Abstract
Retinoblastoma (RB) is one of the most common ophthalmic tumors, and most of the patients have been identified as advanced at the time of diagnosis, which is directly related to high mortality. Recent studies showed that long noncoding RNA (lncRNA) and miRNAs play key roles in the development、progression、or treatment of cancer, such as RB. However, the role of lncRNA -TP73-AS1 in RB remains unclear. In this study, we performed functional and mechanistic investigation of miRNA-874-3p-TP73-AS1 interaction in RB. The experiments results revealed that miRNA-874-3p had anti-oncogenic functions in RB. Moreover, the bioinformatics analysis shown that TP73-AS1 could bind to miRNA-874-3p. TP73-AS1 was inversely correlated with miRNA-874-3p expression. Furthermore, studies confirmed that TP73-AS1 negatively regulated miRNA-874-3p expression via functioning as a ceRNA. In a word, our results suggest that the TP73-AS1/ miRNA-874-3p / TFAP2B (transcription factor activating enhancer-binding protein 2B) pathway contributes to the progression of RB, which may provide novel insights into the function of lncRNA-driven retinoblastogenesis.
Graphical abstract
Keywords: lncRNA-TP73-AS1, miR-874-3p, Transcription factor activating enhancer-binding protein 2B (TFAP2B), Retinoblastoma(RB), WNT/β-catenin pathway
Introduction
Retinoblastoma (RB) is the most common tumor, and it is usually a child’s fatal eye cancer (Shields and Shields 2010). Most patients are diagnosed as advanced at the time of diagnosis, which is associated with a high mortality rate in medulloblastoma (Dimaras et al. 2010). Many retinoblastoma survivors have a risk of blindness and fatal eye loss due to metastasis and drug-resistant chemotherapy or radiation therapy (Fabian et al. 2017, 2018). Therefore, the molecular mechanism of studying the development and progression of retinoblastoma is crucial to find new effective therapeutic targets for retinoblastoma.
LncRNA is a non-protein-encoding transcript of more than 200 nt in length and is involved in a variety of biological functions such as gene transcriptional regulation (Goodrich and Kugel 2006), RNA processing (Gong and Maquat 2011), regulation of apoptosis and invasion (Khaitan et al. 2011), and miRNA host genes (Eis et al. 2005), etc. The regulation of IncRNAs occurs in a variety of human diseases, including nervous system diseases (Quan et al. 2017). In addition, lncRNA can be used not only as a bait for splicing factors (Beltran et al. 2008; Tripathi et al. 2010), but also as a competitive endogenous RNAs (ceRNAs) in some tumors (Fu et al. 2015; Sun et al. 2016). Studies have shown that lncRNAs, including THOR (Chen et al. 2013) and XIST (Zhang et al. 2015), are associated with RB tumorigenesis. Similarly, miRNA-874 has been described as tumor suppressor miRNA in hepatocellular carcinoma cells, thereby inhibiting EMT via DOR/EGFR/ERK signalling pathway (Zhang et al. 2018). However, the specific clinical significance and molecular mechanism of lncRNA-TP73-AS1 in the development of RB tumorigenesis remains unknown.
The role of miRNA-874-3p in the regulation of TP73-AS1 has never been investigated. This study will provide new and important molecular targets for the diagnosis and treatment of RB.
Materials and methods
Clinical tissues
Fifty pairs of RB tissues and tissues adjacent to cancer from the same patients were collected from Tianjin Medical University Eye Hospital (Tianjin, China) and rapid frozen in liquid nitrogen for subsequent Pathological analysis. All protocols of this study were approved by Ethical Oversight Committee of Tianjin Medical University Eye Hospital (Tianjin, China) approved this study. The patients’ clinical characteristics are shown in Table 1.
Table 1.
A correlation analysis between the TP73-AS1 expression level and the clinicopathological characteristics
| Vairables | No. cases(70) | TP73-AS1 expression |
p value | |
|---|---|---|---|---|
| Low (%) | High(%) | |||
| Age | ||||
| <50 years(%) | 30 | 15 | 15 | 0.678 |
| ≥50 years(%) | 40 | 18 | 22 | |
| Gender | ||||
| Male | 31 | 14 | 17 | 0.767 |
| Female | 39 | 19 | 20 | |
| Tumor size | ||||
| ≥5 cm | 33 | 11 | 22 | 0.029 |
| <5 cm | 37 | 22 | 15 | |
| TNM stage | ||||
| I-II | 28 | 18 | 10 | 0.019 |
| III-IV | 42 | 15 | 27 | |
| LNM | ||||
| No | 30 | 20 | 10 | 0.005 |
| Yes | 40 | 13 | 27 | |
| Histological type | ||||
| Squamous | 41 | 19 | 22 | 0.873 |
| Adenocarcinoma | 29 | 14 | 15 | |
aχ 2 test. P-values in bold print indicate statistically significant differences. TNM, Tumor Node Metastasis
Cell culture
The human RB cell lines (Weri-Rb1, SO-RB50, Y79 and HXO-RB44) and a acute retinal pegment epitheliitis (ARPE-19) were purchased from the American Type Culture Collection (Manassas, VA, USA). Cells were cultured in DMEM with 10% FBS, streptomycin (100 μg/mL) and penicillin (100 μg/mL). Cells were maintained at 37 °C with 5% CO2.
Real-time quantitative PCR (RT-qPCR)
Total RNA was purified from tissues or cell lines by Thermo Scientific GeneJET RNA Purification Kit. Quantitative PCR assays were performed via using SYBR-Green PCR Master Mix on a VIAA7 qPCR System (Life Technologies). The primers were as follows:
TFAP2B-For: 5′-TGAAGATGCCAATAACAGCGGCA-3′
and TFAP2B-Rev: 5′-GGAGCAAAACACCTCGCCGGT-3′;
β-catenin-For: 5′-ACTACCACAGCTCCTTCTCT-3′
and β-catenin-Rev: 5′-AAATCCCTGTTCCCACTCATAC-3′;
TCF4-For: 5′-CCACCCATTTCTTTGCTGAAC-3′
and TCF4-Rev: 5′-CCCTGACTCTTAACACCAACTC-3′;
c-myc-For: 5′-TGAGGAGGAACAAGAAGATG-3′
and c-myc-Rev: 5′- ATCCAGACTCTGACCTTTT-3′;
cyclinD1-For: 5′-GGGTTGTGCTACAGATGATAGAG-3′
and cyclinD1-Rev: 5′-AGACGCCTCCTTTGTGTTAAT-3′;
Bcl-2-For: 5′-GGTGGGGTCATGTGTGTGG-3′
and Bcl-2-Rev: 5′-CGGTTCAGGTACTCAGTCATCC-3′;
β-actin-For: 5′-GGGAAATCGTGCGTGACATTAAG-3′
and β-actin-Rev: 5′-GTCAGGCAGCTCGTAGCTCT-3′
Cellular proliferation and colony formation
To investigate cell viability, we performed 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay. Firstly, 3 × 103 Y79 or HXO-RB44 cells that were transfected planted in 96-well plates for 24 h. The cellular proliferation activity rate was detected at 24, 48 and 72 h using a Quant Universal Microplate Spectrophotometer at 490 nm. For the colony formation assay, Y79 or HXO-RB44 cells that were transfected with pcDNA-TP73-AS1 and TP73-AS1-shRNA or their relative controls were planted in 12-well normal attachment plates (300 cells per well) and cultured for 12 days until colonies had formed. Subsequently, the wells of these plates were rinsed out by PBS, fixed with methanol and communities greater than 50 cells were photographed and counted.
