Abstract
Cellular mechanisms causing insulin resistance (IR) in chronic kidney disease (CKD) are poorly understood. One potential mechanism is that CKD-induced inflammation activates the signal transducer and activator of transcription 3 (Stat3) in muscle. We uncovered increased p-Stat3 in muscles of mice with CKD or mice fed high-fat diet (HFD). Activated Stat3 stimulates the expression of Fbxo40, a muscle-specific E3 ubiquitin ligase that stimulates ubiquitin conjugation leading to degradation of insulin receptor substrate 1 (IRS1). Evidence that Stat3 activates Fbxo40 includes 1) potential Stat3 binding sites in Fbxo40 promoters; 2) Stat3 binding to the Fbxo40 promoter; and 3) constitutively active Stat3 stimulating both Fbxo40 expression and its promoter activity. We found that IL-6 activates Stat3 in myotubes, increasing Fbxo40 expression with reduced IRS1 and p-Akt. Knockdown Fbxo40 using siRNA from myotubes results in higher levels of IRS1 and p-Akt despite the presence of IL-6. We treated mice with a small-molecule inhibitor of Stat3 (TTI-101) and found improved glucose tolerance and insulin signaling in skeletal muscles of mice with CKD or fed an HFD. Finally, we uncovered improved glucose tolerance in mice with muscle-specific Stat3 KO versus results in Stat3f/f mice in response to the HFD. Thus Stat3 activation in muscle increases IR in mice. Inhibition of Stat3 by TTI-101 could be developed into clinical strategies to improve muscle insulin signaling in inflammation and other catabolic diseases.
Keywords: CKD, diabetes, E3, Fbxo40, IRS1, insulin resistance, Stat3, and ubiquitin ligases
INTRODUCTION
Insulin resistance (IR) is common in patients with chronic kidney disease (CKD) including patients after successful kidney transplantation, and it is almost universally present in patients with end-stage kidney failure (ESKF) (15, 45). In fact, IR can be detected even in patients with normal levels of glomerular filtration rate (GFR) (16). Development of methods that overcome IR-associated complications of CKD is important because IR is a primary factor responsible for the metabolic syndrome, a collection of cardiometabolic risk factors including obesity, hypertension, and dyslipidemia (37). Importantly, skeletal muscle is the major site for disposal of glucose in healthy individuals. In the IR state, skeletal muscle had decreased insulin-stimulated glucose uptake and impaired insulin signaling and multiple postreceptor intracellular defects including impaired glucose transport, glucose phosphorylation, and reduced glucose oxidation and glycogen synthesis (11, 25, 37). Although the mechanisms accounting for these abnormalities in insulin resistance in skeletal muscle are not fully understood, the presence of inflammation have been shown to play a pivotal role in the development of IR in skeletal muscle (49).
Activation of certain signaling molecules [e.g., IKK-β, transforming growth factor-β1 (TGF-β1), or Smad3] has been linked to the development of IR (2). It also has been determined that the signal transducer and activator of transcription 3 (Stat3) regulates insulin signaling. The influences of Stat3 extend widely because it acts as a transcription factor and is expressed in multiple tissues. Specifically, Stat3 is activated by cytokines, growth factors, and nutrients; these factors induce phosphorylation of Tyr705 and Tyr727 of Stat3. Following its stimulation and nuclear translocation, Stat3 binds to promoters and regulates the expression of genes involved in inflammation, cell development, differentiation, proliferation, survival, and angiogenesis (41). In fact, Stat3 signaling pathways are involved in determining peripheral and hepatic insulin sensitivities. For example, in hepatocarcinoma cell lines, knockdown Stat3 prevents the development of amino acid-induced insulin resistance (26). In rat adipocytes, Stat3 activation is linked to the development of growth hormone-induced insulin resistance (14). Stat3 also affects insulin pathogenesis in skeletal muscles: in cultured skeletal myotubes, Stat3 participates in the development of IL-6-induced insulin resistance in subjects with impaired glucose tolerance (27). We have found that increased Stat3 activation is linked to losses in muscle mass in muscles of mice with CKD or cancer cachexia (43, 53). Stat3 also activates the expression of suppressor of cytokine signaling (SOCS) proteins, which downregulate responses to cytokine signaling (29). The fates of these proteins are relevant because SOCS3 overexpression in turn stimulates the ubiquitin conjugation and degradation of IRS1 leading to defects in intracellular insulin signaling (28). In patients with type 2 diabetes, evaluation of their skeletal muscles revealed constitutive increases in phosphotyrosylated (pY) Stat3 (32). Therefore, Stat3 inhibition could be used to improve skeletal muscle insulin signaling.
Atrogin-1 has been identified as a muscle-specific E3 ubiquitin ligase serving as a marker of the degree of muscle proteolysis in models of skeletal muscle atrophy. It functions as a muscle-specific F-box protein (designated Fbxo32) (7, 18) and is a key component of the SCF (Skp1-Cullin1-F-box protein) complex. The F-box proteins function as adaptors that bind specific substrates and target them for ubiquitin-proteasome-mediated degradation (21, 24). There are over 70 genes encoding F-box-containing proteins, they stimulate E3 ubiquitin ligase activities and participate in the regulation of cell cycle and signal transduction functions. Fbxo40 has been identified as a member of muscle-specific F-box proteins, but its functions have not been fully defined (51). It is reported that Fbxo40 expression is upregulated in skeletal muscles of mice that are subjected to denervation (51). Moreover, mice that cannot express Fbxo40 develop increases in body and muscle weights (42). Thus Fbxo40 could be involved in the development of muscle atrophy.
To identify potential mechanisms that could cause IR, we studied two mouse models of IR, CKD and mice fed a high-fat diet (HFD). We uncovered that activation of Stat3 stimulates Fbxo40 expression leading to IRS1 degradation and IR. To examine Stat3 function in vivo, we administrated TTI-101 (a small-molecule, Stat3 inhibitor to both groups of mice. We also investigated IR that is induced by HFD feeding of mice with muscle-specific Stat3 knockout (KO). Results from these experiments that probed activation of Stat3 using pharmacologic and genetic targeting indicate that Stat3 interruption can improve insulin signaling in mice with CKD or following HFD feeding.
METHODS
Animals and models.
Experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Baylor College of Medicine. Mice were housed in a temperature- and humidity-controlled room with a cycle of 12 h of light/darkness and were allowed free access to water and food. Wild-type (WT) C57BL/6 mice were purchased from Jackson laboratory (Bar Harbor, ME). CKD was created by subtotal nephrectomy, but only male mice were examined as female mice are less prone to proteinuria (23) and less sensitive to ischemic kidney damage (3, 22). The model of CKD was initially created by removal of 60–70% of the left kidney and feeding of 6% protein chow to minimize mortality from uremia. After feeding 6% protein chow for a week, the right kidney was removed and 6% protein chow was continued for 1 wk. Two weeks later, the diet contained 40% protein (Harlan Teklab, Indianapolis, IN) was applied to induce advanced CKD (33). Sham-operated control mice underwent surgery without damaging the kidneys and were fed the same diets. Mice with an average blood urea nitrogen (BUN) ~80 mg/dl were studied: CKD and sham-operated control mice were assigned to one of two subgroups. One subgroup was injected intraperitoneally with TTI-101 [12.5 mg/kg body wt in 5% dextrose (D5W) plus 5.3% DMSO] every other day for 2 wk; another group received an identical volume of D5W plus 5% DMSO for 2 wk. During the 2-wk treatment we pair-fed D5W with D5W/TTI-101 treatment mice. TTI-101 (formerly C1889) is a small molecule, direct Stat3 inhibitor developed by D. J. Tweardy’s group in collaboration with Tvardi Therapeutics, Inc. (Houston, TX). TTI-101 binds Stat3 directly inhibiting its activities. Specifically, TTI-101 inhibits both the recruitment of Stat3 to the tyrosine kinase-containing receptor complexes and receptor homodimerization (5); pharmacokinetic (PK) and toxicology studies in mice, rats, and dogs have demonstrated that TTI-101 administration provides excellent plasma exposures following oral administration (6, 31). TTI-101 is undergoing Phase I testing in patients with solid tumors.
