Abstract
Epigenetic regulation of gene transcription has been shown to coordinate with nutrient availability, yet the mechanisms underlying this coordination remain incompletely understood. Here we show that glucose starvation suppresses histone 2A K119 monoubiquitination (H2Aub), a histone modification that correlates with gene repression. Glucose starvation suppressed H2Aub levels independently of energy stress-mediated AMPK activation and possibly through NADPH depletion and subsequent inhibition of BMI1, an integral component of polycomb repressive complex 1 (PRC1) that catalyzes H2Aub on chromatin. Integrated transcriptomic and epigenomic analyses linked glucose starvation-mediated H2Aub repression to the activation of genes involved in the endoplasmic reticulum (ER) stress response. We further showed that this epigenetic mechanism has a role in glucose starvation-induced cell death and that pharmacologic inhibition of glucose transporter 1 (GLUT1) and PRC1 synergistically promoted ER stress and suppressed tumor growth in vivo. Together, these results reveal a hitherto unrecognized epigenetic mechanism coupling glucose availability to the ER stress response.
Keywords: H2A monoubiquitination, ER stress response, glucose, BMI1, PRC1, tumor development
INTRODUCTION
Sensing and responding to nutrient availability is central to virtually all living organisms (1). To survive and grow under nutrient-limiting environments, eukaryotic cells have evolved various mechanisms to coordinate extracellular nutrient availability with epigenetic control of gene expression in the nucleus (2,3). Common epigenetic mechanisms to control gene expression include modifications of DNA (such as DNA methylation) and histones (such as histone acetylation, methylation, ubiquitination, and phosphorylation) through corresponding chromatin-modifying enzymes (4). Recent studies have shown that metabolic regulation of epigenetics occurs either through intracellular signaling pathways that transduce signals from extracellular nutrients to regulate the activity or expression of epigenetic modifying enzymes or by more direct mechanisms wherein intracellular metabolites serve as the substrates or co-factors of chromatin-modifying enzymes (2,5).
Histone 2A monoubiquitination at lysine 119 (H2Aub) is a histone modification that has been associated with transcriptional repression (6–8). H2Aub levels are balanced by its ubiquitination and de-ubiquitination through corresponding writers and erasers. H2A ubiquitination is catalyzed mainly by its writer polycomb repressive complex 1 (PRC1) (6). The canonical PRC1 contains BMI1, RING1/2, PHC, and CBX proteins (9,10), among which RING1 and 2 are redundant catalytic subunits and BMI1 regulates PRC1 architecture and enzymatic activity toward H2Aub (11). H2A de-ubiquitination is mediated mainly by its eraser BRCA1-associated protein 1 (BAP1) (12). H2Aub dysregulation has been associated with cancer development. For example, BMI1 is upregulated in several types of cancer and has been shown to exert oncogenic functions in various contexts (13–15). Correspondingly, small molecules targeting PRC1, such as PRT-4165, have been developed and tested as potential antitumor agents (16). Conversely, BAP1 is a tumor suppressor that is mutated or downregulated in several cancers (17–22). Although H2Aub regulation by its writers and erasers has been well established, whether and how H2Aub levels and the H2Aub-associated transcriptional network can be modulated by nutrient availability remain largely unknown.
The endoplasmic reticulum (ER) is a dynamic organelle committed to protein folding and trafficking. Accumulation of unfolded proteins in the ER lumen triggers a series of signaling cascades collected called the ER stress response, which initially functions to restore ER homeostasis and to maintain cell survival but ultimately can trigger cell death under excessive stress conditions (23). Disturbance in diverse cellular processes such as redox maintenance, calcium regulation, and nutrient metabolism can lead to an ER stress response. In response to ER stress, various ER membrane‒embedded protein sensors including protein kinase RNA-like endoplasmic reticulum kinase (EIF2AK3/PERK), inositol-requiring enzyme 1 (IRE1), and activating transcription factor 6 (ATF6) are activated and subsequently transduce signaling through distinct downstream transcription factors including ATF4, spliced XBP1 (XBP1s), and cleaved ATF6 (ATF6c), respectively. These transcription factors translocate into the nucleus and regulate gene transcription to either restore ER homeostasis or promote cell death, depending on the duration and severity of the stress (24–26). In contrast to the well-established roles of transcription factors in regulating the ER stress response, the potential epigenetic mechanisms that couple nutrient availability with the ER stress response remain much less well understood. In this study, we identified an H2Aub-involved epigenetic mechanism linking glucose availability to the ER stress response.
MATERIALS AND METHODS
Cell culture studies
The UMRC6 cell line was purchased from Sigma (#08090513, 2017), and other cancer cell lines and the HEK293T embryonic kidney cell line were obtained from the American Type Culture Collection (2011–2017). All cell lines were cultured in DMEM containing penicillin (100 units/mL), streptomycin (100 μg/mL), and 10% (vol/vol) fetal bovine serum (FBS) except for 786-O and H460 cells, which were cultured in RPMI-1640 medium. Cells within eight passages were used for experiments. All cell lines were tested annually to be Mycoplasma-free. For the glucose-deprivation experiments, cells were cultured in glucose-free DMEM with different concentrations of glucose + 10% (vol/vol) dialyzed FBS (27,28). Stable cell lines were generated as previously described (29). Briefly, HEK293T cells were transfected with individual plasmids targeting specific genes together with the psPAX.2 and pMD2.G third-generation lentiviral packaging systems by using Lipofectamine 3000 reagent (ThermoFisher Scientific) according to the manufacturer’s instructions. At 36–48 hours later, lentivirus particles in the medium were collected and filtered before being used to infect cell lines in the presence of 8 μg/mL hexadimethrine bromide (Polybrene; MilliporeSigma, TR1003G). At 24 hours after infection, puromycin was added at a concentration of 2 μg/mL to obtain stable cell lines with successful transduction. To generate CRISPR/Cas9-mediated knockout cells, the single-guide RNA (sgRNA) targeting the specific gene was cloned into the lentiCRISPR v2 (Addgene, #52961). To generate short hairpin RNA (shRNA) -knockdown cell lines, lentiviral transduction with shRNA vectors was conducted as described above. To confirm knockout or knockdown efficiency, expression levels of target genes were determined by immunoblotting with corresponding antibodies.
Constructs and reagents
Knockout of AMPK α1/α2 and ATF4 in UMRC6 cell line was done by using sgRNAs and CRISPR/Cas9 technology as follows. The sgRNAs were cloned into the lentiviral lentiCRISPR v2 vector. A LentiCRISPR v2 vector containing sgRNAs against ATF4 was described in our previous publication (30). All constructs were confirmed by DNA sequencing. pGIPZ-shRNAs against BMI1 were obtained from the Functional Genomics Core at The University of Texas MD Anderson Cancer Center. Other reagents were purchased as follows: 2-deoxy-D-glucose (2DG; D6134), metformin (PHR1084), phenformin (P7045), tunicamycin (T7765), and N-acetyl-cysteine (A7250) from Sigma; A-769662 (LC laboratories, A-1803); 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR; ThermoFisher Scientific, A611700); ISRIB (Apexbio, B3699); brefeldin A (Cayman Chemical, #11861); thapsigargin (Santa Cruz, sc-24017A); PRT-4165 (Abcam, ab146003); BAY-876 (Sigma, SML1774); SP600125 (Selleckchem, S1460); and SB203580 (Selleckchem, S1076).
Cell death/viability assay
Cell death was measured by propidium iodide (PI) staining as described previously (31,32). Briefly, cells were seeded in 12-well plates 24 hours before treatment. After treatment with the appropriate culture medium or drug, cells were trypsinized and collected in a 1.5-mL tube, washed once with phosphate-buffered saline (PBS) and stained with 2 μg/mL PI (Roche) in PBS. Dead (PI-positive) cells were analyzed with a BD Accuri C6 flow cytometer (BD Biosciences). Cell viability was measured as described previously (29). Briefly, 8,000–12,000 cells per well were seeded in 96-well plates and treated, after which the medium in each well was replaced with fresh medium containing Cell Counting Kit-8 (CCK8) reagent (APExBIO, K1018). After incubation for 1 hour at 37°C, plates were read by a FLUOstar Omega microplate reader (BMG Labtech) at an absorbance of 450 nm. Cell viability (%) = [(Absorbance of tested compound minus Absorbance of blank) / (Absorbance of control minus Absorbance of blank)] × 100.
Light microscopy
For light microscopy, cells cultured in 12-well plates were treated as indicated in the figure legends. Phase contrast images were obtained with an EVOSfl (Advanced Microscopy Group) microscope equipped with a 10× phase contrast objective. Crystal violet staining was done as described elsewhere (33). Briefly, cells treated in 12-well plates were washed once in PBS and then stained with 0.5% crystal violet solution containing 20% methanol for 20 min. The plates were then washed in tap water and stored overnight at room temperature to dry. Levels of crystal violet‒stained cells were measured by adding 1 mL of methanol to each well and incubating the plates on a shaker for 30 min. The absorbance (OD570 nm) of each well was measured with a FLUOstar Omega microplate reader (BMG Labtech).
Real-time PCR
Real time PCR was done as previously described (34–36). Briefly, total mRNA was extracted from each sample by using TRIzol reagent (ThermoFisher Scientific) according to the manufacturer’s instructions. Then 2 μg of RNA was subjected to reverse transcription of cDNA by using SuperScript II reverse transcriptase (ThermoFisher Scientific) according to the manufacturer’s instructions. Real-time PCR was performed in triplicate in a 20-μL reaction mixture by using SYBR GreenER qPCR SuperMix Universal (ThermoFisher Scientific, #11762100). β-actin was used to as internal control. The primer sequences used are listed in Table S1.
