Abstract
In the age of a pandemic, such as the ongoing one caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), the world faces a limited supply of tests, personal protective equipment, and factories and supply chains are struggling to meet the growing demands. This study aimed to evaluate the efficacy of specimen pooling for testing of SARS‐CoV‐2 virus, to determine whether costs and resource savings could be achieved without impacting the sensitivity of the testing. Ten previously tested nasopharyngeal and throat swab specimens by real‐time polymerase chain reaction (PCR), were pooled for testing, containing either one or two known positive specimens of varying viral concentrations. Specimen pooling did not affect the sensitivity of detecting SARS‐CoV‐2 when the PCR cycle threshold (Ct) of original specimen was lower than 35. In specimens with low viral load (Ct > 35), 2 of 15 pools (13.3%) were false negative. Pooling specimens to test for Coronavirus Disease 2019 infection in low prevalence (≤1%) areas or in low risk populations can dramatically decrease the resource burden on laboratory operations by up to 80%. This paves the way for large‐scale population screening, allowing for assured policy decisions by governmental bodies to ease lockdown restrictions in areas with a low incidence of infection, or with lower‐risk populations.
Keywords: cost efficiency, COVID‐19, PCR, real‐time PCR, SARS‐CoV‐2, specimen pooling
Highlights
Specimen pooling did not affect the sensitivity of detecting SARS‐CoV.
Pooling specimens to test for COVID‐19 infection can dramatically decrease the resource burden on laboratory operations.
Specimen pooling in samples whose cycle threshold (Ct) value is greater than 35 may yield false‐negative results.
Pooling specimens is especially useful for large‐scale population screening.
1. INTRODUCTION
The ongoing Coronavirus Disease 2019 (COVID‐19) pandemic has highlighted the need for early diagnosis of emerging infectious diseases to better contain an outbreak. Testing for severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), the virus that causes COVID‐19, has been limited due to the considerable strain on global supply chains for reagents, personal protective equipment, and other consumables. 1 , 2 To date, countries that are able to screen patients swiftly have fared better in containing the COVID‐19 outbreak and suppressing the mortality rate associated with the disease. 3 The rapid diagnosis of COVID‐19 in both symptomatic and asymptomatic patients can shed light on transmission patterns and facilitate contact tracing. 2 , 3 Large scale population screening for COVID‐19 infection is generally considered a necessary part of an exit strategy from the coronavirus lockdown.
Specimen pooling is a method of screening a large number of patients for an infection and typically involves combining multiple patient specimens into a single test sample, then testing multiple such samples. This approach has the advantage of cost‐effectiveness and speed, and was used to retrospectively screen for COVID‐19 in specimens that were negative for common respiratory viruses earlier in the course of the pandemic in the United States. 4 Specimen pooling has also been used in screening efforts for several other infectious diseases, including donated blood samples for HIV. 5 , 6 , 7 , 8
Pooling nasopharyngeal and throat swab (NT) specimens would be more economical than individually testing all specimens from low‐risk populations, particularly in limited‐resource settings. 9 It is unclear how pooling biological samples would affect the sensitivity and the false‐negative rate of polymerase chain reaction (PCR) assays. The current study compares laboratory results from pooled testing (10 samples) with individually tested samples using the standard real‐time quantitative PCR (qPCR) to ensure that detection accuracy is not compromised. Additionally, NT specimens with PCR cycle threshold (Ct) greater than 35 were pooled to determine the limit of detection and sensitivity of pooling samples to test for SARS‐CoV‐2.