We created a second model of insulin resistance in wild-type, C57/BL6, male mice by feeding a high fat diet (HFD: 58% kcal from fat, Research Diets, New Brunswick, NJ) for 12 wk. Control mice were fed the regular diet (RD: 11% kcal from fat). In diet-induced examinations of obesity studies, male mice have been studied more frequently because they are more severely affected by diabetes than female mice (5, 47). To study how p-Stat3 affects IR, HFD-fed male mice were randomly assigned to two subgroups: one subgroup was injected intraperitoneally with TTI-101 (12.5 mg/kg body wt in D5W) every other day for 4 wk, while the other subgroup received an identical volume of D5W for 4 wk. The D5W treatment mice were pair fed with D5W/TTI-101 treatment mice.
Muscle-specific Stat3 KO mice were created by crossing female Stat3flox/flox mice with male mice expressing muscle creatine kinase Cre (MCK-Cre) as described previously (53). Genotyping was performed by PCR using genomic DNA isolated from tail tips. Beginning at four weeks after birth, Stat3flox/flox or Stat3 KO male and female mice were fed the HFD for 16 wk.
For glucose tolerance testing (GTT), mice with free access to water were fasted for 16 h and then injected glucose intraperitoneally with 2 mg/g body wt. Tail vein blood was collected at 0, 30, 60, and 120 min following glucose injection; blood glucose concentrations were measured by True Track Glucometer (Nipro Diagnostics, Fort, Lauderdale, FL). An insulin tolerance test was performed in mice fasted for 4 h: mice were injected intraperitoneally with 2 U/kg insulin, and blood was collected after 0, 30, 60, and 120 min to measure blood glucose concentration.
At the end of the experiment, mice were fasted for 6 h and injected with 10 U/kg of insulin. Five minutes later muscles were collected and stored at −70° for future studies. Serum insulin concentration was measured using the rat/mouse insulin ELISA kit (Millipore, Billerica, MA).
Cell culture.
Mouse C2C12 myoblasts were obtained from American Type Culture Collection (ATCC, Manassas, VA). Cells were transfected with Fbxo40 siRNA (Santa Cruz Biotechnology, Dallas, TX) or the control siRNA using the Invitrogen Neon transfection system (Invitrogen Madison, Wisconsin). To induce differentiation, C2C12 myoblasts were grown to 85% confluence and then switched to differentiation media consisting of DMEM plus 2% horse serum (HS) and 1% penicillin-streptomycin (P/S; Invitrogen Madison, Wisconsin). The myotubes were treated with/without 100 ng/ml interleukin 6 (IL-6) (Biolegend, San Diego, CA) for 24 h. Cell lysates were subjected to Western blotting.
Luciferase reporter assays.
The human Fbxo40 promoter was cloned into a Gaussia-luciferase reporter that was obtained from GeneCopoeia, Inc. (Rockville, MD). The 1226 bp Fbxo40 promoter includes 1,062-bp upstream and 163-bp downstream. JASPAR database showed that the consensus Stat3 binding site as 5′-TTCTGGGAA-3′, which is comparable to the sequence in the promoter of Fbxo40 from 520 to 529 bp (see Fig. 3B). Substitution of two cysteine residues within the C-terminal loop of the SH2 domain of Stat3 produces a molecule that dimerizes spontaneously leading to constitutive Stat3 activation (Stat3C) (12). For promoter activity assay, the Fbxo40 promoter clone and plasmid expressing Stat3C or cDNA3 were transfected into C2C12 myoblasts using the Invitrogen Neon transfection system. At 24 h after transfection, the activity of Gaussia luciferase was measured using the Thermo Scientific Pierce Gaussia Luciferase Flash Assay Kit.
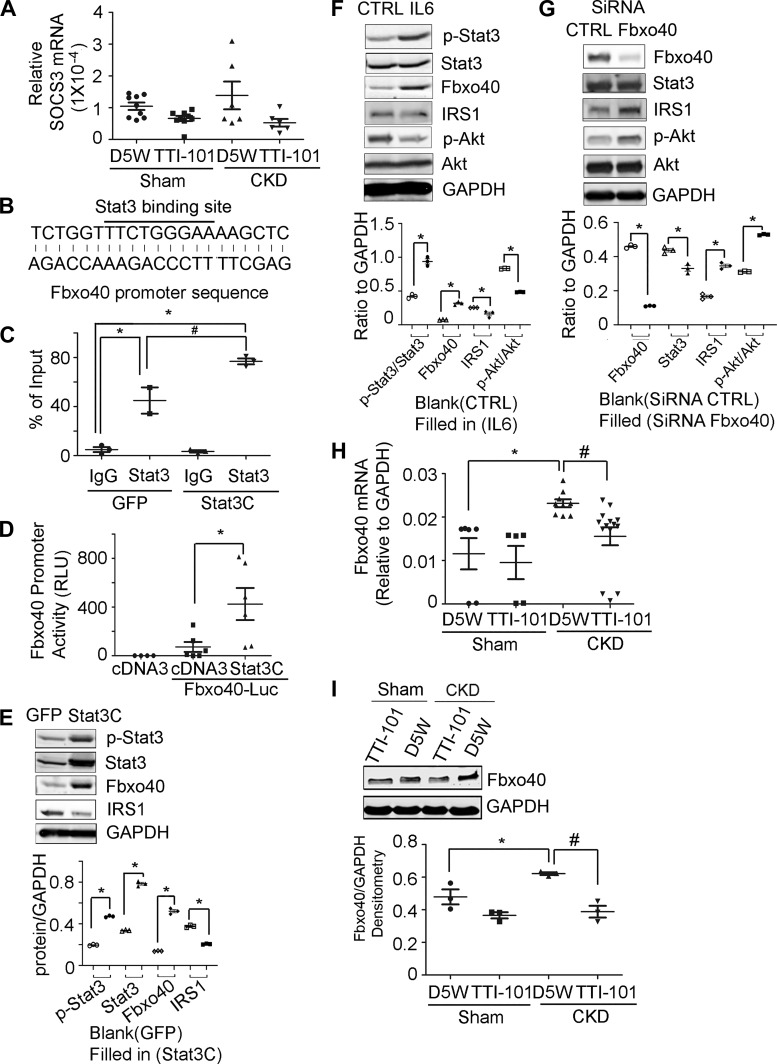
Fig. 3.
