Abstract
The lungs and the immune and nervous systems functionally interact to respond to respiratory environmental exposures and infections. The lungs are innervated by vagal sensory neurons of the jugular and nodose ganglia, fused together in smaller mammals as the jugular-nodose complex (JNC). Whereas the JNC shares properties with the other sensory ganglia, the trigeminal (TG) and dorsal root ganglia (DRG), these sensory structures express differential sets of genes that reflect their unique functionalities. Here, we used RNA sequencing (RNA-seq) in mice to identify the differential transcriptomes of the three sensory ganglia types. Using a fluorescent retrograde tracer and fluorescence-activated cell sorting, we isolated a defined population of airway-innervating JNC neurons and determined their differential transcriptional map after pulmonary exposure to lipopolysaccharide (LPS), a major mediator of acute lung injury (ALI) and acute respiratory distress syndrome (ARDS) after infection with gram-negative bacteria or inhalation of organic dust. JNC neurons activated an injury response program, leading to increased expression of gene products such as the G protein-coupled receptor Cckbr, inducing functional changes in neuronal sensitivity to peptides, and Gpr151, also rapidly induced upon neuropathic nerve injury in pain models. Unique JNC-specific transcripts, present at only minimal levels in TG, DRG, and other organs, were identified. These included TMC3, encoding for a putative mechanosensor, and urotensin 2B, a hypertensive peptide. These findings highlight the unique properties of the JNC and reveal that ALI/ARDS rapidly induces a nerve injury-related state, changing vagal excitability.
Keywords: jugular-nodose complex, lipopolysaccharide, lung inflammation, RNA sequencing
INTRODUCTION
Lung inflammation is often caused by exposure to infectious agents or environmental pro-inflammatory toxicants. The culprits include gram-negative bacteria, which contain lipopolysaccharide (LPS) in their outer membrane, or environmental organic dust contaminated with LPS. Intranasal endotoxin (LPS) administration in mice has been extensively used to study the role of innate immune responses in the pathogenesis of ALI/ARDS in pneumonia (20, 35). LPS stimulates Toll-like receptor (TLR) 4 on mammalian cells, resulting in an innate immune response (53). Structural motifs on bacterial membranes stimulating TLRs not only elicit inflammation but also itch and pain caused by activation of peripheral sensory neurons (49, 50). The immune and nervous systems are functionally interconnected. This interaction is bidirectional. Activation of some nerve types innervating the airways promotes inflammation and elicits mucus secretion, cough, sneezing, and bronchoconstriction (22, 83), whereas activation of other nerve types dampens inflammatory responses (13).
The lungs are innervated by a complex network of sensory neurons (59, 72). The cell bodies of these neurons are located in the neck in the jugular and nodose ganglia of the vagus nerve, of neural crest and placodal origin, respectively (42, 77, 88). In humans and large mammals, these ganglia are anatomically separated and innervate the lungs as well as the heart, stomach, kidneys, and intestines (94). In mice, the ganglia are fused, forming the jugular-nodose complex (JNC). Each mouse JNC contains ∼2,000 neurons tightly surrounded by a large population of satellite glia cells. Although this vagal structure shares similarities with other sensory ganglia, such as the dorsal root ganglia (DRG) and the trigeminal ganglia (TG), there are unique differences based on the different vagal innervation of organs, chemical and physical modalities sensed, and peripheral and central response mechanisms. Previous studies identified transcription factors essential for developmental specification of the JNC and sets of ion channels and G protein-coupled receptors (GPCRs) involved in chemical and mechanical sensing. Some of these, such as the transient receptor potential (TRP) ion channels TRPA1 and TRPV1, are essential to maintain inflammation in asthma and trigger reflex responses such as cough as direct targets of pro-inflammatory environmental exposures (5, 12, 51, 81). However, the repertoire of ion channels, G protein-coupled receptors, and kinases contributing to sensing, signaling, and reflex control by this essential structure remains understudied. Only a fraction of JNC neurons innervate the airways and lungs, with repertoires of expressed genes and functionalities likely different from JNC neurons innervating other organs (70). Hence, it is necessary to implement methods that selectively identify pulmonary sensory neurons to investigate their specific responses following exposure to LPS.
In this study, we generate mouse bulk transcriptome data sets of the three sensory ganglia types, JNC, DRG, and TG, to identify JNC-specific transcripts. We use a fluorescent retrograde tracer to label and isolate airway-innervating neurons of the JNC (34), followed by cell sorting and RNA sequencing, to identify sets of genes differentially regulated upon lung exposure to LPS. Neuronal functional analysis is used to determine whether the observed transcriptional changes impact neuronal signaling and excitability.
MATERIALS AND METHODS
Animals
C57BL/6 mice from Charles River Laboratories (Wilmington, MA) were used (female, 8–12 wk old). Procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Duke University. Mice were housed in standard environmental conditions (12-h light-dark cycle and 23°C) at facilities accredited by the Association for Assessment and Accreditation of Laboratory Animal Care. Food and water were provided ad libitum. All of the animal protocols were submitted to and approved by the Institutional Animal Care and Use Committees of Duke University. All of the research with mice conforms to the ARRIVE guidelines for animal studies (38).
Fast Blue Retrograde Labeling
Under light inhalation anesthesia (sevoflurane, open-drop exposure method), mice received a 40-µL intranasal instillation of 350 µM Fast Blue (Polyscience, Inc.) and 1% DMSO in saline. Mice were euthanized 3–14 days after Fast Blue administration and the jugular-nodose complex were collected (34).
For visualization, JNCs were fixed in 4% PFA overnight and then immersed in 10%, 20%, and 30% sucrose solutions. Tissue was embedded in optimum cutting temperature compound (OCT), sectioned using a Cryostat at 20-µm sections, and collected on slides. Slides were post-fixed in 10% NBF, washed with PBS, and treated with NeuroTrace 530/615 red fluorescence Nissl stain (cat. no. N21482; ThermoFisher) according to the manufacturers’ recommendations. Sections were imaged using a fluorescence Zeiss microscope.
Lipopolysaccharide Airway Exposure
Mice received three intranasal administrations with either lipopolysaccharide (E. Coli, 0111:B4; Calbiochem) or vehicle (saline). Lightly anesthetized mice (sevoflurane) received 8 µg of LPS in 40 µL of saline or saline alone on days 1, 2, and 3. The mice underwent bronchoalveolar lavage and were euthanized for tissue collection 24 h after the last LPS administration.
Bronchoalveolar Lavage and Leukocyte Analysis
Bronchoalveolar lavage (BAL) was performed by cannulation of the trachea and gentle instillation/aspiration (3 times) of 1.0 mL of PBS with 0.1% BSA and protease inhibitor cocktail tablets (Roche, Indianapolis, IN). Animals were then perfused with PBS before tissue collection. The lavage fluid was centrifuged and the supernatant discarded. The cell pellet was treated with red blood cell lysing buffer (BD Biosciences, San Diego, CA), washed, and resuspended in 200 µL of PBS. Total cell counts were determined with a hemocytometer (Scil Vet ABC, Gurnee, IL), centrifuged onto cytoslides (Shandon Cytospin 3 cytocentrifuge, Thermo Electron Corporation), and stained with Diff-Quick (Dade-Behring Inc., Newark, DE). Differential cell counts were obtained using a microscope to count a minimum of 200 cells/slide using standard morphological and staining criteria.
Dissociation of Primary Neurons
JNC, DRG, and/or TG were removed from the animal and immediately placed into 1 mL of neuronal media (Advanced DMEM/F12, GlutaMax, penicillin-streptomycin, 1 M HEPES, N2 supplement, B27). The tissue was washed once with calcium-free PBS and resuspended in digestion media (55 µg Liberase in 1 mL of neuronal media). Tissue was digested at 37°C for 45–60 min, washed, resuspended in 200 µL of neuronal media, and pipetted up and down with a 200-µL tip until tissue was completely disrupted. Cells were passed through a 70-µm cell strainer and then added to a Percoll Gradient (28% over 12.5%). For FAC sorting, and dissociated cells were kept on ice. For calcium imaging, cells were plated in an eight-well chamber precoated with poly-d-lysine and laminin.
FAC Sorting
Flow cytometry was done on a BD FACS Aria II running Diva 8 software (BD Biosciences). For cell sorting, a large nozzle (100 µm) and low pressure (20 psi) were used. To set up the sorting gates, dissociated non-lung-innervating dorsal root ganglia were used. The positive control was dissociated dorsal root ganglia incubated with 350 µM Fast Blue in vitro. Cells were sorted directly into lysis buffer, and RNA was extracted immediately after sorting.
RNA Isolation and Next-Generation Sequencing
Tissue samples were flash-frozen in liquid nitrogen (whole ganglia) or FAC sorted (Fast Blue traced). Then, RNA was isolated with the RNeasy Micro Kit (Qiagen) according to the manufacturer’s protocol, including DNase-I digestion. RNA quality was tested via gel electrophoresis (2200 TapeStation; Agilent Technologies), and each sample was assigned an RNA Integrity Number (RIN) based on the RNA quality. Only samples with a RIN > 7 were used for sequencing. RNA sequencing (RNA-seq) was performed on an Illumina HiSeq 2500 at the Duke Center for Genomic and Computational Biology, using single-end sequencing of 50-bp reads. Samples were pooled four per lane.
For the whole ganglia LPS sequencing data set, there were 10 total samples corresponding to 10 female mice (8–10 wk old); of the 10 mice, five were vehicle (healthy control) and five were LPS (sick). For the Fast Blue tracing RNA sequencing data set, n = 2 vehicle, and n = 2 LPS. Fast Blue positive samples contained only neurons traced from the airways, and the Fast Blue negative samples contained mostly neurons and a fraction of satellite cells. Each sample represents a pool of five mice to yield the minimum cell density required for fluorescence-activated cell sorting and high-quality RNA extraction. The sequencing results were aligned to the genome using TopHat (84), and transcripts were mapped to the reference genome, (mm9, which was mapped by the Genome Bioinformatics Group of the University of California Santa Cruz) using Cufflinks (85). The gene expression values were calculated for each gene in a sample based on the number of fragments mapped per kilobase of transcript per million reads mapped (FPKM). The number of reads aligned was >82% for all samples.
Differential expression was determined in two ways, first by using Cuffdiff and a second manual method, using Matlab with the following criteria: average FPKM > 1, standard error < 33% of the mean, P < 0.05, and at least a onefold change up or down. All of the resulting genes were further filtered with the following criteria; the higher value (vehicle or LPS) was >2 FPKM, greater than twofold increase for upregulated genes or greater than onefold difference for downregulated genes. Based on these stringent restrictions, only the most strongly regulated genes are presented.
