ABSTRACT
As the crucial non-cellular component of tissues, the extracellular matrix (ECM) provides both physical support and signaling regulation to cells. Some ECM molecules provide a fibrillar environment around cells, while others provide a sheet-like basement membrane scaffold beneath epithelial cells. In this Review, we focus on recent studies investigating the mechanical, biophysical and signaling cues provided to developing tissues by different types of ECM in a variety of developing organisms. In addition, we discuss how the ECM helps to regulate tissue morphology during embryonic development by governing key elements of cell shape, adhesion, migration and differentiation.
KEY WORDS: Extracellular matrix, Embryo, Migration, Adhesion, Differentiation, Biophysical
Summary: This Review discusses our current understanding of how the extracellular matrix helps guide developing tissues by influencing cell adhesion, migration, shape and differentiation, emphasizing the biophysical cues it provides.
Introduction
The extracellular matrix (ECM) is essential for metazoan life; without it, we would be merely an amorphous mass of cells. The ECM is the non-cellular component of all tissues, forming the physical environment surrounding cells, and playing both structural and signaling roles (Alberts et al., 2014; Frantz et al., 2010; Hynes and Yamada, 2012; Loganathan et al., 2016). As summarized in this Review, the physical roles of the various types of ECM include anchoring, guiding or restraining cell and tissue movements. For example, epithelial cells are anchored to a basement membrane, but if they become migratory, they can migrate along ECM fibrils or basement membranes.
The physical properties of the ECM (e.g. stiffness) can provide regulatory information to cells (Frantz et al., 2010; Yamada and Sixt, 2019). In addition, the ECM can provide signaling information through its specific biochemical composition and the local concentrations of its constituents (e.g. for gene regulation), and can serve as a reservoir and source of signaling molecules, such as cytokines. The many developmental and cell biological processes regulated or guided by the ECM include: contact guidance-mediated cell directionality (Teixeira et al., 2003), morphogenetic movements of gastrulation and organogenesis (Loganathan et al., 2016; Dzamba and DeSimone, 2018; Wang et al., 2017), stem cell differentiation (Darnell et al., 2018a,b; Smith et al., 2018), anchorage-dependent growth (Huang and Ingber, 1999), and cell survival (anoikis) (Frisch and Ruoslahti, 1997; Reddig and Juliano, 2005).
The importance of the ECM in normal mouse and human development is demonstrated by the many examples of embryonic lethality or functional disorders caused by deficiency or mutation, either experimentally or in a multitude of genetic diseases. Human genetic disorders can result from perturbed ECM structure, dynamics, components and/or interactions (Arseni et al., 2018; Bateman et al., 2009; Lamande and Bateman, 2019; Pozzi et al., 2017). In comparison with studies of the ECM in development, the regulation of developmental processes by transcription and growth factor signaling are much better studied (Chen et al., 2018c; Schweisguth and Corson, 2019; Shahbazi and Zernicka-Goetz, 2018). However, new insights highlight the importance of synergy between biochemical and biophysical signaling in developing tissues. As such, interest in the ECM and the biophysical cues that regulate embryogenesis is growing (Mammoto et al., 2012; Merle and Farge, 2018; Dzamba and DeSimone, 2018).
This Review summarizes key roles of the ECM in cellular processes and tissue morphogenesis during embryonic development. After first introducing principles of cell-ECM interactions, we focus primarily on recently published examples to discuss how the ECM helps to direct developing tissues by influencing cell adhesion, migration, shape and differentiation (Fig. 1A-D). We place particular emphasis on the biophysical properties and signals of the ECM that regulate these processes in a variety of organisms, ranging from humans and mice to Drosophila and Tribolium.
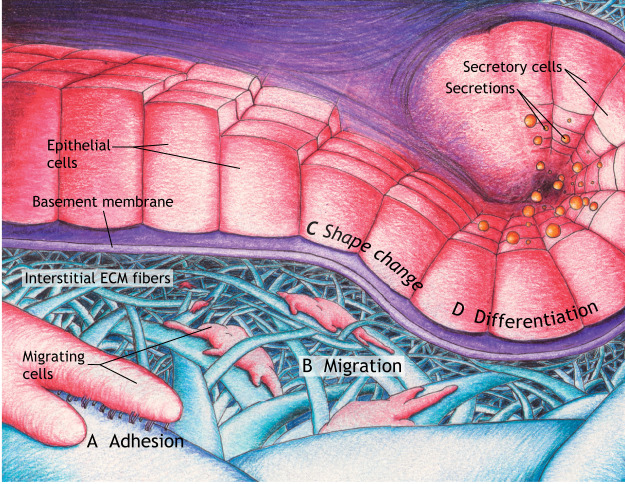
Fig. 1.
A summary of the regulation of developmental processes by the extracellular matrix. Two major forms of ECM are basement membranes and interstitial matrices. These types of ECM help to direct cell and tissue shape during morphogenesis in development by influencing cell adhesion (A), migration (B), morphology (C) and differentiation (D). (A) Cell-ECM adhesion, with cell adhesion complexes between the cell and a fibril substrate shown being mediated by integrins (heterodimeric receptors projecting downward from the closest migrating cell). (B) Migrating cells using oriented protrusions and cell-ECM adhesion complexes to move along interstitial ECM fibers. (C) Epithelial cells undergoing shape change (columnar-to-cuboidal transitions). (D) Epithelial cells differentiating into secretory cells.
Extracellular matrix
The ECM comprises predominantly protein and polysaccharide components (Frantz et al., 2010), but the forms of ECM can be remarkably diverse in biophysical, biochemical and topological properties (Naba et al., 2017; Shao et al., 2020). The precise composition of an ECM is often tissue specific, highly dynamic and responsible for its unique physical properties (e.g. topography, pore size, fiber size, fiber orientation, stiffness/elasticity and ligand density) (Fig. 2A-E) and chemical properties of each tissue (Brown, 2011; Dzamba and DeSimone, 2018; Hynes and Yamada, 2012). Although ECMs can exist in many forms, two major classes are basement membrane and interstitial ECM. Basement membranes are specialized, flat laminar ECMs consisting predominantly of core proteins organized into sheet-like networks of interconnected ECM molecules that include collagen IV, laminins and proteoglycans (e.g. perlecan) (Table 1; Fig. 2F) (Pozzi et al., 2017). Basement membranes underlie epithelia and surround the organs of most metazoans (Pozzi et al., 2017; Sekiguchi and Yamada, 2018). In interstitial matrices, collagens and various non-collagenous proteins (e.g. fibronectin, elastin, laminin and tenascin) contribute to the characteristic fibrous networks of ECMs, while proteoglycans and water contribute to their interstitial spaces (Table 1; Fig. 2G) (Frantz et al., 2010; Hynes and Yamada, 2012).
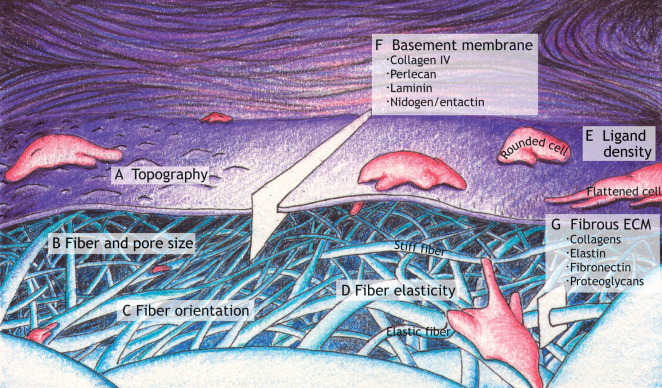
Fig. 2.
Examples of physical properties of the extracellular matrix. (A) Topography encountered by a migrating cell. (B) Examples of varying fiber diameters and sizes of pores between ECM fibers. (C) Examples of fiber orientation: compare oriented fibers near ‘C’ with the other relatively non-oriented fibers. (D) Examples of varying fiber elasticity/stiffness represented as different degrees of fiber deformation as a cell pulls on two fibers using cell processes and cellular contractility. (E) Ligand density (shown as black bristles) affecting the extent of cell spreading. (F) Basement membrane composition: a slice of the basement membrane indicating key molecular components. (G) Fibrous ECM composition: a slice of fibrillar ECM listing several key components.
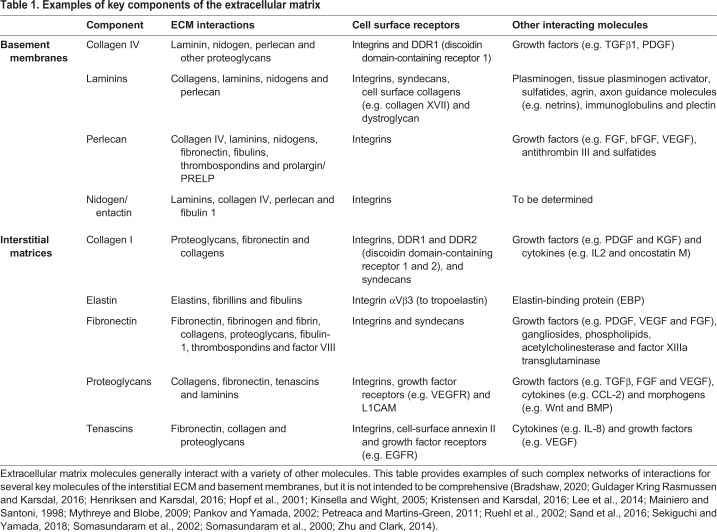
Table 1.
Examples of key components of the extracellular matrix
Cell-ECM and cell-cell adhesions
Cell adhesions are the attachment structures between cells and the ECM, or between cells and other cells. They are essential for the organization of individual cells into three-dimensional tissues. The specific properties of cell-ECM adhesions, such as their distribution, quantity and stability/duration, can vary between organisms, tissues, developmental stages and even neighboring cells (Diaz-de-la-Loza et al., 2018; Etournay et al., 2015; Munster et al., 2019; Ray et al., 2015). Amidst this heterogeneity, the most common cell-ECM adhesions are mediated by integrins, which are linked to the internal cell cytoskeleton (Geiger et al., 2001). Cells use these adhesions to attach directly either to anchoring ligands of the ECM interstitial matrix (e.g. fibrous collagens, fibronectin and vitronectin) or to other glycoproteins in the basement membrane (e.g. laminin or network collagens) (Table 1; Fig. 1A) (Frantz et al., 2010; Hynes and Yamada, 2012). Indeed, several types of integrin-dependent adhesions are involved in crucial developmental processes (Alberts et al., 2014; Gillard et al., 2019; Keller, 2006). The best-characterized integrin-based adhesions are the RhoA-stimulated focal adhesions that anchor the ends of actin stress fibers to the nearby matrix. However, their precursors and variants (e.g. dot-like focal complexes and elongated fibrillar adhesions) are also likely to play roles in developmental events (Davidson et al., 2019; Goodwin et al., 2017; Horton et al., 2016a,b; Lee et al., 2018). Highlighting the importance of integrin-dependent adhesions in development, mutations in various integrin- and integrin-associated-protein family members are implicated in several human developmental diseases, such as those involving renal (Humbert et al., 2014), ocular (Beleggia et al., 2015; Zhang et al., 2016), pulmonary (Yalcin et al., 2015) and dermal/epidermal (Condrat et al., 2018; Mylonas et al., 2019) tissues.
Embryogenesis requires a coordinated balance between cell-ECM and cell-cell dynamics. Cell-cell adhesions mediate tissue cohesion and organization. Through cell-cell adhesions, the ECM can exert physical effects beyond only the first layer of ECM-attached cells into the interior of the tissue/organ. As with cell-ECM adhesions, a variety of cell-cell adhesive contacts are found in the developing embryo, for example various adherens junctions (Halbleib and Nelson, 2006; Letizia et al., 2019), desmosomes (Bharathan and Dickinson, 2019; Garrod and Chidgey, 2008) and tight junctions (Anderson and Van Itallie, 2009; Chan et al., 2019; Eckert and Fleming, 2008). Contacts mediated by the cadherin family of adhesion molecules are particularly important types of cell-cell interactions for maintaining organized solid tissues and transmitting mechanical signals (Balaji et al., 2019; Goodwin and Nelson, 2017; Halbleib and Nelson, 2006; Pinheiro and Bellaiche, 2018; Sumi et al., 2018; Wu and Taneyhill, 2019). As with integrin-mediated adhesions, human mutations in cadherin complexes are implicated in several developmental disorders (Accogli et al., 2019; Cox et al., 2018; Saeidian et al., 2019; Samuelov et al., 2015). For detailed descriptions of the diversity, physiological roles and biochemical properties of the numerous integrins, integrin-associated proteins, cadherins and adhesion complexes, we refer the reader to a number of recent excellent reviews (Bachmann et al., 2019; Green and Brown, 2019; Halbleib and Nelson, 2006; Horton et al., 2016a,b; Hynes, 1992; Hynes, 2002; Takeichi, 2014; Tepass et al., 2000).
In the context of this Review, the ECM uses cellular adhesions to regulate or modulate tissue shaping by anchoring, signal/force transmission and cell migration. Owing to the importance of cell migration in embryogenesis, we devote a separate section to this topic below.
Tissue shaping by anchoring
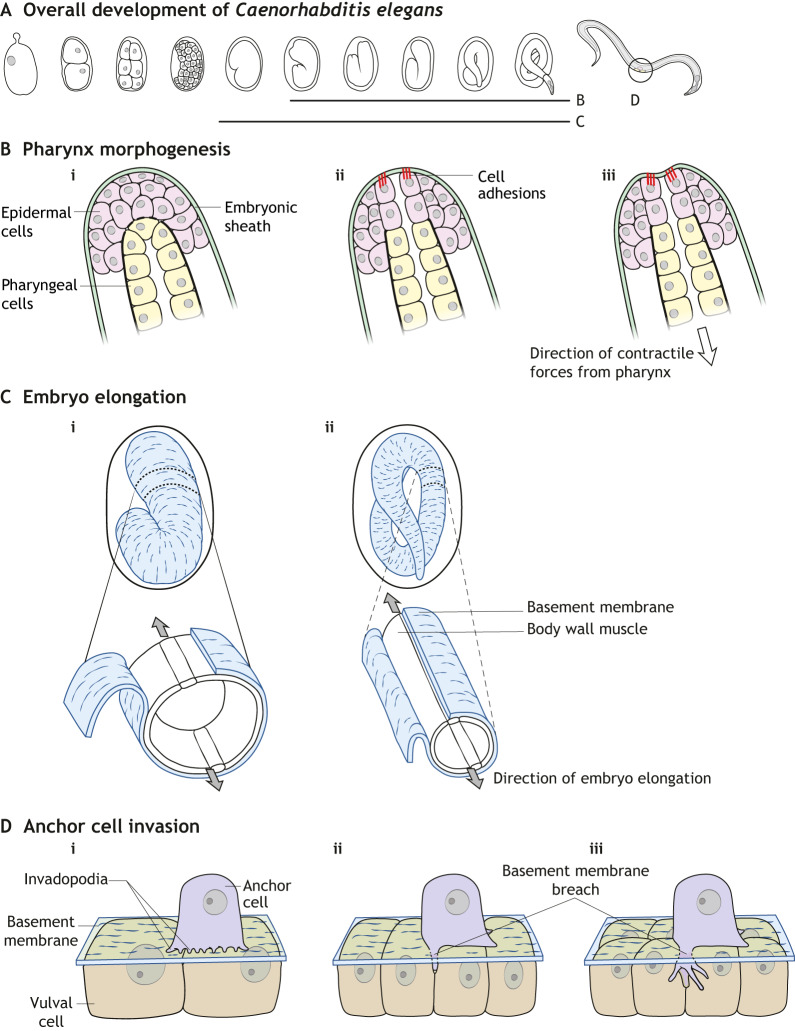
Many developing organisms progress through stages in which a layer of ECM separates embryonic germ layers (Latimer and Jessen, 2010) and/or surrounds a mass of cells. For example, the vitelline envelope surrounds the blastoderm and oocyte of Tribolium, Drosophila and other non-mammalian species (Munster et al., 2019), the cuticle surrounds Drosophila larval and pupal tissues (Ray et al., 2015), and the zona pellucida surrounds the oocyte of humans and other mammals (Bhakta et al., 2019). In these systems, spatiotemporal variations in cell-ECM adhesions during tissue-intrinsic contraction drive tissue shaping. For example, the Tribolium integrin termed ‘inflated’ temporarily mediates adhesion of blastodermal cells to the antero-ventral region of the vitelline envelope. This localized attachment guides unidirectional tissue elongation, because myosin contractile activity causes the non-anchored dorsal tissues to slide along the envelope (Munster et al., 2019). Similarly, in C. elegans, the attachment of epidermal cells to an FBN-1 extracellular fiber meshwork of the embryonic sheath anchors the epidermis to prevent its posterior displacement secondary to pulling forces of the developing pharynx (Fig. 3A,B) (Kelley et al., 2015).
