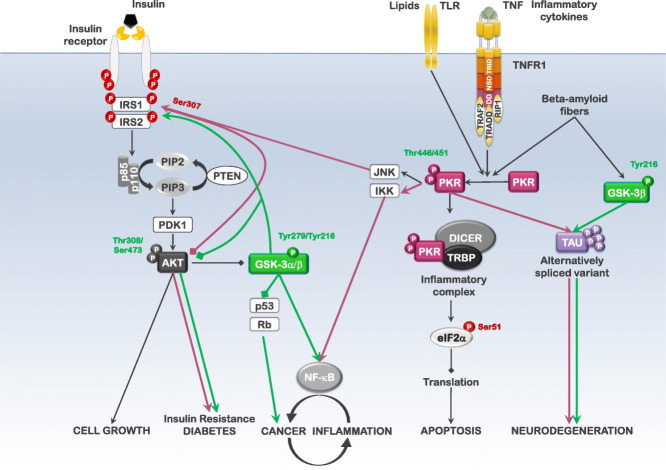
Abstract
Glycogen synthase kinase (GSK)-3α/β and the double-stranded RNA-dependent kinase PKR are two sentinel kinases that carry-out multiple similar yet distinct functions in both the cytosol and the nucleus. While these kinases belong to separate signal transduction cascades, they demonstrate an uncanny propensity to regulate many of the same proteins either through direct phosphorylation or by altering transcription/translation, including: c-MYC, NF-κB, p53 and TAU, as well as each another. A significant number of studies centered on the GSK3 kinases have led to the identification of the GSK3 interactome and a number of substrates, which link GSK3 activity to metabolic control, translation, RNA splicing, ribosome biogenesis, cellular division, DNA repair and stress/inflammatory signaling. Interestingly, many of these same pathways and processes are controlled by PKR, but unlike the GSK3 kinases, a clear picture of proteins interacting with PKR and a complete listing of its substrates is still missing. In this review, we take a detailed look at what is known about the PKR and GSK3 kinases, how these kinases interact to influence common cellular processes (innate immunity, alternative splicing, translation, glucose metabolism) and how aberrant activation of these kinases leads to diseases such as Alzheimer's disease (AD), diabetes mellitus (DM) and cancer.
Abbreviations: 4E-BP, eIF4E-binding protein; AD, Alzheimer's disease; ADAR, adenosine deaminase acting on double-stranded RNA; CDK, cyclin dependent kinase; C/EBP, CCAAT-enhancer binding protein; DM, diabetes mellitus; dsRBD, double-strand RNA binding domain; dsRNA, double-strand RNA; eIF, eukaryotic initiation factor; GCN2, general control non-derepressable 2; GSK3, glycogen synthase kinase; HRI, heme-regulated kinase; IFN, interferon; IGF, insulin-like growth factor; IGF1R, insulin-like growth factor 1 receptor; IκB, inhibitor κB; IKK, inhibitor κB kinase; IR, insulin receptor; IRS, insulin response substrate; JNK, c-Jun N-terminal kinase; mTOR, mammalian target of rapamycin; NEMO, IKK subunit γ; NF-κB, nuclear factor κB; PERK, PKR-related endoplasmic reticulum kinase; PI3K, phosphatidylinositol-3 kinase; PIP, phosphoinositol phosphate; PKR, double-strand RNA-dependent kinas R; PP1, protein phosphatase 1; STAT, signal transducer and activator of transcription; TNFα, tumor necrosis factor-α; TNFR, tumor necrosis factor-α receptor
Keywords: Inflammation, Alternative splicing, Innate immunity, Neurodegenerative disease, Leukemia, Osteosarcoma
Graphical abstract
Highlights
-
•
GSK3α/β and PKR are major regulators of cellular homeostasis and the response to stress/inflammation and infection.
-
•
GSK3α/β and PKR interact with and/or modify many of the same proteins and affect the expression of similar genes.
-
•
A balance between AKT and PKR nuclear signaling may be responsible for regulating the activation of nuclear GSK3β.
-
•
GSK3α/β- and PKR-dependent signaling influence major molecular mechanisms of the cell through similar intermediates.
-
•
Aberrant activation of GSK3α/β and PKR is highly involved in cancer, metabolic disorders, and neurodegenerative diseases.
1. Introduction
Cellular homeostatsis and maintenance are critical for the proper development and function of every tissue and organ system; the activity of specialized cells, stem cell self-renewal, differentiation, cell death and replacement are all tightly controlled. Multiple levels of regulation are in place to control these processes, and when one of these control systems goes awry it puts pressure on the other regulatory systems often resulting in a form of chronic inflammation, as the main cellular response is damage control. Glycogen synthase kinase-3 (GSK-3) and the double-strand RNA-dependent kinase PKR are two major regulators of this response [1,2]. While upstream regulation of these kinases appears to be for the most part independent from one another, GSK-3 responding to metabolic stresses and PKR a more general stress responder, together they co-regulate multiple stress response pathways that result in the inhibition of translation, cell cycle arrest, apoptosis and inflammation; and aberrant activation of either is associated with diverse pathologies including, Alzheimer's disease, diabetes mellitus and cancer [[2], [3], [4]]. In this review, we look closely at the relationship between these two stress-activated kinases, their co-regulation of key mediators of cell homeostasis and their involvement in disease. To limit discussion, we have focused only on those signaling intermediates and processes these kinases have in common, highlighting possible points of therapeutic intervention for the design of novel compounds targeting the signaling of both these sentinel kinases.
1.1. GSK-3
Glycogen synthase kinase-3 is the all-inclusive name given to two kinase isoforms (α and β) that are encoded by different genes. In humans, the α-isoform (GSK3α), a 51 kDa protein, is encoded on Ch.19q13.2, while the β-isoform (GSK3β), a 47 kDa protein, is encoded on Ch.3q13.3; both are ubiquitously expressed with the greatest expression being observed in brain tissue [5,6]. Within the cell, these isoforms have a more restricted localization, with GSK3α localizing throughout the cell (nucleus, cytosol and mitochondria) and GSK3β localizing to mainly the nucleus, cytosol or cell membrane. Other reports have suggested that GSK3α in more important to cytoplasmic signaling, while GSK3β is more important for nuclear signaling [6]. These kinases belong to the CMGC family of serine/threonine kinases and are composed of two domains; a sequence forming seven anti-parallel β-sheets followed by an α-helical region. The catalytic domain lies at the interface of these two domains. The GSK3 proteins are 98% identical, but significant divergence is observed in the C-termini. In addition, GSK3α also contains an extension of the glycine-rich sequence in the amino terminus. These slight differences in structure likely result in the differing cellular localization of these kinases, as well as substrate preference [5,6]. While originally identified and thus named for the ability of what would eventually be designated GSK3β to phosphorylate and inactivate glycogen synthase (GS), these kinases have been found to phosphorylate and regulate proteins involved in glucose homeostasis, WNT/β-catenin pathway, microtubule assembly, apoptosis, inflammation, the cell cycle and translation. Phosphorylation on the part of GSK3α/β is dependent on a priming phosphorylation mediated by a secondary kinase at a site four amino acids C-terminal to the GSK3α/β-dependent site [5,6]. In theory, phosphorylation of a series of GSK3α/β sites, spaced approximately four amino acids apart from one another, would occur sequentially from C-ter to N-ter, following the initial priming phosphorylation mediated by a secondary kinase (ex., β-catenin). As with GS, phosphorylation mediated by GSK3α/β has, with few exceptions, a negative regulatory effect on the substrate [6].
The main mechanism by which the GSK3 kinases are controlled is through the phosphorylation of a key tyrosine residue in the catalytic domain. The GSK3 kinases are constitutively-active at a basal level; phosphorylation at Y279 or Y216 in GSK-3α or -3β, respectively, enhances GSK3 catalytic activity. It has been shown that this site is an autophosphorylation site; thus in this rare case GSK3α/β demonstrate tyrosine kinase ability. In addition, activation of upstream tyrosine kinases such as FYN, SRC, the proline-rich tyrosine kinase, PYK2, and even the dual Ser/Thr and Tyr kinase MEK1 have been shown to phosphorylate the GSK3 proteins at these respective sites when activated [[7], [8], [9], [10]]. In contrast, inactivation of GSK3 activity requires the phosphorylation of a serine residue within the glycine-rich region of the kinase (S21 in GSK3α and S9 in GSK3β). Phosphorylation of this site causes a conformational change in the protein, blocking access of the substrate to the active site. Depending on the GSK3 isoform in question, phosphorylation at this site can occur via AKT1, Aurora kinase, diverse ribosomal S6 kinases (p70S6K1/2, p90RSK and RSK2), serine/threonine protein kinase (SGK)-3, protein kinase A (PKA), inhibitor kappa B kinase (IKK)-ε and various protein kinase C (PKC) isoforms (http://www.phosphosite.org/uniprotAccAction?id=P49840; http://www.phosphosite.org/uniprotAccAction?id=P49841; [6,11,12]). Moreover, additional phosphorylation sites have been demonstrated to be important for GSK3 regulation. In the better studied GSK3β, phosphorylation at S21, T43, Y56, S147, S215, S219, Y221, Y222, T356, S389 and T390 have also been reported. Phosphorylation of S21 was reported in acute leukemia cell lines and in cells following hepatitis C infection, but the significance of phosphorylation at this site has not been determined [13]. Similarly, T43 phosphorylation occurs through ERK-dependent means, but again the exact significance of this modification has not been determined [14]. In contrast, phosphorylation of S147 is carried-out by PKCζ following WNT treatment, resulting in enhanced GSK3α/β kinase activity [15]. While not yet understood, phosphorylation surrounding Y216 (i.e., S215, S219) appears to be inhibitory to the activating phosphorylation of this residue (http://www.phosphosite.org/uniprotAccAction?id=P49841; [12,16]). In mouse tissue, the dual-specificity tyrosine phosphorylation regulated kinase (DYRK) 1A-dependent phosphorylation of T356 was demonstrated to inactivate GSK3β [17]. Moreover, stress-induced phosphorylation of S389 and T390 by p38α has also been suggested to inhibit GSK3β activity [18]. In addition, the phosphorylation of a string of threonine residues in the carboxy-terminus (T392, T395, T402, T417 and T420) appears to regulate the proteolysis of GSK3β (http://www.phosphosite.org/uniprotAccAction?id=P49841; [12]). Although much less studied, similar phosphorylations (T19, S41, S52, S63, S97, S278, S282 and Y284) have been reported in GSK3α. Phosphorylation of the serine residues surrounding Y279 in GSK3α (S278 and S282) appear to mask and inhibit phosphorylation of this Tyr, while the other sites mentioned have been found to be phosphorylated following treatment with various inhibitors: T19 (imatanib, wortmannin), S41 and S97 (MG132 withdrawal; suggesting these sites are involved proteosomal degradation), S52 and S63 (MEK inhibition). Beyond these, other post-translational modifications of both GSK3 isoforms have been documented and include sumoylation (GSK3β only), ubiquitinations, methylations and acetylations; but the significance of these PTMs remains unclear (http://www.phosphosite.org/uniprotAccAction?id=P49840; [12]). Moreover, several reports describe alternately spliced forms of both GSK3 isoforms (discussed below).
1.2. PKR
The double-strand RNA (dsRNA)-dependent kinase PKR is an innate immune, stress/inflammatory kinase originally identified and characterized for its antiviral activity. It belongs to a family of kinases best known for their ability to phosphorylate the eukaryotic initiation factor 2 subunit α (eIF2α). Among this family are PKR, the PKR-related endoplasmic reticulum kinase (PERK), heme-regulated kinase (HRI) and general control nonderepressible (GCN)-2. While PERK, HRI and GCN2 respond to more specific stresses, unfolded protein/endoplasmic reticulum stress, reduced heme concentration or amino acid starvation, respectively, PKR is activated in response to more general stresses, including viral infection, dsRNA, peroxidation, mitochondrial stress, DNA damage, ER stress, inflammatory cytokines, growth factor deprivation and Toll-like receptor activation [19,20], placing it in a unique position as a sensor and first responder to cellular stresses as well as a mediator of inflammation. Together these kinases form a redundant network [[21], [22], [23], [24], [25], [26], [27], [28], [29]]. Interestingly, while all of these kinases are present in the cytoplasm, PKR is the only eIF2α kinase that is also present in the nucleolus and nucleoplasm [25,30]. In humans, the PKR kinase is encoded by the EIF2AK2 gene located on Ch.2p22.2. The gene encodes a 68 kDa protein which is unusual in that the amino terminal end or regulatory region contains two double-strand RNA binding domains (dsRBDs) while the carboxyl terminus contains a protein kinase domain. The EIF2AK2 gene is ubiquitously and constitutively expressed with the highest levels of protein expression being observed in hematopoietic tissue (bone marrow, spleen and thymus) and the brain [[31], [32], [33]]. Moreover, the promoter of EIF2AK2 contains interferon (IFN)-stimulated response elements (ISREs), allowing for enhanced transcription of the gene in cells exposed to Type I (IFNα/β) interferons [34].
The regulation of PKR kinase activity requires the phosphorylation of two primary threonine residues in the catalytic domain. Minimal activation of PKR requires the phosphorylation of T451, while subsequent autophosphorylation of T446 significantly enhances kinase activity. Additional sites of autophosphorylation are observed predominantly in two clusters: S83, T88, T89, T90 and Y101 between dsRBD I and dsRBD II, and Y162, S242, T255, T258 and Y293 situated between dsRBD II and the kinase domain; each of these enhancing PKR enzymatic activity [35,36]. Other than the afore mentioned sites, which have all been biochemically verified, additional sites of phosphorylation have been identified in multiple studies using mass spectrometry: S33, S92, T115, S167, S179, S181, S456 and S542. The exact consequence of phosphorylating these residues though is not known (http://www.phosphosite.org/uniprotAccAction?id=P19525; [12]). PKR is also highly ubiquitinated and sumoylated with a large number of sites within the carboxyl half of the protein. Ubiquitination is predominantly carried-out by the SCF E2 ubiquitinase FBXWII E3 ligase, which targets PKR for proteosomal degradation [37]. In contrast, SUMO1 and SUMO3 sumoylate PKR on K60, K150 and K440 in an enzyme specific manner altering PKR activation, localization and stability [38].
Several mechanisms have been proposed to explain PKR activation. Initially, PKR activation was thought to require only dsRNA, a typical product of viral infection. The binding of PKR to dsRNA through the dsRBDs would facilitate PKR homodimerization and autophosphorylation of T451 followed by T446. Several lines of evidence have suggested that this model was not completely correct: (1) the endogenous PKR activator, PACT/RAX, was demonstrated to promote PKR activation in the absence of dsRNA in in vitro studies [39]; (2) PKR activation following vesicular stomatitis virus (VSV) infection was inhibited in the absence of PACT [40]; (3) T451 phosphorylation is often induced following treatment of cells with the commercial PKR inhibitor [41]; (4) dsRNAs readily available in the cell do not activate PKR, in contrast, the cellular RNA non-coding 886 (nc886) binds to and inhibits PKR activation; and (5) a diverse number of miRNAs bind to the dsRBDs of PKR [42]. These findings also suggested that PKR was not the only kinase capable of phosphorylating T451. Zykova et al. demonstrated that T451 could be phosphorylated by ERK2 and RSK2, likely establishing them as primers of PKR activation [43]. This would also explain PKR activation following Toll-like receptor (TLR) stimulation. During apoptosis, PKR may also be activated through caspase-dependent cleavage at D251, thus removing the regulatory dsRBDs and releasing an active kinase domain [44]. For years, PKR was studied for its ability to phosphorylate the eukaryotic initiation factor (eIF)-2α subunit in the presence of dsRNA (either during viral infection or treatment with poly I:C, a synthetic dsRNA) and was thus analyzed for its ability to block viral replication and/or induce cell death following infection [19]. It is now known that PKR has a much larger role in cell growth and homeostasis than previously thought.
Several seminal studies over the years have begun to shed more light on just how entwined PKR is in normal cellular homeostasis. The simultaneous reporting of PACT and its mouse orthologue RAX as endogenous activators of PKR demonstrated that PKR regulation was more complicated than just the presence or absence of dsRNA [39,45]. The fact that multiple cellular stresses [DNA damage, reactive oxygen species (ROS), and cytotoxic cytokines (IFNγ, TNFα, IL1)] in addition to viral infection could result in PKR activation began to paint a picture of PKR as a sentinel for the cellular stress response. PKR was found to regulate p53, NF-κB, c-MYC, protein phosphatase 2A (PP2A), RNAase A (DHX9), GSK3 and be a component of several cytoplasmic (inflammasome and stress bodies) and nuclear complexes (splicesome, ribosome assembly and DNA repair) [46]. The role of PKR in disease has been controversial, but it is known that PKR is overexpressed and/or activity elevated in multiple solid tumors (breast carcinoma, colon carcinoma, melanoma, osteosarcoma, etc.), hematopoietic malignancies (acute leukemias and myelodysplastic syndromes) and neurodegenerative diseases (Alzheimer's disease, Huntington's corea and Creutzfeldt-Jakob disease) [25,47,48]. In most of these pathologies an increased nuclear presence of PKR has been observed, but while nuclear PKR is known to be associated with splicing factors and components of the assembling ribosome, the significance of PKR nuclear localization in these pathologies remains unclear.
2. PKR-dependent regulation of GSK-3
Regulation of GSK3 by PKR appears to occur at multiple levels. As stated previously, activation of the PI3K-AKT-mTOR pathway leads to AKT-dependent phosphorylation of GSK3α/β on S21/S9 resulting in the inactivation of GSK3 kinase activity. The link between PKR activation and AKT was originally observed in PKR-null MEFs where AKT activation was altered [49]. Baltzis et al. then later reported that both PKR and PERK could lead to the activation of nuclear GSK3β in a manner independent of eIF2α phosphorylation [50]. Blalock et al. subsequently demonstrated that PKR could regulate GSK3α/β through the activation of a phosphatase, but this study stopped short of identifying the phosphatase [41]. The authors demonstrated that inhibition PKR with a small molecule inhibitor resulted in an increase in p-S21/S9 GSK3α/β while at the same time resulting in a slight mobility shift to a faster migrating form of GSK3α/β. In contrast, while use of the general phosphatase inhibitor okadaic acid (OA) resulted in an increase of p-S21/S9 GSK3α/β, it did not result in any mobility shift, suggesting a site specific regulation of GSK3 phosphorylation by PKR [41]. A more recent analysis of the PKR nuclear interactome brought forward the finding that the phosphatase PP1A is associated with active PKR in the nucleus, but the PP1A catalytic subunit is replaced with that of PP1B in the inactive PKR complex [46]. As dephosphorylation of S21/S9 is known to occur through PP1A, it might be presumed that PKR-dependent regulation of GSK3 activity is mediated through PP1A dephosphorylation of S21/S9, which would account for the increase in levels of p-S21/S9 GSK3α/β in the presence of the PKR inhibitor. In the presence of active PKR, PP1A-mediated dephosphorylation at S21/S9 would increase GSK3 activity, thus enhancing Y279/Y216 phosphorylation, increasing GSK3 activity further (Fig. 1A). As PKR also stimulates the p38MAPK, PKR activity may favor additional phosphorylations at S389 and T390 in GSK3β, thus accounting for the significant shift or collapse in bands that occurs in the presence of the PKR inhibitor [18,41]. To date, no clear mechanism for GSK3α/β to regulate PKR activity has been reported, although control at the level of PKR expression would be reasonable to assume taking into consideration the effects that GSK3α/β has on transcription factors, such as MYC and p53, and translation (see below).
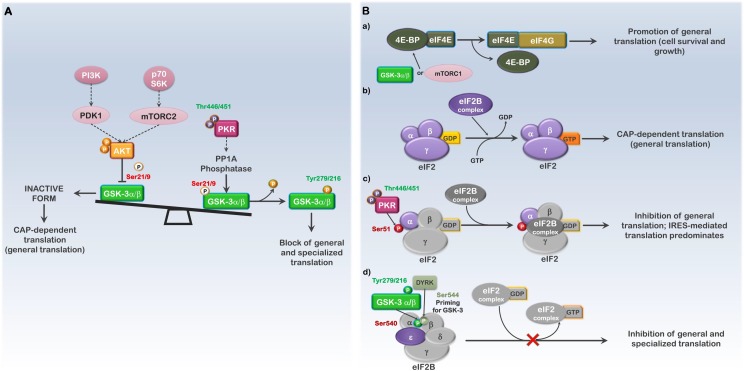
Fig. 1.