Plasmid construction and EGFP reporter assay
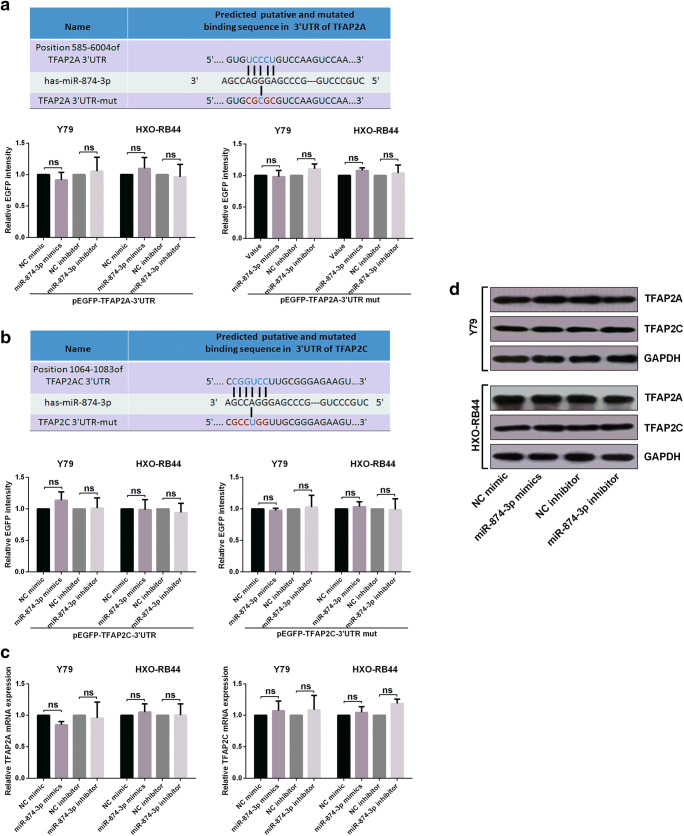
First of all, to confirm the direct target of miR-874-3p, we designed sequences including the two sequences of TFAP2B and two sequences of TP73-AS1 in the Figs. 4a and 5a. The sequences were synthesized as the oligonucleotides. Then the forward and reverse ones were made into double strand DNA fragments. Then we ligated the four DNA fragments into the EGFP reporter plasmids Vector. Sequencing confirmed the ligation was successful. So we successfully inserted the wild type and mutant fragments of the 3’UTR of TFAP2B and TP73-AS1 containing the binding sites of miR-874-3p into the EGFP reporter vector. For miR-874-3p mimics, an overexpression vector containing a miR-874-3p precursor region was amplified from the genomic DNA and inserted into the vector of pcDNA3. The plasmids pEGFP-TFAP2B-3’UTR or pEGFP-TFAP2B-3’UTR-mut were co-transfected into the cells with miR-874-3p mimics, miR-874-3p inhibitor or their relative controls respectively, and then culture for 48 h. The intensities of EGFP fluorescence were determined with a spectrophotometer. TP73-AS1 EGFP reporter assay is the same as above. Then we calculated the ratio of luminescence from the experimental reporter to luminescence from the control reporter, normalized this ratio to the ratio of control wells.
Fig. 4.
TP73-AS1 up-regulated TFAP2B by crosstalk with miR-874-3p in RB cells. A The diagram shows the bioinformatic prediction of miR-874-3p that may target TFAP2B, and the predicted miR-874-3p binding sites in TFAP2B mRNA 3’UTR or the mutational 3’UTR of TFAP2B mRNA were confirmed. B miR-874-3p expression levels in four retinoblastoma cell lines and a normal cell line were analyzed by RT-qPCR assay. C a negative correlations between miR-874-3p and TP73-AS1 was confirmed by a Pearson correlation analysis. D-E RT-qPCR assay confirmed a negative correlations between miR-874-3p and TP73-AS1. F-G EGFP reporter assay showed that TFAP2B is a target of miR-874-3p. H-I TFAP2B mRNA and protein expression levels were measured by RT-qPCR and Western blot. J-K RT-qPCR and Western blot showed that the expression of TFAP2B is affected by miR-874-3p and TP73-AS1
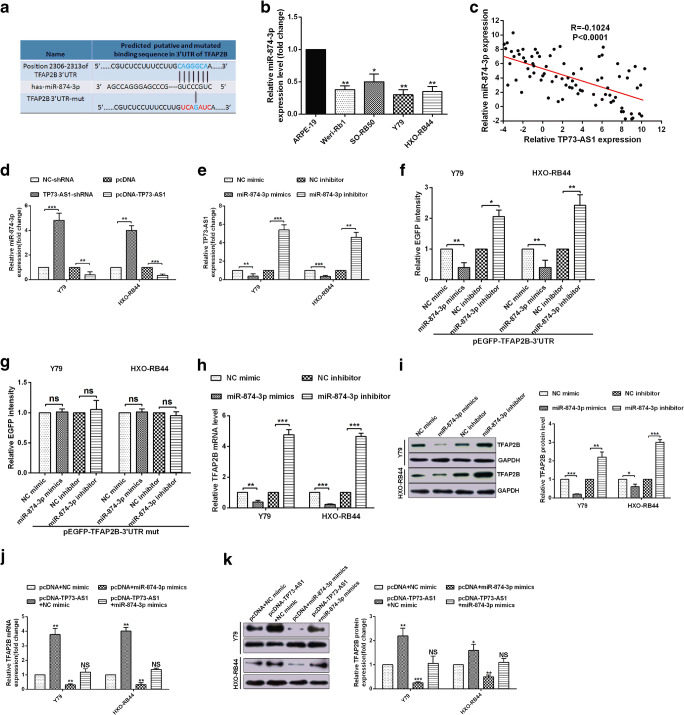
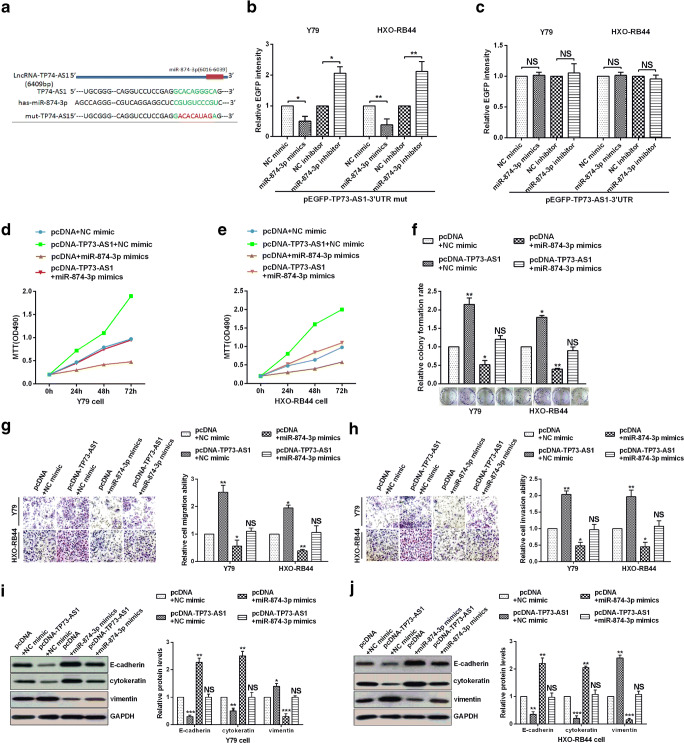
Fig. 5.
