Abstract
There is currently a clear benefit for many countries to utilize wastewater-based epidemiology (WBE) as part of ongoing measures to manage the coronavirus disease 2019 (COVID-19) global pandemic. Since most wastewater virus concentration methods were developed and validated for nonenveloped viruses, it is imperative to determine the efficiency of the most commonly used methods for the enveloped severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Municipal wastewater seeded with a human coronavirus (CoV) surrogate, murine hepatitis virus (MHV), was used to test the efficiency of seven wastewater virus concentration methods: (A–C) adsorption-extraction with three different pre-treatment options, (D–E) centrifugal filter device methods with two different devices, (F) polyethylene glycol (PEG 8000) precipitation, and (G) ultracentrifugation. MHV was quantified by reverse-transcription quantitative polymerase chain reaction and the recovery efficiency was calculated for each method. The mean MHV recoveries ranged from 26.7 to 65.7%. The most efficient methods were adsorption-extraction methods with MgCl2 pre-treatment (Method C), and without pre-treatment (Method B). The third most efficient method used the Amicon® Ultra-15 centrifugal filter device (Method D) and its recovery efficiency was not statistically different from the most efficient methods. The methods with the worst recovery efficiency included the adsorption-extraction method with acidification (A), followed by PEG precipitation (F). Our results suggest that absorption-extraction methods with minimal or without pre-treatment can provide suitably rapid, cost-effective and relatively straightforward recovery of enveloped viruses in wastewater. The MHV is a promising process control for SARS-CoV-2 surveillance and can be used as a quality control measure to support community-level epidemic mitigation and risk assessment.
Keywords: SARS-CoV-2, COVID-19, Murine hepatitis virus, Recovery, Concentration method, Enveloped virus, Untreated wastewater, Filtration
Graphical abstract
Highlights
-
•
Seven virus concentration methods were evaluated to recover CoV from wastewater.
-
•
The mean MHV recoveries ranged from 26.7 to 65.7%.
-
•
Adsorption-extraction with MgCl2 pre-treatment most efficiently concentrated MHV.
-
•
MHV seems to be an appropriate process control.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the etiologic agent of the ongoing global pandemic of coronavirus disease 2019 (COVID-19) (Coronavirus Study Group of the International Committee on Taxonomy of Viruses, 2020). The primary transmission routes of SARS-CoV-2 are inhalation of aerosols/droplets and person-to-person contact (Morawska and Cao, 2020; Yu et al., 2020). There is increasing evidence for the fecal shedding of SARS-CoV-2 and the presence of viral RNA in domestic wastewater (Kitajima et al., 2020; Xiao et al., 2020). Therefore, the presence of SARS-CoV-2 RNA in untreated wastewater suggests that wastewater could be utilized as a tool to monitor for the invasion, prevalence, molecular epidemiology, and potential eradication of SARS-CoV-2 in the community in an approach known as wastewater-based epidemiology (WBE) (Kitajima et al., 2020).
The detection of SARS-CoV-2 RNA in untreated domestic wastewater has been reported in Australia (Ahmed et al., 2020), the Netherlands (Medema et al., 2020), USA (Wu et al., 2020; Nemudryi et al., 2020), France (Wurtzer et al., 2020a, Wurtzer et al., 2020b), China (Zhang et al., 2020), Israel (Bar-Or et al., 2020), Turkey (Kocamemi et al., 2020), Spain (Randazzo et al., 2020a, Randazzo et al., 2020b) and Italy (La Rosa et al., 2020a, La Rosa et al., 2020b). The virus concentration methods used in these studies to recover SARS-CoV-2 RNA from wastewater include ultrafiltration, polyethylene glycol (PEG) precipitation, ultracentrifugation, and filtration with an electronegative membrane. Rapid, efficient (high recovery), and cost-effective virus concentration methods are needed to monitor SARS-CoV-2 and its nucleic acid in untreated wastewater samples for the successful application of WBE for COVID-19 surveillance. Accurate estimates of viral concentration in untreated wastewater require that the concentration observed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) assays be adjusted using the recovery efficiency of a particular combination of virus and concentration method.
The concentration methods used in each of the above studies were originally developed for the detection of nonenveloped enteric viruses, such as adenovirus, norovirus, and enterovirus in water/wastewater samples. Little is known about the recovery efficiencies for an enveloped virus such as SARS-CoV-2. The virus concentration recovery efficiencies of SARS-CoV-2 may be different from those of nonenveloped enteric viruses because of significant structural differences between enveloped viruses and nonenveloped enteric viruses. In a previous study, a head-to-head method comparison of virus recovery efficiencies demonstrated differences between an enveloped virus and a nonenveloped virus in lake water in Japan (Haramoto et al., 2009). Such discrepancies could lead to large errors (i.e., an order of magnitude) in the estimated concentration of SARS-CoV-2 in untreated wastewater.
To date, no information is available regarding the SARS-CoV-2 recovery efficiency for the wastewater concentration methods commonly used (Carducci et al., 2020; Kitajima et al., 2020). Due to the stringent biosafety requirements of working with SARS-CoV-2, a model virus with similar structural and morphological characteristics provides a useful surrogate for estimating the recovery efficiency of SARS-CoV-2 concentration methods. A handful of non-human coronaviruses (CoVs), porcine epidemic diarrhea virus (Randazzo et al., 2020b) and avian infectious bronchitis virus (Kocamemi et al., 2020) have been used to estimate human CoV recoveries. Data suggested that approximately 11% and 3% of the seeded porcine epidemic diarrhea virus were recovered from untreated and treated wastewater, respectively, with aluminum flocculation-based concentration methods (Randazzo et al., 2020b). Interestingly, these CoV recoveries were similar to the recoveries of the nonenveloped mengovirus (Randazzo et al., 2020b), which is often used as a process control for enteric virus detection in environmental samples (da Silva et al., 2007; Sima et al., 2011; Farkas et al., 2018).
Murine hepatitis virus (MHV) is an enveloped and positive-sense single-stranded RNA Betacoronavirus, which belongs to the same genus as SARS-CoV-2 (Coronaviridae Study Group of the International Committee on Taxonomy of Viruses, 2020), and is responsible for a number of diseases in mice and rats (Roth-Cross et al., 2008). MHV has been used as a surrogate for human CoV in recovery and persistence studies (Ye et al., 2016). MHV and other murine viruses (e.g. murine norovirus) have been successfully used as surrogates for many different enveloped and nonenveloped viruses due to their structural and morphological similarities (Casanova et al., 2009; Ye et al., 2016; Patel et al., 2017), and there are no special laboratory requirements for their use since they are non-pathogenic to humans.
In the present study, we evaluated the efficiencies of MHV recovery from wastewater using various virus concentration methods previously used to detect SARS-CoV-2 in wastewater (Ahmed et al., 2020; Medema et al., 2020; Wu et al., 2020). The performance of seven virus concentration methods was estimated and compared by seeding MHV in untreated wastewater samples. RT-qPCR assays were then used to determine MHV concentrations in seeded untreated domestic wastewater samples to identify the relative performance of each method for CoV recovery. The results presented in this study will allow researchers to select an appropriate efficient concentration method(s) for the recovery of SARS-CoV-2 from domestic wastewater for WBE applications.
