ABSTRACT
The specification of organs, tissues and cell types results from cell fate restrictions enacted by nuclear transcription factors under the control of conserved signaling pathways. The progenitor epithelium of the Drosophila compound eye, the eye imaginal disc, is a premier model for the study of such processes. Early in development, apposing cells of the eye disc are established as either retinal progenitors or support cells of the peripodial epithelium (PE), in a process whose genetic and mechanistic determinants are poorly understood. We have identified protein phosphatase 2A (PP2A), and specifically a STRIPAK–PP2A complex that includes the scaffolding and substrate-specificity components Cka, Strip and SLMAP, as a critical player in the retina-PE fate choice. We show that these factors suppress ectopic retina formation in the presumptive PE and do so via the Hippo signaling axis. STRIPAK–PP2A negatively regulates Hippo kinase, and consequently its substrate Warts, to release the transcriptional co-activator Yorkie into the nucleus. Thus, a modular higher-order PP2A complex refines the activity of this general phosphatase to act in a precise specification of cell fate.
KEY WORDS: MST2, PP2A-C (Mts), PP2A-A (PP2A-29B), STRIATIN, TAZ, YAP
Summary: STRIPAK–PP2A promotes peripodial cell fate in the developing Drosophila eye epithelium by antagonizing the Hippo signaling axis, thereby promoting Yorkie activity and suppressing retinal cell fate.
INTRODUCTION
Cell fate specification is a fundamental process of metazoan development that leads to the emergence of distinct body parts, organs, tissues and eventually, highly specialized cell types. Key steps in this process result from the intersecting activity of highly conserved signaling cascades and nuclear factors, which together alter gene expression to restrict developmental potential and drive the acquisition of distinct cellular characteristics. However, the sheer complexity and diversity of the metazoan body plan and its cellular specializations is vast compared with the relatively limited repertoire of developmental factors encoded in the animal genome. Thus, a small set of signaling pathways rely on an array of different regulators and effectors to generate organismal complexity. A deeper understanding of fate choice in development is contingent on ascertaining the divergent and nuanced ways in which these biological factors come together to drive distinct cellular progressions.
The compound eye of Drosophila melanogaster is a premier model for the study of fate acquisition during development, from the initial steps of retina primordium specification in the early second instar larva, to the exquisite choice of photoreceptor neuron subtype in the terminally differentiating eye of the late third instar larva and early pupa. The progenitor epithelium of the eye, the eye-antennal imaginal disc, forms from the embryonic head ectoderm as a flattened sac or vesicle. It eventually gives rise to the compound eye and other adult head structures (Domínguez and Casares, 2005), the emergence of which begin during larval development, making this process easily accessible.
Broad expression of the Pax6-type retinal determination factors Eyeless and Twin-of-eyeless initially prime the disc to give rise to the eye primordium. This event is closely linked to the acquisition of distinct morphological characteristics between the facing portions of the disc epithelium. Cells on one side of the epithelium adopt a columnar morphology and begin to express the retinal determinant Eyes absent (Eya), thus initiating eye formation; cells on the apposed side adopt the squamous and cuboidal morphology typical of the peripodial epithelium (PE) (Bessa et al., 2002; Kenyon et al., 2003). This binary choice between PE and retina developmental potential is a first step in a complex regional specification program that progresses in larvae over a 4-day period while the disc epithelium grows from fewer than 100 cells to more than 50,000 cells (Martin, 1982).
While the development of the eye, from primordium establishment to differentiation of the various retinal cell types, has been extensively studied (Treisman, 2013; Quan et al., 2012), much less is known about PE formation. During metamorphosis, the PE is necessary for the morphogenetic events that transform a pair of apparently simple epithelia into the complex fly head (Agnes et al., 1999; Fristrom and Fristrom, 1993; Milner et al., 1983; Zeitlinger and Bohmann, 1999). However, there is evidence that PE is more broadly required for the development of eye-antennal disc-derived organs, in particular the retina. In fact, the PE is essential for cellular growth in the eye primordium and for retinal neurogenesis; for example, PE ablation during photoreceptor neuron formation results in an arrest of retinogenesis (Cho et al., 2000; Gibson and Schubiger, 2000; Stultz et al., 2006). Notwithstanding these important functions of the PE, very little is known about the genes and molecular mechanisms that specify PE fate. We focus here on the initial step, when retina potential is fostered within a portion of the eye disc epithelium but suppressed elsewhere, in favor of PE formation.
Heterotrimeric protein phosphatase 2A (PP2A) holoenzymes are an abundant group of cellular phosphatases that are highly conserved across Metazoa (Shi, 2009). They consist of A, B and C subunits; the catalytic (C) subunit and adaptor subunit (A) are each encoded by unique genes in Drosophila (mts and PP2A-29B, respectively). In contrast, the interchangeable substrate-specifying and activity-gating B subunits are divided into four families (B, B′, B″ and B‴) and are encoded by numerous genes in flies and vertebrates. Cka encodes the sole atypical B‴ subunit that is orthologous to the mammalian striatins (Chen et al., 2002; Moreno et al., 2000). PP2A holoenzymes containing Cka are found in the striatin-interacting phosphatase and kinase (STRIPAK) complex, which also includes other scaffolding and substrate-associated factors such as Strip (Ribeiro et al., 2010). The contribution of this major cellular phosphatase to developmental fate decisions is yet to be determined.
In this study we demonstrate that STRIPAK–PP2A is essential for PE cell fate specification in the Drosophila eye disc. Loss of function of either Cka or Strip results in the emergence of an ectopic retinal field within the presumptive PE. We show that STRIPAK–PP2A acts by negatively regulating the activity of the Hippo kinase (Hpo) and its effector Warts (Wts), thereby releasing the PE-promoting activity of the transcriptional co-activator Yorkie (Yki). Thus, we elucidate the genetic pathway through which STRIPAK–PP2A promotes PE fate and provide a first example of a STRIPAK–PP2A-controlled cell fate decision in metazoan development.
RESULTS
Loss of Cka results in ectopic retina development within the presumptive PE
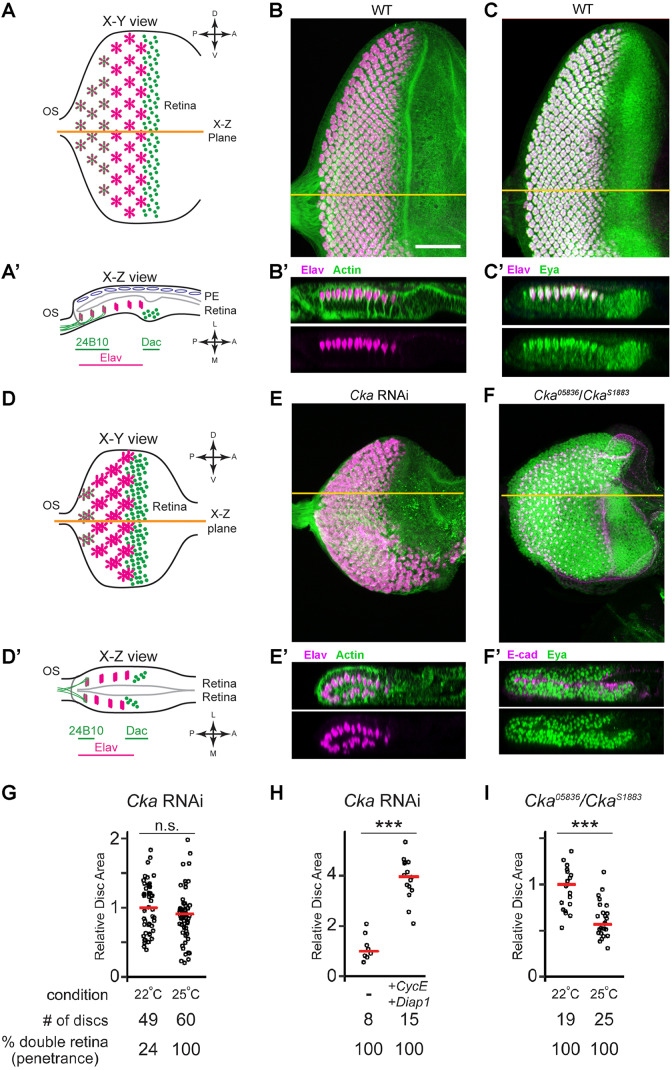
As part of an RNA interference (RNAi) screen to identify genes required for eye disc development, we observed a phenotypic class in which the discs were devoid of non-neuronal PE-like cells, but instead exhibited columnar morphology and expressed the neuronal marker Elav in both apposed cell layers of the epithelium. Cka was identified among this class. Compared with wild-type (WT) eye discs, in which the neural retina develops on one side of the disc and the PE on the other (Fig. 1A–C′), silencing of the Cka locus using RNAi produced discs that were smaller in size, lacked PE, and displayed mirror-image retinal fields within the juxtaposed portions of the disc epithelium (Fig. 1D–E′). A similar phenotype was observed in discs transheterozygous for two mutant alleles of Cka, Cka05836/CkaS1883; expression of the retinal determinant Eya marked both cell layers (compare Fig. 1C,C′ with F,F′; see also Fig. S1A for Elav staining). These analyses of Cka loss-of-function tissue, with mutant alleles and transgenic RNAi, strongly suggested a switch in fate, from PE to retina identity.
Fig. 1.
