Abstract
As a zoonotic disease that has already spread globally to several million human beings and possibly to domestic and wild animals, eradication of coronavirus disease 2019 (COVID‐19) appears practically impossible. There is a pressing need to improve our understanding of the immunology of this disease to contain the pandemic by developing vaccines and medicines for the prevention and treatment of patients. In this review, we aim to improve our understanding on the immune response and immunopathological changes in patients linked to deteriorating clinical conditions such as cytokine storm, acute respiratory distress syndrome, autopsy findings and changes in acute‐phase reactants, and serum biochemistry in COVID‐19. Similar to many other viral infections, asymptomatic disease is present in a significant but currently unknown fraction of the affected individuals. In the majority of the patients, a 1‐week, self‐limiting viral respiratory disease typically occurs, which ends with the development of neutralizing antiviral T cell and antibody immunity. The IgM‐, IgA‐, and IgG‐type virus‐specific antibodies levels are important measurements to predict population immunity against this disease and whether cross‐reactivity with other coronaviruses is taking place. High viral load during the first infection and repeated exposure to virus especially in healthcare workers can be an important factor for severity of disease. It should be noted that many aspects of severe patients are unique to COVID‐19 and are rarely observed in other respiratory viral infections, such as severe lymphopenia and eosinopenia, extensive pneumonia and lung tissue damage, a cytokine storm leading to acute respiratory distress syndrome, and multiorgan failure. Lymphopenia causes a defect in antiviral and immune regulatory immunity. At the same time, a cytokine storm starts with extensive activation of cytokine‐secreting cells with innate and adaptive immune mechanisms both of which contribute to a poor prognosis. Elevated levels of acute‐phase reactants and lymphopenia are early predictors of high disease severity. Prevention of development to severe disease, cytokine storm, acute respiratory distress syndrome, and novel approaches to prevent their development will be main routes for future research areas. As we learn to live amidst the virus, understanding the immunology of the disease can assist in containing the pandemic and in developing vaccines and medicines to prevent and treat individual patients.
Keywords: COVID-19, cytokine storm, immune response, immunologic tests, immunopathology, infections, pandemic, virus
1. INTRODUCTION
Coronaviruses (CoVs) are enveloped, single positive‐strand RNA viruses belonging to the large subfamily Coronavirinae, which can infect mammals and several other animals. 1 Seven CoVs are known to cause human disease and can be divided into low and high pathogenic CoVs. 2 Four low pathogenic CoVs, 229E, NL63, OC43, and HKU1, cause mild diseases and are globally endemic. They are called non–severe acute respiratory syndrome (SARS)‐like CoVs. Three highly pathogenic, novel zoonotic CoVs have emerged during the last 18 years, which can cause outbreaks and lethal human disease. The SARS coronavirus (SARS‐CoV), now named SARS‐CoV‐1, was discovered in November 2002, 1 and the Middle East respiratory syndrome coronavirus (MERS‐CoV) in June 2012, both caused local outbreaks and were contained before causing a pandemic. 3 SARS‐CoV‐2 was identified in December 2019 after sequencing clinical samples from a cluster of patients with pneumonia in Wuhan, China (Figure 1). 4 The disease caused by SARS‐CoV‐2 is named coronavirus disease 2019 (COVID‐19). Since its first report in December 2019 in Wuhan, China, COVID‐19 was declared a pandemic by the World Health Organization (WHO) on March 11, 2020, and continued to aggressively spread across the globe infecting more than 3 million confirmed cases. 5 Three central variants of the current virus that differ in their amino acid sequence have been identified, namely A, B, and C. The ancestral type A and the mutated type C are found in significant proportions outside East Asia, mainly in Europe and in the United States. The B type which has mutated and spread is the most common type in East Asia. 6 Continuous monitoring of the isolated virus mutations by genome sequencing is needed to accurately monitor the development of the pandemic. 1221 (13.3%) out of 9199 persons tested positive for SARS‐CoV‐2 in Iceland. Many of the participants, 86.1%, had visited areas designated as being at high risk by the end of February (China and the Alps mountain regions in Austria, Italy, and Switzerland). The two most common haplotypes in Iceland are A2a3a and A2a2 after sequencing of 643 out of 1221 positive RNA samples. A2a2 haplotype was most commonly seen in travelers coming from Austria to Iceland. 7
Figure 1.
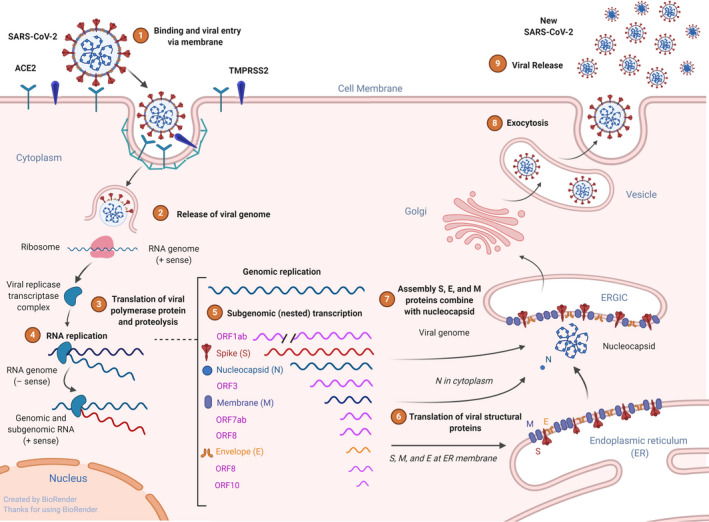
Virus binding, internalization to epithelial cells, and replication. Schematic representation of the genomic and subgenomic organizations of SARS‐CoV and replication. SARS‐CoV‐2 uses receptor ACE2 and transmembrane protease, serine 2 (TMPRSS2) for host cell entry. Following entry to cell cytoplasm, the genomic RNA; the two large open reading frames (ORFs) 1ab are translated into a protein viral transcriptase complex (phosphatase activity and RNA‐dependent RNA polymerase (RdRp) and a helicase). Replication of the genome involves the synthesis of a full‐length negative‐strand RNA and serves as template for full‐length genomic RNA. After translation, structural proteins are localized to the Golgi intracellular membranes, the endoplasmic reticulum Golgi intermediate compartment (ERGIC) that is called site of budding. New virions that are assembled full genome RNA release from the cell
COVID‐19 shows a complex profile with many different clinical presentations. Similar to many other viral infections, the characteristics of currently infected patients and their clinical outcomes may represent the tip of the iceberg (Figure 2). Patients may be asymptomatic; experience mild, moderate, or severe symptoms; and presented with or without pneumonia. 8 , 9 Asymptomatic cases are common but to date there are scarce epidemiological surveys that provide a clear percentage of asymptomatic cases. 8 , 10 Approximately 20% of patients are severe cases that require hospitalization. 11 As of April 2020, the mortality rate is approximately 6% of confirmed cases worldwide, but can vary significantly depending on the quality of healthcare services and hospital capacity. The case fatality ratio of COVID‐19 was estimated substantially higher than recent influenza pandemics (H1N1 influenza in 2009). The mean time from onset to death was 18.8 days in China and 24.7 days out of China. Case fatality ratio was determined as 3.6% (1.9‐7.2). 12
Figure 2.
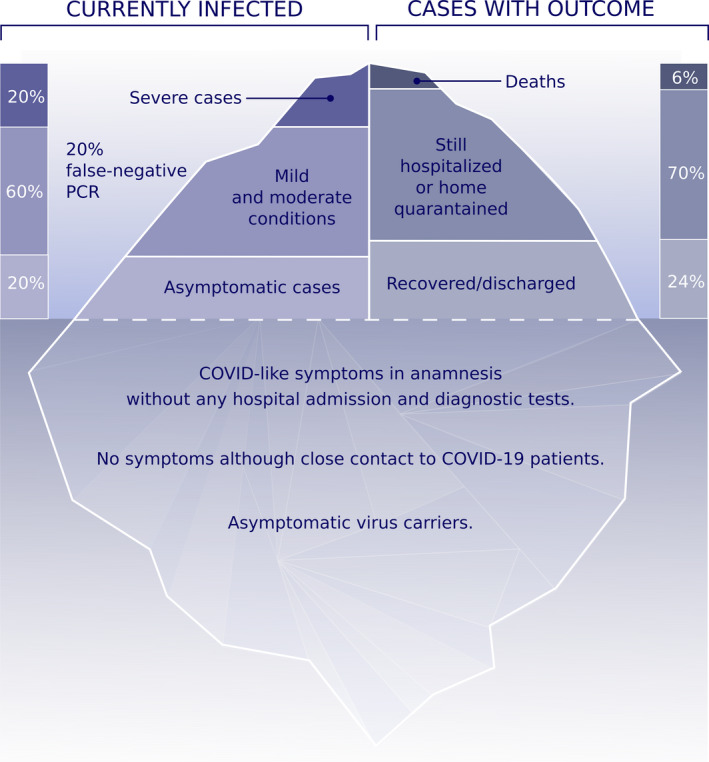
The iceberg of the COVID‐19 pandemic. 10%‐20% of currently diagnosed patients appear with severe cases and 60% with mild to moderate cases. False‐negative viral nucleic acid diagnosis with RT‐PCR should always be considered as 15%‐20% in best experienced hospital conditions, which can be higher in the field. Known asymptomatic cases are diagnosed by random screening of hospital staff and individuals with close contact to COVID‐19 cases in the household. However, there are also a high number of unproven asymptomatic individuals at the bottom of the iceberg with COVID‐like symptoms in the anamnesis without any diagnostic tests and hospital admission. Certain individuals have been reported, who never had symptoms although they had close contact to COVID‐19–positive family members. Overall death rate is 6% and is currently increasing worldwide. The reports on the number of recovered individuals are not currently convincing. Data are collected from https://www.worldometers.info/coronavirus/ and https://www.who.int/health-topics/coronavirus/
Although it is clear that asymptomatic cases exist, 8 the real percentage and how long they carry the virus are not known. This information can be obtained by screening the population for virus‐specific IgM, IgG, and IgA antibodies, which is expected soon in many centers and will be an informative and decisive factor in controlling the pandemic as it is the main indicator of the development of population immunity.
Non–SARS‐CoVs account for 10%‐30% of upper respiratory tract infections in adults and present as a mild common cold disease in humans. 13 Patients with common allergic diseases do not develop additional distinct symptoms or severe outcomes. Allergic children show a mild course similar to nonallergic children. 8 In a study of 1099 cases, 3.8% presented with diarrhea and 5.0% with nausea and vomiting, and these patients were positive for SARS‐CoV‐2 RNA in stool samples. 14 Cases with a previously existing condition of chronic obstructive pulmonary disease or complicated with secondary bacterial pneumonia are more severe and may represent a complex immune pathogenesis. 8 Patients with hypertension, diabetes, obesity, chronic lung disease, active smoking, and old age are more prone to severe disease. 15 The cytokine storm that takes place in severe cases is a major factor for high mortality, multiorgan failure, acute respiratory distress syndrome (ARDS), and disseminated intravascular coagulation.
As it is now a worldwide spread zoonosis, it might be practically impossible to fully eradicate the SARS‐CoV‐2 virus. The containment of the pandemic is challenging as it is very difficult to identify virus carriers due to the nature of this disease. There can be presumably high number of asymptomatic virus carriers, in addition recently infected individuals before the onset of symptoms, clinically recovered individuals who still carry the virus, and many potentially susceptible domestic and wild animals in regular contact with humans (Figure 3). The main question will be to learn how to live together with this virus as humans. There is a pressing need for an improved understanding of its immunopathology, much more than any previous pathogen, as COVID‐19 has become the number one reason of morbidity and mortality in many countries. Even though this pandemic will likely be brought under control in the coming months, unexpected outbreaks and the development of viral resistance to treatments or vaccines are highly possible due to mutations of the virus.
Figure 3.
