Abstract
Activation of the inflammasome-caspase-1 axis in lung endothelial cells is emerging as a novel arm of the innate immune response to pneumonia and sepsis caused by Pseudomonas aeruginosa. Increased levels of circulating autacoids are hallmarks of pneumonia and sepsis and induce physiological responses via cAMP signaling in targeted cells. However, it is unknown whether cAMP affects other functions, such as P. aeruginosa-induced caspase-1 activation. Herein, we describe the effects of cAMP signaling on caspase-1 activation using a single cell flow cytometry-based assay. P. aeruginosa infection of cultured lung endothelial cells caused caspase-1 activation in a distinct population of cells. Unexpectedly, pharmacological cAMP elevation increased the total number of lung endothelial cells with activated caspase-1. Interestingly, addition of cAMP agonists augmented P. aeruginosa infection of lung endothelial cells as a partial explanation underlying cAMP priming of caspase-1 activation. The cAMP effect(s) appeared to function as a priming signal because addition of cAMP agonists was required either before or early during the onset of infection. However, absolute cAMP levels measured by ELISA were not predictive of cAMP-priming effects. Importantly, inhibition of de novo cAMP synthesis decreased the number of lung endothelial cells with activated caspase-1 during infection. Collectively, our data suggest that lung endothelial cells rely on cAMP signaling to prime caspase-1 activation during P. aeruginosa infection.
Keywords: cAMP, caspase-1, Pseudomonas aeruginosa, pulmonary endothelial cells, stress responses
INTRODUCTION
Caspase-1 is an inflammatory member of the caspase family that is activated following sensing of danger signals, such as those elicited during infection and sepsis (12). Canonical caspase-1 activation in immune cells leads to cytokine release and leukocyte recruitment. However, caspase-1 activation in pulmonary microvascular endothelial cells (PMVECs) elicits noncanonical responses regulating maintenance of endothelial cell homeostasis (1). Sensing of danger signals via cytosolic NOD-like receptor proteins (NLRs) triggers assembly of a multiprotein complex, termed the inflammasome, to promote cleavage and maturation of pro-caspase-1 (4). Classically, caspase-1 activation in immune cells leads to IL-1β and IL-18 maturation and gasdermin D-mediated pyroptotic cell death, which together coordinate proinflammatory responses (13). Activation of the inflammasome/caspase-1 axis in experimental models of Pseudomonas aeruginosa-induced pneumonia and sepsis results in potentially confounding outcomes. P. aeruginosa infection of inflammasome knockout mice (5) or infection of mice treated with caspase-1 inhibitors (1) decreased lung parenchymal inflammation but, paradoxically, increased vascular permeability associated with detrimental edema formation. Emerging evidence in experimental models of P. aeruginosa pneumonia and sepsis indicate the involvement of the pulmonary vascular endothelium during P. aeruginosa infection (17). Thus the paradoxical effects of caspase-1 in regulating both inflammation and vascular permeability indicate that caspase-1 activation mediates distinct biological responses that are cell phenotype dependent. Therefore, caspase-1 activation in nonimmune cells is an under-recognized determinant of outcome in the lung during pneumonia and sepsis, for which mechanisms are poorly understood.
During sepsis, circulating prostanoids (16) elicit signaling cascades that result in elevated intracellular cAMP (28). In PMVECs, cellular response is determined by the spatial distribution of cAMP. Specifically, PGE2 elevates cAMP in the near-membrane compartment, which elicits barrier-protective responses that prevent pulmonary edema (3, 30). Conversely, pathogenic adenylyl cyclase (AC) toxins, such as Bacillus anthracis edema factor (32) and the P. aeruginosa exoenzyme Y (25), elevate cAMP in the cytosolic compartment, causing endothelial barrier disruption and pulmonary edema. Thus altered cAMP signals during sepsis may constitute an underappreciated intracellular danger signal. In immune cells, elevation of cAMP has been shown to inhibit caspase-1 activation (15, 27). However, it is unknown whether cAMP signals elicited during sepsis regulate caspase-1 activation in a cell phenotype-dependent manner. Therefore, we sought to explore links between cAMP signaling and caspase-1 activation in PMVECs using our in vitro P. aeruginosa infection model.
MATERIALS AND METHODS
Cell culture methods.
Rat endothelial pulmonary microvascular cells (PMVECs) were isolated and characterized from distal lung parenchyma and obtained through the University of South Alabama Center for Lung Biology cell culture core. PMVECs were routinely cultured on 100-mm cell culture dishes in Dulbecco's modified Eagle's medium, high glucose (DMEM), supplemented with 10% fetal bovine serum (FBS), at 37°C, 5% CO2.
Bacterial strains and preparation for cell culture inoculations.
Frozen stock solutions of P. aeruginosa strain PA103 were stored at −80°C in Difco nutrient broth supplemented with 12.5% glycerol. PA103 was grown on the minimal E salts medium of Vogel and Bonner containing agar (1.5%) at 37°C overnight. PA103 is a nonmotile strain expressing a functional T3SS and the exoenzymes ExoU (a phospholipase A2 cytolysin) and ExoT (a dual-functioning RhoGAP and ADP ribosyltransferase).
Preparation of FLICA to identify cells with activated caspase-1.
Each vial of the fluorescent-labeled inhibitor of caspase-1 (FLICA) reagent (FAM-YVAD-FMK) (ImmunoChemistry Technologies, cat. no. 98) was reconstituted in 50 μL of 100% DMSO to make a 150-fold concentrated stock solution (150× FLICA). 150× FLICA was diluted into sterile phosphate-buffered saline (PBS, pH 7.4) solution to make a 30-fold concentrated solution (30× FLICA). 30× FLICA was diluted into DMEM, high glucose lacking phenol red and FBS, and supplemented with 4 mM GlutaMAX (sfcDMEM) for final working concentration of 0.5× FLICA.
Pharmacological treatments used in this study.
Pharmacological treatments were added to PMVECs in culture medium either before or after infection as follows: forskolin (100 μM) (Sigma-Aldrich, cat no. F3917 or EMD Millipore, cat. no. 24427); rolipram (10 μM) (Alfa Aesar, cat. no. J67177); 8-bromo-cAMP (100–600 μM) (Tocris, cat. no. 1140); 8-bromo-cGMP (100–600 μM) (Tocris, cat. no. 1089); PGE2 (25 μM) (Sigma-Aldrich, cat. no. P5640); butaprost (50 μM) (Sigma-Aldrich, cat. no. B6309); IBMX (500 μM) (Sigma-Aldrich, cat. no. I5879); SQ 22,536 (200 μM) (Sigma-Aldrich, cat. no. S153).
Cell culture infection model.
One day before an experiment, confluent PMVEC monolayers were harvested by treatment with Trypsin-EDTA (0.05%/0.53 mM solution in Hank’s balanced salt solution, without Ca++ and Mg++) and washed by centrifugation (500 g, 4 min, room temperature) into DMEM FBS. Cell suspensions were counted on a Fuchs-Rosenthal ultra-plane counting chamber, seeded in 12-well CellBIND-treated culture dishes at a density of 5.5 × 105 cells in 1 mL DMEM FBS, and incubated overnight at 37°C, 5% CO2. On the day of the experiment (~16 h after seeding), monolayers were visually inspected under the microscope for confluency. Only if monolayers were confluent was the experiment carried out. Duplicate wells were harvested and counted as above to account for any cell growth after seeding and to accurately calculate multiplicity of infection (MOI). Cells in the well were washed in 1 mL of sfcDMEM and serum starved in 1 mL of sfcDMEM for 1 h at 37°C, 5% CO2, before the start of infection. Pharmacological treatments were added either during the serum starvation step or during the infections as indicated in the figure legends. All infections were performed in a 300-µL final volume.
