Abstract
Many drugs that have been proposed for treatment of COVID-19 are reported to cause cardiac adverse events, including ventricular arrhythmias. In order to properly weigh risks against potential benefits, particularly when decisions must be made quickly, mathematical modeling of both drug disposition and drug action can be useful for predicting patient response and making informed decisions. Here we explored the potential effects on cardiac electrophysiology of 4 drugs proposed to treat COVID-19: lopinavir, ritonavir, chloroquine, and azithromycin, as well as combination therapy involving these drugs. Our study combined simulations of pharmacokinetics (PK) with quantitative systems pharmacology (QSP) modeling of ventricular myocytes to predict potential cardiac adverse events caused by these treatments. Simulation results predicted that drug combinations can lead to greater cellular action potential prolongation, analogous to QT prolongation, compared with drugs given in isolation. The combination effect can result from both pharmacokinetic and pharmacodynamic drug interactions. Importantly, simulations of different patient groups predicted that females with pre-existing heart disease are especially susceptible to drug-induced arrhythmias, compared males with disease or healthy individuals of either sex. Overall, the results illustrate how PK and QSP modeling may be combined to more precisely predict cardiac risks of COVID-19 therapies.
Keywords: Adverse events, Drug-induced arrhythmias, Model Informed Drug Development, Pharmacokinetics, Quantitative Systems Pharmacology
INTRODUCTION
The rapid spread of COVID-19 worldwide, in the absence of established therapeutics, has forced clinicians to improvise by treating patients “off label’ with drugs that were approved to treat other diseases. The investigational off label use of such drugs for COVID-19 has led to the initiation of several clinical studies, generally small in scope due to the urgent nature of the health care crisis. Some of these have included drug combinations, such as lopinavir plus ritonavir1 or chloroquine plus azithromycin.2 Under the current pandemic conditions, with clinicians attempting to maintain scientific rigor while delivering treatments quickly, it can be difficult to properly weigh the potential benefits of drugs against the risks of adverse events. It can be especially challenging to determine the risks of drug combinations because specific combinations have often not been examined during preclinical or early clinical safety assessments.
Mathematical models can be used to rapidly predict the physiological effects of drug treatments, and simulations with such models gain outsize importance when time is of the essence. This can become especially true when considering drug combinations due to the complexities that are introduced when drugs are co-administered. These can include both pharmacokinetic (PK) interactions, whereby the presence of one drug alters the concentration of a second drug, and pharmacodynamic interactions, whereby the overall physiological consequences result from the combined biological effects of the two drugs. The former can be studied with pharmacometrics approaches, whereas the latter can be addressed with quantitative systems pharmacology (QSP) models that explicitly incorporate drug mechanisms of action. Although simulation results are always subject to model limitations, they can nonetheless guide decision making. Both PK and QSP simulations are likely to become particularly important for proposed COVID-19 treatments because: (1) adverse cardiac events, including QT prolongation, have been associated with several of these drugs,3, 4, 5 (2) drug disposition has previously been characterized for most of the drugs being considered for treatment;6, 7, 8 and (3) modeling of pharmacological effects on cardiac electrophysiology, including adverse events, is a mature area of research.9, 10, 11
Here we present cellular simulation results that predict an additive drug combination effect on QT prolongation, indicating that these combinations may increase the risk of ventricular arrhythmias in COVID-19 patients. The simulations further highlight the importance of sex differences and the presence of existing comorbidities such that particular drug combinations may be especially dangerous for women with heart failure. Importantly for future studies, the results demonstrate a pipeline for systematic examination and a quantitative methodology that can be used to balance the potential benefits of COVID-19 treatments against the risks of cardiac arrhythmias.
METHODS
Drugs considered and data sources
To begin to understand potential side effects caused by COVID-19 treatments, we considered 4 drugs currently under investigation: lopinavir, ritonavir, chloroquine, and azithromycin, as well as the former two and the latter two drugs in combination. We selected these 4 drugs for initial analysis because their effects on cardiac ionic currents have been assessed under standardized conditions. Crumb et al12 have reported effective free therapeutic plasma concentrations (EFTPCs) for each drug in addition to half maximal inhibitory concentration (IC50) values that indicate the affinity of each drug to block the following cardiac ionic currents: (1) late Na+ current (INaL); (2) transient outward K+ current (Ito); (3) rapid delayed rectifier K+ current (IKr); (4) slow delayed rectifier K+ current (IKs); (5) inward rectifier K+ current (IK1); (6) L-type Ca2+ current (ICaL). This unified data source was exploited for QSP model simulations of drug effects (see below).
