated spectrum and the LCModel fit curve (Fig. 1c). The metabolite concentrations and corresponding line widths measured with SVS-LCM, wbMRSI and hybrid methods, as well as the known concentrations of the phantom are shown in Table 1. Since none of the spectral analysis methods are normalized to result in quantitative, i.e. molar concentrations, all results determined as a ratio to the water signal differed clearly from the known values. In contrast, the concentrations determined as a ratio to tCr were all close to the known values, with slight overestimation or underestimation. These observations showed that the water-referencing procedures with SVS-LCM, wbMRSI and hybrid methods were quite different. The estimated spectral line width was narrower for both of the wbMRSI integrated methods than that obtained for the SVS-LCM method. The CRLB was the lowest by wbMRSI (≤1%), the next by hybrid (1% ≤ CRLB 5%) and highest by SVS-LCM (2% ≤ CRLB ≤ 14%).
Fig. 1.
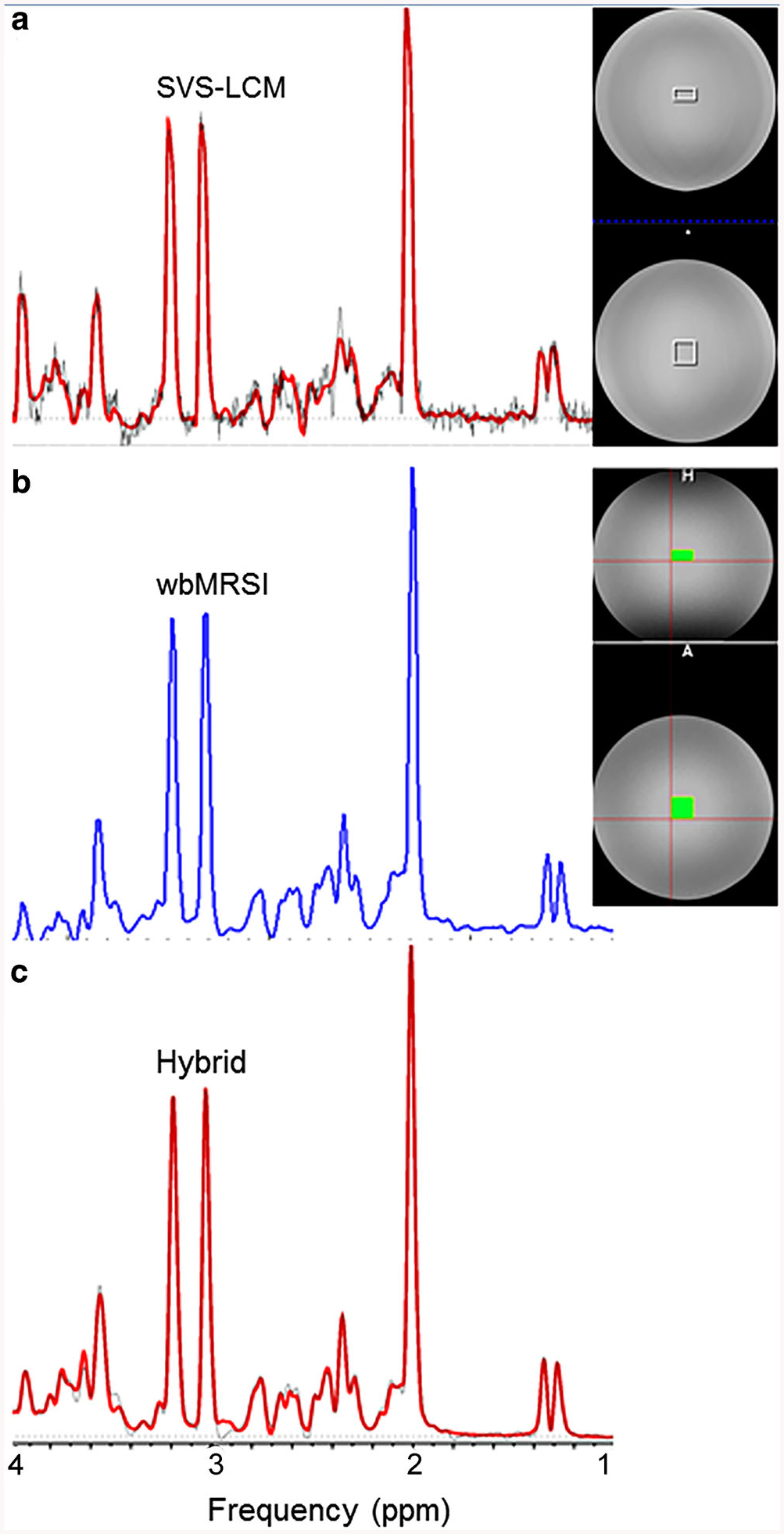
MR spectra and the regions of interest (ROIs) used, shown on the T1-weighted image of the aqueous phantom. a The spectrum of a single voxel spectroscopy (SVS) acquisition and the LCModel fit curve of the ROI are shown as empty squares (SVS-LCM method). b The integrated spectrum from the whole brain MR spectroscopic imaging (wbMRSI) data from the same ROI volume used for the SVS voxel (wbMRSI). c The integrated spectrum and the LCModel fit (hybrid)
Table 1.
