Abstract
The ability of Escherichia coli to tolerate acid stress is important for its survival and colonization in the human digestive tract. Here, we performed adaptive laboratory evolution of the laboratory strain E. coli K-12 MG1655 at pH 5.5 in glucose minimal medium. After 800 generations, six independent populations under evolution had reached 18.0 % higher growth rates than their starting strain at pH 5.5, while maintaining comparable growth rates to the starting strain at pH 7. We characterized the evolved strains and found that: (1) whole genome sequencing of isolated clones from each evolved population revealed mutations in rpoC appearing in five of six sequenced clones; and (2) gene expression profiles revealed different strategies to mitigate acid stress, which are related to amino acid metabolism and energy production and conversion. Thus, a combination of adaptive laboratory evolution, genome resequencing and expression profiling revealed, on a genome scale, the strategies that E. coli uses to mitigate acid stress.
Keywords: adaptive laboratory evolution, acid stress, Escherichia coli, whole genome resequencing, RNA sequencing
Introduction
As a commonly found enteric bacterial species of the human digestive tract, Escherichia coli is known to withstand various levels of acid stress [1–6]. For example, E. coli can survive several hours under pH 2 [1], which is within the range of the extremely acidic stomach (pH 1.5–3) that serves as a barrier for most bacteria [7]. Additionally, E. coli has been shown to grow under mild acid stress [4–6], which is typically found in the human intestinal tract [7, 8]. Such adaptability to low pH environments has raised wide interest in understanding the underlying mechanisms that protect E. coli from acid stress. Furthermore, studying the acid resistance mechanisms of E. coli has important implications in the food and healthcare industries. For example, treatment strategies can be developed to target specific acid resistance mechanisms in a pathogenic E. coli infection.
The acid resistance mechanisms of E. coli have been studied extensively. To maintain intracellular pH homeostasis, E. coli has developed various strategies including cytoplasmic buffering [9], proton-consuming systems [10–13], adjustment of cellular metabolism [14, 15] and physiological responses [16–20]. The buffering capacity of the cytoplasm mainly comes from inorganic phosphates, amino acid side chains, polyphosphates and polyamines [9]. The proton-consuming systems include four types of amino acid decarboxylase systems that function under different pH conditions and formate hydrogen lyase that is active under anaerobic conditions [21]. The metabolic responses under acid stress include the up-regulation of components in the electron transport chain and metabolism of sugar derivatives that have decreased acid production compared to glucose [14, 15]. The physiological responses include the activation of periplasmic chaperones HdeA and HdeB [16], adjustment of membrane lipid compositions [17, 18] and blockage of outer membrane porins [19, 20].
Adaptive laboratory evolution (ALE) is an important experimental approach for understanding the adaptive response of microorganisms to particular environments or after exposure to stresses [22]. During an ALE experiment, the microorganism is cultured under defined conditions for an extended period of time. ALE allows the selection of improved phenotypes, typically growth rates, under certain growth environments. Furthermore, advances in next-generation sequencing technology have made it convenient to obtain the genotypes underlying the favourable traits over the course of evolution [23]. A previous study also investigated the adaptive evolution of E. coli under acid stress, where E. coli K-12 W3110 was evolved in a nutrient-rich environment (LBK medium) buffered at pH 4.6–4.8 for 2000 generations [24, 25]. Here, we are interested in the adaptive evolution of E. coli under acid stress in a nutrient-limited environment, where glucose is the only carbon source.
In this study, we perform ALE on E. coli K-12 MG1655 strain at pH 5.5 in glucose minimal medium. For the evolved strains, we use whole genome sequencing to identify genetic mutations that arise over the course of evolution. Additionally, to examine the change in gene activity after evolution, we perform RNA sequencing (RNA-seq) to characterize the gene expression profile of the evolved endpoints when growing under different pH conditions. We then identify the differentially expressed genes (DEGs) of the evolved endpoints at different pH conditions. We also uncover new cellular processes that emerge over the adaptive evolution under acid stress, using DEGs identified in the starting strain across pH as a reference.