Cardiac cellular QSP modeling of drug effects
The O’Hara et al mathematical model13 of the human endocardial ventricular myocyte was used to simulate the effects of drugs on ventricular action potentials (APs). This model, comprising a system of 41 ordinary differential equations, simulates interactions between ionic currents and Ca2+ cycling in the ventricular myocyte, and drug-induced changes to APs in models such as these are well-correlated with clinically observed changes to electrocardiographic waveforms.9, 10, 11
Block of ionic currents by particular drugs was simulated with a pore block model. With this approach, the conductance (G) of each ionic current is scaled based on drug concentration ([C]) and the IC50 value describing how the drug blocks that current, i.e.
Pharmacokinetic modeling
To link drug concentrations in the QSP simulations with free plasma drug concentrations that are likely to be observed in patients, we used PK models to simulate drug disposition of either azithromycin8 or lopinavir + ritonavir.7 These models, implemented as published, allowed us to simulate temporal changes in either total or free plasma concentrations with different dosing regimens. For lopinavir and ritonavir, we assumed an unbound fraction of 0.01 to relate total drug concentrations to free plasma concentrations that are likely to be present in patients.14 The azithromycin PK model8 calculated free drug concentrations directly. For chloroquine, which has been shown to accumulate in tissues including the heart, we extracted relevant cardiac drug concentrations from a physiologically-based PK model6 and assumed the unbound drug fraction in the heart (0.39) was identical to the fraction in plasma.15
Pharmacodynamic variability between individuals
Two factors that are known to influence the risk of drug-induced arrhythmia are female sex16 and the presence of pre-existing heart failure (HF).17 We simulated differences between male and female and between healthy and failing myocytes using established protocols. These methods, published by Yang et al18 and Gomez et al,19 respectively, consist of scaling values for ionic current maximal conductances based on measured differences in ion transport pathways between male and female hearts, or between healthy and failing myocytes. Applying these scale factors allowed us to create 4 variants of the baseline ventricular myocyte: healthy male (assumed to be the original model), healthy female, HF male, and HF female. Once baseline models for the different patient groups had been created, we generated virtual populations to simulate physiological variability between individuals by randomly varying each baseline model’s maximal conductances, as previously described.20, 21, 22
RESULTS AND DISCUSSION
We performed simulations to predict the effects on human ventricular action potentials (APs) of lopinavir, ritonavir, chloroquine, and azithromycin. Based on a comprehensive study of how various drugs affect cardiac ion channels,12 we calculated that each of the proposed COVID-19 treatments will block a slightly different complement of ionic currents (Fig. 1A). These drug-specific, concentration-dependent alterations to ionic currents were applied to a mathematical model of the human ventricular endocardial myocyte13 to predict drug-induced changes to cardiac APs. Simulations performed with each drug at 10 times the reported12 EFTPC show that all drugs can induce substantial AP prolongation at these high concentrations (Fig. 1B), with lopinavir causing the most dramatic effects. Simulations performed across drug concentrations ranging from 0.3 times to 10 times EFTPC confirm that the drugs prolong AP duration at 90% repolarization (APD90) in a concentration-dependent manner, with the largest effects occurring at the highest drug concentrations (Fig. 1C).
