Abstract
The development of therapeutic agents that specifically target cancer cells while sparing healthy tissue could be used to enhance the efficacy of cancer therapy without increasing its toxicity. Specific targeting of cancer cells can be achieved through the use of pH-low insertion peptides (pHLIPs), which take advantage of the acidity of the tumor microenvironment to deliver cargoes selectively to tumor cells. We developed a pHLIP-peptide nucleic acid (PNA) conjugate as an antisense reagent to reduce expression of the otherwise undruggable DNA double-strand break repair factor, KU80, and thereby radiosensitize tumor cells. Increased antisense activity of the pHLIP-PNA conjugate was achieved by partial mini-PEG sidechain substitution of the PNA at the gamma position, designated pHLIP-αKu80(γ). We evaluated selective effects of pHLIP-αKu80(γ) in cancer cells in acidic culture conditions as well as in two subcutaneous mouse tumor models. Fluorescently labeled pHLIP-αKu80(γ) delivers specifically to acidic cancer cells and accumulates preferentially in tumors when injected intravenously in mice. Furthermore, pHLIP-αKu80(γ) selectively reduced KU80 expression in cells under acidic conditions and in tumors in vivo. When pHLIP-αKu80(γ) was administered to mice prior to local tumor irradiation, tumor growth was substantially reduced compared with radiation treatment alone. Furthermore, there was no evidence of acute toxicity associated with pHLIP-αKu80(γ) administration to the mice. These results establish pHLIP-αKu80(γ) as a tumor-selective radiosensitizing agent.
Keywords: Radiosensitization, non-homologous end joining (NHEJ), KU80, pH-low insertion peptides (pHLIPs), peptide nucleic acids (PNAs)
Introduction
As most current cancer therapies are toxic to healthy tissue, they often result in undesirable side effects that hinder their clinical use. For example, the dosing of radiation therapy, which is estimated by the American Society of Radiation Oncology to treat approximately two-thirds of all cancer patients, is limited by toxicities including inflammation, vascular damage, and fibrosis. The development of radiation sensitizers that specifically target cancer cells while sparing healthy tissue could be used to enhance the efficacy of this widely used treatment without increasing its toxicity, thus providing an important advance in the field.
Because DNA repair is crucial for tumor cell survival after exposure to DNA damaging agents, such as ionizing radiation (IR), it represents a promising target for cancer therapy. Furthermore, upregulation of DNA repair occurs after exposure to DNA damaging agents, and so may underlie resistance to these therapies (1–3). Thus, inhibiting DNA repair renders cells more sensitive to IR, reducing cell survival after IR exposure (radiosensitization). As radiation-induced DNA double strand breaks (DSBs) are lethal threats to genomic integrity, repair of DSBs by homology-directed repair (HDR) or non-homologous end-joining (NHEJ) is particularly important for cellular survival. The NHEJ pathway, which is responsible for repairing 90% of DSBs, requires the DNA-dependent protein kinase (DNA-PK) complex, of which KU80 is an indispensible factor (4,5). In mice, KU80 deficiency is associated with impaired DSB repair and radiosensitization (6,7). While KU80 lacks a known enzyme activity and therefore is not considered a classically “druggable” protein, inhibition of KU80 using antisense has been shown to radiosensitize cells in culture (8,9). However this approach has not yet been tested in vivo, likely due to the limitations of traditional antisense delivery methods.
We sought to overcome these limitations in targeting KU80 for use as a radiosensitizer by using peptide nucleic acids (10) with (R)-diethylene glycol at the γ position (MPγPNA) (11) to develop a molecule that potently reduces KU80 expression via an antisense mechanism (αKu80(γ)). The γ position substitution confers helical pre-organization on the PNA oligomer that substantially improves binding affinity to complementary DNAs or RNAs (11). We further improved the efficacy of αKu80(γ) using a tumor-specific delivery system, pH-low insertion peptides (pHLIPs), which take advantage of the acidity of the tumor microenvironment to deliver cargo specifically to tumors in vivo. Our group has previously used pHLIPs to deliver PNA antisense agents against an oncogenic miRNA in a mouse lymphoma model, causing substantial anti-tumor effects without systemic toxicity (12). Here, we utilized similar technologies to develop a tumor-specific radiosensitizer by conjugating a MPγPNA antisense targeting KU80 to pHLIP (pHLIP-αKu80(γ)). In cell culture, pHLIP-αKu80(γ) delivers to cancer cells and suppresses Ku80 expression with pH-specificity. When delivered systemically to mouse models in vivo, pHLIP-αKu80(γ) targets to tumors and causes knockdown of KU80 expression in tumors but not in non-malignant tissues. Furthermore, systemic treatment with pHLIP-αKu80(γ) causes tumor radiosensitization in vivo but does not appear to cause normal tissue toxicity. These results establish pHLIP-αKu80(γ) as a tumor-selective radiosensitizing agent.
Materials and methods
PNA synthesis
Boc-protected PNA monomers, including MPγ monomers, were obtained from ASM Research Chemicals. All oligomers were synthesized on a solid-support resin using standard Boc chemistry procedures. To facilitate linkage of pHLIP to PNAs, cysteine was conjugated to the C-terminus of PNAs using a Boc-miniPEG-3 linker (11-Amino-3,6,9-Trioxaundecanoic Acid, DCHA, denoted in the sequences by -ooo-) obtained from Peptide International. Single isomer 5-Carboxytetramethylrhodamine (TAMRA, ThermoFisher) was also conjugated to the N-terminus of PNAs using a Boc-miniPEG-3 linker. PNA oligomers were cleaved from the resin using a cocktail solution consisting of m-cresol:thioanisole:TFMSA:TFA (1:1:2:6), and were precipitated with ether. PNAs were then purified by high performance liquid chromatography (HPLC) using a C18 column (Waters) and a mobile phase gradient of water and acetonitrile with 0.1% trifluoroacetic acid. Purified PNAs were further characterized using matrix-assisted laser desorption/ionization– time of flight (MALDI–TOF). PNA stock solutions were prepared using nanopure water and the concentrations were determined on a Nanodrop Spectrophotometer (Thermo Scientific) at 260nm using the following extinction coefficients: 13,700 M−1 cm−1 (A), 6,600 M−1 cm−1 (C), 11,700 M−1 cm−1 (G), and 8,600 M−1 cm−1 (T).
The following PNA sequences were used (underline indicates MPγ monomer):
αKu80: CCCATGAAGAATCTTCTCTG-ooo-Cys
αKu80-TAMRA: TAMRA-ooo-CCCATGAAGAATCTTCTCTG-ooo-Cys
αKu80(γ): CCCATGAAGAATCTTCTCTG-ooo-Cys
αKu80(γ)-TAMRA: TAMRA-ooo-CCCATGAAGAATCTTCTCTG-ooo-Cys
scr(γ): ATATCGTTCACGCACGTATC-ooo-Cys
pHLIP conjugations
To facilitate conjugation of pHLIP to thiolated PNAs, a cysteine group derivatized with 3-nitro-2-pyridinesulphenyl (NPys) was incorporated to the C-terminus of pHLIP (AAEQNPIYWARYADWLFTTPLLLLDLALLVDADEGT(CNPys)G, New England Peptide). To generate pHLIP-PNA conjugates, pHLIP-Cys(NPys) and PNAs were reacted overnight at a 1:1 ratio in the dark in a mixture of DMSO:DMF:0.1mM KH2PO4 pH 4.5 (3:1:1). pHLIP-PNA conjugates were then purified and characterized using HPLC and MALDI-TOF, respectively, and concentrations were determined as described above. For all in vitro and in vivo studies, pHLIP-PNAs were heated at 55°C for 5 mins to prevent aggregation.
