Abstract
Extensive virological testing is central to SARS-CoV-2 containment, but many settings face severe limitations on testing. Group testing offers a way to increase throughput by testing pools of combined samples; however, most proposed designs have not yet addressed key concerns over sensitivity loss and implementation feasibility. Here, we combine a mathematical model of epidemic spread and empirically derived viral kinetics for SARS-CoV-2 infections to identify pooling designs that are robust to changes in prevalence, and to ratify losses in sensitivity against the time course of individual infections. Using this framework, we show that prevalence can be accurately estimated across four orders of magnitude using only a few dozen pooled tests without the need for individual identification. We then exhaustively evaluate the ability of different pooling designs to maximize the number of detected infections under various resource constraints, finding that simple pooling can identify up to 20 times as many positives compared to individual testing with a given budget. We illustrate how pooling affects sensitivity and overall detection capacity during an epidemic and on each day post infection, finding that sensitivity loss is mainly attributed to individuals sampled at the end of infection. Crucially, we confirm that our theoretical results can be accurately translated into practice using pooled human nasopharyngeal specimens. Our results show that accounting for variation in sampled viral loads provides a nuanced picture of how pooling affects sensitivity to detect epidemiologically relevant infections. Using simple, practical group testing designs can vastly increase surveillance capabilities in resource-limited settings.
Introduction
The ongoing pandemic of SARS-CoV-2, a novel coronavirus, has caused over 24 million reported cases of coronavirus disease 2019 (COVID-19) and 800,000 reported deaths between December 2019 and August 2020. (1) Although wide-spread virological testing is essential to inform disease status and where outbreak mitigation measures should be targeted or lifted, sufficient testing of populations with meaningful coverage has proven difficult. (2–7) Disruptions in the global supply chains for testing reagents and supplies, as well as on-the-ground limitations in testing throughput and financial support, restrict the usefulness of testing–both for identifying infected individuals and to measure community prevalence and epidemic trajectory. While these issues have been at the fore in even the highest-income countries, the situation is even more dire in low income regions of the world. Cost barriers alone mean it is often simply not practical to prioritize community testing in any useful way, with the limited testing that exists necessarily reserved for the healthcare setting. These limitations urge new, more efficient, approaches to testing to be developed and adopted both for individual diagnostics and to enable public health epidemic control and containment efforts.
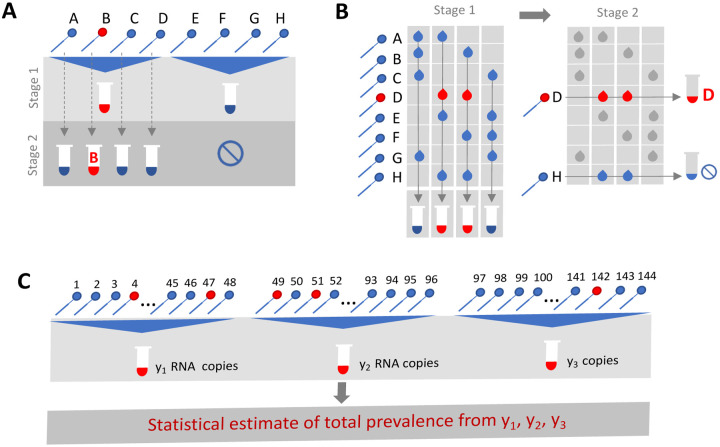
Group or pooled testing offers a way to increase efficiency by combining samples into a group or pool and testing a small number of pools rather than all samples individually. (8–10) For classifying individual samples, including for diagnostic testing, the principle is simple: if a pool tests negative, then all of the constituent samples are assumed negative. If a pool tests positive, then the constituent samples are putatively positive and must be tested again individually (Fig. 1A). Further efficiency gains are possible through combinatorial pooling, where, instead of testing every sample in every positive pool, each sample can instead be represented across multiple pools and potential positives are identified based on the pattern of pooled results (Fig. 1B). (9,10)
Fig. 1. Group testing designs for sample identification or prevalence estimation.
In group testing, multiple samples are pooled and tests are run on one or more pools. The results of these tests can be used for identification of positive samples (A, B) or to estimate prevalence (C). (A) In the simplest design for sample identification, samples are partitioned into non-overlapping pools. In stage 1 of testing, a negative result (Pool 2) indicates each sample in that pool was negative, while a positive result (Pool 1) indicates at least one sample in the pool was positive. These putatively positive samples are subsequently individually tested in stage 2 to identify positive results. (B) In a combinatorial design, samples are included in multiple pools as shown in stage 1. All samples that were included in negative pools are identified as negative, and the remaining putatively positive samples that were not included in any negative test are tested individually in stage 2. (C) In prevalence estimation, samples are partitioned into pools. The pool measurement will depend on the number and viral load of positive samples, and the dilution factor. The (quantitative) results from each pool can be used to estimate the fraction of samples that would have tested positive, had they been tested individually.
Simple pooling designs can also be used to assess prevalence without individual specimen identification (Fig. 1C). It has already been shown that the frequency of positive pools can allow estimation of the overall prevalence. (11) Crucially however, we show here that prevalence estimates can be greatly honed by considering quantitative viral loads measured in each positive pool, rather than simply using binary results (positive / negative). In short, the viral (RNA) load measurement from a pool is proportional to the sum of the (diluted) viral loads from each positive sample in the pool. Thus, here we show how evaluating the viral loads greatly improves potential efficiency gains in prevalence estimates by providing crucial information on the estimated number of positive samples in the pool – when the expected distribution of viral loads across specimens is known, which is easily measured empirically in a given lab. (12,13) Although this approach requires more complex statistical methods, the efficiency gains for public health surveillance can be large, and simplifying templates can be produced to improve ease of use and access to these types of analyses. The outcome is a highly efficient method for estimating population prevalence and enabling robust public health surveillance where it was previously out of reach.
Whilst the literature on theoretically optimized pooling designs for COVID-19 testing has grown rapidly, formal incorporation of biological variation (i.e., viral loads) and incorporation of general position along the epidemic curve, has received little attention. (14–17) Test sensitivity for example is not a fixed value, but depends on viral load, which can vary by many orders of magnitudes across individuals and over the course of an infection. (18–20) This large variation within a single infection affects sensitivity to detect infections at different points in the disease course, which has implications for appropriate intervention and the interpretation of a viral load measurement from a sample pool.
