The genomes of two newly emerged Newcastle disease virus strains, chicken/Indonesia/Mega/001WJ/2013 and chicken/Indonesia/Cimanglid/002WJ/2015, from disease outbreaks in chickens in Indonesia are reported. Phylogenetic analysis of different genotypes of Newcastle disease virus using the F gene coding sequences suggests that these two strains belong to genotype VII.2, in class II of avian paramyxoviruses.
ABSTRACT
The genomes of two newly emerged Newcastle disease virus strains, chicken/Indonesia/Mega/001WJ/2013 and chicken/Indonesia/Cimanglid/002WJ/2015, from disease outbreaks in chickens in Indonesia are reported. Phylogenetic analysis of different genotypes of Newcastle disease virus using the F gene coding sequences suggests that these two strains belong to genotype VII.2, in class II of avian paramyxoviruses.
ANNOUNCEMENT
Newcastle disease (ND) is one of the most severe infectious diseases of chickens. The causative agent, ND virus (NDV), is a member of the avian genus Orthoavulavirus (subfamily Avulavirinae, family Paramyxoviridae) (1). NDV is a single-stranded, nonsegmented, negative-sense, and enveloped RNA virus with six major structural proteins in the order 3′-NP-P-M-F-HN-L-5′ (2, 3). NDV has been divided into two classes; class I represents avirulent strains, while class II represents virulent and nonvirulent strains (4–6). Recent ND outbreaks have appeared in commercial chickens, even vaccinated flocks, leading to mortality rates of 70 to 80%, and are caused mainly by highly virulent genotype VII NDVs (6, 7).
The two virus strains in this study were collected from two brain samples from vaccinated chickens in two different NDV outbreaks in Indonesia in 2013 and 2015. The samples were processed based on the OIE guidelines for laboratory procedures for isolating the virus (8) and then were inoculated into embryonated chicken eggs, followed by collection of allantoic fluid. RNA purification was performed using the viral RNA minikit (Qiagen, USA). cDNA libraries were prepared using random hexamers with the stranded mRNA-Seq kit (Kapa Biosystems, USA) according to the manufacturer’s instructions. The resulting cDNAs were sequenced using the Illumina MiSeq platform 600-cycle kit v3, generating 2 × 300-nucleotide reads, and the library size was checked on a Bioanalyzer 2100 using the high-sensitivity DNA kit (Agilent Technologies, Germany). Adaptors were removed using Trimmomatic v0.36 (9); 616,471 and 794,856 reads for samples 1 and 2, respectively, were de novo assembled using Unicycler v0.4.4 with default parameters (10) and visualized with Bandage (11). Assembled contigs from each sample were compared to the NCBI nonredundant/nucleotide collection using BLASTn (12). Two NDV contigs with a genome GC content of 46% and a length of 15,192 nucleotides were identified for the chicken/Indonesia/Mega/001WJ/2013 (Mega/001WJ) and chicken/Indonesia/Cimanglid/002WJ/2015 (Cimanglid/002WJ) strains. The Mega/001WJ and Cimanglid/002WJ contigs had average coverage of 573× and 1,837×, respectively, and 97.97% and 98% identity to the Sukorejo strain (GenBank accession number HQ697255), respectively. A gap in the sequence of Mega/001WJ was closed using reverse transcriptase PCR (Qiagen) and Sanger sequencing (13), and BioEdit v7.2 was used to merge sequences (14). Geneious release 10.1.3 and ORFfinder were used to annotate genes and to confirm open reading frames, respectively (12).