On the day of an experiment, bacterial lawns were scraped with an inoculation loop into 10 mL of sterile 0.9% saline solution (saline). Bacteria were washed by centrifugation (5,000 g, 10 min, at room temperature). The bacterial pellet was suspended in 1 mL of saline and the optical density at 600 nm (OD600) of the bacterial suspension was measured on a ThermoFisher Evolution 300 spectrophotometer. For P. aeruginosa strain PA103, the OD600 to colony-forming unit (CFU) ratio was previously established by serial dilution and plating to be OD600 0.007:1 × 107 CFU/mL. This ratio was used to calculate the OD600 required for the desired MOI, and a 20-fold concentrated (20×) infection stock solution was prepared in saline.
The sfcDMEM medium was aliquoted into sterile microcentrifuge tubes. Bacterial inoculum (1/20 dilution of the 20× stock solution) or saline was pipetted into the media. Pharmacological agents or vehicle controls were pipetted into the media to the final concentrations indicated in the figures. FLICA was pipetted into media to a final 0.5× concentration. Final volume was 300 μL. Media from cells was discarded. Infection inoculum was mixed by pipetting and then gently added to the well. Pharmacological treatments were present or added as indicated in the figure legends. Infections were incubated at 37°C, 5% CO2.
For determination of bacterial growth during infection, culture medium was collected at 3 h after infection, serially diluted, and plated onto Luria-Bertani (LB) agar growth medium. After overnight incubation at 37°C, colonies were counted to determine the number of colony-forming units (CFUs). For assessment of bacterial attachment to host cells during infection, the culture medium was removed at 3 h after infection, monolayers suspended, and washed three times in 1 mL of sfcDMEM (by centrifugation). The final cell suspension was then serially diluted and plated onto LB agar to determine CFU counts.
ExoU PLA2 functional assay as a reporter of PMVEC infection.
ExoU activity was measured using the PED6 fluorogenic phospholipase A2 (PLA2) substrate [N-((6-(2,4-dinitrophenyl)amino)hexanoyl)-2-(4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-pentanoyl)-1-hexadecanoyl-sn-glycero-3-phosphoethanolamine, triethylammonium salt, ThermoFisher, D23739]. A PED6 stock compound was solubilized in 100% DMSO and diluted in sfcDMEM to a final concentration of 14.85 μM (100 μL final volume). The PED6 suspension was sonicated (50% duty cycle, output = 2, pulses = 5 with icing for 30 s between pulses, using a Heat System Ultrasonics, Inc Sonicator Cell Disruptor Model W-375) and then added to a confluent PMVEC monolayer and incubated for 1 h at 37°C, 5% CO2. Monolayers were washed once in 0.5 mL of sfcDMEM and then inoculated with saline solution or infected with PA103 as described above. Media samples were removed over time (see legend to Fig. 2D) and the PLA2-mediated release of BODIPY-labeled free fatty acid (FFA) was measured using a NanoDrop 3300 Fluorospectrometer (ThermoFisher). Note that the basal rate of FFA release from saline-treated PMVECs was subtracted as a background correction at each time point (reported as adjusted relative fluorescent units, RFUs).
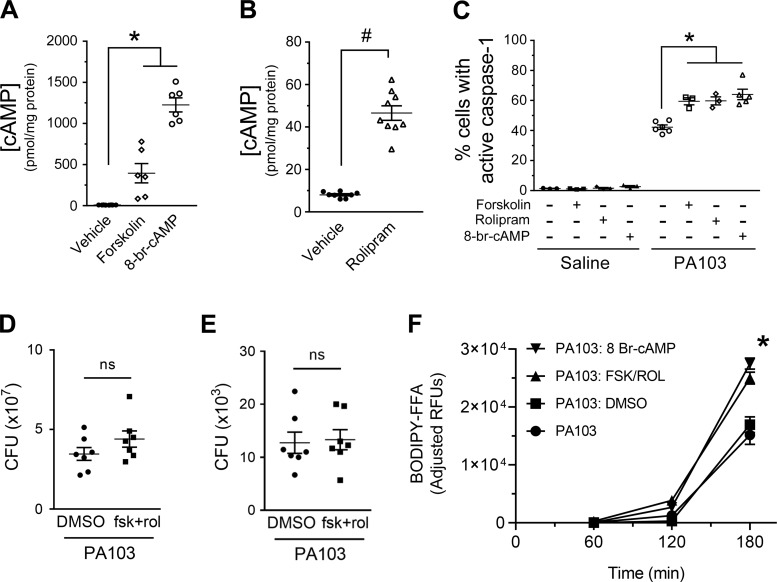
Fig. 2.
Addition of forskolin and rolipram or 8-br cAMP increases the number of pulmonary microvascular endothelial cells (PMVECs) with active caspase-1 during PA103 infection. A and B: PMVECs were serum starved for 60 min. The cAMP-elevating compounds were added to the culture medium 20 min before harvesting for measurement of global cAMP by ELISA. For all conditions, n = 3. *Significant difference from saline and rolipram, P < 0.0001 (one-way ANOVA with Bonferroni’s post hoc analysis); #significant difference from saline, P < 0.0001 (unpaired t test). C: PMVECs were serum starved for 60 min. The cAMP-elevating compounds were added to the culture medium 30 min before addition of fluorescent-labeled inhibitor of caspase-1 (FLICA) reagent and PA103 [multiplicity of infection (MOI) 40:1]. Saline-treated PMVEC monolayers served as negative controls. PMVECs were harvested at 180 min after infection and analyzed for activated caspase-1 by flow cytometry. For all conditions, n = 3–4. *Significant differences compared with infection alone, P = 0.0003 (interaction among groups), P < 0.0001 (between different infection groups), P = 0.0001 (between different treatment groups) (two-way ANOVA with Tukey’s post hoc analysis). D: PMVECs were serum starved for 60 min followed by treatment with DMSO (vehicle control) or a combination of forskolin and rolipram for an additional 30 min. Monolayers were then infected with PA103 (MOI 40:1). At 3 h after infection, culture medium was collected, and bacterial growth was assessed by serial dilution and plating to determine colony-forming units (CFUs). For all conditions, n = 5, and no differences were noted (unpaired t test, P = 0.1754). E: PMVECs were treated and infected as in D, except that culture medium was discarded, and bacterial attachment to cells was assessed by plating. For all conditions, n = 5, and no differences were noted (unpaired t test, P = 0.8385). F: PMVECs were serum starved for 60 min and simultaneously labeled by the addition of PED6. Cells were washed and then inoculated with saline or with PA103 at an MOI of 40:1. The cAMP-elevating compounds or its vehicle control (DMSO) were added to the culture medium 30 min before infection. Culture media was collected over time and assayed for release of BODIPY-labeled free fatty acid (FFA) as an indicator of PLA2 activity. Note that the basal rate of FFA release from saline-treated PMVECs was subtracted as a background correction at each time point (reported as adjusted relative fluorescent units, RFUs). For all conditions, n = 6. *Significant differences at the 3-h time point, P < 0.0001 (two-way ANOVA with Tukey’s post hoc analysis). PA103 versus the PA103: DMSO and PA103: FSK/ROL versus PA103: 8 Br-cAMP were the only groups that did not show significant differences.
Western blotting.
At indicated times after inoculation, cell lysates were collected and assayed for pro-caspase-1 and IL-1β. To harvest cell lysates, culture dishes were placed on ice, and PMVECs were lysed by scraping into a 50-μL cold buffer solution consisting of PBS (pH 7.4), 1% Triton X-100, 0.5% SDS, 1× Roche protein inhibitor cocktail. Whole cell lysates were sonicated (50% duty cycle, output = 2, pulses = 5, using a Heat System Ultrasonics, Inc Sonicator Cell Disruptor Model W-375) and clarified by centrifugation (18,000 g, 10 min, 4°C). Protein concentrations were determined using Bio-Rad’s DC protein assay, and 90 μg of protein was mixed in 2× Laemmli dye. Total protein was resolved by SDS-PAGE on 4–12% Bis-Tris gels in MOPS-SDS running buffer at 200 V for 50 min. Protein was transferred to 0.2-μm nitrocellulose membranes in transfer buffer with 20% methanol on ice at 100 V for 50 min. Ponceau S staining was used to verify transfer followed by washing in 0.5 N NaOH to remove the Ponceau S. Membranes were probed by Western blotting as previously described (1). The α-caspase-1 antibody was a mouse monoclonal antibody (Adipogen, AG-20B-0042-C100, at 1:10,000), the α-IL-1β was polyclonal rabbit (Novus, NB600-633, at 0.001 µg/mL), and the α-β-actin antibody was monoclonal mouse (Santa Cruz Biotechnology, sc-81178, at 0.0001 µg/mL). All secondary antibodies were horseradish peroxidase (HRP) conjugates used at a dilution of 1:2,000. Blots were developed on a Bio-Rad Chemidoc XRS using Supersignal West Femto HRP reagent. Blots were subsequently stripped and reprobed with antibody to β-actin to normalize for loading as previously described (1).