2. Materials and methods
2.1. Murine hepatitis virus (MHV) concentrations in fecal suspension
The enveloped MHV stock was obtained from 20 fecal samples of naturally infected mice. Each MHV positive-fecal sample (approximately 250 mg) was suspended into 1 mL of phosphate buffered saline (1 × PBS) and then the samples were pooled together to produce a homogeneous fecal slurry. The slurry was centrifuged at 1000 g for 10 min. The pellet was discarded, and the supernatant was stored at −80 °C for three days. This supernatant, containing MHV, is referred to as the MHV suspension. The MHV RNA concentration in the fecal suspension was determined using an MHV RT-qPCR assay (see below for detailed methodologies). Briefly, RNA was directly extracted on three occasions from triplicate, 200 μL aliquots of MHV suspension using the RNeasy PowerMicrobiome Kit according to manufacturer instructions with a minor modification (Qiagen, Valencia, CA, USA). The glass beads in the bead tube were replaced with garnet beads which have sharp cutting edges.
2.2. Wastewater sample preparation
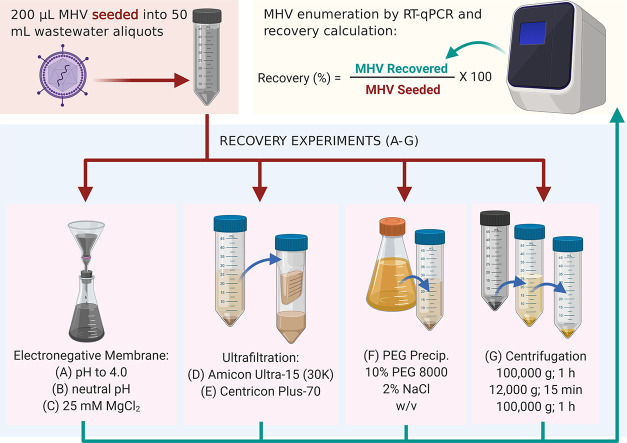
A sample of untreated wastewater (2 L) was collected from a metropolitan wastewater treatment plant (WWTP) in Brisbane, Australia and transported to laboratory on ice. The wastewater sample was kept at 4 °C for 24 h. The WWTP treats domestic wastewater from approximately 325,000 people, as well as industrial wastewater. The treatment process consists of primary treatment, a secondary treatment (activated sludge), and disinfection with chlorine and UV. While stormwater also enters the WWTP, the study catchment did not receive any precipitation 24 h prior to the wastewater sampling. A 200-μL volume of MHV suspension was added to a 50-mL aliquot of untreated wastewater and subjected to each virus concentration method.
2.3. Virus concentration methods
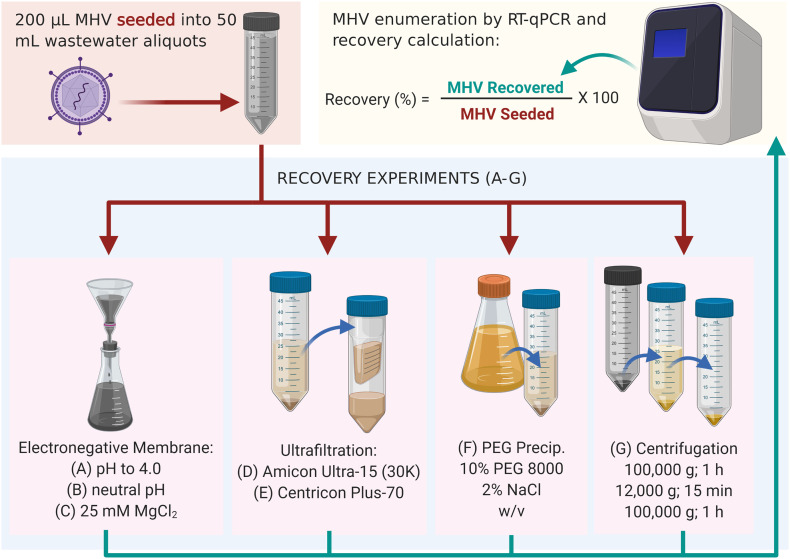
Viruses were concentrated from MHV seeded (n = 3) and unseeded (n = 1) domestic wastewater samples using each of the seven methods as shown in Fig. 1 . The unseeded wastewater samples were used to identify background MHV concentrations in wastewater. Methods A, B and C were derived from virus adsorption extraction methods commonly used to concentrate enteric viruses from water/wastewater (Symonds et al., 2014; Ahmed et al., 2015; Ahmed et al., 2020). Method A began with acidification of sample to pH 4 using 2 N HCl. Method B did not pre-treat the sample; it began with measuring the pH of the sample (pH = 6.9). Method C began with the addition of MgCl2 to the sample to obtain a final concentration of 25 mM MgCl2. For Methods A, B and C, samples were then passed through 0.45-μm pore-size, 47-mm diameter electronegative membranes (HAWP04700; Merck Millipore Ltd., Sydney, Australia) via a magnetic filter funnel (Pall Corporation) and filter flask (Merck Millipore Ltd.) (Ahmed et al., 2015). The membrane was immediately inserted into a 2 mL-bead beating tube (followed by RNA extraction described below).
Fig. 1.
Virus concentration methods used in this study.
Methods D and E were ultrafiltration methods that use centrifugal devices that have previously been used to concentrate viruses from (waste)water (Symonds et al., 2009; Ikner et al., 2011). Both methods began with the centrifugation of the sample at 4500 g for 10 min at 4 °C to obtain a supernatant. For Method D, the supernatant was concentrated using an Amicon® Ultra-15 (molecular weight cut-off 30 kDa) centrifugal filter device (Merck Millipore Ltd.), which was centrifuged at 4750 g for 10 min at 4 °C. This centrifugal concentration step was repeated three times to pass through the entire supernatant volume (Symonds et al., 2009; Ikner et al., 2011; Ahmed et al., 2015). The concentrated sample (400 μL) was collected from the sample reservoir with a pipette and transferred into a 2 mL-bead beating tube. For Method E, the supernatant was further centrifuged at 3500 g for 30 min at 4 °C through the Centricon Plus-70 centrifugal filter device with a molecular weight cut-off of 10 kDa (Merck Millipore). The concentrated sample (300 μL) was collected from the concentrate collection cup with a pipette and mixed with 100 μL of DNase and RNase free water and transferred into a 2 mL-bead beating tube (Ahmed et al., 2020; Medema et al., 2020).
