Abstract
Accumulating evidence suggests that metabolic reprogramming has a critical role in carcinogenesis and tumor progression. The usefulness of formalin-fixed paraffin-embedded (FFPE) tissue material for metabolomics analysis as compared with fresh frozen tissue material remains unclear. Liquid chromatography tandem mass spectrometry-based metabolomics analysis was performed on 11 pairs of matched tumor and normal tissues in both FFPE and fresh frozen tissue materials from colorectal carcinoma patients. Permutation t-test was applied to identify metabolites with differential abundance between tumor and normal tissues. A total of 200 metabolites were detected in the FFPE samples and 536 in the fresh frozen samples. The preservation of metabolites in FFPE samples was diverse according to classes and chemical characteristics, ranging from 78% (energy) to 0% (peptides). Compared with the normal tissues, 34 (17%) and 174 (32%) metabolites were either accumulated or depleted in the tumor tissues derived from FFPE and fresh frozen samples, respectively. Among them, 15 metabolites were common in both FFPE and fresh frozen samples. Notably, branched chain amino acids were highly accumulated in tumor tissues. Using KEGG pathway analyses, glyoxylate and dicarboxylate metabolism, arginine and proline, glycerophospholipid, and glycine, serine and threonine metabolism pathways distinguishing tumor from normal tissues were found in both FFPE and fresh frozen samples. This study demonstrates that informative data of metabolic profiles can be retrieved from FFPE tissue materials.
Keywords: colorectal neoplasms, metabolites, metabolic reprogramming, molecular pathological epidemiology, tumor microenvironment
Introduction
Recent development of analytical techniques, such as chromatography and mass spectrometry (MS), have empowered metabolomics as an important field of omics studies. Metabolites are the downstream products of genomic, transcriptomic, and proteomic variability, thus providing useful information on biological condition (1–4). Accumulating evidence indicates that cancer cells may reprogram cellular metabolism according to external or internal stimuli, such as nutritional starvation and extra- or intracellular stress, to survive in an adverse tissue microenvironment (5–9). Reprogrammed cancer metabolisms have also been suggested to facilitate nutrient production, including nucleotides, amino acids, and lipids, for cancer proliferation and adjustment to disadvantageous environment such as hypoxia and malnutrition. Metabolomics capture snapshots of various combinations of molecular alterations and complex interactions with host cells in the tumor microenvironment (10). Thus, through identifying metabolites and metabolic pathways with differential abundance between cancer and normal tissues, metabolomics could provide an insight into the establishment of novel biomarkers and the development of effective therapeutic strategies.
Tissue based metabolomics research has mainly been conducted using fresh frozen materials (1,11–17). However, a major downside of using fresh frozen materials is that many established cohort lack sufficient fresh frozen samples. This limitation could be overcome by using formalin-fixed paraffin-embedded (FFPE) tissue materials, as these samples are obtained routinely for pathological diagnostic processes. Since FFPE tissues can be easily archived for decades with the corresponding patient data for retrospective assessment, FFPE tissues constitute widely available resources of clinical information, which is essential for retrospective molecular studies. Because of widespread availability and long-term stability of FFPE tissues, accurate profiling of their metabolite content would greatly increase the potential for discovery and validation of clinically useful biomarkers. In this study, we aim to evaluate the precision and performance characteristics of comprehensive metabolomics study using FFPE and matched fresh frozen samples of colorectal cancer by liquid chromatography tandem mass spectrometry.
Materials and Methods
Human colorectal cancer tissue
Eleven samples from radical colectomies were used in the study. All samples were collected with written informed consent and by approval from Brigham and Women’s Hospital Institutional Review Board (Boston, MA). Matched FFPE and fresh frozen samples were collected from each colectomy. Tissue blocks were sectioned at 4 μm and were stained with hematoxylin and eosin (H&E) to identify tumor and normal area in each specimen. Sections of 20 μm were used for the metabolomics analysis. Histopathologic evaluation was performed to assess the tumor cell percentage in each sample.
Metabolome extraction
The metabolite extraction from FFPE and fresh frozen samples was performed, as previously described (18,19). Briefly, the metabolites from fresh frozen samples were extracted by incubating in 1 ml of 80% methanol at room temperature for 4 hours. After centrifugation at 14,000 g for 10 minutes, the supernatants were collected and stored at −80 C° until metabolic analyses. The metabolites from FFPE samples were extracted by incubating in 1ml of 80% methanol at 70 C° for 45 minutes in 1.5 ml micro centrifuge tubes without any de-paraffinization procedure. The samples were placed on ice for 15 min and centrifuged at 14,000 g for 10 minutes at 4 C°. The supernatants were transferred into new 1.5 ml micro centrifuge tubes and placed on ice for 10 minutes, followed by centrifugation at 14,000 g for 5 minutes at 4 C°. The supernatants were collected and stored at −80 C° until metabolic analyses.