RNA sequencing was loaded into Qlucore Omics Explorer (version 1; Lund Sweden) for Principal Component Analysis (PCA) and heat map generation. PCA was used to visualize the data set in three-dimensional space after our variables were filtered with low overall variance to reduce the impact of noise and the remaining variables were centered and scaled to zero mean and unit variance. The projection score was used to determine the optimal filtering threshold, based on P value.
RNA Isolation and Real-Time quantitative PCR
Tissue samples (n = 4, unless otherwise stated) were surgically removed and snap-frozen by immersion in liquid nitrogen. Total RNA was isolated with the RNeasy Micro Kit (Qiagen) according to the manufacturer’s protocol, including DNase-I digestion. RNA quality was tested via gel eletrophoresis (TapeStation; Aligent Technologies), and each sample was assigned an RNA Integrity Number (RIN) according the RNA quality. Only samples with a RIN > 7 were used for quantitative PCR (qPCR). Concentration was determined using the NanoDrop 1000. For real-time qPCR, cDNA was made using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems) based on the manufacturer’s protocol. Real-time qPCR was performed on a LightCycler 480 (Roche, Indianapolis, IN). The expression of genes of interest was determined using the following TaqMan Gene Expression Assays from Applied Biosystems (Foster City, CA): Actb (Mm00607939_s1), Lcn2 (Mm01324470_m1), Atf3 (Mm00476032_m1), Sprr1a (Mm01962902_s1), Gpr151 (Mm00808987_s1), Cyp26a1 (Mm00514486_m1), Cckbr (Mm00432329_m1), and Npy1r (Mm00650798_g1). The threshold cycle (CT) value of each well was determined using the LightCycler 480 software, and the average of the triplicates was calculated. The relative quantification was calculated as 2−∆CT, using 18S and Act-b as housekeeping genes (52). All the qPCR data conform to the MIQE guidelines (11).
Calcium Imaging
Calcium imaging was performed 2 h after ganglia dissociation and culture. Cells were washed once with calcium free PBS then incubated for 45 min at 37°C with 5 µM Fura-2 AM, cell permeant (Life Technologies, Carlsbad, CA) and 0.1% Pluronic F-127 (Life Technologies) in imaging buffer (120 mM NaCl, 3 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 10 mM HEPES, 10 mM glucose). The loading buffer was removed and cells were washed two more times and placed in the dark for 15 min to reach room temperature. Radiometric calcium imaging was done using an Olympus IX51 microscope with a Polychrome V monochromator (Till Photonics, Gräfelfing, Germany) light source and a PCO Cooke Sensical QE camera running Imaging Workbench 6 software (INDEC BioSystems, Santa Clara, CA). Fura-2 emission images were obtained with of 50 ms at 340-nm followed by 30 ms at 380-nm excitation wavelengths. Intracellular calcium changes were measured from the F340/F380 ratio. Ratiometric images were generated using ImageJ software (https://imagej.nih.gov/ij/).
Supplemental Material
See Supplemental Data for complete details. All Supplemental material is available at https://doi.org/10.6084/m9.figshare.10311557.v2; the data are available in Gene Expression Omnibus (GEO) accession number GSE141400.
Statistics
Data are given as means ± SE. Statistical analyses were carried out using Matlab and Excel. Student’s t-test was used for a single comparison between two groups. Results of P values <0.05 were considered significant.
RESULTS
Identification of Jugular-Nodose Complex-Specific Genes
To determine the genes uniquely expressed in the JNC, we compared the JNC transcriptome with that of the other sensory ganglia, the lumbar DRG and the TG. Bulk transcriptome data sets were generated by RNA-seq of whole dissected mouse ganglia, containing all cell types, in triplicate (n = 3). We used next-generation sequencing (NGS, Illumina HiSeq 2500) to quantify mRNA expression. NGS utilizes parallel processing by creating cDNA read fragments where many reads can be recorded at the same time (79). Each sample yielded >29 million, 50 bp, reads.
Principal component analysis (PCA) revealed that components one and two were sufficient to describe 42% and 23% of variability between the different ganglia (Fig. 1A). We then performed a two-group comparison of each ganglia group with the other two groups and identified the genes that were enriched or depleted. Transcripts were filtered by using the P value P ≤ 0.0001. Applying these criteria, we identified 337 unique genes selectively transcribed at high levels in DRG, 1,754 genes in the TG, and 791 genes in the JNC. Gene expression differences are displayed as heatmaps in Fig. 1B. A full list of these genes with their transcript FPKM numbers are shown in Supplemental Data S1 (Gene List-Ganglia).
Fig. 1.
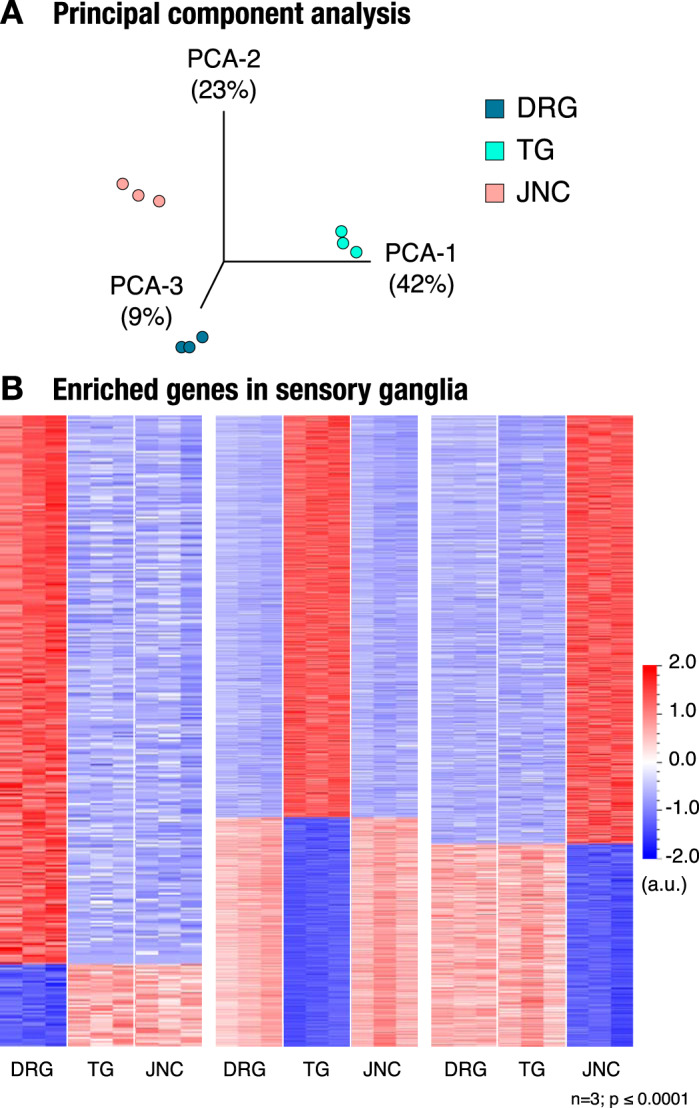
Transcriptional analysis of sensory ganglia. A: principal component analysis (PCA) shows 3 groups associated with different sensory ganglia. B: heat maps show a group of genes that are enriched in dorsal root ganglia (DRG; 337 genes; left), trigeminal ganglia (TG; 1,754 genes; middle), and jugular-nodose complex (JNC; 791 genes; right); n = 3, P ≤ 0.0001.
Genes Selectively Expressed in the Jugular-Nodose Complex
Next, we identified highly expressed genes in the JNC that had little to no expression in the DRG and TG (Table 1). Leading the list were two known JNC marker genes, Phox2b and P2rx2. Phox2b encodes for a unique transcription factor specifying the development of the vagal ganglia (18, 19). Expression of P2rx2, encoding for a purinergic receptor ion channel, allows us to distinguish between nodose and jugular neurons (14), which are fused into the JNC structure in the mouse. Previous studies confirm the lack of expression of these two genes in DRG and TG (86). These findings validate our approach, demonstrating that bulk sequencing of whole ganglia provides sufficient resolution to identify ganglia-specific selective gene expression.
Table 1.
Enriched genes in the JNC
| FPKM |
||||
|---|---|---|---|---|
| Gene | DRG | TG | JNC | Description |
| Phox2b | 0.06 ± 0.02 | 0.13 ± 0.02 | 90.98 ± 2.29 | Paired-like homeobox 2b |
| P2rx2 | 1.24 ± 0.10 | 0.45 ± 0.16 | 90.37 ± 3.04 | P2X purinoceptor 2 |
| Tmem255b | 0.58 ± 0.16 | 0.13 ± 0.04 | 28.78 ± 4.00 | Transmembrane protein 255b |
| Gpr65 | 0.42 ± 0.01 | 1.05 ± 0.13 | 26.62 ± 0.42 | G protein-coupled receptor 65 |
| Uts2b | 0.44 ± 0.16 | 0.12 ± 0.06 | 34.77 ± 5.09 | Urotensin 2b |
| Tmc3 | 0.97 ± 0.13 | 0.61 ± 0.05 | 59.74 ± 7.26 | Transmembrane channel 3 |
| Actb | 565.80 ± 35.50 | 349.87 ± 14.84 | 647.77 ± 31.67 | β-Actin |
Values are means ± SE. These genes are highly expressed in the JNC but have very little to no expression in DRG and TG. Genes were identified as having a high FPKM in the JNC and FPKMs < 2 in both the DRG and the TG. Actb is included as a measure of total RNA. DRG, dorsal root ganglia; FPKM, fragments mapped per kilobase of transcript per million reads mapped; JNC, jugular-nodose complex; TG, trigeminal ganglia.
We then used real-time qPCR for validation of JNC-specific expression of the genes identified by RNA-seq and to compare expression in other neuronal tissues and major organs (Fig. 2). Tmem255b (Fig. 2A) and Gpr65 (data not shown) were found to also be expressed in other tissues. However, Uts2b and Tmc3 (Fig. 2, B and C) were found to be selectively expressed in only JNC and not in any other ganglia or organs, with Tmc3 being particularly highly expressed (2,714.50 ± 485.41 FPKM). The Tmc3 gene belongs to the Tmc (transmembrane channel like) family of genes, with eight genes known in the human and rodent genomes (36). We used qPCR to further examine the expression of all eight Tmc genes in sensory ganglia and other tissues (Supplemental Fig. S1). Tmc3 was the only Tmc gene selectively expressed in the JNC and not in TG and DRG, whereas Tmc6 was highly expressed in all three types of sensory ganglia.
Fig. 2.