Here, we comprehensively evaluate designs for pooled testing of SARS-CoV-2 whilst accounting for epidemic dynamics and variation in viral loads arising from viral kinetics and extraneous features such as sampling variation. We demonstrate efficient, logistically feasible pooling designs for individual identification (i.e., diagnostics) and prevalence estimation (i.e., population surveillance). To do this, we use realistic simulated viral load data at the individual level over time, representing the entire time course of an epidemic to generate synthetic data that reflects the true distribution of viral loads in the population at any given time of the epidemic. We then used these data to derive optimal pooling strategies for different use cases and resource constraints in-silico. Finally, we demonstrate the approach using discarded de-identified human nasopharyngeal swabs initially collected for diagnostic and surveillance purposes.
Results
Modelling a synthetic population to assess pooling designs
To identify optimal pooling strategies for distinct scenarios, we required realistic estimates of viral loads across epidemic trajectories. We developed a population-level mathematical model of SARS-CoV-2 transmission that incorporates empirically measured within-host virus kinetics, and used these simulations to generate population-level viral load distributions representing real data sampled from population surveillance, either using nasopharyngeal swab or sputum samples (Fig. 2). Full details are provided in Materials and Methods and Supplementary Material 1, sections 1–4. These simulations generated a synthetic, realistic epidemic with a peak daily per incidence of 19.5 per 1000 people, and peak daily prevalence of RNA positivity (viral load greater than 100 virus RNA copies per ml) of 265 per 1000 (Fig. 2E). We used these simulation data to evaluate optimal group testing strategies at different points along the epidemic curve for diagnostic as well as public health surveillance, where the true viral loads in the population is known fully.
Fig. 2: Viral kinetics model fits, simulated infection dynamics and population-wide viral load kinetics.
(A) Schematic of the viral kinetics and infection model. Individuals begin susceptible with no viral load, acquire the virus from another infectious individual (exposed), experience an increase in viral load and possibly develop symptoms (infected), and finally either recover following viral waning or die (removed). This process is simulated for many individuals. (B) Model fits to time-varying viral loads in swab samples. The black dots show observed log10 RNA copies per swab; solid lines show posterior median estimates; dark shaded regions show 95% credible intervals (CI) on model-predicted latent viral loads; light shaded regions show 95% CI on simulated viral loads with added observation noise. The blue region shows viral loads before symptom onset and red region shows time after symptom onset. The horizontal dashed line shows the limit of detection. (C) Distribution of positive viral loads from 10,000 individuals sampled at day 140. (D) 25 simulated viral loads over time. The heatmap shows the viral load in each individual over time. (E) Simulated infection incidence and prevalence of virologically positive individuals from the SEIR model. Incidence was defined as the number of new infections per day divided by the population size. Prevalence was defined as the number of individuals with viral load > 100 (log10 viral load > 2) in the population divided by the population size on a given day. (F) As in (D), but for 500 individuals. The distribution of viral loads reflects the increase and subsequent decline of prevalence. We simulated from inferred distributions for the viral load parameters, thereby propagating substantial individual-level variability through the simulations.
Improved testing efficiency for estimating prevalence
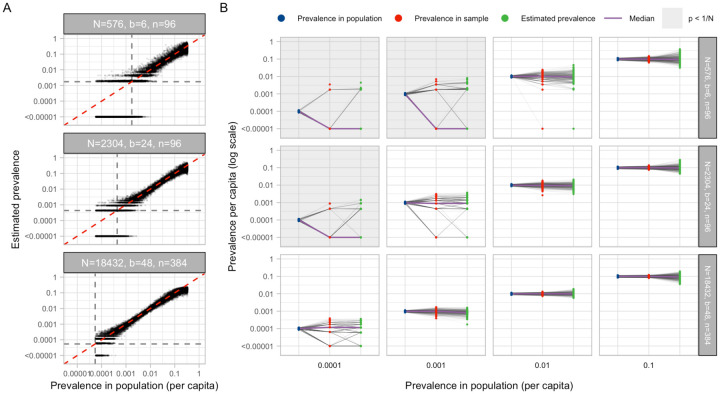
We developed a statistical method to estimate prevalence of SARS-CoV-2 based on cycle threshold (Ct) values measured from pooled samples (Materials and Methods), potentially using far fewer tests than would be required to assess prevalence based on number of positive samples identified. We used our synthetic viral load data to assess inferential accuracy under a range of sample availabilities and pooling designs. Across the spectrum of simulated pools and tests we found that simple pooling allows accurate estimates of prevalence across at least four orders of magnitude, ranging from 0.02% to 20%, with up to 400-times efficiency gains (i.e., 400 times fewer tests) than would be needed without pooling (Fig. 3). For example, in a population prevalence study that collects ~2,000 samples, we accurately estimated infection prevalences as low as 0.05% by using only 24 total qPCR tests (i.e., 24 pools of 96 samples each; Fig. 3A; Fig. S1). Importantly, because the distribution of Ct values may differ depending on the sample type (sputum vs. swab), the instrument, and the phase of the epidemic (growth vs. decline, Fig. S2), in practice, the method should be calibrated to viral load data (i.e., Ct values) specific to the laboratory and instrument (which can differ from one laboratory to the next) and the population under investigation.
Fig. 3: Estimating prevalence from a small number of pooled tests.
In prevalence estimation, a total of N individuals are sampled and partitioned into b pools (with n=N/b samples per pool). The true prevalence in the entire population (x-axis in A) varies over time with epidemic spread. Population prevalences shown here are during the epidemic growth phase. (A) Estimated prevalence against true population prevalence using 100 independent trials sampling N individuals at each day of the epidemic. Each facet shows a different pooling design (more pooling designs shown in Fig. S1). Dashed grey lines show one divided by the sample size, N. (B) For a given true prevalence (x-axis, blue points), estimation error is introduced both through binomial sampling of positive samples (red points) and inference on the sampled viral loads (green points). Sampling variation is a bigger contributor at low prevalence and low sample sizes. When prevalence is less than one divided by N (grey boxes), inference is less accurate due to the high probability of sampling only negative individuals or inclusion of false positives.
Estimation error arises in two stages: sample collection effects, and as part of the inference method (Fig. 3B). Error from sampling collection became less important with increasing numbers of positive samples, which occurred with increasing population prevalence or by increasing the total number of tested samples (Fig. 3B; Fig. S2). At very low prevalence, small sample sizes (N) risk missing positives altogether or becoming biased by false positives. We found that accuracy in prevalence estimation is greatest when population prevalence is greater than 1/N and that when this condition was met, partitioning samples into more pools always improves accuracy (Fig. S2). In summary, very accurate estimates of prevalence can be attained using only a small fraction of the tests that would be needed in the absence of pooling.