Figure 1:
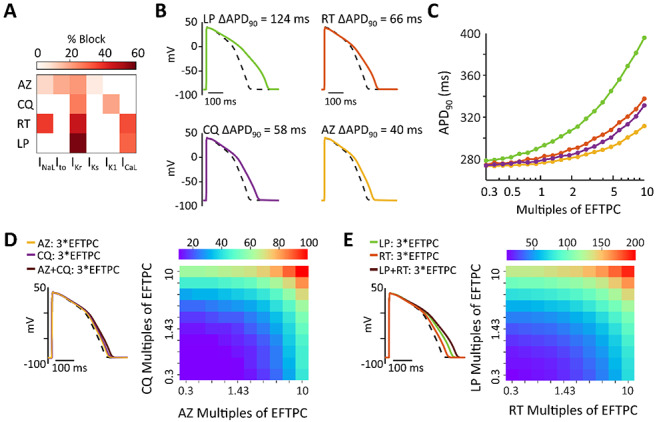
(A) Heatmap illustrating the extent to which azithromycin (AZ), chloroquine (CQ), ritonavir (RT), and lopinavir (LP) inhibit 6 important cardiac ionic currents, as previously measured.12 Reference 11 reported effective free therapeutic plasma concentration (EFTPC) of each drug, in addition to IC50 values that indicated how much each drug influenced 6 cardiac ionic currents (see Methods for abbreviations). Block of currents by particular drugs at 10*EFTPC was calculated based on drug concentration and IC50 values using a simple pore block model. (B) Simulations with the baseline myocyte model demonstrating how each simulated at 10*EFTPC are predicted to influence ventricular action potentials (APs). (C) Concentration-response curves illustrating how the 4 drugs influence APD90, the duration between the action potential upstroke (maximal rate of change of voltage) and 90% repolarization. Drug concentrations tested ranged from 0.3 times to 10 times EFTPC, with logarithmically-spaced increments. (D) Predicted AP prolongation (ΔAPD90) for chloroquine + azithromycin. (E) Predicted ΔAPD90 for lopinavir + ritonavir. Combination therapy causes greater AP prolongation than drugs applied individually, as shown in both heatmaps illustrating ΔAPD90 over a range of drug concentrations, and in example AP traces showing effects at 3*EFTPC.
Because recently-published clinical studies on COVID-19 treatments have delivered combination therapy to at least some patients,1, 2 we next simulated the combined effects of lopinavir + ritonavir1 and chloroquine + azithromycin.2 Importantly, the simulations predict that combination therapy causes a greater increase in APD90 (ΔAPD90) than does either drug in isolation. For instance (Fig.1D), at 3 times EFTPC, lopinavir and ritonavir individually produce ΔAPD90 of 50.5 and 25.2 ms, respectively, whereas ΔAPD90 is 73.9 ms for combination therapy. Similarly (Fig.1E), ΔAPD90 for chloroquine + azithromycin is larger (32.4 ms) than that produced by either drug in isolation (19.8 and 12.6 ms, respectively).
Although the mechanistic simulations indicate the possibility for pharmacodynamic interactions during combination therapy, drug concentrations in patients may not reach the levels assumed in the simulations. To more accurately incorporate clinical drug concentrations, we examined quantitative PK studies on these drugs.6, 7, 8 Simulations with a model developed for lopinavir + ritonavir therapy,7 shown in Fig 2A, predict higher lopinavir concentrations with combination therapy (400 mg/100 mg twice daily) than with lopinavir therapy alone (400 mg twice daily). This occurs due to PK interactions whereby ritonavir inhibits lopinavir clearance. Simulations were next performed in a virtual population to predict how drug plasma concentrations vary between individuals. These results suggest that, between extreme individuals in a population, peak concentrations of either drug may differ by more than 10-fold. Importantly, the results predict that plasma concentrations for lopinavir (Fig. 2B) approach IC50 values for cardiac ionic currents in some individuals whereas plasma concentrations of ritonavir (Fig. 2C) remain considerably lower than IC50 values. Predictions of chloroquine PK were less straightforward to simulate, given uncertainty in the literature about both protein binding15 and accumulation of drug in target tissues, including the heart.6 For chloroquine + azithromycin combination therapy, we therefore combined predictions of a PK model for azithromycin8 (daily dosing, 500 mg) with published concentrations from a physiologically-based PK model for chloroquine that incorporated drug accumulation in different tissues.6 These simulations (Fig. 2D) predict that azithromycin concentrations generally remain well below reported cardiac ionic current IC50 levels whereas chloroquine concentrations in the heart can exceed IC50 values and block ion channels, due to drug accumulation. To link the predictions of the PK models with QSP model simulations, we calculated the mean peak concentration of the 5% of the patients with the highest drug concentrations (highlighted in red in Fig. 2) and used those values as an input.