Fig. 3.
Schematic diagrams of C. elegans model systems discussed in this Review. (A) Overview of C. elegans development indicating stages involved in the following panels. (B) Pharynx morphogenesis. Epidermal cells adhering via cell adhesions to the surrounding embryonic sheath, which prevents deformation of the epidermis by pulling forces from the developing pharynx (pharyngeal cells in yellow). (C) Embryo elongation. The basement membrane serves as a ‘molecular corset’, acting in conjunction with muscle contractions to elongate the embryo. (D) Anchor cell invasion. Anchor cells use invadopodia to produce initial focal sites of basement membrane degradation (i). Upon breaching the basement membrane (ii), further invadopodia formation ceases, a large invasive protrusion forms and the anchor cell inserts itself between underlying vulval cells (iii).
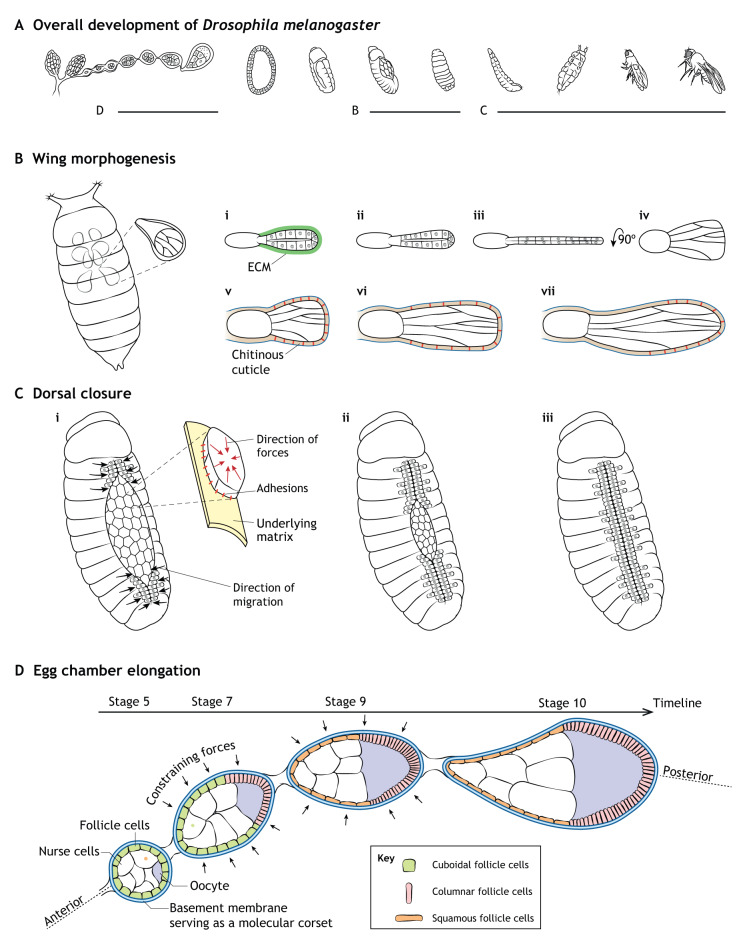
Turning to insights provided by Drosophila systems, the apical ECM protein Dumpy (Dp) anchors distal epithelial cells of the Drosophila pupal wing to the surrounding chitinous cuticle in a patterned manner (Fig. 4A,B) (Ray et al., 2015). This Dp-mediated attachment resists tissue retraction that would otherwise result in the truncated wings, legs and antennae observed in dp loss-of-function mutants (Ray et al., 2015). Several Drosophila systems, including Dp-regulated limb morphogenesis, have been characterized by computational models that simulate the ability of cellular interactions to resist or transmit forces to drive oriented tissue growth during development (Etournay et al., 2015; Sui et al., 2018; Tozluoglu et al., 2019). In addition to force resistance and transmission, these cell-matrix interactions allow the ECM to dissipate forces exerted on cells during tissue morphogenesis. This buffering role of the ECM occurs during formation of the Drosophila leg disc (Proag et al., 2019). In early stages of this process, the peripodial epithelium remains in a relaxed state because tensile forces caused by leg elongation are borne by the attached ECM. At latter stages, however, cell-matrix interactions are lost, retractile forces are transferred to the cell monolayer and the peripodial epithelium opens and retracts (Proag et al., 2019). Embryogenesis requires cooperation between the physical cell-adhesion mechanisms discussed above and various signaling processes that transfer mechanical information between cells and tissues.
Fig. 4.
Schematics of Drosophila model systems discussed in this Review. (A) Overview of Drosophila development indicating stages involved in the following panels. (B) Wing morphogenesis. (i-iv) Removal of the ECM initiates wing elongation secondary to cell columnar-to-cuboidal shape changes. (v-vii) Dynamic patterned attachment of pupal wing epithelial cells to the chitinous cuticle shapes the developing wing. (C) Early (i), middle (ii) and late (iii) dorsal closure. Contracting cells adhering to underlying matrix along with lateral epidermal cells migrating towards the dorsal midline as the amniosera contracts and ingresses. (D) Egg chamber elongation. The basement membrane promotes cuboidal (green)-to-squamous (orange) transitions of anterior follicle cells and cuboidal-to-columnar (pink) transitions of posterior follicle cells; the basement membrane provides constraining forces as a ‘molecular corset’ to elongate the egg chamber.
Force and mechanical signal transmission
Appreciation of the roles of mechanical forces in developing tissues has grown from initial observations more than one century ago that documented load-induced bone remodeling (Churchill, 1970), to recent elaborate investigations using advanced biophysical techniques that include cell migration simulators, in vivo embryo remodeling quantification systems and others (Hou et al., 2019; Lardennois et al., 2019; Roca-Cusachs et al., 2017). The ability of a cell to sense and transduce mechanical signals (termed mechanosensation and mechanotransduction, respectively; Box 1) is fundamental to biophysically guiding tissue morphogenesis (Merle and Farge, 2018; Wozniak and Chen, 2009). Coordination of this signaling between cells and their physical environment during development depends on ECM biophysical properties (Fig. 2A-D) [e.g. geometry, alignment and elasticity (Humphries et al., 2017; Ma et al., 2013; Piotrowski-Daspit et al., 2017; Sopher et al., 2018; Yamada and Sixt, 2019)], cell-matrix adhesion (Fig. 1A) and intercellular adhesions.
Box 1. Mechanotransduction and the ECM.
Cells not only synthesize and remodel the ECM, but also respond to mechanical information coming from the ECM. Cells sense physical stimuli from their microenvironment, such as ECM topography, composition and stiffness. These external signals are converted into cellular responses in the process of mechanotransduction. Research into the multiple mechanisms of mechanotransduction is rapidly expanding (reviewed by Chighizola et al., 2019; Doyle and Yamada, 2016; Jansen et al., 2017, 2015; Ringer et al., 2017).
The following are key terms in this rapidly evolving field:
Mechanobiology: characterizing how cells and tissues respond to mechanical/physical stimuli through integrating biology, engineering and physics.
Mechanosensitive: describing a molecule that undergoes structural changes in response to mechanical stimuli.
Mechanosensing: the process of detecting mechanical stimuli.
Mechanotransduction: the overall process of sensing a mechanical signal and converting it to an intracellular response.
Mechanosignaling: intracellular signaling events induced by extracellular mechanical stimuli.
Among the wide variety of mechanisms of mechanotransduction [e.g. those involving cell-cell adherens junctions (Qin et al., 2020), mechanosensitive ion channels (Barzegari et al., 2020a,b; Jin et al., 2020) and others], integrin-based responses of cells to the extracellular matrix have recently been characterized in particular detail. As depicted schematically in a simplified form in Fig. 1A, the integrin cell-surface receptors for ECM molecules are heterodimers that bind to ECM molecules. Intracellular proteins bind to the cytoplasmic tails of integrins and organize multimolecular cell-matrix adhesion complexes in order to transmit external signals into the cell (Kechagia et al., 2019; Seetharaman and Etienne-Manneville, 2018; Sun et al., 2016). A number of excellent reviews discuss the molecular composition of integrin-associated and other force-stimulated (e.g. non-integrin focal adhesion, stretch-sensitive ion channel, etc.) protein complexes (Barzegari et al., 2020a,b; Doyle and Yamada, 2016; Jansen et al., 2015; Michael and Parsons, 2020).
Upon sensing mechanical stimuli, mechanosensitive molecules can initiate or modulate a wide variety of specific intracellular signaling pathways, including the Rho GTPase-related (Rafiq et al., 2019), Hippo (Barzegari et al., 2020a,b), TGFβ (Nguyen et al., 2020) and YAP/TAZ (Pocaterra et al., 2020) signaling pathways. This signaling can lead to diverse series of functional and/or structural responses in the involved cells and tissues – the most extensively investigated of these are highlighted in each section of this Review (e.g. shape changes, migratory events and differentiation) (Fig. 1).
For example, Drosophila dorsal closure relies on integrin-mediated cell-matrix adhesions for the transmission of intercellular tensile forces generated by cell constriction (Goodwin et al., 2016). When the number of these integrin-mediated focal adhesion-like structures is modified, intercellular apical force transmission is perturbed. The resulting abnormal contraction and ingression of the amniosera prevents opposing embryonic lateral epidermal cells from normally migrating toward the dorsal midline (Fig. 4C) (Goodwin et al., 2016). Besides these integrin-mediated cell-ECM adhesions, cadherin-mediated cell-cell adhesions are also crucial for proper force transmission across cells that make up the amniosera. Changes in cadherin localization and stability result in a similar failure of dorsal closure (Goodwin et al., 2017). In fact, cell-cell and cell-matrix adhesions functionally interact to promote proper mechanical signaling in these developing tissues (Goodwin et al., 2016, 2017).
Many morphogenetic events in embryogenesis are coordinated by muscle contractions, such as embryo elongation in C. elegans (Fig. 3C) and skeletogenesis in zebrafish, mouse, chick and other vertebrates (Shwartz et al., 2012). During these processes, mechanical information from muscle fibers is relayed to local environments via matrix-muscle adhesions and transmitted throughout tissues by cell-matrix and cell-cell junctions. Moreover, elongating C. elegans embryos rely on the ECM to coordinate communication between muscle, lateral epidermal and dorsal/ventral epidermal tissues (Gillard et al., 2019). In this system, mechanical signals generated by muscle contractions are transmitted to and between epidermal cells via cell-matrix molecular tendons and adherens junctions, respectively. When functional mutations alter matrix-muscle adhesion proteins (e.g. altered NOAH-1 and NOAH-2), muscle contractions cannot convey signals through molecular tendons, intracellular actin fibers fail to polarize, cells orient inadequately along the anterior-posterior axis of the embryo and mid-elongation arrest ensues (Vuong-Brender et al., 2016, 2017). These studies, together with others investigating cell adhesion complexes in developing tissues, indicate that cell-matrix interactions can regulate tissue shape using anchoring by relaying mechanical signals between cells and tissues, and by coordinating migratory events.
Migration
Cell migration is crucial for embryogenesis: cells undergo initial specification during gastrulation and can then migrate separately or as collective assemblies guided by environmental cues to reach their destinations (Friedl and Gilmour, 2009; Scarpa and Mayor, 2016; Yamada and Sixt, 2019). These cues can be biochemical, such as diffusible or substrate-bound ligands (known as chemotaxis or haptotaxis, respectively) or physical, mediated by substrate composition, topography (e.g. contact guidance) and stiffness (durotaxis), which can regulate migration and differentiation (Fig. 1B,D; Fig. 2). Advances in live-cell/tissue imaging, tunable biomaterials and in vitro models have revealed mechanisms through which the ECM can regulate a large repertoire of cell migration modalities (Feng et al., 2019; Li et al., 2017; Trappmann et al., 2017; Wang et al., 2019a,b; Yamada et al., 2019; Yamada and Sixt, 2019). In developing tissues, the ECM provides paths that can provide both directional and stop signals for coordinating cell migration.
Numerous model systems have provided insights into cell migration during embryogenesis (Scarpa and Mayor, 2016). Among these, studies of neural crest cell (NCC) development have substantially enhanced our understanding of complex ECM-cell interactions that govern migration. Triggered by cues such as substrate stiffness changes and transcription factors (Barriga et al., 2018; Hockman et al., 2019; Shellard and Mayor, 2019), NCCs display multiple migration modes [e.g. as organized groups, chains, sheets and/or relatively unorganized masses (Rozario and DeSimone, 2010; Shellard and Mayor, 2019; Theveneau and Mayor, 2011)]. In addition, NCC migration is regulated through mechanisms that include contact inhibition of migration (Bahm et al., 2017; Li et al., 2019b; Roycroft et al., 2018; Yoon et al., 2018), durotaxis (Barriga et al., 2018; Chevalier et al., 2016), and chemotaxis (Bajanca et al., 2019; Shellard et al., 2018; Szabo and Mayor, 2018).
Several recent reviews discuss coordinated ECM-NCC interactions during NCC migration in chick, mouse and Xenopus systems (Kechagia et al., 2019; Szabo and Mayor, 2018; Yamada and Sixt, 2019). We instead focus on recent studies and concepts discussing how ECM-cell interactions drive tissue formation in other systems, such as cells of the zebrafish ectoderm and mesendoderm, Xenopus mesendoderm and neurons and Drosophila myotubes, as well as in embryonic cell invasion.
Roads and maps
Embryonic cell migration can include amoeboid, mesenchymal or lobopodial 3D modes of cell migration [the multiple modes of 3D cell migration are reviewed by Yamada and Sixt (2019)]. Mesenchymal and lobopodial migration involve extensive integrin-mediated adhesion to surrounding ECM substrates, whereas amoeboid migration can involve non-specific interactions with ECM. Mesenchymal migration is characterized by cells using actin-driven lamellipodial or filopodial protrusions to adhere to, produce force against and migrate in or on the ECM (Caswell and Zech, 2018; Ghiglione et al., 2018; Plutoni et al., 2019; Sharma et al., 2018; Zeledon et al., 2019). ECM biochemical and physical properties can regulate these leading edge protrusions and the resulting directed locomotion (Love et al., 2018; Plutoni et al., 2019).
The interplay between ECM constituents and cellular migration machinery is remarkably complex, requiring various membrane-bound and secreted cellular proteins to interact functionally at the cell-ECM interface to mediate or modulate embryonic cell migration (Bjerke et al., 2014; Cheng et al., 2019; Sánchez-Sánchez et al., 2017). As portrayed in the following paragraphs, the identity and quantity of expressed proteins determine the positions of cell protrusions, the dimensions and stability of cell-ECM interfaces, and the ability of cells to sense and respond to their microenvironments.
An early step in organized migration is the formation of oriented protrusions. For example, zebrafish prechordal plate cells secrete Cthrc1a (collagen triple helix repeat containing 1a) to generate polarized protrusions that interact functionally with fibronectin, and undergo extensive directed cell migration during axis extension and head formation (Cheng et al., 2019). Depletion of cthrc1a results in failed epiboly, diminished anteroposterior axis elongation and head defects (Cheng et al., 2019). Furthering this concept, ectodermal and mesodermal cells express the membrane-bound planar cell polarity protein Vangl2 (Vang-like 2) to form oriented actin-rich protrusions, achieve proper mediolateral alignment and elongation, and establish planar cell polarity in the zebrafish gastrula. Migrating vangl2 mutant cells lack directionality, but increasing the fibronectin content of the surrounding ECM can repolarize the large protrusions of vangl2 mutants and restore directional migration (Love et al., 2018).