Pooled testing for individual identification
We next analyzed effectiveness of group testing for identifying individual sample results at different points along the epidemic curve with the aim of identifying simple, efficient pooling strategies that are robust to a range of prevalences (Fig. 1A&B). Using the simulated viral load data described in Materials and Methods, we evaluated a large array of pooling designs in silico (Table S1). Based on our models of viral kinetics and given a PCR limit of detection of 100 viral copies per ml, we first estimated a baseline sensitivity of conventional (non-pooled) PCR testing of 85% during the epidemic growth phase (i.e., 15% of the time we sample an infected individual with a viral load greater than 1 but below the LOD of 100 viral copies per mL, Fig. 4A), which largely agrees with reported estimates. (21,22) This reflects sampling during the latent period of the virus (after infection but prior to significant viral growth) or in the relatively long duration of low viral titers during viral clearance.
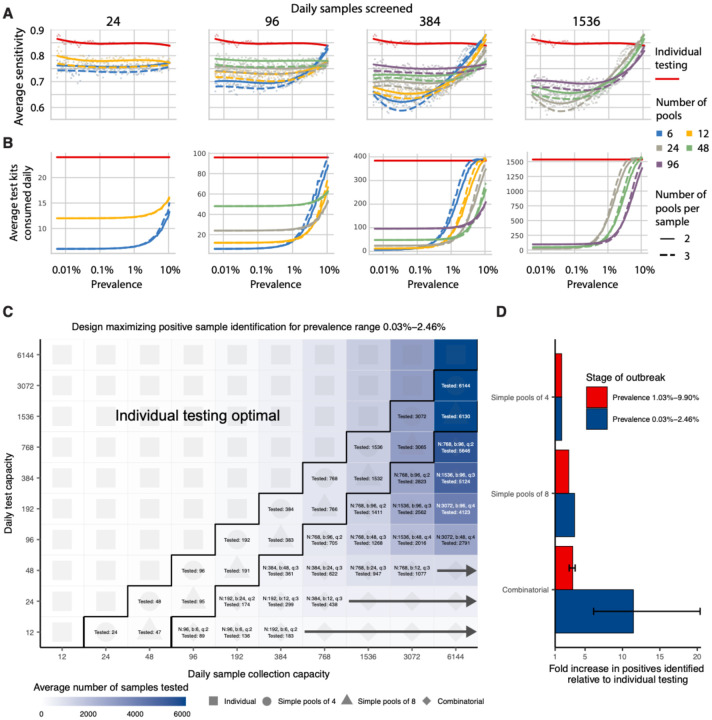
Fig. 4: Group testing for sample identification.
We evaluated a variety of group testing designs for sample identification (Table S1) on the basis of sensitivity (A), efficiency (B), total number of positive samples identified (C) and the fold increase in positive samples identified relative to individual testing (D). (A and B) The average sensitivity (A, y-axis, individual points and spline) and average number of tests needed to identify individual positive samples (B, y-axis) using different pooling designs (individual lines) were measured over days 20–110 in our simulated population, with results plotted against prevalence (x-axis, log-scale). Results show the average of 200,000 trials, with individuals selected at random on each day in each trial. Pooling designs are separated by the number of samples tested on a daily basis (individual panels); the number of pools (color); and the number of pools into which each sample is split (dashed versus solid line). Solid red line indicates results for individual testing. (C) Every design was evaluated under constraints on the maximum number of samples collected (columns) and average number of reactions that can be run on a daily basis (rows) over days 40–90. Text in each box indicates the optimal design for a given set of constraints (number of samples per batch (N), number of pools (b), number of pools into which each sample is split (q), average number of total samples screened per day). Color indicates the average number of samples screened on a daily basis using the optimal design. Arrows indicate that the same pooling design is optimal at higher sample collection capacities due to testing constraints. (D) Fold increase in the number of positive samples identified relative to individual testing with the same resource constraints. Error bar shows range amongst optimal designs.
Sensitivity of pooled tests, relative to individual testing, is affected by the dilution factor of pooling and by the population prevalence - with lower prevalence resulting in generally lower sensitivity as positives are diluted into many negatives (Fig. 4A). The decrease in sensitivity is roughly linear with the log of the dilution factor employed, which largely depends on the number and size of the pools and, for combinatorial pooling, the number of pools that each sample is placed into (Fig. S3A–C).
There is a less intuitive relationship between sensitivity and prevalence as it changes over the course of the epidemic. Early in an epidemic there is an initial dip in sensitivity for both individual and pooled testing (Fig. 4A). Early during exponential growth of an outbreak, a random sample of infected individuals will be sampled closer to their peak viral load, while later on there is an increasing mixture of newly infected with individuals with lower viral loads at the tail end of their infection. We found that this means at peak prevalence, sensitivity of pooled testing increases as samples with lower viral loads, which would otherwise be missed due to dilution, are more likely to be ‘rescued’ by coexisting in the same pool with high viral load samples and thus get individually retested (at their undiluted concentration) during the validation stage. During epidemic decline, fewer new infections arise over time and therefore a randomly selected infected individual is more likely to be sampled during the recovery phase of their infection, when viral loads are lower (Fig. S4D). Overall sensitivity is therefore lower during epidemic decline, as more infected individuals have viral loads below the limit of detection; during epidemic growth (up to day 108), overall sensitivity of RT-PCR for individual testing is 85%, whereas during epidemic decline (from day 168 onward) it is 60% (Fig. S5A). Sensitivity of RT-PCR for individual testing was ~75% across the whole epidemic. We note that in practice, sensitivity is likely higher than estimated here, because individuals are not sampled entirely at random. Together, these results describe how sensitivity is affected by the combination of epidemic dynamics, viral kinetics, and pooling design when individuals are sampled randomly from the population.
We find that on average the majority of false negatives arise from individuals sampled seven days or more after their peak viral loads, or around seven days after what is normally considered symptom onset (~75% in swab samples during epidemic growth; ~96% in swab samples during epidemic decline; ~68% in sputum during epidemic growth). Importantly, only ~3% of false negative swab samples arose from individuals tested during the first week following peak viral load during epidemic growth, and only ~1% during epidemic decline - (peak titers usually coincide with symptom onset) – and thus most false negatives are from individuals with the least risk of onward transmission (Fig. S3D&E).