Cka suppresses ectopic retina development in the presumptive PE, independent of imaginal disc size. (A,A′) Schematic illustrating the orientation (posterior to the left, dorsal up) and wild-type expression patterns of retinal markers (Elav: neurons, magenta; Dac: neural progenitors, green dots; 24B10 (Chaoptin): mature neurons, green outlines) within a typical X-Y projection of a third instar Drosophila eye imaginal disc (OS, optic stalk). Throughout, horizontal orange lines indicate the plane of X-Z images shown below the given X-Y projection. The peripodial epithelium (PE, blue ovals) is continuous with the columnar retinal epithelium (as indicated) in the eye disc. A lumen, outlined by a gray line indicating the apical aspects of the apposed cell layers, is present in the disc. The PE and lumen are exaggerated in the diagram, whereas they are often obfuscated by tissue compression during specimen preparation. (B,B′) Wild-type (WT) eye imaginal disc showing the developing retinal field of Elav-positive neurons (magenta), co-stained for actin (green) (B, n>100). The X–Z section reveals the single neural columnar epithelium (B′). (C,C′) WT eye imaginal disc co-stained for Elav (magenta) and Eya (neurons and progenitors, green) (C, n>100). The X–Z section reveals the restriction of Eya-positive progenitors to the retinal epithelium (C′). (D,D′) Schematic illustrating the mutant phenotype observed in Cka-loss-of-function discs; orientation and markers are as in panels A and A′. Discs are smaller and have superimposed retinal fields that obscure the normal crystalline array of ommatidial clusters in the X-Y projection (D). The two retinal layers are resolved in the X–Z view (D′). (E,E′) Disc-wide expression of Cka-RNAi results in a smaller eye disc (neurons, magenta; actin, green) (E, n=60). The X–Z section reveals two retinal epithelia, with mirrored Elav-positive neurons (E′). (F,F′) Eye disc from a Cka05836/CkaS1883 transheterozygote stained for Eya-positive retinal progenitor cells (green) and E-cadherin (magenta) (F, n=20). The X–Z section shows evidence of retinal progenitor cells in both layers (F′). (G–I) Quantification of relative disc sizes. There was no significant difference in the size of Cka RNAi-expressing discs cultivated at 22°C versus 25°C (two-tailed t-test, P=0.197); however, there was incomplete penetrance of the phenotype at the lower temperature (G). Co-expression of CycE and DIAP1 increased the size of Cka RNAi-expressing discs cultured at 30°C (one-tailed t-test, P=2.14×10−9), and did not affect penetrance of the mutant phenotype (H). Higher cultivation temperature (25°C versus 22°C) decreased the size of Cka05836/CkaS1883 transheterozygous discs (one-tailed t-test, P=2.87×10−6), without affecting penetrance of the phenotype (I). Scale bar (for all panels), 50 µm.
To confirm this, we explored the relationship between the developing retinae and disc size. In Cka-RNAi discs, cultivation temperature independently affected both disc size and the penetrance of the double retina phenotype (Fig. S1B). Notably, disc size was not significantly different in animals cultivated at 25°C versus 22°C, but the penetrance of the phenotype dropped from 100% to less than 25%, respectively (Fig. 1G). As an alternative way to increase the size of Cka-RNAi discs, we overexpressed exogenous pro-proliferation or anti-apoptosis factors, alone or in combination. We found that co-expression of CycE and DIAP1 provided the greatest rescue of size in Cka-RNAi discs, but did not affect phenotype penetrance (Fig. 1H; Fig. S1C). Furthermore, cultivation temperature had a significant effect on disc size in Cka05836/CkaS1883 transheterozygotes, yet the double-retina phenotype remained fully penetrant (Fig. 1I; Fig. S1B). The lack of correlation between disc size and cell fate strongly suggests that Cka is required for PE cell fate independently of its role in cell proliferation and/or survival.
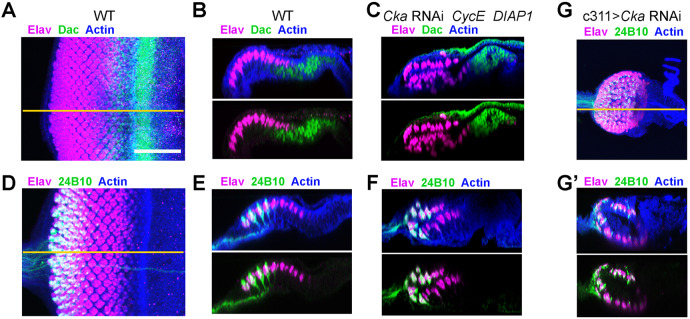
To assess whether neurogenesis in the ectopic retina of Cka-mutant discs was largely normal, we relied on the markers Dac and Chaoptin/24B10 (Shen and Mardon, 1997; Van Vactor et al., 1988). In the developing WT eye disc, eye morphogenesis begins early in the third instar larval stage and continues into the early pupal stage. During this process, progenitor cells first give rise to neurons and then to accessory cells as a posterior-to-anterior wave of differentiation sweeps across the disc epithelium, leaving in its wake rows of developing neuronal clusters, or ommatidia. Thus, a third instar disc offers a snapshot of retinal development with progressively more mature or ‘older’ ommatidial clusters towards the disc's posterior margin (Fig. 1A). Dac expression in the eye disc is largely restricted to the retinal epithelium, where it is expressed transiently in late retinal progenitor cells and precursors of the photoreceptor neurons just ahead of emerging, ‘young’ ommatidia (Shen and Mardon, 1997). As eye morphogenesis continues, Dac expression is lost while expression of Elav marks the developing photoreceptor neurons (Figs 1A,A′ and 2A,B). In the mirrored retinae of size-rescued (CycE+DIAP1-expressing) Cka-RNAi discs, we observed the expected anterior-to-posterior sequence of first Dac- and then Elav-expressing cells (Figs 1D,D′ and 2C). Conversely, the 24B10 antibody detects the membrane protein Chaoptin along the cell bodies and axons of ‘older’ photoreceptor neurons (Van Vactor et al., 1988). Thus in WT discs, ‘older’ neuronal clusters at the disc posterior express both Elav and Chaoptin (Figs 1A,A′ and 2D,E). As above, in size-rescued (CycE+DIAP1-expressing) Cka-RNAi discs, we observed the normal anterior-to-posterior distribution of Elav and Elav+Chaoptin neuronal clusters on both sides of the disc epithelium (Figs 1D,D′ and 2F). Thus, retinal neurogenesis proceeds in the expected pattern in both layers of the disc upon reduction of Cka function.
Fig. 2.
Cka functions within the PE to suppress ectopic neurogenesis. (A–C) Dac expression (green) labels neural progenitor cells anterior to the morphogenetic furrow and Elav-positive cells (magenta) in WT eye discs (A, X–Y; B, X–Z); actin (blue). X–Z sections of Cka RNAi-expressing eye discs exhibit Dac staining anterior to Elav-positive cells in both epithelial layers (C). (D–F) 24B10/Chaoptin (green) labels the membrane and processes of mature photoreceptor neurons, overlapping with the oldest clusters of Elav-positive cells (magenta) in WT discs (D, X–Y; E, X–Z); actin (blue). X–Z sections of Cka RNAi-expressing eye discs exhibit 24B10 staining posterior to Elav-positive single-labeled cells in both epithelial layers (F). (G,G′) c311-GAL4 was used to drive expression of Cka-RNAi only in the presumptive PE, resulting in a small disc phenotype (G, X–Y, n=16), with 24B10 staining (green) posterior to Elav-positive single-labeled cells (magenta) in both epithelial layers (G′, X–Z). Overlap of the Elav and Chaoptin signals, due to image projection, appears as white. Scale bar (for all panels), 50 µm. Orange lines in A, D and G represent the plane of the X-Z slices shown in B, E and G', respectively.
Lastly, given the ubiquitous expression of Cka within the eye disc (Grubbs et al., 2013), we sought to establish its function in the presumptive PE. Using the PE-specific driver c311-GAL4 (Gibson and Schubiger, 2000) to limit Cka RNAi expression to the PE, we observed discs with mirror image retinae. These discs had Elav+Chaoptin-positive cells posterior to Elav-positive cells in both layers (Fig. 2G,G′). These findings support a model whereby the activity of Cka is specifically required in the presumptive PE to suppress retinogenesis.
In conclusion, the loss of Cka results in the conversion of the presumptive PE portion of the eye disc into a second retina that exhibits morphology and posterior-to-anterior neurogenesis akin to that of the normal retinal epithelium.
STRIPAK–PP2A controls PE fate
As mentioned in the introduction, Cka encodes the Drosophila striatin, a B‴ substrate-specificity subunit of the PP2A complex. Therefore, we sought to establish whether the change in PE cell fate induced by Cka loss reflected dysfunction of this major cellular phosphatase. We did so by investigating the loss-of-function phenotypes of other subunits of PP2A complexes.
We first assessed loss of function for the other B-type substrate-specifying subunits (Tws, Wdb, Wrd), which are mutually exclusive to Cka in conventional heterotrimeric PP2A complexes (Slupe et al., 2011). Silencing of Tws, Wdb or Wrd by RNAi revealed less severe phenotypes in the eye disc that were inconsistent with fate changes (data not shown). These results strongly suggested a primary and specific role for Cka-containing PP2A complexes in establishing cell fate in the eye disc.
