Abstract
Relapse has become the major cause of treatment failure after allogeneic stem cell transplantation. Outcome of patients with clinical relapse after transplantation generally remains poor, but intervention prior to florid relapse improves outcome for certain hematologic malignancies. To detect early relapse or minimal residual disease, sensitive methods such as molecular genetics, tumor specific molecular primers, fluorescence in situ hybridisation (FISH), and multiparameter flow cytometry (MFC) are commonly used after allogeneic stem cell transplantation to monitor patients, but not all of them are included in the commonly employed disease-specific response criteria. The highest sensitivity and specificity can be achieved by molecular monitoring of tumor- or patient-specific markers measured by polymerase chain reaction-based techniques, but not all diseases have such targets for monitoring. Similar high sensitivity can be achieved by determination of donor chimerism, but its specificity regarding detection of relapse is low and differs substantially among diseases. Here we summarize the current knowledge about the utilization of such sensitive monitoring techniques based on tumor specific markers and donor cell chimerism and how these methods might augment the standard definitions of post-transplant remission, persistence, progression, relapse and the prediction of relapse. Critically important is the need for standardization of the different residual disease techniques and to assess the clinical relevance of minimal residual disease and chimerism surveillance in individual diseases, which in turn must be followed by studies to assess the potential impact of specific interventional strategies.
Keywords: allogeneic stem cell transplantation, minimal residual disease, chimerism, acute leukemia, myelodysplastic syndrome
INTRODUCTION
Methodological and technological advances allow sensitive detection of minimal residual disease (MRD) and early recognition of recurrence of hematologic malignancies after allogeneic hematopoietic stem cell transplantation (alloHSCT). Importantly, intervention prior to florid relapse improves outcome for certain hematologic malignancies [1, 2]. This manuscript by the Workshop Committee on Disease-Specific Methods and Strategies for Monitoring Relapse following Allogeneic Stem Cell Transplantation is divided into two parts and reviews disease-specific detection methods and available data with the use of such after alloHSCT. Given the critical importance to the goals of this Workshop, standard disease-specific response and relapse criteria are summarized. Outside of the alloHSCT setting, international working groups have developed standard diagnostic criteria that are widely employed in the definition of relapse for the different hematologic malignancies [3]. These are based on morphologic investigations of peripheral blood and/or bone marrow, imaging, and/or specific laboratory findings. Given their critical importance to the goals of this Workshop, these criteria are summarized here. However, after alloHSCT, more sensitive methods such as molecular genetics, tumor specific molecular primers, fluorescence in situ hybridisation (FISH), multiparameter flow cytometry (MFC), and/or chimerism are commonly used to monitor patients with respect to relapse. While some of these have clearly been shown to be predictive of outcome in specific diseases (e.g., chronic myeloid leukemia), and have become part of standard criteria, assays for monitoring of disease status after alloHSCT have not yet been incorporated into relapse definitions across all hematologic malignancies. Recommendations for the utilization of sensitive monitoring techniques to augment the standard definitions of post-transplant remission, persistence, progression, relapse, and in the prediction of relapse are proposed, whenever possible based on current evidence. It is anticipated that sensitive MRD detection will allow for earlier therapeutic intervention, and it is hoped that treatment prior to overt relapse may improve outcome of alloHSCT for hematologic malignancies. Critically important is the need to assess the clinical relevance of MRD surveillance in individual diseases, which in turn must be followed by studies to assess the potential impact of specific interventional strategies. From the point of view of this Workshop, the use of these proposed definitions and methods should facilitate future studies of the natural history of relapse (Committee on Epidemiology and Natural History of Relapse), therapeutic interventions to prevent clinical relapse (Committee on Strategies/Therapies Used to Prevent Relapse), and the treatment of relapse (Committee on Disease-Specific Treatment of Relapse). Finally, major deficits and important questions for further clinical research will be addressed. In this first part we focus on methods to detect and monitor disease response, persistence, progression, and relapse in acute leukemias and myelodysplastic syndrome, while in the forthcoming second part disease specific monitoring for chronic leukemias, myeloproliferative neoplasms and lymphoid malignancies will be reviewed.
METHODS TO DETECT AND MONITOR DISEASE RESPONSE, PERSISTENCE, PROGRESSION, AND RELAPSE
A wide variety of techniques are available to monitor residual disease after therapy, including in the post-transplant setting (Table 1), although the applicability varies by the specific disease subtype and the predictive value of each method is currently not well defined for most diseases. Some of these techniques are difficult to standardize, which is essential to the conduct of multi-center studies to assess the utility in the prediction and possible prevention of overt relapse.
Table 1:
Diagnostic methods to monitor residual disease and relapse of hematologic malignancies after alloHSCT
| TUMOR MARKER | CHIMERISM | ||||||
|---|---|---|---|---|---|---|---|
| METHOD | Chromo some Banding | FISH | Flow cytometry | Antigen receptor PCR | Translocation or other RT-PCR | XY FISH | qPCR / STR- PCR |
| APPLICABILITY | Subset of all types | Subset of all types | ALL; most AML; CLL; myeloma | ALL; lymphoma; CLL | CML; Subset of ALL; subset of AML; subset of lymphoma | Sex mismatched alloHSCT | All types with differences in donor/ recipient polymorphisms |
| SENSITIVITY | 10−1 | 10−2 | 10−3–10−4 | 10−4–10−5 | 10−3–10−6 | 10−2 | 10−3–10−6 |
ALL: acute lymphoblastic leukemia; AML: acute myeloid leukemia; CLL: chronic lymphocytic leukemia; CML: chronic myelogenous leukemia; PCR: polymerase chain reaction; qPCR: quantitative real-time PCR; STR:short tandem repeats.
Broadly, post-transplant monitoring of disease status is assured by two different methodologies: specific MRD detection and characterization of hematopietic chimerism. The latter characterizes the origin of post-transplant hematopoiesis, whereas MRD detection measures the malignant clone directly. For each approach, a variety of techniques is available, though in general there have been more studies looking directly at markers of residual malignancy than of chimerism. Issues of applicability, standardization, sensitivity and specificity are discussed separately for each technique.
Chromosome Banding Analysis
Classical chromosome banding analysis still plays an essential role in the biological characterization and for prognostic predictions in many hematologic malignancies. Whenever possible, cultivation of metaphases is performed from bone marrow instead of peripheral blood due to the higher proliferation rates and higher proportions of malignant cells in the setting of leukemia. In vitro proliferation of cells is supported by specific cytokines. Application of colchicine leads to cell cycle arrest shortly before metaphase preparation. Staining of chromosomes is performed with Giemsa- (G-), Quinacrin (Q-), or reverse (R-) banding techniques. Analysis of metaphases is improved by digital picture capture systems. The International System of Cytogenetic Nomenclature (ISCN) is used for the presentation of karyotypes. Analysis of 20–25 metaphases is required for valid results. With chromosome banding analysis, the whole karyotype can be illustrated within a single investigation. However, chromosomal banding techniques are hampered by insufficient quality of bone marrow samples (e.g., myelofibrosis, recent history of total body irradiation) as vital cells are required for metaphase culture. Moreover, chromosome banding analysis is limited to the detection of microscopically visible abnormalities. Sensitivity is further limited as a maximum of 25 metaphases is usually evaluated within a single analysis. Further, full clarification of the chromosomal abnormalities often requires the combination with molecular cytogenetic methods using the FISH technique.
Fluorescence in situ Hybridization
Diverse fluorescence in situ hybridization (FISH) techniques are available for the detection of submicroscopic alterations or for clarification of complex chromosomal changes including interphase FISH (IP-FISH), whole chromosome painting (WCP-FISH), 24-color- FISH (M-FISH or SKY), and comparative genomic hybridization (CGH). These techniques are all based on the hybridization of fluorescence-tagged probes to a specific chromosomal region. WCP-FISH and 24-color FISH can only be performed on metaphases. In contrast, FISH with loci specific probes or with probes for the centromeric regions can be performed on metaphases or interphase nuclei and thus do not require dividing cells. As interphase-FISH provides a higher sensitivity (100–200 cells can easily be evaluated) [4], the technique can be used for MRD [5], albeit not at a sensitivity approached with other techniques. Moreover, analysis is limited to the probes that have been chosen for the individual case [6].
Multiparameter Flow Cytometry
The principle of detection of MRD by multiparameter flow cytometry (MFC) rests on the property that leukemic cells have phenotypic properties that while broadly similar to those of normal cells, often show subtle differences in antigen expression. Considerable effort has been put into characterizing the patterns of antigen expression in normal and regenerating hematopoietic differentiation. Comparison of specimens of leukemia to these normal templates has shown that most cases have “leukemia-associated immunophenotypes” (LAIPs). Typically, antigens normally expressed at a particular stage of maturation are over- or underexpressed or underexpressed in leukemic cells. Thus, with design of appropriate combinations of antigens in MFC analysis, leukemic cells can be recognized because they occupy areas of “empty space” on correlated plots of antigen expression [7, 8, 9, 10]. By analyzing large numbers of cells (500,000 or more) with modern high speed flow cytometers it is possible to identify clusters of as few as 10 to 20 leukemic cells in a background of normal cells, allowing for a theoretical sensitivity approaching 10−5 cells. In practice, however sensitivities are often limited to about 10−4 and may not even reach that in some cases.
Sensitivity of detection of residual leukemic cells varies between subtypes and depends upon the magnitude of the differences seen between leukemic cells and normal hematopoietic cells. Morever, the proportion of patients with informative phenotypes varies among different diseases. Studying larger panels of markers, either in higher order (more colors) flow cytometry or by adding additional tubes, will typically increase the ability to detect MRD. In B-lineage acute lymphoblastic leukemia (ALL), greater than 90% of patients can be monitored with a relatively simple panel of antibodies [11, 12], and similarly in B- chronic lymphocytic leukemia (CLL) a simple antibody combination is informative for detecting MRD at high sensitivity in nearly all cases [13]. By contrast, MRD detection in acute myeloid leukemia (AML) is more complicated because leukemic signatures are often unique to an individual case, and custom panels are often designed to monitor particular patients [14, 15, 16, 17]. Complicating detection of MRD by flow cytometry in acute leukemias is the fact that there may be phenotypic shifts following therapy, so looking only for those cells with the leukemia specific phenotype may underestimate, or even fail to identify residual leukemic cells [18, 19, 20]. Therefore, application of comprehensive antibody panels has been suggested for follow-up monitoring in AML patients. MRD in ALL can appear to undergo maturation following steroid therapy, and unless one is aware of this, MRD can be missed [21]. Phenotypic changes in AML are even more common, and the complexity of leukemic maturational patterns seen makes it difficult to clearly quantify leukemic populations even when some leukemic cells are identified. CLL and myeloma are clonal B-cell diseases that show light chain restriction, but in the MRD setting, identifying changes in the kappa/lambda ratio, or using other markers to identify subsets of cells that express only one light chain is generally not as sensitive as methods that employ the same kinds of aberrant antigen markers that are useful in MRD monitoring of acute leukemias [13, 22].
Flow cytometric detection of MRD has a number of advantages over other methods. It is often relatively inexpensive compared to highly sensitive molecular methods, and results are obtained rapidly, often within hours or at most a day or two, allowing for the design of studies that have an early therapeutic intervention based on MRD results. The method is particularly suited not only to detect, but also to quantify disease burden. This is of major importance to evaluate disease kinetics. A major disadvantage to MRD studies by flow in all diseases except CLL and multiple myeloma is that no international consensus has been reached on standardized panel design and data interpretation. Different panels of antibodies are used by different investigators and in different studies, and highly specialized experience is necessary for accurate interpretation of MRD results. There is even variablility in specimen processing, with some laboratories using ficolled separated cells [12] and others using cell lysis techniques [11, 26] some but not all of the latter techniques express MRD results as a percentage of mononuclear cells [11] to better match results obtained with ficolled samples. In contrast to molecular methods, there are not well established protocols for demonstrating interlaboratory reproducibility. In B-lineage ALL, which is perhaps the best studied area in this regard, reproducibility is relatively high early in therapy when MRD burden is high and when there are the smallest number of contaminating normal B-cell precursors. However, obtaining reproducibility at sensitivities of 10−5 or even 10−4 is very difficult [23]. Nevertheless, the clinical importance of MRD at such a low level has been well established in a variety of studies performed at one or few reference labs [11, 24, 25, 26], suggesting that it is possible to measure clinically relevant MRD with that sensitivity.
Molecular Methods of Residual Disease Detection
The polymerase chain reaction (PCR) technique has proven to be an extremely robust method of high sensitivity and specificity with a wide range of applicability to residual disease detection depending upon the target chosen. PCR can be performed at the genomic level to look for DNA-based alterations, or alternatively reverse transcription PCR (RT-PCR) can be used to detect either structural or quantitative abnormalities in mRNA. Sensitivity and specificity of PCR can be improved by using “nested” PCR, in which a first reaction using primers directed at consensus sequences is followed by a second reaction using internal primers to further amplify DNA that was amplified in the first round. However, this “nested” PCR approach often yields unreliable quantitative results. Finally, PCR can either be performed using a qualitative or endpoint assay, or, now more commonly, quantitative real-time PCR (qPCR) can be used to quantify targets over a range of many orders of magnitude.
Minimal residual disease detection by antigen receptor PCR in lymphoid malignancies
Approaches for monitoring MRD in B- or T-lymphoid malignant diseases comprise techniques based on PCR in order to quantify tumor burden by disease-specific T cell receptor (TCR) or immunoglobulin (Ig) gene rearrangements [27]. Unique fingerprint-like sequences, assumed to be specific for each lymphoid cell, can be detected in the junctional regions of rearranged Ig and TCR genes. Cells of a lymphoid malignancy have a common clonal origin, with each cell having an identically rearranged Ig or TCR gene. Cells of a lymphoid malignancy have a common clonal origin with identically rearranged Ig or TCR genes. Consequently, these gene rearrangements, located in the junctional regions of Ig and TCR genes, serve as specific targets for each leukemia and can be used for analysis of MRD [28, 29]. At diagnosis, patient-and leukemia-specific clonal rearrangements are amplified by PCR. After this the various disease specific rearrangements can be identified and selected [30]. After selection, PCR products are used for sequencing of the junctional regions. This specific sequence forms the basis for the design of junctional region specific oligonucleotides (allele specific oligonucleotides [ASO]), required for leukemia, lymphoma and myeloma specific sensitive qPCR-based MRD analysis.
There are a number of potential pitfalls in the use of Ig and TCR clonality in MRD monitoring. Ig and TCR gene rearrangements in hematologic malignancies might be susceptible to subclone formation. Furthermore, secondary gene rearrangements could occur between diagnosis and relapse, which might cause false-negative MRD results. In order to reduce the number of false-negative MRD analyses in ALL, at least two Ig/TCR targets are used in clinical MRD studies [28, 31]. Kreyenberg et al. demonstrated that Ig and TCR gene rearrangements are stable markers for MRD in ALL after alloHSCT [32]. Importantly, isolated false positive results in Ig-based qPCR systems have been observed at the time of intense B cell regeneration [33].
In order to assure comparability of MRD results between different laboratories, quality control and standardization are essential. A standardized approach for MRD analysis by qPCR has been implemented by the European Study Group on MRD detection in ALL (ESG-MRD-ALL), consisting of 30 laboratories worldwide. Furthermore, guidelines for the interpretation of qPCR-based MRD data have been developed. Application of these guidelines allows identical interpretation of MRD data between different laboratories of the same MRD-based clinical protocol. Method and guidelines for its interpretation are precise to avoid both false-negative and false-positive results [31].
Fusion gene specific targets and mRNA expression
Quantitative PCR can be used to monitor specific fusion gene transcripts as a sensitive indicator of MRD. Experience is most extensive in the setting of BCR-ABL1 transcript monitoring for individuals with chronic myeloid leukemia (CML). The methodology used for identifying BCR-ABL1 transcripts has evolved over the years. Initially to BCR-ABL transcripts were detected by single-step amplification or a 2-step “nested” amplification. In 1993 Cross et al. developed a competitive semi-quantitative assay that expressed BCR-ABL1 transcripts as a ratio (%) compared with normal ABL (BCR-ABL1/ABL1 × 100) [34]. This method was adapted for qPCR when this technology became available [35, 36]. Subsequently Hughes et al. [37] analyzing patients in the IRIS study introduced the concept of logarithmic (log) reduction using as a baseline the laboratory-specific median BCR-ABL1 transcript level taken from 30 patients.
For valid PCR data, it is imperative to optimize each stage of the procedure, including sample collection, RNA extraction and quantitative PCR. The quality of the RNA is extremely important for reproducible data, and consistency in sample collection, tissue type, transportation, and storage conditions will maximize the accuracy and reliability of analysis. Standard protocols for BCR-ABL1 PCR employ peripheral blood for MRD monitoring instead of bone marrow aspirates.
It is highly desirable that a standardized international scale for measuring transcript levels be established and it is probably preferable to move away from log reduction. An international collaborative group recently published recommendations whereby the standardized baseline would be determined using a reference material and the absolute BCR- ABL1 value representing a major molecular response would be standardized at 0.1%. A value of 1.0% would be approximately equivalent to complete cytogenetic remission [38, 39].
MRD monitoring by PCR techniques has also been utilized in the assessment of treatment response and for the early detection of relapse in other leukemias with sensitivities of 10−4 to 10−6 [40, 41, 42, 43, 44, 45, 46, 47, 48]. Specific reciprocal gene rearrangements (e.g., PML/RARa, RUNX1-RUNX1T1=AML1-ETO, CBFB-MYH11), intragenic duplications (e.g., MLL-PTD, FLT3-ITD) and other mutation types (e.g., alterations of the NPM1 gene) can be used for monitoring MRD in AML [49]. However, reciprocal gene fusions are applicable in a minority of AML patients only. NPM1 mutations represent ideal targets for MRD monitoring by qPCR in adults [50, 51], but have a low prevalence in pediatric AML (8–10%). Thus, MRD monitoring based on molecular markers has so far been realized only for subgroups of patients with AML. In 50% of patients with myelofibrosis and in 5–10% of patients with myelodysplastic syndromes (MDS), the JAK2V617F mutation can be monitored by sensitive qPCR [52]. Furthermore, the MPLW515L/K mutation can be detected and monitored in about 5% of myelofibrosis patients [53].