The stability and dimensionality of cell-ECM interfaces influence cell migration. Drosophila embryonic hemocytes require prolonged, stable cell-ECM interactions to migrate along the ventral nerve cord. Hemocytes achieve stability of lamellipodia and prolonged cell-ECM adhesion states through autocrine deposition of laminin, a major basement membrane structural glycoprotein, in a Rab8-regulated manner (Sánchez-Sánchez et al., 2017). Exemplifying the importance of cell-ECM adhesive contact area, Xenopus mesendodermal cells increase expression of focal adhesion kinase during directional migration (Bjerke et al., 2014; Hens and DeSimone, 1995). Antisense morpholino oligonucleotide knockdown of focal adhesion kinase reduces the area of focal adhesion contacts, causes aberrant actin organization and uncoordinated cell protrusions, and, most notably, reduces spreading/traction forces and migration speed (Bjerke et al., 2014). At the tissue level, this focal adhesion kinase reduction results in defective neurulation, axial elongation and somitogenesis (Bjerke et al., 2014).
To reach their destination and circumvent and/or break down potential ECM barriers, cells must sense and react to their microenvironment. Recent reviews describe how mechanosensitive retinal ganglion cell axons in the developing Xenopus brain traffic along stiffness gradients to achieve proper anatomic distribution (Buchsbaum and Cappello, 2019; Long and Huttner, 2019). The concept of ECM stiffness and viscoelasticity regulating cell migration has been characterized in a variety of NCC systems (Barriga et al., 2018; Barriga and Mayor, 2019; Chaudhuri et al., 2015; Wang et al., 2019a).
Stop signs and road blocks
Recent publications illustrate the intricate ECM-cell exchanges of information that can either prevent initiation of migration or ensure that migrating cells halt at their intended destination. Common mechanisms through which the ECM regulates these events include providing zones of uniform cytokine/growth factor concentration in place of gradients during chemotaxis or haptotaxis (Colak-Champollion et al., 2019; Malhotra et al., 2018; Sieg et al., 2000), forming physical barriers to restrict cell movement (Renkawitz et al., 2019; Zanotelli et al., 2019), and providing signaling cues to alter the cellular machinery responsible for protrusion, adhesion and/or traction force generation (Richier et al., 2018; Sekine et al., 2012; Sieg et al., 2000; Yamada and Sixt, 2019). These ECM signals cooperate with other forms of signaling during development.
A well-characterized example of attractant-guided cell migration involves the Cxcl12-Cxcr4/Cxcr7 signaling pathway. During embryogenesis, migrating mouse, chick, zebrafish and human primordial cells express the chemokine receptor Cxcr4. Stationary somatic cells express both the chemoattractant Cxcl12 to guide trafficking primordial cells, as well as the chemokine receptor Cxcr7 to endocytose excess Cxcl12 (Boldajipour et al., 2008; Breau et al., 2012; Colak-Champollion et al., 2019; Dalle Nogare et al., 2014; Friedl and Gilmour, 2009; Lei et al., 2019; Neelathi et al., 2018; Zheng et al., 2018). In these systems, the concentration of Cxcl12 serves as either a ‘green light’ or a ‘stop sign’ for migrating primordial cells. This chemokine signaling pathway regulates several cellular events not only in development, but also in disease (Del Molino Del Barrio et al., 2018; Pluchino et al., 2018; Teicher and Fricker, 2010; Zheng et al., 2019). In addition, numerous cytokines/chemokines regulate processes in cell migration and embryogenesis that are beyond the scope of this Review, but are discussed elsewhere (Devreotes and Horwitz, 2015; Haeger et al., 2015).
To navigate through physical barriers, cells remodel either their cytoskeleton or the surrounding ECM. The cell nucleus is a relatively large and stiff cytoplasmic organelle that limits the capacity of migrating cells to squeeze through barriers, such as ECM pores that can serve as road blocks to migrating cells (Fig. 2B) (Denais et al., 2016; Harada et al., 2014; Yamada and Sixt, 2019). The structural proteins lamins A and C are major contributors to nucleoskeletal stiffness, and their expression correlates with the ability of a cell to navigate through spaces and pores in the ECM (Bone and Starr, 2016; Chen et al., 2018a,b; Das et al., 2019; Harada et al., 2014; Renkawitz et al., 2019). Alternatively, cells can either proteolytically or non-proteolytically deform their microenvironment (Gifford and Itoh, 2019; van Helvert et al., 2018; Wang et al., 2019c; Wolf and Friedl, 2011). This proteolytic mechanism is nicely portrayed by dorsally migrating endodermal cells during zebrafish gastrulation (Hu et al., 2018). These cells regulate Mmp14a/b (matrix metalloproteinase 14) levels through expression of Gpc4 (glypican 4). In gpc4 zebrafish mutants, loss of functional Gpc4 impairs cell migration due to increased amounts of ECM fibronectin and laminin caused by diminished proteolytic degradation (Hu et al., 2018).
A particularly important determinant of cell trafficking is the presence and activation of functional cellular migration machinery. For example, modifying the cellular contractile apparatus can have an even greater effect on cell migration than altering the surrounding ECM microenvironment, as demonstrated during contact guidance of cells migrating in 3D collagen matrices (Nuhn et al., 2018). The complex interplay between signaling, adhesions and matrix assembly is exemplified by the transcription factor Pitx2 (paired-like homeodomain 2) and its downstream activities. Classically characterized by its involvement in left-right patterning during asymmetric morphogenesis, recent insights suggest that Pitx2c serves an additional key role in chemokine-ECM-integrin-dependent mesendodermal migration in early embryogenesis (Collins et al., 2018). Using pitx2c-deletion mutant zebrafish embryos, Pitx2c expression has been shown to promote mesendodermal cell migration by coordinating Cxcl12b chemokine signaling, integrin β1 expression and ECM fibronectin assembly (Collins et al., 2018). Pitx2 is not only crucial for zebrafish embryogenesis, but also for mouse (Mitiku and Baker, 2007), Xenopus (Ding et al., 2017), chicken (Torlopp et al., 2014) and human development (Hendee et al., 2018; Yin et al., 2014; Zhang et al., 2019).
Further demonstrating the ability of the ECM to modify the cellular migration machinery, ECM cues can actively suppress sensory actin-rich filipodia in an integrin-dependent manner (Richier et al., 2018). This role is observed in elongating Drosophila myotube tips that probe the ECM to locate ‘stop signs’ (the matrix overlying tendon cells) and establish sites of tendon attachment during lateral transverse muscle development. Exploratory and sensory behavior of cellular protrusions, cell-substrate adhesion and cell traction-force generation involve a multitude of signaling mechanisms that contribute to cell migration during embryogenesis (Devreotes and Horwitz, 2015; Doyle and Yamada, 2016; Lauffenburger and Horwitz, 1996; Ridley et al., 2003).
Conversely, a cell that is initially restrained by a barrier such as the underlying basement membrane can use multiple strategies to breach it. A particularly striking developmental example – reminiscent of human cancer cell invasion – is used by the C. elegans anchor cell for vulval invasion (Fig. 3D). This cell initially produces focal points of degradation of the basement membrane using invadopodia, which then proceeds to a large breach using a combination of matrix metalloproteinase degradation and forces generated by actin polymerization driven by Arp2/3 (Cáceres et al., 2018; Kelley et al., 2019; Naegeli et al., 2017). Interestingly, even if protease function is inhibited, the local deformation forces fueled by local mitochondrial enrichment and expanded by lysosomal fusion to form a large protrusion are ultimately sufficient to breach the basement membrane barrier to permit invasion (Sherwood and Plastino, 2018).
Morphology and polarity
In addition to its well-known role as a scaffold (Frantz et al., 2010) the ECM can regulate morphological properties of cells and tissues via a variety of mechanical cues. Cells drive morphogenesis through a series of changes in three-dimensional shape (Fig. 1C), orientation and position [e.g. columnar-to-cuboidal (Diaz-de-la-Loza et al., 2018), cuboidal-to-columnar (Balaji et al., 2019), polarity, intercalation (Chen et al., 2019), etc.] to provide a diverse toolbox for shaping the developing embryo.
The ECM uses this toolbox to help coordinate development. Matrix physical properties, e.g. stiffness, elasticity, density and fiber orientation (Fig. 2) (Chen et al., 2019; Chlasta et al., 2017; Diaz-de-la-Loza et al., 2018), influence local cell and tissue shape and polarity.
Flattening
Recent advances in 3D cell culture techniques have revealed how cell shape changes can drive morphogenesis (Diaz-de-la-Loza et al., 2018; Doyle and Yamada, 2016; Yamada and Sixt, 2019). Cell shape change, along with oriented cell division (Godard and Heisenberg, 2019) and polarized cell intercalation (Huebner and Wallingford, 2018), contribute to driving epithelial elongation in development.
In Drosophila wing and leg elongation, after the peripodial layer is removed, ECM remodeling is responsible for initiating wing elongation (Fig. 4B). Triggered by this matrix remodeling, neighboring cells flatten (completing a columnar-to-cuboidal transition) to drive lateral tissue expansion (Diaz-de-la-Loza et al., 2018).
Furthermore, both cell and tissue shape are influenced by mechanical properties of the basement membrane in the developing Drosophila follicle (Chlasta et al., 2017). A TGFβ-driven decrease in basement membrane stiffness is associated with flattening of anterior follicle cells, which contributes to the final elongated morphology of the egg chamber (Fig. 4D) (Chlasta et al., 2017). In other species, several investigations have identified that similar cell flattening events are responsible for embryonic morphogenetic changes in zebrafish (Bruce, 2016; Dasgupta et al., 2018; Delile et al., 2017) and Xenopus (Kloc and Kubiak, 2014).
Constraining
While the ECM promotes flattening of the anterior Drosophila follicle cells, it simultaneously constrains the posterior follicle cells to induce a cuboidal-to-columnar shape transition (Balaji et al., 2019; Chlasta et al., 2017). Specifically, between stages 6 and 9 of egg chamber development, the basement membrane physically constrains the underlying cells at the posterior pole. In conjunction with medial myosin II contraction and adherens junction remodeling, follicle cells undergo a resulting cuboidal-to-columnar transition (Fig. 4D) (Balaji et al., 2019).
Beyond its effects on individual cells, fibrillar structures of the ECM provide anisotropic constraining forces to drive and orient morphogenetic events at the tissue level (Isabella and Horne-Badovinac, 2016; Vuong-Brender et al., 2017). This is classically illustrated by the polarized fibrillar basement membrane serving as a ‘molecular corset’ surrounding the growing Drosophila egg chamber (Gutzeit et al., 1991; Isabella and Horne-Badovinac, 2016; Ramos-Lewis and Page-McCaw, 2019). In this case, the basement membrane physically constrains outward expansion of the egg chamber to force growth to occur along the anterior-posterior axis (Fig. 4D) (Chen et al., 2019; Isabella and Horne-Badovinac, 2016; Ramos-Lewis and Page-McCaw, 2019). A similar phenomenon is observed in C. elegans, in which the ECM not only constrains the shape of the embryo, but also provides crucial attachment sites for contracting muscle fibers (Fig. 3C) (Vuong-Brender et al., 2016, 2017).
Polarizing
The ECM provides information regulating cell orientation and polarity. For example, stiffness cues provided by the basement membrane of the developing Drosophila follicle regulate polarized reorientation of anterior follicle cells (Chen et al., 2019). When these cues are compromised, Src tyrosine kinase-driven remodeling of cell-cell junctions is altered, anterior follicle cells randomly orient along the anterior-posterior axis and the organ fails to achieve its appropriate shape (Chen et al., 2019).
Polarity and orientation of cells and tissues is closely regulated by several factors, including adhesion complexes, actin organization, actomyosin contraction and ECM signals (Gillard et al., 2019). For example, the ECM surrounding the elongating C. elegans embryo is essential for establishing bipolar planar polarity of the apical PAR module (a protein complex responsible for organizing cell junctions at the apical cell surface) of lateral epidermal cells (Gillard et al., 2019). The resulting planar organization of actin helps to orient cell-shape changes and polarize the developing embryo. Indeed, genetic depletion of the ECM protein perlecan results in altered actin planar polarity and cell orientation (Gillard et al., 2019).
Many of the investigations characterizing the ability of the ECM to regulate polarity and orientation in development are limited to the Drosophila and C. elegans models, as described in this section. This is probably because of the complexity of comprehensively analyzing 3D in vivo embryogenic events in mammals (Chan et al., 2017; Herrera-Perez and Kasza, 2019; Shahbazi et al., 2019). Investigations in this field may soon rapidly expand as emerging techniques provide the ability to manipulate in vivo mechanical signals directly in the developing embryo (Chan et al., 2017; Stooke-Vaughan and Campàs, 2018).
Differentiation
Specific ECM microenvironmental niches, biochemical cues and mechanical signals are intriguing candidate factors for guiding the differentiation of pluripotent embryonic stem cells or induced pluripotent stem cells, as well as fate-restricted adult stem cells (e.g. mesenchymal, hematopoietic, neural or epithelial) (Harvey et al., 2019; Liu et al., 2019; Smith et al., 2018; Zhu and Huangfu, 2013). The ECM regulates stem cells through a complex mixture of mechanical cues that include matrix geometry and chirality (Chen et al., 2018a; Dong et al., 2019; von Erlach et al., 2018; Wei et al., 2019), rigidity (Gerardo et al., 2019), ligand density (Lee et al., 2019) and topography (Fig. 2) (Abagnale et al., 2015; Hulshof et al., 2017). A number of recent reviews have addressed the broad topic of ECM mechanical properties that can regulate stem cell fate (Kumar et al., 2017; Kumari et al., 2018; Smith et al., 2018; Vining and Mooney, 2017). Consequently, this Review provides a brief overview of key concepts in this field and refers readers to recent relevant literature for additional information.
Stem cell fate specification
In addition to promoting stem cell support within a specific microenvironmental niche, the mechanical properties of the ECM can strongly influence a variety of stem cell behaviors including maintenance, self-renewal, proliferation and differentiation (Fig. 1D) (Gattazzo et al., 2014). For example, manipulating substrate elasticity/stiffness alters mesenchymal stem cell (MSC) fate (Darnell et al., 2018a,b; Gerardo et al., 2019; Lee et al., 2019; Engler et al., 2006) and embryonic stem cell (ESC) fate (Przybyla et al., 2016), driving cell lineage commitment towards tissues with similar physical properties, e.g. towards soft adipose or stiff osseous tissue. ECM topographical cues, including geometric chirality (Dong et al., 2019; Wei et al., 2019) and ligand density (Fig. 2E) (Darnell et al., 2018a,b; Lee et al., 2019), as well as matrix stress-relaxation cues (Darnell et al., 2018a,b), influence MSC differentiation. For example, transcriptional changes occur in mouse MSCs when cultured on substrates of varying adhesion ligand density, stiffness and stress relaxation rate, which drive osteogenic or hematopoietic differentiation (Darnell et al., 2018a,b). Similar studies with ESCs characterize the role of surface roughness of the substrate (Jaggy et al., 2015) and the size of landscape features (Fig. 2A) (Lapointe et al., 2013; Macgregor et al., 2017; Reimer et al., 2016) in modulating ESC differentiation potential. Various molecular pathways (e.g. Notch and Wnt/β-catenin signaling, among others) are hypothesized to serve prominent roles in regulating the responses of stem cells to substrate mechanics (Harvey et al., 2019; Kumari et al., 2018; Przybyla et al., 2016; Smith et al., 2018; Totaro et al., 2017). Consequently, the biophysical properties of the ECM regulate stem cell differentiation through a coordinated balance of multiple physical mechanisms, the complexity and mechanisms of which are only starting to be characterized (Harvey et al., 2019; Muncie and Weaver, 2018; Smith et al., 2018; Wen et al., 2014).
Potential applications
Compared with adult stem cells, the body of literature describing biophysical contributions of the ECM to pluripotent ESC differentiation for potential clinical application is currently strikingly sparse (Kumari et al., 2018), with many investigations focusing on the topographical features described above. Such topographical studies move us closer to possible future use of ESCs in clinical tissue regeneration, but a complicating feature is that ESCs are known to respond to a wide range of both biomechanical and biochemical cues (Dogan, 2018). Consequently, even though some adult stem cells, such as hematopoietic stem cells, have achieved clear success in regenerative medicine (Iida et al., 2019), the clinical use of ESCs remains controversial for both ethical and practical reasons (Prentice, 2019). To further our understanding of embryonic development and to continue making progress towards potential clinical applications of embryonic stem cells, the complex interplay between the ECM and ESCs should be better characterized in terms of ECM roles and mechanisms for providing specific microenvironmental niches and biomechanical regulatory mechanisms that can guide cell fate.