ATP level measurement
Levels of ATP in cells in the different drug treatment conditions were measured with the CellTiter-Glo luminescent assay kit (Promega, G7570) according to the manufacturer’s instructions (33,37). Briefly, 10,000 cells/well were seeded in 96-well plates 24 hours before treatment. On the day of treatment, the appropriate drugs and 100 μL of CellTiter-Glo reagent were added to each well, and the plates were incubated for 10 min at room temperature on a shaker. The luminescence of each well was then measured with a Gen5 microplate reader (BIOTEK).
Western blotting
Protein levels were determined by immunoblotting as previously described (38–40). The primary antibodies and concentrations used for western blotting were: H3K27me3 (1:5000, MilliporeSigma, 07–449), tubulin (1:5000; Cell Signaling Technology, #2144), H2Aub (1:5000; MilliporeSigma, 05–678), H2A (1:5000; MilliporeSigma, ABE327), vinculin (1:10000; Sigma, V4505), AMPK (1:1000; Cell Signaling Technology, #5832) phospho-AMPK (1:1000; Cell Signaling Technology, #2535), AMPKα1 (1:1000; Cell Signaling Technology, #2795), AMPKα2 (1:1000; Cell Signaling Technology, #2757), phospho-eIF2α (1:1000; Cell Signaling Technology, #3398), eIF2α (1:1000; Cell Signaling Technology, #9722), ATF4 (1:1000; Cell Signaling Technology, #11815), phospho-JNK (1:1000; Cell Signaling Technology, #4668), JNK (1:1000; Cell Signaling Technology, #9252), XBP1s (1:1000; Cell Signaling Technology, #12782), ATF3 (1:1000; Cell Signaling Technology, #33593), DDIT3 (1:1000; Cell Signaling Technology, #2895), BMI1 (1:1000; Cell Signaling Technology, #6964), RING1A (1:1000; Cell Signaling Technology, #13069) and RING1B (1:1000; Cell Signaling Technology, #5694),.
Nuclear/cytosol fractionation
Cellular fractionation was performed as described before (37). UMRC6 cells cultured in 10-cm dish with or without glucose for 4 hours followed by nuclear and cytosol fractionation. Nuclear and cytosol proteins were analyzed by Western blotting. H2A and vinculin were used as loading control for nuclear and cytosol proteins respectively.
Protein dephosphorylation assay
Cell lysates were collected in NP-40 lysis buffer with no added inhibitors. After protein concentration was determined, the protein sample was combined with water to a total volume of 40 μL and incubated with 5 μL of 10X NEBuffer for protein metallophosphatases, 5 μL of 10 mM MnCl2, and 1 μL of lambda protein phosphatase (New England Biolabs, P0753) for 30 min at 30 °C. Protein samples were then analyzed by western blotting.
ROS level measurement
To measure levels of reactive oxygen species (ROS), treated cells cultured in 12-well plates were incubated with fresh medium containing 2 μM cell-permeant 2’,7’-dichlorodihydrofluorescein diacetate (H2DCFDA; ThermoFisher Scientific, D399) for 30 minutes, washed once with PBS, and subjected to fluorescence-activated cell sorting (FACS) analysis. Fluorescence in channel 1 was captured and plotted by using FlowJo_V10 software.
NADP+ and NADPH measurement
NADP+ and NADPH levels were measured based on a modified method as previously described (PMID: 22660331). UMRC6 cells were cultured in 6-cm dish with or without glucose combined with vehicle or 10 mM 2DG for 4 hours. Cell lysates were collected in 300 μL extraction buffer (20 mM nicotinamide, 20 mM NaHCO3, 100 mM Na2CO3) followed by centrifugation. To measure total NADP, 20 μL cell supernatant was added into a 96-well plate and mixed with 80 μL of NADP-cycling buffer (100 mM Tris-HCl pH8.0, 0.5 mM thiazolyl blue (MTT), 2mM phenazine ethosulfate, 5 mM EDTA) containing 0.75 U of G6PD enzyme (Sigma, #G4134). After incubating in the dark for 1 min at 30 °C and adding 20 μL of 10 mM glucose 6-phosphate to the mixture, absorbance at 570 nm was measured at 30 °C with Gen5 microplate reader (BIOTEK). To measure NADPH, 150 μL supernatant from each lysate was incubated at 60 °C for 30 min (destroyed NADP+ without affecting NADPH), followed by measuring absorbance at 570 nm. Then NADP+ level was calculated by subtracting [NADPH] from [total NADP] and NADP+/NADPH ration was plotted.
Glucose uptake assay
Cells were seeded in 12-well plates 24 hours before each experiment. Then, fresh medium with or without 10 μM of the glucose transporter-1 (GLUT1) inhibitor BAY-876 was added to each well. After treatment for the periods indicated in the figure legends, medium in each well was replaced with glucose uptake medium containing 25 μM glucose and 0.2 μCi 2-[1-14C] labeled 2DG. Plates were then incubated for the indicated times followed by washing with PBS and lysing in 0.1 mM NaOH. Radioactivity was measured with a Tri-Carb Liquid Scintillation Analyzer (PerKinElmer, Model 4810TR) according to the manufacturer’s instructions. All experiments were carried out with three independent replicates.
Xenograft model
Tumor xenograft models were created and tested as described in our previous studies (41) and in accordance with a protocol approved by the Institutional Animal Care and Use Committee of The University of Texas MD Anderson Cancer Center. Female 4- to 6-week-old athymic nude mice (Foxn1nu/Foxn1nu) were obtained from the Experimental Radiation Oncology Breeding Core Facility at MD Anderson Cancer Center and housed in the Animal Care Facility at the Department of Veterinary Medicine and Surgery at MD Anderson. H460 lung cancer cells were resuspended in PBS and 1.5 × 106 cells were injected into mice subcutaneously. Tumor volumes were monitored by bi-dimensional tumor measurements 2 to 3 times a week until the specified endpoint and calculated according to the equation v = length × width2 × 0.5. For drug treatment experiments, nude mice were treated with vehicle, 10 mg/kg PRT-4165, or 5 mg/kg BAY-876 by intraperitoneal injection daily beginning at 12 days after tumor inoculation until the specified endpoint.
ChIP-qPCR
Chromatin immunoprecipitation (ChIP) qPCR experiments were performed as previously described (42). Briefly, cells were cultured and treated in 15-cm dishes and analyzed with a SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling Technology, 9003) per the manufacturer’s instructions. After DNA pulldown with primary antibody against H2Aub (Cell Signaling Technology, #8240), H3K27me3 (Cell Signaling Technology, #9733) or BMI1 (Cell Signaling Technology, #6964) and purification, specific primers targeting different gene promoters were used for RT-PCR. The signal relative to input was evaluated using a formula from the manufacturer’s protocol as follows: percent input = 2% × 2(C[T]2% input sample – C[T]IP sample), where C[T] = the threshold cycle of PCR reaction. The sequences of primers used for ChIP-qPCR are listed in Table S1.
ChIP-seq
Chromatin immunoprecipitation coupled with high-throughput sequencing (ChIP-seq) was done with the SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling Technology, 9003) per the manufacturer’s instructions. To pull down associated DNA, 20 μg of chromatin was incubated overnight with 5 μL H2Aub antibody (Cell Signaling Technology, 8240). During incubation, 100 ng fly chromatin and 0.5 μg H2Av antibody (ActiveMotif, 61686) were added to each tube as spike-in controls. DNA was purified and prepared for ChIP-seq in the Sequencing and Microarray Facility at MD Anderson Cancer Center. The ChIP-seq data (GSE137885) were deposited in the public genomics data repository Gene Expression Omnibus.
ChIP-seq analysis
The raw reads (single-end, 50 bp) were aligned to the human reference genome hg19 by using bowtie (v1.1.0), allowing up to one mismatch. Before peak calling, we used spike-in normalization for sample size correction as previously described (43). For simplicity, the reads were downsampled to keep the same spike-in read counts in different samples. Peak calling was done with model-based analysis of ChIP-Seq (MACS v1.4.2) (44). with a cutoff of P ≤ 1 × 10-8. Clonal reads were automatically removed by MACS. Promoter regions were defined as regions 5 kb upstream to 5 kb downstream of the transcription start site (TSS). The H2Aub occupancies at promoter regions were normalized as reads per kilobase per million reads (RPKM). The average profile and heatmap around the TSS were obtained by generating a data matrix with deepTools (45) and visualized with R.
Gene expression profiling
RNA sequencing was done as described elsewhere (46) and complete datasets were deposited in GEO (GSE95097). Genes that were expressed differently upon glucose starvation were defined by a false discovery rate (FDR) cutoff of < 0.05 with at least a 2-fold change. Gene set enrichment was analyzed with the GSEA method (Broad Institute). Gene ontology (GO) analysis was done with the DAVID online service (http://david.ncifcrf.gov), with P values adjusted by Bonferroni correction. Expression data on BMI1, ATF3, and DDIT3 genes were obtained from The Cancer Genome Atlas (TCGA). All of these data were generated by using the UCSC Xena Browser (http://xena.ucsc.edu/).
Statistics
For all statistical analyses, significance was determined at *P < 0.05, **P < 0.01, ***P < 0.001, or ****P < 0.0001. Comparisons between two conditions or groups were done with two-tailed Student’s t tests in GraphPad Prism (GraphPad Software, Inc.). Two-way analysis of variance was used to calculate differences among three or more groups. Data are expressed as mean ± S.D., with at least 3 independent replicates in each group.