The transcription factor WT1, originally described as a tumor suppressor gene, is a key molecule for neoplastic proliferation in a large number of hematologic malignancies, making it suitable both as an universal MRD maker and as a potential target for therapeutic strategies (e.g., vaccination) [45, 54, 55, 56, 57, 58]. WT1 has been shown to be expressed in the majority of adult and pediatric cases of acute leukemias, CML-blast phase, MDS, and both T- and B-lineage non-hodgkin’s lymphoma (NHL), though the 3–4 log10 overexpression necessary to make it a reasonable MRD marker is only seen in a subset of cases [59, 60, 61, 62, 63, 64, 65]. The frequency and degree of WT1 expression has been shown to correlate with MDS disease progression [66]. WT1 expression, quantified by qPCR, has been evaluated as a marker for risk stratification and for MRD detection in AML [44, 45, 46, 47, 48, 66, 67, 68, 69]. However, there are contradictory reports about the utility of WT1 overexpression to monitor MRD during frontline chemotherapy and after alloHSCT [45, 46, 56, 57, 70, 71].
Chimerism
Analysis of chimerism allows monitoring of hematopoietic cells from donor and recipient origin after alloHSCT to determine engraftment as well as detection of imminent graft rejection and may also serve as an indicator for recurrence of the underlying malignancy. Since chimerism analyses were first performed, many different methods have been developed and implemented, all following the same basic principle using differences in polymorphic genetic markers and their products. These methods include cytogenetics [72, 73], red cell or HLA phenotyping [74, 75, 76], restriction fragment length polymorphism (RFLP) analysis [77, 78, 79, 80], and FISH of sex chromosomes [81, 82, 83, 84, 85, 86]. A major limitation of these different techniques is that they are time consuming and not applicable to all patients.
The breakthrough for clinical applicability came when the PCR technique was developed [87] and also utilized for investigation of chimerism [88, 89, 90, 91, 92, 93, 94]. During the 1990s, these analyses were mainly performed by the amplification of variable number of tandem repeats (VNTR) or by the characterization of short tandem repeat (STR) markers. Fluorescent labeling of the primers and resolution of PCR products by capillary electrophoresis allowed accurate quantification of the degree of mixed chimerism. Semi-automated PCR analyses using the appropriate hardware permitted a high sample throughput [93, 95, 96, 97, 98] facilitiating the study of chimerism in all patients in short time intervals starting early after transplantation. Thus, accurate monitoring of engraftment as well as surveillance of impending graft rejection and imminent relapse has become possible.
Furthermore, a qPCR assay for the analysis of the SRY gene on the Y chromosome has been established, which allows the identification of male DNA in the background of female DNA at very low levels [99]. This method is able to detect one male cell in the background of 100,000 female cells [100, 101] increasing sensitivity enormously.
Over the past years qPCR-based chimerism techniques aiming at the amplification of sequence polymorphisms (SPs) were established. SPs represent the most frequent genetic variation, occur on an average of 1.3 kilobases in the human genome, are mostly biallelic and differ predominantly only in one single nucleotide, denoted by single nucleotide polymorphisms (SNP) [102, 103, 104]. Alizadeh et al. were the first to report a set of 11 biallelic polymorphisms using qPCR amplification for chimerism analysis [105]. The limit of detection for the minor cell population (0.1%) was higher than in STR-PCR. This study was followed by others demonstrating the possibility of accurate characterization of chimerism by qPCR based on polymorphisms [106, 107]. In contrast to STR-PCR, where virtually all recipient/donor pairs could be characterized, only 90% of donor/recipients could be discriminated by this assay. Furthermore, this qPCR technique showed less quantitative accuracy with a variation coefficient of 30–50% for higher autologous signals [105, 106] compared to a variation coefficient of 5% when applying STR-PCR systems [95, 97, 108, 109, 110, 111]. However, this did not hamper analysis when only low levels of mixed chimerism were needed to be quantified. Large prospective trials utilizing this qPCR method are needed to determine whether the clinical impact of chimerism analysis can be improved. For the time being, fluorescence-based PCR amplification of STR seems to be the gold standard method for post-transplant chimerism surveillance and the great majority of the major studies published to date have used this technique. Despite the increasing sensitivity of methods to determine chimerism, the clinical utility of this approach is limited. In most hematologic malignancies, chimerism is not a tumor-specific marker, and finding recipient cells does not necessarily indicate disease recurrence. The persistence or reappearance of recipient cells, deteced by molecular methods, could reflect the survival of leukemic blasts, survival of normal host hematopoiesis or a combination of both phenomena. Surviving host derived hematopoietic cells could promote reemergence of the malignant clone by inhibiting immunocompetent donor derived effectors.
Hematologic relapse was preceded by reappearing host-derived hematopoiesis in the mononuclear cell fraction in CML patients [112]. In consequence, the graft-versus-leukemia (GVL) effect could be weakend by a state of mixed chimerism [113]. In several early reports it remained unclear if a state of mixed chimerism in patients with an acute leukemia was associated with an increased risk of relapse. The dynamic process of the evolution of chimerism was described in the 1990s, and it was concluded that monitoring of chimerism should be performed serially and in short intervals of time. The specificity in this regard is higher in diseases that originate from a stem- or progenitor cell (e.g., CML), in contrast to malignancies that originate from a later cell stage of development (e.g., CLL or myeloma), in which case the usefulness of chimerism to detect MRD or relapse is low. This lack of specificity might be overcome by performing chimerism on specific subsets of cells in some diseases (see below) [114, 115]. Here chimerism is performed on cell subtypes after isolation by cell sorting or enrichment (e.g., CD138 for myeloma, CD34 for AML and MDS). Finding chimerism in a population enriched for tumor cells may increase the likelihood that it is a true marker of residual disease. Chimerism analysis may also have value as it provides information about the alloreactivity and/or tolerance induction of the graft and thus may serve as a prognostic factor independent of its role as a marker for MRD.
Imaging
Imaging studies are not generally useful as MRD markers in leukemias or myeloprolferative neoplasms, but they play a role in lymphomas and multiple myeloma. In myeloma, lytic lesions are generally diagnosed by radiographic analysis. One weakness of radiographic detection is that it may reveal lytic disease only when over 30% of the trabecular bone is lost [116]. Due to these limitations, computed tomography (CT) or magnetic resonance imaging (MRI) have been used to increase the sensitivity and specificity. Computed tomography is currently the most commonly used means for re-staging patients with lymphoma [117, 118]. However, CT lacks functional information, which impedes identification of disease in normal-sized organs. 18-F-fluoro-2-deoxyglucose positron emission tomography may be an alternative to CT [119]. MRI is also utilized for bone and soft tissue re-evaluation in specific situations.
Computed Tomography
The mainstay of imaging studies for lymphoma has always been CT scans performed at various intervals. With current CT scanners, lymph nodes of 5 mm or less in diameter can be detected. Extranodal involvement is typically demonstrated on a CT by organomegaly, abnormal masses, structural changes, or abnormal contrast enhancement. One problem with CT scans is that they have limited specificity, as many different processes can produce enlarged lymph nodes, even in patients with lymphoma [120], and a biopsy may be necessary to confirm residual disease. For use with multiple myeloma patients, CT imaging is superior to that of standard radiography and reveals more lesions as compared with conventional radiology [121].
Magnetic Resonance Imaging
MRI has high spatial resolution and excellent soft-tissue contrast which makes it an ideal tool for the detection of parenchymal and osseous lesions. However, because of long imaging time, limited availability, and extensive costs, MRI was previously used only as a tool to image limited anatomical areas of the body. Recent improvements in MRI technology have resulted in the availability of sufficiently fast and diagnostic sequences for whole body (WB) MRI. However, there is no standard WB-MRI protocol for staging malignant lymphoma at this time. MRI provides superior soft-tissue contrast to CT. As with CT, assessment of the nodal involvement is based on size criteria, and biopsy is needed for final confirmation of relapse. The MRI sequences that are most informative for detection of bone lesions which may be observed in myeloma and lymphomas, are the T1-weighted, the T1-weighted with fat suppression and gadolinium contrast, the T2-weighted with fat suppression, and the short-time inversion recovery (STIR) images. The STIR images are particularly sensitive in detecting myelomatous lesions [122].
Positron emission tomography
Positron emission tomography (PET) is based on the use of positron-emitting radiopharmaceuticals and the detection in coincidence of the two nearly collinear 511-keV photons emitted following positron annihilation with an electron. The increased glycolytic rate of malignant cells is the rationale behind the common use of 18-F-fluoro-2-deoxyglucose (FDG) as a radiotracer [123]. More recently FDG-PET and CT scans have been integrated which provides both functional and anatomic information [124]. FDG-PET positivity is not restricted to malignancy. For example, uptake can be seen with inflammation and infection resulting in “false-positive” results in the setting of cancer evaluations. FDG-PET scanning has been integrated into standard staging criteria for lymphomas [125]. FDG-PET/CT can detect Richter’s transformation of CLL to diffuse large B-cell lymphoma (DLBCL) with a high sensitivity and a high negative predictive value [126]. Standardized FDG-PET and PET/CT imaging procedure guidelines have been published to improve image quality, reporting, and common imaging standards may enable the use of semiquantitative techniques such as the SUV (standardized uptake value) in assessing response to therapy among centers worldwide [127]. PERCIST 1.0 is a proposed FDG-PET/CT response imaging criteria in solid tumors. This proposes imaging methology and image analysis to assess response semiquantiatively or quantitatively. It is intended as a starting point for use FDG-PET in clinical trials and structured quantitative reporting, which may be particularly useful when assessing the activity of newer therapies that stabilize disease [128]. The standardization of imaging and response criteria could be a good step in establishing the use or lack of utility of FDG-PET/CT in early/interim (after 1–3 cycles of therapy) response monitoring or early PET response adapted therapy trials [129]. Multiple novel alternative clinical PET tracers have been developed, but the greatest experience outside of FDG-PET has been with 3’-deoxy-3’- [18F] fluorothymidine (FLT) which is an analogue for thymidine used to image tumor proliferation. Early pilot studies using FLT-PET/CT appear to be very promising in early response evaluation in lymphoma [130].
DISEASE-SPECIFIC DEFINITIONS AND MONITORING OF RELAPSE AFTER ALLOGENEIC HEMATOPOIETIC STEM CELL TRANSPLANTATION
Standard diagnostic criteria have been established to define response and relapse for the hematologic malignancies. These criteria have historically been based on morphological bone marrow investigations (e.g., blast count in acute leukemias), imaging methods (e.g., occurrence of new lymph nodes on FDG-PET scans for NHL), and/or specific laboratory findings (e.g., increased paraprotein by immunofixation and electrophoresis in multiple myeloma). Recently, more sensitive methods have been utilized to assess patients for disease response. Some, but not all of these approaches, have been integrated into response criteria definitions for various hematologic malignancies. Herein we propose criteria for incorporation of currently available methodologies in the definitions for disease response, persistence, progression, relapse, and the prediction of relapse after alloHSCT.
Acute Myeloid Leukemia
According to the International Working Group for Diagnosis, Standardization of Response Criteria, Treatment Outcomes, and Reporting Standards for Therapeutic Trials in Acute Myeloid Leukemia, remission criteria in AML are not solely based on morphology, but also include cyto- and molecular genetic as well as flow cytometric data. Cytomorphology and hematologic parameters retain a central position (Table 2) [131]. However, very few follow-up studies have been performed to specifically assess these criteria in the post- transplant period.
Table 2:
Response criteria in AML according to an international working group [131]
| Criteria | Complete Remission | Relapse |
|---|---|---|
| Morphologic and Hematologic | BM blasts <5%; thrombocytes ≥100 × 109/L; neutrophils ≥1.0 × 109/L |
Reappearance of blasts after CR (BM: ≥5%; PB) |
| Cytogenetic | Disappearance of previous cytogenetic alteration | Reappearance of cytogenetic alteration |
| Molecular | Disappearance of molecular mutation | Reappearance of molecular mutation |
| Flow Cytometric | Disappearance of cells with previously determined LAIP | Reappearance of cells with LAIP |
BM: bone marrow; CR: complete remission; PB: peripheral blood; LAIP: leukemia associated immunophenotype
These criteria could also be integrated into the assessment of remission in the post- transplant period. Due to the limited sensitivity of chromosomal banding [132], interphase FISH seems to be a more suitable parameter for the assessment of the cytogenetic remission status. There are only few studies that assess the prognostic value of interphase FISH after AML therapy [5], and even fewer studies addressing this issue in the post-transplant period. In pediatric cases with MDS or AML Fuehrer et al. showed that clonal cytogenetic markers, as assessed by interphase FISH (e.g., monosomy 7), reappeared at relapse after alloHSCT and could be followed after second transplant [133]. Metaphase FISH proved useful for MRD diagnostics following standard chemotherapy or alloHSCT in an analysis of 22 AML patients performed by El-Rifei et al., as all but one patient with persisting or increasing levels of abnormal cells relapsed [134].
Chimerism
Both in AML and MDS, several studies demonstrated the relevance of chimerism, and especially its kinetics, for the prediction of relapse. Using a semiquantive STR method in 81 pediatric alloHSCT recipients with AML, Bader et al. found a relapse rate of 47% in those with increasing mixed chimerism (i.e., increase of recipient cells) in contrast to 13% in patients with complete or decreasing mixed chimerism (p<0.005) [135]. In the study of Husiman et al., chimerism status was monitored by STR-PCR in T- and non-T-cell subsets in 96 patients with AML after myeloablative (MAC) or reduced-intensity conditioning (RIC). Stable complete donor chimerism was detected in 56% of patients of the MAC group in contrast to 12% in the RIC group. In samples taken between 1–6 months post-transplant, complete donor chimerism or decreasing mixed chimerism (i.e., decrease of recipient cells) was significantly associated with a lower relapse risk (31% vs. 83%) and mortality (38% vs. 83%, p<0.001) when compared to increasing mixed chimerism [136]. Similar results were shown by Zeiser et al. who performed STR-PCR both with conventional and CD34-specific chimerism methods in 168 patients with AML or MDS after MAC. With CD34+-specific chimerism, decrease of donor alleles was detected at a median of 30 days before the clinical manifestation of relapse. The relapse rate was 89% in patients with mixed chimerism in contrast to 6% in those with complete donor chimerism [137].
Molecular genetics
Molecular mutations
A variety of genetic markers are available for MRD studies and have been investigated within a standard chemotherapy setting for their potential to guide therapeutic decisions. In this setting, MRD measurement is well established for the reciprocal rearrangements t(15;17)/PML-RARA, inv(16)/CBFB-MYH11, and t(8;21)/RUNX1-RUNX1T1. These are found in approximately 20% of de novo AML cases and confer a favorable prognosis for patients treated with standard chemotherapy. Ratios of aberrant gene expression after consolidation therapy and at diagnosis were demonstrated to correlate significantly with prognosis [138], and distinct thresholds of transcript copy numbers were determined to be associated with an increased relapse risk [42, 139]. After standard chemotherapy, the interval from the increase of fusion transcripts to morphological manifestation of relapse was 3–6 months. With respect to the post-transplant period, Elmaagacli et al. followed eight patients with inv(16)/CBFB-MYH11 with reverse transcription (RT-) PCR. The CBFB-MYH11 transcript was not detectable in six of seven patients who remained in stable remission post-alloHSCT, while relapse occurred in one of two patients who had been positive three months after alloHSCT [140].
In patients with de novo AML, the prognostically favorable nucleophosmin (NPM1) mutations are detectable in approximately 35% of overall AML cases. In normal karyotype AML, frequencies about 55% were reported. qPCR or high melting resolution PCR (“HRM”) can be used to detect the most frequent NPM1 mutation subtypes A, B, and D (Figure 1). Some studies demonstrated a correlation of the mutation load before and after chemotherapy with outcome [141, 142, 143]. In most studies in the standard therapeutic setting, NPM1 mutations provided high stability during follow-up [50, 141, 142], while one study described loss of the mutation in two of 21 relapsing patients [144]. In an analysis involving 13 stem cell recipients with a history of NPM1 mutated AML, the achievement of PCR negativity after alloHSCT was a precondition for stable remission [49], whereas relapse after alloHSCT was preceded by an increase of the NPM1 mutation level with mean intervals of 24 days (range, 12 – 38 days) in all but one case. The short interval between MRD detection and relapse in NPM1 mutated AML implies that very frequent monitoring (e.g., weekly intervals) would be necessary if this marker were to be used for early therapeutic intervention to prevent hematologic relapse. The more favorable prognosis of patients with isolated NPM1 mutations and a normal karyotype suggests that lower frequencies of this mutation might be expected in the alloHSCT setting. Post-transplant MRD studies for this marker would likely be more important in patients with coincident NPM1 mutations and the prognostically unfavorable internal tandem duplication of the FLT3 gene (FLT3-ITD) [145, 146].
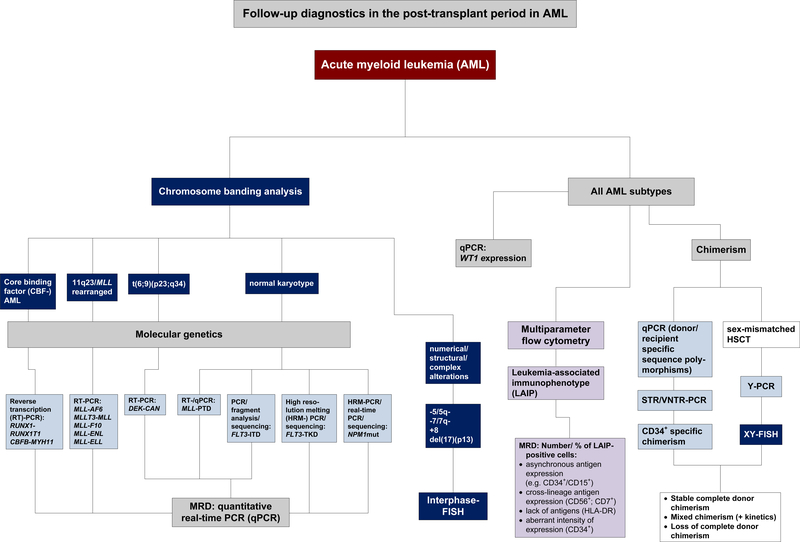
Figure 1:
Proposed algorithm for follow-up diagnostics in the post-transplantperiod in AML
FLT3-ITDs are seen in 20–25% of all AML cases and in approximately 40% of cases with normal karyotypes [147]. FLT3-ITD have variable lengths and different starting points; thus qPCR monitoring requires the design of patient-specific primers [148]. Some studies have indicated that the FLT3-ITD mutation was unstable, with loss of the mutation in nearly a fifth of relapsed patients, limiting the validity of this marker for MRD monitoring [149]. However, others have reported instability rates of <5% [150]. Integration of FLT3-ITD in post-transplantation monitoring has been suggested [151], but is still controversial due to the potential for instability of the mutation [152]. Consequently, a combination with a second MRD strategy (e.g., NPM1 mutations [49, 153]), or with MFC, might be recommended for FLT3 mutated AML.