Metabolite concentrations and spectral line widths of the aqueous phantom as well as the Cramer-Rao lower bound for spectral fits derived with three different methods as indicated
| Methoda | Metabolite concentrationb | Line width (Hz) | |||||
|---|---|---|---|---|---|---|---|
| NAA | Cho | Cr | Glu | MI | Lactate | ||
| In relation to water signal (i. u.) | |||||||
| SVS-LCM | 5.40 | 1.66 | 4.35 | 5.88 | 3.07 | 1.65 | 5.30 |
| wbMRSI | 5.57 | 1.36 | 4.36 | 5.38 | 2.64 | 2.15 | 4.73 |
| Hybrid | 26.10 | 7.31 | 20.60 | 27.70 | 17.00 | 10.20 | 4.32 |
| Known concentration (mM) | 12.5 | 3.0 | 10.0 | 12.5 | 7.5 | 5.0 | – |
| In relation to Cr | |||||||
| SVS-LCM | 1.24 | 0.38 | 1.00 | 1.35 | 0.71 | 0.38 | – |
| wbMRSI | 1.28 | 0.31 | 1.00 | 1.23 | 0.60 | 0.49 | – |
| Hybrid | 1.27 | 0.36 | 1.00 | 1.35 | 0.83 | 0.50 | – |
| Known concentration (mM) | 1.25 | 0.30 | 1.00 | 1.25 | 0.75 | 0.50 | – |
| Cramer-Rao lower bound | |||||||
| SVS-LCM | 2.0 | 3.0 | 3.0 | 5.0 | 4.0 | 14.0 | – |
| wbMRSI | 0.5 | 0.7 | 0.5 | 0.9 | 1.0 | 1.3 | – |
| Hybrid | 1.0 | 1.0 | 1.0 | 2.0 | 2.0 | 5.0 | – |
SVS-LCM: spectral data were scanned with single voxel spectroscopy (SVS) and analyzed with LCModel whole brain MR spectroscopic imaging (wbMRSI): data were scanned with spin-echo planar spectroscopic imaging (EPSI) and analyzed with Metabolic Imaging and Data Analysis System (MIDAS). Hybrid: data were scanned with EPSI and analyzed with LCModel
None of the spectral analysis methods were normalized to result in molar concentrations. All results determined as ratio to the water signal in institutional units (i.u.) or to total creatine (tCr)
In Vivo Measurements
For measurements on volunteers, metabolite values estimated as a ratio to tCr were reported for direct comparison. Fig. 2 shows the ROI locations of SVS scans in basal ganglia, pWM and oGM overlaid on the T1-weighted images (32 years old, female) (Fig. 2a, empty squares), and the same ROIs for wbMRSI measurements that were mapped by extracting the SVS voxel locations from the raw data headers from the SVS acquisitions (Fig. 2b, filled squares). The corresponding spectra of the ROIs are shown in Fig. 3. The mean number of basic voxels excluded by obtaining integrated spectra from wbMRSI data (due to spectral line width larger than 12Hz) in each ROI was very small in pWM (1% of 49 ± 4 voxels) and oGM (2% of 47 ± 4 voxels), and slightly larger in BG (18% of 51 ± 4 voxels).
Fig. 2.
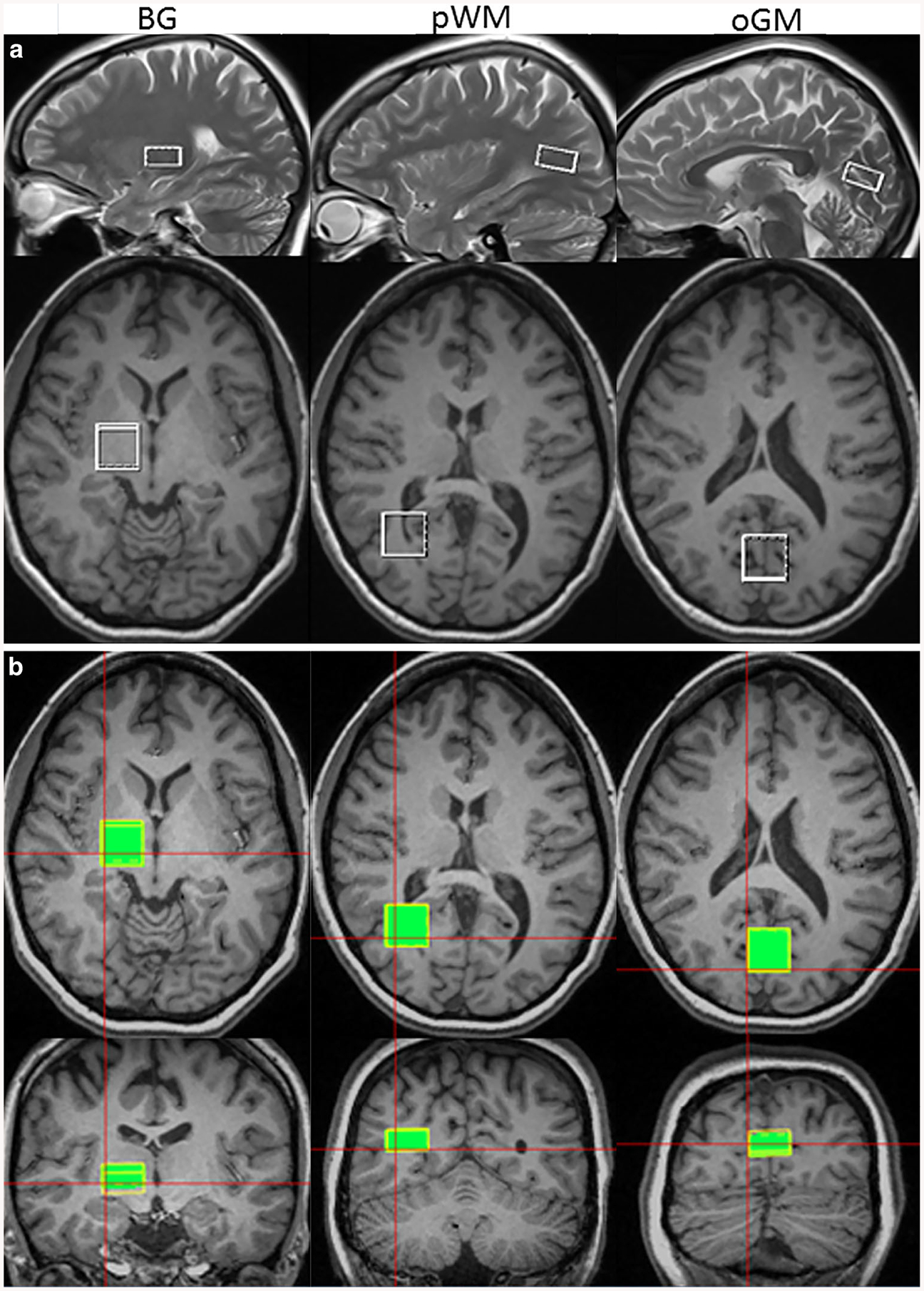
Locations of the ROIs in basal ganglia (BG), parietal white matter (pWM) and occipital grey matter (oGM) drawn on T1-weighted images in axial (2nd and 3rd rows), sagittal (upper row), and coronar (lower row) sections, shown for a the SVS acquisition and b the integrated wbMRSI measurement
Fig. 3.