RESULTS
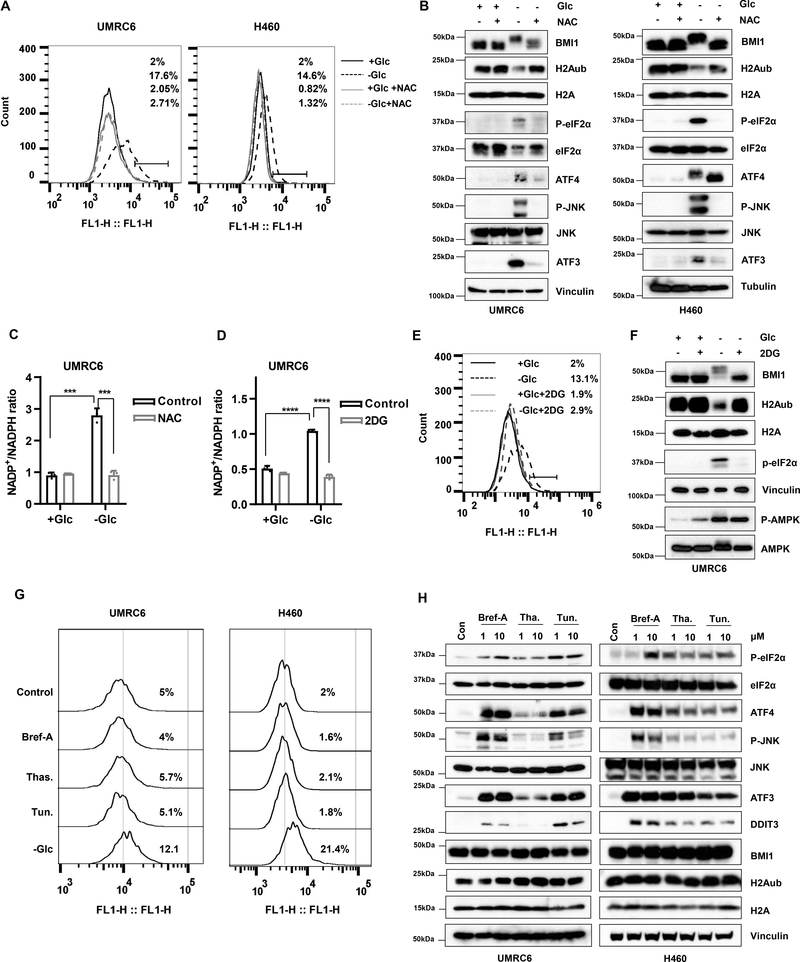
Glucose starvation decreases H2Aub independent of energy stress and AMPK
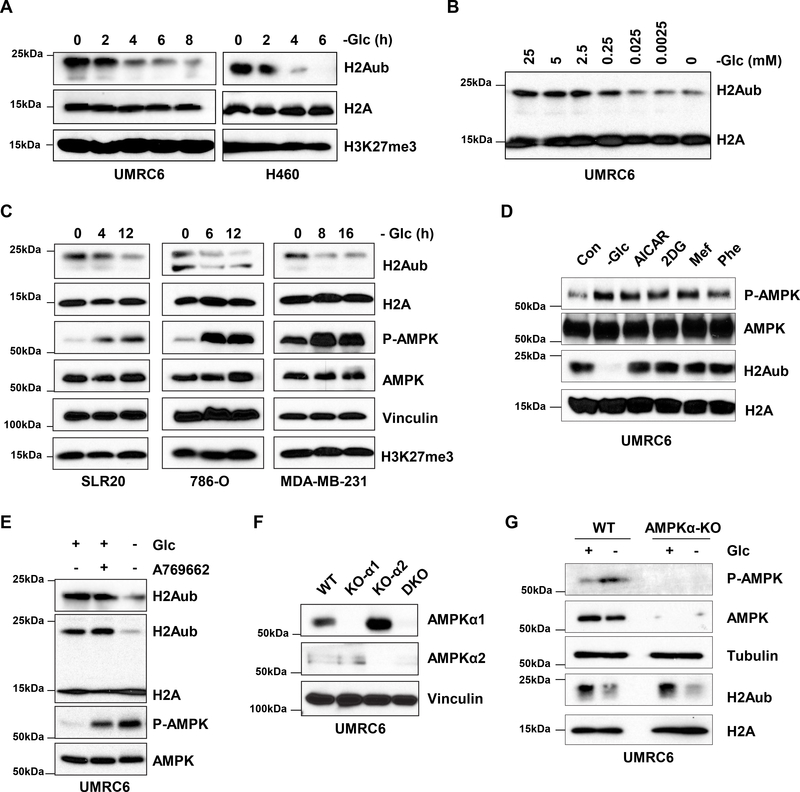
In studying whether H2Aub levels could be modulated by changes in nutrient availability, we found that glucose starvation potently decreased global H2Aub levels in a dose- and time-dependent manner (Fig. 1A–1B). We further confirmed this observation in a variety of cell lines (Fig. 1A–1C). Although previous studies suggested cross-talk between H2Aub and H3K27me3 regulation (47), we found that glucose starvation significantly reduced H2Aub levels without obviously affecting H3K27me3 levels (Fig. 1C), suggesting a specific regulation of H2Aub by glucose starvation.
Figure 1.
Glucose starvation decreases H2Aub independent of energy stress and AMPK. A, Western blots show that glucose deprivation decreases H2A monoubiquitination (H2Aub) levels in UMRC6 and H460 tumor cell lines in a time-dependent manner. B, Western blots show that glucose deprivation for 4 hours decreases H2Aub levels in UMRC6 tumor cells in a concentration-dependent manner. C, Western blots show that glucose deprivation decreases H2Aub levels in additional tumor cell lines in a time-dependent manner. D, Western blots show H2Aub levels in UMRC6 tumor cells after a 4-hour incubation in glucose-free medium or a 16-hour incubation with 1 mM 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR), 10 mM 2-deoxy-D-glucose (2DG), 2 mM metformin (Mef), or 2 mM phenformin (Phe). E, Western blots show H2Aub levels in UMRC6 cells after a 4-hour incubation in glucose-free medium or a 16-hour incubation with 200 μM A769662. F, Western blots show AMPKα1 or AMPKα2 expression in CRISPR/Cas9-mediated knockout (KO) cells. G, Western blots show H2Aub and phosphorylated AMPKα levels in wild-type (WT) or AMPKα1/2 double-knockout (AMPKα-KO) cells after 4 hours of glucose withdrawal.
Glucose starvation is well known to induce energy stress (with decreased cellular ATP and increased AMP levels) and to activate AMP-activated protein kinase (AMPK), a critical cellular energy sensor, by promoting its phosphorylation at Thr172 (48). Consistent with this, we found that the decrease in H2Aub levels was accompanied by increased AMPK Thr172 phosphorylation under glucose starvation conditions (Fig. 1C). Surprisingly, H2Aub levels were not obviously affected by other treatments that either induce or mimic energy stress, including the AMP analog AICAR, the glycolysis inhibitor 2DG, and the mitochondrial respiratory complex I inhibitors metformin and phenformin (Fig. S1), even though all of these treatments induced AMPK phosphorylation (Fig. 1D). Further, treatment with the AMPK activator A769662 induced AMPK phosphorylation to a level comparable to that induced by glucose starvation but did not affect H2Aub levels (Fig. 1E). To study the potential role of AMPK in regulating H2Aub levels under glucose starvation, we generated AMPK α1 and α2 double-knockout cells by using CRISPR/Cas9-mediated gene editing technology (Fig. 1F). As shown in Fig. 1G, glucose starvation decreased the H2Aub level in AMPK double-knockout cells to a level similar to that in wild-type cells. Taken together, these findings suggest that glucose deprivation decreases H2Aub levels independently of energy stress and AMPK signaling.
Genome-wide analyzes link H2Aub regulation to ER stress upon glucose deprivation
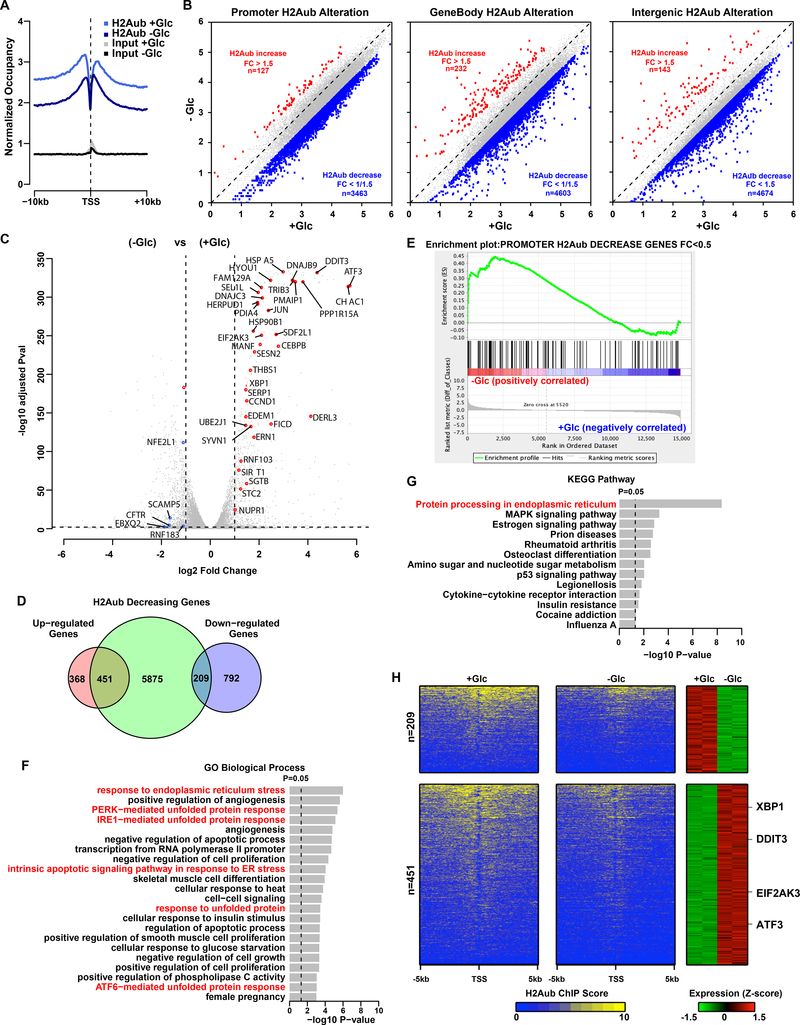
The aforementioned findings prompted us to conduct genome-wide analyses to characterize glucose starvation–associated H2Aub occupancy and corresponding transcriptional alterations in the genome. To this end, we performed H2Aub ChIP-seq analyses in UMRC6 cells cultured in medium with or without glucose, with 2 independent replicates. Both replicates showed consistent global patterns in terms of glucose starvation–induced H2Aub occupancy changes in the genome (Fig. S2A and 2B). Averaged H2Aub profiles from these two replicates clearly showed a genome-wide downregulation of H2Aub levels upon glucose starvation (Fig. 2A), and identified 3463, 4603, and 4674 genes exhibiting decreased H2Aub occupancy at their promoter, gene body, and intergenic regions, respectively; in contrast, only 127, 232, and 143 genes showed increased H2Aub occupancy at these regions (Fig. 2B). Further analysis identified 5875 genes with decreased H2Aub occupancy at their promoters, gene bodies, or both upon glucose starvation. Overall, our H2Aub ChIP-seq findings are consistent with our H2Aub western blotting findings from the same cell line (Fig. 1A).