Ultrahigh Performance Liquid Chromatography-Tandem Mass Spectroscopy
Metabolite profiling was conducted by Metabolon Inc (Durham, NC) as previously described (20). All methods utilized Waters ACQUITY ultra-performance liquid chromatography (UPLC) and a Thermo Scientific Q-Exactive high resolution/accurate MS interfaced with a heated electrospray ionization source and Orbitrap mass analyzer operated at 35,000 mass resolution.
Statistical analysis
All statistical analyses were conducted using R software (version 3.5.3), and all P values were two-sided. Our primary aim was to assess the deferentially abundant metabolites (DAM) between tumor and normal tissues and associated pathways enrichment. Principal component analysis was performed using the detected metabolites in FFPE and fresh frozen samples, individually. Two-dimensional plots constructed using first and second principal components as well as first and third principal components were used as visual aids for assessing sample quality. Permutation test was used for the identification of DAM whereby the null distribution was constructed using all the possible combination of randomly assigned tumor and normal labels to the 20 (or 22) FFPE (or fresh frozen) samples. Metabolites with P < 0.05 were considered significant and metabolites with P < 0.10 were used in metabolic pathways enrichment analysis. The pathway analysis module in MetaboAnalyst 4.0 was employed and hypergeometric test was used to test metabolites involved in a particular pathway are enriched compared to random hits. False discovery rate (FDR) was applied to reduce false-positive discovery in metabolic pathway enrichment analysis. The module also measures an impact score on each pathway based upon the position of metabolite(s), i.e. a centralized position that is connected to many other entities is considered more impactful. As such, we considered pathways satisfying the conditions of number of hits > 1, and either -ln FDR > 2, or impact score > 0.1 as influential pathway.
Results
Comparison of detected metabolites in FFPE and fresh frozen samples according to the class of metabolites
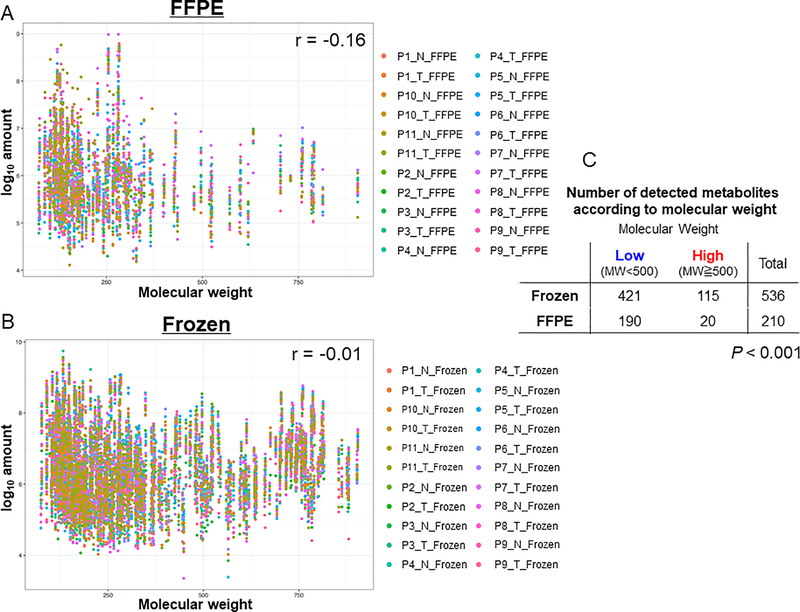
We first examined and compare the metabolite profiles of the FFPE and fresh frozen samples using untargeted UPLC-MS; an overview of the experiment design is depicted in Figure 1A. A total of 539 metabolites were detected from samples. Among them, 197 metabolites were identified in both FFPE and fresh frozen samples, 339 in only fresh frozen samples, and 3 in only FFPE samples (Figure 1B). We next categorized the identified metabolites according to the Human Metabolome Database. As shown in Figure 1C, the ratio of detected metabolites in FFPE samples to that in fresh frozen samples varied according to the class of the metabolites. In general, similar metabolite profile was observed in both FFPE and fresh frozen samples, except for the classes of peptide and energy (Figure 1D). There was no peptide detected in FFPE samples, whereas metabolites associated with energy were highly preserved in FFPE samples (78%) (Figure 1C). In order to investigate if the preservation of metabolites in FFPE samples according to molecular weight of metabolites, we examined the correlation between metabolites molecular weight and amount detected in FFPE and fresh frozen samples (Figure 2A and 2B). There was a stronger negative correlation between metabolites molecular weight and amount detected in FFPE samples (r = −0.16) compared with that in fresh frozen samples (r = −0.014). The detection rate of metabolites with larger than molecular weight of 500 in FFPE samples was 9.5% whereas that in fresh frozen samples was 21.5% (P < 0.001, Figure 2C). Approximately 37% of the metabolites detected in fresh frozen samples were detected in FFPE samples. We visualized in a heat-map that the abundance profile of the 197 common metabolites, which could be detected in both FFPE and fresh frozen samples, tended to be depleted in FFPE samples, as compared with that derived from fresh frozen samples (Figure 1E). Similar analyses for the component sub-classes, specifically 97 lipids, 53 amino acids, 18 nucleotides, 8 carbohydrates, 7 xenobiotics, 7 cofactors and vitamins, and 7 energy metabolites shared by FFPE and fresh frozen samples, are provided in Supplementary Figure 1A–C and Supplementary Figure S2A–D. These results suggest that the reproducibility of metabolite detection in FFPE samples compared to fresh frozen samples varies according to the class of the metabolites.
Figure 1.
Comparison of detected metabolites in formalin-fixed paraffin-embedded (FFPE) and fresh frozen samples according to the classes of metabolites. (A): Schematic overview of the protocol used to prepare FFPE and fresh frozen samples. (B): Venn diagram showing the number of detected metabolites and the intersection between FFPE and fresh frozen samples in the experimental settings. (C): Bar chart showing the percentage of detected metabolites derived from FFPE samples compared with fresh frozen samples according to the class of metabolites. (D): Pie chart showing the distribution of different classes of metabolites detected in FFPE and fresh frozen samples. (E): Heat-map of metabolites grouped by sample types and then tissue types. Heat-map colors represent z-score values of raw detection amount. FFPE, formalin-fixed paraffin-embedded; N, normal; T, tumor; UPLC/MS, ultra-performance liquid chromatography/mass spectrometry.
Figure 2.
(A, B): Scatter plots showing the correlations between metabolite molecular weight (X-axis) and amount (Y-axis) detected in FFPE (A) and fresh frozen (B) samples. Each dot represents one metabolite and the colors represent the sample identities. The r values were Spearman’s correlation coefficients. (C): Table shows the number of detected metabolites in FFPE and frozen samples whereby the metabolites were grouped into high and low molecular weight. P value was calculated by chi-square test. FFPE, formalin-fixed paraffin-embedded; MW, molecular weight; N, normal; T, tumor.
Differentially abundant metabolites between tumor and normal FFPE tissues
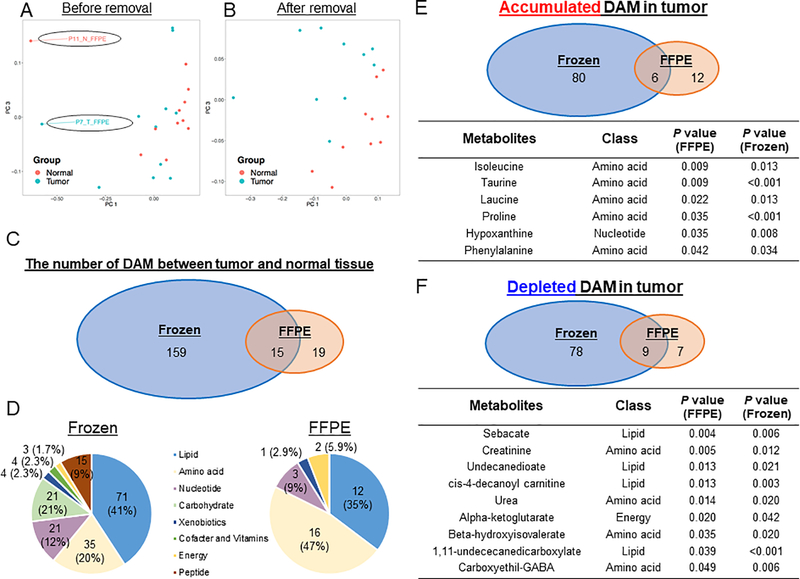
We next investigated the difference of metabolite profiles by comparing the metabolite abundance between tumor and normal tissues in FFPE samples. As a quality assessment, we performed principal component analysis (PCA) using the 200 metabolites detected in FFPE samples (Figure 3A and Supplementary Figure S3A) and 536 metabolites detected in fresh frozen samples (Supplementary Figure S3C and S3D). While the metabolic profiling derived from the tumor fresh frozen tissues were separated from the ones from normal fresh frozen tissues in the PC1-PC2 and PC1-PC3 space, the ones from two FFPE samples (P7_T_FFPE and P11_N_FFPE) exhibited different metabolite profiles compared with the ones from the others. Better separation between tumor and normal tissues within