CRISPR-Cas9 directed mutation of caspase-1 in PMVECs.
CRISPR small-guide sgRNA vectors targeting Rattus norvegicus caspase-1 exon sequences were purchased from GeneCopoeia. Targeting constructs express a single sgRNA, an mCherry reporter, a puromycin resistance gene, and lentivirus packaging sequences (pCRISPR-LvSG02–3-B, no. RCP249089). Replication-incompetent pseudo-lentivirus was generated via CaCl2 transfection of cultured HEK 293FT cells (grown in DMEM FBS), and for safety purposes the lentiviral packaging genes were provided on two different plasmids in trans by cotransfection. Lentivirus-laden culture medium was collected at 48 h after transfection, filtered (0.2 μm SFCA filter), and stored at –80°C. All pseudo-lentivirus stocks were empirically tested for transduction efficiency by diluting different amounts of virus-laden medium into DMEM FBS and incubating the mixture with 3 × 105 PMVECs seeded in six-well cluster plates. At 72 h after transduction, cells were harvested and analyzed on a BD FACS Aria II flow cytometer to determine conditions appropriate to achieve >90% transduction efficiency (i.e., mCherry-expressing cells). Also note that nonmutated sgRNA-only-expressing transductant cells generated under optimized conditions served as parent control PMVECs in subsequent experiments.
One of the resulting PMVEC transductants, expressing a sgRNA targeted to caspase-1 exon 4, was subsequently transfected with a mixture of two plasmids expressing the Cas9 enzyme [pX330-U6-Chimeric_BB-CBh-hSpCas9 was from Feng Zhang (Addgene plasmid no. 42230)] (6) and expressing Cas9-2A-GFP [pSpCas9(BB)-2A-GFP (PX458) was from Feng Zhang (Addgene plasmid # 48138)] (23), using Clontech’s Xfect transfection reagent under conditions optimized per the manufacturer’s instructions. At 24–48 h after transfection, PMVECs were harvested into a single cell suspension and sorted on a BD FACSAria II Flow Cytometry System to collect cells coexpressing mCherry and GFP. A diluted suspension of double-positive cells was seeded on a 150-mm culture dish and grown in DMEM FBS until isolated, individual colonies were observable. Individual colonies were lifted from the culture dish using a trypsin-EDTA-soaked filter paper and transferred to fresh DMEM FBS in a 24-well cluster plate. When cultures reached confluence, they were harvested for low-density reseeding, and a portion was extracted to screen for indel mutations using a previously described multiplex thermocycling-based strategy (33), with three primers that will generate either short DNA fragments (amplification of wild-type alleles) or long DNA fragments (amplification of alleles with indel mutations). Primers for screening were as follows: Cas1Exon4 Forward (5′–ATGTGTAAACACAGTAGAAGGTGA); Cas1Exon4 Reverse Long (5′–GCAGATTGAATGAAGAACCTTGTTA); and Cas1Exon4 Reverse Short (5′– TTGAGGGAACCACTCGGTC). Clones identified as possible indel mutants were further screened by PA103 infection and FLICA assay. Clones that were identified as FLICA-negative (caspase-1 indel mutants) were ultimately screened by Western blotting using anti-caspase-1 antibody. Upon confirmation, the population of caspase-1 indel mutant PMVECs was subjected to a second round of single cell sorting on a BD FACS Aria II Flow Cytometry System, and the resulting single cell-generated clone bank was rescreened as described above. This final sorting step yielded a clonal mutant population, which was confirmed by screening and DNA sequencing to harbor stable indel mutations in the caspase-1 gene (designated mutCasp1). During this final confirmation process, DNA fragments generated during the multiplex thermocycling screen step were cloned into a plasmid shuttle vector (Zero Blunt TOPO PCR cloning kit, Invitrogen), and at least 20 individual fragments were sent for DNA sequencing (Eurofins Genomics) in both orientations to identify the nature of the indel mutations. The resulting sequence analysis revealed the presence of a 2-base pair deletion resulting in substantial mutation and truncation of the caspase-1 protein; a 3-base pair deletion resulting in a single amino acid deletion; and a 5-base pair deletion resulting in substantial mutation and truncation of the caspase-1 protein. Interestingly, Western blotting analysis of the clone indicated that the combined effect of the mutations is sufficient to reduce caspase-1 protein to undetectable levels.
Flow cytometric and FlowJo software analysis to identify PMVEC populations with activated caspase-1.
At indicated times postinoculation, the infection/treatment medium was removed and held in reserve in a 1.5-mL microcentrifuge tube. Adherent cell monolayers were gently washed in 1 mL of wash buffer (provided with the FLICA reagent, contains mammalian proteins to stabilize cells stained with FLICA). The first wash was discarded, and PMVECs were gently scraped into 1 mL of fresh wash buffer. Cells were triturated to make single cell suspension, and the suspension was transferred to a 1.5-mL microcentrifuge tube. At this point, if monolayers were severely damaged by the infection, any floating cells in the reserved culture medium were collected by centrifugation (600 g, 5 min, room temperature) and combined back with the cells harvested from the culture dish. Cells were then washed two additional times by centrifugation (1 mL wash buffer, 600 g, 5 min, room temperature). After the last wash, cell pellets were suspended in 750 μL of wash buffer and transferred to 5 mL flow cytometry tubes containing 250 μL 10% formalin. These fixed cell suspensions were either analyzed immediately or stored at 4°C protected from light.
Fixed PMVEC suspensions were analyzed on a BD FACS Canto II Flow Cytometry System using the FITC laser to detect FAM-YVAD-FMK. Event analysis stopping gate was set to 20,000, and data were analyzed using TreeStar’s FlowJo software v10 (FlowJo, LLC). All events were first displayed as a side scatter area (SSC-A) versus forward scatter area (FSC-A) scatter plot, which indicate cell granulosity and cell size, respectively. A polygonal gate was drawn outlining the events indicative of intact PMVECs. Events within this gate were then displayed in an FSC height versus FSC width scatter plot, and, based on cell height and width, a square gate was drawn around single cell PMVECs (excluding doublets, triplets etc.). Single cell events within this gate were then displayed in a green fluorescence area (FITC-A) versus FSC-A scatter plot to analyze PMVEC fluorescence. Axes were adjusted to center population in the scatter plot. Uninfected, FLICA-treated and washed PMVECs were used to determine nonspecific background FLICA binding and establish the best possible trapezoid gate to delineate FLICA-negative from FLICA-positive events. Delineation accuracy of the gate was checked with infected PMVECs and adjusted accordingly. After satisfactory delineation between FLICA-positive and FLICA-negative cells was established, all gates were copied to all experimental conditions tested to ensure uniformity of all gates.
Global cAMP measurements in whole cell lysates using ELISA.
PMVECs were treated with indicated pharmacological agents for 20–30 min. Monolayers were washed in 1 mL cold PBS, and cells were lysed in 300 µL 1 N HCl, followed by neutralization in 350 µL 1 N NaOH. Lysates were stored at 4°C before cAMP measurement by ELISA according to manufacturer's instructions (Cayman Chemical). Acetylated standards and samples were analyzed following manufacturer's instructions. Protein assay was performed to calculate the concentration of protein in each sample and used to normalize the cAMP concentration.
Statistical analysis.
GraphPad Prism V6 or V8 was used for analysis. Data are reported as mean ± SE. Unpaired t test was used to compare two groups. One-way ANOVA was used to compare three or more groups, and Bonferroni's post hoc analysis was used as necessary. Two-way ANOVA was used to compare two or more groups over time or with multiple treatments, and Tukey's multiple comparison test was applied as necessary. Differences with a P value <0.05 were considered significant.