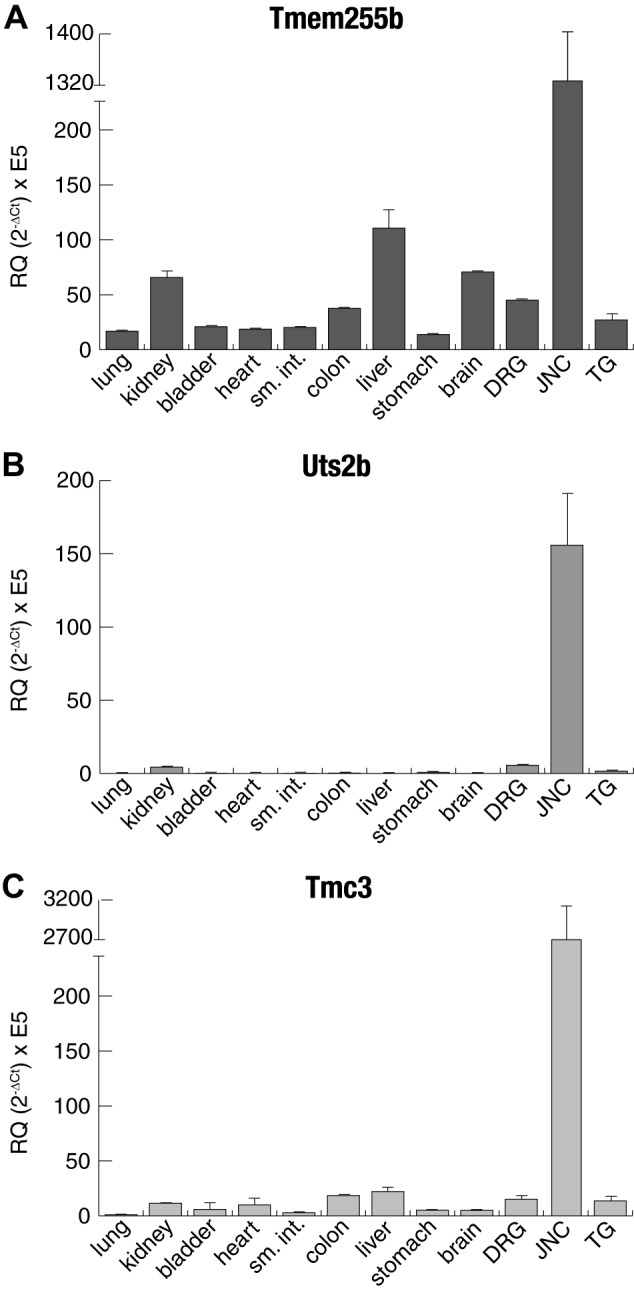
Tissue quantitative PCR (qPCR) expression of jugular-nodose complex (JNC) highly expressed genes. Real-time qPCR was used to determine the expression of Tmem255b (A), Uts2b (B), and Tmc3 (C) in different tissues. Actb was used as the housekeeping gene; n = 4. Values are means ± SE and were calculated as 2−∆CT × E5. DRG, dorsal root ganglia; RQ, relative quantification; sm. int., small intestine; TG, trigeminal ganglia.
Less stringent selectivity criteria identified additional genes with enriched expression in the JNC. These included Chrna3 and Chrnb4, encoding for nicotinic receptor subunits, with DRG and TG showing some baseline expression (Supplemental Fig. S2).
Transcriptomic Responses in Whole JNC and Retrogradely Traced Lung-Innervating JNC Neurons to Lipopolysaccharide-Induced Pulmonary Inflammation
For induction of pulmonary inflammation, mice were given intranasal instillations of 8 µg of LPS in 40 µL of saline, once daily on 3 consecutive days. The vehicle control group was given 40 µL of saline only following the same schedule. On the 4th day, the bronchoalveolar lavage was performed to collect pulmonary cells, followed by immediate dissection of lungs and neuronal tissues. The retrograde tracer Fast Blue was used to label and identify JNC neurons innervating the lungs; 40 µL of 350 µM Fast Blue was administered intranasally, 1 day before the first LPS (or vehicle) administration (Fig. 3A). Fast Blue-labeled neurons were clearly identifiable in JNC cryosections and whole mount preparations (34) (Fig. 3A and Supplemental Video S1). One day after the third LPS administration, JNCs were dissected and sorted by FACS into Fast Blue positive (lung innervating) or negative pools. Sorted cell pools were processed for RNA-seq. Each sample had 29–30 million successful reads and >80% read alignment (Supplemental Table S1). In both Fast Blue-positive and Fast Blue-negative populations, the highly enriched JNC genes (Supplemental Table S2) were similarly expressed, further supporting that the enriched genes we describe are global JNC markers.
Fig. 3.
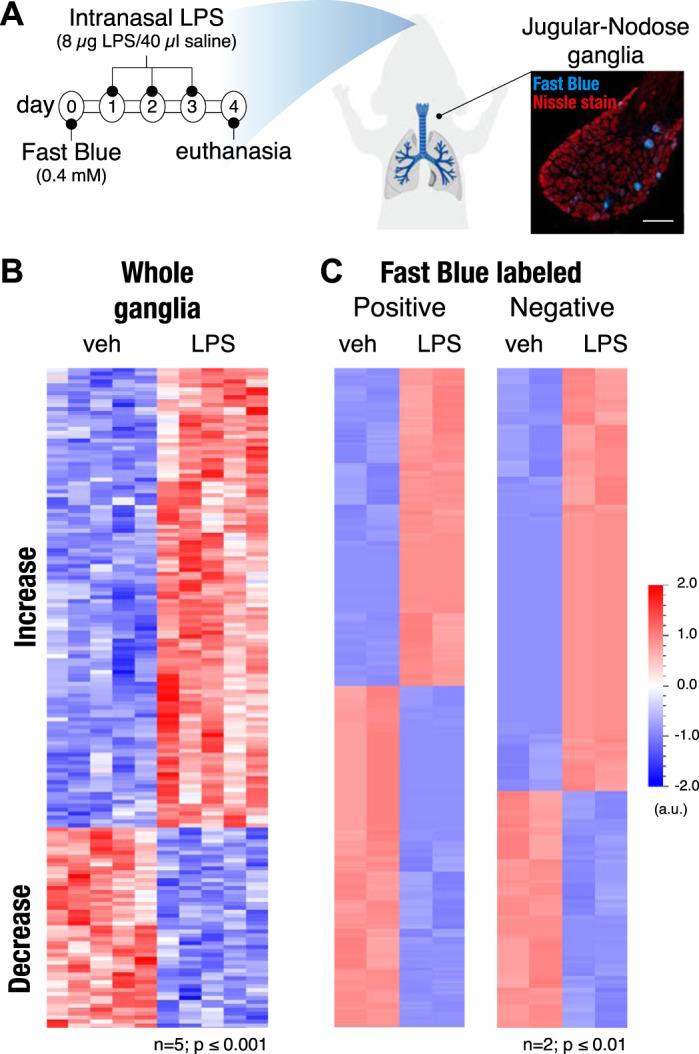
Lipopolysaccharide (LPS) lung exposure changes transcriptional profile of airway-innervating jugular-nodose complex (JNC) neurons. A: timeline for exposure to Fast Blue tracing dye and LPS (8 µg/40 µL saline); vehicle mice were just given saline. B: heat map showing 169 genes that are enriched in whole JNC ganglia with and without LPS-induced inflammation; n = 5, P ≤ 0.001. C: heat map of genes enriched in sorted JNC neurons positive for Fast Blue (255 genes; left) or negative for Fast Blue (164 genes; right); n = 2 (each n consists of 5 mice pooled together; a total of 10 vehicle and 10 LPS mice were used, P ≤ 0.01).
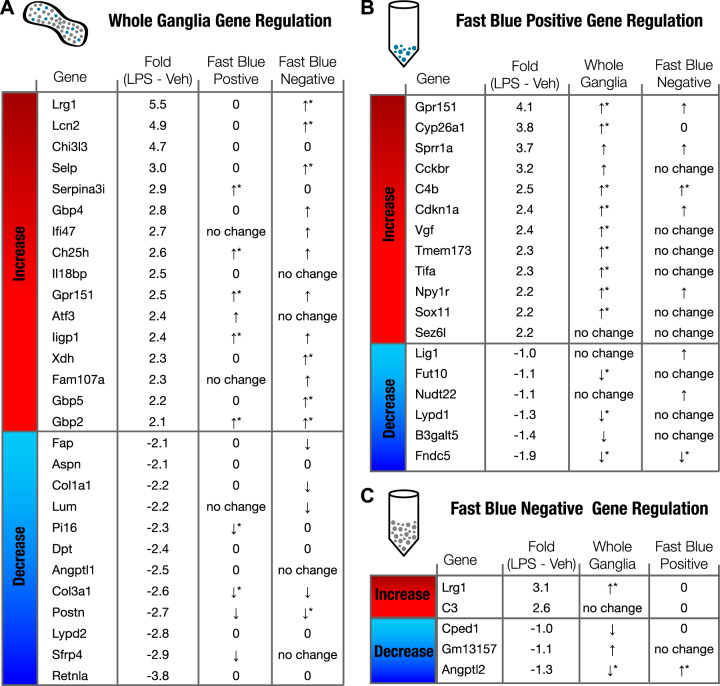
In addition to the Fast Blue-positive and -negative cells, we also created transcriptome data sets for whole, undissociated ganglia from LPS- and vehicle-treated mice. These data sets contain information from all ganglia cell types, both neurons and supporting satellite cells, including immune cell populations located within the JNC. Because of the restriction of Fast Blue to neuronal cells, the Fast Blue-positive population represents the pure lung-innervating population of JNC neurons, whereas the Fast Blue-negative population contains both the non-lung-innervating JNC neurons and satellite cells. Heat maps illustrate the populations of up- and downregulated genes in whole ganglia and the sorted Fast Blue-positive and -negative cell populations (Fig. 3, B and C). The full list of genes with expression altered following LPS administration and their FPKM values can be found in Supplemental Data S2 (Gene List-FB-vehLPS). A full list of genes differentially regulated in lung-innervating JNC neurons was generated by comparing transcriptome data sets from Fast Blue-positive and -negative populations of mice treated with vehicle or LPS (Supplemental Data S3; Gene List-FB-veh). Figure 4 ranks the most strongly regulated genes found in the whole ganglia (Fig. 4A) and in both Fast Blue-positive (Fig. 4B) and Fast Blue-negative populations (Fig. 4C). Genes that are significantly upregulated in the whole ganglia transcriptome data set and also upregulated in the Fast Blue-negative population but have an FPKM value of 0 in the Fast Blue-positive population most likely indicate expression changes in the satellite cells, immune cells, or neurons that innervate other tissue (Fig. 4A). These genes include Lrg1, Lcn2, Selp, Gbp4, Xdh, and Gbp5. Of the upregulated genes in the whole ganglia, Lrg1, Lcn2, Gbp2, Chi3l3, and Ch25h were shown to also be upregulated locally in lung tissue after LPS exposure (9). These genes are likely associated with the innate immune response and may be regulated in immune cells in both the lungs and in JNC.