GSK3α/β and PKR regulate translation initiation. The diagram shows the effects of GSK3α/β and PKR activity on translation initiation and the various potential outcomes of kinase-dependent modifications on translation. (A) Regulation of the PI3K-AKT-mTOR and PKR pathways can influence translation initiation through modulation of GSK3 activity. (B) Activation of the PI3K-AKT (mTOR or GSK3α/β) and PKR pathways affect the eIF4 and eIF2 translation initiation complexes. a; GSK3α/β- or mTOR- mediated phosphorylation of 4E-BP inhibits its association with eIF4E, promoting general translation. b; The eIF2B GDP-GTP exchange factor complex (eIF2B complex) must replace GDP bound to the eIF2 translation initiation complex (eIF2) with GTP in order to initiate each new round of CAP-dependent (general) translation. c; Active PKR (phospho-T446/T451) results in eIF2α phosphorylation on S51 and the inhibition of general translation by stabilizing the eIF2:eIF2B-GDP interaction; IRES-mediated translation of stress-response genes is favored. d; Active GSK3α/β (phospho-Y279/216) phosphorylates the ε subunit of the eIF2B complex on S540, resulting in inhibition of both general and specialized (IRES-mediated) translation. While active PKR is denoted as phosphorylated on T446 and T451, only T451 is required for activity. Phosphorylation of T446 as well as numerous additional sites influences the degree of activation. Phosphorylation sites labeled in green (ex., Tyr216-GSK3β) indicate activating or stabilizing modifications; phosphorylation sites labeled in red (ex., Ser51-eIF2α) indicate modifications that are inhibitory to the substrates activity.
3. Common signaling intermediates of GSK3 and PKR
The extent of interaction between GSK3α/β and PKR signaling goes beyond that of phosphatase regulation; these kinases influence many of the same downstream intermediates. In some cases, these intermediates are directly phosphorylated by both PKR and GSK3; in others, regulation of the downstream protein occurs through phosphorylation by one of the kinases and an alternate mechanism with the other. In the following sections, we discuss some of the common targets of GSK3 and PKR and the influence that these kinases have on these downstream targets (Table 1 ).
Table 1.
Common substrates and/or interacting proteins of GSK3α/β and PKR.
| Acc. # | Gene name | Description | Function | Association and modification |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GSK3α | GSK3β | pGSKα/β | PKR+ | PKR- | pPKR | PKR | ||||
| Q9NYF8 | BCLAF1 | Bcl-2-associated transcription factor 1 | Death-promoting transcriptional repressor; interacts with BCL2-related proteins and adenovirus E1B protein | X | X | X(S531) | X | |||
| Q16543 | CDC37 | Hsp90 co-chaperone Cdc37 | Co-chaperone; mediates interaction with the HSP90 complex | X | X | |||||
| Q8TDD1 | DDX54 | ATP-dependent RNA helicase DDX54 | Represses the transcriptional activity of nuclear receptors | X | X | |||||
| Q99848 | EBNA1BP2 | Probable rRNA-processing protein EBP2 | Required for the processing of the 27S pre-rRNA; interacts with Epstein-Barr virus (EBV) EBNA1 protein; required for stable EBV episome segregation | X | X | X | ||||
| Q15029 | EFTUD2 | 116 kDa U5 small nuclear ribonucleoprotein component | Required for pre-mRNA splicing; spliceosome component | X | X | |||||
| Q15717 | ELAVL1 | ELAV-like protein 1 | Ribonucleoprotein complex; involved in 3′ UTR AU-rich element (ARE) dependent MYC, FOS and IL3 stabilization | X | X | |||||
| Q01844 | EWSR1 | RNA-binding protein EWS | Transcriptional repressor; translocated in many cancers | X | X | X | ||||
| Q9P2P5 | HECW2 | E3 ubiquitin-protein ligase HECW2 | Mediates ubiquitination of p73; involved in metaphase/anaphase transition | X | X | |||||
| Q14103 | HNRNPD | Heterogeneous nuclear ribonucleoprotein D0 | Binds 3′ AU-rich elements (AREs) of mRNA and is involved in translationally coupled mRNA turnover; can bind ssDNA and act as a transcription factor; may play a role in telomere elongation | X | X(S83) | X | X | |||
| P07900 | HSP90AA1 | Heat shock protein HSP 90-alpha | Molecular chaperone involved in multiple cellular processes | X | X | X(T725, S726) | X | |||
| P35568 | IRS1 | Insulin receptor substrate 1 | Regulates insulin signaling; when phosphorylated by the insulin receptor it binds to SH2 containing proteins, including the PI3K p85 subunit | X | X(S337) | X(S312) | X | |||
| P52292 | KPNA2 | Importin subunit alpha-2 | Functions in nuclear protein import as an adapter protein for nuclear receptor KPNB1 | X | X | |||||
| P10636 | MAPT | Microtubule-associated protein tau | Promotes microtubule assembly and stability; establishes neuronal polarity | X | X | X(seeTable 2) | X(S262, S356) | X | ||
| P01106 | MYC | Myc proto-oncogene protein | Transcription factor; involved in cell growth, ribosome biogenesis, proliferation and apoptosis | X | X | X (T58) | X | |||
| P49116 | NR2C2 | Nuclear receptor subfamily 2 group C member 2 | Orphan nuclear receptor that can act as a transcriptional activator or repressor; involved in early embryonic development and spermatogenesis; represses estrogen receptor, Vit D3 receptor and retinoic acid receptor mediated transcription | X | X | |||||
| Q13310 | PABPC4 | Polyadenylate-binding protein 4 | Binds poly A tail of mRNAs; involved in cytoplasmic regulation of mRNA metabolism | X | X | |||||
| P62136 | PPP1CA | Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | Protein phosphatase 1 (PP1) is essential for cell division, and participates in the regulation of glycogen metabolism, muscle contractility and protein synthesis, cell migration; interacts with HHV1 ICP34.5 | X | X | X | ||||
| P62140 | PPP1CB | Serine/threonine-protein phosphatase PP1-beta catalytic subunit | Component of the PTW/PP1 phosphatase complex, which plays a role in the control of chromatin structure and cell cycle progression during the transition from mitosis into interphase | X | X | X | ||||
| P36873 | PPP1CC | Serine/threonine-protein phosphatase PP1-gamma catalytic subunit (1,2) | Component of the PTW/PP1 phosphatase complex, which plays a role in the control of chromatin structure and cell cycle progression during the transition from mitosis into interphase | X | X | X | X | |||
| O94761 | RECQL4 | ATP-dependent DNA helicase Q4 | DNA-dependent ATPase; may regulate chromosome segregation | X | X | |||||
| P27635 | RPL10 | 60S ribosomal protein L10 | Component of the large ribosomal subunit | X | X | |||||
| P62910 | RPL32 | 60S ribosomal protein L32 | Not reported | X | X | |||||
| P15880 | RPS2 | 40S ribosomal protein S2 | Small subunit ribosomal protein involved in initiation | X | X | |||||
| P61247 | RPS3A | 40S ribosomal protein S3a | May play a role in erythropoiesis by regulating the DDIT3 transcription factor | X | X | |||||
| P62854 | RPS26 | 40S ribosomal protein S26 | Not reported | X | X | |||||
| P08621 | SNRNP70 | U1 small nuclear ribonucleoprotein 70 kDa | Component of the spliceosomal U1 snRNP; involved in pre-mRNA 5′ splice site selection and spliceosome assembly | X | X | |||||
| Q8IYB3 | SRRM1 | Serine/arginine repetitive matrix protein 1 | Part of pre- and post-splicing multiprotein mRNP complexes | X | X | |||||
| P42224 | STAT1 | Signal transducer and activator of transcription 1-alpha/beta | Transcription factor; mediates the cellular response to interferons (IFNs) | X | X | |||||
| P04637 | TP53 | Cellular tumor antigen p53 | Transcription factor; acts as a tumor suppressor; mediates the cellular response to stress; involved in DNA-damage and repair | X | X(S376) | X(S392) | X | |||
| Q12933 | TRAF2 | TNF receptor-associated factor 2 | Has ubiquitin ligase activity; involve in the activation of NF-κB and JNK following TNFα treatment | X | X | |||||
| Q9Y4K3 | TRAF6 | TNF receptor-associated factor 6 | E3 ubiquitin ligase; involved in the activation of NF-κB and JNK following TNFα treatment; plays a role in TNFα receptor, interleukin (IL)-1 receptor and IL-17 receptor; polyubiquinates IKBKG/NEMO, IRAK1 and AKT1/2 | X | X | |||||
| Q71U36 | TUBA1A | Tubulin alpha-1A chain | Constituent of microtubules | X | X | |||||
| Q01081 | U2AF1 | Splicing factor U2AF 35 kDa subunit | Constitutive and enhancer-dependent splicing by mediating protein-protein interactions and protein-RNA interactions required for accurate 3′-splice site selection | X | X | X | ||||
| P26368 | U2AF2 | Splicing factor U2AF 65 kDa subunit | Splicing of pre-mRNA; required for mRNA export; regulates troponin exon 5 inclusion; represses TAU exon 10 splicing | X | X | |||||
| P67809 | YBX1 | Nuclease-sensitive element-binding protein 1 | Pre-mRNA alternative splicing regulation; regulates transcription of MDR1, HLA class II genes and promotes MYC mRNA stability | X | X | X | ||||
Proteins are listed in alphabetical order.
Acc. # refers to the identifier in the UniProtKB-SwissProt database.
Gene names are those used by the UniProtKB-SwissProt database.
Functions were retrieved from the UniProtKB database.
Proteins are identified as to whether they were found in association with GSK3α, GSK3β, active PKR (PKR+), inactive PKR (PKR-) or with PKR in an unknown state of activation (PKR).
Proteins are identified as to whether they are a direct substrate of GSK3α/β (pGSK3α/β) or PKR (pPKR).
Information was obtained from the following:
1. UniProtKB database.
2. Gene database-National Center of Biotechnological Information (NCBI), National Institutes of Health (NIH).
3. PhosphoSite Plus (www.phosphosite.org).
4. Blalock et al. (2014) J Cell Physiol. Ref # [46].
3.1. eIF2 complex and translation
Protein synthesis is the most energy consuming process of the cell, requiring both ribosome biogenesis and the subsequent translation of mRNA. From prokaryotes to mammals, these processes are highly regulated in response to the surrounding environment to limit energy expenditure under conditions that are unfavorable for growth, as well as limit the possibility of producing mutant proteins. The most efficient and rapid point to regulate protein synthesis under cell stressing conditions is during the initiation phase of translation, thereby limiting the amount of cellular energy and resources that are fruitlessly utilized. This point also allows for the rapid synthesis of proteins once the stress is alleviated. Many upstream kinases converge on translation initiation, which is the rate limiting step. Recruitment of the mRNA to the 40S ribosomal subunit represents one level of control and is carried-out by the eIF4F complex, which requires the eIF4E subunit to directly bind the 5′ CAP of the mRNA [51]. Under resting conditions, eIF4E is bound by the eIF4E-binding protein, 4E-BP, which sequesters it and inhibits its association with the mRNA. Activation of the PI3K-AKT-mTOR axis promotes metabolism and protein synthesis through mTORC2-mediated phosphorylation of 4E-BP and other substrates [51]. As previously stated, active AKT phosphorylates GSK3α/β on S21/S9, inhibiting GSK3 catalytic activity. Under stress conditions where reduced flux through PI3K-AKT-mTOR pathway no longer leads to mTOR-mediated phosphorylation 4E-BP, active GSK3β is able to maintain CAP-dependent translation initiation via the eIF4F complex translation initiation but alter general translation (Fig. 1A and B, a) [51].
In addition to the eIF4F translation initiation complex, two additional initiation factors represent major points of translation control, eIF2 and eIF2B (Fig. 1B). These complexes bare both the GTP and the initiator Met-tRNA necessary for the formation of the pre-initiation complex (PIC) and translation initiation, as well as the proteins for the GDP to GTP exchange required to initiate the next round of translation (Fig. 1B,b) [52]. Regulation of the eIF2-GDP/GTP exchange is via phosphorylation of the α-subunit (eIF2α) by one of four different kinases (PKR, PERK, GCN2 or HRI) and is probably the best understood mechanism regulating translation initiation. Phosphorylation of eIF2α on S51 results in eIF2 being locked in the GDP bound state with the GDP/GTP exchange complex, eIF2B, unable to catalyze the initiation of protein synthesis. As the eIF2 complex is limited compared to eIF2B, it does not take much phosphorylated eIF2α to soon result in a complete block of general translation (Fig. 1B, c) [53].
Phosphorylation of eIF2α, which was originally thought to be strictly pro-apoptotic, actually results in a shut-down of general CAP-dependent translation while at the same time, allowing for efficient translation of upstream open reading frames (uORFs) in mRNAs that contain complex secondary structure at the 5′ end and an internal ribosome entry site (IRES) element upstream [[54], [55], [56]]. Short-term inhibition of general translation through eIF2α phosphorylation establishes a pro-survival state by allowing for cellular repair and time for the cell to adjust following a particular stress [57]; although, if this stress cannot be resolved and general translation remains inhibited the cell will likely die through apoptotic means. In contrast, under other conditions phosphorylation of eIF2α has been shown to inhibit IRES-mediated translation [58]. These differences may be due in part to specific regulator proteins that differ between IRES elements as well as additional elements inherent to the mRNA (Fig. 1B, c). Many of the mRNAs translated under conditions where eIF2α is phosphorylated encode inflammatory cytokines such as TNFα, IL-1, FGF, VEGF, IL-6 and ATF4 [[59], [60], [61], [62], [63], [64]].
In contrast to PKR which targets eIF2, GSK3α/β target eIF2B. The function of eIF2B is to exchange GTP for the eIF2 complex bound GDP [65]. The eIF2B GTP exchange factor is composed of α, β, γ, δ and ε subunits, of which the 82 kDa ε-subunit is the most critical to eIF2B regulation and is enzymatically responsible for the GDP/GTP exchange. The α-, β-, and δ-subunits associate with Ser51 phosphorylated eIF2α and inhibit eIF2Bε activity, locking eIF2B with eIF2 in the GDP-bound state. The γ-subunit, which is phosphorylated and regulated by casein kinase (CK)-II, promotes the activity of the ε-subunit. Phosphorylation of eIF2Bε on Ser540 by GSK-3β, following amino acid starvation, inhibits eIF2B activity [25,65,66]. As GSK3 requires a priming phosphorylation, eIF2Bε must first be phosphorylated on Ser544 by one of the DYRK family kinases (Fig. 1B, d) [67]. The phosphorylation of these residues results in translation of a set of mRNAs that is different from those translated when only eIF2α is phosphorylated; thus regulation of translation can give rise to proteins that are most efficiently translated under one of three (or more) different conditions (Fig. 1A and B) [25,66,68]. Additional information on alternate translation can be found in the following reviews [59,[69], [70], [71]].
3.2. TP53
Glycogen synthase kinase-3 and PKR have been shown to influence the cell cycle both positively and negatively depending on the context. Among the cell cycle targets that have been demonstrated to be directly phosphorylated by the GSK3 kinases are cyclins D and E (GSK3α and GSK3β) and the dual cyclin phosphatase, CDC25A (GSK3β); each of these phosphorylations favoring ubiquitination and degradation of the target protein, thus resulting in cell cycle arrest [72]. In contrast, while PKR was originally thought of as a tumor suppressor, it was observed that PKR-null mouse embryo fibroblasts (MEFs) grew slower than normal MEFs and expression of dominant-negative PKR (K296R) in the human glioblastoma cell line T98G resulted in a longer G1 phase, suggesting some role for PKR activity in normal cell proliferation, but direct targets were missing [73]. As will be discussed later both GSK3 and PKR influence the activity of diverse transcription factors which promote transcription of both pro-growth and cell cycle inhibiting factors. Among the common targets of GSK3 and PKR is the MDM2-p53 regulatory complex.
Most studies examining GSK3 and p53 have centered on GSK3β, likely a result of the fact that it is this GSK3 isoform that is prominent in the nucleus. Both positive and negative p53 regulation have been associated with GSK3β, and these discrepancies may be related to the cellular localization of the GSK3β in question. In ovarian cancer, GSK3β represses let-7 miRNA through activating p53 [74]. It is interesting that active GSK3β would target the repression of let-7, a miRNA that targets the AKT kinase family members among other proteins. This might be suggestive of a possible feedback regulation: low AKT leads to increased active GSK3β which subsequently stimulates p53 and the repression of let-7, allowing for increased expression of AKT. Enhancement of p53 through GSK3β activation has been shown to occur through acetylation of p53 on K120, resulting in stabilization of p53 [75]. The reported mechanism was dependent on GSK3-dependent phosphorylation of the histone acetylase Tip60 on S86 and increased acetylase activity toward p53. In addition, GSK3β-dependent phosphorylation of S376 leads to enhanced p53 activity (Fig. 2A, a) [76]. In contrast, GSK3β has also been shown to result in p53 degradation. The E3 ligase MDM2 (HDM2) is responsible for the ubiquitination of p53, resulting in its subsequent degradation by the proteosome. Phosphorylation of S240 and S254 in murine cell lines was reported to lead to MDM2 stabilization and p53 degradation, a process that was instrumental for promoting apoptosis following ionizing radiation exposure [77]. Loss of GSK3β or use of an isoform specific inhibitor was shown to result in p53 accumulation and protection from radiation-induced damage; thus loss of GSK3 activity is protective against radiation-induced DNA damage [77,78]. Under physiological conditions it is usually AKT activation that results in the inhibition of GSK3 following ionizing radiation exposure (Fig. 2A, b).
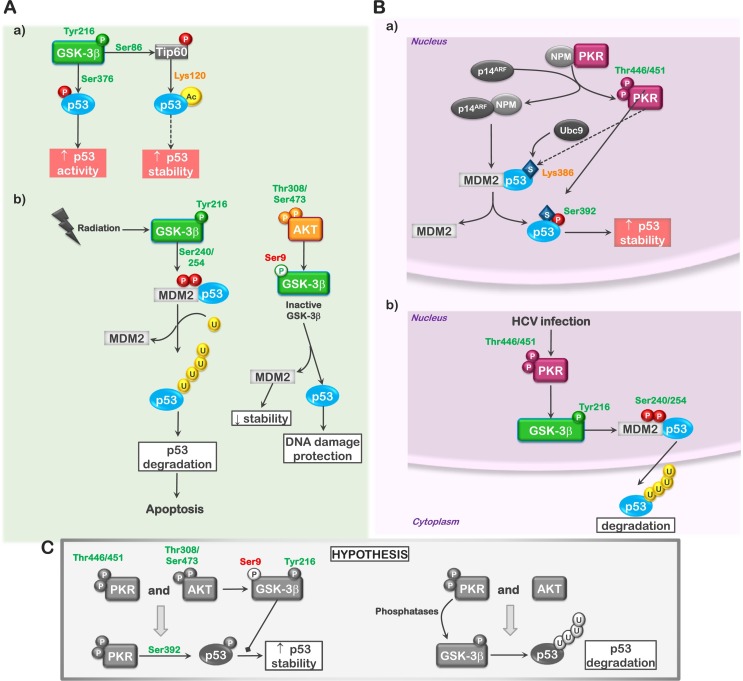
Fig. 2.
The role of GSK3β and PKR in regulating p53. The diagram demonstrates how p53 is regulated by GSK3β and PKR. (A) a; GSK3β can enhance p53 stability and transcriptional activity by inducing acetylation of p53 through Tip60 and phosphorylation of S376. b; In contrast, active, nuclear GSK3β can enhance the stability of MDM2 following DNA damaging stress, through phosphorylation of S240 and S254, resulting in the expulsion of p53 from the nucleus and proteosomal degradation. Activation of AKT inhibits this process. (B) a; PKR can both phosphorylate (S392) as well as promote sumoylation of p53, enhancing its stability. b; On the other hand, PKR can stimulate the activity of nuclear GSK3β through a not completely understood mechanism, resulting in the stabilization of MDM2 and the degradation of p53. (C) The regulation of GSK3β by PKR is believed to be through the PP1A phosphatase. In the presence of AKT, GSK3β would remain inactive. In contrast, low AKT would favor the activating effects of PKR on GSK3β. While active PKR is denoted as phosphorylated on T446 and T451, only T451 is required for activity. Phosphorylation of T446 as well as numerous additional sites influences the degree of activation. Phosphorylation sites labeled in green (ex., Tyr216-GSK3β) indicate activating or stabilizing modifications; phosphorylation sites labeled in red (ex., Ser9-GSK3β) indicate modifications that are inhibitory to the substrates activity. Lines with arrowhead termini represent promoting effects on the downstream signaling; lines with boxed termini represent inhibitory effects on downstream signaling.
