Abstract
Sepsis is characterized as an uncontrolled host response to infection, and it represents a serious health challenge, causing excess mortality and morbidity worldwide. The discovery of sepsis-related epigenetic and molecular mechanisms could result in improved diagnostic and therapeutic approaches, leading to a reduced overall risk for affected patients. Accumulating data show that microRNAs, non-coding RNAs, and exosomes could all be considered as novel diagnostic markers for sepsis patients. These biomarkers have been demonstrated to be involved in regulation of sepsis pathophysiology. However, epigenetic modifications have not yet been widely reported in actual clinical settings, and further investigation is required to determine their importance in intensive care patients. Further studies should be carried out to explore tissue-specific or organ-specific epigenetic RNA-based biomarkers and their therapeutic potential in sepsis patients.
Keywords: sepsis, microRNA, long non-coding RNA, exosomes, biomarker, diagnosis, septic shock, critical care
Graphical Abstract
Non-coding RNAs and exosomes present an opportunity for early diagnosis as well as an ability to interact with key points of the biological mechanisms, suggesting that measurement of non-coding RNAs and exosomes are a promising approach for intensive care patients.
Main Text
Sepsis is a potentially life-threatening condition, which occurs because of an uncontrolled response of the body to an infection.1,2 Sepsis can result in different types of organ dysfunction and the occurrence of secondary infections. The condition called “septic shock” can cause up to 40% mortality in hospitalized patients.2,3 More than 80% of the primary genetic elements have been changed in the critically ill based on genome-wide analysis of gene expression.4
In 1992, the concepts of “systemic inflammatory response syndrome” (SIRS), sepsis, severe sepsis, and septic shock were originally defined by the American College of Chest Physicians and the Society of Critical Care Medicine (SCCM). In SIRS, two or more of the following symptoms are required: temperature >38°C or <36°C, heart rate >90 beats/min, respiratory rate >20 breaths/min or arterial CO2 tension <32 mm Hg or a need for mechanical ventilation, or white blood cell count >12,000/mm3 or <4,000/mm3. SIRS arising from an infection is defined as “sepsis,” whereas “severe sepsis” includes evidence of acute organ dysfunction (e.g., hypotension, lactic acidosis, thrombocytopenia). Sepsis with persistent hypotension after fluid resuscitation is called “septic shock.”5 After much research into the concept of sepsis, and analysis of the clinical evolution of the disease, it has been concluded that SIRS, sepsis, severe sepsis, and septic shock occur along a continuum of disease severity, rather than as discrete clinical entities. Finally, in 2016, new consensus definitions of sepsis were published by the European Society of Intensive Care Medicine and the SCCM. The revised definitions included the following: the terms SIRS and severe sepsis were omitted; sepsis is now defined as life-threatening organ dysfunction caused by a dysregulated host response to infection; and septic shock is defined as a subset of sepsis in which underlying circulatory, metabolic, or cellular abnormalities are sufficiently severe to substantially increase mortality rates.5
Non-coding RNAs are responsible for the regulation of many cell signaling pathways. These molecules include long non-coding RNAs (lncRNAs), circular RNAs (circRNAs), and microRNAs (miRNAs), which are involved in several biological processes such as apoptosis, mitochondrial dysfunction, and innate immunity.6, 7, 8, 9, 10
lncRNAs are transcripts containing more than 200 nt, whose mechanisms of formation and function have been recently investigated.11,12 According to some experimental studies, the differential expression of some lncRNAs has been found in cardiomyocytes, monocytes, and human tubular epithelial cells, when subjected to the addition of plasma obtained from septic patients, or, alternatively, the addition of pure lipopolysaccharide (LPS).13, 14, 15 However, the function of these lncRNAs in sepsis still remains unclear. Human umbilical vein endothelial cells were screened for lncRNAs, and the results showed 28- to 70-fold increases in the expression of these non-coding RNAs after LPS exposure.16 These changes could be involved in the modulation of inflammatory responses. For example, the lncRNA called lnc-IL7R can interact with the gene coding for the human interleukin 7 (IL-7) receptor α subunit to reduce the proinflammatory responses induced by LPS.17 Additionally, the lncRNA HOTAIR has been shown to modulate tumor necrosis factor (TNF)-α synthesis in cardiomyocytes in a murine sepsis model via the nuclear factor κB (NF-κB) pathway.14
circRNAs are known as a class of lncRNAs,18 which are stabilized by their circular structure, and have been proposed to function as biomarkers for various conditions. Despite the limited availability of data concerning any proven association between these molecules and the condition of sepsis, in vitro knockdown of the circRNA known as RasGEF1B suggested a complicated relationship between several cellular pathways occurring in sepsis.19
All cells (both diseased and healthy) are able to synthesize 21- to 25-nt-long RNA molecules called miRNAs. These miRNAs bind to complementary sequences in the untranslated 3′ end of their target mRNAs, and they are able to post-transcriptionally regulate the expression of numerous genes.20 Somewhat confusingly, they can have protective or detrimental effects in different immune-related disorders and can affect the levels of the pro-inflammatory cytokines TNF-α and IL-1β through signaling pathways involving p38 mitogen-activated protein kinase (MAPK) and MAPK phosphatase 1 (MKP-1).6, 7, 8, 9,21, 22, 23 The results of case-control studies showed that there was differential expression of some miRNAs in septic patients compared to control patients. These authors suggested that miRNAs could be biomarkers for diagnosis or prognostic stratification, and they could even possibly be therapeutic targets in sepsis treatment.7,23, 24, 25, 26, 27, 28, 29, 30
A complete understanding of the exact molecular mechanisms involved in sepsis requires further research.31 Researchers have suggested that the pathophysiology of sepsis is reportedly associated with the dysregulation of oxidative stress and excessive inflammatory responses.32 Recent studies have detected extracellular vesicles (EVs) in the circulation of sepsis patients, and they have suggested that these EVs could originate from various cells affected by the pathophysiology of sepsis.33, 34, 35 Exosomes are a specific kind of EVs derived from platelets (as well as other cell types), which can trigger the synthesis of superoxide and the induction of apoptosis in vascular cells in sepsis patients through redox and inflammatory mechanisms.36,37
In one study, a correlation between exosome abundance and cardiac dysfunction was observed. Cardiac dysfunction was proposed to be due to myocardial depression caused by exosomes in the isolated heart preparation and in papillary muscle cells, probably due to myocardial nitric oxide production.38
Exosomes are reported to be active in cell-to-cell communication, both in normal tissues and in diseased conditions such as cancer, due to their contents of functional messenger RNAs (mRNAs) and also miRNAs.39, 40, 41 Although miRNAs have been reported in plasma from septic patients,42,43 only limited data are available on the nucleic acid contents of exosomes during sepsis.30,44,45
Based on clinical and experimental studies, we herein summarize the current knowledge about the role of miRNAs, lncRNAs, and exosomes in the pathophysiology of sepsis.
lncRNA Biogenesis
The family of lncRNAs characterized as 200-nt non-protein-coding transcripts,46 contains more than 60,000 individually cataloged members based on the database LNCipedia for annotation of human lncRNA transcript structures and sequences.47 Poorly conserved lncRNAs might be compensated due to the presence of strong structural conservation.48 The conserved genomic areas of DNA can transcribe lncRNAs (Figure 1),49 and lncRNAs can also be produced by back-splicing of exons from circRNAs.50,51 The lncRNAs possess multiple functions and activities, such as regulation of gene expression, rearrangement of chromatin, modification of histones, and alternative splicing of genes, suggesting their possible role in the pathogenesis of different diseases and conditions.52,53
Figure 1.
Long Non-coding RNA (lncRNAs) Biogenesis
(A) Sense: the lncRNA transcript overlaps with exons of the other transcript in the same strand. (B) Antisense: the lncRNA transcript overlaps with exons of the other transcript in the opposite strand. (C) Intronic: the lncRNA is entirely derived from the intron of a different transcript. (D) Intergenic: the lncRNA sequence is located between the two genes coding for proteins. (E) Bidirectional or divergent: the lncRNA is located in the opposite strand from a protein coding gene; therefore, they are co-regulated. (F) Enhancer: the lncRNA, also known as eRNA, is transcribed from the enhancer region.
circRNAs comprise a large class of non-coding RNAs that are primarily derived from protein-coding gene exons, but they are not produced by the normal RNA splicing model.54 The main features of circRNAs are their covalent loop structures due to their 3′ and 5′ ends being joined together by intron-exon circularization.55 Despite initial reports that circRNAs had only a low abundance, further research showed they were indeed more widespread and had a substantial presence in various transcriptomes.50,51,55
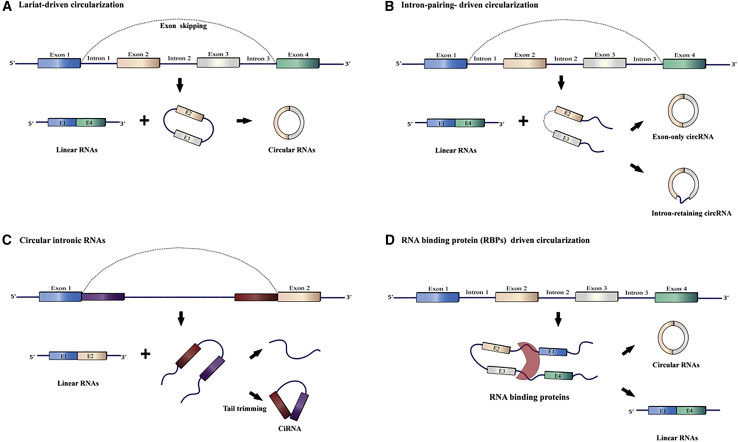
According to Jeck et al.,51 circRNAs can be produced by several mechanisms, including exon-intron pairing-driven and lariat-driven circularization, as shown in Figure 2. As shown in Figure 2C, the introns between the exons are removed after generating a circular structure to form an exon-only circRNA named EIciRNA, or else they can be retained to produce an intron-retaining circRNA.51,56 The circularization of two flanking intronic sequences can also generate circRNAs (Figure 2).57,58 Many different circRNAs have been shown to be specific for different organs or tissues, and some of them have been associated with diseases, suggesting that circRNAs are not merely byproducts caused by splicing or mis-splicing errors.59,60
Figure 2.
Circular RNA (circRNA) Biogenesis Pathways
(A) Lariat-driven circularization. (B) Intron pairing-driven circularization. (C) Circular intronic RNAs. (D) RNA-binding protein (RBP)-driven circularization.
lncRNAs and Sepsis
It has been previously suggested that dysregulation of lncRNAs is associated with different diseases, and many lncRNAs have been detected to play significant roles in developmental and biological processes.12 However, there have only been a limited number of studies aiming to assess any correlation between sepsis and lncRNAs (Table 1). Until now, numerous reports have found that the expression of lncRNAs is altered in human monocytes, cardiomyocytes, and tubular epithelial cells during the sepsis process or after exposure to LPS.13, 14, 15 Moreover, it was recently shown in human umbilical vein endothelial cells that several lncRNAs were downregulated (with a maximum downregulation of about 28-fold), whereas other lncRNAs were upregulated (with a maximum upregulation of about 70-fold).16 Additionally, Cui et al.17 reported that the lncRNA lnc-IL7R (which overlaps with the 3′ UTR of the α subunit gene of IL-7 receptor) was considerably upregulated and could be involved in inflammatory signaling in LPS-treated cells. However, the role of lncRNAs has not been sufficiently explored in sepsis, and further studies are needed to discover how lncRNAs affect sepsis, and to unravel the precise molecular mechanisms.
Table 1.
Selected lncRNAs Involved in Sepsis
| lncRNAs | Expression in Sepsis | Target Gene | Effect(s) | Model | No. of Samples | Ref. |
|---|---|---|---|---|---|---|
| MALAT1 | up | miR-125b | MALAT1 interacts with p38 MAPK/NF-κB and miR-125b, thereby aggravating cardiac inflammation and dysfunction in sepsis | mice | – | 216 |
| NEAT1 | up | miR-204 | the upregulation of NEAT1 was related to the severity of acute kidney injury (AKI) in sepsis patients | humans in vitro | 55 | 217 |
| NEAT1 | up | circulating lncRNA NEAT1 was related to severity, increased risk, and unfavorable prognosis in sepsis patients | humans | 152 | 218 | |
| NEAT1 | up | NEAT1 induced brain injury in septic mice via positively regulating NF-κB | Mice | – | 219 | |
| NEAT1 | up | – | – | humans | 59 | 220 |
| lnc-ANRIL | up | – | – | humans | 26 | 221 |
| lnc-ANRIL/miR-125a axis | up | lnc-ANRIL/miR-125a axis could serve as a biomarker for prognosis, severity, and inflammation in sepsis patients | humans | 26 | 221 | |
| HOTAIR | up | HOTAIR upregulation leads to HK-2 cell apoptosis in kidney injury via the miR-22/HMGB1 pathway | rats in vitro | – | 222 | |
| HOTAIR | – | HOTAIR overexpression can reduce AKI in septic rats by suppressing the apoptosis of kidney tissues via downregulating the miR-34a/Bcl-2 signaling pathway | Rats | – | 223 | |
| lncRNA H19 | down | – | humans in vitro | 69 | 224 | |
| lncRNA ITSN1-2 | up | high expression of ITSN1-2 is associated with disease severity and inflammation in sepsis patients. | Humans | 309 | 225 | |
| HULC | up | – | upregulation of lncRNA HULC is required for the pro-inflammatory response during LPS induced sepsis. | mice in vitro | – | 226 |
| UCA1 | upregulation of UCA1 is needed for the response of pro-inflammatory immune cells during LPS-induced sepsis | |||||
| TUG1 | down | decreased TUG1 expression may induce sepsis-related AKI by modulating the NF-κB pathway and regulating the miR-142-3p/SIRT1 axis | humans in vitro | 28 | 227 | |
| TapSAKI | up | TapSAKI promoted the inflammatory response and HK-2 cell apoptosis through the miR-22/PTEN/TLR4/NF-κB pathway | rats in vitro | – | 228 | |
| HOTAIR | up | - | in vivo studies showed that silencing of HOTAIR could improve cardiac function of septic mice and significantly reduce TNF-α production | Mice | 14 | |
| MALAT1 | up | – | IL-6 induced upregulation of MALAT1 in LPS-treated cardiomyocytes, and MALAT1 could promote the expression of TNF-α at least partly by SAA3 in response to LPS treatment in cardiomyocytes | mice in vitro | – | 229 |
| MALAT1 and EZH2 | up | – | upregulation of MALAT1 and EZH2 were found in the hearts of rats with sepsis | rats in vitro | – | 230 |
Several circRNAs have been recently observed to function as “miRNA sponges” that can remove specific miRNAs, thereby modulating gene expression.50,59,61 In sepsis, many miRNAs are differentially expressed, suggesting that some corresponding circRNAs may also be implicated. It has been recently found that mcircRasGEF1B, as an LPS-inducible circRNA, could modulate the stability of mature intercellular adhesion molecule (ICAM)-1 mRNAs, and might be able to protect cells against microbial infection.19 Improved experimental characterization and identification of circRNAs, as well as some related molecules, including miRNAs, are needed to obtain new insights into sepsis pathogenesis and to identify possible novel biomarkers.
miRNA Biogenesis
miRNAs have been shown to be endogenous non-coding RNA molecules with lengths of 19–22 nt,62 whose biogenesis has been elucidated in previous studies.63,64 Briefly, RNA polymerase II transcribes the DNA to produce a primary miRNA known as pri-miRNA having 500–3,000 nt65 (Figure 3). Subsequently, the pri-miRNA is cleaved into a premature miRNA known as pre-miRNA having 70–80 nt. This cleavage is caused by the microprocessor complex consisting of the DiGeorge syndrome critical region 8 (DGCR) protein and RNase III Drosha enzyme.66 In the next step, the pre-miRNA moves into the cytoplasm using the exportin-5-mediated nuclear export system, which processes the approximately 22-nt miRNA duplex by the interaction of the co-factor double-stranded transactivation-responsive RNA-binding protein with RNase III endonuclease Dicer protein.67 Afterward, the integration of miRNA duplex occurs with the RNA-induced silencing complex (RISC) following binding to glycine tryptophan repeat-containing protein and argonaute protein, which bind to full or partial sequences in the 3′ or 5′ UTR of the target mRNA.20,68 The miRNAs are involved in a comprehensive gene regulation network in the pathophysiology of numerous disease conditions.69,70
Figure 3.
MicroRNA (miRNA) Biogenesis
RNA polymerase II transcribes the DNA to produces a pri-miRNA. The pri-miRNA is cleaved into a pre-miRNA by the microprocessor complex consisting of the DiGeorge syndrome critical region 8 (DGCR8) protein and RNase III Drosha enzyme. The pre-miRNA moves into the cytoplasm using the exportin-5-mediated nuclear export system, which processes the approximately 22-nt miRNA duplex by the interaction of the co-factor double-stranded, transactivation-responsive RBP with RNase III endonuclease Dicer protein. The integration of the miRNA duplex occurs with the RNA-induced silencing complex (RISC) following binding to glycine tryptophan repeat-containing protein and Argonaute protein, which bind to full or partial sequences in the 3′ or 5′ UTR of the target mRNA.
miRNAs and Sepsis
miRNAs play significant roles in the regulation of adaptive and innate immune responses during the pathogenesis of diseases such as sepsis (Table 2).71 These biomarkers are able to regulate the development of regulatory T cells and T helper cells, which are essential to optimizing the host responses to infectious disease.72,73 During sepsis, the immune system of the host appears to be both immunosuppressed, while at the same time being in an accentuated pro-inflammatory state.74,75 In this pro-inflammatory state, numerous cytokines are overexpressed, including TNF-α. miRNAs can control the production of TNF-α at both translational and transcriptional levels. For instance, one investigation carried out by Dan et al.76 found that miR-181 upregulation enhanced the degradation of TNF-α mRNA. Likewise, another study showed that miR-125b was considerably reduced in correlation with higher expression of TNF-α in neonatal monocytes after LPS stimulation.77 Furthermore, miRNAs can also target the TNF pathway and mediate the inflammatory response. miR-511 was suggested to act as a regulator of TNF receptor 1 protein and to affect the sensitivity of cells to TNF, thus possibly protecting against TNF-dependent endotoxic shock syndrome.78 Moreover, it has been also shown that other inflammatory cytokines, such as IL-6, are considerably increased in patients with sepsis.79 In fact, Zhou et al.80 showed that miR-146a downregulation was correlated with increased concentrations of IL-6 in sepsis patients.