Methods
Culture medium
The M9 glucose minimal medium was prepared by adding the following to Milli-Q water: 0.1 mM CaCl2, 0.2 mM MgSO4, 1× trace elements solution, 1× M9 salt solution and 4 g d-glucose l−1. Trace elements solution (4000×) was prepared in concentrated HCl with (per litre) 27 g FeCl3 · 6H2O, 1.3 g ZnCl2, 2 g CoCl2 · 6H2O, 2 g Na2MoO4 · 2H2O, 0.75 g CaCl2, 0.91 g CuCl2 · 2H2O and 0.5 g H3BO3. M9 salt solution (10×) was prepared by dissolving (per litre) 68 g Na2HPO2, 30 g KH2PO4, 5 g NaCl and 10 g NH4Cl in Milli-Q water. It is of note that the concentration of MgSO4 is 10 times lower than used previously [26], as higher concentrations of magnesium ions led to precipitation issues. To maintain the pH around 5.5 during cell culture, the culture medium was supplemented with 150 mM MES buffer from a 500 mM stock prepared in Milli-Q water. After mixing all components of the medium, the pH was adjusted using 2 M H2SO4 and 4 M KOH. All stock solutions as well as the final medium were sterile filtered through a 0.22 µm PVDF membrane.
Adaptive laboratory evolution process
Cultures were initiated from isolated colonies of an E. coli K-12 MG1655 strain (ATCC 47076), which had previously been evolved for approximately 1013 cumulative cell divisions (CCDs) on M9 minimal medium supplemented with glucose at 4 g l−1 [26]. The cultures were first grown overnight and then placed in 35 ml tubes on a platform that performed passage automatically. The tubes allowed free gas exchange with the external environment. The working culture volume was 15 ml, and the culture temperature was maintained at 37 °C. The culture medium was magnetically stirred at 1100 r.p.m. to ensure a well-mixed and aerobic growth environment.
From the start of the culture to the next passage, on average four samples of 100 μl culture medium were taken and optical density measurements at a wavelength of 600 nm (OD600) were made in a spectrophotometer (Tecan Sunrise). To maintain the cells in exponential growth phase, the culture medium containing E. coli (100 μl) was passaged to a tube containing fresh medium when the OD600 of the original medium approached 0.3. The growth rate was determined for each culture using a least-squares fit on ln(OD600) versus time. Growth trajectories were generated by fitting a monotonically increasing cubic-interpolating-spline to the calculated growth rate values versus CCDs, as described previously [26]. Glycerol stocks of the cultures were taken periodically by mixing 800 µl of sterile 50 % glycerol with 800 µl of culture and storing at −80 °C.
Throughout the course of the evolution, the culture medium pH was constantly measured to ensure proper buffering (Table S1, available in the online version of this article). Specifically, the pH values of the fresh medium and the culture medium before the next passage were measured. The culture medium was filtered through 0.22 µm membranes, and the pH was measured using a meter (Fisher Scientific Accumet AB15). Additionally, OD600 measurements of the culture medium were taken before the next passage to assess the possible effect of cell density on culture medium pH.
Whole genome sequencing and analysis of genetic mutations
Genomic DNA was isolated using bead agitation as described previously [27]. Whole genome DNA sequencing libraries were generated using a Kapa HyperPlus library prep kit (Kapa Biosystems). The libraries were then run on an Illumina HiSeq 4000 platform with a HiSeq SBS kit and 150/150 paired-end reads. The raw DNA sequencing reads in fastq format were processed using the breseq computational pipeline v0.33 [28]. Specifically, the workflow includes quality control [29], alignment to the E. coli genome (NCBI accession NC_000913.3) to identify mutations and annotation of the mutations. Note that genomic DNA was extracted for individual clones taken at different CCDs of the evolution and at the end of the evolution. The mutations identified in the clones at the end of the evolution were reported and those found at earlier stages were used to track how different mutations emerge or disappear throughout the course of the evolution.