Likewise, PKR has been demonstrated to both positively and negatively influence p53. Originally, activation of PKR was shown to be associated with p53 expression and transcriptional transactivation of p53 associated genes [50,79]. Studies in a PKR-null background also demonstrated that S18 phosphorylation was diminished [79]. Later studies by Cuddihy et al. indicated that PKR could directly phosphorylate p53 on S392 resulting in its stability [80]. Most recently, Bennett et al. reported that activation of the PACT/PKR pathway resulted in the association of the SUMO E2 ligase Ubc9 with p53 and the subsequent sumoylation of lysine 386 [81]. Sumoylation at this site appeared to be a prerequisite for S392 phosphorylation, as this site was not phosphorylated in the sumoylation deficient p53 mutant (K386R) (Fig. 2B, a). In unpublished experiments our group also noticed that treatment of diverse acute leukemia cell lines with the PKR specific inhibitor [PI; 8-(1h-imidazol-4-ylmethylene)-6,8-dihydrothiazolo[5,4-e]indol-7-one] at a concentration of 1 μM resulted in the rapid loss of p53 expression (unpublished results). Interestingly, PKR activation during hepatitis C (HCV) viral infection has been reported to block the activation of p53 [82]. Similarly, Baltzis et al. reported that the eIF2α kinases, PKR and PERK, influenced GSK3β activity in a manner opposed to that of AKT. The activation of both kinases was shown to result in the activation of nuclear GSK3β, the subsequent expulsion of p53 from the nucleus, followed by its degradation in the cytosol. This effect was not dependent on the phosphorylation of eIF2α, suggesting a mechanism independent of translational regulation and believed to involve the phosphorylation of Y216 (Fig. 2B, b) [50]. This may indeed be the phosphatase-dependent effect that was observed in acute leukemia cells [41]. A critical factor that remains to be determined is what dictates whether these kinases promote or oppose p53 activation; this may center on the status AKT activity. PKR activation in the presence of AKT activity may allow PKR to enhance p53 stability and transactivation, while PKR activation in the absence of AKT activity may promote GSK3-mediated p53 decay (Fig. 2C). The potential of this interplay and the influence that cellular localization of the associated protein complexes plays remains to be well defined.
3.3. TAU/MAPT
The microtubule-associated protein TAU, the product of mapt gene which is expressed almost exclusively in neurons, serves the purpose of facilitating microtubule assembly and links axonal microtubule components to those in the plasma membrane, thus defining the polarity of the neuron [83]. At least nine different isoforms of TAU, which are produced by alternative splicing, have been reported, with some isoforms restricted to development [84]. The largest isoform, PNS-TAU, is expressed in the peripheral nervous system, whereas the other isoforms are found in the central nervous system. TAU is a highly modified protein that is a target of at least twelve different kinases, including GSK3α/β and PKR [32,[83], [84], [85], [86]]. Analysis of the Phosphosite Plus database indicates that while GSK3α has been shown to phosphorylate a number of TAU isoforms (isoforms 2, 5, 6 and 8) both GSK3β and PKR have been demonstrated to phosphorylate namely isoform 8 (Table 2 ) [[87], [88], [89], [90], [91], [92], [93], [94], [95], [96], [97], [98], [99], [100], [101]]. With the exception of two sites phosphorylated by GSK3β, the majority of the sites phosphorylated by the GSK3 and PKR kinases lie near to or within the C-ter tubulin binding repeat (aa 560-690). In general, phosphorylation of these residues results in the detachment of TAU from the microtubule complex and disassembly. Normal neuronal development will see the interplay between O-GlcNacylation, phosphorylation and dephosphorylation by phosphatases. As phosphorylation and O-GlcNacylation are mutually exclusive in this regulation, the interplay between these mechanisms in simple terms either leads to O-GlcNacylation and microtubule assembly or phosphorylation and microtubule disassembly. In multiple degenerative diseases such as Alzheimer's and Parkinson's a loss of O-GlcNacylation with an associated hyperphosphorylation is observed with TAU [83,84]. These hyperphosphorylated TAU molecules become seeds for the development of neural filamentous tangles which lead to progressive neuronal cell death and disease. As will be discussed later, chronic inflammatory signaling leading to the constitutive elevated activation of both GSK3β and PKR has been demonstrated to be intricately associated with neurodegeneration.
Table 2.
Sites of GSK3α/β- and PKR-mediated phosphorylation in TAU (MAPT).
| Isoform | Phosphorylation site | Kinase |
Ref. # | ||
|---|---|---|---|---|---|
| GSK3α | GSK3β | PKR | |||
| TAU isoform 2 | S307 (S713) | X | [87] | ||
| TAU isoform 2 | S315 (S721) | X | [87] | ||
| TAU isoform 5 | T181 (T498) | X | [88] | ||
| TAU isoform 5 | S184 (S501) | X | [88] | ||
| TAU isoform 5 | S195 (S512) | X | [88] | ||
| TAU isoform 5 | S198 (S515) | X | [88] | ||
| TAU isoform 5 | S199 (S516) | X | [88] | ||
| TAU isoform 5 | S202 (S519) | X | [88] | ||
| TAU isoform 5 | T205 (T522) | X | [88] | ||
| TAU isoform 5 | T231 (T548) | X | [87,88] | ||
| TAU isoform 5 | S235 (S552) | X | [87,88] | ||
| TAU isoform 5 | S262 (S575) | X | [88] | ||
| TAU isoform 5 | S325 (S673) | X | [88] | ||
| TAU isoform 5 | S365 (S713) | X | [87] | ||
| TAU isoform 5 | S369 (S717) | X | [88] | ||
| TAU isoform 5 | S373 (S721) | X | [87,88] | ||
| TAU isoform 6 | T173 (T548) | X | [87,89] | ||
| TAU isoform 6 | S177 (S552) | X | [87] | ||
| TAU isoform 6 | S338 (S713) | X | [87,89] | ||
| TAU isoform 6 | S346 (S721) | X | [87,89] | ||
| TAU isoform 8 | S46 (S46) | X | [90] | ||
| TAU isoform 8 | T50 (T50) | X | [90] | ||
| TAU isoform 8 | T153 (T470) | X | [91] | ||
| TAU isoform 8 | T175 (T492) | X | [91] | ||
| TAU isoform 8 | T181 (T498) | X | [[92], [93], [94]] | ||
| TAU isoform 8 | S195 (S512) | X | [95] | ||
| TAU isoform 8 | S199 (S516) | X | [95,96] | ||
| TAU isoform 8 | S202 (S519) | X | [92,96] | ||
| TAU isoform 8 | T205 (T522) | X | [95,96] | ||
| TAU isoform 8 | S210 (S527) | X | [95] | ||
| TAU isoform 8 | T212 (T529) | X | [92,96] | ||
| TAU isoform 8 | S214 (S531) | X | [93,95] | ||
| TAU isoform 8 | T217 (T534) | X | [92,96] | ||
| TAU isoform 8 | T231 (T548) | X | X | [87,92,[97], [98], [99]] | |
| TAU isoform 8 | S235 (S552) | X | X | [87,91,98] | |
| TAU isoform 8 | S262 (S579) | X | X | X | [93,[96], [97], [98],101] |
| TAU isoform 8 | S356 (S673) | X | [101] | ||
| TAU isoform 8 | S396 (S713) | X | X | [87,95,96] | |
| TAU isoform 8 | S400 (S717) | X | [95,100] | ||
| TAU isoform 8 | S404 (S721) | X | X | [87,95,96] | |
| TAU isoform 8 | S409 (S726) | X | [93] | ||
| TAU isoform 8 | S422 (S739) | X | [93] | ||
Information was obtained from the PhosphoSitePlus Database under the substrate page for the following:
3.4. c-Myc
The MYC transcription factor is a critical proto-oncogene for cellular growth and differentiation. In addition, to being indispensible for the first steps in ribosome biogenesis, the synthesis of ribosomal RNAs, and protein synthesis, MYC is also critical to cellular differentiation and the regulation of apoptosis [102]. While MYC is influenced by the PI3K-AKT-mTOR pathway through multiple mechanisms, it is also subject to direct control by GSK3 through phosphorylation. Phosphorylation of MYC on T58, by GSK3α/β, and on S62, by GSK3α/β and other kinases (DYRK2, CDK2 and CDK5) enhances the transcriptional transactivating activity of MYC but at the same time functions to prime a negative regulatory mechanism [[103], [104], [105]]. Phosphorylation of T58 in particular allows for the Fbw7 F-box proteins of the SCF-type ubiquitin ligase to associate with MYC. Association of MYC with Fbw7α and the ubiquitin protease Ubc28 in the nucleus favor MYC stability and transcriptional activity. In contrast, in the nucleolus, MYC phosphorylated on T58 associates with Fbw7γ and is degraded, thus providing a control mechanism over MYC-induced transcription, which is dependent on Fbw7 isoform expression [104]. As is observed with TAU, the interplay between phosphatase-dependent dephosphorylation (T58 and S62), O-GlcNacylation (T58) and phosphorylation (T58 and S62) plays a major role in MYC activity (Fig. 3A) [103,104,106,107].
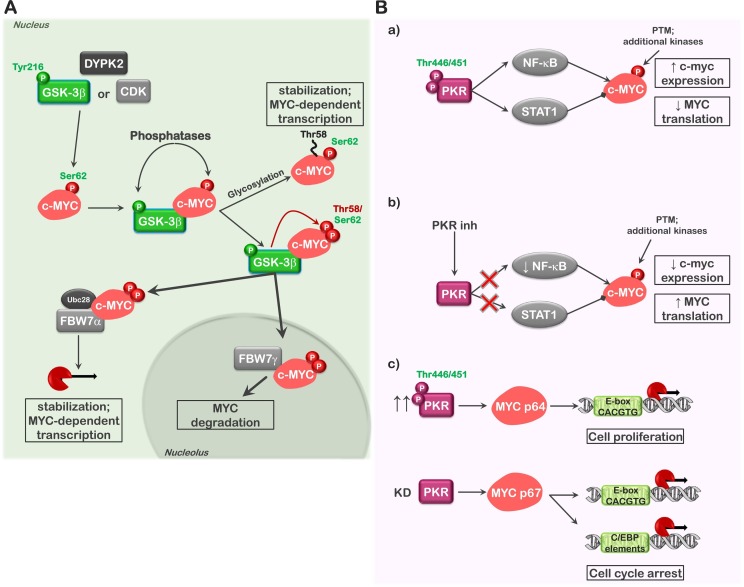
Fig. 3.
GSK3α/β and PKR regulate MYC stability and synthesis. The diagram demonstrates what is known concerning GSK3α/β- and PKR-dependent MYC regulation. (A) MYC, previously phosphorylated on Ser62 by one of a number of kinases, is phosphorylated on T58 by GSK3β (also by GSK3α), stabilizing the association of MYC with Fbw7 F-box proteins (Fbw7α and Fbw7γ) of the SCF-type ubiquitin ligase. Association of Fbw7α and subsequent binding of Ubc28, an ubiquitin-specific protease, promote MYC stability in the nucleus and transcriptional activity. In contrast, association with Fbw7γ in the nucleolus promotes MYC degradation. Glycosylation of T58 inhibits phosphorylation at this site, blocking the association of MYC with Fbw7 family members, resulting in MYC protein stability. (B) PKR regulates the synthesis of MYC through the stimulation of the transcription factors (NF-κB and STAT1) and translation initiation. PKR activation leads to enhanced c-myc expression through NF-κB, which is counterbalanced by activation of STAT1-mediated suppression of c-myc. At the same time active PKR favors the synthesis of the p64 form of MYC through a translational mechanism (a and c). Inhibition of PKR leads to decreased c-myc expression and enhanced translation with the synthesis of p67 MYC favored (b and c). While active PKR is denoted as phosphorylated on T446 and T451, only T451 is required for activity. Phosphorylation of T446 as well as numerous additional sites influences the degree of activation. Phosphorylation sites labeled in green (ex., Tyr216-GSK3β) indicate activating or stabilizing modifications; phosphorylation sites labeled in red (ex., Thr58-MYC) indicate modifications that can result in protein degradation. Lines with arrowhead termini represent promoting effects on the downstream signaling; lines with boxed termini represent inhibitory effects on downstream signaling.
Similarly, PKR also regulates the expression of MYC, but not through direct phosphorylation. Diverse groups have demonstrated that PKR influences the expression of c-myc through the stimulation of nuclear factor (NF)-κB and signal transducer and activator of transcription (STAT) transcription factors (Fig. 3B, a) [49,108]. Previous work examining the effects inhibiting PKR kinase activity in acute leukemia cells with a small molecule inhibitor demonstrated that loss of PKR activity could both enhanced MYC expression and influence the isoform of MYC expressed. Overexpression of PKR was shown to favor p64 MYC expression while siRNA-mediated knock-down of PKR favored p67 MYC expression (Fig. 3B, b and c) [46]. This is an interesting point as the p64 isoform of MYC is initiated from a standard AUG start codon; while in contrast, the p67 isoform is produced from a non-canonical CUG start codon and encodes an additional 15 amino acids at the amino terminus. Both MYC isoforms, p64 and p67, target E-box sites in MYC responsive promoters, but only p67 can also target C/EBP elements, thus leading to the transcription of an additional set of responsive genes (Fig. 3B, c) [46,109]. It has been suggested that the ratio p64/p67 dictates whether MYC expression favors growth and proliferation or stimulates the expression of pro-apoptotic factors, with p64 favoring proliferation and p67 favoring growth arrest [46,109]. As the difference is these isoforms appears to be an effect of altered translation initiation, it might be assumed that PKR activity can influence additional aspects of translation initiation beyond that of simply eIF2α phosphorylation. Moreover, this might also suggest that the loss of PKR may stimulate a feedback control through MYC isoform expression to limit growth and proliferation of cells that do not have the necessary safeguards in place to monitor translation initiation.
3.5. NF-κB
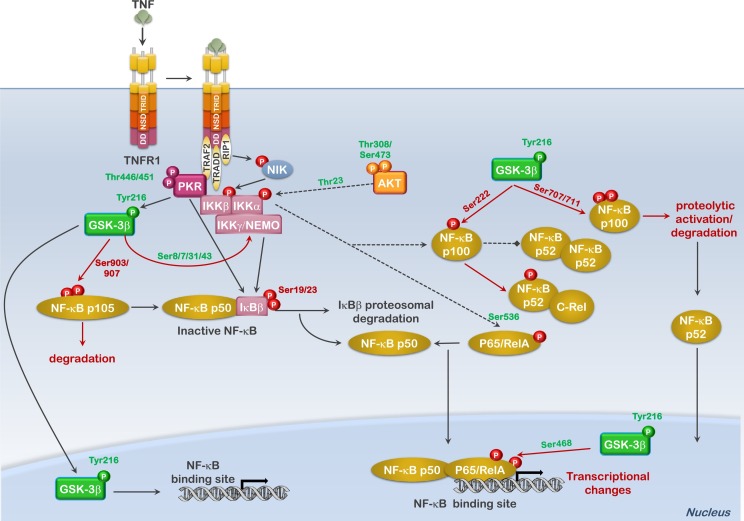
Nuclear factor-κB (NF-κB) consists of hetero- or homodimers derived from five different subunits: p65/RelA, p50 (processed from p105), p52 (processed from p100), RelB and c-Rel. The genes preferentially induced differ according to the hetero- or homodimer present. The NF-κB family regulates the expression of numerous genes involved in cell cycle progression (cyclin D1 and c-MYC), apoptosis inhibition (c-IAP1/2, BCL-XL and A20) oxidation-reduction regulation and inflammation (COX2, GADD45β, HO-1, IL-6. iNOS, MMP-9, MnSOD and RANTES) [110]. Under most situations the regulation of NF-κB relies on subunit binding to the inhibitory proteins IκB-α and -β. These proteins maintain NF-κB localized to the cytoplasm. Following stimulation, IκB proteins are phosphorylated by the IκB kinase or IKK complex. This phosphorylation leads to dissociation between IκB and NF-κB, the targeting of IκB to the proteosome for degradation, and translocation of the NF-κB dimer to the nucleus (Fig. 4 ) [110]. In addition to this level of regulation, the NF-κB proteins can also be post-translationally modified bringing about changes in their activity and stability.
Fig. 4.
Nuclear factor-κB is regulated on multiple levels by both GSK3β and PKR. The schematic diagram shows how cytotoxic cytokine signaling or stress, resulting in GSK3β and PKR activation, can affect NF-κB transcription factors. GSK3β activation regulates the proteosomal degradation of the NF-κB p100 and NF-κB p105, determining whether these are processed to NF-κB p52 and NF-κB p50, respectively. In addition, nuclear GSK3β activity alters p65/RELA transcriptional activation through phosphorylation of S468. PKR in contrast targets both the inhibitor κB (IκB) and the IKKγ/NEMO subunit to promote IκB phosphorylation and degradation. PKR promotes the activation and translocation of NF-κB proteins as well as the transactivating phosphorylation (S536) of p65/RELA through the IKK complex. While active PKR is denoted as phosphorylated on T446 and T451, only T451 is required for activity. Phosphorylation of T446 as well as numerous additional sites influences the degree of activation. Phosphorylation sites labeled in green (ex., Tyr216-GSK3β) indicate activating or stabilizing modifications; phosphorylation sites labeled in red (ex., Ser222-NF-κBp100) indicate modifications that can result in protein degradation/proteolysis. Lines with arrowhead termini represent promoting effects on the downstream signaling; lines with boxed termini represent inhibitory effects on downstream signaling.
NF-κB homo- and heterodimer activation by PI3K-AKT-mTOR signaling occurs on many levels, including AKT-dependent modification of IKKα on Thr23, which, rather than stimulating IκB degradation, seems to stimulate the transactivating domain of p65/RelA [111,112]. The effect of AKT on IKKα also regulates the processing of NF-κB p100 to p52 [113,114]. Downstream targets of AKT can also influence NF-κB activity (Fig. 4). The mTOR complex can stimulate IKK activity; IKK in-turn has been demonstrated to feedback on mTOR activity through the phosphorylation of the mTOR inhibitor proteins TSC1/2 [[115], [116], [117], [118], [119]]. Additionally, AKT also has the ability to inhibit the formation and activation of certain NF-κB transcriptional complexes through its ability to modify GSK3α/β. As mentioned previously, GSK3-α and -β are often activated in response to inflammatory/stress signaling. GSK3β activity has not only been shown to be required for processing of p105 NF-κB, but also for NF-κB transcriptional activity in the nucleus; the main reason GSK3β−/− mice are embryonic lethal and have a phenotype similar to IKK−/− mice (Fig. 4) [[120], [121], [122], [123], [124], [125], [126], [127], [128]]. This regulation again appears to occur at two levels, phosphorylation of the NEMO subunit of the IKK complex and phosphorylation of p65 or the NF-κB precursor proteins p100 and p105. Phosphorylation of S8, S17, S31 and S43 of NEMO results in protein stabilization favoring kinase activity and downstream activation of NF-κB [129]. However, GSK3β-dependent phosphorylations of NF-κB p65, p100 and p105 have very differing effects on the transcriptional outcome. GSK3β-dependent phosphorylation of p65/RelA on S468 primarily occurs in the nucleus and alters the transcriptional activity, favoring the expression of some responsive genes while suppressing others [130]. In contrast, phosphorylation of NF-κBp100 on S222 suppresses homodimerization of p52 favoring p52/c-REL heterodimers, while phosphorylation of S707 and 711 leads to p100 degradation by the Fbxw7α E3 ubiquitin ligase, liberating non-canonical NF-κB-mediated transcription [131,132]. Phosphorylation of NF-κBp105 on S903 and S907 inhibits the proteolysis of p105 to p50 and primes p105 for degradation, thus reducing p50-mediated transcription (Fig. 4) [120,133]. Although data involving the activation status of NF-κB in GSK3α−/− mice have not been reported nor has GSK3α-dependent phosphorylation of NF-κB transcription factors, some reports have suggested GSK3α activity may also play a role in NF-κB nuclear functions, with its loss having severe consequences on survival and proliferation (Fig. 4) [134].