Table 2.
Selected MicroRNAs Involved in Sepsis
| MicroRNAs | Expression in Sepsis | Key Points of Investigation | Model (In Vitro, In Vivo, Human) | Sample Type | Sample Size | Ref. |
|---|---|---|---|---|---|---|
| miR-150 | down | miR-150 levels in both leukocytes and plasma correlated with the severity of sepsis and could be used as a marker of early sepsis | humans | plasma, leukocytes | 8 | 140 |
| plasma ratio of levels of miR-150/IL-18 could be used for assessing the severity of sepsis | ||||||
| miR-125b | down | polymicrobial sepsis (CLP) decreased miR-125b levels in circulation and in myocardium | mice | heart | – | 231 |
| miR-205b | down | serum miR-205b concentrations were decreased in the LPS group compared to control group, but in lungs, spleen, and liver the decrease was not significant | mice | serum, organs | – | 232 |
| miR-23a-5p | down | serum miR-23a-5p was increased after LPS injection | rats | serum, lung tissues | – | 102 |
| miR-21-3p | up | miR-21-3p controls sepsis-related cardiac dysfunction through modulating SORB | in vivo | plasma | 46 | 233 |
| miR-23b | down | LPS downregulates miR-23b expression in human vascular endothelial cells (VECs); upregulation of miR-23b inhibited the expression of NF-κB, TNF-α, IL-6, ICAM-1, E-selectin and vascular cell adhesion molecule-1 (VCAM-1) | in vitro | vascular endothelial cells | – | 234 |
| miR-375 | down | miR-375 can block the JAK2-STAT3 pathway and modulate the level of miR-21 involved in the regulation of late-stage sepsis | in vivo | blood | 33 | 235 |
| miR-342-5p | down | – | humans | plasma, leukocytes | 8 | 140 |
| miR-182 | down | – | humans | plasma, leukocytes | 8 | 140 |
| miR-146a | down | – | humans | serum | 50 + 30 (SIRS) | 24 |
| miR-223 | down | – | humans | serum | 50 + 30 (SIRS) | 24 |
| miR-130a | down | – | humans | PBMCs | 60 | 149 |
| miR-31 | down | miR-31 downregulation in CD4+ T cells contributes to immunosuppression in sepsis patients via promoting TH2 skewing | humans | T cells | 23 | 236 |
| miR-15a | down | – | humans | plasma | 62 | 30 |
| miR-27a | down | miR-34a, miR-15a, and miR-27a are correlated with shock development in severe sepsis patients; they also target cell cycle regulation, apoptosis, cell layer permeability, and inflammatory pathways | humans | plasma | 62 | 30 |
| miR-25 | down | a correlation between levels of miR-25 and the severity of sepsis was observed; surviving patients had higher levels of this biomarker compared with non-surviving subjects; decreased levels of miR-25 were associated with the concentrations of oxidative stress indicators in sepsis | humans | blood | 70 + 30 (SIRS) | 28 |
| miR-146a, miR-181a, miR-584 | down | – | humans | PBMCs | 32 | 80 |
| miR-146a | down | miR-146a gene polymorphism rs2910164 is associated with the risk of severe sepsis | humans | PBMCs | 226 | 237 |
| miR-146a | down | – | humans | plasma | 14 + 14 (SIRS) | 238 |
| miR-499-5p | down | – | humans | serum | 166 | 26 |
| miR-122 | down | – | humans | serum | 166 | 26 |
| miR-193b | down | – | humans | serum | 166 | 26 |
| miR-150 | down | – | humans | PBMCs | 23 + 22 | 29 |
| miR-342 | down | – | humans | PBMCs | 23 + 22 | 29 |
| miR-3173-5p | down | – | humans | PBMCs | 23 + 22 | 29 |
| miR-181b | down | miR-181b regulates NF-κB-mediated endothelial cell activation and vascular inflammation in response to pro-inflammatory stimuli | in vitro | plasma | 26 SP | 139 |
| in vivo | 36 SP + ARDS | |||||
| let-7a | down | – | mice | lung | – | 239 |
| miR-129-5p | down | – | mice | lung | – | 239 |
| miR-218 | down | – | mice | lung | – | 239 |
| miR-21 | up | miR-21 is involved in the regulation of late sepsis | in vivo | blood | 33 | 235 |
| miR-155 | up | miR-155 induced an elevated percentage of CD39+ regulatory T cells, leading to immunosuppression | in vivo | blood | 60 | 97 |
| miR-15a | up | upregulated miR-15a downregulated the LPS-induced inflammatory pathway | humans | serum | 46 | 136 |
| miR-16 | up | upregulated miR-16 downregulated the LPS-induced inflammatory pathway | humans | serum | 46 | 136 |
| miR-15a | up | serum miR-15a/miR-16 levels were significantly elevated in sepsis/SIRS patients when compared to healthy controls | humans | serum | 166 + 32 | 27 |
| miR-16 | ||||||
| miR-574-5p | up | serum level was correlated with the death of sepsis patients | humans | serum | 142 | 103 |
| the combined analysis of miR-574-5p, SOFA scores, and the sepsis stage on the day of diagnosis provided a good predictor for sepsis prognosis | ||||||
| miR-297 | up | serum miR-297 level was higher in survivors than non-survivors among septic patients | humans | serum | 142 | 103 |
| miR-223 | up | – | humans | serum | 166 | 26 |
| miR-19a | up | – | in vitro | B cells | 38 + 26 (SIRS) | 240 |
| miR-486 | up | – | humans | plasma, leukocytes | 8 | 140 |
| miR-34a | up | – | humans | plasma | 62 | 30 |
| miR-145 | up | – | humans | PBMCs | 32 | 80 |
| miR-143 | up | – | humans | PBMCs | 32 | 80 |
| miR-182 | up | – | humans | PBMCs | 32 | 80 |
| miR-486 | up | – | humans | PBMCs | 32 | 80 |
| miR-1308 | up | – | humans | PBMCs | 32 | 80 |
| miR-4772 | up | – | humans | blood | 23 +22 | 29 |
| miR-143 | up | serum miR-143 levels were significantly higher in sepsis than in SIRS and healthy controls | humans | serum | 103 | 241 |
| 95 | ||||||
| miR-133a | up | there was a correlation between the levels of miR-133a and sepsis severity | humans, mice | serum | 138 | 137 |
| miR-4772 | up | – | humans | blood | 23 + 22 | 29 |
| let-7d | up | the levels of these biomarkers were elevated in whole blood after LPS injection | mice | blood, organs | – | 242 |
| miR-15b | up | the levels of these biomarkers were elevated in whole blood after LPS injection | mice | blood, organs | – | 242 |
| miR-16 | up | the levels of these biomarkers were elevated in whole blood after LPS injection | mice | blood, organs | – | 242 |
| miR-25 | up | the levels of these biomarkers were elevated in whole blood after LPS injection | mice | blood, organs | – | 242 |
| miR-92a | up | the levels of these biomarkers were elevated in whole blood after LPS injection | mice | blood, organs | – | 242 |
| miR-103 | up | the levels of these biomarkers were elevated in whole blood after LPS injection | mice | blood, organs | – | 242 |
| miR-107 | up | the levels of these biomarkers were elevated in whole blood after LPS injection | mice | blood, organs | – | 242 |
| miR-451 | up | the levels of these biomarkers were elevated in whole blood after LPS injection | mice | blood, organs | – | 242 |
| miR-15a | up | the levels of these biomarkers were elevated in lung | mice | blood, organs | – | 242 |
| miR-16 | up | the levels of these biomarkers were elevated in lung | mice | blood, organs | – | 242 |
| miR-21 | up | the levels of these biomarkers were elevated in lung | mice | blood, organs | – | 242 |
| miR-146a | up | the levels of these biomarkers were elevated in lung | mice | blood, organs | – | 242 |
| miR-155 | up | the levels of these biomarkers were elevated in lung | mice | blood, organs | – | 242 |
| miR-223 | up | the levels of these biomarkers were elevated in lung | mice | blood, organs | – | 242 |
| miR-195 | up | miR-195 was elevated in lung and liver of sepsis-induced mice | mice | lung, liver | – | 7 |
| miR-27a | up | miR-27a induces an inflammatory response in sepsis | mice | lung | – | 239 |
| miR-143 | up | mice | lung | – | 239 | |
| miR-153 | up | mice | lung | – | 239 | |
| miR-21 | up | miR-21 increased in early sepsis and showed a sustained increase until late sepsis | mice | bone marrow | – | 96 |
| miR-181b | up | miR-181b increased in early sepsis and showed a sustained increase until late sepsis | mice | bone marrow | – | 96 |
| miR-16 | up | miRNAs upregulated after CLP | mice | blood | – | 243 |
| miR-17 | up | miRNAs upregulated after CLP | mice | blood | – | 243 |
| miR-20a/b | up | miRNAs upregulated after CLP | mice | blood | – | 243 |
| miR-26a/b | up | miRNAs upregulated after CLP | mice | blood | – | 243 |
| miR-106a/b | up | miRNAs upregulated after CLP | mice | blood | – | 243 |
| miR-195 | up | miRNAs upregulated after CLP | mice | blood | – | 243 |
| miR-451 | up | miRNAs upregulated after CLP | mice | blood | – | 243 |
| miR-29a | – | miR-29a induces apoptosis of monocytes via targeting STAT3 during sepsis | mice | THP-1 cells | – | 101 |
| miR-30a | – | miR-30a inhibits MD-2 expression by targeting STAT1 in human monocytes | mice | THP-1 cells | – | 244 |
| miR-146 | – | the transfection of miR-146a reduces the sepsis-induced cardiac dysfunction through inhibiting inflammatory cell infiltration, NF-κB activation, and inflammatory cytokine production by targeting TRAF6 and IRAK | mice | H9C2 and J774 cells | – | 83 |
| miR-150 | – | reduced miR-150 serum concentrations are associated with an unfavorable outcome in patients with critical illness, independent of the presence of sepsis | humans | serum | 138 | 42 |
| miR-223 | – | it has been found that this biomarker does not appropriate in predicting sepsis | humans, mice | serum | 137 | 43 |
| miR-122 | – | serum miR-122 levels correlated with short-term mortality in sepsis patients and are a potential biomarker for sepsis and ARDS | humans | serum | 232 | 104 |
| miR-15a | – | the expression levels of miR-193b, miR-122, miR-483-5p, and miR-15a in the sepsis non-survivors were significantly higher than those in the sepsis survivors, and the levels of miR-223 and miR-16 were significantly lower | humans | serum | 214 | 245 |
| miR-16 | ||||||
| miR-122 | ||||||
| miR-193b | ||||||
| miR-483-5p | ||||||
| miR-223 |
Numerous investigations have reported that miRNAs are capable of regulating inflammatory responses via targeting the Toll-like receptor (TLR) signaling pathway (Figure 4). TLR-induced signaling primarily activates NF-κB, a key transcription factor modulating the expression of pro-inflammatory and immunoregulatory factors.81, 82, 83 Some miRNAs, such as miR-155, miR-125, and miR-146a, were found to play essential roles in the negative modulation of the TLR/NF-κB-induced inflammatory cascade and in innate immunity.84, 85, 86 The expression of miR-155 and numerous other miRNAs is NF-κB-dependent.87 For instance, it was found that using LPS to stimulate monocytic THP-1 cells induced expression of both miR-146a and miR-146b.88 miR-146a directly regulates the expression of TNF receptor-associated factor 6 (TRAF6) and IL-1 receptor-associated kinase 1 (IRAK1), which are crucial adaptor molecules in the TLR signaling pathway.81,82,89 In addition, miR-146a plays an important role in the endotoxin tolerance of monocytic cells in vitro,90,91 and this tolerance could be reversed by suppressing miR-146a.91 It has recently been reported that the NF-κB/DICER signaling pathway inhibited TNF-α expression by producing mature forms of miR-130a and miR-125b, thus regulating TNF-α mRNA.92 It is known that the TLR-induced signaling pathway is critical in sepsis.93
Figure 4.
The Association of lncRNAs and miRNAs with the Pathophysiology of Sepsis
eNOS, endothelial nitric oxide synthase; HuR, human antigen R; CISH, cytokine-inducible SH2-containing protein; MDSC, myeloid-derived suppressor cell; IRAK, IL-1 receptor-associated kinase; JNK, c-Jun N-terminal kinase; TRAF6, TNF receptor-associated factor 6; MyD88, myeloid differentiation primary response gene 88; BMPR2, bone morphogenetic protein receptor type II; NF-kB, nuclear factor κB; IkB, inhibitor of κB; SOX6, sex-determining region Y box 6; Sirt1, sirtulin 1; Pim1, proto-oncogene serine/threonine-protein kinase; BMAL1, brain and muscle ARNT-like 1; PDCD4, programmed cell death 4; PGC1A, PPARγ (peroxisome proliferator-activated receptor γ) co-activator 1A; PRKC, primary rat kidney cell.
As the sepsis condition progresses, the immune response is reprogrammed into a more immunosuppressed state.75,94 This immunosuppression is partly induced by miRNAs, which mediate the polarization of immune cells and alter cellular immunity95, 96, 97 (Figure 3). miR-181b and miR-21 were upregulated in myeloid-derived suppressor cells isolated from septic mice. This expression prevents the differentiation of precursors into dendritic cells and macrophages via affecting the transcription factors CCAAT/enhancer-binding protein β (C/EBPβ) and signal transducer and activator of transcription 3 (Stat3).96,98 Moreover, a temporal modulation was found in myeloid cells, with miR-155 inversely correlated with LPS-stimulated levels of brain and muscle ARNT-like 1 (BMAL1).99 Additionally, it has also been demonstrated by other investigators that different miRNAs, including miR-29a, miR-23b, and mi-R210, inhibit IL-6 and NF-κB expression during sepsis by affecting various immune cells.65,100,101 These studies have suggested a role for miRNA involvement in sepsis pathophysiology.
Non-coding RNAs as Therapeutic Targets
Experimental investigations have suggested that the measurement of non-coding RNAs and the use of miRNA modulators could be of some benefit in the fight against sepsis. However, in order to translate these biomarkers into real clinical applications, considerable challenges must be overcome. In fact, the expression pattern of miRNAs is tissue-dependent, which brings into question the suitability of systemic delivery of miRNA mimics or antagonists, as has been mostly conducted in experimental studies. Targeted delivery systems for drugs are often considered to be alternative approaches; however, this approach would be further complicated by the heterogeneous clinical manifestations in patients with sepsis, as well as non-optimized delivery technology.