To confirm that Cka acts via PP2A in the control of PE fate in the eye disc, we focused directly on the core C and A subunits of the enzyme complex, encoded by mts and PP2A-29B, respectively. Loss of function of mts or of PP2A-29B (by RNAi) produced grossly abnormal discs (Fig. 3A,B), whose developmental state could not be assessed. This was not surprising, as PP2A holoenzymes are known to target a multitude of substrates involved in diverse cellular processes, including proliferation and cell survival (Shi, 2009). We therefore leveraged the temperature sensitivity of the Cka-RNAi induced phenotype (Fig. 1G) for genetic interaction testing. At 22°C, the Cka-RNAi phenotype is incompletely penetrant, offering a sensitized background in which to assess a potential role for Mts. Indeed, we found that two different recessive loss-of-function alleles of mts, mts02496 and mtsS5286, dominantly enhanced the Cka-RNAi phenotype (Fig. 3C–E). Together with mts02496, the Cka RNAi phenotype increased from 24 to 83% penetrance, whereas it became fully penetrant in combination with mtsS5286. These genetic interaction data strongly support a model whereby Cka functions via its regulation of PP2A activity in the control of PE fate.
Fig. 3.
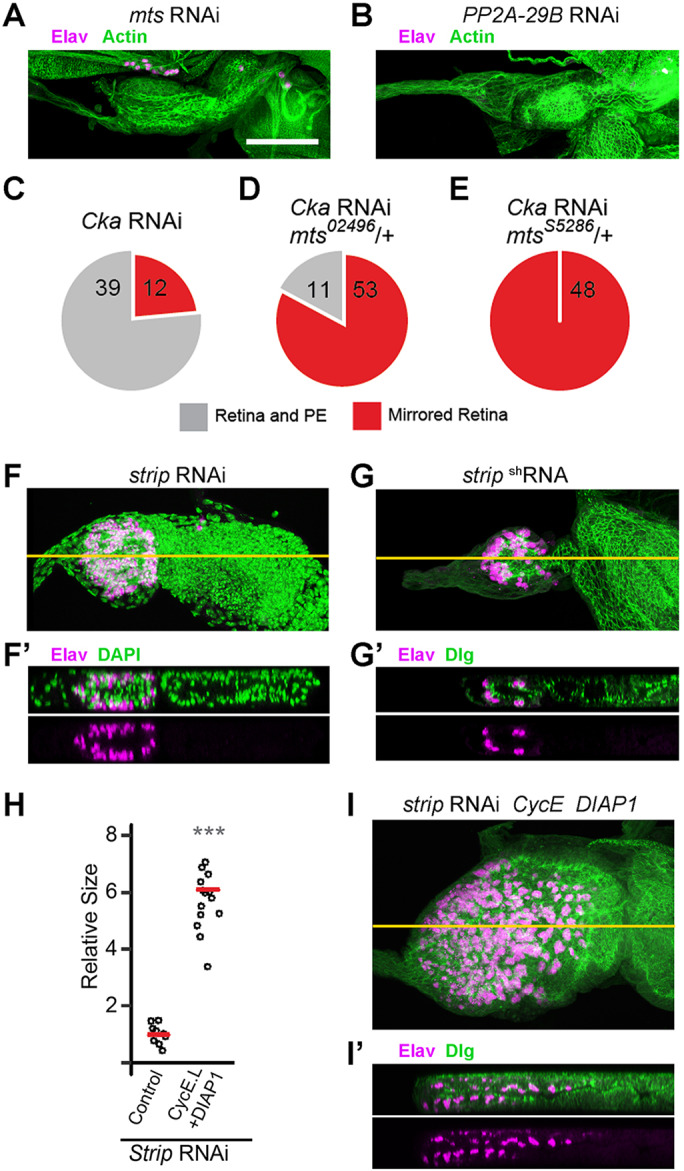
Cka links STRIPAK–PP2A to PE fate determination. (A,B) Loss of function of the PP2A catalytic subunit (A, Mts, n=24) or of the adaptor subunit (B, PP2A-29B, n=16) blocks eye disc development; residual actin (green) is present but neurons (magenta) are not detected in X-Y projections (C–E) Cka and mts exhibit a genetic interaction with respect to the double retina phenotype. At 22°C, the Cka-RNAi phenotype (red) is not fully penetrant, leaving some unaffected discs (gray) (C). The penetrance of the phenotype increases when a loss-of-function allele, mts02496, is present (D); it becomes fully penetrant in the context of an alternate loss-of-function allele, mtsS5286 (E). (F–G′) Strip loss-of-function (F,F′, RNAi, n=18, 9/18 had no neurons; G,G′, shRNA, n=18, 15/18 had no neurons) has a severe effect on disc size (F,G, X–Y sections); nonetheless, X–Z sections (F′,G′) reveal the presence of neurons (magenta) in both cell layers, counterstained with DAPI or Dlg (green). (H–I′) Grown at 30°C, CycE+DIAP1 co-expression significantly rescues Strip-RNAi disc size (H, one-tailed t-test, P=8.14×10−11). These discs remain abnormal (I, X–Y, n=15) and X–Z sections (I′) reveal the continued presence of neurons (magenta) in both cell layers of the size-rescued disc, counterstained with Dlg (green). Scale bar (for all panels), 50 µm. Orange lines in F, G and I represent the plane of the X–Z slices shown in F′, G′ and I′, respectively.
In addition, Cka is known to link PP2A to a larger multi-protein complex known as STRIPAK (Goudreault et al., 2009; Ribeiro et al., 2010). We reasoned that decreased function of other components of STRIPAK should also specifically impair the activity of this subtype of PP2A holoenzyme. Hence, we focused on the scaffolding component Strip and targeted it for gene silencing within the eye disc by transgenic-RNAi. Discs expressing two independent RNAi reagents against Strip, one of which was previously validated (Sakuma et al., 2016), were greatly reduced in size and contained Elav-positive cell clusters in both the retinal and putative PE side of the eye disc epithelium (Fig. 3F–G′). Co-expression of CycE and DIAP1 significantly increased disc size but did not rescue PE fate, as shown by the presence of two retinal fields in CycE+DIAP1-expressing Strip-RNAi discs (Fig. 3H–I′; Fig. S1C). Thus, loss of function of Strip recapitulates the Cka-RNAi mutant phenotype.
Together, this evidence specifically implicates STRIPAK–PP2A in the PE versus retina fate choice.
Loss of Yki results in ectopic retina development within the presumptive PE
The only other regulator of PE fate in the eye disc thus far identified is the transcriptional co-activator Yki, which appears to promote PE fate over retina fate in conjunction with its DNA-binding partners, Sd and Hth. We previously reported that silencing of the genes encoding these factors by transgenic RNAi leads to development of an ectopic mirror-image retina, concomitant with PE loss (Zhang et al., 2011). However, these findings were not consistent with previously published loss-of-function analyses of yki mutant clones (Huang et al., 2005; Peng et al., 2009; Wu et al., 2008). Hence, before testing for a possible link between STRIPAK–PP2A and Yki in PE fate, we sought to solidify our earlier findings using both size rescue and a mutant allele (Bennett and Harvey, 2006; Emoto et al., 2006; Oh and Irvine, 2008).
First, we confirmed that, as for the loss of Cka and Strip, diminished proliferation and cell survival did not play a major role in inducing the duplicated retina phenotype in yki loss-of-function discs. As previously reported, yki-RNAi eye discs are small and feature Elav-positive neuronal clusters on both sides of the epithelium (Fig. 4A,A′; Zhang et al., 2011). We were able to achieve a substantial rescue of disc size by co-expression of CycE and DIAP1 (Fig. 4B,C; Fig. S1C); nonetheless, these discs displayed Elav-positive neuronal clusters in both layers of the disc (Fig. 4B′). Furthermore, analyses of Dac and Chaoptin expression in these size-rescued discs provided evidence that retinogenesis proceeded as expected in both cell layers (Fig. 4D,E).
Fig. 4.
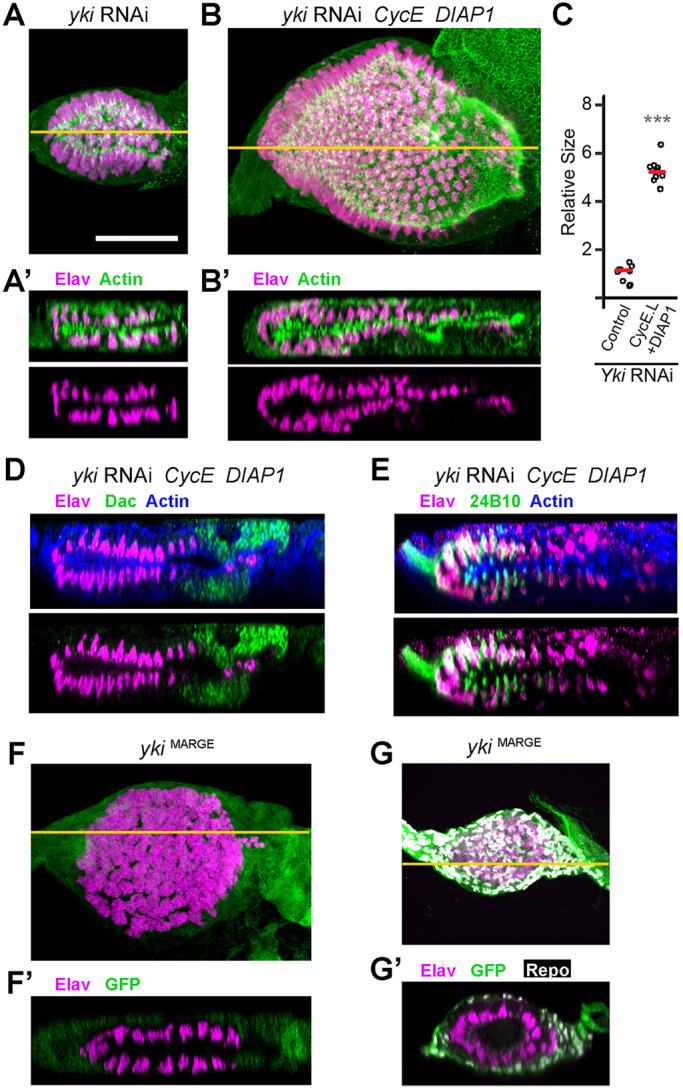
Yki is a bona fide regulator of PE cell fate in the eye disc. (A,A′) yki-RNAi expressing discs are small (A, X–Y projections, n=9) and contain two layers of Elav-positive neurons (magenta) as seen in the X–Z slice (A′); actin counterstain (green). (B,B′) Co-expression of CycE and DIAP1 with yki-RNAi produces larger discs with correspondingly larger retinal neuron fields (B, X-Y projection, n=9). X–Z sections reveal the continued presence of neurons (magenta) in both cell layers of the size-rescued disc (B′). (C) Grown at 30°C, CycE+DIAP1 co-expression significantly rescues yki-RNAi disc size (one-tailed t-test, P=7.79×10−13). (D,E) X–Z sections of size-rescued yki-RNAi eye discs stained for Elav (magenta) and either Dac (green, D) or 24B10/Chaoptin (green, E). Upper images include actin counterstaining (blue), and overlap of the Elav and Chaoptin signals, due to image projection, appears as white. Note that Dac staining appears anterior to Elav staining in both cell layers (D) and that 24B10 co-labels Elav-positive cells located in the posterior portion of the disc in both layers (E). (F–G′) Yki loss-of-function (ykiB5/ykiB5) was induced using the MARGE technique (Zhou et al., 2016), resulting in a small disc phenotype with mirrored retinae (F, X–Y, n=13). Residual yki+-expressing cells co-express GFP (green) while mutant cells do not (F,F′). An X–Z section reveals two layers of Elav-positive neurons (magenta, F′), similar to the phenotype induced by yki-RNAi. The GFP-positive (yki+) cells are immunoreactive for Repo, a glial marker (G, X–Y projection, and G′, X–Z slicen=12); Repo-positive glia infiltrate the disc via the optic stalk, after neurogenesis has initiated (Hummel et al., 2002). Scale bar (for all panels), 50 µm. Orange lines in A, B, F and G represent the plane of the X–Z slices shown in A′ B′, F′ and G′, respectively.
