Abstract
Immunotherapies targeting programmed cell death protein 1 (PD-1) and programmed cell death 1 ligand 1 (PD-L1) immune checkpoints represent a major breakthrough in cancer treatment. PD-1 is an inhibitory receptor expressed on the surface of activated T-cells that dampens T-cell receptor (TCR)/CD28 signaling by engaging with its ligand PD-L1 expressed on cancer cells. Despite the clinical success of PD-1 blockade using monoclonal antibodies, most patients do not respond to the treatment, and the underlying regulatory mechanisms of PD-1 remain incompletely defined. Here we show that PD-1 is extensively N-glycosylated in T cells and the intensities of its specific glycoforms are altered upon TCR activation. Glycosylation was critical for maintaining PD-1 protein stability and cell surface localization. Glycosylation of PD-1, especially at the N58 site, was essential for mediating its interaction with PD-L1. The monoclonal antibody STM418 specifically targeted glycosylated PD-1, exhibiting higher binding affinity to PD-1 than FDA-approved PD-1 antibodies, potently inhibiting PD-L1/PD-1 binding, and enhancing anti-tumor immunity. Together these findings provide novel insights into the functional significance of PD-1 glycosylation and offer a rationale for targeting glycosylated PD-1 as a potential strategy for immunotherapy.
Keywords: Cancer biology, immunotherapy, immune checkpoint, PD-1, post-translational modification
Introduction
T-cell activation is an intricately regulated process that involves peptide-major histocompatibility complex (MHC) engagement of the T-cell receptor (TCR) and costimulatory signals (1). Upon T cell activation, co-inhibitory immune checkpoint proteins, such as programmed cell death protein 1 (PD-1), are induced and act as brakes on the activation (2). Cancer cells utilized the immune checkpoint proteins to avoid and suppress anti-tumor immune responses. The engagement of PD-1 with its ligand programmed death 1 ligand 1 (PD-L1), expressed on the surface of cancer cells, leads to T-cell exhaustion and immunosuppression (3). The inhibitory function of PD-l is primarily mediated by the tyrosine phosphatase SHP-2, which is recruited to the cytoplasmic domain of PD-1 and dephosphorylates the signaling molecules downstream of the TCR/CD28 (4,5). PD-1 transcriptional regulation has been extensively studied, and several transcription factors, including nuclear factor of activated T cells 1 (NFATC1), forkhead box protein O1 (FOXO1), T-bet (also known as T-box transcription factor TBX21), and B lymphocyte-induced maturation protein 1 (BLIMP1) have been reported to regulate PD-1 expression (6). However, the post-translational regulation of PD-1 (7,8) remains less clear.
Glycosylation is an important post-translational modification that covalently attaches carbohydrate moieties to proteins (9). N-linked glycosylation, in which N-acetylglucosamine is linked to the amide side chain, is regulated by the programmed remodeling of glycosyltransferases and glycosidases (10). The resulting abundant repertoire of glycans plays diverse essential roles in a large number of biological events. Recent studies on protein glycosylation (11,12) revealed that glycans associated with the cell surface receptors substantially modify the structure and function of proteins through steric influences, and directly modulate protein turnover and intermolecular interactions. There is considerable evidence showing that T-cell glycosylation is crucial for immune regulation (13,14). Although PD-1 is known to be a glycoprotein, the details of its glycan structure and functional significance of its glycosylation in regulating T-cell activation are not well understood.
Therapeutic blockade of the PD-1 pathway by monoclonal antibodies restores T-cell function and has demonstrated success in promoting long-lasting antitumor immune response (15). This led to the approval of two PD-1 (nivolumab and pembrolizumab) and three PD-L1 (atezolizumab, avelumab, and durvalumab) antibodies by the U.S. Food and Drug Administration (FDA) to treat multiple types of cancers, including melanoma, non-small-cell lung cancer, and head and neck squamous cell carcinoma. Although immune checkpoint blockade has revolutionized cancer treatment for some patients with promising results, most patients do not show complete response, and some cancers were even shown to be completely refractory to checkpoint blockade (16). Furthermore, toxicity and immune-related adverse events have been documented (6). These clinical findings underscore the need for a better mechanistic understanding of PD-1 regulation to identify more effective approaches to potently block the PD-1 pathway and boost the immune response for sustained patient benefits.
Here, we report that PD-1 is heavily glycosylated in T cells, and its glycan structure is altered upon TCR activation. PD-1 glycosylation is critical for its biological functions. Of note, we found that the glycosylation of PD-1, at N58 in particular, is essential for mediating the interaction with PD-L1. Based on those findings, we generated a monoclonal antibody, STM418, which targets glycosylated PD-1 and primarily recognizes N58 of PD-1. STM418 exhibits higher binding affinity to PD-1 than the FDA-approved nivolumab and pembrolizumab PD-1 antibodies, significantly neutralizes PD-L1/PD-1 binding, and enhances anti-tumor immunity. Together, our findings suggest that the glycosylation of PD-1 contributes to the intricate regulatory mechanisms of PD-1 in T cells and provide a rationale for targeting glycosylated PD-1 for immunotherapy.
Materials and Methods
Cell Lines, Antibodies, and Chemicals
All cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA), independently validated by short tandem repeat (STR) DNA fingerprinting at MD Anderson Cancer Center (Houston, TX), and negative for mycoplasma infection. These cells were cultured in DMEM medium or RPMI 1640 media (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific) at 37 °C in a humidified atmosphere with 5% CO2. Antibodies against PD-1, PD-L1 and ubiquitin were purchased from Cell Signaling Technology (Danvers, MA, USA). Antibodies against actin, tubulin and FLAG, phytohemagglutinin (PHA), tunicamycin (TM), cycloheximide (CHX), and MG132 were purchased from Sigma-Aldrich (St. Louis, MO, USA). Biotin-labeled lectins were purchased from Vector Laboratories (Burlingame, CA, USA).
Plasmids
The PD-1 gene was purchased from Origene (Rockville, MD, USA), and cloned into pCDH lentiviral expression vectors. pCDH-puro-PD-1-FLAG expression vector was used as a template to generate the PD-1-FLAG 1 NQ mutants, i.e., N49Q, N58Q, N74Q, and N116Q; 3NQ mutants, i.e., N49 (N58,74,116Q), N58 (N49,74,116Q), N74 (N49,58,116Q), and N116 (N49,58,74Q); and a 4NQ mutant (N49,58,74,116Q) by performing site-directed mutagenesis. All constructs were confirmed using enzyme digestion and DNA sequencing.
Generation of Stable Cells by Lentiviral Infection
To generate PD-1 expressing stable cells, 293T cells were co-transfected with pCDH-puro-PD-1-FLAG expression constructs (4 μg) with pCMV-dR8.2 (3 μg) and pCMV-VSVG (1 μg) helper constructs using Lipofectamine 2000 reagent (Life Technologies, Carlsbad, CA, USA). Viral stocks were harvested from the culture medium after 2 days and then filtered to remove non-adherent 293T cells. To select cells that stably express the PD-1 constructs, cells were plated at subconfluent densities and infected with a cocktail of 1 ml of virus-containing medium, 1 ml of regular medium and 8 μg/ml polybrene, and then selected in 1 μg/ml of puromycin (InvivoGen, San Diego, CA, USA) 48 hours after lentivirus infection.
Immunoblot Analysis and Immunoprecipitation
Immunoblot analysis was performed as described previously (17). Briefly, proteins were resolved by SDS-PAGE and transferred onto polyvinylidene difluoride membranes (Millipore, Burlington, MA, USA). The membranes were blocked in Tris-buffered saline containing 0.2% Tween 20 and 5% fat-free dry milk and incubated first with primary antibodies and then with horseradish peroxidase-conjugated secondary antibodies. Specific proteins were visualized with enhanced chemiluminescence detection reagent according to the manufacturer’s instructions (Pierce Biotechnology, Waltham, MA, USA). Image acquisition and quantification of band intensity were performed using a ChemiDoc imaging system (Bio-Rad, Hercules, CA, USA). For immunoprecipitation, the cells were lysed in lysis buffer (50 mM Tris-HCl [pH7.4], 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid and 1% Triton X-100) and centrifuged at 16,000 × g for 30 min to remove debris. Cleared lysates were subjected to immunoprecipitation by incubating with anti-FLAG M2 agarose (Sigma-Aldrich, St. Louis, MO, USA) at 4 °C. After 2 hours of incubation, the resin was washed three times with TBS and eluted with SDS-PAGE sample buffer.