Recent experimental investigations have revealed that miRNA expression levels are correlated with septic complications and therapeutic interventions in sepsis patients.43,78,102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113 The upregulation of miR-511 induced by glucocorticoid administration inhibited the TNF receptor TNFR1, and therefore decreased its sensitivity to TNF-α in SPRET/Ei mice.78 Likewise, in an LPS-induced sepsis model, dexamethasone administration downregulated miR-155 expression in the liver and reduced the production of pro-inflammatory cytokines.105,114 miRNAs have also been implicated in cortisol non-responsiveness, which may occur during the course of treatment. This phenotype of steroid resistance is associated with changed expression of the glucocorticoid receptor α, an isoform of the cortisol receptor. The mentioned isoform was remarkably downregulated in patients with sepsis, which may be related to miR-124, which was elevated 3-fold upon exposure to glucocorticoids.106
After exposure to systemic LPS, the body switches to a state of tolerance, which prevents prolonged pro-inflammatory reactions. During induction of LPS tolerance, translation and transcription of acute inflammatory genes can be silenced, which is mediated by different miRNAs in vitro.115 Modulation of this tolerance process can be disturbed by miR-146a overexpression in sepsis models.116 Differential expression of sepsis-related miRNAs has implicated a variety of miRNAs in the alterations produced in several organs. The alteration in miRNAs in endotoxin tolerance is observed predominantly in macrophages and involves miR-155 and miR-146a.117,118 Recent evidence has suggested that miRNA modulation during LPS tolerance involves the TLR-NF-κB-cytokine pathways.119, 120, 121
Some therapies that have been tested for sepsis involve the regulation of miRNAs. These miRNAs have been mostly correlated with pathways involved in innate immunity in animals122,123 and cellular models.124,125 Flavonoids are well-studied anti-inflammatory compounds. It has been recently shown that the flavonoid apigenin inhibits LPS-mediated expression of miR-155 in macrophages, resulting in upregulation of the anti-inflammatory modulators MAD-related protein 2 and forkhead box O3a in vitro.125 In a murine model of sepsis, similar protective effects were shown when an apigenin-rich diet significantly decreased TNF-α and miR-155 expression in the mouse lungs.125 Recently, stem cell therapy has attracted further attention for sepsis. In vivo investigations showed that mesenchymal stem cells (MSCs) prolong survival in mice with sepsis induced by cecal ligation and puncture (CLP) through downregulating miR-143.124,126 In septic mice treated with MSCs, microarray analysis revealed that there were 77 miRNAs with more than 1.5-fold differential expression.122 This was accompanied by a decreased inflammatory response as well as less apoptosis.122 Experimental sepsis models that were treated with the 20-hydroxyeicosatetraenoic acid (20-HETE) analog N-(20-hydroxyeicosa-5Z,14Z-dienoyl)glycine showed effects on the expression of miR-297, miR-223, and miR-150.123 It was suggested that these miRNAs were downregulated, resulting in inhibition of the myeloid differentiation primary response gene 88 (MyD88)/NF-κB pathway.127
Assessment of the ability of miRNAs to function as therapeutic agents in sepsis has mainly employed two approaches: use of miRNA mimics or antagomirs. An antagomir is a small synthetic RNA that is perfectly complementary to the specific miRNA target and inhibits Ago2-mediated cleavage. Among all miRNAs, miR-146a has been the most widely investigated. Through targeting TRAF6 and IRAK1, miR-146a reduced cardiac dysfunction in septic mice.83 In vivo delivery of a miR-146a antagomir using jetPEI instillation into the airways of septic mice suppressed the generation of pro-inflammatory cytokines and ameliorated the injury to lung tissue.128 It was recently reported that this miRNA also inhibited the in vitro differentiation of human CD4+ T lymphocytes via affecting protein kinase C (PRKC).129 Other miRNAs, such asmiR-195, miR-142-3p, and miR-124, have also been reported to be beneficial in preventing apoptosis, inflammation, and organ injury in sepsis models.7,116,130,131 The induction of miR-126 expression using CTEC-0214, a stromal cell-derived factor 1α analog, preserved the endothelial cell barrier integrity and decreased pulmonary vascular leakage.132
Non-coding RNAs as Biomarkers in Sepsis
The detection of non-coding RNAs within various organs or tissues may be useful as biomarkers for sepsis, as well as sepsis-associated organ dysfunction.13, 14, 15, 16, 17,133, 134, 135 Currently, blood is the most useful clinical sample from sepsis patients for detecting non-coding RNAs, because it contains leucocytes, plasma, and serum. The existence of extracellular miRNAs within blood can be rapidly quantified in a clinical setting, unlike microbial culture, which is more time-consuming.63 Therefore, this ease of use has encouraged more studies of whether miRNAs could be diagnostic biomarkers for sepsis. As mentioned earlier, various miRNAs are expressed differentially in sepsis. For instance, miR133a, miR122, and miR-16 were elevated in the serum of patients with sepsis,136, 137, 138 while other miRNAs, including miR-181b and miR-25, were reduced.28,139 Additionally, one recently performed study indicated that the clinical accuracy of the measurement of miRNA-25 for the diagnosis of sepsis was better than C-reactive protein (CRP) and procalcitonin (PCT) (area under the receiver operating characteristic [ROC] curve = 0.806, 0.676, and 0.726 for miRNA-25, CRP, and PCT, respectively).28 The downregulation of miRNA-25 levels was associated with the severity of sepsis (Sequential Organ Failure Assessment [SOFA] score), CRP, as well as PCT levels.28 Moreover, a similar investigation revealed that at a cutoff point set at −1.89, miR-233 provided a sensitivity of 80% and a specificity of 100%, while at a cutoff point set at −2.98, miR-146a provided a sensitivity of 63.3% and a specificity of 100%.24 With this high sensitivity and specificity, these miRNAs could be used as novel biomarkers for sepsis.24 miRNAs have also been shown to be correlated with the prognosis of sepsis patients. Vasilescu et al.140 reported that miR-150 concentrations were significantly decreased in plasma samples from patients with sepsis, and they were related to the severity of the disease as assessed by the SOFA scores. miR-150 was shown to control the expression of c-Myb in vivo in a dose-dependent manner over a narrow range of c-Myb concentrations, and this influenced lymphocyte responses and differentiation,141 which supports the importance of miR-150 downregulation in patients with sepsis. A study carried out by Wang et al.103 revealed that the combination of sepsis SOFA scores and miR-574-5p expression levels could predict the mortality of patients with sepsis with 91.84% specificity and 78.13% sensitivity. Thus, miRNAs may play a crucial role in improving sepsis prognosis and diagnosis in affected individuals.
Cardiac dysfunction induced by sepsis is characterized by a reduced ejection fraction and disturbed myocardial contractility.142 However, the pathophysiology of myocardial dysfunction induced by sepsis is not completely understood, and there is no effective medication to reverse sepsis-mediated myocardial dysfunction.143 Numerous studies have suggested that miRNAs may play an important role in sepsis-mediated cardiac dysfunction. For instance, in a experimental model of severe sepsis, miR-233 was repressed, leading to inflammatory responses at several levels, and resulting in the induction of myocardial dysfunction.144 In addition, miR-155 inhibition protected against LPS-mediated cardiac dysfunction.145 Moreover, miR-146a was able to decrease sepsis-mediated cardiac dysfunction through preventing the activation of NF-κB, as well as decreasing the infiltration of inflammatory cells.83 It has been recently shown that LPS inhibited the expression of miR-499, resulting in cardiomyocyte apoptosis via the Bcl-2 family pathway.146 lncRNAs have been shown to be implicated in the sepsis-induced cardiac dysfunction process. The lncRNA HOTAIR was observed to be considerably upregulated and it positively modulated NF-kB activation and p65 phosphorylation in cardiomyocytes taken from septic mice.14 Therefore, several non-coding RNAs have been shown to induce sepsis-mediated cardiac dysfunction. These agents may deserve to be considered as diagnostic biomarkers for patients with sepsis-mediated cardiac dysfunction.
Thrombocytopenia is an important abnormality that occurs in severe sepsis patients.147,148 It has been recently shown that septic patients with thrombocytopenia had elevated concentrations of IL-18 and some miRNAs, as well as reduced miR-130a expression, suggesting that miRNAs could be implicated in sepsis-related thrombocytopenia and thus function as a biomarker.149 Additionally, a study by Wang et al.138 indicated that levels of miR-122 were higher in the abnormal coagulation group compared to normal coagulation patients. Overall, it is unclear how miRNAs influence coagulation, and further investigation is required to understand whether miRNAs can be coagulopathy indicators.
Inflammation of the endothelium has an essential role in sepsis pathogenesis.150 It was shown that LPS-mediated endothelial dysfunction was induced by the Slit2-Robo4 pathway, and Slit2 downregulation decreased miR-218 expression.151 Furthermore, miR-147b has been implicated in endothelial protection via the degradation of the mRNA for ADAM15 (a regulator of endothelial permeability).152 Altogether, the data suggest that different miRNAs could be considered to be novel indicators for sepsis-related cardiac and endothelial dysfunction.
Exosome Biogenesis
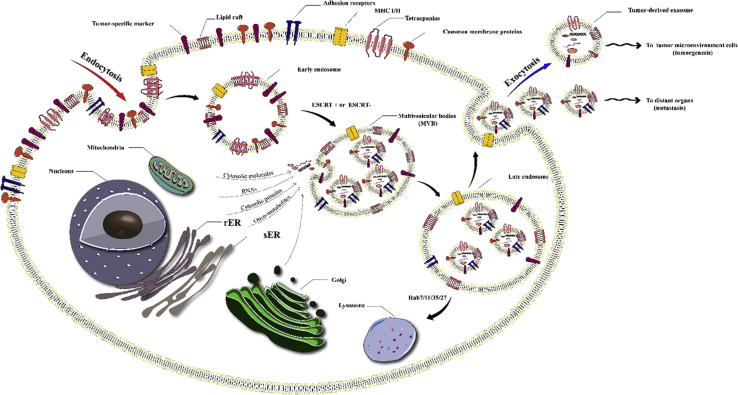
Multivesicular bodies (MVBs) are late-stage endosomes containing numerous intraluminal endosomal vesicles, some of which are targeted for lysosomal degradation, and others for cell membrane integration followed by the extracellular release of the vesicles. The smaller vehicles (30–120 nm in diameter) are known as exosomes, while the larger vehicles (120–1,000 nm in diameter) are called microvesicles.153, 154, 155 The exosomes contain ubiquitinated proteins that have been loaded by an “endosomal sorting complex required for transport” (ESCRT) or other ESCRT-independent mechanisms (Figure 5).156 Electron microscopy images exhibit a “cup-shaped” morphology for exosomes isolated by differential ultracentrifugation.157
Figure 5.
Exosome Biogenesis
Endocytosis at a lipid raft occurs by either a clathrin-dependent pathway or a clathrin-independent pathway. The endocytic vesicles contain signaling proteins, growth factor receptors, and oncoproteins, together with normal membrane proteins, including tetraspanins (e.g., CD9, CD63, and CD81), MHC I and II, and adhesion molecules (e.g., cadherins, integrins). Exosome biogenesis occurs via the endosomal network in the endosomal sorting complexes needed for ESCRT-independent or ESCRT-dependent pathways. Inward budding of multi-vesicular bodies (MVBs) produces intra-luminal vesicles (exosomes). Several cytoplasmic molecules (e.g., heat shock proteins, ubiquitin-related proteins, mRNAs, miRNAs, cytoskeleton proteins) and nuclear molecules (e.g., lncRNAs, transcription factors, DNAs) can be loaded into MVBs via stage-specific pathways, some of which are more or less specific for the state of sepsis. Moreover, plasma membrane fusion of MVBs leads to release of exosomes by exocytosis. Numerous Rab GTPases (such as Rab11/35, Rab7, and Rab27) are contained in secreted exosomes. rER, rough endoplasmic reticulum; sER, smooth endoplasmic reticulum.
The composition of exosomes has been found to be uniquely disparate and complex, so that 194 lipids, 4,563 proteins, 764 miRNAs, and 1,639 mRNAs have been cataloged to be present inside exosomes isolated from various organisms, based on the database ExoCarta (version 4; http://www.exocarta.org).158 The exosomal proteins mainly belong to the families of heat shock proteins (heat shock cognate 70 [HSC70]), membrane transporters, and fusion proteins (annexins, flotillin, and GTPases), MVB biogenesis proteins (tumor suppressor gene 101 [TSG101] and alix), tetraspanins (CD81, CD63, and CD9) lipid-related proteins, and phospholipases.157,159 There are certain mRNAs commonly found inside exosomes that code for proteins, importantly including CD81, CD63, and tetraspanins. Exosomal lipids are mainly sphingolipids, cholesterol, bisphosphates, and phospholipids, which have been detected in exosomes secreted by different cells, such as mast cells and dendritic cells,160 reticulocytes,161 and B lymphocytes.162 Some specific exosomal lipid components, including phosphatidylserine (PS)163 and prostaglandins,164 have been reported to be involved in the biological function of exosomes. Exosomal miRNAs and mRNAs play a role as carriers for genetic information. Despite the degradation of most exosomal RNAs into <200-nt-long RNA fragments, several full-length RNAs can be taken up into cells through an endocytosis process and influence their protein expression. Exosomal miRNA expression has been associated with some diseases, including cancer, indicating the potential of exosomal miRNAs to act as biomarkers for cancer diagnosis.165, 166, 167 miRNAs have been frequently reported to be present in exosomes isolated from body fluids such as saliva and urine,168 thereby encouraging the use of exosomal miRNAs as novel biomarkers.
Exosomes and EVs in Sepsis
Efficient intercellular communication is necessary in order to coordinate the inflammatory responses of immune cells. Chemokines, cytokines, and cell surface receptors are all involved in communications between immune cells. Mounting data indicate that immune cells are able to signal through the release of exosomes that carry various biological molecules, and that can be taken up by recipient cells, thereby altering their function.41,169, 170, 171, 172, 173, 174, 175, 176, 177, 178
Ying et al.179 showed that it was the miRNAs within exosomes that were primarily responsible for the biological effects.41,180 Furthermore, some beneficial immunomodulatory effects of plasma-derived exosomal miRNAs have been recently demonstrated in patients with sepsis (Table 3; Figure 6).181 Therefore, exosomal miRNAs have been proposed to be responsible for the anti-apoptotic effects observed in T lymphocytes by addition of sepsis exosomes (Sepsis-Exos). However, there are still complicated relationships operating between miRNAs and their mRNA targets. Theoretically, one miRNA is capable of targeting more than one mRNA, and vice versa, and there could be more than one miRNA able to target any individual mRNA sequence.182 Hence, bioinformatics analysis of the Sepsis-Exos differentially expressed (DE) miRNAs was carried out, suggesting that the apoptosis-associated guanosine monophosphate-protein kinase G (cGMP-PKG) pathway was involved.
Table 3.
Exosomes and Their Cargos in Sepsis
| Cargo | Detection Methods | Model | Note | Ref. |
|---|---|---|---|---|
| Protein (ATF3) | differential centrifugation + western blot | humans (urine), mice | urinary exosomal ATF3 is an early diagnostic biomarker for sepsis-induced acute kidney injury | 185 |
| Protein (SPTLC3) | one-step ultracentrifugation using OptiPrep + mass spectrometry using Q Exactive Plus | humans (plasma) | SPTLC3 is involved in sphingolipid metabolism, with a negative correlation with the progression of sepsis | 187 |
| Proteins | ExoQuick exosome precipitation | humans (plasma) | proteomic profile analysis of sepsis-derived exosomes and LPS-stimulated, monocyte-derived exosomes exhibited downregulation of several important protein networks, including immune response | 188 |
| miRNA (miR-126) | centrifugation + ExoQuick | mice (serum) | levels of miR-126 in serum exosomes isolated from HSPA12B-deficient (HSPA12B−/−) septic mice were significantly lower than those for wild-type septic mice | 186 |
| felivery of miR-126 containing exosomes significantly improved cardiac function and vascular permeability in HSPA12B−/− septic mice | ||||
| miRNA (miRNA-125b) | – | mice (serum) | miRNA-125b in endothelial progenitor cell-derived exosomes was also downregulated during sepsis | 45 |
| miRNA (miR-155 and miR-146a) | differential centrifugation | mice (serum) | miR-146a inhibited while miR-155 promoted endotoxin-induced inflammation | |
| miRNA (miR-34a, miR-27a, and miR-15a) | ExoQuick exosome precipitation solution | humans (plasma) | miR-34a, miR-27a, and miR-15a in the endothelial progenitor cell-derived exosomes had different expression levels in sepsis patients | 30 |
| miRNA (miR-223) | centrifuged + filtering through 0.2 μm + Tris/EDTA | mice (serum) | exosomal miR-223 plays an essential role for MSC-induced cardioprotection in sepsis | 183 |
| miRNA (miR-126-3p, miR-122-5p, miR-146a-5p, miR-145-5p, miR-26a-5p, miR-150-5p, miR-222-3p, and miR-181a-5p) | differential ultracentrifugation | mice (serum) | EVs of septic animals play an important role in inflammation, and EV-associated miRNAs likely mediate the cytokine production via TLR7-MyD88 signaling | 189 |
| miRNA (miR-122-5p, miR-125b-5p, miR-1260a, miR-1262, miR-127-3p, miR-1290, miR-1298-5p, miR-146a-5p, miR-151a-3p, miR-16-5p, miR-1825, miR-192-5p, miR-193a-5p, miR-221-3p, miR-25-3p, miR-26a-5p, miR-301a-3p, miR-320b, miR-339-3p, miR-340-5p, miR-532-3p, miR-720, miR-744-5p, miR-885-5p, miR-92a-3p) | ultracentrifugation + western blot + nanoparticle-tracking analysis device + nano flow cytometry | humans (plasma) | – | 181 |
| mRNA (myeloperoxidase [MPO], PRDX3, SOD2, FOXM1, SELS, and GLRX2) | ultracentrifugation + western blot + nanoparticle-tracking analysis device + nano flow cytometry | humans (plasma) | – | 181 |
| mRNA (DNMT1, DNMT3A, DNMT3B) | centrifugation | humans (plasma) | EV-DNMT mRNAs load, when coupled with total plasma EV number, may be a novel method to diagnose septic shock | 190 |
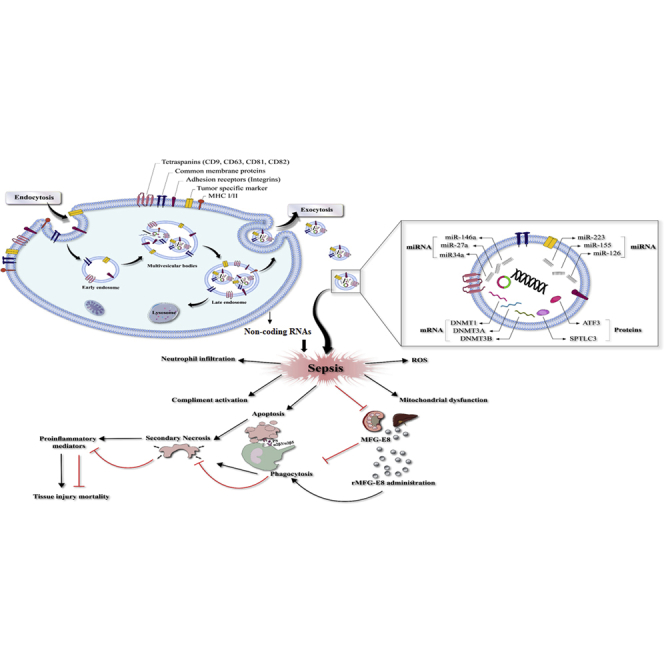
Figure 6.