Conclusions and future perspectives
Over the past decade, classical and innovative research approaches and techniques (Box 2) have identified many diverse biophysical and mechanical roles for the ECM during morphogenesis of many organisms and model systems, including zebrafish, Drosophila, C. elegans and Xenopus. Considerably less is known about the biophysical regulation of embryos developing in utero, although in ovo studies in avian embryos (Gandhi and Bronner, 2018) and new intrauterine methods for mammalian embryos (Beronja et al., 2010; Iwashita et al., 2014) are providing new opportunities to overcome the technical problems of smaller sample size, inaccessibility and long gestational periods.
Box 2. Techniques for analyzing roles of ECM in development.
Gene ablation and overexpression altering ECM composition (George et al., 1993; Liu et al., 2020; Oh et al., 2013; Schinzel et al., 2019; Terajima et al., 2019; Wang et al., 2019c)
Crosslinking of ECM constituents (Deo et al., 2020; Petrie et al., 2012; Piersma et al., 2020; Vallet and Ricard-Blum, 2019)
Alteration in alignment, pore size or other physical parameters (Paul et al., 2019; Wolf et al., 2013; Yamada and Sixt, 2019)
Experimentally induced individual protein degradation (Cavanaugh et al., 2020; Li et al., 2019a; Reynders et al., 2020)
Specific antibodies, pharmacological agents and other inhibitors (Afasizheva et al., 2016; Kapoor et al., 2020; Lu et al., 2020; Valiente-Alandi et al., 2018)
Optogenetic activation or depletion of proteins (Baaske et al., 2019; Liu et al., 2016; Reynders et al., 2020)
3D tissue and organ culture (Clevers, 2016; Yamada and Cukierman, 2007)
Atomic force microscopy (AFM) and microrheology (Alcaraz et al., 2018; Staunton et al., 2019; Viji Babu et al., 2019)
Laser ablation (Balcioglu et al., 2016; Goodwin et al., 2016; Ilina et al., 2011)
Force-sensing molecules [e.g. chimeras with vinculin, talin or peptides (Brockman et al., 2018; Cost et al., 2019; Curry et al., 2018; Grashoff et al., 2010; Rothenberg et al., 2018)]
Local force determination via analyzing droplet deformation (Campas et al., 2014; Serwane et al., 2017)
Local application of force [e.g. by magnetic beads or optical tweezers (Herath et al., 2014; Honarmandi et al., 2011; Jones et al., 2015; Roca-Cusachs et al., 2009)]
Adding to the complexity of species differences in development, the ECM is not static during embryogenesis. Developing organs and tissues interact with similarly dynamically changing matrices throughout embryogenesis (Loganathan et al., 2016). To address some of these hurdles, in vitro bioengineered models recapitulating key milestones of mammalian development provide preliminary insights into the mechanisms of ECM mechanical regulation (Vianello and Lutolf, 2019). However, such models merely skim the surface of the intricate mechanical and molecular signaling systems in embryogenesis.
What controls the changing biophysical properties of the ECM at progressive developmental stages? What molecular mechanisms allow the ECM and the cells that synthesize ECM to sense and respond to cues from cells and tissues? Are these mechanisms consistent between different tissue types? Are such mechanisms conserved between different organisms? These and many other unanswered questions, combined with rapidly emerging new techniques to explore these topics, make this an exciting time for the field of ECM developmental biology. The field is likely to move toward increasingly quantitative approaches involving directly quantifying changing ECM composition and physical parameters as development proceeds, combined with mathematical modeling to characterize mechanisms and generate new testable hypotheses. ECM molecules continue to be identified as therapeutic and prognostic targets in disease (Theocharis et al., 2019). Approaches to precisely control synthetic ECM forces and properties (van Oosten et al., 2019; Wu et al., 2018) are emerging and tissue engineering strategies focusing on biophysical properties of the ECM (Petersen et al., 2018) are rapidly progressing. Novel methods to produce completely autologous implants (Edri et al., 2019) are being explored. Besides ultimately gaining a satisfyingly deep mechanistic understanding of the roles of ECM in development, we can hope to begin to link the basic biophysics of ECM embryology to the clinical field of regenerative medicine.
Acknowledgements
The authors apologize for not being able to cite many other excellent publications due to space limitations. We thank Rei Sekiguchi and Shaohe Wang for helpful comments.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
Research in the authors’ laboratory is supported by the Intramural Research Program of the National Institutes of Health, National Institute of Dental and Craniofacial Research. Deposited in PMC for immediate release.
References
- Abagnale G., Steger M., Nguyen V. H., Hersch N., Sechi A., Joussen S., Denecke B., Merkel R., Hoffmann B., Dreser A. et al. (2015). Surface topography enhances differentiation of mesenchymal stem cells towards osteogenic and adipogenic lineages. Biomaterials 61, 316-326. 10.1016/j.biomaterials.2015.05.030 [DOI] [PubMed] [Google Scholar]
- Accogli A., Calabretta S., St-Onge J., Boudrahem-Addour N., Dionne-Laporte A., Joset P., Azzarello-Burri S., Rauch A., Krier J., Fieg E. et al. (2019). De Novo pathogenic variants in N-cadherin cause a syndromic neurodevelopmental disorder with corpus collosum, axon, cardiac, ocular, and genital defects. Am. J. Hum. Genet. 105, 854-868. 10.1016/j.ajhg.2019.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afasizheva A., Devine A., Tillman H., Fung K. L., Vieira W. D., Blehm B. H., Kotobuki Y., Busby B., Chen E. I. and Tanner K. (2016). Mitogen-activated protein kinase signaling causes malignant melanoma cells to differentially alter extracellular matrix biosynthesis to promote cell survival. BMC Cancer 16, 186 10.1186/s12885-016-2211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts B., Johnson A., Lewis J., Morgan D., Raff M., Roberts K. and Walter P. (2014). Molecular Biology of the Cell. New York, NY: Garland Science. [Google Scholar]
- Alcaraz J., Otero J., Jorba I. and Navajas D. (2018). Bidirectional mechanobiology between cells and their local extracellular matrix probed by atomic force microscopy. Semin. Cell Dev. Biol. 73, 71-81. 10.1016/j.semcdb.2017.07.020 [DOI] [PubMed] [Google Scholar]
- Anderson J. M. and Van Itallie C. M. (2009). Physiology and function of the tight junction. Cold Spring Harb. Perspect. Biol. 1, a002584 10.1101/cshperspect.a002584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arseni L., Lombardi A. and Orioli D. (2018). From structure to phenotype: impact of collagen alterations on human health. Int. J. Mol. Sci. 19, 1407 10.3390/ijms19051407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baaske J., Mühlhäuser W. W. D., Yousefi O. S., Zanner S., Radziwill G., Horner M., Schamel W. W. A. and Weber W. (2019). Optogenetic control of integrin-matrix interaction. Commun. Biol. 2, 15 10.1038/s42003-018-0264-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann M., Kukkurainen S., Hytönen V. P. and Wehrle-Haller B. (2019). Cell adhesion by integrins. Physiol. Rev. 99, 1655-1699. 10.1152/physrev.00036.2018 [DOI] [PubMed] [Google Scholar]
- Bahm I., Barriga E. H., Frolov A., Theveneau E., Frankel P. and Mayor R. (2017). PDGF controls contact inhibition of locomotion by regulating N-cadherin during neural crest migration. Development 144, 2456-2468. 10.1242/dev.147926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajanca F., Gouignard N., Colle C., Parsons M., Mayor R. and Theveneau E. (2019). In vivo topology converts competition for cell-matrix adhesion into directional migration. Nat. Commun. 10, 1518 10.1038/s41467-019-09548-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaji R., Weichselberger V. and Classen A. K. (2019). Response of Drosophila epithelial cell and tissue shape to external forces in vivo. Development 146, dev171256 10.1242/dev.171256 [DOI] [PubMed] [Google Scholar]
- Balcioglu H. E., van de Water B. and Danen E. H. (2016). Tumor-induced remote ECM network orientation steers angiogenesis. Sci. Rep. 6, 22580 10.1038/srep22580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barriga E. H. and Mayor R. (2019). Adjustable viscoelasticity allows for efficient collective cell migration. Semin. Cell Dev. Biol. 93, 55-68. 10.1016/j.semcdb.2018.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barriga E. H., Franze K., Charras G. and Mayor R. (2018). Tissue stiffening coordinates morphogenesis by triggering collective cell migration in vivo. Nature 554, 523-527. 10.1038/nature25742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzegari A., Gueguen V., Omidi Y., Ostadrahimi A., Nouri M. and Pavon-Djavid G. (2020a). The role of Hippo signaling pathway and mechanotransduction in tuning embryoid body formation and differentiation. J. Cell. Physiol. 235, 5072-5083. 10.1002/jcp.29455 [DOI] [PubMed] [Google Scholar]
- Barzegari A., Omidi Y., Ostadrahimi A., Gueguen V., Meddahi-Pelle A., Nouri M. and Pavon-Djavid G. (2020b). The role of Piezo proteins and cellular mechanosensing in tuning the fate of transplanted stem cells. Cell Tissue Res. (in press) 10.1007/s00441-020-03191-z [DOI] [PubMed] [Google Scholar]
- Bateman J. F., Boot-Handford R. P. and Lamande S. R. (2009). Genetic diseases of connective tissues: cellular and extracellular effects of ECM mutations. Nat. Rev. Genet. 10, 173-183. 10.1038/nrg2520 [DOI] [PubMed] [Google Scholar]
- Beleggia F., Li Y., Fan J., Elcioglu N. H., Toker E., Wieland T., Maumenee I. H., Akarsu N. A., Meitinger T., Strom T. M. et al. (2015). CRIM1 haploinsufficiency causes defects in eye development in human and mouse. Hum. Mol. Genet. 24, 2267-2273. 10.1093/hmg/ddu744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beronja S., Livshits G., Williams S. and Fuchs E. (2010). Rapid functional dissection of genetic networks via tissue-specific transduction and RNAi in mouse embryos. Nat. Med. 16, 821-827. 10.1038/nm.2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhakta H. H., Refai F. H. and Avella M. A. (2019). The molecular mechanisms mediating mammalian fertilization. Development 146, dev176966 10.1242/dev.176966 [DOI] [PubMed] [Google Scholar]
- Bharathan N. K. and Dickinson A. J. G. (2019). Desmoplakin is required for epidermal integrity and morphogenesis in the Xenopus laevis embryo. Dev. Biol. 450, 115-131. 10.1016/j.ydbio.2019.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerke M. A., Dzamba B. J., Wang C. and DeSimone D. W. (2014). FAK is required for tension-dependent organization of collective cell movements in Xenopus mesendoderm. Dev. Biol. 394, 340-356. 10.1016/j.ydbio.2014.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldajipour B., Mahabaleshwar H., Kardash E., Reichman-Fried M., Blaser H., Minina S., Wilson D., Xu Q. and Raz E. (2008). Control of chemokine-guided cell migration by ligand sequestration. Cell 132, 463-473. 10.1016/j.cell.2007.12.034 [DOI] [PubMed] [Google Scholar]
- Bone C. R. and Starr D. A. (2016). Nuclear migration events throughout development. J. Cell Sci. 129, 1951-1961. 10.1242/jcs.179788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw A. D. (2020). Regulation of cell behavior by extracellular proteins. In Principles of Tissue Engineering (Fifth Edition) (ed. Lanza R., Langer R., Vacanti J. P. and Atala A.), pp. 205-215. Cambridge: Academic Press. [Google Scholar]
- Breau M. A., Wilson D., Wilkinson D. G. and Xu Q. (2012). Chemokine and Fgf signalling act as opposing guidance cues in formation of the lateral line primordium. Development 139, 2246-2253. 10.1242/dev.080275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockman J. M., Blanchard A. T., Pui-Yan V. M., Derricotte W. D., Zhang Y., Fay M. E., Lam W. A., Evangelista F. A., Mattheyses A. L. and Salaita K. (2018). Mapping the 3D orientation of piconewton integrin traction forces. Nat. Methods 15, 115-118. 10.1038/nmeth.4536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N. H. (2011). Extracellular matrix in development: insights from mechanisms conserved between invertebrates and vertebrates. Cold Spring Harb. Perspect. Biol. 3, a005082 10.1101/cshperspect.a005082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce A. E. (2016). Zebrafish epiboly: spreading thin over the yolk. Dev. Dyn. 245, 244-258. 10.1002/dvdy.24353 [DOI] [PubMed] [Google Scholar]
- Buchsbaum I. Y. and Cappello S. (2019). Neuronal migration in the CNS during development and disease: insights from in vivo and in vitro models. Development 146, dev163766 10.1242/dev.163766 [DOI] [PubMed] [Google Scholar]
- Cáceres R., Bojanala N., Kelley L. C., Dreier J., Manzi J., Di Federico F., Chi Q., Risler T., Testa I., Sherwood D. R. et al. (2018). Forces drive basement membrane invasion in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 115, 11537-11542. 10.1073/pnas.1808760115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campàs O., Mammoto T., Hasso S., Sperling R. A., O'Connell D., Bischof A. G., Maas R., Weitz D. A., Mahadevan L. and Ingber D. E. (2014). Quantifying cell-generated mechanical forces within living embryonic tissues. Nat. Methods 11, 183-189. 10.1038/nmeth.2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell P. T. and Zech T. (2018). Actin-based cell protrusion in a 3D matrix. Trends Cell Biol. 28, 823-834. 10.1016/j.tcb.2018.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh K. E., Oakes P. W. and Gardel M. L. (2020). Optogenetic control of RhoA to probe subcellular mechanochemical circuitry. Curr. Protoc. Cell Biol. 86, e102 10.1002/cpcb.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C. J., Heisenberg C. P. and Hiiragi T. (2017). Coordination of morphogenesis and cell-fate specification in development. Curr. Biol. 27, R1024-R1035. 10.1016/j.cub.2017.07.010 [DOI] [PubMed] [Google Scholar]
- Chan C. J., Costanzo M., Ruiz-Herrero T., Mönke G., Petrie R. J., Bergert M., Diz-Muñoz A., Mahadevan L. and Hiiragi T. (2019). Hydraulic control of mammalian embryo size and cell fate. Nature 571, 112-116. 10.1038/s41586-019-1309-x [DOI] [PubMed] [Google Scholar]
- Chaudhuri O., Gu L., Darnell M., Klumpers D., Bencherif S. A., Weaver J. C., Huebsch N. and Mooney D. J. (2015). Substrate stress relaxation regulates cell spreading. Nat. Commun. 6, 6364 10.1038/ncomms7365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Kumar G., Co C. C. and Ho C. C. (2018a). Author correction: geometric control of cell migration. Sci. Rep. 8, 15257 10.1038/s41598-018-33004-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Jiang F., Qiao Y., Li H., Wei Z., Huang T., Lan J., Xia Y. and Li J. (2018b). Nucleoskeletal stiffness regulates stem cell migration and differentiation through lamin A/C. J. Cell. Physiol. 233, 5112-5118. 10.1002/jcp.26336 [DOI] [PubMed] [Google Scholar]