Glycosylation Analysis of PD-1
To validate the glycosylation of PD-1 proteins, cell lysates were treated with PNGase F (New England BioLabs, Ipswich, MA, USA) as described by the manufacturer. Purified PD-1 proteins were stained using the Glycoprotein Staining Kit (Thermo Fisher Scientific) as described by the manufacturer.
Flow Cytometry
Cells were suspended in Cell Staining Buffer (CSB; BioLegend, San Diego, CA, USA) at 2 × 106 cells/mL. Cells (50 μL) were aliquoted into a 96-well round-bottom plate, to which 50 μL of 20 μg/mL primary antibody was added, followed by gentle mixing and incubation at 4 °C for 1 hour in the dark. Cells were washed with CSB and incubated with anti-mouse IgG-PE conjugate (10 μg/mL) at 4 °C in the dark. Cells were washed, and data were acquired using a Guava easyCyte HT (Millipore, Burlington, MA, USA) or FACS Celesta (Becton Dickinson, Franklin Lakes, NJ, USA) flow cytometer.
Analysis of PD-1/PD-L1 Binding by Live-Cell Imaging Assay
To measure PD-1/PD-L1 interaction, PD-1- expressing 293T cells were incubated with recombinant human PD-L1-Fc chimera protein (R&D Systems) for 1 hour and then anti-human Alexa Fluor 488 dye conjugate (Life Technologies). The fluorescence intensity of Alexa Fluor 488 dye was then monitored using a real-time IncuCyte live-cell analysis system (Essen BioScience, Ann Arbor, ML, USA) according to the manufacturer’s instructions.
Analysis of PD-1/PD-L1 Interaction by In vitro Plate-based Binding Assay
To measure PD-1/PD-L1 interaction, recombinant His-tagged PD-1 was incubated with or without Rapid PNGase F (New England BioLabs, Ipswich, MA, USA) in non-reducing buffer for 30 min at 50 °C and then coated on a nickel-coated 96-well plate. The plate was then incubated with recombinant PD-L1 Fc fusion protein for 1 hour. After that, Alexa Fluor 488 dye-conjugated anti-human IgG, Fc specific antibodies (Life Technologies) was added. The fluorescence intensity of Alexa Fluor 488 dye was determined using a microplate reader (Synergy Neo; BioTeK, Winooski, VT, USA) and normalized.
Detection of PD-1/PD-L1 Binding by Immunoprecipitation
Jurkat cells expressing PD-1 WT or various NQ mutants were washed in PBS and lysed in PTY buffer (50 mM HEPES, 50 mM NaCl, 5 mM EDTA, 1% Triton X-100, 50 mM NaF, 10 mM Na4P2O7. pH to 7.4) containing protease inhibitors. Lysates were incubated with 1 μg/ml mouse IgG2a (control) or PD-L1.Fc (mouse IgG2a) fusion protein and 8 × 106 Dynabeads Pan Mouse IgG for 2 hours. Beads were washed 3 times with 5× TBS-T buffer and once with 1× TBS-T buffer. Protein were eluted in 2× SDS-sample buffer and subjected to Western blot with rabbit anti-PD-1 antibody (Cell Signaling Technology).
Flow Cytometric Analysis of PD-1/PD-L1 Binding
Jurkat cells expressing PD-1 WT or various NQ mutants were incubated with 1 μg/ml hIgG1.Fc or PD-L1.Fc (human IgG1) fusion protein in incubation/wash buffer (PBS containing 0.5% BSA and 0.02% NaN3) for 1 h. Cells were then washed and incubated with APC-labeled anti-human PD-1 (BioLegend) or AF-488-labeled anti-human IgG.Fc antibody for 30 min. Cells were washed and fixed with 1.6% paraformaldehyde in PBS and subjected to analysis by BD FACSCanto II flow cytometer.
NFAT Luciferase Reporter Assay
PD-1-overexpressing Jurkat T cells were transfected with pNFAT-Luc and TK-Renilla reporter construct. After 12 hours, cells were stimulated with α-CD3/CD28 or α-CD3/CD28/PD-L1 conjugated beads in the presence or absence of STM418 (20 μg/mL). Cell lysates were harvested and then subjected to a Dual Luciferase assay (Promega, Madison, WI, USA). Luciferase activity was measured using a TD20/20 luminometer (Turner Designs Inc, Sunnyvale, CA, USA) according to the manufacturer’s instructions. Normalized luciferase activity was presented as the ratio of firefly luciferase activity to Renilla luciferase activity.
Production of PD-1 Antibodies
PD-1 antibodies were produced as described previously (12). Hybridomas producing monoclonal antibodies generated against glycosylated human PD-1 were obtained by the fusion of SP2/0 murine myeloma cells with spleen cells isolated from human PD-1-immunized BALB/c mice (n = 6) (Antibody Solutions, Inc., Sunnyvale, CA, USA) according to the standardized protocol. Before fusion, sera from the immunized mice were validated for binding to the PD-1 immunogen using FACS analysis. mAb-producing hybridomas were generated. The hybridomas that produced antibodies were again tested for specificity. To this end, over 100 candidate MAb-producing hybridomas were selected, grown in ADCF media, and their monoclonal antibody-containing supernatant was concentrated and purified. The purified mAbs were tested for their ability to neutralize or inhibit the interaction between PD-1 and PD-L1 (PD-1/PD-L1 binding interaction) with a live-cell imaging assay. The isotype of STM418 is mouse IgG2b which was determined by ELISA based isotyping kit (Mouse Monoclonal Antibody Isotyping Reagents, Sigma ISO2)
Dot Blot Assay
PD-1-His proteins (500 ng) were immobilized onto the nitrocellulose membrane in a 96-well dot blot apparatus (Bio-Dot, BioRad, Hercules, CA, USA), then detected with either 100 μl hybridoma supernatant or 100 ng purified mouse monoclonal antibody for 12 hours at 4 °C, 1:5000 anti-mouse-IgG-HRP secondary antibody (Jackson Labs, Bar Harbor, ME, USA), SuperSignal West Femto Substrate (Thermo Fisher Scientific), and Chemidoc imager (Bio-Rad).
Antibody Sequencing
Hybridoma cell pellets were frozen at –80°C and total RNA isolated with an RNeasy Plus Mini Kit (Qiagen, Hilden, DE) and quantitated using a NanoDrop 2000 spectrophotometer(Thermo Fisher Scientific). cDNA synthesis and rapid amplification of cDNA ends (RACE) were performed using a SMARTer RACE 5´/3´ Kit following the manufacturer’s instructions (Clontech, Mountain View, CA, USA). PCR primers were purchased in a Novagen Mouse Ig-Primer set (Merck KGaA, Darmstadt, DE, Germany) for Ig-specific PCR amplification. Heavy chain primer sets (A–F), light (kappa) chain primer sets (A–G), and template RNA 0.5 μg per 25 μL reaction volume were used. After gel separation, the PCR product was ligated into a pCR 2.1 vector, transformed into competent cells, and selected on 100 μg/mL ampicillin with 50 μL X-gal/IPTG agar plates. The white colonies were picked after 24 hours. After culturing, plasmid DNA was isolated and subjected to sequencing using M13 forward and reverse primers.
KD Determination
High-throughput KD screening was performed as described previously (12). Antibody ligand was loaded to the sensor of an Octet Red96 system (ForteBio, Fremont, CA, USA) in a 20 nM solution. A baseline was established in PBS containing 1 mg/ml bovine serum albumin (assay buffer), the association step was performed by submerging the sensors in a single concentration of an analyte in assay buffer. Dissociation was performed and monitored in fresh assay buffer. All experiments were performed with sensor shaking at 1,000 rpm. ForteBio data analysis software was used to fit the data to a 1:1 binding model to extract an association rate (Ka) and dissociation rate (Kd). The dissociation constant (KD) was calculated using the ratio Kd/Ka. In a typical epitope binning assay, the antigen, PD-1-His (10 nM), was preincubated with the second antibody (10 nM) for 1 hour at room temperature. Control antibody (20 nM) was loaded onto AMC sensors (ForteBio), and the remaining Fc-binding sites on the sensor were blocked with a whole mouse IgG antibody (Jackson ImmunoResearch, West Grove, PA, USA). The sensors were exposed to preincubated antigen-second antibody mixture. Raw data were processed using ForteBio’s Data Analysis Software version 7.0 to analyze the competitive binding of the antibody.
KD determination was performed via surface plasmon resonance using a Biacore X100 instrument (GE Healthcare, Uppsala, Sweden). Mouse IgG1 was immobilized on a research-grade CM5 chip using standard procedures, and antibody was flown over the chip at 2 μg/mL in HBS-EP+ buffer. Next, six concentrations of PD-1, each a 2-fold dilution, were passed over the chip. Sensorgram data were analyzed by Biacore X100 evaluation software version 2.0.1 with 1:1 binding kinetics.