Signal transducer and activator of transcription 3 (Stat3) activation induces Fbxo40 transcription. Mice with chronic kidney disease (CKD) and sham-operated, pair-fed control mice were injected with TTI-101 or its diluent (D5W) for 2 wk: A: the relative mRNA values of suppressor of cytokine signaling 3 (SOCS3) in gastrocnemius muscles of mice (n ≥ 6). B: Stat3 binding sequences in promoter of mouse Fbxo40. C: chromatin immunoprecipitation (ChIP) assay indicates the enrichment of Stat3 in promoters of Fbxo40 [*P < 0.05 vs. green fluorescent protein (GFP)-IgG; #P < 0.05 vs. GFP-Stat3; n = 3 repeat]. D: Fbxo40 promoter activity (*P < 0.05 vs. cDNA3-Fbxo40-Luc; n ≥ 4 repeat). E: the representative immunoblots of cell lysates from C2C12 cells transfected with Stat3C. Image quantification are shown at bottom (*P < 0.05 vs. GFP-C2C12 cells; n = 4 repeat). F: the representative immunoblots of cell lysates from C2C12 myotubes treated with IL-6. Image quantification is shown at bottom (*P < 0.05 vs. nontreated C2C12 cells; n = 4 repeat). G: C2C12 cells were transfected with siRNA to Fbxo40 or control and then treated with 100 ng/ml IL-6. Cell lysates were subjected to Western blotting. Image quantification is shown at bottom [*P < 0.05 vs. siRNA-control (CTRL); n = 3 repeats]. H: mRNAs of Fbxo40 in tibialis anterior (TA) muscles of mice (*P < 0.05 vs. Sham-D5W; #P < 0.05 vs. CKD-D5W; n ≥ 5 mice). I: muscle lysates were subjected to Western blotting to evaluate protein levels of Fbxo40 in skeletal muscle. Quantification of images is shown at bottom (*P < 0.05 vs. Sham-D5W; #P < 0.05 vs. CKD-D5W n ≥ 3 mice). RLU, relative Luminescence units; IRS1, insulin receptor substrate 1.
Chromatin immunoprecipitation assays.
C2C12 myoblasts were transfected with plasmids expressing Stat3C or green fluorescent protein (GFP) using the Invitrogen Neon transfection system. C2C12 cells were differentiated for 24 h before being treated with 1% formadehyde (Sigma-Aldrich, St. Louis, MO) for 10 min. Cells were washed three times with ice-cold PBS containing a protease inhibitor (Sigma-Aldrich, St. Louis, MO). Myotubes were then lysed vortexed and sonicated according to Millipore Kit manufacturer’s instructions (43). After centrifugation, the protein-DNA lysate was diluted 10-fold in chromatin immunoprecipitation (ChIP) buffer and precleared using salmon sperm DNA and protein A/G agarose beads for 1 h at 4°C. Each 100 μL of the protein-DNA lysate was used as an input control. Cellular protein-DNA lysates were immunoprecipitated overnight at 4°C with antibodies against Stat3, p-Stat3, or rabbit IgG (Santa Cruz Biotechnology, Dallas, TX). Subsequently, lysates were incubated with protein A/G Agarose beads (SCBT) for 1 h at 4°C. The complexes were washed as described by the manufacturer before they were subjected to reverse cross linking at 65°C for 4 h in the presence of 0.2 M NaCl; the mixture was purified by mixing with phenol/chloroform/isoamyl alcohol. A total of 5 μl of the purified DNA was subjected to PCR amplification using primers that cover the Stat3 binding sites in the mouse Fbxo40 promoter. The primers were purchased from Sabiosciences [Frederick, MD; cat. no. GPM1046740 (+) 09A]. The fold enrichment of Stat3 or p-Stat3 relative to IgG was calculated.
RNA extraction and quantitative real-time PCR.
RNAs were isolated using the RNeasy kit (Qiagen, Valencia, CA) as instructed by the company. We performed RT-PCR to obtain relative gene expressions by calculating cycle threshold (Ct) values using GAPDH as an internal control [relative expression = 2(sample Ct − GAPDH Ct)] (52). Sequences of primers will be provided upon request.
Antibodies.
The primary antibodies of rabbit monoclonal antibody (mAb) to p-Akt (Ser473) (D9E) no. 4060, mouse mAb to Akt (40D4) no. 2920, rabbit mAb to p-Stat3 (Tyr705) (D3A7) no. 9145, and mouse mAb to Stat3 (124H6) no. 9139 were from Cell Signaling Technology (Beverly, MA). A rabbit monoclonal antibody to Fbxo40 [EPR15005] no. ab190688 was from Abcam (Cambridge, MA). An antibody against rat mAb to IRS1 no. 611395 was obtained from BD Biosciences (San Jose, CA), and the anti-GAPDH polyclonal antibody no. PA1-987 was from Thermo Fisher Scientific.
IRS1 ubiquitinization assay.
TA muscles from mice were lysed using immunoprecipitation (IP) lysis buffer (Pierce no. 87787). Lysates (1 mg protein) were incubated with anti-IRS1 antibody for overnight at 4°C before they were incubated with protein A/G plus agarose beads. Beads were washed three times with 1 mL of IP lysis buffer and before then boiled in 30 μL of 2× sample buffer. The blots were incubated with anti-ubiquitin and IRS1.
Statistical analysis.
Student’s t test was used when two experimental groups were compared and ANOVA when data from three or four groups were studied. After ANOVA analyses, pair wise comparisons were made by the Student-Newman-Keuls test. The data are presented as means ± SE.
RESULTS
CKD induces insulin resistance in mice.
Two weeks following subtotal nephrectomy and high protein diets, mice with CKD had average BUN values of ~80 mg/dL (see methods) compared with an average BUN of ~20 mg/dL in pair-fed, control mice (Fig. 1A). At 2 mo of CKD, mice had loss of body weight and higher values of fasting blood glucose (Fig. 1, B and C). Glucose tolerance testing revealed that CKD mice had higher glucose levels after glucose injection versus results present in pair-fed, sham-operated, control mice (Fig. 1D). Consistent with the levels of blood glucose, we uncovered higher insulin levels in serum of CKD mice when fasting (Fig. 1E) or during glucose tolerance testing (GTT; Fig. 1F). Notably, the level of p-Stat3 was increased and p-Akt was decreased in muscles of CKD mice versus responses of control mice (Fig. 1G). However, the level of SOCS3 proteins in muscles of CKD mice was not significantly different from values in control mice (Fig. 1G). Interestingly, we found low protein level and high ubiquitin conjugated IRS1 in muscles of CKD mice (Fig. 1, H and I). Thus CKD causes high p-Stat3 in muscle and glucose intolerance in mice.
Fig. 1.
Insulin resistance is present in mice with chronic kidney disease (CKD). A: 2 wk after nephrectomy mice fed with high-protein diet (CKD mice), the blood urea nitrogen (BUN) level was significantly increased vs. that in sham-operated control mice. B–F: 2 mo of CKD mice vs. that in sham control mice, body weights decreased (B), fasting blood glucose levels increased (C), glucose tolerance was impaired (D), and insulin levels (E) and insulin levels were high following glucose tolerance test (GTT) in connection with D (F). G: representative immunoblots from lysates of gastrocnemius muscles of mice with CKD vs. sham-operated, control mice. Quantification of values is shown at bottom. H and I: low protein level and high ubiquitin (Ub)-conjugated insulin receptor substrate (IRS1) in muscles of CKD mice. *P < 0.05 vs. sham; n > 3 mice in each group. IB, immunoblot; IP immunoprecipitated. Stat3, signal transducer and activator of transcription 3; SOCS3, suppressor of cytokine signaling 3.