TP73-AS1 decoyed miR-874-3p to promote TFAP2B-mediated proliferation, migration, invasion and EMT in RB cells. A Sequence alignment of TP73-AS1 with potential wild type and mutant type of miR-874-3p targeting sites. B-C EGFP reporter assay showed that TP73-AS1 is a target of miR-874-3p. D-E Y79 and HXO-RB44 cellular viabilities were confirmed by MTT assay. F Colony formation ability of Y79 and HXO-RB44 cells were detected by colony formation assay. G-H The migration and invasion ability of Y79 and HXO-RB44 cells were confirmed by transwell assays. I E-cadherin, cytokeratin and Vimentin protein levels in Y79 and HXO-RB44 cells were confirmed by Western blot assays
Flow cytometry
Y79 and HXO-RB44 cells after transfected were incubated for 48 h, and were harvested for cell cycle distribution analysis. The cells were washed washed with ice-cold PBS for four times. Subsequently, the cells were washed again after fixation, incubated in a dark room with RNase A and treated with PI stain (Beyotime Biotech).Then, the cells were sorted by A BD FACSCanto™ II flow cytometry system and the cell cycle were analysed by ModFit LT software package. The percentage of cells undergoing apoptosis was evaluated by annexin V-FITC.
Transwell assays
In the transwell assays, Y79 and HXO-RB44 cells transfected with plasmids for 24 h were collected and resuspended in serum-free medium. Subsequently, 1.5–2 × 105 of Y79 and HXO-RB44 cells were seeded in the upper chambers(8 μm pore size, Corning, USA) inserts with or without matrigel (BD Biosciences, USA). Meanwhile,medium containing 30% FBS was added in the lower chamber and incubated at 37 °C for 48 h.At the end of incubation, the cells in the upper surface of the membrane were removed with a cotton swab.Cells in lower chamber were fixed with methanol.Cell migration and invasion were determined by bright-field microscopy.
Western blot
RIPA lysis buffer lysed the transfected cells for total protein extracts, including protease inhibitor cocktail (Roche, Mannheim, Germany). The concentrations of total protein were detected by the BCA method (Beyotime). Protein extracts were separated by 10% SDS-PAGE, and then transferred to PVDF membrane. The PVDF membranes with proteins were blocked with 5% skim fat dry milk in 0.1% Tween-20 in Tris-buffered saline (TBS) for 1 h to block the non-specific sites on the blots, and then incubated with primary antibodies at 4 °C overnight. Following washing, the membranes were incubated with HRP-linked secondary antibodies (Jackson, USA). Western blot data were quantified by Alpha Innotech (San Leandro, CA, USA) imaging software. The antibodies used in the present study were E-cadherin (1:1000; cat. no. 3195; Cell Signaling Technology, Inc.), cytokeratin (1:2000; cat no. sc-15,367; Santa Cruz Biotechnology, Inc.), Vimentin (1:1000; cat. no. 5741 Cell Signaling Technology, Inc.), GAPDH (1:1000; sc-47,724; Santa Cruz Biotechnology, Inc.), Bcl-2 (1:500; SAB4300340; Sigma-Aldrich; Merck KGaA), TFAP2B (1:1000, Abcam, Cambridge, UK, ab18114), β-catenin (1:2000, Abcam, Cambridge, UK, ab16051), TCF4 (1:1000, Abcam, Cambridge, UK, ab185736), c-myc (1:1000, Abcam, Cambridge, UK, ab39688), cyclinD1 (1:1000, Abcam, Cambridge, UK, ab194972), P-β-catenin (1:1000, Abcam, Cambridge, UK, ab27798), H2A.X (1:2000, Abcam, Cambridge, UK, ab188819) and a horseradish peroxidase-conjugated secondary antibody (1:2000, cat. no. 7074; Cell Signaling Technology, Inc.).
Statistical analysis
All data shown are expressed as the mean ± SD from at least three independent experiments. Statistical analysis was demonstrated by unpaired two-sided Student’s t-tests. Fisher’s and chi-square tests were employed to examine differences between the two groups in clinicopathological characteristics. P-values less than 0.05 were considered significant.(*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; NS, not significant).
Results
TP73-AS1 is up-regulated in human retinoblastoma tissues and cells
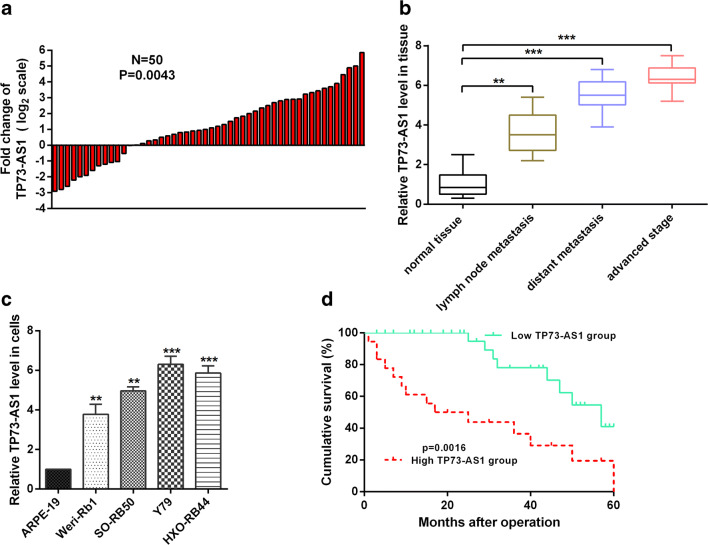
LncRNAs play a certain role in the regulation of tumors (Fu et al. 2015; Sun et al. 2016). Does TP73-AS1 play a regulatory role in the development of RB? Therefore, we examined the levels of TP73-AS1 in RB tissue and adjacent tissues by RT-qPCR. The expression of TP73-AS1 was significantly up-regulated in 38 of the 50 RB tissues (76%) compared to the adjacent tissues (Fig. 1a). Additionally, there is a potential correlation between expression levels of TP73-AS1 and clinicopathological features. We found that the TP73-AS1 expression level was significantly higher in patients with different clinicopathological stage (Fig. 1b). Moreover, we also examined the expression levels of TP73-AS1 in RB cells and ARPE-19 cell. The results confirmed that the expression of TP73-AS1 was significantly up-regulated in RB cells compared to ARPE-19 cells (Fig. 1c). The expression of TP73-AS1 was significantly increased in RB tissues and RB cells. Is the expression level of TP73-AS1 correlated with survival rate? We obtained a longer overall survival rate in RB patients with low expression of TP73-AS1 by Kaplan-Meier survival analysis (p < 0.05; Fig. 1d). Taken together, these data indicate that the expression level of TP73-AS1 is positively correlated with RB malignancies.