RESULTS
PMVEC caspase-1 is activated in a dose- and time-dependent manner during P. aeruginosa infection.
Canonical approaches to assess caspase-1 activation employ analysis of whole cell homogenates and/or secretion products by Western blotting with α-caspase-1 antibody (4). However, we considered that the whole cell lysate approach analyzes the average response of all cells in a population but may misrepresent responses that occur within distinct cell populations. Thus we tested a membrane-permeable, irreversible fluorescent labeled inhibitor of CAspase-1 (FLICA, FAM-YVAD-FMK) using flow cytometry to elucidate the kinetics of P. aeruginosa-induced caspase-1 activation at the single cell level. In uninfected PMVECs, where caspase-1 is inactive, the FLICA compound is freely diffusible and readily washed out of the cell (Fig. 1A and Supplemental Fig. S1A, top; see https://doi.org/10.6084/m9.figshare.11867901). We first determined optimal FLICA concentration and washing conditions to reduce background (Supplemental Fig. S1, A and B; see https://doi.org/10.6084/m9.figshare.11866500). Inoculation of PMVECs with PA103 generated enzymatically active caspase-1, which irreversibly bound to FLICA, trapping it inside the cell, thus producing a distinct population of FLICA-positive cells (Fig. 1A and Supplemental Fig. S1C, bottom; see https://doi.org/10.6084/m9.figshare.11867949). We verified that PA103 infection alone in the absence of FLICA did not spuriously generate green fluorescent cells (Supplemental Fig. S1D; see https://doi.org/10.6084/m9.figshare.11867952), and the FLICA response was abolished when bacteria were heat inactivated (Supplemental Fig. S1E; see https://doi.org/10.6084/m9.figshare.11867955).
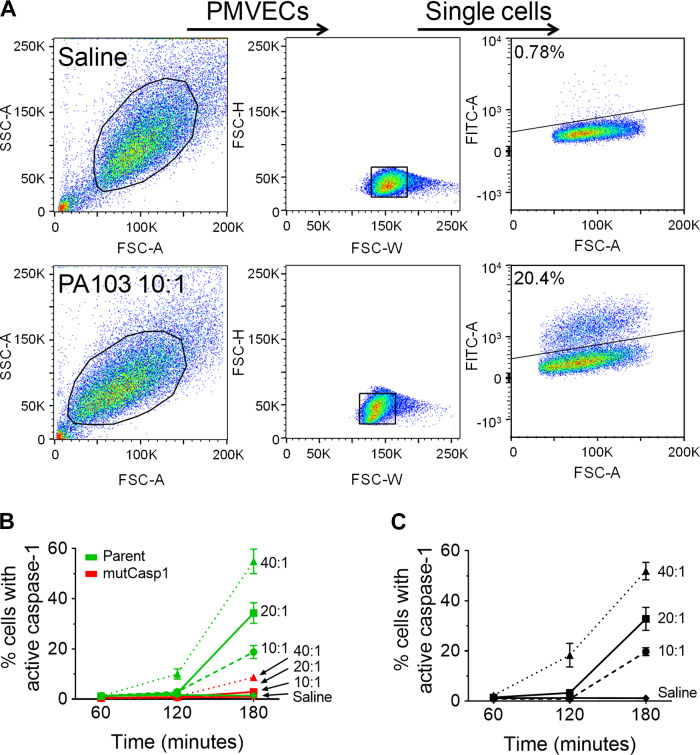
Fig. 1.
PA103 infection of pulmonary microvascular endothelial cells (PMVECs) activates caspase-1 in a dose- and time-dependent manner. PMVECs were serum starved for 60 min, then inoculated with saline or with PA103 at a multiplicity of infection (MOI) of 10:1, 20:1, or 40:1, and cells were collected for analysis at 60, 120, or 180 min after inoculation. A: detection of caspase-1-positive PMVECs was assessed by loading PMVECs with fluorescent-labeled inhibitor of caspase-1 (FLICA) and treating with saline (top) or infecting with PA103 at an MOI of 10:1 (bottom) for 180 min. PMVECs were gated according to size and granulosity (left) and single cells (middle) using light scatter before analysis for green fluorescence (FITC) signal to identify FLICA-positive cells (right). Uninfected control conditions (saline) were used to set the gating threshold to identify the FLICA-positive population. B: the FLICA assay was validated using PMVECs with mutated caspase-1 alleles as a negative control. We compared the effects of dose and time in a parent PMVEC culture expressing only the targeting caspase-1 sgRNAs against effects in mutCasp1 PMVECs lacking caspase-1. C: the kinetics of caspase-1 activation as assessed by FLICA in wild-type PMVECs. n = 3–6, P < 0.0001 (interaction among groups), P < 0.0001 (between different MOI groups), P < 0.0001 (between different time points) (two-way ANOVA). SSC-A, side scatter area; FSC-H, forward scatter height; FSC-A, forward scatter area; FITC-A, green fluorescence area; FSC-W, forward scatter width.
As our next step to rigorously validate the specificity of the FLICA assay, we used CRISPR-Cas9 genome engineering to generate a PMVEC strain harboring deletion mutations in the caspase-1 alleles (designated as mutCasp1, mutations verified by DNA sequencing). To examine the extent of pro-caspase-1 deficiency in the mutCasp1 PMVECs, we used Western blotting for pro-caspase-1 protein and compared the mutCasp1 PMVECs to wild-type PMVECs under control and PA103-infected conditions (Supplemental Fig. S1F; see https://doi.org/10.6084/m9.figshare.11867958, compare lanes 1 and 2 with lanes 5 and 6). Indeed, the mutCasp1 PMVECs produce very little pro-caspase-1 protein compared with wild-type control PMVECs. Furthermore, we demonstrated that the mutCasp1 cells were deficient for activation of IL-1β as further verification that the caspase-1 alleles were mutated (Supplemental Fig. S1G; see https://doi.org/10.6084/m9.figshare.11867961). Together, these data suggest the CRISPR-Cas9-edited mutCasp1 PMVECs are deficient for production and activation of caspase-1 during PA103 infection.
The mutCasp1 PMVECs were used subsequently to validate the FLICA assay. We first compared the effects of infectious dose and time on the number of PMVECs with active caspase-1 during PA103 infection (Fig. 1B). In these assays, we compared the mutCasp1 PMVECs to a culture of parent PMVEC expressing only the sgRNA guides (the parent PMVECs were used to generate the mutCasp1 PMVECs, see materials and methods). The mutCasp1 PMVECs were highly deficient for FLICA staining in response to PA103 infection, indicating the specificity of the assay for measuring caspase-1 activation.
Using the FLICA assay, we next tested the effects of infectious dose and time on caspase-1 activation in wild-type PMVECs. While the intensity of the FLICA signal remained unchanged, the percentage of PMVECs that activated caspase-1 during infection increased in an infectious dose- and time-dependent manner (Fig. 1C). The highly sensitive cell-by-cell analysis of caspase-1 activation in PMVECs using FLICA showed that small populations of caspase-1-activated PMVECs could be detected as early as 120 min after inoculation (Fig. 1C).
Elevation of cAMP increases the number of caspase-1-active PMVECs during P. aeruginosa infection.