Method F employed PEG precipitation, which is commonly used to concentrate viruses from water matrices (Mull and Hill, 2012; Gyawali et al., 2019; Wu et al., 2020). In this study, it began with sample centrifugation at 10,000 g for 20 min at 4 °C to remove larger particles and debris. The resulting supernatant was transferred to a fresh centrifuge tube and stored at 4 °C, while MHV was recovered from the pellet. The pellet was re-suspended in beef extract (3% w/v) in 0.05 M glycine (pH 9.0) at a ratio of 1:5. The pellet was agitated on a shaking incubator at 200 rpm for 30 min at room temperature. The pellet suspension was then centrifuged at 10,000 g for 10 min at 4 °C and the supernatant was transferred into the centrifuge tube containing supernatant from the initial centrifugation step. The pH of the supernatant mixture was neutralized by the addition of 2 M HCl. PEG 8000 and NaCl were added to the supernatant at ratios of 10% and 2% w/v, respectively. The centrifuge tubes were then incubated at 4 °C for 2 h on an orbital shaker set to 120 rpm. Following incubation, the sample was centrifuged at 10,000 g for 30 min at 4 °C to obtain a pellet. The supernatant was discarded, and the pellet was resuspended in 800 μL Trizol (Sigma-Aldrich, Sydney, NSW, Australia). Finally, 400 μL of the concentrated sample was transferred to a 2-mL bead beating tube.
Method G used ultracentrifugation, which is frequently used to concentrate viruses from (waste)water (Fumian et al., 2010; Ye et al., 2016). In this study, it began with sample centrifugation at 100,000 g for 1 h at 4 °C. Supernatant was removed carefully, and the pellet was suspended in 3.5 mL of 0.25 N glycine buffer (pH 9.5). The sample was incubated on ice for 30 min. The sample was neutralized by the addition of 3 mL of 2 × PBS (pH 7.2). The supernatant was clarified by centrifugation (12,000 g for 15 min at 4 °C). The virus was recovered by ultracentrifugation at 100,000 g for 1 h at 4 °C (Fumian et al., 2010). The pellet was resuspended in 400 μL of 1 × PBS (pH 7.2) and transferred to a 2-mL bead beating tube.
2.4. Viral RNA extraction
Viral RNA was extracted using the RNeasy PowerMicrobiome Kit with a slight modification (Qiagen). The glass beads in the bead tube were replaced with garnet beads which have sharp cutting edges. In brief, a 650 μL of buffer PM1 and 6.5 μL of β-Mercaptoethanol (Sigma-Aldrich) were added into each bead beating tube. Bead beating tubes were homogenized using a Precellys Evolution 24 tissue homogenizer (Bertin Technologies, Montigny-le-Bretonneux, France) at conditions 3 × 20 s at 10,000 rpm at a 10 s interval. Tubes were further centrifuged at 10,000 g for 5 min to pellet the filter debris and beads. 450 μL of sample lysate from the bead beating tube was transferred into rotor adapter (Qiagen) using a QIAcube Connect platform (Qiagen) to obtain a final elution volume of 100 μL of RNA. All RNA samples were stored at −80 °C and subjected to RT-qPCR analysis within the same day of RNA extraction to avoid losses associated with storing, as well as freezing and thawing RNA extracts.
2.5. MHV RT-qPCR analyses
A previously published TaqMan-based RT-qPCR assay was used for MHV quantification in the wastewater samples (Besselsen et al., 2002). For the MHV RT-qPCR assay, gBlocks gene fragments (double-stranded DNA), containing the 108 bp assay amplicon, were purchased from Integrated DNA Technologies (Coralville, IA, USA). The gBlock containing the MHV assay amplicon was used as a positive control and to generate the standard curve. MHV RT-qPCR analyses were performed in 25 μL reaction mixtures using iTaq™ Universal Probes One-Step Reaction Mix (Bio-Rad Laboratories, Richmond, CA). The MHV RT-qPCR mixture contained 12.5 μL of Supermix, 300 nM of forward primer (5′-GGA ACT TCT CGT TGG GCA TTA TAC T-3′), 300 nM of reverse primer (5′-ACC ACA AGA TTA TCA TTT TCA CAA CAT A-3′), 400 nM of probe (5′-FAM-ACA TGC TAC GGC TCG TGT AAC CGA ACT GT-BHQ-3′), 0.625 μL of iScript RT enzyme and 5 μL of template RNA. The RT-qPCR assays were performed using a Bio-Rad CFX96 thermal cycler (Bio-Rad Laboratories, Richmond, CA), using automatic settings for threshold and baseline. Thermal cycling conditions consisted of RT at 50 °C for 10 min, denaturation and Taq polymerase activation at 95 °C for 5 min, and 40 cycles of 95 °C for 15 s followed by 60 °C for 1 min (data collection). RT-qPCR reactions were performed in triplicate for each sample and the sample quantification cycle (Cq) mean was used for further analyses. Samples were always analyzed with corresponding positive (standards) and negative controls (DNase-and RNase-free water).
Separately, three instrument runs were executed on different days to analyze a six-point, ten-fold serial dilution of the MHV assay gBlock (5 × 105 to 5 copies/reaction) in triplicate. A standard curve was generated for each instrument run from the log10-linear regression of triplicate Cq values. The lowest number of diluted standards detected in triplicate assays was considered the qPCR assay limit of detection (ALOD). The MHV estimated copy numbers for each virus wastewater concentrate sample were corrected for the difference between the double-stranded standard curve material and the single-stranded MHV virus (i.e., divided by 2).
2.6. Inhibition test
The presence of PCR inhibition in virus wastewater concentrate sample RNA was assessed using the Sketa22 qPCR assay (Haugland et al., 2005) after spiking RNA samples with a known copy number (104/reaction) of Oncorhynchus keta (O. keta) DNA as described previously (Ahmed et al., 2020). In order to determine PCR inhibition, O. keta DNA was also added to DNase- and RNase-free water and the mean Cq value was used to set-up a reference point. Subsequently, Sketa22 qPCR assay (for O. keta) was performed in 25 μL reaction mixtures using Bio-Rad CFX96 thermal cycler (Bio-Rad Laboratories), using automatic settings for threshold and baseline. The qPCR assay mixtures contained 12.5 μL of iQ Supermix (Bio-Rad Laboratories), 300 nM of forward primer (5′-GGT TTC CGC AGC TGG G-3′), 300 nM of reverse primer (5’-CCG AGC CGT CCT GGT CTA-3′) 400 nM of probe (FAM-5′AGT CGC AGG CGG CCA CCG T-3′-BHQ), 3 μL of template DNA and a known copy number (104/reaction) of O. keta DNA. All samples were analyzed alongside three no template controls. If the Cq value of the RNA sample was >2-Cq values compared to the reference Cq value for distilled water, the sample was considered to have PCR inhibitors (Staley et al., 2012; Ahmed et al., 2018). All wastewater RNA samples were within the 2-Cq values of the reference Cq value; thus, no qPCR inhibition was identified.