CKD induces insulin resistance in mice via Stat3 activation.
Earlier, we found that mice with either CKD or cancer cachexia develop Stat3 activation leading to muscle wasting (43, 53). In those experiments, we demonstrated that inhibition of p-Stat3 using a small molecule inhibitor of Stat3, TTI-101 (formerly C188-9), blocked muscle wasting even in models of CKD or cancer cachexia. In the current experiments, we administered TTI-101 to mice with CKD every other day, and 2 wk later, body weight was increased and blood glucose concentrations decreased (Fig. 2, A and B). These outcomes were confirmed when we examined glucose tolerance in mice (Fig. 2C). To evaluate the effectiveness of TTI-101, we performed Western blotting of muscle lysates and found that treatment with TTI-101 decreased levels of p-Stat3 while increasing p-Akt and IRS1 proteins in muscle (Fig. 2D). TTI-101 treatment also increased levels of glucose transporter Glut4 mRNA and its muscle protein expression in mice with CKD (Fig. 2, E and F). These results indicate there is a CKD-initiated pathway in mice that activates Stat3 in muscles and causes IR. The next step is to determine how Stat3 causes IR.
Fig. 2.
Signal transducer and activator of transcription 3 (Stat3) activation in skeletal muscles of mice with chronic kidney disease (CKD) induces insulin resistance (IR). Mice with CKD and sham-operated, pair-fed control mice were injected with TTI-101 or its diluent (D5W) for 2 wk. A: body weights (BW; *P < 0.05 vs. Sham-D5W; #P < 0.05 vs. CKD-D5W; n = 4 mice). B: blood glucose (*P < 0.05 vs. Sham-D5W; #P < 0.05 vs. CKD-D5W; n ≥ 4 mice). C: glucose tolerance test (*P < 0.05 vs. CKD-D5W; n ≥ 5 mice). D: gastrocnemius muscle lysates were subjected to Western blotting of p-Stat3 and p-Akt. Image quantification is shown at bottom (*P < 0.05 vs. Sham-D5W; #P < 0.05 vs. CKD-D5W; n ≥ 4 mice). E and F: glucose transporter Glut4 protein (E) and mRNA (F) levels in muscles of mice (*P < 0.05 vs. CKD-D5W; n ≥ 4 mice). IRS1, insulin receptor substrate 1.
Stat3 activation induces Fbxo40 expression.
To explore mechanisms that link p-Stat3 to changes in insulin signaling in mice with CKD, we examined SOCS3, a downstream target of Stat3. This interaction was examined because SOCS3 reportedly blocks insulin signaling via a pathway that is linked to ubiquitin conjugation and degradation of IRS1 and IRS2 (40). However, we found that SOCS3 levels in muscles of CKD mice are not directly associated with responses of p-Stat3 (Fig. 1E and Fig. 3A). In searching for the target genes of Stat3, we found one putative Stat3 binding site in the promoter of Fbxo40 (1,062-bp upstream of the ATG start codon) (Fig. 3B). Identification of interactions between Stat3 and Fbxo40 was preceded because Fbxo40 reportedly is involved in the ubiquitination and degradation of IRS1 (42, 51). First, we examined whether Stat3 binds to the Fbxo40 promoter and stimulates its expression. We performed ChIP assays by transfecting C2C12 muscle cells with plasmids expressing constitutively active Stat3 (Stat3C); cells transfected with GFP served as controls. Second, we obtained chromatins from both types of cells and immunoprecipitated them with IgG, anti-Stat3, or anti-p-Stat3 antibodies. DNA isolated from immunocomplexes was subjected to PCR analysis using primers derived from sequences in the Fbxo40 promoter near the Stat3 binding sites. Notably, the relative enrichment of Stat3 over IgG in cells indicates that Stat3 binds to the promoter of the Fbxo40 genes (Fig. 3C). Third, we measured Fbxo40 promoter activities by transfecting C2C12 myoblasts with Fbxo40-promoter-luciferase plasmids plus plasmids expressing Stat3C. Fbxo40-promoter-luciferase plus cDNA3 were used as controls. After 24 h, cells were lysed in passive lysis buffer and Gaussia-luciferase activity was measured (see methods). The presence of Stat3C significantly increased Fbxo40 promoter activity (Fig. 3D). Western blotting also demonstrated the presence of increases in Fbxo40 proteins in cells that had been transfected with Stat3C. Notably, this response was accompanied by decreases in the levels of IRS1 protein (Fig. 3E).
Fourth, we examined IL-6-mediated responses to Stat3 activation. This was undertaken because IL-6 stimulates Stat3 activation in muscle (52, 53). We treated C2C12 myotubes with IL-6 for 24 h and found that IL-6 increased both levels of p-Stat3 and Fbxo40 but decreased IRS1 and p-Akt (Fig. 3F). In addition, knockdown of Fbxo40 in C2C12 cells increased protein levels of IRS1 and p-Akt even in myotubes that had been treated with IL-6 (Fig. 3G). This is relevant because the SCF-Fbxo40 complex reportedly induces IRS1 conjugation to ubiquitin in skeletal muscles; this results in limited IGF1 signaling (42). We conclude that Stat3 activation stimulates Fbxo40 expression and impairs insulin signaling in muscle cells.
Consistent with the increase in p-Stat3 in muscles of mice with CKD, there were increases in the levels of Fbxo40 mRNAs in skeletal muscles of mice with CKD (Fig. 3H). When we inhibited Stat3 activation in CKD mice by treating them with TTI-101, we found that the level of Fbxo40 mRNA was suppressed (Fig. 3H). In consistence with mRNA, Western blotting of muscle lysates from the tibialis anterior (TA) muscles revealed that TTI-101 treatment of CKD mice significantly decreased Fbxo40 protein (Fig. 3I). We conclude that activation of STAT3 induces insulin resistance by a pathway that includes Fbxo40-stimulated IRS1 degradation.
Stat3 inhibition improves HFD-induced IR in mice.
To determine whether the Stat3-activated insulin resistance includes a mechanism that is stimulated by CKD, we studied mice with another type of insulin resistance, namely feeding of a high-fat diet (HFD). After 2 wk of feeding the HFD, muscle levels of p-Stat3 and Fbxo40 were increased but IRS1 protein levels were decreased when compared with mice fed standard chow (Fig. 4A). Notably, mice fed the HFD also had increases in the muscle expression of the Fbxo40 mRNA (Fig. 4B). We extended these results by feeding the HFD to mice for 12 wk (Fig. 4C); these mice developed glucose intolerance (Fig. 4D). Subsequently, these mice were divided into two groups: one group received TTI-101 intraperitoneally; the other group received the diluent intraperitoneally. Both groups of mice continued eating the HFD for another 4 wk. We found that TTI-101 treatment resulted in lower fasting blood glucose levels versus responses in mice treated with the diluent (Fig. 4E). Notably, TTI-101 administration improved both glucose and insulin tolerances in mice fed the HFD (Fig. 4, F and G). Interestingly, Western blotting revealed that the TTI-101 treatment of HFD fed mice led to decreases in p-Stat3 and Fbxo40. Instead, there were higher levels of both IRS1 and p-Akt in muscles versus results from mice treated with the diluent (Fig. 4H). We conclude that inhibition of p-Stat3 in mice that were fed an HFD increases insulin signaling pathway in muscles.