Fig. 1.
TP73-AS1 was up-regulated in RB tissues and cells. A RT-qPCR assay was used to detect TP73-AS1 expression levels in retinoblastoma and normal tissues. B TP73-AS1 expression was prominently increased in patients with advanced stage by RT-qPCR assay. C RT-qPCR assay was used to detect TP73-AS1 expression levels in RB cell lines and corresponding normal cell line. D Kaplan-Meier survival curves showed that TP73-AS1 expression in retinoblastoma patients
TP73-AS1 acts AS an oncogene in RB cells
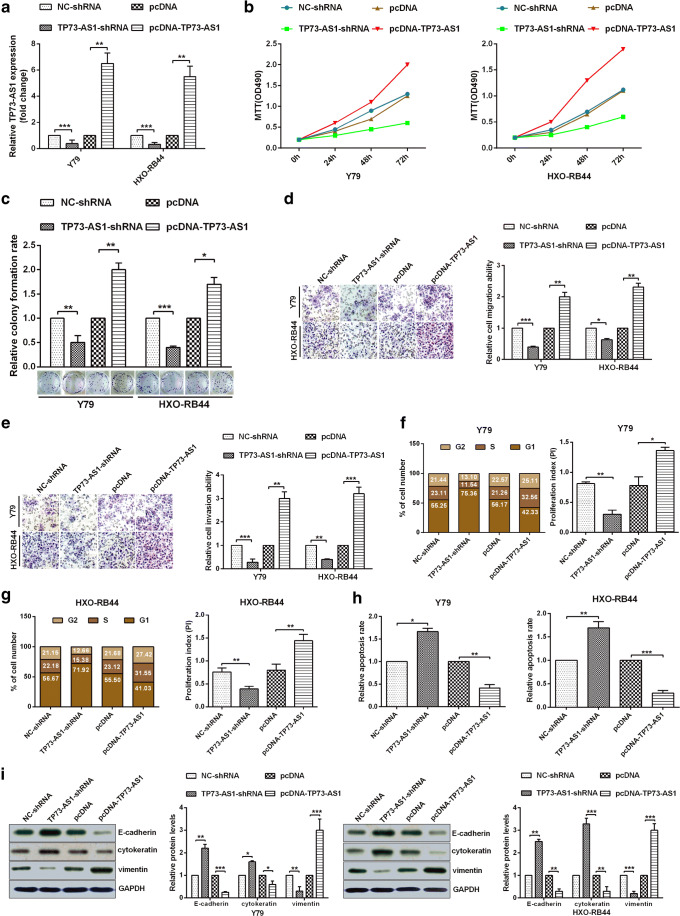
To demonstrate that TP73-AS1 is able to promote RB progression, we first transfected Y79 and HXO-RB44 cells with the relevant plasmids and verified the validity of the plasmid (Fig. 2a). The TP73-AS1 over-expression dramatically promoted the activity and the proliferation of the cells, and TP73-AS1 knockdown significantly inhibited the cells activity and proliferation in Y79 and HXO-RB44 cells (Fig. 2b, c). To confirmed that TP73-AS1 can affect the migration and invasion of HXO-RB44 and Y79 cells, Transwell experiments demonstrated that overexpression of TP73-AS1 significantly increased cell migration and invasion, and downregulation of TP73-AS1 significantly reduced cell migration and invasion ability (Fig. 2d, e). To demonstrate that TP73-AS1 can affect the proliferation and apoptosis of Y79 and HXO-RB44 cells, flow cytometry results showed that overexpression of TP73-AS1 could block cell cycle and inhibit cell apoptosis, while knockdown of TP73-AS1 could extend cell cycle and promote apoptosis (Fig. 2f-h). Finally, the key markers of EMT were examined by Western bolt experiments. The results showed that the expression levels of E-cadherin and cytokeratin were significantly decreased after overexpression of TP73-AS1, while the expression level of vimentin was significantly increased (Fig. 2i). In contrast, the expression levels of E-cadherin, cytokeratin and vimentin after knockdown of TP73-AS1 were opposite to those above (Fig. 2i). Decreased expression of e-cadherin and cytokeratin and increased expression of vimentin indicate EMT activation in RB cells. Therefore, TP73-AS1 played a tumour activator role by regulating cell EMT, cycle and apoptosis in RB cells.
Fig. 2.