Schematic Representation of Exosome Functions in Sepsis
Exosomes and their contents (i.e., miRNAs, mRNAs, and proteins) could be involved in the pathogenesis of sepsis. In this regard, these nanovesicles and their cargos could increase apoptotic cell death in various cells and tissues, and reduce apoptotic cell clearance by phagocytes, such as macrophages and dendritic cells. Disturbance of the phagocytic removal of apoptotic cells has the potential to release toxic and proinflammatory contents due to secondary necrosis, and it could increase tissue injury and mortality.
A few reports have confirmed the presence of miRNAs in exosomes isolated during sepsis. miRNA-125b was detected in endothelial progenitor cell-derived exosomes in a mouse model, and its expression level was also downregulated during sepsis.45 It was recently shown that some miRNAs in endothelial progenitor cell-derived exosomes (miR-34a, miR-27a, and miR-15a) had different expression levels in the plasma of sepsis patients.30
In one study, Real et al.181 assessed the mRNAs and miRNAs associated with inflammatory responses, as well as redox metabolism in the exosomes of septic shock patients. In comparison to healthy subjects, exosomes from sepsis patients had significant alterations in 65 different exosomal miRNAs. Twenty-eight miRNAs were expressed differentially, both at recruitment and after 7 days, with similar kinetics (10 miRNAs downregulated and 18 upregulated). In comparison to control individuals, the pathways enriched by the miRNAs of sepsis patients were correlated with inflammatory responses. The comparison of miRNA levels in sepsis patients with their length of survival in the hospital implicated pathways associated with cell cycle modulation. At recruitment, sepsis was correlated with elevations in mRNA expression associated with redox metabolism (myeloperoxidase, 64-fold; peroxiredoxin 3 [PRDX3], 2.6-fold; superoxide dismutase 2 [SOD2], 2.2-fold) and redox-responsive genes (forhead box protein M1 [FOXM1], 21-fold; SELS, 16-fold; glutaredoxin 2 [GLRX2], 3.4-fold). Myeloperoxidase mRNA expression remained elevated after 7 days (65-fold). Exosomes from septic shock patients contain mRNAs and miRNAs associated with pathogenic processes, such as cell cycle modulation, oxidative stress, and inflammatory response. Overall, exosomal transport represents a novel pathway for intercellular communication during sepsis.
Studies have suggested that exosomal miRNAs could modulate inflammation by regulating target proteins in different signaling pathways. miR-223 directly targets STAT3 to regulate pro-inflammatory cytokines. Alternatively, IL-6 triggered by stimulation of TLR/NF-κB could promote miR-223 downregulation; therefore, IL-6 could form a positive regulatory loop for the production of pro-inflammatory cytokines.183 As a result, when exosomes containing miR-223, which were derived from MSCs, were administered to animal models of sepsis, they could downregulate the expression of its target genes and suppress inflammatory response in macrophages. It has been shown that pro-inflammatory and anti-inflammatory miRNAs, which are both present in exosomes, could regulate the inflammatory responses. Alexander et al.184 showed that dendritic cells released exosomes containing miR-155 and miR-146a, and that could be taken up by other recipient dendritic cells, thereby reprogramming the cellular response to endotoxin. Exosome-delivered miR-155 increased the expression of inflammatory genes, whereas miR-146a decreased it. They showed that miR-155 and miR-146a could pass between immune cells in vivo, and that exosomal miR-146a inhibited endotoxin-induced inflammation in mice, while, alternatively, miR-155 promoted the inflammatory response.
In another study, Wang et al.183 evaluated the effect of treating sepsis-induced mice with miR-223-knockout (KO) exosomes obtained from knockout mice, and they showed that the mice had higher levels of Sema3A and STAT3 (targets of miR-223) than did mice treated with wild-type (WT) exosomes. These exosomal proteins could be transferred to cardiomyocytes, leading to increased inflammation and more cell death. In contrast, WT exosomes, which had higher levels of miR-223, could be delivered to cardiomyocytes, causing downregulation of Sema3A and Stat3. Therefore, they concluded that exosomal miR-223 could play a role in MSC-induced cardioprotection in sepsis patients.
Panich et al.185 investigated possible biomarkers in urine samples from patients with early sepsis-induced acute kidney injury. They found that urinary exosomal ATF3 could be an interesting early diagnostic biomarker for sepsis-induced acute kidney injury.
Zhang et al.186 investigated the cardioprotective mechanism of HSPA12B in sepsis-induced cardiovascular dysfunction. They found that the levels of miR-126 in serum exosomes isolated from HSPA12B−/− septic mice were lower than those from WT septic mice. Interestingly, delivery of miR-126 containing exosomes improved cardiac function, decreased immune cell infiltration in the myocardium, and improved vascular permeability in HSPA12B−/− septic mice. Therefore, they showed that HSPA12B could improve endothelial function in sepsis and suggested that exosomes containing miR-126 could play a role in the cardiovascular protective mechanisms in polymicrobial sepsis.
Xu et al.187 used a one-step ultracentrifugation process using an OptiPrep density gradient medium to extract exosomes from human plasma samples with volumes as small as 300 μL. They obtained the exosomal proteomic profile from septic patient blood samples at six time points after diagnosis. Among the 238 proteins, the protein SPTLC3, which is involved in sphingolipid metabolism, showed a negative correlation with disease progression measured by body temperature and CRP levels. The results showed that SPTLC3 may be involved in the development of sepsis and could be a biomarker to monitor the clinical progression of sepsis.
Wisler et al.188 analyzed the proteomic pathways in monocyte-derived exosomes isolated from patients suffering from surgical sepsis to identify possible immunoregulatory functions. They compared the proteomic networks in exosomes derived from LPS-stimulated monocytes with those isolated from patients with surgical sepsis. Proteomic analysis showed that 17 proteins were downregulated, while 1 was upregulated in sepsis-derived exosomes, whereas 14 were downregulated and 1 was upregulated in the LPS-stimulated exosomes. Functional enrichment analysis showed that the downregulated processes included biogenesis, localization, and metabolic and cellular pathways, in addition to those involved in the immune system. In LPS-stimulated macrophages, similar metabolic and cellular processes were downregulated. Human monocytes treated with sepsis-derived exosomes, or exosomes from LPS-stimulated monocytes, suggested that circulating exosomes could be involved in systemic signaling and immunomodulation during sepsis.
Xu et al.189 demonstrated that circulating plasma EVs isolated from septic mice could increase inflammation and cytokine production. They found that the effects of the EVs were not affected by polymyxin B (an endotoxin inhibitor), but were significantly inhibited by anti-miR inhibitors designed against miR-34a, miR-122, and miR-146a. The cytokine production induced by septic EVs was lower in TLR7−/− or MyD88−/− cells. Therefore, EV-associated miRNAs were suggested to mediate cytokine production via TLR7-MyD88 signaling. DNA methyltransferases (DNMTs) are conserved enzymes that are epigenetic regulators of gene expression. The balance between DNMTs involved in DNA repair (DNMT1) and the de novo DNMTs (DNMT3A and DNMT3B) may change during serious diseases. Dakhlallah et al.190 investigated the mRNAs coding for DNMTs inside EVs as prognostic markers for septic shock. Comparison of EV-DNMT1 (maintenance methylation) with EV-DNMT3A+DNMT3B (de novo methylation) correlated with the severity of sepsis, EVs obtained from septic shock patients carried more total DNMT mRNAs and more DNMT3A+DNMT3B mRNAs than did control EVs. Total circulating (plasma) EVs also correlated with sepsis severity. Therefore, the EV-DNMT mRNA load, coupled with total number of plasma EVs, could be a method to diagnose septic shock in patients at intensive care unit (ICU) admission, and could offer opportunities to more precisely intervene with standard therapy or test more advanced approaches.
Human umbilical cord MSC-derived exosomes (hucMSC-Exs) have been investigated for the repair of acute kidney injury (AKI) caused by cisplatin or ischemia/reperfusion injury. Zhang et al.191 investigated the benefits of hucMSC-Exs in sepsis. They found that treatment with hucMSC-Exs ameliorated morphological damage and inhibited renal tubular cell apoptosis. Treatment with hucMSC-Exs improved the survival of mice with sepsis. The effects of hucMSC-Exs were proposed to be via the inhibition of NF-κB signaling and the lessening of pro-inflammatory responses.
Although the triggering of thrombotic pathways is thought to inhibit bacterial dissemination throughout the body by shutting down blood flow, thrombosis may also result in excessive consumption and activation of coagulation components. Because EVs have shown a pro-coagulant activity, they may be implicated in the pathophysiology of disseminated intravascular coagulation (DIC) in patients with sepsis.192 DIC is recognized to be an extreme symptom in sepsis, and its onset is correlated with increased mortality. DIC is characterized by the massive accumulation of thrombi within the microcirculation, as well as excessive consumption of platelets and coagulation components. Approximately 35% of patients with sepsis develop DIC.193 At present, the management of sepsis, which is complicated by DIC, requires treatment of the underlying infection with broad-spectrum antibiotics and surgical control of the infection source,194 accompanied by support for organ function. Despite many attempts, other therapeutic approaches have not yet shown any statistical success in large-scale clinical trials. An early quantification of the risk for developing DIC would be highly useful in patients who present in the initial stages of sepsis. Therefore, EV assays may contribute to this risk assessment, and also aid in identification of novel targets for therapy. EVs also exhibit direct pro-coagulant activities due to the exposure of PS on their surface. PS is a cell membrane phospholipid that helps the function of tissue factor (TF) and coagulation enzymes, and is a major initiator of the coagulation cascade.195,196 In sepsis, pro-coagulant EVs are principally secreted by monocytes (MEVs), endothelial cells (EEVs), and platelets (PEVs).197 TF serves as a primary initiator of the coagulation cascade, while PS is a catalyst for the activation of coagulation components. Both functions make this EV subset a likely factor for the dissemination of the pro-coagulant phenotype during sepsis.198,199 As mentioned above, PEVs are the major source of TF in plasma.200 In addition to upregulation of TF in monocytes and endothelial cells, and the activation of fibrinolytic and prothrombotic pathways, blood-borne TF-positive EVs are also partly responsible for the overall prothrombotic milieu underlying DIC.195,201
Numerous studies have suggested an indirect role of EVs as “carrier vehicles” linking coagulation and inflammation. It has been previously proposed that the release of EVs acts as a danger signal, leading to the transition into a pro-thrombotic phenotype.202 IL-33 has also been implicated in the increased activity of TF in EEVs.203 Budding and remodeling processes within the multivesicular bodies, or at the cell membrane, are induced by highly conserved entities such as the ESCRT.204 An increase in intracellular calcium leads to activation of scramblase and calpain enzymes as well as the loss of symmetry in the phospholipid bilayer leading to activation of inflammatory cells.205,206
Patients suffering from sepsis frequently require mechanical ventilation, leading to possible lung damage. EVs may be involved in sepsis-related lung injuries, as well as other lung-associated conditions in ICU patients. For instance, elevated EVs have been correlated with a decreased risk of acute respiratory distress syndrome (ARDS).207 Moreover, after post-operative extubation, Mutschler et al.208 studied the effects of mechanical ventilation on the expression of EVs in the bronchoalveolar lavage fluid taken from both ventilated human patients and laboratory pigs. It was found that EVs bound to neutrophil granulocytes at the pulmonary air-blood interface, suggesting that EVs could be used as a biomarker to guide the mechanical ventilation strategy. For instance, a predefined level of EVs could help to make the decision whether patients with ARDS require temporary extracorporeal membrane oxygenation to prevent possible lung injuries being caused by mechanical ventilation.
A stretched model of pathological pulmonary endothelial cells (which mimics extreme tidal volume ventilation) triggered the release of EVs, which could be counteracted by the caspase inhibitor Z-VAD209 and was unaffected by co-treatment with calpeptin (a calpain inhibitor), and the Rho kinase inhibitor Y-27632, LPS, and thrombin. It was recently shown that high tidal volume ventilation in healthy mice increased the systemic concentration of EVs derived from pulmonary endothelial cells (CD31+) and reduced the expression of CD31 in the lung tissue. Utilizing a stretched model of human pulmonary endothelial cells, the same mechanism was proposed to explain the shedding of CD31 and annexin V-positive EEVs.203 After intratracheal instillation of EEVs into healthy animals, lung inflammation was increased. According to proteomic analysis, several factors shared by the various EEV populations (such as CD31) have been detected.203 As mentioned above, the disintegration of the pulmonary endothelial barrier triggers the release of EVs, which express numerous adhesion molecules and other factors that modulate inflammatory events.210 It has recently been reported that an increased level of EVs in plasma was inversely correlated with the risk for ARDS development in a group of critically ill patients with different underlying pathologies. Further post hoc analysis showed that this correlation was true in septic patients but not correct in non-septic subjects.207
Moreover, systemic and endothelial activation caused by LPS and thrombin/CD40L increased the proportion of EVs carrying CD40L and matrix metalloproteinase 10 (MMP10) both in vivo and in vitro. Increases in circulating CD40L and MMP10 were correlated with increased mortality in patients with sepsis who had higher thrombin levels.211 It was found that immunization of healthy rats with septic rat-derived EVs decreased their arterial blood pressure.212 Additionally, inhibition of calpains seems to have a therapeutic impact. In another investigation, the role of calpains in DIC development was assessed in an experimental sepsis model. After suppressing calpain activity by overexpressing its inhibitor calpastatin, they found an increased survival benefit and attenuation of DIC.209
EVs could be useful biomarkers for the occurrence of DIC in sepsis patients. For instance, in patients with meningococcal sepsis with organ dysfunction, the circulating EVs were mostly platelet derived, and in comparison with healthy subjects, their level was almost 15-fold higher.213 In addition, EVs with exposed TF were reduced in sepsis patients with organ dysfunction.197 However, it was recently reported that, in meningococcal septic shock patients compared to non-shock affected subjects, the EVs possessed increased pro-coagulant activity.214 Overall, patients with sepsis could significantly benefit from early detection of biomarkers predicting the onset of DIC, leading to early and more efficient management and therapy. CD31+ EVs and CD105+ EVs are candidates for these biomarkers that warrant further validation.195,215
Conclusions
Despite many studies, sepsis remains a complex clinical condition whose pathophysiology is incompletely understood. Non-coding RNAs present an opportunity for early diagnosis as well as an ability to interact with key points of the biological mechanisms, suggesting that measurement of the expression of miRNAs is a promising approach for intensive care patients. The well-investigated markers are miR-4772-5p, miR-4722-5p-iso, miR-4772-3p, miR-576-5p, miR-146a, miR-133a, and miR-150, which all have higher concentrations in patients with sepsis. Alternatively, miR-122, miR-181b, and miR-223 have lower concentrations in sepsis patients. Furthermore, in critically ill polytrauma patients, these biomarkers can be detected in various body fluids, including blood, plasma, and serum, widening the options for the epigenetic detection of sepsis. Until now, epigenetic interactions have not been widely documented in sepsis, and more investigations are required to determine the role of the expression of miRNAs.
Exosomes may be able to prevent the occurrence of excess inflammation, thereby reducing mortality as well as lessening morbidity. Since the majority of these investigations have been performed in experimental models, further studies are required to translate the findings into clinical applications.
Exosomes isolated from urine, cerebrospinal fluid, saliva, and plasma can serve as prognostic and diagnostic markers for liver injury, kidney disease, neurodegenerative diseases, and cancer. Clinical studies of exosomes, including their use as drug-delivery systems, have shown some promising results. It is accepted that any treatment that interacts with basic biological processes carries a considerable risk of causing undesirable side effects. Drug development and drug delivery investigations based on exosomes, which are intended for future therapeutic applications, must undergo a comprehensive and strict risk and safety analysis before these agents can be tested in human studies.
The future perspective of miRNAs as an emerging therapeutic option is not only due to the fact that miRNAs contain an entirely known sequence that is often completely conserved among species, but they are also relatively easily synthesized and can be manufactured in bulk. Moreover, there still needs to be more knowledge about which specific circulating miRNAs are deregulated in sepsis in order to develop sensitive and specific screening biomarkers. Fortunately, miRNA profiling data can be obtained from multiple pooled samples as a promising way to identify potential miRNA biomarkers. Moreover, miRNA profiling combined with sensitive, accurate, and flexible detection systems, such as small RNA sequencing, quantitative PCR methods for miRNAs in circulating blood, and analysis of complex datasets by powerful bioinformatics/biostatistical algorithms, will work together to produce innovative miRNA-based diagnostic and prognostic biomarkers.
Author Contributions
H.M. and M.R.H. contributed to conception, design, statistical analysis, and drafting of the manuscript. S.M.H., M.H.P., S.F., and A.A.V. contributed in data collection and manuscript drafting. All authors approved the final version of the manuscript for submission.
Conflicts of Interest
M.R.H. declares the following potential conflicts of interest. Scientific Advisory Boards: Transdermal Cap, Cleveland, OH, USA; BeWell Global, Wan Chai, Hong Kong; Hologenix, Santa Monica, CA, USA; LumiThera, Poulsbo, WA, USA; Vielight, Toronto, ON, Canada; Bright Photomedicine, Sao Paulo, Brazil; Quantum Dynamics, Cambridge, MA, USA; Global Photon, Bee Cave, TX, USA; Medical Coherence, Boston MA, USA; NeuroThera, Newark, DE, USA; JOOVV, Minneapolis-St. Paul, MN, USA; AIRx Medical, Pleasanton, CA, USA; FIR Industries, Ramsey, NJ, USA; UVLRx Therapeutics, Oldsmar, FL, USA; Ultralux UV, Lansing, MI, USA; Illumiheal & Petthera, Shoreline, WA, USA; MB Lasertherapy, Houston, TX, USA; ARRC LED, San Clemente, CA, USA; Varuna Biomedical, Incline Village, NV, USA; and Niraxx Light Therapeutics, Boston, MA, USA. Consulting: Lexington International, Boca Raton, FL, USA; USHIO Corporation, Japan; Merck, KGaA, Darmstadt, Germany; Philips Electronics Nederland, B.V. Eindhoven, the Netherlands; Johnson & Johnson, Philadelphia, PA, USA; and Sanofi-Aventis Deutschland, GmbH, Frankfurt am Main, Germany. Stock holdings: Global Photon, Bee Cave, TX, USA; Mitonix, Newark, DE, USA. The remaining authors declare no competing interests.