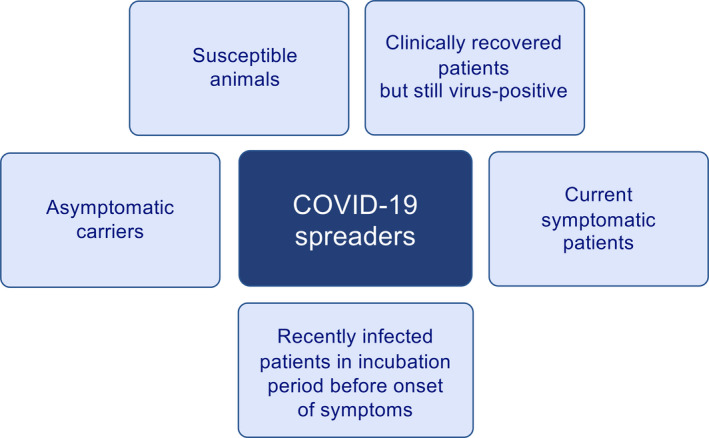
SARS‐CoV‐2 carriers and spreaders. There are different types of virus carriers in COVID‐19, an important factor for the containment of the pandemic. They are listed as asymptomatic virus carriers, current symptomatic patients, recently infected patients in window period before the onset of symptoms, clinically recovered patients who remain SARS‐CoV‐2‐positive, and susceptible domestic and wild animals. All of these have specific immunological mechanisms that are discussed in this review
This article aims to comprehensively review the current knowledge on the immunological changes observed in COVID‐19 infection. All of the previously described clinical manifestations are expected to show distinct local or systemic immune profiles. Most of the immunological studies to date have been reported on hospitalized severe cases, and almost no longitudinal follow‐up studies currently exist. However, it is clear that immune responses to SARS‐CoV‐2 infections include all arms of the immune system, such as tissue barriers, innate and adaptive cells, and mediators. Our combined knowledge and experiences with combating all existing infectious agents is now needed to fight this virus.
2. EPITHELIAL INFECTION AND VIRAL CYCLE
Coronaviruses are named for their large spikes projecting from the envelope giving the virus a crown‐like shape of approximately 100 nm. The envelope consists of a lipid bilayer derived from the cell membrane of the host and four structural proteins, spike (S), envelope (E), membrane (M), and nucleoprotein (N), as well as a variable number of nonstructural proteins. SARS‐CoV‐2 recognizes angiotensin‐converting enzyme 2 (ACE2) to attach to cells, particularly respiratory epithelial cells of the host (Figure 1). 16 , 17 This process is dependent on the host serine protease transmembrane protease serine 2 (TMPRSS2), which cleaves viral spike protein at the S1/S2. S2 subunit allows for fusion of viral and cellular membranes. 18 SARS‐CoV‐2 replicates efficiently in cats and ferrets and poorly in dogs, pigs, chickens, and ducks. It was recently demonstrated that cats are susceptible to experimental airborne infection. 19 The infection of ferrets and cats may be via the same receptor mediated by the transmembrane spike (S). 17 Following receptor binding, the virus can enter the cell cytoplasm via endocytosis. 20 Translation and budding process in vitro and in vivo have been studied in SARS‐CoV‐1 and MERS viruses. 21 Infection with SARS‐CoV‐1 is detected by various intercellular sensors such as RIG I/MDA5/MAVS/TRAF3/IRF3/IRF7 and various TLRs/TRIF/MyD88/IkB/NF‐kB/MAPK/AP‐1 pathways. 22 Importantly, SARS proteins can inhibit antiviral response by blocking RIG I and IRF3/7 pathway on different levels, which leads to inefficient production of type 1 interferons and impaired antiviral response, while increasing NF‐kB activation, pro‐inflammatory cytokine production, and necroptosis. 22 All of these signaling events may lead to increased cellular death, hyperinflammation, and cytokine storm.
3. TRAINED IMMUNITY AND INNATE IMMUNE RESPONSE
Immunological memory in innate immune system is called “trained immunity” and may affect the spread and intensity of certain infections. During the COVID‐19 pandemics, it was hypothesized that general BCG vaccination policies adopted by different countries might have impacted the transmission patterns and/or COVID‐19–associated morbidity and mortality. 23 , 24 Role of the abandonment of BCG vaccination within the last couple of decades in several countries should be considered and investigated, especially by its impact on immune responses to viral infections of non–BCG‐vaccinated young populations. The pathogenicity of COVID‐19 is complex, and the virulence and pathogenicity of the disease are additionally associated with viral activation of cytoplasmic NOD‐like receptor family, pyrin domain containing 3 (NLRP3) inflammasome (Table 1). Inflammasome activation in macrophages, epithelial cells, and maybe even endothelial cells releases pro‐inflammatory cytokines, interleukin (IL)‐1β and IL‐18, which contribute to the pathogenic inflammation responsible for the severity of symptoms of COVID‐19. 25 , 26 In addition, sensing viral RNA by toll‐like receptor (TLR)3, TLR7, TLR8, and TLR9 activates the NF‐κB pathway and a high number of pro‐inflammatory cytokines with a major role in initiating virus‐induced inflammation. 27 There is limited knowledge on the innate immune response, other than elevated levels of acute‐phase reactants and cytokine storm. Most of the reports to date have focused on severe outcomes and adaptive immune responses.
Table 1.
Summary of immunologic characteristics of COVID‐19
| Immunologic changes | COVID‐19 |
|---|---|
| T‐cell responses | Lymphopenia in severe cases (<20%). Initial lymphopenia is predictive of severe disease. |
| CD8+ T cells | Severe lymphopenia (<5%) is observed in CD8+ T cells and can be a predictor of severe disease. |
| Th1‐Th2 responses | Normal antiviral immunity requires a CD4 and CD8 Th1 response. Severe disease shows a systemic severe inflammatory response with a cytokine storm. Cytokine storm response is mainly Th1 and inflammatory. It can also have a major role in inflammasome activation. |
| Eosinophils | Decreased circulating eosinophil numbers in 50%‐80% of the hospitalized patients. |
| Specific antibody levels | In the acute phase, virus‐specific IgM increases followed by virus‐specific IgG during convalescence. |
| Cytokine storm | Innate and adaptive cytokines are released in high amounts linked to severe disease. |
| Acute‐phase reactants | High in severe cases. Initially high values are predictive of severe disease. |
Several innate immune signaling proteins are targeted by SARS‐CoV‐2 viral proteins. The interferon (IFN) pathway is targeted by Nsp13, Nsp15, and open reading frame (ORF)9b, and the NF‐κB pathway is targeted by Nsp13 and Orf9c. SARS‐CoV‐2 Orf6 impedes NUP98‐RAE1, an interferon‐inducible mRNA nuclear export complex. Orf3b and Orf9c of SARS‐CoV‐2 are canonical for replication. As above mentioned, these data will shed light for the development of antiviral against to SARS‐Cov‐2. One of the antiviral drugs is remdesivir, a nucleoside analog RNA‐dependent RNA polymerase (RdRP) inhibitor for treatment of COVID‐19. 28 , 29
4. T‐ AND B‐CELL RESPONSES
Human SARS‐CoV‐2 infection has a classical respiratory virus‐like clinical course in more than 80% of patients with a mild to moderate and self‐limiting course. It involves all of the so far known aspects of innate immune response and T‐ and B‐cell immunity and antiviral neutralizing antibody response (Table 1, Figure 4). Interestingly, SARS‐CoV‐2 infects human T‐cell lines via a novel route through CD147 spike protein, present on the surface of T lymphocytes. 30 CD147 (also known as basigin or EMMPRIN) is expressed in many tissues and cells and plays a role of cell proliferation, apoptosis, tumor cell migration, metastasis, and differentiation, especially under hypoxic conditions. A second isoform of CD147, called CD147 Ig0‐Ig1–Ig2, has also been characterized. Dysregulation of CD147 has been associated with nearly every type of cancer and regulates matrix metalloproteinase (MMP) and vascular endothelial growth factor production or signals for tumor cell invasion and metastasis. 31 To block CD147 protein by meplazumab, preventing SARS‐CoV‐2 spike binding and subsequent infection may also have beneficial effects on COVID‐19 treatment. 32
Figure 4.
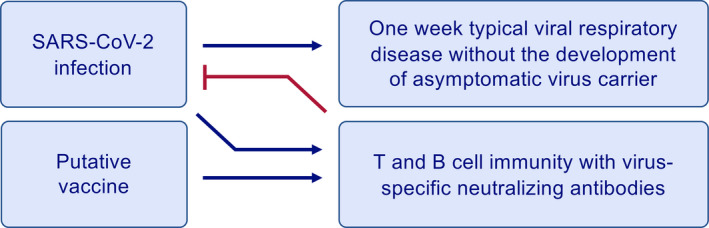
Ideal immune response to SARS‐CoV‐2 infection or a vaccine. Acute infection and the vaccine are expected to develop same type of an immune response with T‐ and B‐cell immunity and development of virus‐specific neutralizing antibodies. The normal immune response is characterized with one‐week typical viral respiratory disease without the development of asymptomatic virus carriers. This type of immune response is observed in almost all mild cases
4.1. T‐ and B‐cell epitopes
Upon entry into the host, a virus attaches to and invades cells expressing its specific receptor in order to replicate. Once the virus is inside the tissue cells, such as respiratory epithelial cells in the case of SARS‐CoV‐2, viral peptides are presented through class I major histocompatibility complex (MHC) proteins to CD8+ cytotoxic T cells. 33 CD8+ T cells become activated and start to divide and show clonal expansion and develop virus‐specific effector and memory T cells. CD8+ cytotoxic T cells lyse the virus‐infected tissue cells (Figure 5). For a short time, whole virus and viral particles are recognized by professional antigen‐presenting cells, which are mainly dendritic cells and macrophages and present viral peptides to CD4+ T cells through MHC‐Class‐II molecules. 33 B cells can directly recognize the viruses and get activated by them and also interact with CD4+ T cells. The IgM isotype primary virus‐specific antibody response is observed within the first week following symptoms. 34 IgG isotype antibodies follow the early IgM response that mostly retain a lifelong immunity (Figure 6).
Figure 5.
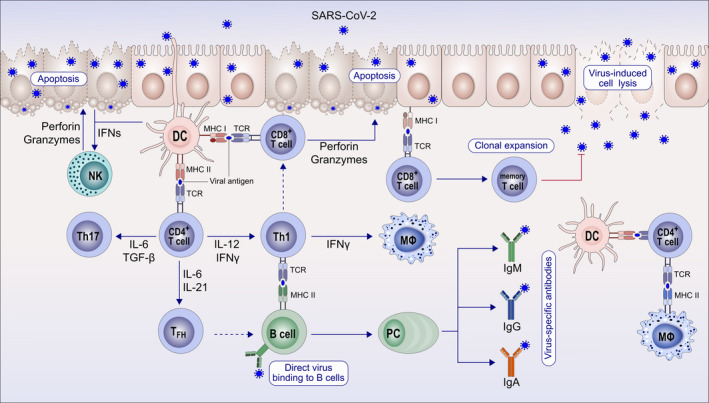
Immune response to coronaviruses and other respiratory viruses. After the epithelium is infected with SARS‐CoV‐2, the replicating virus can cause cell lysis and direct damage to the epithelium. The epithelium presents virus antigens to CD8+ T cells. With their perforin and granzymes, CD8+ T cells and natural killer (NK) cells can show cytotoxicity to virus‐infected epithelial cells and induce apoptosis. Subepithelial dendritic cells (DC) recognize virus antigens and present them to CD4 T cells and induce the differentiation of these T cells toward memory Th1, Th17, and memory T follicular helper (FH). TFH helps B cells to develop into plasma cells (PC) and promotes the production of IgM, IgA, and IgG isotype virus‐specific antibodies. Tissue macrophages (MΦ) and DCs also present viral antigens to CD4+ T cells
Figure 6.