TP73-AS1 promotes tumorigenesis in RB cell lines. A The expression of pcDNA-TP73-AS1 or TP73-AS1-shRNA was analyzed via RT-qPCR. B Cellular viabilities were confirmed by MTT assay. C Colony formation ability of Y79 and HXO-RB44 cells were confirmed by colony formation assay. D-E TP73-AS1 promoted cell migration ability and cell invasion ability were demonstrated via transwell migration assays or transwell invasion assays. F-G Flow cytometry assay showed the cell cycle in Y79 and HXO-RB44 cells, and proliferation index was counted via PI = (S + G2)/G1. H The apoptosis rate was demonstrated via Flow cytometry or AV/PI double staining in Y79 and HXO-RB44 cells. I Western blot assays showed that The protein levels of E-cadherin, cytokeratin, and Vimentin in Y79 and HXO-RB44 cells
TP73-AS1 promoted proliferation, migration and invasion via up-regulation of TFAP2B in RB cells
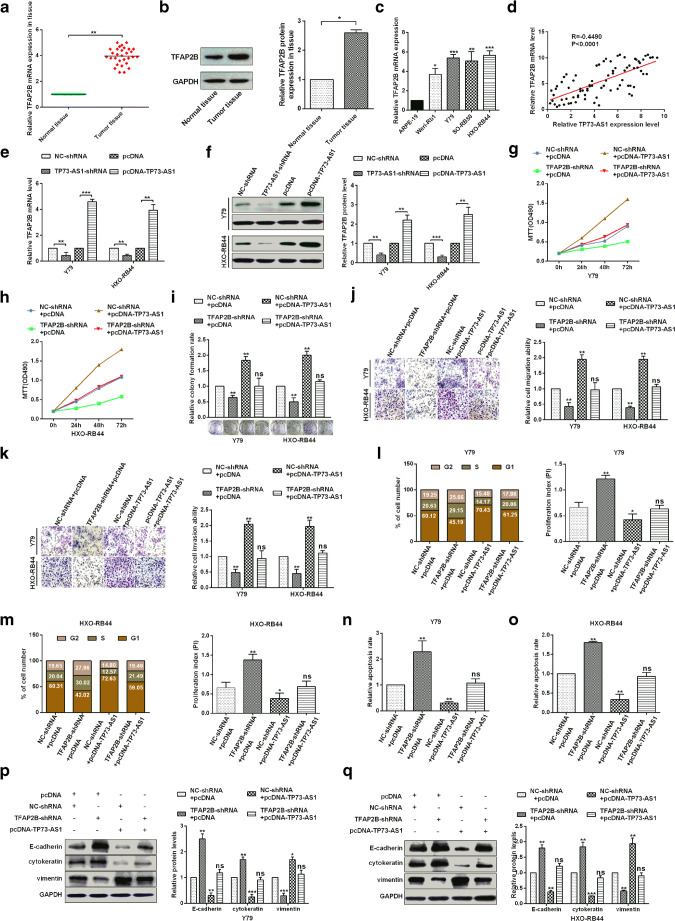
Many studies have shown that lncRNAs exerted multiple functions by regulating various downstream genes. According to other previous researches had confirmed that TFAP2B is involved in tumorigenesis and is a cancer-promoting factor that is closely related to tumor progression (Fu et al. 2014; Raap et al. 2018). Therefore, we sought to determine whether TP73-AS1 regulates RB cell proliferation, migration and invasion by regulating TFAP2B expression. Firstly, the levels of TFAP2B mRNA were detected in 30 paired RB tissues by RT-qPCR assay. The results showed that expression levels of TFAP2B mRNA were significantly up-regulated in RB tissues compared with adjacent tissues (Fig. 3a). The levels of TFAP2B protein were also detected in normal tissues and RB tissues by western blot, and the results showed that expression levels of TFAP2B protein were significantly up-regulated in RB tissues compared with adjacent tissues (Fig. 3b). In addition, the expression levels of TFAP2B mRNA were also up-regulated in RB cells compared with those in ARPE-19(Fig. 3c). Then we studied the correlation between TFAP2B and TP73-AS1, suggesting that TFAP2B and TP73-AS1 were positively correlated(Fig. 3d). To investigate whether TP73-AS1 affects TFAP2B expression in Y79 and HXO-RB44 cells, RT-qPCR was used to detect TFAP2B mRNA levels, and western blot was used to detect TFAP2B protein levels (Figs. 3e, f). The results showed that the overexpression of TP73-AS1 significantly up-regulated the expression of TFAP2B, while the knock down of TP73-AS1 significantly down-regulated the expression of TFAP2B. In addition, to investigate whether TP73-AS1 affects RB progression by regulating TFAP2B, TP73-AS1 over-expression significantly promoted the cells activity and proliferation and TFAP2B knockdown dramatically inhibited the cells activity and proliferation, whereas TFAP2B knockdown rescued the promotion of over-expression of TP73-AS1 on tumor cells activity and proliferation (Fig. 3g-i)、migration and invasion (Fig. 3j, k). Flow cytometry results showed that overexpression of TP73-AS1 promoted the cell cycle of Y79 and HXO-RB44 and inhibited their apoptosis. However, knockdown of TFAP2B could reversed the above results in Y79 and HXO-RB44 cells (Fig. 3l-o). Finally, Western bolt results showed that overexpression of TP73-AS1 inhibited E-cadherin or cytokeratin expression and promoted vimentin expression in cells (Fig. 3p, q). In contrast, TFAP2B knockdown could upregulate E-cadherin or cytokeratin expression and downregulate vimentin expression in Y79 and HXO-RB44 cells (Fig. 3p, q). Taken together, these results indicate that TP73-AS1 plays a carcinogenic role in RB cells by up-regulating the expression of TFAP2B.
Fig. 3.
TP73-AS1 promoted proliferation, migration and invasion via up-regulation of TFAP2B in RB cells. A Relative expression levels of TFAP2B mRNA in normal tissues and retinoblastoma tissues were analyzed via RT-qPCR. B Relative expression levels of TFAP2B protein in normal tissues and retinoblastoma tissues were analyzed via western blot. C TFAP2B expression levels in four retinoblastoma cell lines and a normal cell line were analyzed by RT-qPCR assay. D A positive correlations between TP73-AS1 and TFAP2B was confirmed by a Pearson correlation analysis. E-F qRT-PCR and western blotting were used to measure the expression of TFAP2B after TP73-AS1 regulation. G-H Analysis of TP73-AS1 and TFAP2B in Y79 and HXO-RB44 cellular viabilities were confirmed by MTT assay. I Colony formation ability of Y79 and HXO-RB44 cells were detected by colony formation assay. J-K TP73-AS1 promoted cell migration and cell invasion ability and TFAP2B inhibited cell migration and cell invasion ability, which were confirmed by transwell assays. L-M Flow cytometry assay detected the cell cycle in Y79 and HXO-RB44 by transfectting with pcDNA-TP73-AS1 and TFAP2B-shRNA and respective controls, and the proliferation index was counted via PI = (S + G2)/G1. N-O The apoptosis rate was demonstrated via Flow cytometry and AV/PI double staining in Y79 and HXO-RB44. P-Q. E-cadherin, cytokeratin and Vimentin protein levels in Y79 and HXO-RB44 cells were confirmed by Western blot assays
TP73-AS1 up-regulated TFAP2B by crosstalk with miR-874-3p in RB cells
To speculate whether TP73-AS1 might interact with miRNAs in retinoblastoma, We predicted potential miRNAs that interact with TP73-AS1 via lncRNA prediction software (DIANA-LncBase). MiR-874-3p was studied because TP73-AS1 and TFAP2B shared the similar miRNA response elements for miR-874-3p (Fig. 4a). The miR-874-3p expression in RB cells were determined by qRT-PCR (Fig. 4b). Moreover, inverse correlations of TP73-AS1 and miR-874-3p was presented by Pearson correlation analysis (Fig. 4c). We confirmed that increase or decrease TP73-AS1 negatively regulated miR-874-3p (Fig. 4d). Conversely, up-regulated and down-regulated of miR-874-3p also inversely affected in expression level of TP73-AS1 (Fig. 4e). In addition, we also confirmed that TFAP2B was a direct target of miR-874-3p and that miR-874-3p could negatively regulate TFAP2B mRNA expression (Fig. 4f-i). Finally, miR-874-3p were used to detect the effect that they might execute on TP73-AS1-induced TFAP2B elevation. As we expected, opposite to NC mimic, miR-874-3p mimics attenuated TP73-AS1 induced TFAP2B elevation (Fig. 4j, k). The above results indicate that TP73-AS1 up-regulated TFAP2B by crosstalk with miR-874-3p in RB cells.