Acknowledgments
This study was supported by the Kashan University of Medical Sciences, Kashan, Iran. M.R.H. was supported by NIH grants R01AI050875 and R21AI121700.
Contributor Information
Hamed Mirzaei, Email: h.mirzaei2002@gmail.com.
Michael R. Hamblin, Email: hamblin@helix.mgh.harvard.edu.
References
- 1.Angus D.C., van der Poll T. Severe sepsis and septic shock. N. Engl. J. Med. 2013;369:840–851. doi: 10.1056/NEJMra1208623. [DOI] [PubMed] [Google Scholar]
- 2.Singer M., Deutschman C.S., Seymour C.W., Shankar-Hari M., Annane D., Bauer M., Bellomo R., Bernard G.R., Chiche J.D., Coopersmith C.M. The Third International Consensus Definitions for sepsis and septic shock (Sepsis-3) JAMA. 2016;315:801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seymour C.W., Liu V.X., Iwashyna T.J., Brunkhorst F.M., Rea T.D., Scherag A., Rubenfeld G., Kahn J.M., Shankar-Hari M., Singer M. Assessment of clinical criteria for sepsis: for the Third International Consensus Definitions for sepsis and septic shock (Sepsis-3) JAMA. 2016;315:762–774. doi: 10.1001/jama.2016.0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao W., Mindrinos M.N., Seok J., Cuschieri J., Cuenca A.G., Gao H., Hayden D.L., Hennessy L., Moore E.E., Minei J.P., Inflammation and Host Response to Injury Large-Scale Collaborative Research Program A genomic storm in critically injured humans. J. Exp. Med. 2011;208:2581–2590. doi: 10.1084/jem.20111354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gotts J.E., Matthay M.A. Sepsis: pathophysiology and clinical management. BMJ. 2016;353:i1585. doi: 10.1136/bmj.i1585. [DOI] [PubMed] [Google Scholar]
- 6.Precone V., Stornaiuolo G., Amato A., Brancaccio G., Nardiello S., Gaeta G.B. Different changes in mitochondrial apoptotic pathway in lymphocytes and granulocytes in cirrhotic patients with sepsis. Liver Int. 2013;33:834–842. doi: 10.1111/liv.12169. [DOI] [PubMed] [Google Scholar]
- 7.Zheng D., Yu Y., Li M., Wang G., Chen R., Fan G.-C., Martin C., Xiong S., Peng T. Inhibition of microRNA 195 prevents apoptosis and multiple-organ injury in mouse models of sepsis. J. Infect. Dis. 2016;213:1661–1670. doi: 10.1093/infdis/jiv760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brudecki L., Ferguson D.A., McCall C.E., El Gazzar M. Mitogen-activated protein kinase phosphatase 1 disrupts proinflammatory protein synthesis in endotoxin-adapted monocytes. Clin. Vaccine Immunol. 2013;20:1396–1404. doi: 10.1128/CVI.00264-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brudecki L., Ferguson D.A., McCall C.E., El Gazzar M. MicroRNA-146a and RBM4 form a negative feed-forward loop that disrupts cytokine mRNA translation following TLR4 responses in human THP-1 monocytes. Immunol. Cell Biol. 2013;91:532–540. doi: 10.1038/icb.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morris K.V., Mattick J.S. The rise of regulatory RNA. Nat. Rev. Genet. 2014;15:423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wapinski O., Chang H.Y. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. [Google Scholar]
- 12.Dey B.K., Mueller A.C., Dutta A. Long non-coding RNAs as emerging regulators of differentiation, development, and disease. Transcription. 2014;5:e944014. doi: 10.4161/21541272.2014.944014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin J., Zhang X., Xue C., Zhang H., Shashaty M.G., Gosai S.J., Meyer N., Grazioli A., Hinkle C., Caughey J. The long noncoding RNA landscape in hypoxic and inflammatory renal epithelial injury. Am. J. Physiol. Renal Physiol. 2015;309:F901–F913. doi: 10.1152/ajprenal.00290.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu H., Liu J., Li W., Liu G., Li Z. lncRNA-HOTAIR promotes TNF-α production in cardiomyocytes of LPS-induced sepsis mice by activating NF-κB pathway. Biochem. Biophys. Res. Commun. 2016;471:240–246. doi: 10.1016/j.bbrc.2016.01.117. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y., Ferguson J.F., Xue C., Ballantyne R.L., Silverman I.M., Gosai S.J., Serfecz J., Morley M.P., Gregory B.D., Li M., Reilly M.P. Tissue-specific RNA-seq in human evoked inflammation identifies blood and adipose lincRNA signatures of cardiometabolic diseases. Arterioscler. Thromb. Vasc. Biol. 2014;34:902–912. doi: 10.1161/ATVBAHA.113.303123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh K.K., Matkar P.N., Muhammad S., Quan A., Gupta V., Teoh H., Al-Omran M., Verma S. Investigation of novel LPS-induced differentially expressed long non-coding RNAs in endothelial cells. Mol. Cell. Biochem. 2016;421:157–168. doi: 10.1007/s11010-016-2797-8. [DOI] [PubMed] [Google Scholar]
- 17.Cui H., Xie N., Tan Z., Banerjee S., Thannickal V.J., Abraham E., Liu G. The human long noncoding RNA lnc-IL7R regulates the inflammatory response. Eur. J. Immunol. 2014;44:2085–2095. doi: 10.1002/eji.201344126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L.-L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 19.Ng W.L., Marinov G.K., Liau E.S., Lam Y.L., Lim Y.-Y., Ea C.-K. Inducible RasGEF1B circular RNA is a positive regulator of ICAM-1 in the TLR4/LPS pathway. RNA Biol. 2016;13:861–871. doi: 10.1080/15476286.2016.1207036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 21.Ho P.-C., Chang K.-C., Chuang Y.-S., Wei L.-N. Cholesterol regulation of receptor-interacting protein 140 via microRNA-33 in inflammatory cytokine production. FASEB J. 2011;25:1758–1766. doi: 10.1096/fj.10-179267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.How C.-K., Hou S.-K., Shih H.-C., Huang M.-S., Chiou S.-H., Lee C.-H., Juan C.C. Expression profile of microRNAs in gram-negative bacterial sepsis. Shock. 2015;43:121–127. doi: 10.1097/SHK.0000000000000282. [DOI] [PubMed] [Google Scholar]
- 23.Tili E., Michaille J.-J., Cimino A., Costinean S., Dumitru C.D., Adair B., Fabbri M., Alder H., Liu C.G., Calin G.A., Croce C.M. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-α stimulation and their possible roles in regulating the response to endotoxin shock. J. Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 24.Wang J.F., Yu M.L., Yu G., Bian J.J., Deng X.M., Wan X.J., Zhu K.M. Serum miR-146a and miR-223 as potential new biomarkers for sepsis. Biochem. Biophys. Res. Commun. 2010;394:184–188. doi: 10.1016/j.bbrc.2010.02.145. [DOI] [PubMed] [Google Scholar]
- 25.Huang J., Sun Z., Yan W., Zhu Y., Lin Y., Chen J., Shen B., Wang J. Identification of microRNA as sepsis biomarker based on miRNAs regulatory network analysis. BioMed Res. Int. 2014;2014:594350. doi: 10.1155/2014/594350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang H.J., Zhang P.J., Chen W.J., Feng D., Jia Y.H., Xie L.X. Four serum microRNAs identified as diagnostic biomarkers of sepsis. J. Trauma Acute Care Surg. 2012;73:850–854. doi: 10.1097/TA.0b013e31825a7560. [DOI] [PubMed] [Google Scholar]
- 27.Wang H., Zhang P., Chen W., Feng D., Jia Y., Xie L.X. Evidence for serum miR-15a and miR-16 levels as biomarkers that distinguish sepsis from systemic inflammatory response syndrome in human subjects. Clin. Chem. Lab. Med. 2012;50:1423–1428. doi: 10.1515/cclm-2011-0826. [DOI] [PubMed] [Google Scholar]
- 28.Yao L., Liu Z., Zhu J., Li B., Chai C., Tian Y. Clinical evaluation of circulating microRNA-25 level change in sepsis and its potential relationship with oxidative stress. Int. J. Clin. Exp. Pathol. 2015;8:7675–7684. [PMC free article] [PubMed] [Google Scholar]
- 29.Ma Y., Vilanova D., Atalar K., Delfour O., Edgeworth J., Ostermann M., Hernandez-Fuentes M., Razafimahatratra S., Michot B., Persing D.H. Genome-wide sequencing of cellular microRNAs identifies a combinatorial expression signature diagnostic of sepsis. PLoS ONE. 2013;8:e75918. doi: 10.1371/journal.pone.0075918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goodwin A.J., Guo C., Cook J.A., Wolf B., Halushka P.V., Fan H. Plasma levels of microRNA are altered with the development of shock in human sepsis: an observational study. Crit. Care. 2015;19:440. doi: 10.1186/s13054-015-1162-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salomao R., Brunialti M.K.C., Rapozo M.M., Baggio-Zappia G.L., Galanos C., Freudenberg M. Bacterial sensing, cell signaling, and modulation of the immune response during sepsis. Shock. 2012;38:227–242. doi: 10.1097/SHK.0b013e318262c4b0. [DOI] [PubMed] [Google Scholar]
- 32.Azevedo L., Janiszewski M., Soriano F., Laurindo F. Redox mechanisms of vascular cell dysfunction in sepsis. Endocr. Metab. Immune Disord. Drug Targets. 2006;6:159–164. doi: 10.2174/187153006777442431. [DOI] [PubMed] [Google Scholar]
- 33.Reid V.L., Webster N.R. Role of microparticles in sepsis. Br. J. Anaesth. 2012;109:503–513. doi: 10.1093/bja/aes321. [DOI] [PubMed] [Google Scholar]
- 34.Terrasini N., Lionetti V. Exosomes in critical illness. Crit. Care Med. 2017;45:1054–1060. doi: 10.1097/CCM.0000000000002328. [DOI] [PubMed] [Google Scholar]
- 35.Azevedo L.C., Pedro M.A., Laurindo F.R. Circulating microparticles as therapeutic targets in cardiovascular diseases. Recent Pat. Cardiovasc. Drug Discov. 2007;2:41–51. doi: 10.2174/157489007779606121. [DOI] [PubMed] [Google Scholar]
- 36.Janiszewski M., Do Carmo A.O., Pedro M.A., Silva E., Knobel E., Laurindo F.R. Platelet-derived exosomes of septic individuals possess proapoptotic NAD(P)H oxidase activity: a novel vascular redox pathway. Crit. Care Med. 2004;32:818–825. doi: 10.1097/01.ccm.0000114829.17746.19. [DOI] [PubMed] [Google Scholar]
- 37.Gambim M.H., do Carmo Ade.O., Marti L., Veríssimo-Filho S., Lopes L.R., Janiszewski M. Platelet-derived exosomes induce endothelial cell apoptosis through peroxynitrite generation: experimental evidence for a novel mechanism of septic vascular dysfunction. Crit. Care. 2007;11:R107. doi: 10.1186/cc6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Azevedo L.C.P., Janiszewski M., Pontieri V., Pedro Mde.A., Bassi E., Tucci P.J.F., Laurindo F.R. Platelet-derived exosomes from septic shock patients induce myocardial dysfunction. Crit. Care. 2007;11:R120. doi: 10.1186/cc6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lv L.-L., Cao Y.-H., Ni H.-F., Xu M., Liu D., Liu H., Chen P.S., Liu B.C. MicroRNA-29c in urinary exosome/microvesicle as a biomarker of renal fibrosis. Am. J. Physiol. Renal Physiol. 2013;305:F1220–F1227. doi: 10.1152/ajprenal.00148.2013. [DOI] [PubMed] [Google Scholar]
- 40.Hunter M.P., Ismail N., Zhang X., Aguda B.D., Lee E.J., Yu L., Xiao T., Schafer J., Lee M.L., Schmittgen T.D. Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J.J., Lötvall J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 42.Roderburg C., Luedde M., Vargas Cardenas D., Vucur M., Scholten D., Frey N., Koch A., Trautwein C., Tacke F., Luedde T. Circulating microRNA-150 serum levels predict survival in patients with critical illness and sepsis. PLoS ONE. 2013;8:e54612. doi: 10.1371/journal.pone.0054612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benz F., Tacke F., Luedde M., Trautwein C., Luedde T., Koch A., Roderburg C. Circulating microRNA-223 serum levels do not predict sepsis or survival in patients with critical illness. Dis. Markers. 2015;2015:384208. doi: 10.1155/2015/384208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reithmair M., Buschmann D., Märte M., Kirchner B., Hagl D., Kaufmann I., Pfob M., Chouker A., Steinlein O.K., Pfaffl M.W., Schelling G. Cellular and extracellular miRNAs are blood-compartment-specific diagnostic targets in sepsis. J. Cell. Mol. Med. 2017;21:2403–2411. doi: 10.1111/jcmm.13162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fan H., Goodwin A.J., Chang E., Zingarelli B., Borg K., Guan S., Halushka P.V., Cook J.A. Endothelial progenitor cells and a stromal cell-derived factor-1α analogue synergistically improve survival in sepsis. Am. J. Respir. Crit. Care Med. 2014;189:1509–1519. doi: 10.1164/rccm.201312-2163OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 47.Volders P.-J., Helsens K., Wang X., Menten B., Martens L., Gevaert K., Vandesompele J., Mestdagh P. LNCipedia: a database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Res. 2013;41:D246–D251. doi: 10.1093/nar/gks915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnsson P., Lipovich L., Grandér D., Morris K.V. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim. Biophys. Acta. 2014;1840:1063–1071. doi: 10.1016/j.bbagen.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boon R.A., Jaé N., Holdt L., Dimmeler S. Long noncoding RNAs: from clinical genetics to therapeutic targets? J. Am. Coll. Cardiol. 2016;67:1214–1226. doi: 10.1016/j.jacc.2015.12.051. [DOI] [PubMed] [Google Scholar]
- 50.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 51.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fan R., Cao C., Zhao X., Shi Q., Zhao J., Xu S. Downregulated long noncoding RNA ALDBGALG0000005049 induces inflammation in chicken muscle suffered from selenium deficiency by regulating stearoyl-CoA desaturase. Oncotarget. 2017;8:52761–52774. doi: 10.18632/oncotarget.17187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu G., Dong B., Zhang J., Zhai W., Xie T., Huang B., Huang C., Yao X., Zheng J., Che J., Xu Y.F. The long noncoding RNA HOTAIR activates the Hippo pathway by directly binding to SAV1 in renal cell carcinoma. Oncotarget. 2017;8:58654–58667. doi: 10.18632/oncotarget.17414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salzman J., Gawad C., Wang P.L., Lacayo N., Brown P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 57.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 58.Conn S.J., Pillman K.A., Toubia J., Conn V.M., Salmanidis M., Phillips C.A., Roslan S., Schreiber A.W., Gregory P.A., Goodall G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 59.Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B., Luo Y., Lyu D., Li Y., Shi G. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang K., Long B., Liu F., Wang J.-X., Liu C.-Y., Zhao B., Zhou L.Y., Sun T., Wang M., Yu T. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 2016;37:2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 61.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 62.Krol J., Loedige I., Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 63.Essandoh K., Fan G.-C. Role of extracellular and intracellular microRNAs in sepsis. Biochim. Biophys. Acta. 2014;1842:2155–2162. doi: 10.1016/j.bbadis.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ho J., Chan H., Wong S.H., Wang M.H., Yu J., Xiao Z., Liu X., Choi G., Leung C.C., Wong W.T. The involvement of regulatory non-coding RNAs in sepsis: a systematic review. Crit. Care. 2016;20:383. doi: 10.1186/s13054-016-1555-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Benz F., Roy S., Trautwein C., Roderburg C., Luedde T. Circulating microRNAs as biomarkers for sepsis. Int. J. Mol. Sci. 2016;17:78. doi: 10.3390/ijms17010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Rådmark O., Kim S., Kim V.N. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 67.Lund E., Güttinger S., Calado A., Dahlberg J.E., Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 68.Macfarlane L.-A., Murphy P.R. MicroRNA: biogenesis, function and role in cancer. Curr. Genomics. 2010;11:537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li S., Lee C., Song J., Lu C., Liu J., Cui Y., Liang H., Cao C., Zhang F., Chen H. Circulating microRNAs as potential biomarkers for coronary plaque rupture. Oncotarget. 2017;8:48145–48156. doi: 10.18632/oncotarget.18308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Q., Su J., Wang Z., Qi H., Ge Z., Li Z., Chen W.D., Wang Y.D. MicroRNA-149∗ suppresses hepatic inflammatory response through antagonizing STAT3 signaling pathway. Oncotarget. 2017;8:65397–65406. doi: 10.18632/oncotarget.18541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.O’Connell R.M., Rao D.S., Baltimore D. MicroRNA regulation of inflammatory responses. Annu. Rev. Immunol. 2012;30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- 72.Xiao C., Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 73.O’Connell R.M., Kahn D., Gibson W.S., Round J.L., Scholz R.L., Chaudhuri A.A., Kahn M.E., Rao D.S., Baltimore D. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–619. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Delano M.J., Scumpia P.O., Weinstein J.S., Coco D., Nagaraj S., Kelly-Scumpia K.M., O’Malley K.A., Wynn J.L., Antonenko S., Al-Quran S.Z. MyD88-dependent expansion of an immature GR-1+CD11b+ population induces T cell suppression and Th2 polarization in sepsis. J. Exp. Med. 2007;204:1463–1474. doi: 10.1084/jem.20062602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gentile L.F., Cuenca A.G., Efron P.A., Ang D., Bihorac A., McKinley B.A., Moldawer L.L., Moore F.A. Persistent inflammation and immunosuppression: a common syndrome and new horizon for surgical intensive care. J. Trauma Acute Care Surg. 2012;72:1491–1501. doi: 10.1097/TA.0b013e318256e000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dan C., Jinjun B., Zi-Chun H., Lin M., Wei C., Xu Z., Ri Z., Shun C., Wen-Zhu S., Qing-Cai J., Wu Y. Modulation of TNF-α mRNA stability by human antigen R and miR181s in sepsis-induced immunoparalysis. EMBO Mol. Med. 2015;7:140–157. doi: 10.15252/emmm.201404797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang H.C., Yu H.R., Huang L.T., Huang H.C., Chen R.F., Lin I.C., Ou C.Y., Hsu T.Y., Yang K.D. miRNA-125b regulates TNF-α production in CD14+ neonatal monocytes via post-transcriptional regulation. J. Leukoc. Biol. 2012;92:171–182. doi: 10.1189/jlb.1211593. [DOI] [PubMed] [Google Scholar]