As mentioned above, the lower sensitivity of pooled testing is counterbalanced by gains in efficiency. When prevalence is low, efficiency is roughly the number of samples divided by the number of pools, since there are rarely putative positives to test individually. However, the number of validation tests required will increase as prevalence increases, and designs that are initially more efficient will lose efficiency (Fig. 4B). In general, we find that at very low population prevalence the use of fewer pools each with larger numbers of specimens offers relative efficiency gains compared to larger numbers of pools, as the majority of pools will test negative. However, as prevalence increases, testing a greater number of smaller pools pays off as more validations will be performed on fewer samples overall (Fig. 4B). For combinatorial designs with a given number of total samples and pools, splitting each sample across fewer pools results in a modest efficiency gains (dashed versus solid lines in Fig. 4B).
To address realistic resource constraints, we integrated our analyses of sensitivity and efficiency with limits on daily sample collection and testing capacity to maximize the number of positive individuals identified (see Materials and Methods). We analyzed the total number of samples screened and the fold increase in the number of positive samples identified relative to individual testing for a wide array of pooling designs evaluated over a period of 50 days during epidemic spread (days 40–90 where point prevalence reaches ~2.5%; Fig. 4C&D). Because prevalence changes over time, the number of validation tests may vary each day despite constant pooling strategies. Thus, tests saved on days requiring fewer validation tests can be stored for days where more validation tests are required.
Across all resource constraints considered, we found that effectiveness ranged from one (when testing every sample individually is optimal) to 20 (i.e., identifying 20x more positive samples on a daily basis compared with individual testing within the same budget; Fig. 4D). As expected, when capacity to collect samples exceeds capacity to test, group testing becomes increasingly effective. Simple pooling designs are most effective when samples are in slight excess of testing capacity (2–8x), whereas we find that increasingly complex combinatorial designs become the most effective when the number of samples to be tested greatly exceeds testing capacity. Additionally, when prevalence is higher (i.e., sample prevalence from 1.03% to 9.90%), the optimal pooling designs shift towards combinatorial pooling, and the overall effectiveness decreases - but still remains up to 4x more effective than individual testing (Fig. S6). Our results were qualitatively unchanged when evaluating the effectiveness of pooling sputum samples, and the optimal pooling designs under each set of sample constraints were either the same or very similar (Fig. S7). Furthermore, we evaluated the same strategies during a 50-day window of epidemic decline (days 190–250) and found that similar pooling strategies were optimally effective, despite lower overall sensitivity as described above (Fig. S5).
Pilot and validation experiments
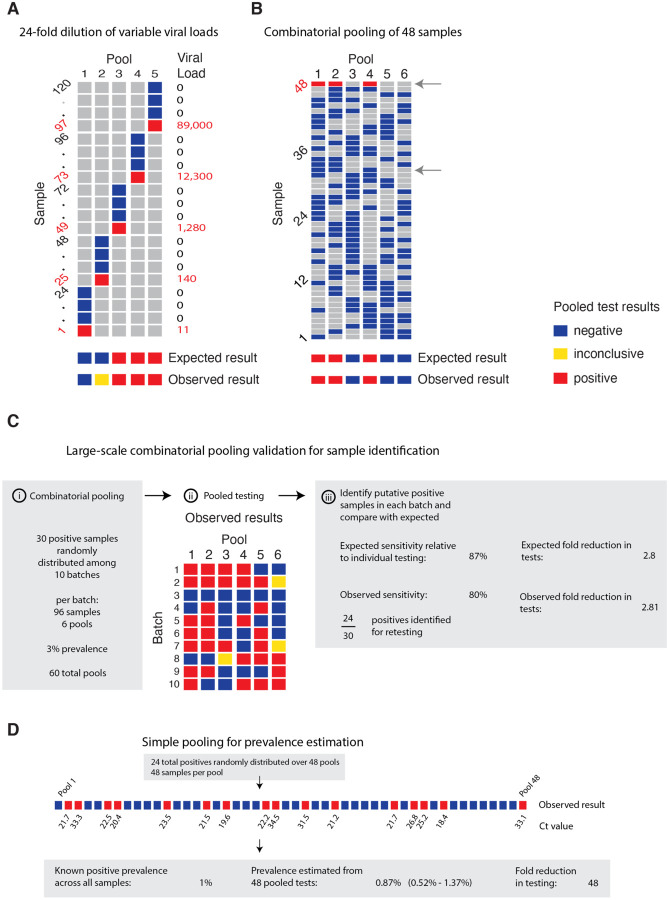
We validated our pooling strategies using anonymized clinical nasopharyngeal swab specimens. To evaluate simple pooling across a range of inputs, we diluted 5 nasopharyngeal clinical swab samples with viral loads of 89000, 12300, 1280, 140 and 11 viral copies per ml, respectively, into 23 negative nasopharyngeal swab samples (pools of 24). Further details are provided in Materials and Methods, and Supplementary Material 1, sections 7&8. The results matched the simulated sampling results: the first three pools were all positive, the fourth was inconclusive (negative on N1, positive on N2), and the remaining pool was negative (Fig. 5A, Table S2). These results are as expected because the EUA approved assay used has a limit of detection of ~100 virus copies per ml, such that the last two specimens fall below the limit of detection given a dilution factor of 24 (i.e. 0.46 and 5.8 virus copies per ml once pooled).
Fig. 5: Validation of simple and combinatorial pooling.
Pooled testing of samples was validated experimentally. (A) Five pools (columns of matrix), each consisting of 24 nasopharyngeal swab samples (rows of matrix; 23 negative samples per pool and 1 positive, with viral load indicated in red on right) were tested by viral extraction and RT+qPCR. Pooled results indicated as: negative (blue), inconclusive (yellow), or positive (red). (B) Six combinatorial pools (columns) of 48 samples (rows; 47 negative and 1 positive with viral load of 12,300) were tested as above. Pools 1, 2, and 4 tested positive. Arrows indicate two samples that were in pools 1, 2, and 4: sample 32 (negative), and sample 48 (positive). (C) Previously tested de-identified samples were pooled using a combinatorial design with 96 samples, 6 pools, and 2 pools per sample. 30 positive samples were randomly distributed across 10 batches of the design. Viral RNA extraction and RT-qPCR were performed on each pool, with the results used to identify potentially positive samples. (D) Samples were pooled according to a simple design (48 pools with 48 samples per pool). 24 positive samples were randomly distributed among the pools (establishing a 1% prevalence). The pooled test results were used with an MLE procedure to estimate prevalence (0.87%), and bootstrapping was used to estimate a 95% confidence interval (0.52% – 1.37%).