FFPE samples was achieved after excluding these two outlier samples (Figure 3B and Supplementary Figure S3B). As such, P7_T_FFPE and P11_N_FFPE were not included in downstream differentially abundant metabolites (DAM) and metabolic pathways analysis. A total of 193 metabolites were differentially abundant (P < 0.05) between tumor and normal tissues in either FFPE and fresh frozen samples (Figure 3C). Among them, 15 metabolites were identified in both FFPE and fresh frozen samples, while 159 were exclusive to fresh frozen samples and 19 were only detected in FFPE samples. These 34 DAM belong to different classes of metabolites in which higher proportions of amino acid and energy related metabolites were observed in FFPE samples (Figure 3D). Among tumor-accumulated DAM, 6 metabolites, including branched chain amino acids (BCAA) isoleucine and leucine, were identified from both FFPE and fresh frozen samples (Figure 3E). Among tumor-depleted DAM, 9 metabolites were identified in both FFPE and fresh frozen samples (Figure 3F). Therefore, metabolite profile aberrations caused by tumorigenesis as captured in DAM were partially preserved in FFPE samples as compared with fresh frozen samples.
Figure 3.
Differentially abundant metabolites (DAM) between tumor and normal tissues, in both FFPE and fresh frozen samples. (A, B): Principal component analysis plot showing the distribution of the tumor and normal tissues in FFPE samples in the first and third principal components space before (A) and after (B) removal of 2 samples; whereby the analysis was conducted using the Spearman correlation between the tumor and normal tissues on the 200 detected metabolites. (C): Venn diagram comparing the DAM of tumor and normal tissues between FFPE and fresh frozen samples (P < 0.05). (D): Pie charts showing the distribution of different classes of DAM of tumor and normal tissues, in FFPE and fresh frozen samples, respectively. (E): Upper Venn diagram showing the number of overlapping DAM accumulated in tumor against normal tissues between FFPE and fresh frozen samples. Lower table listing the types and P values of the 6 DAM found commonly in FFPE and fresh frozen samples. (F): Upper Venn diagram showing the number of overlapping DAM depleted in tumor against normal tissues between FFPE and fresh frozen samples. Lower table listing the types and P values of the 9 DAM found commonly in FFPE and fresh frozen samples. DAM, differentially abundant metabolites; FFPE, formalin-fixed paraffin-embedded; PC, principal component.
Metabolic pathways enrichment analysis using differentially abundant metabolites between tumor and normal tissues
In addition to assessing individual DAM, we further examined the metabolite pathways enriched in the DAM. Our interpretation focused on the influential pathways which were selected based on 3 conditions, i.e. -ln FDR > 2, impact scores > 0.1, and hit > 1. There was one pathway, glyoxylate and dicarboxylate metabolism pathway, enriched in tumor-depleted DAM between FFPE and fresh frozen samples (Figure 4A and 4B) and 3 pathways which were enriched in tumor-accumulated DAM detected simultaneously in both FFPE and fresh frozen samples (Figure 4C and 4D). These included arginine and proline, glycerophospholipid, and glycine, serine and threonine metabolism pathways. Hence, difference of profiling metabolic pathways between tumor and normal tissue in FFPE samples were, in part, similar to that in fresh frozen samples.
Figure 4.