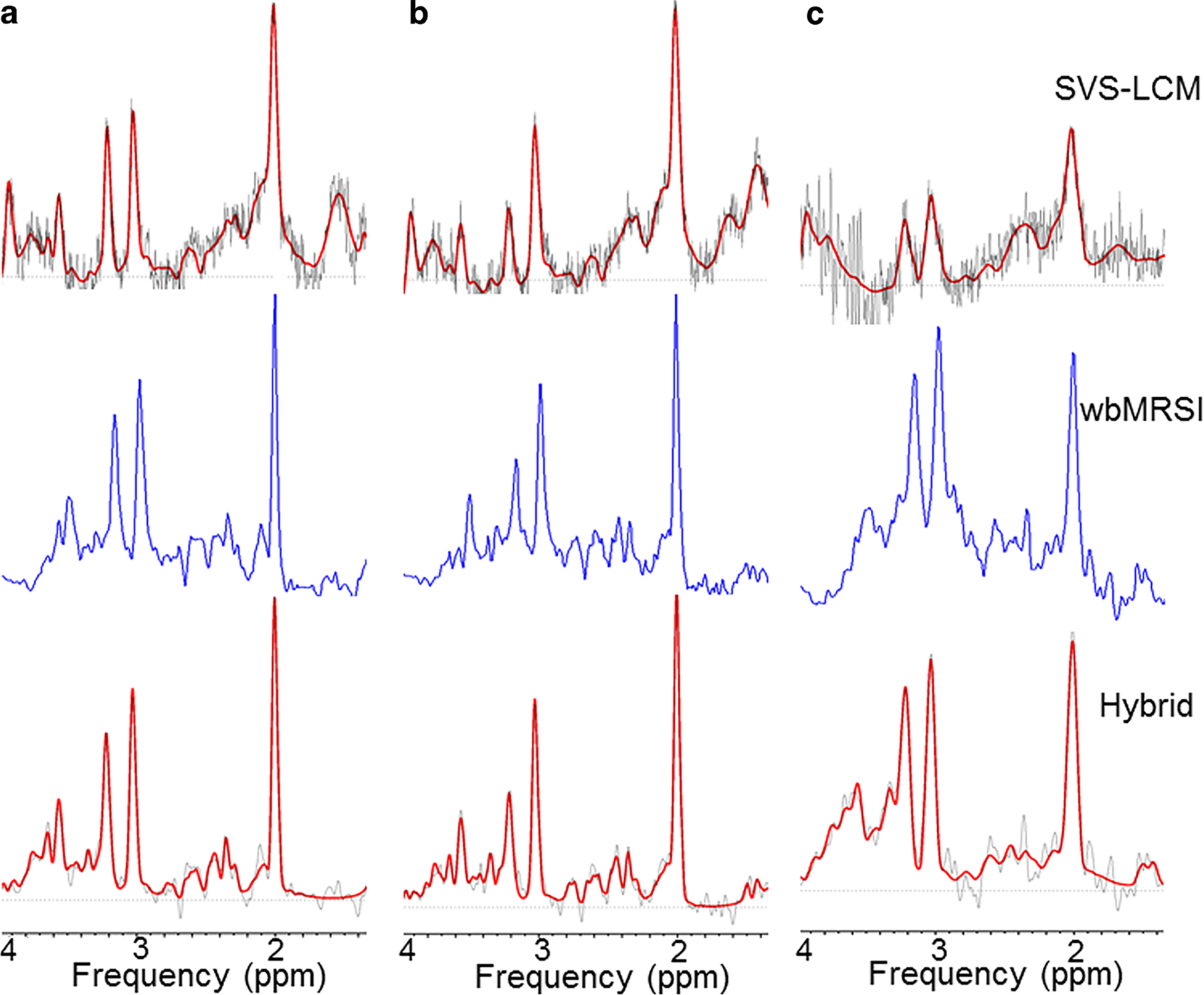
Spectra and fit results for SVS scans with the LCModel fit (first row), wbMRSI measurements (second row) and wbMRSI measurements with LCModel fit (third row). Data are shown for parietal white matter (column a), occipital grey matter (column b) and basal ganglia (column c)
No statistically significant differences were found between metabolite concentrations or spectral line widths of males and females using two-sided t-tests, different to that observed by Maudsley et al. in a retrospective work with wbMRSI. The discrepancy is thought to be caused by the fact that these authors also included data from volunteers with a BMI larger than 30 that is reported to impact measured metabolite values [9]. Therefore, male and female data in this study were combined for further analysis. Regional concentrations of NAA/tCr, tCho/tCr, Gln/tCr, Glu/tCr, and MI/tCr of all volunteers at each ROI measured with SVS-LCM, wbMRSI, and the hybrid method as well as the corresponding spectral line widths are shown in Fig. 4. The metabolite values and the spectral line widths obtained with three methods were relatively close to each other except in basal ganglia, where the metabolite values measured with SVS-LCM revealed a wide spread and the spectral line width was much broader than those estimated with other methods, as shown in Fig. 4.
Fig. 4.