Fig. 4.
LPS-induced gene regulation. Fold changes are shown as base 2 for LPS – vehicle (Veh) [fold-change = log2 (LPS FPKM) – log2 (Veh FPKM)]. A: whole ganglia gene regulation fold changes compared Fast Blue-positive and -negative sequencing data. B: Fast Blue-positive gene regulation fold changes compared with whole ganglia and Fast Blue negative data. C: Fast Blue negative gene regulation fold changes compared with whole ganglia and Fast Blue positive data. No change indicates a significance value of P > 0.2. An arrow (no star) indicates 0.2 < P < 0.05 and an arrow with a star indicates P < 0.05. A “0” means FPKM < 1.
LPS lung exposure only affected transcription of very few genes in the Fast Blue-negative (non-lung-innervating) population (Fig. 4C). This is an important observation suggesting that JNC nerve populations respond selectively to changes in the pathophysiological states of their target organ, whereas neighboring populations innervating other organs remain unaffected. The two genes upregulated in the FB population, Lrg1 and C3, are both associated with innate immunity (65, 78). The selective regulation of these genes suggests that either immune cells are maturing in the JNC (45) or satellite cells are taking on immune cell characteristics (33, 66). Satellite glia cell activation may help maintain neuronal changes during inflammation. Genes upregulated in the Fast Blue-positive population of lung-innervating JNC neurons (Fig. 4B) include Gpr151, Cckbr, both encoding for G protein-coupled receptors (GPCRs) and Sprr1a, Sox11, Sez6l, Vgf, and the transcription factor Atf3 (25, 47, 73).
Changes in transcript levels of the above genes in the JNC following LPS exposure were validated by real time qPCR and compared with levels in cDNA generated from lumbar DRG of the same animals (Fig. 5A). This analysis confirmed their strong upregulation in JNC, but only minimal expression changes in DRG (Fig. 5A). Two genes, Atf3 and Gpr151, were moderately upregulated in DRG compared with controls, suggesting that these are highly sensitive to beginning systemic changes.
Fig. 5.
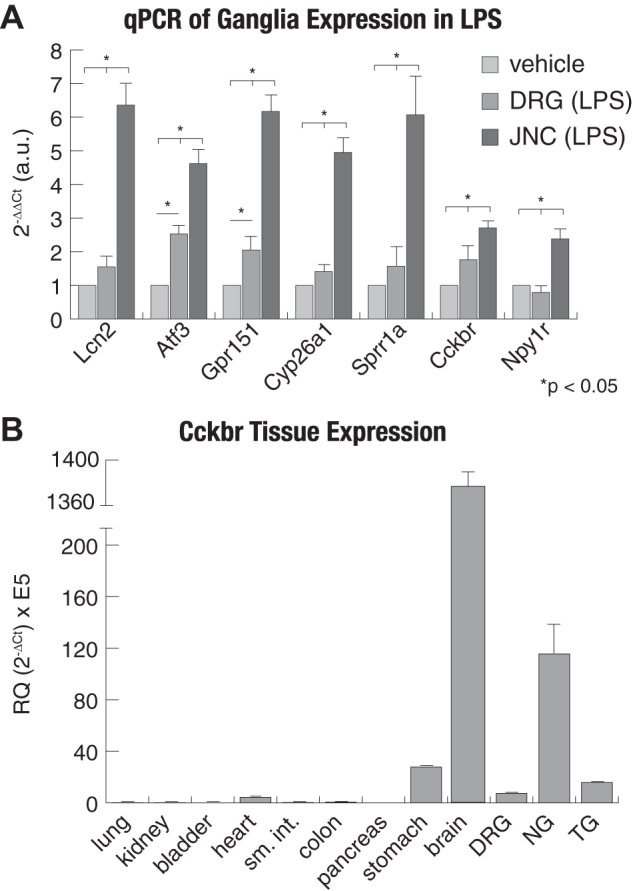
Real-time quantitative PCR (qPCR) confirmation of gene expression. A: real-time qPCR was used to validate RNA-sequencing findings. The ∆∆CT measurement was used to calculate fold change of each gene compared with vehicle. B: tissue specificity of Cckbr expression shows that the jugular-nodose complex (JNC) is enriched for the gastrin/CCK receptor. For each group, n = 4–6, and Actb was used as the housekeeping gene. Values are reported as means ± SE. DRG, dorsal root ganglia; NG, nodose ganglia; TG, trigeminal ganglia.
Correlation of Transcriptome and Functional Changes in JNC Neurons After LPS Treatment
The gene encoding for the cholecystokinin B receptor (CCKBR), Cckbr, was among the most strongly LPS-induced genes in the JNC. Whereas the majority of the identified LPS-induced gene products are orphan receptors or not targetable, highly selective agonists of CCKBR are available to examine whether the observed changes in JNC transcriptome levels resulted in neuronal functional changes. Real-time qPCR analysis was performed to compare Cckbr transcript levels in JNC with those in other sensory ganglia and other tissues (Fig. 5B). Cckbr transcript levels are highest in brain, followed by JNC. As shown previously, the stomach shows moderate expression (44, 63, 64). Cckbr transcript levels were higher in JNC compared with DRG and TG (Fig. 5B).
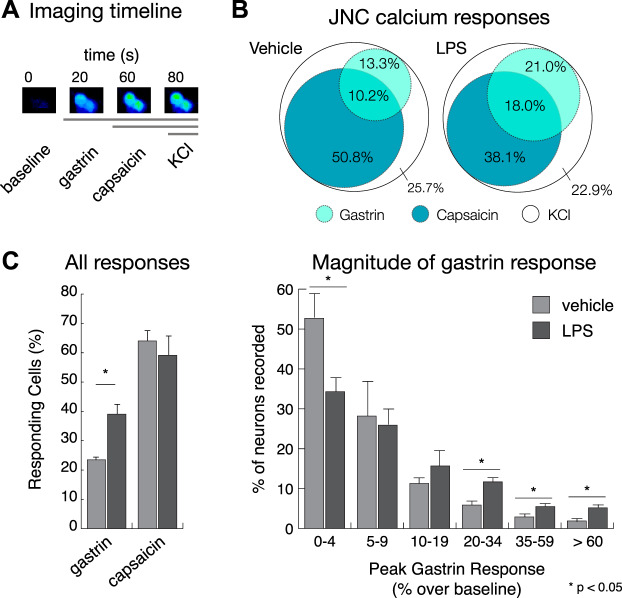
To examine whether LPS exposure-induced Cckbr transcription resulted in functional changes, JNC neurons were dissociated 1 day after the last of three pulmonary LPS treatments. Neurons were analyzed by fluorescent calcium imaging 2 h after plating. Gastrin (1 nM), a CCKBR-specific agonist, was used for stimulation following recording of baselines (30). Subsequently, JNC neurons were superfused with capsaicin (100 nM) to excite TRPV1-expressing neurons (C-fibers), followed by KCl (30 mM) as a strong depolarizing stimulus identifying all neurons (Fig. 6A).
Fig. 6.
Cholecystokinin B receptor (CCKBR) neuronal activity increases after LPS-induced lung inflammation. A: imaging timeline for stimuli (gastrin: 1 nM; capsaicin: 100 nM; KCl: 30 mM) application. B: a Venn diagram showing the overlap between the gastrin and capsaicin responding cells. C, left: %cells that responded to gastrin in the vehicle (healthy) vs. LPS (sick) mice significantly increased. C, right: magnitude of the gastrin response over baseline in vehicle (healthy) vs. LPS (sick) neurons also significantly increased. The capsaicin response remained constant after treatment. The total no. of neurons recorded was 1,542 vehicle and 1,270 LPS neurons. A positive calcium response was defined as a >10% increase, over baseline, of the ratiometric signal (F340/380); n = 5 mice for each group. Data are reported as means ± SE.
JNC neurons dissociated from LPS-treated animals contained larger numbers of neurons responding to gastrin with calcium influx than neurons from vehicle-treated animals. The percentage of gastrin-responding cells significantly increased from 23.5% ± 1.2 in the vehicle group to 39.0% ± 4.8 in the LPS-treated group (Fig. 6, B and C). The number of capsaicin-sensitive neurons was not significantly different between groups. When we sorted the gastrin-responsive neurons recorded for Fig. 6C according to their peak calcium amplitudes, we saw a significant increase in the larger calcium responses (>20% increase over baseline) and a decrease in the number of neurons that did not respond (<4% increase over baseline) (Fig. 6D). This implies that more neurons express functional CCKB receptor and are responding more strongly after LPS-induced lung inflammation.
DISCUSSION
Sensory ganglia innervate different regions of the body and different organs. As such, their molecular profiles are expected to differ based on target organs and functional needs. Our comparison of sensory ganglia transcriptome data sets revealed specialized molecular markers expressed in the murine vagal sensory neurons of the JNC. We purified the airway-innervating JNC nerve cell population using a retrograde tracer and identified a set of neuronal genes differentially regulated in response to pulmonary LPS exposure.
Comparison of Whole Sensory Ganglia Transcriptomes
Comparison of the whole transcriptomes of the JNC, DRG, and TG identified several transcripts uniquely expressed in JNC and absent in DRG and TG. Some of these were exclusively found in the JNC, with little expression in other nervous tissues or other organs:
Tmc3.
Tmc3 is a gene of the Tmc (transmembrane channel like) family of eight mammalian genes (43). Although the function of TMC3 in JNC neurons is not known, TMC1 and TMC2 were identified as potential components of the auditory hair cell mechanotransduction complex. Mutations in Tmc1 and Tmc2 were linked to deafness in multiple human populations (26, 36). These findings suggest that some Tmc proteins may contribute to mechanical sensing in other sensory systems. In C. elegans, tmc-1 was shown to be a sodium-sensitive ion channel, implicating a role in homeostatic ion sensing or, potentially, osmotic sensing (17). A longitudinal genome-wide study linked a polymorphism near the IL6/STARD5/TMC3 locus on chromosome 15 to a higher rate of lung function decline in humans (80). However, it is not clear which of the three genes in this locus are differentially regulated due to the polymorphisms. The highly selective expression of Tmc3 in JNC, and not in other sensory ganglia or other organs, warrants further studies examining its role in JNC development, target organ innervation, and sensory mechanisms.