Second, we validated the RNAi-based findings and confirmed the role of Yki in establishing the PE using the lethal mutant allele ykiB5. To achieve this goal, we employed the Mutant Analysis by Rescue Gene Excision (MARGE) technique (Zhou et al., 2016), in which tissue-specific FLP-mediated excision is used to eliminate a genomic WT rescue transgene (yki+) in the context of a homozygous mutant animal (ykiB5/ykiB5). Thus, we produced ykiB5 eye discs using ey-FLP, in the context of an otherwise yki+-rescued viable organism. MARGE ykiB5/ykiB5 discs revealed a phenotype very similar to that observed using yki-RNAi (compare Fig. 4F,F′ with A,A′), thus conclusively demonstrating the validity of the phenotype induced by the transgenic-RNAi reagent. We did, however, observe GFP immunoreactivity in cells surrounding the field of neuronal clusters, suggesting the presence of cells in which yki+ transgene was still expressed. Indeed, the GFP-positive cells co-stain for the glial marker Repo (Fig. 4G,G′). These glial cells are known to migrate from the brain and through the optic stalk to spread out along the basal surface of the retina epithelium as neurogenesis proceeds (Hummel et al., 2002).
In conclusion, Yki is indeed necessary to promote PE fate in the developing eye disc and it does so independently of its extensively documented role in proliferation and cell survival.
STRIPAK–PP2A promotes PE fate via the protein kinase Hpo
Given the similar effects of loss of function for Yki and STRIPAK–PP2A components in the eye disc, we explored a possible mechanistic connection between these two players. The Hippo (Hpo) kinase is a well-known negative regulator of Yki (Ma et al., 2018; Misra and Irvine, 2018; Wu et al., 2003). Interestingly, Hpo was shown to be genetically downstream of Cka in the regulation of host survival and anti-microbial peptide expression in the fly immune system; in cell culture, Hpo phosphorylation increased when the Cka locus was silenced (Liu et al., 2016). Hence, we decided to test whether Hpo could also be under STRIPAK–PP2A control in the specification of PE fate within the eye disc.
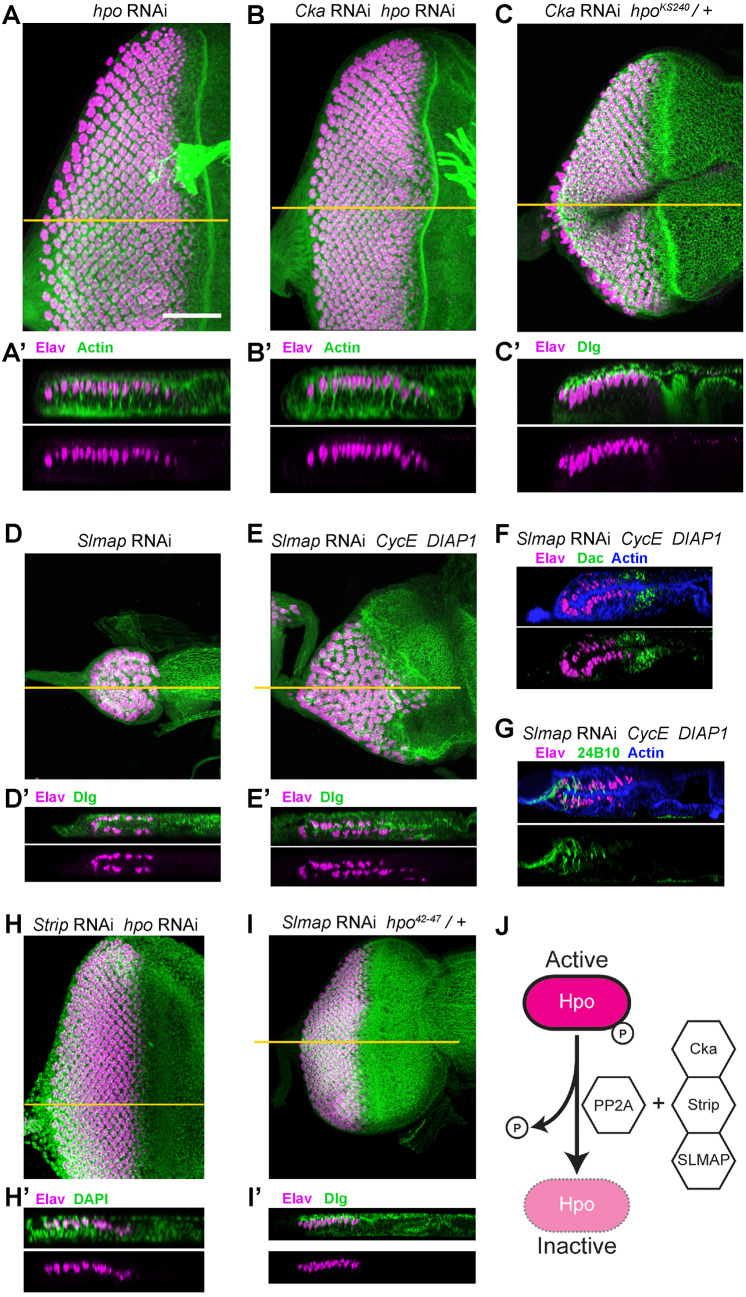
We thus investigated whether a reduction in hpo could mitigate the effect of loss of STRIPAK–PP2A function, by inducing a hpo knockdown condition in the Cka-RNAi background, using either a hpo-RNAi reagent or hpo loss-of-function allele (hpoKS240). Strikingly, expression of a mild hpo-RNAi reagent (showing little to no effect on WT discs; Fig. 5A), or introduction of one loss-of-function mutant allele of hpo (hpoKS240/+) rescued both size and PE fate in Cka-RNAi animals, producing discs that were largely indistinguishable from WT (Fig. 5B,B′; Cka-RNAi+hpo-RNAi) or with mild defects in disc morphology (Fig. 5C,C′; Cka-RNAi+hpoKS240). These findings support the hypothesized relationship between STRIPAK–PP2A and Hpo in the control of cell fate within the developing eye disc of Drosophila.
Fig. 5.
STRIPAK–PP2A functions genetically upstream of Hpo. (A–I′) Discs of the indicated genotypes stained with Elav (neurons; magenta) alone, or together with counterstaining (actin, Dlg, Dac, 24B10 or DAPI; green, as indicated). (A,A′) Expression of hpo-RNAi in the eye disc results in a slight increase in disc size (A, X–Y, n=12) but does not perturb the organization of the retina or PE (A′, X–Z). (B,B′) Co-expression of hpo-RNAi with Cka-RNAi produces eye discs that are largely indistinguishable from WT (B, X–Y, n=18), restoring the PE cell layer and single retinal layer expressing Elav (B′, X–Z). (C,C′) Cka-RNAi expressing animals that are heterozygous for hpoKS240, a loss-of-function allele, exhibit some morphological defects (C, X–Y, n=19), but contain only a single retinal layer (C′, X–Z). (D,D′) Transgenic expression of Slmap-RNAi in the eye disc phenocopies Cka- and Strip-loss-of-function. Discs are small (D, X–Y, n=20, 8/20 had no neurons) and exhibit two layers of Elav-positive cells (D′, X–Z). (E,E′) Co-expression of CycE and DIAP1 with Slmap-RNAi produces larger discs with correspondingly larger retinal neuron fields (E, X–Y, n=16). X–Z sections reveal the continued presence of neurons (magenta) in both cell layers of the size-rescued disc (E′). (F,G) X–Z sections of size-rescued Slmap-RNAi eye discs stained for Elav (magenta) and either Dac (green, F) or 24B10/Chaoptin (green, G). Upper images include actin counterstaining (blue), and overlap of the Elav and Chaoptin signals, due to image projection, appears as white. Note that Dac staining appears anterior to Elav staining in both cell layers (F) and that 24B10 co-labels Elav-positive cells located in the posterior portion of the disc in both layers (G). (H,H′) Co-expression of hpo-RNAi with Strip-RNAi produces eye discs that are largely indistinguishable from WT (H, X–Y, n=18), restoring the PE cell layer and single retinal layer expressing Elav (H′, X–Z). (I,I′) Slmap-RNAi expressing animals that are heterozygous for hpo42-47, a loss-of-function allele, exhibit some morphological defects (I, X–Y, n=19), but contain only a single retinal layer (I′, X–Z, 8/19 show WT phenotype). (J) Schematic illustrating the dephosphorylation of Hpo by PP2A, together with the STRIPAK components Cka, Strip and SLMAP, converting Hpo from its active state (bright magenta) to its inactive state (faint magenta, dashed border). Scale bar (for all panels), 50 µm. Orange lines in A–E, H and I represent the plane of the X–Z slices shown in A′– E′, H′ and I′, respectively.