Gel shift assays
pHLIP-PNAs were incubated at a 2:1 ratio with a DNA sequence corresponding to the predicted binding site of PNAs to KU80 mRNA and allowed to anneal at 37°C overnight. 125mM TCEP was added to a subset of samples to assess for disulfide reduction. Samples were run on a polyacrylamide gel, and SYBR Gold (Life Technologies) was used to visualize KU80 (pHLIP and free PNA are not detected by this stain).
The following DNA sequences were used:
Human KU80: 5’-AGGTTCAGAGAAGATTCTTCATGGGAAATC-3’
Mouse Ku80: 5’-AGGTTCATAGAAGATTCTTCATGGGACATC3’
Cell culture
A549 cells were obtained from ATCC. DLD1-BRCA2KO cells were obtained from Horizon Discovery. EMT6 cells were provided by Dr. Sara Rockwell. Human cells were authenticated using STR profiling (ATCC). All cell lines were regularly tested for mycoplasma contamination (MycoAlert, Lonza). Cells were grown in F12K (A549), RPMI (DLD1-BRCA2KO), or DMEM (EMT6) media containing 10% fetal bovine serum without antibiotics. For pH-specificity experiments, Leibovitz L-15 media was titrated to pH=6.2 using HCl. Cells were treated with pHLIP-PNAs in pH-titrated media for 24 hours, followed by a series of several washes with phosphate buffered saline. Cells were then immediately processed for flow cytometry or collected 24 hours later for Western blot analysis.
Animals
All studies were approved by the Yale University Institutional Animal Care and Use Committee. For in vivo xenograft studies. 1×106 human DLD1-BRCA2KO cells were implanted subcutaneously in 100μl of media bilaterally in the flanks of athymic nu/nu mice (Harlan). For in vivo mouse tumor studies, 2×105 mouse EMT6 or EMT6-GFP cells were implanted subcutaneously in 100μl of media in the flanks of BALBc/Rw mice. Three times a week, tumors were measured using calipers to determine tumor volume by the following formula: V = 1/2(4π/3)(length/2)(width/2)(height). Mice were treated when tumors reached an average size of 100mm3, and irradiation (7 or 10Gy) was applied locally to tumors using a Seimen’s Stalipan.
Ex vivo fluorescence imaging
Mice were euthanized, subcutaneous EMT6 tumors and selected organs were dissected, and ex vivo fluorescence imaging was immediately performed on an IVIS Spectrum System (Caliper) using TAMRA filter sets.
Tumor collection and fluorescence Activated Cell Sorting (FACS)
Tumors were dissected and dissociated to single cell suspension using dispase/collagenase followed by trypsinization. Red blood cells were removed by lysis in ammonium chloride solution. A mouse cell depletion kit (Miltenyi Biotech) was used to isolate human tumor cells from DLD1-BRCA2KO tumors. EMT6-GFP tumors were sorted into GFP+ and GFP- fractions using a FACSAria cell sorter (BD Biosciences).
Western blot
Organs collected from treated mice were flash-frozen in liquid nitrogen, homogenized, and lysed with AZ lysis buffer (50 mmol/L Tris, 250 mmol/L NaCl, 1% Igepal, 0.1% SDS, 5 mmol/L EDTA, 10 mmol/L Na2P2O7, 10 mmol/L NaF) supplemented with Protease Inhibitor Cocktail (Roche) and Phospho-STOP (Sigma). Cells were also lysed in protease and phosphatase inhibitor supplemented AZ lysis buffer. KU80 expression was detected using antibody #611360 (BD). Expression of the DNA damage marker γH2Ax was measured using antibody #9718 (Cell Signaling Technologies). Apoptotis was evaluated using antibodies targeting cleaved caspase-3 (CST, #9664) and cleaved PARP (CST, #9544). GAPDH (Santa Cruz, sc-365062) was used as an endogenous loading control. Where shown, band intensities were quantified using ImageJ64 software.
siRNA-mediated knockdown
Cells were treated with siRNA for KU80 (Dharmacon SmartPOOL) at final concentration of 0.001–10nmol/L for 72 hours. A non-targeting scrambled siRNA (Dharmacon SmartPOOL) was used as a control. Extent of knockdown was confirmed using Western blot.
Clonogenic survival assay
A549 or DLD1-BRCA2KO cells were seeded at a density of 1×105 cells per well in 6-well plates. For siRNA experiments, cells were treated with control non-targeting or KU80 siRNA (0.01nmol/L or 1 nmol/L) for 72 hours. For pHLIP-PNA experiments, cells were treated with pHLIP-scr(γ) or pHLIP-αKu80(γ) (1 μmol/L) 72 hours and 24 hours prior to irradiation. Cells were then reseeded at a density of 500 cells per well in a 6-well plate and irradiated with 2 or 4 Gy using an X-RAD 320 X-Ray Biological Irradiator (Precision X-Ray Inc). Non-irradiated controls were handled in parallel but kept outside of the irradiator during treatment. Approximately 1.5 weeks later, cells were permeabilized in 0.9% saline solution and stained with crystal violet in 80% methanol. Colonies were counted manually.
Peripheral blood analysis
For peripheral blood collection, all mice were anesthetized using open-drop 30% w/v isoflurane in propylene glycol. Blood was collected retro-orbitally using heparinized micro-hematocrit capillary tubes (Fisher Scientific). For complete blood counts, 50μl blood was evacuated into heparinized coated tubes containing 10μl 0.5 M EDTA acid, and analysis was performed using a Hemavet 950FS (Drew Scientific) according to the manufacturer’s protocol. For blood chemistry analysis, blood from vehicle or pHLIP-αKu80(γ) (5mg/kg, i.v.) treated mice was collected 72 hours after treatment, serum separated using lithium heparin, and sent to Antech Diagnostics for analysis. For peripheral cytokine analysis, serum was isolated by centrifugation (1500 rcf for 10 minutes) from blood collected from vehicle or pHLIP-αKu80(γ) (5mg/kg, i.v.) treated mice 72 hours after treatment. Serum was submitted to the CytoPlex Core Facility at Yale University for luminex-based cytokine detection and quantification using the Bio-Plex Pro Mouse Cytokine 23-Plex (Bio-Rad).
Bone marrow
Bone marrow cells were harvested by flushing femurs and tibias from mice 72 hours after treatment with vehicle or pHLIP-αKu80(γ) (5mg/kg, i.v.). Cells were automatically counted using a Cellometer Auto T4 (Nexcelom). For relative quantification of cell types, cells were stained with fluorescently tagged antibodies to different lineage markers (KIT: #12–1171-82, SCA1: #45–5981-82, TER-119 #11–5921-82, GR1 #11–5931-82, CD45 #11–0452-82; Thermo Scientific), washed with PBS, and analyzed via flow cytometry.
Statistical analysis
Statistical analysis was performed in GraphPad Prism. Unless otherwise stated, data is presented as mean±SEM, with n=3 replicates.