Mutations of the tyrosine kinase domain of the FLT3 gene (FLT3-TKD) are less frequent than the FLT3-ITD in AML. They occur in 6%–8% of all AML cases with normal karyotypes [154, 155, 156, 157]. The impact on prognosis remains incompletely defined [157, 158]. Scholl et al. demonstrated that quantitative assessment during follow-up with qPCR is feasible as the FLT3-TKD mutations are represented by single nucleotide base exchanges [148]. Quantitative follow-up of the FLT3-ITD and the FLT3-TKD was performed with qPCR in four stem cell recipients with a history of FLT3 mutated AML and showed a significant association of relapse to post-transplant PCR positivity, whereas disease-free survival (DFS) was associated with attainment of PCR negativity [159].
Partial tandem duplications of the MLL gene (MLL-PTD) represent another promising marker for post-transplant monitoring in AML. These prognostically unfavorable gain-of- function mutations [160] occur in 5–10% of patients with a normal karyotype [161, 162, 163, 164]. MRD studies with qPCR demonstrated that the amount of reduction of mutated cells was prognostically relevant in the standard therapeutic setting. Molecular relapse preceded clinical manifestations by several weeks [165]. To date, no study has been performed with this marker specifically in the post-transplant period.
Single gene expression
Monitoring of the expression of the WT1 gene represents another option for post- transplant MRD detection in patients with AML [46, 152]. Using qPCR in patients with AML, ALL, or advanced phases of CML, Ogawa et al. were able to determine thresholds above which relapse was predictable within 40 days after alloHSCT. There was a 100% relapse probability at an expression level of 1–5 × 10−2, while the relapse risk was less than 1% with expression levels less than 4 × 10−4. Relapsed patients who responded to adoptive immunotherapy by donor lymphocyte infusion (DLI) or withdrawal of immunosuppression showed a significantly longer doubling time of WT1 expression as compared to patients who were refractory to these approaches. Doubling time of WT1 transcripts less than 13 days was predictive for failure of treatment [166]. Candoni et al. described full concordance between WT1 expression levels and the remission status of AML before and after alloHSCT [167].
Jacobsohn et al. evaluated the prognostic significance of pre-transplant WT1 gene expression as a marker of MRD in 36 pediatric patients with AML. Elevated WT1 gene expression before alloHSCT was predictive of post-transplant relapse and lower 5-year event-free survival (EFS). After alloHSCT, 76% of patients with high pre-transplant WT1 expression relapsed, in contrast none of those with low WT1 expression. [168].
Multicenter studies have been initiated to develop standardization of WT1 measurement (e.g., by qRT-PCR normalized to the ABL gene) [58]. However, the physiological background expression of WT1 in healthy individuals has to be considered. Barragan et al. compared WT1 expression and monitoring of the NPM1 mutation load in 24 AML patients and found a strong correlation between both markers (p<0.001), but with different disappearance kinetics. WT1 expression showed rapid decrease after induction therapy, but residual levels were maintained during complete remission (CR), whereas NPM1 mutations showed a mild reduction after induction, but they were undetectable in long-term survivors. Thus, assessment by qPCR of the NPM1 mutation load was superior to the assay for WT1 quantification due to the specificity for leukemic cells and higher levels of expression at presentation [143].
Expression of other genes such as BAALC (brain and acute leukemia, cytoplasmic) [169] might be investigated for post-transplant monitoring in normal karyotype AML as well, though to date there are no data on their usefulness for this specific purpose. Thus, a variety of additional genetic markers should be investigated for their potential utility after alloHSCT, particularly those with an adverse impact on prognosis as they are likely to be overrepresented in the transplant setting. Efforts should continue to develop algorithms for monitoring patients with AML in the post-transplant period (Figure 1).
Multiparameter flow cytometry
The value of immunophenotyping by MFC for the determination of the remission status in AML has been confirmed in many studies in the setting of standard therapy [131]. The decrease of cells with the specific leukemia-associated immunophenotype (LAIP) from diagnosis to the end of induction therapy showed significant correlation with the remission rates and long-term prognosis [170, 171, 172], and the reduction of aberrant cells to a threshold of less than 0.1% after chemotherapy was associated with improved survival rates [173]. Only very few studies [174] have addressed the value of MFC specifically in the post-transplantation period, but it was shown that MFC was able to discriminate leukemic cells from regeneration blasts after alloHSCT [175]. In an individual patient with AML, withdrawal of immunosuppression was guided by the results of MFC, as there was an increase of cells with the LAIP two months post-transplant. This was followed by a subsequent reduction of aberrant cells [176]. Perez-Simon et al. performed MFC in 13 patients with AML or MDS who had received RIC-alloHSCT. Persistence of cells with the LAIP within 21–56 days post-transplant was associated with a relapse risk close to 60%, whereas all patients with a negative MRD status in this period achieved stable remission [177]. Diez-Campelo et al. performed MRD monitoring by MFC in 41 alloHSCT recipients with AML or MDS. A cut-off level of 10−3 malignant cells at 100 days post-transplant was able to discriminate different risk populations. The 4-year overall survival was 73% in patients with a low MRD level versus 25% among patients with high MRD level at this time point, and EFS after 4 years was 74% versus 17% (p = 0.01) for the same two groups. The authors thus proposed that MRD monitoring with MFC has relevance for the therapeutic decisions in the early post-transplant period [178].
It should be emphasized that one advantage of MFC is the option to perform this technique in AML patients without evidence of a specific genetic marker. However, the frequency of changes of the LAIP in patients relapsing after alloHSCT is unknown, while relapse after standard therapy was associated with loss of the previous LAIP in approximately 25% of cases [179]. Thus, further evaluation is needed to evaluate the status of MFC for AML patients in the post-transplant period.
Myelodysplastic Syndromes
As in AML, standard criteria for the assessment of remission in MDS have been developed and revised by an international committee [180]. These new criteria integrate clinical, morphological, cytogenetic and hematologic parameters (Table 3). Few studies have evaluated MRD diagnostics in patients with MDS in the post-transplant period.
Table 3:
Revised criteria for assessment of remission in MDS according to an international working group [180].
| Criteria | Response |
|---|---|
| Morphologic and Hematologic |
Complete Remission (CR): bone marrow blasts <5% without dysplasia, hemoglobin ≥11 g/dL, platelets ≥100 × 109/L, neutrophils ≥1.5 × 109/L. Partial Remission (PR): reduction of blasts by at least 50% or achievement of lower risk category than prior to treatment. |
| Cytogenetic |
Major Cytogenetic Response: disappearance of a cytogenetic abnormality. Minor Cytogenetic Response: ≥50% reduction of abnormal metaphases. |
Molecular markers
WT1 expression was shown to increase during leukemic transformation of MDS and to decrease following chemotherapy or alloHSCT [66]. In a study from Cilloni et al. that included 131 patients with MDS, 65% of bone marrow and 78% of peripheral blood samples from patients with refractory anemia (RA) showed WT1 overexpression, while 100% of samples from patients with either refractory anemia with excess blasts (RAEB) or secondary AML (sAML) samples WT1 transcript amounts greater than the level observed in healthy volunteers. Thus, there was significant correlation between the WT1 expression level and the IPSS score [181]. Similar results were reported by Patmasiriwat et al. who described increased WT1 levels in advanced MDS cases, but not in RA [182]. Bader et al. found significantly higher mean WT1 expression levels in pediatric patients with RAEB and juvenile myelomonocytic leukemia (JMML) when compared to expression levels of healthy donors (p<0.001). However, single samples showed overlap between patients and healthy individuals [69].
Tamura et al. reported on a patient of 14 years of age who had a diagnosis of RAEB with an increase of WT1 expression to 16,000 copies/μg RNA in peripheral blood after alloHSCT. Withdrawal of immunosuppression was followed by transient and self-limiting skin graft-versus-host disease (GVHD) and gradual decrease of WT1 expression during the next months. Sixteen months after alloHSCT, the patient was in CR while WT1 expression was below the detection level [183]. However, because it is difficult to differentiate clearly between phyisological and aberrant gene expression, the usefulness of WT1 expression for post-transplant monitoring of myeloid malignancies remains controversial.
Only a few studies have investigated molecular mutations in MDS. Most frequent are intragenic mutations of the RUNX1/AML1 gene [184, 185]. FLT3-ITD occurs in 3–5% of MDS patients [186, 187, 188]. Mutations of the NRAS protooncogene were identified in 6–10% of all MDS cases. In contrast to AML, NPM1 mutations are rarely if ever seen in MDS [189]. Thus, the proportion of MDS patients who can be characterized with molecular markers is lower than in AML. However, as FLT3-ITD or NRAS mutations were demonstrated to increase during leukemic transformation [187, 188], the selection of high- risk MDS cases for alloHSCT might be associated with an overrepresentation of these markers when compared to standard therapy cohorts. So far no study has specifically determined the frequency of these molecular markers in the allo-transplant setting.
Multiparameter flow cytometry
Several studies have demonstrated that MFC has potential diagnostic utility in MDS. Frequently, the side-scatter (SSC) signal is reduced due to lower granularity of granulocytes. Aberrant expression patterns of CD13 and CD16 on granulocytes or lack of CD71 on erythrocytes can also be characteristic [190, 191]. The progression of MDS to higher stages was shown to be accompanied by an increase of the flow cytometric scores [192], which correlated with the International Prognostic Scoring System and prognosis [193]. At this time, no flow cytometric study has been performed specifically in the post-transplant period in MDS patients. Whether the frequent finding of bone marrow dysplasia in the post- transplant period will be an obstacle for this approach, remains to be clarified.
In conclusion, considering that molecular MRDs markers are available or under development for specific AML subgroups only, and the lack of MRD markers in most MDS patients, surveillance of chimerism represents the most broadly applicable molecular tool in patients with these disorders. Monitoring of the kinetics of chimerism in short intervals is clearly superior to the interpretation of a single measurement. In addition, the potential of CD34+ specific chimerism for myeloid disorders seems worthy of further evaluation.
Acute Lymphoblastic Leukemia (ALL)
Standard definitions for response and relapse are based on clinicopathologic evaluation of peripheral blood, bone marrow, cerebrospinal fluid, and potential extramedullary sites of involvement. Response criteria rely most commonly on blood, marrow, and CSF cell counts and morphology (Table 4). From the standpoint of long-term outcome, CR has historically been considered the only meaningful goal and endpoint. However, criteria for CR with incomplete blood count recovery (CRi), partial response, and stable disease are sometimes utilized in the context of early phase clinical trials (Table 4). Importantly, the standard definition for relapse in patients with CR1 (greater than 25% bone marrow blasts) is insensitive for use after allogeneic haemaotopoietic stem cell transplantation , as detailed below.
Table 4a.
Response criteria in ALL
|
Complete Remission (CR): A CR requires that all of the following be recorded concurrently: • < 5% marrow leukemia blast cells (M1). • No circulating blasts. • Absolute neutrophil count (neutrophils and bands) ≥ 1.0 × 109/L. • Platelet count ≥ 100 × 109/L. • Adequate bone marrow cellularity with trilineage hematopoiesis. • Absence of extramedullary manifestations of disease (e.g., CNS1). • No evidence of recurrence of ALL for at least 4 weeks. |
| *Morphologic CR with incomplete blood count recovery (CRi): Above CR criteria without specified blood counts. |
| Cytogenetic CR (CRc): In addition to above CR criteria, reversion to normal karyotype for those with previously detected cytogenetic abnormality. |
| Molecular CR (CRm): In addition to above CRc criteria, normalization of previously detected molecular cytogenetic abnormality. |
|
*Partial Response (PR): Requires that all of the following be recorded concurrently: • Decrease in the percentage of marrow blasts, absolute peripheral blast count, and extramedullary disease by at least 50%. • ≤ 25% marrow leukemia blast cells. • Absolute neutrophil count (neutrophils and bands) ≥ 1.0 × 109/L. • Platelet count ≥ 100 × 109/L. |
|
*Stable Disease (SD) • Criteria not met for CR, PR, or progression. |
|
Progressive Disease • An increase of at least 25% in the absolute number of circulating or bone marrow leukemic blasts or extramedullary disease burden; or • Development of new extramedullary disease. |
|
Relapsed Disease • The reappearance of leukemia blast cells in the blood or the bone marrow (≥ 25%) or in any other extramedullary site after a CR with confirmation of lymphoid blasts by morphology and flow cytometry, PCR for antigen receptor loci or fusion genes, or cytogenetics/FISH; or • Increase to > 25% blasts in the marrow after a PR. • Importantly, isolated extramedullary relapses (e.g., CNS) are considered relapse from a diagnostic standpoint, although these are commonly approached differently in terms of therapy. |
Additional definitions sometimes employed in the context of clinical trial
Prognostic significance of MRD in non-transplant studies
Multiple studies support the independent prognostic value of MRD measurements in pediatric and adult patients with B- and T-lineage ALL [11, 194, 195, 196, 197, 198, 199, 200, 201, 202, 203, 204, 205, 206]. It has been well demonstrated that MRD positivity (MFC and PCR-based molecular methods) after induction [204, 205, 207] and consolidation [208, 209, 210] correlates with the risk of relapse. Most studies have employed bone marrow and in general this is the preferred sample source; however, in T-ALL, in contrast to B-ALL, levels of MRD in blood and marrow correlate closely so that peripheral blood may be utilized in MRD detection [211, 212]. However, measurement in peripheral blood may also provide useful prognostic information in B-ALL [211], especially early in therapy during periods of marrow aplasia (i.e. day 8) [11]. Furthermore, the risk of relapse appears to be proportional to the level of MRD, which in some studies was found to be the most powerful prognostic factor for relapse in multivariate analyses [25, 194, 195, 196, 197, 198, 206, 209, 213]. The kinetics of the decrease in MRD level has been shown to be more predictive than MRD analyzed at one time point [25, 197,]. Consequently, MRD detection has been utilized in the stratification for several ALL treatment protocols [199, 200, 209] and to evaluate the effectiveness of alloHSCT or alternative treatment approaches [197].
Prognostic significance of MRD at the time of transplant
Pediatric studies
Numerous studies have shown on a retrospective basis that the MRD status prior to conditioning was the strongest predictor for post-transplant relapse. [214, 215, 216]. A British pediatric study in Bristol reported that an MRD load of greater than 10−3 pre-transplant was associated with poor survival. In contrast, a portion of patients with low level disease burden (<10−3) revealed a more favorable outcome and MRD negative children had the best chance of survival [213].
A retrospective study in Germany including 45 children reconfirmed these results, applying the same semiquantitative PCR approach. Those patients, entering transplantation with high-level MRD (>10−3) were rarely cured. Interestingly, residual disease could be eradicated by post-transplant immunotherapy in some of these high-risk patients [209].
Recently Bader et al. published prospective data on MRD analysis in 91 children with relapsed ALL in second or higher remission prior to alloHSCT within the ALL-REZ BFM Study Group. The probability of EFS and cumulative incidence of relapse (CIR) in patients with MRD equal to or greater than 10−4 were 0.27 and 0.57, respectively, as compared to 0.60 and 0.13 in 46 patients with MRD less than 10−4 (EFS: p = 0.004; CIR: p <0.001). Multivariate Cox regression analysis revealed MRD as the only independent parameter predictive for EFS. These findings proved the clinical significance of MRD analysis prior to alloHSCT as a predictor for post-transplant outcome in a large prospective study. Based on this, new strategies with modified alloHSCT procedures will be evaluated within the ALL- BFM trials [217].
Adult studies
There are few studies examining the prognostic significance of MRD detection prior to and following alloHSCT for adults with standard risk B-cell ALL. In several older studies [201, 202, 203, 204], semi-quantitative PCR analysis failed to detect a strong association of MRD in CR1 prior to alloHSCT with post-transplant DFS. In one of these studies [204], there was no significant difference in pre-transplant MRD detection for the 14 patients who were in continuous CR after transplant; eight were MRD positive and six were MRD negative prior to alloHSCT. However, there was an association with pre-transplant MRD and risk of relapse for a cohort of patients who underwent autologous HSCT in CR. Twenty one of 23 patients (91%) who were MRD-negative prior to autologous HSCT remained in long-term remission following transplant. In contrast, six of seven patients (86%) who tested MRD positive prior to autologous HSCT relapsed following the transplant (p = 0.005). In this same study, post- transplant MRD detection was strongly associated with relapse-free-survival (RFS) for the 19 patients undergoing alloHSCT. There were no relapses in patients who remained MRD negative following alloHSCT, whereas both patients with detectable MRD following transplant relapsed.
A more recent study using qPCR examined the clinical impact of MRD detection in 43 adults with high-risk ALL who underwent alloHSCT [218]. The group was heterogeneous in regard to disease subtype and status, conditioning, and donor type. Overall survival at 36 months was 48%. For patients who were PCR-negative prior to transplant, 80% were alive at 36 months in contrast to 49% survival for PCR-positive patients (p = 0.17). For the same patients, the cumulative incidence of relapse was 0% for PCR-negative and 46% for PCR- positive patients (p = 0.03). The relapse rate calculated according to the molecular results at 100 days following alloHSCT confirmed that the achievement or maintenance of a PCR- negative status within three months following transplantation was significantly associated with a lower incidence of relapse at 36 months post-transplant (7% vs. 80%, p = 0.0006). In a multivariate analysis to investigate the role of clinical findings including disease status prior to transplant, cytogenetics, immunophenotype, age and pre-transplant MRD level, only molecular CR before conditioning proved to be a significant predictor of the achievement of MRD negativity at 100 days after transplantation.
