Abstract
In nature, plants are always under the threat of pests and diseases. Pathogenic bacteria are one of the major pathogen types to cause diseases in diverse plants, resulting in negative effects on plant growth and crop yield. Chemical bactericides and antibiotics have been used as major approaches for controlling bacterial plant diseases in the field or greenhouse. However, the appearance of resistant bacteria to common antibiotics and bactericides as well as their potential negative effects on environment and human health demands bacteriologists to develop alternative control agents. Bacteriophages, the viruses that can infect and kill only target bacteria very specifically, have been demonstrated as potential agents, which may have no negative effects on environment and human health. Many bacteriophages have been isolated against diverse plant-pathogenic bacteria, and many studies have shown to efficiently manage the disease development in both controlled and open conditions such as greenhouse and field. Moreover, the specificity of bacteriophages to certain bacterial species has been applied to develop detection tools for the diagnosis of plant-pathogenic bacteria. In this paper, we summarize the promising results from greenhouse or field experiments with bacteriophages to manage diseases caused by plant-pathogenic bacteria. In addition, we summarize the usage of bacteriophages for the specific detection of plant-pathogenic bacteria.
Keywords: bacteriophages, disease management, plant-pathogenic bacteria
Plants, in particular, crops play essential roles for daily human diet as well as food security. In nature, they face with threats of diverse pests and diseases very often. There are several types of plant pathogens such as fungi, viruses, and bacteria to cause devastating diseases in crops, resulting in major economic losses in agriculture (Agrios, 2005). Therefore, the efficient disease management is critical for stable income to growers and stable food supply to consumers. However, the bacterial disease management for agricultural crops is a difficult challenge. The main challenges are only a few effective bactericides, pathogen variability, high mutation probability or high rate of gene transfer among pathogens, and high build-up population rate during optimal conditions for disease development (Balogh et al., 2010).
Conventionally, chemical-based bactericides such as antibiotics (e.g., oxytetracycline and streptomycin) and copper have been extensively used to combat many bacterial plant diseases. After Second World War, antibiotics have been incorporated for the control of plant pathogens (McManus et al., 2002). However, the results of abuse or overuse of antibiotics or copper, resistant strains against those chemicals have been consistently reported in many plant-pathogenic bacteria, and these issues have become a significant concern. Moreover, the uncontrolled use of antibiotics negatively affects the environment and human health (McManus et al., 2002; Svircev et al., 2018). These led scientists to develop eco-friendly and sustainable methods to control bacterial diseases in plants. So far, many agents in this category such as antagonists themselves, antagonist-originated natural chemicals, and agents to boost up plant immunity have been developed and widely used in agricultural crops, including ones in organic farming (Calvo-Garrido et al., 2014; Carisse et al., 2000; Compant et al., 2005; Dennis and Webster, 1971; Wiesel et al., 2014). Among those agents, the use of bacteriophages, which are viruses to infect and destroy only host bacteria as an antimicrobial agent, so called phage therapy, is a promising and reemerging approach for management of bacterial diseases in plants.
The use of bacteriophages as antimicrobials has two major advantages. One is that bacteriophages carry narrow host-range, implying that they specifically infect and eliminate only target bacteria without damaging others (Loc-Carrillo and Abedon, 2011). They are self-replicating in the existence of host bacteria, but at the same time, self-limiting without host bacteria. Therefore, they can minimize effects on the microbial systems in soil as well as treated environment. In addition, this feature could make bacteriophages being used for detection of bacterial pathogens (Farooq et al., 2018; Javed et al., 2013; Wittmann et al., 2010). The other is that, until now, there are no evidence of negative effects of bacteriophage to the eukaryotic cells in both plants and animals (Greer, 2005). This feature could make bacteriophages being used not only to control plant diseases (Bae et al., 2012; Chae et al., 2014; Fujiwara et al., 2011; Nagai et al., 2017), but also to prevent human diseases (Abedon et al., 2011; Hagens and Loessner, 2007; Leverentz et al., 2003; Weber-Dabrowska et al., 2001).
In this review, we summarize the usage of bacteriophages as antimicrobial agents to control bacterial diseases in plants only under greenhouse or field conditions and also as tools to detect certain bacterial pathogens in plants with high specificity.
The General Characteristics and Life Cycles of Bacteriophages
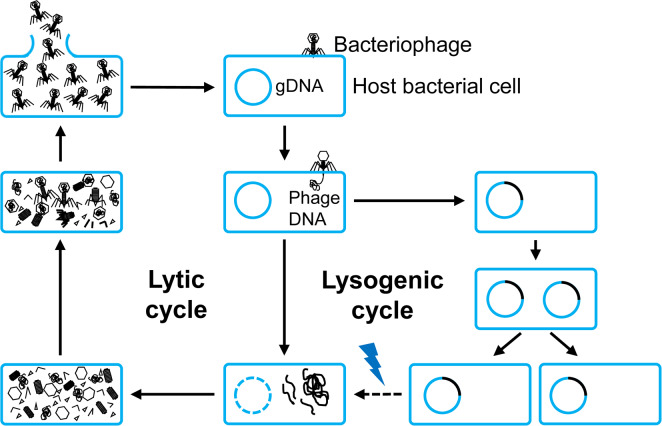
Bacteriophages were discovered more than a century ago and estimated to the most diverse and abundant biological entities on earth. They play an important role in controlling bacterial community (Gómez and Buckling, 2011; Koskella and Brockhurst, 2014), nutrient cycling (Weinbauer, 2004; Wilhelm and Suttle, 1999) and bacteria genome evolution (Howard-Varona et al., 2017; Morgan et al., 2010). Although the billions of bacteriophages infecting different classes of bacteria exist in the environment, most characterized bacteriophages are the members of the Caudovirales, which are either virulent or temperate bacteriophages, depending on their types of infection: lytic and lysogenic (Dy et al., 2018). The infection cycle of bacteriophages is begun by the adsorption of bacteriophages with the special receptors located in the cell surface of susceptible bacteria. Upon irreversible attachment, they inject their genomic DNA (gDNA) into the host cell cytoplasm (Fig. 1). In terms of lytic replication cycle, after gDNA injection, bacteriophages utilize ribosomes of the host bacterium to manufacture phage proteins. The bacterium also provides resources for bacteriophage genome replication and production of virion-related protein components (Dy et al., 2018). At the late stage of the lytic replication cycle, bacteriophages encode holins and lysins, known as endolysins, to lyse the bacterium for release of the phage progenies. In contrast to the lytic cycle, the lysogenic replication cycle has been known as the integration of bacteriophage genome into the bacterial cell chromosome—termed as prophage—or existence as an episomal element, and then they replicate and transfer to daughter bacterial cell (Fortier and Sekulovic, 2013). Prophages can spontaneously alter to a lytic cycle and kill their host spontaneously (Nanda et al., 2015) due to certain environmental stresses, metabolic condition of host bacteria, or antibiotic treatment (Davies et al., 2016). In general, lytic bacteriophages have been used for disease management of bacterial diseases because they can directly kill pathogens, resulting in protecting plants from those pathogens, while temperate bacteriophages may be used for bacterial-pathogen diagnosis because of their genome integration with bacterial chromosome. If the various fusion proteins, including green fluorescent protein can be used in lytic bacteriophages, bacterial pathogen with phage proteins can be easily detected.
Fig. 1.
The general life cycles of bacteriophages. They begin with the adherence of a bacteriophage into the host bacterial cell, then the translocation of its genetic material (phage DNA). Based on the destination of the genetic material, a bacteriophage comes to either lytic or lysogenic cycles. In the case of a lytic cycle, a bacteriophage multiplies and lyses the host cell, while in the case of a lysogenic cycle, its genetic material is integrated into bacterial genomic DNA (gDNA). Under certain conditions (indicated by the lightning), a lysogenic cycle can be converted to a lytic cycle.
Early History and Rehabilitation of Interest of Bacteriophages for Bacterial Disease Management in Plants
The pioneer in phage therapy was Ferick Twort (1915) and Felix d’Hellex (1921), who observed some small agents parasitizing bacteria in growing culture and named as “bacteriophage”. It was recognized as potential antimicrobial agents soon after that (Hermoso et al., 2007). However, the interest in phage therapy was rapidly reversed and displaced by the discovery into new broad-spectrum antibiotics in the 1940s. Bacteriophages, in terms of association with plant pathogenic bacteria, were first discovered by Mallmann and Hemstreet, who demonstrated the inhibited growth of Xanthomonas campestris pv. campestris, by treatment with filtered decomposed cabbage (Mallmann and Hemstreet, 1924). Subsequently, Kotila and Coons suggested that the isolated bacteriophages could prevent soft rot on slices of potato tuber and carrot caused by Pectobacterium atrosepticum and Pectobacterium carotovorum subsp. carotovorum, respectively (Coons and Kotila, 1925; Kotila and Coons, 1925). Bacteriophage effects on plant bacterial pathogen under open conditions were first shown that bacteriophage-treated corn seeds displayed a reduced incidence of Stewart’s wilt disease incited by Pantoea stewartii by 16.5% (Thomas, 1935). However, early scientists found that bacteriophage treatment in the field to control bacterial diseases in plants was less effective than newly discovered broad-spectrum antibiotics (Goto, 2012). Due to this reason, the interest in phage therapy was rapidly faded, despite their promising results for disease management.