Additionally, the RNA extraction-RT-qPCR process was tested to investigate whether RT-qPCR inhibitors were present using Methods C and D. A 200 μL of MHV suspension was added to a 50-mL aliquot of distilled water and subjected to virus concentration Methods C and D. The reference Cq values obtained for MHV seeded distilled water (for Methods C and D) were compared with the Cq values of the MHV seeded untreated wastewater to obtain information on potential RT-PCR inhibition. Wastewater RNA samples processed with Methods C and D were within the 1 Cq value of the reference Cq value; thus, no evidence of inhibition was identified in the RT-PCR process.
2.7. MHV recovery efficiency
The MHV recovery efficiency of each replicate for each concentration method was calculated based upon the copies quantified per by RT-qPCR as follows:
The mean and standard deviation for each concentration method was calculated.
2.8. Quality control
To minimize qPCR contamination, RNA extraction and RT-qPCR set up were performed in separate laboratories. A method blank was included for each concentration method. A reagent blank was also included during nucleic acid extraction to account for any contamination during extraction. All method and extraction blanks were negative for MHV.
2.9. Statistical analysis
The one-way analysis of variance (ANOVA) was used to determine whether there was a difference in MHV recovery among the concentration methods tested and also MHV concentrations in fecal suspensions. Tukey's honest significant difference (HSD) test was used for post-hoc evaluation of group pairings that were significantly different (α = 0.05) using GraphPad software (Prism 8.3, La Jolla, CA, USA).
3. Results
3.1. MHV qPCR assay performance
MHV RT-qPCR standard curves had a dynamic linear range of quantification from 5 × 105 to 5 copies/reaction. The slope of the standard curves ranged from −3.245 to −3.334 (Table 1 ). The amplification efficiencies ranged from 98.7 to 103% and the correlation coefficient (r 2) ranged from 0.992 to 0.995. The ALOQ (i.e., lowest copy number detected 100% of the time) was 5 copies/reaction.
Table 1.
Murine hepatitis virus (MHV) RT-qPCR performance characteristics.
| MHV assay run | Performance characteristic (range) |
|||
|---|---|---|---|---|
| Efficiency (E) (%) | Linearity (r2) | Slope | Y-intercept | |
| 1 | 103.3 | 0.995 | −3.245 | 39.323 |
| 2 | 102.4 | 0.993 | −3.256 | 39.564 |
| 3 | 98.7 | 0.992 | −3.334 | 40.124 |
3.2. MHV concentrations in untreated wastewater and fecal suspension
RNA was extracted from seven unseeded wastewater virus concentrates to determine the background concentrations of MHV. None of the samples were positive for MHV. The MHV concentration was 5.46 ± 0.20 log10 copies/200 μL of fecal suspension as determined by RT-qPCR assay and was used to determine the recovery efficiency of the seven concentration methods examined. The fecal suspension’s MHV concentration was not significantly different among replicate analyses (p > .05).
3.3. MHV recovery efficiency from untreated wastewater
The mean log10 copies of MHV recovered/RT-qPCR reaction for each concentration method used in this study are shown in Table 2 . For the 50 mL untreated wastewater samples seeded with MHV, Method C (adsorption-extraction method, supplemented with MgCl2) provided the highest mean MHV recovery of 65.7 ± 23.0% (Table 2). The second highest mean recovery was for Method B the adsorption-extraction method without pre-treatment. Interestingly, a slightly modified version of this adsorption-extraction method (Method A, adjustment sample pH to 4) recovered less (26.7 ± 15.3%) MHV. Overall, Method D (Amicon Ultra-15 centrifugal filter device) yielded the third-highest mean recovery (56.0 ± 32.3%) of MHV. Method E (Centricon Plus-70 ultrafilter centrifugal device) was similar to Method D, but produced approximately 50% less recovery (28.0 ± 9.10%) of MHV from untreated wastewater samples compared to Method D. Method F (PEG precipitation) provided greater recoveries (44.0 ± 27.7%) than Methods A and E. Methods B and C recovery efficiencies were significantly different (p < .05) than Methods A, E, F and G. Method D recovery efficiency also significantly (p < .05) different than Methods A, E and G.
Table 2.
Mean (±SD) of murine hepatitis virus (MHV) recovered through each concentration method and recovery efficiency of MHV using seven different virus concentration methods (A-G) from untreated wastewater.
| Concentration methods | Mean ± SD MHV concentration (log10 copies of MHV recovered) | Mean ± SD of % recovery of MHV |
|---|---|---|
| Method A | 4.85 ± 0.20 | 26.7 ± 15.3 |
| Method B | 5.24 ± 0.08 | 60.5 ± 22.2 |
| Method C | 5.28 ± 0.09 | 65.7 ± 23.8 |
| Method D | 5.13 ± 0.33 | 56.0 ± 32.3 |
| Method E | 4.91 ± 0.05 | 28.0 ± 9.10 |
| Method F | 5.07 ± 0.21 | 44.0 ± 27.7 |
| Method G | 4.98 ± 0.09 | 33.5 ± 12.1 |
5.46 ± 0.20 log10 copies were seeded; SD: Standard deviation.
4. Discussion
There is a need for the development of efficient methods to concentrate and detect SARS-CoV-2 from wastewater, which has been identified as a key research need for WBE (Kitajima et al., 2020, La Rosa et al., 2020b). Furthermore, it is important to determine the SARS-CoV-2 concentration method recovery efficiency. First, it can enable the user to select the appropriate method to recover SARS-CoV-2 from wastewater with optimal efficiency. Additionally, characterizing method performance will allow more detailed assessments of the actual SARS-CoV-2 load present in wastewater influent, as well as any given water source receiving (un)treated wastewater. A thorough understanding of SARS-CoV-2 concentration method efficiencies will enable accurate measurements in wastewater and allow public health officials to develop appropriate mitigation strategies needed on a community level. Due to the inherent risks associated with laboratory work with infectious SARS-CoV-2 (WHO, 2020), we used a human CoV surrogate, MHV, belonging to the same genus as SARS-CoV-2 to evaluate concentration method performance. In this study, seven concentration methods were evaluated for their recovery of MHV from wastewater. The benefits and limitations of these methods are summarized in Table 3 . These methods have been previously used to recover nonenveloped enteric viruses from various water matrices but have not been evaluated for their ability to detect and concentrate enveloped virus to date except for a few instances (Shi et al., 2017; Ahmed et al., 2020; Medema et al., 2020; Wu et al., 2020). When reported, recovery efficiencies for nonenveloped viruses varied widely depending on the matrix and the virus (Haramoto et al., 2018), but analogous information for enveloped viruses is not available.
Table 3.
