Abstract
Purpose
A Renal Genetics Clinic (RGC) was established to optimize diagnostic testing, facilitate genetic counseling, and direct clinical management.
Methods
Retrospective review of patients seen over a two-year period in the RGC.
Results
One hundred eleven patients (mean age: 39.9 years) were referred to the RGC: 65 for genetic evaluation, 19 for management of a known genetic disease, and 18 healthy living kidney donors (LKDs) and their 9 related transplant candidates for screening. Forty-three patients underwent genetic testing with a diagnosis in 60% of patients including 9 with Alport syndrome, 7 with autosomal dominant polycystic kidney disease (ADPKD), 2 with genetic focal segmental glomerulosclerosis (FSGS), 2 with PAX2-mediated CAKUT, and 1 each with autosomal recessive polycystic kidney disease (ARPKD), Dent, Frasier, Gordon, Gitelman, and Zellweger syndromes. Four of 18 LKDs were referred only for APOL1 screening. For the remaining 14 LKDs, their transplant candidates were first tested to establish a genetic diagnosis. Five LKDs tested negative for the familial genetic variant, four were positive for their familial variant. In five transplant candidates, a genetic variant could not be identified.
Conclusion
An RGC that includes genetic counseling enhances care of renal patients by improving diagnosis, directing management, affording presymptomatic family focused genetic counseling, and assisting patients and LKDs to make informed decisions.
Keywords: genetic screening, genetic counseling, presymptomatic testing, comprehensive renal panel, next-generation sequencing
INTRODUCTION
Kidney diseases encompass a spectrum of disorders that may be limited to a subset of tubular transport pathways or are associated with a more global decline in kidney function. In turn, diseases associated with an overall decline in kidney function may arise from abnormalities in kidney and urinary tract development, specific defects in ciliary structure or function, or disorders that begin as tubulointerstitial or glomerular disease processes. The vast and varied phenotypic spectrum of these disorders includes several monogenic renal diseases as well as acquired disorders that can, at least superficially, resemble different genetic conditions.
With mapping of the human genome and advances in genetic sequencing technology, an increasing number of genetic causes of renal disease have been identified.1 It has become evident that many genetic diseases are present in individuals without an apparent family history of kidney disease, often reflecting the nonrecognition of affected family members, possibly from variable or subtle disease expression or nonpenetrance. In addition, occasionally family history cannot be ascertained (adoptees) or be misleading (misattributed paternity). Finally, genetic diseases from pathogenic variants that arise de novo or with autosomal recessive inheritance are less likely to have a family history. However, once a genetic diagnosis is confirmed this information can substantially enhance the management of patients and their families by affording the ability to screen family members at risk for disease and offering them targeted genetic counseling and testing including prenatal counseling and assessment. Providing preconception counseling also becomes possible.
While genetic disorders are acknowledged as the most common cause of renal diseases in children, their role in adults, except for those with polycystic kidney disease, is frequently overlooked.2 Even when considered, the dearth of certified genetic counselors (CGCs) who can provide timely counseling appointments can become an impediment to optimal patient management. Furthermore, the pressure of a busy clinic load with the inability to routinely extend the time available for evaluation and management of more complicated cases, especially those that have been on a diagnostic odyssey, often limits the ability to provide disease-focused or genotype-directed care. Additionally, in the field of living related renal transplantation identifying the genetic basis of kidney disease in the patient with end-stage renal disease (ESRD) can assist in the screening of the related living donor.3 Testing living donors of recent African ancestry for the APOL1 risk alleles G1 and G2 is also a consideration, given the emerging data on the risk of kidney disease.4,5
To help with a perceived need within our own practice, we established a Renal Genetics Clinic (RGC) with physicians who had a strong interest and clinical expertise in a broad range of renal genetic disorders and a CGC whose practice was limited to nephrology and cardiology. The renal genetics practice consisted of two adult nephrologists, one focused on polycystic kidney disease and another who saw all the remaining patients. The RGC worked closely with a pediatric nephrologist with an interest in developmental disorders and a clinical genetics provider with an interest in lysosomal storage disorders. The dedicated RGC operated weekly and all new referrals for a genetic diagnosis met the nephrologist and the CGC for clinical review and formulation of a differential diagnosis. Patients who underwent genetic testing returned to the RGC at least once for counseling and discussion of test results, and if appropriate, returned to their referring nephrologist. The goals of the clinic were:
To offer genetic counseling and testing services for patients, presymptomatic family members, and health-care providers
To assess the need for and facilitate genetic testing for kidney disease suspected to have an inherited basis
To offer consultative and ongoing management advice for inherited tubulopathies, ciliopathies, familial renal stone disease, and genetic glomerular diseases
To oversee the management of rare multisystem inherited diseases with a renal component such as Fabry disease, cystinosis, and tuberous sclerosis, and coordinate specialist consultative services
To help with the transition to an adult nephrology practice for patients with genetic renal disease who have been referred by their pediatric nephrology providers
To evaluate kidney transplant candidates and their related asymptomatic living donors for inherited renal disease to increase the safety and informed choice of living donation
MATERIALS AND METHODS
We performed a retrospective review of all patients who were evaluated in the RGC between January 2017 and December 2018. We collected information, including demographic data, the clinical features of the disease, and family history (a family member in this context is an individual genetically related to the patient). The study was approved by the University of Iowa Institutional Review Board (IRB 201810847). The study adheres to the Declaration of Helsinki and informed consent was waived by the IRB.
For the purposes of this analysis, we classified patients by the following four broad phenotypes: congenital anomalies of the kidney and urinary tract (CAKUT), ciliopathy and tubulointerstitial diseases, tubular transport disorders including nephrolithiasis and nephrocalcinosis, and glomerular diseases. We classified genetic variants using criteria developed by the American College of Medical Genetics and Genomics (ACMG) as benign (B), likely benign (LB), variant of unknown significance (VUS), likely pathogenic (LP), and pathogenic (P).6 We assessed the effect of genetic testing, when performed, on establishing or confirming a diagnosis and attempted to determine the impact of the genetic test results on patient management.
RESULTS
In total, 111 patients from 88 pedigrees were referred to the RGC for consultation with a renal genetics physician and/or the CGC during this time period. Fifty-two patients were male and 59 were female ranging in age from 1 to 79 years, with 16 patients under the age of 25 and a mean of 39.9 years. In this cohort, 65 patients from 57 pedigrees were referred for assistance with establishing a genetic diagnosis. Of these referrals, 39 came from internal nephrologists (pediatric nephrology: 5 and adult nephrology: 34), 15 referrals were from external nephrologists, and 11 were self-referrals. Nineteen patients with a known genetic disease were referred for continued management of their condition and 18 living donors and 9 related recipient candidates were referred for assessment of living donor genetic risk. The clinical features of the patients referred for consideration of a genetic diagnosis are shown in Supplementary Table S1.