2. MATERIALS AND METHODS
This study is an evaluation of laboratory techniques using archived clinical specimens and was exempted from Chulalongkorn University Institutional Review Board review. NT specimens used in this study had been collected from patients under investigation (PUI) for COVID‐19 infection at King Chulalongkorn Memorial Hospital, placed in 2.0 mL viral transport media (VTM) and sent to the Thai Red Cross Emerging Infectious Diseases Health Science Centre Laboratory for testing between 1 February and 31 March 2020. All specimens had been stored at −80°C. A total of 50 leftover specimens that had tested negative for SARS‐CoV‐2 by qPCR amplifying the ORF1ab gene (BGI, Shenzhen, China) were combined into a single sample, which was then used as the negative portion of all pooling tests. The purpose of homogenized negative pooled specimen was to standardize and eliminate the possibility of variations between pools, which could have potentially affected our results. This pooled negative NT‐VTM was retested for SARS‐CoV‐2 using qPCR to confirm the negative result before pooling with selected positive specimens.
This study used the Boom method's magnetic extraction‐based assay (NucliSens, easyMag, bioMérieux, Marcy‐l’Étoile, France) to extract DNA and RNA, which allows a maximum specimen volume of 1.0 mL. 10 By using magnetic beads to capture DNA and RNA during the extraction step, pooling 10 specimens of 0.1 mL each (total of 1.0 mL extraction sample) can result in the same extraction capability as 0.1 mL if the elution volume at the end is equal and there is no PCR interference from the specimen such as lipid, protein or cell debris.
Two pooling ratios were evaluated in this study, termed 1X and 2X. In the 1X ratio, 0.1 mL of NT‐VTM from one SARS‐CoV‐2 positive specimen was combined with 0.9 mL pooled negative NT‐VTM, thus modeling a 10% infection rate. Correspondingly, in the 2X ratio, 0.1 mL of NT‐VTM each from two SARS‐CoV‐2 positive specimens were pooled with 0.8 mL pooled negative NT‐VTM, thus modeling a 20% infection rate (see Figure 1). All pooled samples (1.0 mL each) were added to 2.0 mL of lysis buffer (total 3.0 mL) and processed for nucleic acid extraction using the NucliSens easyMAG instrument (bioMérieux). In addition, 0.1 mL of the same positive specimens that were used in the pooled samples were retested individually for sensitivity comparison using a separate extraction system (EZ1, Qiagen, Hilden, Germany). Real‐time PCR (qPCR) for detection of SARS‐CoV‐2 was performed using a commercial kit that targets the ORF1ab gene as per the manufacturer's protocol (BGI, Shenzhen, China). The protocol's stated limit of detection of ORF1ab real‐time PCR was 100 copies/mL and the cutoff PCR cycle threshold (Ct) was 38.
Figure 1.
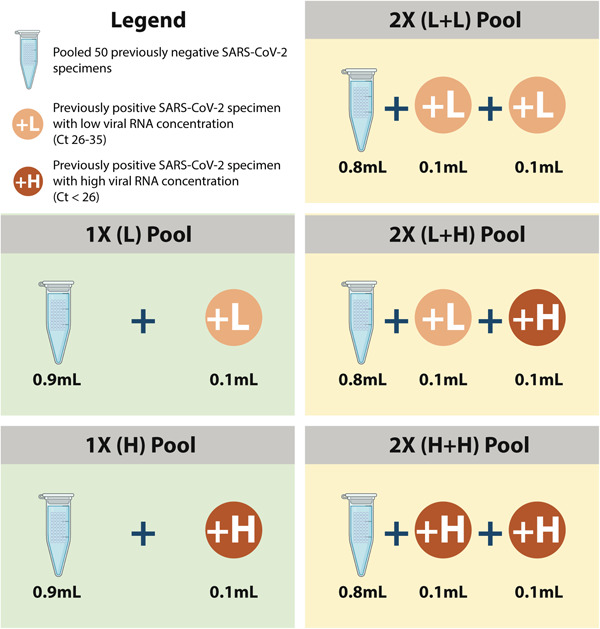
Illustrates the experimental design of the pooling strategies tested in this study
Previously positive specimens with high and low‐concentrations of RNA, as determined by PCR Ct values at the time of detection, were selected to determine the effect of viral load on pooling to ensure that the sensitivity and accuracy of the assay were maintained (Table 1). Low Ct values indicate the presence of higher amounts of viral RNA and vice versa. In this study, specimens with Ct values between 26 and 35 were considered to have low concentrations of viral RNA, while those with Ct values lower than 26 were considered to have of high‐concentrations viral RNA. Ct values higher than 35 were considered weakly positive. As per the laboratory's protocol, samples that test weakly positive are retested for confirmation. The experimental design is summarized in Figure 1.