Figure 2.
Genome-wide analyzes link H2Aub regulation to endoplasmic reticulum (ER) stress upon glucose deprivation. A, Average profiles of H2Aub in the indicated conditions. Glc, glucose; TSS, transcription start site. B, Scatter plots show changes in H2Aub occupancy due to glucose deprivation at the promoter, genebody, and intergenic regions. H2Aub occupancy was calculated as log2 kilobase per million read (RPKM) value. FC, fold change. C, Volcano plot shows changes in gene expression due to glucose deprivation, with ER stress genes highlighted in red. P values were adjusted by Bonferroni correction. D, Venn diagram shows overlap between 6,535 genes with decreased H2Aub occupancies and 1,820 differentially expressed genes (fold change > 2, false discovery rate < 0.05) upon glucose deprivation in UMRC6 cells. E, Gene set enrichment analysis showing that the 324 genes with >2-fold reduction in H2Aub were positively enriched in glucose deprivation upregulated genes. F, Gene ontology (GO) and G, Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses of the 451 H2Aub decreased and upregulated genes. P values were determined by Fisher’s exact test and adjusted by Bonferroni correction. H, Left 2 panels, heatmaps show the H2Aub profile around the TSS of 209 downregulated and 451 upregulated genes from overlap analysis in D, in decreasing order. Right panel shows the expression levels of the corresponding genes.
We also used RNA-seq analysis to profile changes in glucose starvation–associated gene expression in UMRC6 cells, which identified 1,820 differentially expressed genes, including 819 upregulated and 1001 downregulated genes, upon glucose starvation (fold change > 2, FDR < 0.05); notably, glucose starvation induced more pronounced expression changes in upregulated genes than in downregulated genes (Fig. 2C). Integration of both H2Aub ChIP-seq and RNA-seq data sets identified 660 genes that exhibited differential expression and decreased H2Aub occupancy upon glucose starvation, including 451 upregulated genes and 209 downregulated genes (Fig. 2D). Gene set enrichment analysis (GSEA) revealed that upregulated genes were significantly enriched in these 660 genes with decreased H2Aub occupancy (Fig. 2E; normalized enrichment score = 1.80, FDR = 0), which is consistent with the prevailing model that H2Aub is generally associated with transcriptional repression (6–8).
To identify the biological processes associated with the genes identified in our genome-wide analyses, we used Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses for these 451 upregulated and 209 downregulated genes (Fig. 2F–2G, S2C–2D). Notably, although no biological process or pathway was significantly enriched in the downregulated genes (Fig. S2C–2D), both the GO and KEGG pathway analyses identified a striking enrichment of ER stress‒related biological processes or pathways in upregulated genes (Fig. 2F–2G). Further analysis revealed that numerous genes involved in the ER stress response, such as EIF2AK3 (also known as PERK), DDIT3 (also known as CHOP), ATF3, and XBP1, were drastically upregulated (Fig. 2C) with decreased H2Aub occupancy upon glucose starvation (Fig. 2H). Together, our genome-wide analyses reveal that glucose starvation–mediated H2Aub repression is mainly associated with transcriptional activation, with a particular enrichment in genes involved in the ER stress response.
Glucose starvation decreases H2Aub binding on ER stress genes and induces their expression
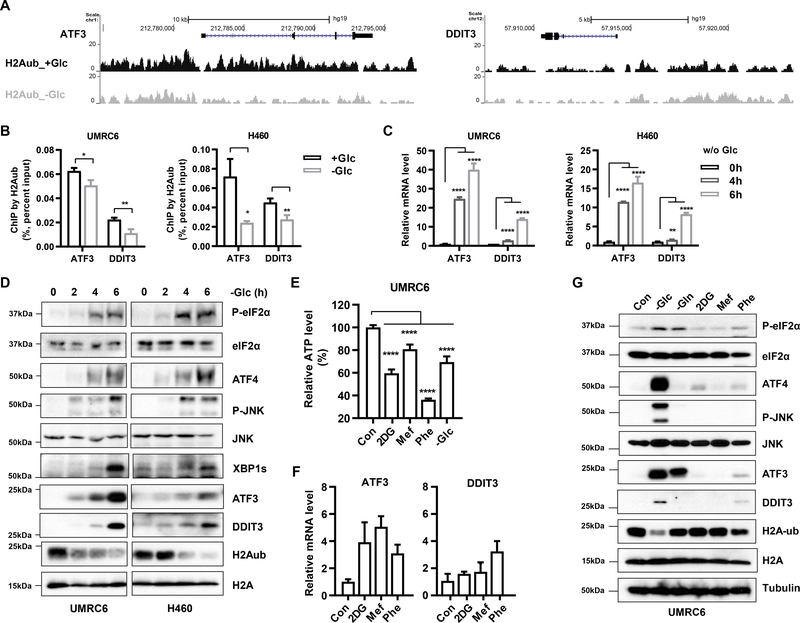
Given the significant enrichment of ER stress pathways in our genome-wide analyses, we focused on ER stress genes in our subsequent analyses. Analysis of our H2Aub ChIP-seq data revealed that glucose starvation significantly decreased H2Aub occupancy at both the promoter and body of the genes EIF2AK3, DDIT3, ATF3, and XBP1 (Figs. 3A and S3A). H2Aub ChIP assays confirmed decreased binding of H2Aub on the promoters of DDIT3 and ATF3 genes, two critical transcription factors and effectors in the ER stress response (Figs. 3B and S3B). Consistent with our observation in Fig. 1A, glucose starvation did not significantly affect H3K27me3 levels on the ATF3 and DDIT3 gene promoters in UMRC6 cells (Fig. S3C). Correspondingly, glucose starvation markedly increased the expression levels of DDIT3 and ATF3 (Fig. 3C), suggesting that glucose starvation decreases H2Aub binding on ER stress genes, leading to their increased expression. Glucose starvation also drastically increased other ER stress surrogates such as eIF2α phosphorylation, XBP1s, and ATF4 induction, which correlated with the H2Aub reduction during glucose starvation (Figs. 3D and S3D).
Figure 3.
Glucose starvation decreases H2Aub binding on ER stress genes and induces their expression. A, H2Aub ChIP–seq occupancy profiles at the ATF3 and DDIT3 loci in UMRC6 cells treated or not treated with glucose. B, ChIP-qPCR analysis of H2Aub levels on ATF3 and DDIT3 gene promoters upon glucose starvation in UMRC6 and H460 tumor cells. C, RT-PCR analysis of mRNA levels of ATF3 and DDIT3 genes upon glucose starvation for indicated times in UMRC6 and H460 cells. D, Western blots show activation of ER stress signaling in UMRC6 and H460 cells upon glucose starvation for the indicated times. E, UMRC6 cells were treated with 10 mM 2-deoxyglucose (2DG), 2 mM meformin, or 2 mM phenformin for 6 hours, or glucose-free medium for 3 hours, after which ATP levels were measured in the treated cells and normalized to levels in untreated cells. F, UMRC6 cells were treated as described in E followed by RT-PCR analysis for mRNA levels of ATF3 and DDIT3 genes. G, Western blots show H2Aub levels and ER stress signaling in UMRC6 cells after a 16-hour incubation with 10 mM 2DG, 2 mM meformin, or 2 mM phenformin, a 3-hour incubation in glucose-free medium, or a 6-hour incubation in glutamine-free medium. Error bars are mean ± s.d., n=3 (B, C, F) or 5 (E) independent repeats. Two-tailed unpaired Student’s t tests were used to compare two groups. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
As noted earlier, we found that other energy stress inducers, such as 2DG, metformin, and phenformin, did not regulate H2Aub levels (Fig. 1D). We therefore studied their effects on inducing the ER stress response. Under treatment conditions that 2DG, metformin, and phenformin treatment decreased cellular ATP to levels similar to or even lower than those caused by glucose starvation (Fig. 3E), their effects on inducing DDIT3 and ATF3 expression or other ER stress markers were significantly weaker than glucose starvation (Fig. 3F–3G). Further time course experiments revealed that in general the effects of these other energy stress inducers on ER stress markers were significantly weaker than glucose starvation at different time points (except for DDIT3 induction at 4-hour treatment with phenformin) (Fig. S3E). Amino acid deprivation is also well known to induce an integrated stress response (49). Consistent with this, we found that glutamine deprivation induced some surrogates involved in the ER stress response, such as eIF2α phosphorylation; however, unlike glucose deprivation, glutamine deprivation did not obviously affect H2Aub levels (Fig. 3G). Therefore, although numerous mechanisms are likely involved in inducing ER stress in response to different stress stimuli, our findings suggest that H2Aub regulation of ER stress gene expression serves as an unique mechanism linking glucose starvation to the ER stress response, which is also consistent with the more potent ER stress induced by glucose starvation relative to that induced by other stress stimuli we examined.