Of the patients referred for a genetic diagnosis, 58 were considered candidates for genetic testing and 43 proceeded to counseling and testing. In 41 patients genetic testing was performed with a 264-gene (Supplementary Table S2) renal panel (KidneySeq™ V3, Iowa Institute of Human Genetics [IIHG], https://medicine.uiowa.edu/humangenetics/kidneyseq). With KidneySeq™, targeted capture of coding exons and splice sites of renal disease genes are performed with RNA baits and then subjected to massively parallel sequencing and bioinformatic processing as previously described.3,7 For one patient, testing for Fabry disease (OMIM 301500) was performed at Mount Sinai, New York, and for another patient, testing was limited to APOL1 risk alleles, G1 and G2 (IIHG). Outcomes of patients who underwent genetic testing with positive results are shown in Table 1. In 26 of 43 patients (60%), we identified pathogenic or likely pathogenic variants and were thus able to confirm a genetic diagnosis. Positive diagnoses included nine with Alport disease (four autosomal dominant: OMIM 104200 and five X-linked: OMIM301050), seven with autosomal dominant polycystic kidney disease (ADPKD) (OMIM 173900 and 613095), two with genetic focal segmental glomerulosclerosis (FSGS) (TRPC6: OMIM 603965, APOL1: OMIM 612551), two with PAX2-mediated CAKUT (OMIM 120330), and one each with autosomal recessive polycystic kidney disease (ARPKD) (OMIM 263200), Dent (OMIM 300009), Frasier (OMIM 136680), Gordon (OMIM 614495), Gitelman (OMIM 263800), and Zellweger spectrum (OMIM 601539) syndromes. In an additional five patients, the identified variants corresponded to the phenotype but did not meet criteria to be classified as pathogenic or likely pathogenic. These included two patients with suspected ADPKD, each of whom carried a VUS in PKD1. It is possible that the VUSs in these two patients are disease causing and may be reclassifiable as LP/P if segregation analysis can be performed in these families or if functional studies on these missense variants are completed. The remaining three patients each carried a single heterozygous pathogenic variant in an autosomal recessive disease (SCL12A3 in possible Gitelman: OMIM 263800; PKHD1 in a possible ARPKD: OMIM 263200 and CYP11B1 in a possible congenital adrenal hyperplasia: OMIM 202010). In 16 patients genetic testing was deferred: 4 because of limited genetic testing elsewhere, 4 due to insurance decline, 6 due to patient choice or expected out of pocket expense, 1 due to presumed genetic diagnosis based on family screening, and 1 was deferred by the RGC for later consideration. A summary of testing results is illustrated in Figure 1.
Table 1.
Patients (n = 31) with positive genetic variants are shown arranged by phenotype categories.
| Subject | Clinical features | FH | Sex/age/ethnicity | Genetic testing | ACMG criteria | Genetic diagnosis | Impact of genetic diagnosis |
|---|---|---|---|---|---|---|---|
| Tubular transport | |||||||
| 15 | Hypokalemia, nephrocalcinosis | N | M/40/EUR | SLC12A3 p.Gly741Arg—homozygous | Pathogenic (PS1, PS3, PM2, PM3, PP3) | Gitelman syndrome | Altered clinical diagnosis |
| 24 | Nephrocalcinosis | Y | M/41/EUR | CLCN5 p.Arg704Ter | Pathogenic (PVS1, PS1, PM2, PP3) | X-linked Dent disease | Establish a diagnosis, cascade screening, recurrence risk |
| 33 | Resistant HTN childhood onset | Y | F/23/EUR | CYP11B1 p.Phe79Ile | Likely pathogenic (PS1, PM2, PP3) | Carrier for 11 β-hydroxylase deficiency | Exclude other monogenic forms of resistant hypertension |
| 55 | Hyperkalemia | Y | M/56/EUR | KLHL3 p.Arg496His; SLC3A1 p. Glu321Valfs*6 | Likely pathogenic (PS1, PM2, PP2, PP3, PP5) | Gordon syndrome | Establish a diagnosis, cascade screening, initiated thiazides |
| 57 | Hypokalemia | N | F/62/EUR | SLC12A3 p.Cys994Tyr | Likely pathogenic (PS1, PP2, PP3, PP5) | Carrier for Gitelman syndrome | Diagnosis not confirmed; treat as Gitelman |
| Glomerular | |||||||
| 3 | Microscopic hematuria | Y | M/31/EUR | COL4A5 p.Gly1415Asp | Likely pathogenic (PM1, PM2, PP2, PP3, PP5) | X-linked Alport nephropathy | Initiated ACEI, discuss prognosis |
| 25 | Proteinuria, ESRD, kidney transplant | Y | M/59/EUR | COL4A5 p.Gly1143Ser | Likely pathogenic (PM1, PM2, PM5, PP3, PP5) | X-linked Alport nephropathy | Cascade screening |
| 29–1 | Alport syndrome (biopsy) | Y | F/59/EUR | COL4A3 p.Gly934Arg | Likely pathogenic (PM1, PM2, PP1, PP3) | Autosomal dominant Alport nephropathy | Cascade screening |
| 38 | Deafness, retinal dystrophy, podocyte myeloid bodies | Y | M/54/EUR | PEX1 p.Gln1192Stop; PEX1 p.Ile989Thr; PEX1 p.Pro674Leu | Pathogenic (PVS1, PM2, PP3, PP5) VUS (PM2, PP3, PP5) VUS (PM2, PP3) | Zellweger spectrum disorder | Establish a diagnosis, treatment |
| 39–2 | Chronic kidney disease | Y | M/33/EUR | COL4A5 p.Cys1521Ser | Likely pathogenic (PM1, PM2, PP1, PP3, PP5) | X-linked Alport nephropathy | Establish a diagnosis, cascade screening, recurrence risk |
| 39–3 | Microhematuria, proteinuria | Y | M/28/EUR | COL4A5 p.Cys1521Ser | Likely pathogenic (PM1, PM2, PP1, PP3, PP5) | X-linked Alport nephropathy | Establish a diagnosis, initiate ARB, prognostic information |