Bassan et al. recently demonstrated the utility of risk-adapted therapy based on MRD measurements for adults with ALL [209]. MRD measurements taken at weeks 10, 16, and 22 of therapy using qPCR were used to stratify treatment for adults receiving intensive post-remission chemotherapy for ALL in CR1. Of 280 patients registered to the trial, 112 were evaluable for MRD at the end of consolidation therapy. Of these, 58 had no measurable MRD and 54 were MRD-positive. Five-year overall survival was 75% and DFS was 72% for the MRD-negative group in comparison with 33% overall survival and 14% DFS (p = 0.001) for the MRD-positive group (regardless of pre-treatment risk assignment based on clinical, immunophenotypic, and cytogenetic features). They further stratified post-remission treatment based on MRD. Those patients who were MRD-positive at weeks 16 through 22 were assigned to receive alloHSCT in first complete remission if an available donor was identified, or in the absence of a donor, to an intensification of post-remission chemotherapy. Those who were MRD-negative went on to receive traditional maintenance therapy. Thirty-six of 54 (67%) MRD-positive cases proceeded to alloHSCT or to the intensified chemotherapy phase. There was a clear improvement in 4-year DFS for these 36 patients estimated at 33% in comparison to no survivors among the 18 MRD-positive patients who did not receive either intensified treatment approach. Notably, MRD status after the intensified treatment further defined prognosis for this group. For the patients who achieved a MRD-negative state following alloHSCT or intensified chemotherapy, the DFS was 51%; for those who were remained MRD-positive, there were no long term survivors at 4 years. These data suggest that allocation to standard maintenance therapy for patients with MRD-negative status following consolidation results in excellent DFS and that intensification therapy with alloHSCT or repeated cycles of intensification chemotherapy can salvage some very high risk patients (MRD-positive at the end of consolidation) who would otherwise relapse with near certainty.
Pre-transplant MRD monitoring in Philadelphia chromosome positive ALL
MRD detection prior to transplant for Philadelphia chromosome positive (Ph+) ALL is a good predictor of relapse-free survival after transplant [219, 220, 221, 222, 223]. The addition of targeted tyrosine kinase inhibition with imatinib to frontline therapy of Ph+ ALL has resulted in the ability to significantly reduce or eradicate MRD during the first months of treatment using sensitive qRT-PCR assays [224, 225, 226, 227]. A number of recent studies have demonstrated that achievement of a molecular CR as assessed by qPCR immediately prior to the time of transplant correlates with improved DFS and overall survival after alloHSCT. In one of the larger studies, when imatinib was added to front-line therapy, 82% of the patients became PCR negative and the majority was able to proceed to transplant [228]. With a median follow-up of 25 months following transplant, outcome of these patients appeared to be improved with DFS of 78% in comparison to studies in the pre-imatinib era where DFS following alloHSCT for adults with Ph+ ALL was in the 30–50% range. The kinetics of MRD eradication prior to alloHSCT may also be an important prognostic feature. Lee et al. [229] studied MRD in 41 patients with Ph+ ALL who were treated with combination chemotherapy induction followed by four weeks of imatinib therapy. They found that 36 of 41 patients achieved at least a 3-log reduction of MRD following the first four weeks of treatment with imatinib. This early reduction in MRD was identified as the most powerful predictor of lower relapse rate following alloHSCT. For patients with at least a 3-log reduction in MRD, the relapse rate following alloHSCT was 12% in comparison to the other patients who had a relapse rate of 45% (p = 0.011) [230]. These data, similar to the pediatric data presented above, suggest that pre-transplant MRD detection in adults with ALL may be an important guide to early post-transplant treatment to attempt to avert clinical relapse.
Role of MRD monitoring and chimerism assessment following alloHSCT for ALL
Pediatric and adult studies (Table 5)
Table 5:
Published studies on the use of chimerism and MRD in alloHSCT for ALL
| Chimerism | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Late 1980s to mid 1990s | |||||||||
| Author | Patients [n] | Graft [n] | T-cell depleted | Interval of investigations | Methods | State of chimerism | Relapses | ||
| MC | CC | MC | CC | ||||||
| Schattenberg et al., Blood 1989 [74] | 29 | 29 | yes | 6, 12 months | Red cell phenotype Cytogenetics RFLP |
19/29 | 3/19 | ||
| Van Leeuwen et al. Blood 1993 [248] | 53 | 46 7 | yes no |
2, 6, 12 months | VNTR | 23/53 | 30/53 | 8/23 | 10/30 |
| Roy et al. Blood 1990 [249] | 43 | 43 | yes | 0.5, 1, 3, 6, 9, 12, 18, 24 months | RFLP | 22/43 | 2¼3 | 1½2 | 6/21 |
| Lawler et al. Blood 1991 [89] | 32 | 12 20 |
yes no |
not given | VNTR and STR | 10/12 8/20 |
2/12 12/20 |
4/10 2/8 |
0/2 0/12 |
| Bader et al. BMT 1998 [250] | 55 | 55 | no | Weekly | VNTR and STR | 18/36 1 autologous recovery | Increasing MC: relapse | ||
| Chimerism with Intervention | |||||||||
| 2003 to 2008 | |||||||||
| Author | Patients [n] | Diagnosis | Interval of investigations | Methods | Relapses | ||||
| Formankova et al. Haematologica 2003 [232] | 54 | ALL, AML, CML and MDS Children | weekly to +100; monthly | STR | MC associated with rejection and relapse, immunotherapy was possible | ||||
| Gorczynska et al. BMT 2004 [233] | 14 | ALL, AML Children | weekly to +100; monthly | STR | Increasing MC could be converted by immunotherapy to CC | ||||
| Bader et al. JCO 2004 [238] | 163 | ALL Children | weekly to +100; monthly | STR | MC associated with rejection and relapse, immunotherapy was possible | ||||
| Horn et al. BMT 2008 [251] | 20 | ALL, AML Children | 1, 3, 6, 12 months; in MC bi-weekly | STR | MC associated with relapse, immunotherapy was not possible | ||||
| Pre-Transplant MRD | |||||||||
| Retrospective Studies | |||||||||
| Knechtli et al. Blood 1998 [216] | 64 | ALL | prior to conditioning | Ig / TCR PCR |
high level positive – 0% low level positive – 36% negative – 73% |
||||
| Bader et al. Leukemia 2002 [214] | 41 | ALL | prior to conditioning | Ig / TCR PCR |
high level positive – 23% low level poitive – 48% negative – 78% |
||||
| Uzunel et al. Blood 2001 [252] | 30 | ALL | prior to conditioning | Ig / TCR PCR |
high level positive – 47% low level positive – 50% negative – 100% |
||||
| Sramkova et al. Ped Blood Cancer 2007 [253] | 25 | ALL | prior to conditioning | Ig / TCR PCR |
positive – 0% negative – 94% |
||||
| Prospective Studies | |||||||||
| Bader et al. JCO 2009 [217] | 91 | ALL | prior to conditioning | Ig / TCR PCR |
MRD ≥10−4: EFS 27%; CIR 57% MRD <10−4: EFS 60%; CIR 13% |
||||
| Post-Transplant MRD | |||||||||
| Retrospective Studies | |||||||||
| Knechtli et al. BJH 1998 [240] | 68 | ALL | up to 24 months post- transplant | Ig / TCR PCR |
relapse – 88% positive remission – 22% positive |
||||
| Uzunel et al. BJH 2003 [242] | 23 | ALL | 24 months | Ig / TCR PCR |
MRD positive associated with relapse | ||||
| Sanchez et al. BJH 2002 [241] | 40 | ALL | day 30, 60,90, every 2 to 3 months |
Flow cytometry | positive – 33% negative – 74% |
||||
| Prospective Studies | |||||||||
| Bader et al. BMT 2009 [243] | 92 | ALL | day 30, 60,100, 200, 365 post- transplant | Ig / TCR PCR |
MRD ≥10−4: EFS 9%; RFS 11% MRD <10−4: EFS 48%; RFS 62% |
||||
| Mortuza et al. JCO 2002 [204] | 19 | ALL (B-lineage) | 1 to 20 months | Ig / TCR PCR (semi-quant.) |
positive – 0% negative – 100% |
||||
| Spinelli et al. Haematologica 2007 [218] | 37 | ALL | day 100 | Ig / TCR or fusion gene PCR (quantitative) |
positive >10−4 – 20% negative – 93% |
||||
| Bassan et al. Blood 2009 [209] | 18 | ALL (All patients were PCR positive prior to transplant) | not defined | Ig / TCR PCR |
positive > 10 −4 – 0% negative – 50% |
||||
Legend: ALL: acute lymphoblastic leukemia; AML: acute myeloid leukemia; CIR, cumulative incidence of relapse; CC, complete chimerism; CML: chronic myelogenous leukemia; EFS, event-free survival; Ig, immunoglobulin; MC, mixed chimerism; MDS: myelodysplastic syndromes; PCR, polymerase chain reaction; RFLP, restriction fragment length polymorphism; RFS, relapse free survival; STR, short tandem repeats; TCR, T-cell receptor; VNTR, variable number tandem repeats.
Assessment of chimerism using the semiquantitative STR-PCR approach, which analyzed microsatellite regions, has demonstrated that ALL patients with rapid increase of mixed chimerism were at the highest risk of relapse post-alloHSCT [113, 231, 232, 233, 234]. The analysis of cell subpopulations in acute leukemia patients revealed a potential difference between children and adults. Guimond et al. demonstrated that mixed chimerism in T and natural killer (NK) cells was frequently detected in children with relapsed leukemia but not in pediatric patients, who remained in remission. Furthermore this phenomenon was not observed in relapsed adult patients [235].
The clinical relevance of a state of increasing mixed chimerism as a risk factor for relapse in patients with acute leukemia was demonstrated by Barrios et al. in 2003 [236]. However, a group in Germany showed a more complicated relationship between chimerism and relapse. [237]. They showed that the persistence of mixed chimerism in the early phase post-transplant as predominantly caused by normal recipient derived hematopoiesis. The reappearance of leukemic blasts was preceded by an increase of this recipient derived normal hematopiesis, denoted by increasing recipient chimerism. These results promoted the hypothesis that mixed chimerism weakened the clinical GVL effect, which was thought to be mediated by donor-derived alloreactive effectors in acute leukemia patients. These results formed the basis for several clinical trials to study the efficacy of relapse prevention by pre-emptive immunotherapeutic intervention based on the analysis of chimerism in acute leukemia patients [232, 233].
By applying STR-based analysis of chimerism at defined regular intervals post-transplant, In 2004, Bader et al. defined a subset of children with impending relapse [238], applying STR-based analysis of chimerism at defined regular intervals post-transplant. Beyond that, prevention of overt relapse was possible in a portion of patients by pre-emptive immunotherapeutic intervention (withdrawal of immunosuppression or administration of DLI), though impending relapse was not diagnosed in all patients because of the short time interval between the conversion of chimerism and the occurrence of relapse in some patients.
Importantly, analysis of chimerism by conventional STR-PCR does not incorporate the potential to detect MRD because of its limited sensitivity (approximately 1%). Thus, the decision to start immunotherapy should not be based exclusively on the state of chimerism, since this might result in a delay of onset of beneficial interventional therapy.
A German retrospective analysis including patients who had received DLI based on a state of increasing mixed chimerism also measured the MRD burden, applying qPCR (Ig/TCR rearrangements). It was demonstrated that those patients with an MRD burden of greater than10−3 did not benefit from interventional therapy, whereas patients with an MRD load <10−3 at initiation of immunotherapy showed a survival rate just below 40% at two years [239].
The significance of MRD analysis post-transplant has only been investigated in a few prospective studies. In 1998, Knechtli et al. showed that persistent MRD positivity represented an unfavorable predictor for EFS. Here, MRD was assessed by a semiquantitative dot blot hypridization approach [240]. These initial results could be confirmed later in a smaller cohort of patients by immunophenotyping of antigens associated with leukemia [241] and by molecular qPCR based approaches [218, 177, 242]. These trials all showed MRD to be a predictor for poor post-transplant outcome.
Recently, the ALL-REZ BFM Study Group monitored MRD in 92 pediatric ALL patients after alloHSCT in order to highlight patients with highest risk for relapse to whom pre-emptive treatment might be offered. The probability of EFS and RFS for MRD-negative patients was 0.55 and 0.77, respectively, as compared to 0.48 and 0.62 in MRD-positive patients with less than 10−4 and compared to 0.09 and 0.11 in MRD positive patients with equal or greater than 10−4, respectively (EFS: p<0.005; RFS: p<0.001). Patients who remained persistently MRD-negative showed a pRFS of 0.78 as compared to 0.5 in patients who became MRD-negative, and compared to only 0.1 in patients who developed an increase of MRD equal to or greater than 10−4 and of 0 in patients with MRD levels remaining permanently greater than 10−4 (p<0.001). Consequently, patients with reappearing or persisting MRD load at a level of ≥10−4 faced the highest risk for subsequent relapse and may therefore be candidates for further interventional therapy [243].
MRD by molecular Ig/TCR rearrangements is impossible in patients lacking a disease-specific marker. In these cases, the status of chimerism can be used as a surrogate marker for MRD if it is analysed in specific cell subpopulations. Thiede et al. demonstrated that a state of mixed chimerism in the CD34+ cell fraction predicted relapse in peripheral blood in ALL and AML patients [244]. In 1998, Winiarski et al. characterized chimerism in cell subsets, which carried the leukemic phenotype. A remarkable correlation between the presence of MRD and a state of mixed chimerism in the respective cell subpopulation was detected [245]. However, large prospective trials proving the predictive value of mixed chimerism in cell subsets have not been published yet.
Taken together, post-transplant serial monitoring of chimerism by conventional STR-PCR in peripheral blood allowed for the identification of patients with impending relapse. Therefore, frequent chimerism analyses are indispensable within the first 200 post-transplant days, when most relapses occur.
The combination of characterization of chimerism and analysis of MRD ensures the documentation of engraftment and surveillance of post-transplant state of remission. These approaches provide a rational basis for individual immunotherapeutic intervention in order to prevent the recurrence of the underlying malignancy. However, it should be noted that the clinical importance of MRD in ALL patients after allogeneic stem cell transplantation could not be verified in some studies [213].
Post-transplant MRD monitoring in Philadelphia chromosome positive ALL
Beginning in the late 1990’s studies to assess MRD status following alloHSCT have provided intriguing information about the risk of relapse in Ph+ ALL [219, 220, 221, 222, 246]. First, using qualitative RT-PCR testing with sensitivities reported in the 10−5 to 10−6 range, a number of studies found that patients who were consistently PCR negative following alloHSCT were unlikely to relapse. Conversely, patients in whom MRD was detected after transplant were at very high risk of relapse. In the largest published series, Radich et al. found that the relative risk (RR) of relapse was significantly higher for patients with detectable MRD following transplantation than for those without detectable BCR-ABL1 transcripts (RR: 5.7; p = 0.025) [221, 222]. The prognostic significance of the PCR assay remained after controlling for other clinical variables (e.g., stem cell source, GVHD) that could influence relapse risk. Interestingly, the genetic context of MRD may also be of relevance. In this study, Radich et al. noted that the risk of relapse was greatest for MRD- positive patients with a p190 BCR-ABL1 transcript in comparison to patients who had detectable p210 BCR-ABL1 transcripts after transplant.
Use of imatinib after alloHSCT appears to be useful for eradication of MRD in some patients. Wassmann et al. showed that initiation of imatinib for MRD-positive patients after alloHSCT was associated with a rapid complete molecular remission and prolonged DFS in 14/27 treated patients [247]. There have also been anecdotal reports of initiation of targeted tyrosine kinase inhibitor therapy at the time of clinical relapse with re-institution of donor chimersim and achievement of prolonged remission with or without DLI.
Summary and future directions: Risk-adapted therapy of ALL based on MRD
Taken together, all the studies suggest that MRD measurements in patients with ALL taken in the first 4 – 20 weeks of post-remission therapy have strong prognostic significance and can identify patients with a both a good and poor prognosis. Similarly, detection of pre-transplant MRD in pediatric and some adult studies is highly predictive of relapse following alloHSCT and, coupled with post-transplant MRD evaluation, may guide early post- transplant intervention such as early withdrawal of immunosuppression, administration of DLI, or addition of post-transplant maintenance therapy (e.g., targeted tyrosine kinase inhibiation for Ph+ ALL).
Thus, there are ample data demonstrating that MRD monitoring is a powerful prognostic tool in pediatric and adult ALL and may be used to risk-adapt treatment. The introduction of novel treatment strategies to eradicate MRD early during treatment should be a goal of future trials and may eventually obviate the need for alloHSCT. Characterization of MRD before transplantation defines those patients at highest risk for relapse after alloHSCT. Importantly, while it appears that MRD detection can facilitate the decision to advise an allogeneic transplant in CR1, the outcome for MRD+ positive patients, even with the most aggressive approaches currently available is still relatively poor. After alloHSCT, MRD measurements can be used to guide novel therapeutic interventions and assess their efficacy. MRD monitoring allows the detection of impending relapse in a substantial percentage of children undergoing transplantation for ALL. These analyses have recently served as the basis for treatment intervention to avoid graft rejection, maintain engraftment and treat imminent relapse.
SUMMARY, CONCLUSIONS AND FUTURE RESEARCH
Because the intention of alloHSCT is to cure the underlying hematologic malignancy, and because there is increasing evidence that minimal disease after alloHSCT may be eradicated by immunotherapeutic approaches such as DLI, monitoring of disease is of great importance. The current definitions of remission and relapse utilized to evaluate most hematologic malignancies during upfront therapy lack sufficient sensitivity for use after alloHSCT. Flow cytometry, molecular methods and new imaging modalities have been investigated in recent years and can substiantially increase the sensitivity of disease detection up to 10−4 to 10−6. The highest sensitivity and specificity can be achieved by molecular monitoring of tumor- or patient-specific markers measured by PCR, but not all diseases have such targets for monitoring. Similar sensitivity can be achieved by determination of donor chimerism, but its specificity regarding detection of relapse is low and differs substantially among diseases. A higher specificity might be obtained by lineage-specific donor chimerism, but there are only a few such studies with limited number of patients.