Metabolic pathways enrichment analysis results obtained from MetaboAnalyst 4.0, using differentially abundant metabolites (DAM) between tumor and normal tissues, in (A, C) FFPE and (B, D) fresh frozen samples. (A-D) display the -ln FDR (vertical axis) and impact scores (horizontal axis) of individual pathways involved by DAM. Influential pathways which satisfied the conditions of -ln FDR > 2, impact score > 0.1, and number of hits > 1 as an influential pathway were highlighted and labelled with pathway names. One pathway enriched in tumor-depleted DAM and three pathways enriched in tumor-accumulated DAM, which were highlighted in boxes, were found in both FFPE and fresh frozen samples. *1) shows Valine, leucine and isoleucine biosynthesis pathway; *2) shows Cyanoamino acid metabolism pathway; *3) shows Phenylalanine, tyrosine and tryptophan biosynthesis pathway; *4) shows Alanine, aspartate and glutamate metabolism pathway; and *5) shows Amino sugar and nucleotide sugar metabolism pathway. DAM, differentially abundant metabolites; FFPE, formalin-fixed paraffin-embedded; FDR, false discovery rate.
Discussion
Experimental evidence indicates that genetic and epigenetic alterations occurring during carcinogenesis alter metabolic programming in favor of cancer cell survival and growth in tumor microenvironment (4,7,8,16,17,21–24). The detection of changes in metabolic profiles has the potential to advance early detection of cancer, as well as the prediction of prognosis or drug sensitivity (14,25–28). In view of the availability of FFPE archival samples compared to fresh frozen samples, we aimed to assess the feasibility of utilizing the FFPE samples, in addition to fresh frozen samples, for metabolomics analysis. Our data demonstrated tumorigenesis-related metabolic profiling in FFPE samples and is, thus, suggestive of the potential of utilizing FFPE samples for metabolomics studies, particularly for small molecular metabolites, although fresh frozen sample-derived metabolite profile was only partially recapitulated in the one from FFPE samples. Our data gives insight into feasibility of metabolomics analyses for molecular pathological epidemiology studies (29–31).
Accumulating evidence supports differential metabolic programming between tumor and normal tissues and the biological importance of metabolites (25,27,32). One remarkable example is the activation of glycolysis in cancer tissue, known as Warburg effect (33,34). Interestingly, activated glycolysis in cancer cells appear to be beneficial to cancer cell survival and growth by several mechanisms, such as increased nucleotide production utilizing activating pentose phosphate pathway (21,22). Consequently, depleted citric acid is replaced by utilizing alpha-ketoglutarate which is metabolized to citric acid by IDH (isocitrate dehydrogenase) and ACO (aconitase) (35). The current study detected the depletion of alpha-ketoglutarate, which is one of the metabolites in tricarboxylic acid cycle, in tumor tissues. Alpha-ketoglutarate is also decomposed into 2-hydroxyglutarate by IDH which often harbors point mutation in several cancers (4,23). The depletion of alpha-ketoglutarate in this study may indicate the activation of citric acid synthesizing and IDH pathways. In addition to energy metabolites, the depletion and accumulation of amino acid in tumor tissues were robustly detected in both FFPE and fresh frozen samples, and this study suggests that BCAA including isoleucine and leucine are accumulated in tumor tissues. Since these essential ketogenic BCAAs are degraded directly into acetyl-CoA and succinyl-CoA in human mitochondria, their accumulation reiterates mitochondrial dysfunction in tumor cells to completely oxidize ketogenic substrates into carbon dioxide that is accompanied by inherent transfer of protons with less deuterium from nutrients to metabolic matrix water (36,37). The fundamental mitochondrial function of curtailing deuterium oncoisotope accumulation in intermediary metabolites and nucleotides to prevent cell transformation has been argued as a surrogate marker for predicting response in personalized treatment and as a prognostic marker for patient survival in several clinical cancer studies (38,39). Our study thus ingeminates again the clinical importance of insufficient mitochondrial deuterium depletion with resulting oncogenic transformation that can be unearthed from paraffin embedded tissue samples via metabolic profiling. Moreover, BCAT1 (BCAA transaminase 1) plays a role in maintaining the stemness of cancer stem cells in producing and accumulating BCAA, whereby BCAA enhances cancer progression (4,7,23–25). Therefore, current study demonstrated the profiling of amino acid and energy was partially recapitulated in FFPE samples. Further investigations using larger cohorts are required to clarify the exact mechanisms that underlie the relationship between metabolic alterations and carcinogenesis in colorectal cancer.
Conventionally, a fresh frozen tissue material, frozen immediately after surgical resection, is believed to have much better preserve metabolites than a FFPE sample, which undergo the fixation. In the current study, the detectable metabolites in FFPE samples were approximately 40% of that in fresh frozen samples. Of note, large molecular metabolites, such as peptides, were undetected in FFPE samples. Our results are consistent with a recent prostate cancer study, which reported that FFPE samples have approximately 45% of metabolites compared with fresh frozen samples, but small molecular amino acids and a part of lipids are relatively-well preserved and detected in FFPE materials compared with that in fresh frozen tissue materials (19). Therefore, these findings support the potential utility of FFPE samples for comprehensive analyses targeting small molecular metabolites.