Glucose starvation promotes BMI1 phosphorylation and decreases its binding to ER stress genes
Next, we sought to study the underlying mechanisms by which glucose starvation regulates H2Aub levels. H2Aub levels are balanced by its writers and erasers. PRC1, the major H2Aub writer, monoubiquitinates H2A at K119 (9), whereas BAP1, the major H2Aub eraser, functions to remove monoubiquitin from H2A at K119 (12). Therefore, we hypothesized that glucose starvation modulates H2Aub levels by regulating PRC1, BAP1, or both. Notably, glucose starvation potently decreased H2Aub levels in UMRC6 cells (Figs. 1–2), a BAP1-deficient cancer cell line (18,21). We further showed that restoration of BAP1 wild-type, but not its catalytically dead mutant C91A, in UMRC6 cells drastically decreased H2A levels (Fig. S4A), and that glucose starvation was still capable of reducing H2Aub levels and inducing ER stress in BAP1 C91A-expressing UMRC6 cells (Fig. S4B), indicating that glucose starvation decreases H2Aub levels independent of BAP1 and its deubiquitinase activity.
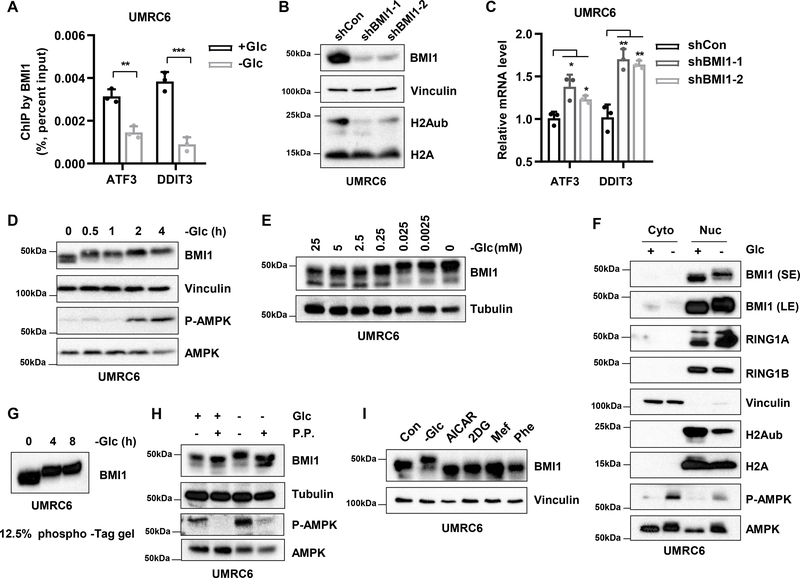
We then examined the potential relevance of PRC1 to glucose starvation–mediated H2Aub reduction. We showed that glucose starvation decreased the binding of BMI1, a critical component in PRC1, on the ATF3 and DDIT3 promoters (Fig. 4A). Further, similar to glucose starvation, knocking down BMI1 decreased H2Aub levels (Fig. 4B) and increased ATF3 and DDIT3 expression (Fig. 4C). Consistent with this, analyses of TCGA data sets revealed that BMI1 expression inversely correlated with that of ATF3 (or DDIT3) in a variety of cancer types, including testicular germ cell tumor, breast invasive carcinoma, renal clear cell carcinoma, renal papillary cell carcinoma, prostate adenocarcinoma, and uterine corpus endometrial carcinoma (Fig. S4C), suggesting that BMI1 regulation of ATF3 and DDIT3 expression likely has relevance in human cancers.
Figure 4.
Glucose starvation promotes BMI1 phosphorylation and decreases its binding to ER stress genes. A, ChIP-qPCR analysis of BMI1 binding on ATF3 and DDIT3 gene promoters in UMRC6 cells after 8 hours of glucose (Glc) deprivation. B, Western blots show decreased H2Aub levels upon BMI1 knockdown in UMRC6 cells. C, RT-PCR analysis of mRNA levels of ATF3 and DDIT3 in UMRC6 cells after BMI1 knockdown. D, Western blots show protein levels in UMRC6 cells cultured in glucose-free medium for the indicated times. E, Western blots show protein levels in UMRC6 cells cultured for 6 hours in medium with the indicated glucose concentrations. F, UMRC6 cultured in media with or without glucose for 4 hours followed by fractionation and Western blotting analysis. Vinculin, a marker for cytosol (Cyto); H2A, a marker for nucleus (Nuc). G, Phosphorylation of BMI1 in UMRC6 cells upon glucose deprivation was analyzed by Phos-tag sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). H, UMRC6 cells were cultured with or without glucose for 4 hours, after which cell lysates were subjected to lambda phosphatase (P.P.) treatment followed by western blotting. I, Western blots show protein levels in UMRC6 cells after the treatments described in Fig. 1D. Error bars are mean ± s.d., n=3 independent repeats. Two-tailed unpaired Student’s t tests were used to compare two groups. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
We further noted that glucose starvation induced a potent mobility shift of BMI1 protein in several cell lines (Fig. 4D–4E and S4D), suggesting that glucose starvation induces post-translational modifications to BMI1. Of note, glucose starvation did not affect BMI1 nuclear localization, nor affected RING1A or 1B protein levels (or its protein mobility or nuclear localization) (Fig. 4F). We also showed that glucose starvation similarly induced retarded migration of BMI protein on phos-tag gels (in which phosphorylated proteins bind to phos-tag and migrate more slowly) (Fig. 4G) and that protein phosphatase treatment abolished the glucose starvation–induced BMI1 mobility shift (Fig. 4H), confirming that glucose starvation induces BMI1 phosphorylation. Importantly, glucose starvation, but not AICAR, 2DG, metformin, or phenformin treatment, induced BMI1 phosphorylation (Fig. 4I).
Together, these findings suggest a model in which under glucose-replete conditions, PRC1 binds on the promoters of ER stress genes and represses their expression by promoting H2Aub levels on these genes; glucose starvation dissociates BMI1-containing PRC1 from ER stress gene promoters, potentially through BMI1 phosphorylation, leading to decreased H2Aub levels and de-repression of ER stress genes. Further elucidation of this model would require identifying the upstream kinase(s) that mediate glucose starvation–induced BMI phosphorylation and the corresponding phosphorylation site(s) on BMI1. However, at this time technical issues have prevented us from identifying such phosphorylation sites on BMI1. We did examine the potential involvement of several glucose starvation–activated kinases, including AMPK, JNK, and p38, in glucose starvation–induced BMI1 phosphorylation. However, our results showed that genetic or pharmacologic inactivation of these kinases did not affect glucose starvation–induced BMI1 phosphorylation (Fig. S4E–S4G), suggesting that some other kinase(s) might mediate BMI1 phosphorylation in response to glucose starvation.
Glucose starvation regulates BMI1 phosphorylation and H2Aub likely through NADPH depletion
Our aforementioned findings next led us to question how glucose starvation modulates BMI1 phosphorylation and H2Aub levels. Once glucose is taken up into cells, it is mainly shunted into glycolysis to generate ATP (as well as other important metabolic intermediates) (50). Consistent with the role of 2DG as a glycolysis inhibitor, both glucose starvation and 2DG treatment depleted ATP (see Fig. 3E). However, glucose starvation, but not 2DG treatment, affected H2Aub levels (see Fig. 1D); therefore, it seems unlikely that glucose starvation modulates H2Aub levels through glycolysis or ATP generation. Glucose can also be shunted into the pentose phosphate pathway (PPP) to generate NADPH, a reducing power to maintain redox homeostasis and to suppress ROS (50,51); glucose starvation is well known to induce ROS (51). Indeed, glucose starvation but none of other energy stress inducers significantly induced ROS (Fig. S5A), consistent with their effects on H2Aub deubiquitination (Fig. 3G and Fig. S5B). Therefore, we first tested whether treatment with ROS scavengers N-acetyl cysteine (NAC), Tempol or Trolox can reverse glucose starvation–induced BMI1 phosphorylation and H2A deubiquitination. Our analyses showed that NAC abolished glucose starvation–induced ROS (Fig. 5A) and BMI1 phosphorylation, restored H2Aub levels, significantly dampened ER stress induction under glucose starvation conditions (Fig. 5B). In contrast, Tempol or Trolox did not exert any rescuing effect on glucose starvation-induced cell death, BMI1 phosphorylation, H2A deubiquitination, or ER stress marker induction (Fig. S5C–5D); however, in these experiments we also noticed that Tempol or Trolox could significantly, but not completely, suppressed DCFDA staining under glucose starvation (Fig. S5E), making it challenging to draw a definitive conclusion. Conversely, we tested whether ROS-inducing conditions, such as H2O2 treatment, can mimic glucose starvation to induce BMI1 phosphorylation and H2A deubiquitination. As shown in Fig. S5F–S5G, H2O2 treatment potently induced ROS but did not obviously affect BMI1 phosphorylation or H2Aub levels, suggesting that ROS induced by H2O2 is not sufficient to induce these effects.
Figure 5.