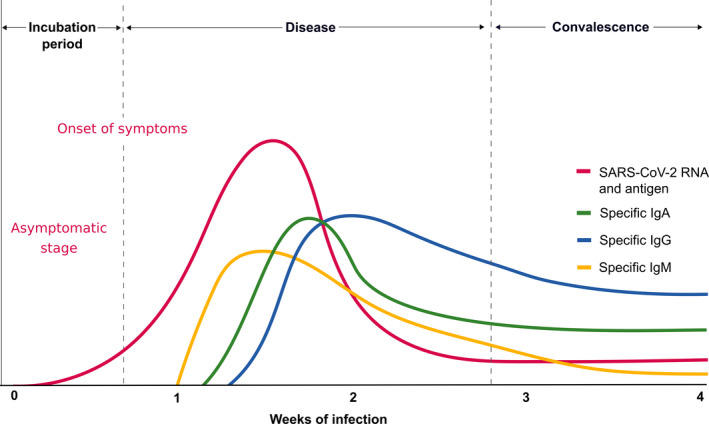
Specific antibody response to SARS‐CoV‐2. The incubation period of COVID‐19 is relatively long and has been reported to be 5‐10 d. A specific IgM response is the early antibody response that starts and peaks within 7 d. IgM continues as long as the acute phase of the disease continues. Specific IgA and IgG antibodies develop several days after IgM and do not decrease to undetectable levels and are assumed to continue lifelong as protective antibodies. This research requires an international consensus on the usage of correct methodology and antigens of SARS‐CoV‐2
T‐ and B‐cell response to viral determinants and recognition of virus‐infected cells prove to be central to our ultimate understanding of several major phenomena about antiviral immune responses. Coronavirus spike protein harbors the S1 and S2 subunits, which are cleaved at the S1/S2 boundary and the S2′ cleavage site. SARS‐CoV‐2 spike protein harbors a polybasic furin cleavage sequence (PRRARS) with an insertion of four amino acid residue distinct from SARS‐CoV and other SARS‐like viruses. Indicated immunodominant T‐ and B‐cell epitopes have been reported. 35 As SARS‐CoV‐2 is the third virus of its kind making an outbreak, due to similarities with the previous 2 viruses, SARS‐CoV‐1 and the MERS‐CoV, it is an important question whether the existing population immunity from 2002 and 2012 outbreaks is protective and whether vaccine development efforts of those times may continue from where they stopped because there were limited numbers of cases to make clinical trials for the developed vaccines. These efforts are fully dependent on the similarity of the T‐ and B‐cell epitopes. Based on the similarity of the SARS‐CoV‐2 to SARS‐CoV potential T‐cell and B‐cell epitopes, has been predicted using bioinformatic approach. 36 Three SARS‐recovered individuals, 9 and 11 years postinfection, were analyzed for T‐cell responses against a total of 550 peptides from SARS‐CoV structural proteins that may also show cross‐reactivity to MERS‐CoV. SARS‐specific memory T cells persist at 9 and 11 years post‐SARS in the absence of antigen. All memory T cells detected were specific against SARS‐CoV structural S, N, and M proteins, but lacked cross‐reactivity to MERS. 37 Structural proteins of SARS‐CoV‐2 are genetically similar to SARS‐CoV, but not to MERS‐CoV. Only 23% and 16% of known SARS‐CoV T‐ and B‐cell epitopes map identically to SARS‐CoV‐2, respectively. 38 Clearly recognizable T‐ and B‐cell epitopes represent a potential for eliciting a robust T‐cell or antibody response in SARS‐CoV‐2 or in response to its mutual vaccine. The observation that many B‐ and T‐cell epitopes are highly conserved between SARS‐CoV and SARS‐CoV‐2 is important. Vaccination strategies designed to target the immune response toward these conserved epitope regions could generate immunity that is not only cross‐protective across multiple coronaviruses, but also relatively resistant to ongoing virus evolution to protect against mutated future virus strains. 36
5. LYMPHOPENIA IN T CELLS
Lymphopenia associated with COVID‐19 is an important pathological finding and severity criteria, which serves as like a biomarker and a possible target for intervention to minimize the risk of severe disease. Some infectious agents, such as human immunodeficiency virus (HIV), can cause lymphopenia in humans. Although it is not a part of the natural course and expected type of immune response, it can be observed in influenza, tuberculosis, malaria, and sepsis. 39 The changes in peripheral lymphocyte counts and the transition of lymphocyte subgroups may suggest possible mechanisms in the pathogenesis of SARS‐CoV‐2 infection. Limited numbers of studies have focused on severe COVID‐19 cases, which have a relatively distinct profile of decreased memory T cells and cytotoxic CD8+ T cells. Flow cytometric analyses showed that the percentage of CD4+ naïve T cells (CD3+CD4+CD45RA+) increased and memory helper T cells (CD3+CD4+CD45RO+) decreased in peripheral blood. 40 The percentage of CD3+CD8+CD28+ cytotoxic T cells also decreased in severe cases. However, there was no significant difference in activated total T cells (CD3+HLADR+) and activated cytotoxic T cells (CD3+CD8+HLA−DR+). These severe patients also presented with lower levels of regulatory T (Treg) cells (CD3+CD4+CD25+CD127low+). 40 In addition, the overactivation of T cells, manifested by an increase in T‐helper (Th)17 and the high cytotoxicity of CD8+ T cells, partially accounts for the severe immune injury. 41 In a similar study, the levels of peripheral lymphocyte subsets were evaluated in 60 patients with COVID‐19 by flow cytometry during the course of disease. Decrease in total lymphocytes, CD4+ T‐cell, CD8+ T‐cell, B‐cell, and natural killer (NK)‐cell counts were observed. The authors suggested that lymphopenia in CD8+ T cells could be an independent predictor for COVID‐19 severity and treatment efficacy. 42 In another earlier study analyzing laboratory indexes, twelve patients with COVID‐19 showed a decreased percentage of lymphocytes and CD8 cell count. 43 In 286 patients with COVID‐19, severe cases had higher leukocyte and neutrophil and lower lymphocyte counts, with a high neutrophil‐to‐lymphocyte ratio, as well as lower percentages of monocytes, eosinophils, and basophils. The mean T‐ and NK‐cell counts were below normal levels, while B‐cell values were at the low end of the normal range. T cells were shown to be more affected by SARS‐CoV‐2 as the T‐cell count was nearly half the lower reference limit and tend to be more reduced in severe cases. 40 Lymphocyte subset analysis revealed a decreased percentage of CD16+ CD56+ lymphocytes (4 out of 8 patients) and increased percentage of CD3+ (2 out of 8 patients), CD4+ (4 out of 8 patients), and CD8+ (1 out of 8 patients) lymphocytes. 44 In a study analyzing critically ill patients with SARS‐CoV‐2 pneumonia, 85% of the patients showed lymphopenia. 45 Intensive care unit (ICU) patients suffering with this infection had a median lymphocyte count of 800 cells/microL (normal values 1000‐4000 cells per 1 µL). 45 , 46 It was observed that lymphocytes in patients with COVID‐19 gradually decrease as the disease progresses. The older patients with lower counts of lymphocytes and platelets were at higher risk of severe disease and increased length of hospitalization. 47 A significant reduction in lymphocyte counts and their rapid depletion may play a role in the pathogenesis and contribute to the progression to severe COVID‐19. Accordingly, drugs targeting lymphocyte proliferation or preventing apoptosis, such as interleukin IL‐2, IL‐7, or programmed cell death protein 1 (PD1/PD‐L1) inhibitors, could help to prevent lymphopenia or restore lymphocyte counts in severe patients. 47 The mean values of the three main subsets of lymphocytes generally decreased in severe patients with COVID‐19, with T and NK cells were below normal values, while B cells were at the lower end of the normal range. 40
In view of the above results, lymphocyte percentage was suggested as a predictive biomarker for severity or recovery (Table 2). 48 At the first time point, 10‐12 days after symptom onset, patients with lymphocyte percentage greater than 20% were classified as mild to moderate and recovered quickly. Patients with less than 20% lymphocytes at the time of diagnosis were initially classified as severe. At the 2nd time point, 17‐19 days after the onset of symptoms, patients with more than 20% lymphocytes are accepted as being in recovery, and patients with a lymphocyte % between 5% and 20% are still in an at‐risk group, while patients with a lymphocyte % less than 5% are critically ill with a high mortality rate.
Table 2.
The percentage of lymphocytes is decisive for prognosis at the first and second visits 48
| Blood lymphocyte percentage |
1st time point 10‐12 d after symptom onset |
2nd time point 17‐19 d after symptom onset |
|---|---|---|
| >20% | Mild/Moderate | Recovering |
| 5%‐20% | Severe | Risky |
| <5% | Severe | Critically ill |
The mechanisms by which there is a significant lymphocyte reduction in severe cases remain unclear. The type of lymphocyte death should be extensively studied in COVID‐19. It may represent a therapeutic modality for severe cases, if its mechanisms are better understood. The apoptotic process of lymphocytes can be regulated in the cell by exogenous and/or intrinsic factors and can be modulated with the role of pro‐ and anti‐apoptotic molecules. 49 , 50 Lymphopenia has been found in SARS and MERS, which were reported to induce apoptosis in the liver, lung, and T lymphocytes. 51 To investigate the role of apoptosis in lymphopenia patients with SARS, fifteen patients were investigated for plasma soluble Fas‐ligand levels and altered cleaved caspase‐3 activation. SARS patients were found to have higher plasma Fas‐ligand levels, associated with higher intracellular cleaved caspase‐3–positive CD4 and CD8 lymphocytes in the acute phase of SARS. 52 It was proposed that apoptosis of lymphocytes induces lymphopenia in critically ill patients with SARS‐CoV‐2 infection. 53 In addition, pyroptosis induced by IL‐1β has been proposed. 54 It has to be noted here that evidence is accumulating on direct infection of T cells with SARS‐CoV‐2, which may also cause cytopathic effect on infected T cells. 30 Other mechanisms of lymphopenia that remain to be studied are bone marrow suppression during cytokine storm, and sequestration in the lungs during extensive bilateral pneumonia.
High‐ versus low‐dose virus exposure of SARS‐CoV‐2 may also influence lymphocyte responses. High‐dose exposure is the case with healthcare workers and individuals who have currently sick or asymptomatic virus spreading family members. Whereas low‐dose infection may lead to appropriate effector T‐ and B‐cell response, neutralizing antibodies, and rapid viral clearance, high‐dose exposure may cause severe disease and delayed viral clearance. This may be due to lymphopenia leading to inefficient T‐ and B‐cell immunity, subsequently the cytokine storm and destructive tissue inflammation (Figure 7).
Figure 7.