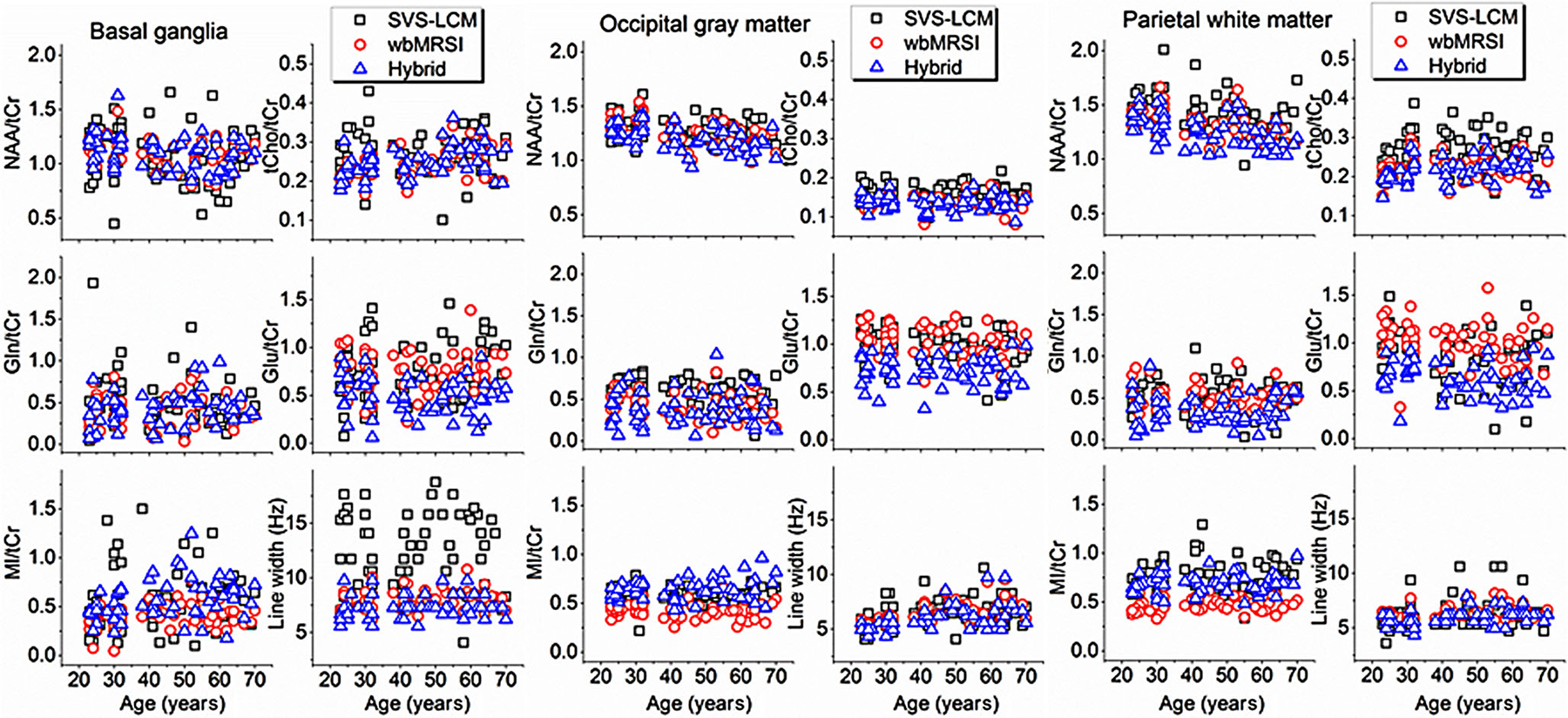
Regional metabolite concentrations of NAA/tCr, tCho/tCr, Gln/tCr, Glu/tCr, and MI/tCr, and spectral line widths at each ROI plotted against age, which were measured with SVS and analyzed with the LCModel (indicated as SVS-LCM), wbMRSI and analyzed with MIDAS (indicated as wbMRSI), and wbMRSI and analyzed with LCModel (indicated as hybrid)
Mean values averaged among the volunteers for metabolite concentrations and spectral line widths are summarized in Table 2. Paired metabolite values from different methods in same brain structure showing significant differences from each other, as estimated by using rANOVA with Bonferroni post hoc test, are indicated by the same letter suffixed a, b, or c behind the paired values. Note that not all data were sampled due to limitations for line width and CRLB according to quality criteria, as indicated by the number of sampled subjects given in Table 2. This is particularly notable for values at the ROI basal ganglia, where more spectra were excluded for the SVS-LCM method and the COVs were all larger in comparison to those estimated using both of the wbMRSI integrated methods. The mean CRLBs obtained by each method are shown in Table 3, where for all three methods the CRLB of the metabolites with small spectral signals (Glu, Gln, mI) were larger than those of the prominent singlet resonances (NAA, tCho, tCr), and the CRLB derived with SVSL-LCM were mainly larger than those with other two methods.
Table 2.
Mean values of brain metabolite concentrations and coefficients of variance (COV) obtained by averaging corresponding values among the volunteers that were estimated with the methods as indicated
| Metabolite | Methodd | BGf | oGMf | pWMf | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ne | Mean | COV (%) | N | Mean | COV (%) | N | Mean | COV (%) | ||
| NAA/tCr | SVS-LCM | 32 | 1.17 | 18.3 | 59 | 1.30a | 8.3 | 59 | 1.43a,b | 12.6 |
| wbMRSI | 59 | 1.09 | 12.5 | 59 | 1.26b | 9.3 | 59 | 1.33a,c | 10.1 | |
| Hybrid | 59 | 1.10 | 13.7 | 59 | 1.21a,b | 9.5 | 59 | 1.26b,c | 11.2 | |
| tCho/tCr | SVS-LCM | 32 | 0.29a,b | 21.3 | 59 | 0.16a,b | 13.0 | 59 | 0.27a,b | 16.6 |
| wbMRSI | 59 | 0.24a | 16.1 | 59 | 0.14a | 15.3 | 59 | 0.22a | 15.1 | |
| Hybrid | 59 | 0.25b | 15.6 | 59 | 0.13b | 14.2 | 59 | 0.22b | 14.6 | |
| mIns/tCr | SVS-LCM | 25 | 0.70a | 47.3 | 59 | 0.58a,b | 15.2 | 59 | 0.76a,b | 21.5 |
| wbMRSI | 57 | 0.41a,b | 26.3 | 59 | 0.42a,c | 17.9 | 59 | 0.47a,c | 15.4 | |
| Hybrid | 58 | 0.56b | 35.7 | 59 | 0.63b,c | 16.8 | 59 | 0.68b,c | 14.6 | |
| Gln/tCr | SVS-LCM | 14 | 0.63 | 30.2 | 57 | 0.62a,b | 19.5 | 42 | 0.59a | 24.5 |
| wbMRSI | 29 | 0.51 | 30.5 | 47 | 0.46a | 29.0 | 58 | 0.51b | 27.8 | |
| Hybrid | 47 | 0.48 | 33.8 | 43 | 0.45b | 40.8 | 46 | 0.40a,b | 37.6 | |
| Glu/tCr | SVS-LCM | 23 | 0.97a | 25.5 | 59 | 1.00a | 18.1 | 53 | 0.92a,b | 22.3 |
| wbMRSI | 55 | 0.77b | 25.0 | 59 | 1.03b | 15.1 | 58 | 1.01a,c | 18.2 | |
| Hybrid | 50 | 0.55a,b | 28.3 | 59 | 0.71a,b | 20.2 | 58 | 0.63b,c | 24.9 | |
| Linewidth (Hz) | SVS-LCM | 32 | 10.07a,b | 20.2 | 59 | 6.43 | 19.9 | 59 | 6.17 | 23.2 |
| wbMRSI | 59 | 7.91a,c | 12.1 | 59 | 6.38a | 15.6 | 59 | 6.35a | 9.9 | |
| Hybrid | 59 | 7.32b,c | 14.1 | 59 | 5.93a | 18.6 | 59 | 5.90a | 11.3 | |
The suffixed letter a, b, or c indicate the paired metabolite values of different methods in same brain structure were significantly different that estimated by using rANOVA with Bonferroni post hoc test
The suffixed letter a, b, or c indicate the paired metabolite values of different methods in same brain structure were significantly different that estimated by using rANOVA with Bonferroni post hoc test
The suffixed letter a, b, or c indicate the paired metabolite values of different methods in same brain structure were significantly different that estimated by using rANOVA with Bonferroni post hoc test
SVS-LCM spectral data were scanned with SVS and analyzed with LCModel, wbMRSI data were scanned with EPSI and analyzed with MIDAS, Hybrid data were scanned with EPSI and analyzed with LCModel
N represents the number of sampled subjects. The data with line width >14Hz, Cramer-Rao lower bound >20% for NAA, tCho, and tCr, and >30% for Gln, Glu and MI were not sampled
BG represents the ROI in basal ganglia, oGM the ROI in occipital grey matter, and pWM the ROI in parietal white matter
Table 3.