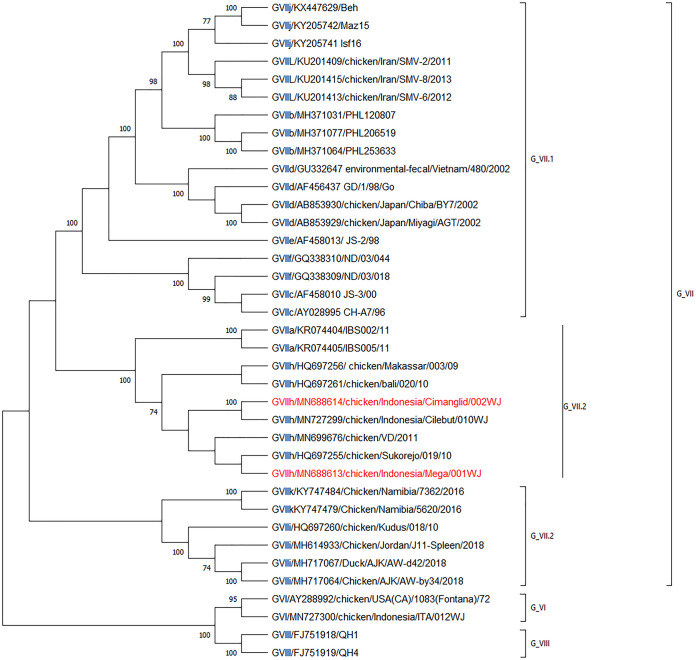
Sequence analysis revealed that the two strains differed in the amino acid sequence at the C terminus of the F protein cleavage site, which is a key determinant of NDV pathogenicity (15, 16). The Cimanglid/002WJ strain encodes the amino acid sequence motif 112RRRKRF117, while the Mega/001WJ strain encodes 112RRQKRF117. Phylogenetic analysis carried out on F gene sequences using MEGA X (17) suggests that these circulating strains belong to NDV genotype VII.2 (Fig. 1), a cause of many recent disease outbreaks in Indonesia. The amino acid identities of NP, P, M, F, HN, and L proteins between the current virus strains and the LaSota vaccine strain that is most commonly used in Indonesia are 92%, 81%, 88%, 89%, 85%, and 94%, respectively. These differences could contribute to poor protection against these strains by NDV vaccines, which highlights the need for a vaccine development strategy against newly emerged NDV strains in Southeast Asia.
FIG 1.
Phylogenetic analysis based on the full-length fusion protein gene of representative NDV isolates. The evolutionary history was inferred by using the maximum likelihood method and general time-reversible model in MEGA X. The tree with the highest log likelihood (−7,196.50) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial trees for the heuristic search were obtained automatically by applying neighbor-joining and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood (MCL) approach and then selecting the topology with a superior log likelihood value. A discrete gamma distribution was used to model evolutionary rate differences among sites (five categories [+G; parameter, 1.0951]). The rate variation model allowed for some sites to be evolutionarily invariable (+I; 33.02% of sites). This analysis involved 37 nucleotide sequences. Codon positions included were first, second, third, and noncoding. All positions with <95% site coverage were eliminated, i.e., fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position (partial deletion option). There were a total of 1,647 positions in the final data set. The vertical lines show genotype VII, with subgenotypes VII.1 and VII.2, genotype VI, and genotype VIII. The two virulent strains of subgenotype VII.2 in this study are highlighted in red.
Data availability.
The genome sequences for Mega/001WJ and Cimanglid/002WJ were deposited in GenBank with accession numbers MN688613 and MN688614, respectively. The raw sequence data were deposited in the NCBI Sequence Read Archive (SRA) under BioProject number PRJNA613298.
ACKNOWLEDGMENTS
This work was supported by Australian Centre for International Agricultural Research grant AH/2015/003.
We thank Indrawati Sendow (manager of biosafety level 3 facilities at Bbalitvet), Harimurti Nuradji and staff of the virology unit, and Unieq Syafitrie (communication manager from the Indonesian Research Center for Veterinary Science) (Bbalitvet) for assistance in sample preparation and animal experiments.