| 42–1 | Microscopic hematuria | Y | M/4/EUR | COL4A4 p.Gly622Argfs*35 | Likely pathogenic (PVS1, PM2, PP1, PP3) | Autosomal dominant Alport nephropathy | Establish a diagnosis, cascade screening, prognosis |
| 42–2 | Microhematuria, proteinuria, and TBMD | Y | F/36/EUR | COL4A4 p.Gly622Argfs*35 | Likely pathogenic (PVS1, PM2, PP1, PP3) | Autosomal dominant Alport nephropathy | Prognostic information |
| 42–3 | ESRD, kidney transplant | Y | M/78/EUR | COL4A4 p.Gly622Argfs*35 | Likely pathogenic (PVS1, PM2, PP1, PP3) | Autosomal dominant Alport nephropathy | Establish a diagnosis |
| 53 | CKD 5, global glomerulosclerosis | Y | F/30/AFR | APOL1 G1/G2 | Risk alleles | APOL1 associated nephropathy | Siblings may be at risk |
| 59 | Microhematuria, ESRD | Y | M/34/EUR | COL4A5 p.Gln1234Ter | Pathogenic (PVS1, PS1, PM2, PM4, PP3) | X-linked Alport nephropathy | Establish a diagnosis, family planning, cascade screening |
| 65 | Focal segmental glomerulosclerosis | N | F/26/EUR | TRPC6 p. Asn125Ser | Likely pathogenic (PS1, PP3, PP5) | Autosomal dominant FSGS | Establish a diagnosis, avoid steroids |
| 69 | ESRD, delayed female puberty | N | F/15/EUR | WT1 c.745+5G>A | Likely pathogenic (PS1, PM2, PP3, PP5) | Frasier syndrome | Establish a diagnosis, prophylactic gonadectomy |
| Ciliopathy | |||||||
| 4–2 | Bilateral renal cysts | Y | M/33/EUR | PKD1 p.Tyr2622Ter | Likely pathogenic (PVS1, PM2) | Autosomal dominant polycystic kidney disease | Family planning, prognostic information, tolvaptan |
| 19–1 | Bilateral renal cysts, liver cysts | Y | F/38/EUR | PKD2 p.Tyr311Leufs*2 | Pathogenic (PVS1, PM2, PP1, PP3, PP5) | Autosomal dominant polycystic kidney disease | Prognostic information, cascade screening |
| 19–2 | Bilateral renal cysts, liver cysts | Y | F/20/EUR | PKD2 p.Tyr311Leufs*2 | Pathogenic (PVS1, PM2, PP1, PP3, PP5) | Autosomal dominant polycystic kidney disease | Prognosis, cascade screening, family planning |
| 21 | Cystic kidney disease | N | F/21/EUR | PKD1 p.Thr2250Met | VUS (PM1, PP3) | Not established | Family screening and segregation analysis |
| 35–1 | Bilateral renal cysts, enlarged kidneys | N | F/12/EUR | PKHD1 c.390+1G>T; PKD1 p.Leu1106Val | Pathogenic (PVS1, PM2, PP3); likely benign (BP4, BP6) | Carrier for ARPKD | Likely ARPKD based on cascade screening |
| 40–1 | Asymptomatic | Y | F/28/EUR | PKD1 p.Tyr2004Thrfs*112 | Pathogenic (PVS1, PM2, PP3) | Autosomal dominant polycystic kidney disease | Establish a diagnosis, family planning, treatment |
| 48 | CKD 3, congenital hepatic fibrosis | Y | F/27/EUR | PKHD1 p.Arg375Trp; PKHD1 p.Ile222Leu | Likely pathogenic (PS1, PM1, PM2, PP3); likely pathogenic: PM1, PM2, PM5, PP2, PP3 | Autosomal recessive polycystic kidney disease | Establish a diagnosis |
| 58 | Renal and liver cysts, ESRD, HTN | Adopted | M/50/EUR | PKD1 p.Ala2332Trpfs*7 | Pathogenic (PVS1, PS1, PM2, PP3) | Autosomal dominant polycystic kidney disease | Recurrence risk, cascade screening, prognosis, treatment |
| 60 | Bilateral renal cysts | Y | F/41/EUR | PKD1 p.Val1105Gly | VUS (PM1, PM2, PP3) | Not established | Family screening and segregation analysis |
| 68 | Bilateral renal cysts | N | M/23/EUR | PKD1 p.Val1144Alafs*68 | Pathogenic (PVS1, PM2, PP3) | Autosomal dominant polycystic kidney disease | Establish a diagnosis, cascade screening, prognosis, tolvaptan |
| 70 | Bilateral renal cysts, liver cysts | N | F/43/EUR | PKD2 p.Leu409Argfs*42 | Pathogenic (PVS1, PM2, PP3) | Autosomal dominant polycystic kidney disease | Establish a diagnosis, prognosis, cascade screening |
| CAKUT | |||||||
| 12 | Unilateral atrophy versus hypoplasia | Y | F/48/EUR | PAX2 p.Arg140Leu | Likely pathogenic (PM1, PM2, PP1, PP2, PP3) | PAX2-mediated CAKUT | Establish a diagnosis, evaluate for eye disease, recurrence risk |
| 52 | Multicystic dysplastic kidney | Y | F/7/EUR | PAX2 p.His62Tyr | Likely pathogenic (PM1, PM2, PP2, PP3) | PAX2-mediated CAKUT | Cascade screening, recurrence risk |
ACEI angiotensin-converting enzyme inhibitor, ACMG American College of Medical Genetics and Genomics, AFR African/African American, AJ Ashkenazi Jewish, ARB Angiotensin 2 receptor blocker, ARPKD autosomal recessive polycystic kidney disorder, CAKUT congenital anomaly of the kidney or urinary tract, CKD chronic kidney disease, EA East Asian, ESRD end-stage renal disease, EUR Caucasian, FH family history, FSGS focal segmental glomerulosclerosis, HTN hypertension, LAT Hispanic or Latino, NA American Indian or Alaska Native, TBMD thin basement membrane disease, VUS variant of unknown significance.
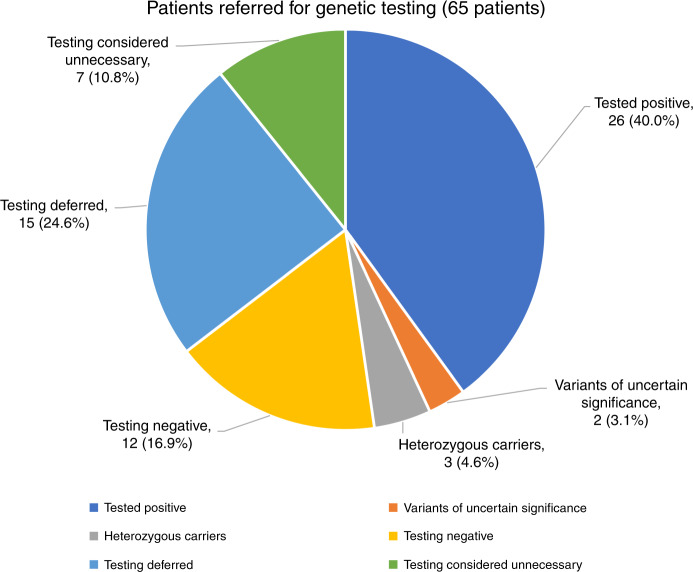
Fig. 1. Outcome of genetic clinic visit in 65 patients referred for genetic evaluation.
See Table 1. Forty-three patients were tested and of those tested, 58% had a genetic diagnosis established (tested positive). Actual numbers and percentages (in parentheses) for each category are shown. Three patients had one pathogenic variant for an autosomal recessive disorder (heterozygous carrier) that fit the phenotype. In seven patients, the preliminary evaluation concluded that genetic testing would not be informative (testing considered unnecessary).