RNA sequencing
RNA-seq data were generated from cell cultures under exponential growth phase at pH 5.5 and pH 7. The culture conditions at pH 5.5 were the same as used in ALE experiments mentioned above. The culture conditions at pH 7 were the same as the regular M9 glucose minimal medium [26], with no additional modifications to the medium. Cells were stabilized with Qiagen RNA-protect Bacteria Reagent. Cell pellets were stored at −80 °C before RNA extraction. Frozen cell pellets were then thawed and incubated with lysozyme, protease K, SuperaseIN and 20 % SDS for 30 min at 4 °C. Total RNA was isolated and purified using Qiagen’s RNeasy Plus Mini Kit based on the manufacturer's protocol. Total RNA quality was checked using the RNA 6000 Nano kit from Agilent Bioanalyzer. For gram-negative bacteria, rRNA was removed using the Ribo-Zero rRNA removal kit from Epicentre. Single-end, strand-specific RNA-seq libraries were generated using KAPA RNA HyperPrep Kit from Kapa Biosystem. RNA-seq libraries were run on an Illumina NextSeq platform using a 75 cycle mid-output kit.
Analysis of DEGs on RNA-seq data
Raw sequencing reads in fastq format were first mapped to the reference genome (NCBI accession NC_000913.3) using bowtie v1.2.2 [30]. The abundance of the transcript was obtained using the summarizeOverlaps function from the GenomicAlignments package in R [31]. From the transcript abundance, the DEGs between two conditions were identified through the DESeq2 package in Bioconductor [32]. The output for the DEGs include log2(fold change) and the corresponding P-values [false discovery rate (FDR)-adjusted]. DEGs with log2(fold change) >1 and P-value <0.01 were considered to be significantly changed between the two conditions compared.
Enrichment analysis for cluster of orthologous group (COG) categories
The set of DEGs between two different conditions were annotated using COG categories. The hypergeometric test was then performed for the set of upregulated genes and downregulated genes, respectively. To calculate the enrichment of each COG category in the gene set, four values were obtained to perform the test: the total number of genes mapped in RNA-seq data, the number of genes in the current set, the number of genes with the current COG category out of all genes, and the number of genes with the COG category out of the current gene set. The FDR correction was applied to the P-values of the COG categories in the gene set. COG category with corrected P-value <0.05 was considered enriched in the gene set.
Data availability
The genomic sequence data have been deposited in the NCBI Sequence Read Archive (SRA) under accession number PRJNA546056. The RNA-seq data have been deposited in the NCBI SRA under accession number PRJNA546062.
Results
Laboratory evolution and acid-adapted endpoint strains
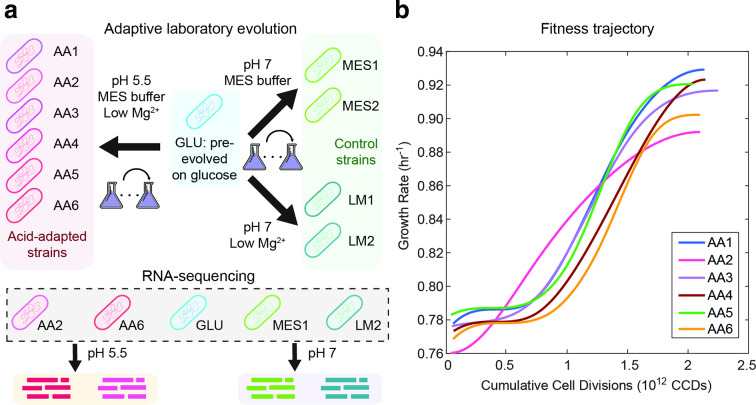
We used wild-type E. coli K-12 MG1655 that had been previously evolved on M9 glucose minimal medium as the starting strain for evolution and refer to it as the GLU strain [26]. We used GLU as the starting strain to isolate changes due to adaptation to acid stress from those caused by adaptation to the culture medium. The genetic mutations of the GLU strain against E. coli K-12 MG1655 are also documented in ALEdb (aledb.org) under the experiment name GLU. Six independent cultures were established under pH 5.5 in glucose minimal medium, buffered with 150 mM MES (pK a=6.1) [33]. In addition, we lowered the magnesium (Mg) concentration in the media to 0.2 mM to minimize precipitation. We refer to six acid-adapted strains as AA1 to AA6, respectively. To account for possible effects due to changes in media composition, we also set up two independent cultures under pH 7 with 150 mM MES buffer (MES1, MES2) and lowered the Mg concentration (LM1, LM2), respectively. All strains used in this study and their relationships are detailed in Fig. 1(a).
Fig. 1.