- Chen Q., Shi J., Tao Y. and Zernicka-Goetz M. (2018c). Tracing the origin of heterogeneity and symmetry breaking in the early mammalian embryo. Nat. Commun. 9, 1819 10.1038/s41467-018-04155-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D. Y., Crest J., Streichan S. J. and Bilder D. (2019). Extracellular matrix stiffness cues junctional remodeling for 3D tissue elongation. Nat. Commun. 10, 3339 10.1038/s41467-019-10874-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X. N., Shao M. and Shi D. L. (2019). Collagen triple helix repeat containing 1a (Cthrc1a) regulates cell adhesion and migration during gastrulation in zebrafish. Exp. Cell Res. 381, 112-120. 10.1016/j.yexcr.2019.04.033 [DOI] [PubMed] [Google Scholar]
- Chevalier N. R., Gazguez E., Bidault L., Guilbert T., Vias C., Vian E., Watanabe Y., Muller L., Germain S., Bondurand N. et al. (2016). How tissue mechanical properties affect enteric neural crest cell migration. Sci. Rep. 6, 20927 10.1038/srep20927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chighizola M., Dini T., Lenardi C., Milani P., Podesta A. and Schulte C. (2019). Mechanotransduction in neuronal cell development and functioning. Biophys. Rev. 11, 701-720. 10.1007/s12551-019-00587-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlasta J., Milani P., Runel G., Duteyrat J.-L., Arias L., Lamiré L.-A., Boudaoud A. and Grammont M. (2017). Variations in basement membrane mechanics are linked to epithelial morphogenesis. Development 144, 4350-4362. 10.1242/dev.152652 [DOI] [PubMed] [Google Scholar]
- Churchill F. B. (1970). Hertwig, weismann, and the meaning of reduction division circa 1890. Isis 61, 429-457. 10.1086/350680 [DOI] [PubMed] [Google Scholar]
- Clevers H. (2016). Modeling development and disease with organoids. Cell 165, 1586-1597. 10.1016/j.cell.2016.05.082 [DOI] [PubMed] [Google Scholar]
- Colak-Champollion T., Lan L., Jadhav A. R., Yamaguchi N., Venkiteswaran G., Patel H., Cammer M., Meier-Schellersheim M. and Knaut H. (2019). Cadherin-mediated cell coupling coordinates chemokine sensing across collectively migrating cells. Curr. Biol. 29, 2570-2579.e2577. 10.1016/j.cub.2019.06.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins M. M., Maischein H. M., Dufourcq P., Charpentier M., Blader P. and Stainier D. Y. (2018). Pitx2c orchestrates embryonic axis extension via mesendodermal cell migration. Elife 7, e34880 10.7554/eLife.34880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condrat I., He Y., Cosgarea R. and Has C. (2018). Junctional epidermolysis bullosa: allelic heterogeneity and mutation stratification for precision medicine. Front. Med. 5, 363 10.3389/fmed.2018.00363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cost A. L., Khalaji S. and Grashoff C. (2019). Genetically encoded FRET-based tension sensors. Curr. Protoc. Cell Biol. 83, e85 10.1002/cpcb.85 [DOI] [PubMed] [Google Scholar]
- Cox L. L., Cox T. C., Moreno Uribe L. M., Zhu Y., Richter C. T., Nidey N., Standley J. M., Deng M., Blue E., Chong J. X. et al. (2018). Mutations in the Epithelial Cadherin-p120-Catenin Complex Cause Mendelian Non-Syndromic Cleft Lip with or without Cleft Palate. Am. J. Hum. Genet. 102, 1143-1157. 10.1016/j.ajhg.2018.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curry E. J., Ke K., Chorsi M. T., Wrobel K. S., Miller A. N. III, Patel A., Kim I., Feng J., Yue L., Wu Q. et al. (2018). Biodegradable piezoelectric force sensor. Proc. Natl. Acad. Sci. USA 115, 909-914. 10.1073/pnas.1710874115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalle Nogare D., Somers K., Rao S., Matsuda M., Reichman-Fried M., Raz E. and Chitnis A. B. (2014). Leading and trailing cells cooperate in collective migration of the zebrafish posterior lateral line primordium. Development 141, 3188-3196. 10.1242/dev.106690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell M., Gu L. and Mooney D. (2018a). RNA-seq reveals diverse effects of substrate stiffness on mesenchymal stem cells. Biomaterials 181, 182-188. 10.1016/j.biomaterials.2018.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell M., O'Neil A., Mao A., Gu L., Rubin L. L. and Mooney D. J. (2018b). Material microenvironmental properties couple to induce distinct transcriptional programs in mammalian stem cells. Proc. Natl. Acad. Sci. USA 115, E8368-E8377. 10.1073/pnas.1802568115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A., Barai A., Monteiro M., Kumar S. and Sen S. (2019). Nuclear softening is essential for protease-independent migration. Matrix Biol. 82, 4-19. 10.1016/j.matbio.2019.01.001 [DOI] [PubMed] [Google Scholar]
- Dasgupta A., Merkel M., Clark M. J., Jacob A. E., Dawson J. E., Manning M. L. and Amack J. D. (2018). Cell volume changes contribute to epithelial morphogenesis in zebrafish Kupffer's vesicle. Elife 7, e30963 10.7554/eLife.30963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson C. D., Wang W. Y., Zaimi I., Jayco D. K. P. and Baker B. M. (2019). Cell force-mediated matrix reorganization underlies multicellular network assembly. Sci. Rep. 9, 12 10.1038/s41598-018-37044-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Molino Del Barrio I., Wilkins G. C., Meeson A., Ali S. and Kirby J. A. (2018). Breast cancer: an examination of the potential of ACKR3 to modify the response of CXCR4 to CXCL12. Int. J. Mol. Sci. 19, 3592 10.3390/ijms19113592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delile J., Herrmann M., Peyrieras N. and Doursat R. (2017). A cell-based computational model of early embryogenesis coupling mechanical behaviour and gene regulation. Nat. Commun. 8, 13929 10.1038/ncomms13929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denais C. M., Gilbert R. M., Isermann P., McGregor A. L., te Lindert M., Weigelin B., Davidson P. M., Friedl P., Wolf K. and Lammerding J. (2016). Nuclear envelope rupture and repair during cancer cell migration. Science 352, 353-358. 10.1126/science.aad7297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo K. A., Singh K. A., Peak C. W., Alge D. L. and Gaharwar A. K. (2020). Bioprinting 101: design, fabrication, and evaluation of cell-laden 3D bioprinted scaffolds. Tissue Eng. Part A 26, 318-338. 10.1089/ten.tea.2019.0298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devreotes P. and Horwitz A. R. (2015). Signaling networks that regulate cell migration. Cold Spring Harb. Perspect. Biol. 7, a005959 10.1101/cshperspect.a005959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-de-la-Loza M. D., Ray R. P., Ganguly P. S., Alt S., Davis J. R., Hoppe A., Tapon N., Salbreux G. and Thompson B. J. (2018). Apical and basal matrix remodeling control epithelial morphogenesis. Dev. Cell 46, 23-39.e25. 10.1016/j.devcel.2018.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Colozza G., Zhang K., Moriyama Y., Ploper D., Sosa E. A., Benitez M. D. J. and De Robertis E. M. (2017). Genome-wide analysis of dorsal and ventral transcriptomes of the Xenopus laevis gastrula. Dev. Biol. 426, 176-187. 10.1016/j.ydbio.2016.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogan A. (2018). Embryonic stem cells in development and regenerative medicine. Adv. Exp. Med. Biol. 1079, 1-15. 10.1007/5584_2018_175 [DOI] [PubMed] [Google Scholar]
- Dong L., Gong J., Wang Y., He J., You D., Zhou Y., Li Q., Liu Y., Cheng K., Qian J. et al. (2019). Chiral geometry regulates stem cell fate and activity. Biomaterials 222, 119456 10.1016/j.biomaterials.2019.119456 [DOI] [PubMed] [Google Scholar]
- Doyle A. D. and Yamada K. M. (2016). Mechanosensing via cell-matrix adhesions in 3D microenvironments. Exp. Cell Res. 343, 60-66. 10.1016/j.yexcr.2015.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzamba B. J. and DeSimone D. W. (2018). Extracellular matrix (ECM) and the sculpting of embryonic tissues. Curr. Top. Dev. Biol. 130, 245-274. 10.1016/bs.ctdb.2018.03.006 [DOI] [PubMed] [Google Scholar]
- Eckert J. J. and Fleming T. P. (2008). Tight junction biogenesis during early development. Biochim. Biophys. Acta 1778, 717-728. 10.1016/j.bbamem.2007.09.031 [DOI] [PubMed] [Google Scholar]
- Edri R., Gal I., Noor N., Harel T., Fleischer S., Adadi N., Green O., Shabat D., Heller L., Shapira A. et al. (2019). Personalized hydrogels for engineering diverse fully autologous tissue implants. Adv. Mater. 31, e1803895 10.1002/adma.201803895 [DOI] [PubMed] [Google Scholar]
- Engler A. J., Sen S., Sweeney H. L. and Discher D. E. (2006). Matrix elasticity directs stem cell lineage specification. Cell 126, 677-689. 10.1016/j.cell.2006.06.044 [DOI] [PubMed] [Google Scholar]
- Etournay R., Popovic M., Merkel M., Nandi A., Blasse C., Aigouy B., Brandl H., Myers G., Salbreux G., Julicher F. et al. (2015). Interplay of cell dynamics and epithelial tension during morphogenesis of the Drosophila pupal wing. Elife 4, e07090 10.7554/eLife.07090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Levine H., Mao X. and Sander L. M. (2019). Cell motility, contact guidance, and durotaxis. Soft Mat. 15, 4856-4864. 10.1039/C8SM02564A [DOI] [PubMed] [Google Scholar]
- Frantz C., Stewart K. M. and Weaver V. M. (2010). The extracellular matrix at a glance. J. Cell Sci. 123, 4195-4200. 10.1242/jcs.023820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P. and Gilmour D. (2009). Collective cell migration in morphogenesis, regeneration and cancer. Nat. Rev. Mol. Cell Biol. 10, 445-457. 10.1038/nrm2720 [DOI] [PubMed] [Google Scholar]
- Frisch S. M. and Ruoslahti E. (1997). Integrins and anoikis. Curr. Opin. Cell Biol. 9, 701-706. 10.1016/S0955-0674(97)80124-X [DOI] [PubMed] [Google Scholar]
- Gandhi S. and Bronner M. E. (2018). Insights into neural crest development from studies of avian embryos. Int. J. Dev. Biol. 62, 183-194. 10.1387/ijdb.180038sg [DOI] [PubMed] [Google Scholar]
- Garrod D. and Chidgey M. (2008). Desmosome structure, composition and function. Biochim. Biophys. Acta 1778, 572-587. 10.1016/j.bbamem.2007.07.014 [DOI] [PubMed] [Google Scholar]
- Gattazzo F., Urciuolo A. and Bonaldo P. (2014). Extracellular matrix: a dynamic microenvironment for stem cell niche. Biochim. Biophys. Acta 1840, 2506-2519. 10.1016/j.bbagen.2014.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger B., Bershadsky A., Pankov R. and Yamada K. M. (2001). Transmembrane crosstalk between the extracellular matrix and the cytoskeleton. Nat. Rev. Mol. Cell Biol. 2, 793-805. 10.1038/35099066 [DOI] [PubMed] [Google Scholar]
- George E. L., Georges-Labouesse E. N., Patel-King R. S., Rayburn H. and Hynes R. O. (1993). Defects in mesoderm, neural tube and vascular development in mouse embryos lacking fibronectin. Development 119, 1079-1091. [DOI] [PubMed] [Google Scholar]
- Gerardo H., Lima A., Carvalho J., Ramos J. R. D., Couceiro S., Travasso R. D. M., Pires das Neves R. and Graos M. (2019). Soft culture substrates favor stem-like cellular phenotype and facilitate reprogramming of human mesenchymal stem/stromal cells (hMSCs) through mechanotransduction. Sci. Rep. 9, 9086 10.1038/s41598-019-45352-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiglione C., Jouandin P., Cérézo D. and Noselli S. (2018). The Drosophila insulin pathway controls Profilin expression and dynamic actin-rich protrusions during collective cell migration. Development 145, dev161117 10.1242/dev.161117 [DOI] [PubMed] [Google Scholar]
- Gifford V. and Itoh Y. (2019). MT1-MMP-dependent cell migration: proteolytic and non-proteolytic mechanisms. Biochem. Soc. Trans. 47, 811-826. 10.1042/BST20180363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillard G., Nicolle O., Brugière T., Prigent S., Pinot M. and Michaux G. (2019). Force transmission between three tissues controls bipolar planar polarity establishment and morphogenesis. Curr. Biol. 29, 1360-1368.e1364. 10.1016/j.cub.2019.02.059 [DOI] [PubMed] [Google Scholar]
- Godard B. G. and Heisenberg C. P. (2019). Cell division and tissue mechanics. Curr. Opin. Cell Biol. 60, 114-120. 10.1016/j.ceb.2019.05.007 [DOI] [PubMed] [Google Scholar]
- Goodwin K., Ellis S. J., Lostchuck E., Zulueta-Coarasa T., Fernandez-Gonzalez R. and Tanentzapf G. (2016). Basal cell-extracellular matrix adhesion regulates force transmission during tissue morphogenesis. Dev. Cell 39, 611-625. 10.1016/j.devcel.2016.11.003 [DOI] [PubMed] [Google Scholar]
- Goodwin K., Lostchuck E. E., Cramb K. M. L., Zulueta-Coarasa T., Fernandez-Gonzalez R. and Tanentzapf G. (2017). Cell-cell and cell-extracellular matrix adhesions cooperate to organize actomyosin networks and maintain force transmission during dorsal closure. Mol. Biol. Cell 28, 1301-1310. 10.1091/mbc.e17-01-0033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin K. and Nelson C. M. (2017). Generating tissue topology through remodeling of cell-cell adhesions. Exp. Cell Res. 358, 45-51. 10.1016/j.yexcr.2017.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grashoff C., Hoffman B. D., Brenner M. D., Zhou R., Parsons M., Yang M. T., McLean M. A., Sligar S. G., Chen C. S., Ha T. et al. (2010). Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature 466, 263-266. 10.1038/nature09198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green H. J. and Brown N. H. (2019). Integrin intracellular machinery in action. Exp. Cell Res. 378, 226-231. 10.1016/j.yexcr.2019.03.011 [DOI] [PubMed] [Google Scholar]
- Guldager Kring Rasmussen D. and Karsdal M. A. (2016). Laminins. In Biochemistry of Collagens, Laminins and Elastin (ed. Karsdal M. A.), pp. 163-196. Cambridge: Academic Press. [Google Scholar]
- Gutzeit H. O., Eberhardt W. and Gratwohl E. (1991). Laminin and basement membrane-associated microfilaments in wild-type and mutant Drosophila ovarian follicles. J. Cell Sci. 100, 781-788. [DOI] [PubMed] [Google Scholar]
- Haeger A., Wolf K., Zegers M. M. and Friedl P. (2015). Collective cell migration: guidance principles and hierarchies. Trends Cell Biol. 25, 556-566. 10.1016/j.tcb.2015.06.003 [DOI] [PubMed] [Google Scholar]
- Halbleib J. M. and Nelson W. J. (2006). Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev. 20, 3199-3214. 10.1101/gad.1486806 [DOI] [PubMed] [Google Scholar]
- Harada T., Swift J., Irianto J., Shin J.-W., Spinler K. R., Athirasala A., Diegmiller R., Dingal P. C., Ivanovska I. L. and Discher D. E. (2014). Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival. J. Cell Biol. 204, 669-682. 10.1083/jcb.201308029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey A., Caretti G., Moresi V., Renzini A. and Adamo S. (2019). Interplay between metabolites and the epigenome in regulating embryonic and adult stem cell potency and maintenance. Stem Cell Rep. 13, 573-589. 10.1016/j.stemcr.2019.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendee K. E., Sorokina E. A., Muheisen S. S., Reis L. M., Tyler R. C., Markovic V., Cuturilo G., Link B. A. and Semina E. V. (2018). PITX2 deficiency and associated human disease: insights from the zebrafish model. Hum. Mol. Genet. 27, 1675-1695. 10.1093/hmg/ddy074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksen K. and Karsdal M. A. (2016). Type I Collagen. In Biochemistry of Collagens, Laminins and Elastin (ed. Karsdal M. A.), pp. 1-11. Cambridge: Academic Press. [Google Scholar]
- Hens M. D. and DeSimone D. W. (1995). Molecular analysis and developmental expression of the focal adhesion kinase pp125FAK in Xenopus laevis. Dev. Biol. 170, 274-288. 10.1006/dbio.1995.1214 [DOI] [PubMed] [Google Scholar]
- Herath S. C., Yue D., Hui S., Kim M. C., Wang D. A., Wang Q., Van Vliet K. J., Asada H. and Chen P. C. (2014). Quantification of magnetically induced changes in ECM local apparent stiffness. Biophys. J. 106, 332-341. 10.1016/j.bpj.2013.11.4459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera-Perez R. M. and Kasza K. E. (2019). Manipulating the patterns of mechanical forces that shape multicellular tissues. Physiology 34, 381-391. 10.1152/physiol.00018.2019 [DOI] [PubMed] [Google Scholar]