Due to the control efficacy, antibiotics and bactericidal chemicals have been major components of management strategies for controlling bacterial plant diseases for decades (Agrios, 2005). However, their negative effects on environment and human health were rapidly emerged (Hermoso et al., 2007). Moreover, the prevalence of antibiotic-, pesticideor copper-resistant bacteria such as Erwinia amylovora (Manulis et al., 2000; Stall, 1962), Pseudomonas syringae (Hwang et al., 2005; Masami et al., 2004; Mellano and Cooksey, 1988), Xanthomonas campestris pv. juglandis (Lee et al., 1994), Xanthomonas citri spp. citri and Xanthomonas alfalfa spp. citrumelonis (Behlau et al., 2011), as well as very slow development of new effective antibiotics, twisted scientists’ attention toward other potential biocontrol agents for control of bacterial plant diseases. Combined with consumers’ demand for organic and antibiotic-free products resulted in the rehabilitation of phage therapy applications in plant disease management.
As stated above, only lytic bacteriophages were given a priority for this aspect with immediate lysis ability as well as avoiding the probable negative effects of lysogenic bacteriophages such as enhancing the virulence of their host bacteria (Frobisher and Brown, 1927; Groman, 1953, 1955), promoting the ecological fitness of bacteria during infection (Flockhart et al., 2012; Fortier and Sekulovic, 2013), and protecting host bacteria from the infection of other lytic phages (Davies et al., 2016). Stonier et al. (1967) reported that fewer than 10 bacteriophage particles showing the clear plaques present at the beginning of 21-h induction period were able, at times, to inhibit completely tumor induction by highly virulent Agrobacterium tumefaciens corn strain B6. In the following years, other bacteriophages also found to target some other plant pathogenic bacteria such as Xanthomonas campestris pv. pruni (Civerolo, 1973; Civerolo and Keil, 1969; Saccardi et al., 1993), A. tumefaciens (Boyd et al., 1971), Xanthomonas oryzae (Kuo et al., 1971), Pseudomonas solanacearum currently renamed as Ralstonia solanacearum (Tanaka et al., 1990), and E. amylovora (Schnabel et al., 1998).
Recent Application of Bacteriophages for Bacterial Disease Management in Plants
Application of individual bacteriophages
Nowadays, almost publications of bacteriophage-based researches for bacterial plant pathogens were restricted in isolation and characterizations, but some of the isolated bacteriophages proved the potential for phage therapy (Frampton et al., 2014; Rahimi-Midani et al., 2018; Yin et al., 2019; Yu et al., 2016). In 2012, top 10 scientifically and economically important bacterial plant pathogens were generated by a survey of bacterial pathologists in the journal Molecular Plant Pathology. The most important plant bacterial pathogen was account for P. syringae pathovars causing diseases on a variety of plants from monocots, herbaceous dicots to woody dicots throughout the world (Mansfield et al., 2012). Two parallel field trials on three locations were carried out with a bacteriophage cocktail of six isolated bacteriophages as a biocontrol for bacterial blight in leek incited by P. syringae pv. porri (Rombouts et al., 2016). Although the result exhibited variable effects, one of these trials showed the attenuation of symptom development. These results indicated that the bacteriophage mixture may have the therapeutic potential for control of bacterial blight disease (Rombouts et al., 2016). In recent years, several studies have been published on biocontrol efficacy with bacteriophages for management of the other important bacterial plant pathogens in the top 10 list, with many considerable results.
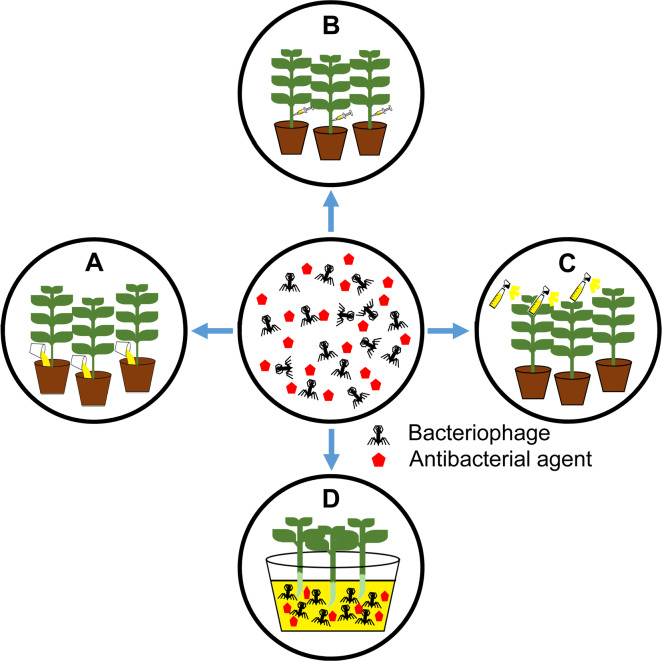
Bacteriophages have been treated in various ways in the greenhouse and field conditions, such as soil drenching, foliar spraying, infiltration, and immersion in the case of seed treatments (Fig. 2). To control R. solanacearum—a soilborne pathogen, direct phage application into rhizosphere via soil drenching showed high suppression in wilting development in tomato (Elhalag et al., 2018). Using foliar spraying, the application of phage effectively decreased disease incidence caused by X. campestris pv. campestris (Nagai et al., 2017), Xanthomonas euvesicatoria (Gašić et al., 2018), and P. carotovorum subsp. carotovorum (Lim et al., 2013). Those examples were listed in Table 1. Recently, the potential of filamentous phages in plant disease control has also been reported. The infection of the filamentous ΦRSM-type phages—ΦRSM3—enhanced the expression of pathogenesis-related genes in tomato plants and led to the loss of virulence of R. solanacearum (Addy et al., 2012). Likewise, several negative changes in extracellular polysaccharide production, mobility, growth rate or virulence were observed in X. axonopodis pv. citri strains along with the filamentous phage XacF1 infection (Ahmad et al., 2014).
Fig. 2.
Treatment methods of bacteriophages in a greenhouse or field. A, soil drench: phage solution was directly added into the base of the plant; B, direct infiltration into plant tissues; C, foliar application by spraying; D, immersion: the seedlings or seeds can be immersed into phage solution before planting. Red pentagons indicate antibacterial agents like antibiotics and systemic acquired resistance inducers.
Table 1.
Application of bacteriophages in the greenhouse or field conditions, which have been conducted since 2010 to present
| Target diseases and pathogens | Bacteriophages | Supplements | Hosts | Treatment methods | Test conditions | Control efficacy (reduction of disease incidence, %) | Reference |
|---|---|---|---|---|---|---|---|
| Bacterial wilt: Ralstonia solanacearum | Single (RsPod1EGY) | None | Tomato | Soil drench | Greenhouse | 100 | Elhalag et al. (2018) |
| Cocktail (NJ-P3, NB-P21, NC-P34, NN-P42) | None | Tomato | Soil drench | Greenhouse | 80 | Wang et al. (2019) | |
| Field | 80 | Wang et al. (2019) | |||||
| Cocktail (M5, M8) | None | Banana | Soil treatment | Greenhouse | 100 | Ramírez et al. (2020) | |
| Bacterial blight: Xanthomonas oryzae pv. oryzae | Cocktail (P4L, P43M, P23M1) | Skim milk | Rice | Spray | Field | 50.8 | Chae et al. (2014) |
| Black rot: X. campestris pv. campestris | Single (XcpSFC211) | None | Broccoli | Spray | Greenhouse | 60 | Nagai et al. (2017) |
| Nonpathogenic Xanthomonas sp. strain | Broccoli | Spray | Field | 16.7-55 | Nagai et al. (2017) | ||
| Bacterial spot: X. axonopodis | Cocktail (X. phage -1, X. phage -2) | None | Pepper | Spray | Greenhouse | 65 | Tewfike and Desoky (2015) |
| Skim milk or corn flour | Pepper | Spray | Field | 53.3, 66 | Tewfike and Desoky (2015) | ||
| Bacterial spot: X. euvesicatoria | Single (KФ1) | None | Pepper | Spray | Greenhouse | 50-67 | Gašić et al. (2018) |
| Canker: X. citri subsp. citri | Cocktail | ASM | Citrus | Spray and soil drench | Greenhouse | 56.9 | Ibrahim et al. (2017) |
| ASM | Citrus | Spray and soil drench | Field | 42.6-56.9 | Ibrahim et al. (2017) | ||
| ASM + Skim milk | Citrus | Spray and soil drench | Field | 81.2-86.1 | Ibrahim et al. (2017) | ||
| Soft rot: Pectobacterium carotovorum subsp. carotovorum | Single (PP1) | None | Cabbage | Spray | Greenhouse | 80 | Lim et al. (2013) |
| Cocktail (φMA11, φMA12, φMA13, φMA14) | None | Onion | Immersion and spray | Field | 2.5-15 | Zaczek-Moczydłowska et al. (2020) | |
| Soft rot: Dickeya solani | Single (vB_DsoMLIMEstone1) | None | Potato | Spray | Field | 5 | Adriaenssens et al. (2012) |
ASM, acibenzolar-S-methyl.