Critically important is the need for standardization of the different residual disease techniques. Table 6 summarizes the different methods of MRD detection in the monitoring of ALL, AML, and MDS and the relative pros and cons of use of these methods after alloHSCT. Further clinical trials to assess the utility of these techniques in each disease entity are also mandatory. The predictive value of post-transplant MRD and chimerism, and the optimal timepoints to assess kinetic changes in these measurements remains to be determined across all hematologic malignancies. Subsequent studies must then be performed in order to evaluate the efficacy of MRD- and chimerism-guided therapeutic interventions designed to prevent overt relapse. Thus, critical objectives for future studies in this area should include the following:
Table 6:
Response and relapse definitions after alloHSCT - Application of monitoring methodologies
| Disease | Definition of Complete Remission | Definition of Relapse | Molecular Marker | Cytogenetics | Chimerism | Imaging | Flow Cytometry |
|---|---|---|---|---|---|---|---|
| AML/MDS | IWG | IWG | Molecular mutations | Chromosome banding analysis, FISH | PCR or VNTR/STR | 4–8 color flow | |
| Applicable | All patients | All patients | Subgroups | Subgroups | All patients | Not applicable | All patients |
| Comment | Well established. | Well established, but less sensitive. | Expansion of MRD marker panel for post-transplant monitoring in AML (e.g., NPM1 mutations) or MDS (e.g., RUNX1/AML1 mutations).* | No standardization for MRD monitoring, useful for specific aberrations.* | Well-established, lack of specificity: investigation of CD34+ specific chimerism*; and standardization of techniques. | Few studies.* | |
| ALL | Less than 5% blasts in BM | More than 5% blasts in BM | TCR- and Ig- Gene rearrangement | Chromosome banding analysis, FISH | PCR or VNTR/STR | 4–6 color flow | |
| Applicable | All patients | All patients | 90% of all patients | Subgroups | All patients | Not applicable | > 95% of patients |
| Comment | ASO primer: 80–90% of patients. Ig VDJ: Most patients. BCR-ABL1: All Ph+ ALL. |
Clinical not important for MRD assessment. | Gold standard: Singleplex PCR with fluorescent labelled STR primers. Importantly: product resolution using capillary electrophoresis. | Sensitivity in B- lineage ALL is limited after SCT because of large numbers of hematogones. | |||
ALL: acute lymphoblastic leukemia; AML: acute myeloid leukemia; Flow: multiparameter flow cytometry; MDS: myelodysplastic syndromes; qPCR: quantitative real-time PCR; STR: short tandem repeats; VNTR: variable number tandem repeat;
Further studies needed.
Standardize measurement of molecular markers for each hematologic malignancy for which alloHSCT is employed.
Determine the optimal frequency for monitoring MRD and chimerism after alloHSCT.
Define kinetic changes in MRD and chimerism that occur after alloHSCT and establish criteria for remission and relapse that incorporate measurement of these molecular markers in the definition of response and remission after alloHSCT.
Assess the efficacy of interventional strategies based changes in MRD and/or chimerism to prevent clinical relapse.
Table 4b.
Bone marrow classification
| Blast Percentage (at least 200 nucleated cells counted) | |
|---|---|
| M1 | < 5% |
| M2 | 5 – 25% |
| M3 | > 25% |
Table 4c.
Central nervous system (CNS) classification
| Cerebrospinal Fluid Cell Count and Cytology | |
|---|---|
| CNS1 | Absence of blasts on cytospin preparation. |
| CNS2 | < 5 white blood cells/μl and cytospin positive for blasts. |
| CNS3 | ≥ 5 white blood cells/μl and cytospin positive for blasts. |
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES:
- 1.Raanani P, Dazzi F, Sohal J, et al. : The rate and kinetics of molecular response to donor leucocyte transfusions in chronic myeloid leukaemia patients treated for relapse after allogeneic bone marrow transplantation. Br J Haematol 99:1997:945–950. [DOI] [PubMed] [Google Scholar]
- 2.Bader P, Klingebiel T, Schaudt A, et al. : Prevention of relapse in pediatric patients with acute leukemias and MDS after allogeneic SCT by early immunotherapy initiated on the basis of increasing mixed chimerism: a single center experience of 12 children. Leukemia 1999; 13:2079–2086. [DOI] [PubMed] [Google Scholar]
- 3.Baccarani M, Cortes J, Pane, F, et al. ; European LeukemiaNet. Chronic myeloid leukemia: an update of concepts and management recommendations of European LeukemiaNet. J Clin Oncol. 2009;27:6041–6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haferlach T, Bacher U, Kern W, et al. Diagnostic pathways in acute leukemias: a proposal for a multimodal approach. Ann.Hematol 2007; 86,:311–327. [DOI] [PubMed] [Google Scholar]
- 5.Bacher U, Kern W, Schoch C, et al. Evaluation of complete disease remission in acute myeloid leukemia: a prospective study based on cytomorphology, interphase fluorescence in situ hybridization, and immunophenotyping during follow-up in patients with acute myeloid leukemia. Cancer 2006;106:839–847. [DOI] [PubMed] [Google Scholar]
- 6.Fuehrer M, Gerusel-Bleck M, Konstantopoulos N, et al. FISH analysis of native smears from bone marrow and blood for the monitoring of chimerism and clonal markers after stem cell transplantation in children. Int.J.Mol.Med.2005, 15:291–297. [PubMed] [Google Scholar]
- 7.Dworzak MN, Fritsch G, Panzer-Grumayer ER, Mann G, Gadner H: Detection of residual disease in pediatric B-cell precursor acute lymphoblastic leukemia by comparative phenotype mapping: method and significance. Leuk Lymphoma 2000; 38(3–4):295–308. [DOI] [PubMed] [Google Scholar]
- 8.Lucio P, Gaipa G, van Lochem EG, et al. : BIOMED-I concerted action report: flow cytometric immunophenotyping of precursor B-ALL with standardized triple-stainings. BIOMED-1 Concerted Action Investigation of Minimal Residual Disease in Acute Leukemia: International Standardization and Clinical Evaluation. Leukemia 2001; 15:1185–1192. [DOI] [PubMed] [Google Scholar]
- 9.Porwit-MacDonald A, Bjorklund E, Lucio P, et al. : BIOMED-1 concerted action report: flow cytometric characterization of CD7+ cell subsets in normal bone marrow as a basis for the diagnosis and follow-up of T cell acute lymphoblastic leukemia (T-ALL). Leukemia 2000; 14:816–825. [DOI] [PubMed] [Google Scholar]
- 10.Weir EG, Cowan K, LeBeau P, Borowitz MJ: A limited antibody panel can distinguish B-precursor acute lymphoblastic leukemia from normal B precursors with four color flow cytometry: implications for residual disease detection. Leukemia 1999; 13:558–567. [DOI] [PubMed] [Google Scholar]
- 11.Borowitz MJ, Devidas M, Hunger SP, et al. : Clinical significance of minimal residual disease in childhood acute lymphoblastic leukemia and its relationship to other prognostic factors: a Children’s Oncology Group study. Blood 2008; 111:5477–5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campana D, Coustan-Smith E: Minimal residual disease studies by flow cytometry in acute leukemia. Acta Haematol 2004; 112:8–15. [DOI] [PubMed] [Google Scholar]
- 13.Rawstron AC, Villamor N, Ritgen M, et al. : International standardized approach for flow cytometric residual disease monitoring in chronic lymphocytic leukaemia. Leukemia. 2007;21:956–964. [DOI] [PubMed] [Google Scholar]
- 14.Al-Mawali A, Gillis D, Lewis I: The role of multiparameter flow cytometry for detection of minimal residual disease in acute myeloid leukemia. Am J Clin Pathol. 2009;131:16–26. [DOI] [PubMed] [Google Scholar]
- 15.Kern W, Danhauser-Riedl S, Ratei R, et al. : Detection of minimal residual disease in unselected patients with acute myeloid leukemia using multiparameter flow cytometry for definition of leukemia- associated immunophenotypes and determination of their frequencies in normal bone marrow. Haematologica. 2003; 88:646–653. [PubMed] [Google Scholar]
- 16.Macedo A, Orfao A, Gonzalez M, et al. : Immunological detection of blast cell subpopulations in acute myeloblastic leukemia at diagnosis: implications for minimal residual disease studies. Leukemia. 1995; 9:993–998. [PubMed] [Google Scholar]
- 17.San Miguel JF, Martinez A, Macedo A, et al. : Immunophenotyping investigation of minimal residual disease is a useful approach for predicting relapse in acute myeloid leukemia patients. Blood. 1997; 90:2465–2470. [PubMed] [Google Scholar]
- 18.Borowitz MJ, Pullen DJ, Winick N, et al. : Comparison of diagnostic and relapse flow cytometry phenotypes in childhood acute lymphoblastic leukemia: implications for residual disease detection: a report from the children’s oncology group. Cytometry B Clin Cytom. 2005; 68:18–24. [DOI] [PubMed] [Google Scholar]
- 19.Tomova A, Babusikova O: Shifts in expression of immunological cell markers in relapsed acute leukemia. Neoplasma. 2001; 48:164–168. [PubMed] [Google Scholar]
- 20.Zeleznikova T, Babusikova O: The impact of cell heterogeneity and immunophenotypic changes on monitoring minimal residual disease in acute myeloid leukemia. Neoplasma. 2006; 53:500–506. [PubMed] [Google Scholar]
- 21.Gaipa G, Basso G, Maglia O, et al. : Drug-induced immunophenotypic modulation in childhood ALL: implications for minimal residual disease detection. Leukemia 2005; 19:49–56. [DOI] [PubMed] [Google Scholar]
- 22.Rawstron AC, Orfao A, Beksac M, et al. : Report of the European Myeloma Network on multiparametric flow cytometry in multiple myeloma and related disorders. Haematologica 2008; 93:431–438. [DOI] [PubMed] [Google Scholar]
- 23.Dworzak MN, Gaipa G, Ratei R et al. : Standardization of flow cytometric minimal residual disease evaluation in acute lymphoblastic leukemia: Multicentric assessment is feasible. Cytometry B Clin Cytom 2008; 74:331–340. [DOI] [PubMed] [Google Scholar]
- 24.Bjorklund E, Mazur J, Soderhall S, et al. : Flow cytometric follow-up of minimal residual disease in bone marrow gives prognostic information in children with acute lymphoblastic leukemia. Leukemia 2003; 17:138–148. [DOI] [PubMed] [Google Scholar]
- 25.Coustan-Smith E, Sancho J, Hancock ML, et al. : Clinical importance of minimal residual disease in childhood acute lymphoblastic leukemia. Blood 2000; 96:2691–2696. [PubMed] [Google Scholar]
- 26.Dworzak MN, Froschl G, Printz D, et al. : Prognostic significance and modalities of flow cytometric minimal residual disease detection in childhood acute lymphoblastic leukemia. Blood 2002; 99:1952–58. [DOI] [PubMed] [Google Scholar]
- 27.Gabert J, Beillard E, van der Velden V, et al. Standardization and quality control studies of ‘real-time’ quantitative reverse transcription transcription polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia - a Europe Against Cancer program. Leukemia 2003;17:2318–2357. [DOI] [PubMed] [Google Scholar]
- 28.van der Velden VH, Hochhaus A, Cazzaniga G, et al. Detection of minimal residual disease in hematologic malignancies by real-time quantitative PCR: principles, approaches, and laboratory aspects. Leukemia 2003;17:1013–1034. [DOI] [PubMed] [Google Scholar]
- 29.van der Velden V, van Dongen JJ. MRD detection in acute lymphoblastic leukemia patients using Ig/TCR gene rearrangements as targets for real-time quantitative PCR. Methods Mol Biol 2009;538:115–50. [DOI] [PubMed] [Google Scholar]
- 30.Pongers-Willemse MJ, Seriu T, Stolz F, et al. Primers and protocols for standardized detection of minimal residual disease in acute lymphoblastic leukemia using immunoglobulin and T cell receptor gene rearrangements and TAL1 deletions as PCR targets: report of the BIOMED-1 CONCERTED ACTION: investigation of minimal residual disease in acute leukemia. Leukemia 1999;13:110–118. [DOI] [PubMed] [Google Scholar]
- 31.van der Velden VH, Cazzaniga G, Schrauder A, et al. Analysis of minimal residual disease by Ig/TCR gene rearrangements: guidelines for interpretation of real-time quantitative PCR data. Leukemia 2007;21:604–611. [DOI] [PubMed] [Google Scholar]
- 32.Kreyenberg H, Eckert C, Yarkin Y, et al. Immunoglobulin and T-cell receptor gene rearrangements as PCR-based targets are stable markers for monitoring minimal residual disease in acute lymphoblastic leukemia after stem cell transplantation. Leukemia 2009; 23:1355–1358. [DOI] [PubMed] [Google Scholar]
- 33.Fronkova E, Muzikova K, Mejstrikova E, et al. B-cell reconstitution after allogeneic SCT impairs minimal residual disease monitoring in children with ALL. Bone Marrow Transplant 2008;42:187–196. [DOI] [PubMed] [Google Scholar]
- 34.Cross NC, Hughes TP, Feng L, et al. Minimal residual disease after allogeneic bone marrow transplantation for chronic myeloid leukaemia in first chronic phase: correlations with acute graft- versus-host disease and relapse. Br J Haematol. 1993;84:67–74. [DOI] [PubMed] [Google Scholar]
- 35.Hochhaus A, Lin F, Reiter A, et al. Quantification of residual disease in chronic myelogenous leukemia patients on interferon-alpha therapy by competitive polymerase chain reaction. Blood 1996;87:1549–1555. [PubMed] [Google Scholar]
- 36.Cross NC, Feng L, Chase A, et al. Competitive polymerase chain reaction to estimate the number of BCR-ABL transcripts in chronic myeloid leukemia patients after bone marrow transplantation. Blood 1993;82:1929–1936. [PubMed] [Google Scholar]
- 37.Hughes TP, Kaeda J, Branford S, et al. Frequency of Major Molecular Responses to Imatinib or Interferon Alfa plus Cytarabine in Newly Diagnosed Chronic Myeloid Leukemia. N Engl J Med 2003;349:1423–1432. [DOI] [PubMed] [Google Scholar]
- 38.Muller MC, Saglio G, Lin F, et al. An international study to standardize the detection and quantitation of BCR-ABL transcripts from stabilized peripheral blood preparations by quantitative RT-PCR. Haematologica 2007;92:970–973. [DOI] [PubMed] [Google Scholar]
- 39.Cross NCP, Hughes TP, Hochhaus A, et al. International standardisation of quantitative real-time RT- PCR for BCR-ABL. Leukemia Research 2008;32:505–506. [DOI] [PubMed] [Google Scholar]
- 40.Gallagher RE, Yeap BY, Bi W, et al. Quantitative real-time RT-PCR analysis of PML-RAR alpha mRNA in acute promyelocytic leukemia: assessment of prognostic significance in adult patients from intergroup protocol 0129. Blood 2003;101:2521–2528. [DOI] [PubMed] [Google Scholar]
- 41.Marcucci G, Caligiuri MA, Dohner H, et al. Quantification of CBFbeta/MYH11 fusion transcript by real time RT-PCR in patients with INV(16) acute myeloid leukemia. Leukemia 2001;15:1072–1080. [DOI] [PubMed] [Google Scholar]
- 42.Krauter J, Gorlich K, Ottmann O, et al. Prognostic value of minimal residual disease quantification by real-time reverse transcription transcription polymerase chain reaction in patients with core binding factor leukemias. J Clin Oncol 2003;21:4413–4422. [DOI] [PubMed] [Google Scholar]
- 43.Schnittger S, Weisser M, Schoch C, et al. New score predicting for prognosis in PML-RARA+, AML1- ETO+, or CBFBMYH11+ acute myeloid leukemia based on quantification of fusion transcripts. Blood 2003;102:2746–2755. [DOI] [PubMed] [Google Scholar]
- 44.Cilloni D, Saglio G. WT1 as a universal marker for minimal residual disease detection and quantification in myeloid leukemias and in myelodysplastic syndrome. Acta Haematol 2004;112:79–84. [DOI] [PubMed] [Google Scholar]
- 45.Lapillonne H, Renneville A, Auvrignon A, et al. High WT1 expression after induction therapy predicts high risk of relapse and death in pediatric acute myeloid leukemia. J Clin Oncol 2006;24:1507–1515. [DOI] [PubMed] [Google Scholar]
- 46.Weisser M, Kern W, Rauhut S, et al. Prognostic impact of RT-PCR-based quantification of WT1 gene expression during MRD monitoring of acute myeloid leukemia. Leukemia 2005;19:1416–1423. [DOI] [PubMed] [Google Scholar]
- 47.Barragan E, Cervera J, Bolufer P, et al. Prognostic implications of Wilms’ tumor gene (WT1) expression in patients with de novo acute myeloid leukemia. Haematologica 2004;89:926–933. [PubMed] [Google Scholar]
- 48.Steinbach D, Schramm A, Eggert A, et al. Identification of a set of seven genes for the monitoring of minimal residual disease in pediatric acute myeloid leukemia. Clin Cancer Res 2006;12:2434–2441. [DOI] [PubMed] [Google Scholar]
- 49.Bacher U, Badbaran A, Fehse B, et al. Quantitative monitoring of NPM1 mutations provides a valid minimal residual disease parameter following allogeneic stem cell transplantation. Exp Hematol 2009; 37:135–1342. [DOI] [PubMed] [Google Scholar]
- 50.Schnittger S, Kern W, Tschulik C, et al. Minimal residual disease levels assessed by NPM1 mutation-specific RQ-PCR provide important prognostic information in AML. Blood 2009;114:2220–2231. [DOI] [PubMed] [Google Scholar]