Logistical and theoretical advantages and disadvantages of the virus concentration methods evaluated in this study.
| Concentration method | Advantages | Disadvantages | Potential refinement |
|---|---|---|---|
| Methods A, B and C | - Rapid (<40 min to process a sample) - Concentrate viruses from both solid and liquid phases. - Easy to upscale using 90-mm membranes. - Up to 200 mL of sample can be processed, depending on the filter size and turbidity of the sample. - Can be undertaken in the field. - Only a filtration unit and a pump are required. - Multiple samples can be processed at a time if multiple filtration units are available. - Easy to store and transport membrane. - Relatively inexpensive supplies and generally routine microbial laboratory equipment are required (similar to fecal indicator bacteria membrane filtration methods). |
- Requires washing and cleaning filtration units. - pH adjustment is required (Method A only). - Addition of MgCl2 is required (Method C only) - Clogging may occur due to high turbidity. - 90-mm filtration units are expensive. - Ideally, a bead-beating system, which is expensive, should be used; however, the RNeasy PowerWater kit (Qiagen) involves a sample homogenizing step that is undertaken with an adaptor and vortex. |
- Electropositive membrane can also be used. - Pre-filter sample to eliminate debris and lower turbidity, which will allow more sample to pass through the filter. - Use membrane with a larger pore size (0.8 μm) to process larger volume of wastewater sample. - Pre-treating membrane with MgCl2 or AlCl3 could further increase recovery (Method C only). |
| Methods D and E | - Rapid (1 h depending on the turbidity of the sample). - The main equipment required is a centrifuge (up to 4,750 g). - Can process up to 70 mL of sample at a time (Method E only). |
- Concentrate viruses only from liquid fraction. - Ultrafiltration centrifugal unit is expensive. - Method D can only process up to 15 mL of sample at a time. - Multiple centrifugal units may be needed for high turbidity samples (both Methods D and E). - Clogging occurs when turbidity is high. - Cannot be used in the field. - Viruses adsorb to the membrane, which decreases recovery. - Co-concentrates PCR inhibitors. - A large, benchtop centrifuge is required, which is expensive. |
- Use centrifugal unit with 100 kDa filter to speed up the process and reduce clogging. - Centriprep (Merck, Millipore) may be an alternative when Centricon is not available. - Similar devices can be sourced from other vendors, such as Pall Corporation. |
| Method F | - The only equipment required is a centrifuge (up to 10,000 g). - Concentrate viruses from both solid and liquid phases. - Relatively inexpensive. - Large volume (e.g., 1 L) of wastewater can be processed. |
- Time consuming (4-6 h). - Requires handling of hazardous chemical (Trizol). - Cannot be used in the field. - Only a portion of viral concentrate is used to extract RNA, which prohibits the inclusion of all viruses in the sample. |
- Elute pellet in PBS to reduce hazardous chemical usage. |
| Method G | - Concentrate viruses from both solid and liquid phase. - The cost per sample is low. |
- Time consuming (3 h). - Only a small number (n = 6) of samples can be processed at a time. - The sample volume that can be processed is limited (e.g., 50 mL). - Requires expensive equipment (ultracentrifuge), which may not be available in a routine microbiology laboratory. - Cannot be used in the field. - Requires training to operate ultracentrifuge. |
- Higher centrifugation speeds. |
Of the methods tested, two of the adsorption-extraction methods had the most optimal MHV recovery for wastewater samples (Methods B and C) and their mean recovery efficiencies were not significantly different. Adsorption-extraction methods use electronegative membranes with a 0.45-μm pore-size to concentrate viruses; these methods have been used to concentrate nonenveloped viruses in wastewater and environmental waters (as recently reviewed in Haramoto et al., 2018; Bofill-Mas and Rusinol, 2020) and only recently for SARS-CoV-2 (Nemudryi et al., 2020). The best mean MHV recovery was achieved by Method C, which involved the addition of MgCl2 to the wastewater sample prior to filtration. The second highest mean recovery was achieved using Method B, which did not receive any pre-treatment prior to filtration. It is likely that the addition of MgCl2 increased MHV adsorption to the filter occurred in Method C. Increased virus adsorption with high MgCl2 concentrations was previously described and attributed to salt-bridging (Wallis et al., 1979; Lukasik et al., 2000; Villar et al., 2006; Ikner et al., 2012). Additionally, RT-qPCR inhibitors were not significantly co-concentrated by Method C per comparing the MHV-seeded wastewater and distilled water results.
It is important to note that the adsorption-extraction Methods B and C do not include pre-filtration or centrifugation; thus, they concentrated viruses from the liquid and solid fractions of the wastewater sample. Previous studies have suggested that considerable portions of MHV (and other CoV) may be bound to particulate matter in wastewater because upwards of 26% MHVs were adsorbed to organic matter within the sample (Gundy et al., 2009; Ye et al., 2016). Since the most efficient MHV concentration methods in this study were those that equally concentrated viruses from the solid and liquid fractions, the present study validates the observation about MHV adsorption to organic matter. Furthermore, the results of the present study highlight the need to concentrate both liquid and solid fractions of wastewater samples. Interestingly, Method A (adsorption-extraction, with acidification pre-treatment) also concentrated both fractions of the sample, but unlike Methods B and C, it required acidification of the sample to pH 4 and yielded the lowest recoveries. It has been reported that sample acidification might affect virus integrity and infectivity (Abdelzaher et al., 2008; Sabatino and Maier, 1980). This suggests that, unlike other enteric viruses, CoV may be sensitive to low pH. Further cross-comparison may be required to identify the impacts of acidification and other pre-treatment on the recovery of SARS-CoV-2 from untreated wastewater samples.
We found that ultrafiltration using centrifugal concentration devices may also be suitable for the recovery of MHV; however, recovery efficiency varied greatly based upon the centrifugal concentration device utilized (Methods D and E). Ultrafiltration using centrifugal concentration devices concentrate viruses based on size exclusion rather than electrostatic interactions between negatively charged viruses and electronegative or electropositive membranes. In this method, molecules smaller than the molecular weight cut-off are passed through the membrane by centrifugation; thus, the viruses are collected in the retentate fraction. These methods have been previously used to concentrate viruses from (waste)water (Symonds et al., 2009; Ikner et al., 2011) and recently used for SARS-CoV-2 RNA detection in untreated wastewater (Ahmed et al., 2020; Medema et al., 2020; Kocamemi et al., 2020). Overall, Method D (Amicon Ultra-15 centrifugal filter device) yielded the third-highest recovery, while Method E (Centricon Plus-70 centrifugal concentration device) had the second worst MHV recovery. The significantly different (p < .05) MHV recovery efficiencies between the two different types of centrifugal concentration devices demonstrate that not all centrifugal devices can effectively concentrate CoV from wastewater.
The difference in recovery efficiency could be due to differences in design. The Centricon plus-70 unit has a greater surface area for filtration than the Amicon Ultra-15 centrifugal filter device; thus, more MHV could have been lost via adsorption to the membrane via van der Waals interactive forces and/or hydrophobic bonding (Ikner et al., 2012). Ikner et al. (2011) used 30 kDa Centricon ultrafiltration units in a secondary concentration step following electropositive NanoCeram cartridge filters. The Centricon recovered 75% of MS2 coliphage, 61% of Echovirus 1, 95% of Poliovirus 1, and 109% of Coxsackievirus B5; however, only 33% of adenovirus 2 was recovered. Adenovirus recovery was similar to that obtained for MHV in our study. Such results suggest that a given centrifugal concentration device may also yield variable recoveries for different classes of viruses.