ALE of E. coli under acid stress. (a) Schematic of the ALE process and strains used for RNA-seq. Starting from the GLU strain, we performed ALE at pH 5.5 to obtain six acid-adapted (AA) strains and at pH 7 to obtain four control strains. Two control strains were adapted in glucose minimal medium with lowered magnesium concentration (LM) and two control strains were adapted in glucose minmal medium with MES buffer (MES). Using RNA-seq, we obtained the gene expression profiles of five selected strains at pH 5.5 and 7. (b) Smoothed fitness trajectories of six acid-adapted strains (raw data in Fig. S1). Raw data for the fitness change of the control strains can be found in Fig. S2. We show here the change in growth rate over cumulative cell divisions through the evolution process. The average growth rate improvement is 18 %.
We performed ALE using an automated system, which tracked culture growth rates and passed the cells to fresh media when OD600 measurements reached 0.3 to ensure selection at exponential-phase growth. Additionally, we periodically measured the pH of the clean media and recently passaged cultures to ensure proper buffering (Table S1). The culture pH remained relatively stable for strains evolved in MES buffer under pH 5.5 and 7. For strains evolved under lowered Mg concentration and with only phosphate buffers (pK a=7.2) in glucose minimal medium, the culture pH dropped significantly at the end of the culture, probably due to the secretion of organic acids during growth. The laboratory evolution process lasted for 35 days for strains AA1 to AA6 under pH 5.5, corresponding to 800 generations and 2.1×1012 CCDs. The fitness trajectories of the evolved strains are shown in Fig. 1(b). We observed that growth rates continuously improved with CCDs and approached stable values at the end of the evolution. Overall, we found the evolved endpoints to have an average of 18.0 % improvement in growth rate (from 0.77±0.01 to 0.91±0.01 h−1) over their starting strain values.
To evaluate the fitness of acid-adapted strains against the starting GLU strain, we obtained the growth rates of the strains at pH 5.5 and 7 in a separate experiment. The acid-adapted strains showed increased fitness at pH 5.5, with growth rate of 0.83±0.01 h−1 compared to the growth rate of 0.67±0.02 h−1 of the GLU strain. Growth rate under pH 7 for acid-adapted strains was 1.00±0.01 h−1, slightly higher than that of the GLU strain at 0.94±0.03 h−1.
Genetic mutations of the evolved strains
To understand the genetic basis of the observed phenotypic change, we performed whole genome sequencing on individual clones picked from acid-adapted strains AA1 to AA6, as well as the control strains MES1, MES2, LM1 and LM2. We identified the genetic mutations of the evolved strains by comparing them to the reference genome using breseq computational pipeline v0.33 (see Methods) [28]. The converged mutations of acid-adapted strains are reported in Table 1. Converged mutations are mutations on the same gene identified across multiple strains from independent cultures. All mutations identified in acid-adapted strains and those of control strains are presented in Tables S2 and S3.
Table 1.
Converged mutations identified in the clones of acid-adapted strains at pH 5.5
Flask number stands for the number of cell passages during the evolution. Numbers in bold represent the final flask number (and thus the endpoint) in the evolution process of the specific acid-adapted strain. Non-bold numbers are intermediate flasks during the evolution where samples were selected for whole genome sequencing.
|
Gene |
Mutation |
Protein change |
Flask number |
AA1 |
AA2 |
AA3 |
AA4 |
AA5 |
AA6 |
|---|---|---|---|---|---|---|---|---|---|
|
rho |
C→A |
R102S (CGC→AGC) |
87 |
X |
|||||
|
rho |
C→T |
R102C (CGC→TGC) |
111 |
|
X |
||||
|
rpoC |
C→A |
A397E (GCG→GAG) |
111 |
|
X |
||||
|
rpoC |
G→C |
G444A (GGT→GCT) |
88, 118 |
|
X |
||||
|
rpoC |
Δ7 bp |
coding (4106 −4112/4224 nt) |
113 |
|
X |
||||
|
rpoC |
Δ1 bp |
coding (4111/4224 nt) |
111 |
|
X |
||||
|
rpoC |
C→T |
S539F (TCT→TTT) |
111 |
|
X |
||||
|
nagA |
G→C |
S90* (TCA→TGA) |
84 |
|
X |
||||
|
nagA |
C→T |
R149H (CGT→CAT) |
83 |
|
X |
Overall, we found a total of 22 mutations in all acid-adapted strains, including those of clones picked from the endpoints (Tables 1 and S2, bold flask number) and midpoints of the evolution (Tables 1 and S2, non-bold flask number). Notably, we observed that mutations in rpoC appeared in five of the six endpoint clones. The rpoC gene encodes a subunit of RNA polymerase, which is known to act as a global regulator of gene expression [34, 35]. The mutations in rpoC include both SNPs and deletions. These mutations were located on the interaction interfaces of the protein product. Specifically, mutation with protein change A397E (Table 1) is located at the exit gate of the newly synthesized RNA strand. The region with mutation G444A interacts with the rpoB subunit and the region with mutation S539F interacts with the rpoA subunit (Table 1). The two deletion mutations are located at the interaction interface with the rpoZ subunit (Table 1). Mutations in rpoC in E. coli have been found in several previous ALE experiments, covering a variety of experimental conditions or perturbations (e.g. high temperature, alternating substrate, gene knockouts [36–38]). Several studies have suggested mutations in rpoC to be mainly associated with improvement in metabolic efficiency and growth rate [39–41].