Fig. 4.
Signal transducer and activator of transcription 3 (Stat3) inhibition improves high-fat diet (HFD)-induced insulin resistance (IR) in mice. A and B: C56/BL6 mice were fed with HFD for 2 wk, and the representative Western blotting is shown with image quantification at bottom (A) and the Fbxo40 mRNA (B) [*P < 0.05 vs. regular diet (RD), n ≥ 4 mice]. C and D: 12 wk of HFD feeding induced obesity (C) and glucose intolerance (D) in mice (*P < 0.05 vs. RD; n ≥ 5 mice). E: TTI-101 treatment of HFD mice decreased their fasting glucose level [*P < 0.05 vs. HFD-diluent (D5W); n ≥ 7 mice]. F: TTI-101 treatment of HFD-fed mice improved their glucose tolerance (*P < 0.05 vs. HFD-D5W; n = 10 mice). G: TTI-101 treatment of HFD-fed mice improved their insulin tolerance (*P < 0.05 vs. HFD-D5W; n = 10 mice). H: the representative Western blotting is shown with image quantification at bottom (*P < 0.05 vs. RD; #P < 0.05 vs. HFD-D5W; n = 3 mice for Western). BW, body weight; GTT, glucose tolerance test; ITT, insulin tolerance test; IRS1, insulin receptor substrate 1.
Stat3 KO in muscles suppresses HFD-induced IR in mice.
To examine whether p-Stat3 causes IR genetically, we studied mice with muscle-specific Stat3 KO; they were created by crossing transgenic mice that express floxed-Stat3 with MCK-cre mice (43, 53). In muscles of Stat3 KO mice, we only found a decrease in p-Stat3 but not complete knockout; the low level of Stat3 could be from residual protein within the myofiber. Alternatively (or in addition), it may be from other cell types such as fibroblasts and blood vessels. After 16 wk of feeding these mice plus control mice with HFD, we found significant increases in body weight and adipocytes but decreased muscle weights compared with results of mice fed the regular diet (Fig. 5, A–E). In comparing results obtained from Stat3 KO mice and the control, floxed-Stat3 mice, there were no significant differences in body or muscle weights or in masses of adipose tissues. However, mice with muscle-specific, Stat3 KO, there were substantial decreases in fasting blood glucose (Fig. 5F) even though mice were fed the HFD. There also were improvements in glucose tolerance (Fig. 5G). In muscles of Stat3 KO mice fed the HFD, there were decreases in p-Stat3 and Fbxo40 but increases in p-Akt and IRS1 (Fig. 5H). We conclude that Stat3 activation in muscles of mice impairs insulin functions while inhibition of Stat3 activation blocks insulin actions even when the mice were being fed an HFD.
Fig. 5.
Signal transducer and activator of transcription 3 (Stat3)-specific knockout (KO) in mouse muscles suppressed insulin resistance (IR) responded to feeding a high-fat diet (HFD). A: body weights (BW) during 12 wk of HFD feeding [*P < 0.05 vs. regular diet (RD) Stat3flox/flox (F/F) mice; n ≥ 5 mice]. B and C: muscle weights in mice after 16 wk HFD (*P < 0.05 vs. RD Stat3flox/flox; n ≥ 5 mice). TA, tibialis anterior. D and E: adipose tissues mass after 16 wk of HFD (*P < 0.05 vs. RD Stat3flox/flox; n ≥ 5 mice). F: HFD STAT3 KO mice decreased fasting glucose level in mice (*P < 0.05 vs. RD Stat3flox/flox; #P < 0.05 vs. HFD Stat3flox/flox; n ≥ 7 mice). G: glucose tolerance testing in mice after a 16-wk HFD (*P < 0.05 vs. HFD Stat3flox/flox; n = 6 mice). H: the representative Western blots from muscle of mice are shown with image quantification at bottom (*P < 0.05 vs. RD- Stat3flox/flox; #P < 0.05 vs. HFD; Stat3flox/flox; n ≥ 7 mice).
DISCUSSION
We have uncovered certain complications of CKD, including an increase in the rate of muscle protein degradation plus impairment in protein synthesis producing loss of muscle mass (52, 54). We also examined if the increase in muscle protein degradation was mediated via stimulation of the ubiquitin-proteasome system (UPS) (34). Specifically, we found that the two muscle-specific E3 ubiquitin ligases (Atrogin-1 and MuRF-1) were elevated in muscles of rodents with CKD (30) and that p-Akt levels were suppressed resulting in the development of IR (4). In the present experiments, we confirmed that Stat3 activation in muscles is associated with an increase in the expression of another ubiquitin E3 ligase, Fbxo40, that causes ubiquitin conjugation and degradation of IRS1 (42) leading to the development of IR (Fig. 6). This conclusion was supported pharmacologically and genetically. Notably, we found improved insulin signaling in muscle when we inhibited Stat3 with the small molecule inhibitor TTI-101 in mice with CKD or HFD or when we knocked out Stat3 specifically from muscles of mice in the presence of feeding an HFD.
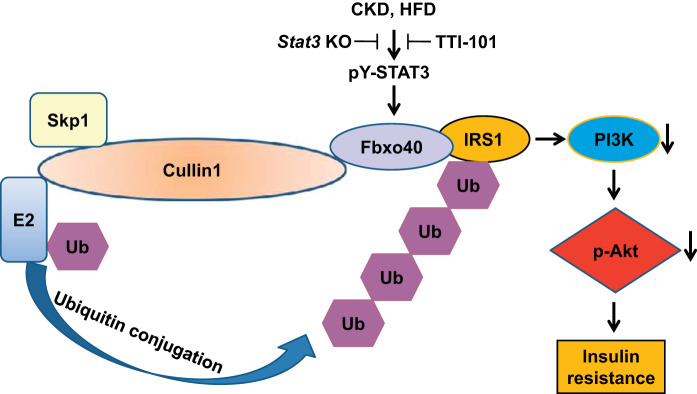
Fig. 6.
Chronic kidney disease (CKD) stimulates signal transducer and activator of transcription 3 (Stat3) activation leading to insulin resistance (IR). Inflammation developing during CKD or prolonged feeding of a high-fat diet (HFD) leads to stimulation of Stat3, and its activation induces the expression of Fbxo40, which leads to ubiquitin conjugation and degradation of insulin receptor substrate 1 (IRS1). The low level of IRS1 impairs phosphatidylinositol 3-kinase (PI3K) and p-Akt resulting in insulin resistance. This response is blocked by administration of a small molecule Stat3 inhibitor or elimination of Stat3 expression in muscle.