Uts2b.
Similar to Tmc3, the gene Uts2b proved to be selectively expressed in the JNC, with minimal expression in TG, DRG, and kidney, but undetectable in other major organs. Uts2b encodes for the peptide urotensin II, a potent vasoconstrictor involved in blood pressure regulation (55). Subcutaneous injection of urotensin II in monkeys caused cardiac dysfunction (3). Receptors for urotensin II were targeted for drug development with the aim to generate antihypertensive treatments. Some neuronal expression of Uts2b was reported in in motor neurons of the spinal cord (8). The kidney was discussed as the major systemic source of urotensin II (16, 75). Our data suggest that, in fact, urotensin II is a vagal sensory neuropeptide, potentially released by JNC neurons peripherally upon vagal nerve stimulation.
Tmem255b.
Expression of Tmem255b was confirmed to be high in the JNC; however, it also has basal low-level expression in all other tissues we tested. Not much is known about TMEM255B. In humans, Tmem255b expression is increased in colon cancer (23), and a mutation was found in an obese child (87). These findings do not establish a functional role or pattern for this gene encoding for a hypothetical membrane protein. Its function in vagal development and sensing has yet to be determined.
Gpr65.
The G protein-coupled receptor Gpr65 was originally identified as a receptor for certain lipid species and as a proton receptor activated by acidity (27, 29, 89). Although expression of Gpr65 in JNC neurons was reported previously (14), the authors did not observe any respiratory effects of Gpr65 gene ablation in mice. This is consistent with our data that did not detect significant Gpr65 expression in the airway-innervating population of JNC neurons. Instead, this gene likely to contributes to sensing and control of other visceral organs. Indeed, a polymorphism associated with the Gpr65 gene was shown to determine the severity of inflammatory bowel disease (IBD) (82).
Chrna3 and Chrnb4.
These genes encode for the α3- and β4-subunits of nicotinic acetylcholine receptors (nAChRs). α3β4-containing nAChRs are associated with nicotine’s effects in the brain, including reward mechanisms, addiction, and withdrawal. Nicotine is known to directly excite vagal sensory neurons and is a major mediator of tobacco smoke-induced respiratory irritation and cough (37, 46). Immunoprecipitation studies identified α3- and β4-subunits as the most prevalent nAChRs subunits in nodose sensory neurons (57). Although not exclusive to the JNC, α3- and β4-nAChR subunit transcripts were expressed at much higher levels in the JNC compared with the other sensory ganglia (TG, DRG; Supplemental Fig. S2). The role of these receptors in nicotine’s adverse respiratory effects, especially in the context of e-cigarette use, needs to be explored further (2).
Comparison With Recent Single-Cell Transcriptome Studies
A recent single-cell transcriptome analysis of vagal sensory nerves categorized neuronal subclasses according to certain unique marker genes (42). Whereas that study also inferred lung innervating neuronal populations, no validations were performed through retrograde tracing or other means. Our study, identifying transcripts specifically enriched in lung-innervating neurons retrogradely traced with Fast Blue, enables a more accurate assignment of cell categories. Certain markers used for categorization in the single-cell study were strongly enriched in the retrogradely traced lung-innervating neurons. These included populations labeled by Tmem233, Slc17a7, Piezo2, and Tac1 (encoding for the neuropeptide, Substance P) and Aqp1 that innervate the lung (Supplemental Fig. S3).
The single-cell study also identified a neuronal population positive for urotensin II (Utsb2b) and hypothesizes that this population contributes to digestive tract innervation (42). Uts2b transcript levels were comparably low in airway-innervating JNC neurons in our study, suggesting a minor role in control of respiratory function (Supplemental Fig. S3). A urotensin II antagonist has been tested as an oral treatment in human asthmatic patients but failed to produce positive bronchodilation effects, suggesting that urotensin II plays only a limited role in pulmonary inflammatory conditions (68).
A Neuropathic Injury Transcriptome Response in Lung-Innervating JNC Neurons Triggered By Pulmonary LPS Exposure
Use of the retrograde fluorescent tracer, Fast Blue, enabled us to identify a population of murine JNC sensory neurons that innervates the airways and lungs. This approach may not have labeled all lung-innervating sensory nerves, since some JNC neurons may not extend to airway surfaces within the lung, where Fast Blue uptake occurs after aspiration. Nevertheless, a substantial number of neurons was labeled, enabling us to gather transcriptome data from pooled Fast Blue-positive neurons and to identify LPS-induced genes. The transcriptome data set is most probably from a heterogeneous lung-innervating neuronal population and needs to be further analyzed using single-cell approaches that so far have not used organ-specific retrograde tracing methods (10). Our approach was sensitive enough to identify the major LPS-induced neuronal genes and to detect essential differences between data sets produced from other whole sensory ganglia (TG, DRG). The following genes were induced after LPS exposure, with information provided about their expression site (lung-innervating nerves, nerves innervating other organs, satellite cells) and potential functions.
Gpr151.
This gene is the most strongly LPS-induced gene in lung-innervating JNC neurons, encoding for a G protein-coupled receptor that was first described as a galanin receptor like receptor (GALRL) in 2004 (28). However, this receptor is sensitive to galanin only concentrations by far exceeding physiological levels. GPR151 may have other endogenous agonists that remain to be identified. Intriguingly, Gpr151 is strongly induced in DRG after neuropathic injury in pain models, described in several recent studies (25, 32, 58, 69). Similar to GPR65, GPR151 was shown to be activated by acidity; however, its role in pain signaling may be stimulus dependent (25, 32, 58, 69). The induction of this gene in JNC after LPS exposure suggests that JNC neurons experience neuropathic injury and activate a response program with partial similarities to injury-induced gene sets in DRG neurons.
Lcn2.
The gene Lcn2 encodes the protein lipocalin-2, which is associated with innate immune responses to bacteria activated downstream of TLR4 activation by LPS. Lipocalin-2 is found in neutrophils as well as endothelial cells, astrocytes, and microglia (33). In the brain and spinal cord, inflammation activates microglia. Once activated, they secrete LCN2, which is toxic to neurons (6, 31). Upregulation of the Lcn2 gene following LPS exposure was observed in the bulk whole ganglia data sets but not in the Fast Blue-positive (lung-innervating) neurons, which implies that this gene is induced by LPS in the satellite cells or neurons that innervate other tissue.
Atf3/Sprr1a.
Activated transcription factor 3 (ATF3) plays multiple roles in gene expression regulation in different cell types. In Atf3−/− mice, neutrophils are unresponsive to chemotaxic gradients and unable to find their target (86). In DRG sensory neurons, however, Atf3 expression is associated with nerve injury and promotes nerve growth and regeneration by turning on Sprr1a expression (9, 36). SPRR1A associates with F-actin to promote nerve growth (7). This activated pathway in neurons may promote nerve growth and repair of JNC neurons after LPS exposure.
Cyp26a1.
CYP26A1 is a cytochrome enzyme that breaks down retinoic acid, preventing excessive retinoic acid signaling. Retinoic acid plays an important role in cell proliferation neuronal and limb development (21). The Cyp26a1−/− mouse is embryonic lethal (1). Activated microglia has also been shown to produce CYP26A1 (24). The role of Cyp26a1 expression in JNC neurons after LPS exposure is unclear but may contribute to nerve regeneration or growth and sprouting following nerve injury.
Npy1r.
NPY-R1 is a receptor for neuropeptide Y (NPY), a neuropeptide upregulated in the lungs during inflammation (56). Its receptor can be found on immune cells and neurons. In DRG neurons, NPY-R1 is found mostly in pain-transducing neurons, and its activation leads to an inhibitory current (54, 93). This presents a complex scenario in which some neurons may become sensitized by inflammation and other inhibititions due to increased sensitivity to NPY.
Cckbr.
Our unbiased approach revealed that the expression of Cckbr, encoding for the cholecystokinin B receptor, is upregulated in the JNC neurons that innervate the airways following LPS exposure. CCKBR, a G protein-coupled receptor, is the only known target of gastrin, a peptide produced in the pancreas and other digestive organs, first discovered in 1993 (30). CCKBR is also activated by cholecystokinin (CCK), a peptide implicated in pain signaling in neuropathic pain within the spinal cord and, potentially, in the periphery.
The analysis of nocifensive behavioral responses of Cckbr−/− mice in the chronic constriction model of nerve injury observed a lack of allodynia, demonstrating that this gene fulfills a role in pain perception (40). Interestingly, the authors reported increased expression of TLR4 in the brain of Cckbr−/− mice compared with wild-type (40). This suggests that Cckbr may regulate neuronal sensitivity to innate immune stimuli. Cckbr expression was also upregulated in DRG neurons in a burn injury model, and inhibition of CCKBR was shown to have an analgesic effect (91).
Whereas the expression of gastrin is restricted to digestive tissues, CCK peptides are expressed within the lung by pulmonary neuroendocrine cells (PNECs) and other cell types (4, 67, 90). A CCK peptide was shown to induce bronchoconstriction in humans and guinea pigs (76). Exogenously administered CCK peptides were shown to alter cytokine expression and counteracted inflammatory responses in an LPS-induced mode rat model of endotoxic shock (48, 62). CCK signaling has also been implicated in lung injury due to pancreatitis, and CCKBR is also involved in proliferative responses such as cell growth in the stomach lining and in aberrant proliferative responses such as pancreatic cancer cells (15), colon cancer (71), and small-cell lung cancer (74). In the digestive tract, the CCK/gastrin/CCKBR system responds to bacterial infection by Helicobacter pylori (H. pylori), gram-negative bacteria, initiating an immune response and increasing gastrin secretion (39).
To infer the function of the CCKBR receptor in neurons, it is helpful to compare with the function of the other genes induced by LPS in the in Fast Blue-positive neurons. Some of these genes, including Atf3 and Sprr1a, are also induced in DRG neurons following neuropathic injury in pain models and may either initiate a nerve repair program or stimulate pathological nerve branching and sprouting, potentially leading to increased sensitivity and chronic pain perception (10, 69). CCKBR may contribute to these mechanisms, as it contributes to proliferative and growth mechanisms in other cell types.