The other converged mutations are found in rho (transcription regulation) and nagA (metabolism of N-acetyl-d-glucosamine) (Table 1). The mutations in these two genes are all SNPs. Unlike rpoC in which mutations are found in endpoint clones, mutations in rho appear in the midpoint clone of strain AA1 and endpoint clone of strain AA6. Mutations in nagA appear in midpoint clones of strains AA4 and AA6 but are not found in the endpoint clones of these two strains. For the rest of the mutations observed in acid-adapted strains, each appears only in a single strain (Table S2). These mutations appeared in both the coding regions and intergenic regions. The types of mutations include SNPs, deletions and insertion elements.
We found distinct patterns when examining mutations in control strains evolved under different conditions (Table S3). SNPs on the oxyR gene were the converged mutations in strains LM1 and LM2. On the other hand, the converged mutations for strains MES1 and MES2 are found in the intergenic region of ilvL and ilvX. Notably, the same mutation between ilvL and ilvX is also found in the endpoint clone of acid-adapted strain AA5 (Table S2), confirming the possible effect of the MES buffer during the evolution process.
Differential gene expression of the evolved endpoints at different pH
To understand how the mutations can affect gene products in evolved strains, we used RNA-seq to examine the gene expressions of ALE endpoints. We selected two acid-adapted strains for RNA-seq, strain AA2 which has a single mutation in rpoC and strain AA6 which has the largest number of mutations among all AA strains (Table S2). We selected strains MES1 and LM2 for different control conditions. We performed RNA-seq and obtained gene expression profiles of the selected strains as well as the starting GLU strain grown at pH 7 and pH 5.5 (Fig. 1a; see Methods). We then analysed the expression profiles at the level of individual genes and their related cellular processes. Specifically, we performed statistical tests to identify DEGs of the same strain grown at pH 5.5 compared to pH 7 (see Methods).
To ensure the DEGs identified for acid-adapted strains across different pH are only due to the effect of adaptive evolution under acid stress, we first need to understand the response to acid stress of the starting strain and also control for possible variations in culture medium during the evolution process. Therefore, we examined DEGs across pH values for the GLU strain, as well as the MES1 strain and LM2 strain to account for the possible effects due to MES buffer and lowered magnesium concentration. We found significant overlap of upregulated genes involved in cell wall/membrane biogenesis and translation processes among the acid-adapted strains and the control strains, implicating these two cellular processes as the common acid resistance mechanisms in E. coli .