- 51.Hollink IH, Zwaan CM, Zimmermann M, et al. Favorable prognostic impact of NPM1 gene mutations in childhood acute myeloid leukemia, with emphasis on cytogenetically normal AML. Leukemia 2009;23:262–270. [DOI] [PubMed] [Google Scholar]
- 52.Kröger N, Badbaran A, Holler E, et al. Monitoring of the JAK2-V617F mutation by highly sensitive quantitative real-time PCR after allogeneic stem cell transplantation in patients with myelofibrosis. Blood. 2007;109:1316–1321. [DOI] [PubMed] [Google Scholar]
- 53.Alchalby H, Badbaran A, Bacher U, et al. Screening and monitoring of MPL W515L mutation with real-time PCR in patients with myelofibrosis undergoing allogeneic stem cell transplantation. Bone Marrow Transplantation 2010. January 11 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 54.Keilholz U, Menssen HD, Gaiger A, et al. Wilms’ tumour gene 1 (WT1) in human neoplasia. Leukemia 2005;19:1318–1323. [DOI] [PubMed] [Google Scholar]
- 55.Yang L, Han Y, Suarez SF, et al. A tumor suppressor and oncogene: the WT1 story. Leukemia 2007;21:868–76. [DOI] [PubMed] [Google Scholar]
- 56.Trka J, Kalinova M, Hrusak O, et al. Real-time quantitative PCR detection of WT1 gene expression in children with AML: prognostic significance, correlation with disease status and residual disease detection by flow cytometry. Leukemia 2002;16:1381–1389. [DOI] [PubMed] [Google Scholar]
- 57.Cilloni D, Gottardi E, De Micheli D, et al. Quantitative assessment of WT1 expression by real time quantitative PCR may be a useful tool for monitoring minimal residual disease in acute leukemia patients. Leukemia 2002;16:2115–2121. [DOI] [PubMed] [Google Scholar]
- 58.Willasch AM, Gruhn B, Coliva T, et al. Standardization of WT1 mRNA quantitation for minimal residual disease monitoring in childhood AML and implications of WT1 gene mutations: a European multicenter study. Leukemia 2009;23: 1472–1479. [DOI] [PubMed] [Google Scholar]
- 59.Inoue K, Ogawa H, Sonoda Y, et al. : Aberrant overexpression of the Wilms tumor gene (WT1) in human leukemia. Blood 1997;89:1405–1412. [PubMed] [Google Scholar]
- 60.Menssen HD, Renkl HJ, Rodeck U, et al. : Presence of Wilms’ tumor gene (wt1) transcripts and the WT1 nuclear protein in the majority of human acute leukemias. Leukemia 1995;9:1060–67. [PubMed] [Google Scholar]
- 61.Rosenfeld C, Cheever MA, Gaiger A: WT1 in acute leukemia, chronic myelogenous leukemia and myelodysplastic syndrome: therapeutic potential of WT1 targeted therapies. Leukemia 2003;17:1301–1312. [DOI] [PubMed] [Google Scholar]
- 62.Elisseeva OA, Oka Y, Tsuboi A, et al. : Humoral immune responses against Wilms’ tumor gene (WT1) product in patients with hematopoietic malignancies. Blood 2002;99:3272–3279. [DOI] [PubMed] [Google Scholar]
- 63.Algar E: A review of the Wilms’ tumor 1 gene (WT1) and its role in hematopoiesis and leukemia. J Hematother Stem Cell Res 2002;11:589–599. [DOI] [PubMed] [Google Scholar]
- 64.Menssen HD, Siehl JM, Theil E: Wilms’ tumor gene (wt1) expression as a panleukemia marker. Int J Hematol 2002;76:103–109. [DOI] [PubMed] [Google Scholar]
- 65.Drakos E, Rassidakis GZ, Tsioli P, Lai et al. Differential expression of WT1 gene product in non- Hodgkin lymphomas. Appl Immunohistochem Mol Morphol 2005;13:132–137. [DOI] [PubMed] [Google Scholar]
- 66.Tamaki H, Ogawa H, Ohyashiki K, et al. : The Wilms’ tumor gene WT1 is a good marker for diagnosis of disease progression of myelodysplastic syndromes. Leukemia 1999;13:393–399. [DOI] [PubMed] [Google Scholar]
- 67.Ostergaard M, Olesen LH, Hasle H, et al. WT1 gene expression: an excellent tool for monitoring minimal residual disease in 70% of acute myeloid leukaemia patients - results from a single-centre study. Br J Haematol 2004;125:590–600. [DOI] [PubMed] [Google Scholar]
- 68.Osborne D, Frost L, Tobal K, et al. Elevated levels of WT1 transcripts in bone marrow harvests are associated with a high relapse risk in patients autografted for acute myeloid leukaemia. Bone Marrow Transplantation 2005;36:67–70. [DOI] [PubMed] [Google Scholar]
- 69.Bader P, Niemeyer C, Weber G, et al. WT1 gene expression: useful marker for minimal residual disease in childhood myelodysplastic syndromes and juvenile myelo-monocytic leukemia? Eur J Haematol 2004;73:25–28. [DOI] [PubMed] [Google Scholar]
- 70.Cilloni D, Gottardi E, Messa F, et al. Significant correlation between the degree of WT1 expression and the International Prognostic Scoring System Score in patients with myelodysplastic syndromes. J Clin Oncol 2003;21:1988–1995. [DOI] [PubMed] [Google Scholar]
- 71.Hamalainen MM, Kairisto V, Juvonen V, et al. Wilms tumour gene 1 overexpression in bone marrow as a marker for minimal residual disease in acute myeloid leukaemia. Eur J Haematol 2008;80:201–207. [DOI] [PubMed] [Google Scholar]
- 72.Offit K, Burns JP, Cunningham I, et al. Cytogenetic analysis of chimerism and leukemia relapse in chronic myelogenous leukemia patients after T cell-depleted bone marrow transplantation. Blood 1990;75:1346–1355. [PubMed] [Google Scholar]
- 73.Sparkes R Cytogenetic analysis in human bone marrow transplantation. Cancer Genet Cytogenet 1981;4:345–352. [DOI] [PubMed] [Google Scholar]
- 74.Schattenberg A, de Witte T, Salden M, et al. Mixed hematopoietic chimerism after allogeneic transplantation with lymphocyte-depleted bone marrow is not associated with a higher incidence of relapse. Blood 1989;73:1367–1372. [PubMed] [Google Scholar]
- 75.Schaap N, Schattenberg A, Bar B, et al. Red blood cell phenotyping is a sensitive technique for monitoring chronic myeloid leukaemia patients after T-cell-depleted bone marrow transplantation and after donor leucocyte infusion. Br J Haematol 2000;108:116–125. [DOI] [PubMed] [Google Scholar]
- 76.Schumm M, Feuchtinger T, Pfeiffer M, et al. Flow cytometry with anti HLA-antibodies: a simple but highly sensitive method for monitoring chimerism and minimal residual disease after HLA-mismatched stem cell transplantation. Bone Marrow Transplant. 2007;39:767–773. [DOI] [PubMed] [Google Scholar]
- 77.Casarino L, Carbone C, Capucci MA, Izzi T, Ferrara GB. Analysis of chimerism after bone marrow transplantation using specific oligonucleotide probes. Bone Marrow Transplant 1992;10:165–170. [PubMed] [Google Scholar]
- 78.Knowlton RG, Brown VA, Braman JC, et al. Use of highly polymorphic DNA probes for genotypic analysis following bone marrow transplantation. Blood 1986;68:378–385. [PubMed] [Google Scholar]
- 79.Yam PY, Petz LD, Knowlton RG, et al. Use of DNA restriction fragment length polymorphisms to document marrow engraftment and mixed hematopoietic chimerism following bone marrow transplantation. Transplantation 1987;43:399–407. [DOI] [PubMed] [Google Scholar]
- 80.Brunet S, Casals T, Madoz P, et al. DNA polymorphisms as implant markers in allogeneic bone marrow transplantation. Preliminary evaluation. Med Clin (Barc) 1989;93:765–771. [PubMed] [Google Scholar]
- 81.Kogler G, Wolf HH, Heyll A, et al. Detection of mixed chimerism and leukemic relapse after allogeneic bone marrow transplantation in subpopulations of leucocytes by fluorescent in situ hybridization in combination with the simultaneous immunophenotypic analysis of interphase cells. Bone Marrow Transpl 1995;15:41–48. [PubMed] [Google Scholar]
- 82.Petz LD, Yam P, Wallace RB, et al. Mixed hematopoietic chimerism following bone marrow transplantation for hematologic malignancies. Blood 1987;70:1331–1337. [PubMed] [Google Scholar]
- 83.Bielorai B, Trakhtenbrot L, Amariglio N, et al. Multilineage hematopoietic engraftment after allogeneic peripheral blood stem cell transplantation without conditioning in SCID patients. Bone Marrow Transplant 2004;34:317–320. [DOI] [PubMed] [Google Scholar]
- 84.Thiele J, Wickenhauser C, Kvasnicka HM, et al. Mixed chimerism of erythro- and megakaryopoiesis following allogeneic bone marrow transplantation. Acta Haematol 2003;109:176–183. [DOI] [PubMed] [Google Scholar]
- 85.Thiele J, Wickenhauser C, Kvasnicka HM, et al. Dynamics of lineage-restricted mixed chimerism following sex-mismatched allogeneic bone marrow transplantation. Histol Histopathol 2003;18:557–574. [DOI] [PubMed] [Google Scholar]
- 86.Seong CM, Giralt S, Kantarjian H, et al. Early detection of relapse by hypermetaphase fluorescence in situ hybridization after allogeneic bone marrow transplantation for chronic myeloid leukemia. J Clin Oncol 2000;18:1831–1836. [DOI] [PubMed] [Google Scholar]
- 87.Mulli KB. The unusual origin of the polymerase chain reaction. Sci Am 1990;262:56–61. [DOI] [PubMed] [Google Scholar]
- 88.Lawler M, McCann SR, Conneally E, et al. Chimaerism following allogeneic bone marrow transplantation: detection of residual host cells using the polymerase chain reaction. Br J Haematol 1989;73:205–210. [DOI] [PubMed] [Google Scholar]
- 89.Lawler M, Humphries P, McCann S. Evaluation of mixed chimerism by in vitro amplification of dinucleotide repeat sequences using the polymerase chaim reaction. Blood 1991;77:2504–2514. [PubMed] [Google Scholar]
- 90.Suttorp M, Schmitz N, Dreger P, et al. Monitoring of chimerism after allogeneic bone marrow transplantation with unmanipulated marrow by use of DNA polymorphisms. Leukemia 1993;7:679–687. [PubMed] [Google Scholar]
- 91.van Leeuwen JE, van Tol MJ, Bodzinga BG, et al. Detection of mixed chimaerism in flow-sorted cell subpopulations by PCR-amplified VNTR markers after allogeneic bone marow transplantation. Br J Haematol 1991;79:218–225. [DOI] [PubMed] [Google Scholar]
- 92.Bertheas MF, Lafage M, Blaise D, et al. Mixed chimerism after allogeneic bone marrow transplantation for leukemias. Bone Marrow Transplant 1990;6(Suppl 1):61–63. [PubMed] [Google Scholar]
- 93-.Scharf SJ, Smith AG, Hansen JA, et al. Quantitative determination of bone marrow transplant engraftment using fluorescent polymerase chain reaction primers for human identity markers. Blood 1995;85:1954–1963. [PubMed] [Google Scholar]
- 94.Oberkircher AR, Strout MP, Herzig GP, et al. Description of an efficient and highly informative method for the evaluation of hematopoietic chimerism following allogeneic bone marrow transplantation. Bone Marrow Transplant 1995;16:695–702. [PubMed] [Google Scholar]
- 95.Kreyenberg H, Holle W, Mohrle S, et al. Quantitative analysis of chimerism after allogeneic stem cell transplantation by PCR amplification of microsatellite markers and capillary electrophoresis with fluorescence detection: the Tuebingen experience. Leukemia 2003;17:237–240. [DOI] [PubMed] [Google Scholar]
- 96.Hancock JP, Goulden NJ, Oakhill A, et al. Quantitative analysis of chimerism after allogeneic bone marrow transplantation using immunomagnetic selection and fluorescent microsatellite PCR. Leukemia 2003;17:247–251. [DOI] [PubMed] [Google Scholar]
- 97.Acquaviva C, Duval M, Mirebeau D, et al. Quantitative analysis of chimerism after allogeneic stem cell transplantation by PCR amplification of microsatellite markers and capillary electrophoresis with fluorescence detection: the Paris-Robert Debre experience. Leukemia 2003;17:241–246. [DOI] [PubMed] [Google Scholar]
- 98.Chalandon Y, Vischer S, Helg C, et al. Quantitative analysis of chimerism after allogeneic stem cell transplantation by PCR amplification of microsatellite markers and capillary electrophoresis with fluorescence detection: the Geneva experience. Leukemia 2003;17:228–231. [DOI] [PubMed] [Google Scholar]
- 99.Lo YM, Tein MS, Lau TK, et al. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am J Hum Genet 1998;62:768–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Thiede C, Kellermann T, Schwerdtfeger R, et al. Real-time PCR for the SRY-gene allows sensitive and quantiative chimerism analysis after allogeneic blood stem cell transplantation: Clinical results in 43 patients. Bone Marrow Transplant 2003;31(Suppl. 1):23.12621503 [Google Scholar]
- 101.Fehse B, Chukhlovin A, Kühlcke K, et al. Real-time quantitative Y chromosome-specific PCR (QYCS- PCR) for monitoring hematopoietic chimerism after sex-mismatched allogeneic stem cell transplantation. J Hematother Stem Cell Res. 2001;10:419–425. [DOI] [PubMed] [Google Scholar]
- 102.Sachidanandam R, Weissman D, Schmidt SC, et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 2001;409:928–933. [DOI] [PubMed] [Google Scholar]
- 103.Wang DG, Fan JB, Siao CJ, et al. Large-scale identification, mapping, and genotyping of single- nucleotide polymorphisms in the human genome. Science 1998;280:1077–1082. [DOI] [PubMed] [Google Scholar]
- 104.Hochberg EP, Miklos DB, Neuberg D, et al. A novel rapid single nucleotide polymorphism (SNP)- based method for assessment of hematopoietic chimerism after allogeneic stem cell transplantation. Blood 2003;101:363–369. [DOI] [PubMed] [Google Scholar]
- 105.Alizadeh M, Bernard M, Danic B, et al. Quantitative assessment of hematopoietic chimerism after bone marrow transplantation by real-time quantitative polymerase chain reaction. Blood 2002;99(:4618–4625. [DOI] [PubMed] [Google Scholar]
- 106.Maas F, Schaap N, Kolen S, et al. Quantification of donor and recipient hemopoietic cells by real-time PCR of single nucleotide polymorphisms. Leukemia 2003;17:621–629. [DOI] [PubMed] [Google Scholar]
- 107.Fredriksson M, Barbany G, Liljedahl U, et al. Assessing hematopoietic chimerism after allogeneic stem cell transplantation by multiplexed SNP genotyping using microarrays and quantitative analysis of SNP alleles. Leukemia 2004;18:255–266. [DOI] [PubMed] [Google Scholar]
- 108.Thiede C, Florek M, Bornhauser M, et al. Rapid quantification of mixed chimerism using multiplex amplification of short tandem repeat markers and fluorescence detection. Bone Marrow Transplant 1999;23:1055–1060. [DOI] [PubMed] [Google Scholar]
- 109.de Weger RA, Tilanus MG, Scheidel KC, et al. Monitoring of residual disease and guided donor leucocyte infusion after allogeneic bone marrow transplantation by chimaerism analysis with short tandem repeats. Br J Haematol 2000;110:647–653. [DOI] [PubMed] [Google Scholar]
- 110.Lion T, Daxberger H, Dubovsky J, et al. Analysis of chimerism within specific leukocyte subsets for detection of residual or recurrent leukemia in pediatric patients after allogeneic stem cell transplantation. Leukemia 2001;15:307–310. [DOI] [PubMed] [Google Scholar]
- 111.Willasch A, Schneider G, Reincke BS, et al. Sequence polymorphism systems for quantitative real-time polymerase chain reaction to characterize hematopoietic chimerism-high informativity and sensitivity as well as excellent reproducibility and precision of measurement. Lab Hematol 2007;13:73–84. [DOI] [PubMed] [Google Scholar]
- 112.Roux E, Abdi K, Speiser D, et al. Characterization of mixed chimerism in patients with chronic myeloid leukemia transplanted with T-cell-depleted bone marrow: involvement of different hematologic lineages before and after relapse. Blood 1993;81,243–248. [PubMed] [Google Scholar]
- 113.Fernández-Avilés F, Urbano-Ispizua A, Aymerich M,. et al. Serial quantification of lymphoid and myeloid mixed chimerism using multiplex PCR amplification of short tandem repeat-markers predicts graft rejection and relapse, respectively, after allogeneic transplantation of CD34+ selected cells from peripheral blood. Leukemia 2003; 17,613–620. [DOI] [PubMed] [Google Scholar]
- 114.Kröger N, Zagrivnaja M, Schwartz S, et al. Kinetics of plasma-cell chimerism after allogeneic stem cell transplantation by highly-sensitive real-time PCR based on sequence polymorphism and its value to quantify minimal residual disease in patients with multiple myeloma. Exp Hematol. 2006;34:688–694. [DOI] [PubMed] [Google Scholar]
- 115.Antin JH, Childs R, Filipovich AH, et al. Establishemnt of complete and mixed donor chimerism after allogeneic lymphohematopoietic transplantation. Biol Blood Marrow Transplant 2001;7:473–485. [DOI] [PubMed] [Google Scholar]
- 116.Edelstyn GA, Gillespie PJ, Grebbell FS: The radiological demonstration of oseous metastases. Experimental observations. Clin Radiol 1967;18:158–162. [DOI] [PubMed] [Google Scholar]
- 117.Armitage JO: Staging non-Hodgkin’s lymphoma. CA Cancer J Clin 2005; 55:368–376. [DOI] [PubMed] [Google Scholar]
- 118.Lister TA, Crowther D, Sutcliffe SB, et al. Report of a committee convened to discuss the evaluation and staging of patients with Hodgkin’s disease: Coswolds meeting. J Clin Oncol 1989; 7:1630–1636. [DOI] [PubMed] [Google Scholar]
- 119.Jhanwar YS, Straus DJ. The role of PET in lymphoma. J Nucl Med 2006;47:1326–1334. [PubMed] [Google Scholar]
- 120.Vinnicombe SJ, Reznek RH. Computerised tomography in the staging of Hodgkin’s disease and non- Hodgkin’s lymphoma. Eur J Nucl Med Mol Imaging. 2003;30:S42–S55. [DOI] [PubMed] [Google Scholar]