In models of CKD or streptozotocin-induced acute diabetes or cancer cachexia, we found high level of p-Stat3 present in skeletal muscle leading to muscle wasting via Stat3/CCAAT/enhancer binding protein δ (CEBP)/myostatin signaling pathway (43, 53). In the present studies, we found that Stat3 activation mediates the development of IR in mice with CKD and in mice fed an HFD. Our results are supported by Mashili et al., who found that Stat3 is constitutively phosphorylated in muscles of patients with type 2 diabetes (32). In addition, the investigators found that Stat3 activation is present in myotubes and involved in lipid-induced IR. Our results differ from those of White et al. (48), who studied mice with muscle-specific Stat3 KO plus feeding an HFD for 20 days and found both Stat3 KO and control mice exhibited similar phenotypes and there were no significant differences in metabolic responses. They concluded that muscle-specific Stat3 KO does not prevent HFD-induced IR (48). One potential explanation for the differences between our two groups is that we studied mice fed an HFD for a more prolonged feeding of the HFD.
The development of IR and of type II diabetes is stimulated by low levels of IRS1 (13). Genetic disruption of IRS1 can impair insulin-stimulated glucose disposal in vivo plus glucose transport in vitro (46). These responses are relevant because IRS proteins activate phosphatidylinositol 3-kinase (PI3K), which recruits Akt to the plasma membrane where it is activated. Activated Akt is required for translocation of glucose transporter GLUT4 to the plasma membrane, which results in glucose uptake and its transfer into muscle or adipocytes. What causes low levels of IRS1? Cell-based mechanisms have been proposed including phosphotyrosine-dephosphorylation, serine-threonine phosphorylation, and, ultimately, IRS1 degradation (1, 36, 38). In fact, investigators have reported that specific ubiquitin E3 ligases can interact with IRS1 leading to its degradation by the UPS. For example, the presence of inflammation reportedly stimulates activity of E3 ubiquitin ligases, SOCS1 and SOCS3, where these E3 ligases interact with IRS1 or IRS2 resulting in their degradation (39, 40). Alternatively, the E3 ubiquitin ligase Cbl-b can activate proteolysis of IRS1 producing muscle atrophy (35). Interestingly, Cbl-b activation can induce the development of IR that occurs following HFD feeding. Finally, the cullin 7 enzyme complex containing the E3 ubiquitin ligase Fbxw8 is activated by a mammalian target of rapamycin (mTOR)-dependent, negative feedback mechanism. The result stimulates the degradation of IRS1, resulting in IR (50). It is tempting to conclude that E3 ubiquitin ligases are selective in terms of advancing degradation of different proteins. For example, Shi et. al reported that Fbxo40 induces ubiquitin conjugation and UPS-mediated degradation of IRS1 in skeletal muscle cells and only in response to IGF1 stimulation (42). Our discovery differs from that of Shi et. al. because we find that inflammation stimulates Fbxo40 expression activating IR.
We did not find increases of SOCS3 in muscles of CKD mice even when p-Stat3 was stimulated. This is not surprising because the role of SOCS3 in the development of muscle atrophy or IR is not settled. SOCS3 expression increases in both muscle hypertrophy (19) and exercise training (44) and decreases in unloading-induced muscle atrophy (17). In addition, the level of SOCS3 is not correlated with the degradation of IRS-1 proteins in adipocytes (20). SOCS3 expression was reported in experimental cancer-induced muscle wasting in mice (10). Interestingly, in this model, the SOCS3 protein level in cachectic muscles was unchanged even when there is persistent Stat3 signaling in muscle. In this case, Stat3 is the primary mediator of muscle wasting (9). These data suggest that persistent Stat3 signaling may play an important pathophysiologic role in the development of muscle wasting in some disease states not through SOCS3 but through other mechanisms, possibly Fbxo40.
We have utilized two models of inhibited Stat3 to evaluate how it affects HFD-induced IR: pharmacological inhibition with TTI-101 and a genetic KO of Stat3 but only in mouse muscles. Even though we found similar results for a Stat3 influence in IR, pharmaceutical inhibition of Stat3 model achieved more response than the genetic model. The potential explanation for this difference is that HFD model affected Stat3 not only in muscle but also other tissues (e.g., adipocytes and liver). Consequently, when we used the inhibitor in mice fed the HFD, we expected to find a lower degree of p-Sstat3 in other organs that could contribute to insulin signaling in muscle (14, 27). In contrast, when we fed HFD to mice with Stat3 KO in muscle, there still is an increase in adipocytes that leads to lower response of muscle Stat3 to regulate IR.
We have uncovered a novel signaling pathway beginning with Stat3 activation that stimulates UPS-mediated IRS1 degradation and hence causes IR. We provide evidence that the mediator of these changes is an upregulation of the ubiquitin E3 ligase Fbxo40. It is tempting to speculate that the pathway we have uncovered is a general mechanism that depends on Stat3 to influence systemic disorders such as CKD, type II diabetes, obesity, and cardiovascular diseases. Thus our results provide a rationale for developing a clinical strategy that targets Stat3 as a means of treating IR arising in response to complex disorders such as inflammation.
GRANTS
This work was supported by National Institutes of Health Grants R01-DK-37175 (to W. E. Mitch) and R01-HL-147108 (and to L. Zhang with P. I. X. Wehrens as Co-Investigator). We also acknowledge the generous support of Dr. H. Selzman. L. Zhang, Investigator for the Norman S. Coplon extramural research grant from Satellite Health, from American Diabetic Association (1-11-BS-194) and from pilot/feasibility award of the Diabetes Research Center (P30-DK-079638).
DISCLOSURES
D.J.T. is a co-founder, President, and Director of Tvardi. The other authors have no conflicts of interest, financial or otherwise.
AUTHOR CONTRIBUTIONS
L.Z., D.J.T., and W.E.M. conceived and designed research; L.Z., Z.C., and Y.W. performed experiments; L.Z. analyzed data; L.Z., D.J.T., and W.E.M. interpreted results of experiments; L.Z. prepared figures; L.Z. drafted manuscript; L.Z. and W.E.M. edited and revised manuscript; L.Z., Z.C., Y.W., D.J.T., and W.E.M. approved final version of manuscript.