- 78.Puimège L., Van Hauwermeiren F., Steeland S., Van Ryckeghem S., Vandewalle J., Lodens S., Dejager L., Vandevyver S., Staelens J., Timmermans S. Glucocorticoid-induced microRNA-511 protects against TNF by down-regulating TNFR1. EMBO Mol. Med. 2015;7:1004–1017. doi: 10.15252/emmm.201405010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mera S., Tatulescu D., Cismaru C., Bondor C., Slavcovici A., Zanc V., Carstina D., Oltean M. Multiplex cytokine profiling in patients with sepsis. APMIS. 2011;119:155–163. doi: 10.1111/j.1600-0463.2010.02705.x. [DOI] [PubMed] [Google Scholar]
- 80.Zhou J., Chaudhry H., Zhong Y., Ali M.M., Perkins L.A., Owens W.B., Morales J.E., McGuire F.R., Zumbrun E.E., Zhang J. Dysregulation in microRNA expression in peripheral blood mononuclear cells of sepsis patients is associated with immunopathology. Cytokine. 2015;71:89–100. doi: 10.1016/j.cyto.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Medzhitov R., Preston-Hurlburt P., Janeway C.A., Jr. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 82.Zhang G., Ghosh S. Toll-like receptor-mediated NF-κB activation: a phylogenetically conserved paradigm in innate immunity. J. Clin. Invest. 2001;107:13–19. doi: 10.1172/JCI11837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gao M., Wang X., Zhang X., Ha T., Ma H., Liu L., Kalbfleisch J.H., Gao X., Kao R.L., Williams D.L., Li C. Attenuation of cardiac dysfunction in polymicrobial sepsis by microRNA-146a is mediated via targeting of IRAK1 and TRAF6 expression. J. Immunol. 2015;195:672–682. doi: 10.4049/jimmunol.1403155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.O’Connell R.M., Rao D.S., Chaudhuri A.A., Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 85.Quinn S.R., O’Neill L.A. A trio of microRNAs that control Toll-like receptor signalling. Int. Immunol. 2011;23:421–425. doi: 10.1093/intimm/dxr034. [DOI] [PubMed] [Google Scholar]
- 86.O’Neill L.A., Sheedy F.J., McCoy C.E. MicroRNAs: the fine-tuners of Toll-like receptor signalling. Nat. Rev. Immunol. 2011;11:163–175. doi: 10.1038/nri2957. [DOI] [PubMed] [Google Scholar]
- 87.Monk C.E., Hutvagner G., Arthur J.S.C. Regulation of miRNA transcription in macrophages in response to Candida albicans. PLoS ONE. 2010;5:e13669. doi: 10.1371/journal.pone.0013669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Taganov K.D., Boldin M.P., Chang K.-J., Baltimore D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Quinn E.M., Wang J.H., O’Callaghan G., Redmond H.P. MicroRNA-146a is upregulated by and negatively regulates TLR2 signaling. PLoS ONE. 2013;8:e62232. doi: 10.1371/journal.pone.0062232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nahid M.A., Satoh M., Chan E.K. MicroRNA in TLR signaling and endotoxin tolerance. Cell. Mol. Immunol. 2011;8:388–403. doi: 10.1038/cmi.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.El Gazzar M., Church A., Liu T., McCall C.E. MicroRNA-146a regulates both transcription silencing and translation disruption of TNF-α during TLR4-induced gene reprogramming. J. Leukoc. Biol. 2011;90:509–519. doi: 10.1189/jlb.0211074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Guan Y., Yao H., Wang J., Sun K., Cao L., Wang Y. NF-κB-DICER-miRs axis regulates TNF-α expression in responses to endotoxin stress. Int. J. Biol. Sci. 2015;11:1257–1268. doi: 10.7150/ijbs.12611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Williams D.L., Ha T., Li C., Kalbfleisch J.H., Schweitzer J., Vogt W., Browder I.W. Modulation of tissue Toll-like receptor 2 and 4 during the early phases of polymicrobial sepsis correlates with mortality. Crit. Care Med. 2003;31:1808–1818. doi: 10.1097/01.CCM.0000069343.27691.F3. [DOI] [PubMed] [Google Scholar]
- 94.Ho J., Yu J., Wong S.H., Zhang L., Liu X., Wong W.T., Leung C.C., Choi G., Wang M.H., Gin T. Autophagy in sepsis: degradation into exhaustion? Autophagy. 2016;12:1073–1082. doi: 10.1080/15548627.2016.1179410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li Y., Dalli J., Chiang N., Baron R.M., Quintana C., Serhan C.N. Plasticity of leukocytic exudates in resolving acute inflammation is regulated by microRNA and proresolving mediators. Immunity. 2013;39:885–898. doi: 10.1016/j.immuni.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.McClure C., Brudecki L., Ferguson D.A., Yao Z.Q., Moorman J.P., McCall C.E., El Gazzar M. MicroRNA 21 (miR-21) and miR-181b couple with NFI-A to generate myeloid-derived suppressor cells and promote immunosuppression in late sepsis. Infect. Immun. 2014;82:3816–3825. doi: 10.1128/IAI.01495-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu J., Shi K., Chen M., Xu L., Hong J., Hu B., Yang X., Sun R. Elevated miR-155 expression induces immunosuppression via CD39+ regulatory T-cells in sepsis patient. Int. J. Infect. Dis. 2015;40:135–141. doi: 10.1016/j.ijid.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 98.McClure C., McPeak M.B., Youssef D., Yao Z.Q., McCall C.E., El Gazzar M. Stat3 and C/EBPβ synergize to induce miR-21 and miR-181b expression during sepsis. Immunol. Cell Biol. 2017;95:42–55. doi: 10.1038/icb.2016.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Curtis A.M., Fagundes C.T., Yang G., Palsson-McDermott E.M., Wochal P., McGettrick A.F., Foley N.H., Early J.O., Chen L., Zhang H. Circadian control of innate immunity in macrophages by miR-155 targeting Bmal1. Proc. Natl. Acad. Sci. USA. 2015;112:7231–7236. doi: 10.1073/pnas.1501327112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Qi J., Qiao Y., Wang P., Li S., Zhao W., Gao C. MicroRNA-210 negatively regulates LPS-induced production of proinflammatory cytokines by targeting NF-κB1 in murine macrophages. FEBS Lett. 2012;586:1201–1207. doi: 10.1016/j.febslet.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 101.Song X., Wang C.T., Geng X.H. MicroRNA-29a promotes apoptosis of monocytes by targeting STAT3 during sepsis. Genet. Mol. Res. 2015;14:13746–13753. doi: 10.4238/2015.October.28.37. [DOI] [PubMed] [Google Scholar]
- 102.Liu S., Liu C., Wang Z., Huang J., Zeng Q. MicroRNA-23a-5p acts as a potential biomarker for sepsis-induced acute respiratory distress syndrome in early stage. Cell. Mol. Biol. 2016;62:31–37. [PubMed] [Google Scholar]
- 103.Wang H., Meng K., Chen W.j., Feng D., Jia Y., Xie L. Serum miR-574-5p: a prognostic predictor of sepsis patients. Shock. 2012;37:263–267. doi: 10.1097/SHK.0b013e318241baf8. [DOI] [PubMed] [Google Scholar]
- 104.Wang H., Yu B., Deng J., Jin Y., Xie L. Serum miR-122 correlates with short-term mortality in sepsis patients. Crit. Care. 2014;18:704. doi: 10.1186/s13054-014-0704-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang Z.H., Liang Y.B., Tang H., Chen Z.B., Li Z.Y., Hu X.C., Ma Z.F. Dexamethasone down-regulates the expression of microRNA-155 in the livers of septic mice. PLoS ONE. 2013;8:e80547. doi: 10.1371/journal.pone.0080547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ledderose C., Möhnle P., Limbeck E., Schütz S., Weis F., Rink J., Briegel J., Kreth S. Corticosteroid resistance in sepsis is influenced by microRNA-124-induced downregulation of glucocorticoid receptor-α. Crit. Care Med. 2012;40:2745–2753. doi: 10.1097/CCM.0b013e31825b8ebc. [DOI] [PubMed] [Google Scholar]
- 107.Acosta-Herrera M., Lorenzo-Diaz F., Pino-Yanes M., Corrales A., Valladares F., Klassert T.E., Valladares B., Slevogt H., Ma S.F., Villar J., Flores C. Lung transcriptomics during protective ventilatory support in sepsis-induced acute lung injury. PLoS ONE. 2015;10:e0132296. doi: 10.1371/journal.pone.0132296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang Q., Zhao C., Cai Q., Zhu H. [Expression of microRNA-155 and regulative T cell in sepsis patients and their relationship] Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2014;26:179–183. doi: 10.3760/cma.j.issn.2095-4352.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 109.Caserta S., Kern F., Cohen J., Drage S., Newbury S.F., Llewelyn M.J. Circulating plasma microRNAs can differentiate human sepsis and systemic inflammatory response syndrome (SIRS) Sci. Rep. 2016;6:28006. doi: 10.1038/srep28006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yang W., Wu H., Zhang H., Liu H., Wei Y., Shi B. [Prognostic value of Picco monitoring combined with plasma microRNA-150 detection in septic shock patients] Zhejiang Da Xue Xue Bao Yi Xue Ban. 2015;44:659–664. doi: 10.3785/j.issn.1008-9292.2015.11.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang H.J., Wang B.Z., Zhang P.J., Deng J., Zhao Z.R., Zhang X., Xiao K., Feng D., Jia Y.H., Liu Y.N., Xie L.X. Identification of four novel serum protein biomarkers in sepsis patients encoded by target genes of sepsis-related miRNAs. Clin. Sci. (Lond.) 2014;126:857–867. doi: 10.1042/CS20130301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schlosser K., McIntyre L.A., White R.J., Stewart D.J. Customized internal reference controls for improved assessment of circulating microRNAs in disease. PLoS ONE. 2015;10:e0127443. doi: 10.1371/journal.pone.0127443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Leelahavanichkul A., Somparn P., Panich T., Chancharoenthana W., Wongphom J., Pisitkun T., Hirankarn N., Eiam-Ong S. Serum miRNA-122 in acute liver injury induced by kidney injury and sepsis in CD-1 mouse models. Hepatol. Res. 2015;45:1341–1352. doi: 10.1111/hepr.12501. [DOI] [PubMed] [Google Scholar]
- 114.Wang X., Zhao Q., Matta R., Meng X., Liu X., Liu C.-G., Nelin L.D., Liu Y. Inducible nitric-oxide synthase expression is regulated by mitogen-activated protein kinase phosphatase-1. J. Biol. Chem. 2009;284:27123–27134. doi: 10.1074/jbc.M109.051235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.El Gazzar M., McCall C.E. MicroRNAs distinguish translational from transcriptional silencing during endotoxin tolerance. J. Biol. Chem. 2010;285:20940–20951. doi: 10.1074/jbc.M110.115063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Banerjee S., Meng J., Das S., Krishnan A., Haworth J., Charboneau R., Zeng Y., Ramakrishnan S., Roy S. Morphine induced exacerbation of sepsis is mediated by tempering endotoxin tolerance through modulation of miR-146a. Sci. Rep. 2013;3:1977. doi: 10.1038/srep01977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Doxaki C., Kampranis S.C., Eliopoulos A.G., Spilianakis C., Tsatsanis C. Coordinated regulation of miR-155 and miR-146a genes during induction of endotoxin tolerance in macrophages. J. Immunol. 2015;195:5750–5761. doi: 10.4049/jimmunol.1500615. [DOI] [PubMed] [Google Scholar]
- 118.Nahid M.A., Pauley K.M., Satoh M., Chan E.K. miR-146a is critical for endotoxin-induced tolerance: implication in innate immunity. J. Biol. Chem. 2009;284:34590–34599. doi: 10.1074/jbc.M109.056317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dai Y., Jia P., Fang Y., Liu H., Jiao X., He J.C., Ding X. miR-146a is essential for lipopolysaccharide (LPS)-induced cross-tolerance against kidney ischemia/reperfusion injury in mice. Sci. Rep. 2016;6:27091. doi: 10.1038/srep27091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nahid M.A., Satoh M., Chan E.K. Interleukin 1β-responsive microRNA-146a is critical for the cytokine-induced tolerance and cross-tolerance to Toll-like receptor ligands. J. Innate Immun. 2015;7:428–440. doi: 10.1159/000371517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Androulidaki A., Iliopoulos D., Arranz A., Doxaki C., Schworer S., Zacharioudaki V., Margioris A.N., Tsichlis P.N., Tsatsanis C. The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity. 2009;31:220–231. doi: 10.1016/j.immuni.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wang Y., Tan L., Jin J., Sun H., Chen Z., Tan X., Su Y., Shi C. Non-cultured dermal-derived mesenchymal cells attenuate sepsis induced by cecal ligation and puncture in mice. Sci. Rep. 2015;5:16973. doi: 10.1038/srep16973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sari A.N., Korkmaz B., Serin M.S., Kacan M., Unsal D., Buharalioglu C.K., Sahan Firat S., Manthati V.L., Falck J.R., Malik K.U., Tunctan B. Effects of 5,14-HEDGE, a 20-HETE mimetic, on lipopolysaccharide-induced changes in MyD88/TAK1/IKKβ/IκB-α/NF-κB pathway and circulating miR-150, miR-223, and miR-297 levels in a rat model of septic shock. Inflamm. Res. 2014;63:741–756. doi: 10.1007/s00011-014-0747-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhao X., Liu D., Gong W., Zhao G., Liu L., Yang L., Hou Y. The Toll-like receptor 3 ligand, poly(I:C), improves immunosuppressive function and therapeutic effect of mesenchymal stem cells on sepsis via inhibiting miR-143. Stem Cells. 2014;32:521–533. doi: 10.1002/stem.1543. [DOI] [PubMed] [Google Scholar]
- 125.Arango D., Diosa-Toro M., Rojas-Hernandez L.S., Cooperstone J.L., Schwartz S.J., Mo X., Jiang J., Schmittgen T.D., Doseff A.I. Dietary apigenin reduces LPS-induced expression of miR-155 restoring immune balance during inflammation. Mol. Nutr. Food Res. 2015;59:763–772. doi: 10.1002/mnfr.201400705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ho M.S., Mei S.H., Stewart D.J. The immunomodulatory and therapeutic effects of mesenchymal stromal cells for acute lung injury and sepsis. J. Cell. Physiol. 2015;230:2606–2617. doi: 10.1002/jcp.25028. [DOI] [PubMed] [Google Scholar]
- 127.Shukla K., Sharma A.K., Ward A., Will R., Hielscher T., Balwierz A., Breunig C., Münstermann E., König R., Keklikoglou I., Wiemann S. MicroRNA-30c-2-3p negatively regulates NF-κB signaling and cell cycle progression through downregulation of TRADD and CCNE1 in breast cancer. Mol. Oncol. 2015;9:1106–1119. doi: 10.1016/j.molonc.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhang J., Ding C., Shao Q., Liu F., Zeng Z., Nie C., Qian K. [The protective effects of transfected microRNA-146a on mice with sepsis-induced acute lung injury in vivo] Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2015;27:591–594. doi: 10.3760/cma.j.issn.2095-4352.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 129.Möhnle P., Schütz S.V., van der Heide V., Hübner M., Luchting B., Sedlbauer J., Limbeck E., Hinske L.C., Briegel J., Kreth S. MicroRNA-146a controls Th1-cell differentiation of human CD4+ T lymphocytes by targeting PRKCε. Eur. J. Immunol. 2015;45:260–272. doi: 10.1002/eji.201444667. [DOI] [PubMed] [Google Scholar]