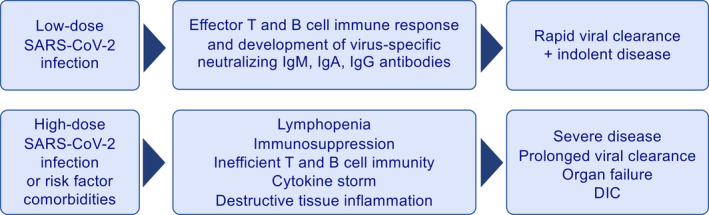
Distinct response to high‐ and low‐dose virus exposure and infection. High‐dose exposure may occur in healthcare workers and individuals who are exposed to currently sick or asymptomatic virus spreading family members. Whereas low‐dose infection may lead to appropriate effector T‐ and B‐cell response, neutralizing antibodies, and rapid viral clearance, high‐dose exposure may cause severe disease and delayed viral clearance. This may be due to lymphopenia leading to inefficient T‐ and B‐cell immunity, subsequently the cytokine storm and destructive tissue inflammation
5.1. Regulatory T cells (Tregs)
Treg cells play a crucial role in suppressing excessive immune responses to pathogens, cancer cells, and transplanted organs and to prevent and control the development of autoimmune and allergic diseases. 55 , 56 Treg cells can play a crucial role in the airway mucosa by suppressing effector cells and tissue‐damaging mechanisms. 57 , 58 In respiratory virus infections, Treg cells can limit pulmonary immunopathology. 59 Tregs can protect from central nervous system immunopathology by controlling CD8 T‐cell responses in West Nile infection. 60 The molecular mechanisms involved in regulating forkhead box P3 (FOXP3) expression and antigen‐specific response of Treg cells in COVID‐19 remain unclear, and further studies are needed to exploit their applicability in the clinical setting. Decreased numbers of circulating Treg cells (CD3+CD4+CD25+CD127low+) as a part of lymphopenia have been reported in COVID‐19. 40
6. EOSINOPENIA
Within many of their functions, eosinophils have been previously suggested as cells with antiviral effects. 61 , 62 Eosinophils contain and produce molecules with antiviral activity and participate in adaptive immunity, serving as antigen‐presenting cells as demonstrated against some respiratory viruses in vitro and in vivo, including respiratory syncytial virus and influenza. 61 In addition to lymphopenia, eosinopenia was observed in 73 out of 138 (52.9%) in hospitalized COVID‐19 cases. Decreased blood eosinophil counts correlate positively with lymphocyte counts in severe (r = .486, P < .001) and nonsevere (r = .469, P < .001) patients after hospital admission. 15 Eosinopenia has been reported in three following studies. It was suggested that eosinophil counts below normal levels could be a viable biomarker for COVID‐19 diagnosis. The changes in peripheral blood leukocyte differential counts in patients with COVID‐19 and patients with other viral pneumonia were compared. Whereas 70% of patients in the COVID‐19 group presented with eosinopenia, only 16.7% of the patients in non–COVID‐19 viral pneumonia group experienced a decline in eosinophil numbers below the normal levels. 63 In a study of the clinical features of 85 fatal cases with COVID‐19 in two hospitals in Wuhan, 81.2% patients had very low eosinophil counts on admission. 64 Accordingly, eosinopenia was suggested to have a diagnostic value or can also be an indicator of disease severity. The observed eosinopenia in particularly in severe COVID‐19 cases could be due to a number of reasons. It can be a result of immune exhaustion; it can represent the high migration rate of eosinophils from the peripheral blood to the infected organ or can be due to continuous and potent type 1 responses that antagonize the type 2 response, including IL‐5 that may promote eosinophil survival and activation. In addition, inhibition of eosinophil egress from the bone marrow, blockade of eosinophilopoiesis, reduced expression of chemokine receptors/adhesion factors, and/or direct eosinophil apoptosis induced by type 1 interferons released during the acute infection can be counted within these mechanisms. No eosinophil enrichment in the lung tissue has been observed in samples from patients at early stages of COVID‐19 or in postmortem analyses. 65 , 66
7. CYTOTOXIC CD8 T‐CELL RESPONSES
Cytotoxic T lymphocytes and NK cells in patients infected with SARS‐CoV‐2 are essential for mounting an appropriate antiviral response. 67 The classical convention is that the T‐cell receptor of the CD8 cytotoxic T cells recognizes the viral peptides presented by MHC‐class‐I molecules of virus‐infected cells and is cytotoxic to virus‐infected cells via multiple mechanisms including perforin and granzymes (Figure 5). 68 CD8 T cells are critical for mediating clearance following many acute viral infections in the lung. In addition, memory CD8 T cells are capable of providing protection against secondary infections. Therefore, the combined induction of virus‐specific CD8 T cells and antibodies may provide optimal protective immunity. 69 In COVID‐19, these cells are strongly affected from the observed lymphopenia and their numbers are decreased. As a part of the lymphopenia, COVID‐19 patients show functional exhaustion of cytotoxic lymphocytes associated with SARS‐CoV‐2 infection. The total number of NK and CD8+ T cells was markedly decreased in patients with SARS‐CoV‐2 infection. In addition, increased expression of NK group 2 member A (NKG2A) was thought to be responsible for the hampered function of exhausted NK and CD8+ T cells. Interestingly, the number of NK and CD8+ T cells was restored in parallel to reduced expression of NKG2A in patients convalescing after therapy. 67 These data suggest that a breakdown of antiviral immunity may play a role in the pathogenesis and severity in COVID‐19. In CD8+ T cells, the frequency of the nonexhausted (PD‐1− CTLA‐4− TIGIT−) subset in the severe group was significantly lower. 70 It was suggested that the virus promotes an initial excessive activation in the beginning of the disease and is followed by subsequent exhaustion of CD8+ T cells.
8. VIRUS‐SPECIFIC ANTIBODY RESPONSES
Similar to many other viral diseases, an increase in virus‐specific IgM in the acute phase followed by an increase in virus‐specific IgG at later phases has been observed in the course of COVID‐19 (Figure 6). 8 This area is fully open to future research; however, it is anticipated that infusion of the convalescent serum obtained from already recovered patients may prevent or limit SARS‐CoV‐2 infection. 71 It was reported that 10 seriously ill patients receiving convalescent serum therapy demonstrated improved lung function, oxygenation, reduced inflammation, and viral load. 72 There are more than one million recovered patients as of today, whose convalescent serum can be used for passive immunization for treatment or prevention. One important point to emphasize convalescent serum treatment is if the patient is at high virus load condition with massive pneumonia, convalescent serum containing IgG1‐, IgG2‐, and IgG3‐type virus‐specific antibodies may activate complement in the lungs and increase tissue destruction. In such cases, virus‐specific IgG4 treatment should be preferentially considered as it does not have any complement activating property. 73 Total immunoglobulins (IgA, IgG, and IgM) and complement proteins (C3 and C4) in patients with COVID‐19 were within normal range. 40 An increase was noted in IgG or IgM antibody levels against SARS‐CoV‐2 nucleoprotein (NP) or receptor‐binding domain (RBD) in most of the patients 10 days or later after onset of symptoms. An early increase in IgM later followed by development of IgG is a normal expected antibody response (Figure 6). However, specific IgG levels can be found at already high levels in serum at the same time or earlier than IgM against SARS‐CoV‐2. 34 In another study, SARS‐CoV‐2 virus‐specific IgG and IgM reached to peak levels 17‐19 days and 20‐22 days after symptom onset, respectively. Another interesting observation was that IgG and IgM titers in the COVID‐19 severe group are higher than the nonsevere group. Different types of seroconversion were reported, such as synchronous seroconversion of IgG and IgM, IgM seroconversion earlier than that of IgG, and IgM seroconversion later than that of IgG. 34 , 74 In a study analyzing specific IgM and IgG by enzyme immunoassay, more patients were found seropositive for IgG than IgM at day 0 and day 5 of hospital admission. A higher proportion of patients in that study also had earlier IgG than IgM seroconversion. 75 These findings suggest cross‐reactivity of the used antigens with previously existing specific IgG due to other coronaviruses and such assays may not be useful for diagnosis of COVID‐19.
The development of mucosal immunity via IgA may be important for preventing SARS‐CoV infections. It has been demonstrated in the context of influenza infection that secretory IgA plays a crucial role in the protection on mucosal surfaces by neutralizing the virus or preventing the attachment of viruses to the mucosal epithelium. Passive immunization was sufficient to prevent infection of the lung, in influenza but did not appear to protect the upper respiratory tract of mice and ferrets against viral infection. 76 Secretory influenza hemagglutinin (HA)‐specific IgA response in the respiratory tract is more effective and more cross‐protective against influenza infections than the systemic immunity induced by parenteral vaccines in human and mice models. 77 , 78 Mice orally vaccinated with influenza viruses have a protective effect in the nose, which is thought to be related to a nasal IgA antibody response. 79 The influenza virus‐specific secretory IgA response has a protective role in the pulmonary fluids of aged mice following oral vaccination with inactivated virus. 80 MERS‐CoV‐S1–specific neutralizing IgA antibodies in the bronchoalveolar lavage fluid were shown to be protective in mice. 81 SARS‐CoV–specific IgA may play a role in protection for the vaccinated mice with SARS. 82 Intranasal vaccination with recombinant adenovirus that have SARS genome induced local IgA and systemic IgG neutralizing antibodies and specific T‐cell responses, which are able to protect against SARS‐CoV infection in animal models. 83 The sensitivity and specificity of current commercial specific antibody kits should be evaluated in independent laboratories, and WHO‐confirmed tests should be made available. The IgA‐based enzyme‐linked immunosorbent assay (ELISA) kit was more sensitive but less specific than the IgG‐based ELISA kit. 84 However, new kits are continuously coming to market and it is possible to develop in‐house kits depending on the quality of the coated SARS‐CoV‐2 antigen to plates. The evaluation of IgM and IgG isotype virus‐specific antibody levels together with RT‐PCR are shown in Table 3. Home‐made indirect ELISA for detecting IgM, IgA, and IgG antibodies against SARS‐CoV‐2 using purified recombinant nucleocapsid (NP) proteins as coating antigens has been studied. The development time for specific antibodies has significantly varied from method to method, and in this study, it was 5 days (range 3‐6 days) for specific IgM and IgA, whereas 14 days (range 10‐18 days) for specific IgG after the onset of symptoms (Figure 6). 85
Table 3.
Proposed immunologic criteria for the diagnosis of the stage of the disease
| Test results | Clinical significance | ||
|---|---|---|---|
| PCR | IgM | IgG | |
| + | − | − |
Proposed immunologic period of infection before the symptoms develop Early days of infection before detectable antibody response Antibody assay may have given false‐negative results |
| + | + | − | Early stage of infection approximately 5‐7 d after the beginning of symptoms |
| + | + | + | Active phase of infection |
| + | − | + |
Late phase or recurrent infection IgM ELISA is false‐negative |
| − | + | − |
Early stage of infection IgG may be false‐negative RT‐PCR result may be false‐negative |
| − | − | + |
Past infection Recovered PCR‐negative patients Cross‐reactivity with other coronaviruses |
| − | + | + |
Recovery stage of infection Early infection with false‐negative RT‐PCR |
9. ACUTE‐PHASE REACTANTS
Acute‐phase reactants are potential diagnostic markers and predictors of disease outcome in human and animals. 86 , 87 Infectious and noninfectious agents can induce an inflammatory response, where acute‐phase proteins increase and play a role in the host defense against infection. The follow‐up of SARS and MERS patients initially demonstrated the importance of acute‐phase reactants for diagnosis and prediction of the prognosis in severe coronavirus infections. It was reported that most patients with SARS had elevated C‐reactive protein (CRP), alanine transaminase (ALT), lactate dehydrogenase (LDH), and creatine kinase levels. 88 Among 45 MERS‐CoV–infected patient, 13 (28.9%) had no pneumonia, 19 patients had pneumonia without respiratory failure (42.2%), and 13 patients had respiratory failures (28.9%). Increased CRP levels represented a predictive factor for pneumonia development and respiratory failure in that study. 89
The overall evaluation of acute‐phase reactants in COVID‐19 correlates high levels to increased disease severity and death (Table 4). In 140 hospitalized COVID‐19 patients, significantly higher levels of D‐dimer, CRP, and procalcitonin (PCT) were associated with severe disease compared to nonsevere disease. Nonsurvivors had higher levels of neutrophils, D‐dimer, blood urea nitrogen, and creatinine compared to recovered patients. 15 Anticoagulation therapy is recommended for COVID‐19 patients, when the D‐dimer value is 4 times higher than the normal upper limit, except for patients with anticoagulant contraindications. 45 , 90 Zhou et al reported elevated levels of ALT, LDH, high‐sensitivity cardiac troponin I, creatine kinase, D‐dimer, serum ferritin, IL‐6, prothrombin time (PT), creatinine, and PCT were associated with mortality. 91 Another group investigated forty‐three (mild N = 28 and severe N = 15) adult patients with COVID‐19. The levels of CRP, IL‐6, PT, fibrinogen, and D‐dimer were significantly higher in the severe group. 92 In a total of 135 patients with COVID‐19, severe patients (N = 40) showed higher plasma levels of PT, activated partial thromboplastin time, D‐dimer, LDH, PCT, albumin, CRP, and aspartate aminotransferase (AST) compared to the mild group (N = 95). Severe patients were significantly older (median age 56 years) and were more likely to have underlying comorbidities, such as diabetes, cardiovascular disease, hypertension, and malignancy. 93 In another study, 113 patients died of COVID‐19 and 161 patients fully recovered and were discharged. D‐dimer concentrations were significantly higher in deceased patients (4.6 μg/mL) compared to recovered patients (0.6 μg/mL). Concentrations of PCT, high‐sensitivity CRP, ferritin, and erythrocyte sedimentation rate were significantly higher in deceased patients. 94 Significantly elevated levels of LDH, creatine kinase, troponin I, PT, D‐dimer, CRP, ferritin, PCT, and IL‐6 were repeatedly reported at the time of initial diagnosis, suggesting the development of a severe outcome (Table 4). Further research, consensus, and standard approach are needed to identify which acute‐phase reactants are essential for early diagnosis, prediction, and follow‐up of patients. 95
Table 4.