Results
Synthesis and characterization of pHLIP-conjugated antisense PNAs targeting KU80
We sought to overcome traditional limitations in targeting KU80 for use as a radiosensitizer by using peptide nucleic acids (PNAs), DNA analogs in which the sugar phosphodiester backbone is replaced with homomorphous achiral N-(2 aminoethyl) glycine units (10). PNAs are a particularly useful tool as they hybridize with RNA and DNA with high affinity in a sequence-specific fashion and their charge neutral structure confers resistance to both protease and nuclease degradation (13). PNA-DNA and PNA-RNA complexes also have enhanced stability from a reduction of the backbone charge repulsion that destabilizes natural double stranded complexes. PNAs have been used to decrease gene expression via an antisense mechanism in numerous targets ,(14–16), and their efficacy can be enhanced by structural modifications and by novel delivery systems (17–19). Using PNA monomers with (R)-diethylene glycol at the γ position (MPγPNA) to increase solubility and nucleic acid binding (11), we synthesized a MPγPNA that we hypothesized would reduce KU80 expression via an antisense mechanism (αKu80(γ)). We first identified a 20-nucleotide sequence with known antisense activity against KU80 (8). Using solid-phase synthesis of Boc-protected monomers, we prepared both an unmodified PNA and a MPγPNA corresponding to this antisense sequence (αKu80 and αKu80(γ), respectively) as well as a scrambled control MPγPNA sequence (scr(γ)) (Fig. 1A). In some cases, the fluorescent dye TAMRA was conjugated to these PNAs to allow visualization in uptake experiments.
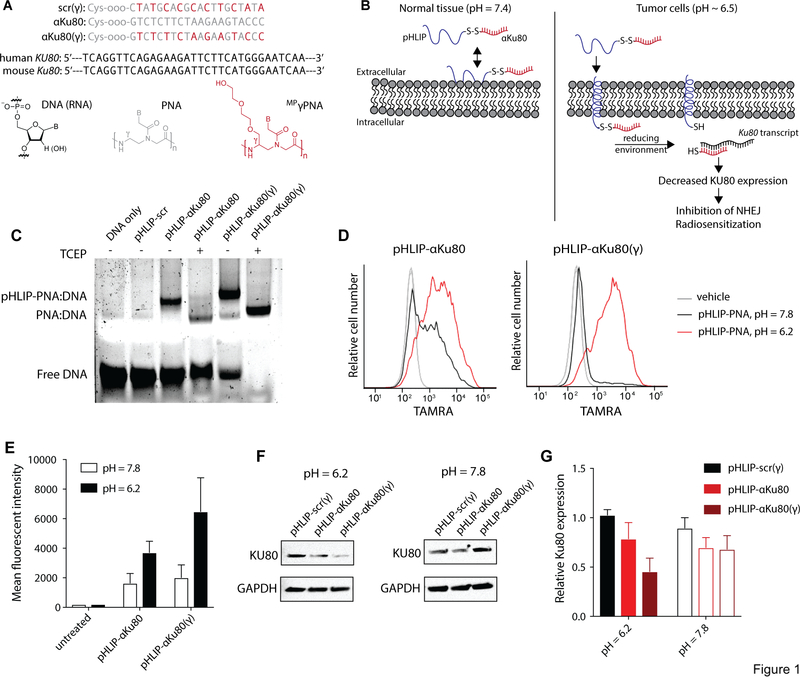
Fig. 1: pHLIP-αKu80 and pHLIP-αKu80 (γ) induce pH-specific suppression of Ku80 expression in cultured cancer cells.
(A) PNA sequences and diagrams of DNA, RNA, PNA, and MP γ PNA monomers. (B) Proposed mechanism of tumor-specific targeting of KU80 by pHLIP-αKu80: at physiologic extracellular pH, pHLIP is unable to deliver antisense targeting Ku80 (αKu80) across cellular membranes. At the acidic pH found at the cell membrane of tumor cells, protonation of Asp residues on pHLIP allow for membrane insertion and delivery of αKu80. The disulfide bond linking αKu80 to pHLIP is reduced under intracellular conditions, freeing the antisense to bind the Ku80 transcript and reduce KU80 expression, thereby inhibiting non-homologous end-joining (NHEJ) and sensitizing cells to ionizing radiation. (C) Gel shift assay indicating specific binding of pHLIP-αKu80 and pHLIP-αKu80(γ) to a sequence corresponding to the predicted binding site of PNAs to human Ku80 mRNA. (D) Representative flow cytometry traces of TAMRA fluorescence in A549 cells after treatment with vehicle or 1 μM pHLIP-αKu80-TAMRA (left), or pHLIP-αKu80(γ)-TAMRA (right) under acidic (pH = 6.2) or neutral (pH = 7.8) culture conditions (n = 3 technical replicates). (E) Quantification of TAMRA fluorescence by flow cytometry (t-test, untreated P = 0.99, pHLIP-αKu80 P = 0.20, pHLIP-αKu80(γ) P < 0.05, n = 3 technical replicates). (F) Western blot of KU80 expression in A549 cells treated with 1 μM pHLIP-scr(γ), pHLIP-αKu80, or pHLIP-αKu80(γ) under acidic (left) or neutral (right) culture conditions (n = 3 technical replicates). (G) Quantification of KU80 expression by Western blot (two-way ANOVA, effect of treatment P < 0.05; pH = 6.2: pHLIP-scr(γ) vs. pHLIP-αKu80 P = 0.24, pHLIP-scr(γ) vs. pHLIP-αKu80(γ) P < 0.05; pH = 7.8: pHLIP-scr(γ) vs. pHLIP-αKu80 P = 0.43, pHLIP-scr(γ) vs. pHLIP-αKu80(γ) P = 0.38; n = 3 technical replicates). Data are represented as means ± SEM.
To deliver PNAs in cell culture and in vivo, we utilized a tumor-specific delivery system, pH-low insertion peptides (pHLIPs), which takes advantage of the acidity at the surfaces of tumor cells. While extracellular pH is tightly regulated to approximately 7.4 in healthy tissue, tumor cell and tumor macrophage surfaces are largely acidic (20), resulting from high rates of glycolytic lactic acid production, surface carbonic anhydrase expression, and the electrochemical gradient (21). Thus, a delivery system with specificity for acidic conditions at cell surfaces facilitates the selective targeting of cargoes to the tumor microenvironment in vivo. pHLIPs are small peptides that insert their C-terminus across cell membranes at acidic extracellular pH (22,23). Cargo, including small molecules, PNAs, and dyes, can be linked to the C-terminus of a pHLIP using a disulfide bond that is cleaved in the intracellular reducing environment (24) (Fig. 1B).
Thiol-disulfide interchange reactions were performed to conjugate PNAs to pHLIP via a disulfide linkage (pHLIP-αKu80, pHLIP-αKu80(γ), and pHLIP-scr(γ)). After purifying and characterizing the pHLIP-PNA conjugates using mass spectrometry and high-performance liquid chromatography, we then confirmed the ability of these conjugates to bind the sequence of interest (using a DNA sequence as a proxy for the RNA target) using a gel-shift assay (Fig. 1C). Both pHLIP-αKu80 and pHLIP-αKu80(γ), but not pHLIP-scr(γ)), bound the KU80 mRNA sequence as indicated by a shift in migration of the detected band (Fig. 1C), confirming the sequence-specific binding of our pHLIP-PNA conjugates. As expected, pHLIP-αKu80(γ) showed enhanced nucleic acid binding as compared with pHLIP-αKu80, which is indicated by a reduced amount of free, unbound DNA. In some samples, the reducing agent TCEP was added to cleave disulfide linkages between pHLIP and PNAs. The addition of TCEP shifted the size of detected bands, confirming cleavage of conjugates and release of the peptide under reducing conditions. Gel shift assays also confirmed the ability of pHLIP-αKu80(γ) to bind the mouse sequence of Ku80 at the corresponding sequence (Fig. S1), which differs from the human sequence by one base pair.
pH-specific delivery and suppression of KU80 using pHLIP
We next confirmed the pH-dependent insertion of pHLIP-αKu80 and pHLIP-αKu80(γ) in cultured cells. To do this, we treated cells with TAMRA-labeled pHLIP-PNA conjugates in acidic (pH=6.2) or neutral (pH=7.8) media followed by several washes with PBS. Flow cytometry was used to quantify TAMRA fluorescence. As demonstrated in Fig. 1D–E, these culture conditions lead to robust pH-dependent delivery of pHLIP-αKu80 and pHLIP-αKu80(γ).