Table 1.
Comparison of specimen pooling and individual testing of nasopharyngeal and throat swab specimen using qPCR threshold cycles from SARS‐CoV‐2 testing
| No. | Pooling patterna | PCR results (Ct) | Ct difference (pooled – individual testingb) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Pooled testing | Individual testing | ||||||||
| Replicate I | Replicate II | Avg Ct | Positive NT 1 | Avg Ct of positive NT 1 | Positive NT 2 | ||||
| Replicate I | Replicate II | ||||||||
| 1 | 1X(L) | 31.63 | na | … | 32.01 | na | … | na | −0.38 |
| 2 | 1X(L) | 33.47 | na | … | 31.81 | na | … | na | +1.66 |
| 3 | 1X(L) | 34.00 | na | … | 33.98 | na | … | na | +0.02 |
| 4 | 1X(L) | 33.06 | na | … | 33.90 | na | … | na | −0.84 |
| 5 | 1X(L) | 34.24 | na | … | 33.50 | na | … | na | +0.74 |
| 6 | 1X(L) | 34.66 | na | … | 34.86 | na | … | na | −0.2 |
| 7 | 1X(L) | 32.47 | na | … | 32.51 | na | … | na | −0.04 |
| 8 | 1X(L) | 33.10 | na | … | 32.99 | na | … | na | +0.11 |
| 9 | 1X(L) | 29.91 | na | … | 29.97 | na | … | na | −0.06 |
| 10 | 1X(L) | 33.48 | na | … | 33.84 | na | … | na | −0.36 |
| 11 | 1X(L) | 33.66 | na | … | 33.73 | na | … | na | −0.07 |
| 12 | 1X(L) | 27.37 | na | … | 27.90 | na | … | na | −0.53 |
| 13 | 1X(L > 35) | 35.22 | 35.93 | 35.58 | 35.49 | 35.23 | 35.36 | na | +0.22 |
| 14 | 1X(L > 35) | 37.07 | 36.48 | 36.78 | 35.48 | 36.80 | 36.14 | na | +0.63 |
| 15 | 1X(L > 35) | 36.48 | Negative | 36.48 | 36.15 | 36.14 | 36.15 | na | +0.34 (One false negative in pooled testing) |
| 16 | 1X(L > 35) | 36.57 | Negative | 36.57 | 36.57 | 36.45 | 36.51 | na | +0.06 (One false negative in pooled testing) |
| 17 | 1X(L > 35) | 36.40 | 35.09 | 35.75 | 35.81 | 36.38 | 36.10 | na | −0.35 |
| 18 | 1X(L > 35) | 37.43 | Negative | 37.43 | 36.68 | 36.83 | 36.76 | na | +0.68 (One false negative in pooled testing) |
| 19 | 1X(L > 35) | 36.96 | 35.85 | 36.41 | 37.00 | 35.83 | 36.42 | na | −0.01 |
| 20 | 1X(L > 35) | 35.35 | 36.56 | 35.96 | 35.60 | 36.41 | 36.01 | na | −0.05 |
| 21 | 1X(L > 35) | 35.99 | 35.50 | 35.75 | 35.16 | 35.17 | 35.17 | na | +0.58 |
| 22 | 1X(L > 35) | Negative | Negative | Negative | 35.30 | Negative | 35.30 | na | … |
| One false negative in individual testing and one false negative in pooled testing | |||||||||
| 23 | 1X(L > 35) | Negative | Negative | Negative | 37.10 | Negative | 37.10 | na | … |
| One false negative in individual testing and one false negative in pooled testing | |||||||||
| 24 | 1X(L > 35) | 36.56 | Negative | 36.56 | 36.87 | 36.79 | 36.83 | na | −0.27 (One false negative in pooled testing) |