We tested nine kidney transplant candidates, seven with a comprehensive genetic test panel (KidneySeq™ V3), one with an atypical hemolytic uremic syndrome (aHUS)/C3G panel (Genetic Renal Panel, Molecular Otolaryngology and Renal Research Labs, University of Iowa), and one for ADTKD (Wake Forest University, Winston-Salem, NC) (Table 2). We identified genetic variants that could account for their kidney disease in five recipient candidates, which permitted the screening of nine related living donor candidates for the familial genetic variant and, in turn, allowed five potential donors in whom the disease-causing variant was excluded to be accepted as living donors. In another five transplant candidates, genetic testing was negative, which nevertheless facilitated the counseling of living donors and their subsequent decision to donate. Four donor candidates who had positive genetic testing following familial screening were offered counseling and did not proceed with organ donation. In two of these, their transplant candidate had a VUS in a relevant gene for FSGS, ARHGAP24, and despite the lack of certainty about its role in the transplant candidate’s kidney disease, it was prudent to counsel the donor candidates, who carried the VUS, against donating. Four living kidney donors (LKDs) were referred only for APOL1 screening with two testing positive for two high-risk alleles. The donors were counseled before and after testing and offered the option of donating after a thorough explanation of the known effects of APOL1 risk status in the general population and in living donors.4,5,8,9
Table 2.
Outcome of testing in kidney transplant recipients (n = 10) and their related (n = 14) living donor candidates.
| Recipients | Living donors | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Clinical features | Subject | FH | Sex/age/ethnicity | Genetic testing | ACMG criteria | Genetic diagnosis | Subject | Sex/age/ethnicity | Relation to recipient | Donor clinical evaluation | Impact of genetic diagnosis |
| Microhematuria, proteinuria, TBMD | 29–1 | Y | F/59/EUR | COL4A3 p.Gly934Arg | LP: PM1, PM2, PP1, PP3 | AD Alport | 29–2 | M/51/EUR | Brother | Microhematuria | Positive for familial variant, not allowed to donate |
| 29–3 | M/54/EUR | Brother | Microhematuria | Positive for familial variant, not allowed to donate | |||||||
| FSGS-ESRD | 76–1 | Y | M/67/EUR | ARHGAP24 p.Gly493Arg | VUS: PM2, PP3 | None | 76–2 | M/25/EUR | Son | Negative | Positive for ARHGAP24 variant, not allowed to donate |
| 76–3 | M/35/EUR | Son | Negative | Positive for ARHGAP24 variant, not allowed to donate | |||||||
| 76–4 | F/33/EUR | Daughter | Negative | Negative for ARHGAP24 variant, allowed to donate | |||||||
| Heavy proteinuria ESRD | 77–1 | N | M/59/EUR | Negative | None | 77–2 | F/19/EUR | Daughter | Negative | Negative for any causal variants, allowed to donate | |
| Interstitial nephritis | 78–1 | Y | M/45/EUR | MUC1 insC in VNTR | LP: PVS1, PS1, PM2, PP3 | ADTKD-MUC1 | 78–2 | F/43/EUR | Sister | Negative | Negative for familial MUC1, allowed to donate |
| 78–3 | M/48/EUR | Brother | Negative | Negative for familial MUC1, allowed to donate | |||||||
| ADPKD | 79–1 | N | M/28/EUR | PKD1 p.Glu2771Lys; | P: PS1, PM1, PM2,PP3, PP5 | ADPKD-PKD1 | 79–2 | F/26/EUR | Sister | Negative | Negative for familial PKD1 variant, allowed to donate |
| C3 glomerulopathy | 80–1 | N | M/25/AFR | CFH p.Ser884Tyr | VUS: PM2, PP3 | None | 80–2 | F/44/EUR | Aunt | Negative | Negative for familial CFH variant; allowed to donate |
| FSGS-ESRD | 81–1 | Y | M/54/EUR | Negative (incl. MUC1) | None | 81–2 | M/22/EUR | Son | Negative | Negative for any causal variants, allowed to donate | |
| Proteinuria, hematuria, ESRD | 82–1 | Y | F/37/EUR,LAT | MYO1E p.Glu154Gln (AR) | VUS: PM2, PP3 | None | 82–2 | M/35/EUR, LAT | Brother | Negative | Negative for causal variants, allowed to donate |
| ESRD, bland urine | 83–1 | Y | F/61/EUR | CLCNKB p.Gly164Arg; NPHP4 p.Phe729Ser | VUS: PM2, PP3; VUS: PM2, PP3; | None | 83–2 | F/40/EUR | Daughter | Negative | Negative for causal variants, allowed to donate |
| Bilateral VUR, MCDK | 84–1 | N | M/1/EUR | Negative | None | 84–2 | M/36/EUR | Father | Few cysts | Affected son negative for causal variants | |
Living donors who underwent testing for APOL1 risk alleles alone are not included. Subject 29–1 is also in Table 1.ACMG American College of Medical Genetics and Genomics, AD autosomal dominant, ADPKD autosomal dominant polycystic kidney disease, ADTKD autosomal dominant tubulointerstitial kidney disease, AFR African/African American, AR autosomal recessive, ESRD end-stage renal disease, EUR Caucasian, FH family history, FSGS focal segmental glomerulosclerosis, LAT Hispanic or Latino, LP likely pathogenic, MCKD medullary cystic kidney disease, P pathogenic, TBMD thin basement membrane disease, VUR vesicoureteric reflux, VUS variant of unknown significance.
Finally, we had 19 patients with a variety of known genetic renal diseases including aHUS, Fabry disease, cystinosis, and tuberous sclerosis who were referred to the clinic for continued management (Table 3). These included the prescription and monitoring of chronic therapies such as eculizumab, cysteamine, agalsidase beta, and everolimus; periodic surveillance for potential or known disease manifestations; and the coordination of specialist care.
Table 3.
Patients with a known genetic disease referred to the clinic for disease management (n = 19).