- Hockman D., Chong-Morrison V., Green S. A., Gavriouchkina D., Candido-Ferreira I., Ling I. T. C., Williams R. M., Amemiya C. T., Smith J. J., Bronner M. E. et al. (2019). A genome-wide assessment of the ancestral neural crest gene regulatory network. Nat. Commun. 10, 4689 10.1038/s41467-019-12687-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honarmandi P., Lee H., Lang M. J. and Kamm R. D. (2011). A microfluidic system with optical laser tweezers to study mechanotransduction and focal adhesion recruitment. Lab. Chip 11, 684-694. 10.1039/C0LC00487A [DOI] [PubMed] [Google Scholar]
- Hopf M., Göhring W., Mann K. and Timpl R. (2001). Mapping of binding sites for nidogens, fibulin-2, fibronectin and heparin to different IG modules of perlecan. J. Mol. Biol. 311, 529-541. 10.1006/jmbi.2001.4878 [DOI] [PubMed] [Google Scholar]
- Horton E. R., Astudillo P., Humphries M. J. and Humphries J. D. (2016a). Mechanosensitivity of integrin adhesion complexes: role of the consensus adhesome. Exp. Cell Res. 343, 7-13. 10.1016/j.yexcr.2015.10.025 [DOI] [PubMed] [Google Scholar]
- Horton E. R., Humphries J. D., James J., Jones M. C., Askari J. A. and Humphries M. J. (2016b). The integrin adhesome network at a glance. J. Cell Sci. 129, 4159-4163. 10.1242/jcs.192054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J. C., Shamsan G. A., Anderson S. M., McMahon M. M., Tyler L. P., Castle B. T., Heussner R. K., Provenzano P. P., Keefe D. F., Barocas V. H. et al. (2019). Modeling distributed forces within cell adhesions of varying size on continuous substrates. Cytoskeleton 76, 571-585. 10.1002/cm.21561 [DOI] [PubMed] [Google Scholar]
- Hu B., Gao Y., Davies L., Woo S., Topczewski J., Jessen J. R. and Lin F. (2018). Glypican 4 and Mmp14 interact in regulating the migration of anterior endodermal cells by limiting extracellular matrix deposition. Development 145, dev163303 10.1242/dev.163303 [DOI] [PubMed] [Google Scholar]
- Huang S. and Ingber D. E. (1999). The structural and mechanical complexity of cell-growth control. Nat. Cell Biol. 1, E131-E138. 10.1038/13043 [DOI] [PubMed] [Google Scholar]
- Huebner R. J. and Wallingford J. B. (2018). Coming to consensus: a unifying model emerges for convergent extension. Dev. Cell 46, 389-396. 10.1016/j.devcel.2018.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulshof F. F. B., Papenburg B., Vasilevich A., Hulsman M., Zhao Y., Levers M., Fekete N., de Boer M., Yuan H., Singh S. et al. (2017). Mining for osteogenic surface topographies: In silico design to in vivo osseo-integration. Biomaterials 137, 49-60. 10.1016/j.biomaterials.2017.05.020 [DOI] [PubMed] [Google Scholar]
- Humbert C., Silbermann F., Morar B., Parisot M., Zarhrate M., Masson C., Tores F., Blanchet P., Perez M. J., Petrov Y. et al. (2014). Integrin alpha 8 recessive mutations are responsible for bilateral renal agenesis in humans. Am. J. Hum. Genet. 94, 288-294. 10.1016/j.ajhg.2013.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries D. L., Grogan J. A. and Gaffney E. A. (2017). Mechanical cell-cell communication in fibrous networks: the importance of network geometry. Bull. Math. Biol. 79, 498-524. 10.1007/s11538-016-0242-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes R. O. (1992). Integrins: versatility, modulation, and signaling in cell adhesion. Cell 69, 11-25. 10.1016/0092-8674(92)90115-S [DOI] [PubMed] [Google Scholar]
- Hynes R. O. (2002). Integrins: bidirectional, allosteric signaling machines. Cell 110, 673-687. 10.1016/S0092-8674(02)00971-6 [DOI] [PubMed] [Google Scholar]
- Hynes R. O. and Yamada K. M. (2012). Extracellular Matrix Biology. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Iida M., Kodera Y., Dodds A., Ho A. Y. L., Nivison-Smith I., Akter M. R., Wu T., Lie A. K. W., Ghavamzadeh A., Kang H. J. et al. (2019). Advances in hematopoietic stem cell transplantation in the Asia-Pacific region: the second report from APBMT 2005-2015. Bone Marrow. Transplant. 54, 1973-1986. 10.1038/s41409-019-0554-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilina O., Bakker G. J., Vasaturo A., Hofmann R. M. and Friedl P. (2011). Two-photon laser-generated microtracks in 3D collagen lattices: principles of MMP-dependent and -independent collective cancer cell invasion. Phys. Biol. 8, 015010 10.1088/1478-3975/8/1/015010 [DOI] [PubMed] [Google Scholar]
- Isabella A. J. and Horne-Badovinac S. (2016). Rab10-mediated secretion synergizes with tissue movement to build a polarized basement membrane architecture for organ morphogenesis. Dev. Cell 38, 47-60. 10.1016/j.devcel.2016.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwashita M., Kataoka N., Toida K. and Kosodo Y. (2014). Systematic profiling of spatiotemporal tissue and cellular stiffness in the developing brain. Development 141, 3793-3798. 10.1242/dev.109637 [DOI] [PubMed] [Google Scholar]
- Jaggy M., Zhang P., Greiner A. M., Autenrieth T. J., Nedashkivska V., Efremov A. N., Blattner C., Bastmeyer M. and Levkin P. A. (2015). Hierarchical micro-nano surface topography promotes long-term maintenance of undifferentiated mouse embryonic stem cells. Nano Lett. 15, 7146-7154. 10.1021/acs.nanolett.5b03359 [DOI] [PubMed] [Google Scholar]
- Jansen K. A., Donato D. M., Balcioglu H. E., Schmidt T., Danen E. H. and Koenderink G. H. (2015). A guide to mechanobiology: where biology and physics meet. Biochim. Biophys. Acta 1853, 3043-3052. 10.1016/j.bbamcr.2015.05.007 [DOI] [PubMed] [Google Scholar]
- Jansen K. A., Atherton P. and Ballestrem C. (2017). Mechanotransduction at the cell-matrix interface. Semin. Cell Dev. Biol. 71, 75-83. 10.1016/j.semcdb.2017.07.027 [DOI] [PubMed] [Google Scholar]
- Jin P., Jan L. Y. and Jan Y. N. (2020). Mechanosensitive ion channels: structural features relevant to mechanotransduction mechanisms. Annu. Rev. Neurosci. (in press) 10.1146/annurev-neuro-070918-050509 [DOI] [PubMed] [Google Scholar]
- Jones C. A., Cibula M., Feng J., Krnacik E. A., McIntyre D. H., Levine H. and Sun B. (2015). Micromechanics of cellularized biopolymer networks. Proc. Natl. Acad. Sci. USA 112, E5117-E5122. 10.1073/pnas.1509663112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A., Chen C. G. and Iozzo R. V. (2020). Endorepellin evokes an angiostatic stress signaling cascade in endothelial cells. J. Biol. Chem. (in press) 10.1074/jbc.RA120.012525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kechagia J. Z., Ivaska J. and Roca-Cusachs P. (2019). Integrins as biomechanical sensors of the microenvironment. Nat. Rev. Mol. Cell Biol. 20, 457-473. 10.1038/s41580-019-0134-2 [DOI] [PubMed] [Google Scholar]
- Keller R. (2006). Mechanisms of elongation in embryogenesis. Development 133, 2291-2302. 10.1242/dev.02406 [DOI] [PubMed] [Google Scholar]
- Kelley M., Yochem J., Krieg M., Calixto A., Heiman M. G., Kuzmanov A., Meli V., Chalfie M., Goodman M. B., Shaham S. et al. (2015). FBN-1, a fibrillin-related protein, is required for resistance of the epidermis to mechanical deformation during C. elegans embryogenesis. Elife 4, e06565 10.7554/eLife.06565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L. C., Chi Q., Cáceres R., Hastie E., Schindler A. J., Jiang Y., Matus D. Q., Plastino J. and Sherwood D. R. (2019). Adaptive F-actin polymerization and localized ATP production drive basement membrane invasion in the absence of MMPs. Dev. Cell. 48, 313-328.e318. 10.1016/j.devcel.2018.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsella M. G. and Wight T. N. (2005). Perlecan: an extracellular matrix heparan sulfate proteoglycan that regulates key events in vascular development and disease. In Chemistry and Biology of Heparin and Heparan Sulfate (ed. Garg H. G. Linhardt R. J. and Hales C. A.), pp. 607-635. Amsterdam: Elsevier Science. [Google Scholar]
- Kloc M. and Kubiak J. Z. (2014). Xenopus Development. Hoboken, New Jersey: Wiley. [Google Scholar]
- Kristensen J. H. and Karsdal M. A. (2016). Elastin. In Biochemistry of Collagens, Laminins and Elastin (ed. Karsdal M. A.), pp. 197-201. Cambridge: Academic Press. [Google Scholar]
- Kumar A., Placone J. K. and Engler A. J. (2017). Understanding the extracellular forces that determine cell fate and maintenance. Development 144, 4261-4270. 10.1242/dev.158469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari S., Vermeulen S., van der Veer B., Carlier A., de Boer J. and Subramanyam D. (2018). Shaping cell fate: influence of topographical substratum properties on embryonic stem cells. Tissue Eng. Part B Rev. 24, 255-266. 10.1089/ten.teb.2017.0468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamande S. R. and Bateman J. F. (2019). Genetic disorders of the extracellular matrix. Anat Rec (Hoboken). (in press) 10.1002/ar.24086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe V. L., Fernandes A. T., Bell N. C., Stellacci F. and Stevens M. M. (2013). Nanoscale topography and chemistry affect embryonic stem cell self-renewal and early differentiation. Adv. Healthc. Mater. 2, 1644-1650. 10.1002/adhm.201200382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardennois A., Pasti G., Ferraro T., Llense F., Mahou P., Pontabry J., Rodriguez D., Kim S., Ono S., Beaurepaire E. et al. (2019). An actin-based viscoplastic lock ensures progressive body-axis elongation. Nature 573, 266-270. 10.1038/s41586-019-1509-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latimer A. and Jessen J. R. (2010). Extracellular matrix assembly and organization during zebrafish gastrulation. Matrix Biol. 29, 89-96. 10.1016/j.matbio.2009.10.002 [DOI] [PubMed] [Google Scholar]
- Lauffenburger D. A. and Horwitz A. F. (1996). Cell migration: a physically integrated molecular process. Cell 84, 359-369. 10.1016/S0092-8674(00)81280-5 [DOI] [PubMed] [Google Scholar]
- Lee P., Bax D. V., Bilek M. M. and Weiss A. S. (2014). A novel cell adhesion region in tropoelastin mediates attachment to integrin alphaVbeta5. J. Biol. Chem. 289, 1467-1477. 10.1074/jbc.M113.518381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Kassianidou E. and Kumar S. (2018). Actomyosin stress fiber subtypes have unique viscoelastic properties and roles in tension generation. Mol. Biol. Cell 29, 1992-2004. 10.1091/mbc.E18-02-0106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Stanton A. E., Tong X. and Yang F. (2019). Hydrogels with enhanced protein conjugation efficiency reveal stiffness-induced YAP localization in stem cells depends on biochemical cues. Biomaterials 202, 26-34. 10.1016/j.biomaterials.2019.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei G. Q., Wu Z. Y., Jiang W. B., Luo J., Xu H., Luo S. F., Peng Z. Y., Wang W., Chen M. and Yu L. L. (2019). Effect of CXCL12/CXCR4 on migration of decidua-derived mesenchymal stem cells from pregnancies with preeclampsia. Am. J. Reprod. Immunol. 82, e13180 10.1111/aji.13180 [DOI] [PubMed] [Google Scholar]
- Letizia A., He D., Astigarraga S., Colombelli J., Hatini V., Llimargas M. and Treisman J. E. (2019). Sidekick is a key component of tricellular adherens junctions that acts to resolve cell rearrangements. Dev. Cell 50, 313-326.e315. 10.1016/j.devcel.2019.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Eyckmans J. and Chen C. S. (2017). Designer biomaterials for mechanobiology. Nat. Mater. 16, 1164-1168. 10.1038/nmat5049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Prasanna X., Salo V. T., Vattulainen I. and Ikonen E. (2019a). An efficient auxin-inducible degron system with low basal degradation in human cells. Nat. Methods 16, 866-869. 10.1038/s41592-019-0512-x [DOI] [PubMed] [Google Scholar]
- Li Y., Vieceli F. M., Gonzalez W. G., Li A., Tang W., Lois C. and Bronner M. E. (2019b). In vivo quantitative imaging provides insights into trunk neural crest migration. Cell Rep. 26, 1489-1500.e1483. 10.1016/j.celrep.2019.01.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Liu Y., Chang Y., Seyf H. R., Henry A., Mattheyses A. L., Yehl K., Zhang Y., Huang Z. and Salaita K. (2016). Nanoscale optomechanical actuators for controlling mechanotransduction in living cells. Nat. Methods 13, 143-146. 10.1038/nmeth.3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Michowski W., Kolodziejczyk A. and Sicinski P. (2019). The cell cycle in stem cell proliferation, pluripotency and differentiation. Nat. Cell Biol. 21, 1060-1067. 10.1038/s41556-019-0384-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Kim Y. S., Richardson C. E., Tom A., Ramakrishnan C., Birey F., Katsumata T., Chen S., Wang C., Wang X. et al. (2020). Genetically targeted chemical assembly of functional materials in living cells, tissues, and animals. Science 367, 1372-1376. 10.1126/science.aay4866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loganathan R., Rongish B. J., Smith C. M., Filla M. B., Czirok A., Benazeraf B. and Little C. D. (2016). Extracellular matrix motion and early morphogenesis. Development 143, 2056-2065. 10.1242/dev.127886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long K. R. and Huttner W. B. (2019). How the extracellular matrix shapes neural development. Open Biol. 9, 180216 10.1098/rsob.180216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love A. M., Prince D. J. and Jessen J. R. (2018). Vangl2-dependent regulation of membrane protrusions and directed migration requires a fibronectin extracellular matrix. Development 145, dev165472 10.1242/dev.165472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Doyle A. D., Shinsato Y., Wang S., Bodendorfer M. A., Zheng M. and Yamada K. M. (2020). Basement membrane regulates fibronectin organization using sliding focal adhesions driven by a contractile winch. Dev. Cell 52, 631-646.e634. 10.1016/j.devcel.2020.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Schickel M. E., Stevenson M. D., Sarang-Sieminski A. L., Gooch K. J., Ghadiali S. N. and Hart R. T. (2013). Fibers in the extracellular matrix enable long-range stress transmission between cells. Biophys. J. 104, 1410-1418. 10.1016/j.bpj.2013.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macgregor M., Williams R., Downes J., Bachhuka A. and Vasilev K. (2017). The role of controlled surface topography and chemistry on mouse embryonic stem cell attachment, growth and self-renewal. Materials 10, 1081 10.3390/ma10091081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainiero F. and Santoni A. (1998). Fibronectin. In Encyclopedia of Immunology (Second Edition) (ed. Delves P. J.), pp. 909-913. Oxford: Elsevier. [Google Scholar]
- Malhotra D., Shin J., Solnica-Krezel L. and Raz E. (2018). Spatio-temporal regulation of concurrent developmental processes by generic signaling downstream of chemokine receptors. Elife 7, e33574 10.7554/eLife.33574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammoto A., Mammoto T. and Ingber D. E. (2012). Mechanosensitive mechanisms in transcriptional regulation. J. Cell Sci. 125, 3061-3073. 10.1242/jcs.093005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merle T. and Farge E. (2018). Trans-scale mechanotransductive cascade of biochemical and biomechanical patterning in embryonic development: the light side of the force. Curr. Opin. Cell Biol. 55, 111-118. 10.1016/j.ceb.2018.07.003 [DOI] [PubMed] [Google Scholar]