- 130.Zhao G., Su Z., Song D., Mao Y., Mao X. The long noncoding RNA MALAT1 regulates the lipopolysaccharide-induced inflammatory response through its interaction with NF-κB. FEBS Lett. 2016;590:2884–2895. doi: 10.1002/1873-3468.12315. [DOI] [PubMed] [Google Scholar]
- 131.Sun Y., Li Q., Gui H., Xu D.-P., Yang Y.-L., Su D.-F., Liu X. MicroRNA-124 mediates the cholinergic anti-inflammatory action through inhibiting the production of pro-inflammatory cytokines. Cell Res. 2013;23:1270–1283. doi: 10.1038/cr.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Guo C., Goodwin A., Buie J.N.J., Cook J., Halushka P., Argraves K., Zingarelli B., Zhang X., Wang L., Fan H. A stromal cell-derived factor 1α analogue improves endothelial cell function in lipopolysaccharide-induced acute respiratory distress syndrome. Mol. Med. 2016;22:115–123. doi: 10.2119/molmed.2015.00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sun C., Xue L., Zhu Z., Zhang F., Yang R., Yuan X., Jia Z., Liu Q. Insights from lncRNAs profiling of MIN6 beta cells undergoing inflammation. Mediators Inflamm. 2016;2016:9275106. doi: 10.1155/2016/9275106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yu X.-J., Zou L.-H., Jin J.-H., Xiao F., Li L., Liu N., Yang J.F., Zou T. Long noncoding RNAs and novel inflammatory genes determined by RNA sequencing in human lymphocytes are up-regulated in permanent atrial fibrillation. Am. J. Transl. Res. 2017;9:2314–2326. [PMC free article] [PubMed] [Google Scholar]
- 135.Wang S.-Y., Fan X.-L., Yu Q.-N., Deng M.-X., Sun Y.-Q., Gao W.-X., Li C.L., Shi J.B., Fu Q.L. The lncRNAs involved in mouse airway allergic inflammation following induced pluripotent stem cell-mesenchymal stem cell treatment. Stem Cell Res. Ther. 2017;8:2. doi: 10.1186/s13287-016-0456-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wang X., Wang X., Liu X., Wang X., Xu J., Hou S., Zhang X., Ding Y. miR-15a/16 are upreuglated in the serum of neonatal sepsis patients and inhibit the LPS-induced inflammatory pathway. Int. J. Clin. Exp. Med. 2015;8:5683–5690. [PMC free article] [PubMed] [Google Scholar]
- 137.Tacke F., Roderburg C., Benz F., Cardenas D.V., Luedde M., Hippe H.-J., Frey N., Vucur M., Gautheron J., Koch A. Levels of circulating miR-133a are elevated in sepsis and predict mortality in critically ill patients. Crit. Care Med. 2014;42:1096–1104. doi: 10.1097/CCM.0000000000000131. [DOI] [PubMed] [Google Scholar]
- 138.Wang H.-J., Deng J., Wang J.-Y., Zhang P.-J., Xin Z., Xiao K., Feng D., Jia Y.H., Liu Y.N., Xie L.X. Serum miR-122 levels are related to coagulation disorders in sepsis patients. Clin. Chem. Lab. Med. 2014;52:927–933. doi: 10.1515/cclm-2013-0899. [DOI] [PubMed] [Google Scholar]
- 139.Sun X., Icli B., Wara A.K., Belkin N., He S., Kobzik L., Hunninghake G.M., Vera M.P., Blackwell T.S., Baron R.M., Feinberg M.W., MICU Registry MicroRNA-181b regulates NF-κB-mediated vascular inflammation. J. Clin. Invest. 2012;122:1973–1990. doi: 10.1172/JCI61495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Vasilescu C., Rossi S., Shimizu M., Tudor S., Veronese A., Ferracin M., Nicoloso M.S., Barbarotto E., Popa M., Stanciulea O. MicroRNA fingerprints identify miR-150 as a plasma prognostic marker in patients with sepsis. PLoS ONE. 2009;4:e7405. doi: 10.1371/journal.pone.0007405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Xiao C., Calado D.P., Galler G., Thai T.-H., Patterson H.C., Wang J., Rajewsky N., Bender T.P., Rajewsky K. miR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 142.Flierl M.A., Rittirsch D., Huber-Lang M.S., Sarma J.V., Ward P.A. Molecular events in the cardiomyopathy of sepsis. Mol. Med. 2008;14:327–336. doi: 10.2119/2007-00130.Flierl. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sluijter J.P., Doevendans P.A. Sepsis-associated cardiac dysfunction is controlled by small RNA molecules. J. Mol. Cell. Cardiol. 2016;97:67–69. doi: 10.1016/j.yjmcc.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 144.Wang X., Huang W., Yang Y., Wang Y., Peng T., Chang J., Caldwell C.C., Zingarelli B., Fan G.C. Loss of duplexmiR-223 (5p and 3p) aggravates myocardial depression and mortality in polymicrobial sepsis. Biochim. Biophys. Acta. 2014;1842:701–711. doi: 10.1016/j.bbadis.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wang H., Bei Y., Huang P., Zhou Q., Shi J., Sun Q., Zhong J., Li X., Kong X., Xiao J. Inhibition of miR-155 protects against LPS-induced cardiac dysfunction and apoptosis in mice. Mol. Ther. Nucleic Acids. 2016;5:e374. doi: 10.1038/mtna.2016.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Jia Z., Wang J., Shi Q., Liu S., Wang W., Tian Y., Lu Q., Chen P., Ma K., Zhou C. SOX6 and PDCD4 enhance cardiomyocyte apoptosis through LPS-induced miR-499 inhibition. Apoptosis. 2016;21:174–183. doi: 10.1007/s10495-015-1201-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Vincent J.-L., Yagushi A., Pradier O. Platelet function in sepsis. Crit. Care Med. 2002;30(5, Suppl):S313–S317. doi: 10.1097/00003246-200205001-00022. [DOI] [PubMed] [Google Scholar]
- 148.Sakr Y. Heparin-induced thrombocytopenia in the ICU: an overview. Crit. Care. 2011;15:211. doi: 10.1186/cc9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cui Y.-L., Wang B., Gao H.-M., Xing Y.-H., Li J., Li H.-J., Lin Z., Wang Y.Q. Interleukin-18 and miR-130a in severe sepsis patients with thrombocytopenia. Patient Prefer. Adherence. 2016;10:313–319. doi: 10.2147/PPA.S95588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Riedemann N.C., Guo R.-F., Ward P.A. Novel strategies for the treatment of sepsis. Nat. Med. 2003;9:517–524. doi: 10.1038/nm0503-517. [DOI] [PubMed] [Google Scholar]
- 151.Zhao H., Anand A.R., Ganju R.K. Slit2-Robo4 pathway modulates lipopolysaccharide-induced endothelial inflammation and its expression is dysregulated during endotoxemia. J. Immunol. 2014;192:385–393. doi: 10.4049/jimmunol.1302021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chatterjee V., Beard R.S., Jr., Reynolds J.J., Haines R., Guo M., Rubin M., Guido J., Wu M.H., Yuan S.Y. MicroRNA-147b regulates vascular endothelial barrier function by targeting ADAM15 expression. PLoS ONE. 2014;9:e110286. doi: 10.1371/journal.pone.0110286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Vlassov A.V., Magdaleno S., Setterquist R., Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta. 2012;1820:940–948. doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 154.Sun D., Zhuang X., Zhang S., Deng Z.-B., Grizzle W., Miller D., Zhang H.G. Exosomes are endogenous nanoparticles that can deliver biological information between cells. Adv. Drug Deliv. Rev. 2013;65:342–347. doi: 10.1016/j.addr.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 155.Bang C., Thum T. Exosomes: new players in cell-cell communication. Int. J. Biochem. Cell Biol. 2012;44:2060–2064. doi: 10.1016/j.biocel.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 156.EL Andaloussi S., Mäger I., Breakefield X.O., Wood M.J. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013;12:347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 157.Conde-Vancells J., Rodriguez-Suarez E., Embade N., Gil D., Matthiesen R., Valle M., Elortza F., Lu S.C., Mato J.M., Falcon-Perez J.M. Characterization and comprehensive proteome profiling of exosomes secreted by hepatocytes. J. Proteome Res. 2008;7:5157–5166. doi: 10.1021/pr8004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mathivanan S., Fahner C.J., Reid G.E., Simpson R.J. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012;40:D1241–D1244. doi: 10.1093/nar/gkr828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Subra C., Grand D., Laulagnier K., Stella A., Lambeau G., Paillasse M., De Medina P., Monsarrat B., Perret B., Silvente-Poirot S. Exosomes account for vesicle-mediated transcellular transport of activatable phospholipases and prostaglandins. J. Lipid Res. 2010;51:2105–2120. doi: 10.1194/jlr.M003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Laulagnier K., Motta C., Hamdi S., Roy S., Fauvelle F., Pageaux J.-F., Kobayashi T., Salles J.P., Perret B., Bonnerot C., Record M. Mast cell- and dendritic cell-derived exosomes display a specific lipid composition and an unusual membrane organization. Biochem. J. 2004;380:161–171. doi: 10.1042/BJ20031594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Olver C., Vidal M. Proteomic analysis of secreted exosomes. Subcell. Biochem. 2007;43:99–131. doi: 10.1007/978-1-4020-5943-8_7. [DOI] [PubMed] [Google Scholar]
- 162.Wubbolts R., Leckie R.S., Veenhuizen P.T., Schwarzmann G., Möbius W., Hoernschemeyer J., Slot J.W., Geuze H.J., Stoorvogel W. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J. Biol. Chem. 2003;278:10963–10972. doi: 10.1074/jbc.M207550200. [DOI] [PubMed] [Google Scholar]
- 163.Zakharova L., Svetlova M., Fomina A.F. T cell exosomes induce cholesterol accumulation in human monocytes via phosphatidylserine receptor. J. Cell. Physiol. 2007;212:174–181. doi: 10.1002/jcp.21013. [DOI] [PubMed] [Google Scholar]
- 164.Xiang X., Poliakov A., Liu C., Liu Y., Deng Z.B., Wang J., Cheng Z., Shah S.V., Wang G.J., Zhang L. Induction of myeloid-derived suppressor cells by tumor exosomes. Int. J. Cancer. 2009;124:2621–2633. doi: 10.1002/ijc.24249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Taylor D.D., Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 166.Rabinowits G., Gerçel-Taylor C., Day J.M., Taylor D.D., Kloecker G.H. Exosomal microRNA: a diagnostic marker for lung cancer. Clin. Lung Cancer. 2009;10:42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 167.Feng D.-Q., Huang B., Li J., Liu J., Chen X.-M., Xu Y.-M., Chen X., Zhang H.B., Hu L.H., Wang X.Z. Selective miRNA expression profile in chronic myeloid leukemia K562 cell-derived exosomes. Asian Pac. J. Cancer Prev. 2013;14:7501–7508. doi: 10.7314/apjcp.2013.14.12.7501. [DOI] [PubMed] [Google Scholar]
- 168.Michael A., Bajracharya S.D., Yuen P.S., Zhou H., Star R.A., Illei G.G., Alevizos I. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 2010;16:34–38. doi: 10.1111/j.1601-0825.2009.01604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Théry C., Zitvogel L., Amigorena S. Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 170.Raposo G., Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Tian T., Wang Y., Wang H., Zhu Z., Xiao Z. Visualizing of the cellular uptake and intracellular trafficking of exosomes by live-cell microscopy. J. Cell. Biochem. 2010;111:488–496. doi: 10.1002/jcb.22733. [DOI] [PubMed] [Google Scholar]
- 172.Raposo G., Nijman H.W., Stoorvogel W., Liejendekker R., Harding C.V., Melief C.J., Geuze H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996;183:1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Sheldon H., Heikamp E., Turley H., Dragovic R., Thomas P., Oon C.E., Leek R., Edelmann M., Kessler B., Sainson R.C. New mechanism for Notch signaling to endothelium at a distance by Delta-like 4 incorporation into exosomes. Blood. 2010;116:2385–2394. doi: 10.1182/blood-2009-08-239228. [DOI] [PubMed] [Google Scholar]
- 174.Chairoungdua A., Smith D.L., Pochard P., Hull M., Caplan M.J. Exosome release of β-catenin: a novel mechanism that antagonizes Wnt signaling. J. Cell Biol. 2010;190:1079–1091. doi: 10.1083/jcb.201002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Li J., Zhang Y., Liu Y., Dai X., Li W., Cai X., Yin Y., Wang Q., Xue Y., Wang C. Microvesicle-mediated transfer of microRNA-150 from monocytes to endothelial cells promotes angiogenesis. J. Biol. Chem. 2013;288:23586–23596. doi: 10.1074/jbc.M113.489302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Aucher A., Rudnicka D., Davis D.M. MicroRNAs transfer from human macrophages to hepato-carcinoma cells and inhibit proliferation. J. Immunol. 2013;191:6250–6260. doi: 10.4049/jimmunol.1301728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bang C., Batkai S., Dangwal S., Gupta S.K., Foinquinos A., Holzmann A., Just A., Remke J., Zimmer K., Zeug A. Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J. Clin. Invest. 2014;124:2136–2146. doi: 10.1172/JCI70577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhou W., Fong M.Y., Min Y., Somlo G., Liu L., Palomares M.R., Yu Y., Chow A., O’Connor S.T., Chin A.R. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25:501–515. doi: 10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ying W., Riopel M., Bandyopadhyay G., Dong Y., Birmingham A., Seo J.B., Ofrecio J.M., Wollam J., Hernandez-Carretero A., Fu W. Adipose tissue macrophage-derived exosomal miRNAs can modulate in vivo and in vitro insulin sensitivity. Cell. 2017;171:372–384.e12. doi: 10.1016/j.cell.2017.08.035. [DOI] [PubMed] [Google Scholar]
- 180.Zhang Y., Kim M.S., Jia B., Yan J., Zuniga-Hertz J.P., Han C., Cai D. Hypothalamic stem cells control ageing speed partly through exosomal miRNAs. Nature. 2017;548:52–57. doi: 10.1038/nature23282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Real J.M., Ferreira L.R.P., Esteves G.H., Koyama F.C., Dias M.V.S., Bezerra-Neto J.E., Cunha-Neto E., Machado F.R., Salomão R., Azevedo L.C.P. Exosomes from patients with septic shock convey miRNAs related to inflammation and cell cycle regulation: new signaling pathways in sepsis? Crit. Care. 2018;22:68. doi: 10.1186/s13054-018-2003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wang X., Gu H., Qin D., Yang L., Huang W., Essandoh K., Wang Y., Caldwell C.C., Peng T., Zingarelli B., Fan G.C. Exosomal miR-223 contributes to mesenchymal stem cell-elicited cardioprotection in polymicrobial sepsis. Sci. Rep. 2015;5:13721. doi: 10.1038/srep13721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Alexander M., Hu R., Runtsch M.C., Kagele D.A., Mosbruger T.L., Tolmachova T., Seabra M.C., Round J.L., Ward D.M., O’Connell R.M. Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nat. Commun. 2015;6:7321. doi: 10.1038/ncomms8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Panich T., Chancharoenthana W., Somparn P., Issara-Amphorn J., Hirankarn N., Leelahavanichkul A. Urinary exosomal activating transcriptional factor 3 as the early diagnostic biomarker for sepsis-induced acute kidney injury. BMC Nephrol. 2017;18:10. doi: 10.1186/s12882-016-0415-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Zhang X. East Tennessee State University; 2016. Endothelial HSPA12B is a novel protein for the preservation of cardiovascular function in polymicrobial sepsis via exosome miR-126. PhD thesis. [Google Scholar]
- 187.Xu Y., Ku X., Wu C., Cai C., Tang J., Yan W. Exosomal proteome analysis of human plasma to monitor sepsis progression. Biochem. Biophys. Res. Commun. 2018;499:856–861. doi: 10.1016/j.bbrc.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 188.Wisler J.R., Singh K., Mccarty A.R., Abouhashem A.S.E., Christman J.W., Sen C.K. Proteomic pathway analysis of monocyte-derived exosomes during surgical sepsis identifies immunoregulatory functions. Surg. Infect. (Larchmt.) 2020;21:101–111. doi: 10.1089/sur.2019.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Xu J., Feng Y., Jeyaram A., Jay S.M., Zou L., Chao W. Circulating plasma extracellular vesicles from septic mice induce inflammation via microRNA- and TLR7-dependent mechanisms. J. Immunol. 2018;201:3392–3400. doi: 10.4049/jimmunol.1801008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Dakhlallah D.A., Wisler J., Gencheva M., Brown C.M., Leatherman E.R., Singh K., Brundage K., Karsies T., Dakhlallah A., Witwer K.W. Circulating extracellular vesicle content reveals de novo DNA methyltransferase expression as a molecular method to predict septic shock. J. Extracell. Vesicles. 2019;8:1669881. doi: 10.1080/20013078.2019.1669881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Zhang R., Zhu Y., Li Y., Liu W., Yin L., Yin S., Ji C., Hu Y., Wang Q., Zhou X. Human umbilical cord mesenchymal stem cell exosomes alleviate sepsis-associated acute kidney injury via regulating microRNA-146b expression. Biotechnol. Lett. 2020;42:669–679. doi: 10.1007/s10529-020-02831-2. [DOI] [PubMed] [Google Scholar]