| Subject number | Sex/age/ ethnicity | FH | Diagnosis | Basis for diagnosis | Genetic testing | Testing lab | ACMG criteria | Reason for referral |
|---|---|---|---|---|---|---|---|---|
| 1 | F/50/EUR | Yes—multiple | Fabry disease | Low α-GAL A; positive family history | GLA p.Arg227Gln | Mount Sinai, New York, NY | LP: PM1, PM2, PP2, PP3, PP5 | Renal biopsy |
| 4–1 | F/56/EUR | Yes—multiple | ADPKD | Cystic kidneys, positive family history | IIHG (Kidneyseq™), Iowa City, IA | CKD f/u | ||
| 6 | M/21/EUR | Yes—multiple | Fabry disease | Low α-GAL A; positive family history | GLA p.Ser297Tyr | Mount Sinai, New York, NY | LP: PM1, PM2, PM5, PP2, PP3 | CKD f/u |
| 7 | M/29/EUR | No | Cystinosis | Fanconi syndrome, renal rickets and corneal crystals in infancy | Not done | Cysteamine Rx, manage disease | ||
| 8 | F/59/EUR | Yes—multiple | Fabry disease | Slit lamp, positive family history | GLA p.Trp204Ter | Mount Sinai, New York, NY | P: PVS1, PM1, PM2, PP3 | CKD f/u |
| 9 | M/54/AFR | Yes—multiple | Fabry disease | Low α-GAL A; positive family history | GLA p.Trp340Ter | Mount Sinai, New York, NY | P: PVS1, PM1, PM2, PP3 | CKD f/u |
| 10 | M/47/EUR | Yes—multiple | Fabry disease | Low α-GAL A; positive family history | GLA p.Ala29GlyfsTer2 | Mount Sinai, New York, NY | P; PVS1, PM1, PM2, PP3 | CKD f/u |
| 11 | F/27/EUR | Yes—sister | Cystinosis | Bone marrow biopsy positive for cystine crystals | Not done | Cysteamine Rx | ||
| 19 | F/23/EUR | No | Tuberous sclerosis | Clinical criteria | Not done | Manage renal AMLs | ||
| 26 | F/32/EUR | No | Tuberous sclerosis + TMA in pregnancy | TSC: clinical criteria | TSC 1c.1029+3A>G; PLG p.Thr200Ala |
CHG, Cambridge, MA; MORL (Genetic Renal Panel), Iowa City, IA |
VUS: PM2, PP3, PP5; VUS: PP3 | Manage tuberous sclerosis |
| 27 | M/18/EUR | Yes—multiple | Fabry disease | Kidney biopsy | GLA p.Cys63Arg | Mount Sinai, New York, NY | LP: PM1, PM2, PM5, PP2, PP3 | CKD f/u |
| 28 | M/34/EUR | No | Fabry disease | Symptoms; positive family history | GLA p.Gly260Glu | Mount Sinai, New York, NY | LP: PM1, PM2, PM5, PP2, PP3 | CKD f/u |
| 31 | M/24/EUR | No | Unilateral renal aplasia | Antenatal and postnatal imaging | Not done | CAKUT f/u | ||
| 37 | M/59/EUR | Yes—multiple | Suspected Fabry, no manifestation | Low α-GAL A; positive family history | GLA p.Ala143Thr | Mount Sinai, New York, NY | LP: PM1, PM5, PP2, PP3, PP5 | Referred for renal biopsy |
| 62 | F/79/EUR | Yes—multiple | Familial hypocalciuric hypercalcemia | Hypercalcemia, positive family history | CaSR p.Pro55Leu | Mayo Medical Lab, Rochester, MN | LP: PM1, PM2, PP2, PP3, PP4, PP5 | Post-test genetic counseling |
| 66 | F/34/EUR | Yes—sister | aHUS | TMA, genetic screening | CFH p.Leu1189Argfs*2 | MORL (Genetic Renal Panel), Iowa City, IA | P: PVS1, PM2, PP3 | aHUS post-transplant f/u |
| 67 | M/30/EUR | No | None | Asymptomatic | Negative for NPHP1 variant | IIHG (Kidneyseq™), Iowa City, IA | Preconception counseling, spouse with NPHP1 deletion | |
| 74 | F/40/EUR | No | aHUS | TMA, genetic screening | CFI p.Tyr369Ser | MORL (Genetic Renal Panel), Iowa City, IA | LP: PM1, PM2, PP3, PP5 | aHUS post-transplant f/u |
| 75 | F/38/EUR | No | aHUS | TMA, genetic screening | CFH p.Glu625Ter | MORL (Genetic Renal Panel), Iowa City, IA | P: PVS1, PM2, PP3 | aHUS post-transplant f/u |
Genetic screening in these patients was performed prior to referral.
ACMG American College of Medical Genetics and Genomics, ADPKD autosomal dominant polycystic kidney disease, AFR African/African American, aHUS atypical hemolytic uremic syndrome, AML angiomyolipoma, CAKUT congenital anomalies of kidney and urinary tract, CHG Center for Human Genetics, CKD chronic kidney disease, EUR Caucasian, f/u follow up, FH family history, IIHG Iowa Institute of Human Genetics, LP likely pathogenic, MORL Molecular Otolaryngology and Renal Research Laboratories, P pathogenic, TMA thrombotic microangiopathy, TSC tuberous sclerosis, VUR vesicoureteric reflux, VUS variant of unknown significance, α-GAL A α-galactosidase A.
DISCUSSION
In the practice of medicine, establishing a clinical or laboratory diagnosis is generally a prerequisite for appropriate management of a patient, including prognostication and guiding therapy. Many diseases that present as one of the distinct renal phenotypes such as glomerular disorders, tubulointerstitial diseases, cystic kidney diseases, tubular transport disorders, and disorders of renal development are single-gene disorders. Genetic renal diseases may be diagnosable with traditional testing methods such as ultrasound imaging in the setting of a positive family history as in the case of ADPKD or a renal biopsy with a low plasma α-galactosidase in a male with suspected Fabry disease. However, genetic testing often provides diagnostic certainty for many other renal diseases, especially when their presentation is nonclassic or when standard studies are merely suggestive but not diagnostic. In the case of progressive chronic kidney diseases with no distinctive imaging finding, a renal biopsy may not offer enough etiologic information and if the presentation is late there is often nonspecific tubulointerstitial fibrosis and glomerulosclerosis obscuring the characteristic findings that may have been present. Genetic testing also offers other advantages as it can be performed at any stage of the disease including presymptomatically and is noninvasive and cost effective.
With the advent of more efficient sequencing technologies with high-throughput bioinformatic processing, there are several options available for the clinician wishing to establish a genetic diagnosis. When a single-gene disorder such as Fabry disease is suspected, the relevant gene can be screened by polymerase chain reaction (PCR)-based sequencing of coding exons, which is available at several clinical testing laboratories.10 More often a genetic diagnosis is suspected but the exact nature of the genetic disease is unknown, or due to considerable locus heterogeneity in disease causation, more broad-based testing is necessary. These broader methods take advantage of next-generation sequencing methods and are either phenotype focused (e.g., FSGS panel, ciliopathy panel, nephrolithiasis panel) or more comprehensive and thereby include all or nearly all genes implicated in monogenic renal diseases (comprehensive targeted gene panel such as KidneySeq™) or even the exome.7,11,12
As stated previously, establishing a diagnosis is a vital part of the management of a patient, even if the diagnosis does not lead to a specific therapy. Some patients with rare genetic disorders have been on a diagnostic odyssey, and having a diagnosis avoids unnecessary testing and continued uncertainty.13,14 Furthermore, with a diagnosis comes the ability to provide a meaningful prognosis and, if appropriate, counseling for family members who may be at risk. In some situations, the recognition that a common renal disease like FSGS may be genetic in origin can also have an impact on management. For example, autosomal dominant FSGS from heterozygous missense or loss-of-function variants in COL4A3 and COL4A4 (cases 29–1 and 42–3) or ADPKD from PKD2 variants (cases 19–1 and 19–2) predicts a slowly progressive disease with very late development of ESRD.15–17 Conversely, the identification of pathogenic null variants in PKD1 with a resulting high ProPKD score may identify ADPKD patients who require frequent follow up and who may benefit from tolvaptan therapy.18 Genetic FSGS can present like primary FSGS with nephrotic syndrome and diffuse foot process effacement on renal biopsy (case 64 FSGS-TRPC6) but is generally unresponsive to steroids.19 Genetic variants that typically cause CAKUT or tubulointerstitial diseases may also be identified in patients with FSGS, likely a manifestation of secondary FSGS in the context of reduced nephron mass, as we report here in at least one family member of case 12.20
Confirming a genetic diagnosis can be helpful in tailoring therapy for a patient. The identification of a likely pathogenic variant in KLHL3 confirmed a diagnosis of Gordon syndrome (case 55) prompting the use of low-dose thiazide diuretic rather than chronic furosemide and sodium polystyrene sulfonate, which up to that point had been ineffective. In Fabry disease, a heterozygous female may have normal plasma α-galactosidase activity and unrecognized end organ disease and identifying the familial genetic variant in a female may therefore prompt cardiac testing or a renal biopsy.21,22 In other situations, specific treatment strategies require knowledge of not only the underlying gene but also the specific genetic variant. For example, while enzyme replacement therapy (agalsidase beta) can be offered to any patient with Fabry disease, specific amenable variants in the GLA gene can be rescued by the pharmacologic chaperone migalastat (case 28).23,24 Similarly, some genetic causes of FSGS (e.g., COQ2, COQ6, ADCK4) may respond to CoQ10, offering a cheap and innocuous treatment.25,26
A genetic diagnosis may also offer specific management strategies in the post-transplant period or predict a risk of recurrent disease following a kidney transplant. Genetic FSGS, except for NPHS1-mediated disease, is considered to have a low risk of recurrence in the post-transplant setting.27 Atypical HUS from CFH and CFI variants requires perioperative and likely lifelong complement blockade to prevent post-transplant recurrence, and based on identified variants, we have continued eculizumab in three cases (cases 74, 75, and 76). In another case diagnosed as possible aHUS when severe thrombotic microangiopathy occurred during pregnancy, genetic testing identified only a VUS in PLG, and we withdrew eculizumab one year after a successful kidney transplant (case 26).