Antibody Neutralizing Assay
To measure the inhibitory effect of antibodies on PD-1 and PD-L1 interaction, 293T cells expressing PD-1 were seeded in 96-well plates and incubated with PD-1 antibodies, recombinant human PD-L1-Fc protein, and anti-human-Fc Alexa Fluor 488 dye conjugate (Life Technologies). The green fluorescent signals were measured every 2 hours and quantified using an IncuCyte Zoom system(Essen BioScience).
Epitope Mapping by Mass Spectrometry
Epitope mapping of the antigen-antibody complex was performed as described previously (12). First, 5 μl of the antigen sample (4 μM) was mixed with 5 μl of the antibody sample (2 μM) to obtain an antibody and antigen mix with a final concentration of 2 μM or 1 μM, respectively. The mixture was incubated at 37 °C for 180 min. Next, 1 mg of d0 crosslinker was mixed with 1 mg of d12 crosslinker. Prepared crosslinkers (2 mg) were mixed with 1 ml of DMF to obtain a 2 mg/ml solution of DSS d0/d12. The antibody/antigen mix (10 μl) prepared previously was mixed with 1 μl of the solution of crosslinker d0/d12 (2 mg/ml). The solution was incubated for 180 min at room temperature to complete the crosslinking reaction. The crosslinked solution (10 μl) was mixed with 40 μl of ammonium bicarbonate (25 mM, pH 8.3) followed by the addition of 2 μl of DTT (500 mM). The mixture was incubated for 1 hour at 55 °C, and then 2 μl of iodoacetamide (1 M) was added for an additional 1-hour incubation at room temperature in the dark. After incubation, a 1:5 dilution was created by adding the buffer used for the proteolysis. The reduced/alkylated antigen was mixed with trypsin (1:100), chymotrypsin (1:200), ASP-N protease (1:200), elastase (1:100) or thermolysin (1:50 dilution; Roche Diagnostic, Risch-Rotkreuz, Switzerland). The proteolytic mixture was incubated overnight at 37 °C. The samples were analyzed by high-mass MALDI analysis immediately after crystallization. The MALDI ToF MS analysis was performed using CovalX’s HM4 interaction module with a standard nitrogen laser and focusing on different mass ranges from 0 to 2,000 kDa. For the analysis, the following parameters were applied: linear and positive mode, ion source 1: 20 kV, ion source 2:17 kV, lens: 12 kV, and pulse ion extraction: 400 ns for mass spectrometer, and gain voltage: 3.14 kV and acceleration voltage: 20 kV for HM4. The crosslinker peptides were analyzed using the xQuest version 2.0 and StavroX 2.1 software.
Mixed Lymphocyte Reaction (MLR)
Dendritic cells (DCs) were induced from PBMCs of the allogeneic donor (Immunospot #CTL-CP1) by culturing the cells in the presence of IL-4 (500 U/mL) and GM-CSF (250 U/mL) for 7 days. DCs were isolated by human pan-DC enrichment kit (#130–100-777; Miltenyi Biotech, San Diego, CA, USA) according to the manufacturer’s recommendation and used to stimulate allogeneic memory or naïve CD4+ T cells. CD4+ T cells were also enriched from PBMCs of another allogeneic donor (Immunospot #CTL-CP1) by CD4 microbeads (#130–045-101; Miltenyi Biotech). DCs (1 × 104 per well) were co-cultured in 96-well flat-bottom plates (Nunc; Thermo Fisher Scientific) with 1 × 105 T cells per well in culture medium in the presence of STM418 (10 μg/mL). T cells were labeled with 5 μM of CellTrace CFSE (Thermo Fisher Scientific) following the manufacturer’s instructions. After 5 days of culturing, the concentrations of IFN-γ and IL-2 in the culture supernatants were determined by a cytokine ELISA kit (BioLegend) according to the manufacturer’s instructions. CFSE staining was measured with a FACSCelesta (Becton Dickinson) flow cytometer.
T-cell Killing Assay
T cells were enriched from a PMBC donor (#CTL-CP1; Immunospot) by culturing cells in the presence of CD3 antibody (#16–0037, eBioscience), 10 ng/mL IL-2 (#200–02, PreproTech) and 20 μg/mL STM418, and then co-cultured with NucLight Red-labeled NCI-H226 cells. The NucLight Red lentivirus reagent was used according to the manufacturer’s instructions (Essen Bioscience). Red fluorescent signals were measured every 2 hours and quantified using an IncuCyte Zoom system (Essen BioScience).
Animal Studies
All procedures using NOD-SCID-IL2Rg- null mice (female, 6 weeks old; The Jackson Laboratory, Bar Harbor, ME, USA) were conducted under guidelines approved by the MD Anderson Institutional Animal Care and Use Committee. Tumorigenesis assays were performed using mouse orthotopic breast cancer models. NOD-SCID-IL2Rg null mice were engrafted with 107 human PBMCs to humanize the immune system. MDA-MB-231 cells (5 × 106) were injected into the mammary fat pad of humanized NOD-SCID-IL2Rg-null mice. Mice were randomly assigned to groups according to mean tumor volume. For antibody-based drug intervention, PD1 antibodies nivolumab (Bristol-Myers Squibb), pembrolizumab (Merck), or STM418 (STCube Pharmaceuticals) or immunoglobulin G (control; Bio X Cell) (5 mg/kg) was injected intraperitoneally every 3 days 1 week after tumor cell inoculation. Tumor sizes were measured using a caliper, twice a week. Tumor volumes were calculated using the formula (length × width2)/2. At the experimental endpoint, mice were killed using CO2 exposure followed by cervical dislocation, and tumors were excised for subsequent histologic analysis or processed immediately for flow cytometric analyses.
Statistical Analysis
Data in bar graphs represent mean fold-change relative to untreated or control groups with standard deviation from three independent experiments. Statistical analyses were performed using SPSS software (version 20, SPSS, Chicago, IL). The correlation between protein expression levels was analyzed using Spearman’s correlation and the Mann-Whitney test. One-way ANOVA with a post hoc Tukey test was used to compare the difference between groups. A P value < 0.05 was considered statistically significant.
Data and Materials Availability
The STM418 and hSTM418 antibodies were developed by the authors and are not commercially available. Small aliquot can be provided by the corresponding author upon requested. A material transfer agreement (MTA) from both MD Anderson Cancer Center and STCube, Inc. is required to release the hSTM418 for industrial development.
Results
PD-1 is heavily glycosylated in T cells
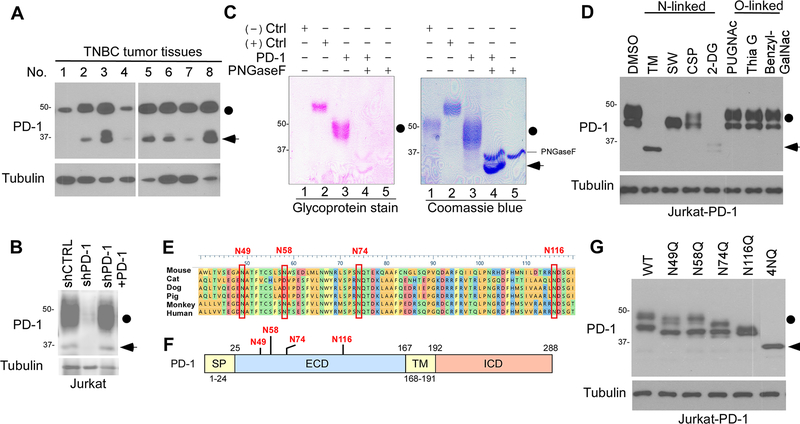
The PD-1 protein (288 amino acids) is expressed on both immune and cancer cells (18). When we analyzed the expression of PD-1 in the tumor tissues from patients with triple-negative breast cancer (TNBC), we detected two bands, 46-kDa and 32-kDa, by Western blot analysis after long exposure (Fig. 1A). Knocking down PD-1 by lentiviral short-hairpin RNA (shRNA) in Jurkat T cells decreased the levels of both forms, and reconstitution of PD-1 restored their expression, suggesting that the two bands indeed represent PD-1 (Fig. 1B). The heterogeneous expression pattern of PD-1 suggested the potential involvement of post-translational modification of PD-1, such as glycosylation. To further validate this concept, we purified PD-1 from Jurkat T cells and then performed glycoprotein staining. As shown in Figure 1C (lane 3 vs. 4), the addition of recombinant glycosidase (peptide-N-glycosidase F; PNGase F) reduced the molecular weight of PD-1 on SDS-PAGE, confirming that the higher molecular weight of PD-1 is the glycosylated form. Furthermore, treatment with the inhibitors of N-linked glycosylation, such as tunicamycin (TM), swainsonine (SW), castanospermine (CSP), or 2-deoxyglucose (2-DG) also decreased the molecular weight (as indicated by the changes in the migration pattern in SDS-PAGE) or expression of PD-1 (Fig. 1D). In contrast, inhibitors blocking O-linked glycosylation, e.g., benzyl-GalNAc, did not have any noticeable effects on the PD-1 expression pattern (Fig. 1D). These results indicated that PD-1 is primarily N-linked glycosylated.