During sepsis, elevated levels of circulating autacoids, prostanoids, and other mediators stimulate intracellular cyclic nucleotide production (e.g., cAMP) to alter host cell physiological responses (16, 19). The importance of cAMP signaling in innate immune cells was previously highlighted by the observation that increased levels of cAMP inhibited NLRP3 inflammasome-mediated caspase-1 activation (15, 27) although this finding is somewhat controversial (14, 22). Thus, we next tested the effects of increased cAMP levels on caspase-1 activation in PMVECs using three different pharmacological agents: 1) forskolin activates particulate adenylyl cyclase (AC); 2) rolipram inhibits phosphodiesterase type 4 (PDE4-)-mediated cAMP degradation; or 3) cell -permeant 8-br-cAMP. Compounds were added before inoculation and maintained throughout the experiment as described in the figure legend. In the absence of infection, all three compounds significantly elevated intracellular cAMP to varying levels (Fig. 2, A and B), but none of the compounds endogenously activated caspase-1 (Fig. 2C, saline). Unexpectedly, all three cAMP-elevating compounds significantly increased the total number of caspase-1-active PMVECs elicited during PA103 infection (Fig. 2C, PA103). We also verified that the effect of 8-br-cAMP caused a dose-dependent increase in the number of caspase-1-active PMVECs (Supplemental Fig. S2A; see https://doi.org/10.6084/m9.figshare.11867964) and that elevation of a different cyclic nucleotide (cGMP levels via addition of 8-br-cGMP) had no effect (Supplemental Fig. 2SB; see https://doi.org/10.6084/m9.figshare.11867967).
To gain insight into possible mechanisms underlying cAMP-mediated increases in the number of caspase-1-active PMVECs elicited during PA103 activation, we assayed for effects of cAMP-elevating agents on bacterial growth (Fig. 2D) and bacterial attachment to host PMVECs (Fig. 2E). Interestingly, changes in host cell cAMP levels had no significant effects on PA103 growth or attachment to host cells. We next examined effects of elevating host cAMP levels on a second aspect of the host-pathogen interaction. Strain PA103 uses the type III secretion system (T3SS) to inject the ExoU phospholipase A2 (PLA2) into host cells during infection; thus we labeled PMVECs with a fluorogenic PLA2 reporter (PED6) and measured free fatty acid (FFA) release as a surrogate measure of infectivity. Interestingly, elevation of cAMP levels by addition of forskolin and rolipram or 8-bromo cAMP increased the amount of FFA released during PA103 infection (Fig. 2F). Together, these data suggest that elevation of intracellular cAMP serves as a priming step to sense P. aeruginosa infection and subsequently activate caspase-1.
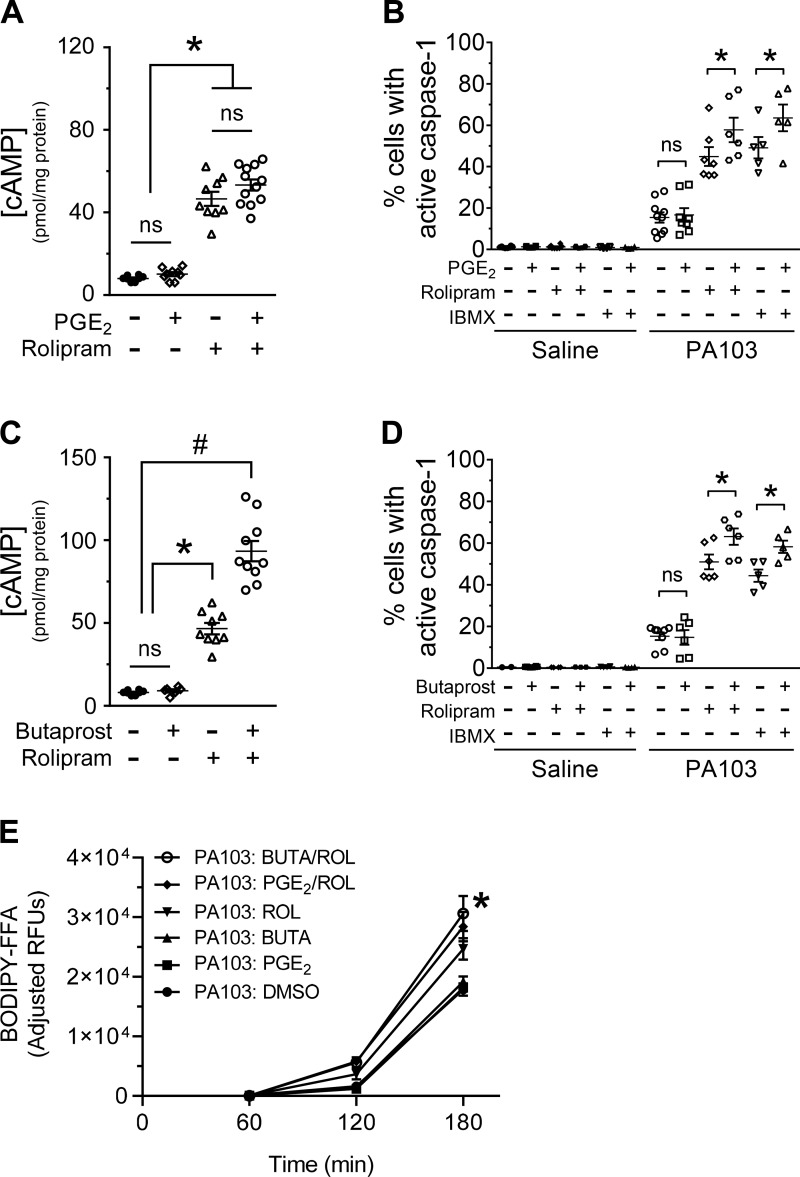
To better approximate the cAMP signals generated during sepsis, we next used PGE2 and the prostaglandin receptor for prostaglandin E2 (EP2) agonist butaprost to elevate cAMP levels through Gαs-mediated AC activation (3). Compounds were added before inoculation and maintained throughout the experiment as described in the figure legend. In the absence of infection, PGE2 did not produce elevation of intracellular cAMP (Fig. 3A) and did not endogenously activate caspase-1 (Fig. 3B, saline). Consistent with the lack of intracellular cAMP elevation, addition of PGE2 to PMVECs did not increase the number of caspase-1-active PMVECs elicited during PA103 infection (Fig. 3B, PA103). Identical trends were observed using the EP2 receptor-specific agonist butaprost (Fig. 3, C and D). These data raised the prospect that a minimal threshold of intracellular cAMP is necessary to prime PMVECs to activate caspase-1 during infection. Thus we combined PGE2 or butaprost with rolipram to inhibit cAMP degradation. Intriguingly, compared with rolipram alone, the combination of PGE2 and rolipram did not further increase intracellular cAMP (Fig. 3A), but the combination treatment significantly increased the number of caspase-1-active PMVECs elicited during infection (Fig. 3B). The combination of butaprost and rolipram significantly increased intracellular cAMP levels (Fig. 3C) and significantly increased the number of caspase-1-active PMVECs elicited during infection (Fig. 3D). To further verify these observations, we showed that combining PGE2 or butaprost with the pan-PDE inhibitor IBMX also significantly increased the number of caspase-1-active PMVECs elicited during infection (Fig. 3, B and D). Moreover, we showed that the EP2 receptor agonist iloprost had no effect on the number of PMVECs with activated caspase-1 during P. aeruginosa infection (data not shown). We next assayed for the effects of PGE2 and butaprost on the host-pathogen interaction using the PED6 PLA2 assay (described in Fig. 2D). Intriguingly, the same combinations that increased the number of caspase-1-active PMVECs elicited during infection also increased FFA release (Fig. 3E).
Fig. 3.