Application of bacteriophage mixtures or cocktails
Although there are some bacteriophages attacking one or more bacterial genera (Ahern et al., 2014; Okabe and Goto, 1963), most bacteriophages can infect restrictively some strains within a single bacterial species. The restriction is mainly derived from the highly specific interaction between host cell surface receptors and phage attachment structures (Sulakvelidze et al., 2001). However, bacteria quickly overcome the infection of bacteriophages via a variety of resistant mechanisms from simple mutation to other more complex resistant mechanisms such as the adsorption-blocking mechanism to avoid phage binding to cellular receptors on the bacterial cell surface (Chopin et al., 2005; Coffey and Ross, 2002; Forde et al., 1999), cleaving phage genetic material via restriction–modification systems or CRISPR-Cas systems (Ranjani et al., 2018; Rezzonico et al., 2011; Semenova et al., 2009). The bacteriophage-resistant R. solanacearum became evident approximately 30 hours after bacteriophage addition to the culture (Fujiwara et al., 2011). Similarly, 16 to 17 h after treated with bacteriophage Xoo-Sp2, resistant bacteria were observed in liquid culture and maintained at a low level after 35 h after infection (Dong et al., 2018).
Using a mixture of lytic bacteriophages known as a bacteriophage cocktail is also a potential approach to compensate for the limitation of host-range of a single bacteriophage as well as prevention of the development of bacteriophage-resistant bacteria (Schmerer et al., 2014; Tewfike and Desoky, 2015). Those examples were included in Table 1. The phage mixtures were used to control a variety of bacterial plant pathogens such as R. solanacearum (Ramírez et al., 2020; Wang et al., 2019; Wei et al., 2017), Xanthomonas sp. (Chae et al., 2014; Ibrahim et al., 2017; Tewfike and Desoky, 2015), and P. carotovorum sp. carotovorum (Zaczek-Moczydłowska et al., 2020).
Furthermore, to overcome bacterial resistance to bacteriophages, a patented process involving preparing a mixture of host-range (h-) mutant bacteriophages was developed. The h-mutant bacteriophages exhibit a wider intraspecific range including bacteria strains that are resistant to parent bacteriophages and maintain specificity toward the wild-type bacteria (Le Roy, 1989). Indeed, a mixture of five hmutant bacteriophages was generated to control bacterial blight in geranium incited by X. campestris pv. pelargonii. These h-mutants enabled to lyse all 21 tested X. campestris pv. pelargonii strains, including the non-host strains of parent bacteriophages (Flaherty et al., 2001). In addition, the daily application of the bacteriophage mixture effectively controlled the spread of bacterial blight on potted and seedling geranium under greenhouse conditions. This research group also used a similar strategy for foliar application as a biological control of bacterial spot caused by X. campestris pv. vesicatoria in tomato (Flaherty et al., 2000). The result showed that the application of bacteriophages consistently reduced the incidence and severity of bacterial spot disease. In addition, the bacteriophage-treated plant also produced significantly higher yields than untreated or copper/mancozeb bactericide-treated plants.
Application of bacteriophages with other antimicrobial agents
The combination of bacteriophages with other antimicrobial agents contributing to a reduction of disease severity such as plant systemic acquired resistance (SAR) inducers and antibiotics has been considerably attended and showed its potential. In 2005, Obradovic et al. evaluated the effects of the combination of SAR inducers with other biocontrol agents on tomato-bacterial spot disease incited by X. campestris pv. vesicatoria in greenhouse experiments. They figured out that the application of bacteriophages in combination with acibenzolar-S-methyl (ASM) suppressed a visible hypersensitive response caused by ASM and provided excellent disease control efficacy (Obradovic et al., 2005). A similar approach was also conducted to control bacterial leaf blight of onion caused by X. axonopodis pv. allii under field conditions. Indeed, the treatment with bacteriophage mixture together with ASM reduced disease severity by 50% compared to 31% of copper hydroxide-mancozeb treatment (Lang et al., 2007). Under greenhouse conditions, the integration of phage KΦ1 and copper hydroxide significantly reduced the lesion number on pepper leaves caused by X. euvesicatoria (approximately 81%, 90%, and 88% for three separate trials) (Table 1). Although there is no statistical difference in the control efficacy of copper hydroxide treatment alone, its treatment together with the bacteriophage reduced the number of lesions significantly (Gašić et al., 2018).
Application of bacteriophage-derived proteins—endolysins—as antimicrobial agents
To release the new progenies from host cells to the outside environment, bacteriophages have evolved two basics strategies: (1) constant liberation of new virions from bacterial cells via extracellular membrane vesicle without lysing them (Russel et al., 1997) often found in filamentous bacteriophages (Borysowski et al., 2006) or (2) release via phage lytic enzymes employed by dsDNA phages (Drulis-Kawa et al., 2015). Unlike the application of the whole-bacteriophages carrying the narrow host specificity and potential to appear phage-resistant strains, the use of lytic enzymes only has some advantages for control of pathogenic bacteria: broader host range and different physicochemical property as well as almost impossible development of resistant strains (Borysowski et al., 2006).
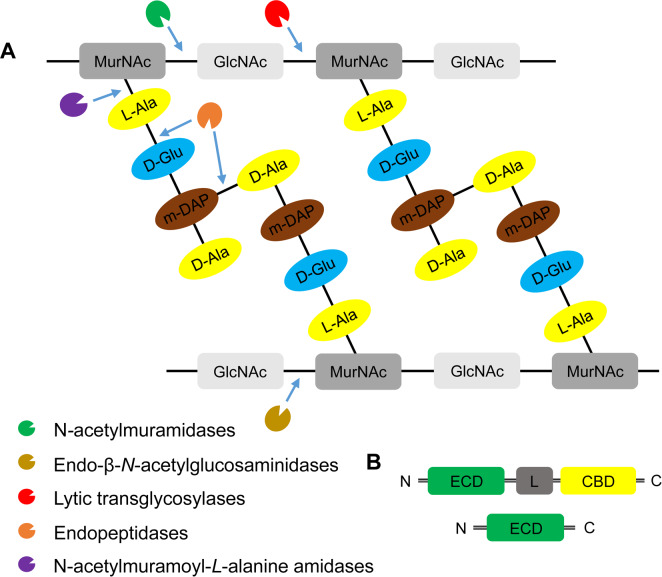
Endolysins and holins are two dsDNA phage enzymes commonly encoded at the late stage of lytic cycle after phage replication and assembly (Borysowski et al., 2006). Endolysins are peptidoglycan (PG) hydrolases, which can be distinguished into five groups, according to their catalytic activity: N-acetylmuramidases (lysozymes or muranidases); endo-β-N-acetylglucosaminidases (glucosamindases), N-acetylmuramoyl-L-alanine amidases, endopeptidases and lytic transglycosylases (Fig. 3A). These enzymes also can be classified based on the targeted bacteria of their original phages: Gram-positive or negative bacteria. Most endolysins from Gram-positive infecting bacteriophages have modular structures (Fig. 3B), carrying an enzymatic catalytic domain (ECD), a cell wall-binding domain, and a linker between two domains (Nelson et al., 2012). By contrast, endolysins with single ECD commonly found in the Gram-negative specificity phages with few exceptions (Pohane and Jain, 2015).
Fig. 3.
The domain structures, diversity, and the mode-of-action of endolysins. (A) Peptidoglycan structure and targets of five classes of endolysins. Peptidoglycan is a heteropolymer of alternating amino sugars N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MucNAc), and tetrapeptide side chains, L-alanine (L-Ala), D-glutamate (D-Glu), meso-diaminopimeric acid (m-DAP) or L-lysine, and D-alanine (D-Ala), attached to the lactyl group of muramic acid. Depending on the enzymatic specificity, endolysins can be divided into five classes, N-acetylmuramindase (lysozymes or muramidases), endo-β-N-acetylglucosaminidase (glucosaminidases), N-acetylmuramoyl-L-alanine amidase (NAM-amidases), endopeptidases and lytic transglycosylases. Bonds cleaved by different endolysins were indicated by blue arrows. (B) Domain structures of bacteriophage endolysins. Ones derived from bacteriophages infecting Grampositive-bacteria include an enzymatic catalytic domain (ECD) and at least one cell-wall binding domain (CBD) and connected by a flexible linker (L) (Nelson et al., 2012). By contrast, endolysins from bacteriophages infecting Gram-negative bacteria are mostly globular structure with single ECD without a specific CBD (Pohane and Jain, 2015). N, N-terminus; C, C-terminus.
In 2010, Wittmann et al. reported the specific bacteriolytic activity of two endolysins encoded by bacteriophages CMP1 and CN77. Although these bacteriophages only lysed Clavibacter michiganensis and C. nebraskensis respectively, their endolysins could specifically lyse related strains within C. michiganensis subspecies without affecting other bacteria in soil (Wittmann et al., 2010). This feature could be useful for specific biocontrol of the plant-pathogenic C. michiganensis. The expression of genes encoding putative peptidoglycan hydrolases (PPH) from lytic bacteriophages Atu_ph02 and Atu_ph03 in A. tumefaciens cells led to a block in cell division and rapidly lysis of host cell (Attai et al., 2017). Additionally, the rapid-cell lysis was observed in the mutation of positively charged residues at the extreme C terminus of PPH. Moreover, bacteriophage-derived endolysins showed a broader bacteriolytic spectrum not only within a species but also other genera. Indeed, a PG hydrolase (gp21 protein) located in the tail of X. oryzae bacteriophage Xop411 showed its ability to kill 3 out of 6 tested species of Xanthomonas as well as other opportunistic Pseudomonas aeruginosa and Stenotrophomonas maltophilla in the plate assay (Weng et al., 2018). These examples indicate that exogenous application of endolysins could be successful to kill bacteria probably by degrading peptidoglycans (Pohane and Jain, 2015).