As mentioned above, PKR activation often leads to increased nuclear GSK3β activity. Although the outcome of PKR-mediated modification of GSK3β on NF-κB associated transcription has not been determined, it is interesting to note that many of the same NF-κB transcribed genes that are repressed in PKR-null fibroblasts or by overexpression of catalytically-dead PKR are also inhibited in GSK3β-null fibroblasts and cells overexpressing kinase defective GSK3β mutants (Fig. 4 and Table 3 ) [57,121,124,127,[135], [136], [137], [138]].
Table 3.
NF-κB induced genes dependent on both GSK3β and PKR.
| Gene/protein | Stimulation | Method used | aFunction | Ref. # |
|---|---|---|---|---|
| A1/Bfl-1 | TNFα stimulation in PKR+/+ and −/− MEFs | Western blot | A1 is an anti-apoptotic BCL-2 family protein that binds BAX thus inhibiting cytochrome c release and caspase 3 activation. | [49,135] |
| TNFα stimulation in GSK3β+/+ and −/− MEFs | Western blot | |||
| BCL-XL | TNFα stimulation in PKR+/+ and −/− MEFs | Western blot | BCL-XL is the major hematopoetic anti-apoptotic BCL-2 family member. BCL-XL is best known for its ability to bind BAX blocking the homodimerization of BAX and the initiation of the caspase-9 mediated apoptotic pathway. | [49,135,136] |
| TNFα stimulation in GSK3β+/+ and −/− MEFs; treatment of pancreatic cancer cell lines with a pharmacological inhibitor of GSK3β siRNA | Western blot | |||
| Cyclin D1 | TNFα stimulation in PKR+/+ and −/− MEFs | Western blot | Controls entry into S-phase of the cell cycle. Cyclin D1 is a target of both MYC and NF-κB. Cyclin D1 in association with CDK4/6 phosphorylate the retinoblastoma protein (Rb) resulting in the release of E2F and the transcription of cell cycle promoting genes. Overexpression of cyclin D1 is associated with enhanced cell proliferation. | [49,135,136] |
| TNFα stimulation in GSK3β+/+ and −/− MEFs; treatment of pancreatic cancer cell lines with a pharmacological inhibitor of GSK3β siRNA | Western blot | |||
| FLIP | TNFα stimulation in PKR+/+ and −/− MEFs | Western blot | FLICE inhibitor protein associates with caspase 8, FADD, caspase 3, TRAF1 and TRAF2. Expression of full-length FLIP (FLIPL) or the C-ter domain is pro-apoptotic while a splice variant lacking the C-ter (FLIPS) is anti-apoptotic in response to death receptor mediated apoptosis. Loss of FLIP is embryonic lethal in mice due to lack of proper heart development. FLIP−/− embryonic fibroblasts were hypersensitive to TNFα- and FAS-mediated apoptosis. | [49,135] |
| TNFα stimulation in GSK3β+/+ and −/− MEFs | Western blot | |||
| c-IAP-1 | Tet-inducible PKR/TNFα stimulation in PKR+/+ and −/− MEFs | cDNA array/Western blot | Inhibitor of apoptosis protein-1, -2 and XIAP block the induction of apoptosis by associating with cytosolic regions of death domain containing receptors to block the induction of associated caspases such as caspase 8. | [49,57,135] |
| TNFα stimulation in GSK3β+/+ and −/− MEFs | Western blot | |||
| c-IAP-2 | Tet-inducible PKR/TNFα stimulation in PKR+/+ and −/− MEFs | cDNA array/Western blot | Inhibitor of apoptosis protein-1, -2 and XIAP block the induction of apoptosis by associating with cytosolic regions of death domain containing receptors to block the induction of associated caspases such as caspase 8. | [57,135] |
| TNFα stimulation in GSK3β+/+ and −/− MEFs | Western blot | |||
| MMP-9 | TNFα stimulation in PKR+/+ and −/− MEFs | Western blot | Matrix metalloproteinase 9 is a member of a family of extracellular proteases that degrade Type IV and V collagens. MMP9 has been shown to be involved in vascularization, tissue remodeling, angiogenesis, and migration. MMP9 was shown to have a significant role in mobilization of hematopoeitic stem cells from the bone marrow induced by IL8. | [49,135] |
| TNFα stimulation in GSK3β+/+ and −/− MEFs | Western blot | |||
| XIAP | TNFα stimulation in PKR+/+ and −/− MEFs | Western blot | Inhibitor of apoptosis protein-1, -2 and XIAP block the induction of apoptosis by associating with cytosolic regions of death domain containing receptors to block the induction of associated caspases such as caspase 8. | [49,124,136] |
| Treatment of CLL patient samples and cell lines and pancreatic cancer cell lines with a pharmacological inhibitor or GSK3β siRNA | RT-PCR/Western blot |
Descriptive information of the genes and proteins above was taken from the Mendelian Inheritance in Man website available through the National Center for Biotechnology Information (NCBI), National Institutes of Health (NIH).
The role of PKR in regulating NF-κB-dependent transcription was first suggested when it was discovered that dsRNA and IFNβ activate NF-κB DNA binding [[139], [140], [141]]. Later, studies reported that expression of DN-PKR blocked dsRNA and IFNβ activation of NF-κΒ [142,143]. Conclusive evidence was finally demonstrated by the inability of TNFα to stimulate NF-κB translocation and DNA binding activity in PKR−/− cells [144,145]. Exactly how PKR induces the activation of NF-κB is somewhat still debated. Originally, PKR was thought to directly phosphorylate inhibitor κB (IκB)-α resulting in its release of NF-κB and subsequent proteosomal degradation [142,146]. This idea was later challenged by the discovery that PKR associated with the IKK complex in order to activate NF-κB transcription [147,148]. Other data challenged whether PKR catalytic activity was even need at all, as truncated forms of PKR consisting of the amino terminus were shown to associate with the IKK complex and stimulate IκBβ phosphorylation [149,150]. Similarly, kinase-deficient mutants of PKR were reported to associate with the IKK complex and induce NF-κB; while other reports suggested that kinase activity was required [151,152]. Much of this discrepancy was solved when Donze et al. published a seminal paper demonstrating that PKR, irregardless of catalytic activity, could induce the transcriptional activation of NF-κB and the synthesis of some NF-κB-dependent transcripts, but the transcriptional activity of NF-κB and the transcription of additional NF-κΒ-dependent genes was greatly potentiated in the presence of active PKR kinase [57]. This suggested that both PKR association with the IKK complex and PKR catalytic activity are important for PKR-mediated effects on NF-κB. It should be stated at this point that PKR and its associated catalytic activity mainly favor p65 NF-κB-containing transcription complexes. To this end the current understanding is that PKR activity is required for the full effects of PKR on NF-κB-mediated transcription, by both influencing IκB expression and phosphorylation/release as well as later points, such as enhancing the nuclear activity of GSK-3β (Fig. 4) [50,147]. A list of NF-κB-induced genes/proteins dependent on PKR and GSK3β is presented in Table 3.
3.6. C/EBP
Similar to MYC, both GSK3α/β and PKR influence CCAAT-enhancer binding transcription factor proteins, C/EBPα and C/EBPβ through phosphorylation and alternative translation, respectively. These transcription factors are essential for the differentiation of hepatocytes, adipocytes, and myeloid progenitors as well as tissues of the lung and placenta [153,154]. Through association with IRE elements, C/EBP transcription factors can regulate gluconeogenesis and lipogenesis programs in the liver to balance metabolic energy demand and storage. Of significant importance is the activation of these transcription factors under stress and inflammation, resulting in cellular differentiation and energy homeostasis. C/EBPα is expressed as either a full-length (p42) or truncated (p30) protein [153]. Although both forms contain sites for interaction with additional transcription factors, only p42 contains the transactivation domain [153]. In the hematopoietic compartment, C/EBPα is required for normal granulocyte differentiation and C/EBPα−/− mice have a loss of myeloid maturation accompanied by blast accumulation [153,155]. Additionally, C/EBPα is mutated or repressed in many other hematologic malignancies [153,156,157]. Interestingly, expression of p30 C/EBPα increases the expression and activity of the Ubc9 ubiquitin ligase toward p42 C/EBPα, resulting in p42 degradation; thus blocking differentiation in CD34+ hematopoietic stem cells, favoring self-renewal [25,158].
In many ways similar to C/EBPα, C/EBPβ is expressed as one of three forms: p38, p33 and p20, and shares many overlapping functions with C/EBPα; but unlike C/EBPα, it promotes the proliferation of a number of cell types, with the noted exception of T-lymphocytes whose proliferation is blocked by C/EBPβ-mediated MYC repression [153,159]. The full-length form is most associated with regulating the expression of genes involved in the immunologic response and inflammation while the p33 isoform is associated with gene transcription in activated macrophages, the CD4+ T-cell response, and diverse aspects of intracellular innate immunity. The shortest form, p20, functions as a dominant-negative factor when bound to p33 and has been best associated with osteoblast differentiation and osteoclastogenesis. Interestingly, whereas the loss of C/EBPα transcriptional activity is associated with tumorigenesis, loss of C/EBPβ is not [25,153,160,161].
In humans, the GSK3 kinases phosphorylate T226, T230 and S234 in C/EBPα and T226 and S231 in C/EBPβ, which is proposed to enhance their transcriptional activity favoring among other things adipogenesis and hematopoietic differentiation [162,163]. In contrast, stimulation of the PI3K-AKT-mTOR pathway following stimulation of insulin-related growth factor receptor (IGF-1R), or other growth factor receptors, leads to GSK3 inhibition through AKT-dependent phosphorylation of S21/S9 and the suppression of differentiation programming with a concatenate increase in glucose uptake/usage and proliferation. Zeng et al. reported that C/EBPα activation can contrast this activity through the induction of miR-122 expression, which represses IGF-1R expression [164]. In contrast to the control mechanisms mediated by GSK3α/β, phosphorylation of eIF2α by PKR in response to inflammatory/stress signaling favors the translation of full-length C/EBPα and β [154].
4. Regulation of alternative splicing
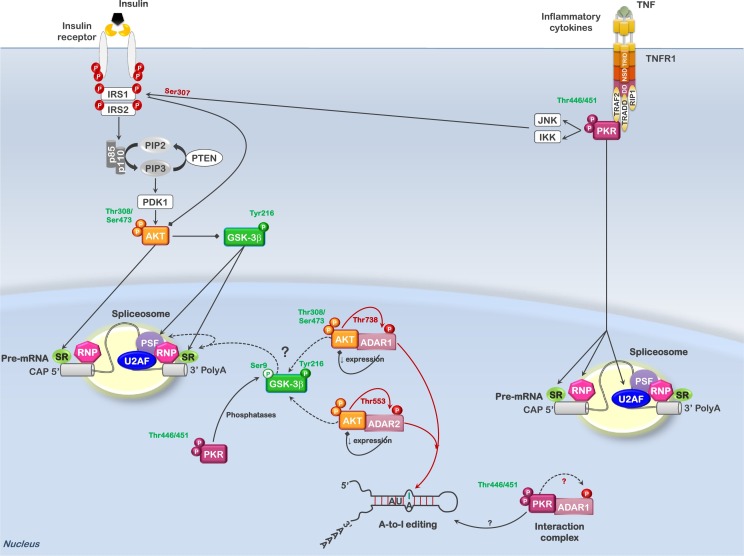
In the past several years proteomic studies examining the interactome and phosphoproteome have begun to associate a diverse number of kinases to cellular processes to which they were previously not associated; this is no truer than for GSK3α/β and PKR. As stated above, both PKR and GSK3α/β are considered major regulators of ribosome biogenesis and translation by directly or indirectly influencing these processes at multiple levels (for more detail information see Ref. [51]). More recently a role for both these kinases in alternative splicing has been documented. Blalock et al. previously reported that multiple proteins involved in RNA processing and alternative splicing were associated with PKR in the nucleus [46]. The study examined which proteins were complexed with nuclear PKR when active and when inactive (in the presence of the PKR inhibitor). Proteins associated with RNA processing/splicing that interacted only with the active PKR complex included the EIF4A3 (or DDX48), HNRNPM, SNRPA1, SON, SRSF2, THOC2, U2AF1, U2AF2, and YBX1. Those associated with the inactive PKR complex included COIL, SNRPD2, SNRPD3, SRRM1 and TRA2B (Table 4 ). Interestingly, while several RNA binding proteins were found to be associated with both active and inactive PKR complexes, nucleophosmin (NPM1) and the adenosine deaminase acting on dsRNA (ADAR)-1 were the only proteins with a known role in splicing/alternative splicing that were identified in both active and inactive PKR complexes [46]. This suggested that PKR activity could significantly affect assembly and composition of the splicing complex, thereby changing the alternatively spliced RNA landscape (Fig. 5 ). A shortcoming of this study was the lack of identifying direct targets of PKR phosphorylation in these complexes. More recently, Shinde et al. reported in a phosphoproteome study examining GSK3α/β-dependent signaling in wild-type and gsk3α/β double-knockout mouse embryonic stem cells (ESCs) that GSK3α/β regulate the phosphorylation of a number of proteins involved in RNA splicing, going on to both define the sites of GSK3α/β-dependent phosphorylation as well as a number of transcripts that are alternatively spliced in the presence of GSK3α/β activity [165]. The proteins whose phosphorylation was reduced in the gsk3α/β double-knockout ESCs and the sites of phosphorylation that were identified are as follows: BCLAF1, CELF1, NPM1, PPP4R2, RBM8A (S166 and/or S168), RBM39 (S129), SRRM1, SRRM2 (S1864), TRA2B (S39 and S83 and/or S85 and/or S87), SRSF9 (S190), and WBP11. While this data does not necessarily indicate direct phosphorylation of all these proteins by GSK3α/β, it does indicate that phosphorylation of these proteins is regulated by GSK3α/β. Additionally, GSK3α and GSK3β have also been demonstrated to interact with U2AF1 (α and β), U2AF2 (α only) and YBX1 (α and β), but phosphorylation of these proteins by GSK3α/β has not been observed (Fig. 5 and Table 4) [165].
Table 4.
Splicing-related proteins shared by GSK3α/β and PKR.
| Acc. # | Gene name | Description | Function | In association with |
||
|---|---|---|---|---|---|---|
| Phosphorylation dependent on GSK3α/β | GSK3α/β | PKR | ||||
| Q9NYF8 | BCLAF1 | Bcl-2-associated transcription factor 1 | Death-promoting transcriptional repressor; interacts with BCL2-related proteins and adenovirus E1B protein. Can repress pre-mRNA processing. | X | Xa | - |
| P06748 | NPM1 | Nucleophosmin | Involved in cellular division, ribosome biogenesis and ribosomal export; substrate for EIF2AK2 and GSK3. Can repress pre-mRNA processing. | X | +/- | |
| Q8IYB3 | SRRM1 | Serine/arginine repetitive matrix protein 1 | Part of pre- and post-splicing multiprotein mRNP complexes | X | Xa | - |
| P62995 | TRA2B | Transformer-2 protein homolog beta | Sequence-specific RNA-binding protein which participates in the control of pre-mRNA splicing; promotes TAU 10 exon inclusion | X | - | |
| Q01081 | U2AF1 | Splicing factor U2AF 35 kDa subunit | Constitutive and enhancer-dependent splicing by mediating protein-protein interactions and protein-RNA interactions required for accurate 3′-splice site selection | Xa,b | + | |
| P26368 | U2AF2 | Splicing factor U2AF 65 kDa subunit | Splicing of pre-mRNA; required for mRNA export; regulates troponin exon 5 inclusion; represses TAU exon 10 splicing | Xa | + | |
| P67809 | YBX1 | Nuclease-sensitive element-binding protein 1 | Pre-mRNA alternative splicing regulation; regulates transcription of MDR1, HLA class II genes and promotes MYC mRNA stability | Xa,b | + | |
Identified proteins are listed in alphabetical order.
Acc. # refers to the identifier in the UniProtKB-SwissProt data base.
Gene name is that used by the UniProtKB-SwissProt data base.
Function and Involvement in disease were retrieved in the UniProtKB data base.
Superscript “a” and “b” indicate whether the protein is known to interact with GSK3α or GSK3β, respectively.
“+” and “-” indicate whether the protein was found complexed with active PKR or inactive PKR.
Data were compiled from the following sources:
1. National Institute for Biotechnological Information (NCBI) National Institutes of Health (NIH) Gene Database.
2. Blalock, WL et al. (2014) J Cell Physiol. Ref # [46].
3. Shinde, MY et al. (2017) J Biol Chem. Ref # [165].
Fig. 5.
GSK3α/β and PKR are both regulators of alternative splicing. The diagram demonstrates the points of GSK3α/β and PKR regulation during mRNA processing and splicing following cytokine stimulation. Cytokine stimulation leads to the activation of both the PI3K-AKT and PKR pathways. AKT, GSK3α/β and PKR interact with diverse splicing proteins (some of them shared) and can regulate their phosphorylation. In addition, ADAR1 and ADAR2 editing activity can alter splice sites, facilitating alternative splicing. AKT activation would inhibit GSK3α/β-mediated changes to splicing associated proteins as well as result in reduced ADAR-dependent editing through phosphorylation of T738 and T553 in ADAR1 and ADAR2, respectively; thus favoring the production of a certain set of mRNA isoforms. Active PKR would favor activation of nuclear GSK3β and potentially enhance ADAR-dependent editing, altering the mRNA repertoire. While active PKR is denoted as phosphorylated on T446 and T451, only T451 is required for activity. Phosphorylation of T446 as well as numerous additional sites influences the degree of activation. Phosphorylation sites labeled in green (ex., Tyr216-GSK3β) indicate activating or stabilizing modifications; phosphorylation sites labeled in red (ex., Thr738-ADAR1) indicate modifications that result in inhibition of activity. Lines with arrowhead termini represent promoting effects on the downstream signaling; lines with boxed termini represent inhibitory effects on downstream signaling. SR = serine/arginine-rich splicing factors; RNP = heteronuclear ribonucleoprotein complex proteins; PSF = splicing factor, proline- and glutamine-rich (SFPQ).
Interestingly, activity of both PKR and GSK3α/β has been associated with hyperphosphorylated TAU, neural filamentous tangles and Alzheimer's disease. In addition, the inclusion or exclusion of TAU exon 10 through alternate splicing results in TAU isoforms containing either 3 or 4 microtubule repeat domains (3R or 4R). In TAU-related pathologies, or TAUopathies, the 3R/4R ratio is disturbed with an increase in the 3R:4R ratio [166]. Of the proteins mentioned above that have a role in TAU exon 10 splicing, U2AF2, which represses exon 10 inclusion, interacts with GSK3α and was found in association with active nuclear PKR [46,165]. In contrast, TRA2B which promotes the inclusion of TAU exon 10, as mention above, is phosphorylated in response to active GSK3α/β and was found to interact with inactive PKR in the nucleus (Table 4) [46,165]. Also associated with inactive nuclear PKR was the serine/arginine-rich splicing factor (SRSF)-6, which also represses the inclusion of TAU exon 10 [46]. Previous studies have demonstrated that GSK3β could phosphorylate SC35, a serine/arginine splicing factor that favors TAU exon 10 inclusion [166]. The GSK3β phosphorylated SC35 is redistributed in the nucleus and TAU exon 10 inclusion is repressed. These data seem to support a combined role for PKR and GSKα/β in the regulation of alternate splicing of TAU transcripts and the production of isoforms that have been linked to neurodegenerative pathologies.
Recent data also suggest that mRNA transcripts of both kinases undergo alternative splicing. In several organisms, GSK3β has been found to be expressed in multiple alternatively spliced forms [167,168]. In humans, two alternatively spliced forms have been observed. The primary form consists of 420 amino acids and has a predicted molecular weight of 46.7 kDa, while the alternative spliced form contains an additional 13 amino acids and has a predicted molecular weight of 48 kDa. Isoform 2, which shows reduced binding to AXIN1 and reduced kinase activity toward TAU, has been predicted to have a role in axon and neurite growth [168]. Similarly, PKR, which previously was not thought to undergo significant alternative splicing, has recently been found to produce more than 10 different alternately spliced forms in osteosarcoma (unpublished data). The significance of these isoforms in the associated pathology is currently under investigation.