Addition of PGE2 and rolipram or butaprost and rolipram increases the number of pulmonary microvascular endothelial cells (PMVECs) with active caspase-1 during PA103 infection. A: PMVECs were serum starved for 60 min. The cAMP-elevating compounds were added to the culture medium 20 min before harvesting for measurement of global cAMP by ELISA. For all conditions, n = 3–8. *Significant difference between designated groups, P < 0.0001 (one-way ANOVA with Bonferroni’s post hoc analysis). B: PMVECs were serum starved for 60 min, at which time the cAMP-elevating compounds, the fluorescent-labeled inhibitor of caspase-1 (FLICA) reagent, and PA103 [multiplicity of infection (MOI) 5:1] were added to the culture medium. Saline-treated PMVEC monolayers served as negative controls. PMVECs were harvested at 180 min after infection and analyzed for activated caspase-1 by flow cytometry. For all conditions, n = 3–5. *Statistical significance between designated groups, P < 0.0001 (interaction among groups), P < 0.0001 (between different infection groups), P < 0.0001 (between different treatment groups) (two-way ANOVA with Tukey’s post hoc analysis). C: PMVECs were serum starved for 60 min. The cAMP-elevating compounds were added to the culture medium 20 min before harvesting for measurement of global cAMP by ELISA. For all conditions, n = 3–8. *Significant difference from saline; #significant difference from all other treatments, P < 0.0001 (one-way ANOVA with Bonferroni’s post hoc analysis). D: PMVECs were serum starved for 60 min, at which time the cAMP-elevating compounds, the FLICA reagent, and PA103 (MOI 5:1) were added to the culture medium. Saline-treated PMVEC monolayers served as negative controls. PMVECs were harvested at 180 min after infection and analyzed for activated caspase-1 by flow cytometry. For all conditions, n = 3–5. *Significant difference between designated groups, P < 0.0001 (interaction among groups), P < 0.0001 (between different infection groups), P < 0.0001 (between different treatment groups) (two-way ANOVA with Tukey’s post hoc analysis). E: PMVECs were serum starved for 60 min and simultaneously labeled by the addition of PED6. Cells were washed and then inoculated with saline or with PA103 at an MOI of 40:1. The cAMP-elevating compounds or its vehicle control (DMSO) were added to the culture medium 30 min before infection. Culture media was collected over time and assayed for release of BODIPY-labeled free fatty acid (FFA) as an indicator of PLA2 activity. Note that the basal rate of FFA release from saline-treated PMVECs was subtracted as a background correction at each time point (reported as adjusted relative fluorescent units, RFUs). For all conditions, n = 6. *Significant differences at the 3-h time point, P < 0.0001 (two-way ANOVA with Tukey’s post hoc analysis) between the following groups: PA103: DMSO versus PA103: PGE2/ROL, PA103: DMSO versus PA103: BUTA/ROL, and PA103: PGE2 versus PA103: BUTA/ROL. ROL, rolipram; BUTA, butaprost.
There is a specific window for cAMP priming of caspase-1 activation in PMVECs during P. aeruginosa infection.
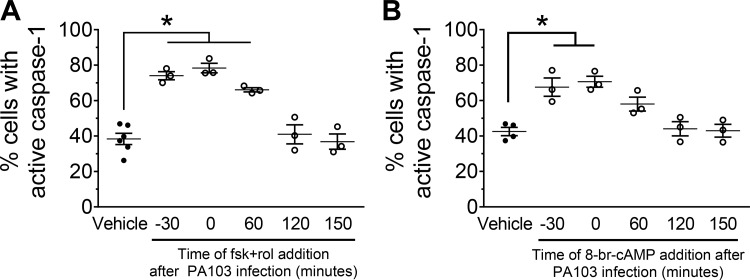
To assess whether temporal dynamics of cAMP elevation influence P. aeruginosa-induced caspase-1 activation, cAMP-elevating agents were added at different time points during PA103 infection. The combination of forskolin and rolipram treatment, or 8-br-cAMP, only increased the number of caspase-1-active PMVECs when added before or during the first hour of PA103 infection (Fig. 4, A and B). Collectively, these data indicate that the temporal dynamics of elevated cAMP signals (i.e., time of cAMP elevation related to time of initial infection) influence P. aeruginosa-induced caspase-1 activation in PMVECs.
Fig. 4.
cAMP-dependent priming of caspase-1 activation in pulmonary microvascular endothelial cells (PMVECs) occurs before or early during the onset of PA103 infection. A: PMVECs were serum starved for 60 min before infection. The cAMP-elevating compounds were added to the culture medium at the time points indicated in the figure, where t = 0 is the time of infection [PA103, multiplicity of infection (MOI) 40:1] and fluorescent-labeled inhibitor of caspase-1 (FLICA) reagent addition. Saline-treated PMVEC monolayers served as a negative control. PMVECs were harvested at 180 min after infection and analyzed for activated caspase-1 by flow cytometry. For all conditions, n = 3. *Significant difference compared with saline, P < 0.0001 (one-way ANOVA with Bonferroni’s post hoc analysis). B: PMVECs were prepared and analyzed as described in A. For all conditions, n = 3. *Significant difference compared with vehicle, P = 0.0003 (one-way ANOVA with Bonferroni’s post hoc analysis).
Inhibiting adenylyl cyclase activity reduces the number of PMVECs that activate caspase-1 during P. aeruginosa infection.
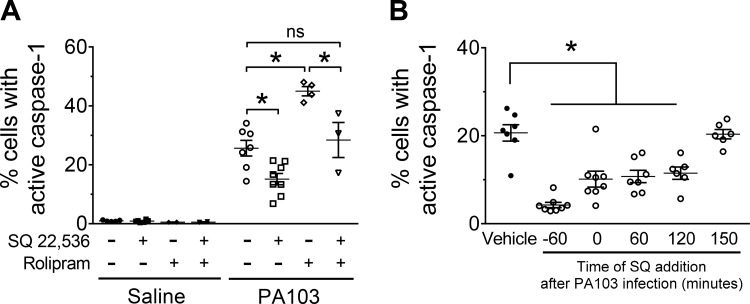
It is well established that basal cAMP signaling in PMVECs is a balance between AC-mediated cAMP synthesis and PDE4-mediated cAMP degradation (8). To assess the effect of basal cAMP signaling on P. aeruginosa-induced caspase-1 activation in PMVECs, we tested the AC inhibitor SQ 22,536 to inhibit de novo cAMP synthesis (7). The SQ 22,536 compound was added before inoculation and maintained throughout the experiment as described in the figure legend. Inhibition of cAMP synthesis by SQ 22,536 treatment decreased the number of PMVECs with active caspase-1 during PA103 infection (Fig. 5A). Moreover, simultaneous treatment with SQ 22,536 and rolipram ameliorated the number of PMVECs that activated caspase-1 during PA103 infection compared with rolipram alone (Fig. 5A). Consistent with temporal dynamics of cAMP signaling shown in Fig. 4, the inhibitory effect of SQ 22,536 was only observed when added before or within the first two hours of infection (Fig. 5B). These data suggest that basal cAMP production primes PMVECs for caspase-1 activation during P. aeruginosa infection.
Fig. 5.
Inhibition of adenylyl cyclase (AC) prevents cAMP-dependent priming of caspase-1 activation in pulmonary microvascular endothelial cells (PMVECs) during PA103 infection. A: PMVECs were serum starved for 60 min, at which time the cAMP-modulating compounds, the fluorescent-labeled inhibitor of caspase-1 (FLICA) reagent, and PA103 [multiplicity of infection (MOI) 10:1] were added to the culture medium. Saline-treated PMVEC monolayers served as negative controls. PMVECs were harvested at 180 min after infection and analyzed for activated caspase-1 by flow cytometry. For all conditions, n = 3. *Significant difference between designated groups, P < 0.0001 (interaction among groups), P < 0.0001 (between different infection groups), P < 0.0001 (between different treatment groups) (two-way ANOVA with Tukey’s post hoc analysis). B: PMVECs were serum starved for 60 min before infection. The cAMP inhibitor was added to the culture medium at the time points indicated in the figure, where t = 0 is the time of infection (PA103, MOI 10:1) and FLICA reagent addition. Saline-treated PMVEC monolayers served as a negative control. PMVECs were harvested at 180 min after infection and analyzed for activated caspase-1 by flow cytometry. For all conditions, n = 3. *Significant difference from vehicle, P < 0.0001 (one-way ANOVA with Bonferroni’s post hoc analysis).SQ, SQ 22,536 compound.
DISCUSSION
In this study, we demonstrated heterogeneous responses of PMVECs to P. aeruginosa infection using a high-throughput, single cell assay. Specifically, we showed that PMVECs activate caspase-1 during PA103 infection in a dose- and time-dependent manner and that cAMP signaling primes PMVECs to activate caspase-1 via a mechanism that appears to render them more susceptible to intoxication by the T3SS. The PMVEC priming mechanism was also induced by sepsis-associated factors that elevate cytosolic cAMP. Intriguingly, inhibiting de novo cAMP production reduced the number of PMVECs with active caspase-1, indicating a requirement for endogenous cAMP signaling. Considering the recent discovery that caspase-1 activation in PMVECs governs responses to P. aeruginosa infection (1), our data suggest that cAMP signaling is a critical priming mechanism of caspase-1 signaling in PMVECs.