- Michael M. and Parsons M. (2020). New perspectives on integrin-dependent adhesions. Curr. Opin. Cell Biol. 63, 31-37. 10.1016/j.ceb.2019.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitiku N. and Baker J. C. (2007). Genomic analysis of gastrulation and organogenesis in the mouse. Dev. Cell 13, 897-907. 10.1016/j.devcel.2007.10.004 [DOI] [PubMed] [Google Scholar]
- Muncie J. M. and Weaver V. M. (2018). The physical and biochemical properties of the extracellular matrix regulate cell fate. Curr. Top. Dev. Biol. 130, 1-37. 10.1016/bs.ctdb.2018.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster S., Jain A., Mietke A., Pavlopoulos A., Grill S. W. and Tomancak P. (2019). Attachment of the blastoderm to the vitelline envelope affects gastrulation of insects. Nature 568, 395-399. 10.1038/s41586-019-1044-3 [DOI] [PubMed] [Google Scholar]
- Mylonas K. S., Hayes M., Ko L. N., Griggs C. L., Kroshinsky D. and Masiakos P. T. (2019). Clinical outcomes and molecular profile of patients with Carmi syndrome: a systematic review and evidence quality assessment. J. Pediatr. Surg. 54, 1351-1358. 10.1016/j.jpedsurg.2018.05.019 [DOI] [PubMed] [Google Scholar]
- Mythreye K. and Blobe G. C. (2009). Proteoglycan signaling co-receptors: roles in cell adhesion, migration and invasion. Cell. Signal. 21, 1548-1558. 10.1016/j.cellsig.2009.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naba A., Pearce O. M. T., Del Rosario A., Ma D., Ding H., Rajeeve V., Cutillas P. R., Balkwill F. R. and Hynes R. O. (2017). Characterization of the extracellular matrix of normal and diseased tissues using proteomics. J. Proteome Res. 16, 3083-3091. 10.1021/acs.jproteome.7b00191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naegeli K. M., Hastie E., Garde A., Wang Z., Keeley D. P., Gordon K. L., Pani A. M., Kelley L. C., Morrissey M. A., Chi Q. et al. (2017). Cell invasion in vivo via rapid exocytosis of a transient lysosome-derived membrane domain. Dev. Cell 43, 403-417.e410. 10.1016/j.devcel.2017.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelathi U. M., Dalle Nogare D. and Chitnis A. B. (2018). Cxcl12a induces snail1b expression to initiate collective migration and sequential Fgf-dependent neuromast formation in the zebrafish posterior lateral line primordium. Development 145, dev162453 10.1242/dev.162453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen J., Massoumi R. and Alliston T. (2020). CYLD, a mechanosensitive deubiquitinase, regulates TGFbeta signaling in load-induced bone formation. Bone 131, 115148 10.1016/j.bone.2019.115148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuhn J. A. M., Perez A. M. and Schneider I. C. (2018). Contact guidance diversity in rotationally aligned collagen matrices. Acta Biomater. 66, 248-257. 10.1016/j.actbio.2017.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh D. J., Kang M. H., Ooi Y. H., Choi K. R., Sage E. H. and Rhee D. J. (2013). Overexpression of SPARC in human trabecular meshwork increases intraocular pressure and alters extracellular matrix. Invest. Ophthalmol. Vis. Sci. 54, 3309-3319. 10.1167/iovs.12-11362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankov R. and Yamada K. M. (2002). Fibronectin at a glance. J. Cell Sci. 115, 3861-3863. 10.1242/jcs.00059 [DOI] [PubMed] [Google Scholar]
- Paul C. D., Hruska A., Staunton J. R., Burr H. A., Daly K. M., Kim J., Jiang N. and Tanner K. (2019). Probing cellular response to topography in three dimensions. Biomaterials 197, 101-118. 10.1016/j.biomaterials.2019.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen A., Princ A., Korus G., Ellinghaus A., Leemhuis H., Herrera A., Klaumunzer A., Schreivogel S., Woloszyk A., Schmidt-Bleek K. et al. (2018). A biomaterial with a channel-like pore architecture induces endochondral healing of bone defects. Nat. Commun. 9, 4430 10.1038/s41467-018-06504-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petreaca M. and Martins-Green M. (2011). Cell-ECM Interactions in Repair and Regeneration. In Principles of Regenerative Medicine, 2nd edn (ed. Atala A., Lanza R., Thomson J. A. and Nerem R.), pp. 19-65. Cambridge: Academic Press. [Google Scholar]
- Petrie R. J., Gavara N., Chadwick R. S. and Yamada K. M. (2012). Nonpolarized signaling reveals two distinct modes of 3D cell migration. J. Cell Biol. 197, 439-455. 10.1083/jcb.201201124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piersma B., Hayward M. K. and Weaver V. M. (2020). Fibrosis and cancer: a strained relationship. Biochim. Biophys. Acta Rev. Cancer 1873, 188356 10.1016/j.bbcan.2020.188356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro D. and Bellaiche Y. (2018). Mechanical force-driven adherens junction remodeling and epithelial dynamics. Dev. Cell 47, 3-19. 10.1016/j.devcel.2018.09.014 [DOI] [PubMed] [Google Scholar]
- Piotrowski-Daspit A. S., Nerger B. A., Wolf A. E., Sundaresan S. and Nelson C. M. (2017). Dynamics of tissue-induced alignment of fibrous extracellular matrix. Biophys. J. 113, 702-713. 10.1016/j.bpj.2017.06.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluchino N., Mamillapalli R., Moridi I., Tal R. and Taylor H. S. (2018). G-protein-coupled receptor CXCR7 is overexpressed in human and murine endometriosis. Reprod. Sci. 25, 1168-1174. 10.1177/1933719118766256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plutoni C., Keil S., Zeledon C., Delsin L. E. A., Decelle B., Roux P. P., Carréno S. and Emery G. (2019). Misshapen coordinates protrusion restriction and actomyosin contractility during collective cell migration. Nat. Commun. 10, 3940 10.1038/s41467-019-11963-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pocaterra A., Romani P. and Dupont S. (2020). YAP/TAZ functions and their regulation at a glance. J. Cell Sci. 133, jcs230425. [DOI] [PubMed] [Google Scholar]
- Pozzi A., Yurchenco P. D. and Iozzo R. V. (2017). The nature and biology of basement membranes. Matrix Biol. 57-58, 1-11. 10.1016/j.matbio.2016.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice D. A. (2019). Adult stem cells. Circ. Res. 124, 837-839. 10.1161/CIRCRESAHA.118.313664 [DOI] [PubMed] [Google Scholar]
- Proag A., Monier B. and Suzanne M. (2019). Physical and functional cell-matrix uncoupling in a developing tissue under tension. Development 146, dev172577 10.1242/dev.172577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przybyla L., Lakins J. N. and Weaver V. M. (2016). Tissue mechanics orchestrate Wnt-dependent human embryonic stem cell differentiation. Cell Stem Cell 19, 462-475. 10.1016/j.stem.2016.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin E. C., Ahmed S. T., Sehgal P., Vu V. H., Kong H. and Leckband D. E. (2020). Comparative effects of N-cadherin protein and peptide fragments on mesenchymal stem cell mechanotransduction and paracrine function. Biomaterials 239, 119846 10.1016/j.biomaterials.2020.119846 [DOI] [PubMed] [Google Scholar]
- Rafiq N. B. M., Nishimura Y., Plotnikov S. V., Thiagarajan V., Zhang Z., Shi S., Natarajan M., Viasnoff V., Kanchanawong P., Jones G. E. et al. (2019). A mechano-signalling network linking microtubules, myosin IIA filaments and integrin-based adhesions. Nat. Mater. 18, 638-649. 10.1038/s41563-019-0371-y [DOI] [PubMed] [Google Scholar]
- Ramos-Lewis W. and Page-McCaw A. (2019). Basement membrane mechanics shape development: Lessons from the fly. Matrix Biol. 75-76, 72-81. 10.1016/j.matbio.2018.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray R. P., Matamoro-Vidal A., Ribeiro P. S., Tapon N., Houle D., Salazar-Ciudad I. and Thompson B. J. (2015). Patterned anchorage to the apical extracellular matrix defines tissue shape in the developing appendages of Drosophila. Dev. Cell 34, 310-322. 10.1016/j.devcel.2015.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddig P. J. and Juliano R. L. (2005). Clinging to life: cell to matrix adhesion and cell survival. Cancer Metastasis Rev. 24, 425-439. 10.1007/s10555-005-5134-3 [DOI] [PubMed] [Google Scholar]
- Reimer A., Vasilevich A., Hulshof F., Viswanathan P., van Blitterswijk C. A., de Boer J. and Watt F. M. (2016). Scalable topographies to support proliferation and Oct4 expression by human induced pluripotent stem cells. Sci. Rep. 6, 18948 10.1038/srep18948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renkawitz J., Kopf A., Stopp J., de Vries I., Driscoll M. K., Merrin J., Hauschild R., Welf E. S., Danuser G., Fiolka R. et al. (2019). Nuclear positioning facilitates amoeboid migration along the path of least resistance. Nature 568, 546-550. 10.1038/s41586-019-1087-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynders M., Matsuura B. S., Bérouti M., Simoneschi D., Marzio A., Pagano M. and Trauner D. (2020). PHOTACs enable optical control of protein degradation. Sci. Adv. 6, eaay5064 10.1126/sciadv.aay5064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richier B., Inoue Y., Dobramysl U., Friedlander J., Brown N. H. and Gallop J. L. (2018). Integrin signaling downregulates filopodia during muscle-tendon attachment. J. Cell Sci. 131, jcs217133 10.1242/jcs.217133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley A. J., Schwartz M. A., Burridge K., Firtel R. A., Ginsberg M. H., Borisy G., Parsons J. T. and Horwitz A. R. (2003). Cell migration: integrating signals from front to back. Science 302, 1704-1709. 10.1126/science.1092053 [DOI] [PubMed] [Google Scholar]
- Ringer P., Colo G., Fassler R. and Grashoff C. (2017). Sensing the mechano-chemical properties of the extracellular matrix. Matrix Biol. 64, 6-16. 10.1016/j.matbio.2017.03.004 [DOI] [PubMed] [Google Scholar]
- Roca-Cusachs P., Gauthier N. C., Del Rio A. and Sheetz M. P. (2009). Clustering of alpha(5)beta(1) integrins determines adhesion strength whereas alpha(v)beta(3) and talin enable mechanotransduction. Proc. Natl. Acad. Sci. USA 106, 16245-16250. 10.1073/pnas.0902818106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca-Cusachs P., Conte V. and Trepat X. (2017). Quantifying forces in cell biology. Nat. Cell Biol. 19, 742-751. 10.1038/ncb3564 [DOI] [PubMed] [Google Scholar]
- Rothenberg K. E., Puranam I. and Hoffman B. D. (2018). Measurement of force-sensitive protein dynamics in living cells using a combination of fluorescent techniques. J. Vis. Exp 141, e58619 10.1242/jcs.230425 [DOI] [PubMed] [Google Scholar]
- Roycroft A., Szabó A., Bahm I., Daly L., Charras G., Parsons M. and Mayor R. (2018). Redistribution of adhesive forces through Src/FAK drives contact inhibition of locomotion in neural crest. Dev. Cell 45, 565-579.e563. 10.1016/j.devcel.2018.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozario T. and DeSimone D. W. (2010). The extracellular matrix in development and morphogenesis: a dynamic view. Dev. Biol. 341, 126-140. 10.1016/j.ydbio.2009.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruehl M., Somasundaram R., Schoenfelder I., Farndale R. W., Knight C. G., Schmid M., Ackermann R., Riecken E. O., Zeitz M. and Schuppan D. (2002). The epithelial mitogen keratinocyte growth factor binds to collagens via the consensus sequence glycine-proline-hydroxyproline. J. Biol. Chem. 277, 26872-26878. 10.1074/jbc.M202335200 [DOI] [PubMed] [Google Scholar]
- Saeidian A. H., Vahidnezhad H., Youssefian L., Sotudeh S., Sargazi M., Zeinali S. and Uitto J. (2019). Hypotrichosis with juvenile macular dystrophy: combination of whole-genome sequencing and genome-wide homozygosity mapping identifies a large deletion in CDH3 initially undetected by whole-exome sequencing-A lesson from next-generation sequencing. Mol. Genet. Genomic Med. 7, e975 10.1002/mgg3.975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelov L., Sprecher E. and Paus R. (2015). The role of P-cadherin in skin biology and skin pathology: lessons from the hair follicle. Cell Tissue Res. 360, 761-771. 10.1007/s00441-015-2114-y [DOI] [PubMed] [Google Scholar]
- Sánchez-Sánchez B. J., Urbano J. M., Comber K., Dragu A., Wood W., Stramer B. and Martin-Bermudo M. D. (2017). Drosophila embryonic hemocytes produce laminins to strengthen migratory response. Cell Rep. 21, 1461-1470. 10.1016/j.celrep.2017.10.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sand J. M. B., Genovese F. and Karsdal M. A. (2016). Type IV Collagen. In Biochemistry of Collagens, Laminins and Elastin (ed. Karsdal M. A.), pp. 31-41. Cambridge: Academic Press. [Google Scholar]
- Scarpa E. and Mayor R. (2016). Collective cell migration in development. J. Cell Biol. 212, 143-155. 10.1083/jcb.201508047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinzel R. T., Higuchi-Sanabria R., Shalem O., Moehle E. A., Webster B. M., Joe L., Bar-Ziv R., Frankino P. A., Durieux J., Pender C. et al. (2019). The Hyaluronidase, TMEM2, Promotes ER Homeostasis and Longevity Independent of the UPR(ER). Cell 179, 1306-1318.e1318. 10.1016/j.cell.2019.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweisguth F. and Corson F. (2019). Self-organization in pattern formation. Dev. Cell 49, 659-677. 10.1016/j.devcel.2019.05.019 [DOI] [PubMed] [Google Scholar]
- Seetharaman S. and Etienne-Manneville S. (2018). Integrin diversity brings specificity in mechanotransduction. Biol. Cell 110, 49-64. 10.1111/boc.201700060 [DOI] [PubMed] [Google Scholar]
- Sekiguchi R. and Yamada K. M. (2018). Basement membranes in development and disease. Curr. Top. Dev. Biol. 130, 143-191. 10.1016/bs.ctdb.2018.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine K., Kawauchi T., Kubo K., Honda T., Herz J., Hattori M., Kinashi T. and Nakajima K. (2012). Reelin controls neuronal positioning by promoting cell-matrix adhesion via inside-out activation of integrin α5β1. Neuron 76, 353-369. 10.1016/j.neuron.2012.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serwane F., Mongera A., Rowghanian P., Kealhofer D. A., Lucio A. A., Hockenbery Z. M. and Campàs O. (2017). In vivo quantification of spatially varying mechanical properties in developing tissues. Nat. Methods 14, 181-186. 10.1038/nmeth.4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazi M. N. and Zernicka-Goetz M. (2018). Deconstructing and reconstructing the mouse and human early embryo. Nat. Cell Biol. 20, 878-887. 10.1038/s41556-018-0144-x [DOI] [PubMed] [Google Scholar]
- Shahbazi M. N., Siggia E. D. and Zernicka-Goetz M. (2019). Self-organization of stem cells into embryos: a window on early mammalian development. Science 364, 948-951. 10.1126/science.aax0164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao X., Taha I. N., Clauser K. R., Gao Y. T. and Naba A. (2020). MatrisomeDB: the ECM-protein knowledge database. Nucleic Acids Res. 48, D1136-D1144. 10.1093/nar/gkz849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Halder S., Felix M., Nisaa K., Deshpande G. and Prasad M. (2018). Insulin signaling modulates border cell movement in Drosophila oogenesis. Development 145, dev166165 10.1242/dev.166165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shellard A. and Mayor R. (2019). Integrating chemical and mechanical signals in neural crest cell migration. Curr. Opin. Genet. Dev. 57, 16-24. 10.1016/j.gde.2019.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shellard A., Szabó A., Trepat X. and Mayor R. (2018). Supracellular contraction at the rear of neural crest cell groups drives collective chemotaxis. Science 362, 339-343. 10.1126/science.aau3301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood D. R. and Plastino J. (2018). Invading, leading and navigating cells in Caenorhabditis elegans: insights into cell movement in vivo. Genetics 208, 53-78. 10.1534/genetics.117.300082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shwartz Y., Farkas Z., Stern T., Aszodi A. and Zelzer E. (2012). Muscle contraction controls skeletal morphogenesis through regulation of chondrocyte convergent extension. Dev. Biol. 370, 154-163. 10.1016/j.ydbio.2012.07.026 [DOI] [PubMed] [Google Scholar]