The ectopic expression of endolysins in plants also confers resistance to bacterial pathogens. The bacteriophage CMP1 endolysin gene (lys) transformed into tomato plants showed significant reduction in disease severity as well as the number of bacterial cells in the xylem sap and leaf extracts in comparison to non-transgenic plants under infection of C. michiganensis (Wittmann et al., 2016). Such effect is not only limited within bacterial pathogens, surprisingly but also pathogenic fungi. Brown patch and gray leaf spot in tall fescue are severe diseases caused by Rhizoctonia solani and Magnaporthe oryzae. The introduction of bacteriophage T4-lysozymes gene into tall fescue showed remarkable results. Particularly, 6 out of 13 transgenic plants showed high resistance to a mixture of two M. oryzae isolates and 3 out of the six plants also exhibited significant resistance to a R. solani isolate (Dong et al., 2008).
The high bactericidal activity of endolysins against Gram-positive bacteria was not observed for Gramnegative bacteria because of their outer membrane protecting the PG from the hydrolysis by phage endolysins from outside (Santos et al., 2018). So far, only few endolysins were reported to affect against Gram-negative bacteria (Lai et al., 2013; Lood et al., 2015). Several strategies have been designed to overcome this disadvantage such as a combination of two or more endolysins fulfilling the limitation of each others to obtain a chimeric protein (Santos et al., 2018). The sensitivity of E. amylovora to a virulent and broad-host-range bacteriophage Y2 dramatically was increased with the recombinant depolymerases enzyme (DopL1) encoded by the T7-like bacteriophage Y1 degrading amylovoran—the main component of extracellular polysaccharide—a physical barrier of bacteriophage Y2 infection (Born et al., 2014). Moreover, the introduction of an N-terminal truncated version of the depolymerase gene (dopL1-C) from phage Y1 into Y2 genome enhanced bactericidal activity toward E. amylovora by clearer plaques and expanded surrounding haloes. It also significantly reduced the number of E. amylovora cells by approximately 6 logs on contaminated flowers (Born et al., 2017). Remarkably, no bacteria could be recovered from 95% of the flowers.
Potential Challenges of Bacteriophages for Bacterial Disease Management in Plants
Even though many laboratory-based assays indicated the significant potential of bacteriophages as biocontrol agents, the applications of bacteriophage for plant disease management in field still need more technical improvement to overcome several limitations or challenges. Bacteriophages have been mostly applied directly to the rhizosphere by soil drench or sprayed to the phyllosphere of plants (Fig. 2). Gill and Abedon (Gill and Abedon, 2003) figured out several factors influencing the success of phage therapy in both environments. In rhizosphere, mostly soil, the available water is essential for free-bacteriophage diffusion. Indeed, the biofilm-trapped bacteriophages or reversibly absorption to particles are commonly found in soil, which lacks a continuous aqueous phase. In this case, bacteriophages become less mobile, sometimes permanently inactivated in low soil pH and may be hard to encounter a suitable host.
The phyllosphere is not a safe place and leads to a sharp reduction of the bacteriophage population (Balogh, 2002; Balogh et al., 2003; Civerolo and Keil, 1969; Iriarte et al., 2007). Many studies in field and laboratory conditions demonstrated that bacteriophages are easily inactivated by exposure to high temperature, high and low pH (Gašić et al., 2018) and sunlight irradiation (especially UV-A and UV-B spectrum) (Gašić et al., 2018; Iriarte et al., 2007). In the open field conditions, the population of bacteriophage mixtures was rapidly decreased and practically eliminated 36-48 h after spraying (Tewfike and Desoky, 2015). The short-period persistence of bacteriophages in the leaf surface is the main limitation of phyllosphere-bacteriophage application. Several approaches have been investigated to improve the efficacy of controlling plant pathogens in the phyllosphere. Encapsulated bacteriophages with some kind of formulation such as corn flour, skim milk, casein, sucrose, congo red, and lignin (Arthurs et al., 2006; Balogh, 2002; Balogh et al., 2003; Behle et al., 1996; Ignoffo et al., 1997); use of carrier bacteria for bacteriophage propagation in the target environment (Balogh, 2006; Boulé et al., 2011; Tanaka et al., 1990) or treatment in the early morning or in the evening (Balogh et al., 2003; Iriarte et al., 2007) showed promising results in bacteriophage-longevity of extension and bacteriophage-treatment efficacy. The control efficacy of bacteriophage-based treatment can be also influenced by many other factors: bacterial flora and bacteriophage density, virion decay rates, bacteriophage enrichment in the target environment and the timing of treatment and the surrounding environment (Gill and Abedon, 2003).
An additional challenge of biological control using bacteriophages in agriculture is a low correlation between bacteriophage characteristics in vitro such as host range, plaque size, and in vitro lytic activity and actual control efficacy in field (Bae et al., 2012; Bhunchoth et al., 2015; Gašić et al., 2018). Indeed, while in vitro bioassays showed the stability in various environmental factors and reduced diseasesymptom on leek leaves against P. syringae pv. porri, no significant difference between bacteriophage-treated and untreated plants in the field trial (Rombouts et al., 2016). The strategical use of ΦRSL1, which did not rapidly kill cells in vitro, compared to other isolated-lytic bacteriophages, which killed rapidly in vitro, showed efficient control the R. solanacearum population as well as movement in tomato root (Fujiwara et al., 2011). Moreover, its protection capability against R. solanacearum remained for 4 months, while other highly lytic bacteriophages did not induce similar plant-protecting effects. Filamentous bacteriophage XacF1, which formed small and turbid plaque during plaque assay, caused significant defects in extracellular polysaccharide production, mobility, growth rate as well as virulence to host bacteria—X. axonopodis pv. citri (Ahmad et al., 2014).
Bacteriophage-Based Biosensors and Bioassay for Detection of Plant-Pathogenic Bacteria
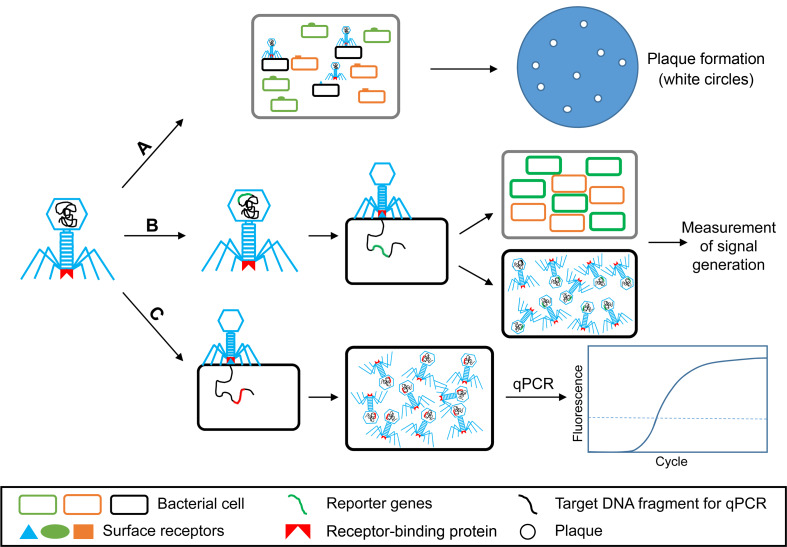
To effectively control and reduce the spread of bacterial diseases, specific and rapid detection is important. The extreme specificity to host bacterial strains, the possibility of massive production and resistance to critical conditions, potential discrimination between live and dead bacteria have led the bacteriophages as potential tools for bacterial detection (Farooq et al., 2018). A number of publications have demonstrated the possibility of bacteriophages as a biosensor for plant pathogen detection during their host infections. Sutton and Katznelson (1953) aimed to isolate bacteriophages for diagnosis and identification of some seed-borne pathogenic bacteria. Four polyvirulent bacteriophages for P. pisi isolated from peas successfully discovered the existence of bacterial blight pathogen out of 14 Pseudomonas species in pea seeds and plant tissues from the infected field (Sutton and Katznelson, 1953). Such way was defined as phage typing schemes, which traditionally screen the sample with the specific phages (A in Fig. 4). However, to isolate a specific phage for the particular pathogen is laborious and time-consuming (Singh et al., 2012).
Fig. 4.
The working models of bacteriophages to detect plant-pathogenic bacteria. A, phage typing: traditionally employed a specific phage for identification and characterization of different pathogenic bacteria based on its lysis activity (Singh et al., 2012); B, reporter phages: engineered phages are used as an importer of marker gene that makes target bacterial cells detectable; C, phage progeny-based detection: using specific bacteriophage to generate rapid amplification of progenies before detection by quantitative PCR (qPCR).
With the possibilities and of genetic engineering techniques, bacteriophages can be engineered as a reporter phage to transfer or insert a reporter gene into target bacteria (B in Fig. 4) (Farooq et al., 2018). The expression of such genes marks the bacteria as a signal marker for bacterial diagnosis (Burnham et al., 2014). Indeed, recombination of luxAB gene encoding the bacterial luciferase was inserted into the bacteriophage PBSPCA1 as a diagnostic for the detection of Pseudomonas cannabina pv. alisalensis (Schofield et al., 2013). PBSPCA1::luxAB was sensitive to detect P. cannabina pv. alisalensis in liquid culture within 20 minutes as well as in diseased plant specimens. The same reporter gene was also introduced into bacteriophage Y2 to detect the fire blight pathogen, E. amylovora. The engineered bacteriophage Y2::luxAB rapidly and effectively detected the pathogen at lower number of viable bacteria, approximately 3.8 × 103 without enrichment in both in vitro or and plant materials (Born et al., 2017).