A genetic diagnosis may also permit the earlier recognition of extrarenal manifestations of disease such as cardiac disease in Fabry disease (case 9, left ventricular hypertrophy [LVH], dilated aortic root), or allow strategies to prevent the development of future severe complications (e.g., prophylactic gonadectomy for future gonadoblastoma risk in an XY female with Frasier syndrome, case 69). In some cases genetic testing may even uncover a new genetic basis for renal disease. Case 38 was referred to us with asymptomatic proteinuria, impaired kidney function, and zebra bodies in podocytes but a normal plasma ɑ-galatosidase. We noted that he also had pigmentary retinal dystrophy, bilateral sensorineural hearing loss, tremors, bradykinesia, myoclonus, and cerebellar signs. He did not have an identifiable genetic variant when tested with KidneySeq™, a targeted renal gene panel with 264 genes, but was found to have compound heterozygous variants in PEX1 when tested with OtoSCOPE™, a multigene panel of hearing loss genes.7,28 Variants in PEX1 cause a peroxisomal biogenesis disorder manifesting as one of a group of conditions called Zellweger spectrum disorder.29 This disease accounts for the auditory, visual, and central nervous system (CNS) manifestations and is thought to explain the renal phenotype. However, proteinuric chronic kidney disease (CKD) with podocyte inclusions has not been recognized previously in this condition although crystalline inclusions have been seen in affected livers.30
Finally, the confirmation of the genetic basis of a disease, which may have been previously suspected to be sporadic, can prompt at-risk family members to be screened for disease. This occurred for case 39–3, the sibling of 39–2, who was prompted to see a physician for the first time following the identification of familial Alport disease and who was discovered to have stage 3 CKD with microscopic hematuria together with the identified familial COL4A5 variant.
With respect to living kidney donors, it is important to ascertain the cause of kidney disease in the related recipient, so that the living donor can be screened for genetic disease if appropriate.31 Unfortunately, too often the cause of ESRD in patients on the transplant waitlist is unknown or assumed and the increased risk of ESRD postdonation in related living donors may reflect a missed genetic disease.32,33 In the case of the gene APOL1, the inheritance of two copies of the risk alleles G1 or G2 substantially increases the risk of ESRD in patients of sub-Saharan African ancestry who have hypertension, HIV, sickle cell disease, or systemic lupus erythematosus (SLE).8,9 Preliminary studies suggest that living donors with two APOL1 risk variants may have a lower estimated glomerular filtration rate (eGFR) and possibly a higher risk of ESRD postdonation.5 In our opinion, all at-risk living donors should be offered genetic counseling and the option of APOL1 genetic testing before they are accepted as living kidney donors.
There are several barriers to genetic testing in the typical nephrology practice. First, many genetic disorders have an incomplete or overlapping phenotype and may phenocopy an acquired disease, thus causing a genetic disease to go unrecognized. Alport gene variants are now considered to be the most common genetic cause of adult onset FSGS and yet many of these patients do not have the typical syndromic manifestations of sensorineural deafness or lenticonus.34 Other classic syndromic conditions such as nail patella syndrome and renal coloboma syndrome may also present with isolated FSGS without skeletal or ocular manifestations, respectively.20,35 Second, although there have been tremendous advances in the ability to rapidly sequence and interpret variants in the human exome and genome, an understanding of the power and limitations of sequencing technology has not reached the practicing clinician. Furthermore, the rapidly changing landscape of testing labs and capabilities makes it challenging for a clinician to determine the best testing approach (i.e., single-gene vs. limited panel vs. comprehensive panel vs. exome) or how and where to have genetic testing done. Since it is considered a new and emerging technology, even though it is cost-efficient, some insurance payers require preauthorization, which can again dampen a clinician’s enthusiasm for ordering these tests. Finally, the genetic test results, especially when scored based on ACMG criteria, can be confusing to the clinician when a series of variants of unknown significance (VUS) are reported in genes that may not even be relevant to the patient’s disease.
A CGC is a vital part of any genetics clinic.36,37 Patients may be referred for consideration of genetic testing and if the indication for testing is not clear, a nephrologist with experience in renal genetic diseases should evaluate the patient to formulate a differential diagnosis and determine whether genetic testing is indicated. If the patient is agreeable to genetic testing, he or she should be seen by a CGC prior to genetic testing. Genetic counseling involves obtaining a detailed medical and family history, performing a risk assessment for the patient, and providing education, psychosocial counseling, and support. The implications of a genetic diagnosis on the patient’s prognosis, medical management, familial risk, and risk of recurrence following kidney transplant are also discussed. Patients are informed of the current policies in place that protect them from discrimination based on a genetic testing result. Currently, the Genetic Information Nondiscrimination Act (GINA) of 2008 protects them from health insurance and employment discrimination. However, it does not cover life insurance, long-term care insurance, or disability insurance and is not protective in companies with fewer than 15 employees. It is important for patients to understand the benefits, risks, and limitations of a genetic test to make an informed decision.