We next tested combinatorial pooling, first using only a modest pooling design. We split 48 samples, including 1 positive, into 6 pools with each sample spread across three different pools. The method correctly identified the three pools containing the positive specimen (Fig. 5B, Table S2). One negative sample was included in the same 3 pools as the positive sample; thus, 8 total tests (6 pools + 2 validations) were needed to accurately identify the status of all 48 samples, a 6x efficiency gain, which matched our expectations from the simulations.
We next performed two larger validation studies (Materials and Methods, and Supplementary Material 1, section 8). To validate combinatorial pooling, we used anonymized samples representing 930 negative and 30 distinct positive specimens (3.1% prevalence), split across 10 batches of 96 specimens each (Table S3). For each batch of 96, we split the specimens into 6 pools and each specimen was spread across 2 pools (Fig. 5C, Table S4). For this combinatorial pooling design and prevalence, our simulations suggest that we would expect to identify 26 out of 30 known positives (87%) and would see a 2.81x efficiency gain - using only 35% of the number of tests compared to no pooling. We identified 24 of the 30 known positives (80%) and, indeed, required 35% fewer tests (341 vs 960, a 2.8x efficiency gain).
To further validate our methods for prevalence estimation, we created a large study representing 2,304 samples with a (true) positive prevalence of 1%. We aimed to determine how well our methods would work to estimate the true prevalence using 1/48th the number of tests compared to testing samples individually. To do this, we randomly assigned 24 distinct positive samples into 48 pools, with each pool containing 48 samples (Table S3; to create the full set of pools, we treated some known negatives as distinct samples across separate pools). By using the measured viral loads detected in each of the pools, our methods estimated a prevalence of 0.87% (compared to the true prevalence of 1%) with a bootstrapped 95% confidence interval of 0.52% – 1.37% (Fig. 5D), and did so using 48x fewer tests than without pooling. This level of accuracy is in line with our expectations from our simulations. Notably, the inference algorithm applied to these data used viral load distributions calibrated from our simulated epidemic, which in turn had viral kinetics calibrated to samples collected and tested on another continent, demonstrating robustness of the training procedure.
Discussion
Our results show that group testing for SARS-CoV-2 can be a highly effective tool to increase surveillance coverage and capacity when resources are constrained. For prevalence testing, we find that fewer than 40 tests can be used to accurately infer prevalences across four orders of magnitude, providing large savings on tests required. For individual identification, we determined an array of designs that optimize the rate at which infected individuals are identified under constraints on sample collection and daily test capacities. These results provide pooling designs that maximize the number of positive individuals identified on a daily basis, while accounting for epidemic dynamics, viral kinetics, viral loads measured from nasopharyngeal swabs or sputum, and practical considerations of laboratory capacity.
While our experiments suggest that pooling designs may be beneficial for SARS-CoV-2 surveillance and identification of individual specimens, there are substantial logistical challenges to implementing theoretically optimized pooling designs. Large-scale testing without the use of pooling already requires managing thousands of specimens per day, largely in series. Pooling adds complexity because samples must be tracked across multiple pools and stored for potential re-testing. These complexities can be overcome with proper tracking software (including simple spreadsheets) and standard operating procedures in place before pooling begins. Such procedures can mitigate the risk of handling error or specimen mix-up. In addition, expecting laboratories to regularly adapt their workflow and optimize pool sizes based on prevalence may not be feasible in some settings. (8,16) A potential solution is to follow a simple, fixed protocol that is robust to a range of prevalences. Supplementary Material 2 provides an example spreadsheet guiding a technician receiving 96 labeled samples to create 6 pools, enter the result of each pool and be provided a list of putative positives to be retested.
Enhancing efficiency at the expense of sensitivity must be considered depending on the purpose of testing. For prevalence testing, accurate estimates can be obtained using very few tests if individual identification is not the aim. For individual testing, although identifying all positive samples that are tested is of course the objective, increasing the number of specimens tested when sacrificing sensitivity may be a crucially important tradeoff. This tradeoff is particularly pertinent because the specimens most likely to be lost due to dilution are those samples with the lowest viral loads already near the limit of detection (Fig. S3D&E). Although there is a chance that the low viral load samples missed are on the upswing of an infection - when identifying the individual would be maximally beneficial - the asymmetric course of viral titers over the full duration of positivity means that most false negatives would arise from failure to detect late-stage, low-titer individuals who are less likely to be infectious. (20) Optimal strategies and expectations of sensitivity should also be considered alongside the phase of the epidemic and how samples are collected, as this will dictate the distribution of sampled viral loads. For example, if individuals are under a regular testing regimen or are tested due to recent exposure or symptom onset, then viral loads at the time of sampling will typically be higher, leading to higher sensitivity in spite of dilution effects.
Testing throughput and staffing resources should also be considered. If a testing facility can only run a limited number of tests per day, it may be preferable to process more samples at a slight cost to sensitivity. Back-logs of individual testing can result in substantial delays in returning individual test results, which can ultimately defeat the purpose of identifying individuals for isolation - potentially further justifying some sensitivity losses. (23,24) Choosing a pooling strategy will therefore depend on target population and availability of resources. For testing in the community or in existing sentinel surveillance populations (e.g., antenatal clinics), point prevalence is likely to be low (<0.1 – 3%), which may favor strategies with fewer pools. (6,25–27) Conversely, secondary attack rates in contacts of index cases may vary from <1% to 17% depending on the setting (e.g., casual vs. household contacts), (28–30) and may be even higher in some instances, (31,32) favoring more pools. These high point prevalence sub-populations may represent less efficient use cases for pooled testing, as our results suggest that pooling for individual identification is inefficient once prevalence reaches 10%. However, group testing may still be useful if testing capacity is severely limited - for example, samples from all members of a household could be tested as a pool and quarantined if tested positive, enabling faster turnaround than testing individuals. This approach may be even more efficient if samples can be pooled at the point of collection, requiring no change to laboratory protocols.