Mean Cramer-Rao lower bounds (CRLB) of the spectral data analysis
| Metabolite | Methoda | BGc | oGMc | pWMc | |||
|---|---|---|---|---|---|---|---|
| Nb | CRLB | N | CRLB | N | CRLB | ||
| NAA | SVS-LCM | 32 | 8% ± 2% | 59 | 5% ± 1% | 59 | 5% ± 1% |
| wbMRSI | 59 | 4% ± 1% | 59 | 4% ± 1% | 59 | 3% ± 1% | |
| Hybrid | 59 | 3% ± 1% | 59 | 2% ± 0% | 59 | 2% ± 0% | |
| tCho | SVS-LCM | 32 | 9% ± 3% | 59 | 9% ± 1% | 59 | 7% ± 1% |
| wbMRSI | 59 | 6% ± 1% | 59 | 9% ± 3% | 59 | 5% ± 1% | |
| Hybrid | 59 | 4% ± 1% | 59 | 6% ± 1% | 59 | 4% ± 1% | |
| tCr | SVS-LCM | 32 | 8% ±3% | 59 | 5% ± 1% | 59 | 6% ± 1% |
| wbMRSI | 59 | 3% ± 1% | 59 | 4% ± 1% | 59 | 3% ± 1% | |
| Hybrid | 59 | 3% ± 0% | 59 | 3% ± 1% | 59 | 3% ± 0% | |
| mI | SVS-LCM | 25 | 14% ± 5% | 59 | 10% ± 3% | 59 | 10% ± 3% |
| wbMRSI | 57 | 11% ± 4% | 59 | 10% ± 3% | 59 | 8% ± 2% | |
| Hybrid | 58 | 8% ± 4% | 59 | 6% ± 1% | 59 | 5% ± 1% | |
| Gln | SVS-LCM | 14 | 22% ± 4% | 57 | 18% ± 4% | 42 | 22% ± 5% |
| wbMRSI | 29 | 22% ± 5% | 47 | 18% ± 6% | 58 | 15% ± 4% | |
| Hybrid | 47 | 18% ± 5% | 43 | 18% ± 6% | 46 | 19% ± 6% | |
| Glu | SVS-LCM | 23 | 18% ± 5% | 59 | 13% ± 4% | 53 | 16% ± 5% |
| wbMRSI | 55 | 13% ± 5% | 59 | 7% ± 3% | 58 | 6% ± 2% | |
| Hybrid | 50 | 15% ± 5 % | 59 | 10% ± 3% | 58 | 11% ± 4% | |
SVS-LCM spectral data were scanned with SVS and analyzed with LCModel, wbMRSI data were scanned with EPSI and analyzed with MIDAS, Hybrid data were scanned with EPSI and analyzed with LCModel
N represents number of sampled subjects. The data with linewidth >14Hz, Cramer-Rao lower bound >20% for NAA, tCho, and tCr, and >30% for Gln, Glu and MI were not sampled
BG represents the ROI in basal ganglia, oGM the ROI in occipital grey matter, and pWM the ROI in parietal white matter
The results of the linear regression analysis of age and metabolite values in ratio to tCr and to internal water measured in oGM and pWM are shown in Table 4 and in Table 5, respectively. As shown in Table 4, age-related changes were mainly observed for NAA/tCr, which significantly deceased with age in pWM and oGM, and the spectral line widths, which increased with age significantly in oGM, both independent of the method used; however, a comparison between methods for the p-values and R-values showed that for all cases where all methods reached significance, both p and R were more confidential for both wbMRSI integrated methods. Similar effects were also observed when values determined in ratio to internal water were used (Table 5). In addition, a significant decrease of Glu/tCr with age in pWM and oGM was observed from the data obtained with wbMRSI, and a significant increase of line width with age in pWM from the data obtained with hybrid. No linear regression analysis was made for metabolites measured in basal ganglia, since many values, especially those obtained with SVS method, were excluded according to data quality criteria.
Table 4.