In addition to not being the most efficient method for MHV recovery, ultrafiltration using centrifugal concentration devices has a variety of drawbacks. One limitation is that a pre-filtration step at low speed is required to remove most of the larger debris and cells before ultrafilter centrifugation. Such pre-filtration can cause the loss of particle-associated viruses in the pellet. The resultant pellets from Methods D and E were analyzed for MHV in our study; we observed ~30% loss of MHV during the pre-filtration step. Furthermore, Method D is time-consuming because the maximum sample volume of these units is 15 mL; hence, repeated centrifugation of 15–20 mL aliquots is required to concentrate a 50-mL sample. While no evidence of RT-qPCR inhibition was identified in this study using Method D, these centrifugal concentration devices may co-concentrate PCR inhibitors. Furthermore, smaller molecular cut-off (i.e., 10 kDa or 30 kDa) device will take significant longer time to process wastewater samples than larger molecular cut-off (i.e., 100 kDa) and also co-concentrate different levels of PCR inhibitors. Finally, centrifugal concentration devices are quite expensive and often not readily available in many countries.
Thus, adsorption-extraction methods (Methods B and C) may be more logistically feasible and efficient for SARS-CoV-2 WBE. Method F (PEG precipitation) appeared to be a promising approach for MHV concentration because it incorporated the concentration of viruses from both the liquid and solid fractions of wastewater, however; it recovered significantly less MHV in comparison to Methods B and C. Different versions of PEG precipitation have been used for the assessment of SARS-CoV-2 in untreated wastewater, but the efficiencies were not reported (Wu et al., 2020; Kocamemi et al., 2020; Zhang et al., 2020; Bar-or et al., 2020). To the best of our knowledge, only one study reported the MHV recovery (~5%) using PEG precipitation (Ye et al., 2016), which was much lower than the value obtained in this study. This is likely because in our study PEG method concentrated MHV from both liquid and solid fractions, whereas in the previous study MHV was concentrated only from liquid phase (Ye et al., 2016). Using PEG precipitation methods, Kocamemi et al. (2020) found 1–1.5 log10-reduction in detection due to RT-qPCR inhibition when using this method and it is likely that the co-concentration of inhibitors with these methods explained their lower MHV recoveries. While qPCR inhibition was not observed in our study, the RT process may have been inhibited.
In this study, Method G (ultracentrifugation) yielded a mean recovery of 33.5%, which was significantly less than the most efficient methods (Methods B and C). Wurtzer et al. (2020a) used ultracentrifugation to recover SARS-CoV-2 from 11 mL of wastewater but did not provide recovery efficiency data. Ultracentrifugation has been used for decades to concentrate viruses from environmental matrices. It has been reported that ultracentrifugation at 100,000 g is required to pellet most macromolecules and viruses (Ammersbach and Bienzle, 2011). For example, Fumian et al. (2010) reported that ultracentrifugation (100,000 g for 1 h at 4 °C) had a mean recovery of 47% (range of 34–60%) of nonenveloped rotavirus A from wastewater samples. Ye et al. (2016) seeded 60 mL of wastewater with MHV and with the nonenveloped MS2 coliphage. Low mean recoveries (~5%) were achieved for both MHV and MS2 using the ultracentrifugation method; however, analyses were culture-based, and authors attributed low MHV and MS2 recoveries to virus inactivation by the high g force of the ultracentrifugation. The method also involves discarding supernatant a few times which may have resulted in loss of MHV. Nevertheless, this method may not be suitable for WBE studies because it requires expensive specialized centrifuges that may not be found in all laboratories.
This study is the first to evaluate established wastewater virus concentration methodologies for their use to concentrate CoV from wastewater, and ultimately, facilitate WBE. Since free viral RNA can degrade within minutes in wastewater (Limsawat and Ohgaki, 1997), most of the RNA copies detected in the wastewater virus concentration methods will be most likely from intact virus particles that may or may not be infectious. While we identified superior performance by the adsorption extraction concentration methods, with MgCl2 pre-treatment and without pre-treatment, it is important to note that the methods performance evaluation was based on small volumes (50 mL) of one municipal WWTP and the use of a surrogate (MHV). It is possible that CoV recovery efficiencies may vary based upon the wastewater matrix characteristics (e.g., concentration of total and dissolved suspended solids).
Frequently, it is possible to filter as much as 100–200 mL of wastewater using the adsorption extraction method used in this study. If these larger volumes were filtered, then the adsorption-extraction methods may become less efficient given the increased co-concentration of inhibitors. Also, centrifugal concentration devices, such as those used in this study, may co-concentrate RT-PCR inhibitors. To obtain information on the RT-PCR inhibitors, 50 mL of distilled water was seeded with MHV and the MHV recovery efficiency was compared to that for the MHV seeded wastewater for Methods C and D. Given that the recovery efficiency was only improved for distilled water by 5–8%, it is unlikely that 50 mL wastewater samples tested in this study had RT-PCR inhibitors. It is likely that the co-concentration of inhibitors from larger volumes will have a minimal influence on CoV recovery but will require further investigation. Furthermore, RNA extraction kit used in this study is equipped with next-generation inhibition removal technology (IRT) which is designed to remove PCR inhibitors effectively for soil and fecal samples.
Several methods identified in this preliminary study can be used in conjunction with large-scale primary concentration procedures (2–100 L, such as dead-end or tangential flow hollow fibre ultrafiltration) as a primary step to reduce filter elution to volumes, which are applicable to molecular detection assays. Furthermore, the focus of future research efforts should be to compare recovery efficiencies of MHV to those of actual SARS-CoV-2, considering the multiple benefits of working with the former as a surrogate. Also, the recovery efficiency of various levels of surrogate CoV or SARS-CoV-2 from large volumes of wastewater needs to be assessed to better understand how these variabilities affect recovery efficiency. Furthermore, the best methods for the recovery of intact virus particles need to be identified. Given the high standard deviations observed in the recoveries of any given method, the use of a process control is highly recommended for assessing and normalizing SARS-CoV-2 concentrations for WBE. MHV is a promising process control, which can be purchased from ATCC® VR-764™ and similar companies; however, it may be difficult to obtain this virus in a timely-manner during the pandemic. Other coronaviruses, and/or other viral surrogates, could also be useful process controls for SARS-CoV-2 surveillance; thus, future studies are needed to assess their use as a process control. The culmination of these efforts will aid public health officials, epidemiologist and modellers in assessing incidences of SARS-CoV-2 infection rates at the community scale using WBE approaches, so that appropriate public health response and intervention strategies are deployed.
Disclaimers
The views expressed in this article are those of the author(s) and do not necessarily represent the views or policies of the U.S. Environmental Protection Agency. The U.S. Environmental Protection Agency through the Office of Research and Development provided technical direction but did not collect, generate, evaluate, or use the environmental data described herein.