Recent data also indicate that the adenosine deaminase acting on double-strand RNA (ADAR) family proteins are significant players in an alternative splicing regulatory network consisting of ADAR1/2, AKT, GSK3α/β and PKR. The ADAR proteins are a family of RNA editing enzymes that carry-out the deamination of specific adenosine (A) residues in areas of complex RNA structure, converting them into inosine (I). As inosine is interpreted by the cells molecular machinery as guanine, the A-to-I conversion can alter amino acid coding, change mRNA stability, and alter splice acceptor/donor sites, favoring alternative splicing of transcripts. The ADAR family, which consists of ADAR1 (DSRAD), ADAR2 (ADARB1 or RED1) and ADAR3 (ADARB2 or RED2), are responsible by far for the majority of A-to-I editing events in the cell. While ADAR1 and ADAR2 are rather ubiquitously expressed, ADAR3 expression is more restricted to the brain. Of the family members, only ADAR1 and ADAR2 demonstrate deaminase activity [169,170]. As these enzymes operate as homo- and heterodimers, ADAR3 may serve to regulate activity of the other two family members. Likewise, the dimer composition has a great influence on substrate selection and rate of catalysis. Moreover, multiple forms of ADAR1 and ADAR2 resulting both from alternate transcriptional initiation and alternative splicing have been documented [[170], [171], [172], [173]]. It was previously established in diverse studies that ADAR1 interacts with PKR and has the ability to suppress PKR activity [174,175]. Our group has also found that nuclear PKR associates with ADAR2. Whether this interaction is direct or mediated through ADAR1 is currently not known, and the full significance of the PKR-ADAR1/2 interaction in the nucleus has not been determined. What is known about this interaction is that ADAR1 and ADAR2 in the nucleus are found either in AKT containing complexes or PKR containing complexes, as no duel AKT-PKR complexes with ADAR1/2 are observed. Recently, Bavelloni et al. reported that AKT could regulate ADAR1 and ADAR2 through the phosphorylation of a conserved threonine residue in the catalytic domain [176,177]. Mimicking phosphorylation by site-directed mutagenesis of this site in either ADAR1 or ADAR2 resulted in a differential decrease in editase activity toward tested substrates (Fig. 5). Conversely, it was observed that phosphorylation of the ADARs also had an effect on AKT expression. To date whether, PKR can phosphorylate ADAR1 or ADAR2 has not been determined; a result of few studies examining the substrate network of this kinase and the lack of a true consensus phosphorylation site. What is now evident is that a regulatory balance is apparently at play whereby AKT- and PKR-dependent signaling influence ADAR-dependent editing and GSK3α/β activity. Both ADAR and GSK3α/β then have a tremendous influence on spliceosome composition and alternative splicing; ADAR though direct editing of splice donor/acceptor sites or alteration of transcripts encoding additional splice regulators and GSK3α/β through the modification of spliceosome components and splicing regulatory proteins. Similarly, both AKT and PKR are known to alter the composition of the spliceosome. Fig. 5 depicts the balance that must be maintained between PKR and AKT to achieve proper mRNA splicing in the cell and how metabolic stimulation of AKT or inflammatory activation of PKR can have a significant influence on alternative splicing.
5. Cancer, neurodegenerative diseases, diabetes/metabolic disorders and viral infection
Chronic inflammation and stress have increasingly become associated with disease. Most degenerative pathologies and cancer each have a significant inflammatory component. What has remained elusive is, much like the chicken or the egg, which comes first. Is chronic inflammation a result of the disease or is the disease a result of chronic inflammation. Insulin/insulin-related growth factor signaling may prove the best example. Normal insulin, signaling through the insulin receptor (IR) or insulin-like growth factor (IGF)-1 signaling through the IGF1 receptor (IGF-1R) results in PI3K-AKT-mTOR pathway activation among others. Stimulation of this pathway and other secondary pathways lead to increased protein synthesis, enhanced glucose uptake and metabolism, glycogen storage, and mitochondrial respiration; thus producing the biomolecules necessary for cell growth and proliferation (amino acids, lipids, nucleic acids, ATP). At the same time, increased metabolism generates reactive oxygen species (ROS) and toxic metabolic by-products that result in the stimulation of stress/inflammatory pathways to protect the cell from subsequent damage. In addition, transient imbalances in synthetic pathways, such as ribosome biogenesis and translation, result in the stimulation of stress response pathways as a mode to re-establish equilibrium [51]. The kinases PKR and GSK3α/β each become activated in response to insulin/IGF1-induced metabolic stress and either directly phosphorylate and/or reduce the synthesis of the insulin receptor substrates (IRS)-1 or IRS2, inhibiting IR and IGF-1R ligand-mediated stimulation of PI3K, thereby making the cells resistant to insulin and IGF1 [[178], [179], [180]]. As insulin resistance is the main feature defining diabetes mellitus, it is not surprising that constitutive activation of PKR and GSK3α/β is observed in obese/diabetic patients [181,182]. In fact, the feeding of high fat diets in mice has been observed to result in constitutively elevated levels of active PKR and GSK3α/β, favoring insulin resistance [180,181]. In addition to IRS1 and IRS2, the active PKR and GSK3α/β kinases target other downstream proteins that tweak additional pathways regulating transcription, cell division, ribosome biogenesis and translation (Fig. 6A).
Fig. 6.
Alteration of GSK3α/β and PKR activity can promote disease. The GSK3α/β and PKR interact to maintain cellular homeostasis, but, under altered conditions where their activity is aberrantly stimulated, they can lead to disease. (A) The diagram indicates the relationship between GSK3β signaling and PKR signaling in homeostasis and diverse pathologies, such as diabetes, cancer and neurodegenerative diseases. (B) Coronaviruses (SARS-CoV and MERS-CoV) utilize GSK3 activity for viral replication while inhibiting the ability of PKR to block protein synthesis. Other PKR-dependent signaling is often left intact, enhancing inflammation. While active PKR is denoted as phosphorylated on T446 and T451, only T451 is required for activity. Phosphorylation of T446 as well as numerous additional sites influences the degree of activation. Phosphorylation sites labeled in green (ex., Tyr216-GSK3β) indicate activating or stabilizing modifications; phosphorylation sites labeled in red (ex., Ser51-eIF2α) indicate modifications that are inhibitory to the substrates activity. Lines with arrowhead termini represent promoting effects on the downstream signaling; lines with boxed termini represent inhibitory effects on downstream signaling. pN = SARS-CoV nucleocapsid protein; p4a = MERS-CoV non-structural (NS) protein 4a.
In the case of Alzheimer's disease, a chronic inflammatory state leads to improper protein folding, activation of the unfolded protein response (UPR), and continuous stimulation of GSK3α/β and PKR kinases, resulting among other things in alternate splicing and phosphorylation of TAU, the formation of fibrogenous neural tangles and neuronal cell death, favoring disease progression [32,183]. Both PKR and GSK3 have become prime candidates for the design of targeted therapies to AD. Those directed toward inhibiting GSK3 have shown some promise in Phase II trials but only at early stages of the disease [184]. In contrast, therapies targeting PKR have thus far shown promising results in in vitro and animal studies, but have yet to enter clinical trials mainly as a result of the lack of critical attention given to PKR in the past [85,185]. This stress signaling circuit has made it clear that AD, like many other inflammatory related diseases may result from chronic systemic stress and not necessarily local stresses [186]. Empirical evidence supports this conclusion as metabolic disorders like diabetes correlate with enhanced risk of Alzheimer's and various cancers (Fig. 6A) [187].
Cancer much like degenerative diseases has a chronic inflammatory component. It has been proposed that the relation between cancer and inflammation/stress is that chronic inflammation and stress lead to a selective pressure on the tissues of an organism with survival dependent on random mutations. This is no more clearly evident than in bone marrow failure disorders, where genetic or idiopathic stresses contribute to the loss of bone marrow progenitors in the early stages. In the later stages this is usually replaced by accumulation of blasts in the periphery and full blown leukemia [25,188]. In many cases, alterations in apoptotic proteins or altered splicing are observed. In cancer, PKR has been suggested to have both a positive and negative role in the disease [25,30,48,189]. What seems evident is that the influence of PKR on diverse cancers is closely associated with its primary cellular location; cytosolic (suppresses tumorigenesis/pro-apoptotic) or nuclear (favors tumorigenesis/progression). Similarly, GSK3α/β have been found to display both tumor promoting and tumor suppressor functions. Enhanced activity, mainly that of GSK3β, has been documented in pancreatic cancer, colorectal cancer and renal cancer while alternative splicing has been reported in leukemia [5]. In contrast, loss of GSK3α/β activity has been associated with skin, lung and breast cancers [5]. What is not clear from many of these studies is the major cellular localization of the GSK3 kinase in question. The importance of this is critical as enhanced nuclear GSK3β activity would be expected to enhance alternative splicing and lead to the degradation of p53, thus favoring genome instability and a pro-tumorigenic climate. On the other hand, active GSK3α/β activity in the cytosol would favor antiproliferative/homeostasis oriented cellular programming (Fig. 6A).
Under the current global circumstances, it would be a failure to ignore the role of PKR and GSK3α/β in viral infection. As modulators of the inflammatory response, both PKR and GSK3α/β might be considered innate immune kinases. PKR has long held that role, possibly too well, as the majority of studies over the years have left it labeled simply as an antiviral kinase that autophosphorylates in the presence of dsRNA, thereby lagging far behind the number of studies involving AKT and GSK3. Both kinases are quick to respond to cellular stresses which include among other things viral infection. PKR is the quintessential antiviral kinase. As stated previously, PKR is both a pathogen recognition receptor (PRR) and interferon response gene (ISG); as such it can be activated by viral dsRNAs in a PACT-dependent manner and its expression induced by interferons [19,25,40]. Activation of PKR under these conditions leads to the phosphorylation of eIF2α, the accumulation of stress granules, the suppression of general protein synthesis, and the induction of interferon response factor (IRF) stimulated genes as well as ifnα/β transcription and mRNA stability; thus, in theory, having a negative effect on viral protein synthesis [19,25]. But a number of viruses encode or have found a means to by-pass this checkpoint while still allowing other arms of PKR-mediated signal transduction to continue, thereby turning the activation and synthesis of PKR into a facilitator of the pathology associated with viral infection, as well as in some cases the viral infection itself (see the following Reviews [19,190,191]). Similarly, GSK3α/β are known mediators of the inflammatory response, and GSK3 activity is modulated by certain viruses. In human immunodeficiency virus (HIV) infection, activation of GSK3β is largely responsible for viral associated neuronal cell death and accounts for the main reason why infection with neurotropic viruses such as HIV and herpesviruses are linked to increased risk of developing neurodegenerative disease [[192], [193], [194]]. For other viruses such as influenza A, GSK3 activation has antiviral activity, while in contrast GSK3β activity promotes hepatitis C viral replication and viral particle production [195,196].
Coronaviruses such as severe acute respiratory syndrome coronavirus (SARS-CoV), Mediterranean emerging respiratory syndrome coronavirus (MERS-CoV) and SARS-CoV-2 (COVID-19) are positive-strand RNA viruses that as a family are known for producing a number of unique proteins that repress the cellular antiviral response [197]. In fact, the major pathological manifestations of an enhanced inflammatory response with these viruses result from a delay in IFN induction following infection; this results in an over abundance of proinflammatory monocytes and macrophages and the release of proinflammatory cytokines in the lung, causing tissue damage [198,199]. Previous, investigations have shown that PKR becomes activated in SARS-CoV and MERS-CoV infections, but this activation was reported to have little antiviral effect while it was still able to contribute to apoptosis of infected cells (Fig. 6B) [[200], [201], [202]]. In MERS-CoV infections, the absence of PKR antiviral effects were found to be mediated by the viral accessory protein p4a, which binds to PKR and inhibits its ability to phosphorylate eIF2α, while at the same time leaving other aspects of PKR signaling largely intact [202]. In fact, the targeting of eIF2α phosphorylation more than PKR activity seems to be paramount to coronavirus infection and replication as other non-human coronaviruses both inhibit PKR-dependent phosphorylation of eIF2α as well as enhance the dephosphorylation of eIF2α S51 through a viral-mediated enhancement of GADD34, a component of the PP1 phosphatase [203]. The exact role of PKR in coronavirus infection and the associated pathology is complex, requiring additional clarifying studies. In addition, the current lack of promising activators or inhibitors of PKR-mediated signaling places specific therapeutic interventions along this line out of immediate reach.
In contrast to PKR, the few studies examining the role of GSK3α/β in coronavirus infection have demonstrated that GSK3 activity has a proviral effect. In SARS-CoV infection, GSK3 activity is required to phosphorylate the viral nucleocapsid protein (N), thus allowing the interaction of the host cell RNA helicase DDX1 with phosphorylated nucleocapsid complexes, which alters the viral polymerase in a manner that favors viral genome replication over viral mRNA transcription [204,205]. Inhibition of GSK3 with a specific inhibitor was shown to block viral replication and reduce the viral titer in vitro [205]. Compounds, targeting GSK3β, such as the orphan drug Tideglusib, which was given orphan status after phase II trials did not reach the set end points in treating Alzheimer's disease, may be promising therapeutics to repurpose as antiviral therapies to combat emerging coronavirus diseases like SARS-CoV-2 (Fig. 6B) [206].
6. Conclusions
The GSK3 and PKR kinases control responses from ribosome biogenesis, translation, cellular division and metabolic regulation placing them at key positions in normal cell homeostasis and pathologies such as cancer, neurodegenerative disease, diabetes mellitus and viral infection. While these kinase share much in common (substrates and signal transduction pathways) as well as have the capability to regulate one another or facilitate/reinforce the signal transduction mediated by the other, there is still a tremendous amount of work to be done to understand the regulatory effects that are specific to each kinase and what upstream signaling dictates the intersection of their mediated signaling. As both kinases have been shown to contribute significantly to diverse inflammatory-related pathologies, the detailed parallel analysis of GSK3α/β and PKR has the possibility to result in the discovery and design of a number of targeted therapies.
CRediT authorship contribution statement
Manuela Piazzi: Visualization, Data curation, Writing - review & editing. Alberto Bavelloni:Writing - review & editing. Irene Faenza:Writing - review & editing. William Blalock:Conceptualization, Funding acquisition, Writing - original draft, Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by grants from the Leukemia Research Foundation (to WLB), the Associazione Italiana per la Ricerca sul Cancro (AIRC)-IG-2015 (Grant number 17137; to WLB).
Footnotes
This article is part of a Special Issue entitled: GSK-3 and related kinases in cancer, neurological and other disorders edited by James McCubrey, Agnieszka Gizak and Dariusz Rakus.
References
- 1.Sud N., Rutledge A.C., Pan K., Su Q. Activation of the dsRNA-activated protein kinase PKR in mitochondrial dysfunction and inflammatory stress in metabolic syndrome. Curr. Pharm. Des. 2016;22:2697–2703. doi: 10.2174/1381612822666160202141845. [DOI] [PubMed] [Google Scholar]
- 2.Beurel E., Grieco S.F., Jope R.S. Glycogen synthase kinase-3 (GSK3): regulation, actions, and diseases. Pharmacol. Ther. 2015;148:114–131. doi: 10.1016/j.pharmthera.2014.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duda P., Wisniewski J., Wojtowicz T., Wojcicka O., Jaskiewicz M., Drulis-Fajdasz D., Rakus D., McCubrey J.A., Gizak A. Targeting GSK3 signaling as a potential therapy of neurodegenerative diseases and aging. Expert Opin. Ther. Targets. 2018;22:833–848. doi: 10.1080/14728222.2018.1526925. [DOI] [PubMed] [Google Scholar]
- 4.Marchal J.A., Lopez G.J., Peran M., Comino A., Delgado J.R., Garcia-Garcia J.A., Conde V., Aranda F.M., Rivas C., Esteban M., Garcia M.A. The impact of PKR activation: from neurodegeneration to cancer. FASEB J. 2014;28:1965–1974. doi: 10.1096/fj.13-248294. [DOI] [PubMed] [Google Scholar]
- 5.Nagini S., Sophia J., Mishra R. Glycogen synthase kinases: moonlighting proteins with theranostic potential in cancer. Semin. Cancer Biol. 2019;56:25–36. doi: 10.1016/j.semcancer.2017.12.010. [DOI] [PubMed] [Google Scholar]
- 6.Mancinelli R., Carpino G., Petrungaro S., Mammola C.L., Tomaipitinca L., Filippini A., Facchiano A., Ziparo E., Giampietri C. Multifaceted roles of GSK-3 in cancer and autophagy-related diseases. Oxidative Med. Cell. Longev. 2017;2017:4629495. doi: 10.1155/2017/4629495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole A., Frame S., Cohen P. Further evidence that the tyrosine phosphorylation of glycogen synthase kinase-3 (GSK3) in mammalian cells is an autophosphorylation event. Biochem. J. 2004;377:249–255. doi: 10.1042/BJ20031259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi-Yanaga F., Shiraishi F., Hirata M., Miwa Y., Morimoto S., Sasaguri T. Glycogen synthase kinase-3beta is tyrosine-phosphorylated by MEK1 in human skin fibroblasts. Biochem. Biophys. Res. Commun. 2004;316:411–415. doi: 10.1016/j.bbrc.2004.02.061. [DOI] [PubMed] [Google Scholar]