- Sieg D. J., Hauck C. R., Ilic D., Klingbeil C. K., Schaefer E., Damsky C. H. and Schlaepfer D. D. (2000). FAK integrates growth-factor and integrin signals to promote cell migration. Nat. Cell Biol. 2, 249-256. 10.1038/35010517 [DOI] [PubMed] [Google Scholar]
- Smith L. R., Cho S. and Discher D. E. (2018). Stem cell differentiation is regulated by extracellular matrix mechanics. Physiology 33, 16-25. 10.1152/physiol.00026.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somasundaram R., Ruehl M., Tiling N., Ackermann R., Schmid M., Riecken E. O. and Schuppan D. (2000). Collagens serve as an extracellular store of bioactive interleukin 2. J. Biol. Chem. 275, 38170-38175. 10.1074/jbc.M006616200 [DOI] [PubMed] [Google Scholar]
- Somasundaram R., Ruehl M., Schaefer B., Schmid M., Ackermann R., Riecken E. O., Zeitz M. and Schuppan D. (2002). Interstitial collagens I, III, and VI sequester and modulate the multifunctional cytokine oncostatin M. J. Biol. Chem. 277, 3242-3246. 10.1074/jbc.M110011200 [DOI] [PubMed] [Google Scholar]
- Sopher R. S., Tokash H., Natan S., Sharabi M., Shelah O., Tchaicheeyan O. and Lesman A. (2018). Nonlinear elasticity of the ECM fibers facilitates efficient intercellular communication. Biophys. J. 115, 1357-1370. 10.1016/j.bpj.2018.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staunton J. R., So W. Y., Paul C. D. and Tanner K. (2019). High-frequency microrheology in 3D reveals mismatch between cytoskeletal and extracellular matrix mechanics. Proc. Natl. Acad. Sci. USA 116, 14448-14455. 10.1073/pnas.1814271116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stooke-Vaughan G. A. and Campas O. (2018). Physical control of tissue morphogenesis across scales. Curr. Opin. Genet. Dev. 51, 111-119. 10.1016/j.gde.2018.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui L., Alt S., Weigert M., Dye N., Eaton S., Jug F., Myers E. W., Julicher F., Salbreux G. and Dahmann C. (2018). Differential lateral and basal tension drive folding of Drosophila wing discs through two distinct mechanisms. Nat. Commun. 9, 4620 10.1038/s41467-018-06497-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumi A., Hayes P., D'Angelo A., Colombelli J., Salbreux G., Dierkes K. and Solon J. (2018). Adherens junction length during tissue contraction is controlled by the mechanosensitive activity of actomyosin and junctional recycling. Dev. Cell 47, 453-463.e453. 10.1016/j.devcel.2018.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Guo S. S. and Fassler R. (2016). Integrin-mediated mechanotransduction. J. Cell Biol. 215, 445-456. 10.1083/jcb.201609037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo A. and Mayor R. (2018). Mechanisms of neural crest migration. Annu. Rev. Genet. 52, 43-63. 10.1146/annurev-genet-120417-031559 [DOI] [PubMed] [Google Scholar]
- Takeichi M. (2014). Dynamic contacts: rearranging adherens junctions to drive epithelial remodelling. Nat. Rev. Mol. Cell Biol. 15, 397-410. 10.1038/nrm3802 [DOI] [PubMed] [Google Scholar]
- Teicher B. A. and Fricker S. P. (2010). CXCL12 (SDF-1)/CXCR4 pathway in cancer. Clin. Cancer. Res. 16, 2927-2931. 10.1158/1078-0432.CCR-09-2329 [DOI] [PubMed] [Google Scholar]
- Teixeira A. I., Abrams G. A., Bertics P. J., Murphy C. J. and Nealey P. F. (2003). Epithelial contact guidance on well-defined micro- and nanostructured substrates. J. Cell Sci. 116, 1881-1892. 10.1242/jcs.00383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepass U., Truong K., Godt D., Ikura M. and Peifer M. (2000). Cadherins in embryonic and neural morphogenesis. Nat. Rev. Mol. Cell Biol. 1, 91-100. 10.1038/35040042 [DOI] [PubMed] [Google Scholar]
- Terajima M., Taga Y., Cabral W. A., Liu Y., Nagasawa M., Sumida N., Kayashima Y., Chandrasekaran P., Han L., Maeda N. et al. (2019). Cyclophilin B control of lysine post-translational modifications of skin type I collagen. PLoS Genet. 15, e1008196 10.1371/journal.pgen.1008196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theocharis A. D., Manou D. and Karamanos N. K. (2019). The extracellular matrix as a multitasking player in disease. FEBS J. 286, 2830-2869. 10.1111/febs.14818 [DOI] [PubMed] [Google Scholar]
- Theveneau E. and Mayor R. (2011). Can mesenchymal cells undergo collective cell migration? The case of the neural crest. Cell. Adh. Migr. 5, 490-498. 10.4161/cam.5.6.18623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torlopp A., Khan M. A., Oliveira N. M., Lekk I., Soto-Jimenez L. M., Sosinsky A. and Stern C. D. (2014). The transcription factor Pitx2 positions the embryonic axis and regulates twinning. Elife 3, e03743 10.7554/eLife.03743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totaro A., Castellan M., Battilana G., Zanconato F., Azzolin L., Giulitti S., Cordenonsi M. and Piccolo S. (2017). YAP/TAZ link cell mechanics to Notch signalling to control epidermal stem cell fate. Nat. Commun. 8, 15206 10.1038/ncomms15206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tozluoglu M., Duda M., Kirkland N. J., Barrientos R., Burden J. J., Munoz J. J. and Mao Y. (2019). Planar differential growth rates initiate precise fold positions in complex epithelia. Dev. Cell. 51, 299-312. 10.1016/j.devcel.2019.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trappmann B., Baker B. M., Polacheck W. J., Choi C. K., Burdick J. A. and Chen C. S. (2017). Matrix degradability controls multicellularity of 3D cell migration. Nat. Commun. 8, 371 10.1038/s41467-017-00418-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiente-Alandi I., Potter S. J., Salvador A. M., Schafer A. E., Schips T., Carrillo-Salinas F., Gibson A. M., Nieman M. L., Perkins C., Sargent M. A. et al. (2018). Inhibiting fibronectin attenuates fibrosis and improves cardiac function in a model of heart failure. Circulation 138, 1236-1252. 10.1161/CIRCULATIONAHA.118.034609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallet S. D. and Ricard-Blum S. (2019). Lysyl oxidases: from enzyme activity to extracellular matrix cross-links. Essays Biochem. 63, 349-364. 10.1042/EBC20180050 [DOI] [PubMed] [Google Scholar]
- van Helvert S., Storm C. and Friedl P. (2018). Mechanoreciprocity in cell migration. Nat. Cell Biol. 20, 8-20. 10.1038/s41556-017-0012-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oosten A. S. G., Chen X., Chin L., Cruz K., Patteson A. E., Pogoda K., Shenoy V. B. and Janmey P. A. (2019). Emergence of tissue-like mechanics from fibrous networks confined by close-packed cells. Nature 573, 96-101. 10.1038/s41586-019-1516-5 [DOI] [PubMed] [Google Scholar]
- Vianello S. and Lutolf M. P. (2019). Understanding the mechanobiology of early mammalian development through bioengineered models. Dev. Cell 48, 751-763. 10.1016/j.devcel.2019.02.024 [DOI] [PubMed] [Google Scholar]
- Viji Babu P. K., Rianna C., Mirastschijski U. and Radmacher M. (2019). Nano-mechanical mapping of interdependent cell and ECM mechanics by AFM force spectroscopy. Sci. Rep. 9, 12317 10.1038/s41598-019-48566-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vining K. H. and Mooney D. J. (2017). Mechanical forces direct stem cell behaviour in development and regeneration. Nat. Rev. Mol. Cell Biol. 18, 728-742. 10.1038/nrm.2017.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Erlach T. C., Bertazzo S., Wozniak M. A., Horejs C. M., Maynard S. A., Attwood S., Robinson B. K., Autefage H., Kallepitis C., Del Rio Hernandez A. et al. (2018). Cell-geometry-dependent changes in plasma membrane order direct stem cell signalling and fate. Nat. Mater. 17, 237-242. 10.1038/s41563-017-0014-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong-Brender T. T., Yang X. and Labouesse M. (2016). C. elegans embryonic morphogenesis. Curr. Top. Dev. Biol. 116, 597-616. 10.1016/bs.ctdb.2015.11.012 [DOI] [PubMed] [Google Scholar]
- Vuong-Brender T. T. K., Suman S. K. and Labouesse M. (2017). The apical ECM preserves embryonic integrity and distributes mechanical stress during morphogenesis. Development 144, 4336-4349. 10.1242/dev.150383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Sekiguchi R., Daley W. and Yamada K. M. (2017). Patterned cell and matrix dynamics in branching morphogenesis. J. Cell Biol. 216, 559-570. 10.1083/jcb.201610048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Shi J., Wei J., Tu X. and Chen Y. (2019a). Fabrication of elastomer pillar arrays with elasticity gradient for cell migration, elongation and patterning. Biofabrication 11, 045003 10.1088/1758-5090/ab21b3 [DOI] [PubMed] [Google Scholar]
- Wang W. Y., Davidson C. D., Lin D. and Baker B. M. (2019b). Actomyosin contractility-dependent matrix stretch and recoil induces rapid cell migration. Nat. Commun. 10, 1186 10.1038/s41467-019-09121-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Rojas-Quintero J., Wilder J., Tesfaigzi Y., Zhang D. and Owen C. A. (2019c). Tissue inhibitor of metalloproteinase-1 promotes polymorphonuclear neutrophil (PMN) pericellular proteolysis by anchoring matrix metalloproteinase-8 and −9 to PMN Surfaces. J. Immunol. 202, 3267-3281. 10.4049/jimmunol.1801466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Jiang S., Si M., Zhang X., Liu J., Wang Z., Cao C., Huang J., Huang H., Chen L. et al. (2019). chirality controls mesenchymal stem cell lineage diversification through mechanoresponses. Adv. Mater. 31, e1900582 10.1002/adma.201900582 [DOI] [PubMed] [Google Scholar]
- Wen J. H., Vincent L. G., Fuhrmann A., Choi Y. S., Hribar K. C., Taylor-Weiner H., Chen S. and Engler A. J. (2014). Interplay of matrix stiffness and protein tethering in stem cell differentiation. Nat. Mater. 13, 979-987. 10.1038/nmat4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K. and Friedl P. (2011). Extracellular matrix determinants of proteolytic and non-proteolytic cell migration. Trends Cell Biol. 21, 736-744. 10.1016/j.tcb.2011.09.006 [DOI] [PubMed] [Google Scholar]
- Wolf K., Te Lindert M., Krause M., Alexander S., Te Riet J., Willis A. L., Hoffman R. M., Figdor C. G., Weiss S. J. and Friedl P. (2013). Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J. Cell Biol. 201, 1069-1084. 10.1083/jcb.201210152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak M. A. and Chen C. S. (2009). Mechanotransduction in development: a growing role for contractility. Nat. Rev. Mol. Cell Biol. 10, 34-43. 10.1038/nrm2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. Y. and Taneyhill L. A. (2019). Cadherin-7 mediates proper neural crest cell-placodal neuron interactions during trigeminal ganglion assembly. Genesis 57, e23264 10.1002/dvg.23264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Li P., Dong C., Jiang H., Bin X., Gao X., Qin M., Wang W., Bin C. and Cao Y. (2018). Rationally designed synthetic protein hydrogels with predictable mechanical properties. Nat. Commun. 9, 620 10.1038/s41467-018-02917-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin E. G., He Y., Orhan D., Pazzagli C., Emiralioglu N. and Has C. (2015). Crucial role of posttranslational modifications of integrin alpha3 in interstitial lung disease and nephrotic syndrome. Hum. Mol. Genet. 24, 3679-3688. 10.1093/hmg/ddv111 [DOI] [PubMed] [Google Scholar]
- Yamada K. M. and Cukierman E. (2007). Modeling tissue morphogenesis and cancer in 3D. Cell 130, 601-610. 10.1016/j.cell.2007.08.006 [DOI] [PubMed] [Google Scholar]
- Yamada K. M. and Sixt M. (2019). Mechanisms of 3D cell migration. Nat. Rev. Mol. Cell Biol. 20, 738-752. 10.1038/s41580-019-0172-9 [DOI] [PubMed] [Google Scholar]
- Yamada K. M., Collins J. W., Cruz Walma D. A., Doyle A. D., Morales S. G., Lu J., Matsumoto K., Nazari S. S., Sekiguchi R., Shinsato Y. et al. (2019). Extracellular matrix dynamics in cell migration, invasion and tissue morphogenesis. Int. J. Exp. Pathol. 100, 144-152. 10.1111/iep.12329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H. F., Fang X. Y., Jin C. F., Yin J. F., Li J. Y., Zhao S. J., Miao Q. and Song F. W. (2014). Identification of a novel frameshift mutation in PITX2 gene in a Chinese family with Axenfeld-Rieger syndrome. J. Zhejiang. Univ. Sci. B. 15, 43-50. 10.1631/jzus.B1300053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J., Hwang Y. S., Lee M., Sun J., Cho H. J., Knapik L. and Daar I. O. (2018). TBC1d24-ephrinB2 interaction regulates contact inhibition of locomotion in neural crest cell migration. Nat. Commun. 9, 3491 10.1038/s41467-018-05924-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotelli M. R., Rahman-Zaman A., VanderBurgh J. A., Taufalele P. V., Jain A., Erickson D., Bordeleau F. and Reinhart-King C. A. (2019). Energetic costs regulated by cell mechanics and confinement are predictive of migration path during decision-making. Nat. Commun. 10, 4185 10.1038/s41467-019-12155-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeledon C., Sun X., Plutoni C. and Emery G. (2019). The ArfGAP drongo promotes actomyosin contractility during collective cell migration by releasing myosin phosphatase from the trailing edge. Cell Rep. 28, 3238-3248.e3233. 10.1016/j.celrep.2019.08.044 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Fan J., Ho J. W., Hu T., Kneeland S. C., Fan X., Xi Q., Sellarole M. A., de Vries W. N., Lu W. et al. (2016). Crim1 regulates integrin signaling in murine lens development. Development 143, 356-366. 10.1242/dev.125591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Peng Y., Ouyang P., Liang Y., Zeng H., Wang N., Duan X. and Shi J. (2019). A novel frameshift mutation in the PITX2 gene in a family with Axenfeld-Rieger syndrome using targeted exome sequencing. BMC Med. Genet. 20, 105 10.1186/s12881-019-0840-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Wang H. and Zhou W. (2018). Modulatory effects of trophoblast-secreted CXCL12 on the migration and invasion of human first-trimester decidual epithelial cells are mediated by CXCR4 rather than CXCR7. Reprod. Biol. Endocrinol. 16, 17 10.1186/s12958-018-0333-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N., Liu W., Chen J., Li B., Liu J., Wang J., Gao Y., Shao J. and Jia L. (2019). CXCR7 is not obligatory for CXCL12-CXCR4-induced epithelial-mesenchymal transition in human ovarian cancer. Mol. Carcinog. 58, 144-155. 10.1002/mc.22916 [DOI] [PubMed] [Google Scholar]
- Zhu J. and Clark R. A. F. (2014). Fibronectin at select sites binds multiple growth factors and enhances their activity: expansion of the collaborative ECM-GF paradigm. J. Invest. Dermatol. 134, 895-901. 10.1038/jid.2013.484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z. and Huangfu D. (2013). Human pluripotent stem cells: an emerging model in developmental biology. Development 140, 705-717. 10.1242/dev.086165 [DOI] [PMC free article] [PubMed] [Google Scholar]