Using the culture conditions described above, we also confirmed pH-dependent antisense activity of pHLIP-αKu80 conjugates in the human lung adenocarcinoma cancer cell line A549 (Fig. 1F–G). Specifically, after treatment with pHLIP-αKu80(γ) at pH=6.2, the expression of Ku80 is knocked down to 45±14% of controls in A549 cells. Treatment with pHLIP-αKu80 caused a more modest reduction in KU80 expression (Fig. 1F–G), consistent with its reduced nucleic acid binding. Importantly, KU80 expression is unaffected in cells treated with pHLIP-PNA at pH=7.8, demonstrating that pHLIP-αKu80(γ) exhibits pH-specific activity against KU80.
Pharmacokinetics and pharmacodynamics of systemically administrated pHLIP-αKu80(γ) in tumor-bearing mice
To inform timing and dosing for subsequent radiosensitization and synthetic lethality experiments, we established the pharmacokinetic and pharmacodynamic properties of systemic pHLIP-αKu80(γ) treatment in mice bearing EMT6 mouse breast cancer tumors. We used the fluorescently labeled pHLIP-αKu80(γ)-TAMRA conjugate to visualize tumor and organ distribution after intravenous (i.v.) administration (via tail vein injection) using IVIS Spectrum. Intratumoral TAMRA fluorescence was detected and quantified at both 24 and 72 hours after a 5 mg/kg i.v. injection of pHLIP-αKu80(γ)-TAMRA (Fig. 2A–B). As expected, pHLIP-αKu80(γ)-TAMRA was also observed in the kidney (Fig. 2A), likely due to excretion of pHLIP by the renal system as well as uptake of pHLIP-αKu80(γ)-TAMRA by acidic regions in the kidney. We also observed a lower level of fluorescence in the liver (Fig. 2A–B). These results confirm prior work showing relative tumor specificity of pHLIP delivery (22) and also establish successful in vivo delivery of MPγPNAs by pHLIP.
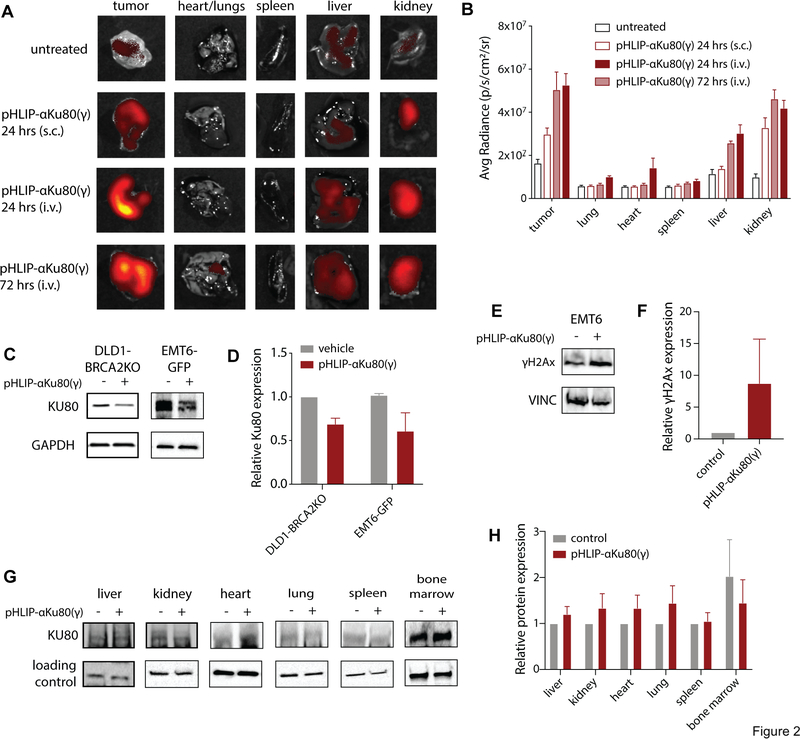
Fig. 2: Tumor-selective suppression of KU80 expression after systemic delivery of pHLIP-αKu80(γ).
(A) TAMRA fluorescence in isolated organs and EMT6 tumors after systemic (i.v.) treatment with pHLIP-αKu80(γ)-TAMRA (5 mg/kg). (B) Quantification of fluorescence data (two-way ANOVA, effect of treatment: P < 0.0001, 24 hrs (s.c.) vs. 24 hrs (i.v.): tumor P < 0.0001, liver P < 0.05, kidney P < 0.05, n = 3 mice/group). (C) Representative Western blot of KU80 expression in DLD1-BRCA2KO (left) and EMT6-GFP (right) tumors after pHLIP-αKu80(γ) treatment (5 mg/kg). (D) Quantification of Western blot data (two-way ANOVA, effect of treatment P < 0.05, n = 3 mice/group). (E) Representative Western blot of γH2Ax expression in EMT6 tumors from control and pHLIP-αKu80(γ) (5 mg/kg) treated mice. (F) Quantification of Western blot data (t-test, P = 0.33, n = 3 mice/group). (G) Representative Western blot of KU80 expression in isolated organs after pHLIP-αKu80(γ) treatment (5 mg/kg). (H) Quantification of Western blot data (two-way ANOVA, effect of treatment P = 0.50, n = 3 to 5 mice/group). Data are represented as means ± SEM.
To assess the pharmacodynamic properties of pHLIP-αKu80(γ), we treated DLD1-BRCA2KO human colon cancer xenograft tumor-bearing mice with pHLIP-αKu80(γ) (5 mg/kg, i.v.), and collected tumors and organs for Western blot analysis. In order to better measure the suppression of KU80 expression within tumor cells, we isolated DLD1-BRCA2KO tumor cells from stromal cells by immune depletion of mouse cells. At 48 hours after treatment, we observed a ~40% decrease in KU80 expression in isolated DLD1-BRCA2KO tumor cells (Fig. 2C–D). We then confirmed these results in EMT6 tumors, using EMT6 cells expressing GFP (EMT6-GFP) in order to isolate tumor cells from stromal and immune cells on the basis of GFP expression using fluorescence-activated cell sorting (FACS) (Fig. S2). Similar to the DLD1-BRCA2KO tumor cells, in EMT6-GFP cells, systemic treatment with pHLIP-αKu80(γ) caused a partial suppression of KU80 expression (Fig. 2C–D). To determine whether this degree of KU80 knockdown was sufficient to induce radiosensitization, we titrated siRNA-mediated knockdown of KU80 in tumor cells in culture and used a clonogenic survival assay to assess radiosensitization. siRNA doses corresponding to a ~40% knockdown were determined to increase cellular sensitivity to irradiation (Fig. S3), indicating that the extent of KU80 knockdown induced by pHLIP-αKu80(γ) in vivo would be sufficient to mediate radiosensitization. Furthermore, as compared with controls, tumors treated with pHLIP-αKu80(γ) also had a trend towards higher expression of γH2Ax, a marker of DNA damage, providing further evidence of the functional effects of pHLIP-αKu80(γ) on DNA repair (Fig. 2E–F).