Kutin et al. (2009) developed a sensitive, effective protocol for detecting R. solanacearum based on the rapid self-replication of bacteriophage with the quantitative PCR (qPCR) (C in Fig. 4). A bacteriophage M_DS1, which specifically infected 61 of 63 R. solanacearum isolates, was selected for the development of the bacteriophage-indirect assay. qPCR indirectly detected R. solanacearum via the detection of a bacteriophage DNA fragment. The method sensitively detected the presence of the pathogen in potted plants, and the detection limit was near 102 cfu/g in 0.1 g of leaf tissues and 103 cfu/ml in drainage water from the pot. In the infected soil, the detection limit was approximately 102 cfu/g (Kutin et al., 2009).
Future Perspectives of Bacteriophage Usage in Plants
Bacteriophages have been explored as potential agents to manage bacterial diseases in plants and also to specifically detect plant-pathogenic bacteria, as highlighted in this paper. Multiple investigations of the potential application of bacteriophages in the case of bacterial disease control have been studied and observed many promising results. However, almost successful applications of bacteriophages were performed in controlled conditions like greenhouses, while agricultural production mainly occurs in an open environment where the environmental factors are constantly changeable and uncontrolled. Therefore, more field trials have to perform to fully implement its efficacy in open conditions. Moreover, even though many promising results were published throughout last decades, there are few commercial bacteriophage-based products that have reached the market for the control of several bacterial plant diseases such as AgriPhages for bacterial spot or speck of tomatoes and peppers and fire blight of apple and pear trees, Erwiphage for fire blight of apple trees, Biolyses for soft rot disease of potato tubers (Buttimer et al., 2017). The efficacy of bacteriophage application is also impaired by several environmental factors so it is also critical to the development of delivery strategies or formulations for commercial purposes.
Developing standard criteria for selecting bacteriophages is also needed more attention for phage therapy. Many evidences demonstrated that current criteria may be effective for several situations but it also remained some exceptions (Ahmad et al., 2014; Fujiwara et al., 2011; Rombouts et al., 2016). Only lytic bacteriophages have been utilized for plant disease management nowadays, but there is still a big question mark over the potential and risk of temperate bacteriophages. Although the natural-temperate bacteriophages were not ideal as biological agents for plant disease control because of their replication cycle, they can be modified to become virulent or work as a delivery vehicle for genetic elements for restoration of antimicrobial susceptibility or virulence-factor disruption (Balogh et al., 2010). Moreover, in the case of phage-based pathogen detection, the engineered phages aim to introduce a marker gene into the target bacterial genome. Therefore, no matter whether reporter phages are lytic or lysogenic, it still potentially detects the targeted bacterial pathogen (Farooq et al., 2018).
The ectopic expression of phage-based proteins in plants exhibited the enhancement of plant resistance to pathogenic bacteria (Dong et al., 2008; Wittmann et al., 2016). However, the use of transgenic plants may present a challenge in certain countries and to consumers. Therefore, it needs more detailed analysis to optimize the efficacy as well as minimize potential side effects.
In conclusion, although agrochemicals such as antibiotics and copper are still mainly used for control of bacterial plant diseases in field, there is a significant potential of bacteriophage usage to reduce the amount of agrochemicals or to replace those agrochemicals for the control of bacterial diseases in plants. For this, more bacteriophages should be collected for diverse bacterial pathogens, and more field trials instead of trials in the controlled conditions are necessary.
Acknowledgments
We are grateful to In Sun Hwang and Yu-Rim Song for critical review. This work was carried out with the support of Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry (IPET) through AgriBio industry Technology Development Program, funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA) (No. 317012-4).
References
- Abedon S. T., Kuhl S. J., Blasdel B. G., Kutter E. M. Phage treatment of human infections. Bacteriophage. 2011;1:66–85. doi: 10.4161/bact.1.2.15845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addy H. S., Askora A., Kawasaki T., Fujie M., Yamada T. Utilization of filamentous phage φRSM3 to control bacterial wilt caused by Ralstonia solanacearum. Plant Dis. 2012;96:1204–1209. doi: 10.1094/PDIS-12-11-1023-RE. [DOI] [PubMed] [Google Scholar]
- Adriaenssens E. M., Van Vaerenbergh J., Vandenheuvel D., Dunon V., Ceyssens P.-J., De Proft M., Kropinski A. M., Noben J.-P., Maes M., Lavigne R. T4-related bacteriophage LIMEstone isolates for the control of soft rot on potato caused by 'Dickeya solani'. PLoS ONE. 2012;7:e33227. doi: 10.1371/journal.pone.0033227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrios G. Plant pathology. 5th ed. Elsevier Academic Press; Burlington, MA, USA: 2005. 952 [Google Scholar]
- Ahern S. J., Das M., Bhowmick T. S., Young R., Gonzalez C. F. Characterization of novel virulent broad-hostrange phages of Xylella fastidiosa and Xanthomonas. J. Bacteriol. 2014;196:459–471. doi: 10.1128/JB.01080-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad A. A., Askora A., Kawasaki T., Fujie M., Yamada T. The filamentous phage XacF1 causes loss of virulence in Xanthomonas axonopodis pv. citri, the causative agent of citrus canker disease. Front. Microbiol. 2014;5:321. doi: 10.3389/fmicb.2014.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthurs S. P., Lacey L. A., Behle R. W. Evaluation of spray-dried lignin-based formulations and adjuvants as solar protectants for the granulovirus of the codling moth, Cydia pomonella (L) J. Invertebr. Pathol. 2006;93:88–95. doi: 10.1016/j.jip.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Attai H., Rimbey J., Smith G. P., Brown P. J. B. Expression of a peptidoglycan hydrolase from lytic bacteriophages Atu_ph02 and Atu_ph03 triggers lysis of Agrobacterium tumefaciens. Appl. Environ. Microbiol. 2017;83:e01498-17. doi: 10.1128/AEM.01498-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae J. Y., Wu J., Lee H. J., Jo E. J., Murugaiyan S., Chung E., Lee S.-W. Biocontrol potential of a lytic bacteriophage PE204 against bacterial wilt of tomato. J. Microbiol. Biotechnol. 2012;22:1613–1620. doi: 10.4014/jmb.1208.08072. [DOI] [PubMed] [Google Scholar]
- Balogh B. University of Florida; Gainesville, FL, USA: 2002. Strategies for improving the efficacy of bacteriophages for controlling bacterial spot of tomato. M.S. thesis. [Google Scholar]
- Balogh B. University of Florida; Gainesville, FL, USA: 2006. Characterization and use of bacteriophages associated with citrus bacterial pathogens for disease control. Ph.D. thesis. [Google Scholar]
- Balogh B., Jones J. B., Iriarte F. B., Momol M. T. Phage therapy for plant disease control. Curr. Pharm. Biotechnol. 2010;11:48–57. doi: 10.2174/138920110790725302. [DOI] [PubMed] [Google Scholar]
- Balogh B., Jones J. B., Momol M. T., Olson S. M., Obradovic A., King P., Jackson L. E. Improved efficacy of newly formulated bacteriophages for management of bacterial spot on tomato. Plant Dis. 2003;87:949–954. doi: 10.1094/PDIS.2003.87.8.949. [DOI] [PubMed] [Google Scholar]
- Behlau F., Canteros B. I., Minsavage G. V., Jones J. B., Graham J. H. Molecular characterization of copper resistance genes from Xanthomonas citri subsp. citri and Xanthomonas alfalfae subsp. citrumelonis. Appl. Environ. Microbiol. 2011;77:4089–4096. doi: 10.1128/AEM.03043-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behle R. W., McGuire M. R., Shasha B. S. Extending the residual toxicity of Bacillus thuringiensis with casein-based formulations. J. Econ. Entomol. 1996;89:1399–1405. doi: 10.1093/jee/89.6.1399. [DOI] [Google Scholar]
- Bhunchoth A., Phironrit N., Leksomboon C., Chatchawankanphanich O., Kotera S., Narulita E., Kawasaki T., Fujie M., Yamada T. Isolation of Ralstonia solanacearum-infecting bacteriophages from tomato fields in Chiang Mai, Thailand, and their experimental use as biocontrol agents. J. Appl. Microbiol. 2015;118:1023–1033. doi: 10.1111/jam.12763. [DOI] [PubMed] [Google Scholar]