Our modelling results have a number of limitations and may be updated as more data become available. First, our simulation results depend on the generalizability of the simulated Ct values, which were based on viral load data from symptomatic patients. Although some features of viral trajectories, such as viral waning, differ between symptomatic and asymptomatic individuals, population-wide data suggest that the range of Ct values do not differ based on symptom status. (20,33) Furthermore, we have assumed a simple hinge function to describe viral kinetics. Different shapes for the viral kinetics trajectory may become apparent as more data become available. Nonetheless, our simulated population distribution of Ct values is comparable to existing data and we propagated substantial uncertainty in viral kinetics parameters to generate a wide range of viral trajectories. For prevalence estimation, the MLE framework requires training on a distribution of Ct values. Such data can be available based on past tests from a given laboratory, but care should be taken to use a distribution appropriate for the population under consideration. For example, training the virus kinetics model on data skewed towards lower viral loads (as would be observed during the tail end of an epidemic curve) may be inappropriate when the true viral load distribution is skewed higher (as might be the case during the growth phase of an epidemic curve). Nevertheless, we used our simulated distribution of Ct, which were fit to virus kinetics in published reports from distinct labs from across the world and obtained highly accurate results throughout. Thus, despite the limitations just mentioned, this shows that the virus kinetics models are quite robust and may not, in practice, require new fitting to the individual laboratory or population. In addition, while we assume that individuals are sampled from the population at random in our analysis, in practice samples that are processed together are also typically collected together, which may bias the distribution of positive samples among pools.
We have shown that simple designs that are straightforward to implement have the potential to greatly improve testing throughput across the time course of the pandemic. These principles likely also hold for pooling of sera for antibody testing, which remains an avenue for future work. There are logistical challenges and additional costs associated with pooling that we do not consider deeply here, and it will therefore be up to laboratories and policy makers to decide where these designs are feasible. Substantial coordination will therefore be necessary to make group testing practical but investing in these efforts could enable community screening where it is currently infeasible and provide epidemiological insights that are urgently needed.
Materials and Methods
Simulation model of infection dynamics and viral load kinetics
We developed a population-level mathematical model of SARS-CoV-2 transmission that incorporates realistic within-host virus kinetics. Full details are provided in Supplementary Material 1, sections 1–4, but we provide an overview here. First, we fit a viral kinetics model to published longitudinally collected viral load data from nasopharyngeal swab and sputum samples using a Bayesian hierarchical model that captures the variation of peak viral loads, delays from infection to peak and virus decline rates across infected individuals (Fig. 5A&B; Fig. S8). (19) By incorporating estimated biological variation in virus kinetics, this model allows random draws each representing distinct within-host virus trajectories. We then simulated infection prevalence during a SARS-CoV-2 outbreak using a deterministic Susceptible-Exposed-Infected-Removed (SEIR) model with parameters reflecting the natural history of SARS-CoV-2 (Fig. 5D). For each simulated infection, we generated longitudinal virus titers over time by drawing from the distribution of fitted virus kinetic curves, using distributions derived using either nasopharyngeal swab or sputum data (Fig. S4). All estimated and assumed model parameters are shown in Table S5, with model fits shown in Fig. S8. Posterior estimates and Markov chain Monte Carlo trace plots are shown in Fig. S9 and Fig. S10. We accounted for measurement variation by: i) transforming viral loads into Ct values under a range of Ct calibration curves, ii) simulating false positives with 1% probability, and, importantly, iii) simulating sampling variation. We assumed a limit of detection (LOD) of 100 RNA copies / ml.
Estimating prevalence from pooled test results
We adapted a statistical (maximum likelihood) framework initially developed to estimate HIV prevalence with pooled antibody tests to estimate prevalence of SARS-CoV-2 using pooled samples. (12,13) The framework accounts for the distribution of viral loads (and uncertainty around them) measured in pools containing a mixture of negative and potentially positive samples. By measuring viral loads from multiple such pools, it is possible to estimate the prevalence of positive samples without individual testing. See Supplementary Material 1, section 5 for full details.
We evaluated prevalence estimation under a range of sample availabilities (N total samples; N=288 to ~18,000) and pooling designs. We varied the pool size of combined specimens (n samples per pool; n=48, 96, 192, or 384) and the number of pools (b=6, 12, 24, or 48). For each combination, we estimated the point prevalence from pooled tests on random samples of individuals drawn during epidemic growth (days 20–120) and decline (days 155–300). Because the data is realistic but simulated, we used ground truth prevalence in the population and, separately, in the specific set of samples collected from the overall population to assess accuracy of our estimates (see for example Fig. 3B). We calculated estimates for 100 entirely distinct epidemic simulations.
Pooled tests for individual sample identification
Using the same simulated population, we evaluated a range of simple and combinatorial pooling strategies for individual positive sample identification (Supplementary Material 1, section 6). In simple pooling designs, each sample is placed in one pool, and each pool consists of some pre-specified number of samples. If a pool tests positive, all samples that were placed in that pool are retested individually (Fig. 1A). For combinatorial pooling, each sample is split into multiple, partially overlapping pools (Fig. 1B). (9,10) Every sample that was placed in any pool that tested negative is inferred to be negative, and the remaining samples are identified as potential positives. Here, we consider a very simple form of combinatorial testing, where identified potential positive samples are individually tested in a validation stage.
A given pooling design is defined by three parameters: the total number of individuals to be tested (N); the total number of pools to test (b); and the number of pools a given sample is included in (q). For instance, if we have 50 individuals (N) to test, we might split the 50 samples into four pools (b) of 25 samples each, where each sample is included in two pools. Note that, by definition, in simple pooling designs each sample is placed in one pool (q=1).
To identify optimal testing designs under different resource constraints, we systematically analyzed a large array of pooling designs under various sample and test kit availabilities. We evaluated different combinations of between 12 and ~6,000 available samples/tests per day. The daily testing capacity shown is the daily average, though we assume that there is some flexibility to use fewer or more tests day to day (i.e., there is a budget for period of time under evaluation).
For each set of resource constraints, we evaluated designs that split N samples between 1 to 96 distinct pools, and with samples included in q=1 (simple pooling), 2, 3, or 4 (combinatorial pooling) pools (Table S1). To ensure robust estimates (especially at low prevalences of less than 1 in 10,000), we repeated each simulated pooling protocol at each time point in the epidemic up to 200,000 times.
In each scenario, we calculated: i) the sensitivity to detect positive samples when they existed in the pool; ii) the efficiency, defined as the total number of samples tested divided by the total number of tests used; iii) the total number of identified true positives (total recall); and iv) the effectiveness, defined as the total recall relative to individual testing.