We acknowledge that there were several limitations in our study. First, as our sample size was relatively small, the statistical power is low. Hence, our findings need to be validated in independent datasets with larger number of samples. Second, since tissue samples include both cancer cells and stromal cells, it would be impossible to test if the detected metabolic changes were exclusively caused by the cancer cells. Third, the clinicopathological data were limited. Since colorectal cancer represents a heterogeneous group of neoplasms, the metabolic profiles of colorectal cancer may differ by tumor location, disease stage, microsatellite instability status, and somatic mutation status. However, as long as both FFPE and fresh frozen tissue materials were collected from the same patient and from adjacent tumor location, the comparison study between FFPE and fresh frozen materials should serve its purpose. Fourth, we have excluded two outliers for DAM analyses since these outliers demonstrated metabolic profiles that were different from the other specimens based on the principal component analysis. These outliers might represent different tumor subtype, such as microsatellite instability-high; therefore, further research is warranted to investigate the association of tumor subtype with metabolic profiling. Fifth, more comprehensive analyses including treatment and clinicopathological features using a larger sample size were not feasible in the current study.
In conclusion, our study suggests the potential utility of FFPE samples in studying small molecule metabolites in colorectal cancer and at the same time cautions its deficiency in preserving larger metabolites such as peptides. These findings need to be validated in additional large populations. Through leveraging the rich sources of FFPE archival samples, our study may help to shape future translational studies through developing strategies for colorectal cancer prevention and treatment through targeting metabolites.
Supplementary Material
Implications.
Our findings suggest potential value of metabolic profiling using FFPE tumor tissues and may help to shape future translational studies through developing treatment strategies targeting metabolites.
Acknowledgements
This work was supported by U.S. National Institutes of Health (NIH) grants (P50 CA127003 to C.S.F.; R01 CA118553 to C.S.F.; R01 CA169141 to C.S.F.; R01 CA137178 to A.T.C.; K24 DK098311 to A.T.C.; R35 CA197735 to S.O.; R01 CA151993 to S.O.; R21 CA230873 to S.O., K07 CA190673 to R.N.); by Nodal Award (2016-02) from the Dana-Farber Harvard Cancer Center (to S.O.); by Stand Up to Cancer Colorectal Cancer Dream Team Translational Research Grant (grant number SU2C-AACR-DT22-17; to C.S.F. and M.Gi.); and by grants from the Project P Fund, The Friends of the Dana-Farber Cancer Institute, Bennett Family Fund, and the Entertainment Industry Foundation through National Colorectal Cancer Research Alliance. Stand Up To Cancer is a division of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the scientific partner of SU2C. K.A. was supported by grants from Overseas Research Fellowship from Japan Society for the Promotion of Science (201860083). K.H. was supported by a fellowship grant from the Uehara Memorial Foundation and the Mitsukoshi Health and Welfare Foundation. K.F. was supported by a fellowship grant from the Uehara Memorial Foundation. A.T.C. is a Stuart and Suzanne Steele MGH Research Scholar. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We thank the supports of the Department of Surgery, Brigham and Women’s Hospital for providing us with tissue specimens.
Disclosure of Potential Conflicts of Interest: A.T.C. previously served as a consultant for Bayer Healthcare and Pfizer Inc.. R.N. is currently employed by Pfizer Inc.; she contributed to this study before she became an employee of Pfizer Inc. J.A.M has also served as an advisor/consultant to Ignyta, Array Pharmaceutical, and Cota. C.S.F. previously served as a consultant for Agios, Bain Capital, Bayer, Celgene, Dicerna, Five Prime Therapeutics, Gilead Sciences, Eli Lilly, Entrinsic Health, Genentech, KEW, Merck, Merrimack Pharmaceuticals, Pfizer Inc, Sanofi, Taiho, and Unum Therapeutics; C.S.F. also serves as a Director for CytomX Therapeutics and owns unexercised stock options for CytomX and Entrinsic Health. M.G. was on an advisory board for AstraZeneca and receives research funding from Bristol-Myers Squibb. This study was not funded by any of these commercial entities. No other conflicts of interest exist. The other authors declare that they have no conflicts of interest.