Changes in acute‐phase reactants and serum biochemistry in COVID‐19
| Acute‐phase reactants and serum biochemistry | Changes in COVID‐19 | References |
|---|---|---|
| C‐reactive protein (CRP) | ↑ | [15, 44, 91, 92, 94, 107, 131, 132, 133, 134, 135, 136] |
| Erythrocyte sedimentation rate (ESR) | ↑ | [94, 135, 136] |
| Procalcitonin (PCT) | ↑ | [15, 44, 91, 94, 107, 131, 134, 137, 138] |
| Interleukin 6 (IL‐6) | ↑ | [27, 44, 91, 92, 139] |
| Urea nitrogen (BUN) | ↑ | [94, 135, 140] |
| Creatinine (CRN) | ↑ | [16, 91, 94, 133, 135] |
| Alanine transaminase (ALT) | ↑ | [44, 91, 94, 132, 133, 139] |
| Aspartate aminotransferase (AST) | ↑ | [94, 107, 133, 141] |
| Albumin (ALB) | ↓ | [94, 107] |
| Lactate dehydrogenase (LDH) | ↑ | [44, 45, 91, 94, 107, 131, 135, 136, 139, 141, 142, 143] |
| Ferritin | ↑ | [91, 94, 139] |
| Fibrinogen (FIB) | ↑ | [92, 94, 144] |
| D‐dimer | ↑ | [15, 45, 91, 92, 94, 107, 131, 135, 137, 139, 145] |
| Serum amyloid A (SAA) | ↑ | [15, 131] |
| Cardiac troponin I (cTnI) | ↑ | [91, 94, 131] |
| Prothrombin time (PT) | ↑ | [91, 92, 94, 145] |
| Activated partial thromboplastin time (APTT) | ↑ | [94, 107] |
10. CYTOKINE STORM
Although most of the COVID‐19 patients recover with mild and moderate disease in one week, some develop to severe pneumonia in the second week followed by cytokine storm, ARDS, multiorgan failure, and disseminated intravascular coagulation (DIC) within the 3rd week of the disease (Figure 8). The cytokine storm is a complex network of severe molecular events unified by a clinical phenotype of systemic inflammation, multiorgan failure, and hyper‐ferritinemia. It is consistently linked with severity and fatal disease outcome. Cytokine storm is induced by the activation of large numbers of white blood cells, including B cells, T cells, NK cells, macrophages, dendritic cells, neutrophils, monocytes, and resident tissue cells, such as epithelial and endothelial cells, which release high amounts of pro‐inflammatory cytokines. 96 Any cells that bloodstream or the virus itself and degraded viral products can reach in the body can contribute to cytokine storm. Cytokine storm can also occur in a number of infectious and noninfectious diseases, such as bacterial sepsis, Ebola and other hemorrhagic fevers, influenza, and blunt trauma, and as a side effect of immune stimulatory drugs. 97 , 98 , 99 , 100 , 101 , 102 Although many cells are involved as mentioned above, multiple pro‐inflammatory cytokines and inflammasome activation appear to play a major role in its pathogenesis. IL‐10 appears to control tissue damage in experimental models. 103 A pro‐inflammatory eicosanoid storm may also accompany the cytokine storm. 104 The balance of the pro‐/anti‐inflammatory and proresolving lipid mediator response during viral infection is often decisive over the clinical outcome. 105 Among the numerous molecules that increase in serum in cytokine storm, complements, IFN‐γ, IL‐1β, IL‐6, IL‐12, IL‐17, and tumor necrosis factor‐α (TNF‐α), are of crucial importance. 106 In 88 SARS patients, IFN‐γ, IL‐18, TGF‐β, IL‐6, IP‐10 (interferon‐γ–inducible protein‐10), monocyte chemoattractant protein‐1 (MCP‐1), monokine induced by IFN‐γ(MIG), and IL‐8, but not TNF‐α, IL‐2, IL‐4, IL‐10, IL‐13, or TNFRI, were highly elevated in the acute‐phase sera of SARS patients. IFN‐γ, IL‐18, MCP‐1, MIG, and IP‐10 were immediately upregulated following the onset of fever in patients with SARS. Furthermore, IL‐18, IP‐10, MIG, and MCP‐1 levels were significantly higher in the deceased patients compared to survivors. 97 In a recent study, the RNA sequencing transcriptional changes in a small number of bronchoalveolar lavage fluid (BALF) and peripheral blood mononuclear cells (PBMC) specimens from COVID‐19 patients. 107 Several immune pathways and pro‐inflammatory cytokines were induced by SARS‐CoV‐2 infection, notably CC chemokine ligand (CCL)2, C‐X‐C motif chemokine ligand(CXCL)2, CCL8, CXCL1, IL33, CCL3L1 in BALF and IP‐10, tumor necrosis factor superfamily (TNFSF)10, tissue inhibitors of metalloproteinases (TIMP)1, C5, IL18, amphiregulin, neuregulin1, and IL10 in PBMC, indicating sustained inflammation and cytokine storm in the patients. Pathway analysis of PBMC transcriptome revealed that patient's lymphopenia may be associated with activation of apoptosis and P53 signaling pathway in lymphocytes suggesting a role for lymphopenia. 107
Figure 8.
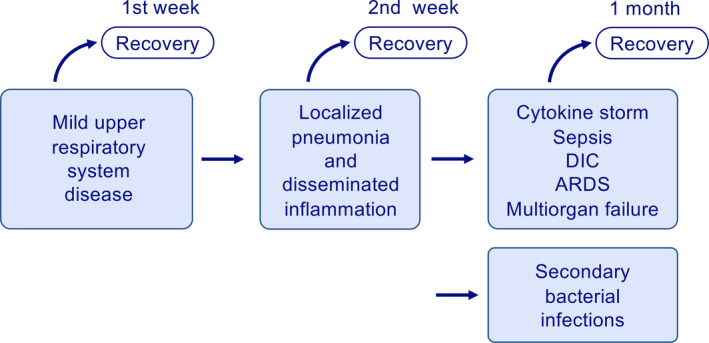
Different clinical phases of COVID‐19. COVID‐19 presents with three stages of diseases. It starts with upper respiratory disease symptoms followed by pneumonia and disseminated inflammation, and in the third‐stage cytokine storm, sepsis‐like syndrome, ARDS, DIC, and multiorgan failure take place. The disease that moves into second phage always carries the risk for secondary bacterial infections. Antiviral immune response and tissue and systemic inflammation are different in all stages
It has been previously reported that the Ebola virus Tim‐1 protein can induce a cytokine storm. 100 SARS‐CoV activates a considerable level of pro‐inflammatory cytokines in vitro, such as TNF‐α, IL‐6, and IL‐12 release via the TLR7 and TLR8, almost twofold higher than a strong TLR7/8 stimulating ssRNA40. 108 Upon influenza infection, high levels of complement C3 and C5, including fragments C3a and C5a, are produced. H5N1 replicates in the lower respiratory tract leading to complement activation, and C5a is responsible for the acute lung injury. 109
Similar to SARS and MERS, cytokine storm is a common feature of severe COVID‐19 cases and a major reason for ARDS and multiorgan failure. Several levels of evidence suggest that mortality in COVID‐19 is due to the virus‐activated “cytokine storm syndrome.” 110 In a study of 41 hospitalized COVID‐19 patients, high levels of pro‐inflammatory cytokines were observed in severe cases, including IL‐2, IL‐7, IL‐10, granulocyte‐colony stimulating factor, IP‐10, MCP‐1, macrophage inflammatory protein 1 alpha, and TNF‐α. These findings are in line with SARS and MERS in that the presence of lymphopenia and “cytokine storm” may play a major role in the pathogenesis of severe COVID‐19. Patients with COVID‐19 had elevated levels of IL1B, IFNγ, IP10, and MCP‐1 suggesting hyperactivation of Th1 cell responses. 45 , 111 Predictors of fatality in a recent retrospective, multicenter study of 150 confirmed COVID‐19 cases in Wuhan, China, included elevated ferritin and IL‐6 suggesting that mortality might be due to virally driven hyperinflammation. 110 Increased circulating cytokines are visible in patients with fever, and they are mostly tolerated in mild cases, however turns into a tissue‐damaging storm condition affecting severe patients. Although with limited efficacy, many agents or methods have been proposed for the treatment of cytokine storm syndromes, such as T‐cell inhibitors, IFN‐γ neutralization, IL‐6 receptor blockade, IL‐6 neutralization, corticosteroids, B‐cell ablation with rituximab, T‐cell ablation with anti‐thymocyte globulin, T cell–directed immunomodulation, blockade of IL‐1 family member cytokines, such as IL‐33 receptor blockade, IL‐1β blockade, and IL‐18 binding protein, and JAK inhibition. 96
11. ACUTE RESPIRATORY DISTRESS SYNDROME IN COVID‐19
Acute respiratory distress syndrome is an acute inflammatory lung injury leading to severe SARS manifesting as acute hypoxemic respiratory failure with bilateral alveolar opacities on chest radiography and decreased lung compliance (Figure 9). 112 Fifteen out of 75 patients (20%) have developed ARDS in 3 weeks following SARS infection. 113 In most cases, the course of COVID‐19 is mild, but life‐threatening ARDS develops in some critically ill patients within 8‐9 days after onset of the disease. 45 , 46 The reported incidence of ARDS and critical illness requiring ICU admissions varies widely across multiple studies. 9 , 45 , 46 , 114 , 115 , 116 , 117 ARDS is more common in older people, those with comorbidities, including hypertension, diabetes, coronary artery disease, bronchitis, ischemic disease of the central nervous system, and Parkinson's disease. 118 Wu et al reported the clinical characteristics and risk factors of patients with COVID‐19 pneumonia with the development of ARDS or death. In their study, older age, comorbidities, high fever, neutrophilia, lymphopenia, elevated end‐organ related indices, elevated high‐sensitivity CRP, serum ferritin, and coagulation function–related indicators (PT and D‐dimer) were significantly associated with higher risk of developing ARDS. 119
Figure 9.
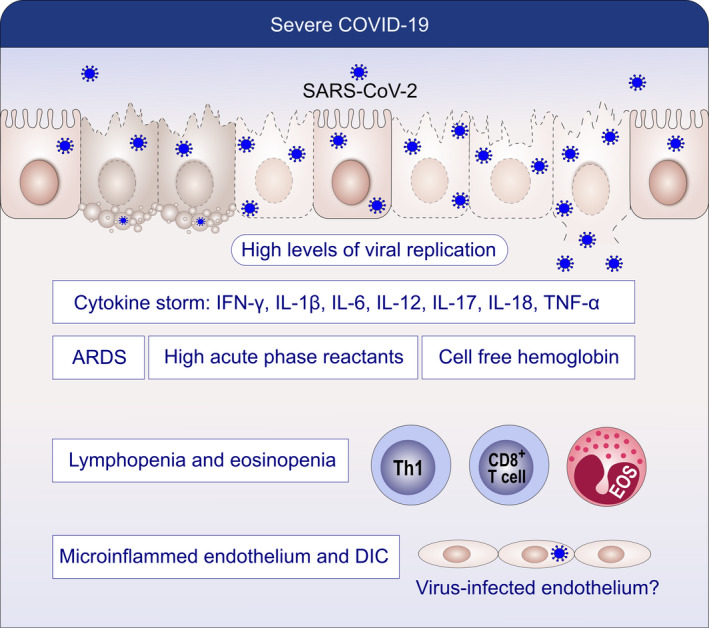
Pathogenesis of severe COVID‐19. Severe COVID‐19 presents with high viral load, cytokine storm, ARDS, cell‐free hemoglobin, high acute‐phase reactants, lymphopenia, eosinopenia, microinflammed endothelium, and DIC. Endothelial damage, cell‐free hemoglobin, cytokine storm, and lymphopenia show links to severe disease
Critically ill patients with ARDS are extremely difficult to oxygenate as their lungs are filled with fluid and cell‐free hemoglobin (CFH) occupying most of the airspace. A study by Shaver et al indicated that the red color observed in the exudates from ARDS patients is not merely a benign sign of edema, but demonstrates the presence of CFH and hemolysis (Figure 9). 120 Secretions from COVID‐19 patients with ARDS are pink in color. CFH in the air space correlates with measures of alveolar‐capillary barrier dysfunction in humans with ARDS (r = .89, P < .001) and in mice with ventilator‐induced acute lung injury (r = .89, P < .001). 120 CFH contributes to the cytokine storm by increasing pro‐inflammatory cytokine expression and paracellular permeability. COVID‐19 ARDS patients are difficult to oxygenate because of systematic destruction of red blood cells resulting in CFH that have oxidized iron ions in the ferric state. Under normal conditions, the iron ions in heme can be reduced by Cytb561.