Pilot experiments
For validation experiments of our simulation-based approach, we used fully de-identified, discarded human nasopharyngeal specimens obtained from the Broad Institute of MIT and Harvard. In each experiment, sample aliquots were pooled before RNA extraction and qPCR and pooled specimens were tested using the Emergency Use Authorization (EUA) approved SARS-CoV-2 assay performed by the Broad Institute CLIA laboratory. The protocol is described in full detail in Supplementary Material 1, section 7. Specifics of each pooling approach is detailed alongside the relevant results and in Supplementary Material 1, section 8.
Supplementary Material
Acknowledgements:
We thank Benedicte Gnangnon, Rounak Dey, Xihong Lin, Edgar Doriban, Rene Niehus and Heather Shakerchi for useful discussions. Work was supported by the Merkin Institute Fellowship at the Broad Institute of MIT and Harvard (B.C.), by the National Institute of General Medical Sciences (#U54GM088558; J.H. and M.M.); and by an NIH DP5 grant (M.M.), and the Dean’s Fund for Postdoctoral Research of the Wharton School and NSF BIGDATA grant IIS 1837992 (D.H.).
Footnotes
Competing interests: A.R. is a co-founder and equity holder of Celsius Therapeutics, an equity holder in Immunitas, and until July 31, 2020, was an SAB member of ThermoFisher Scientific, Syros Pharmaceuticals, Asimov, and Neogene Therapeutics. From August 1, 2020, A.R. is an employee of Genentech.
Code availability: The SEIR model, viral kinetics model and MCMC were implemented in R version 3.6.2. The remainder of the work was performed in Python version 3.7. The code used for the MCMC framework is available at: https://github.com/jameshay218/lazymcmc. All other code and data used are available at: https://github.com/cleary-lab/covid19-group-tests.
References
- 1.Coronavirus disease (COVID-19) Weekly Epidemiological Update Global epidemiological situation. [cited 2020 Aug 28]. Available from: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200824-weekly-epi-update.pdf?sfvrsn=806986d1_4
- 2.World Health Organization (WHO). Critical preparedness, readiness and response actions for COVID-19. World Heal Organ. 2020; [Google Scholar]
- 3.Cheng MP, Papenburg J, Desjardins M, Kanjilal S, Quach C, Libman M, et al. Diagnostic Testing for Severe Acute Respiratory Syndrome–Related Coronavirus-2. Ann Intern Med. 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoo JH, Chung MS, Kim JY, Ko JH, Kim Y, Kim YJ, Kim JM, Chung YS, Kim HM, Han MG KSY. Report on the epidemiological features of coronavirus disease 2019 (covid-19) outbreak in the republic of korea from january 19 to march 2, 2020. J Korean Med Sci. 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee VJ, Chiew CJ, Khong WX. Interrupting transmission of COVID-19: lessons from containment efforts in Singapore. J Travel Med. 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gudbjartsson DF, Helgason A, Jonsson H, Magnusson OT, Melsted P, Norddahl GL, et al. Spread of SARS-CoV-2 in the Icelandic Population. N Engl J Med. 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grassly NC, Pons-Salort M, Parker EPK, White PJ, Ferguson NM, Ainslie K, et al. Comparison of molecular testing strategies for COVID-19 control: a mathematical modelling study. Lancet Infect Dis. 2020. August [cited 2020 Aug 20];0(0). Available from: https://linkinghub.elsevier.com/retrieve/pii/S1473309920306307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorfman R. The Detection of Defective Members of Large Populations. Ann Math Stat. 1943; [Google Scholar]
- 9.Du D, Hwang FK, Hwang F. Combinatorial group testing and its applications. Vol. 12 World Scientific; 2000. [Google Scholar]
- 10.Porat E, Rothschild A. Explicit non-adaptive combinatorial group testing schemes. In: Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). 2008. [Google Scholar]
- 11.Centre for Disease Prevention E. Methodology for estimating point prevalence of SARS-CoV-2 infection by pooled RT-PCR testing. 2020. [cited 2020 Aug 28]. Available from: https://www.ecdc.europa.eu/en/publications-data/methodology-estimating-point-prevalence-sars-cov-2-infection-pooled-rt-pcr
- 12.Zenios SA, Wein LM. Pooled testing for HIV prevalence estimation: Exploiting the dilution effect Vol. 17, Statistics in Medicine. John Wiley & Sons, Ltd; 1998. [cited 2020 Aug 14]. p. 1447–67. Available from: https://onlinelibrary.wiley.com/doi/full/10.1002/%28SICI%291097-0258%2819980715%2917%3A13%3C1447%3A%3AAID-SIM862%3E3.0.CO%3B2-K [DOI] [PubMed] [Google Scholar]
- 13.Wein LM, Zenios SA. Pooled Testing for HIV Screening: Capturing the Dilution Effect Vol. 44, Operations Research. INFORMS; [cited 2020 Aug 14]. p. 543–69. Available from: https://www.jstor.org/stable/171999 [Google Scholar]
- 14.Hogan CA, Sahoo MK, Pinsky BA. Sample Pooling as a Strategy to Detect Community Transmission of SARS-CoV-2. JAMA. 2020. May 19 [cited 2020 Jul 3];323(19):1967 Available from: https://jamanetwork.com/journals/jama/fullarticle/2764364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bilder CR, Iwen PC, Abdalhamid B, Tebbs JM, McMahan CS. Tests in short supply? Try group testing. Significance (Oxford, England). 2020. June [cited 2020 Jul 8];17(3):15–6. Available from: http://www.ncbi.nlm.nih.gov/pubmed/32536952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ben-Ami R, Klochendler A, Seidel M, Sido T, Gurel-Gurevich O, Yassour M, et al. Large-scale implementation of pooled RNA extraction and RT-PCR for SARS-CoV-2 detection. Clin Microbiol Infect. 2020. September 1;26(9):1248–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shental N, Levy S, Wuvshet V, Skorniakov S, Shalem B, Ottolenghi A, et al. Efficient high-throughput SARS-CoV-2 testing to detect asymptomatic carriers. Sci Adv. 2020. August 21 [cited 2020 Aug 28];eabc5961 Available from: https://advances.sciencemag.org/lookup/doi/10.1126/sciadv.abc5961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. New England Journal of Medicine. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020; [DOI] [PubMed] [Google Scholar]