Although LPS is a common stimulator of innate immune pathways leading to acute and chronic inflammation in the lung and other organs, our study revealed that LPS induces a nerve injury transcriptomic response in lung-innervating sensory nerves and not a typical inflammatory response as seen in inflammatory pain states in DRG or TG. Whether similar responses occur in human vagal sensory nerves following LPS exposure is unknown. Whereas in mice and rats the neural crest-derived jugular neurons and the placode-derived nodose neurons are contained within a single anatomical structure, the JNC, in humans and other larger mammals these neurons reside in separate ganglia (60). A recent study revealed that key distinctions in gene expression between rodent nodose and jugular neurons are conserved in the separate nodose and jugular neurons in primates, suggesting that certain anatomic and functional properties are translatable (41).
It remains to be established whether the LPS-induced changes in gene expression in the lung-innervating JNC neurons counteract inflammatory responses or contribute to transient or permanent neuropathic states promoting vagal reflex responses such as cough, glandular secretions, and cardiovascular reflexes (60, 61, 92). Some of the identified receptors and membrane proteins may serve as targets to interfere with exaggerated vagal responses during LPS-induced pulmonary inflammation.
GRANTS
This work was supported by National Heart, Lung, and Blood Institute (NHLBI) Grants R01 HL105635 and National Institute of Environmental Health Sciences (NIEHS) Grant R01 ES029435 of the National Institutes of Health (NIH) to S.E.J. Flow cytometry was performed at the Duke Human Vaccine Institute Research Flow Cytometry Shared Resource Facility (Durham, NC), which received partial support for construction from the National Institutes of Health, National Institute of Allergy and Infectious Diseases (NIAID), Grant UC6-AI058607.
DISCLAIMERS
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
S.-E.J. conceived and designed research; M.M.K. and A.I.C. performed experiments; M.M.K. and A.I.C. analyzed data; M.M.K., A.I.C., and S.-E.J. interpreted results of experiments; M.M.K. and A.I.C. prepared figures; M.M.K. and A.I.C. drafted manuscript; M.M.K., A.I.C., and S.-E.J. edited and revised manuscript; M.M.K., A.I.C., and S.-E.J. approved final version of manuscript.
REFERENCES
- 1.Abu-Abed S, Dollé P, Metzger D, Beckett B, Chambon P, Petkovich M. The retinoic acid-metabolizing enzyme, CYP26A1, is essential for normal hindbrain patterning, vertebral identity, and development of posterior structures. Genes Dev 15: 226–240, 2001. doi: 10.1101/gad.855001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmad S, Zafar I, Mariappan N, Husain M, Wei CC, Vetal N, Eltoum IA, Ahmad A. Acute pulmonary effects of aerosolized nicotine. Am J Physiol Lung Cell Mol Physiol 316: L94–L104, 2019. doi: 10.1152/ajplung.00564.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ames RS, Sarau HM, Chambers JK, Willette RN, Aiyar NV, Romanic AM, Louden CS, Foley JJ, Sauermelch CF, Coatney RW, Ao Z, Disa J, Holmes SD, Stadel JM, Martin JD, Liu WS, Glover GI, Wilson S, McNulty DE, Ellis CE, Elshourbagy NA, Shabon U, Trill JJ, Hay DW, Ohlstein EH, Bergsma DJ, Douglas SA. Human urotensin-II is a potent vasoconstrictor and agonist for the orphan receptor GPR14. Nature 401: 282–286, 1999. [Erratum in: Nature 402: 898, 1999.] doi: 10.1038/45809. [DOI] [PubMed] [Google Scholar]
- 4.Balaguer L, Romano J, Ruiz-Pesini P. Localization of serotonin, cholecystokinin and somatostatin immunoreactivity in the lower respiratory tract of embryonic, foetal and postnatal sheep. Histol Histopathol 7: 703–708, 1992. [PubMed] [Google Scholar]
- 5.Bautista DM, Jordt SE, Nikai T, Tsuruda PR, Read AJ, Poblete J, Yamoah EN, Basbaum AI, Julius D. TRPA1 mediates the inflammatory actions of environmental irritants and proalgesic agents. Cell 124: 1269–1282, 2006. doi: 10.1016/j.cell.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 6.Bi F, Huang C, Tong J, Qiu G, Huang B, Wu Q, Li F, Xu Z, Bowser R, Xia XG, Zhou H. Reactive astrocytes secrete lcn2 to promote neuron death. Proc Natl Acad Sci USA 110: 4069–4074, 2013. doi: 10.1073/pnas.1218497110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonilla IE, Tanabe K, Strittmatter SM. Small proline-rich repeat protein 1A is expressed by axotomized neurons and promotes axonal outgrowth. J Neurosci 22: 1303–1315, 2002. doi: 10.1523/JNEUROSCI.22-04-01303.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bougerol M, Auradé F, Lambert FM, Le Ray D, Combes D, Thoby-Brisson M, Relaix F, Pollet N, Tostivint H. Generation of BAC transgenic tadpoles enabling live imaging of motoneurons by using the urotensin II-related peptide (ust2b) gene as a driver. PLoS One 10: e0117370, 2015. doi: 10.1371/journal.pone.0117370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brass DM, Yang IV, Kennedy MP, Whitehead GS, Rutledge H, Burch LH, Schwartz DA. Fibroproliferation in LPS-induced airway remodeling and bleomycin-induced fibrosis share common patterns of gene expression. Immunogenetics 60: 353–369, 2008. doi: 10.1007/s00251-008-0293-3. [DOI] [PubMed] [Google Scholar]
- 10.Braun A, Appel E, Baruch R, Herz U, Botchkarev V, Paus R, Brodie C, Renz H. Role of nerve growth factor in a mouse model of allergic airway inflammation and asthma. Eur J Immunol 28: 3240–3251, 1998. doi:. [DOI] [PubMed] [Google Scholar]
- 11.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55: 611–622, 2009. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 12.Caceres AI, Brackmann M, Elia MD, Bessac BF, del Camino D, D’Amours M, Witek JS, Fanger CM, Chong JA, Hayward NJ, Homer RJ, Cohn L, Huang X, Moran MM, Jordt SE. A sensory neuronal ion channel essential for airway inflammation and hyperreactivity in asthma. Proc Natl Acad Sci USA 106: 9099–9104, 2009. doi: 10.1073/pnas.0900591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caceres AI, Liu B, Jabba SV, Achanta S, Morris JB, Jordt SE. Transient receptor potential cation channel subfamily M member 8 channels mediate the anti-inflammatory effects of eucalyptol. Br J Pharmacol 174: 867–879, 2017. doi: 10.1111/bph.13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang RB, Strochlic DE, Williams EK, Umans BD, Liberles SD. Vagal sensory neuron subtypes that differentially control breathing. Cell 161: 622–633, 2015. doi: 10.1016/j.cell.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chao C, Hellmich MR. Gastrin, inflammation, and carcinogenesis. Curr Opin Endocrinol Diabetes Obes 17: 33–39, 2010. doi: 10.1097/MED.0b013e328333faf8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charles CJ, Rademaker MT, Richards AM, Yandle TG. Urotensin II: evidence for cardiac, hepatic and renal production. Peptides 26: 2211–2214, 2005. doi: 10.1016/j.peptides.2005.03.043. [DOI] [PubMed] [Google Scholar]
- 17.Chatzigeorgiou M, Bang S, Hwang SW, Schafer WR. tmc-1 encodes a sodium-sensitive channel required for salt chemosensation in C. elegans. Nature 494: 95–99, 2013. doi: 10.1038/nature11845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D’Autréaux F, Coppola E, Hirsch MR, Birchmeier C, Brunet JF. Homeoprotein Phox2b commands a somatic-to-visceral switch in cranial sensory pathways. Proc Natl Acad Sci USA 108: 20018–20023, 2011. doi: 10.1073/pnas.1110416108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dauger S, Pattyn A, Lofaso F, Gaultier C, Goridis C, Gallego J, Brunet JF. Phox2b controls the development of peripheral chemoreceptors and afferent visceral pathways. Development 130: 6635–6642, 2003. doi: 10.1242/dev.00866. [DOI] [PubMed] [Google Scholar]
- 20.de Souza Xavier Costa N, Ribeiro Júnior G, Dos Santos Alemany AA, Belotti L, Zati DH, Frota Cavalcante M, Matera Veras M, Ribeiro S, Kallás EG, Nascimento Saldiva PH, Dolhnikoff M, Ferraz da Silva LF. Early and late pulmonary effects of nebulized LPS in mice: An acute lung injury model. PLoS One 12: e0185474, 2017. doi: 10.1371/journal.pone.0185474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elmasri H, Karaaslan C, Teper Y, Ghelfi E, Weng M, Ince TA, Kozakewich H, Bischoff J, Cataltepe S. Fatty acid binding protein 4 is a target of VEGF and a regulator of cell proliferation in endothelial cells. FASEB J 23: 3865–3873, 2009. doi: 10.1096/fj.09-134882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groneberg DA, Quarcoo D, Frossard N, Fischer A. Neurogenic mechanisms in bronchial inflammatory diseases. Allergy 59: 1139–1152, 2004. doi: 10.1111/j.1398-9995.2004.00665.x. [DOI] [PubMed] [Google Scholar]
- 23.Hawthorn L, Lan L, Mojica W. Evidence for field effect cancerization in colorectal cancer. Genomics 103: 211–221, 2014. doi: 10.1016/j.ygeno.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 24.Hellmann-Regen J, Kronenberg G, Uhlemann R, Freyer D, Endres M, Gertz K. Accelerated degradation of retinoic acid by activated microglia. J Neuroimmunol 256: 1–6, 2013. doi: 10.1016/j.jneuroim.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 25.Holmes FE, Kerr N, Chen YJ, Vanderplank P, McArdle CA, Wynick D. Targeted disruption of the orphan receptor Gpr151 does not alter pain-related behaviour despite a strong induction in dorsal root ganglion expression in a model of neuropathic pain. Mol Cell Neurosci 78: 35–40, 2017. doi: 10.1016/j.mcn.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holt JR, Pan B, Koussa MA, Asai Y. TMC function in hair cell transduction. Hear Res 311: 17–24, 2014. doi: 10.1016/j.heares.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang XP, Karpiak J, Kroeze WK, Zhu H, Chen X, Moy SS, Saddoris KA, Nikolova VD, Farrell MS, Wang S, Mangano TJ, Deshpande DA, Jiang A, Penn RB, Jin J, Koller BH, Kenakin T, Shoichet BK, Roth BL. Allosteric ligands for the pharmacologically dark receptors GPR68 and GPR65. Nature 527: 477–483, 2015. doi: 10.1038/nature15699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ignatov A, Hermans-Borgmeyer I, Schaller HC. Cloning and characterization of a novel G-protein-coupled receptor with homology to galanin receptors. Neuropharmacology 46: 1114–1120, 2004. doi: 10.1016/j.neuropharm.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 29.Im DS. Two ligands for a GPCR, proton vs lysolipid. Acta Pharmacol Sin 26: 1435–1441, 2005. doi: 10.1111/j.1745-7254.2005.00237.x. [DOI] [PubMed] [Google Scholar]