- Born Y., Fieseler L., Klumpp J., Eugster M. R., Zurfluh K., Duffy B., Loessner M. J. The tail-associated depolymerase of Erwinia amylovora phage L1 mediates host cell adsorption and enzymatic capsule removal, which can enhance infection by other phage. Environ. Microbiol. 2014;16:2168–2180. doi: 10.1111/1462-2920.12212. [DOI] [PubMed] [Google Scholar]
- Born Y., Fieseler L., Thöny V., Leimer N., Duffy B., Loessner M. J. Engineering of bacteriophages Y2::dpoL1C and Y2::luxAB for efficient control and rapid detection of the fire blight pathogen, Erwinia amylovora. Appl. Environ. Microbiol. 2017;83:e00341-17. doi: 10.1128/AEM.00341-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borysowski J., Weber-Dabrowska B., Górski A. Bacteriophage endolysins as a novel class of antibacterial agents. Exp. Biol. Med. (Maywood) 2006;231:366–377. doi: 10.1177/153537020623100402. [DOI] [PubMed] [Google Scholar]
- Boulé J., Sholberg P. L., Lehman S. M., O'gorman D. T., Svircev A. M. Isolation and characterization of eight bacteriophages infecting Erwinia amylovora and their potential as biological control agents in British Columbia, Canada. Can. J. Plant Pathol. 2011;33:308–317. doi: 10.1080/07060661.2011.588250. [DOI] [Google Scholar]
- Boyd R. J., Hildebrandt A. C., Allen O. N. Retardation of crown gall enlargement after bacteriophage treatment. Plant Dis. Rep. 1971;55:145–148. [Google Scholar]
- Burnham S., Hu J., Anany H., Brovko L., Deiss F., Derda R., Griffiths M. W. Towards rapid on-site phage-mediated detection of generic Escherichia coli in water using luminescent and visual readout. Anal. Bioanal. Chem. 2014;406:5685–5693. doi: 10.1007/s00216-014-7985-3. [DOI] [PubMed] [Google Scholar]
- Buttimer C., McAuliffe O., Ross R. P., Hill C., O'Mahony J., Coffey A. Bacteriophages and bacterial plant diseases. Front. Microbiol. 2017;8:34. doi: 10.3389/fmicb.2017.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo-Garrido C., Viñas I., Elmer P. A., Usall J., Teixidó N. Suppression of Botrytis cinerea on necrotic grapevine tissues by early-season applications of natural products and biological control agents. Pest Manag. Sci. 2014;70:595–602. doi: 10.1002/ps.3587. [DOI] [PubMed] [Google Scholar]
- Carisse O., Philion V., Rolland D., Bernier J. Effect of fall application of fungal antagonists on spring ascospore production of the apple scab pathogen, Venturia inaequalis. Phytopathology. 2000;90:31–37. doi: 10.1094/PHYTO.2000.90.1.31. [DOI] [PubMed] [Google Scholar]
- Chae J.-C., Hung N. B., Yu S.-M., Lee H. K., Lee Y. H. Diversity of bacteriophages infecting Xanthomonas oryzae pv. oryzae in paddy fields and its potential to control bacterial leaf blight of rice. J. Microbiol. Biotechnol. 2014;24:740–747. doi: 10.4014/jmb.1402.02013. [DOI] [PubMed] [Google Scholar]
- Chopin M.-C., Chopin A., Bidnenko E. Phage abortive infection in lactococci: variations on a theme. Curr. Opin. Microbiol. 2005;8:473–479. doi: 10.1016/j.mib.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Civerolo E. L. Relationship of Xanthomonas pruni bacteriophages to bacterial spot disease in prunus. Phytopathology. 1973;63:1279–1284. doi: 10.1094/Phyto-63-1279. [DOI] [Google Scholar]
- Civerolo E. L., Keil H. L. Inhibition of bacterial spot of peach foliage by Xanthomonas pruni bacteriophage. Phytopathology. 1969;59:1966–1967. [Google Scholar]
- Coffey A., Ross R. P. Bacteriophage-resistance systems in dairy starter strains: molecular analysis to application. Antonie Van Leeuwenhoek. 2002;82:303–321. doi: 10.1007/978-94-017-2029-8_20. [DOI] [PubMed] [Google Scholar]
- Compant S., Duffy B., Nowak J., Clément C., Barka E. A. Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 2005;71:4951–4959. doi: 10.1128/AEM.71.9.4951-4959.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coons G. H., Kotila J. E. The transmissible lytic principle (bacteriophage) in relation to plant pathogens. Phytopathology. 1925;15:357–370. [Google Scholar]
- Davies E. V., Winstanley C., Fothergill J. L., James C. E. The role of temperate bacteriophages in bacterial infection. FEMS Microbiol. Lett. 2016;363:fnw015. doi: 10.1093/femsle/fnw015. [DOI] [PubMed] [Google Scholar]
- Dennis C., Webster J. Antagonistic properties of species-groups of Trichoderma: I. Production of non-volatile antibiotics. Trans. Br. Mycol. Soc. 1971;57:25–39. doi: 10.1016/S0007-1536(71)80077-3. [DOI] [Google Scholar]
- Herelle F. Sur un microbe invisible antagoniste des Bacillies dysentériqué. C. R. Acad. Sci. 1917;165:373–375. [Google Scholar]
- Dong S., Shew H. D., Tredway L. P., Lu J., Sivamani E., Miller E. S., Qu R. Expression of the bacteriophage T4 lysozyme gene in tall fescue confers resistance to gray leaf spot and brown patch diseases. Transgenic Res. 2008;17:47–57. doi: 10.1007/s11248-007-9073-3. [DOI] [PubMed] [Google Scholar]
- Dong Z., Xing S., Liu J., Tang X., Ruan L., Sun M., Tong Y., Peng D. Isolation and characterization of a novel phage Xoo-sp2 that infects Xanthomonas oryzae pv. oryzae. J. Gen. Virol. 2018;99:1453–1462. doi: 10.1099/jgv.0.001133. [DOI] [PubMed] [Google Scholar]
- Drulis-Kawa Z., Majkowska-Skrobek G., Maciejewska B. Bacteriophages and phage-derived proteins: application approaches. Curr. Med. Chem. 2015;22:1757–1773. doi: 10.2174/0929867322666150209152851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dy R. L., Rigano L. A., Fineran P. C. Phage-based biocontrol strategies and their application in agriculture and aquaculture. Biochem. Soc. Trans. 2018;46:1605–1613. doi: 10.1042/BST20180178. [DOI] [PubMed] [Google Scholar]
- Elhalag K., Nasr-Eldin M., Hussien A., Ahmad A. Potential use of soilborne lytic Podoviridae phage as a biocontrol agent against Ralstonia solanacearum. J. Basic Microbiol. 2018;58:658–669. doi: 10.1002/jobm.201800039. [DOI] [PubMed] [Google Scholar]
- Farooq U., Yang Q., Ullah M. W., Wang S. Bacterial biosensing: recent advances in phage-based bioassays and biosensors. Biosens. Bioelectron. 2018;118:204–216. doi: 10.1016/j.bios.2018.07.058. [DOI] [PubMed] [Google Scholar]
- Flaherty J. E., Harbaugh B. K., Jones J. B., Somodi G. C., Jackson L. E. H-mutant bacteriophages as a potential biocontrol of bacterial blight of geranium. HortScience. 2001;36:98–100. doi: 10.21273/HORTSCI.36.1.98. [DOI] [Google Scholar]
- Flaherty J. E., Jones J. B., Harbaugh B. K., Somodi G. C., Jackson L. E. Control of bacterial spot on tomato in the greenhouse and field with H-mutant bacteriophages. HortScience. 2000;35:882–884. doi: 10.21273/HORTSCI.35.5.882. [DOI] [Google Scholar]
- Flockhart A. F., Tree J. J., Xu X., Karpiyevich M., McAteer S. P., Rosenblum R., Shaw D. J., Low C. J., Best A., Gannon V., Laing C., Murphy K. C., Leong J. M., Schneiders T., La Ragione R., Gally D. L. Identification of a novel prophage regulator in Escherichia coli controlling the expression of type III secretion. Mol. Microbiol. 2012;83:208–223. doi: 10.1111/j.1365-2958.2011.07927.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde A., Daly C., Fitzgerald G. F. Identification of four phage resistance plasmids from Lactococcus lactis sub-sp. cremoris HO2. Appl. Environ. Microbiol. 1999;65:1540–1547. doi: 10.1128/AEM.65.4.1540-1547.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortier L.-C., Sekulovic O. Importance of prophages to evolution and virulence of bacterial pathogens. Virulence. 2013;4:354–365. doi: 10.4161/viru.24498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frampton R. A., Taylor C., Moreno A. V. H., Visnovsky S. B., Petty N. K., Pitman A. R., Fineran P. C. Identification of bacteriophages for biocontrol of the kiwifruit canker phytopathogen Pseudomonas syringae pv. actinidiae. Appl. Environ. Microbiol. 2014;80:2216–2228. doi: 10.1128/AEM.00062-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frobisher M., Brown J. H. Transmissible toxicogenicity of streptococci. Bull. Johns Hopkins Hosp. 1927;41:167–173. [Google Scholar]
- Fujiwara A., Fujisawa M., Hamasaki R., Kawasaki T., Fujie M., Yamada T. Biocontrol of Ralstonia solanacearum by treatment with lytic bacteriophages. Appl. Environ. Microbiol. 2011;77:4155–4162. doi: 10.1128/AEM.02847-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gašić K., Kuzmanović N., Ivanović M., Prokić A., Šević M., Obradović A. Complete genome of the Xanthomonas euvesicatoria specific bacteriophage KΦ1, its survival and potential in control of pepper bacterial spot. Front. Microbiol. 2018;9:2021. doi: 10.3389/fmicb.2018.02021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill J., Abedon S. T. Bacteriophage ecology and plants. APSnet Features. 2003 doi: 10.1094/APSnetFeature-2003-1103. doi: 10.1094/APSnetFeature-2003-1103. [DOI] [Google Scholar]