- 121.Mahnken AH, Wildberger JE, Gehbauer G, et al. : Multidetector CT of the spine in multiple myeloma: comparison with MR imaging and radiography. AJR 2002;180:1429–1436. [DOI] [PubMed] [Google Scholar]
- 122.Libshitz HI, Malthouse SR, Cunningham D, et al. : Multiple myeloma: appearnace at MR imaging. Radiology 1992;182:833–837. [DOI] [PubMed] [Google Scholar]
- 123.Rohren EM, Turkington TG, Coleman RE. Clinical applications of PET in oncology. Radiology 2004;231:305–332. [DOI] [PubMed] [Google Scholar]
- 124.Von Schulthess GK, Steinert HC, Hany TF: Integrated PET/CT: current applications and future directions. Radiology 2006;238:405–422. [DOI] [PubMed] [Google Scholar]
- 125.Cheson BD, Pfistner B, Juweid ME, et al. Revised response criteria for malignant lymphoma. J Clin Oncol 2007;25:579–586. [DOI] [PubMed] [Google Scholar]
- 126.Bruzzi JF, Macapinlac H, Apostolia M., et al. Detection of Richter’s Transformation of Chronic Lymphocytic Leukemia by PET/CT. J Nucl Med 2006;47:1267–1273. [PubMed] [Google Scholar]
- 127.Boellaard R, O’Doherty MJ, Weber WA, et al. FDG PET and PET/CT: EANM procedure guidelines for tumour PET imaging: version 1.0. Eur J Nucl Med Mol Imaging. 2010. January;37:181–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wahl RL. Jacene H, Kasamon Y, et al. From RECIST to PERCIST: Evolving Considerations for PET Response Criteria in Solid Tumors. J Nucl Med. 2009;50:5 (Suppl):122S–150S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hutchings M and Barrington SF.: PET/CT for Therapy Response Assessment in Lymphoma. J Nucl Med. 2009;50:5 (Suppl):21S–30S. [DOI] [PubMed] [Google Scholar]
- 130.Hermann K, Weider HA, Buck AK, et al. Early response assessment using 3’-deoxy-3’-[18F] fluorothymidine-positron emission tomography in high-grade non-Hodgkin’s lymphoma. Clin Cancer Res 2007; 5;13:3552–3558. [DOI] [PubMed] [Google Scholar]
- 131.Cheson BD, Bennett JM, Kopecky KJ, et al. Revised recommendations of the International Working Group for Diagnosis, Standardization of Response Criteria, Treatment Outcomes, and Reporting Standards for Therapeutic Trials in Acute Myeloid Leukemia. J Clin Oncol 2003;21:4642–4649. [DOI] [PubMed] [Google Scholar]
- 132.Marcucci G, Mrozek K, Ruppert AS, et al. Abnormal cytogenetics at date of morphologic complete remission predicts short overall and disease-free survival, and higher relapse rate in adult acute myeloid leukemia: results from cancer and leukemia group B study 8461. J Clin Oncol 2004;22:2410–2418. [DOI] [PubMed] [Google Scholar]
- 133.Fuehrer M, Gerusel-Bleck M, Konstantopoulos N, et al. Walther JU. FISH analysis of native smears from bone marrow and blood for the monitoring of chimerism and clonal markers after stem cell transplantation in children. Int J Mol Med 2005;15:291–297. [PubMed] [Google Scholar]
- 134.El-Rifai W, Ruutu T, Elonen E, et al. Prognostic value of metaphase-fluorescence in situ hybridization in follow-up of patients with acute myeloid leukemia in remission. Blood 1997;89:3330–3334. [PubMed] [Google Scholar]
- 135.Bader P, Kreyenberg H, Hoelle W, et al. Increasing mixed chimerism defines a high-risk group of childhood acute myelogenous leukemia patients after allogeneic stem cell transplantation where pre- emptive immunotherapy may be effective. Bone Marrow Transplant 2004;33:815–821. [DOI] [PubMed] [Google Scholar]
- 136.Huisman C, de Weger RA, de Vries L, et al. Chimerism analysis within 6 months of allogeneic stem cell transplantation predicts relapse in acute myeloid leukemia. Bone Marrow Transplant 2007;39:285–291. [DOI] [PubMed] [Google Scholar]
- 137.Zeiser R, Spyridonidis A, Wasch R, et al. Evaluation of immunomodulatory treatment based on conventional and lineage-specific chimerism analysis in patients with myeloid malignancies after myeloablative allogeneic hematopoietic cell transplantation. Leukemia 2005;19:814–821. [DOI] [PubMed] [Google Scholar]
- 138.Leroy H, de Botton S, Grardel-Duflos N, et al. Prognostic value of real-time quantitative PCR (RQ- PCR) in AML with t(8;21). Leukemia 2005;19:367–372. [DOI] [PubMed] [Google Scholar]
- 139.Marcucci G, Caligiuri MA, Dohner H, et al. Quantification of CBFbeta/MYH11 fusion transcript by real time RT-PCR in patients with INV(16) acute myeloid leukemia. Leukemia 2001;15:1072–1080. [DOI] [PubMed] [Google Scholar]
- 140.Elmaagacli AH, Beelen DW, Kroll M, et al. Detection of CBFbeta/MYH11 fusion transcripts in patients with inv(16) acute myeloid leukemia after allogeneic bone marrow or peripheral blood progenitor cell transplantation. Bone Marrow Transplant 1998;21:159–166. [DOI] [PubMed] [Google Scholar]
- 141.Gorello P, Cazzaniga G, Alberti F, et al. Quantitative assessment of minimal residual disease in acute myeloid leukemia carrying nucleophosmin (NPM1) gene mutations. Leukemia 2006;20:1103–1108. [DOI] [PubMed] [Google Scholar]
- 142.Chou WC, Tang JL, Wu SJ, et al. Clinical implications of minimal residual disease monitoring by quantitative polymerase chain reaction in acute myeloid leukemia patients bearing nucleophosmin (NPM1) mutations. Leukemia 2007;21:998–1004. [DOI] [PubMed] [Google Scholar]
- 143.Barragan E, Pajuelo JC, Ballester S, et al. Minimal residual disease detection in acute myeloid leukemia by mutant nucleophosmin (NPM1): comparison with WT1 gene expression. Clin Chim Acta 2008;395:120–123. [DOI] [PubMed] [Google Scholar]
- 144.Papadaki C, Dufour A, Seibl M, et al. Monitoring minimal residual disease in acute myeloid leukaemia with NPM1 mutations by quantitative PCR: clonal evolution is a limiting factor1. Br J Haematol 2009;144:517–523. [DOI] [PubMed] [Google Scholar]
- 145.Falini B, Mecucci C, Tiacci E, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med 2005;352:254–266. [DOI] [PubMed] [Google Scholar]
- 146.Dohner K, Schlenk RF, Habdank M, et al. Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: interaction with other gene mutations. Blood 2005;106:3740–3746. [DOI] [PubMed] [Google Scholar]
- 147.Schnittger S, Schoch C, Dugas M, et al. Analysis of FLT3 length mutations in 1003 patients with acute myeloid leukemia: correlation to cytogenetics, FAB subtype, and prognosis in the AMLCG study and usefulness as a marker for the detection of minimal residual disease. Blood 2002;100:59–66. [DOI] [PubMed] [Google Scholar]
- 148.Scholl S, Loncarevic IF, Krause C, et al. Minimal residual disease based on patient specific Flt3-ITD and -ITT mutations in acute myeloid leukemia. Leuk Res 2005;29:849–853. [DOI] [PubMed] [Google Scholar]
- 149.Cloos J, Goemans BF, Hess CJ, et al. Stability and prognostic influence of FLT3 mutations in paired initial and relapsed AML samples. Leukemia 2006;20(7):1217–1220. [DOI] [PubMed] [Google Scholar]
- 150.Schnittger S, Schoch C, Kern W, et al. FLT3 length mutations as marker for follow-up studies in acute myeloid leukaemia. Acta Haematol 2004;112:68–78. [DOI] [PubMed] [Google Scholar]
- 151.Elmaagacli AH. Molecular methods used for detection of minimal residual disease following hematopoietic stem cell transplantation in myeloid disorders. Methods Mol Med 2007;134:161–178. [DOI] [PubMed] [Google Scholar]
- 152.Shimoni A, Nagler A. Clinical implications of minimal residual disease monitoring for stem cell transplantation after reduced intensity and nonmyeloablative conditioning. Acta Haematol 2004;112: 93–104. [DOI] [PubMed] [Google Scholar]
- 153.Falini B, Nicoletti I, Martelli MF, et al. Acute myeloid leukemia carrying cytoplasmic/mutated nucleophosmin (NPMc+ AML): biologic and clinical features. Blood 2007;109:874–885. [DOI] [PubMed] [Google Scholar]
- 154.Abu-Duhier FM, Goodeve AC, Wilson GA, et al. FLT3 internal tandem duplication mutations in adult acute myeloid leukaemia define a high-risk group. Br J Haematol 2000;111:190–195. [DOI] [PubMed] [Google Scholar]
- 155.Thiede C, Steudel C, Mohr B, et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood 2002;99:4326–4335. [DOI] [PubMed] [Google Scholar]
- 156.Yamamoto Y, Kiyoi H, Nakano Y, et al. Activating mutation of D835 within the activation loop of FLT3 in human hematologic malignancies. Blood 2001;97:2434–2439. [DOI] [PubMed] [Google Scholar]
- 157.Bacher U, Haferlach C, Kern W, et al. Prognostic relevance of FLT3-TKD mutations in AML: the combination matters - an analysis of 3082 patients. Blood 2008;111:2527–2537. [DOI] [PubMed] [Google Scholar]
- 158.Mead AJ, Linch DC, Hills RK, et al. FLT3 tyrosine kinase domain mutations are biologically distinct from and have a significantly more favorable prognosis than FLT3 internal tandem duplications in patients with acute myeloid leukemia. Blood 2007;110:1262–1270. [DOI] [PubMed] [Google Scholar]
- 159.Scholl S, Loncarevic IF, Krause C, et al. Analyses of minimal residual disease based on Flt3 mutations in allogeneic peripheral blood stem cell transplantation. J Cancer Res Clin Oncol 2005;131:279–283. [DOI] [PubMed] [Google Scholar]
- 160.Schlenk RF, Dohner K, Krauter J, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med 2008;358:1909–1918. [DOI] [PubMed] [Google Scholar]
- 161.Schichman SA, Caligiuri MA, Strout MP, et al. ALL-1 tandem duplication in acute myeloid leukemia with a normal karyotype involves homologous recombination between Alu elements. Cancer Res 1994;54(:4277–4280. [PubMed] [Google Scholar]
- 162.Schnittger S, Kinkelin U, Schoch C, et al. Screening for MLL tandem duplication in 387 unselected patients with AML identify a prognostically unfavorable subset of AML. Leukemia 2000;14:796–804. [DOI] [PubMed] [Google Scholar]
- 163.Dohner K, Tobis K, Ulrich R, et al. Prognostic significance of partial tandem duplications of the MLL gene in adult patients 16 to 60 years old with acute myeloid leukemia and normal cytogenetics: a study of the Acute Myeloid Leukemia Study Group Ulm. J Clin Oncol 2002;20:3254–3261. [DOI] [PubMed] [Google Scholar]
- 164.Schoch C, Schnittger S, Klaus M, et al. AML with 11q23/MLL abnormalities as defined by the WHO classification: incidence, partner chromosomes, FAB subtype, age distribution, and prognostic impact in an unselected series of 1897 cytogenetically analyzed AML cases. Blood 2003;102:2395–2402. [DOI] [PubMed] [Google Scholar]
- 165.Weisser M, Kern W, Schoch C, et al. Risk assessment by monitoring expression levels of partial tandem duplications in the MLL gene in acute myeloid leukemia during therapy. Haematologica 2005;90: 881–889. [PubMed] [Google Scholar]
- 166.Ogawa H, Tamaki H, Ikegame K, et al. The usefulness of monitoring WT1 gene transcripts for the prediction and management of relapse following allogeneic stem cell transplantation in acute type leukemia. Blood 2003;101:1698–1704. [DOI] [PubMed] [Google Scholar]
- 167.Candoni A, Tiribelli M, Toffoletti E, et al. Quantitative assessment of WT1 gene expression after allogeneic stem cell transplantation is a useful tool for monitoring minimal residual disease in acute myeloid leukemia. Eur J Haematol 2009;82: 61–68. [DOI] [PubMed] [Google Scholar]
- 168.Jacobsohn DA, Tse WT, Chaleff S, et al. High WT1 gene expression before haematopoietic stem cell transplant in children with acute myeloid leukaemia predicts poor event-free survival. Br J Haematol. 2009;146:669–674. [DOI] [PubMed] [Google Scholar]
- 169.Langer C, Radmacher MD, Ruppert AS, et al. High BAALC expression associates with other molecular prognostic markers, poor outcome, and a distinct gene-expression signature in cytogenetically normal patients younger than 60 years with acute myeloid leukemia: a Cancer and Leukemia Group B (CALGB) study. Blood 2008;111:5371–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Campana D Determination of minimal residual disease in leukaemia patients. Br J Haematol 2003;121: 823–838. [DOI] [PubMed] [Google Scholar]
- 171.Kern W, Voskova D, Schoch C, et al. Determination of relapse risk based on assessment of minimal residual disease during complete remission by multiparameter flow cytometry in unselected patients with acute myeloid leukemia. Blood 2004;104:3078–3085. [DOI] [PubMed] [Google Scholar]
- 172.Griesinger F, Piro-Noack M, Kaib N, et al. Leukaemia-associated immunophenotypes (LAIP) are observed in 90% of adult and childhood acute lymphoblastic leukaemia: detection in remission marrow predicts outcome. Br J Haematol 1999;105:241–255. [PubMed] [Google Scholar]
- 173.Coustan-Smith E, Ribeiro RC, Rubnitz JE, et al. Clinical significance of residual disease during treatment in childhood acute myeloid leukaemia. Br J Haematol 2003;123:243–322. [DOI] [PubMed] [Google Scholar]
- 174.Nagler A, Condiotti R, Rabinowitz R, et al. Detection of minimal residual disease (MRD) after bone marrow transplantation (BMT) by multi-parameter flow cytometry (MPFC). Med Oncol 1999;16:177–187. [DOI] [PubMed] [Google Scholar]
- 175.Wells DA, Sale GE, Shulman HM, et al. Multidimensional flow cytometry of marrow can differentiate leukemic from normal lymphoblasts and myeloblasts after chemotherapy and bone marrow transplantation. Am J Clin Pathol 1998;110:84–94. [DOI] [PubMed] [Google Scholar]
- 176.Ito S, Ishida Y, Murai K, et al. Flow cytometric analysis of aberrant antigen expression of blasts using CD45 blast gating for minimal residual disease in acute leukemia and high-risk myelodysplastic syndrome. Leuk Res 2001;25:205–211. [DOI] [PubMed] [Google Scholar]
- 177.Perez-Simon JA, Caballero D, Diez-Campelo M, et al. Chimerism and minimal residual disease monitoring after reduced intensity conditioning (RIC) allogeneic transplantation. Leukemia 2002;16:1423–1431. [DOI] [PubMed] [Google Scholar]
- 178.Diez-Campelo M, Perez-Simon JA, Perez J, et al. Minimal residual disease monitoring after allogeneic transplantation may help to individualize post-transplant therapeutic strategies in acute myeloid malignancies. Am J Hematol 2009;84:149–152. [DOI] [PubMed] [Google Scholar]
- 179.Voskova D, Schoch C, Schnittger S, et al. Stability of leukemia-associated aberrant immunophenotypes in patients with acute myeloid leukemia between diagnosis and relapse: comparison with cytomorphologic, cytogenetic, and molecular genetic findings. Cytometry B Clin Cytom 2004;62:25–38. [DOI] [PubMed] [Google Scholar]
- 180.Cheson BD, Greenberg PL, Bennett JM, et al. Clinical application and proposal for modification of the International Working Group (IWG) response criteria in myelodysplasia. Blood 2006;108:419–425. [DOI] [PubMed] [Google Scholar]
- 181.Cilloni D, Gottardi E, De Micheli D, et al. Quantitative assessment of WT1 expression by real time quantitative PCR may be a useful tool for monitoring minimal residual disease in acute leukemia patients. Leukemia 2002;16:2115–2121. [DOI] [PubMed] [Google Scholar]
- 182.Patmasiriwat P, Fraizer G, Kantarjian H, et al. WT1 and GATA1 expression in myelodysplastic syndrome and acute leukemia. Leukemia 1999;13:891–900. [DOI] [PubMed] [Google Scholar]
- 183.Tamura K, Kanazawa T, Suzuki M, . Successful rapid discontinuation of immunosuppressive therapy at molecular relapse after allogeneic bone marrow transplantation in a pediatric patient with myelodysplastic syndrome. Am J Hematol 2006;81:139–141. [DOI] [PubMed] [Google Scholar]
- 184.Chen CY, Lin LI, Tang JL, et al. RUNX1 gene mutation in primary myelodysplastic syndrome--the mutation can be detected early at diagnosis or acquired during disease progression and is associated with poor outcome. Br J Haematol 2007;139:405–414. [DOI] [PubMed] [Google Scholar]
- 185.Christiansen DH, Andersen MK, Pedersen-Bjergaard J. Mutations of AML1 are common in therapy- related myelodysplasia following therapy with alkylating agents and are significantly associated with deletion or loss of chromosome arm 7q and with subsequent leukemic transformation. Blood 2004;104:1474–1481. [DOI] [PubMed] [Google Scholar]
- 186.Yanada M, Matsuo K, Suzuki Tet al. Prognostic significance of FLT3 internal tandem duplication and tyrosine kinase domain mutations for acute myeloid leukemia: a meta-analysis. Leukemia 2005;19:1345–1349. [DOI] [PubMed] [Google Scholar]
- 187-.Shih LY, Huang CF, Wang PN, et al. Acquisition of FLT3 or N-ras mutations is frequently associated with progression of myelodysplastic syndrome to acute myeloid leukemia. Leukemia 2004;18:466–475. [DOI] [PubMed] [Google Scholar]
- 188.Bacher U, Haferlach T, Kern W, et al. A comparative study of molecular mutations in 381 patients with myelodysplastic syndrome and in 4130 patients with acute myeloid leukemia. Haematologica 2007;92:744–752. [DOI] [PubMed] [Google Scholar]