- 20.Cevik M, Tate M, Lloyd O, Maraolo AE, Schafers J, Ho A. SARS-CoV-2 viral load dynamics, duration of viral shedding and infectiousness: a living systematic review and meta-analysis. medRxiv. 2020. July 29 [cited 2020 Aug 28];pre-print:2020.07.25.20162107. Available from: http://medrxiv.org/content/early/2020/07/28/2020.07.25.20162107.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams TC, Wastnedge E, McAllister G, Bhatia R, Cuschieri K, Kefala K, et al. Sensitivity of RT-PCR testing of upper respiratory tract samples for SARS-CoV-2 in hospitalised patients: a retrospective cohort study. medRxiv. 2020. June 20 [cited 2020 Aug 28];2020.06.19.20135756. Available from: http://medrxiv.org/content/early/2020/06/20/2020.06.19.20135756.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arevalo-Rodriguez I, Buitrago-Garcia D, Simancas-Racines D, Zambrano-Achig P, del Campo R, Ciapponi A, et al. FALSE-NEGATIVE RESULTS OF INITIAL RT-PCR ASSAYS FOR COVID-19: A SYSTEMATIC REVIEW. medRxiv. 2020. August 13 [cited 2020 Sep 25];2020.04.16.20066787. Available from: 10.1101/2020.04.16.20066787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larremore DB, Wilder B, Lester E, Shehata S, Burke JM, Hay JA, et al. Test sensitivity is secondary to frequency and turnaround time for COVID-19 surveillance. medRxiv. 2020. June 27 [cited 2020 Aug 28];2020.06.22.20136309. Available from: https://www.medrxiv.org/content/10.1101/2020.06.22.20136309v2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paltiel AD, Zheng A, Walensky RP. Assessment of SARS-CoV-2 Screening Strategies to Permit the Safe Reopening of College Campuses in the United States. JAMA Netw open. 2020. July 1 [cited 2020 Aug 28];3(7):e2016818 Available from: https://jamanetwork.com/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riley S, Ainslie K, Eales O, Jeffrey B, Walters C, Atchison C, et al. Community prevalence of SARS-CoV-2 virus in England during May 2020: REACT study. medRxiv. 2020. July 11 [cited 2020 Aug 28];2020.07.10.20150524. Available from: 10.1101/2020.07.10.20150524 [DOI] [Google Scholar]
- 26.Pouwels KB, House T, Robotham J V, Birrell P, Gelman AB, Bowers N, et al. Community prevalence of SARS-CoV-2 in England: Results from the ONS Coronavirus Infection Survey Pilot. medRxiv. 2020. July 7 [cited 2020 Jul 8];2020.07.06.20147348. Available from: https://www.medrxiv.org/content/10.1101/2020.07.06.20147348v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavezzo E, Franchin E, Ciavarella C, Cuomo-Dannenburg G, Barzon L, Del Vecchio C, et al. Suppression of a SARS-CoV-2 outbreak in the Italian municipality of Vo’. Nature. 2020. June 30 [cited 2020 Aug 28];584(7821):425 Available from: 10.1038/s41586-020-2488-1 [DOI] [PubMed] [Google Scholar]
- 28.Jing Q-L, Liu M-J, Zhang Z-B, Fang L-Q, Yuan J, Zhang A-R, et al. Household secondary attack rate of COVID-19 and associated determinants in Guangzhou, China: a retrospective cohort study. Lancet Infect Dis. 2020. June [cited 2020 Aug 28];0(0). Available from: 10.1016/S1473-3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li W, Zhang B, Lu J, Liu S, Chang Z, Cao P, et al. The characteristics of household transmission of COVID-19. Clin Infect Dis. 2020. April 17 [cited 2020 Aug 28]; Available from: /pmc/articles/PMC7184465/?report=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Y-T, Tu Y-K, Lai P-C. Estimation of the secondary attack rate of COVID-19 using proportional meta-analysis of nationwide contact tracing data in Taiwan. J Microbiol Immunol Infect. 2020. June 11 [cited 2020 Jul 8]; Available from: http://www.ncbi.nlm.nih.gov/pubmed/32553448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, Eggo RM, Kucharski AJ. Secondary attack rate and superspreading events for SARS-CoV-2 Vol. 395, The Lancet. Lancet Publishing Group; 2020. [cited 2020 Aug 28]. p. e47 Available from: http://ees.elsevier.com/thelancet/www.thelancet.com [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamner L, Dubbel P, Capron I, Ross A, Jordan A, Lee J, et al. High SARS-CoV-2 Attack Rate Following Exposure at a Choir Practice - Skagit County, Washington, March 2020. MMWR Morb Mortal Wkly Rep. 2020. May 15 [cited 2020 Jul 8];69(19):606–10. Available from: http://www.ncbi.nlm.nih.gov/pubmed/32407303 [DOI] [PubMed] [Google Scholar]
- 33.Lennon NJ, Bhattacharyya RP, Mina MJ, Rehm HL, Hung DT, Smole S, et al. Comparison of viral levels in individuals with or without symptoms at time of COVID-19 testing among 32,480 residents and staff of nursing homes and assisted living facilities in Massachusetts. medRxiv. 2020. July 26 [cited 2020 Aug 14];2020.07.20.20157792. Available from: 10.1101/2020.07.20.20157792 [DOI] [Google Scholar]
- 34.Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020. May 28 [cited 2020 Aug 14];581(7809):465–9. Available from: 10.1038/s41586-020-2196-x [DOI] [PubMed] [Google Scholar]
- 35.Lauer SA, Grantz KH, Bi Q, Jones FK, Zheng Q, Meredith HR, et al. The Incubation Period of Coronavirus Disease 2019 (COVID-19) From Publicly Reported Confirmed Cases: Estimation and Application. Ann Intern Med. 2020. March 10 [cited 2020 Apr 3]; Available from: http://www.ncbi.nlm.nih.gov/pubmed/32150748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.lazymcmc. [cited 2018 Jun 21]. Available from: https://github.com/jameshay218/lazymcmc
- 37.Poon LLM, Chan KH, Wong OK, Cheung TKW, Ng I, Zheng B, et al. Detection of SARS Coronavirus in Patients with Severe Acute Respiratory Syndrome by Conventional and Real-Time Quantitative Reverse Transcription-PCR Assays. Clin Chem. 2004. January [cited 2020 Aug 14];50(1):67–72. Available from: https://pubmed.ncbi.nlm.nih.gov/14709637/ [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.