To test this further, we investigated additional factors that link STRIPAK–PP2A to Hpo. Current models, based on extensive biochemical evidence in mammalian cells, postulate that STRIP1/2 (Strip) and a second STRIPAK component, SLMAP, make principal contacts with the Mst2 (Hpo) substrate (Kück et al., 2019; Tang et al., 2019). For instance, SLMAP has recently been shown to directly bind a ‘linker’ region of the Hpo ortholog Mst2, and mediate its interaction with STRIPAK–PP2A (Bae et al., 2017), a mechanism conserved in Drosophila (Zheng et al., 2017). Based on this model and our finding that a reduction in hpo can restore the PE in Cka-RNAi discs, STRIPAK factors linking the PP2A enzyme and Hpo should affect PE fate, and do so in a manner that is sensitive to hpo gene dosage.
Having already shown that Strip is indeed required for PE fate (Strip-RNAi discs display the double-retina phenotype; Fig. 3F–G′), we investigated the second component, SLMAP. We found that expression of Slmap-RNAi in the developing disc also produces the mirror-image retina phenotype (Fig. 5D,D′) typical of Cka and Strip gene silencing (Figs 1E,E′ and 3F,F′). DIAP1+CycE co-expression partially rescued the size of Slmap-RNAi discs but did not restore PE fate (Fig. 5E–G; Fig. S1C). Moreover, in both developing retinae of size-rescued (CycE+DIAP1-expressing) Slmap-RNAi discs, the markers Dac and Chaoptin displayed the expected anterior–posterior distribution indicating a normal pattern of eye morphogenesis (Fig. 5F,G). We next set out to test the effect of hpo knockdown on the Strip and Slmap loss-of-function phenotypes. Importantly, Strip-RNAi discs were rescued by co-expression of hpo-RNAi (Fig. 5H,H′) and normal disc morphogenesis was restored to Slmap-RNAi discs upon genetic attenuation of hpo (hpo42-47/+; Fig. 5I,I′).
These results strongly support a model whereby STRIPAK–PP2A, with its components Cka, Strip and SLMAP, negatively regulates Hpo in the eye disc to promote PE fate (Fig. 5J).
STRIPAK–PP2A promotes PE fate through the transcriptional regulator Yki
Having established that STRIPAK–PP2A works via Hpo in PE fate, we sought evidence that this relationship reflected a role for STRIPAK–PP2A in the regulation of Yki.
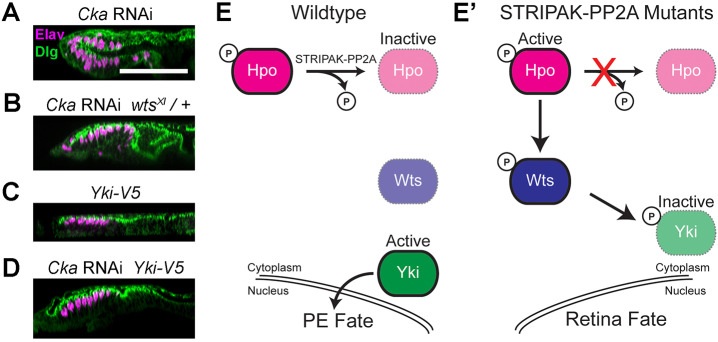
First, we investigated Wts, as a well-known intermediary linking Hpo and Yki. Wts is a kinase whose activity is dependent on direct phosphorylation by Hpo and which, in turn, directly phosphorylates Yki to inhibit its nuclear activity by promoting its cytoplasmic sequestration (Misra and Irvine, 2018). Therefore, we reasoned that reducing Wts activity should be analogous to reducing Hpo activity, and its functional haploidy (wts+/−) should genetically suppress the Cka-RNAi double-retina phenotype. As predicted, in wtsX1-heterozygous animals expressing Cka-RNAi, we consistently observed restoration of the PE cell layer overlying a normal retinal field of Elav-positive neuronal clusters (compare Fig. 6A with 6B). Hence, in agreement with its role in the homonymous pathway, Hippo affects the PE versus retina cell fate choice through its regulation of Wts.
Fig. 6.
STRIPAK–PP2A antagonizes Hpo/Wts signaling to release PE-fate-promoting Yki activity. (A–D) Discs of the indicated genotypes stained with Elav (neurons; magenta) together with counterstaining (Dlg; green). (A) X–Z view of a Cka-RNAi-expressing eye disc exhibiting two layers of Elav-positive cells. (B) Cka-RNAi expressing animals that are heterozygous for wtsX1, a loss-of-function allele, exhibit only a single retinal layer of Elav-positive cells (n=20). (C) Overexpression of a wild-type (WT) yki allele (yki-V5) in WT animals does not disrupt the PE/retinal organization of the eye disc (n=10). (D) Co-expression of yki-V5 with Cka-RNAi blocks the double retina phenotype (n=40, 4/40 WT, 26/40 abnormal with single retina, 10/40 double retina). An example of complete rescue is shown. (E,E′) Schematics illustrating the WT signaling cascade that promotes PE fate in the eye disc (E) and the mechanism by which it is disrupted when STRIPAK–PP2A function is lost (E′). Hyperactive Hpo signaling sequesters Yki in the cytoplasm and prevents it from suppressing retinogenesis in the presumptive PE. Active proteins are in bold colors with continuous borders, while inactive proteins are faint with dashed borders. Scale bar (for panels A–D), 50 µm.
Finally, we reasoned that if the ultimate outcome of STRIPAK–PP2A regulation of Hpo is to maintain a high level of Yki activity, it may be possible to overcome a lack of Cka function by over-expressing Yki. Hence, we elected to use a WT yki transgene (UAS-yki.V5) to over-produce Yki in the developing eye disc and thus boost its function. In a WT background, Yki-V5 produced a slight increase in disc size but discs displayed normal PE-retinal organization (Fig. 6C). In the context of Cka-RNAi, the over-expression of Yki-V5 consistently rescued the mutant phenotype (75%) (Fig. 6D), with 10% of discs being indistinguishable from WT and 65% showing slight morphological abnormalities.
Thus, multiple lines of evidence strongly link Cka to Yki through the suppression of its negative regulators Hpo and Wts, in the PE versus retina cell fate decision of the developing Drosophila eye disc.
DISCUSSION
We document here for the first time a role for PP2A in the control of cell fate during development, and identify the Hpo signaling axis and the nuclear effector Yki as its mechanistic targets. Specifically, we provide genetic evidence in support of a model whereby a STRIPAK–PP2A complex, inclusive of Cka, Strip and SLMAP, suppresses Hpo kinase activity with consequent reduction in Wts kinase function and release of Yki activity to prevent ectopic retina development and promote PE fate (model; Fig. 6E,E′).
STRIPAK–PP2A is a regulator of cell fate in the eye disc
Our results demonstrate a clear role for STRIPAK–PP2A in regulating the adoption of distinct cell fates in the eye disc. PE was absent when Cka function was reduced, and instead we detected the development of a mirror-image ectopic retina within the presumptive PE cell layer (Fig. 1E–F′). We observed similar phenotypes when we induced the loss of function of the STRIPAK–PP2A subunits Strip and SLMAP (Figs 3F–G′ and 5D,D′). Furthermore, we observed a dominant enhancement of the Cka RNAi phenotype by two different loss-of-function alleles of the PP2A catalytic subunit, Mts (Fig. 3C–E). In all tested cases, including temperature modulation and the co-expression of pro-proliferation and anti-apoptotic factors, changes in disc size did not correlate with the PE-to-retina transformation (Figs 1G–I, 3H–I′, 4A–C and 5D–E′) and development of the ectopic retinal field proceeded in an apparently normal pattern (Figs 2E,F, 4D,E and 5F,G).
Interestingly, PE fate was restored by the removal of one wild-type copy of hpo in Cka- or SLMAP-deficient discs, and by hpo RNAi in Strip-deficient discs (Fig. 5B–C′, H,H′ and I,I′), strongly suggesting that the latter factors act genetically upstream of Hpo. Given the direct roles of Strip and SLMAP in the recruitment of Hpo as a PP2A substrate in cell culture (Bae et al., 2017; Kück et al., 2019; Tang et al., 2019; Zheng et al., 2017), our findings strongly point to the Hpo kinase as the target of negative regulation by STRIPAK–PP2A in the control of PE cell fate.