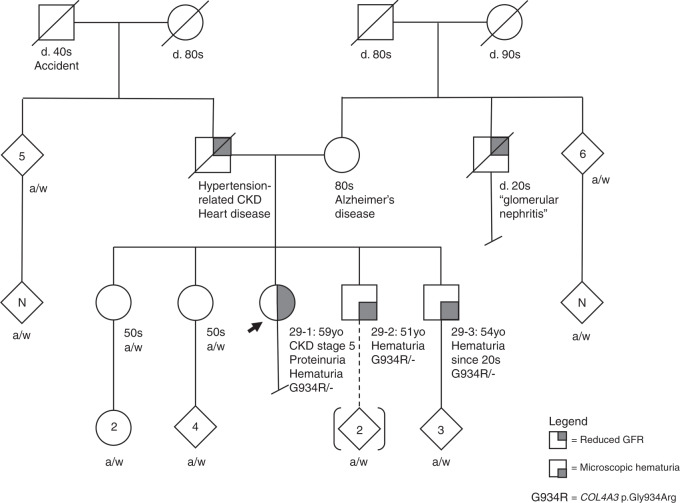
Once the genetic test report is available, the test results are communicated to the patient as soon as practical, ideally with a follow-up visit. Genetic counseling should again be offered at this point to ensure the patient understands the implications of the results. It is important to note that in some cases identified genetic variants may not rise to the level of pathogenicity required to establish causality. Therefore, additional studies including segregation analysis within an affected family can provide further evidence to support pathogenicity of the identified variant. We were able to demonstrate segregation of the identified variant in pedigree 19 and pedigree 29, which helped establish pathogenicity (Figure 2). Sometimes, genetic variants that could explain the phenotype may be difficult to identify, as is the case with large gene segmental deletions or duplications that may not be detected with a standard bioinformatic pipeline.38 In addition, if phenotype-limited gene panels are used the causal gene may not be included in the selected panel chosen such that a negative result would need to be interpreted with caution. Since panels and exomes typically focus on coding exons and their associated splice sites, a negative result may also arise because the causal variant is in an untranslated portion of a gene that is not assessed in the sequencing strategy. This may also be an explanation for the identification of just one causal variant in an autosomal recessive disease. For example, in two large studies, only one pathogenic variant was identified in ~9% of patients with Gitelman syndrome and in 21% of patients with ARPKD.39,40 We were only able to identify a single heterozygous pathogenic variant in each of three patients with suspected autosomal recessive disease (cases 33, 35–1, and 57).
Fig. 2. Pedigree chart for subjects 29–1, 29–2, and 29–3.
The identified heterozygous genetic variant COL4A3 p.Gly934Arg (G934R) was initially classified as a variant of unknown significance (VUS) (American College of Medical Genetics and Genomics [ACMG] criteria: PM1, PM2, PP3) but following segregation analysis was reclassified as likely pathogenic (ACMG: PM1, PM2, PP1, PP3). CKD chronic kidney disease, GFR glomerular filtration rate.
Summary
As a growing number of genes become implicated in kidney diseases associated with a variety of renal phenotypes that present at all ages, an RGC may become increasingly relevant. Ideally, such a clinic would include an adult and pediatric nephrologist with an interest in genetic renal disorders and a CGC. Developing the clinical expertise in the assessment, diagnosis, and treatment of renal genetic diseases offers the potential for specific disease management and/or genotype-directed care. The genetics clinic can serve primarily as a consultative service to determine if genetic testing is necessary and then identify the appropriate testing strategy and facilitate testing where indicated. In other situations, where chronic disease–specific care or coordination of specialist management of a multisystem disease is required, the genetics clinic can provide this long-term care. An RGC can also determine the appropriate screening strategy for presymptomatic testing of related family members, especially living donor candidates where excluding renal disease is of vital importance. In this study, we demonstrate a role for the RGC in these areas with a high rate of genetic confirmation of diagnosis, although it is a single-center study of patients predominantly of European ancestry. Nevertheless, given the obvious benefits, we believe that a specialized RGC will have an increasingly important place in the evaluation and care of patients with a wide variety of kidney diseases.
Supplementary information
Acknowledgements
The study was approved by the University of Iowa Institutional Review Board (IRB number 201810847) for human subject research.
Disclosure
C.P.T. and R.J.S. help develop KidneySeq™ at the University of Iowa as a comprehensive clinical genetic testing panel to evaluate patients with various renal phenotypes. M.H. has received financial support from Sanofi Genzyme for Advisory Board activity. He has received research support from Protalix Biotherapeutics and Idorsia Pharmaceuticals Ltd. The institution received funds to support the research study. The other authors declare no conflicts of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
10/1/2020
The original PDF version of this Article contained an error in Table 3. For subject number 74, the column labelled "Genetic testing" should read CFI p.Tyr369Ser not CFI p.Tyr200Ser. This has now been corrected in both the PDF and HTML versions of the Article.
Change history
10/6/2020
A Correction to this paper has been published: 10.1038/s41436-020-01000-0
Supplementary information
The online version of this article (10.1038/s41436-020-0772-y) contains supplementary material, which is available to authorized users.
References
- 1.Armstrong ME, Thomas CP. Diagnosis of monogenic chronic kidney diseases. Curr Opin Nephrol Hypertens. 2019;28:183–194. doi: 10.1097/MNH.0000000000000486. [DOI] [PubMed] [Google Scholar]
- 2.Harambat J, van Stralen KJ, Kim JJ, Tizard EJ. Epidemiology of chronic kidney disease in children. Pediatr Nephrol. 2012;27:363–373. doi: 10.1007/s00467-011-1939-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas CP, Mansilla MA, Sompallae R, et al. Screening of living kidney donors for genetic diseases using a comprehensive genetic testing strategy. Am J Transplant. 2017;17:401–410. doi: 10.1111/ajt.13970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Locke JE, Sawinski D, Reed RD, et al. Apolipoprotein L1 and chronic kidney disease rsk in young potential living kidney donors. Ann Surg. 2018;267:1161–1168. doi: 10.1097/SLA.0000000000002174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doshi MD, Ortigosa-Goggins M, Garg AX, et al. APOL1 genotype and renal function of black living donors. J Am Soc Nephrol. 2018;29:1309–1316. doi: 10.1681/ASN.2017060658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–423. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mansilla M, Sompallae R, Nishimura C, et al. Targeted broad-based genetic testing by next-generation sequencing informs diagnosis and facilitates management in patients with kidney diseases. Nephrol Dial Transplant. 2019. 10.1093/ndt/gfz173 [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 8.Genovese G, Friedman DJ, Ross MD, et al. Association of trypanolytic ApoL1 variants with kidney disease in African-Americans. Science. 2010;329:841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedman DJ, Pollak MR. Apolipoprotein L1 and kidney disease in African Americans. Trends Endocrinol Metab. 2016;27:204–215. doi: 10.1016/j.tem.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shabbeer J, Yasuda M, Benson SD, Desnick RJ. Fabry disease: identification of 50 novel alpha-galactosidase A mutations causing the classic phenotype and three-dimensional structural analysis of 29 missense mutations. Hum Genomics. 2006;2:297–309. doi: 10.1186/1479-7364-2-5-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Groopman EE, Marasa M, Cameron-Christie S, et al. Diagnostic utility of exome sequencing for kidney disease. N Engl J Med. 2019;380:142–151. doi: 10.1056/NEJMoa1806891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Connaughton DM, Kennedy C, Shril S, et al. Monogenic causes of chronic kidney disease in adults. Kidney Int. 2019;95:914–928. doi: 10.1016/j.kint.2018.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marshall DA, MacDonald KV, Heidenreich S, et al. The value of diagnostic testing for parents of children withrare genetic diseases. Genet Med. 2019;21:2798–2806. doi: 10.1038/s41436-019-0583-1. [DOI] [PubMed] [Google Scholar]