- Gómez P., Buckling A. Bacteria-phage antagonistic coevolution in soil. Science. 2011;332:106–109. doi: 10.1126/science.1198767. [DOI] [PubMed] [Google Scholar]
- Goto M. Fundamentals of bacterial plant pathology. Academic Press; Burlington, MA, USA: 2012. p. 342. [Google Scholar]
- Greer G. G. Bacteriophage control of foodborne bacteria. J. Food Prot. 2005;68:1102–1111. doi: 10.4315/0362-028X-68.5.1102. [DOI] [PubMed] [Google Scholar]
- Groman N. B. Evidence for the induced nature of the change from nontoxigenicity to toxigenicity in Corynebacterium diphtheriae as a result of exposure to specific bacteriophage. J. Bacteriol. 1953;66:184–191. doi: 10.1128/JB.66.2.184-191.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groman N. B. Evidence for the active role of bacteriophage in the conversion of nontoxigenic Corynebacterium diphtheriae to toxin production. J. Bacteriol. 1955;69:9–15. doi: 10.1128/JB.69.1.9-15.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagens S., Loessner M. J. Application of bacteriophages for detection and control of foodborne pathogens. Appl. Microbiol. Biotechnol. 2007;76:513–519. doi: 10.1007/s00253-007-1031-8. [DOI] [PubMed] [Google Scholar]
- Hermoso J. A., García J. L., García P. Taking aim on bacterial pathogens: from phage therapy to enzybiotics. Curr. Opin. Microbiol. 2007;10:461–472. doi: 10.1016/j.mib.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Howard-Varona C., Hargreaves K. R., Abedon S. T., Sullivan M. B. Lysogeny in nature: mechanisms, impact and ecology of temperate phages. ISME J. 2017;11:1511–1520. doi: 10.1038/ismej.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang M. S., Morgan R. L., Sarkar S. F., Wang P. W., Guttman D. S. Phylogenetic characterization of virulence and resistance phenotypes of Pseudomonas syringae. Appl. Environ. Microbiol. 2005;71:5182–5191. doi: 10.1128/AEM.71.9.5182-5191.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim Y. E., Saleh A. A., Al-Saleh M. A. Management of asiatic citrus canker under field conditions in Saudi Arabia using bacteriophages and acibenzolar-S-methyl. Plant Dis. 2017;101:761–765. doi: 10.1094/PDIS-08-16-1213-RE. [DOI] [PubMed] [Google Scholar]
- Ignoffo C. M., Garcia C., Saathoff S. G. Sunlight stability and rain-fastness of formulations of Baculovirus heliothis. Environ. Entomol. 1997;26:1470–1474. doi: 10.1093/ee/26.6.1470. [DOI] [Google Scholar]
- Iriarte F. B., Balogh B., Momol M. T., Smith L. M., Wilson M., Jones J. B. Factors affecting survival of bacteriophage on tomato leaf surfaces. Appl. Environ. Microbiol. 2007;73:1704–1711. doi: 10.1128/AEM.02118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javed M. A., Poshtiban S., Arutyunov D., Evoy S., Szymanski C. M. Bacteriophage receptor binding protein based assays for the simultaneous detection of Campylobacter jejuni and Campylobacter coli. PLoS ONE. 2013;8:e69770. doi: 10.1371/journal.pone.0069770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskella B., Brockhurst M. A. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol. Rev. 2014;38:916–931. doi: 10.1111/1574-6976.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotila J. E., Coons G. H. Investigations on the blackleg disease of the potato. Mich. Agric. Exp. Stn. Tech. Bull. 1925;67:3–29. [Google Scholar]
- Kuo T. T., Chang L. C., Yang C. M., Yang S. E. Bacterial leaf blight of rice plant. IV. Effect of bacteriophage on the infectivity of Xanthomonas oryzae. Acad. Sin. Inst. Bot. Bot. Bull. 1971;12:1–9. [Google Scholar]
- Kutin R. K., Alvarez A., Jenkins D. M. Detection of Ralstonia solanacearum in natural substrates using phage amplification integrated with real-time PCR assay. J. Microbiol. Methods. 2009;76:241–246. doi: 10.1016/j.mimet.2008.11.008. [DOI] [PubMed] [Google Scholar]
- Lai M.-J., Soo P.-C., Lin N.-T., Hu A., Chen Y.-J., Chen L.-K., Chang K.-C. Identification and characterisation of the putative phage-related endolysins through full genome sequence analysis in Acinetobacter baumannii ATCC 17978. Int. J. Antimicrob. Agents. 2013;42:141–148. doi: 10.1016/j.ijantimicag.2013.04.022. [DOI] [PubMed] [Google Scholar]
- Lang J. M., Gent D. H., Schwartz H. F. Management of Xanthomonas leaf blight of onion with bacteriophages and a plant activator. Plant Dis. 2007;91:871–878. doi: 10.1094/PDIS-91-7-0871. [DOI] [PubMed] [Google Scholar]
- Le Roy E. J., inventor. Bacteriophage prevention and control of harmful plant bacteria. U.S. Patent No. US4828999A U.S. Patent and Trademark Office, Washington, DC, USA. 1989
- Lee Y. A., Hendson M., Panopoulos N. J., Schroth M. N. Molecular cloning, chromosomal mapping, and sequence analysis of copper resistance genes from Xanthomonas campestris pv. juglandis: homology with small blue copper proteins and multicopper oxidase. J. Bacteriol. 1994;176:173–188. doi: 10.1128/JB.176.1.173-188.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leverentz B., Conway W. S., Camp M. J., Janisiewicz W. J., Abuladze T., Yang M., Saftner R., Sulakvelidze A. Biocontrol of Listeria monocytogenes on fresh-cut produce by treatment with lytic bacteriophages and a bacteriocin. Appl. Environ. Microbiol. 2003;69:4519–4526. doi: 10.1128/AEM.69.8.4519-4526.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J.-A., Jee S., Lee D. H., Roh E., Jung K., Oh C., Heu S. Biocontrol of Pectobacterium carotovorum subsp. carotovorum using bacteriophage PP1. J. Microbiol. Biotechnol. 2013;23:1147–1153. doi: 10.4014/jmb.1304.04001. [DOI] [PubMed] [Google Scholar]
- Loc-Carrillo C., Abedon S. T. Pros and cons of phage therapy. Bacteriophage. 2011;1:111–114. doi: 10.4161/bact.1.2.14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lood R., Winer B. Y., Pelzek A. J., Diez-Martinez R., Thandar M., Euler C. W., Schuch R., Fischetti V. A. Novel phage lysin capable of killing the multidrug-resistant gramnegative bacterium Acinetobacter baumannii in a mouse bacteremia model. Antimicrob. Agents Chemother. 2015;59:1983–1991. doi: 10.1128/AAC.04641-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallmann W. L., Hemstreet C. Isolation of an inhibitory substance from plants. J. Agric. Res. 1924;28:599–602. [Google Scholar]
- Mansfield J., Genin S., Magori S., Citovsky V., Sriariyanum M., Ronald P., Dow M., Verdier V., Beer S. V., Machado M. A., Toth I., Salmond G., Foster G. D. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 2012;13:614–629. doi: 10.1111/j.1364-3703.2012.00804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manulis S., Kleitman F., Dror O., Shabi E. Isolation of strains of Erwinia amylovora resistant to oxolinic acid. IOBC/WPRS Bull. 2000;23:89–92. [Google Scholar]
- Masami N., Masao G., Katsumi A., Tadaaki H. Nucleotide sequence and organization of copper resistance genes from Pseudomonas syringae pv. actinidiae. Eur. J. Plant Pathol. 2004;110:223–226. doi: 10.1023/B:EJPP.0000015360.88352.a9. [DOI] [Google Scholar]
- McManus P. S., Stockwell V. O., Sundin G. W., Jones A. L. Antibiotic use in plant agriculture. Annu. Rev. Phytopathol. 2002;40:443–465. doi: 10.1146/annurev.phyto.40.120301.093927. [DOI] [PubMed] [Google Scholar]
- Mellano M. A., Cooksey D. A. Nucleotide sequence and organization of copper resistance genes from Pseudomonas syringae pv. tomato. J. Bacteriol. 1988;170:2879–2883. doi: 10.1128/JB.170.6.2879-2883.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan A. D., Bonsall M. B., Buckling A. Impact of bacterial mutation rate on coevolutionary dynamics between bacteria and phages. Evolution. 2010;64:2980–2987. doi: 10.1111/j.1558-5646.2010.01037.x. [DOI] [PubMed] [Google Scholar]
- Nagai H., Miyake N., Kato S., Maekawa D., Inoue Y., Takikawa Y. Improved control of black rot of broccoli caused by Xanthomonas campestris pv. campestris using a bacteriophage and a nonpathogenic Xanthomonas sp. strain. J. Gen. Plant Pathol. 2017;83:373–381. doi: 10.1007/s10327-017-0745-4. [DOI] [Google Scholar]
- Nanda A. M., Thormann K., Frunzke J. Impact of spontaneous prophage induction on the fitness of bacterial populations and host-microbe interactions. J. Bacteriol. 2015;197:410–419. doi: 10.1128/JB.02230-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D. C., Schmelcher M., Rodriguez-Rubio L., Klumpp J., Pritchard D. G., Dong S., Donovan D. M. Endolysins as antimicrobials. Adv. Virus Res. 2012;83:299–365. doi: 10.1016/B978-0-12-394438-2.00007-4. [DOI] [PubMed] [Google Scholar]
- Obradovic A., Jones J. B., Momol M. T., Olson S. M., Jackson L. E., Balogh B., Guven K., Iriarte F. B. Integration of biological control agents and systemic acquired resistance inducers against bacterial spot on tomato. Plant Dis. 2005;89:712–716. doi: 10.1094/PD-89-0712. [DOI] [PubMed] [Google Scholar]