- 30.Ito M, Matsui T, Taniguchi T, Tsukamoto T, Murayama T, Arima N, Nakata H, Chiba T, Chihara K. Functional characterization of a human brain cholecystokinin-B receptor. A trophic effect of cholecystokinin and gastrin. J Biol Chem 268: 18300–18305, 1993. [PubMed] [Google Scholar]
- 31.Jeon S, Jha MK, Ock J, Seo J, Jin M, Cho H, Lee WH, Suk K. Role of lipocalin-2-chemokine axis in the development of neuropathic pain following peripheral nerve injury. J Biol Chem 288: 24116–24127, 2013. doi: 10.1074/jbc.M113.454140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang BC, Zhang WW, Yang T, Guo CY, Cao DL, Zhang ZJ, Gao YJ. Demethylation of G-protein-coupled receptor 151 promoter facilitates the binding of Krüppel-like factor 5 and enhances neuropathic pain after nerve injury in mice. J Neurosci 38: 10535–10551, 2018. doi: 10.1523/JNEUROSCI.0702-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin M, Jang E, Suk K. Lipocalin-2 acts as a neuroinflammatogen in lipopolysaccharide-injected mice. Exp Neurobiol 23: 155–162, 2014. doi: 10.5607/en.2014.23.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaelberer MM, Jordt SE. A method to target and isolate airway-innervating sensory neurons in mice. J Vis Exp (110): 2016. doi: 10.3791/53917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karmpaliotis D, Kosmidou I, Ingenito EP, Hong K, Malhotra A, Sunday ME, Haley KJ. Angiogenic growth factors in the pathophysiology of a murine model of acute lung injury. Am J Physiol Lung Cell Mol Physiol 283: L585–L595, 2002. doi: 10.1152/ajplung.00048.2002. [DOI] [PubMed] [Google Scholar]
- 36.Keresztes G, Mutai H, Heller S. TMC and EVER genes belong to a larger novel family, the TMC gene family encoding transmembrane proteins. BMC Genomics 4: 24, 2003. doi: 10.1186/1471-2164-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kichko TI, Lennerz J, Eberhardt M, Babes RM, Neuhuber W, Kobal G, Reeh PW. Bimodal concentration-response of nicotine involves the nicotinic acetylcholine receptor, transient receptor potential vanilloid type 1, and transient receptor potential ankyrin 1 channels in mouse trachea and sensory neurons. J Pharmacol Exp Ther 347: 529–539, 2013. doi: 10.1124/jpet.113.205971. [DOI] [PubMed] [Google Scholar]
- 38.Kilkenny C, Browne W, Cuthill IC, Emerson M, Altman DG; NC3Rs Reporting Guidelines Working Group . Animal research: reporting in vivo experiments: the ARRIVE guidelines. Br J Pharmacol 160: 1577–1579, 2010. doi: 10.1111/j.1476-5381.2010.00872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim Y, Remacle AG, Chernov AV, Liu H, Shubayev I, Lai C, Dolkas J, Shiryaev SA, Golubkov VS, Mizisin AP, Strongin AY, Shubayev VI. The MMP-9/TIMP-1 axis controls the status of differentiation and function of myelin-forming Schwann cells in nerve regeneration. PLoS One 7: e33664, 2012. doi: 10.1371/journal.pone.0033664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kõks S, Fernandes C, Kurrikoff K, Vasar E, Schalkwyk LC. Gene expression profiling reveals upregulation of Tlr4 receptors in Cckb receptor deficient mice. Behav Brain Res 188: 62–70, 2008. doi: 10.1016/j.bbr.2007.10.020. [DOI] [PubMed] [Google Scholar]
- 41.Kollarik M, Ru F, Undem BJ. Phenotypic distinctions between the nodose and jugular TRPV1-positive vagal sensory neurons in the cynomolgus monkey. Neuroreport 30: 533–537, 2019. doi: 10.1097/WNR.0000000000001231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kupari J, Häring M, Agirre E, Castelo-Branco G, Ernfors P. An atlas of vagal sensory neurons and their molecular specialization. Cell Reports 27: 2508–2523.e4, 2019. doi: 10.1016/j.celrep.2019.04.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kurima K, Yang Y, Sorber K, Griffith AJ. Characterization of the transmembrane channel-like (TMC) gene family: functional clues from hearing loss and epidermodysplasia verruciformis. Genomics 82: 300–308, 2003. doi: 10.1016/S0888-7543(03)00154-X. [DOI] [PubMed] [Google Scholar]
- 44.Lay JM, Jenkins C, Friis-Hansen L, Samuelson LC. Structure and developmental expression of the mouse CCK-B receptor gene. Biochem Biophys Res Commun 272: 837–842, 2000. doi: 10.1006/bbrc.2000.2875. [DOI] [PubMed] [Google Scholar]
- 45.Le DD, Rochlitzer S, Fischer A, Heck S, Tschernig T, Sester M, Bals R, Welte T, Braun A, Dinh QT. Allergic airway inflammation induces the migration of dendritic cells into airway sensory ganglia. Respir Res 15: 73, 2014. doi: 10.1186/1465-9921-15-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee LY, Burki NK, Gerhardstein DC, Gu Q, Kou YR, Xu J. Airway irritation and cough evoked by inhaled cigarette smoke: role of neuronal nicotinic acetylcholine receptors. Pulm Pharmacol Ther 20: 355–364, 2007. doi: 10.1016/j.pupt.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 47.Lindwall C, Dahlin L, Lundborg G, Kanje M. Inhibition of c-Jun phosphorylation reduces axonal outgrowth of adult rat nodose ganglia and dorsal root ganglia sensory neurons. Mol Cell Neurosci 27: 267–279, 2004. doi: 10.1016/j.mcn.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 48.Ling YL, Meng AH, Zhao XY, Shan BE, Zhang JL, Zhang XP. Effect of cholecystokinin on cytokines during endotoxic shock in rats. World J Gastroenterol 7: 667–671, 2001. doi: 10.3748/wjg.v7.i5.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu T, Gao YJ, Ji RR. Emerging role of Toll-like receptors in the control of pain and itch. Neurosci Bull 28: 131–144, 2012. doi: 10.1007/s12264-012-1219-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu T, Han Q, Chen G, Huang Y, Zhao LX, Berta T, Gao YJ, Ji RR. Toll-like receptor 4 contributes to chronic itch, alloknesis, and spinal astrocyte activation in male mice. Pain 157: 806–817, 2016. doi: 10.1097/j.pain.0000000000000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Z, Hu Y, Yu X, Xi J, Fan X, Tse CM, Myers AC, Pasricha PJ, Li X, Yu S. Allergen challenge sensitizes TRPA1 in vagal sensory neurons and afferent C-fiber subtypes in guinea pig esophagus. Am J Physiol Gastrointest Liver Physiol 308: G482–G488, 2015. doi: 10.1152/ajpgi.00374.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 25: 402–408, 2001. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 53.Lu YC, Yeh WC, Ohashi PS. LPS/TLR4 signal transduction pathway. Cytokine 42: 145–151, 2008. doi: 10.1016/j.cyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 54.Macia L, Rao PT, Wheway J, Sierro F, Mackay F, Herzog H. Y1 signalling has a critical role in allergic airway inflammation. Immunol Cell Biol 89: 882–888, 2011. doi: 10.1038/icb.2011.6. [DOI] [PubMed] [Google Scholar]
- 55.MacLean MR, Alexander D, Stirrat A, Gallagher M, Douglas SA, Ohlstein EH, Morecroft I, Polland K. Contractile responses to human urotensin-II in rat and human pulmonary arteries: effect of endothelial factors and chronic hypoxia in the rat. Br J Pharmacol 130: 201–204, 2000. doi: 10.1038/sj.bjp.0703314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Makinde TO, Steininger R, Agrawal DK. NPY and NPY receptors in airway structural and inflammatory cells in allergic asthma. Exp Mol Pathol 94: 45–50, 2013. doi: 10.1016/j.yexmp.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mao D, Yasuda RP, Fan H, Wolfe BB, Kellar KJ. Heterogeneity of nicotinic cholinergic receptors in rat superior cervical and nodose Ganglia. Mol Pharmacol 70: 1693–1699, 2006. doi: 10.1124/mol.106.027458. [DOI] [PubMed] [Google Scholar]
- 58.Mashiko M, Kurosawa A, Tani Y, Tsuji T, Takeda S. GPR31 and GPR151 are activated under acidic conditions. J Biochem 166: 317– 322, 2019. doi: 10.1093/jb/mvz042. [DOI] [PubMed] [Google Scholar]
- 59.Mazzone SB, Canning BJ. Autonomic neural control of the airways. Handb Clin Neurol 117: 215–228, 2013. doi: 10.1016/B978-0-444-53491-0.00018-3. [DOI] [PubMed] [Google Scholar]
- 60.Mazzone SB, Undem BJ. Vagal Afferent Innervation of the Airways in Health and Disease. Physiol Rev 96: 975–1024, 2016. doi: 10.1152/physrev.00039.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McAlexander MA, Gavett SH, Kollarik M, Undem BJ. Vagotomy reverses established allergen-induced airway hyperreactivity to methacholine in the mouse. Respir Physiol Neurobiol 212-214: 20–24, 2015. doi: 10.1016/j.resp.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meng AH, Ling YL, Zhang XP, Zhang JL. Anti-inflammatory effect of cholecystokinin and its signal transduction mechanism in endotoxic shock rat. World J Gastroenterol 8: 712–717, 2002. doi: 10.3748/wjg.v8.i4.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nagata A, Ito M, Iwata N, Kuno J, Takano H, Minowa O, Chihara K, Matsui T, Noda T. G protein-coupled cholecystokinin-B/gastrin receptors are responsible for physiological cell growth of the stomach mucosa in vivo. Proc Natl Acad Sci USA 93: 11825–11830, 1996. doi: 10.1073/pnas.93.21.11825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Noble F, Wank SA, Crawley JN, Bradwejn J, Seroogy KB, Hamon M, Roques BP. International Union of Pharmacology. XXI. Structure, distribution, and functions of cholecystokinin receptors. Pharmacol Rev 51: 745–781, 1999. [PubMed] [Google Scholar]