TP73-AS1 decoyed miR-874-3p to promote TFAP2B-mediated proliferation, migration, invasion and EMT in RB cells
The above results demonstrate that TFAP2B is a target of miR-874-3p. In this section, we first demonstrate that TP73-AS1 is a target of miR-874-3p and that the site of miR-874-3p targeting TP73-AS1 is identical to TFAP2B (Fig. 5a-c). To confirm the role of TP73-AS1 could“crosstalk” with miR-874-3p and promoted TFAP2B-mediated retinoblastoma progression. The over-expression of TP73-AS1 enhanced cells activity and proliferation and up-regulation of miR-874-3p greatly inhibited cells activity and proliferation, whereas over-expression of miR-874-3p rescued the activity and proliferation of RB cells caused by over-expression of TP73-AS1 (Fig. 5d-f). Transwell migration and invasion assays also confirmed that the over-expression of TP73-AS1 obviously promoted migration and invasion of RB cells and up-regulation of miR-874-3p greatly hindered migration and invasion of RB cells, whereas over-expression of miR-874-3p reversed the migration and invasion of RB cells caused by over-expression of TP73-AS1 (Fig. 5g, h). Finally, Western bolt assays demonstrated that the E-cadherin and cytokeratin were down-regulated and vimentin was up-regulated in pcDNA-TP73-AS1 and NC-mimics cotransfected cells, and the E-cadherin or cytokeratin were up-regulated and vimentin weas down-regulated in pcDNA and miR-874-3p mimics cotransfected cells. Conversely, overexpression of miR-874-3p could rescued the up-regulation of E-cadherin or cytokeratin and down-regulation of vimentin caused by TP73-AS1 over-expression in Y79 and HXO-RB44 cells (Fig. 5i, j). All the results showed that TP73-AS1 decoyed miR-874-3p to promote TFAP2B-mediated proliferation, migration, invasion and EMT in RB cells.
TP73-AS1 interacted with miR-874-3p to facilitate RB tumorigenesis through Wnt/β-catenin pathway
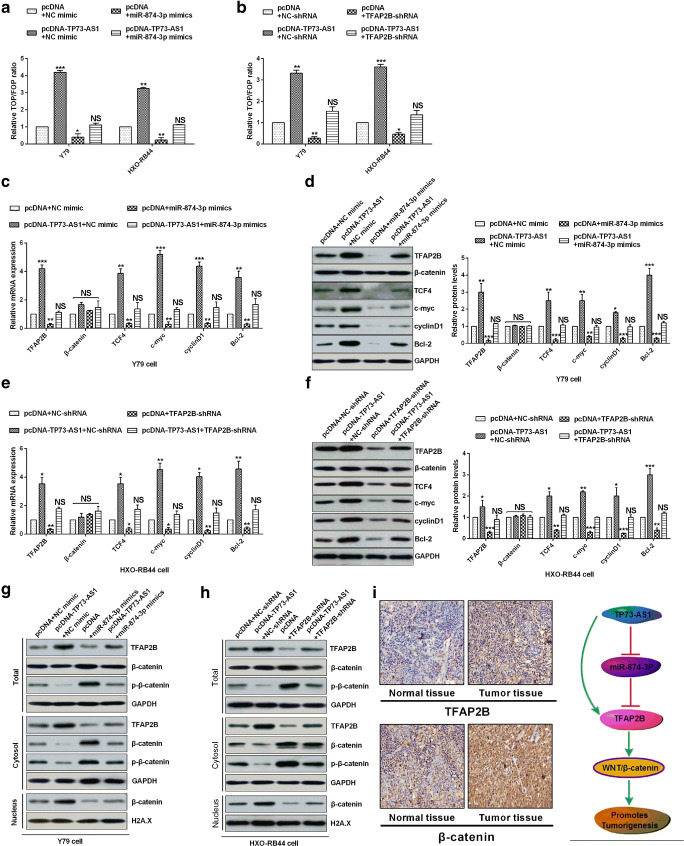
To verify the regulation of Wnt /β-catenin signaling pathway by TP73-AS1, we detected Wnt signaling after overexpressing TP73-AS1 and miR-874-3p in RB cells. As expected, over-expression TP73-AS1 increased Wnt/β-catenin activity in RB cells, whereas over-expression miR-874-3p dramatically inhibited the activity of Wnt/β-catenin caused by over-expression TP73-AS1 in RB cells (Fig. 6a). In addition, we also detected the effect of knockdown of TFAP2B on the endogenous Wnt/β-catenin activity in MB, and we found that TFAP2B knockdown significantly decreased Wnt/β-catenin activity and rescued Wnt/β-catenin activity because of causing TP73-AS1 over-expression in RB cells (Fig. 6b). Over-expression of TP73-AS1 consistently enhanced the mRNA and protein expression levels of TFAP2B, TCF4, Bcl-2, cyclin D1 and c-myc, and miR-874-3p over-expression significantly inhibited those mRNA and protein levels, but they had no effect on β-catenin, whereas miR-874-3p over-expression revoked the effects of TP73-AS1 over-expression on TFAP2B, TCF4, c-myc, cyclin D1 and Bcl-2 in Y79 and HXO-RB44 cells (Fig. 6c, d). TCF4, c-myc, cyclin D1 and bcl-2 are tumorigene-related factors. The overexpression of TP73-AS1 up-regulates the expression of tumorigene-related factors, and the overexpression of mir-874-3p saves the overexpression of tumorigene-related factors caused by the overexpression of TP73-AS1, indicating that the overexpression of TP73-AS1 promotes the occurrence of tumors. In addition, the effect of knockdown of TFAP2B induced the mRNA and protein levels of TFAP2B, TCF4, c-myc, cyclin D1 and Bcl-2 was the same as that miR-874-3p over-expression in HXO-RB44 cells (Fig. 6e, f). The results showed that TP73-AS1 promotes TFAP2B-mediated tumorigenesis in retinoblastoma via decoying of miRNA-874-3p. In the canonical Wnt pathway (Kim et al. 2013), β-catenin phosphorylation was repressed and β-catenin protein stability was increased and allowed to translocate to the nucleus in activity of Wnt/β-catenin. Nuclear and cytoplasmic extracts analysis had shown that over-expression of TP73-AS1 decreased the β-catenin phosphorylation levels in the cytoplasm and markedly promoted its nuclear translocation (Fig. 6g, h). On the contrary, miR-874-3p over-expression and knockdown of TFAP2B increased the β-catenin phosphorylation levels in the cytoplasm and supressed nuclear translocation and retrieved the effect of Wnt/β-catenin activity caused by TP73-AS1 over-expression (Fig. 6g, h). However, the total expression levels of β-catenin were not affected by TP73-AS1, miR-874-3p and TFAP2B (Fig. 6g, h). The results showed that TP73-AS1 activated the Wnt signaling pathway by promoting the expression of TFAP2B in the RB. In addition, we have also shown that Wnt signal activation in RB and normal tissues by IHC in Fig. 6i. The expression level of TFAP2B in RB tissues was significantly higher than that in normal tissues, and the expression level of β-catenin in the nucleus of RB tissues was significantly higher than that in normal tissues, indicating the activation of Wnt signaling pathway. To sum up, proposed model showed that TP73-AS1 promotes TFAP2B over-expression by interacting with miR-874-3p and promoting WNT/β-catenin pathway activation to promote tumorigenesis (Fig. 6i). These results showed that TP73-AS1 interacted with miR-874-3p to facilitate RB tumorigenesis through Wnt/β-catenin Pathway.