| 25 | 1X(L > 35) | 37.00 | 35.63 | 36.32 | 36.40 | 36.34 | 36.37 | na | −0.06 |
| 26 | 1X(L > 35) | 34.65 | 34.95 | 34.80 | 35.27 | 35.50 | 35.39 | na | −0.59 |
| 27 | 1X(L > 35) | 37.00 | 37.11 | 37.06 | 36.91 | Negative | 36.91 | na | +0.15 (One false negative in individual testing) |
| 28 | 1X(H) | 22.40 | na | … | 23.76 | na | … | na | −1.36 |
| 29 | 1X(H) | 19.22 | na | … | 18.00 | na | … | na | +1.22 |
| 30 | 1X(H) | 23.76 | na | … | 23.69 | na | … | na | +0.07 |
| 31 | 1X(H) | 23.87 | na | … | 23.57 | na | … | na | +0.30 |
| 32 | 2X(L+L) | 31.84 | na | … | 31.73 | na | … | 33.57 | +0.11 |
| 33 | 2X(L+L) | 29.82 | na | … | 29.26 | na | … | 35.48 | +0.56 |
| 34 | 2X(L+L) | 31.67 | na | … | 31.32 | na | … | 35.52 | +0.35 |
| 35 | 2X(L+L) | 34.73 | na | … | 33.98 | na | … | 35.52 | +0.75 |
| 36 | 2X(L+L) | 35.75 | na | … | 34.16 | na | … | 35.49 | +1.59 |
| 37 | 2X(H+H) | 13.04 | na | … | 12.91 | na | … | 25.65 | +0.13 |
| 38 | 2X(H+H) | 15.02 | na | … | 15.34 | na | … | 23.57 | −0.32 |
| 39 | 2X(H+H) | 19.83 | na | … | 18.32 | na | … | 22.95 | +1.51 |
| 40 | 2X(H+H) | 18.26 | na | … | 19.06 | na | … | 22.01 | −0.8 |
| 41 | 2X(H+H) | 20.19 | na | … | 21.91 | na | … | 24.17 | −1.72 |
| 42 | 2X(H+L) | 21.99 | na | … | 23.44 | na | … | 33.41 | −1.45 |
| 43 | 2X(H+L) | 18.70 | na | … | 18.47 | na | … | 29.38 | +0.23 |
| 44 | 2X(H+L) | 20.36 | na | … | 20.33 | na | … | 33.31 | +0.03 |
| 45 | 2X(H+L) | 24.07 | na | … | 23.69 | na | … | 27.21 | +0.38 |
| 46 | 2X(H+L) | 24.32 | na | … | 23.57 | na | … | 31.27 | +0.75 |
| 47 | 2X(H+L) | 18.43 | na | … | 18.32 | na | … | 26.51 | +0.11 |
| 48 | 2X(H+L) | 20.87 | na | … | 19.06 | na | … | 29.09 | +1.81 |
| 49 | 2X(H+L) | 24.10 | na | … | 24.17 | na | … | 27.90 | −0.07 |
Note: Avg, Average; Ct, PCR cycle threshold (lower values = higher viral load); na, not available; Positive NT, nasopharyngeal and throat swab specimen positive for SARS‐CoV‐2; 1 X, one positive specimen in pool of 10; 2 X , two positive specimens in pool of 10; L, low‐concentration of viral RNA (PCR Ct between 26‐35); L > 35, Low‐concentration of viral RNA (PCR Ct>35); H, high‐concentration of viral RNA (PCR Ct < 26)
Abbreviations: qPCR, quantitative polymerase chain reaction; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
See Figure 1.
Negative and positive values of Ct indicate higher and lower sensitivity of pooling, respectively. Positive NT 1 (lower Ct) was used to calculate Ct difference in 2X ratio.