- 9.McCubrey J.A., Steelman L.S., Bertrand F.E., Davis N.M., Sokolosky M., Abrams S.L., Montalto G., D'Assoro A.B., Libra M., Nicoletti F., Maestro R., Basecke J., Rakus D., Gizak A., Demidenko Z.N., Cocco L., Martelli A.M., Cervello M. GSK-3 as potential target for therapeutic intervention in cancer. Oncotarget. 2014;5:2881–2911. doi: 10.18632/oncotarget.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sayas C.L., Ariaens A., Ponsioen B., Moolenaar W.H. GSK-3 is activated by the tyrosine kinase Pyk2 during LPA1-mediated neurite retraction. Mol. Biol. Cell. 2006;17:1834–1844. doi: 10.1091/mbc.E05-07-0688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forde J.E., Dale T.C. Glycogen synthase kinase 3: a key regulator of cellular fate. Cell. Mol. Life Sci. 2007;64:1930–1944. doi: 10.1007/s00018-007-7045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hornbeck P.V., Zhang B., Murray B., Kornhauser J.M., Latham V., Skrzypek E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lerat H., Imache M.R., Polyte J., Gaudin A., Mercey M., Donati F., Baudesson C., Higgs M.R., Picard A., Magnan C., Foufelle F., Pawlotsky J.M. Hepatitis C virus induces a prediabetic state by directly impairing hepatic glucose metabolism in mice. J. Biol. Chem. 2017;292:12860–12873. doi: 10.1074/jbc.M117.785030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding Q., Xia W., Liu J.C., Yang J.Y., Lee D.F., Xia J., Bartholomeusz G., Li Y., Pan Y., Li Z., Bargou R.C., Qin J., Lai C.C., Tsai F.J., Tsai C.H., Hung M.C. Erk associates with and primes GSK-3beta for its inactivation resulting in upregulation of beta-catenin. Mol. Cell. 2005;19:159–170. doi: 10.1016/j.molcel.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 15.Tejeda-Munoz N., Gonzalez-Aguilar H., Santoyo-Ramos P., Castaneda-Patlan M.C., Robles-Flores M. Glycogen synthase kinase 3beta is positively regulated by protein kinase Czeta-mediated phosphorylation induced by Wnt agonists. Mol. Cell. Biol. 2015;36:731–741. doi: 10.1128/MCB.00828-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wrede C.E., Dickson L.M., Lingohr M.K., Briaud I., Rhodes C.J. Protein kinase B/Akt prevents fatty acid-induced apoptosis in pancreatic beta-cells (INS-1) J. Biol. Chem. 2002;277:49676–49684. doi: 10.1074/jbc.M208756200. [DOI] [PubMed] [Google Scholar]
- 17.Song W.J., Song E.A., Jung M.S., Choi S.H., Baik H.H., Jin B.K., Kim J.H., Chung S.H. Phosphorylation and inactivation of glycogen synthase kinase 3beta (GSK3beta) by dual-specificity tyrosine phosphorylation-regulated kinase 1A (Dyrk1A) J. Biol. Chem. 2015;290:2321–2333. doi: 10.1074/jbc.M114.594952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thornton T.M., Pedraza-Alva G., Deng B., Wood C.D., Aronshtam A., Clements J.L., Sabio G., Davis R.J., Matthews D.E., Doble B., Rincon M. Phosphorylation by p38 MAPK as an alternative pathway for GSK3beta inactivation. Science. 2008;320:667–670. doi: 10.1126/science.1156037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia M.A., Gil J., Ventoso I., Guerra S., Domingo E., Rivas C., Esteban M. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol. Mol. Biol. Rev. 2006;70:1032–1060. doi: 10.1128/MMBR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams B.R. PKR; a sentinel kinase for cellular stress. Oncogene. 1999;18:6112–6120. doi: 10.1038/sj.onc.1203127. [DOI] [PubMed] [Google Scholar]
- 21.Hamanaka R.B., Bennett B.S., Cullinan S.B., Diehl J.A. PERK and GCN2 contribute to eIF2alpha phosphorylation and cell cycle arrest after activation of the unfolded protein response pathway. Mol. Biol. Cell. 2005;16:5493–5501. doi: 10.1091/mbc.E05-03-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito T., Warnken S.P., May W.S. Protein synthesis inhibition by flavonoids: roles of eukaryotic initiation factor 2alpha kinases. Biochem. Biophys. Res. Commun. 1999;265:589–594. doi: 10.1006/bbrc.1999.1727. [DOI] [PubMed] [Google Scholar]
- 23.Pervin S., Tran A.H., Zekavati S., Fukuto J.M., Singh R., Chaudhuri G. Increased susceptibility of breast cancer cells to stress mediated inhibition of protein synthesis. Cancer Res. 2008;68:4862–4874. doi: 10.1158/0008-5472.CAN-08-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wek R.C., Jiang H.Y., Anthony T.G. Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- 25.Blalock W.L., Bavelloni A., Piazzi M., Faenza I., Cocco L. A role for PKR in hematologic malignancies. J. Cell. Physiol. 2010;223:572–591. doi: 10.1002/jcp.22092. [DOI] [PubMed] [Google Scholar]
- 26.Kimball S.R., Clemens M.J., Tilleray V.J., Wek R.C., Horetsky R.L., Jefferson L.S. The double-stranded RNA-activated protein kinase PKR is dispensable for regulation of translation initiation in response to either calcium mobilization from the endoplasmic reticulum or essential amino acid starvation. Biochem. Biophys. Res. Commun. 2001;280:293–300. doi: 10.1006/bbrc.2000.4103. [DOI] [PubMed] [Google Scholar]
- 27.Shi Y., Vattem K.M., Sood R., An J., Liang J., Stramm L., Wek R.C. Identification and characterization of pancreatic eukaryotic initiation factor 2 alpha-subunit kinase, PEK, involved in translational control. Mol. Cell. Biol. 1998;18:7499–7509. doi: 10.1128/mcb.18.12.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mounir Z., Krishnamoorthy J.L., Wang S., Papadopoulou B., Campbell S., Muller W.J., Hatzoglou M., Koromilas A.E. Akt determines cell fate through inhibition of the PERK-eIF2alpha phosphorylation pathway. Sci. Signal. 2011;4 doi: 10.1126/scisignal.2001630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J.J. Regulation of protein synthesis by the heme-regulated eIF2alpha kinase: relevance to anemias. Blood. 2007;109:2693–2699. doi: 10.1182/blood-2006-08-041830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blalock W.L., Bavelloni A., Piazzi M., Tagliavini F., Faenza I., Martelli A.M., Follo M.Y., Cocco L. Multiple forms of PKR present in the nuclei of acute leukemia cells represent an active kinase that is responsive to stress. Leukemia. 2011;25:236–245. doi: 10.1038/leu.2010.264. [DOI] [PubMed] [Google Scholar]
- 31.Ward S.V., Samuel C.E. The PKR kinase promoter binds both Sp1 and Sp3, but only Sp3 functions as part of the interferon-inducible complex with ISGF-3 proteins. Virology. 2003;313:553–566. doi: 10.1016/s0042-6822(03)00347-7. [DOI] [PubMed] [Google Scholar]
- 32.Bose A., Mouton-Liger F., Paquet C., Mazot P., Vigny M., Gray F., Hugon J. Modulation of tau phosphorylation by the kinase PKR: implications in Alzheimer’s disease. Brain Pathol. 2011;21:189–200. doi: 10.1111/j.1750-3639.2010.00437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X., Bennett R.L., Cheng X., Byrne M., Reinhard M.K., May W.S., Jr. PKR regulates proliferation, differentiation, and survival of murine hematopoietic stem/progenitor cells. Blood. 2013;121:3364–3374. doi: 10.1182/blood-2012-09-456400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuhen K.L., Samuel C.E. Isolation of the interferon-inducible RNA-dependent protein kinase Pkr promoter and identification of a novel DNA element within the 5′-flanking region of human and mouse Pkr genes. Virology. 1997;227:119–130. doi: 10.1006/viro.1996.8306. [DOI] [PubMed] [Google Scholar]
- 35.Su Q., Wang S., Baltzis D., Qu L.K., Wong A.H., Koromilas A.E. Tyrosine phosphorylation acts as a molecular switch to full-scale activation of the eIF2alpha RNA-dependent protein kinase. Proc. Natl. Acad. Sci. U. S. A. 2006;103:63–68. doi: 10.1073/pnas.0508207103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Romano P.R., Garcia-Barrio M.T., Zhang X., Wang Q., Taylor D.R., Zhang F., Herring C., Mathews M.B., Qin J., Hinnebusch A.G. Autophosphorylation in the activation loop is required for full kinase activity in vivo of human and yeast eukaryotic initiation factor 2alpha kinases PKR and GCN2. Mol. Cell. Biol. 1998;18:2282–2297. doi: 10.1128/mcb.18.4.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mudhasani R., Tran J.P., Retterer C., Kota K.P., Whitehouse C.A., Bavari S. Protein kinase R degradation is essential for Rift Valley fever virus infection and is regulated by SKP1-CUL1-F-box (SCF)FBXW11-NSs E3 ligase. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maarifi G., El Asmi F., Maroui M.A., Dianoux L., Chelbi-Alix M.K. Differential effects of SUMO1 and SUMO3 on PKR activation and stability. Sci. Rep. 2018;8:1277. doi: 10.1038/s41598-018-19683-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ito T., Yang M., May W.S. RAX, a cellular activator for double-stranded RNA-dependent protein kinase during stress signaling. J. Biol. Chem. 1999;274:15427–15432. doi: 10.1074/jbc.274.22.15427. [DOI] [PubMed] [Google Scholar]
- 40.Bennett R.L., Blalock W.L., Abtahi D.M., Pan Y., Moyer S.A., May W.S. RAX, the PKR activator, sensitizes cells to inflammatory cytokines, serum withdrawal, chemotherapy, and viral infection. Blood. 2006;108:821–829. doi: 10.1182/blood-2005-11-006817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blalock W.L., Grimaldi C., Fala F., Follo M., Horn S., Basecke J., Martinelli G., Cocco L., Martelli A.M. PKR activity is required for acute leukemic cell maintenance and growth: a role for PKR-mediated phosphatase activity to regulate GSK-3 phosphorylation. J. Cell. Physiol. 2009;221:232–241. doi: 10.1002/jcp.21848. [DOI] [PubMed] [Google Scholar]
- 42.Treiber T., Treiber N., Plessmann U., Harlander S., Daiss J.L., Eichner N., Lehmann G., Schall K., Urlaub H., Meister G. A compendium of RNA-binding proteins that regulate MicroRNA biogenesis. Mol. Cell. 2017;66:270–284. doi: 10.1016/j.molcel.2017.03.014. (e213) [DOI] [PubMed] [Google Scholar]
- 43.Zykova T.A., Zhu F., Zhang Y., Bode A.M., Dong Z. Involvement of ERKs, RSK2 and PKR in UVA-induced signal transduction toward phosphorylation of eIF2alpha (Ser(51)) Carcinogenesis. 2007;28:1543–1551. doi: 10.1093/carcin/bgm070. [DOI] [PubMed] [Google Scholar]
- 44.Saelens X., Kalai M., Vandenabeele P. Translation inhibition in apoptosis: caspase-dependent PKR activation and eIF2-alpha phosphorylation. J. Biol. Chem. 2001;276:41620–41628. doi: 10.1074/jbc.M103674200. [DOI] [PubMed] [Google Scholar]
- 45.Patel R.C., Sen G.C. PACT, a protein activator of the interferon-induced protein kinase, PKR. EMBO J. 1998;17:4379–4390. doi: 10.1093/emboj/17.15.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blalock W.L., Piazzi M., Bavelloni A., Raffini M., Faenza I., D’Angelo A., Cocco L. Identification of the PKR nuclear interactome reveals roles in ribosome biogenesis, mRNA processing and cell division. J. Cell. Physiol. 2014;229:1047–1060. doi: 10.1002/jcp.24529. [DOI] [PubMed] [Google Scholar]
- 47.Gal-Ben-Ari S., Barrera I., Ehrlich M., Rosenblum K. PKR: a kinase to remember. Front. Mol. Neurosci. 2018;11:480. doi: 10.3389/fnmol.2018.00480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee Y.S., Kunkeaw N., Lee Y.S. Protein kinase R and its cellular regulators in cancer: an active player or a surveillant? Wiley Interdiscip. RNA. 2020;11 doi: 10.1002/wrna.1558. [DOI] [PubMed] [Google Scholar]
- 49.Takada Y., Ichikawa H., Pataer A., Swisher S., Aggarwal B.B. Genetic deletion of PKR abrogates TNF-induced activation of IkappaBalpha kinase, JNK, Akt and cell proliferation but potentiates p44/p42 MAPK and p38 MAPK activation. Oncogene. 2007;26:1201–1212. doi: 10.1038/sj.onc.1209906. [DOI] [PubMed] [Google Scholar]
- 50.Baltzis D., Pluquet O., Papadakis A.I., Kazemi S., Qu L.K., Koromilas A.E. The eIF2alpha kinases PERK and PKR activate glycogen synthase kinase 3 to promote the proteasomal degradation of p53. J. Biol. Chem. 2007;282:31675–31687. doi: 10.1074/jbc.M704491200. [DOI] [PubMed] [Google Scholar]
- 51.Piazzi M., Bavelloni A., Gallo A., Faenza I., Blalock W.L. Signal transduction in ribosome biogenesis: a recipe to avoid disaster. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20112718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kimball S.R., Fabian J.R., Pavitt G.D., Hinnebusch A.G., Jefferson L.S. Regulation of guanine nucleotide exchange through phosphorylation of eukaryotic initiation factor eIF2alpha. Role of the alpha- and delta-subunits of eiF2b. J. Biol. Chem. 1998;273:12841–12845. doi: 10.1074/jbc.273.21.12841. [DOI] [PubMed] [Google Scholar]
- 53.Krishnamoorthy T., Pavitt G.D., Zhang F., Dever T.E., Hinnebusch A.G. Tight binding of the phosphorylated alpha subunit of initiation factor 2 (eIF2alpha) to the regulatory subunits of guanine nucleotide exchange factor eIF2B is required for inhibition of translation initiation. Mol. Cell. Biol. 2001;21:5018–5030. doi: 10.1128/MCB.21.15.5018-5030.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fernandez J., Yaman I., Merrick W.C., Koromilas A., Wek R.C., Sood R., Hensold J., Hatzoglou M. Regulation of internal ribosome entry site-mediated translation by eukaryotic initiation factor-2alpha phosphorylation and translation of a small upstream open reading frame. J. Biol. Chem. 2002;277:2050–2058. doi: 10.1074/jbc.M109199200. [DOI] [PubMed] [Google Scholar]
- 55.Yaman I., Fernandez J., Liu H., Caprara M., Komar A.A., Koromilas A.E., Zhou L., Snider M.D., Scheuner D., Kaufman R.J., Hatzoglou M. The zipper model of translational control: a small upstream ORF is the switch that controls structural remodeling of an mRNA leader. Cell. 2003;113:519–531. doi: 10.1016/s0092-8674(03)00345-3. [DOI] [PubMed] [Google Scholar]
- 56.Gerlitz G., Jagus R., Elroy-Stein O. Phosphorylation of initiation factor-2 alpha is required for activation of internal translation initiation during cell differentiation. Eur. J. Biochem. 2002;269:2810–2819. doi: 10.1046/j.1432-1033.2002.02974.x. [DOI] [PubMed] [Google Scholar]
- 57.Donze O., Deng J., Curran J., Sladek R., Picard D., Sonenberg N. The protein kinase PKR: a molecular clock that sequentially activates survival and death programs. EMBO J. 2004;23:564–571. doi: 10.1038/sj.emboj.7600078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanz M.A., Redondo N., Garcia-Moreno M., Carrasco L. Phosphorylation of eIF2alpha is responsible for the failure of the picornavirus internal ribosome entry site to direct translation from Sindbis virus replicons. J. Gen. Virol. 2013;94:796–806. doi: 10.1099/vir.0.049064-0. [DOI] [PubMed] [Google Scholar]
- 59.Stoneley M., Willis A.E. Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23:3200–3207. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- 60.Aggarwal B.B., Shishodia S., Takada Y., Jackson-Bernitsas D., Ahn K.S., Sethi G., Ichikawa H. TNF blockade: an inflammatory issue. Ernst Schering Res. Found Workshop. 2006:161–186. doi: 10.1007/3-540-37673-9_10. [DOI] [PubMed] [Google Scholar]
- 61.Aggarwal B.B., Shishodia S., Sandur S.K., Pandey M.K., Sethi G. Inflammation and cancer: how hot is the link? Biochem. Pharmacol. 2006;72:1605–1621. doi: 10.1016/j.bcp.2006.06.029. [DOI] [PubMed] [Google Scholar]
- 62.Koschmieder S., D'Alo F., Radomska H., Schoneich C., Chang J.S., Konopleva M., Kobayashi S., Levantini E., Suh N., Di Ruscio A., Voso M.T., Watt J.C., Santhanam R., Sargin B., Kantarjian H., Andreeff M., Sporn M.B., Perrotti D., Berdel W.E., Muller-Tidow C., Serve H., Tenen D.G. CDDO induces granulocytic differentiation of myeloid leukemic blasts through translational up-regulation of p42 CCAAT enhancer binding protein alpha. Blood. 2007;110:3695–3705. doi: 10.1182/blood-2006-11-058941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee Y.Y., Cevallos R.C., Jan E. An upstream open reading frame regulates translation of GADD34 during cellular stresses that induce eIF2alpha phosphorylation. J. Biol. Chem. 2009;284:6661–6673. doi: 10.1074/jbc.M806735200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van den Beucken T., Magagnin M.G., Savelkouls K., Lambin P., Koritzinsky M., Wouters B.G. Regulation of Cited2 expression provides a functional link between translational and transcriptional responses during hypoxia. Radiother. Oncol. 2007;83:346–352. doi: 10.1016/j.radonc.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 65.Proud C.G. eIF2 and the control of cell physiology. Semin. Cell Dev. Biol. 2005;16:3–12. doi: 10.1016/j.semcdb.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 66.Wang X., Paulin F.E., Campbell L.E., Gomez E., O’Brien K., Morrice N., Proud C.G. Eukaryotic initiation factor 2B: identification of multiple phosphorylation sites in the epsilon-subunit and their functions in vivo. EMBO J. 2001;20:4349–4359. doi: 10.1093/emboj/20.16.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Woods Y.L., Cohen P., Becker W., Jakes R., Goedert M., Wang X., Proud C.G. The kinase DYRK phosphorylates protein-synthesis initiation factor eIF2Bepsilon at Ser539 and the microtubule-associated protein tau at Thr212: potential role for DYRK as a glycogen synthase kinase 3-priming kinase. Biochem. J. 2001;355:609–615. doi: 10.1042/bj3550609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kubica N., Jefferson L.S., Kimball S.R. Eukaryotic initiation factor 2B and its role in alterations in mRNA translation that occur under a number of pathophysiological and physiological conditions. Prog. Nucleic Acid Res. Mol. Biol. 2006;81:271–296. doi: 10.1016/S0079-6603(06)81007-X. [DOI] [PubMed] [Google Scholar]
- 69.Willis A.E. Translational control of growth factor and proto-oncogene expression. Int. J. Biochem. Cell Biol. 1999;31:73–86. doi: 10.1016/s1357-2725(98)00133-2. [DOI] [PubMed] [Google Scholar]
- 70.van den Beucken T., Koritzinsky M., Wouters B.G. Translational control of gene expression during hypoxia. Cancer Biol. Ther. 2006;5:749–755. doi: 10.4161/cbt.5.7.2972. [DOI] [PubMed] [Google Scholar]
- 71.Spriggs K.A., Bushell M., Mitchell S.A., Willis A.E. Internal ribosome entry segment-mediated translation during apoptosis: the role of IRES-trans-acting factors. Cell Death Differ. 2005;12:585–591. doi: 10.1038/sj.cdd.4401642. [DOI] [PubMed] [Google Scholar]
- 72.Ryves W.J., Harwood A.J. The interaction of glycogen synthase kinase-3 (GSK-3) with the cell cycle. Prog. Cell Cycle Res. 2003;5:489–495. [PubMed] [Google Scholar]
- 73.Zamanian-Daryoush M., Der S.D., Williams B.R. Cell cycle regulation of the double stranded RNA activated protein kinase, PKR. Oncogene. 1999;18:315–326. doi: 10.1038/sj.onc.1202293. [DOI] [PubMed] [Google Scholar]