Results of linear regression analysis of brain metabolite concentrations (in ratio to tCr) to age
| Metabolite | Methoda | oGMc | pWMc | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Nb | Rd | p | αe | βe | N | R | p | α | β | ||
| NAA/tCr | SVS-LCM | 59 | −0.27 | 0.039 | 1.39 | −0.0020 | 59 | −0.39 | 0.002 | 1.64 | −0.0048 |
| wbMRSI | 59 | −0.54 | 0.000 | 1.45 | −0.0042 | 59 | −0.55 | 0.000 | 1.55 | −0.0050 | |
| Hybrid | 59 | −0.51 | 0.000 | 1.38 | −0.0040 | 59 | −0.49 | 0.000 | 1.47 | −0.0047 | |
| tCho/tCr | SVS-LCM | 59 | 0.02 | 0.867 | – | – | 59 | 0.07 | 0.575 | – | – |
| wbMRSI | 59 | −0.07 | 0.582 | – | – | 59 | 0.03 | 0.811 | – | – | |
| Hybrid | 59 | −0.11 | 0.425 | – | – | 59 | 0.23 | 0.074 | – | – | |
| mIns/tCr | SVS-LCM | 59 | 0.04 | 0.763 | – | – | 59 | 0.16 | 0.216 | – | – |
| wbMRSI | 59 | −0.03 | 0.851 | – | – | 59 | 0.06 | 0.655 | – | – | |
| Hybrid | 59 | 0.18 | 0.163 | – | – | 59 | 0.14 | 0.294 | – | – | |
| Gln/tCr | SVS-LCM | 57 | −0.13 | 0.341 | – | – | 42 | −0.10 | 0.535 | – | – |
| wbMRSI | 47 | −0.10 | 0.514 | – | – | 58 | 0.00 | 0.978 | – | – | |
| Hybrid | 43 | 0.07 | 0.653 | – | – | 46 | −0.04 | 0.816 | – | – | |
| Glu/tCr | SVS-LCM | 59 | −0.25 | 0.054 | – | – | 53 | −0.14 | 0.331 | – | – |
| wbMRSI | 59 | −0.35 | 0.006 | 1.20 | −0.0037 | 58 | −0.35 | 0.008 | 1.20 | −0.0043 | |
| Hybrid | 59 | −0.04 | 0.786 | – | – | 58 | −0.26 | 0.051 | – | – | |
| Linewidth | SVS-LCM | 59 | 0.29 | 0.026 | 5.33 | 0.0249 | 59 | 0.17 | 0.186 | – | – |
| wbMRSI | 59 | 0.58 | 0.000 | 4.67 | 0.0388 | 59 | 0.25 | 0.060 | – | – | |
| Hybrid | 59 | 0.53 | 0.000 | 4.18 | 0.0396 | 59 | 0.32 | 0.012 | 5.25 | 0.0146 | |
Only numbers in Bold print signifity, while all numbers of p vlaues were in italic
SVS-LCM: spectral data were scanned with SVS and analyzed with LCModel. wbMRSI: data were scanned with EPSI and analyzed with MIDAS. Hybrid: data were scanned with EPSI and analyzed with LCModel
N represents number of sampled subjects. The data with line width >14Hz, Cramer-Rao lower bounds >20% for NAA, tCho, and tCr, and >30% for Gln, Glu and MI were not sampled
BG represents the ROI in basal ganglia, oGM the ROI in occipital grey matter, and pWM the ROI in parietal white matter
R is Pearson’s correlation coefficient
Intercept (α) and slope (β) were calculated only in cases with significance value p< 0.05
Table 5.
Results of linear regression analysis of brain metabolite concentrations (in reference to internal water) to age
| Metabolite | Methoda | oGMc | pEMc | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Nb | Rd | p | αe | βe | N | R | p | α | β | ||
| NAA | SVS-LCM | 59 | −0.10 | 0.461 | – | – | 59 | −0.12 | 0.365 | – | – |
| wbMRSI | 59 | −0.48 | 0.000 | 14.15 | −0.0384 | 59 | −0.39 | 0.002 | 12.04 | −0.0299 | |
| Hybrid | 59 | −0.28 | 0.029 | 29.67 | −0.0580 | 59 | −0.45 | 0.000 | 26.60 | −0.0818 | |
| tCho | SVS-LCM | 59 | 0.13 | 0.333 | – | – | 59 | 0.40 | 0.002 | 1.06 | 0.0045 |
| wbMRSI | 59 | −0.04 | 0.752 | – | – | 59 | 0.17 | 0.209 | – | – | |
| Hybrid | 59 | 0.02 | 0.901 | – | – | 59 | 0.29 | 0.025 | 3.46 | 0.0111 | |
| tCr | SVS-LCM | 59 | 0.19 | 0.139 | – | – | 59 | 0.35 | 0.006 | 4.11 | 0.0130 |
| wbMRSI | 59 | 0.04 | 0.735 | – | – | 59 | 0.16 | 0.241 | – | – | |
| Hybrid | 59 | 0.15 | 0.260 | – | – | 59 | 0.02 | 0.854 | – | – | |
| mIns | SVS-LCM | 59 | 0.12 | 0.355 | – | – | 59 | 0.33 | 0.010 | 2.74 | 0.0187 |
| wbMRSI | 59 | −0.01 | 0.921 | – | – | 59 | 0.17 | 0.205 | – | – | |
| Hybrid | 59 | 0.26 | 0.050 | – | – | 59 | 0.18 | 0.174 | – | – | |
| Gln | SVS-LCM | 57 | −0.04 | 0.755 | – | – | 42 | 0.05 | 0.752 | – | – |
| wbMRSI | 47 | −0.10 | 0.483 | – | – | 58 | 0.04 | 0.756 | – | – | |
| Hybrid | 43 | 0.15 | 0.332 | – | – | 46 | −0.04 | 0.797 | – | – | |
| Glu | SVS-LCM | 59 | −0.17 | 0.207 | – | – | 53 | 0.05 | 0.722 | – | – |
| wbMRSI | 59 | −0.29 | 0.028 | 11.73 | −0.0340 | 58 | −0.31 | 0.016 | 9.31 | −0.2700 | |
| Hybrid | 59 | 0.05 | 0.713 | – | – | 58 | −0.24 | 0.070 | – | – | |
SVS-LCM: spectral data were scanned with SVS and analyzed with LCModel. wbMRSI: data were scanned with EPSI and analyzed with MIDAS. Hybrid: data were scanned with EPSI and analyzed with LCModel
N represents number of sampled subjects. The data with line width >14Hz, Cramer-Rao lower bounds >20% for NAA, tCho, and tCr, and >30% for Gln, Glu and MI were not sampled
oGM represents the ROI in occipital grey matter, and pWM the ROI in parietal white matter
R is Pearson’s correlation coefficient
Intercept (α) and slope (β) were calculated only in cases with significance value p< 0.05
Discussion
This study compared three ways for obtaining regional proton metabolite measurements using in vitro and in vivo studies in the brain and compared performance for evaluation of age-dependent changes in metabolite concentrations in the brains of 55 healthy volunteers. The study presents a comparison of SVS, where a STEAM sequence was used to keep the TE (20ms) as close as possible to that used in wbMRSI (17.6ms), and wbMRSI measurements, together with a comparison of two spectral analysis programs for analysis of the wbMRSI results.