Forty‐nine PCR positive NT specimens yielding Ct ranging from 12.91 to 37.10 were selected for the study in five pooling ratios (Figure 1). Thirty‐one of these had a 1X pooling ratio and 18 had 2X ratios. Among the 1X ratio, 12 had low viral concentrations, (L, Ct values from 27.90 to 34.86), 15 had weakly positive viral concentrations (L > 35, Ct values from 35.23 to 37.10), and 4 had high viral concentrations (H, Ct values from 18.00 to 23.76).
The fifteen 1X(L > 35) pools were tested by performing duplicate (replicates I and II) qPCR assays to determine the limit of detection of specimen pooling when compared to individual testing. The 2X ratio pools had two positive specimens each (Positive NT 1 and 2 in Table 1), with viral concentrations as follows: five pools had two low‐concentration specimens (L+L, Ct values from 29.82 to 35.52), five pools had two high concentration specimens (H+H, Ct values from 12.91 to 25.56), and eight pools had one high and one low concentration specimens (H+L, Ct values from 18.47 to 33.41).
The sensitivity of viral RNA detection for each pool was compared with the sensitivity of qPCR results for the individually tested positive specimen in that pool. For 2X ratio pools, the positive specimen with the lower Ct value (Positive NT 1), when individually tested, was used for comparison.
3. RESULTS
All 1X ratio pools (Ct < 35) were positive, with Ct value difference within a range of −1.36 to +1.66 when compared to individual (non‐pooled) testing. All 2X ratio pools were positive, with Ct value difference within a range of −1.72 to +1.81 when compared to individual testing (Table 1). Statistical paired t test was calculated to compare the Ct value differences between pooled (including all patterns in Figure 1) and individual tests. The result showed no significant difference in all comparisons including individual vs 1X L ratio pool (P = .853), or individual vs 1X H ratio pool (P = 0.921). The 2X pooling ratio showed similar results. There were no significant difference between the Ct values of individual testing vs ratio pools 2X L+L, 2X H+L, or 2X H+H (P = .063, .507, and .6766, respectively). Thus, sensitivity was not affected by pooling specimens, while accuracy was maintained.
In pooled testing of 1X L > 35, 13 of 15 of either replicate pools tested positive for SARS‐CoV‐2. Of the 13 positive pools, 4 pools had only 1 replicate that tested positive. The two false‐negative pooled samples tested positive in only 1 replicate when individually tested (numbers 22 and 23, with Ct 35.3 and 37.10, respectively, Table 1). During the individual testing, 3 samples (out of 15) had 1 undetected result.
Cost‐effectiveness of the pooling strategy was calculated, based on varying disease prevalence rates (0.1‐10%) (Table 2). Pooling appears most cost‐effective when testing among populations with lower COVID‐19 prevalence. Estimated laboratory costs were reduced from $35 per patient to $3.85, $6.85, $17.54, and $26.30 at prevalences of COVID‐19 in the tested population of 0.1%, 1%, 5%, and 10%, respectively. By this estimation, pooled‐specimen testing of 10 00 000 subjects in a population with 1% COVID‐19 prevalence would save approximately $28.15 million, assuming evenly distributed positive specimens in each pool (Table 2).
Table 2.
Cost comparison for specimen pooling using real‐time polymerase chain reaction at four different prevalence rates
| Total population | 10 00 000 samples | |||
|---|---|---|---|---|
| % infection | 0.10% | 1.00% | 5.00% | 10.00% |
| % of the noninfected samples | 99.90% | 99.00% | 95.00% | 90.00% |
| Number of samples per pool | 10 | 10 | 10 | 10 |
| Total number of pool | 10 0000 | 10 0000 | 10 0000 | 10 0000 |
| % of pool with no infectiona | 99.00% | 90.44% | 59.87% | 34.87% |
| Total number of pool without an infection | 99 004 | 90 438 | 59 874 | 34 868 |
| Total number of pool with an infection | 996 | 9562 | 40 126 | 65 132 |
| Number of samples that need to be tested individually after pooled qPCR | 9955 | 95 618 | 40 1263 | 65 1322 |
| Total number of tests that need to be performed | 10 9955 | 19 5618 | 50 1263 | 75 1322 |
| Cost per test (USD) | $35.00 | $35.00 | $35.00 | $35.00 |
| Total cost of individual testing | $35 000 000.00 | $35 000 000.00 | $35 000 000.00 | $35 000 000.00 |
| Total cost of specimen pooling | $38 48 429.19 | $68 46 627.37 | $17 544 207.13 | $26 296 254.60 |
| % discount | 89.00% | 80.44% | 49.87% | 24.87% |
| Cost per patient | $3.85 | $6.85 | $17.54 | $26.30 |
Abbreviation: qPCR, quantitative polymerase chain reaction.