CRediT authorship contribution statement
Warish Ahmed: Investigation, Resources, Writing - original draft, Writing - review & editing. Paul M. Bertsch: Writing - original draft, Writing - review & editing. Aaron Bivins: Resources. Kyle Bibby: Resources. Kata Farkas: Writing - original draft, Writing - review & editing. Amy Gathercole: Resources. Eiji Haramoto: Writing - original draft, Writing - review & editing. Pradip Gyawali: Resources. Asja Korajkic: Writing - original draft, Writing - review & editing. Brian R. McMinn: Writing - original draft, Writing - review & editing. Jochen F. Mueller: Resources. Stuart L. Simpson: Writing - original draft, Writing - review & editing. Wendy J.M. Smith: Investigation. Erin M. Symonds: Writing - original draft, Writing - review & editing, Resources. Kevin V. Thomas: Writing - original draft, Writing - review & editing. Rory Verhagen: Resources. Masaaki Kitajima: Writing - original draft, Writing - review & editing, Resources.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
We thank CSIRO Land and Water for strategic funding to complete this research project. We thank Drs. Sonja Toft, Jason Dwyer, and Paul Sherman (Urban Utilities) for providing untreated wastewater samples. E.M. Symonds was partly funded by the US National Science Foundation grant OCE-1566562. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the US National Science Foundation. This study was partly supported by the JST-Mirai Program Grant Number JPMJMI18DB, Japan.
Editor: Damia Barcelo
References
- Abdelzaher A.M., Solo-Gabriele H.M., Wright M.E., Palmer C.J. Sequential concentration of bacteria and viruses from marine waters using a dual membrane system. J. Environ. Qual. 2008;37(4):1648–1655. doi: 10.2134/jeq2007.0238. [DOI] [PubMed] [Google Scholar]
- Ahmed W., Harwood V.J., Gyawali P., Sidhu J.P., Toze S. Comparison of concentration methods for quantitative detection of sewage-associated viral markers in environmental waters. Appl. Environ. Microbiol. 2015;81(6):2042–2049. doi: 10.1128/AEM.03851-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Payyappat S., Cassidy M., Besley C., Power K. Novel crAssphage marker genes ascertain sewage pollution in a recreational lake receiving urban stormwater runoff. Water Res. 2018;145:769–778. doi: 10.1016/j.watres.2018.08.049. [DOI] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., Brien J.W.O., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. 138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Or I., Yaniv K., Shagan M., Ozer E., Erster O., Mendelson E., Mannasse B., Shirazi R., Kramarsky-Winter E., Nir O., Abu-Ali H., Ronen Z., Rinott E., Lewis Y.E., Friedler E., Bitkover E., Paitan Y., Berchenko Y., Kushmaro A. Regressing SARS-CoV-2 sewage measurements onto COVID-19 burden in the population: a proof-of-concept for quantitative environmental surveillance. medRxiv. 2020 doi: 10.1101/2020.04.26.20073569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besselsen D.G., Wagner A.M., Loganbill J.K. Detection of rodent coronaviruses by use of fluorogenic reverse transcriptase-polymerase chain reaction analysis. Comp. Med. 2002;52(2):111–116. [PubMed] [Google Scholar]
- Bofill-Mas S., Rusinol M. Recent trends on methods for the concentration of viruses from water samples. Curr. Opin. Environ. Sci. Health. 2020;16:7–13. doi: 10.1016/j.coesh.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carducci A., Federigi I., Liu D., Thompson J.R., Varni M. Making waves: coronavirus detection, persistence and persistence in the water environment: state of the art and knowledge needs for public health. Water Res. 2020;179 doi: 10.1016/j.watres.2020.115907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova L., Rutala W.A., Weber D.J., Sobsey M.D. Survival of surrogate 561 coronaviruses in water. Water Res. 2009;43(7):1893–1898. doi: 10.1016/j.watres.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva A.K., Le Saux J.-C., Parnaudeau S., Pommepuy M., Elimelech M., Le Guyader F.S. Evaluation of removal of noroviruses during wastewater treatment, using real-time reverse transcription-PCR: different behaviors of genogroups I and II. Appl. Environ. Microbiol. 2007;73(24):7891–7897. doi: 10.1128/AEM.01428-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas K., Cooper D.M., McDonald J.E., Malham S.K., Rougemont A., Jones D.L. Seasonal and spatial dynamics of enteric viruses in wastewater and in riverine and estuarine receiving waters. Sci. Total Environ. 2018;634:1174–1183. doi: 10.1016/j.scitotenv.2018.04.038. [DOI] [PubMed] [Google Scholar]
- Fumian T.M., Leite J.P., castello A.A., Gaggero A., Caillou M.S., Miagostovich M.P. Detection of rotavirus A in sewage samples using multiplex qPCR and an evaluation of the ultracentrifugation and adsorption-elution methods for virus concentration. J. Virol. Methods. 2010;170(1–2):42–46. doi: 10.1016/j.jviromet.2010.08.017. [DOI] [PubMed] [Google Scholar]
- Gundy P.M., Gerba C.P., Pepper I.L. Survival of coronaviruses in water and wastewater. Food Environ. Virol. 2009;1(1):10. [Google Scholar]
- Gyawali P., Croucher D., Ahmed W., Devane M., Hewitt J. Evaluation of pepper mild mottle virus as an indicator of human faecal pollution in shellfish and growing waters. Water Res. 2019;154:370–376. doi: 10.1016/j.watres.2019.02.003. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M., Katayama H., Ito T., Ohgaki S. Development of virus concentration methods for detection of koi herpesvirus in water. J. Fish Dis. 2009;32:297–300. doi: 10.1111/j.1365-2761.2008.00977.x. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M., Hata A., Torrey J.R., Masago Y., Sano D., Katayama H. A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Res. 2018;135:168–186. doi: 10.1016/j.watres.2018.02.004. [DOI] [PubMed] [Google Scholar]
- Haugland R.A., Siefring S.C., Wymer L.J., Brenner K.P., Dufour A.P. Comparison of Enterococcus measurements in freshwater at two recreational beaches by quantitative polymerase chain reaction and membrane filter culture analysis. Water Res. 2005;39(4):559–568. doi: 10.1016/j.watres.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Ikner L.A., Soto-Beltran M., Bright K.R. New method using a positively charged microporous filter and ultrafiltration for concentration of viruses from tap water. Appl. Environ. Microbiol. 2011;77(10):3500–3506. doi: 10.1128/AEM.02705-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikner L.A., Gerba C.P., Bright K.R. Concentration and recovery of viruses from water: a comprehensive review. Food Environ. Virol. 2012;4(2):41–67. doi: 10.1007/s12560-012-9080-2. [DOI] [PubMed] [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.139076. 139076, In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocamemi B.A., Kurt H., Hacıo S., Yaralı C., Saatci A.M., Pakdemirli B. First data-set on SARS-CoV-2 detection for Istanbul wastewaters in Turkey. medRxiv. 2020 doi: 10.1101/2020.05.03.20089417. [DOI] [Google Scholar]