We also assessed for the effects of pHLIP-αKu80(γ) treatment on KU80 expression in non-malignant tissue. Importantly, we did not observe any change in KU80 expression in any of the organs assessed at this time point, including liver, kidney, heart lung, and spleen (Fig. 2G–H). Together these results indicate that pHLIP-αKu80(γ) causes a selective suppression of KU80 expression within tumors.
Systemic administration of pHLIP-αKu80 causes tumor radiosensitization
We first assessed for potential tumor radiosensitizing effects of pHLIP-αKu80(γ) in subcutaneous mouse tumor xenografts of the mouse EMT6 breast cancer cell line. In this immune-competent model of breast cancer, pHLIP-αKu80(γ) treatment alone did not affect tumor growth or mouse survival (Fig. 3A–B). However, when administered with local tumor irradiation (1 × 15 Gy), pHLIP-αKu80(γ) significantly delayed tumor growth and prolonged survival (Fig. 3A–B). Other treatment protocols combining pHLIP-αKu80(γ) with local tumor irradiation (2 or 3 i.v. administrations of vehicle alone or pHLIP-αKu80(γ), combined with 1 × 10 Gy or 2 × 7 Gy) had similar effects on tumor growth and mouse survival in the EMT6 tumor model (Fig. S4).
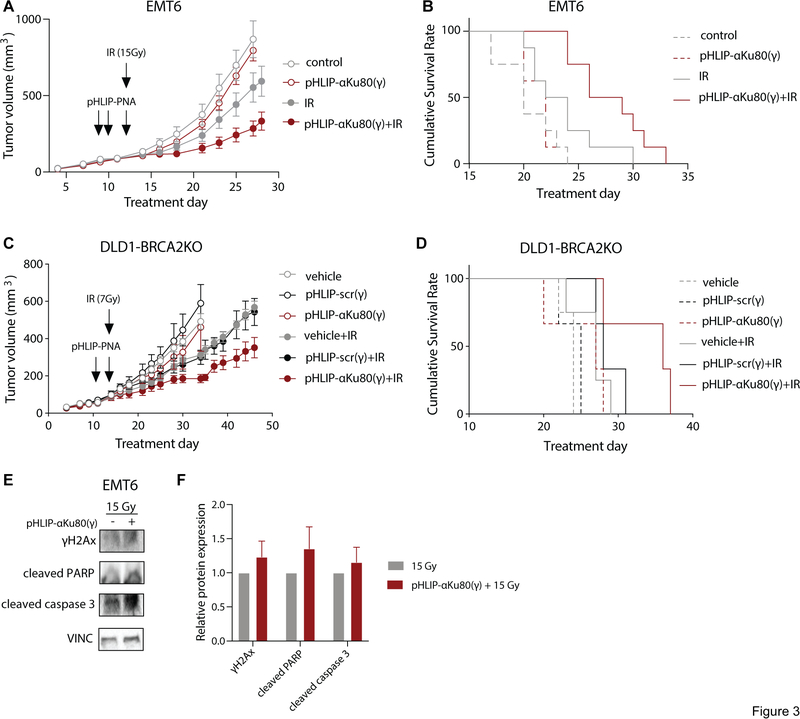
Fig. 3: pHLIP-αKu80(γ) causes tumor radiosensitization in vivo.
(A) Tumor growth curves in control or pHLIP-αKu80(γ) treated EMT6 tumors (5 mg/kg) alone (left) or with local tumor irradiation (1 × 15 Gy, right) (two-way RM-ANOVA, effect of treatment: unirradiated: P = 0.40, irradiated: P < 0.05, n = 8 mice/group). (B) Survival curves in control or pHLIP-αKu80(γ) treated EMT6 tumors (5 mg/kg) with or without local tumor irradiation (1 × 15 Gy) (Log-rank (Mantel-Cox) test, unirradiated: P = 0.54, irradiated: P < 0.05, n = 8 mice/group). (C) Tumor growth curves in DLD1-BRCA2KO tumors treated with vehicle, pHLIP-scr(γ) (5 mg/kg), or pHLIP-αKu80(γ) (5 mg/kg) alone or with local tumor irradiation (1 × 7 Gy) (two-way RM-ANOVA, unirradiated: effect of treatment P = 0.63, n = 3–4 mice/group; irradiated: effect of treatment P < 0.05, vehicle vs. pHLIP-αKu80(γ) P < 0.05, vehicle vs. pHLIP-scr(γ) P = 0.88, n = 3–4 mice/group). (D) Survival curves in DLD1-BRCA2KO tumors treated with vehicle, pHLIP-scr(γ) (5 mg/kg), or pHLIP-αKu80(γ) (5 mg/kg) alone or with local tumor irradiation (1 × 7 Gy) (Log-rank (Mantel-Cox) test, unirradiated: P = 0.27, n = 3–4 mice/group; irradiated: P = 0.08, n = 3–4 mice/group). (E) Representative Western blot of DNA damage and apoptotic markers in control or pHLIP-αKu80(γ) treated EMT6 tumors with local tumor irradiation (1 × 15 Gy) (n = 4 mice/group). (F) Quantification of Western blot data (two-way ANOVA, effect of treatment: P = 0.15, n = 3–4 tumors/group). Data are represented as means ± SEM.
We then tested for radiosensitization by pHLIP-αKu80(γ) in tumor xenografts using the human DLD1-BRCA2KO colon cancer cell line in immune-deficient athymic nude mice. These cells are BRCA2-deficient, conferring deficits in the HDR pathway of DNA double-strand break repair. There is some evidence that HDR deficiency may confer increased susceptibility to NHEJ inhibition (25), and so we first assessed for effects of pHLIP-αKu80(γ) monotherapy on DLD1-BRCA2KO tumor growth and survival (as defined by tumor volume 4X that of initial treatment). We found that pHLIP-αKu80(γ) treatment did not alter tumor growth or survival as compared with vehicle treated controls (Fig. 3C–D). However, when pHLIP-αKu80(γ) was administered in combination with local tumor irradiation (7Gy), it caused a significant tumor growth delay compared to irradiation alone along with a trend (P = 0.07) towards prolonged survival (Fig. 3C–D), similar to the results observed using the EMT6 model. Importantly, pHLIP-scr(γ) did not affect tumor growth or survival when administered alone or in combination with local tumor irradiation (Fig. 3C–D). Together, these results indicate that systemic administration of pHLIP-αKu80(γ) treatment induces radiosensitization in tumors.
We also tested for the effects of in vitro pHLIP-αKu80(γ) treatment on cell survival using clonogenic survival assays in DLD1-BRCA2KO and A549 cells treated with pHLIP-conjugated scr(γ) or αKu80(γ) PNAs under neutral and acidic conditions, with or without irradiation. In DLD1-BRCA2KO cells treated under acidic conditions, pHLIP-αKu80(γ) caused significant radiosensitization as compared with pHLIP-scr(γ) treatment (Fig S5). No radiosensitization was observed in cells treated under neutral conditions (Fig S5). Similar results were observed in A549 cells, with pHLIP-αKu80(γ) treatment causing a trend (P = 0.06) towards radiosensitization under acidic but not neutral conditions (Fig S5).