Figure 1. PD-1 is heavily glycosylated in T cells.
(A) Immunoblot of PD-1 expression in human TNBC tumor tissues. Black circle, glycosylated PD-1; arrowhead, non-glycosylated PD-1. (B) Immunoblot of PD-1 expression in shCTRL, shPD-1, and PD-1-re-expressing Jurkat T cells stimulated by PHA overnight. (C) Glycoprotein staining and Coomassie blue staining of PNGase F-treated purified PD-1. Horseradish peroxidase (HRP) and soybean trypsin inhibitor (STI) served as positive and negative control, respectively. (D) Immunoblot of PD-1 in PD-1-expressing Jurkat (Jurkat-PD-1) T cells treated with inhibitors blocking N-linked or O-linked glycosylation as indicated. (E) Schematic diagram of PD-1 amino acid sequence alignment among different species. The four putative NXT motifs are shown in red. (F) Schematic diagram of full-length PD-1. ECD, extracellular domain; ICD, intracellular domain; SP, signal peptide; TM, transmembrane domain. Four putative NXT motifs in the ECD domain are labeled in red. The numbers indicate the amino acid positions. (G) Immunoblot of the protein expression pattern of PD-1 WT and indicated NQ mutants overexpressed in Jurkat T cells.
To define the glycosylation sites, we searched for evolutionarily conserved NXT motifs in the PD-1 amino acid sequences from different species (Fig. 1E). Consistent with previous predictions (8), we identified four conserved NXT motifs (N49, N58, N74, and N116) in the extracellular domain of PD-1 (Fig. 1F). Substitution of asparagine (N) with glutamine (Q) at each or all of the 4 sites (N49Q, N58Q, N74Q, N116Q, or 4NQ) reduced the molecular weight of the PD-1 mutants compared with their WT counterpart (Fig. 1G). These results together suggested that PD-1 is N-linked glycosylated primarily at N49, N58, N74, and N116 in T cells.
The specific glycoforms of PD-1 are altered upon T cell activation
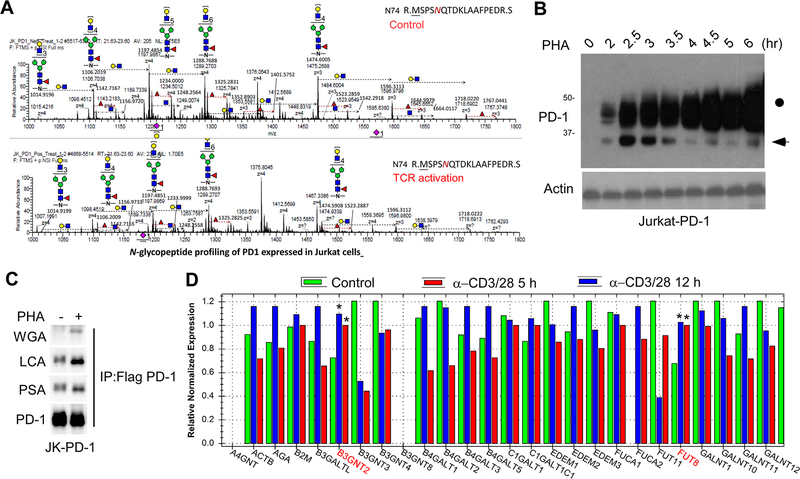
To decipher the detailed glycan structure of PD-1 and investigate whether it is affected by TCR activation, we analyzed the tryptic glycopeptides of PD-1 purified from Jurkat T cells by nanoscale liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS). Interestingly, we found that TCR activation by phytohemagglutinin (PHA) induced dynamic changes in the intensity of PD-1 specific glycoforms (Fig. 2A). Moreover, T-cell activation led to an increase in the levels of the glycosylated form of PD-1 (46 kDa) but a decrease in that of the non-glycosylated form (32 kDa) in a time-dependent manner (Fig. 2B). To determine the changes in specific glycans, we performed lectin immunoblotting by using biotin-labeled plant lectins, such as wheat germ agglutinin (WGA) and Lens culinaris agglutinin (LCA). We found that PD-1 purified from activated T cells exhibited higher levels of poly-N-acetyl-lactosamine (poly-LacNAc; bound by WGA) and core fucose (bound by LCA) (Fig. 2C). Altogether, these data showed that TCR activation alters the intensity of PD-1 specific glycoforms, such as poly-LacNAc and core fucose.
Figure 2. TCR activation induces alterations of specific glycoforms of PD-1.
(A) LC-MS/MS-based analysis of N-glycopeptides containing N74-glycans. PD-1 was purified from Jurkat-PD-1 FLAG cells treated with or without PHA overnight. The cartoon symbols used for the glycans conform to the standard representation recommended by the Consortium for Functional Glycomics. (B) Immunoblot of PD-1 in Jurkat T cells treated with PHA for the indicated time. (C) Lectin immunoblot of PD-1 purified from Jurkat-PD-1 FLAG treated with or without PHA for 5 hours. (D) Quantification of mRNA levels of various glycosyltransferases by real-time PCR in Jurkat T cells stimulated with α−CD3/CD28 for the indicated time points. All data represent mean ± SD from at least three independent experiments. *, P < 0.05.
Next, to identify the glycosyltransferases that might be involved in regulating PD-1 glycosylation, we performed a qPCR array, which includes more than 80 N- or O-linked glycosyltransferases. The result showed that T-cell activation altered the mRNA expression levels of those glycosyltransferases (Supplementary Fig. 1A). Of note, two glycosyltransferases responsible for initiating the synthesis of poly-LacNAc and core fucose, B3GNT2 and FUT8, respectively, were upregulated upon T cell activation (Fig. 2D), suggesting the potential involvement of these enzymes in regulating PD-1 glycosylation. We analyzed the GSE78220 dataset to determine the correlation between the expression of B3GNT2 or FUT8 and the outcomes of PD-1 therapy. As shown in the Supplementary Figure 1B, the progressive disease (PD) group had higher expression of B3GNT2 or FUT8 compared with the partial response/complete response (PRCR) group. These results indicated that the level of B3GNT2 or FUT8 is inversely correlated with the outcome of PD-1 therapy.
Glycosylation is critical for maintaining PD-1 stability and membrane expression
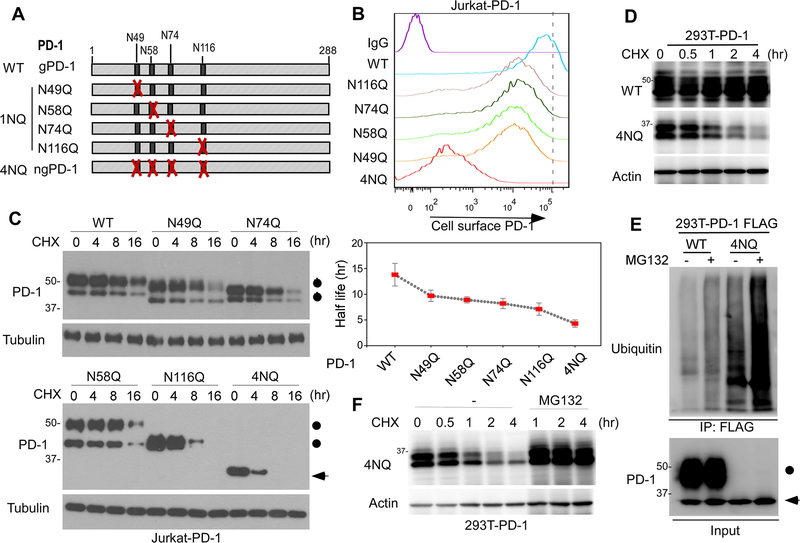
To determine the biological functions of PD-1 glycosylation, we evaluated the cell surface levels of PD-1 WT and the indicated mutants (Fig. 3A) and found that the membranous levels of PD-1 mutants decreased compared with their WT counterpart by fluorescence-activated cell sorting (FACS; Fig. 3B). Similarly, blocking N-linked glycosylation by TM treatment significantly reduced the levels of cell surface PD-1 (Supplementary Fig. 2A). Next, to determine whether the reduction in cell surface PD-1 mutants is attributed to decreased protein stability, we treated Jurkat T cells overexpressing WT or various PD-1 N/Q mutants with protein synthesis inhibitor cycloheximide (CHX) at specific time points. The half-lives of the PD-1 mutants, in particular the 4NQ mutant, were shorter than that of PD-1 WT (Fig. 3C). Similar results were observed in PD-1-overexpressing 293T cells (Fig. 3D). To deduce whether the ubiquitin-proteasome pathway is involved in PD-1 degradation, we treated cells with proteasome inhibitor MG132 and showed that the non-glycosylated PD-1 (4NQ) was substantially more ubiquitinated (Fig. 3E), and its expression was restored in the presence of MG132 (Fig. 3F). These data suggested that glycosylation is important for maintaining PD-1 stability and membrane expression.