REFERENCES
- 1.Aguirre V, Werner ED, Giraud J, Lee YH, Shoelson SE, White MF. Phosphorylation of Ser307 in insulin receptor substrate-1 blocks interactions with the insulin receptor and inhibits insulin action. J Biol Chem 277: 1531–1537, 2002. doi: 10.1074/jbc.M101521200. [DOI] [PubMed] [Google Scholar]
- 2.Arkan MC, Hevener AL, Greten FR, Maeda S, Li ZW, Long JM, Wynshaw-Boris A, Poli G, Olefsky J, Karin M. IKK-beta links inflammation to obesity-induced insulin resistance. Nat Med 11: 191–198, 2005. doi: 10.1038/nm1185. [DOI] [PubMed] [Google Scholar]
- 3.Aufhauser DD Jr, Wang Z, Murken DR, Bhatti TR, Wang Y, Ge G, Redfield RR 3rd, Abt PL, Wang L, Svoronos N, Thomasson A, Reese PP, Hancock WW, Levine MH. Improved renal ischemia tolerance in females influences kidney transplantation outcomes. J Clin Invest 126: 1968–1977, 2016. doi: 10.1172/JCI84712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey JL, Zheng B, Hu Z, Price SR, Mitch WE. Chronic kidney disease causes defects in signaling through the insulin receptor substrate/phosphatidylinositol 3-kinase/Akt pathway: implications for muscle atrophy. J Am Soc Nephrol 17: 1388–1394, 2006. doi: 10.1681/ASN.2004100842. [DOI] [PubMed] [Google Scholar]
- 5.Bégin-Heick N. Beta-adrenergic receptors and G-proteins in the ob/ob mouse. Int J Obes Relat Metab Disord 20, Suppl 3: S32–S35, 1996. [PubMed] [Google Scholar]
- 6.Bharadwaj U, Eckols TK, Xu X, Kasembeli MM, Chen Y, Adachi M, Song Y, Mo Q, Lai SY, Tweardy DJ. Small-molecule inhibition of STAT3 in radioresistant head and neck squamous cell carcinoma. Oncotarget 7: 26307–26330, 2016. doi: 10.18632/oncotarget.8368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294: 1704–1708, 2001. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 9.Bonetto A, Aydogdu T, Jin X, Zhang Z, Zhan R, Puzis L, Koniaris LG, Zimmers TA. JAK/STAT3 pathway inhibition blocks skeletal muscle wasting downstream of IL-6 and in experimental cancer cachexia. Am J Physiol Endocrinol Metab 303: E410–E421, 2012. doi: 10.1152/ajpendo.00039.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonetto A, Aydogdu T, Kunzevitzky N, Guttridge DC, Khuri S, Koniaris LG, Zimmers TA. STAT3 activation in skeletal muscle links muscle wasting and the acute phase response in cancer cachexia. PLoS One 6: e22538, 2011. doi: 10.1371/journal.pone.0022538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bouzakri K, Koistinen HA, Zierath JR. Molecular mechanisms of skeletal muscle insulin resistance in type 2 diabetes. Curr Diabetes Rev 1: 167–174, 2005. doi: 10.2174/1573399054022785. [DOI] [PubMed] [Google Scholar]
- 12.Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE Jr. Stat3 as an oncogene. Cell 98: 295–303, 1999. doi: 10.1016/S0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 13.Carvalho E, Jansson PA, Axelsen M, Eriksson JW, Huang X, Groop L, Rondinone C, Sjöström L, Smith U. Low cellular IRS 1 gene and protein expression predict insulin resistance and NIDDM. FASEB J 13: 2173–2178, 1999. doi: 10.1096/fasebj.13.15.2173. [DOI] [PubMed] [Google Scholar]
- 14.de Castro Barbosa T, de Carvalho JE, Poyares LL, Bordin S, Machado UF, Nunes MT. Potential role of growth hormone in impairment of insulin signaling in skeletal muscle, adipose tissue, and liver of rats chronically treated with arginine. Endocrinology 150: 2080–2086, 2009. doi: 10.1210/en.2008-1487. [DOI] [PubMed] [Google Scholar]
- 15.DeFronzo RA, Alvestrand A, Smith D, Hendler R, Hendler E, Wahren J. Insulin resistance in uremia. J Clin Invest 67: 563–568, 1981. doi: 10.1172/JCI110067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fliser D, Pacini G, Engelleiter R, Kautzky-Willer A, Prager R, Franek E, Ritz E. Insulin resistance and hyperinsulinemia are already present in patients with incipient renal disease. Kidney Int 53: 1343–1347, 1998. doi: 10.1046/j.1523-1755.1998.00898.x. [DOI] [PubMed] [Google Scholar]
- 17.Giger JM, Bodell PW, Zeng M, Baldwin KM, Haddad F. Rapid muscle atrophy response to unloading: pretranslational processes involving MHC and actin. J Appl Physiol (1985) 107: 1204–1212, 2009. doi: 10.1152/japplphysiol.00344.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci USA 98: 14440–14445, 2001. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haddad F, Adams GR. Aging-sensitive cellular and molecular mechanisms associated with skeletal muscle hypertrophy. J Appl Physiol (1985) 100: 1188–1203, 2006. doi: 10.1152/japplphysiol.01227.2005. [DOI] [PubMed] [Google Scholar]
- 20.He F, Stephens JM. Induction of SOCS-3 is insufficient to confer IRS-1 protein degradation in 3T3-L1 adipocytes. Biochem Biophys Res Commun 344: 95–98, 2006. doi: 10.1016/j.bbrc.2006.03.142. [DOI] [PubMed] [Google Scholar]
- 21.Ho MS, Tsai PI, Chien CT. F-box proteins: the key to protein degradation. J Biomed Sci 13: 181–191, 2006. doi: 10.1007/s11373-005-9058-2. [DOI] [PubMed] [Google Scholar]
- 22.Hu H, Wang G, Batteux F, Nicco C. Gender differences in the susceptibility to renal ischemia-reperfusion injury in BALB/c mice. Tohoku J Exp Med 218: 325–329, 2009. doi: 10.1620/tjem.218.325. [DOI] [PubMed] [Google Scholar]
- 23.Ishola DA Jr, van der Giezen DM, Hahnel B, Goldschmeding R, Kriz W, Koomans HA, Joles JA. In mice, proteinuria and renal inflammatory responses to albumin overload are strain-dependent. Nephrol Dial Transplant 21: 591–597, 2006. doi: 10.1093/ndt/gfi303. [DOI] [PubMed] [Google Scholar]
- 24.Jackson PK, Eldridge AG. The SCF ubiquitin ligase: an extended look. Mol Cell 9: 923–925, 2002. doi: 10.1016/S1097-2765(02)00538-5. [DOI] [PubMed] [Google Scholar]
- 25.Karlsson HK, Zierath JR. Insulin signaling and glucose transport in insulin resistant human skeletal muscle. Cell Biochem Biophys 48: 103–113, 2007. doi: 10.1007/s12013-007-0030-9. [DOI] [PubMed] [Google Scholar]
- 26.Kim JH, Yoon MS, Chen J. Signal transducer and activator of transcription 3 (STAT3) mediates amino acid inhibition of insulin signaling through serine 727 phosphorylation. J Biol Chem 284: 35425–35432, 2009. doi: 10.1074/jbc.M109.051516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim TH, Choi SE, Ha ES, Jung JG, Han SJ, Kim HJ, Kim DJ, Kang Y, Lee KW. IL-6 induction of TLR-4 gene expression via STAT3 has an effect on insulin resistance in human skeletal muscle. Acta Diabetol 50: 189–200, 2013. doi: 10.1007/s00592-011-0259-z. [DOI] [PubMed] [Google Scholar]
- 28.Krebs DL, Hilton DJ. A new role for SOCS in insulin action. Suppressor of cytokine signaling. Sci STKE 2003: PE6, 2003. [DOI] [PubMed] [Google Scholar]
- 29.Krebs DL, Hilton DJ. SOCS: physiological suppressors of cytokine signaling. J Cell Sci 113: 2813–2819, 2000. [DOI] [PubMed] [Google Scholar]