% of pool with no infection = (% of noninfected samples in one pool)^number of samples per pools.
4. DISCUSSION
This study demonstrates that specimen pooling (either 1X or 2X pooling ratios) does not compromise the sensitivity of detecting SARS‐CoV‐2 provided the Ct value of the individually tested sample is lower than 35. In 2X ratio pooling, qPCR testing detected higher viral concentrations (lower PCR Ct) compared to those of the corresponding positive specimens when tested individually. This suggests that specimen pooling did not lower the sensitivity of PCR testing but actually increased the viral concentration when more than one positive sample was present in the same pool which combined the viral amount from 2 samples in the same extraction tube.
Inconsistencies were noted in the Ct values between the two PCR runs (duplicate testing) in either pooled or individually tested specimens of Ct> 35. Two of 15 (13.3% false‐negative rate) pools were negative for both replicates and 4 pools were negative in one replicate. PCR testing of COVID‐19 patients with low viral load (Ct > 35) may yield false‐negative results when the pooling ratio is 1X. This result is similar to the study from Spain where false positives were found in samples with Ct values greater than 35.7 and 35.8 for RdRp and E gene PCR, respectively. 11
It was previously demonstrated that when the prevalence of COVID‐19 is 1%, the optimal specimen pool size is 11 with an overall increase in testing efficiency calculated at 400%. 9 In this study, a 10‐specimen pool size (0.1 mL each specimen) was chosen based on the capacity of the RNA extraction system in the laboratory where this study was performed, and the result was similar to five samples pooling. 9 , 11 The capability of the extraction protocol can affect the sensitivity of pooled testing. In this study, the maximum volume of specimen for extraction was 1.0 mL (0.1 mL × 10 samples). The sensitivity of the assay can be improved if ratio of one to five (0.2 mL of each specimen) is used. It can also be improved by collecting specimens directly in 1.0 mL of lysis buffer (extraction buffer), where maximum of 0.3 mL of 10 samples can be pooled (NucliSens easyMAG or miniMAG, bioMérieux), instead of 2.0 mL of VTM. The nucleic acid can therefore be directly extracted without diluting with VTM. Additionally, the lysis buffer inactivates the virus, making it safer to handle. Further, similar PCR Ct values (within ± 2; statistically not significant) between pooled and individually tested specimens indicated there was no interference of PCR inhibitor from 1.0 mL pooled specimens in one extraction tube.
Beyond maintaining accuracy, specimen pooling will almost certainly reduce cost. For example, if 1% of the population is infected, pooling 10 specimens can reduce the cost of laboratory operation by about 80% (Table 2). However, in the case of 10% prevalence, specimen pooling will only save 24.87%, as positive pooled samples will need to be individually tested. Therefore, pooling samples is especially useful in areas with low prevalence rates, or when conducting proactive surveillance in areas of low infection rate. Proactive surveillance, particularly in asymptomatic cases, remains a challenge to surmount to exit lockdown, as screening on a large scale is required.
A limitation of this study is the maximum number of two positive specimens in the 10‐specimen pool. In theory, more positive specimens in a pool could decrease the sensitivity of qPCR as it would result in too many viral copies, causing an insufficiency of PCR enzyme and other reagents in the mix to amplify all the viral copies. Practically, however, this does not affect the overall testing results, since positive pools would require individual testing in any case.