During sepsis, dysregulated host responses to infection increase circulating mediators that act as telecrine danger signals (26). Notable among these mediators is PGE2 (16) that selectively binds to the G protein-coupled receptors EP2 and EP4, eliciting activation of plasma membrane-bound ACs, resulting in cAMP synthesis. In PMVECs, the effects of PGE2-mediated signaling are balanced by cAMP synthesis via ACs and cAMP turnover via PDE4, which create cAMP gradients and/or thresholds within the cell that differentially affect cellular responses (25, 29). Our experiments using PGE2, the EP2 receptor agonist butaprost, and rolipram provided critical information regarding the cAMP signals required for priming of PMVECs to activate caspase-1 during infection. Consistent with the concept of cAMP signaling, PGE2 addition at a concentration that did not increase global cAMP levels measured in whole cell lysates did not change the number of PMVECs with active caspase-1. In contrast, addition of the PDE4 inhibitor rolipram increased global cAMP levels, which concomitantly increased the number of PMVECs with active caspase-1. In addition, we showed a temporal component to cAMP priming, whereby cAMP-elevating agents primed PMVEC caspase-1 activation only when given 30 min before or up to 60 min after infection. These data suggest that a minimal cAMP threshold exists within an activation window and defines the priming response required to increase the number of PMVECs with active caspase-1 during P. aeruginosa infection. The MOI and pharmacological agent concentrations used in our experimental model were chosen to elicit maximal responses (24). Future studies will be necessary to uncover the mechanism of cAMP thresholding that regulates caspase-1 activation.
P. aeruginosa flagellin, the ExlA exolysin, and the T3SS are known activators of the inflammasome/caspase-1 axis, and these P. aeruginosa virulence factors are sensed by the NLRC4/Ipaf inflammasome (2, 5, 9, 18, 31, 34). In addition, P. aeruginosa strains expressing the T3SS deliver exoenzyme effectors (Exo) into host cells, among which ExoU and ExoS are known to inhibit/delay the onset of inflammasome activation (1, 10, 11, 31). Our studies using a single cell caspase-1 assay and P. aeruginosa strain PA103 expressing ExoU suggest that the elevation of PMVEC cAMP signaling may partially overcome the inhibition/delay of caspase-1 activation mediated by ExoU. Indeed, inhibition of endogenous cAMP production decreased the number of PMVECs that activate caspase-1 during PA103 infection, indicating that cAMP signaling is necessary for priming. In addition, treatment of PMVECs with agonists that increase cAMP signaling rendered the PMVECs more susceptible to P. aeruginosa infection, as measured by increased activity of the ExoU PLA2. Together, these data suggest a novel regulatory connection between both the T3SS and/or ExoU PLA2 activity and host cAMP signaling that should be explored as part of future studies. Adding to this fascinating interplay between host and pathogen exoenzymes, some naturally occurring P. aeruginosa strains express ExoY, a promiscuous nucleotidyl cyclase known to elevate intracellular cGMP, cAMP, cUMP, and cCMP (20, 21). While wild-type PA103 does not naturally express ExoY, infection of PMVECs with recombinant PA103 strains engineered to ectopically express ExoY has been shown to primarily elevate cGMP over a time course similar to the one used in our infection studies. In addition, ExoY-mediated production of cAMP is not observed until after ~6 h after infection (20, 21). Considering that our data showed that cGMP had no effect on caspase-1 activation in PMVECs, it remains an outstanding question as to whether ExoU and ExoY activities produce antagonistic effects on caspase-1 activation during infection with P. aeruginosa strains naturally expressing both exoenzymes.
Elevated cAMP was previously associated with inhibition of the NLRP3 inflammasome and decreases caspase-1 activation in macrophages (15, 27). In contrast, our findings indicate that elevated cAMP promotes caspase-1 activation in discreet PMVEC populations. These paradoxical observations raise interesting questions regarding the nature and regulation of inflammasome/caspase-1 axis activation in different cell phenotypes in response to different stimuli. Experiments are ongoing to identify the nature of the inflammasome in PMVECs and how it is regulated by cAMP and whether these observations are generic to all Pseudomonas species. It will be important to determine whether cAMP and/or downstream effectors such as PKA directly affect inflammasome assembly/activation. It is also possible that cAMP and/or downstream effectors such as PKA elicit a heretofore unrecognized effect on the P. aeruginosa T3SS and/or its exoenzyme effectors.
Although paradoxical, the divergent effects of cAMP signaling on caspase-1 activation in immune versus lung endothelial cells ultimately result in salutary effects limiting hyperinflammation by decreasing interleukin release from immune cells, while strengthening the endothelial barrier and preventing edema development (1). In conclusion, we present a paradigm in which cAMP signaling impacts caspase-1-dependent lung endothelial signaling during sepsis to diminish dysregulated host responses and reestablish homeostasis.
GRANTS
This work was supported by National Institutes of Health National Heart, Lung, and Blood Institute Grant HL118334 to D.F.A. and J.P.A., HL118334-S1 to K.S.H., and HL066299 Core B to D.F.A. and T.S. This work was also supported by S10RR027535 (Nikon A1R spectral confocal microscopy system).
DISCLAIMERS
The funders had no role in study design, data collection, and interpretation or the decision to submit this work for publication.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
P.R., J.P.A., and D.F.A. conceived and designed research; P.R., K.S.H., N.H., G.D., N.A., A.B., and D.S. performed experiments; P.R., S.L., T.R., J.P.A., and D.F.A. analyzed data; P.R., S.L., T.R., J.P.A., and D.F.A. interpreted results of experiments; P.R., J.P.A., and D.F.A. prepared figures; P.R., J.P.A., and D.F.A. drafted manuscript; P.R., S.L., T.R., J.P.A., and D.F.A. edited and revised manuscript; P.R., K.S.H., N.H., N.A., A.B., D.S., S.L., T.R., J.P.A., and D.F.A. approved final version of manuscript.
ACKNOWLEDGMENTS
The authors thank Drs. Troy Stevens and Gabriel Nuñez for insightful recommendations during manuscript preparation.
Present address of D. F. Alvarez: Sam Houston State University, College of Osteopathic Medicine, 925 City Central Avenue, Conroe, TX 77304.