Abbreviations
- BCAA
branched chain amino acid
- DAM
differentially abundant metabolites
- FDR
false discovery rate
- FFPE
formalin-fixed paraffin-embedded
- KEGG
Kyoto encyclopedia of genes and genomes
- MS
mass spectrometry
- PCA
principal component analysis
- UPLC
ultra-performance liquid chromatography
Footnotes
Ethics approval and consent to participate: The research was approved by the Institutional Review Board of Brigham and Women’s Hospital.
Consent of publication: All subjects have written informed consent.
Use of standardized official symbols: We use HUGO (Human Genome Organization)-approved official symbols (or root symbols) for genes and gene products, including ACO, BCAT1 and IDH; all of which are described at www.genenames.org. Gene symbols are italicized whereas symbols for gene products are not italicized.
References
- 1.Suhre K, Shin SY, Petersen AK, Mohney RP, Meredith D, Wagele B, et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature 2011;477(7362):54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sullivan LB, Gui DY, Vander Heiden MG. Altered metabolite levels in cancer: implications for tumour biology and cancer therapy. Nat Rev Cancer 2016;16(11):680–93. [DOI] [PubMed] [Google Scholar]
- 3.Newgard CB. Metabolomics and Metabolic Diseases: Where Do We Stand? Cell Metab 2017;25(1):43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McBrayer SK, Mayers JR, DiNatale GJ, Shi DD, Khanal J, Chakraborty AA, et al. Transaminase Inhibition by 2-Hydroxyglutarate Impairs Glutamate Biosynthesis and Redox Homeostasis in Glioma. Cell 2018;175(1):101–16 e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, Metabolism, and Cancer. Cancer Discov 2015;5(10):1024–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kerr EM, Gaude E, Turrell FK, Frezza C, Martins CP. Mutant Kras copy number defines metabolic reprogramming and therapeutic susceptibilities. Nature 2016;531(7592):110–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fontana L, Cummings NE, Arriola Apelo SI, Neuman JC, Kasza I, Schmidt BA, et al. Decreased Consumption of Branched-Chain Amino Acids Improves Metabolic Health. Cell Rep 2016;16(2):520–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silvente-Poirot S, Dalenc F, Poirot M. The Effects of Cholesterol-Derived Oncometabolites on Nuclear Receptor Function in Cancer. Cancer Res 2018;78(17):4803–8. [DOI] [PubMed] [Google Scholar]
- 9.Arima K, Ohmuraya M, Miyake K, Koiwa M, Uchihara T, Izumi D, et al. Inhibition of 15-PGDH causes Kras-driven tumor expansion through prostaglandin E2-ALDH1 signaling in the pancreas. Oncogene 2019;38(8):1211–24. [DOI] [PubMed] [Google Scholar]
- 10.Priolo C, Pyne S, Rose J, Regan ER, Zadra G, Photopoulos C, et al. AKT1 and MYC induce distinctive metabolic fingerprints in human prostate cancer. Cancer Res 2014;74(24):7198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chinnaiyan P, Kensicki E, Bloom G, Prabhu A, Sarcar B, Kahali S, et al. The metabolomic signature of malignant glioma reflects accelerated anabolic metabolism. Cancer Res 2012;72(22):5878–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diamond DL, Krasnoselsky AL, Burnum KE, Monroe ME, Webb-Robertson BJ, McDermott JE, et al. Proteome and computational analyses reveal new insights into the mechanisms of hepatitis C virus-mediated liver disease posttransplantation. Hepatology 2012;56(1):28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ganti S, Taylor SL, Abu Aboud O, Yang J, Evans C, Osier MV, et al. Kidney tumor biomarkers revealed by simultaneous multiple matrix metabolomics analysis. Cancer Res 2012;72(14):3471–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Budhu A, Roessler S, Zhao X, Yu Z, Forgues M, Ji J, et al. Integrated metabolite and gene expression profiles identify lipid biomarkers associated with progression of hepatocellular carcinoma and patient outcomes. Gastroenterology 2013;144(5):1066–75 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wettersten HI, Hakimi AA, Morin D, Bianchi C, Johnstone ME, Donohoe DR, et al. Grade-Dependent Metabolic Reprogramming in Kidney Cancer Revealed by Combined Proteomics and Metabolomics Analysis. Cancer Res 2015;75(12):2541–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zand B, Previs RA, Zacharias NM, Rupaimoole R, Mitamura T, Nagaraja AS, et al. Role of Increased n-acetylaspartate Levels in Cancer. J Natl Cancer Inst 2016;108(6):djv426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu J, Reznik E, Lee HJ, Gundem G, Jonsson P, Sarungbam J, et al. Abnormal oxidative metabolism in a quiet genomic background underlies clear cell papillary renal cell carcinoma. Elife 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan M, Breitkopf SB, Yang X, Asara JM. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat Protoc 2012;7(5):872–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cacciatore S, Zadra G, Bango C, Penney KL, Tyekucheva S, Yanes O, et al. Metabolic Profiling in Formalin-Fixed and Paraffin-Embedded Prostate Cancer Tissues. Mol Cancer Res 2017;15(4):439–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem 2009;81(16):6656–67. [DOI] [PubMed] [Google Scholar]