Rapid identification of SARS‐CoV‐2 infection is crucial to curbing the COVID‐19 pandemic. The present gold standard for testing SARS‐CoV‐2 is qPCR, which requires resources that are currently limited, along with specialized equipment and technically skilled labor. Shortage of testing reagents and equipment may result in delays in testing and result in reduced effectiveness in containing the outbreak. Pooled specimen testing would enable substantial savings in reagent costs, technical burden and time to generate laboratory results.
ACKNOWLEDGMENTS
This study was supported by a research grant from the King Chulalongkorn Memorial Hospital's Excellent Center Program (Grant No. EC‐63‐30101‐29), National Research Council of Thailand (NRCT), and the Biological Threat Reduction Program (BTRP) of the US Defense Threat Reduction Agency (DTRA) (Grant No. HDTRA 1‐17‐C‐0004). The authors would also like to acknowledge Dr. Paul Gaudio (Yale University) for his kind assistance in the critical editing of this manuscript.
Wacharapluesadee S, Kaewpom T, Ampoot W, et al. Evaluating the efficiency of specimen pooling for PCR‐based detection of COVID‐19. J Med Virol. 2020;92:2193–2199. 10.1002/jmv.26005
REFERENCES
- 1. Tanne JH, Hayasaki E, Zastrow M, Pulla P, Smith P, Rada AG. Covid‐19: how doctors and healthcare systems are tackling coronavirus worldwide. BMJ. 2020;368:m1090. 10.1136/bmj.m1090 [DOI] [PubMed] [Google Scholar]
- 2. Emanuel EJ, Persad G, Upshur R, et al. Fair allocation of scarce medical resources in the time of Covid‐19. N Engl J Med. 2020;382(21):2049–2055. 10.1056/NEJMsb2005114 [DOI] [PubMed] [Google Scholar]
- 3. Cohen J, Kupferschmidt K. Countries test tactics in ‘war'against COVID‐19. Science. 2020;367(6484):1287‐1288. 10.1126/science.367.6484.1287 [DOI] [PubMed] [Google Scholar]
- 4. Hogan CA, Sahoo MK, Pinsky BA. Sample pooling as a strategy to detect community transmission of SARS‐CoV‐2. JAMA. 2020;323(19):1967–1969. 10.1001/jama.2020.5445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stramer SL, Glynn SA, Kleinman SH, et al. Detection of HIV‐1 and HCV infections among antibody‐negative blood donors by nucleic acid–amplification testing. N Engl J Med. 2004;351(8):760‐768. [DOI] [PubMed] [Google Scholar]
- 6. Pilcher CD, Fiscus SA, Nguyen TQ, et al. Detection of acute infections during HIV testing in North Carolina. N Engl J Med. 2005;352(18):1873‐1883. [DOI] [PubMed] [Google Scholar]
- 7. Klausner JD, Grant RM, Kent CK. Detection of acute HIV infections. N Engl J Med. 2005;353(6):631‐633. [DOI] [PubMed] [Google Scholar]
- 8. Pilcher CD, Eron JJ, Galvin S, Gay C, Cohen MS. Acute HIV revisited: new opportunities for treatment and prevention. J Clin Invest. 2004;113(7):937‐945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Abdalhamid B, Bilder CR, McCutchen EL, Hinrichs SH, Koepsell SA, Iwen PC. Assessment of specimen pooling to conserve SARS CoV‐2 testing resources. Am J Clin Pathol. 2020;153:715‐718. 10.1093/ajcp/aqaa064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boom RC, Sol CJ, Salimans MM, Jansen CL, Wertheim‐van Dillen PM, Van der Noordaa JP. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28(3):495‐503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Torres I, Albert E, Navarro D. Pooling of nasopharyngeal swab specimens for SARS‐CoV‐2 detection by RT‐PCR. J Med Virol. 2020. 10.1002/jmv.25971 [DOI] [PMC free article] [PubMed] [Google Scholar]