- 192.Oehmcke S., Westman J., Malmström J., Mörgelin M., Olin A.I., Kreikemeyer B., Herwald H. A novel role for pro-coagulant microvesicles in the early host defense against streptococcus pyogenes. PLoS Pathog. 2013;9:e1003529. doi: 10.1371/journal.ppat.1003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Levi M., van der Poll T. Coagulation and sepsis. Thromb. Res. 2017;149:38–44. doi: 10.1016/j.thromres.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 194.Rivers E., Nguyen B., Havstad S., Ressler J., Muzzin A., Knoblich B., Peterson E., Tomlanovich M., Early Goal-Directed Therapy Collaborative Group Early goal-directed therapy in the treatment of severe sepsis and septic shock. N. Engl. J. Med. 2001;345:1368–1377. doi: 10.1056/NEJMoa010307. [DOI] [PubMed] [Google Scholar]
- 195.Delabranche X., Boisramé-Helms J., Asfar P., Berger A., Mootien Y., Lavigne T., Grunebaum L., Lanza F., Gachet C., Freyssinet J.M. Microparticles are new biomarkers of septic shock-induced disseminated intravascular coagulopathy. Intensive Care Med. 2013;39:1695–1703. doi: 10.1007/s00134-013-2993-x. [DOI] [PubMed] [Google Scholar]
- 196.Zafrani L., Ince C., Yuen P.S. Microparticles during sepsis: target, canary or cure? Intensive Care Med. 2013;39:1854–1856. doi: 10.1007/s00134-013-3047-0. [DOI] [PubMed] [Google Scholar]
- 197.Joop K., Berckmans R.J., Nieuwland R., Berkhout J., Romijn F.P., Hack C.E., Sturk A. Microparticles from patients with multiple organ dysfunction syndrome and sepsis support coagulation through multiple mechanisms. Thromb. Haemost. 2001;85:810–820. [PubMed] [Google Scholar]
- 198.Wang J.G., Manly D., Kirchhofer D., Pawlinski R., Mackman N. Levels of microparticle tissue factor activity correlate with coagulation activation in endotoxemic mice. J. Thromb. Haemost. 2009;7:1092–1098. doi: 10.1111/j.1538-7836.2009.03448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Zhang Y., Meng H., Ma R., He Z., Wu X., Cao M., Yao Z., Zhao L., Li T., Deng R. Circulating microparticles, blood cells, and endothelium induce procoagulant activity in sepsis through phosphatidylserine exposure. Shock. 2016;45:299–307. doi: 10.1097/SHK.0000000000000509. [DOI] [PubMed] [Google Scholar]
- 200.Martínez de Lizarrondo S., Roncal C., Calvayrac O., Rodríguez C., Varo N., Purroy A., Lorente L., Rodríguez J.A., Doeuvre L., Hervás-Stubbs S. Synergistic effect of thrombin and CD40 ligand on endothelial matrix metalloproteinase-10 expression and microparticle generation in vitro and in vivo. Arterioscler. Thromb. Vasc. Biol. 2012;32:1477–1487. doi: 10.1161/ATVBAHA.112.248773. [DOI] [PubMed] [Google Scholar]
- 201.Matsumoto H., Yamakawa K., Ogura H., Koh T., Matsumoto N., Shimazu T. Enhanced expression of cell-specific surface antigens on endothelial microparticles in sepsis-induced disseminated intravascular coagulation. Shock. 2015;43:443–449. doi: 10.1097/SHK.0000000000000331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Holnthoner W., Bonstingl C., Hromada C., Muehleder S., Zipperle J., Stojkovic S., Redl H., Wojta J., Schöchl H., Grillari J. Endothelial cell-derived extracellular vesicles size-dependently exert procoagulant activity detected by thromboelastometry. Sci. Rep. 2017;7:3707. doi: 10.1038/s41598-017-03159-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Letsiou E., Sammani S., Zhang W., Zhou T., Quijada H., Moreno-Vinasco L., Dudek S.M., Garcia J.G. Pathologic mechanical stress and endotoxin exposure increases lung endothelial microparticle shedding. Am. J. Respir. Cell Mol. Biol. 2015;52:193–204. doi: 10.1165/rcmb.2013-0347OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Beer K.B., Wehman A.M. Mechanisms and functions of extracellular vesicle release in vivo-What we can learn from flies and worms. Cell Adhes. Migr. 2017;11:135–150. doi: 10.1080/19336918.2016.1236899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Yuana Y., Sturk A., Nieuwland R. Extracellular vesicles in physiological and pathological conditions. Blood Rev. 2013;27:31–39. doi: 10.1016/j.blre.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 206.Lee S.K., Yang S.-H., Kwon I., Lee O.-H., Heo J.H. Role of tumour necrosis factor receptor-1 and nuclear factor-κB in production of TNF-α-induced pro-inflammatory microparticles in endothelial cells. Thromb. Haemost. 2014;112:580–588. doi: 10.1160/TH13-11-0975. [DOI] [PubMed] [Google Scholar]
- 207.Shaver C.M., Woods J., Clune J.K., Grove B.S., Wickersham N.E., McNeil J.B., Shemancik G., Ware L.B., Bastarache J.A. Circulating microparticle levels are reduced in patients with ARDS. Crit. Care. 2017;21:120. doi: 10.1186/s13054-017-1700-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Mutschler D.K., Larsson A.O., Basu S., Nordgren A., Eriksson M.B. Effects of mechanical ventilation on platelet microparticles in bronchoalveolar lavage fluid. Thromb. Res. 2002;108:215–220. doi: 10.1016/s0049-3848(03)00005-7. [DOI] [PubMed] [Google Scholar]
- 209.Vion A.C., Birukova A.A., Boulanger C.M., Birukov K.G. Mechanical forces stimulate endothelial microparticle generation via caspase-dependent apoptosis-independent mechanism. Pulm. Circ. 2013;3:95–99. doi: 10.4103/2045-8932.109921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Cabrera-Benítez N.E., Valladares F., García-Hernández S., Ramos-Nuez Á., Martín-Barrasa J.L., Martínez-Saavedra M.-T., Rodríguez-Gallego C., Muros M., Flores C., Liu M. Altered profile of circulating endothelial-derived microparticles in ventilator-induced lung injury. Crit. Care Med. 2015;43:e551–e559. doi: 10.1097/CCM.0000000000001280. [DOI] [PubMed] [Google Scholar]
- 211.Müller I., Klocke A., Alex M., Kotzsch M., Luther T., Morgenstern E., Zieseniss S., Zahler S., Preissner K., Engelmann B. Intravascular tissue factor initiates coagulation via circulating microvesicles and platelets. FASEB J. 2003;17:476–478. doi: 10.1096/fj.02-0574fje. [DOI] [PubMed] [Google Scholar]
- 212.Boisramé-Helms J., Delabranche X., Degirmenci S.-E., Zobairi F., Berger A., Meyer G., Burban M., Mostefai H.A., Levy B., Toti F., Meziani F. Pharmacological modulation of procoagulant microparticles improves haemodynamic dysfunction during septic shock in rats. Thromb. Haemost. 2014;111:154–164. doi: 10.1160/TH13-04-0313. [DOI] [PubMed] [Google Scholar]
- 213.Nieuwland R., Berckmans R.J., McGregor S., Böing A.N., Romijn F.P.T.M., Westendorp R.G., Hack C.E., Sturk A. Cellular origin and procoagulant properties of microparticles in meningococcal sepsis. Blood. 2000;95:930–935. [PubMed] [Google Scholar]
- 214.Hellum M., Øvstebø R., Brusletto B.S., Berg J.P., Brandtzaeg P., Henriksson C.E. Microparticle-associated tissue factor activity correlates with plasma levels of bacterial lipopolysaccharides in meningococcal septic shock. Thromb. Res. 2014;133:507–514. doi: 10.1016/j.thromres.2013.12.031. [DOI] [PubMed] [Google Scholar]
- 215.Delabranche X., Quenot J.-P., Lavigne T., Mercier E., François B., Severac F., Grunebaum L., Mehdi M., Zobairi F., Toti F., on behalf to the Clinical Research in Intensive Care and Sepsis Network Early detection of disseminated intravascular coagulation during septic shock: a multicenter prospective study. Crit. Care Med. 2016;44:e930–e939. doi: 10.1097/CCM.0000000000001836. [DOI] [PubMed] [Google Scholar]
- 216.Chen H., Wang X., Yan X., Cheng X., He X., Zheng W. lncRNA MALAT1 regulates sepsis-induced cardiac inflammation and dysfunction via interaction with miR-125b and p38 MAPK/NFκB. Int. Immunopharmacol. 2018;55:69–76. doi: 10.1016/j.intimp.2017.11.038. [DOI] [PubMed] [Google Scholar]
- 217.Chen Y., Qiu J., Chen B., Lin Y., Chen Y., Xie G., Qiu J., Tong H., Jiang D. Long non-coding RNA NEAT1 plays an important role in sepsis-induced acute kidney injury by targeting miR-204 and modulating the NF-κB pathway. Int. Immunopharmacol. 2018;59:252–260. doi: 10.1016/j.intimp.2018.03.023. [DOI] [PubMed] [Google Scholar]
- 218.Huang Q., Huang C., Luo Y., He F., Zhang R. Circulating lncRNA NEAT1 correlates with increased risk, elevated severity and unfavorable prognosis in sepsis patients. Am. J. Emerg. Med. 2018;36:1659–1663. doi: 10.1016/j.ajem.2018.06.008. [DOI] [PubMed] [Google Scholar]
- 219.Liu W.Q., Wang Y.J., Zheng Y., Chen X. Effects of long non-coding RNA NEAT1 on sepsis-induced brain injury in mice via NF-κB. Eur. Rev. Med. Pharmacol. Sci. 2019;23:3933–3939. doi: 10.26355/eurrev_201905_17822. [DOI] [PubMed] [Google Scholar]
- 220.Huang S., Qian K., Zhu Y., Huang Z., Luo Q., Qing C. Diagnostic value of the lncRNA NEAT1 in peripheral blood mononuclear cells of patients with sepsis. Dis. Markers. 2017;2017:7962836. doi: 10.1155/2017/7962836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Gui F., Peng H., Liu Y. Elevated circulating lnc-ANRIL/miR-125a axis level predicts higher risk, more severe disease condition, and worse prognosis of sepsis. J. Clin. Lab. Anal. 2019;33:e22917. doi: 10.1002/jcla.22917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Shen J., Zhang J., Jiang X., Wang H., Pan G. lncRNA HOX transcript antisense RNA accelerated kidney injury induced by urine-derived sepsis through the miR-22/high mobility group box 1 pathway. Life Sci. 2018;210:185–191. doi: 10.1016/j.lfs.2018.08.041. [DOI] [PubMed] [Google Scholar]
- 223.Jiang Z.J., Zhang M.Y., Fan Z.W., Sun W.L., Tang Y. Influence of lncRNA HOTAIR on acute kidney injury in sepsis rats through regulating miR-34a/Bcl-2 pathway. Eur. Rev. Med. Pharmacol. Sci. 2019;23:3512–3519. doi: 10.26355/eurrev_201904_17717. [DOI] [PubMed] [Google Scholar]
- 224.Fang Y., Hu J., Wang Z., Zong H., Zhang L., Zhang R., Sun L. lncRNA H19 functions as an Aquaporin 1 competitive endogenous RNA to regulate microRNA-874 expression in LPS sepsis. Biomed. Pharmacother. 2018;105:1183–1191. doi: 10.1016/j.biopha.2018.06.007. [DOI] [PubMed] [Google Scholar]
- 225.Zeng Q., Wu J., Yang S. Circulating lncRNA ITSN1-2 is upregulated, and its high expression correlates with increased disease severity, elevated inflammation, and poor survival in sepsis patients. J. Clin. Lab. Anal. 2019;33:e22836. doi: 10.1002/jcla.22836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Rüdebusch J., Benkner A., Nath N., Kaderali L., Klingel K., Eckstein G., Meitinger T., Fielitz J., Grube K., Felix S.B. Soluble guanylate cyclase as a therapeutic target in heart failure: myocardial gene expression in response to sGC stimulation in pressure overload. Eur. Heart J. 2019;40(Suppl 1) ehz748.0373. [Google Scholar]
- 227.Liu X., Hong C., Wu S., Song S., Yang Z., Cao L., Song T., Yang Y. Downregulation of lncRNA TUG1 contributes to the development of sepsis-associated acute kidney injury via regulating miR-142-3p/sirtuin 1 axis and modulating NF-κB pathway. J. Cell. Biochem. 2019;120:11331–11341. doi: 10.1002/jcb.28409. [DOI] [PubMed] [Google Scholar]
- 228.Shen J., Liu L., Zhang F., Gu J., Pan G. lncRNA TapSAKI promotes inflammation injury in HK-2 cells and urine derived sepsis-induced kidney injury. J. Pharm. Pharmacol. 2019;71:839–848. doi: 10.1111/jphp.13049. [DOI] [PubMed] [Google Scholar]
- 229.Zhuang Y.T., Xu D.Y., Wang G.Y., Sun J.L., Huang Y., Wang S.Z. IL-6 induced lncRNA MALAT1 enhances TNF-α expression in LPS-induced septic cardiomyocytes via activation of SAA3. Eur. Rev. Med. Pharmacol. Sci. 2017;21:302–309. [PubMed] [Google Scholar]
- 230.Yu Z., Rayile A., Zhang X., Li Y., Zhao Q. Ulinastatin protects against lipopolysaccharide-induced cardiac microvascular endothelial cell dysfunction via downregulation of lncRNA MALAT1 and EZH2 in sepsis. Int. J. Mol. Med. 2017;39:1269–1276. doi: 10.3892/ijmm.2017.2920. [DOI] [PubMed] [Google Scholar]
- 231.Ma H., Wang X., Ha T., Gao M., Liu L., Wang R., Yu K., Kalbfleisch J.H., Kao R.L., Williams D.L., Li C. MicroRNA-125b prevents cardiac dysfunction in polymicrobial sepsis by targeting TRAF6-mediated nuclear factor κB activation and p53-mediated apoptotic signaling. J. Infect. Dis. 2016;214:1773–1783. doi: 10.1093/infdis/jiw449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Zhou W., Wang J., Li Z., Li J., Sang M. MicroRNA-205-5b inhibits HMGB1 expression in LPS-induced sepsis. Int. J. Mol. Med. 2016;38:312–318. doi: 10.3892/ijmm.2016.2613. [DOI] [PubMed] [Google Scholar]
- 233.Wang H., Bei Y., Shen S., Huang P., Shi J., Zhang J., Sun Q., Chen Y., Yang Y., Xu T. miR-21-3p controls sepsis-associated cardiac dysfunction via regulating SORBS2. J. Mol. Cell. Cardiol. 2016;94:43–53. doi: 10.1016/j.yjmcc.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 234.Wu M., Gu J.T., Yi B., Tang Z.Z., Tao G.C. MicroRNA-23b regulates the expression of inflammatory factors in vascular endothelial cells during sepsis. Exp. Ther. Med. 2015;9:1125–1132. doi: 10.3892/etm.2015.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Sheng B., Zhao L., Zang X., Zhen J., Chen W. miR-375 ameliorates sepsis by downregulating miR-21 level via inhibiting JAK2-STAT3 signaling. Biomed. Pharmacother. 2017;86:254–261. doi: 10.1016/j.biopha.2016.11.147. [DOI] [PubMed] [Google Scholar]
- 236.van der Heide V., Möhnle P., Rink J., Briegel J., Kreth S. Down-regulation of microRNA-31 in CD4+ T cells contributes to immunosuppression in human sepsis by promoting TH2 skewing. Anesthesiology. 2016;124:908–922. doi: 10.1097/ALN.0000000000001031. [DOI] [PubMed] [Google Scholar]
- 237.Shao Y., Li J., Cai Y., Xie Y., Ma G., Li Y., Chen Y., Liu G., Zhao B., Cui L., Li K. The functional polymorphisms of miR-146a are associated with susceptibility to severe sepsis in the Chinese population. Mediators Inflamm. 2014;2014:916202. doi: 10.1155/2014/916202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Wang L., Wang H.-C., Chen C., Zeng J., Wang Q., Zheng L., Yu H.D. Differential expression of plasma miR-146a in sepsis patients compared with non-sepsis-SIRS patients. Exp. Ther. Med. 2013;5:1101–1104. doi: 10.3892/etm.2013.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Wang Z., Ruan Z., Mao Y., Dong W., Zhang Y., Yin N., Jiang L. miR-27a is up regulated and promotes inflammatory response in sepsis. Cell. Immunol. 2014;290:190–195. doi: 10.1016/j.cellimm.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 240.Jiang Y., Zhou H., Ma D., Chen Z.K., Cai X. MicroRNA-19a and CD22 comprise a feedback loop for B cell response in sepsis. Med. Sci. Monit. 2015;21:1548–1555. doi: 10.12659/MSM.894321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Han Y., Dai Q.-C., Shen H.-L., Zhang X.-W. Diagnostic value of elevated serum miRNA-143 levels in sepsis. J. Int. Med. Res. 2016;44:875–881. doi: 10.1177/0300060516645003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Hsieh C.-H., Rau C.-S., Jeng J.C., Chen Y.-C., Lu T.-H., Wu C.-J., Wu Y.C., Tzeng S.L., Yang J.C. Whole blood-derived microRNA signatures in mice exposed to lipopolysaccharides. J. Biomed. Sci. 2012;19:69. doi: 10.1186/1423-0127-19-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Wu S.-C., Yang J.C.-S., Rau C.-S., Chen Y.-C., Lu T.-H., Lin M.-W., Tzeng S.L., Wu Y.C., Wu C.J., Hsieh C.H. Profiling circulating microRNA expression in experimental sepsis using cecal ligation and puncture. PLoS ONE. 2013;8:e77936. doi: 10.1371/journal.pone.0077936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Wang Y., Li T., Wu B., Liu H., Luo J., Feng D., Shi Y. STAT1 regulates MD-2 expression in monocytes of sepsis via miR-30a. Inflammation. 2014;37:1903–1911. doi: 10.1007/s10753-014-9922-1. [DOI] [PubMed] [Google Scholar]
- 245.Wang H., Zhang P., Chen W., Feng D., Jia Y., Xie L. Serum microRNA signatures identified by Solexa sequencing predict sepsis patients’ mortality: a prospective observational study. PLoS ONE. 2012;7:e38885. doi: 10.1371/journal.pone.0038885. [DOI] [PMC free article] [PubMed] [Google Scholar]