To better understand the mechanism underlying pHLIP-αKu80(γ)-induced radiosensitization, we analyzed the expression of DNA damage and apoptosis markers in control and pHLIP-αKu80(γ)-treated tumors with or without local tumor irradiation (15 Gy). Tumors treated with pHLIP-αKu80(γ) plus irradiation showed a trend towards increased expression of γH2Ax, a marker of DNA damage, as well as cleaved PARP and cleaved caspase 3, markers of apoptosis (Fig. 3E–F).
Systemic administration of pHLIP-αKu80 is not toxic
After administration of pHLIP-αKu80(γ), no gross toxicity was noted in treated mice, including weight loss (Fig. 4A), skin reactions, or behavioral changes. Furthermore, an evaluation of blood chemistries did not reveal any electrolytes abnormalities (Fig. S6) or any signs of renal or hepatic damage (Fig. 4B).
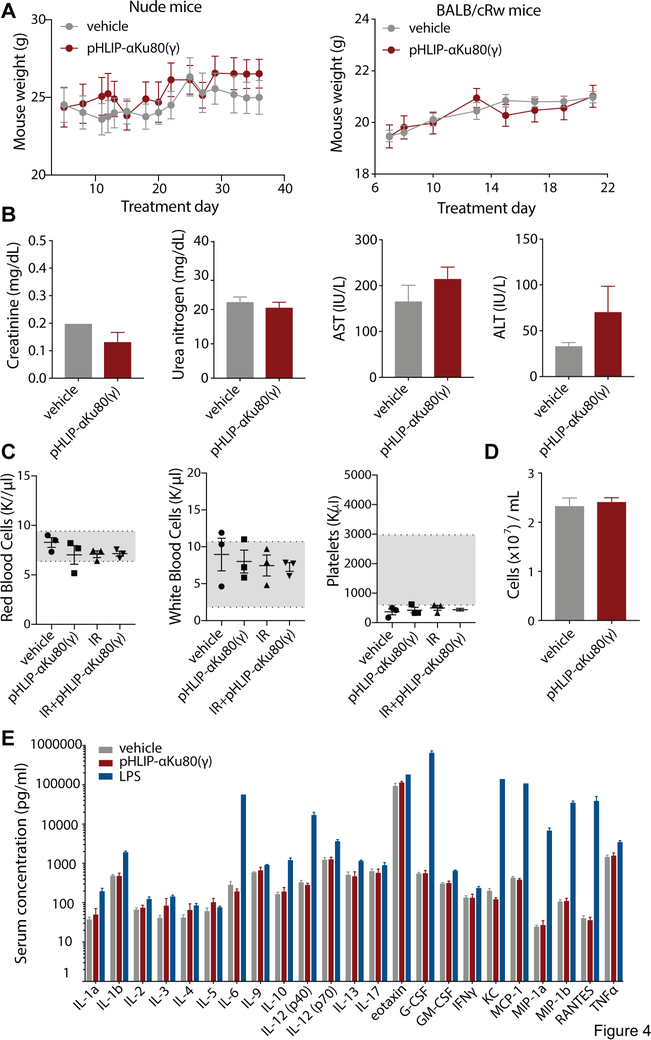
Fig. 4: No observable toxicity after pHLIP-αKu80(γ) treatment.
(A) Body weights of control and pHLIP-αKu80(γ)-treated nude (left) and BALBc/Rw (right) (nude mice: two-way RM ANOVA, effect of treatment P = 0.60, n = 3 mice/group; BALBc/Rw mice: two-way RM ANOVA, effect of treatment, P = 0.88, n = 8 mice/group). (B) Serum chemistry analysis in control and pHLIP-αKu80(γ)-treated mice (unpaired t-tests, creatinine: P = 0.12, urea nitrogen: P = 0.45, AST: P = 0.31, ALT: P = 0.25, n = 3 mice/group). (C) Peripheral blood cell analysis in control and pHLIP-αKu80(γ)-treated mice (1-way ANOVA, white blood cells P = 0.74, red blood cells: P = 0.40, platelets: P = 0.75, n = 3 mice/group). (D) Bone marrow cellularity in control and pHLIP-αKu80(γ)-treated mice (unpaired t-test, P = 0.64, n = 3 mice/group). (E) Serum cytokine levels in mice treated with vehicle, pHLIP-αKu80(γ) (5 mg/kg), or lipopolysaccharide (LPS, 10 mg/kg) (two-way ANOVA, effect of treatment P < 0.0001; Tukey’s multiple comparisons test, vehicle vs. pHLIP-αKu80(γ): P = 0.88. vehicle vs. LPS: P < 0.0001; n = 3 mice/group). Data are represented as mean±SEM.
As many cancer therapeutics display bone marrow toxicity, we also evaluated the effects of pHLIP-αKu80(γ) on bone marrow. Analysis of mature blood cells in the periphery did not indicate any bone marrow toxicity, as the number of red blood cells, white blood cells, and platelets were comparable between control and pHLIP-αKu80(γ) treated mice (Fig 4C). Furthermore, treatment with pHLIP-αKu80(γ) did not significantly change bone marrow cellularity (Fig. 4D) or the relative percentages of different bone marrow cell types as assessed by flow cytometry (Fig. S7).
We also evaluated the immunogenicity of pHLIP-αKu80(γ). To do this, we tested the serum levels of 23 different cytokines in the serum of mice treated with pHLIP-αKu80(γ). pHLIP-αKu80(γ) treatment did not alter the expression of peripheral cytokines (Fig. 4E), consistent with a lack of an immune response. Together these results do not indicate any normal tissue toxicity with pHLIP-αKu80(γ) treatment.
Discussion
In this report, we utilize new technologies to target a clinically important pathway for which therapeutic targeting has been historically challenging. While several small molecule inhibitors targeting the NHEJ pathway have been identified (26,27), the clinical potential of these molecules is limited by their lack of specificity and in vivo toxicity (28,29). Additionally, while antisense suppression and siRNA-mediated inhibition of NHEJ factor expression have been demonstrated in cell culture (8,9,30–34), they have not been reported in vivo (with the exception of Li, et.al., in which intratumoral injections of an adenovirus vector, requiring tumoral heat shock (42ºC), achieved in vivo radiosensitization). Here we report the in vivo use of antisense-based technology via systemic (i.v.) administration to target the non-enzymatic NHEJ factor KU80, which, lacking a known enzyme acivity, is not a classically “druggable” target.
Furthermore, we confer tumor selectivity on the antisense PNA molecule using a pH-based delivery system, pHLIP. We demonstrate that in culture, pHLIP-αKu80(γ) delivers the PNA to cancer cells and suppresses KU80 expression with pH-specificity. Furthermore, in vivo, pHLIP-αKu80(γ) targets to tumors and causes selective suppression of KU80 expression in tumors. This study builds upon prior work identifying pHLIPs as a potential delivery tool for anticancer therapeutics (12,35–39), and represents the first delivery of MPγPNA using pHLIPs as well as the first usage of pHLIP-PNA technology to reduce protein expression via an antisense mechanism.
Importantly, we show that pHLIP-αKu80(γ) increases the sensitivity of tumors to radiation therapy, causing a substantial delay in tumor growth and prolonging host survival when administered in combination with local tumor irradiation. Interestingly, while prior work has indicated that inhibition of NHEJ may be toxic in the setting of BRCA deficiency (25), we do not find pHLIP-αKu80(γ) to be effective as a monotherapy in DLD1-BRCA2KO tumor xenografts. This discrepancy may be explained by the fact that pHLIP-αKu80(γ) only induces a partial, and not complete, suppression of KU80 expression, and therefore treated tumors likely retain partial NHEJ capacity. Complete suppression of NHEJ may be required for the previously identified synthetic lethal interactions.