- Okabe N., Goto M. Bacteriophages of plant pathogens. Annu. Rev. Phytopathol. 1963;1:397–418. doi: 10.1146/annurev.py.01.090163.002145. [DOI] [Google Scholar]
- Pohane A. A., Jain V. Insights into the regulation of bacteriophage endolysin: multiple means to the same end. Microbiology. 2015;161:2269–2276. doi: 10.1099/mic.0.000190. [DOI] [PubMed] [Google Scholar]
- Rahimi-Midani A., Lee Y. S., Kang S.-W., Kim M.-K., Choi T.-J. First isolation and molecular characterization of bacteriophages infecting Acidovorax citrulli, the causal agent of bacterial fruit blotch. Plant Pathol. J. 2018;34:59–64. doi: 10.5423/PPJ.NT.08.2017.0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez M., Neuman B., Ramírez C. A. Bacteriophages as promising agents for the biological control of moko disease (Ralstonia solanacearum) of banana. Biol. Control. 2020 doi: 10.1016/j.biocontrol.2020.104238. doi: 10.1016/j.biocontrol.2020.104238. [DOI] [Google Scholar]
- Ranjani P., Gowthami Y., Gnanamanickam S. S., Palani P. Bacteriophages: a new weapon for the control of bacterial blight disease in rice caused by Xanthomonas oryzae. Microbiol. Biotechnol. Lett. 2018;46:346–359. doi: 10.4014/mbl.1807.07009. [DOI] [Google Scholar]
- Rezzonico F., Smits T. H., Duffy B. Diversity, evolution, and functionality of clustered regularly interspaced short palindromic repeat (CRISPR) regions in the fire blight pathogen Erwinia amylovora. Appl. Environ. Microbiol. 2011;77:3819–3829. doi: 10.1128/AEM.00177-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombouts S., Volckaert A., Venneman S., Declercq B., Vandenheuvel D., Allonsius C. N., Van Malderghem C., Jang H. B., Briers Y., Noben J. P., Klumpp J., Van Vaerenbergh J., Maes M., Lavigne R. Characterization of novel bacteriophages for biocontrol of bacterial blight in leek caused by Pseudomonas syringae pv. porri. Front. Microbiol. 2016;7:279. doi: 10.3389/fmicb.2016.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel M., Linderoth N. A., Šali A. Filamentous phage assembly: variation on a protein export theme. Gene. 1997;192:23–32. doi: 10.1016/S0378-1119(96)00801-3. [DOI] [PubMed] [Google Scholar]
- Saccardi A., Gambin E., Zaccardelli M., Barone G., Mazzucchi U. Xanthomonas campestris pv. pruni control trials with phage treatments on peaches in the orchard. Phytopathol. Mediterr. 1993;32:206–210. [Google Scholar]
- Santos S. B., Costa A. R., Carvalho C., Nóbrega F. L., Azeredo J. Exploiting bacteriophage proteomes: the hidden biotechnological potential. Trends Biotechnol. 2018;36:966–984. doi: 10.1016/j.tibtech.2018.04.006. [DOI] [PubMed] [Google Scholar]
- Schmerer M., Molineux I. J., Bull J. J. Synergy as a rationale for phage therapy using phage cocktails. PeerJ. 2014;2:e590. doi: 10.7717/peerj.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel E. L., Fernando W. G. D., Meyer M. P., Jones A. L., Jackson L. E. Bacteriophage of Erwinia amylovora and their potential for biocontrol. Acta Hortic. 1998;489:649–654. doi: 10.17660/ActaHortic.1999.489.116. [DOI] [Google Scholar]
- Schofield D. A., Bull C. T., Rubio I., Wechter W. P., Westwater C., Molineux I. J. "Light-tagged" bacteriophage as a diagnostic tool for the detection of phytopathogens. Bioengineered. 2013;4:50–54. doi: 10.4161/bioe.22159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenova E., Nagornykh M., Pyatnitskiy M., Artamonova I. I., Severinov K. Analysis of CRISPR system function in plant pathogen Xanthomonas oryzae. FEMS Microbiol. Lett. 2009;296:110–116. doi: 10.1111/j.1574-6968.2009.01626.x. [DOI] [PubMed] [Google Scholar]
- Singh A., Arutyunov D., Szymanski C. M., Evoy S. Bacteriophage based probes for pathogen detection. Analyst. 2012;137:3405–3421. doi: 10.1039/c2an35371g. [DOI] [PubMed] [Google Scholar]
- Stall R. E. Streptomycin resistance of the bacterial spot pathogen and control with streptomycin. Plant Dis. Rep. 1962;46:389–392. [Google Scholar]
- Stonier T., McSharry J., Speitel T. Agrobacterium tumefaciens Conn IV. Bacteriophage PB21 and its inhibitory effect on tumor induction. J. Virol. 1967;1:268–273. doi: 10.1128/JVI.1.2.268-273.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulakvelidze A., Alavidze Z., Morris J. G. Bacteriophage therapy. Antimicrob. Agent Chemother. 2001;45:649–659. doi: 10.1128/AAC.45.3.649-659.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton M. D., Katznelson H. Isolation of bacteriophages for the detection and identification of some seedborne pathogenic bacteria. Can. J. Bot. 1953;31:201–205. doi: 10.1139/b53-019. [DOI] [Google Scholar]
- Svircev A., Roach D., Castle A. Framing the future with bacteriophages in agriculture. Viruses. 2018;10:E218. doi: 10.3390/v10050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H., Negishi H., Maeda H. Control of tobacco bacterial wilt by an avirulent strain of Pseudomonas solanacearum M4S and its bacteriophage. Ann. Phytopathol. Soc. Jpn. 1990;56:243–246. doi: 10.3186/jjphytopath.56.243. [DOI] [Google Scholar]
- Tewfike T. A., Desoky S. M. Biocontrol of Xanthomonas axonopodis causing bacterial spot by application of formulated phage. Ann. Agric. Sci. Moshtohor. 2015;53:615–624. [Google Scholar]
- Thomas R. A bacteriophage in relation to Stewart's disease of corn. Phytopathology. 1935;25:371–372. [Google Scholar]
- Twort F. W. An Investigation on the nature of ultra-microscopic viruses. Lancet. 1915;186:1241–1243. doi: 10.1016/S0140-6736(01)20383-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wei Z., Yang K., Wang J., Jousset A., Xu Y., Shen Q., Friman V.-P. Phage combination therapies for bacterial wilt disease in tomato. Nat. Biotechnol. 2019;37:1513–1520. doi: 10.1038/s41587-019-0328-3. [DOI] [PubMed] [Google Scholar]
- Weber-Dabrowska B., Mulczyk M., Górski A. Bacteriophage therapy for infections in cancer patients. Clin. Appl. Immunol. Rev. 2001;1:131–134. doi: 10.1016/S1529-1049(01)00015-0. [DOI] [Google Scholar]
- Wei C., Liu J., Maina A. N., Mwaura F. B., Yu J., Yan C., Zhang R., Wei H. Developing a bacteriophage cocktail for biocontrol of potato bacterial wilt. Virol. Sin. 2017;32:476–484. doi: 10.1007/s12250-017-3987-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinbauer M. G. Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 2004;28:127–181. doi: 10.1016/j.femsre.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Weng S.-F., Fu Y.-C., Lin J.-W., Tseng T.-T. Identification of a broad-spectrum peptidoglycan hydrolase associated with the particle of Xanthomonas oryzae phage Xop411. J. Mol. Microbiol. Biotechnol. 2018;28:78–86. doi: 10.1159/000488678. [DOI] [PubMed] [Google Scholar]
- Wiesel L., Newton A. C., Elliott I., Booty D., Gilroy E. M., Birch P. R. J., Hein I. Molecular effects of resistance elicitors from biological origin and their potential for crop protection. Front. Plant Sci. 2014;5:655. doi: 10.3389/fpls.2014.00655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm S. W., Suttle C. A. Viruses and nutrient cycles in the sea: viruses play critical roles in the structure and function of aquatic food webs. Bioscience. 1999;49:781–788. doi: 10.2307/1313569. [DOI] [Google Scholar]
- Wittmann J., Brancato C., Berendzen K. W., Dreiseikelmann B. Development of a tomato plant resistant to Clavibacter michiganensis using the endolysin gene of bacteriophage CMP1 as a transgene. Plant Pathol. 2016;65:496–502. doi: 10.1111/ppa.12417. [DOI] [Google Scholar]
- Wittmann J., Eichenlaub R., Dreiseikelmann B. The endolysins of bacteriophages CMP1 and CN77 are specific for the lysis of Clavibacter michiganensis strains. Microbiology. 2010;156:2366–2373. doi: 10.1099/mic.0.037291-0. [DOI] [PubMed] [Google Scholar]
- Yin Y., Ni P., Deng B., Wang S., Xu W., Wang D. Isolation and characterisation of phages against Pseudomonas syringae pv. actinidiae. Acta. Agric. Sect. B Soil Plant Sci. 2019;69:199–208. doi: 10.1080/09064710.2018.1526965. [DOI] [Google Scholar]
- Yu J.-G., Lim J.-A., Song Y.-R., Heu S., Kim G. H., Koh Y. J., Oh C.-S. Isolation and characterization of bacteriophages against Pseudomonas syringae pv. actinidiae causing bacterial canker disease in kiwifruit. J. Microbiol. Biotechnol. 2016;26:385–393. doi: 10.4014/jmb.1509.09012. [DOI] [PubMed] [Google Scholar]
- Zaczek-Moczydłowska M. A., Young G. K., Trudgett J., Fleming C. C., Campbell K., O'Hanlon R. Genomic characterization, formulation and efficacy in planta of a Siphoviridae and Podoviridae protection cocktail against the bacterial plant pathogens Pectobacterium spp. Viruses. 2020;12:150. doi: 10.3390/v12020150. [DOI] [PMC free article] [PubMed] [Google Scholar]