- 74.Guo R., Abdelmohsen K., Morin P.J., Gorospe M. Novel microRNA reporter uncovers repression of Let-7 by GSK-3beta. PLoS One. 2013;8 doi: 10.1371/journal.pone.0066330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Charvet C., Wissler M., Brauns-Schubert P., Wang S.J., Tang Y., Sigloch F.C., Mellert H., Brandenburg M., Lindner S.E., Breit B., Green D.R., McMahon S.B., Borner C., Gu W., Maurer U. Phosphorylation of Tip60 by GSK-3 determines the induction of PUMA and apoptosis by p53. Mol. Cell. 2011;42:584–596. doi: 10.1016/j.molcel.2011.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Qu L., Huang S., Baltzis D., Rivas-Estilla A.M., Pluquet O., Hatzoglou M., Koumenis C., Taya Y., Yoshimura A., Koromilas A.E. Endoplasmic reticulum stress induces p53 cytoplasmic localization and prevents p53-dependent apoptosis by a pathway involving glycogen synthase kinase-3beta. Genes Dev. 2004;18:261–277. doi: 10.1101/gad.1165804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kulikov R., Boehme K.A., Blattner C. Glycogen synthase kinase 3-dependent phosphorylation of Mdm2 regulates p53 abundance. Mol. Cell. Biol. 2005;25:7170–7180. doi: 10.1128/MCB.25.16.7170-7180.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kwon O., Kim K.A., He L., Kim S.O., Kim M.S., Cha E.Y., Yoon B.D., Sok D.E., Jung M., Ahn J.S., Kim B.Y. Ionizing radiation can induce GSK-3beta phosphorylation and NF-kappaB transcriptional transactivation in ATM-deficient fibroblasts. Cell. Signal. 2008;20:602–612. doi: 10.1016/j.cellsig.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 79.Cuddihy A.R., Li S., Tam N.W., Wong A.H., Taya Y., Abraham N., Bell J.C., Koromilas A.E. Double-stranded-RNA-activated protein kinase PKR enhances transcriptional activation by tumor suppressor p53. Mol. Cell. Biol. 1999;19:2475–2484. doi: 10.1128/mcb.19.4.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cuddihy A.R., Wong A.H., Tam N.W., Li S., Koromilas A.E. The double-stranded RNA activated protein kinase PKR physically associates with the tumor suppressor p53 protein and phosphorylates human p53 on serine 392 in vitro. Oncogene. 1999;18:2690–2702. doi: 10.1038/sj.onc.1202620. [DOI] [PubMed] [Google Scholar]
- 81.Bennett R.L., Pan Y., Christian J., Hui T., May W.S., Jr. The RAX/PACT-PKR stress response pathway promotes p53 sumoylation and activation, leading to G(1) arrest. Cell Cycle. 2012;11:407–417. doi: 10.4161/cc.11.2.18999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mitchell J.K., Midkiff B.R., Israelow B., Evans M.J., Lanford R.E., Walker C.M., Lemon S.M., McGivern D.R. Hepatitis C virus indirectly disrupts DNA damage-induced p53 responses by activating protein kinase R. mBio. 2017;8 doi: 10.1128/mBio.00121-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Balaji V., Kaniyappan S., Mandelkow E., Wang Y., Mandelkow E.M. Pathological missorting of endogenous MAPT/Tau in neurons caused by failure of protein degradation systems. Autophagy. 2018;14:2139–2154. doi: 10.1080/15548627.2018.1509607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Park S.A., Ahn S.I., Gallo J.M. Tau mis-splicing in the pathogenesis of neurodegenerative disorders. BMB Rep. 2016;49:405–413. doi: 10.5483/BMBRep.2016.49.8.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hugon J., Mouton-Liger F., Dumurgier J., Paquet C. PKR involvement in Alzheimer’s disease. Alzheimers Res. Ther. 2017;9:83. doi: 10.1186/s13195-017-0308-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Llorens-Martin M., Jurado J., Hernandez F., Avila J. GSK-3beta, a pivotal kinase in Alzheimer disease. Front. Mol. Neurosci. 2014;7:46. doi: 10.3389/fnmol.2014.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Singh T.J., Grundke-Iqbal I., Wu W.Q., Chauhan V., Novak M., Kontzekova E., Iqbal K. Protein kinase C and calcium/calmodulin-dependent protein kinase II phosphorylate three-repeat and four-repeat tau isoforms at different rates. Mol. Cell. Biochem. 1997;168:141–148. doi: 10.1023/a:1006807105059. [DOI] [PubMed] [Google Scholar]
- 88.Wang J.Z., Wu Q., Smith A., Grundke-Iqbal I., Iqbal K. Tau is phosphorylated by GSK-3 at several sites found in Alzheimer disease and its biological activity markedly inhibited only after it is prephosphorylated by A-kinase. FEBS Lett. 1998;436:28–34. doi: 10.1016/s0014-5793(98)01090-4. [DOI] [PubMed] [Google Scholar]
- 89.Mailliot C., Podevin-Dimster V., Rosenthal R.E., Sergeant N., Delacourte A., Fiskum G., Buee L. Rapid tau protein dephosphorylation and differential rephosphorylation during cardiac arrest-induced cerebral ischemia and reperfusion. J. Cereb. Blood Flow Metab. 2000;20:543–549. doi: 10.1097/00004647-200003000-00013. [DOI] [PubMed] [Google Scholar]
- 90.Illenberger S., Zheng-Fischhofer Q., Preuss U., Stamer K., Baumann K., Trinczek B., Biernat J., Godemann R., Mandelkow E.M., Mandelkow E. The endogenous and cell cycle-dependent phosphorylation of tau protein in living cells: implications for Alzheimer’s disease. Mol. Biol. Cell. 1998;9:1495–1512. doi: 10.1091/mbc.9.6.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schneider A., Biernat J., von Bergen M., Mandelkow E., Mandelkow E.M. Phosphorylation that detaches tau protein from microtubules (Ser262, Ser214) also protects it against aggregation into Alzheimer paired helical filaments. Biochemistry. 1999;38:3549–3558. doi: 10.1021/bi981874p. [DOI] [PubMed] [Google Scholar]
- 92.Liu F., Li B., Tung E.J., Grundke-Iqbal I., Iqbal K., Gong C.X. Site-specific effects of tau phosphorylation on its microtubule assembly activity and self-aggregation. Eur. J. Neurosci. 2007;26:3429–3436. doi: 10.1111/j.1460-9568.2007.05955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu F., Liang Z., Shi J., Yin D., El-Akkad E., Grundke-Iqbal I., Iqbal K., Gong C.X. PKA modulates GSK-3beta- and cdk5-catalyzed phosphorylation of tau in site- and kinase-specific manners. FEBS Lett. 2006;580:6269–6274. doi: 10.1016/j.febslet.2006.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu F., Iqbal K., Grundke-Iqbal I., Gong C.X. Involvement of aberrant glycosylation in phosphorylation of tau by cdk5 and GSK-3beta. FEBS Lett. 2002;530:209–214. doi: 10.1016/s0014-5793(02)03487-7. [DOI] [PubMed] [Google Scholar]
- 95.Leroy A., Landrieu I., Huvent I., Legrand D., Codeville B., Wieruszeski J.M., Lippens G. Spectroscopic studies of GSK3{beta} phosphorylation of the neuronal tau protein and its interaction with the N-terminal domain of apolipoprotein E. J. Biol. Chem. 2010;285:33435–33444. doi: 10.1074/jbc.M110.149419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Qian W., Shi J., Yin X., Iqbal K., Grundke-Iqbal I., Gong C.X., Liu F. PP2A regulates tau phosphorylation directly and also indirectly via activating GSK-3beta. J. Alzheimers Dis. 2010;19:1221–1229. doi: 10.3233/JAD-2010-1317. [DOI] [PubMed] [Google Scholar]
- 97.Sengupta A., Novak M., Grundke-Iqbal I., Iqbal K. Regulation of phosphorylation of tau by cyclin-dependent kinase 5 and glycogen synthase kinase-3 at substrate level. FEBS Lett. 2006;580:5925–5933. doi: 10.1016/j.febslet.2006.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sengupta A., Kabat J., Novak M., Wu Q., Grundke-Iqbal I., Iqbal K. Phosphorylation of tau at both Thr 231 and Ser 262 is required for maximal inhibition of its binding to microtubules. Arch. Biochem. Biophys. 1998;357:299–309. doi: 10.1006/abbi.1998.0813. [DOI] [PubMed] [Google Scholar]
- 99.Moszczynski A.J., Gohar M., Volkening K., Leystra-Lantz C., Strong W., Strong M.J. Thr175-phosphorylated tau induces pathologic fibril formation via GSK3beta-mediated phosphorylation of Thr231 in vitro. Neurobiol. Aging. 2015;36:1590–1599. doi: 10.1016/j.neurobiolaging.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 100.Tatebayashi Y., Planel E., Chui D.H., Sato S., Miyasaka T., Sahara N., Murayama M., Kikuchi N., Yoshioka K., Rivka R., Takashima A. c-Jun N-terminal kinase hyperphosphorylates R406W tau at the PHF-1 site during mitosis. FASEB J. 2006;20:762–764. doi: 10.1096/fj.05-4362fje. [DOI] [PubMed] [Google Scholar]
- 101.Azorsa D.O., Robeson R.H., Frost D., hoovet B. Meec, Brautigam G.R., Dickey C., Beaudry C., Basu G.D., Holz D.R., Hernandez J.A., Bisanz K.M., Gwinn L., Grover A., Rogers J., Reiman E.M., Hutton M., Stephan D.A., Mousses S., Dunckley T. High-content siRNA screening of the kinome identifies kinases involved in Alzheimer's disease-related tau hyperphosphorylation. BMC Genomics. 2010;11:25. doi: 10.1186/1471-2164-11-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nepal R.M., Martin A. Unmasking the mysteries of MYC. J. Immunol. 2019;202:2517–2518. doi: 10.4049/jimmunol.1900186. [DOI] [PubMed] [Google Scholar]
- 103.Gregory M.A., Qi Y., Hann S.R. Phosphorylation by glycogen synthase kinase-3 controls c-myc proteolysis and subnuclear localization. J. Biol. Chem. 2003;278:51606–51612. doi: 10.1074/jbc.M310722200. [DOI] [PubMed] [Google Scholar]
- 104.Welcker M., Orian A., Jin J., Grim J.E., Harper J.W., Eisenman R.N., Clurman B.E. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Seo H.R., Kim J., Bae S., Soh J.W., Lee Y.S. Cdk5-mediated phosphorylation of c-Myc on Ser-62 is essential in transcriptional activation of cyclin B1 by cyclin G1. J. Biol. Chem. 2008;283:15601–15610. doi: 10.1074/jbc.M800987200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kamemura K., Hayes B.K., Comer F.I., Hart G.W. Dynamic interplay between O-glycosylation and O-phosphorylation of nucleocytoplasmic proteins: alternative glycosylation/phosphorylation of THR-58, a known mutational hot spot of c-Myc in lymphomas, is regulated by mitogens. J. Biol. Chem. 2002;277:19229–19235. doi: 10.1074/jbc.M201729200. [DOI] [PubMed] [Google Scholar]
- 107.Arnold H.K., Sears R.C. Protein phosphatase 2A regulatory subunit B56alpha associates with c-myc and negatively regulates c-myc accumulation. Mol. Cell. Biol. 2006;26:2832–2844. doi: 10.1128/MCB.26.7.2832-2844.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ramana C.V., Grammatikakis N., Chernov M., Nguyen H., Goh K.C., Williams B.R., Stark G.R. Regulation of c-myc expression by IFN-gamma through Stat1-dependent and -independent pathways. EMBO J. 2000;19:263–272. doi: 10.1093/emboj/19.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hann S.R., Dixit M., Sears R.C., Sealy L. The alternatively initiated c-Myc proteins differentially regulate transcription through a noncanonical DNA-binding site. Genes Dev. 1994;8:2441–2452. doi: 10.1101/gad.8.20.2441. [DOI] [PubMed] [Google Scholar]
- 110.Mitchell S., Vargas J., Hoffmann A. Signaling via the NFkappaB system. Wiley Interdiscip. Rev. Syst. Biol. Med. 2016;8:227–241. doi: 10.1002/wsbm.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ozes O.N., Mayo L.D., Gustin J.A., Pfeffer S.R., Pfeffer L.M., Donner D.B. NF-kappaB activation by tumour necrosis factor requires the Akt serine-threonine kinase. Nature. 1999;401:82–85. doi: 10.1038/43466. [DOI] [PubMed] [Google Scholar]
- 112.Sizemore N., Lerner N., Dombrowski N., Sakurai H., Stark G.R. Distinct roles of the Ikappa B kinase alpha and beta subunits in liberating nuclear factor kappa B (NF-kappa B) from Ikappa B and in phosphorylating the p65 subunit of NF-kappa B. J. Biol. Chem. 2002;277:3863–3869. doi: 10.1074/jbc.M110572200. [DOI] [PubMed] [Google Scholar]
- 113.Gustin J.A., Korgaonkar C.K., Pincheira R., Li Q., Donner D.B. Akt regulates basal and induced processing of NF-kappaB2 (p100) to p52. J. Biol. Chem. 2006;281:16473–16481. doi: 10.1074/jbc.M507373200. [DOI] [PubMed] [Google Scholar]
- 114.Barre B., Perkins N.D. A cell cycle regulatory network controlling NF-kappaB subunit activity and function. EMBO J. 2007;26:4841–4855. doi: 10.1038/sj.emboj.7601899. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 115.Dan H.C., Adli M., Baldwin A.S. Regulation of mammalian target of rapamycin activity in PTEN-inactive prostate cancer cells by I kappa B kinase alpha. Cancer Res. 2007;67:6263–6269. doi: 10.1158/0008-5472.CAN-07-1232. [DOI] [PubMed] [Google Scholar]
- 116.Lee D.F., Kuo H.P., Chen C.T., Hsu J.M., Chou C.K., Wei Y., Sun H.L., Li L.Y., Ping B., Huang W.C., He X., Hung J.Y., Lai C.C., Ding Q., Su J.L., Yang J.Y., Sahin A.A., Hortobagyi G.N., Tsai F.J., Tsai C.H., Hung M.C. IKK beta suppression of TSC1 links inflammation and tumor angiogenesis via the mTOR pathway. Cell. 2007;130:440–455. doi: 10.1016/j.cell.2007.05.058. [DOI] [PubMed] [Google Scholar]
- 117.Yen C.J., Izzo J.G., Lee D.F., Guha S., Wei Y., Wu T.T., Chen C.T., Kuo H.P., Hsu J.M., Sun H.L., Chou C.K., Buttar N.S., Wang K.K., Huang P., Ajani J., Hung M.C. Bile acid exposure up-regulates tuberous sclerosis complex 1/mammalian target of rapamycin pathway in Barrett’s-associated esophageal adenocarcinoma. Cancer Res. 2008;68:2632–2640. doi: 10.1158/0008-5472.CAN-07-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dan H.C., Cooper M.J., Cogswell P.C., Duncan J.A., Ting J.P., Baldwin A.S. Akt-dependent regulation of NF-{kappa}B is controlled by mTOR and Raptor in association with IKK. Genes Dev. 2008;22:1490–1500. doi: 10.1101/gad.1662308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dan H.C., Baldwin A.S. Differential involvement of IkappaB kinases alpha and beta in cytokine- and insulin-induced mammalian target of rapamycin activation determined by Akt. J. Immunol. 2008;180:7582–7589. doi: 10.4049/jimmunol.180.11.7582. [DOI] [PubMed] [Google Scholar]
- 120.Demarchi F., Bertoli C., Sandy P., Schneider C. Glycogen synthase kinase-3 beta regulates NF-kappa B1/p105 stability. J. Biol. Chem. 2003;278:39583–39590. doi: 10.1074/jbc.M305676200. [DOI] [PubMed] [Google Scholar]
- 121.Steinbrecher K.A., Wilson W., 3rd, Cogswell P.C., Baldwin A.S. Glycogen synthase kinase 3beta functions to specify gene-specific, NF-kappaB-dependent transcription. Mol. Cell. Biol. 2005;25:8444–8455. doi: 10.1128/MCB.25.19.8444-8455.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schwabe R.F., Brenner D.A. Role of glycogen synthase kinase-3 in TNF-alpha-induced NF-kappaB activation and apoptosis in hepatocytes. Am. J. Physiol. Gastrointest. Liver Physiol. 2002;283:G204–G211. doi: 10.1152/ajpgi.00016.2002. [DOI] [PubMed] [Google Scholar]
- 123.Haefner B. A model for NF-kappa B regulation by GSK-3 beta. Drug Discov. Today. 2003;8:1062–1063. doi: 10.1016/s1359-6446(03)02898-8. [DOI] [PubMed] [Google Scholar]
- 124.Ougolkov A.V., Fernandez-Zapico M.E., Savoy D.N., Urrutia R.A., Billadeau D.D. Glycogen synthase kinase-3beta participates in nuclear factor kappaB-mediated gene transcription and cell survival in pancreatic cancer cells. Cancer Res. 2005;65:2076–2081. doi: 10.1158/0008-5472.CAN-04-3642. [DOI] [PubMed] [Google Scholar]
- 125.Martin M., Rehani K., Jope R.S., Michalek S.M. Toll-like receptor-mediated cytokine production is differentially regulated by glycogen synthase kinase 3. Nat. Immunol. 2005;6:777–784. doi: 10.1038/ni1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chen H., Yang S., Yang Z., Ma L., Jiang D., Mao J., Jiao B., Cai Z. Inhibition of GSK-3beta decreases NF-kappaB-dependent gene expression and impairs the rat liver regeneration. J. Cell. Biochem. 2007;102:1281–1289. doi: 10.1002/jcb.21358. [DOI] [PubMed] [Google Scholar]
- 127.Ougolkov A.V., Bone N.D., Fernandez-Zapico M.E., Kay N.E., Billadeau D.D. Inhibition of glycogen synthase kinase-3 activity leads to epigenetic silencing of nuclear factor kappaB target genes and induction of apoptosis in chronic lymphocytic leukemia B cells. Blood. 2007;110:735–742. doi: 10.1182/blood-2006-12-060947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hoeflich K.P., Luo J., Rubie E.A., Tsao M.S., Jin O., Woodgett J.R. Requirement for glycogen synthase kinase-3beta in cell survival and NF-kappaB activation. Nature. 2000;406:86–90. doi: 10.1038/35017574. [DOI] [PubMed] [Google Scholar]
- 129.Medunjanin S., Schleithoff L., Fiegehenn C., Weinert S., Zuschratter W., Braun-Dullaeus R.C. GSK-3beta controls NF-kappaB activity via IKKgamma/NEMO. Sci. Rep. 2016;6:38553. doi: 10.1038/srep38553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Buss H., Dorrie A., Schmitz M.L., Frank R., Livingstone M., Resch K., Kracht M. Phosphorylation of serine 468 by GSK-3beta negatively regulates basal p65 NF-kappaB activity. J. Biol. Chem. 2004;279:49571–49574. doi: 10.1074/jbc.C400442200. [DOI] [PubMed] [Google Scholar]
- 131.Arabi A., Ullah K., Branca R.M., Johansson J., Bandarra D., Haneklaus M., Fu J., Aries I., Nilsson P., Den Boer M.L., Pokrovskaja K., Grander D., Xiao G., Rocha S., Lehtio J., Sangfelt O. Proteomic screen reveals Fbw7 as a modulator of the NF-kappaB pathway. Nat. Commun. 2012;3:976. doi: 10.1038/ncomms1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Busino L., Millman S.E., Scotto L., Kyratsous C.A., Basrur V., O’Connor O., Hoffmann A., Elenitoba-Johnson K.S., Pagano M. Fbxw7alpha- and GSK3-mediated degradation of p100 is a pro-survival mechanism in multiple myeloma. Nat. Cell Biol. 2012;14:375–385. doi: 10.1038/ncb2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fujimoto K., Yasuda H., Sato Y., Yamamoto K. A role for phosphorylation in the proteolytic processing of the human NF-kappa B1 precursor. Gene. 1995;165:183–189. doi: 10.1016/0378-1119(95)00507-3. [DOI] [PubMed] [Google Scholar]
- 134.Wilson W., 3rd, Baldwin A.S. Maintenance of constitutive IkappaB kinase activity by glycogen synthase kinase-3alpha/beta in pancreatic cancer. Cancer Res. 2008;68:8156–8163. doi: 10.1158/0008-5472.CAN-08-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Takada Y., Fang X., Jamaluddin M.S., Boyd D.D., Aggarwal B.B. Genetic deletion of glycogen synthase kinase-3beta abrogates activation of IkappaBalpha kinase, JNK, Akt, and p44/p42 MAPK but potentiates apoptosis induced by tumor necrosis factor. J. Biol. Chem. 2004;279:39541–39554. doi: 10.1074/jbc.M403449200. [DOI] [PubMed] [Google Scholar]
- 136.Mamaghani S., Patel S., Hedley D.W. Glycogen synthase kinase-3 inhibition disrupts nuclear factor-kappaB activity in pancreatic cancer, but fails to sensitize to gemcitabine chemotherapy. BMC Cancer. 2009;9:132. doi: 10.1186/1471-2407-9-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Giannopoulou M., Dai C., Tan X., Wen X., Michalopoulos G.K., Liu Y. Hepatocyte growth factor exerts its anti-inflammatory action by disrupting nuclear factor-kappaB signaling. Am. J. Pathol. 2008;173:30–41. doi: 10.2353/ajpath.2008.070583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gong R., Ge Y., Chen S., Liang E., Esparza A., Sabo E., Yango A., Gohh R., Rifai A., Dworkin L.D. Glycogen synthase kinase 3beta: a novel marker and modulator of inflammatory injury in chronic renal allograft disease. Am. J. Transplant. 2008;8:1852–1863. doi: 10.1111/j.1600-6143.2008.02319.x. [DOI] [PubMed] [Google Scholar]
- 139.Visvanathan K.V., Goodbourn S. Double-stranded RNA activates binding of NF-kappa B to an inducible element in the human beta-interferon promoter. EMBO J. 1989;8:1129–1138. doi: 10.1002/j.1460-2075.1989.tb03483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lenardo M.J., Fan C.M., Maniatis T., Baltimore D. The involvement of NF-kappa B in beta-interferon gene regulation reveals its role as widely inducible mediator of signal transduction. Cell. 1989;57:287–294. doi: 10.1016/0092-8674(89)90966-5. [DOI] [PubMed] [Google Scholar]
- 141.Gromkowski S.H., Mama K., Yagi J., Sen R., Rath S. Double-stranded RNA and bacterial lipopolysaccharide enhance sensitivity to TNF-alpha-mediated cell death. Int. Immunol. 1990;2:903–908. doi: 10.1093/intimm/2.9.903. [DOI] [PubMed] [Google Scholar]
- 142.Kumar A., Haque J., Lacoste J., Hiscott J., Williams B.R. Double-stranded RNA-dependent protein kinase activates transcription factor NF-kappa B by phosphorylating I kappa B. Proc. Natl. Acad. Sci. U. S. A. 1994;91:6288–6292. doi: 10.1073/pnas.91.14.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Koromilas A.E., Cantin C., Craig A.W., Jagus R., Hiscott J., Sonenberg N. The interferon-inducible protein kinase PKR modulates the transcriptional activation of immunoglobulin kappa gene. J. Biol. Chem. 1995;270:25426–25434. doi: 10.1074/jbc.270.43.25426. [DOI] [PubMed] [Google Scholar]