Endothelial damage and capillary endothelial cell permeability in the lungs leading to fluid leakage into the pulmonary parenchyma in the presence of an acute inflammatory response together with neutrophils and cytokines describe the pathogenesis of ARDS. 121 Protein‐ and CFH‐rich fluid floods the alveolar spaces through the tight junctions of the epithelial cells. Neutrophils play an important role in the development of pulmonary edema associated with acute lung injury or ARDS. Endothelial injury appears within minutes to hours after acute lung injury, and the endothelial cell gaps allow the permeability of fluid, neutrophils and cytokines into the pulmonary parenchymal space. Both epithelial and endothelial barrier integrity is damaged by barrier opening cytokines in the presence of parenchymal lymphocyte infiltration and inflammations that are all important in the pathogenesis.
Although there are conflicting results regarding corticosteroid treatment in different studies, their use is generally not recommended. 122 , 123 Shen et al reported the results of 5 patients who were critically ill with COVID‐19 and ARDS and were treated with convalescent plasma. Administration of convalescent plasma containing neutralizing antibodies was followed by clearance of the virus and improvement in the patients’ clinical status. These results require further evaluation in clinical trials because of the limited sample size and study design. 124 Low tidal volume mechanical ventilation, positive end‐expiratory pressure, and fluid management guidelines have improved outcomes for patients with ARDS. Extracorporeal membrane oxygenation is used in specialized centers for severe cases. Prone positioning has recently proven to have significantly decreased days in the ventilator and in the intensive care unit. Studies on mesenchymal stem cell therapy, partial fluid ventilation, TIP peptide, cytokine inhibitors, and several pharmaceutical drugs are being pursued. The lectin‐like domain of TNF, mimicked by a circular seventeen amino acid peptide, TIP peptide, is responsible for the reduction in lung edema clearance in murine models of noncardiogenic pulmonary edema. 125
12. AUTOPSY FINDINGS
The most common complications frequently observed in patients who died of COVID‐19 were sepsis (100%) followed by respiratory failure (98%), ARDS (93%), septic shock (70%), acute cardiac injury (59%), heart failure (52%), coagulopathy (50%), secondary infection (50%), and acute kidney injury (50%). 91 Autopsy features of COVID‐19 are very similar to SARS and MERS findings. 126 , 127 The macroscopic features of COVID‐19 are likely to be observed in the chest and may include pleurisy, pericarditis, lung consolidation, and pulmonary edema. A secondary bacterial infection may be superimposed on the viral infection that can lead to purulent inflammation. 128 Early histopathological features were reported in two patients who underwent surgical resections for lung adenocarcinoma but were later found to be SARS‐CoV‐2–positive at the time of the operation. These patients were asymptomatic with respect to COVID‐19 at the time of the operation. Nonspecific findings included edema, pneumocyte hyperplasia, focal inflammation, and multinucleated giant cell formation, while no hyaline membranes were seen. These findings may reflect early changes of acute lung injury in the infection. 129 Histopathological findings were noted in a 50‐year‐old man who died from severe COVID‐19 infection. The microscopic findings included diffuse alveolar damage with exudates. 41 A lymphocytic inflammation was predominant suggesting that one of the mechanisms for peripheral lymphopenia could be the sequestration of lymphocytes in the lungs. In addition, multinucleated giant cells and large atypical pneumocytes were observed. The histopathologic changes seen on postmortem transthoracic needle biopsies from another COVID‐19 patient with respiratory failure and radiographic bilateral ground‐glass opacities showed diffuse alveolar damage, denudation of alveolar lining cells, the presence of reactive type II pneumocyte hyperplasia, intra‐alveolar fibrinous exudates, with chronic inflammatory infiltrates, injured and desquamated epithelial cells, and intra‐alveolar organized fibrin. 130 An antibody to the Rp3 NP protein of SARS‐CoV‐2 was used for virus immunostaining which revealed prominent expression of the virus on alveolar epithelial cells. Viral proteins were minimally detectable on blood vessels. Apparently, more detailed pathological studies characterizing the immune infiltrate that can be linked to the cytokine storm and ARDS are needed. In one of the two recently reported autopsy cases, there were no eosinophils or neutrophils identified in the lungs. 66 Immunological staining of the lymphocyte infiltrate showed a sparse infiltrate of CD3‐positive T lymphocytes within the alveolar septa with CD8‐positive T cells slightly higher than CD4‐positive T cells. There were only rare CD20‐positive B‐lymphocytes and CD68‐positive macrophages. The second case had increased numbers of neutrophils, macrophages, histiocytes, secondary bacterial infection, and aspirated food particles. So far, the postmortem cases analyzed to date show pathological changes due to viral pneumonia but also changes related to individual outcomes, such as the development of ARDS and secondary bacterial infection.
13. CONCLUSIONS AND KEY MESSAGES
SARS‐CoV‐2 and its disease COVID‐19 are associated with significant morbidity and mortality globally, probably becoming one of the biggest health and economic burden of the last 100 years. COVID‐19 became a major fights of humans in modern times, emphasizing the importance and need of a global strategy and solidarity among nations. As a zoonotic disease that has already spread globally to several millions of humans and probably animals, it will be practically impossible to eradicate COVID‐19. We have to learn how to live together with the virus and disease.
The SARS‐CoV‐2 showed an unexpected high speed of transmission and global spreadability because of long infectious incubation period before the onset of symptoms, asymptomatic virus carriers, super spreaders, and the extent of globalization with individual traveling. Due to the nature of the disease, it is difficult to control COVID‐19 spreaders, such as asymptomatic virus carriers, recently infected patients in incubation period, clinically recovered but still virus‐positive cases, and virus‐carrying domestic animals.
COVID‐19 represents clinical forms from asymptomatic to mild, moderate, and very severe cases, and there are still many unknowns. The immunology of all these cases is different and becomes more complex when the patients progress to severe disease. Lymphopenia and its monitoring is important and represents a key mechanism in the pathogenesis and biomarker of severity development and a target for treatment. High viral load during the first infection and repeated exposure to virus especially in healthcare workers can be an important factor for severity and should be strictly avoided. Better understanding the pathogenesis of lymphopenia, cytokine storm, and ARDS and novel ways to prevent their development will be main routes for future research to avoid severe disease.
The know‐how generated at the time of SARS and MERS is extremely valuable. Because of T‐ and B‐cell cross‐reactivities, the vaccines and drugs developed at that time may be helpful. Important immune therapies are awaited in the next months, such as antiviral therapies to boost immunity and use of biologics to dampen or prevent the cytokine storm. Chemical antiviral therapies and effective vaccines and their worldwide usage will help to control the disease. Specific antibody detection is an important add‐on method for diagnosis and for better understanding the population immunity. Until present, an internationally approved antibody detection method remains an unmet need. Determining the broad response of the innate and adaptive immune system to SARS‐CoV‐2 will be also essential in preparations to the next zoonotic disease, which may happen in the future.
There are many unknowns in COVID‐19. Worldwide efforts to contain the pandemic can be split into three different domains: research, public education, and clinical care. A global strategy to reduce the burden of COVID‐19 is needed. It is well known that these action plans can only be successful by the combination of efforts of all stakeholders: WHO, governments, researchers, physicians, patient organizations, economists, pharmacists, industry, and policy makers. Global and regional COVID‐19 guidelines in every aspect of this disease should be developed and implemented. In this fight against COVID‐19, we need international solidarity and prompt sharing of accurate scientific information. The implementation of open data concept for all COVID‐19 and SARS‐CoV‐2 studies should be obligatory.
CONFLICT OF INTERESTS
LM reports personal fees from AHL and grants from GSK. KN reports grants and/or personal fees and/or other from NIAID, Novartis, Regeneron, FARE, EAT, Sanofi, Astellas, Nestle, BeforeBrands, Alladapt, ForTra, Genentech, AImmune Therapeutics, DBV Technologies, AstraZeneca, ImmuneWorks, Cour Pharmaceuticals, AllerGenis, Ukko Pharma, AnaptysBio, Adare Pharmaceuticals, Stallergenes‐Greer, NHLBI, NIEHS, EPA, WAO Center of Excellence, Iggenix, Probio, Vedanta, Centocor, Seed, Immune Tolerance Network, and NIH. In addition, KN has pending patents on Inhibition of Allergic Reaction to Peanut Allergen using an IL‐33 Inhibitor, Special Oral Formula for Decreasing Food Allergy Risk and Treatment for Food Allergy, Basophil Activation Based Diagnostic Allergy Test, Granulocyte‐based methods for detecting and monitoring immune system disorders, Methods and Assays for Detecting and Quantifying Pure Subpopulations of White Blood Cells in Immune System Disorders, Mixed Allergen Compositions and Methods for Using the Same, and Microfluidic Device and Diagnostic Methods for Allergy Testing Based on Detection of Basophil Activation. CA reports grants from Allergopharma, Idorsia, Swiss National Science Foundation, Christine Kühne‐Center for Allergy Research and Education, European Commission's Horizon's 2020 Framework Programme, Cure, Novartis Research Institutes, AstraZeneca, SciBase, and advisory role in Sanofi/Regeneron. All other authors have no conflict of interest within the scope of the submitted work.
ACKNOWLEDGMENT
The authors are grateful to Anna Globinska for graphics art and Laura Alberch for critical reading of the manuscript.
Azkur AK, Akdis M, Azkur D, et al. Immune response to SARS‐CoV‐2 and mechanisms of immunopathological changes in COVID‐19. Allergy. 2020;75:1564–1581. 10.1111/all.14364
Azkur and Akdis are the first co‐authors.