Figure 3. PD-1 glycosylation is critical for maintaining its stability and membrane expression.
(A) Schematic diagram of various PD-1 NQ mutants. The numbers indicate the amino acid positions on PD-1. (B) Flow cytometric analysis of cell surface PD-1 WT or the indicated NQ mutants over-expressed in Jurkat T cells. (C) Protein half-life of PD-1 WT or the indicated NQ mutants over-expressed in Jurkat T cells. Cells were treated with CHX for the indicated time, and PD-1 levels were examined by immunoblotting. (D) Protein half-life of PD-1 WT or 4NQ overexpressed in 293T cells. Cells were treated with CHX for the indicated time and PD-1 levels were examined by immunoblotting. (E) Ubiquitination of PD-1 WT or 4NQ purified from 293T cells treated with or without MG132. (F) Immunoblot of PD-1 4NQ in 293T cells treated with CHX for the indicated time in the presence or absence of MG132.
PD-1 glycosylation is essential for mediating the interaction with PD-L1
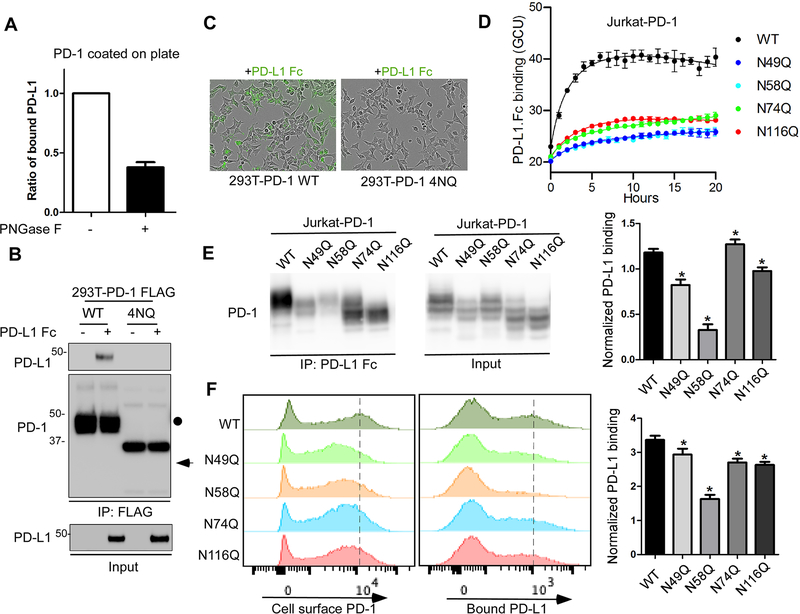
Next, we asked whether the integrity of PD-1 glycans is important for PD-L1 binding. We found that the removal of PD-1 glycans by PNGase F treatment reduced the binding of purified PD-1 with PD-L1 Fc as shown by an in vitro plate-based binding assay (Fig. 4A). To validate that, we incubated PD-L1 Fc with the lysates from 293T cells expressing FLAG-tagged PD-1 WT or PD-1 4NQ and then analyzed PD-L1/PD-1 interaction by immunoprecipitation followed by Western blotting. As shown in Figure 4B, only PD-1 WT but not the PD-1 4NQ mutant bound to PD-L1. Similar results were obtained from the live-cell imaging assay by incubating fluorescent-labeled PD-L1 Fc with 293 T cells expressing PD-1 WT or 4NQ (Fig. 4C and Supplementary Fig. 2B). Next, we sought to map the glycosylated sites that are critical for PD-L1 binding by live-cell imaging assay. Substitution of each asparagine (N) with glutamine (Q) led to a reduction in the binding affinity with PD-L1 compared with their WT counterpart (Fig. 4D). We further validated the result by IP-Western and showed that the N58Q mutant exhibited a significant decrease in binding with PD-L1 Fc (Fig. 4E). Similar results were observed by FACS (Fig. 4F). Thus, the glycosylation of PD-1, especially at the N58 site, plays an important role in mediating PD-L1 interaction.
Figure 4. The glycosylation of PD-1 is essential for its interaction with PD-L1.
(A) In vitro plate-based binding analysis of purified PD-1 with PD-L1 Fc in the absence or presence of PNGase F. (B) Analysis of PD-1/PD-L1 binding by immunoprecipitation. The lysates of 293T cells overexpressing FLAG-tagged PD-1 WT or 4NQ mutant were incubated with PD-L1 Fc and anti-FLAG M2 agarose, and then subjected to IP-Western. (C) Time-lapse microscopy of the dynamic interaction between green fluorescent-labeled PD-L1-Fc and PD-1 WT or 4NQ expressed in 293T cells. (D) Time-lapse evaluation and quantitation of the dynamic interaction between green fluorescent-labeled PD-L1-Fc and PD-1 WT or the indicated mutants expressed in Jurkat cells. (E) Mapping of the glycosylated sites critical for binding with PD-L1 by immunoprecipitation. The lysates of Jurkat cells overexpressing PD-1 WT or the indicated NQ mutants were incubated with pan mouse IgG dynabeads and Fc (mouse IgG) fused human PD-L1, and then subjected to IP-Western. The band intensity was quantified and normalized to show PD-L1 binding levels (right panel). (F) Flow cytometric analysis of the binding of PD-L1 Fc with cell surface PD-1 by using Jurkat T cells expressing PD-1 WT or the indicated NQ mutants. Median Fluorescence Intensity was quantified and normalized to show PD-L1 binding levels (right panel). All data represent mean ± SD from at least three independent experiments. *, P < 0.05.
Given that PD-1/PD-L1 interaction is the key to initiate immunosuppressive signaling in T cells, we further asked whether PD-1 immunosuppressive function is affected by glycosylation. The activity of NFAT, a crucial transcription factor in regulating cytokine interleukin-2 (IL-2) expression, is used as a marker for T cell activation. By using the NFAT luciferase reporter assay, we showed that the engagement of WT PD-1, but not 4NQ, with PD-L1-Fc conjugated beads, significantly inhibited anti-CD3/CD28-induced T-cell activation in Jurkat T cells (Supplementary Fig. 2C), suggesting that the glycosylation of PD-1 is essential for its immunosuppressive function.
Production and characterization of antibodies targeting glycosylated PD-1
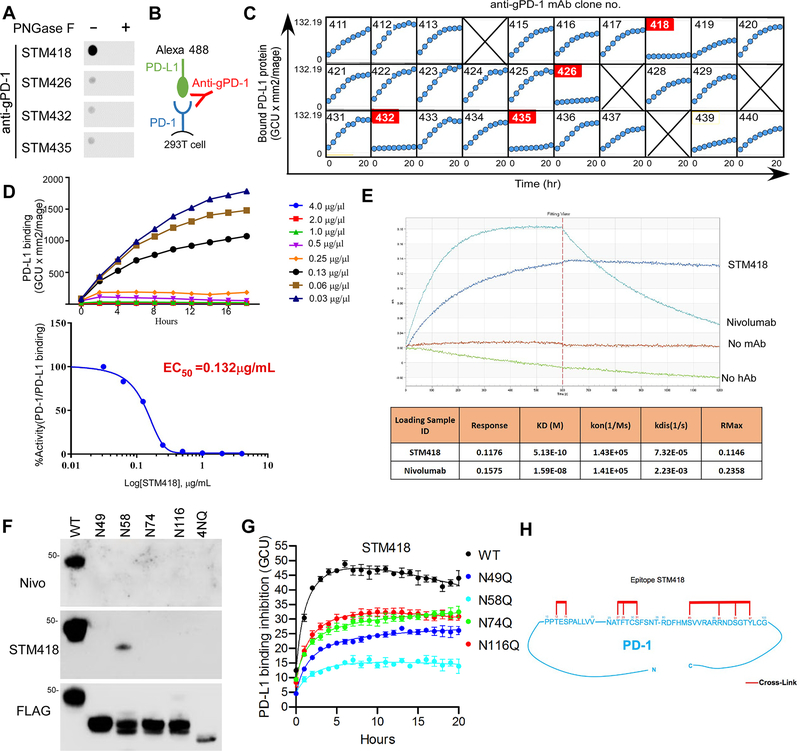
The results showing that glycosylation is critical for PD-L1/PD-1 interaction prompted us to generate monoclonal antibodies (mAbs) against glycosylated PD-1 (gPD-1). We purified gPD-1 from 293T cells overexpressing heavily glycosylated PD-1, and over 100 hybridoma supernatants were screened in a dot blot assay (Supplementary Fig. 3A). Several of the tested antibodies, including STM418, STM426, STM432, and STM435, exhibited glyco-specific binding activities (Figure 5A). Antibody sequencing by 5ŕapid amplification of cDNA ends and subsequent gene-specific PCR revealed the same protein sequence for STM426, STM432, and STM435 whereas STM418 had a unique sequence in its variable region.