Finally, we provide evidence that pHLIP-αKu80(γ) is safe and non-toxic when administered in vivo, as assessed by several end-points. These indications of safety are in line with previous work that has established PNAs and pHLIPs as potential therapeutic agents with favorable side effect profiles (12,19,40). We expect relatively low off-target effects from pHLIP- αKu80(γ) treatment, as uptake of the pHLIP-PNA is primarily limited to tumor cells, and the αKu80(γ) PNA has 100% complementarity to the KU80 mRNA only (based on nucleotide BLAST search). While PNA-nucleotide binding is sequence specific, and mismatches in PNA sequences have been shown to significantly reduce nucleotide binding (41), it is possible that within cancer cells, the αKu80(γ) PNA may cause off-target effects by binding to sequences of partial complementarity (i.e. PCMTD2, KCNIP4).
While the suppression of KU80 expression and radiosensitization induced by pHLIP-αKu80(γ) treatment is modest, efficacy could be improved through a number of methods. A more detailed evaluation of dosing and timing protocols could optimize the treatment regimen. An alternative approach could be simultaneous delivery of multiple antisense PNAs targeting KU80. Additionally, serine modified PNAs (SγPNAs) could be used in place of MPγPNAs. Similar to MPγPNAs, SγPNAs also preorganize into a helical structure and demonstrate increased solubility and nucleic acid binding as compared with unmodified PNAs (42). Furthermore, synthesis of SγPNA monomers is simpler than MPγPNA monomers. Efforts could also be made to conjugate small molecule inhibitors of DNA-PK to pHLIPs. However, as these small molecules do not have available thiol groups, this approach would require the development of additional pHLIP-cargo linkage techniques to ensure that the small molecules could be inserted across the cell membrane and cleaved from pHLIP while still retaining activity against DNA-PK.
Together, these results establish pHLIP-αKu80(γ) as a tumor-specific radiosensitizer. The development of compounds like pHLIP-αKu80(γ) may eventually help lead to novel treatment regimens for cancer therapy that have both high specificity and low toxicity.
Supplementary Material
Implications.
This study describes a novel agent, pHLIP-αKu80(γ), which combines peptide nucleic acid (PNA) antisense and pH-low insertion peptide (pHLIP) technologies to selectively reduce the expression of the DNA repair factor KU80 in tumors and confer tumor-selective radiosensitization.
Acknowledgments
This work was supported by grants from the NIH: R35CA197574 to P.M.G., R01GM073857 to D.M.E., and F30CA221065 to A.R.K. Support for this research was also provided by NIH Medical Scientist Training Program Training grant T32GM007205. We thank D. Hegan, A. Gupta, and A. Dhawan for assistance.
Footnotes
Competing Interests
P.M.G is a consultant to and has equity interests in Cybrexa Therapeutics Inc. of New Haven, CT, which has licensed IP related to the topic of this work. D.M.E. participates and has a financial interest in pHLIP,Inc. Neither company was involved in this work.
References
- 1.Long XH, Zhao ZQ, He XP, Wang HP, Xu QZ, An J, et al. Dose-dependent expression changes of early response genes to ionizing radiation in human lymphoblastoid cells. Int J Mol Med 2007;19(4):607–15. [PubMed] [Google Scholar]
- 2.Shan B, Xu J, Zhuo Y, Morris CA, Morris GF. Induction of p53-dependent activation of the human proliferating cell nuclear antigen gene in chromatin by ionizing radiation. J Biol Chem 2003;278(45):44009–17 doi 10.1074/jbc.M302671200. [DOI] [PubMed] [Google Scholar]
- 3.Fritz G, Tano K, Mitra S, Kaina B. Inducibility of the DNA repair gene encoding O6-methylguanine-DNA methyltransferase in mammalian cells by DNA-damaging treatments. Mol Cell Biol 1991;11(9):4660–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weinstock DM, Jasin M. Alternative pathways for the repair of RAG-induced DNA breaks. Mol Cell Biol 2006;26(1):131–9 doi 10.1128/MCB.26.1.131-139.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gottlieb TM, Jackson SP. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell 1993;72(1):131–42. [DOI] [PubMed] [Google Scholar]
- 6.Nussenzweig A, Sokol K, Burgman P, Li L, Li GC. Hypersensitivity of Ku80-deficient cell lines and mice to DNA damage: the effects of ionizing radiation on growth, survival, and development. Proc Natl Acad Sci U S A 1997;94(25):13588–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ouyang H, Nussenzweig A, Kurimasa A, Soares VC, Li X, Cordon-Cardo C, et al. Ku70 is required for DNA repair but not for T cell antigen receptor gene recombination In vivo. J Exp Med 1997;186(6):921–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belenkov AI, Paiement JP, Panasci LC, Monia BP, Chow TY. An antisense oligonucleotide targeted to human Ku86 messenger RNA sensitizes M059K malignant glioma cells to ionizing radiation, bleomycin, and etoposide but not DNA cross-linking agents. Cancer Res 2002;62(20):5888–96. [PubMed] [Google Scholar]
- 9.Marangoni E, Le Romancer M, Foray N, Muller C, Douc-Rasy S, Vaganay S, et al. Transfer of Ku86 RNA antisense decreases the radioresistance of human fibroblasts. Cancer Gene Ther 2000;7(2):339–46 doi 10.1038/sj.cgt.7700111. [DOI] [PubMed] [Google Scholar]
- 10.Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science 1991;254(5037):1497–500. [DOI] [PubMed] [Google Scholar]
- 11.Sahu B, Sacui I, Rapireddy S, Zanotti KJ, Bahal R, Armitage BA, et al. Synthesis and characterization of conformationally preorganized, (R)-diethylene glycol-containing gamma-peptide nucleic acids with superior hybridization properties and water solubility. J Org Chem 2011;76(14):5614–27 doi 10.1021/jo200482d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng CJ, Bahal R, Babar IA, Pincus Z, Barrera F, Liu C, et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature 2015;518(7537):107–10 doi 10.1038/nature13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demidov VV, Potaman VN, Frank-Kamenetskii MD, Egholm M, Buchard O, Sonnichsen SH, et al. Stability of peptide nucleic acids in human serum and cellular extracts. Biochem Pharmacol 1994;48(6):1310–3. [DOI] [PubMed] [Google Scholar]