REFERENCES
- 1. Coronaviridae Study Group of the International Committee on Taxonomy of V . The species Severe acute respiratory syndrome‐related coronavirus: classifying 2019‐nCoV and naming it SARS‐CoV‐2. Nat Microbiol. 2020;5(4):536‐544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. To KK, Hung IF, Chan JF, Yuen KY. From SARS coronavirus to novel animal and human coronaviruses. J Thorac Dis. 2013;5(Suppl 2):S103‐108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367(19):1814‐1820. [DOI] [PubMed] [Google Scholar]
- 4. Zhu N, Zhang D, Wang W, et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382(8):727‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. WHO . Coronavirus disease (COVID‐2019) situation reports. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports. Accessed April 9
- 6. Forster P, Forster L, Renfrew C, Forster M. Phylogenetic network analysis of SARS‐CoV‐2 genomes. Proc Natl Acad Sci USA 2020;117(17):9241‐9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gudbjartsson DF, Helgason A, Jonsson H, et al. Spread of SARS‐CoV‐2 in the Icelandic Population. N Engl J Med. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dong X, Cao YY, Lu XX, et al. Eleven faces of coronavirus disease 2019. Allergy 2020;75:1699‐1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of Coronavirus disease 2019 in China. N Engl J Med. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bai Y, Yao L, Wei T, et al. Presumed asymptomatic carrier transmission of COVID‐19. JAMA. 2020;323(14):1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. https://www.worldometers.info/coronavirus/
- 12. Verity R, Okell LC, Dorigatti I, et al. Estimates of the severity of coronavirus disease 2019: a model‐based analysis. Lancet Infect Dis 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Paules CI, Marston HD, Fauci AS. Coronavirus infections‐more than just the common cold. JAMA. 2020;323(8):707. [DOI] [PubMed] [Google Scholar]
- 14. Hindson J. COVID‐19: faecal‐oral transmission? Nat Rev Gastroenterol Hepatol. 2020;17(5):259‐259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang JJ, Dong X, Cao YY, et al. Clinical characteristics of 140 patients infected with SARS‐CoV‐2 in Wuhan, China. Allergy. 2020;75:1730‐1741. [DOI] [PubMed] [Google Scholar]
- 16. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim YI, Kim SG, Kim SM, et al. Infection and rapid transmission of SARS‐CoV‐2 in ferrets. Cell Host Microbe 2020;27(5):704‐709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hoffmann M, Kleine‐Weber H, Schroeder S, et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and Is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271‐280. e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shi J, Wen Z, Zhong G, et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS‐coronavirus 2. Science 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ou X, Liu Y, Lei X, et al. Characterization of spike glycoprotein of SARS‐CoV‐2 on virus entry and its immune cross‐reactivity with SARS‐CoV. Nat Commun. 2020;11(1):1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stertz S, Reichelt M, Spiegel M, et al. The intracellular sites of early replication and budding of SARS‐coronavirus. Virology. 2007;361(2):304‐315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Siu KL, Kok KH, Ng MH, et al. Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3.TANK.TBK1/IKKepsilon complex. J Biol Chem. 2009;284(24):16202‐16209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ozdemir C, Kucuksezer UC, Tamay Z. Is BCG vaccination effecting the spread and severity of COVID‐19. Allergy 2020;75:1815‐1819. [DOI] [PubMed] [Google Scholar]
- 24. Gursel M, Gursel I. Is global BCG vaccination‐induced trained immunity relevant to the progression of the SARS.CoV‐2 pandemic? Allergy. 2020;75:1815‐1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Deftereos SG, Siasos G, Giannopoulos G, et al. The Greek study in the effects of colchicine in COvid‐19 complications prevention (GRECCO‐19 study): Rationale and study design. Hellenic J Cardiol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shneider A, Kudriavtsev A, Vakhrusheva A. Can melatonin reduce the severity of COVID‐19 pandemic? Int Rev Immunol. 2020;1‐10. [DOI] [PubMed] [Google Scholar]
- 27. Conti P, Ronconi G, Caraffa A, et al. Induction of pro‐inflammatory cytokines (IL‐1 and IL‐6) and lung inflammation by Coronavirus‐19 (COVI‐19 or SARS‐CoV‐ 2): anti‐inflammatory strategies. J Biol Regul Homeost Agents. 2020;34(2). [DOI] [PubMed] [Google Scholar]
- 28. Gordon DE, Jang GM, Bouhaddou M, et al. A SARS‐CoV‐2 protein interaction map reveals targets for drug repurposing. Nature. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dai W, Zhang B, Su H, et al. Structure‐based design of antiviral drug candidates targeting the SARS‐CoV‐2 main protease. Science 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang X, Xu W, Hu G, et al. SARS‐CoV‐2 infects T lymphocytes through its spike protein‐mediated membrane fusion. Cell Mol Immunol 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xiong L, Edwards CK 3rd, Zhou L. The biological function and clinical utilization of CD147 in human diseases: a review of the current scientific literature. Int J Mol Sci. 2014;15(10):17411‐17441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ulrich H, Pillat MM. CD147 as a target for COVID‐19 treatment: suggested effects of azithromycin and stem cell engagement. Stem Cell Rev Rep. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jansen JM, Gerlach T, Elbahesh H, Rimmelzwaan GF, Saletti G. Influenza virus‐specific CD4+ and CD8+ T cell‐mediated immunity induced by infection and vaccination. J Clin Virol. 2019;119:44‐52. [DOI] [PubMed] [Google Scholar]
- 34. Long QX, Liu BZ, Deng HJ, et al. Antibody responses to SARS‐CoV‐2 in patients with COVID‐19. Nat Med. 2020. [DOI] [PubMed] [Google Scholar]
- 35. Karamloo F, König R. SARS‐CoV‐2 immunogenecity at the crossroads. Allergy. 2020;75:1822‐1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Grifoni A, Sidney J, Zhang Y, Scheuermann RH, Peters B, Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS‐CoV‐2. Cell Host Microbe. 2020;27(4):671‐680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ng OW, Chia A, Tan AT, et al. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post‐infection. Vaccine. 2016;34(17):2008‐2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ahmed SF, Quadeer AA, McKay MR. Preliminary identification of potential vaccine targets for the COVID‐19 Coronavirus (SARS‐CoV‐2) based on SARS‐ CoV immunological studies. Viruses. 2020;12(3):254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Herbinger KH, Hanus I, Beissner M, et al. Lymphocytosis and lymphopenia induced by imported infectious diseases: a controlled cross‐sectional study of 17,229 diseased German travelers returning from the tropics and subtropics. Am J Trop Med Hyg. 2016;94(6):1385‐1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Qin C, Zhou L, Hu Z, et al. Dysregulation of immune response in patients with COVID‐19 in Wuhan, China. Clin Infect Dis. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xu Z, Shi L, Wang Y, et al. Pathological findings of COVID‐19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8(4):420‐422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang F, Nie J, Wang H, et al. Characteristics of peripheral lymphocyte subset alteration in COVID‐19 pneumonia. J Infect Dis. 2020;221(11):1762‐1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu Y, Yang Y, Zhang C, et al. Clinical and biochemical indexes from 2019‐nCoV infected patients linked to viral loads and lung injury. Sci China Life Sci. 2020;63(3):364‐374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sun D, Li H, Lu XX, et al. Clinical features of severe pediatric patients with coronavirus disease 2019 in Wuhan: a single center's observational study. World J Pediatr. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel Coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bermejo‐Martin JF, Almansa R, Menendez R, Mendez R, Kelvin DJ, Torres A. Lymphopenic community acquired pneumonia as signature of severe COVID‐19 infection. J Infect. 2020;80(5):e23‐e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tan L, Wang Q, Zhang D, et al. Lymphopenia predicts disease severity of COVID‐19: a descriptive and predictive study. Signal Transduct Target Ther. 2020;5:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang HP, Sun YX, Lin Z, et al. CARsomes inhibit airway allergic inflammation in mice by inducing antigen‐specific Th2 cell apoptosis. Allergy. 2020. 75(5):1205‐1216. [DOI] [PubMed] [Google Scholar]
- 50. Aksoy E, Azkur AK. Schmallenberg virus induces apoptosis in Vero cell line via extrinsic and intrinsic pathways in a time and dose dependent manner. J Vet Med Sci. 2019;81(2):204‐212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fung SY, Yuen KS, Ye ZW, Chan CP, Jin DY. A tug‐of‐war between severe acute respiratory syndrome coronavirus 2 and host antiviral defence: lessons from other pathogenic viruses. Emerg Microbes Infect. 2020;9(1):558‐570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chen RF, Chang JC, Yeh WT, et al. Role of vascular cell adhesion molecules and leukocyte apoptosis in the lymphopenia and thrombocytopenia of patients with severe acute respiratory syndrome (SARS). Microbes Infect. 2006;8(1):122‐127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Qu R, Ling Y, Zhang YH, et al. Platelet‐to‐lymphocyte ratio is associated with prognosis in patients with Corona Virus Disease‐19. J Med Virol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tay MZ, Poh CM, Renia L, MacAry PA, Ng LFP. The trinity of COVID‐19: immunity, inflammation and intervention. Nat Rev Immunol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Palomares O, Akdis M, Martin‐Fontecha M, Akdis CA. Mechanisms of immune regulation in allergic diseases: the role of regulatory T and B cells. Immunol Rev. 2017;278(1):219‐236. [DOI] [PubMed] [Google Scholar]
- 56. Akdis CA, Akdis M. Mechanisms and treatment of allergic disease in the big picture of regulatory T cells. J Allergy Clin Immunol. 2009;123(4):735‐746. quiz 747–738. [DOI] [PubMed] [Google Scholar]
- 57. Suvas S, Azkur AK, Kim BS, Kumaraguru U, Rouse BT. CD4+CD25+ regulatory T cells control the severity of viral immunoinflammatory lesions. J Immunol. 2004;172(7):4123‐4132. [DOI] [PubMed] [Google Scholar]
- 58. Lan F, Zhang N, Bachert C, Zhang L. Stability of regulatory T cells in T helper 2‐biased allergic airway diseases. Allergy. 2020; 10.1111/all.14257 [DOI] [PubMed] [Google Scholar]
- 59. Loebbermann J, Durant L, Thornton H, Johansson C, Openshaw PJ. Defective immunoregulation in RSV vaccine‐augmented viral lung disease restored by selective chemoattraction of regulatory T cells. Proc Natl Acad Sci USA. 2013;110(8):2987‐2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lanteri MC, O'Brien KM, Purtha WE, et al. Tregs control the development of symptomatic West Nile virus infection in humans and mice. J Clin Invest. 2009;119(11):3266‐3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Flores‐Torres AS, Salinas‐Carmona MC, Salinas E, Rosas‐Taraco AG. Eosinophils and respiratory viruses. Viral Immunol. 2019;32(5):198‐207. [DOI] [PubMed] [Google Scholar]
- 62. Jesenak M, Schwarze J. Lung eosinophils‐A novel "virus sink" that is defective in asthma? Allergy. 2019;74(10):1832‐1834. [DOI] [PubMed] [Google Scholar]
- 63. Li YX, Wu W, Yang T, et al. Characteristics of peripheral blood leukocyte differential counts in patients with COVID‐19. Zhonghua Nei Ke Za Zhi. 2020;59:E003. [PubMed] [Google Scholar]
- 64. Du Y, Tu L, Zhu P, et al. Clinical features of 85 fatal cases of COVID‐19 from Wuhan: A retrospective observational study. Am J Respir Crit Care Med. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lindsley AW, Schwartz JT, Rothenberg ME. Eosinophil responses during COVID‐19 infections and Coronavirus vaccination. J Allergy Clin Immunol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Barton LM, Duval EJ, Stroberg E, Ghosh S, Mukhopadhyay S. COVID‐19 autopsies, Oklahoma, USA. Am J Clin Pathol. 2020;153(6):725‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zheng M, Gao Y, Wang G, et al. Functional exhaustion of antiviral lymphocytes in COVID‐19 patients. Cell Mol Immunol. 2020;17(5):533‐535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Retamal‐Diaz A, Covian C, Pacheco GA, et al. Contribution of resident memory CD8(+) T cells to protective immunity against respiratory syncytial virus and their impact on vaccine design. Pathogens. 2019;8(3):147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Schmidt ME, Varga SM. The CD8 T cell response to respiratory virus infections. Front Immunol. 2018;9:678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zheng HY, Zhang M, Yang CX, et al. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID‐19 patients. Cell Mol Immunol. 2020;17(5):541‐543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Casadevall A, Pirofski LA. The convalescent sera option for containing COVID‐19. J Clin Invest. 2020;130(4):1545‐1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cunningham AC, Goh HP, Koh D. Treatment of COVID‐19: old tricks for new challenges. Crit Care. 2020;24(1):91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. van de Veen W, Akdis M. Tolerance mechanisms of AIT. Allergy. 2020;75:1071‐1018. [DOI] [PubMed] [Google Scholar]
- 74. Lee YL, Liao CH, Liu PY, et al. Dynamics of anti‐SARS‐Cov‐2 IgM and IgG antibodies among COVID‐19 patients. J Infect. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. To KK, Tsang OT, Leung WS, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS‐CoV‐ 2: an observational cohort study. Lancet Infect Dis. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Renegar KB, Small PA Jr, Boykins LG, Wright PF. Role of IgA versus IgG in the control of influenza viral infection in the murine respiratory tract. J Immunol. 2004;173(3):1978‐1986. [DOI] [PubMed] [Google Scholar]