Glucose starvation regulates BMI1 phosphorylation and H2Aub likely through NADPH depletion. A, Relative ROS levels of UMRC6 and H460 cells in glucose-free medium with or without 5 mM N-acetyl cysteine for 4 hours were measured by H2DCFDA staining followed by FACS analysis. B, Western blotting analyzes of protein levels in UMRC6 and H460 cells treated as described in A. C, NADP+ and NADPH levels in UMRC6 cells treated cultured in medium with or without glucose combined with vehicle or NAC (5mM) for 4 hours. NADP+ /NADPH ratio was plotted. D, NADP+ and NADPH levels in UMRC6 cells cultured in medium with or without glucose combined with vehicle or 2DG (10mM) for 4 hours were measured and plotted as NADP+/NADPH ratio. E, Relative ROS levels in UMRC6 cells treated as described in D followed by H2DCFDA staining and FACS analysis. F, UMRC6 cells were treated as described in D followed by Western blotting analysis. G, Relative ROS levels in UMRC6 and H460 cells after a 16-hour incubation with 10 μM brefeldin A (Bref-A), thapsigargin (Thas.), or tunicamycin (Tun.) or a 4-hour incubation with glucose-free medium were measured by H2DCFDA staining followed by FACS analysis. H, Western blots show H2Aub levels and ER stress signaling in URMC6 and H460 cells treated with the indicated drugs at the concentrations described in G. Error bars are mean ± s.d., n=3 independent repeats. Two-tailed unpaired Student’s t tests were used to compare two groups. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
As noted above, glucose supplies NADPH through the PPP. We showed that glucose starvation decreased NADPH levels (shown as increased NADP+/NADPH ratio), and NAC, but not Tempol or Trolox, restored NADPH levels under glucose starvation (Fig. 5C and S5H). We therefore took another approach to further examine the role of NADPH depletion on glucose starvation–induced BMI1 phosphorylation or H2A deubiquitination. Specifically, although 2DG cannot enter the glycolysis pathway downstream of the phosphoglucose isomerase reaction, it can still be shunted to the PPP to supply NADPH (52). Consistent with this, we found that 2DG significantly restored NADPH level, reversed ROS, BMI1 phosphorylation, H2A deubiquitination, and eIF2α phosphorylation under glucose starvation (Fig. 5D–5F). Correspondingly, 2DG also significantly rescued cell death upon glucose starvation (Fig. S5I). These data suggest that glucose starvation induces BMI1 phosphorylation and H2A deubiquitination likely through NADPH depletion.
Finally, we showed that treatment with ER stress inducers, including brefeldin A, thapsigargin, and tunicamycin, potently induced ER stress surrogates but did not affect ROS, BMI1 phosphorylation, or H2Aub levels (Fig. 5G–5H). Conversely, blocking PERK-eIF2-ATF4 signaling by ISRIB treatment abolished ATF4 induction upon glucose starvation as expected (53), but had no effect on BMI1 phosphorylation or H2Aub level under glucose starvation (Fig. S5J), suggesting that BMI1 phosphorylation and H2A deubiquitination under glucose starvation occur independent of ER stress induction.
PRC1 inactivation cooperates with ATF4 induction to promote cell death under glucose starvation
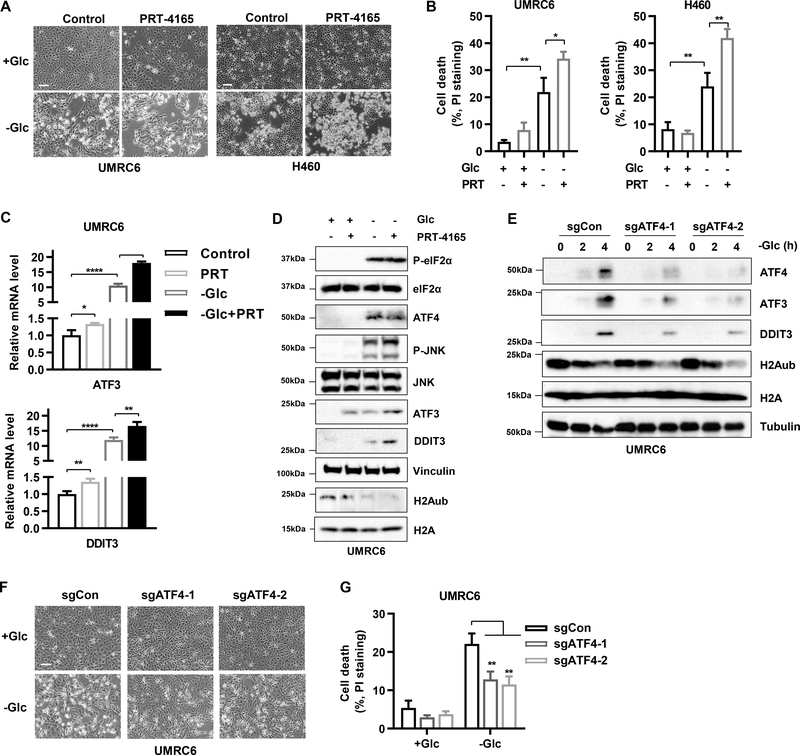
Glucose starvation induces rapid cell death in many cancer cell lines. We therefore studied whether PRC1 has a role in glucose starvation–induced cell death. As expected, treatment with the PRC1 inhibitor PRT-4165 decreased global H2Aub levels as well as H2Aub binding on the ATF3 and DDIT3 promoters (Fig. S6A–S6B). We also found that although PRT-4165 treatment alone is not sufficient to induce obvious cell death, it did potentiate glucose starvation–induced cell death (Fig. 6A–6B). Likewise, BMI1 knockdown also promoted cell death upon glucose starvation (Fig. S6C). We also found PRT-4165 treatment synergized with glucose starvation to induce ATF3 and DDIT3 expression (Figs. 6C–6D and S6D). These findings suggest that upon glucose starvation, PRC1 inactivation–induced H2A de-ubiquitination needs to cooperate with another glucose starvation–induced parallel signaling event to induce ER stress response and promote cell death.
Figure 6.
PRC1 inactivation cooperates with ATF4 induction to promote cell death under glucose starvation. A, Representative photos of UMRC6 and H460 cells pretreated or not pretreated with 50 μM PRT-4165 for 16 hours followed by culturing in glucose-free medium for 4 hours. Scale bar, 100 μm. B, Propidium iodide staining of cells treated as described in A followed by FACS analysis. C, RT-PCR analysis of ATF3 and DDIT3 mRNA levels in UMRC6 cells pretreated or not pretreated with 50 μM PRT-4165 for 16 hours followed by culturing in glucose-free medium for 4 hours. D, Western blots show protein levels in UMRC6 cells pretreated with 50 μM PRT-4165 and then cultured in medium with or without glucose. E, Western blots show protein levels in CRISPR/Cas9 mediated ATF4-knockout cells cultured in glucose-free medium for 4 hours. F, Representative photos of the treated cells in D. G, Propidium iodide staining of cells treated as described in E followed by FACS analysis. Error bars are mean ± s.d., n=3 independent repeats. Two-tailed unpaired Student’s t tests were used to compare two groups. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
We considered ATF4 transcription factor activation as one such signaling event, given that (i) ATF4 is a master transcription factor involved in ER stress response (49); (ii) glucose starvation is known to induce ATF4 levels (30,54); and (iii) stress-induced ATF4 typically involves enhanced ATF4 mRNA translation but not transcription (55) and therefore is most likely independent of PRC1- and H2Aub-mediated epigenetic mechanisms; therefore, PRC1 and ATF4 could act in a parallel way to regulate ER stress gene expression. Consistent with this, glucose starvation markedly increased ATF4 protein levels (Fig. 6E), but analyses of our RNA-seq and H2Aub ChIP-seq data revealed that glucose starvation did not significantly increase ATF4 mRNA levels or H2Aub binding on the ATF4 promoter (Fig. 3C and S6E). Moreover, PRT-4165 treatment did not affect ATF4 levels under either glucose-replete or glucose-deprivation conditions (Fig. 6D). We further showed that ATF4 deletion by CRISPR-Cas9 largely abolished glucose starvation–induced ATF3 and DDIT3 expression (without affecting H2Aub levels) (Fig. 6E), and significantly protected cells from glucose starvation–induced cell death (Fig. 6F–6G). Finally, PRT-4165-induced ATF3 and DDIT3 expression was largely abolished in ATF4 knockout cells (Fig. S6F), suggesting that ATF4 is required for PRC1-mediated epigenetic regulation of ATF3 and DDIT3 gene expression. Together, these findings suggest that under glucose starvation conditions, PRC1 inactivation–mediated H2Aub reduction cooperates with ATF4 induction to promote ER stress and cell death.