- 21.Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, Aburatani H, et al. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell 2012;22(1):66–79. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto T, Takano N, Ishiwata K, Ohmura M, Nagahata Y, Matsuura T, et al. Reduced methylation of PFKFB3 in cancer cells shunts glucose towards the pentose phosphate pathway. Nat Commun 2014;5:3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hattori A, Tsunoda M, Konuma T, Kobayashi M, Nagy T, Glushka J, et al. Cancer progression by reprogrammed BCAA metabolism in myeloid leukaemia. Nature 2017;545(7655):500–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delphan M, Lin T, Liesenfeld DB, Nattenmuller J, Bohm JT, Gigic B, et al. Associations of branched-chain amino acids with parameters of energy balance and survival in colorectal cancer patients: Results from the ColoCare Study. Metabolomics 2018;2018(14):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayers JR, Wu C, Clish CB, Kraft P, Torrence ME, Fiske BP, et al. Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nat Med 2014;20(10):1193–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daemen A, Peterson D, Sahu N, McCord R, Du X, Liu B, et al. Metabolite profiling stratifies pancreatic ductal adenocarcinomas into subtypes with distinct sensitivities to metabolic inhibitors. Proc Natl Acad Sci U S A 2015;112(32):E4410–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kon S, Ishibashi K, Katoh H, Kitamoto S, Shirai T, Tanaka S, et al. Cell competition with normal epithelial cells promotes apical extrusion of transformed cells through metabolic changes. Nat Cell Biol 2017;19(5):530–41. [DOI] [PubMed] [Google Scholar]
- 28.Li J, Condello S, Thomes-Pepin J, Ma X, Xia Y, Hurley TD, et al. Lipid Desaturation Is a Metabolic Marker and Therapeutic Target of Ovarian Cancer Stem Cells. Cell Stem Cell 2017;20(3):303–14 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogino S, Chan AT, Fuchs CS, Giovannucci E. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut 2011;60(3):397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogino S, Nowak JA, Hamada T, Phipps AI, Peters U, Milner DA, Jr., et al. Integrative analysis of exogenous, endogenous, tumour and immune factors for precision medicine. Gut 2018;67(6):1168–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogino S, Nowak JA, Hamada T, Milner DA Jr., Nishihara R Insights into Pathogenic Interactions Among Environment, Host, and Tumor at the Crossroads of Molecular Pathology and Epidemiology. Annu Rev Pathol 2019;14:83–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munoz-Pinedo C, El Mjiyad N, Ricci JE. Cancer metabolism: current perspectives and future directions. Cell Death Dis 2012;3:e248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Warburg O On respiratory impairment in cancer cells. Science 1956;124(3215):269–70. [PubMed] [Google Scholar]
- 34.Warburg O On the origin of cancer cells. Science 1956;123(3191):309–14. [DOI] [PubMed] [Google Scholar]
- 35.Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 2011;481(7381):380–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boros LG, D’Agostino DP, Katz HE, Roth JP, Meuillet EJ, Somlyai G. Submolecular regulation of cell transformation by deuterium depleting water exchange reactions in the tricarboxylic acid substrate cycle. Med Hypotheses 2016;87:69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boros LG, Collins TQ, Somlyai G. What to eat or what not to eat-that is still the question. Neuro Oncol 2017;19(4):595–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hao D, Sarfaraz MO, Farshidfar F, Bebb DG, Lee CY, Card CM, et al. Temporal characterization of serum metabolite signatures in lung cancer patients undergoing treatment. Metabolomics 2016;12:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gros SJ, Holland-Cunz SG, Supuran CT, Braissant O. Personalized Treatment Response Assessment for Rare Childhood Tumors Using Microcalorimetry-Exemplified by Use of Carbonic Anhydrase IX and Aquaporin 1 Inhibitors. Int J Mol Sci 2019;20(20). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.