- 77. Liew FY, Russell SM, Appleyard G, Brand CM, Beale J. Cross‐protection in mice infected with influenza A virus by the respiratory route is correlated with local IgA antibody rather than serum antibody or cytotoxic T cell reactivity. Eur J Immunol. 1984;14(4):350‐356. [DOI] [PubMed] [Google Scholar]
- 78. Tamura S, Funato H, Hirabayashi Y, et al. Functional role of respiratory tract haemagglutinin‐specific IgA antibodies in protection against influenza. Vaccine. 1990;8(5):479‐485. [DOI] [PubMed] [Google Scholar]
- 79. Takase H, Murakami Y, Endo A, Ikeuchi T. Antibody responses and protection in mice immunized orally against influenza virus. Vaccine. 1996;14(17–18):1651‐1656. [DOI] [PubMed] [Google Scholar]
- 80. Chen KS, Quinnan GV Jr. Secretory immunoglobulin A antibody response is conserved in aged mice following oral immunization with influenza virus vaccine. J Gen Virol. 1989;70(Pt 12):3291‐3296. [DOI] [PubMed] [Google Scholar]
- 81. Kim MH, Kim HJ, Chang J. Superior immune responses induced by intranasal immunization with recombinant adenovirus‐based vaccine expressing full‐length Spike protein of Middle East respiratory syndrome coronavirus. PLoS One. 2019;14(7):e0220196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. See RH, Zakhartchouk AN, Petric M, et al. Comparative evaluation of two severe acute respiratory syndrome (SARS) vaccine candidates in mice challenged with SARS coronavirus. J Gen Virol. 2006;87(Pt 3):641‐650. [DOI] [PubMed] [Google Scholar]
- 83. Zheng BJ, Du LY, Zhao GY, et al. Studies of SARS virus vaccines. Hong Kong Med J. 2008;14(Suppl 4):39‐43. [PubMed] [Google Scholar]
- 84. Okba NMA, Muller MA, Li W, et al. Severe acute respiratory syndrome Coronavirus 2−specific antibody responses in Coronavirus disease 2019 patients. Emerg Infect Dis. 2020;26(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Guo L, Ren L, Yang S, et al. Profiling early humoral response to diagnose novel Coronavirus disease (COVID‐19). Clin Infect Dis. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Perez L. Acute phase protein response to viral infection and vaccination. Arch Biochem Biophys. 2019;671:196‐202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Herbinger KH, Hanus I, Schunk M, et al. Elevated Values of C‐reactive protein induced by imported infectious diseases: a controlled cross‐sectional study of 11,079 diseased german travelers returning from the tropics and subtropics. Am J Trop Med Hyg. 2016;95(4):938‐944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Wang JT, Sheng WH, Fang CT, et al. Clinical manifestations, laboratory findings, and treatment outcomes of SARS patients. Emerg Infect Dis. 2004;10(5):818‐824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ko JH, Park GE, Lee JY, et al. Predictive factors for pneumonia development and progression to respiratory failure in MERS‐CoV infected patients. J Infect. 2016;73(5):468‐475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Lin L, Lu L, Cao W, Li T. Hypothesis for potential pathogenesis of SARS‐CoV‐2 infection‐a review of immune changes in patients with viral pneumonia. Emerg Microbes Infect. 2020;9(1):727‐732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID‐19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054‐1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Gao Y, Li T, Han M, et al. Diagnostic utility of clinical laboratory data determinations for patients with the severe COVID‐19. J Med Virol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wan S, Xiang Y, Fang W, et al. Clinical features and treatment of COVID‐19 patients in Northeast Chongqing. J Med Virol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Chen T, Wu D, Chen H, et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 2020;368:m1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Favaloro EJ, Lippi G. Recommendations for minimal laboratory testing panels in patients with COVID‐19: potential for prognostic monitoring. Semin Thromb Hemost. 2020;46(03):379‐382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Behrens EM, Koretzky GA. Review: cytokine storm syndrome: looking toward the precision medicine Era. Arthritis Rheumatol. 2017;69(6):1135‐1143. [DOI] [PubMed] [Google Scholar]
- 97. Huang KJ, Su IJ, Theron M, et al. An interferon‐gamma‐related cytokine storm in SARS patients. J Med Virol. 2005;75(2):185‐194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Channappanavar R, Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol. 2017;39(5):529‐539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Liu Q, Zhou YH, Yang ZQ. The cytokine storm of severe influenza and development of immunomodulatory therapy. Cell Mol Immunol. 2016;13(1):3‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Younan P, Iampietro M, Nishida A, et al. Ebola virus binding to Tim‐1 on T lymphocytes induces a cytokine storm. MBio 2017;8(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Eastwood D, Bird C, Dilger P, et al. Severity of the TGN1412 trial disaster cytokine storm correlated with IL‐2 release. Br J Clin Pharmacol. 2013;76(2):299‐315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Simon‐Loriere E, Lin RJ, Kalayanarooj SM, et al. High anti‐dengue virus activity of the OAS gene family is associated with increased severity of dengue. J Infect Dis. 2015;212(12):2011‐2020. [DOI] [PubMed] [Google Scholar]
- 103. Savarin C, Bergmann CC. Fine tuning the cytokine storm by IFN and IL‐10 following neurotropic coronavirus encephalomyelitis. Front Immunol. 2018;9:3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Dennis EA, Norris PC. Eicosanoid storm in infection and inflammation. Nat Rev Immunol. 2015;15(8):511‐523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Tam VC, Quehenberger O, Oshansky CM, et al. Lipidomic profiling of influenza infection identifies mediators that induce and resolve inflammation. Cell. 2013;154(1):213‐227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Chousterman BG, Swirski FK, Weber GF. Cytokine storm and sepsis disease pathogenesis. Semin Immunopathol. 2017;39(5):517‐528. [DOI] [PubMed] [Google Scholar]
- 107. Xiong Y, Liu Y, Cao L, et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID‐19 patients. Emerg Microbes Infect. 2020;9(1):761‐770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Li Y, Chen M, Cao H, Zhu Y, Zheng J, Zhou H. Extraordinary GU‐rich single‐strand RNA identified from SARS coronavirus contributes an excessive innate immune response. Microbes Infect. 2013;15(2):88‐95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Wang R, Xiao H, Guo R, Li Y, Shen B. The role of C5a in acute lung injury induced by highly pathogenic viral infections. Emerg Microbes Infect. 2015;4(5):e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Ruan Q, Yang K, Wang W, Jiang L, Song J. Clinical predictors of mortality due to COVID‐19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Prompetchara E, Ketloy C, Palaga T. Immune responses in COVID‐19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol. 2020;38(1):1‐9. [DOI] [PubMed] [Google Scholar]
- 112. Fan E, Brodie D, Slutsky AS. Acute respiratory distress syndrome: advances in diagnosis and treatment. JAMA. 2018;319(7):698‐710. [DOI] [PubMed] [Google Scholar]
- 113. Peiris JS, Chu CM, Cheng VC, et al. Clinical progression and viral load in a community outbreak of coronavirus‐associated SARS pneumonia: a prospective study. Lancet. 2003;361(9371):1767‐1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Arentz M, Yim E, Klaff L, et al. Characteristics and Outcomes of 21 Critically Ill Patients With COVID‐19 in Washington State. JAMA. 2020;323(16):1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507‐513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Yang X, Yu Y, Xu J, et al. Clinical course and outcomes of critically ill patients with SARS‐CoV‐2 pneumonia in Wuhan, China: a single‐centered, retrospective, observational study. Lancet Respir Med. 2020;8(5):475‐481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Sun P, Qie S, Liu Z, Ren J, Li K, Xi J. Clinical characteristics of hospitalized patients with SARS‐CoV‐2 infection: A single arm meta‐analysis. J Med Virol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Wujtewicz M, Dylczyk‐Sommer A, Aszkielowicz A, Zdanowski S, Piwowarczyk S, Owczuk R. COVID‐19 ‐ what should anaethesiologists and intensivists know about it? Anaesthesiol Intensive Ther. 2020;52(1):34‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Wu C, Chen X, Cai Y, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with Coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Shaver CM, Upchurch CP, Janz DR, et al. Cell‐free hemoglobin: a novel mediator of acute lung injury. Am J Physiol Lung Cell Mol Physiol. 2016;310(6):L532‐541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Gonzales JN, Lucas R, Verin AD. The acute respiratory distress syndrome: mechanisms and perspective therapeutic approaches. Austin J Vasc Med. 2015;2(1). [PMC free article] [PubMed] [Google Scholar]
- 122. Poston JT, Patel BK, Davis AM. Management of critically ill adults with COVID‐19. JAMA. 2020. [DOI] [PubMed] [Google Scholar]
- 123. Matthay MA, Aldrich JM, Gotts JE. Treatment for severe acute respiratory distress syndrome from COVID‐19. Lancet Respir Med. 2020;8(5):433‐434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Shen C, Wang Z, Zhao F, et al. Treatment of 5 s COVID‐19 with convalescent plasma. JAMA. 2020;323(16):1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Hartmann EK, Boehme S, Duenges B, et al. An inhaled tumor necrosis factor‐alpha‐derived TIP peptide improves the pulmonary function in experimental lung injury. Acta Anaesthesiol Scand. 2013;57(3):334‐341. [DOI] [PubMed] [Google Scholar]
- 126. Ding Y, Wang H, Shen H, et al. The clinical pathology of severe acute respiratory syndrome (SARS): a report from China. J Pathol. 2003;200(3):282‐289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Ng DL, Al Hosani F, Keating MK, et al. Clinicopathologic, immunohistochemical, and ultrastructural findings of a fatal case of middle east respiratory syndrome Coronavirus infection in the United Arab Emirates, April 2014. Am J Pathol. 2016;186(3):652‐658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Hanley B, Lucas SB, Youd E, Swift B, Osborn M. Autopsy in suspected COVID‐19 cases. J Clin Pathol. 2020;73(5):239‐242. [DOI] [PubMed] [Google Scholar]
- 129. Tian S, Hu W, Niu L, Liu H, Xu H, Xiao SY. Pulmonary pathology of early‐phase 2019 novel Coronavirus (COVID‐19) pneumonia in two patients with lung cancer. J Thorac Oncol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Zhang H, Zhou P, Wei Y, et al. Histopathologic Changes and SARS‐CoV‐2 Immunostaining in the Lung of a Patient With COVID‐19. Ann Intern Med 2020. [DOI] [PubMed] [Google Scholar]
- 131. Li X, Wang L, Yan S, et al. Clinical characteristics of 25 death cases with COVID‐19: a retrospective review of medical records in a single medical center, Wuhan, China. Int J Infect Dis. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Zhang Y, Zheng L, Liu L, Zhao M, Xiao J, Zhao Q. Liver impairment in COVID‐19 patients: a retrospective analysis of 115 cases from a single center in Wuhan city, China. Liver Int. 2020. [DOI] [PubMed] [Google Scholar]
- 133. Deng Y, Liu W, Liu K, et al. Clinical characteristics of fatal and recovered cases of coronavirus disease 2019 (COVID‐19) in Wuhan, China: a retrospective study. Chin Med J (Engl). 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Lippi G, Plebani M. Procalcitonin in patients with severe coronavirus disease 2019 (COVID‐19): A meta‐analysis. Clin Chim Acta. 2020;505:190‐191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Cheng Y, Luo R, Wang K, et al. Kidney disease is associated with in‐hospital death of patients with COVID‐19. Kidney Int. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Wang Z, Yang B, Li Q, Wen L, Zhang R. Clinical features of 69 cases with Coronavirus disease 2019 in Wuhan, China. Clin Infect Dis. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Wu P, Duan F, Luo C, et al. Characteristics of ocular findings of patients with Coronavirus disease 2019 (COVID‐19) in Hubei Province, China. JAMA Ophthalmol. 2020;138(5):575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Xia W, Shao J, Guo Y, Peng X, Li Z, Hu D. Clinical and CT features in pediatric patients with COVID‐19 infection: Different points from adults. Pediatr Pulmonol 2020;55(5):1169‐1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Chen G, Wu D, Guo W, et al. Clinical and immunologic features in severe and moderate Coronavirus Disease 2019. J Clin Invest. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Zhang H, Chen Y, Yuan Q, et al. Identification of kidney transplant recipients with Coronavirus disease 2019. Eur Urol 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Zheng F, Tang W, Li H, Huang YX, Xie YL, Zhou ZG. Clinical characteristics of 161 cases of corona virus disease 2019 (COVID‐19) in Changsha. Eur Rev Med Pharmacol Sci. 2020;24(6):3404‐3410. [DOI] [PubMed] [Google Scholar]
- 142. Goh KJ, Choong MC, Cheong EH, et al. Rapid progression to acute respiratory distress syndrome: review of current understanding of critical illness from COVID‐19 infection. Ann Acad Med Singapore. 2020;49(3):1‐9. [PubMed] [Google Scholar]
- 143. Ji D, Zhang D, Xu J, et al. Prediction for progression risk in patients with COVID‐19 pneumonia: the CALL score. Clin Infect Dis. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Han H, Yang L, Liu R, et al. Prominent changes in blood coagulation of patients with SARS‐CoV‐2 infection. Clin Chem Lab Med 2020. [DOI] [PubMed] [Google Scholar]
- 145. Ji HL, Zhao R, Matalon S, Matthay MA. Elevated plasmin(ogen) as a common risk factor for COVID‐19 susceptibility. Physiol Rev. 2020;100(3):1065‐1075. [DOI] [PMC free article] [PubMed] [Google Scholar]