For measurements in phantoms, the comparison of metabolite values determined as a ratio to water signal and as a ratio to tCr, by reference to the known concentrations, revealed that all three methods used different methods for metabolite signal normalization using the internal water signal. This primarily reflects differences in magnitude of the water reference signal, since the EPSI measurement uses an interleaved low flip angle measurement of the water signal whereas the SVS measurement used a 90° excitation. There are also differences in the signal normalization methods, since there is a clear difference in estimation of internal water content between both wbMRSI integrated methods (LCModel vs. MIDAS) [6, 14]. The spectra obtained with both methods that used the wbMRSI data resulted in a narrower line width than the SVS method. This arises from the acquisition of a smaller nominal voxel size with the wbMRSI method, which is augmented by the use of spatial oversampling in the implementation of the EPSI sequence used for this study [21], followed by correction for B0 inhomogeneities before ROI signal integration. The difference in line widths between the wbMRSI and hybrid methods was very small (0.4Hz), indicating that the line width result from MIDAS/FITT spectral analysis is comparable to that from LCmodel, even though these programs use different line shape models, i.e. LCModel did not use a fixed line shape but a linear combination of model spectra of metabolite solutions [6], while FITT used a Gaussian line shape. The observation that CRLB was the lowest by wbMRSI, then hybrid and the highest by SVS-LCM showed that CRLB was strongly dependent on the spectral line width. Overall, for the in vitro measurements using tCr as internal reference, SVS-LCM, wbMRSI, and hybrid methods provided comparable results with an acceptable spectral resolution and satisfactory CRLB for spectral fits.
As shown in Fig. 4, the concentrations of the metabolites measured in vivo as a ratio to tCr in BG, oGM and pWM determined with SVS-LCM, wbMRSI and hybrid methods were close to each other and lay within the ranges previously reported in adult human brain [22–24]; however, significant differences between most of the group mean values derived with different methods in the same brain structures were still found with rANOVA (Table 2). The COVs (Table 2) and CRLBs (Table 3) of MI/tCr, Glu/tCr and Gln/tCr derived by the three methods were all larger than those of main metabolites NAA/tCr, tCho/tCr, which reflects the lower signal amplitude and more complex spectral patterns of MI, Glu and Gln. The comparison of COVs of all five metabolites derived with three methods did not reveal a clear advantage of any method. In contrast, the COVs of the line width derived with wbMRSI was the smallest, that with hybrid was slightly larger, and that with SVS-LCM was the largest, which further indicates that the multivoxel ROI integration procedure results in better spectral resolution than SVS. This is also indicated by the CRLB values in Table 3, where the CRLBs derived with SVS-LCM were larger or equal to those derived with wbMRSI and hybrid. The fact that the spectral line widths in basal ganglia derived with three methods were all broader than those of the other two brain structures, may be explained by accumulated iron deposition in the brain structure, which is consistent with the knowledge of basal ganglia being a preferable brain structure for pathological iron deposition in neurodegeneration [25]. The broader line width and wide spread of the metabolite values in basal ganglia estimated with SVS-LCM in comparison to those with other methods (Fig. 4) reflects difficulties with shimming for SVS in basal ganglia.
As presented in Table 4, significant decreases of NAA/tCr with age were observed in pWM and oGM, and increases of spectral line widths with age were observed in oGM for all the three methods, which are consistent with previous reports [1, 9]. The decrease of NAA/tCr has been attributed to age-related reduction of neuronal volume/density and metabolic activity in the brain tissue [1], and the positive correlations of the spectral line width to age to increased brain iron concentrations and shorter metabolite T2 relaxation times in older subjects [9, 26–28]. While negative Pearson’s coefficients were observed for Glu/tCr with age in pWM and oGM with all three methods, the accompanying significances were divergent depending on which method was used, i.e. varying from p< 0.05 for data in pWM and oGM obtained with wbMRSI to p ≈ 0.05 for data in oGM obtained with SVS+ LCM, and in pWM obtained with hybrid, and to p> 0.05 for data in oGM obtained with hybrid, and in pWM obtained with SVS+ LCM. Similar variation was observed also for line width in pWM. The regression analysis of metabolite values in reference to internal water also revealed differences between the results of SVS and wbMRSI (Table 5), where decreases of NAA with age without age-related changes of tCr reached the significance level only for data obtained with wbMRSI and hybrid methods, while these observations proved that the observed decreases of NAA/tCr with age with these two methods was mainly caused by the changes of NAA. These divergent observations may result mainly from differences of spectral line widths of the data as already discussed.
For a direct comparison of the wbMRSI and SVS acquisition approaches the same data acquisition parameters should ideally be used; however, the two acquisition methods in this study used different TE, excitation pulse angle, RF pulse shapes, and acquisition sequence (spin echo vs. stimulated echo, MRSI vs. SVS). These differences reflect basic considerations of the sequence design and implementation; therefore, this study has compared two available methods with the aim of evaluating the resultant reproducibility for in vivo studies. Several other differences between these acquisition methods were not considered, including the relative acquisition times (16min for the wbMRSI vs. 5min for one SVS measurement), sensitivity to subject motion, and integration of the sequences into the clinical workflow. The choice of acquisition method must also consider the purpose for the measurement. While SVS measurements can be conveniently implemented and are of diagnostic value for selected pathologies, clearly an advantage of wbMRSI is that it is possible to estimate brain metabolites in an image format or in multiple ROIs using post-processing [10].
Limitations of the in vivo measurements presented in this study include that the metabolite concentrations were measured and compared in only selected brain areas, and as a ratio with tCr, meaning that the changes of tCr with age were not considered, as well as corrections of T1/T2 relaxations of the metabolites. The subject selection also did not consider body weight, which may influence age-related changes of brain metabolites as reported previously [9].
In conclusion, this study demonstrated that the wbMRSI technique, which uses the EPSI data acquisition technique and processing using MIDAS, could be used to estimate metabolites in aging human brain with the same reliability as using a conventional SVS measurement with the commonly used LCModel software for data analysis, with the main advantage being that metabolite concentrations can be measured in multiple brain structure simultaneously.
Acknowledgements
We would like to thank the research volunteers.
Funding This work was partially supported by the Deutsche For-schungsgemeinschaft. Additional support was provided under NIH grant R01 EB016064 (AAM).
Footnotes
Conflict of interest H. Maghsudi, B. Schmitz, A.A. Maudsley, S. Sheriff, P. Bronzlik, M. Schütze, H. Lanfermann and X. Ding declare that they have no competing interests.