Furthermore, the loss-of-Cka phenotype was also suppressed by genetic reduction of Wts (Fig. 6B) and by overexpression of Yki (Fig. 6D). As the Wts kinase is a direct intermediate between Hpo and Yki, this evidence links STRIPAK–PP2A to the eye disc PE fate-promoting factor Yki. Therefore, we postulate that during the induction of PE fate, high levels of Yki activity are maintained in the presumptive PE via the negative regulation of the Hpo-Wts kinase axis by a STRIPAK–PP2A complex containing Cka, Strip and SLMAP (Fig. 6E). Notably, the ubiquitous role of the Hpo-Yki signaling axis in regulating tissue size appears to be in parallel to, and genetically separable from, its role in determining PE fate. Hence, STRIPAK–PP2A represents one effector of the developmental regulation of Yki activity in fate specification.
PE fate is specified by distinct mechanisms in different imaginal discs
PE fate specification was first characterized in the wing imaginal disc, where the factor Bowl is a driver of PE fate (Nusinow et al., 2008). In addition, ectopic activation of Wg signaling or expression of Lines within the presumptive PE domain suppress PE fate by repressing the accumulation of Bowl (Hatini et al., 2005).
We previously demonstrated that the transcription co-activator Yki, with its DNA-binding partners Sd and Hth, appeared to be necessary for the establishment of PE in the eye disc, based on phenotypes induced by RNAi-mediated gene silencing (Zhang et al., 2011). We confirm this here by showing that ykiB5/ykiB5 mutant eye discs generated by MARGE are nearly identical to yki-RNAi discs (compare Fig. 4F–G′ with 4A,A′), and also demonstrate that Yki's role in the PE-retina cell fate choice is independent of its nearly universal role in proliferation and cell survival (Fig. 4B,B′).
Interestingly, these mechanisms of PE versus non-PE fate specification are not conserved between the eye and wing discs. Whereas a similar role for Wg has been proposed for the leg and haltere PE, based on the induction of columnar cell morphology by ectopic Wg in the PE, the same effect is not observed in the eye antennal disc (Baena-Lopez et al., 2003). Lines overexpression within the eye disc PE also has no effect on cell fate, whereas in the retinal progenitor region of the disc, Lines repression of Bowl initiates precocious eye morphogenesis (Bras-Pereira et al., 2006). Thus, Wg, Lines and Bowl do not appear to be required for PE specification in the eye disc. Conversely, extensive analyses of Yki and Sd function in the wing imaginal disc have not revealed roles for Yki in the PE versus wing-proper fate choice (Genevet et al., 2009; Huang et al., 2005; Oh and Irvine, 2008; Thompson and Cohen, 2006), consistent with the specific role for Yki in suppressing ectopic retina formation that we document here.
Thus, the acquisition of PE fate results from different genetic determinants in the eye and wing discs, and is mechanistically distinct.
PP2A holoenzymes as regulators of cell fate
Given the plethora of cellular kinases that regulate myriad biological processes, it has been appreciated that cellular phosphatases might play equally important and diverse biological roles (Bononi et al., 2011; Mishra et al., 2017). However, there are far fewer cellular phosphatases, and each is known to target a broad range of cellular substrates (Shi, 2009). PP2A is no exception to this; it is essential for cell proliferation and viability (Cygnar et al., 2005; Silverstein et al., 2002; Ugi et al., 2002). Hence, it is difficult to investigate potential roles of this phosphatase in more specific biological processes, such as cell fate specification. We report here the involvement of STRIPAK–PP2A in regulating Hippo-Yki signaling in the PE fate choice within the developing eye disc. We were able to reveal this role of STRIPAK–PP2A by targeted silencing of complex-specific PP2A co-factors (Cka-B‴, Strip, SLMAP), and we confirmed the link to PP2A by genetic interaction testing using alleles of the vital catalytic subunit (mts).
Few studies in metazoans have identified contexts in which PP2A complexes may play a role in fate specification during development. Screens in Drosophila and Caenorhabditis elegans have identified mutant alleles of fly mts (PP2A-C) and worm sur-6 (a PP2A-B-proper subunit) as modifiers of the MAPK pathway in the R7 photoreceptor and vulval fate choices, respectively (Karim et al., 1996; Sieburth et al., 1999; Wassarman et al., 1996). In the fly, mts mutant alleles dominantly enhanced the effects of activated Ras signaling and dominantly suppressed the effects of activated Raf signaling (Karim et al., 1996; Wassarman et al., 1996), whereas in the worm, sur-6 mutant alleles dominantly suppressed the effects of activated Ras signaling (Sieburth et al., 1999). Yet, homozygous loss-of-function conditions for these PP2A components could not be shown to affect either fate in an otherwise wild-type background.
Two additional phenotypes suggestive of potential changes in cell fate have also been linked to loss-of-function alleles of tws, which encodes the sole Drosophila PP2A-B-proper subunit (Shiomi et al., 1994; Uemura et al., 1993). First, loss of tws led to the overgrowth and duplication of the pouch in the developing wing imaginal disc (Uemura et al., 1993), and second, hypomorphic alleles of tws were shown to yield flies with atypical sensory bristles that display supernumerary shaft and socket cells with coincident loss of their single neuronal and sheath cells (Shiomi et al., 1994). However, neither study substantiated a true switch in cell fate vis-à-vis aberrant proliferation or reorganization of patterning cues, nor did they establish a connection between the PP2A catalytic subunit and the putative fate-switch phenotypes. The molecular substrates of PP2A-Tws also remain to be determined in these contexts. Nonetheless, these data hint at the possibility that specific PP2A holoenzymes, STRIPAK–PP2A (shown here) and PP2A-Tws/Sur-6 (cited above), may direct different cell fate choices and do so via the modulation of distinct signaling pathways.
Does STRIPAK–PP2A function in other Yki (YAP/TAZ) regulated cell fate decisions?
Whereas there is a lack of evidence in the literature for PP2A- and/or STRIPAK-regulated fate decisions, Hippo (MST1/2) signaling and Yki (YAP/TAZ) have been implicated in fate decisions in different developmental contexts (Ma et al., 2018; Misra and Irvine, 2018; Moya and Halder, 2016). Arguably the best studied example also involves the Drosophila eye, where Hpo-Yki-Sd signaling has been shown to instruct the choice between Rh5- versus Rh6-expressing R8 photoreceptor subtypes (Jukam et al., 2013; Jukam and Desplan, 2011; Mikeladze-Dvali et al., 2005).
Perhaps more intriguing is that, during the development of the vertebrate eye, a fate paradigm strikingly similar to the PE/retina fate choice has been described to involve the Yki orthologs YAP and TAZ. In vertebrates, the primordia of the retina and the retinal pigment epithelium (RPE) are specified at the optic vesicle stage, when the vesicle epithelium is partitioned into two fields, one that generates neural retina and one that develops into non-neural RPE in the optic cup. Miesfeld and colleagues demonstrated that YAP and its paralog TAZ play important roles in promoting RPE fate in zebrafish, as loss of these factors leads to transdifferentiation of RPE into retina-like cells (Miesfeld et al., 2015). A similar role for YAP in RPE fate specification was subsequently demonstrated in mice (Kim et al., 2016), where loss of YAP produces optic cups with mirror-image retinae, consisting of a pseudostratified epithelium that expresses markers of retinal progenitor cells in the presumptive RPE.
The contribution of STRIPAK–PP2A to cell fate decisions in these models has not been investigated. However, given the similar fate-determining roles for Yki and YAP/TAZ, particularly in the latter example, it will be of great interest to determine whether STRIPAK–PP2A-mediated regulation of YAP and/or TAZ activity through the MST2 (Hpo)-LATS1/2 (Wts) kinase cascade is equally important in the patterning of the vertebrate optic vesicle, and possibly in other Yki/YAP/TAZ-linked cell fate decisions.
MATERIALS AND METHODS
Drosophila genetics
Precise genotypes and temperature conditions for all samples shown are listed in Table S1. All stocks used and their origin are listed in Table S2. Transgene expression in the eye-antennal imaginal disc was driven by the constitutive ‘flip-out’ method, using ey-FLP with Act5C>CD2>GAL4 (Pignoni and Zipursky, 1997). Excision of the yki+ transgene from ykiB5/ykiB5 mutant eye-antennal discs was carried out by the Mutant Analysis by Rescue Gene Excision (MARGE) method (Zhou et al., 2016).
Immunohistology
Standard protocols were used to dissect and immunostain eye-antennal discs. Primary antibodies were from the Developmental Studies Hybridoma Bank [1:200 rat α-Elav (clone 7E8A10), 1:50 rat α-dE-cadherin (clone DCAD2), 1:200 mouse α-Dlg (clone 4F3), 1:200 mouse α-Eya (clone eya10H6), 1:500 mouse α-Dac (clone mAbdac1-1), 1:200 mouse α-Chaoptin (clone 24B10), 1:100 mouse α-Repo (clone 8D12)] or Invitrogen [1:104 rabbit α-GFP (catalog no. A-6455)]. Cy2-, Cy3- and Cy5-conjugated goat α-mouse, rat or rabbit secondary antibodies were used at 1:250 (Jackson ImmunoResearch). Phalloidin Alexa 546 conjugate (1:50; Molecular Probes) and/or Hoechst 33342 (1:105; Invitrogen) or 4′,6-diamidino-2-phenylindole (DAPI) were included in either the secondary antibody solution or in the mounting medium as counterstains, where indicated.