Figure 2:
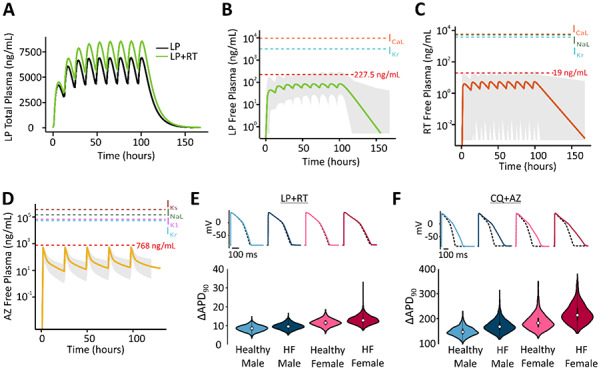
(A) Simulations show that plasma concentrations of lopinavir are greater with co-administration of ritonavir than when the former drug is given alone. (B) Predicted free plasma concentrations of lopinavir under standard dosing regimen with ritonavir, for median individual (thick line) within a virtual population of 1000 individuals (gray shaded area). Red dashed line indicates mean peak concentration of the 5% of the patients with highest drug concentrations. Additional dashed lines indicate IC50 values for cardiac ion channel inhibition. (C) Predicted free plasma concentrations of ritonavir under standard dosing regimen with lopinavir, displayed as in (B). (D) Predicted free plasma concentrations of azithromycin, displayed as in (B). (E) Distributions of ΔAPD90 caused by clinical concentrations of lopinavir + ritonavir, in 4 virtual patient populations, as indicated. (F) Distributions of ΔAPD90 caused by clinical concentrations of chloroquine + azithromycin, in 4 virtual patient populations, as indicated. The free chloroquine concentration used as an input was 3900 ng/ml based on the original publication results (total chloroquine concentrations in the heart around 10,000 ng/ml multiplied by the unbound fraction of 39%).
Finally we performed simulations to predict how sex differences and pre-existing cardiac disease may influence the AP prolongation caused by COVID-19 therapies. To do this, we created virtual populations of ventricular myocytes representing 4 patient groups: healthy males, healthy females, males with heart failure, and females with heart failure. Simulations of both lopinavir + ritonavir (Fig. 2E) and chloroquine + azithromycin (Fig. 2F), using drug concentrations from the PK simulations, predict that AP prolongation is greatest in the female heart failure group. For lopinavir + ritonavir therapy, median AP prolongation is below 20 ms in all groups, but can reach as high as 30 ms for individual cells in the female heart failure population. Chloroquine + azithromycin therapy, which produced greater AP prolongation in general, caused arrhythmic dynamics in many more cells in the female heart failure population (63 cells from a population of 1000) compared with other groups (1 cell in male healthy, 13 cells in male heart failure, 8 cells in female healthy).
Overall the simulation results indicate that proposed treatments for COVID-19 do indeed carry cardiac risk, and special caution should be exercised when developing combination therapies. With standard dosing regimens, the simulations suggest that AP prolongation is primarily driven by either lopinavir or chloroquine in the two combination therapy regimens examined, with chloroquine causing the most pronounced effects. This result is consistent with a recent small clinical trial in Brazil, which was terminated due to increased mortality in the high-dose chloroquine arm of the study.2 An improved safety profile may be a reason for clinicians to favor the chloroquine derivative hydroxychloroquine for COVID-19,23 but this drug is also associated with cardiac adverse events.3 The simulations of virtual populations suggest that women with pre-existing heart disease will be most at greatest risk of arrhythmia during treatment with these drugs. This increased vulnerability of women, combined with the generally worse prognosis for men with COVID-19,24 suggests that sex differences should be considered when weighing risks against benefits. Overall, our study indicates how simulations of both drug disposition and drug mechanisms can be used to identify patient groups at risk of adverse events, thereby helping to guide treatment decisions during this rapidly-evolving pandemic.
Acknowledgments
Sources of Funding
Research in Dr. Sobie's laboratory is supported by the National Institutes of Health (U01 HL136297, U54 HG008098), the National Science Foundation (MCB 1615677) and the US Food and Drug Administration (75F40119C10021). Meera Varshneya is supported by a fellowship from the National Heart Lung and Blood institute (F31 HL149358) and has previously been supported by a training grant from the National Institute of General Medical Sciences (T32 GM062754).