- 30.Lee SW, Dai G, Hu Z, Wang X, Du J, Mitch WE. Regulation of muscle protein degradation: coordinated control of apoptotic and ubiquitin-proteasome systems by phosphatidylinositol 3 kinase. J Am Soc Nephrol 15: 1537–1545, 2004. doi: 10.1097/01.ASN.0000127211.86206.E1. [DOI] [PubMed] [Google Scholar]
- 31.Lewis KM, Bharadwaj U, Eckols TK, Kolosov M, Kasembeli MM, Fridley C, Siller R, Tweardy DJ. Small-molecule targeting of signal transducer and activator of transcription (STAT) 3 to treat non-small cell lung cancer. Lung Cancer 90: 182–190, 2015. doi: 10.1016/j.lungcan.2015.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mashili F, Chibalin AV, Krook A, Zierath JR. Constitutive STAT3 phosphorylation contributes to skeletal muscle insulin resistance in type 2 diabetes. Diabetes 62: 457–465, 2013. doi: 10.2337/db12-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.May RC, Kelly RA, Mitch WE. Mechanisms for defects in muscle protein metabolism in rats with chronic uremia. Influence of metabolic acidosis. J Clin Invest 79: 1099–1103, 1987. doi: 10.1172/JCI112924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitch WE, Goldberg AL. Mechanisms of muscle wasting. The role of the ubiquitin-proteasome pathway. N Engl J Med 335: 1897–1905, 1996. doi: 10.1056/NEJM199612193352507. [DOI] [PubMed] [Google Scholar]
- 35.Nakao R, Hirasaka K, Goto J, Ishidoh K, Yamada C, Ohno A, Okumura Y, Nonaka I, Yasutomo K, Baldwin KM, Kominami E, Higashibata A, Nagano K, Tanaka K, Yasui N, Mills EM, Takeda S, Nikawa T. Ubiquitin ligase Cbl-b is a negative regulator for insulin-like growth factor 1 signaling during muscle atrophy caused by unloading. Mol Cell Biol 29: 4798–4811, 2009. doi: 10.1128/MCB.01347-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pederson TM, Kramer DL, Rondinone CM. Serine/threonine phosphorylation of IRS-1 triggers its degradation: possible regulation by tyrosine phosphorylation. Diabetes 50: 24–31, 2001. doi: 10.2337/diabetes.50.1.24. [DOI] [PubMed] [Google Scholar]
- 37.Roberts CK, Hevener AL, Barnard RJ. Metabolic syndrome and insulin resistance: underlying causes and modification by exercise training. Compr Physiol 3: 1–58, 2013. doi: 10.1002/cphy.c110062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rui L, Aguirre V, Kim JK, Shulman GI, Lee A, Corbould A, Dunaif A, White MF. Insulin/IGF-1 and TNF-α stimulate phosphorylation of IRS-1 at inhibitory Ser307 via distinct pathways. J Clin Invest 107: 181–189, 2001. doi: 10.1172/JCI10934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rui L, Fisher TL, Thomas J, White MF. Regulation of insulin/insulin-like growth factor-1 signaling by proteasome-mediated degradation of insulin receptor substrate-2. J Biol Chem 276: 40362–40367, 2001. doi: 10.1074/jbc.M105332200. [DOI] [PubMed] [Google Scholar]
- 40.Rui L, Yuan M, Frantz D, Shoelson S, White MF. SOCS-1 and SOCS-3 block insulin signaling by ubiquitin-mediated degradation of IRS1 and IRS2. J Biol Chem 277: 42394–42398, 2002. doi: 10.1074/jbc.C200444200. [DOI] [PubMed] [Google Scholar]
- 41.Schindler C, Levy DE, Decker T. JAK-STAT signaling: from interferons to cytokines. J Biol Chem 282: 20059–20063, 2007. doi: 10.1074/jbc.R700016200. [DOI] [PubMed] [Google Scholar]
- 42.Shi J, Luo L, Eash J, Ibebunjo C, Glass DJ. The SCF-Fbxo40 complex induces IRS1 ubiquitination in skeletal muscle, limiting IGF1 signaling. Dev Cell 21: 835–847, 2011. doi: 10.1016/j.devcel.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 43.Silva KA, Dong J, Dong Y, Dong Y, Schor N, Tweardy DJ, Zhang L, Mitch WE. Inhibition of Stat3 activation suppresses caspase-3 and the ubiquitin-proteasome system, leading to preservation of muscle mass in cancer cachexia. J Biol Chem 290: 11177–11187, 2015. doi: 10.1074/jbc.M115.641514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spangenburg EE, Brown DA, Johnson MS, Moore RL. Exercise increases SOCS-3 expression in rat skeletal muscle: potential relationship to IL-6 expression. J Physiol 572: 839–848, 2006. doi: 10.1113/jphysiol.2005.104315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spoto B, Pisano A, Zoccali C. Insulin resistance in chronic kidney disease: a systematic review. Am J Physiol Renal Physiol 311: F1087–F1108, 2016. doi: 10.1152/ajprenal.00340.2016. [DOI] [PubMed] [Google Scholar]
- 46.Tamemoto H, Kadowaki T, Tobe K, Yagi T, Sakura H, Hayakawa T, Terauchi Y, Ueki K, Kaburagi Y, Satoh S, Sekihara H, Yoshioka S, Horikoshi H, Furuta Y, Ikawa Y, Kasuga M, Yazaki Y, Aizawa S. Insulin resistance and growth retardation in mice lacking insulin receptor substrate-1. Nature 372: 182–186, 1994. doi: 10.1038/372182a0. [DOI] [PubMed] [Google Scholar]
- 47.Wang CY, Liao JK. A mouse model of diet-induced obesity and insulin resistance. Methods Mol Biol 821: 421–433, 2012. doi: 10.1007/978-1-61779-430-8_27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White AT, LaBarge SA, McCurdy CE, Schenk S. Knockout of STAT3 in skeletal muscle does not prevent high-fat diet-induced insulin resistance. Mol Metab 4: 569–575, 2015. doi: 10.1016/j.molmet.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu H, Ballantyne CM. Skeletal muscle inflammation and insulin resistance in obesity. J Clin Invest 127: 43–54, 2017. doi: 10.1172/JCI88880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu X, Sarikas A, Dias-Santagata DC, Dolios G, Lafontant PJ, Tsai SC, Zhu W, Nakajima H, Nakajima HO, Field LJ, Wang R, Pan ZQ. The CUL7 E3 ubiquitin ligase targets insulin receptor substrate 1 for ubiquitin-dependent degradation. Mol Cell 30: 403–414, 2008. doi: 10.1016/j.molcel.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ye J, Zhang Y, Xu J, Zhang Q, Zhu D. FBXO40, a gene encoding a novel muscle-specific F-box protein, is upregulated in denervation-related muscle atrophy. Gene 404: 53–60, 2007. doi: 10.1016/j.gene.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 52.Zhang L, Du J, Hu Z, Han G, Delafontaine P, Garcia G, Mitch WE. IL-6 and serum amyloid A synergy mediates angiotensin II-induced muscle wasting. J Am Soc Nephrol 20: 604–612, 2009. doi: 10.1681/ASN.2008060628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang L, Pan J, Dong Y, Tweardy DJ, Dong Y, Garibotto G, Mitch WE. Stat3 activation links a C/EBPδ to myostatin pathway to stimulate loss of muscle mass. Cell Metab 18: 368–379, 2013. doi: 10.1016/j.cmet.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang L, Rajan V, Lin E, Hu Z, Han HQ, Zhou X, Song Y, Min H, Wang X, Du J, Mitch WE. Pharmacological inhibition of myostatin suppresses systemic inflammation and muscle atrophy in mice with chronic kidney disease. FASEB J 25: 1653–1663, 2011. doi: 10.1096/fj.10-176917. [DOI] [PMC free article] [PubMed] [Google Scholar]