Image acquisition and processing
Image stacks were recorded in Leica LASX software using a Leica DM5500Q microscope with SPEII confocal head. Stacks were rotated, projected, adjusted and/or 3D-rendered using LASX. Laser power, gain and post-processing were adjusted for best presentation; no quantitative inferences were made with respect to fluorescence intensities. Figures were assembled in Adobe Photoshop and Adobe Illustrator. Two-dimensional images for disc size analysis were acquired using a Leica DFC-7000 camera on the above microscope, or using an Accu-Scope on a Nikon E600 compound microscope.
Quantification of disc size and statistical analyses
Quantification of disc size was conducted in either Fiji or Adobe Photoshop software by manually tracing the outline of each disc and calculating the area. t-tests were performed to identify differences in the mean disc area among genotypes; data were normalized to the mean or median disc size of the leftmost category, as indicated. Individual data points are plotted.
Reproducibility
Representative images are shown throughout. Crosses were repeated at least once, and imaginal discs were visually scored on a compound fluorescent microscope for each experiment; at least three representative discs were scanned by confocal microscopy. Precise numbers of discs observed are given in the relevant figure captions.
Supplementary Material
Acknowledgements
Stocks were obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537), the Vienna Drosophila Resource Center, or kindly provided by Duojia Pan and Takahiro Chihara. The authors thank Nisveta Jusić for administrative support and Drosophila maintenance, and Dana DeSantis for critical comments on the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.J.N., Q.Z., F.P.; Methodology: S.J.N., Q.Z., F.P.; Validation: S.J.N., Q.Z.; Formal analysis: S.J.N., Q.Z., F.P.; Investigation: S.J.N., Q.Z.; Writing - original draft: S.J.N., F.P.; Writing - review & editing: S.J.N., Q.Z., F.P.; Visualization: S.J.N., Q.Z.; Supervision: F.P.; Project administration: F.P.; Funding acquisition: F.P.
Funding
This work was supported by a grant from the National Institutes of Health to F.P. (NEI 5R01EY028221), and by unrestricted grants to the Department of Ophthalmology and Visual Sciences from Lions Clubs International Foundation (Lions 20-Y) and from Research to Prevent Blindness. S.J.N. was partially supported by a postdoctoral grant from Fight for Sight. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.237834.supplemental
References
- Agnes F., Suzanne M. and Noselli S. (1999). The Drosophila JNK pathway controls the morphogenesis of imaginal discs during metamorphosis. Development 126, 5453-5462. [DOI] [PubMed] [Google Scholar]
- Bae S. J., Ni L., Osinski A., Tomchick D. R., Brautigam C. A. and Luo X. (2017). SAV1 promotes Hippo kinase activation through antagonizing the PP2A phosphatase STRIPAK. eLife 6, e30278 10.7554/eLife.30278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-Lopez L. A., Pastor-Pareja J. C. and Resino J. (2003). Wg and Egfr signalling antagonise the development of the peripodial epithelium in Drosophila wing discs. Development 130, 6497-6506. 10.1242/dev.00884 [DOI] [PubMed] [Google Scholar]
- Bennett F. C. and Harvey K. F. (2006). Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr. Biol. 16, 2101-2110. 10.1016/j.cub.2006.09.045 [DOI] [PubMed] [Google Scholar]
- Bessa J., Gebelein B., Pichaud F., Casares F. and Mann R. S. (2002). Combinatorial control of Drosophila eye development by eyeless, homothorax, and teashirt. Genes Dev. 16, 2415-2427. 10.1101/gad.1009002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bononi A., Agnoletto C., De Marchi E., Marchi S., Patergnani S., Bonora M., Giorgi C., Missiroli S., Poletti F., Rimessi A. et al. (2011). Protein kinases and phosphatases in the control of cell fate. Enzyme Res. 2011, 329098 10.4061/2011/329098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bras-Pereira C., Bessa J. and Casares F. (2006). Odd-skipped genes specify the signaling center that triggers retinogenesis in Drosophila. Development 133, 4145-4149. 10.1242/dev.02593 [DOI] [PubMed] [Google Scholar]
- Chen H.-W., Marinissen M. J., Oh S.-W., Chen X., Melnick M., Perrimon N., Gutkind J. S. and Hou S. X. (2002). CKA, a novel multidomain protein, regulates the JUN N-terminal kinase signal transduction pathway in Drosophila. Mol. Cell. Biol. 22, 1792-1803. 10.1128/MCB.22.6.1792-1803.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho K.-O., Chern J., Izaddoost S. and Choi K.-W. (2000). Novel signaling from the peripodial membrane is essential for eye disc patterning in Drosophila. Cell 103, 331-342. 10.1016/S0092-8674(00)00124-0 [DOI] [PubMed] [Google Scholar]
- Cygnar K. D., Gao X., Pan D. and Neufeld T. P. (2005). The phosphatase subunit tap42 functions independently of target of rapamycin to regulate cell division and survival in Drosophila. Genetics 170, 733-740. 10.1534/genetics.104.039909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez M. and Casares F. (2005). Organ specification-growth control connection: new in-sights from the Drosophila eye-antennal disc. Dev. Dyn. 232, 673-684. 10.1002/dvdy.20311 [DOI] [PubMed] [Google Scholar]
- Emoto K., Parrish J. Z., Jan L. Y. and Jan Y.-N. (2006). The tumour suppressor Hippo acts with the NDR kinases in dendritic tiling and maintenance. Nature 443, 210-213. 10.1038/nature05090 [DOI] [PubMed] [Google Scholar]
- Fristrom D. and Fristrom J. W. (1993). The metamorphic development of the adult epidermis. In The Development of Drosophila Melanogaster (ed. Bate M. and Arias A. M.), pp. 843-897 Cold Spring Harbor Laboratory Press. [Google Scholar]
- Genevet A., Polesello C., Blight K., Robertson F., Collinson L. M., Pichaud F. and Tapon N. (2009). The Hippo pathway regulates apical-domain size independently of its growth-control function. J. Cell Sci. 122, 2360-2370. 10.1242/jcs.041806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson M. C. and Schubiger G. (2000). Peripodial cells regulate proliferation and patterning of Drosophila imaginal discs. Cell 103, 343-350. 10.1016/S0092-8674(00)00125-2 [DOI] [PubMed] [Google Scholar]
- Goudreault M., D'Ambrosio L. M., Kean M. J., Mullin M. J., Larsen B. G., Sanchez A., Chaudhry S., Chen G. I., Sicheri F., Nesvizhskii A. I. et al. (2009). A PP2A phosphatase high density interaction network identifies a novel Striatin-interacting phosphatase and kinase complex linked to the cerebral cavernous malformation 3 (CCM3) protein. Mol. Cell. Proteomics 8, 157-171. 10.1074/mcp.M800266-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubbs N., Leach M., Su X., Petrisko T., Rosario J. B. and Mahaffey J. W. (2013). New components of Drosophila leg development identified through genome wide association studies. PLoS ONE 8, e60261 10.1371/journal.pone.0060261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatini V., Green R. B., Lengyel J. A., Bray S. J. and Dinardo S. (2005). The Drumstick/Lines/Bowl regulatory pathway links antagonistic Hedgehog and Wingless signaling inputs to epidermal cell differentiation. Genes Dev. 19, 709-718. 10.1101/gad.1268005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Wu S., Barrera J., Matthews K. and Pan D. (2005). The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell 122, 421-434. 10.1016/j.cell.2005.06.007 [DOI] [PubMed] [Google Scholar]
- Hummel T., Attix S., Gunning D. and Zipursky S. L. (2002). Temporal control of glial cell migration in the Drosophila eye requires gilgamesh, hedgehog, and eye specification genes. Neuron 33, 193-203. 10.1016/S0896-6273(01)00581-5 [DOI] [PubMed] [Google Scholar]
- Jukam D. and Desplan C. (2011). Binary regulation of Hippo pathway by Merlin/NF2, Kibra, Lgl, and Melted specifies and maintains postmitotic neuronal fate. Dev. Cell 21, 874-887. 10.1016/j.devcel.2011.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukam D., Xie B., Rister J., Terrell D., Charlton-Perkins M., Pistillo D., Gebelein B., Desplan C. and Cook T. (2013). Opposite feedbacks in the Hippo pathway for growth control and neural fate. Science 342, 1238016 10.1126/science.1238016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim F. D., Chang H. C., Therrien M., Wassarman D. A., Laverty T. and Rubin G. M. (1996). A screen for genes that function downstream of Ras1 during Drosophila eye development. Genetics 143, 315-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon K. L., Ranade S. S., Curtiss J., Mlodzik M. and Pignoni F. (2003). Coordinating proliferation and tissue specification to promote regional identity in the Drosophila head. Dev. Cell 5, 403-414. 10.1016/S1534-5807(03)00243-0 [DOI] [PubMed] [Google Scholar]
- Kim J. Y., Park R., Lee J. H. J., Shin J., Nickas J., Kim S. and Cho S.-H. (2016). Yap is essential for retinal progenitor cell cycle progression and RPE cell fate acquisition in the developing mouse eye. Dev. Biol. 419, 336-347. 10.1016/j.ydbio.2016.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kück U., Radchenko D. and Teichert I. (2019). STRIPAK, a highly conserved signaling complex, controls multiple eukaryotic cellular and developmental processes and is linked with human diseases. Biol. Chem. 400, 1005-1022. 10.1515/hsz-2019-0173 [DOI] [PubMed] [Google Scholar]