- 189.Zhang Y, Zhang M, Yang L, et al. NPM1 mutations in myelodysplastic syndromes and acute myeloid leukemia with normal karyotype. Leuk Res 2007;31:109–111. [DOI] [PubMed] [Google Scholar]
- 190.Stetler-Stevenson M, Arthur DC, Jabbour N, et al. Diagnostic utility of flow cytometric immunophenotyping in myelodysplastic syndrome. Blood 2001;98:979–987. [DOI] [PubMed] [Google Scholar]
- 191.Della Porta MG, Malcovati L, Invernizzi R, et al. Flow cytometry evaluation of erythroid dysplasia in patients with myelodysplastic syndrome. Leukemia 2006;20:549–555. [DOI] [PubMed] [Google Scholar]
- 192.Van de Loosdrecht A, Alhan C, Bene MC, et al. Standardization of flow cytometry in myelodysplastic syndromes: Report from the first ELNet working conference on flow cytometry in MDS. Haematologica 2009; 94(:1124–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Wells DA, Benesch M, Loken MR et al. Myeloid and monocytic dyspoiesis as determined by flow cytometric scoring in myelodysplastic syndrome correlates with the IPSS and with outcome after hematopoietic stem cell transplantation. Blood 2003;102:394–403. [DOI] [PubMed] [Google Scholar]
- 194.Brisco MJ, Condon J, Hughes E, et al. Outcome prediction in childhood acute lymphoblastic leukaemia by molecular quantification of residual disease at the end of induction. Lancet 1994;343:196–200. [DOI] [PubMed] [Google Scholar]
- 195.Cavé H, van der Werff ten Bosch J, Suciu S,. et al. Clinical significance of minimal residual disease in childhood acute lymphoblastic leukemia. European Organization for Research and Treatment of Cancer--Childhood Leukemia Cooperative Group. N Engl J Med 1998;339:591–598. [DOI] [PubMed] [Google Scholar]
- 196.Coustan-Smith E, Behm FG, Sanchez J,. et al. Immunological detection of minimal residual disease in children with acute lymphoblastic leukaemia. Lancet 1998;351:550–554.. [DOI] [PubMed] [Google Scholar]
- 197.Szczepanski T, Orfao A, van der Velden VH, et al. Minimal residual disease in leukaemia patients. Lancet Oncol 2001;2:409–417. [DOI] [PubMed] [Google Scholar]
- 198.an Dongen JJ, Seriu T, Panzer-Grümayer ER, et al. Prognostic value of minimal residual disease in acute lymphoblastic leukaemia in childhood. Lancet 1998;352:731–1738. [DOI] [PubMed] [Google Scholar]
- 199.Pui CH and Campana D New definition of remission in childhood acute lymphoblastic leukemia. Leukemia 2000;14:783–785. [DOI] [PubMed] [Google Scholar]
- 200.Schrappe M, Beier R, Burger B New treatment strategies in childhood acute lymphoblastic leukaemia. Best Pract Res Clin Haematol 2002; 15:729–740. [DOI] [PubMed] [Google Scholar]
- 201.Gameiro P, Mortuza FY, Hoffbrand, et al. Minimal residual disease monitoring in adult T-cell acute lymphoblastic leukemia: a molecular based approach using T-cell receptor G and D gene rearrangements. Haematologica 2002;87:1126–1134. [PubMed] [Google Scholar]
- 202.Specchia G, Liso A, Pannunzio A, et al. Molecular detection of minimal residual disease is associated with early relapse in adult acute lymphoblastic leukemia. Haematologica 2004;89:1271–1273. [PubMed] [Google Scholar]
- 203.Toubai T, Tanaka J, Ota S,. et al. Minimal residual disease (MRD) monitoring using rearrangement of T-cell receptor and immunoglobulin H gene in the treatment of adult acute lymphoblastic leukemia patients. Am J Hematol 2005;80:181–187. [DOI] [PubMed] [Google Scholar]
- 204.Mortuza FY, Papaioannou M, Moreira IM, et al. Minimal residual disease tests provide an independent predictor of clinical outcome in adult acute lymphoblastic leukemia. J Clin Oncol 2002;20:1094–1104. [DOI] [PubMed] [Google Scholar]
- 205.Brüggemann M, Raff T, Flohr T, et al. Clinical significance of minimal residual disease quantification in adult patients with standard-risk acute lymphoblastic leukemia. Blood 2006;107:1116–1123. [DOI] [PubMed] [Google Scholar]
- 206.Krampera M, Vitale A, Vincenzi C, et al. Outcome prediction by immunophenotypic minimal residual disease detection in adult T-cell acute lymphoblastic leukaemia. Br J Haematol 2003;120:74–79. [DOI] [PubMed] [Google Scholar]
- 207.Holowiecki J, Krawczyk-Kulis M, Giebel S, et al. Status of minimal residual disease after induction predicts outcome in both standard and high-risk Ph-negative adult acute lymphoblastic leukaemia. The Polish Adult Leukemia Group ALL 4–2002 MRD Study. Br J Haematol 2008;142:227–237. [DOI] [PubMed] [Google Scholar]
- 208.Raff T, Gökbuget N, Lüschen S, et al. Molecular relapse in adult standard-risk ALL patients detected by prospective MRD monitoring during and after maintenance treatment: data from the GMALL 06/99 and 07/03 trials. Blood 2007;109:910–915. [DOI] [PubMed] [Google Scholar]
- 209.Bassan R, Spinelli O, Oldani E, et al. Improved risk classification for risk-specific therapy based on the molecular study of minimal residual disease (MRD) in adult acute lymphoblastic leukemia (ALL). Blood 2009;113:4153–4162. [DOI] [PubMed] [Google Scholar]
- 210.Bruggemann M, Droese, J, Raff T. the prognostic significance of minimal residual disease in adult standard risk patients with acute lymphoblastic leukemia. Blood 2001;98:14a. [DOI] [PubMed] [Google Scholar]
- 211.Coustan-Smith E, Sancho J, Hancock ML, et al. Use of peripheral blood instead of bone marrow to monitor residual disease in children with acute lymphoblastic leukemia. Blood 2002;100:2399–402. [DOI] [PubMed] [Google Scholar]
- 212.van der Velden VH, Jacobs DC, Wijkhuijs AJ, et al. Minimal residual disease levels in bone marrow and peripheral blood are comparable in children with T cell acute lymphoblastic leukemia (ALL), but not in precursor-B-ALL. Leukemia 2002;16:1432–1436. [DOI] [PubMed] [Google Scholar]
- 213.Imashuku S, Terui K, Matsuyama T, et al. Multi-institutional collaborative study in Japan. Lack of clinical utility of minimal residual disease detection in allogeneic stem cell recipients with childhood acute lymphoblastic leukemia: multi-institutional collaborative study in Japan. Bone Marrow Transpant 2003;31(12):1127–1135. [DOI] [PubMed] [Google Scholar]
- 214.Bader P, Hancock J, Kreyenberg H, et al. Minimal residual disease (MRD) status prior to allogeneic stem cell transplantation is a powerful predictor for post-transplant outcome in children with ALL. Leukemia 2002;16:1668–1672. [DOI] [PubMed] [Google Scholar]
- 215.Campana D Minimal residual disease in acute lymphoblastic leukemia. Semin Hematol 2009;46: 100–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Knechtli CJ, Goulden NJ, Hancock JP, et al. Minimal residual disease status before allogeneic bone marrow transplantation is an important determinant of successful outcome for children and adolescents with acute lymphoblastic leukemia. Blood 1998;92:4072–4079. [PubMed] [Google Scholar]
- 217.Bader P, Kreyenberg H, Henze GH, Eckert C, et al. Prognostic value of minimal residual disease quantification before allogeneic stem-cell transplantation in relapsed childhood acute lymphoblastic leukemia: the ALL-REZ BFM Study Group. J Clin Oncol 2009;27:377–384. [DOI] [PubMed] [Google Scholar]
- 218.Spinelli O, Peruta B, Tosi M, et al. Clearance of minimal residual disease after allogeneic stem cell transplantation and the prediction of the clinical outcome of adult patients with high-risk acute lymphoblastic leukemia. Haematologica 2007;92:612–618. [DOI] [PubMed] [Google Scholar]
- 219.Gehly GB, Bryant EM, Lee AM, et al. Chimeric BCR-abl messenger RNA as a marker for minimal residual disease in patients transplanted for Philadelphia chromosome-positive acute lymphoblastic leukemia. Blood 1991;78: 458–465. [PubMed] [Google Scholar]
- 220.Miyamura K, Tanimoto M, Morishima Y, et al. Detection of Philadelphia chromosome-positive acute lymphoblastic leukemia by polymerase chain reaction: possible eradication of minimal residual disease by marrow transplantation. Blood 1992;79:1366–1370. [PubMed] [Google Scholar]
- 221.Radich J, Gehly G, Lee A, et al. Detection of bcr-abl transcripts in Philadelphia chromosome-positive acute lymphoblastic leukemia after marrow transplantation. Blood 1997;89:2602–2609. [PubMed] [Google Scholar]
- 222.Radich J, Ladne P, Gooley T. Polymerase chain reaction-based detection of minimal residual disease in acute lymphoblastic leukemia predicts relapse after allogeneic BMT. Biol Blood Marrow Transplant 1995;1:24–31. [PubMed] [Google Scholar]
- 223.Dombret H, Gabert J, Boiron JM, et al. Outcome of treatment in adults with Philadelphia chromosome- positive acute lymphoblastic leukemia--results of the prospective multicenter LALA-94 trial. Blood 2002;100:2357–2366. [DOI] [PubMed] [Google Scholar]
- 224.Lee S, Kim YJ, Min CK, et al. The effect of first-line imatinib interim therapy on the outcome of allogeneic stem cell transplantation in adults with newly diagnosed Philadelphia chromosome-positive acute lymphoblastic leukemia. Blood 2005;105:3449–3457. [DOI] [PubMed] [Google Scholar]
- 225.Yanada M, Takeuchi J, Sugiura I, et al. High complete remission rate and promising outcome by combination of imatinib and chemotherapy for newly diagnosed BCR-ABL-positive acute lymphoblastic leukemia: a phase II study by the Japan Adult Leukemia Study Group. J Clin Oncol 2006;24:460–466. [DOI] [PubMed] [Google Scholar]
- 226.Ottmann OG, Wassmann B, Pfeifer H, et al. Imatinib compared with chemotherapy as front-line treatment of elderly patients with Philadelphia chromosome-positive acute lymphoblastic leukemia (Ph+ALL). Cancer 2007;109:2068–2076. [DOI] [PubMed] [Google Scholar]
- 227.Thomas DA, Faderl S, Cortes J, et al. Treatment of Philadelphia chromosome-positive acute lymphocytic leukemia with hyper-CVAD and imatinib mesylate. Blood 2004;103:4396–4407. [DOI] [PubMed] [Google Scholar]
- 228.Towatari M, Yanada M, Usui N, et al. Combination of intensive chemotherapy and imatinib can rapidly induce high-quality complete remission for a majority of patients with newly diagnosed BCR- ABL-positive acute lymphoblastic leukemia. Blood 2004;104:3507–3512. [DOI] [PubMed] [Google Scholar]
- 229.Lee S, Kim DW, Kim YJ, et al. Minimal residual disease-based role of imatinib as a first-line interim therapy prior to allogeneic stem cell transplantation in Philadelphia chromosome-positive acute lymphoblastic leukemia. Blood 2003;102:3068–3070. [DOI] [PubMed] [Google Scholar]
- 230.Lee S, Kim YJ, Chung NG, et al. The extent of minimal residual disease reduction after the first 4- week imatinib therapy determines outcome of allogeneic stem cell transplantation in adults with Philadelphia chromosome-positive acute lymphoblastic leukemia. Cancer 2009;115: 561–570. [DOI] [PubMed] [Google Scholar]
- 231.Bader P, Hölle W, Klingebiel T, et al. Mixed hematopoietic chimerism after allogeneic bone marrow transplantation: the impact of quantitative PCR analysis for prediction of relapse and graft rejection in children. Bone Marrow Transplant 1997;19: 697–702. [DOI] [PubMed] [Google Scholar]
- 232.Formánková R, Sedlácek P, Krsková L, et al. Chimerism-directed adoptive immunotherapy in prevention and treatment of post-transplant relapse of leukemia in childhood. Haematologica 2003;88:117–118. [PubMed] [Google Scholar]
- 233.Gorczyñska E, Turkiewicz D, Toporski J, et al. Prompt initiation of immunotherapy in children with an increasing number of autologous cells after allogeneic HCT can induce complete donor-type chimerism: a report of 14 children. Bone Marrow Transplant 2004;33:211–217. [DOI] [PubMed] [Google Scholar]
- 234.Ramírez M, Díaz MA, García-Sánchez F, et al. Chimerism after allogeneic hematopoietic cell transplantation in childhood acute lymphoblastic leukemia. Bone Marrow Transplant 1996;18:1161–1165. [PubMed] [Google Scholar]
- 235.Guimond M, Busque L, Baron C, et al. Relapse after bone marrow transplantation: evidence for distinct immunological mechanisms between adult and paediatric populations. Br J Haematol 2000;109:130–137. [DOI] [PubMed] [Google Scholar]
- 236.Barrios M, Jiménez-Velasco A, Román-Gómez J, et al. Chimerism status is a useful predictor of relapse after allogeneic stem cell transplantation for acute leukemia. Haematologica 2003;88:801–810. [PubMed] [Google Scholar]
- 237.Bader P, Stoll K, Huber A, et al. Characterization of lineages specific chimerism in patients with acute leukaemia and myelodysplastic syndrome after allogeneic stem cell transplantation before and after relapse. Br J Haematology 2000;108:761–768. [DOI] [PubMed] [Google Scholar]
- 238.Bader P, Kreyenberg H, Hoelle W, et al. Increasing mixed chimerism is an important prognostic factor for unfavorable outcome in children with acute lymphoblastic leukemia after allogeneic stem-cell transplantation: possible role for pre-emptive immunotherapy? J Clin Oncol 2004;22:1696–1705. [DOI] [PubMed] [Google Scholar]
- 239.Pulsipher MA, Bader P, Klingebiel T, et al. Allogeneic transplantation for pediatric acute lymphoblastic leukemia: the emerging role of peritransplantation minimal residual disease/chimerism monitoring and novel chemotherapeutic, molecular, and immune approaches aimed at preventing relapse. Biol Blood Marrow Transplant 2008;15:62–71. [DOI] [PubMed] [Google Scholar]
- 240.Knechtli CJ, Goulden NJ, Hancock JP, et al. Minimal residual disease status as a predictor of relapse after allogeneic bone marrow transplantation for children with acute lymphoblastic leukaemia. Br J Haematol 1998;102:860–871. [DOI] [PubMed] [Google Scholar]
- 241.Sánchez J, Serrano J, Gómez P, et al. Clinical value of immunological monitoring of minimal residual disease in acute lymphoblastic leukaemia after allogeneic transplantation. Br J Haematol 2002;116: 686–694. [DOI] [PubMed] [Google Scholar]
- 242.Uzunel M, Jaksch M, Mattsson J et al. Minimal residual disease detection after allogeneic stem cell transplantation is correlated to relapse in patients with acute lymphoblastic leukaemia. Br J Haematol 2003;122:788–794. [DOI] [PubMed] [Google Scholar]
- 243.Bader P Predictive value of MRD monitoring after allogeneic SCT in relapsed childhood acute lymphoblastic leukaemia: analysis of the ALL-REZ BFM Group. Bone Marrow Transplant 2009;43: 40. [Google Scholar]
- 244.Ch Thiede, Lutterbeck K, Oelschlägel U, et al. Detection of relapse by sequential monitoring of chimerism in circulating CD34+ cells. Ann Hematol 2002;81 (Suppl 2):,S27–S28. [PubMed] [Google Scholar]
- 245.Winiarski J, Mattsson J, Gustafsson A, et al. Engraftment and chimerism, particularly of T- and B- cells, in children undergoing allogeneic bone marrow transplantation. Pediatr Transplant 1998;2: 150–156. [PubMed] [Google Scholar]
- 246.Sierra J, Radich J, Hansen JA, et al. Marrow transplants from unrelated donors for treatment of Philadelphia chromosome-positive acute lymphoblastic leukemia. Blood 1997;90: 1410–1414. [PubMed] [Google Scholar]
- 247.Wassmann B, Pfeifer H, Stadler M, et al. Early molecular response to posttransplantation imatinib determines outcome in MRD+ Philadelphia-positive acute lymphoblastic leukemia (Ph+ ALL). Blood 2005;106:458–463. [DOI] [PubMed] [Google Scholar]
- 248.van Leeuwen JE, van Tol MJ, Joosten AM, et al. Mixed T-lymphoid chimerism after allogeneic bone marrow transplantation for hematologic malignancies of children is not correlated with relapse. Blood 1993;82:1921–1928. [PubMed] [Google Scholar]
- 249.Roy DC, Tantravahi R, Murray C, et al. Natural history of mixed chimerism after bone marrow transplantation with CD6-depleted allogeneic marrow: a stable equilibrium. Blood 1990;75:296–304. [PubMed] [Google Scholar]
- 250.Bader P, Beck j, Frey A, et al. Serial and quantitative analysis of mixed hematopoietic chimerism by PCR in patients with acute leukemias allows the prediction of relapse after allogeneic BMT. Bone Marrow Transplant 1998;21:487–495. [DOI] [PubMed] [Google Scholar]
- 251.Horn B, Soni S, Khan S, et al. Feasibility study of preemptive withdrawal of immunosuppression based on chimerism testing in children undergoing myeloablative allogeneic transplantation for hematologic malignancies. Bone Marrow Transplant 2009;43:469–476. [DOI] [PubMed] [Google Scholar]
- 252.Uzunel M, Mattsson J, Jaksch M, et al. The significance of graft-versus-host disease and pretransplantation minimal residual disease status to outcome after allogeneic stem cell transplantation in patients with acute lymphoblastic leukemia. Blood 2001;98:1982–1984. [DOI] [PubMed] [Google Scholar]
- 253.Sramkova L, Muzikova K, Fronkova E, et al. Detectable minimal residual disease before allogeneic hematopoietic stem cell transplantation predicts extremely poor prognosis in children with acute lymphoblastic leukemia. Pediatr Blood Cancer 2007;48:93–100. [DOI] [PubMed] [Google Scholar]