REFERENCES
- 1.Alvarez DF, Housley N, Koloteva A, Zhou C, O’Donnell K, Audia JP. Caspase-1 activation protects lung endothelial barrier function during infection-induced stress. Am J Respir Cell Mol Biol 55: 500–510, 2016. doi: 10.1165/rcmb.2015-0386OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basso P, Wallet P, Elsen S, Soleilhac E, Henry T, Faudry E, Attrée I. Multiple Pseudomonas species secrete exolysin-like toxins and provoke Caspase-1-dependent macrophage death. Environ Microbiol 19: 4045–4064, 2017. doi: 10.1111/1462-2920.13841. [DOI] [PubMed] [Google Scholar]
- 3.Birukova AA, Zagranichnaya T, Fu P, Alekseeva E, Chen W, Jacobson JR, Birukov KG. Prostaglandins PGE2 and PGI2 promote endothelial barrier enhancement via PKA- and Epac1/Rap1-dependent Rac activation. Exp Cell Res 313: 2504–2520, 2007. doi: 10.1016/j.yexcr.2007.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broz P, Monack DM. Measuring inflammasome activation in response to bacterial infection. Methods Mol Biol 1040: 65–84, 2013. doi: 10.1007/978-1-62703-523-1_6. [DOI] [PubMed] [Google Scholar]
- 5.Cohen TS, Prince AS. Activation of inflammasome signaling mediates pathology of acute P. aeruginosa pneumonia. J Clin Invest 123: 1630–1637, 2013. doi: 10.1172/JCI66142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823, 2013. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fabbri E, Brighenti L, Ottolenghi C. Inhibition of adenylate cyclase of catfish and rat hepatocyte membranes by 9-(tetrahydro-2-furyl)adenine (SQ 22536). J Enzyme Inhib 5: 87–98, 1991. doi: 10.3109/14756369109069062. [DOI] [PubMed] [Google Scholar]
- 8.Feinstein WP, Zhu B, Leavesley SJ, Sayner SL, Rich TC. Assessment of cellular mechanisms contributing to cAMP compartmentalization in pulmonary microvascular endothelial cells. Am J Physiol Cell Physiol 302: C839–C852, 2012. doi: 10.1152/ajpcell.00361.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franchi L, Stoolman J, Kanneganti TD, Verma A, Ramphal R, Núñez G. Critical role for Ipaf in Pseudomonas aeruginosa-induced caspase-1 activation. Eur J Immunol 37: 3030–3039, 2007. doi: 10.1002/eji.200737532. [DOI] [PubMed] [Google Scholar]
- 10.Galle M, Jin S, Bogaert P, Haegman M, Vandenabeele P, Beyaert R. The Pseudomonas aeruginosa type III secretion system has an exotoxin S/T/Y independent pathogenic role during acute lung infection. PLoS One 7: e41547, 2012. doi: 10.1371/journal.pone.0041547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galle M, Schotte P, Haegman M, Wullaert A, Yang HJ, Jin S, Beyaert R. The Pseudomonas aeruginosa Type III secretion system plays a dual role in the regulation of caspase-1 mediated IL-1beta maturation. J Cell Mol Med 12, 5A: 1767–1776, 2008. doi: 10.1111/j.1582-4934.2007.00190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gross O, Thomas CJ, Guarda G, Tschopp J. The inflammasome: an integrated view. Immunol Rev 243: 136–151, 2011. doi: 10.1111/j.1600-065X.2011.01046.x. [DOI] [PubMed] [Google Scholar]
- 13.He WT, Wan H, Hu L, Chen P, Wang X, Huang Z, Yang ZH, Zhong CQ, Han J. Gasdermin D is an executor of pyroptosis and required for interleukin-1β secretion. Cell Res 25: 1285–1298, 2015. doi: 10.1038/cr.2015.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He Y, Hara H, Núñez G. Mechanism and regulation of NLRP3 inflammasome activation. Trends Biochem Sci 41: 1012–1021, 2016. doi: 10.1016/j.tibs.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee GS, Subramanian N, Kim AI, Aksentijevich I, Goldbach-Mansky R, Sacks DB, Germain RN, Kastner DL, Chae JJ. The calcium-sensing receptor regulates the NLRP3 inflammasome through Ca2+ and cAMP. Nature 492: 123–127, 2012. doi: 10.1038/nature11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee S, Nakahira K, Dalli J, Siempos II, Norris PC, Colas RA, Moon JS, Shinohara M, Hisata S, Howrylak JA, Suh GY, Ryter SW, Serhan CN, Choi AMK. NLRP3 inflammasome deficiency protects against microbial sepsis via increased lipoxin B4 synthesis. Am J Respir Crit Care Med 196: 713–726, 2017. doi: 10.1164/rccm.201604-0892OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindsey AS, Sullivan LM, Housley NA, Koloteva A, King JA, Audia JP, Alvarez DF. Analysis of pulmonary vascular injury and repair during Pseudomonas aeruginosa infection-induced pneumonia and acute respiratory distress syndrome. Pulm Circ 9: 2045894019826941, 2019. doi: 10.1177/2045894019826941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miao EA, Ernst RK, Dors M, Mao DP, Aderem A. Pseudomonas aeruginosa activates caspase 1 through Ipaf. Proc Natl Acad Sci USA 105: 2562–2567, 2008. doi: 10.1073/pnas.0712183105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitaka C, Hirata Y, Nagura T, Sakanishi N, Tsunoda Y, Amaha K. Plasma alpha-human atrial natriuretic peptide concentration in patients with acute lung injury. Am Rev Respir Dis 146: 43–46, 1992. doi: 10.1164/ajrccm/146.1.43. [DOI] [PubMed] [Google Scholar]
- 20.Morrow KA, Frank DW, Balczon R, Stevens T. The Pseudomonas aeruginosa exoenzyme Y: a promiscuous nucleotidyl cyclase edema factor and virulence determinant. Handb Exp Pharmacol 238: 67–85, 2017. doi: 10.1007/164_2016_5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morrow KA, Seifert R, Kaever V, Britain AL, Sayner SL, Ochoa CD, Cioffi EA, Frank DW, Rich TC, Stevens T. Heterogeneity of pulmonary endothelial cyclic nucleotide response to Pseudomonas aeruginosa ExoY infection. Am J Physiol Lung Cell Mol Physiol 309: L1199–L1207, 2015. doi: 10.1152/ajplung.00165.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muñoz-Planillo R, Kuffa P, Martínez-Colón G, Smith BL, Rajendiran TM, Núñez G. K+ efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity 38: 1142–1153, 2013. doi: 10.1016/j.immuni.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8: 2281–2308, 2013. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sayner SL, Alexeyev M, Dessauer CW, Stevens T. Soluble adenylyl cyclase reveals the significance of cAMP compartmentation on pulmonary microvascular endothelial cell barrier. Circ Res 98: 675–681, 2006. doi: 10.1161/01.RES.0000209516.84815.3e. [DOI] [PubMed] [Google Scholar]
- 25.Sayner SL, Frank DW, King J, Chen H, VandeWaa J, Stevens T. Paradoxical cAMP-induced lung endothelial hyperpermeability revealed by Pseudomonas aeruginosa ExoY. Circ Res 95: 196–203, 2004. doi: 10.1161/01.RES.0000134922.25721.d9. [DOI] [PubMed] [Google Scholar]
- 26.Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche JD, Coopersmith CM, Hotchkiss RS, Levy MM, Marshall JC, Martin GS, Opal SM, Rubenfeld GD, van der Poll T, Vincent JL, Angus DC. The third international consensus definitions for sepsis and septic shock (sepsis-3). JAMA 315: 801–810, 2016. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sokolowska M, Chen LY, Liu Y, Martinez-Anton A, Qi HY, Logun C, Alsaaty S, Park YH, Kastner DL, Chae JJ, Shelhamer JH. Prostaglandin E2 inhibits NLRP3 inflammasome activation through EP4 receptor and intracellular cyclic AMP in human macrophages. J Immunol 194: 5472–5487, 2015. doi: 10.4049/jimmunol.1401343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spindler V, Waschke J. β-adrenergic stimulation contributes to maintenance of endothelial barrier functions under baseline conditions. Microcirculation 18: 118–127, 2011. doi: 10.1111/j.1549-8719.2010.00072.x. [DOI] [PubMed] [Google Scholar]
- 29.Stevens T, Creighton J, Thompson WJ. Control of cAMP in lung endothelial cell phenotypes. Implications for control of barrier function. Am J Physiol Lung Cell Mol Physiol 277: L119–L126, 1999. doi: 10.1152/ajplung.1999.277.1.L119. [DOI] [PubMed] [Google Scholar]
- 30.Stevens T, Nakahashi Y, Cornfield DN, McMurtry IF, Cooper DM, Rodman DM. Ca2+-inhibitable adenylyl cyclase modulates pulmonary artery endothelial cell cAMP content and barrier function. Proc Natl Acad Sci USA 92: 2696–2700, 1995. doi: 10.1073/pnas.92.7.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sutterwala FS, Mijares LA, Li L, Ogura Y, Kazmierczak BI, Flavell RA. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J Exp Med 204: 3235–3245, 2007. doi: 10.1084/jem.20071239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang WJ, Guo Q. The adenylyl cyclase activity of anthrax edema factor. Mol Aspects Med 30: 423–430, 2009. doi: 10.1016/j.mam.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu C, Zhang Y, Yao S, Wei Y. A PCR based protocol for detecting indel mutations induced by TALENs and CRISPR/Cas9 in zebrafish. PLoS One 9: e98282, 2014. doi: 10.1371/journal.pone.0098282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao Y, Yang J, Shi J, Gong YN, Lu Q, Xu H, Liu L, Shao F. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature 477: 596–600, 2011. doi: 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]