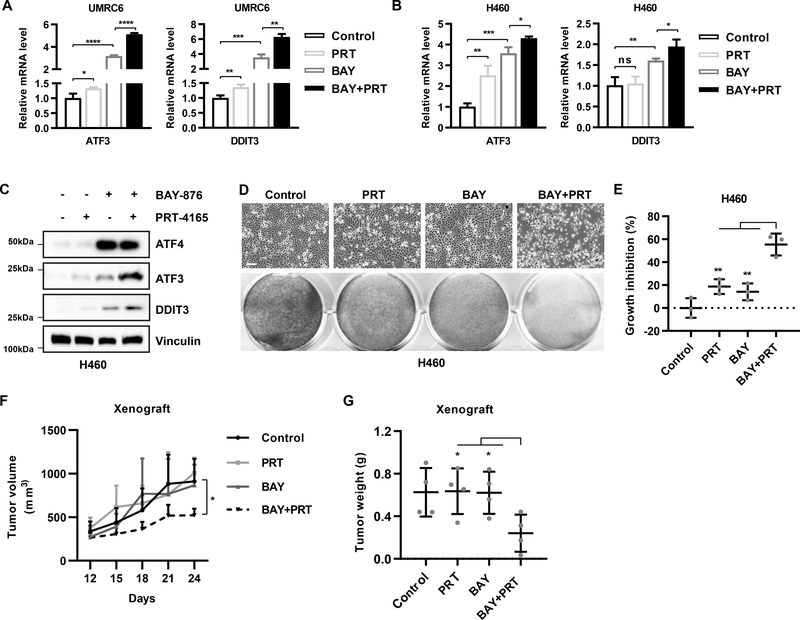
Combined inhibition of GLUT1 and PRC1 synergistically promotes ER stress and suppresses tumor growth
Considerable interest has been expressed in targeting GLUTs in cancer therapies (51). On the basis of our findings that PRC1 inhibition potentiated glucose starvation–induced ER stress and cell death (Fig. 6A–6B), we tested whether PRC1 inhibition would similarly synergize with pharmacologic blockade of glucose uptake to induce ER stress and cell death. To this end, we tested BAY-876, a potent GLUT1 inhibitor recently developed by Bayer (56). We confirmed that treatment with BAY-876 potently inhibited glucose uptake in UMRC6 and H460 cells (Fig. S7A–S7B); of note, residual glucose uptake still remained in the BAY-876-treated cells, likely because BAY-876 cannot completely block GLUT1-mediated glucose transport, or because glucose uptake can also be mediated by other glucose transporters (such as GLUT3 or sodium-dependent glucose transporters) in cancer cells. Correspondingly, we observed a more moderate induction of ATF3 and DDIT3 expression by BAY-876 treatment than by glucose starvation (compare Fig. 7A–7B and Figs. 6C and S6C). Importantly, PRT-4165 treatment significantly synergized with BAY-876 treatment to induce ATF3 and DDIT3 expression (Fig. 7A–7C) as well as cell death and growth inhibition (Fig. 7D–7E). Finally, using an H460 xenograft model, we showed that although PRT-4165 or BAY-876 treatment alone did not affect tumor growth, combined treatment with PRT-4165 and BAY-876 significantly inhibited tumor growth in vivo (Fig. 7F–7G). Notably, we did not observe any obvious weight loss from treatment with these inhibitors (either individually or in combination) in our animal studies (Fig. S7C), indicating that this combination treatment was well tolerated in vivo. Together, these findings show that combined inhibition of GLUT1 and PRC1 synergistically promotes ER stress and suppresses tumor growth, thereby suggesting that combining GLUT1 and PRC1 inhibitors may be a novel anticancer therapeutic strategy.
Figure 7.
Combined inhibition of GLUT1 and PRC1 synergistically promotes ER stress and suppresses tumor growth. A, RT-PCR analysis of ATF3 and DDIT3 mRNA levels in URMC6 cells treated with 2 μM BAY-876, 50 μM PRT-4165, or both for 16 hours. B, RT-PCR analysis of ATF3 and DDIT3 mRNA levels in H460 cells treated with 5 μM BAY-876, 50 μM PRT-4165, or both for 16 hours. C, Western blots show protein levels in H460 cells treated with 5 μM BAY-876, 50 μM PRT-4165, or both for 24 hours. D, Representative photos of H460 cells treated with 5 μM BAY-876, 50 μM PRT-4165, or both for 48 hours. Bottom row shows cells stained with crystal violet solution. E, Growth inhibition after drug treatments was calculated after quantification of crystal violet staining for H460 cells as described in D. F, Tumor volume of xenografts generated from H460 cells after the indicated treatments, which were given as daily injections. n=4. G, Tumor weights at the end of treatment in each treatment group. n=4. Error bars are mean ± s.d., n=3 (A, B, E) or 4 (G) independent repeats. Two-tailed unpaired Student’s t tests were used to compare two groups. *P < 0.05; **P < 0.01; ***P < 0.001; ****, P < 0.0001.
DISCUSSION
Regulation of H2Aub-mediated gene transcription is critical for numerous biological processes, including development, stem cell pluripotency, hematopoiesis, DNA damage, and ferroptosis, and its dysregulation has been associated with various human diseases such as cancer (9,18,57–59). However, whether H2Aub levels and transcription of its associated gene can be regulated by nutrient availability and cellular metabolism was unknown until now. Here we showed that glucose availability regulates H2Aub levels and ER stress response in cancer cells. Our study suggests a model in which glucose starvation inhibits PRC1 function potentially through glucose starvation–induced BMI phosphorylation and thereby decreases H2Aub levels, which leads to the de-repression of ER stress gene expression and contributes to the ER stress response and glucose starvation–induced cell death (Fig. S8). Combining glucose starvation (or pharmacologic inhibition of GLUT1) with PRC1 inhibition precipitated ER stress–induced cell death and synergistically suppressed tumor growth.
We showed that glucose starvation, but not other energy stress inducers or mimetics, induced BMI1 phosphorylation, decreased H2Aub levels, and increased ER stress surrogates. Importantly, treatment with NAC and 2DG restored NADPH levels upon glucose starvation, and largely rescued the effects of glucose starvation on BMI1 phosphorylation, H2Aub levels, and ER stress. Therefore, our study supports a critical role of NADPH depletion, but not energy stress response, in mediating glucose starvation–induced H2Aub reduction and ER stress response.
Currently, how glucose starvation–induced NADPH depletion decreases H2Aub levels remains unclear. Our findings with BAP1-deficient cells strongly suggest that this regulation occurs through BAP1-independent mechanisms. Because PRC1-deficient cells already exhibited significantly lower H2Aub levels under basal conditions (see Fig. 5B for BMI1-knockdown cells), it would be challenging to assess a further reduction of H2Aub levels under glucose starvation conditions in PRC1-deficient cells, thereby precluding us from using this approach to study whether PRC1 is involved in glucose starvation regulation of H2Aub. Rather, we found that glucose starvation induced BMI1 phosphorylation and decreased BMI1 binding on ATF3 and DDIT3 promoters (Fig. 5A and 5D–5H). Because both glucose starvation and BMI1 knockdown increased ATF3 and DDIT3 expression, our findings suggest that glucose starvation–induced BMI1 phosphorylation likely inhibits BMI1 binding on ATF3 and DDIT3 promoters and BMI1-mediated repression of ATF3 and DDIT3 expression. Further testing this hypothesis would require identifying BMI1 phosphorylation site(s) induced by glucose starvation and characterization of corresponding phosphomimetic and non-phosphorylatable mutants of BMI1. However, technical issues have thus far prevented us from identifying such BMI1 phosphorylation site(s) by mass spectrometry. We also attempted to identify the upstream kinase(s) that might mediate glucose starvation–induced BMI1 phosphorylation. However, our findings ruled out the involvement of several kinases known to be activated by glucose starvation in this regulation (Fig. S5E). Further studies will be required to dissect the underlying mechanisms by which glucose starvation regulates BMI1 function and H2Aub levels.
In summary, by using integrated ChIP-seq and RNA-seq analyses, we uncovered a signaling coupling between glucose availability and epigenetic regulation of ER stress response. Mechanistically, glucose starvation represses H2Aub occupancy on ER stress genes, potentially through NADPH depletion–induced BMI1 phosphorylation, resulting in de-repression of ER stress gene expression and increases in glucose starvation–induced cell death. Therapeutically, we showed that combined inhibition of both GLUT1 and PRC1 promotes cell death and suppresses tumor growth, providing a potential novel therapeutic anticancer strategy.
Supplementary Material
Significance.
Findings link glucose deprivation and H2A ubiquitination to regulation of the ER stress response in tumor growth and demonstrate pharmacologic susceptibility to inhibition of polycomb and glucose transporters.
Acknowledgements
We thank the members of the Gan laboratory for their advice and technical assistance. We thank Christine F. Wogan at the Department of Radiation Oncology of MD Anderson Cancer Center for manuscript editing. This research was supported by Bridge Funding from The University of Texas MD Anderson Cancer Center, an NGS allowance from the Center for Cancer Epigenetics at MD Anderson Cancer Center, a Career Enhancement Award from UT SPORE in Lung Cancer NIH/NCI 5P50CA070907, grant KC180131 from Department of Defense Kidney Cancer Research Program, grant R01CA181196 from the National Institutes of Health (to B.G.), grant R01HG007538, R01CA193466 and R01CA228140 from the National Institutes of Health (to W.L.), and by the Cancer Center Support (Core) Grant P30CA016672 from the National Cancer Institute, National Institutes of Health. Y.Z. and P.K. were Scholars at the Center for Cancer Epigenetics at MD Anderson. P.K. is also supported by Cancer Prevention and Research Institute of Texas (CPRIT) Research Training Grant RP170067 and by the Dr. John J. Kopchick Research Award from the MD Anderson UTHealth Graduate School of Biomedical Sciences.
Footnotes
Conflict of interest statement: No potential conflicts of interest were disclosed.