References
- 1.Ding XQ, Maudsley AA, Sabati M, Sheriff S, Schmitz B, Schütze M, Bronzlik P, Kahl KG, Lanfermann H. Physiological neuronal decline in healthy aging human brain - An in vivo study with MRI and short echo-time whole-brain (1)H MR spectroscopic imaging. Neuroimage. 2016;137:45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Griffith HR, den Hollander JA, Okonkwo OC, O’Brien T, Watts RL, Marson DC. Brain metabolism differs in Alzheimer’s disease and Parkinson’s disease dementia. Alzheimers Dement. 2008;4:421–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hall H, Cuellar-Baena S, Dahlberg C, In’t Zandt R, Denisov V, Kirik D. Magnetic resonance spectroscopic methods for the assessment of metabolic functions in the diseased brain. Curr Top Behav Neurosci. 2012;11:169–98. [DOI] [PubMed] [Google Scholar]
- 4.Frahm J, Bruhn H, Gyngell ML, Merboldt KD, Hänicke W, SauterR. Localized high-resolution proton NMR spectroscopy using stimulated echoes: initial applications to human brain in vivo. Magn Reson Med. 1989;9:79–93. [DOI] [PubMed] [Google Scholar]
- 5.Naressi A, Couturier C, Devos JM, Janssen M, Mangeat C, de Beer R, Graveron-Demilly D. Java-based graphical user interface for the MRUI quantitation package. MAGMA. 2001;12:141–52. [DOI] [PubMed] [Google Scholar]
- 6.Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med. 1993;30: 672–9. [DOI] [PubMed] [Google Scholar]
- 7.Sabati M, Sheriff S, Gu M, Wei J, Zhu H, Barker PB, Spielman DM, Alger JR, Maudsley AA. Multivendor implementation and comparison of volumetric whole-brain echo-planar MR spectroscopic imaging. Magn Reson Med. 2015;74:1209–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding XQ, Lanfermann H. Whole brain (1)H-spectroscopy: a developing technique for advanced analysis of cerebral metabolism. Clin Neuroradiol. 2015;25(Suppl 2):245–50. [DOI] [PubMed] [Google Scholar]
- 9.Maudsley AA, Govind V, Arheart KL. Associations of age, gender and body mass with 1H MR-observed brain metabolites and tissue distributions. NMR Biomed. 2012;25:580–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eylers VV, Maudsley AA, Bronzlik P, Dellani PR, Lanfermann H, Ding XQ. Detection of normal aging effects on human brain metabolite concentrations and microstructure with whole-brain MR spectroscopic imaging and quantitative MR imaging. AJNR Am J Neuroradiol. 2016;37:447–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding XQ, Maudsley AA, Sabati M, Sheriff S, Dellani PR, Lanfermann H. Reproducibility and reliability of short-TE whole-brain MR spectroscopic imaging of human brain at 3T. Magn Reson Med. 2015;73:921–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Taub E, Salibi N, Uswatte G, Maudsley AA, Sheriff S, Womble B, Mark VW, Knight DC. Comparison of reproducibility of single voxel spectroscopy and whole-brain magnetic resonance spectroscopy imaging at 3T. NMR Biomed. 2018;31:e3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maudsley AA, Darkazanli A, Alger JR, Hall LO, Schuff N, Studholme C, Yu Y, Ebel A, Frew A, Goldgof D, Gu Y, Pagare R, Rousseau F, Sivasankaran K, Soher BJ, Weber P, Young K, Zhu X. Comprehensive processing, display and analysis for in vivo MR spectroscopic imaging. NMR Biomed. 2006;19:492–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maudsley AA, Domenig C, Govind V, Darkazanli A, Studholme C, Arheart K, Bloomer C. Mapping of brain metabolite distributions by volumetric proton MR spectroscopic imaging (MRSI). Magn Reson Med. 2009;61:548–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steer RA, Clark DA, Beck AT, Ranieri WF. Common and specific dimensions of self-reported anxiety and depression: the BDI-II versus the BDI-IA. Behav Res Ther. 1999;37:183–90. [DOI] [PubMed] [Google Scholar]
- 16.Kalbe E, Kessler J, Calabrese P, Smith R, Passmore AP, Brand M, Bullock R. DemTect: a new, sensitive cognitive screening test to support the diagnosis of mild cognitive impairment and early dementia. Int J Geriatr Psychiatry. 2004;19:136–43. [DOI] [PubMed] [Google Scholar]
- 17.Haupt CI, Schuff N, Weiner MW, Maudsley AA. Removal of lipid artifacts in 1H spectroscopic imaging by data extrapolation. Magn Reson Med. 1996;35:678–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM, Matthews PM. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23(Suppl 1):S208–19. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging. 2001;20:45–57. [DOI] [PubMed] [Google Scholar]
- 20.Goryawala MZ, Sheriff S, Maudsley AA. Regional distributions of brain glutamate and glutamine in normal subjects. NMR Biomed. 2016;29:1108–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ebel A, Maudsley AA. Improved spectral quality for 3D MR spectroscopic imaging using a high spatial resolution acquisition strategy. Magn Reson Imaging. 2003;21:113–20. [DOI] [PubMed] [Google Scholar]
- 22.Grachev ID, Apkarian AV. Chemical heterogeneity of the living human brain: a proton MR spectroscopy study on the effects of sex, age, and brain region. Neuroimage. 2000;11(5 Pt 1):554–63. [DOI] [PubMed] [Google Scholar]
- 23.Pouwels PJ, Brockmann K, Kruse B, Wilken B, Wick M, Hanefeld F, Frahm J. Regional age dependence of human brain metabolites from infancy to adulthood as detected by quantitative localized proton MRS. Pediatr Res. 1999;46:474–85. [DOI] [PubMed] [Google Scholar]
- 24.Natt O, Bezkorovaynyy V, Michaelis T, Frahm J. Use of phased array coils for a determination of absolute metabolite concentrations. Magn Reson Med. 2005;53:3–8. [DOI] [PubMed] [Google Scholar]
- 25.Boelmans K, Holst B, Hackius M, Finsterbusch J, Gerloff C, Fiehler J, Münchau A. Brain iron deposition fingerprints in Parkinson’s disease and progressive supranuclear palsy. Mov Disord. 2012;27:421–7. [DOI] [PubMed] [Google Scholar]
- 26.Kirov II, Fleysher L, Fleysher R, Patil V, Liu S, Gonen O. Age dependence of regional proton metabolites T2 relaxation times in the human brain at 3T. Magn Reson Med. 2008;60:790–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marjanska M, Emir UE, Deelchand DK, Terpstra M. Faster metabolite (1)H transverse relaxation in the elder human brain. PLoS ONE. 2013;8:e77572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitsumori F, Watanabe H, Takaya N. Estimation of brain iron concentration in vivo using a linear relationship between regional iron and apparent transverse relaxation rate of the tissue water at 4.7T. Magn Reson Med. 2009;62:1326–30. [DOI] [PubMed] [Google Scholar]