Fig. 6.
TP73-AS1 interacted with miR-874-3p to facilitate RB tumorigenesis through Wnt/β-catenin Pathway. A Luciferase reporter assays. B TOP-flash assay shown that the rescue effect of TFAP2B knockdown on WNT/β-catenin pathway caused by TP73-AS1. C-F RT-qPCR and Western blot analysis showed that TP73-AS1 and miR-874-3p could regulate the expression levels of TFAP2B and protein levels of β-catenin downstream targets. G-H Western blot analysis shown that cytoplasm or nuclear β-catenin protein levels in the Y79 or HXO-RB44 cells. I Proposed model showing that TP73-AS1 promotes TFAP2B over-expression by interacting with miR-874-3p and promoting WNT/β-catenin pathway activation to promote tumorigenesis
Discussion
It has been demonstrated that lncRNAs are emerging as functional regulators in various cancer processes, including proliferation, invasion, and metastasis (Hirata et al. 2015; Huarte 2015; Raveh et al. 2015). LncRNAs can act as a miRNA sponge and promote the expression of miRNA target genes by interacting with miRNAs, thereby promoting the development of cancer (Chang et al. 2016).
In this paper, we investigated the role of TP73-AS1 in RB and its regulatory mechanisms during RB development. Initially, in order to observe the function of TP73-AS1 in RB, we performed in vitro cell experiments, such as cell proliferation assays, apoptosis assays, and EMT assays. The results confirmed that inhibition of TP73-AS1 expression can inhibit the deterioration of RB, and promotion of TP73-AS1 expression can promote the progression of RB, resulting in TP73-AS1 as an oncogene in RB, which is consistent with previous reports (Zang et al. 2016).
Although TP73-AS1 is considered to be an oncogene in breast cancer, the biological molecular mechanisms involved in RB remain unclear. (Zhang et al. 2017) have indicated that lncRNA-CCAT1 up-regulated may promote EMT and decrease apoptosis in RB through negative regulation of miR-218-5p. Therefore, we also hypothesized that TP73-AS1 promotes the development of RB by interacting with miRNAs. Recently, (Zhang et al. 2018) has reported that miRNA-874 can suppress tumor EMT via targeting the DOR/EGFR/ERK signalling pathway in HCC. Combined these, miRNA-874-3p may server as a tumor suppressor in RB. Therefore, we examined the expression of miRNA-874-3p in RB tissues and cells by RT-qPCR assay, and the results showed that the expression of miRNA-874-3p was down-regulated in RB tissues and cell lines. After knockdown of TP73-AS1, the expression of miRNA-874-3p was detected by RT-qPCR. The results showed that the expression of miRNA-874-3p was up-regulated after knockdown of TP73-AS1. Furthermore, in order to determine the targeting relationship between TP73-AS1 and miRNA-874-3p, we demonstrated that miRNA-874-3p targets TP73-AS1 by luciferase assay. At the same time, in order to demonstrate whether the function of TP73-AS1 in RB cells is achieved by mediating miRNA-874-3p. By inhibiting the expression of miRNA-874-3p and then detecting the effect of TP73-AS1 knockdown on RB cells, we confirmed that miRNA-874-3p knockdown can partially reverse the effect of knockdown of TP73-AS1 on RB cells. Taken together, these results are consistent with our previous hypothesis, demonstrating that TP73-AS1 as an oncogene in RB is achieved by down-regulating miRNA-874-3p.
TP73-AS1 can induce miRNA-874-3p inhibition, then whether TP73-AS1 can affect the expression of potential target genes by inducing miRNA-874-3p, so we further study the target gene TFAP2B of miRNA-874-3p. Clear studies have shown that TFAP2B is a cancer-promoting factor for many cancers and can be used as a potential diagnostic marker and therapeutic target for cancer, such as lung adenocarcinoma (Fu et al. 2014) and neuroblastoma (Ikram et al. 2016). Firstly, we studied the expression of TFAP2B in RB tissues and obtained a significant increase in the expression of TFAP2B in RB tissues. Furthermore, the expression of TFAP2B was significantly inhibited by transfection of TP73-AS1 shRNA vector in vitro. The inhibition of TFAP2B expression was partially reversed by co-transfection of TP73-AS1 shRNA vector with miRNA-874-3p inhibitor. Taken together, it is shown that the TP73-AS1 / miRNA-874-3p / TFAP2B axis may be important signaling pathways involved in RB progression.
TFAP2B can promote breast cancer progression by activating the WNT/β-catenin pathway (Cizkova et al. 2010). We suspect that TP73-AS1 also activated WNT/β-catenin pathway in RB, and we proved that the related proteins of WNT/β-catenin pathway. Those results showed that TP73-AS1 significantly activated WNT/β-catenin pathwayby ceRNA in RB. RT-qPCR and Western blot assay confirmed that TFAP2B was the upstream signal of WNT/β-catenin pathway in RB cells. Therefore, TP73-AS1 was up-regulated in RB and cells. It promoted tumourigenesis by enhancing TFAP2B expression and facilitating the WNT/β-catenin pathway activation in RB.
Funding information
National Natural Science Foundation Project (NO. 81800813).
Compliance with ethical standards
Conflict of interest
There are none.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article has been retracted. Please see the retraction notice for more detail: https://doi.org/10.1007/s12079-023-00774-7"
Change history
6/13/2023
This article has been retracted. Please see the Retraction Notice for more detail: 10.1007/s12079-023-00774-7
Contributor Information
Lina Wang, Email: 30819312@nankai.edu.cn.
Fengyuan Sun, Email: ydyk2016@126.com.