- La Rosa G., Bonadonna L., Lucentini L., Kenmoe S., Suffredini E. Coronavirus in water environments: occurrence, persistence and concentration methods — a scoping review. Water Res. 2020;179 doi: 10.1016/j.watres.2020.115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Bonanno G., Ferraro G.B., Veneri C., Bonadonna L., Lucentini L., Suffredini E. Vol. 736. Sci. Total Environ; 2020. First Detection of SARS-CoV-2 in Untreated Wastewater in Italy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limsawat S., Ohgaki S. Fate of liberated viral RNA in wastewater determined by PCR. Appl. Environ. Microbiol. 1997;63(7):2932–2933. doi: 10.1128/aem.63.7.2932-2933.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukasik J., Scott T.M., Andryshak D., Farrah S.R. Influence of salts on virus adsorption to microporous filters. Appl. Environ. Microbiol. 2000;66(7):2914–2920. doi: 10.1128/aem.66.7.2914-2920.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020 doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Morawska L., Cao J. Airborne transmission of SARS-CoV-2: the world should face the reality. Environ. Int. 2020;139 doi: 10.1016/j.envint.2020.105730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mull B., Hill V.R. Recovery of diverse microbes in high turbidity surface water samples using dead-end ultrafiltration. J. Microbiol. Methods. 2012;91(3):429–433. doi: 10.1016/j.mimet.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemudryi A., Nemudraia A., Surya K., Wiegand T., Buyukyoruk M., Wilkinson R., Wiedenheft B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. medRxiv. 2020 doi: 10.1101/2020.04.15.20066746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S.J., Zhao G., Penna V.R., Park E., lauron E.J., Harvey I.B., Beatty W.L., Plougastel-Douglas B., Poursine-Laurent J., Fremont D.H., Wang D., Yokoyama W.M. A murine herpesvirus closely related to ubiquitous human herpesvirus causes T-cell depletion. J. Virol. 2017;91(9) doi: 10.1128/JVI.02463-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Cuevas-Ferrando E., Sanjuan R., Domingo-Calap P., Sanchez G. 2020. Metropolitan Wastewater Analysis for COVID-19 Epidemiological Surveillance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Ferranfo E.C., Simon P., Allende A., Sanchez G. SARS-CoV-2 RNA titers in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth-Cross J.K., Bender S.J., Weiss S.R. Murine coronavirus mouse hepatitis virus is recognized by MDA5 and induces type I interferon in brain macrophages/microgila. J. Virol. 2008;82(20):9829–9838. doi: 10.1128/JVI.01199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatino C.M., Maier S. Differential inactivation of three bacteriophages by acid and alkaline pH used in the membrane adsorption-elution method of virus recovery. Can. J. Microbiol. 1980;26(12):1403–1407. doi: 10.1139/m80-233. [DOI] [PubMed] [Google Scholar]
- Shi H., Pasco E.V., Tarabara V.V. Membrane-based methods of virus concentration from water: a review of process parameters and their effects on virus recovery. Environ. Sci: Water Res. Technol. 2017;3(5):778–792. [Google Scholar]
- Sima L.C., Schaeffer J., Le Saux J.-C., Parnaudeau S., Elimelch M., Le Guyader F.S. Calicivirus removal in a membrane bioreactor wastewater treatment plant. Appl. Environ. Microbiol. 2011;77(15):5170–5177. doi: 10.1128/AEM.00583-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley C., Gordon K.V., Schoen M.E., Harwood V.J. Performance of two quantitative PCR methods for microbial source tracking of human sewage and implications for microbial risk assessment in recreational waters. Appl. Environ. Microbiol. 2012;78(20):7317–7326. doi: 10.1128/AEM.01430-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonds E.M., Griffin D.W., Breitbart M. Eukaryotic viruses in wastewater samples from the United States. Appl. Environ. Microbiol. 2009;75(5):1402–1409. doi: 10.1128/AEM.01899-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonds E.M., Verbyla M.E., Lukasik J.O., Kafle R.C., Breitbart M., Mihelcic J.R. A case study of enteric virus removal and insights into the associated risk of water reuse for two wastewater treatment pond systems in Bolivia. Water Res. 2014;65:257–270. doi: 10.1016/j.watres.2014.07.032. [DOI] [PubMed] [Google Scholar]
- Villar L.M., de Paula V.S., Diniz-Mendes L., Lampe E., Gaspar A.M.C. Evaluation of methods used to concentrate and detect hepatitis A virus in water samples. J. Virol. Methods. 2006;137(2):169–176. doi: 10.1016/j.jviromet.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Wallis C., Melnick J.L., Gerba C.P. Concentration of viruses from water by membrane chromatography. Annu. Rev. Microbiol. 1979;33:413–437. doi: 10.1146/annurev.mi.33.100179.002213. [DOI] [PubMed] [Google Scholar]
- WHO . Interim Guidance, 13 May 2020. World Health Organization; Geneva: 2020. Laboratory biosafety guidance related to coronavirus disease (COVID-19) [Google Scholar]
- Wu F., Xiao A., Zhang J., Gu X., Lee W., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Moniz K., Erickson T., Chai P., Thompson J., Alm E. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. medRxiv. 2020 doi: 10.1101/2020.04.05.20051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J., Maday Y., Teyssou R., Richard E., Almayrac J., Moulin L. Evaluation of lockdown impact on SARS-CoV-2 dynamics through viral genome quantification in Paris wastewaters. medRxiv. 2020 doi: 10.1101/2020.04.12.20062679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J., Moulin L. Time course quantitative detection of SARS-CoV-2 in Parisian wastewaters correlates with COVID-19 confirmed cases. medRxiv. 2020:10–13. doi: 10.1101/2020.04.12.20062679. [DOI] [Google Scholar]
- Xiao F., Sun J., Xu Y., Li F., Huang X., Li F., Huang X., Li H., Zhao J., Hunag J., Zhao J. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg. Infect. Dis. 2020;26(8) doi: 10.3201/eid2608.200681. Aug 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50:5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Yu P., Zhu J., Zhang Z., Han Y. A familial cluster of infection associated with the 2019 novel coronavirus indicating possible person-to-person transmission during the incubation period. J. Infect. Dis. 2020;221(11):757–1761. doi: 10.1093/infdis/jiaa077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Ling H., Huang X., Li J., Li W., Yi C., Jiang Y., He Y., Deng S., Zhang X., Liu Y., Li G., Qu J., Sciences E., Qu P.J. 2020. Potential Spreading Risks and Disinfection Challenges of Medical Wastewater by the Presence of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Viral RNA in Septic Tanks of Fangcang Hospital; p. 86. [DOI] [PMC free article] [PubMed] [Google Scholar]