REFERENCES
- 1.Efeyan A, Comb WC, Sabatini DM. Nutrient-sensing mechanisms and pathways. Nature 2015;517:302–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu C, Thompson CB. Metabolic regulation of epigenetics. Cell Metab 2012;16:9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharma U, Rando OJ. Metabolic Inputs into the Epigenome. Cell Metab 2017;25:544–58 [DOI] [PubMed] [Google Scholar]
- 4.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 2003;33 Suppl:245–54 [DOI] [PubMed] [Google Scholar]
- 5.Campbell SL, Wellen KE. Metabolic Signaling to the Nucleus in Cancer. Mol Cell 2018;71:398–408 [DOI] [PubMed] [Google Scholar]
- 6.Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature 2004;431:873–8 [DOI] [PubMed] [Google Scholar]
- 7.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell 2005;20:845–54 [DOI] [PubMed] [Google Scholar]
- 8.Scheuermann JC, Gutierrez L, Muller J. Histone H2A monoubiquitination and Polycomb repression: the missing pieces of the puzzle. Fly (Austin) 2012;6:162–8 [DOI] [PubMed] [Google Scholar]
- 9.Gil J, O’Loghlen A. PRC1 complex diversity: where is it taking us? Trends Cell Biol 2014;24:632–41 [DOI] [PubMed] [Google Scholar]
- 10.Blackledge NP, Rose NR, Klose RJ. Targeting Polycomb systems to regulate gene expression: modifications to a complex story. Nat Rev Mol Cell Biol 2015;16:643–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gray F, Cho HJ, Shukla S, He S, Harris A, Boytsov B, et al. BMI1 regulates PRC1 architecture and activity through homo- and hetero-oligomerization. Nat Commun 2016;7:13343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheuermann JC, de Ayala Alonso AG, Oktaba K, Ly-Hartig N, McGinty RK, Fraterman S, et al. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 2010;465:243–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yong KJ, Basseres DS, Welner RS, Zhang WC, Yang H, Yan B, et al. Targeted BMI1 inhibition impairs tumor growth in lung adenocarcinomas with low CEBPalpha expression. Sci Transl Med 2016;8:350ra104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ganaie AA, Beigh FH, Astone M, Ferrari MG, Maqbool R, Umbreen S, et al. BMI1 Drives Metastasis of Prostate Cancer in Caucasian and African-American Men and Is A Potential Therapeutic Target: Hypothesis Tested in Race-specific Models. Clin Cancer Res 2018;24:6421–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bansal N, Bartucci M, Yusuff S, Davis S, Flaherty K, Huselid E, et al. BMI-1 Targeting Interferes with Patient-Derived Tumor-Initiating Cell Survival and Tumor Growth in Prostate Cancer. Clin Cancer Res 2016;22:6176–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ismail IH, McDonald D, Strickfaden H, Xu Z, Hendzel MJ. A small molecule inhibitor of polycomb repressive complex 1 inhibits ubiquitin signaling at DNA double-strand breaks. J Biol Chem 2013;288:26944–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carbone M, Yang H, Pass HI, Krausz T, Testa JR, Gaudino G. BAP1 and cancer. Nat Rev Cancer 2013;13:153–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Shi J, Liu X, Feng L, Gong Z, Koppula P, et al. BAP1 links metabolic regulation of ferroptosis to tumour suppression. Nature cell biology 2018;20:1181–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bott M, Brevet M, Taylor BS, Shimizu S, Ito T, Wang L, et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat Genet 2011;43:668–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiesner T, Obenauf AC, Murali R, Fried I, Griewank KG, Ulz P, et al. Germline mutations in BAP1 predispose to melanocytic tumors. Nat Genet 2011;43:1018–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pena-Llopis S, Vega-Rubin-de-Celis S, Liao A, Leng N, Pavia-Jimenez A, Wang S, et al. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet 2012;44:751–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiao Y, Pawlik TM, Anders RA, Selaru FM, Streppel MM, Lucas DJ, et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet 2013;45:1470–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Senft D, Ronai ZA. UPR, autophagy, and mitochondria crosstalk underlies the ER stress response. Trends Biochem Sci 2015;40:141–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu C, Bailly-Maitre B, Reed JC. Endoplasmic reticulum stress: cell life and death decisions. J Clin Invest 2005;115:2656–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hetz C The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol 2012;13:89–102 [DOI] [PubMed] [Google Scholar]
- 26.Iurlaro R, Munoz-Pinedo C. Cell death induced by endoplasmic reticulum stress. FEBS J 2016;283:2640–52 [DOI] [PubMed] [Google Scholar]
- 27.Lin A, Yao J, Zhuang L, Wang D, Han J, Lam EW, et al. The FoxO-BNIP3 axis exerts a unique regulation of mTORC1 and cell survival under energy stress. Oncogene 2014;33:3183–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X, Xiao ZD, Han L, Zhang J, Lee SW, Wang W, et al. LncRNA NBR2 engages a metabolic checkpoint by regulating AMPK under energy stress. Nature cell biology 2016;18:431–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Koppula P, Gan B. Regulation of H2A ubiquitination and SLC7A11 expression by BAP1 and PRC1. Cell Cycle 2019:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koppula P, Zhang Y, Shi J, Li W, Gan B. The glutamate/cystine antiporter SLC7A11/xCT enhances cancer cell dependency on glucose by exporting glutamate. J Biol Chem 2017;292:14240–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu X, Gan B. lncRNA NBR2 modulates cancer cell sensitivity to phenformin through GLUT1. Cell Cycle 2016;15:3471–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chauhan AS, Liu X, Jing J, Lee H, Yadav RK, Liu J, et al. STIM2 interacts with AMPK and regulates calcium-induced AMPK activation. FASEB J 2019;33:2957–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gan B, Hu J, Jiang S, Liu Y, Sahin E, Zhuang L, et al. Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature 2010;468:701–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gan B, Sahin E, Jiang S, Sanchez-Aguilera A, Scott KL, Chin L, et al. mTORC1-dependent and -independent regulation of stem cell renewal, differentiation, and mobilization. Proc Natl Acad Sci U S A 2008;105:19384–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gan B, Lim C, Chu G, Hua S, Ding Z, Collins M, et al. FoxOs enforce a progression checkpoint to constrain mTORC1-activated renal tumorigenesis. Cancer Cell 2010;18:472–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee H, Dai F, Zhuang L, Xiao ZD, Kim J, Zhang Y, et al. BAF180 regulates cellular senescence and hematopoietic stem cell homeostasis through p21. Oncotarget 2016;7:19134–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee H, Zandkarimi F, Zhang Y, Meena JK, Kim J, Zhuang L, et al. Energy-stress-mediated AMPK activation inhibits ferroptosis. Nature cell biology 2020;22:225–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao ZD, Han L, Lee H, Zhuang L, Zhang Y, Baddour J, et al. Energy stress-induced lncRNA FILNC1 represses c-Myc-mediated energy metabolism and inhibits renal tumor development. Nat Commun 2017;8:783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gan B, Yoo Y, Guan JL. Association of focal adhesion kinase with tuberous sclerosis complex 2 in the regulation of s6 kinase activation and cell growth. J Biol Chem 2006;281:37321–9 [DOI] [PubMed] [Google Scholar]
- 40.Gan B, Melkoumian ZK, Wu X, Guan KL, Guan JL. Identification of FIP200 interaction with the TSC1-TSC2 complex and its role in regulation of cell size control. The Journal of cell biology 2005;170:379–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lei G, Zhang Y, Koppula P, Liu X, Zhang J, Lin SH, et al. The role of ferroptosis in ionizing radiation-induced cell death and tumor suppression. Cell Res 2020;30:146–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin A, Piao HL, Zhuang L, Sarbassov DD, Ma L, Gan B. FoxO transcription factors promote AKT Ser473 phosphorylation and renal tumor growth in response to pharmacological inhibition of the PI3K-AKT pathway. Cancer Res 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen K, Hu Z, Xia Z, Zhao D, Li W, Tyler JK. The Overlooked Fact: Fundamental Need for Spike-In Control for Virtually All Genome-Wide Analyses. Mol Cell Biol 2015;36:662–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008;9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010;26:136–8 [DOI] [PubMed] [Google Scholar]
- 46.Dai F, Lee H, Zhang Y, Zhuang L, Yao H, Xi Y, et al. BAP1 inhibits the ER stress gene regulatory network and modulates metabolic stress response. Proc Natl Acad Sci U S A 2017;114:3192–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kalb R, Latwiel S, Baymaz HI, Jansen PW, Muller CW, Vermeulen M, et al. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat Struct Mol Biol 2014;21:569–71 [DOI] [PubMed] [Google Scholar]
- 48.Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol 2012;13:251–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM. The integrated stress response. EMBO Rep 2016;17:1374–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 2009;324:1029–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hay N Reprogramming glucose metabolism in cancer: can it be exploited for cancer therapy? Nat Rev Cancer 2016;16:635–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang D, Li J, Wang F, Hu J, Wang S, Sun Y. 2-Deoxy-D-glucose targeting of glucose metabolism in cancer cells as a potential therapy. Cancer Lett 2014;355:176–83 [DOI] [PubMed] [Google Scholar]
- 53.Albert AE, Adua SJ, Cai WL, Arnal-Estape A, Cline GW, Liu Z, et al. Adaptive Protein Translation by the Integrated Stress Response Maintains the Proliferative and Migratory Capacity of Lung Adenocarcinoma Cells. Mol Cancer Res 2019;17:2343–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rozpedek W, Pytel D, Mucha B, Leszczynska H, Diehl JA, Majsterek I. The Role of the PERK/eIF2alpha/ATF4/CHOP Signaling Pathway in Tumor Progression During Endoplasmic Reticulum Stress. Curr Mol Med 2016;16:533–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci U S A 2004;101:11269–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siebeneicher H, Cleve A, Rehwinkel H, Neuhaus R, Heisler I, Muller T, et al. Identification and Optimization of the First Highly Selective GLUT1 Inhibitor BAY-876. ChemMedChem 2016;11:2261–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vidal M, Starowicz K. Polycomb complexes PRC1 and their function in hematopoiesis. Exp Hematol 2017;48:12–31 [DOI] [PubMed] [Google Scholar]
- 58.Kuznetsov JN, Aguero TH, Owens DA, Kurtenbach S, Field MG, Durante MA, et al. BAP1 regulates epigenetic switch from pluripotency to differentiation in developmental lineages giving rise to BAP1-mutant cancers. Sci Adv 2019;5:eaax1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van HT, Santos MA. Histone modifications and the DNA double-strand break response. Cell Cycle 2018;17:2399–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.