Figure 5. Production and characterization of gPD-1 antibodies.
(A) Dot blot analysis of PD-1 antibodies using purified PD-1 treated with or without PNGase F. (B) Schematic diagram of the live-cell imaging PD-1/PD-L1 binding assay in the presence of gPD-1 antibody. (C) Measurement of the neutralizing activity of gPD-1 antibodies using an IncuCyte system. 293T-PD-1 cells were incubated with fluorescent-labeled PD-L1-Fc fusion protein in the presence of gPD-1 antibodies. The binding of PD-1 and PD-L1 was quantified by counting fluorescent objects per mm2 using the Incucyte™ Zoom system every 2 hours. (D) EC50 calculation of STM418 by GraphPad Prism software. Experiments were conducted as described in (C) in the presence of indicated doses of STM418. (E) KD determination of STM418 or nivolumab. (F) Glyco-specificity evaluation of STM418 by immunoblot analysis of PD-1 WT or the indicated NQ mutants over-expressed in 293T cells. (G) Time-lapse evaluation and quantitation of the dynamic interaction between green fluorescent-labeled PD-L1-Fc and PD-1 WT or the indicated mutants expressed in Jurkat cells in the absence of presence of STM418. PD-L1 binding inhibition is represented by the fluorescence intensity ratio of the STM418 to IgG treatment in each cell line. (H) Epitope mapping of STM418 by high-mass MALDI mass spectrometry (CovalX). Sequences on the PD-1 protein shown in red represent the STM418 antibody binding sites.
Next, we carried out a live-cell imaging assay to test the purified mAbs for their ability to neutralize or inhibit the interaction between PD-1 and PD-L1 (Figure 5B, C). Among those tested, STM418 potently blocked the PD-1 and PD-L1 interaction with a neutralizing activity (decreased half-maximal effective concentrations [EC50]) of 0.132 μg/mL (Fig. 5D). We further compared the binding affinity (decreased equilibrium dissociation constant [KD] values) between STM418 and the FDA-approved PD-1 antibody nivolumab using an Octet binding assay. The data showed a KD of 0.513 nM for STM418, indicating a strong binding affinity for PD-1 compared with a KD of 15.9 nM for nivolumab (Fig. 5E). Next, we humanized STM418 (hSTM418) by using the Composite Human Antibody™ (Abzena) to more accurately compare its neutralizing activity (Supplementary Fig. 3B) and binding affinity with nivolumab and pembrolizumab. Biacore analysis further revealed a KD value of 7.13 nM, 23 nM, and 30.5 nM for hSTM418, nivolumab, and pembrolizumab, respectively (Supplementary Fig. 3C), indicating a much stronger binding affinity of hSTM418 for PD-1 than either nivolumab or pembrolizumab. These findings suggested that STM418 may be an ideal therapeutic candidate, and therefore, STM418 was chosen for functional characterization in the subsequent experiments.
By immunoblot analysis of various PD-1 3NQ mutants overexpressed in 293 T cells, we found that the major glycosylated site that STM418 recognized was N58 (Fig. 5F), the key site mediating PD-1/PD-L1 interaction. Similar results were observed in Jurkat T cells overexpressing different PD-1 1NQ mutants (Supplementary Fig. 4A). Results from live-cell imaging assay indicated that STM418 effectively disrupted PD1/PDL1-Fc binding in Jurkat-PD-1 WT cells but not in Jurkat-PD-1 N58Q cells (Fig. 5G), suggesting that N58 is important for STM418 recognition and function. The cross-species reactivity of STM418 was examined by flow cytometry, which showed high affinity for human and rhesus monkey but not for mouse PD-1 expressed in 293T cells (Supplementary Fig. 4B). The specificity of STM418 was validated by excluding its binding with other CD28 or B7 family proteins, such as ICOS, CD86, etc. (Supplementary Fig. 4C, D). In addition, epitope mapping indicated that STM418 crosslinked with amino acids at 34–44, 49–59, and 104–124 (34PPTESPALLVV44…49NATFTCSFSNT59…104RDFHMSVVRARRNDSGTYLCG124) with binding contacts underlined at positions 36, 38, 51, 53, 55, 109, 115, 118, and 121 of PD-1 ( Fig. 5H). These epitopes were different from those identified in nivolumab and pembrolizumab (Supplementary Fig. 4E). Collectively, the STM418 antibody, which targets glycosylated PD-1 and primarily recognizes N58, demonstrates high affinity for PD-1 and strong ability to block PD-L1/PD-1 binding.
STM418 enhances T-cell activity in vitro
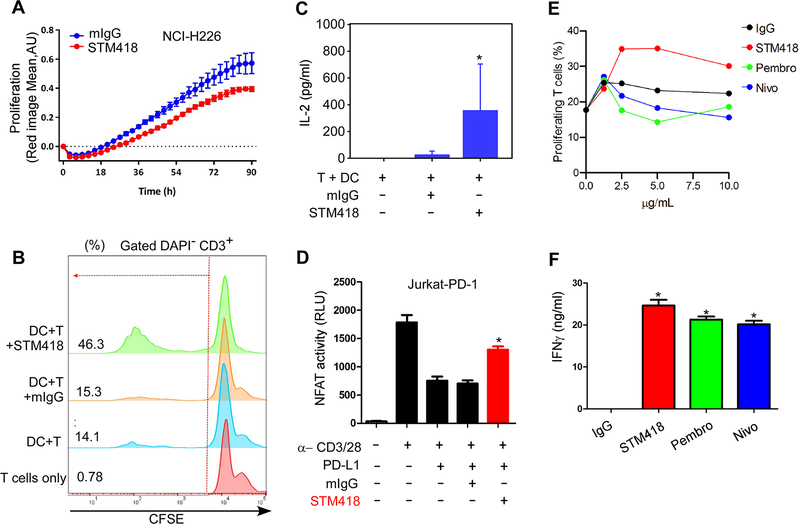
To evaluate the efficacy of STM418 in vitro, we utilized a T-cell-mediated killing assay by co-culturing T cells isolated from peripheral blood mononuclear cells (PBMCs) with NCI-H226 human lung carcinoma cells expressing high levels of PD-L1. Compared with the isotype control (mIgG2b), STM418 treatment enhanced T-cell mediated killing of NCI-H226 cells (Fig. 6A). To further validate its efficacy, we carried out mixed lymphocyte reaction (MLR) assays to evaluate the in vitro response of carboxyfluorescein succinimidyl ester (CFSE)-labeled peripheral T cells isolated from PBMCs upon cell contact-dependent antigenic stimulation (19). Control T cells exhibited low levels of proliferation when co-cultured with allogeneic dendritic cells. Treatment with STM418, but not the isotype control, greatly increased the proliferative response of T cells (Fig. 6B). Likewise, we observed significantly increased secretion of IL-2 (Fig. 6C) and IFN-γ (Supplementary Fig. 5A), which are markers of T cell activation, in the presence of STM418. To further validate the efficacy of STM418, we measured NFAT activity (Fig. 6D) and IL-2 secretion (Supplementary Fig. 5B) in PD-1-overexpressing Jurkat T cells after stimulation with α-CD3/CD28/IgG or α-CD3/CD28/PD-L1 conjugated beads. The results indicated that STM418 enhanced T-cell activation in the setting of PD-1/PD-L1 engagement. Next, we performed an in vitro MLR assay to compare the efficacy of STM418, nivolumab, and pembrolizumab. In this donor T-cell/DC pair, enhanced T cell proliferation was observed only in the presence of STM418 but not nivolumab or pembrolizumab (Fig 6E, Supplementary Fig. 5C). In addition, STM418 induced higher level of IFN-γ secretion compared with that of nivolumab and pembrolizumab (Fig 6F). In total, the results from the various in vitro functional assays demonstrated that STM418 potently promotes T cell proliferation and restores T-cell activities.