REFERENCES
- 1.Cao B, et al. A Trial of Lopinavir-Ritonavir in Adults Hospitalized with Severe Covid-19. N Engl J Med. (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borba MGS, et al. Effect of High vs Low Doses of Chloroquine Diphosphate as Adjunctive Therapy for Patients Hospitalized With Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infection: A Randomized Clinical Trial. JAMA Netw Open 3 e208857 (2020) [DOI] [PubMed] [Google Scholar]
- 3.Smith TB J; LeClaire A; Prosser T. Elsevier Clinical Solutions: COVID-19 Drug Therapy. Elsevier; website Updated April 30, 2020 https://www.elsevier.com/__data/assets/pdf_file/0007/988648/COVID-988619-Drug-Therapy_March-982020.pdf. (2020) [Google Scholar]
- 4.Saleh M, et al. The Effect of Chloroquine, Hydroxychloroquine and Azithromycin on the Corrected QT Interval in Patients with SARS-CoV-2 Infection. Circ Arrhythm Electrophysiol. (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y, Xu Q, Sun Z, Zhou L. Current targeted therapeutics against COVID-19: Based on first-line experience in China. Pharmacol Res 157 104854 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui C, et al. Dose selection of chloroquine phosphate for treatment of COVID-19 based on a physiologically based pharmacokinetic model. Acta Pharmaceutica Sinica B. (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang K, et al. Integrated population pharmacokinetic/viral dynamic modelling of lopinavir/ritonavir in HIV-1 treatment-naive patients. Clin Pharmacokinet 53 361–371. (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng S, Matzneller P, Zeitlinger M, Schmidt S. Development of a population pharmacokinetic model characterizing the tissue distribution of azithromycin in healthy subjects. Antimicrob Agents Chemother 58 6675–6684. (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lancaster MC, Sobie EA. Improved Prediction of Drug-Induced Torsades de Pointes Through Simulations of Dynamics and Machine Learning Algorithms. Clin Pharmacol Ther 100 371–379. (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Z, et al. Assessment of an In Silico Mechanistic Model for Proarrhythmia Risk Prediction Under the CiPA Initiative. Clin Pharmacol Ther 105 466–475. (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Passini E, et al. Human In Silico Drug Trials Demonstrate Higher Accuracy than Animal Models in Predicting Clinical Pro-Arrhythmic Cardiotoxicity. Front Physiol 8 668 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crumb WJ Jr., Vicente J, Johannesen L, Strauss DG. An evaluation of 30 clinical drugs against the comprehensive in vitro proarrhythmia assay (CiPA) proposed ion channel panel. J Pharmacol Toxicol Methods 81 251–262. (2016) [DOI] [PubMed] [Google Scholar]
- 13.O'Hara T, Virag L, Varro A, Rudy Y. Simulation of the undiseased human cardiac ventricular action potential: model formulation and experimental validation. PLoS Comput Biol 7 e1002061 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patterson KB, et al. Protein binding of lopinavir and ritonavir during 4 phases of pregnancy: implications for treatment guidelines. J Acquir Immune Defic Syndr 63 51–58. (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker O, Birkett DJ, Alvan G, Gustafsson LL, Sjoqvist F. Characterization of chloroquine plasma protein binding in man. Br J Clin Pharmacol 15 375–377. (1983) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Regitz-Zagrosek V, Kararigas G. Mechanistic Pathways of Sex Differences in Cardiovascular Disease. Physiol Rev 97 1–37. (2017) [DOI] [PubMed] [Google Scholar]
- 17.Kannankeril P, Roden DM, Darbar D. Drug-induced long QT syndrome. Pharmacol Rev 62 760–781. (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang PC, et al. A multiscale computational modelling approach predicts mechanisms of female sex risk in the setting of arousal-induced arrhythmias. J Physiol 595 4695–4723. (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomez JF, Cardona K, Romero L, Ferrero JM Jr., Trenor B. Electrophysiological and structural remodeling in heart failure modulate arrhythmogenesis. 1D simulation study. PLoS One 9 e106602 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Devenyi RA, Sobie EA. There and back again: Iterating between population-based modeling and experiments reveals surprising regulation of calcium transients in rat cardiac myocytes. J Mol Cell Cardiol 96 38–48. (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varshneya M, Devenyi RA, Sobie EA. Slow Delayed Rectifier Current Protects Ventricular Myocytes From Arrhythmic Dynamics Across Multiple Species. Circ Arrhythm Electrophysiol 11 e006558 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gong JQX, Sobie EA. Population-based mechanistic modeling allows for quantitative predictions of drug responses across cell types. NPJ Syst Biol Appl 4 11 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gautret P, et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents 105949 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helmy YA, et al. The COVID-19 Pandemic: A Comprehensive Review of Taxonomy, Genetics, Epidemiology, Diagnosis, Treatment, and Control. J Clin Med 9 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]