- Liu B., Zheng Y., Yin F., Yu J., Silverman N. and Pan D. (2016). Toll receptor-mediated Hippo signaling controls innate immunity in Drosophila. Cell 164, 406-419. 10.1016/j.cell.2015.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S., Meng Z., Chen R. and Guan K. L. (2018). The Hippo pathway: biology and pathophysiology. Annu. Rev. Biochem. 88, 577-604. 10.1146/annurev-biochem-013118-111829 [DOI] [PubMed] [Google Scholar]
- Martin P. F. (1982). Direct determination of the growth rate of Drosophila imaginal discs. J. Exp. Zool. 222, 97-102. 10.1002/jez.1402220113 [DOI] [Google Scholar]
- Miesfeld J. B., Gestri G., Clark B. S., Flinn M. A., Poole R. J., Bader J. R., Besharse J. C., Wilson S. W. and Link B. A. (2015). Yap and Taz regulate retinal pigment epithelial cell fate. Development 142, 3021-3032. 10.1242/dev.119008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikeladze-Dvali T., Wernet M. F., Pistillo D., Mazzoni E. O., Teleman A. A., Chen Y.-W., Cohen S. and Desplan C. (2005). The growth regulators Warts/lats and melted interact in a bistable loop to specify opposite fates in Drosophila R8 photoreceptors. Cell 122, 775-787. 10.1016/j.cell.2005.07.026 [DOI] [PubMed] [Google Scholar]
- Milner M. J., Bleasby A. J. and Pyott A. (1983). The role of the peripodial membrane in the morphogenesis of the eye-antennal disc of Drosophila melanogaster. Wilehm Roux. Arch. Dev. Biol. 192, 164-170. 10.1007/BF00848686 [DOI] [PubMed] [Google Scholar]
- Mishra A., Oules B., Pisco A. O., Ly T., Liakath-Ali K., Walko G., Viswanathan P., Tihy M., Nijjher J., Dunn S. J. et al. (2017). A protein phosphatase network controls the temporal and spatial dynamics of differentiation commitment in human epidermis. eLife 6, e27356 10.7554/eLife.27356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra J. R. and Irvine K. D. (2018). The Hippo signaling network and its biological functions. Annu. Rev. Genet. 52, 65-87. 10.1146/annurev-genet-120417-031621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno C. S., Park S., Nelson K., Ashby D., Hubalek F., Lane W. S. and Pallas D. C. (2000). WD40 repeat proteins Striatin and S/G2 nuclear autoantigen are members of a novel family of calmodulin-binding proteins that associate with protein phosphatase 2A. J. Biol. Chem. 275, 5257-5263. 10.1074/jbc.275.8.5257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya I. M. and Halder G. (2016). The Hippo pathway in cellular reprogramming and regeneration of different organs. Curr. Opin. Cell Biol. 43, 62-68. 10.1016/j.ceb.2016.08.004 [DOI] [PubMed] [Google Scholar]
- Nusinow D., Greenberg L. and Hatini V. (2008). Reciprocal roles for bowl and lines in specifying the peripodial epithelium and the disc proper of the Drosophila wing primordium. Development 135, 3031-3041. 10.1242/dev.020800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H. and Irvine K. D. (2008). In vivo regulation of Yorkie phosphorylation and localization. Development 135, 1081-1088. 10.1242/dev.015255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H. W., Slattery M. and Mann R. S. (2009). Transcription factor choice in the Hippo signaling pathway: homothorax and Yorkie regulation of the microRNA bantam in the progenitor domain of the Drosophila eye imaginal disc. Genes Dev. 23, 2307-2319. 10.1101/gad.1820009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignoni F. and Zipursky S. L. (1997). Induction of Drosophila eye development by decapentaplegic. Development 124, 271-278. [DOI] [PubMed] [Google Scholar]
- Quan X.-J., Ramaekers A. and Hassan B. A. (2012). Transcriptional control of cell fate specification: lessons from the fly retina. Curr. Top. Dev. Biol. 98, 259-276. 10.1016/B978-0-12-386499-4.00010-0 [DOI] [PubMed] [Google Scholar]
- Ribeiro P. S., Josué F., Wepf A., Wehr M. C., Rinner O., Kelly G., Tapon N. and Gstaiger M. (2010). Combined functional genomic and proteomic approaches identify a PP2A complex as a negative regulator of Hippo signaling. Mol. Cell 39, 521-534. 10.1016/j.molcel.2010.08.002 [DOI] [PubMed] [Google Scholar]
- Sakuma C., Saito Y., Umehara T., Kamimura K., Maeda N., Mosca T. J., Miura M. and Chihara T. (2016). The strip-Hippo pathway regulates synaptic terminal formation by modulating actin organization at the Drosophila neuromuscular synapses. Cell Rep. 16, 2289-2297. 10.1016/j.celrep.2016.07.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W. and Mardon G. (1997). Ectopic eye development in Drosophila induced by directed dachshund expression. Development 124, 45-52. [DOI] [PubMed] [Google Scholar]
- Shi Y. (2009). Serine/threonine phosphatases: mechanism through structure. Cell 139, 468-484. 10.1016/j.cell.2009.10.006 [DOI] [PubMed] [Google Scholar]
- Shiomi K., Takeichi M., Nishida Y., Nishi Y. and Uemura T. (1994). Alternative cell fate choice induced by low-level expression of a regulator of protein phosphatase 2A in the Drosophila peripheral nervous system. Development 120, 1591-1599. [DOI] [PubMed] [Google Scholar]
- Sieburth D. S., Sundaram M., Howard R. M. and Han M. (1999). A PP2A regulatory subunit positively regulates Ras-mediated signaling during Caenorhabditis elegans vulval induction. Genes Dev. 13, 2562-2569. 10.1101/gad.13.19.2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein A. M., Barrow C. A., Davis A. J. and Mumby M. C. (2002). Actions of PP2A on the MAP kinase pathway and apoptosis are mediated by distinct regulatory subunits. Proc. Natl. Acad. Sci. USA 99, 4221-4226. 10.1073/pnas.072071699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupe A. M., Merrill R. A. and Strack S. (2011). Determinants for substrate specificity of protein phosphatase 2A. Enzyme Res. 2011, 398751 10.4061/2011/398751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stultz B. G., Lee H., Ramon K. and Hursh D. A. (2006). Decapentaplegic head capsule mutations disrupt novel peripodial expression controlling the morphogenesis of the Drosophila ventral head. Dev. Biol. 296, 329-339. 10.1016/j.ydbio.2006.05.034 [DOI] [PubMed] [Google Scholar]
- Tang Y., Chen M., Zhou L., Ma J., Li Y., Zhang H., Shi Z., Xu Q., Zhang X., Gao Z. et al. (2019). Architecture, substructures, and dynamic assembly of STRIPAK complexes in Hippo signaling. Cell Discov. 5, 3 10.1038/s41421-018-0077-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B. J. and Cohen S. M. (2006). The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell 126, 767-774. 10.1016/j.cell.2006.07.013 [DOI] [PubMed] [Google Scholar]
- Treisman J. E. (2013). Retinal differentiation in Drosophila. Wiley Interdiscip. Rev. Dev. Biol. 2, 545-557. 10.1002/wdev.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T., Shiomi K., Togashi S. and Takeichi M. (1993). Mutation of twins encoding a regulator of protein phosphatase 2A leads to pattern duplication in Drosophila imaginal discs. Genes Dev. 7, 429-440. 10.1101/gad.7.3.429 [DOI] [PubMed] [Google Scholar]
- Ugi S., Imamura T., Ricketts W. and Olefsky J. M. (2002). Protein phosphatase 2A forms a molecular complex with Shc and regulates Shc tyrosine phosphorylation and downstream mitogenic signaling. Mol. Cell. Biol. 22, 2375-2387. 10.1128/MCB.22.7.2375-2387.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vactor D. Jr, Krantz D. E., Reinke R. and Zipursky S. L. (1988). Analysis of mutants in chaoptin, a photoreceptor cell-specific glycoprotein in Drosophila, reveals its role in cellular morphogenesis. Cell 52, 281-290. 10.1016/0092-8674(88)90517-X [DOI] [PubMed] [Google Scholar]
- Wassarman D. A., Solomon N. M., Chang H. C., Karim F. D., Therrien M. and Rubin G. M. (1996). Protein phosphatase 2A positively and negatively regulates Ras1-mediated photoreceptor development in Drosophila. Genes Dev. 10, 272-278. 10.1101/gad.10.3.272 [DOI] [PubMed] [Google Scholar]
- Wu S., Huang J., Dong J. and Pan D. (2003). Hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with Salvador and Warts. Cell 114, 445-456. 10.1016/S0092-8674(03)00549-X [DOI] [PubMed] [Google Scholar]
- Wu S., Liu Y., Zheng Y., Dong J. and Pan D. (2008). The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev. Cell 14, 388-398. 10.1016/j.devcel.2008.01.007 [DOI] [PubMed] [Google Scholar]
- Zeitlinger J. and Bohmann D. (1999). Thorax closure in Drosophila: involvement of Fos and the JNK pathway. Development 126, 3947-3956. [DOI] [PubMed] [Google Scholar]
- Zhang T., Zhou Q. and Pignoni F. (2011). Yki/YAP, Sd/TEAD and Hth/MEIS control tissue specification in the Drosophila eye disc epithelium. PLoS ONE 6, e22278 10.1371/journal.pone.0022278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Liu B., Wang L., Lei H., Pulgar Prieto K. D. and Pan D. (2017). Homeostatic control of Hpo/MST kinase activity through autophosphorylation-dependent recruitment of the STRIPAK PP2A phosphatase complex. Cell Rep. 21, 3612-3623. 10.1016/j.celrep.2017.11.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Neal S. J. and Pignoni F. (2016). Mutant analysis by rescue gene excision: new tools for mosaic studies in Drosophila. Genesis 54, 589-592. 10.1002/dvg.22984 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.