Figure 6. STM418 enhances T-cell proliferation and activation in vitro.
(A) Quantification of the proliferation of NCI-H226 cells co-cultured with T cells isolated from PMBCs in the presence or absence of STM418 (20 μg/mL). (B) Flow cytometric analysis of the proliferation of CFSE-labeled T cells co-cultured with dendritic cells (DCs) in the presence or absence of STM418 (10 μg/mL). (C) Quantification of IL-2 levels by ELISA. Experiments were conducted as described in (B). Supernatants were collected on day 5 and subjected to ELISA. (D) Normalized luminescence of PD-1 expressing Jurkat T cells transiently transfected with an NFAT-Luc reporter construct and stimulated with α-CD3/CD28/IgG or α-CD3/CD28/PD-L1 in the presence or absence of STM418 (20 μg/mL). (E) Flow cytometric analysis of the proliferation of CFSE-labeled T cells co-cultured with dendritic cells (DCs) in the presence of the indicated dose of IgG, STM418, nivolumab, or pembrolizumab. (F) Quantification of IFNγ levels by ELISA. Experiments were conducted as described in (E). Supernatants were collected on day 5 and subjected to ELISA. All data represent mean ± SD from at least three independent experiments. *, P < 0.05.
STM418 induces strong anti-tumor immunity in vivo
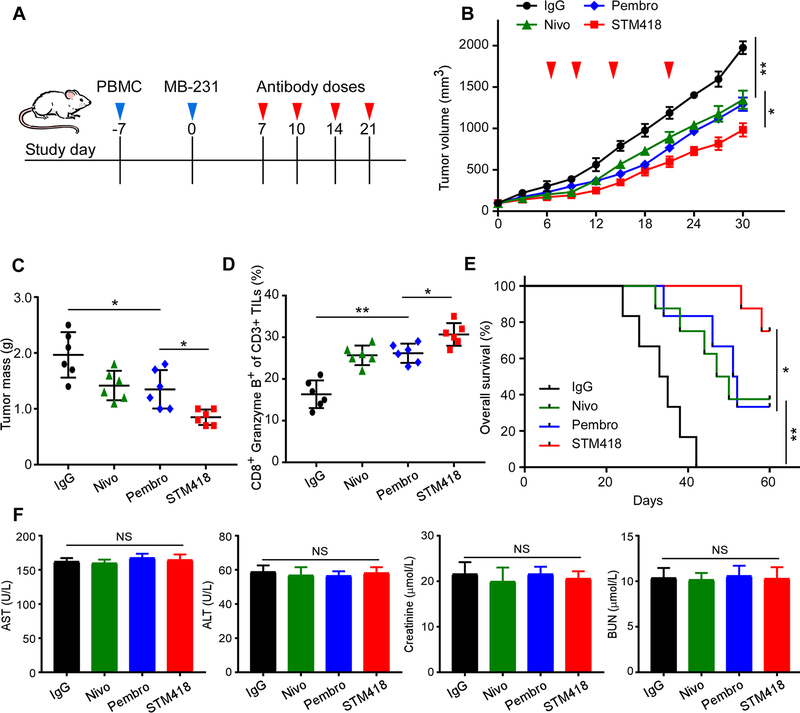
We next examined whether targeting gPD-1 with STM418 induces stronger tumor-infiltrating lymphocyte (TIL) expansion and promotes anti-tumor immunity in vivo. To this end, we utilized a humanized TNBC animal model to examine the efficacy of STM418. NOD-SCID-IL2Rg null mice were engrafted with human PBMCs to remodel and humanize the immune system, and then MDA-MB-231 cells were implanted into the mammary fat pad followed by administration of nivolumab, pembrolizumab, or STM418 (Fig. 7A). Our data indicated that STM418 impaired tumor growth (Fig. 7B) and decreased tumor burden (Fig. 7C) more effectively in mice than did nivolumab or pembrolizumab under the same doses. Similarly, hSTM418 also demonstrated higher efficacy in inhibiting tumor growth (Supplementary Fig. 6A). Importantly, MDA-MB-231 tumor-bearing mice treated with STM418 exhibited higher levels of PD-1 expression (Supplementary Fig. 6B), activated tumor-infiltrating CD8+/granzyme B+ T cells (Fig. 7D), and longer overall survival (Fig. 7E) than those treated with nivolumab or pembrolizumab. These results indicated STM418 induced stronger anti-tumor immunity in mice than did nivolumab or pembrolizumab in the model used.
Figure 7. STM418 induces potent anti-tumor immunity in a humanized TNBC animal model.
(A) Schematic diagram of a drug intervention protocol for anti-PD-1 antibodies (5 mg/kg) in a humanized animal model. (B) The growth of orthotopic MDA-MB-231 tumors in PD-1 antibody-treated SCID-PBMC mice. Tumors were measured until tumors reached > 100 mm3 in volume at the indicated time points. Nivo, nivolumab; Pembro, pembrolizumab. (C) The weight of tumors at the drug intervention endpoint. (D) Intracellular cytokine staining of CD8+ and granzyme B+ cells in CD3+ T-cell populations from isolated tumor-infiltrating lymphocytes. Results are presented as the mean ± standard deviation from a representative experiment. (E) Kaplan Meier survival curve for mice bearing MDA-MB-231 tumors after the drug intervention with anti-PD-1 antibodies. Significance was determined using the log-rank test. (F) Quantitative analysis of indicated biochemistry indices for liver and kidney function after the experiments. All error bars represent mean ± SD. *P < 0.05; NS, not significant.
Next, to investigate the safety of the therapeutic doses used in the experiments, we compared the biochemical indicators of liver and kidney functions, including aspartate transaminase (AST), alanine aminotransferase (ALT), creatinine, and blood urea nitrogen (BUN) levels, of mice before and over the course of the STM418 treatment. As shown in Figure 7F, there were no substantial changes between the experimental and control groups. Similar results were observed for the hSTM418 treatment, as demonstrated by the body weight (Supplementary Fig. 6C). Collectively, our findings in humanized breast tumor models illustrated the potent therapeutic benefits of targeting the glycosylated PD-1 by STM418 with relative safety profile.
Discussion
The T-cell function is primarily determined by the precise timing of gene expression and cytokine production. Although post-translational modifications, such as phosphorylation, have been extensively studied in T cells, glycosylation has largely been overlooked. There is increasing evidence demonstrating the importance of the glycans associated with T-cell surface proteins in regulating immunity. In the current study, we showed that PD-1, a well-established immune checkpoint, is heavily glycosylated at N49, N58, N74, and N116 in T cells. Enzymatic removal of N-linked glycans reduced the molecular weight of PD-l by nearly 15 kDa. These carbohydrate chains on PD-1 may provide a binding domain for the glycan-binding proteins, such as galectins, a family of animal lectins that play important roles in diverse immune processes (20). Interestingly, our LC-MS/MS analysis revealed differential intensities among the various N-glycoforms on PD-1 in response to T-cell activation. Of note, by using lectin blot, we found that the level of poly-LacNAc, which is synthesized by the glycosyltransferases, such as B3GNT2, was increased upon TCR activation. Consistent with the previous report (8), we also showed that PD-1 is highly fucosylated. Our findings that TCR activation alters the intensity of PD-1 specific glycoforms suggest that, in addition to transcriptionally upregulating PD-1 expression, T-cell activation also likely affects PD-1 functions via post-translational modifications, e.g., glycosylation.
Recently, Zak et al. solved the crystal structure of the hPD-1/hPD-L1 complex and provided the molecular details of their interaction (21). However, the structure was determined by using non-glycosylated proteins purified from bacteria. Here, we provide evidence that the integrity of PD-1 glycans, especially the glycosylation of N58, is essential for PD-L1 interaction. Given that glycosylation of PD-L1 is also critical for binding to PD-1 (12), it would be important to determine the structure of PD-1/PD-L1 binding using the glycosylated forms of these proteins in the future. Thus, our study reveals the functional significance of PD-1 glycosylation and helps to better understand the interaction between PD-1 and PD-L1.