- 65.O’Donnell LC, Druhan LJ, Avalos BR. Molecular characterization and expression analysis of leucine-rich alpha2-glycoprotein, a novel marker of granulocytic differentiation. J Leukoc Biol 72: 478–485, 2002. [PubMed] [Google Scholar]
- 66.Ohara PT, Vit JP, Bhargava A, Romero M, Sundberg C, Charles AC, Jasmin L. Gliopathic pain: when satellite glial cells go bad. Neuroscientist 15: 450–463, 2009. doi: 10.1177/1073858409336094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Polak JM, Bloom SR. Regulatory peptides and neuron-specific enolase in the respiratory tract of man and other mammals. Exp Lung Res 3: 313–328, 1982. doi: 10.3109/01902148209069660. [DOI] [PubMed] [Google Scholar]
- 68.Portnoy A, Kumar S, Behm DJ, Mahar KM, Noble RB, Throup JP, Russ SF. Effects of Urotensin II Receptor Antagonist, GSK1440115, in Asthma. Front Pharmacol 4: 54, 2013. doi: 10.3389/fphar.2013.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reinhold AK, Batti L, Bilbao D, Buness A, Rittner HL, Heppenstall PA. Differential transcriptional profiling of damaged and intact adjacent dorsal root ganglia neurons in neuropathic pain. PLoS One 10: e0123342, 2015. doi: 10.1371/journal.pone.0123342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reznikov LR, Meyerholz DK, Abou Alaiwa M, Kuan SP, Liao YJ, Bormann NL, Bair TB, Price M, Stoltz DA, Welsh MJ. The vagal ganglia transcriptome identifies candidate therapeutics for airway hyperreactivity. Am J Physiol Lung Cell Mol Physiol 315: L133–L148, 2018. doi: 10.1152/ajplung.00557.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schubert ML, Peura DA. Control of gastric acid secretion in health and disease. Gastroenterology 134: 1842–1860, 2008. doi: 10.1053/j.gastro.2008.05.021. [DOI] [PubMed] [Google Scholar]
- 72.Scott GD, Blum ED, Fryer AD, Jacoby DB. Tissue optical clearing, three-dimensional imaging, and computer morphometry in whole mouse lungs and human airways. Am J Respir Cell Mol Biol 51: 43–55, 2014. doi: 10.1165/rcmb.2013-0284OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Seijffers R, Mills CD, Woolf CJ. ATF3 increases the intrinsic growth state of DRG neurons to enhance peripheral nerve regeneration. J Neurosci 27: 7911–7920, 2007. doi: 10.1523/JNEUROSCI.5313-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sethi T, Herget T, Wu SV, Walsh JH, Rozengurt E. CCKA and CCKB receptors are expressed in small cell lung cancer lines and mediate Ca2+ mobilization and clonal growth. Cancer Res 53: 5208–5213, 1993. [PubMed] [Google Scholar]
- 75.Shenouda A, Douglas SA, Ohlstein EH, Giaid A. Localization of urotensin-II immunoreactivity in normal human kidneys and renal carcinoma. J Histochem Cytochem 50: 885–889, 2002. doi: 10.1177/002215540205000702. [DOI] [PubMed] [Google Scholar]
- 76.Stretton CD, Barnes PJ. Cholecystokinin-octapeptide constricts guinea-pig and human airways. Br J Pharmacol 97: 675–682, 1989. doi: 10.1111/j.1476-5381.1989.tb12003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Surdenikova L, Ru F, Nassenstein C, Tatar M, Kollarik M. The neural crest- and placodes-derived afferent innervation of the mouse esophagus. Neurogastroenterol Motil 24: e517–e525, 2012. doi: 10.1111/nmo.12002. [DOI] [PubMed] [Google Scholar]
- 78.Suresh M, Molina H, Salvato MS, Mastellos D, Lambris JD, Sandor M. Complement component 3 is required for optimal expansion of CD8 T cells during a systemic viral infection. J Immunol 170: 788–794, 2003. doi: 10.4049/jimmunol.170.2.788. [DOI] [PubMed] [Google Scholar]
- 79.Talbot S, Abdulnour RE, Burkett PR, Lee S, Cronin SJ, Pascal MA, Laedermann C, Foster SL, Tran JV, Lai N, Chiu IM, Ghasemlou N, DiBiase M, Roberson D, Von Hehn C, Agac B, Haworth O, Seki H, Penninger JM, Kuchroo VK, Bean BP, Levy BD, Woolf CJ. Silencing nociceptor neurons reduces allergic airway inflammation. Neuron 87: 341–354, 2015. doi: 10.1016/j.neuron.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tang W, Kowgier M, Loth DW, Soler Artigas M, Joubert BR, Hodge E, Gharib SA, Smith AV, Ruczinski I, Gudnason V, Mathias RA, Harris TB, Hansel NN, Launer LJ, Barnes KC, Hansen JG, Albrecht E, Aldrich MC, Allerhand M, Barr RG, Brusselle GG, Couper DJ, Curjuric I, Davies G, Deary IJ, Dupuis J, Fall T, Foy M, Franceschini N, Gao W, Gläser S, Gu X, Hancock DB, Heinrich J, Hofman A, Imboden M, Ingelsson E, James A, Karrasch S, Koch B, Kritchevsky SB, Kumar A, Lahousse L, Li G, Lind L, Lindgren C, Liu Y, Lohman K, Lumley T, McArdle WL, Meibohm B, Morris AP, Morrison AC, Musk B, North KE, Palmer LJ, Probst-Hensch NM, Psaty BM, Rivadeneira F, Rotter JI, Schulz H, Smith LJ, Sood A, Starr JM, Strachan DP, Teumer A, Uitterlinden AG, Völzke H, Voorman A, Wain LV, Wells MT, Wilk JB, Williams OD, Heckbert SR, Stricker BH, London SJ, Fornage M, Tobin MD, O’Connor GT, Hall IP, Cassano PA. Large-scale genome-wide association studies and meta-analyses of longitudinal change in adult lung function. PLoS One 9: e100776, 2014. doi: 10.1371/journal.pone.0100776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Taylor-Clark TE, McAlexander MA, Nassenstein C, Sheardown SA, Wilson S, Thornton J, Carr MJ, Undem BJ. Relative contributions of TRPA1 and TRPV1 channels in the activation of vagal bronchopulmonary C-fibres by the endogenous autacoid 4-oxononenal. J Physiol 586: 3447–3459, 2008. doi: 10.1113/jphysiol.2008.153585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tcymbarevich IV, Eloranta JJ, Rossel JB, Obialo N, Spalinger M, Cosin-Roger J, Lang S, Kullak-Ublick GA, Wagner CA, Scharl M, Seuwen K, Ruiz PA, Rogler G, de Vallière C, Misselwitz B; Swiss IBD Cohort Study Group . The impact of the rs8005161 polymorphism on G protein-coupled receptor GPR65 (TDAG8) pH-associated activation in intestinal inflammation. BMC Gastroenterol 19: 2, 2019. doi: 10.1186/s12876-018-0922-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tränkner D, Hahne N, Sugino K, Hoon MA, Zuker C. Population of sensory neurons essential for asthmatic hyperreactivity of inflamed airways. Proc Natl Acad Sci USA 111: 11515–11520, 2014. doi: 10.1073/pnas.1411032111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111, 2009. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578, 2012. [Erratum in Nat Protoc 9: 2513, 2014.] doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Usoskin D, Furlan A, Islam S, Abdo H, Lönnerberg P, Lou D, Hjerling-Leffler J, Haeggström J, Kharchenko O, Kharchenko PV, Linnarsson S, Ernfors P. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci 18: 145–153, 2015. doi: 10.1038/nn.3881. [DOI] [PubMed] [Google Scholar]
- 87.Vuillaume ML, Naudion S, Banneau G, Diene G, Cartault A, Cailley D, Bouron J, Toutain J, Bourrouillou G, Vigouroux A, Bouneau L, Nacka F, Kieffer I, Arveiler B, Knoll-Gellida A, Babin PJ, Bieth E, Jouret B, Julia S, Sarda P, Geneviève D, Faivre L, Lacombe D, Barat P, Tauber M, Delrue MA, Rooryck C. New candidate loci identified by array-CGH in a cohort of 100 children presenting with syndromic obesity. Am J Med Genet A 164A: 1965–1975, 2014. doi: 10.1002/ajmg.a.36587. [DOI] [PubMed] [Google Scholar]
- 88.Wang J, Kollarik M, Ru F, Sun H, McNeil B, Dong X, Stephens G, Korolevich S, Brohawn P, Kolbeck R, Undem B. Distinct and common expression of receptors for inflammatory mediators in vagal nodose versus jugular capsaicin-sensitive/TRPV1-positive neurons detected by low input RNA sequencing. PLoS One 12: e0185985, 2017. doi: 10.1371/journal.pone.0185985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang JQ, Kon J, Mogi C, Tobo M, Damirin A, Sato K, Komachi M, Malchinkhuu E, Murata N, Kimura T, Kuwabara A, Wakamatsu K, Koizumi H, Uede T, Tsujimoto G, Kurose H, Sato T, Harada A, Misawa N, Tomura H, Okajima F. TDAG8 is a proton-sensing and psychosine-sensitive G-protein-coupled receptor. J Biol Chem 279: 45626–45633, 2004. doi: 10.1074/jbc.M406966200. [DOI] [PubMed] [Google Scholar]
- 90.Wang YY, Cutz E. Localization of cholecystokinin-like peptide in neuroendocrine cells of mammalian lungs: a light and electron microscopic immunohistochemical study. Anat Rec 236: 198–205, 1993. doi: 10.1002/ar.1092360124. [DOI] [PubMed] [Google Scholar]
- 91.Yin K, Deuis JR, Lewis RJ, Vetter I. Transcriptomic and behavioural characterisation of a mouse model of burn pain identify the cholecystokinin 2 receptor as an analgesic target. Mol Pain 12: 1744806916665366, 2016. doi: 10.1177/1744806916665366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zaccone EJ, Undem BJ. Airway vagal neuroplasticity associated with respiratory viral infections. Hai 194: 25–29, 2016. doi: 10.1007/s00408-015-9832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang X, Bao L, Xu ZQ, Kopp J, Arvidsson U, Elde R, Hökfelt T. Localization of neuropeptide Y Y1 receptors in the rat nervous system with special reference to somatic receptors on small dorsal root ganglion neurons. Proc Natl Acad Sci USA 91: 11738–11742, 1994. doi: 10.1073/pnas.91.24.11738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhuo H, Ichikawa H, Helke CJ. Neurochemistry of the nodose ganglion. Prog Neurobiol 52: 79–107, 1997. doi: 10.1016/S0301-0082(97)00003-8. [DOI] [PubMed] [Google Scholar]