References
- Beltran M, Puig I, Peña C, García JM, Alvarez AB, Peña R, Bonilla F, de Herreros AG. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22(6):756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YN, Zhang K, Hu ZM, Qi HX, Shi ZM, Han XH, Han YW, Hong W. Hypoxia-regulated lncRNAs in cancer. Gene. 2016;575(1):1–8. doi: 10.1016/j.gene.2015.08.049. [DOI] [PubMed] [Google Scholar]
- Chen FJ, Sun M, Li SQ, Wu QQ, Ji L, Liu ZL, Zhou GZ, Cao G, Jin L, Xie HW. Upregulation of the long non-coding rna hotair promotes esophageal squamous cell carcinoma metastasis and poor prognosis. Mol Carcinog. 2013;52(11):908–915. doi: 10.1002/mc.21944. [DOI] [PubMed] [Google Scholar]
- Cizkova M, Cizeronclairac G, Vacher S, Susini A, Andrieu C, Lidereau R, Bièche I. Gene Expression Profiling Reveals New Aspects of PIK3CA Mutation in ERalpha-Positive Breast Cancer: Major Implication of the Wnt Signaling Pathway. PLoS One. 2010;5(12):e15647. doi: 10.1371/journal.pone.0015647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimaras H, Dimba EAO, Gallie BL. Challenging the global retinoblastoma survival disparity through a collaborative research effort. Br J Ophthalmol. 2010;94(11):1415–1416. doi: 10.1136/bjo.2009.174136. [DOI] [PubMed] [Google Scholar]
- Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci U S A. 2005;102(10):3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian ID,Puccinelli F,Gaillard MC,Beck-Popovic M, Munier FL (2017) Diagnosis and management of secondary epipapillary retinoblastoma. Br J Ophthalmol. bjophthalmol-2016-309899 [DOI] [PubMed]
- Fabian ID, Onadim Z, Karaa E, Duncan C, Chowdhury T, Scheimberg I, Ohnuma SI, Reddy MA, Sagoo MS (2018) The management of retinoblastoma. Oncogene 37(12) [DOI] [PubMed]
- Fu L, Ke S, Wang J, Chen W, Shi D, Yun T, Wei G, Yu W, Xiao X, Kang T. TFAP2B overexpression contributes to tumor growth and a poor prognosis of human lung adenocarcinoma through modulation of ERK and VEGF/PEDF signaling. Mol Cancer. 2014;13(1):89. doi: 10.1186/1476-4598-13-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu WM, Zhu X, Wang WM, Lu YF, Hu BG, Wang H, Liang WC, Wang SS, Ko CH, Waye MM. Hotair mediates hepatocarcinogenesis through suppressing miRNA-218 expression and activating P14 and P16 signaling. J Hepatol. 2015;63(4):886–895. doi: 10.1016/j.jhep.2015.05.016. [DOI] [PubMed] [Google Scholar]
- Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3' UTRs via Alu elements. Nature. 2011;470(7333):284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JA, Kugel JF. Non-coding-RNA regulators of RNA polymerase II transcription. Nat Rev Mol Cell Biol. 2006;7(8):612–616. doi: 10.1038/nrm1946. [DOI] [PubMed] [Google Scholar]
- Hirata H, Hinoda Y, Shahryari V, Deng G, Nakajima K, Tabatabai ZL, Ishii N, Dahiya R. Long Noncoding RNA MALAT1 Promotes Aggressive Renal Cell Carcinoma through Ezh2 and Interacts with miR-205. Cancer Res. 2015;75(7):1322. doi: 10.1158/0008-5472.CAN-14-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- Ikram F, Ackermann S, Kahlert Y, Volland R, Roels F, Engesser A, Hertwig F, Kocak H, Hero B, Dreidax D. Transcription factor activating protein 2 beta (TFAP2B) mediates noradrenergic neuronal differentiation in neuroblastoma. Mol Oncol. 2016;10(2):344–359. doi: 10.1016/j.molonc.2015.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitan D, Dinger ME, Mazar J, Crawford J, Smith MA, Mattick JS, Perera RJ. The Melanoma-Upregulated Long Noncoding RNA SPRY4-IT1 Modulates Apoptosis and Invasion. Cancer Res. 2011;71(11):3852. doi: 10.1158/0008-5472.CAN-10-4460. [DOI] [PubMed] [Google Scholar]
- Kim S-E, Huang H, Zhao M, Zhang X, Zhang A, Semonov MV, MacDonald BT, Zhang X, Garcia Abreu J, Peng L, He X. Wnt stabilization of β-catenin reveals principles for morphogen receptor-scaffold assemblies. Science (New York, NY) 2013;340(6134):867–870. doi: 10.1126/science.1232389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan Z, Zheng D, Hong Q. Regulatory Roles of Long Non-Coding RNAs in the Central Nervous System and Associated Neurodegenerative Diseases. Front Cell Neurosci. 2017;11:175. doi: 10.3389/fncel.2017.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raap M, Gronewold M, Christgen H, Glage S, Bentires-Alj M, Koren S, Derksen PW, Boelens M, Jonkers J, Lehmann U, Feuerhake F, Kuehnle E, Gluz O, Kates R, Nitz U, Harbeck N, Kreipe HH, Christgen M. Lobular carcinoma in situ and invasive lobular breast cancer are characterized by enhanced expression of transcription factor AP-2beta. Lab Investig. 2018;98(1):117–129. doi: 10.1038/labinvest.2017.106. [DOI] [PubMed] [Google Scholar]
- Raveh E, Matouk IJ, Gilon M, Hochberg A. The H19 Long non-coding RNA in cancer initiation, progression and metastasis – a proposed unifying theory. Mol Cancer. 2015;14(1):184. doi: 10.1186/s12943-015-0458-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields CL, Shields JA. Retinoblastoma management: advances in enucleation, intravenous chemoreduction, and intra-arterial chemotherapy. Curr Opin Ophthalmol. 2010;21(3):203–212. doi: 10.1097/ICU.0b013e328338676a. [DOI] [PubMed] [Google Scholar]
- Sun C, Li S, Zhang F, Xi Y, Wang L, Bi Y, Li D. Long non-coding RNA NEAT1 promotes non-small cell lung cancer progression through regulation of miR-377-3p-E2F3 pathway. Oncotarget. 2016;7(32):51784–51814. doi: 10.18632/oncotarget.10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39(6):925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang W, Wang T, Wang Y, Chen X, Du Y, Sun Q, Li M, Dong Z, Zhao G. Knockdown of long non-coding RNA TP73-AS1 inhibits cell proliferation and induces apoptosis in esophageal squamous cell carcinoma. Oncotarget. 2016;7(15):19960. doi: 10.18632/oncotarget.6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Tian L, Ma P, Sun Q, Zhang K, Liu H, Xu B. Potential role of differentially expressed lncRNAs in the pathogenesis of oral squamous cell carcinoma. Arch Oral Biol. 2015;60(10):1581–1587. doi: 10.1016/j.archoralbio.2015.08.003. [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhong J, Bian Z, Fang X, Peng Y, Hu Y. Long non-coding RNA CCAT1 promotes human retinoblastoma SO-RB50 and Y79 cells through negative regulation of miR-218-5p. Biomed Pharmacother. 2017;87:683–691. doi: 10.1016/j.biopha.2017.01.004. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wei Y, Li X, Liang X, Wang L, Song J, Zhang X, Zhang C, Niu J, Zhang P. microRNA-874 suppresses tumor proliferation and metastasis in hepatocellular carcinoma by targeting the DOR/EGFR/ERK pathway. Cell Death Dis. 2018;9(2):130. doi: 10.1038/s41419-017-0131-3. [DOI] [PMC free article] [PubMed] [Google Scholar]