- 14.Gainotti S, Mascalzoni D, Bros-Facer V, et al. Meeting patients’ right to the correct diagnosis: ongoing international initiatives on undiagnosed rare diseases and ethical and social issues. Int J Environ Res Public Health. 2018;15:2072. doi: 10.3390/ijerph15102072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pierides A, Voskarides K, Athanasiou Y, et al. Clinico-pathological correlations in 127 patients in 11 large pedigrees, segregating one of three heterozygous mutations in the COL4A3/ COL4A4 genes associated with familial haematuria and significant late progression to proteinuria and chronic kidney disease from focal segmental glomerulosclerosis. Nephrol Dial Transplant. 2009;24:2721–2729. doi: 10.1093/ndt/gfp158. [DOI] [PubMed] [Google Scholar]
- 16.Hwang YH, Conklin J, Chan W, et al. Refining genotype-phenotype correlation in autosomal dominant polycystic kidney disease. J Am Soc Nephrol. 2016;27:1861–1868. doi: 10.1681/ASN.2015060648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cornec-Le Gall E, Audrezet MP, Renaudineau E, et al. PKD2-related autosomal dominant polycystic kidney disease: prevalence, clinical presentation, mutation spectrum, and rrognosis. Am J Kidney Dis. 2017;70:476–485. doi: 10.1053/j.ajkd.2017.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cornec-Le Gall E, Audrezet MP, Rousseau A, et al. The PROPKD score: a new algorithm to predict renal survival in autosomal dominant polycystic kidney disease. J Am Soc Nephrol. 2016;27:942–951. doi: 10.1681/ASN.2015010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao T, Udwan K, John R, et al. Integration of genetic testing and pathology for the diagnosis of adults with FSGS. Clin J Am Soc Nephrol. 2019;14:213–223. doi: 10.2215/CJN.08750718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barua M, Stellacci E, Stella L, et al. Mutations in PAX2 associate with adult-onset FSGS. J Am Soc Nephrol. 2014;25:1942–1953. doi: 10.1681/ASN.2013070686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Germain DP, Arad M, Burlina A, et al. The effect of enzyme replacement therapy on clinical outcomes in female patients with Fabry disease—a systematic literature review by a European panel of experts. Mol Genet Metab. 2019;126:224–235. doi: 10.1016/j.ymgme.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 22.Ortiz A, Germain DP, Desnick RJ, et al. Fabry disease revisited: management and treatment recommendations for adult patients. Mol Genet Metab. 2018;123:416–427. doi: 10.1016/j.ymgme.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 23.Benjamin ER, Della Valle MC, Wu X, et al. The validation of pharmacogenetics for the identification of Fabry patients to be treated with migalastat. Genet Med. 2017;19:430–438. doi: 10.1038/gim.2016.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCafferty EH, Scott LJ. Migalastat: a review in Fabry disease. Drugs. 2019;79:543–554. doi: 10.1007/s40265-019-01090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ozaltin F. Primary coenzyme Q10 (CoQ 10) deficiencies and related nephropathies. Pediatr Nephrol. 2014;29:961–969. doi: 10.1007/s00467-013-2482-z. [DOI] [PubMed] [Google Scholar]
- 26.Lovric S, Ashraf S, Tan W, Hildebrandt F. Genetic testing in steroid-resistant nephrotic syndrome: when and how? Nephrol Dial Transplant. 2016;31:1802–1813. doi: 10.1093/ndt/gfv355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patrakka J, Ruotsalainen V, Reponen P, et al. Recurrence of nephrotic syndrome in kidney grafts of patients with congenital nephrotic syndrome of the Finnish type: role of nephrin. Transplantation. 2002;73:394–403. doi: 10.1097/00007890-200202150-00013. [DOI] [PubMed] [Google Scholar]
- 28.Shearer AE, DeLuca AP, Hildebrand MS, et al. Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc Natl Acad Sci USA. 2010;107:21104–21109. doi: 10.1073/pnas.1012989107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steinberg S, RaymondG. Braverman N, Moser A. Zellweger spectrum disorder. In: Adam MP, et al., editors. GeneReviews. Seattle, WA: University of Washington; 2017.
- 30.Hughes JL, Poulos A, Robertson E, et al. Pathology of hepatic peroxisomes and mitochondria in patients with peroxisomal disorders. Virchows Arch A Pathol Anat Histopathol. 1990;416:255–264. doi: 10.1007/BF01678985. [DOI] [PubMed] [Google Scholar]
- 31.Kuppachi S, Smith RJH, Thomas CP. Evaluation of genetic renal diseases in potential living kidney donors. Curr Transplant Rep. 2015;2:1–14. doi: 10.1007/s40472-014-0042-5. [DOI] [Google Scholar]
- 32.Layton JB, Hogan SL, Jennette CE, et al. Discrepancy between medical evidence form 2728 and renal biopsy for glomerular diseases. Clin J Am Soc Nephrol. 2010;5:2046–2052. doi: 10.2215/CJN.03550410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Massie AB, Muzaale AD, Luo X, et al. Quantifying postdonation risk of ESRD in living kidney donors. J Am Soc Nephrol. 2017;28:2749–2755. doi: 10.1681/ASN.2016101084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gast C, Pengelly RJ, Lyon M, et al. Collagen (COL4A) mutations are the most frequent mutations underlying adult focal segmental glomerulosclerosis. Nephrol Dial Transplant. 2016;31:961–970. doi: 10.1093/ndt/gfv325. [DOI] [PubMed] [Google Scholar]
- 35.Boyer O, Woerner S, Yang F, et al. LMX1B mutations cause hereditary FSGS without extrarenal involvement. J Am Soc Nephrol. 2013;24:1216–1222. doi: 10.1681/ASN.2013020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mallett A, Fowles LF, McGaughran J, Healy H, Patel C. A multidisciplinary renal genetics clinic improves patient diagnosis. Med J Aust. 2016;204:58–59. doi: 10.5694/mja15.01157. [DOI] [PubMed] [Google Scholar]
- 37.Alkanderi S, Yates LM, Johnson SA, Sayer JA. Lessons learned from a multidisciplinary renal genetics clinic. QJM. 2017;110:453–457. doi: 10.1093/qjmed/hcx030. [DOI] [PubMed] [Google Scholar]
- 38.Fromer M, Moran JL, Chambert K, et al. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am J Hum Genet. 2012;91:597–607. doi: 10.1016/j.ajhg.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vargas-Poussou R, Dahan K, Kahila D, et al. Spectrum of mutations in Gitelman syndrome. J Am Soc Nephrol. 2011;22:693–703. doi: 10.1681/ASN.2010090907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Melchionda S, Palladino T, Castellana S, et al. Expanding the mutation spectrum in 130 probands with ARPKD: identification of 62 novel PKHD1 mutations by sanger sequencing and MLPA analysis. J Hum Genet. 2016;61:811. doi: 10.1038/jhg.2016.58. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.