To further explore the therapeutic potential of our findings, we focused on the development of a specific neutralizing monoclonal antibody targeting glycosylated PD-1 and isolated one particular clone, STM418, among 100 hybridomas screened. The binding mode and blocking mechanism of STM418 appears to be distinct from that of nivolumab, pembrolizumab or other recently reported anti-PD-1 antibodies, which do not require the glycans to bind with PD-1 (22–25). The major glycosylated site that STM418 recognizes is N58, the one that plays a key role in mediating PD-L1 binding, suggesting that STM418 binds to this site to inhibit PD-1/PD-L1 interaction. Interestingly, we showed that STM418 exhibited higher affinity for PD-1 compared with nivolumab. We speculated that this may be because the heavy glycan moieties of PD-1 hinder its recognition by PD-1 antibodies, such as nivolumab, which does not recognize glycosylation. This notion is consistent with our recent findings that the removal of PD-L1 glycosylation enhances PD-L1 detection by PD-L1 antibodies (26). Therefore, our current study offers potential strategies in designing therapeutic antibodies that can target particular immune checkpoints that are heavily glycosylated. Based on the MLR assay results, STM418 significantly promoted T-cell proliferation; however, neither nivolumab nor pembrolizumab had any appreciable effects on T cell proliferation. Although the lack of improved T-cell proliferation by nivolumab seemed unexpected, it has indeed been observed previously (27). We speculated that this may be due to donor variability. More investigations are warranted in the future to further clarify the underlying mechanisms. Overall, our findings shed lights on functional significance of PD-1 post-translational modification, specifically glycosylation, in T cells and highlight the rationale of targeting glycosylated PD-1 as a potential approach to improve immunotherapy.
Supplementary Material
Significance.
Findings demonstrate that glycosylation of PD-1 is functionally significant and targeting glycosylated PD-1 may serve as a means to improve immunotherapy response.
Acknowledgements
We would like to thank the Department of Scientific Publications at MD Anderson for editorial assistance. This work was funded in part by the following: National Natural Science Foundation of China (81972186 and 31301160 to L. Sun) and China Scholarship Council (to L. Sun); Natural Science Foundation of Tianjin (16JCYBJC24400 to L. Sun); Cancer Prevention & Research Institute of Texas (MIRA grant RP160710; to M.-C. Hung); National Breast Cancer Foundation, Inc.; Breast Cancer Research Foundation (BCRF-17-069; to M.-C. Hung); Patel Memorial Breast Cancer Endowment Fund (to M.-C. Hung); The University of Texas MD Anderson-China Medical University and Hospital Sister Institution Fund (to M.-C. Hung); U.S. National Institutes of Health Cancer Center Support Grant (CCSG CA016672; to M.-C. Hung) and T32 Training Grant (5T32CA186892 to H.-H. Lee and L.-C. Chan); YingTsai Young Scholar Award (CMU108-YTY-02 to J.-M. Hsu); Ministry of Education (Taiwan) Joint of International Talent Training Program (1040082029B to Y.-H. Wang); and China Scholarship Council (to Y. Qiu).
Footnotes
Competing interests: M.-C.H. received a sponsored research agreement from STCube Pharmaceuticals, Inc. through MD Anderson Cancer Center. E.M.C., Y.-S.K., A.H.P., and S.S.Y. are employees of STCube Pharmaceuticals, Inc. C.-W.L. and M.-C.H. are inventors on patent applications under review. The remaining authors declare no conflicts of interest.
References
- 1.Chen L, Flies DB. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nature reviews Immunology 2013;13:227–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, et al. Engagement of the Pd-1 Immunoinhibitory Receptor by a Novel B7 Family Member Leads to Negative Regulation of Lymphocyte Activation. The Journal of Experimental Medicine 2000;192:1027–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okazaki T, Honjo T. PD-1 and PD-1 ligands: from discovery to clinical application. Int Immunol 2007;19:813–24 [DOI] [PubMed] [Google Scholar]
- 4.Riley JL. PD-1 signaling in primary T cells. Immunol Rev 2009;229:114–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hui E, Cheung J, Zhu J, Su X, Taylor MJ, Wallweber HA, et al. T cell costimulatory receptor CD28 is a primary target for PD-1-mediated inhibition. Science 2017;355:1428–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharpe AH, Pauken KE. The diverse functions of the PD1 inhibitory pathway. Nature reviews Immunology 2018;18:153–67 [DOI] [PubMed] [Google Scholar]
- 7.Meng X, Liu X, Guo X, Jiang S, Chen T, Hu Z, et al. FBXO38 mediates PD-1 ubiquitination and regulates anti-tumour immunity of T cells. Nature 2018;564:130–5 [DOI] [PubMed] [Google Scholar]
- 8.Okada M, Chikuma S, Kondo T, Hibino S, Machiyama H, Yokosuka T, et al. Blockage of Core Fucosylation Reduces Cell-Surface Expression of PD-1 and Promotes Anti-tumor Immune Responses of T Cells. Cell Rep 2017;20:1017–28 [DOI] [PubMed] [Google Scholar]
- 9.Moremen KW, Tiemeyer M, Nairn AV. Vertebrate protein glycosylation: diversity, synthesis and function. Nat Rev Mol Cell Biol 2012;13:448–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer 2015;15:540–55 [DOI] [PubMed] [Google Scholar]
- 11.Li CW, Lim SO, Xia W, Lee HH, Chan LC, Kuo CW, et al. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat Commun 2016;7:12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li CW, Lim SO, Chung EM, Kim YS, Park AH, Yao J, et al. Eradication of Triple-Negative Breast Cancer Cells by Targeting Glycosylated PD-L1. Cancer cell 2018;33:187–201 e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marth JD, Grewal PK. Mammalian glycosylation in immunity. Nature reviews Immunology 2008;8:874–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Kooyk Y, Rabinovich GA. Protein-glycan interactions in the control of innate and adaptive immune responses. Nat Immunol 2008;9:593–601 [DOI] [PubMed] [Google Scholar]
- 15.Boussiotis VA, Longo DL. Molecular and Biochemical Aspects of the PD-1 Checkpoint Pathway. New England Journal of Medicine 2016;375:1767–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei SC, Duffy CR, Allison JP. Fundamental Mechanisms of Immune Checkpoint Blockade Therapy. Cancer Discov 2018;8:1069–86 [DOI] [PubMed] [Google Scholar]
- 17.Sun LL, Yang RY, Li CW, Chen MK, Shao B, Hsu JM, et al. Inhibition of ATR downregulates PD-L1 and sensitizes tumor cells to T cell-mediated killing. Am J Cancer Res 2018;8:1307–16 [PMC free article] [PubMed] [Google Scholar]
- 18.Kleffel S, Posch C, Barthel SR, Mueller H, Schlapbach C, Guenova E, et al. Melanoma Cell-Intrinsic PD-1 Receptor Functions Promote Tumor Growth. Cell 2015;162:1242–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bach FH, Voynow NK. One-way stimulation in mixed leukocyte cultures. Science 1966;153:545–7 [DOI] [PubMed] [Google Scholar]
- 20.Rabinovich GA, Toscano MA. Turning ‘sweet’ on immunity: galectin-glycan interactions in immune tolerance and inflammation. Nature reviews Immunology 2009;9:338–52 [DOI] [PubMed] [Google Scholar]
- 21.Zak KM, Grudnik P, Magiera K, Domling A, Dubin G, Holak TA. Structural Biology of the Immune Checkpoint Receptor PD-1 and Its Ligands PD-L1/PD-L2. Structure 2017;25:1163–74 [DOI] [PubMed] [Google Scholar]
- 22.Tan S, Zhang H, Chai Y, Song H, Tong Z, Wang Q, et al. An unexpected N-terminal loop in PD-1 dominates binding by nivolumab. Nat Commun 2017;8:14369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horita S, Nomura Y, Sato Y, Shimamura T, Iwata S, Nomura N. High-resolution crystal structure of the therapeutic antibody pembrolizumab bound to the human PD-1. Scientific reports 2016;6:35297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu H, Guo L, Zhang J, Zhou Y, Zhou J, Yao J, et al. Glycosylation-independent binding of monoclonal antibody toripalimab to FG loop of PD-1 for tumor immune checkpoint therapy. mAbs 2019;11:681–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen D, Tan S, Zhang H, Wang H, He W, Shi R, et al. The FG Loop of PD-1 Serves as a “Hotspot” for Therapeutic Monoclonal Antibodies in Tumor Immune Checkpoint Therapy. iScience 2019;14:113–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee HH, Wang YN, Xia W, Chen CH, Rau KM, Ye L, et al. Removal of N-Linked Glycosylation Enhances PD-L1 Detection and Predicts Anti-PD-1/PD-L1 Therapeutic Efficacy. Cancer cell 2019;36:168–78 e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang C, Thudium KB, Han M, Wang XT, Huang H, Feingersh D, et al. In vitro characterization of the anti-PD-1 antibody nivolumab, BMS-936558, and in vivo toxicology in non-human primates. Cancer immunology research 2014;2:846–56 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.