- 144.Yang Y.L., Reis L.F., Pavlovic J., Aguzzi A., Schafer R., Kumar A., Williams B.R., Aguet M., Weissmann C. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 1995;14:6095–6106. doi: 10.1002/j.1460-2075.1995.tb00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kumar A., Yang Y.L., Flati V., Der S., Kadereit S., Deb A., Haque J., Reis L., Weissmann C., Williams B.R. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role of IRF-1 and NF-kappaB. EMBO J. 1997;16:406–416. doi: 10.1093/emboj/16.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.DeLuca C., Roulston A., Koromilas A., Wainberg M.A., Hiscott J. Chronic human immunodeficiency virus type 1 infection of myeloid cells disrupts the autoregulatory control of the NF-kappaB/Rel pathway via enhanced IkappaBalpha degradation. J. Virol. 1996;70:5183–5193. doi: 10.1128/jvi.70.8.5183-5193.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zamanian-Daryoush M., Mogensen T.H., DiDonato J.A., Williams B.R. NF-kappaB activation by double-stranded-RNA-activated protein kinase (PKR) is mediated through NF-kappaB-inducing kinase and IkappaB kinase. Mol. Cell. Biol. 2000;20:1278–1290. doi: 10.1128/mcb.20.4.1278-1290.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gil J., Alcami J., Esteban M. Activation of NF-kappa B by the dsRNA-dependent protein kinase, PKR involves the I kappa B kinase complex. Oncogene. 2000;19:1369–1378. doi: 10.1038/sj.onc.1203448. [DOI] [PubMed] [Google Scholar]
- 149.Bonnet M.C., Weil R., Dam E., Hovanessian A.G., Meurs E.F. PKR stimulates NF-kappaB irrespective of its kinase function by interacting with the IkappaB kinase complex. Mol. Cell. Biol. 2000;20:4532–4542. doi: 10.1128/mcb.20.13.4532-4542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bonnet M.C., Daurat C., Ottone C., Meurs E.F. The N-terminus of PKR is responsible for the activation of the NF-kappaB signaling pathway by interacting with the IKK complex. Cell. Signal. 2006;18:1865–1875. doi: 10.1016/j.cellsig.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 151.Ishii T., Kwon H., Hiscott J., Mosialos G., Koromilas A.E. Activation of the I kappa B alpha kinase (IKK) complex by double-stranded RNA-binding defective and catalytic inactive mutants of the interferon-inducible protein kinase PKR. Oncogene. 2001;20:1900–1912. doi: 10.1038/sj.onc.1204267. [DOI] [PubMed] [Google Scholar]
- 152.Gil J., Rullas J., Garcia M.A., Alcami J., Esteban M. The catalytic activity of dsRNA-dependent protein kinase, PKR, is required for NF-kappaB activation. Oncogene. 2001;20:385–394. doi: 10.1038/sj.onc.1204109. [DOI] [PubMed] [Google Scholar]
- 153.Nerlov C. The C/EBP family of transcription factors: a paradigm for interaction between gene expression and proliferation control. Trends Cell Biol. 2007;17:318–324. doi: 10.1016/j.tcb.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 154.Calkhoven C.F., Muller C., Leutz A. Translational control of C/EBPalpha and C/EBPbeta isoform expression. Genes Dev. 2000;14:1920–1932. [PMC free article] [PubMed] [Google Scholar]
- 155.Zhang D.E., Zhang P., Wang N.D., Hetherington C.J., Darlington G.J., Tenen D.G. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 1997;94:569–574. doi: 10.1073/pnas.94.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Akasaka T., Balasas T., Russell L.J., Sugimoto K.J., Majid A., Walewska R., Karran E.L., Brown D.G., Cain K., Harder L., Gesk S., Martin-Subero J.I., Atherton M.G., Bruggemann M., Calasanz M.J., Davies T., Haas O.A., Hagemeijer A., Kempski H., Lessard M., Lillington D.M., Moore S., Nguyen-Khac F., Radford-Weiss I., Schoch C., Struski S., Talley P., Welham M.J., Worley H., Strefford J.C., Harrison C.J., Siebert R., Dyer M.J. Five members of the CEBP transcription factor family are targeted by recurrent IGH translocations in B-cell precursor acute lymphoblastic leukemia (BCP-ALL) Blood. 2007;109:3451–3461. doi: 10.1182/blood-2006-08-041012. [DOI] [PubMed] [Google Scholar]
- 157.Chapiro E., Russell L., Radford-Weiss I., Bastard C., Lessard M., Struski S., Cave H., Fert-Ferrer S., Barin C., Maarek O., Della-Valle V., Strefford J.C., Berger R., Harrison C.J., Bernard O.A., Nguyen-Khac F. Overexpression of CEBPA resulting from the translocation t(14;19)(q32;q13) of human precursor B acute lymphoblastic leukemia. Blood. 2006;108:3560–3563. doi: 10.1182/blood-2006-03-010835. [DOI] [PubMed] [Google Scholar]
- 158.Geletu M., Balkhi M.Y., Peer Zada A.A., Christopeit M., Pulikkan J.A., Trivedi A.K., Tenen D.G., Behre G. Target proteins of C/EBPalphap30 in AML: C/EBPalphap30 enhances sumoylation of C/EBPalphap42 via up-regulation of Ubc9. Blood. 2007;110:3301–3309. doi: 10.1182/blood-2007-01-071035. [DOI] [PubMed] [Google Scholar]
- 159.Berberich-Siebelt F., Berberich I., Andrulis M., Santner-Nanan B., Jha M.K., Klein-Hessling S., Schimpl A., Serfling E. SUMOylation interferes with CCAAT/enhancer-binding protein beta-mediated c-myc repression, but not IL-4 activation in T cells. J. Immunol. 2006;176:4843–4851. doi: 10.4049/jimmunol.176.8.4843. [DOI] [PubMed] [Google Scholar]
- 160.Zhu S., Yoon K., Sterneck E., Johnson P.F., Smart R.C. CCAAT/enhancer binding protein-beta is a mediator of keratinocyte survival and skin tumorigenesis involving oncogenic Ras signaling. Proc. Natl. Acad. Sci. U. S. A. 2002;99:207–212. doi: 10.1073/pnas.012437299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sterneck E., Zhu S., Ramirez A., Jorcano J.L., Smart R.C. Conditional ablation of C/EBP beta demonstrates its keratinocyte-specific requirement for cell survival and mouse skin tumorigenesis. Oncogene. 2006;25:1272–1276. doi: 10.1038/sj.onc.1209144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Liu H.K., Perrier S., Lipina C., Finlay D., McLauchlan H., Hastie C.J., Hundal H.S., Sutherland C. Functional characterisation of the regulation of CAAT enhancer binding protein alpha by GSK-3 phosphorylation of Threonines 222/226. BMC Mol. Biol. 2006;7:14. doi: 10.1186/1471-2199-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Tang Q.Q., Gronborg M., Huang H., Kim J.W., Otto T.C., Pandey A., Lane M.D. Sequential phosphorylation of CCAAT enhancer-binding protein beta by MAPK and glycogen synthase kinase 3beta is required for adipogenesis. Proc. Natl. Acad. Sci. U. S. A. 2005;102:9766–9771. doi: 10.1073/pnas.0503891102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zeng C., Wang R., Li D., Lin X.J., Wei Q.K., Yuan Y., Wang Q., Chen W., Zhuang S.M. A novel GSK-3 beta-C/EBP alpha-miR-122-insulin-like growth factor 1 receptor regulatory circuitry in human hepatocellular carcinoma. Hepatology. 2010;52:1702–1712. doi: 10.1002/hep.23875. [DOI] [PubMed] [Google Scholar]
- 165.Shinde M.Y., Sidoli S., Kulej K., Mallory M.J., Radens C.M., Reicherter A.L., Myers R.L., Barash Y., Lynch K.W., Garcia B.A., Klein P.S. Phosphoproteomics reveals that glycogen synthase kinase-3 phosphorylates multiple splicing factors and is associated with alternative splicing. J. Biol. Chem. 2017;292:18240–18255. doi: 10.1074/jbc.M117.813527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Chen K.L., Yuan R.Y., Hu C.J., Hsu C.Y. Amyloid-beta peptide alteration of tau exon-10 splicing via the GSK3beta-SC35 pathway. Neurobiol. Dis. 2010;40:378–385. doi: 10.1016/j.nbd.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 167.Schaffer B., Wiedau-Pazos M., Geschwind D.H. Gene structure and alternative splicing of glycogen synthase kinase 3 beta (GSK-3beta) in neural and non-neural tissues. Gene. 2003;302:73–81. doi: 10.1016/s0378-1119(02)01092-2. [DOI] [PubMed] [Google Scholar]
- 168.Wood-Kaczmar A., Kraus M., Ishiguro K., Philpott K.L., Gordon-Weeks P.R. An alternatively spliced form of glycogen synthase kinase-3beta is targeted to growing neurites and growth cones. Mol. Cell. Neurosci. 2009;42:184–194. doi: 10.1016/j.mcn.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 169.Gallo A., Vukic D., Michalik D., O’Connell M.A., Keegan L.P. ADAR RNA editing in human disease; more to it than meets the I. Hum. Genet. 2017;136:1265–1278. doi: 10.1007/s00439-017-1837-0. [DOI] [PubMed] [Google Scholar]
- 170.George C.X., Wagner M.V., Samuel C.E. Expression of interferon-inducible RNA adenosine deaminase ADAR1 during pathogen infection and mouse embryo development involves tissue-selective promoter utilization and alternative splicing. J. Biol. Chem. 2005;280:15020–15028. doi: 10.1074/jbc.M500476200. [DOI] [PubMed] [Google Scholar]
- 171.George C.X., Samuel C.E. Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible. Proc. Natl. Acad. Sci. U. S. A. 1999;96:4621–4626. doi: 10.1073/pnas.96.8.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Filippini A., Bonini D., Giacopuzzi E., La Via L., Gangemi F., Colombi M., Barbon A. Differential enzymatic activity of Rat ADAR2 splicing variants is due to altered capability to interact with RNA in the deaminase domain. Genes. 2018;9 doi: 10.3390/genes9020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kawahara Y., Ito K., Ito M., Tsuji S., Kwak S. Novel splice variants of human ADAR2 mRNA: skipping of the exon encoding the dsRNA-binding domains, and multiple C-terminal splice sites. Gene. 2005;363:193–201. doi: 10.1016/j.gene.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 174.Chung H., Calis J.J.A., Wu X., Sun T., Yu Y., Sarbanes S.L., Dao Thi V.L., Shilvock A.R., Hoffmann H.H., Rosenberg B.R., Rice C.M. Human ADAR1 prevents endogenous RNA from triggering translational shutdown. Cell. 2018;172:811–824. doi: 10.1016/j.cell.2017.12.038. (e814) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Okonski K.M., Samuel C.E. Stress granule formation induced by measles virus is protein kinase PKR dependent and impaired by RNA adenosine deaminase ADAR1. J. Virol. 2013;87:756–766. doi: 10.1128/JVI.02270-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bavelloni A., Focaccia E., Piazzi M., Raffini M., Cesarini V., Tomaselli S., Orsini A., Ratti S., Faenza I., Cocco L., Gallo A., Blalock W.L. AKT-dependent phosphorylation of the adenosine deaminases ADAR-1 and -2 inhibits deaminase activity. FASEB J. 2019;33:9044–9061. doi: 10.1096/fj.201800490RR. [DOI] [PubMed] [Google Scholar]
- 177.Piazzi M., Bavelloni A., Gallo A., Blalock W.L. AKT-dependent phosphorylation of ADAR1p110 and ADAR2 represents a new and important link between cell signaling and RNA editing. DNA Cell Biol. 2020;39:343–348. doi: 10.1089/dna.2020.5351. [DOI] [PubMed] [Google Scholar]
- 178.Liberman Z., Eldar-Finkelman H. Serine 332 phosphorylation of insulin receptor substrate-1 by glycogen synthase kinase-3 attenuates insulin signaling. J. Biol. Chem. 2005;280:4422–4428. doi: 10.1074/jbc.M410610200. [DOI] [PubMed] [Google Scholar]
- 179.Sharfi H., Eldar-Finkelman H. Sequential phosphorylation of insulin receptor substrate-2 by glycogen synthase kinase-3 and c-Jun NH2-terminal kinase plays a role in hepatic insulin signaling. Am. J. Physiol. Endocrinol. Metab. 2008;294:E307–E315. doi: 10.1152/ajpendo.00534.2007. [DOI] [PubMed] [Google Scholar]
- 180.Nakamura T., Furuhashi M., Li P., Cao H., Tuncman G., Sonenberg N., Gorgun C.Z., Hotamisligil G.S. Double-stranded RNA-dependent protein kinase links pathogen sensing with stress and metabolic homeostasis. Cell. 2010;140:338–348. doi: 10.1016/j.cell.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Henriksen E.J. Dysregulation of glycogen synthase kinase-3 in skeletal muscle and the etiology of insulin resistance and type 2 diabetes. Curr. Diabetes Rev. 2010;6:285–293. doi: 10.2174/157339910793360888. [DOI] [PubMed] [Google Scholar]
- 182.Carvalho B.M., Oliveira A.G., Ueno M., Araujo T.G., Guadagnini D., Carvalho-Filho M.A., Geloneze B., Lima M.M., Pareja J.C., Carvalheira J.B., Saad M.J. Modulation of double-stranded RNA-activated protein kinase in insulin sensitive tissues of obese humans. Obesity. 2013;21:2452–2457. doi: 10.1002/oby.20410. [DOI] [PubMed] [Google Scholar]
- 183.Amin J., Paquet C., Baker A., Asuni A.A., Love S., Holmes C., Hugon J., Nicoll J.A., Boche D. Effect of amyloid-beta (Abeta) immunization on hyperphosphorylated tau: a potential role for glycogen synthase kinase (GSK)-3beta. Neuropathol. Appl. Neurobiol. 2015;41:445–457. doi: 10.1111/nan.12205. [DOI] [PubMed] [Google Scholar]
- 184.Lovestone S., Boada M., Dubois B., Hull M., Rinne J.O., Huppertz H.J., Calero M., Andres M.V., Gomez-Carrillo B., Leon T., del Ser T., A. investigators A phase II trial of tideglusib in Alzheimer's disease. J. Alzheimers Dis. 2015;45:75–88. doi: 10.3233/JAD-141959. [DOI] [PubMed] [Google Scholar]
- 185.Hwang K.D., Bak M.S., Kim S.J., Rhee S., Lee Y.S. Restoring synaptic plasticity and memory in mouse models of Alzheimer's disease by PKR inhibition. Mol. Brain. 2017;10:57. doi: 10.1186/s13041-017-0338-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Morris G., Berk M., Maes M., Puri B.K. Could Alzheimer’s disease originate in the periphery and if so how so? Mol. Neurobiol. 2019;56:406–434. doi: 10.1007/s12035-018-1092-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Zhang Y., Huang N.Q., Yan F., Jin H., Zhou S.Y., Shi J.S., Jin F. Diabetes mellitus and Alzheimer’s disease: GSK-3beta as a potential link. Behav. Brain Res. 2018;339:57–65. doi: 10.1016/j.bbr.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 188.Blalock, M. W.L.P., Gallo A., Bavelloni A., Focaccia E., Faenza I. RNA processing and RNA ribosome biogenesis in bone marrow failure disorders. RNA Dis. 2017;4:e1531. [Google Scholar]
- 189.Follo M.Y., Finelli C., Mongiorgi S., Clissa C., Bosi C., Martinelli G., Blalock W.L., Cocco L., Martelli A.M. PKR is activated in MDS patients and its subcellular localization depends on disease severity. Leukemia. 2008;22:2267–2269. doi: 10.1038/leu.2008.122. [DOI] [PubMed] [Google Scholar]
- 190.Yim H.C., Williams B.R. Protein kinase R and the inflammasome. J. Interf. Cytokine Res. 2014;34:447–454. doi: 10.1089/jir.2014.0008. [DOI] [PubMed] [Google Scholar]
- 191.Pfaller C.K., Li Z., George C.X., Samuel C.E. Protein kinase PKR and RNA adenosine deaminase ADAR1: new roles for old players as modulators of the interferon response. Curr. Opin. Immunol. 2011;23:573–582. doi: 10.1016/j.coi.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Nguyen T.B., Lucero G.R., Chana G., Hult B.J., Tatro E.T., Masliah E., Grant I., Achim C.L., Everall I.P., H.I.V.N.R. Group Glycogen synthase kinase-3beta (GSK-3beta) inhibitors AR-A014418 and B6B3O prevent human immunodeficiency virus-mediated neurotoxicity in primary human neurons. J. Neurovirol. 2009;15:434–438. doi: 10.1080/13550280903168131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Piacentini R., Li Puma D.D., Ripoli C., Marcocci M.E., De Chiara G., Garaci E., Palamara A.T., Grassi C. Herpes Simplex Virus type-1 infection induces synaptic dysfunction in cultured cortical neurons via GSK-3 activation and intraneuronal amyloid-beta protein accumulation. Sci. Rep. 2015;5:15444. doi: 10.1038/srep15444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Dewhurst S., Maggirwar S.B., Schifitto G., Gendelman H.E., Gelbard H.A. Glycogen synthase kinase 3 beta (GSK-3 beta) as a therapeutic target in neuroAIDS. J. NeuroImmune Pharmacol. 2007;2:93–96. doi: 10.1007/s11481-006-9051-1. [DOI] [PubMed] [Google Scholar]
- 195.Hirata N., Suizu F., Matsuda-Lennikov M., Edamura T., Bala J., Noguchi M. Inhibition of Akt kinase activity suppresses entry and replication of influenza virus. Biochem. Biophys. Res. Commun. 2014;450:891–898. doi: 10.1016/j.bbrc.2014.06.077. [DOI] [PubMed] [Google Scholar]
- 196.Saleh M., Ruschenbaum S., Welsch C., Zeuzem S., Moradpour D., Gouttenoire J., Lange C.M. Glycogen synthase kinase 3beta enhances hepatitis C virus replication by supporting miR-122. Front. Microbiol. 2018;9:2949. doi: 10.3389/fmicb.2018.02949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kikkert M. Innate immune evasion by human respiratory RNA viruses. J. Innate Immun. 2020;12:4–20. doi: 10.1159/000503030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Zhou J., Chu H., Li C., Wong B.H., Cheng Z.S., Poon V.K., Sun T., Lau C.C., Wong K.K., Chan J.Y., Chan J.F., K.K. To, Chan K.H., Zheng B.J., Yuen K.Y. Active replication of Middle East respiratory syndrome coronavirus and aberrant induction of inflammatory cytokines and chemokines in human macrophages: implications for pathogenesis. J. Infect. Dis. 2014;209:1331–1342. doi: 10.1093/infdis/jit504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Frieman M.B., Chen J., Morrison T.E., Whitmore A., Funkhouser W., Ward J.M., Lamirande E.W., Roberts A., Heise M., Subbarao K., Baric R.S. SARS-CoV pathogenesis is regulated by a STAT1 dependent but a type I, II and III interferon receptor independent mechanism. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Comar C.E., Goldstein S.A., Li Y., Yount B., Baric R.S., Weiss S.R. Antagonism of dsRNA-induced innate immune pathways by NS4a and NS4b accessory proteins during MERS coronavirus infection. mBio. 2019;10 doi: 10.1128/mBio.00319-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Nakagawa K., Narayanan K., Wada M., Makino S. Inhibition of stress granule formation by middle east respiratory syndrome coronavirus 4a accessory protein facilitates viral translation, leading to efficient virus replication. J. Virol. 2018;92 doi: 10.1128/JVI.00902-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Rabouw H.H., Langereis M.A., Knaap R.C., Dalebout T.J., Canton J., Sola I., Enjuanes L., Bredenbeek P.J., Kikkert M., de Groot R.J., van Kuppeveld F.J. Middle East respiratory coronavirus accessory protein 4a inhibits PKR-mediated antiviral stress responses. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wang X., Liao Y., Yap P.L., Png K.J., Tam J.P., Liu D.X. Inhibition of protein kinase R activation and upregulation of GADD34 expression play a synergistic role in facilitating coronavirus replication by maintaining de novo protein synthesis in virus-infected cells. J. Virol. 2009;83:12462–12472. doi: 10.1128/JVI.01546-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Wu C.H., Yeh S.H., Tsay Y.G., Shieh Y.H., Kao C.L., Chen Y.S., Wang S.H., Kuo T.J., Chen D.S., Chen P.J. Glycogen synthase kinase-3 regulates the phosphorylation of severe acute respiratory syndrome coronavirus nucleocapsid protein and viral replication. J. Biol. Chem. 2009;284:5229–5239. doi: 10.1074/jbc.M805747200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Wu C.H., Chen P.J., Yeh S.H. Nucleocapsid phosphorylation and RNA helicase DDX1 recruitment enables coronavirus transition from discontinuous to continuous transcription. Cell Host Microbe. 2014;16:462–472. doi: 10.1016/j.chom.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Mathuram T.L., Reece L.M., Cherian K.M. GSK-3 inhibitors: a double-edged sword? - an update on Tideglusib. Drug Res. 2018;68:436–443. doi: 10.1055/s-0044-100186. [DOI] [PubMed] [Google Scholar]