- 14.Thomas SM, Sahu B, Rapireddy S, Bahal R, Wheeler SE, Procopio EM, et al. Antitumor effects of EGFR antisense guanidine-based peptide nucleic acids in cancer models. ACS Chem Biol 2013;8(2):345–52 doi 10.1021/cb3003946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delgado E, Bahal R, Yang J, Lee JM, Ly DH, Monga SP. beta-Catenin knockdown in liver tumor cells by a cell permeable gamma guanidine-based peptide nucleic acid. Curr Cancer Drug Targets 2013;13(8):867–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bahal R, McNeer NA, Ly DH, Saltzman WM, Glazer PM. Nanoparticle for delivery of antisense gammaPNA oligomers targeting CCR5. Artif DNA PNA XNA 2013;4(2):49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McNeer NA, Anandalingam K, Fields RJ, Caputo C, Kopic S, Gupta A, et al. Nanoparticles that deliver triplex-forming peptide nucleic acid molecules correct F508del CFTR in airway epithelium. Nat Commun 2015;6:6952 doi 10.1038/ncomms7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McNeer NA, Schleifman EB, Cuthbert A, Brehm M, Jackson A, Cheng C, et al. Systemic delivery of triplex-forming PNA and donor DNA by nanoparticles mediates site-specific genome editing of human hematopoietic cells in vivo. Gene Ther 2013;20(6):658–69 doi 10.1038/gt.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bahal R, Ali McNeer N, Quijano E, Liu Y, Sulkowski P, Turchick A, et al. In vivo correction of anaemia in beta-thalassemic mice by gammaPNA-mediated gene editing with nanoparticle delivery. Nat Commun 2016;7:13304 doi 10.1038/ncomms13304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaupel P, Kallinowski F, Okunieff P. Blood flow, oxygen and nutrient supply, and metabolic microenvironment of human tumors: a review. Cancer Res 1989;49(23):6449–65. [PubMed] [Google Scholar]
- 21.Ivanov S, Liao SY, Ivanova A, Danilkovitch-Miagkova A, Tarasova N, Weirich G, et al. Expression of hypoxia-inducible cell-surface transmembrane carbonic anhydrases in human cancer. Am J Pathol 2001;158(3):905–19 doi 10.1016/S0002-9440(10)64038-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andreev OA, Dupuy AD, Segala M, Sandugu S, Serra DA, Chichester CO, et al. Mechanism and uses of a membrane peptide that targets tumors and other acidic tissues in vivo. Proc Natl Acad Sci U S A 2007;104(19):7893–8 doi 10.1073/pnas.0702439104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson M, Moshnikova A, Engelman DM, Reshetnyak YK, Andreev OA. Probe for the measurement of cell surface pH in vivo and ex vivo. Proc Natl Acad Sci U S A 2016;113(29):8177–81 doi 10.1073/pnas.1608247113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reshetnyak YK, Andreev OA, Lehnert U, Engelman DM. Translocation of molecules into cells by pH-dependent insertion of a transmembrane helix. Proc Natl Acad Sci U S A 2006;103(17):6460–5 doi 10.1073/pnas.0601463103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dietlein TS, Lappas A, Rosentreter A. [Clinical value of ab-interno trabecular surgery in the management of glaucoma]. Klin Monbl Augenheilkd 2014;231(11):1097–102 doi 10.1055/s-0034-1368609. [DOI] [PubMed] [Google Scholar]
- 26.Pospisilova M, Seifrtova M, Rezacova M. Small molecule inhibitors of DNA-PK for tumor sensitization to anticancer therapy. J Physiol Pharmacol 2017;68(3):337–44. [PubMed] [Google Scholar]
- 27.Davidson D, Amrein L, Panasci L, Aloyz R . Small Molecules, Inhibitors of DNA-PK, Targeting DNA Repair, and Beyond. Front Pharmacol 2013;4:5 doi 10.3389/fphar.2013.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta AK, Cerniglia GJ, Mick R, Ahmed MS, Bakanauskas VJ, Muschel RJ, et al. Radiation sensitization of human cancer cells in vivo by inhibiting the activity of PI3K using LY294002. Int J Radiat Oncol Biol Phys 2003;56(3):846–53. [DOI] [PubMed] [Google Scholar]
- 29.Nutley BP, Smith NF, Hayes A, Kelland LR, Brunton L, Golding BT, et al. Preclinical pharmacokinetics and metabolism of a novel prototype DNA-PK inhibitor NU7026. Br J Cancer 2005;93(9):1011–8 doi 10.1038/sj.bjc.6602823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Omori S, Takiguchi Y, Suda A, Sugimoto T, Miyazawa H, Takiguchi Y, et al. Suppression of a DNA double-strand break repair gene, Ku70, increases radio- and chemosensitivity in a human lung carcinoma cell line. DNA Repair (Amst) 2002;1(4):299–310. [DOI] [PubMed] [Google Scholar]
- 31.Peng Y, Zhang Q, Nagasawa H, Okayasu R, Liber HL, Bedford JS. Silencing expression of the catalytic subunit of DNA-dependent protein kinase by small interfering RNA sensitizes human cells for radiation-induced chromosome damage, cell killing, and mutation. Cancer Res 2002;62(22):6400–4. [PubMed] [Google Scholar]
- 32.Sak A, Stuschke M, Wurm R, Schroeder G, Sinn B, Wolf G, et al. Selective inactivation of DNA-dependent protein kinase with antisense oligodeoxynucleotides: consequences for the rejoining of radiation-induced DNA double-strand breaks and radiosensitivity of human cancer cell lines. Cancer Res 2002;62(22):6621–4. [PubMed] [Google Scholar]
- 33.Li GC, He F, Shao X, Urano M, Shen L, Kim D, et al. Adenovirus-mediated heat-activated antisense Ku70 expression radiosensitizes tumor cells in vitro and in vivo. Cancer Res 2003;63(12):3268–74. [PubMed] [Google Scholar]
- 34.Collis SJ, Swartz MJ, Nelson WG, DeWeese TL. Enhanced radiation and chemotherapy-mediated cell killing of human cancer cells by small inhibitory RNA silencing of DNA repair factors. Cancer Res 2003;63(7):1550–4. [PubMed] [Google Scholar]
- 35.An M, Wijesinghe D, Andreev OA, Reshetnyak YK, Engelman DM. pH-(low)-insertion-peptide (pHLIP) translocation of membrane impermeable phalloidin toxin inhibits cancer cell proliferation. Proc Natl Acad Sci U S A 2010;107(47):20246–50 doi 10.1073/pnas.1014403107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moshnikova A, Moshnikova V, Andreev OA, Reshetnyak YK. Antiproliferative effect of pHLIP-amanitin. Biochemistry 2013;52(7):1171–8 doi 10.1021/bi301647y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burns KE, McCleerey TP, Thevenin D. pH-Selective Cytotoxicity of pHLIP-Antimicrobial Peptide Conjugates. Sci Rep 2016;6:28465 doi 10.1038/srep28465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burns KE, Robinson MK, Thevenin D. Inhibition of cancer cell proliferation and breast tumor targeting of pHLIP-monomethyl auristatin E conjugates. Mol Pharm 2015;12(4):1250–8 doi 10.1021/mp500779k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Antosh MP, Wijesinghe DD, Shrestha S, Lanou R, Huang YH, Hasselbacher T, et al. Enhancement of radiation effect on cancer cells by gold-pHLIP. Proc Natl Acad Sci U S A 2015;112(17):5372–6 doi 10.1073/pnas.1501628112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ricciardi AS, Quijano E, Putman R, Saltzman WM, Glazer PM. Peptide Nucleic Acids as a Tool for Site-Specific Gene Editing. Molecules 2018;23(3) doi 10.3390/molecules23030632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ratilainen T, Holmen A, Tuite E, Nielsen PE, Norden B. Thermodynamics of sequence-specific binding of PNA to DNA. Biochemistry 2000;39(26):7781–91 doi 10.1021/bi000039g. [DOI] [PubMed] [Google Scholar]
- 42.Dragulescu-Andrasi A, Rapireddy S, Frezza BM, Gayathri C, Gil RR, Ly DH. A simple gamma-backbone modification preorganizes peptide nucleic acid into a helical structure. J Am Chem Soc 2006;128(31):10258–67 doi 10.1021/ja0625576. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.