REFERENCES
- 1.International Committee on Taxonomy of Viruses. 2019. Genus: Orthoavulavirus. In Virus taxonomy: 2018b release. International Committee on Taxonomy of Viruses; London, United Kingdom: https://talk.ictvonline.org/ictv-reports/ictv_online_report/negative-sense-rna-viruses/mononegavirales/w/paramyxoviridae/1193/genus-orthoavulavirus. [Google Scholar]
- 2.Miller PJ, Koch G. 2013. Newcastle disease, p 89–138. In Swayne DE, Glisson JR, McDougald LR, Nolan LK, Suarez DL, Nair V (ed), Diseases of poultry, 13th ed. Wiley-Blackwell, Hoboken, NJ. [Google Scholar]
- 3.Xiao S, Paldurai A, Nayak B, Samuel A, Bharoto EE, Prajitno TY, Collins PL, Samal SK. 2012. Complete genome sequences of Newcastle disease virus strains circulating in chicken populations of Indonesia. J Virol 86:5969–5970. doi: 10.1128/JVI.00546-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller PJ, Decanini EL, Afonso CL. 2010. Newcastle disease: evolution of genotypes and the related diagnostic challenges. Infect Genet Evol 10:26–35. doi: 10.1016/j.meegid.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 5.Orynbayev MB, Fereidouni S, Sansyzbai AR, Seidakhmetova BA, Strochkov VM, Nametov AM, Sadikaliyeva SO, Nurgazieva A, Tabynov KK, Rametov NM, Sultankulova KT. 2018. Genetic diversity of avian avulavirus 1 (Newcastle disease virus genotypes VIg and VIIb) circulating in wild birds in Kazakhstan. Arch Virol 163:1949–1954. doi: 10.1007/s00705-018-3815-9. [DOI] [PubMed] [Google Scholar]
- 6.Dimitrov KM, Abolnik C, Afonso CL, Albina E, Bahl J, Berg M, Briand FX, Brown IH, Choi KS, Chvala I, Diel DG, Durr PA, Ferreira HL, Fusaro A, Gil P, Goujgoulova GV, Grund C, Hicks JT, Joannis TM, Torchetti MK, Kolosov S, Lambrecht B, Lewis NS, Liu H, Liu H, McCullough S, Miller PJ, Monne I, Muller CP, Munir M, Reischak D, Sabra M, Samal SK, Servan de Almeida R, Shittu I, Snoeck CJ, Suarez DL, Van Borm S, Wang Z, Wong F. 2019. Updated unified phylogenetic classification system and revised nomenclature for Newcastle disease virus. Infect Genet Evol 74:103917. doi: 10.1016/j.meegid.2019.103917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hemmatzadeh F, McAlister M, Ebrahimie E, Tarigon S, Cahyono MI. 2016. Molecular characterisation of newly emerged Newcastle disease viruses in Indonesia. Australian Centre for International Agricultural Research, Canberra, Australia. [Google Scholar]
- 8.World Organisation for Animal Health. 2012. Chapter 2.3.14: Newcastle disease, p 555–573. In OIE terrestrial manual 2012: manual of diagnostic tests and vaccines for terrestrial animals. World Organisation for Animal Health, Paris, France. [Google Scholar]
- 9.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31:3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Edgar R, Federhen S, Feolo M, Geer LY, Helmberg W, Kapustin Y, Khovayko O, Landsman D, Lipman DJ, Madden TL, Maglott DR, Miller V, Ostell J, Pruitt KD, Schuler GD, Shumway M, Sequeira E, Sherry ST, Sirotkin K, Souvorov A, Starchenko G, Tatusov RL, Tatusova TA, Wagner L, Yaschenko E. 2008. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 36:D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sabra M, Dimitrov KM, Goraichuk IV, Wajid A, Sharma P, Williams-Coplin D, Basharat A, Rehmani SF, Muzyka DV, Miller PJ, Afonso CL. 2017. Phylogenetic assessment reveals continuous evolution and circulation of pigeon-derived virulent avian avulaviruses 1 in Eastern Europe, Asia, and Africa. BMC Vet Res 13:291. doi: 10.1186/s12917-017-1211-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98. [Google Scholar]
- 15.Panda A, Huang Z, Elankumaran S, Rockemann DD, Samal SK. 2004. Role of fusion protein cleavage site in the virulence of Newcastle disease virus. Microb Pathog 36:1–10. doi: 10.1016/j.micpath.2003.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Leeuw OS, Koch G, Hartog L, Ravenshorst N, Peeters BP. 2005. Virulence of Newcastle disease virus is determined by the cleavage site of the fusion protein and by both the stem region and globular head of the haemagglutinin-neuraminidase protein. J Gen Virol 86:1759–1769. doi: 10.1099/vir.0.80822-0. [DOI] [PubMed] [Google Scholar]
- 17.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequences for Mega/001WJ and Cimanglid/002WJ were deposited in GenBank with accession numbers MN688613 and MN688614, respectively. The raw sequence data were deposited in the NCBI Sequence Read Archive (SRA) under BioProject number PRJNA613298.