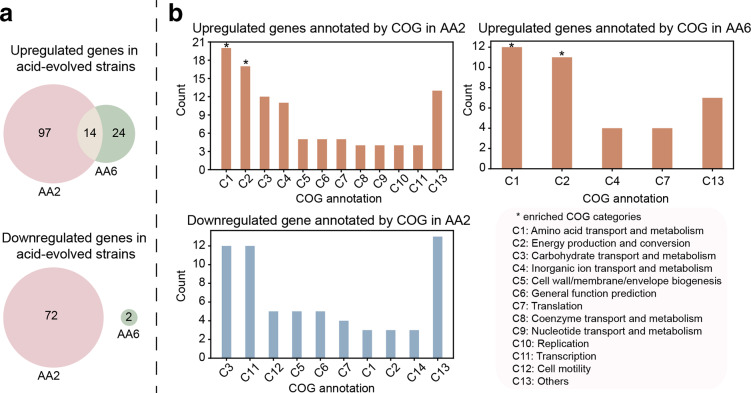
We then examined DEGs in the acid-adapted strains at pH 5.5 compared to pH 7, after removing DEGs also found in GLU and the control strains. We found 183 genes to be differentially expressed for strain AA2 (111 upregulated and 72 downregulated) and 40 genes for strain AA6 (38 upregulated and two downregulated) (Tables S4 and S5). Of these, we found 14 upregulated genes that appeared in both acid-adapted strains (Fig. 2a). Based on COG annotation [42], the 14 genes were found to be mainly involved in energy production and conversion (e.g. TCA cycle, respiratory chain, ATP synthase) and amino acid transport and metabolism (e.g. biosynthesis of glutamate).
Fig. 2.
DEGs of acid-adapted (AA) strains at different pH conditions. The DEGs are determined for the same strain by comparing its gene expression profiles when growing at pH 5.5 and pH 7. (a) Number of upregulated (top panel) and downregulated genes (bottom panel) in acid-adapted strains AA2 and AA6. AA2 and AA6 shared 14 upregulated genes. (b) COG categories in the upregulated and downregulated genes in acid-adapted strains. An asterisk above the bar means that the COG category is enriched, as calculated using a hypergeometric test (Methods). COG categories for downregulated genes in strain AA6 are not shown because there were only two. The upregulated and downregulated genes for strains AA2 and AA6 are listed in Tables S4 and S5.
We next examined the specific COG categories of the DEGs across pH in each acid-adapted strain. For strain AA2, the upregulated genes are associated with more than ten COG categories (Fig. 2b), with the largest number of genes related to amino acid transport and metabolism (e.g. biosynthesis of histidine, threonine), energy production and conversion (e.g. nitrite/nitrate reductase, succinate dehydrogenase, TCA cycle), carbohydrate transport and metabolism (e.g. glycolysis), and inorganic ion transport and metabolism (e.g. transport of iron, zinc, nitrite, nitrate). Among the processes identified, the upregulated genes are enriched in amino acid transport and metabolism and energy production and conversion based on a hypergeometric test (Fig. 2b, asterisks) (Methods). The downregulated genes of strain AA2 are found mostly in carbohydrate transport and metabolism (e.g. secondary carbon sources such as xylose and arabinose) and transcription. No COG categories are found to be enriched in these downregulated genes. For the other acid-adapted strain, AA6, the upregulated genes are mainly enriched in amino acid transport and metabolism (e.g. biosynthesis of glutamate, arginine) and energy production and conversion (e.g. TCA cycle, respiratory chain, ATP synthase) based on a hypergeometric test (Fig. 2b, asterisks). Again, no COG categories are enriched in the downregulated genes in strain AA6.
Overall, we observed the DEGs to be enriched in similar general COG categories for two acid-adapted strains, AA2 and AA6. However, the specific underlying cellular processes still differ, indicating different strategies developed by E. coli over the course of evolution under acid stress. Such differences cover different processes that are upregulated in amino acid biosynthesis (e.g. histidine for AA2 and glutamate for AA6) and energy production and conversion (e.g. anaerobic respiration found in AA2 but not in AA6) between two acid-adapted strains. Additionally, we found the downregulated genes to be involved in different COG processes between these two strains.
Discussion
In this study, we have used ALE to investigate the adaptation of E. coli K-12 MG1655 under acid stress in glucose minimal medium. Using whole genome sequencing, we identified mutations on rpoC, rho and nagA to be the converged mutations in acid-adapted strains. We then used RNA-seq to examine the gene expression profiles of acid-adapted strains across different pH values and compared them to those of the starting GLU strain and the control strains. Through analysis of DEGs, we identified cellular processes acquired by E. coli through adaptive evolution under acid stress.
A previous study by Harden et al. also investigated the adaptive evolution of E. coli K-12 W3110 under acid stress [24]. Before comparing the outcome of their ALE experiments with ours, it is important to note that their study used significantly different experimental conditions from those in the present study. First, Harden et al. used LBK as the culture medium, while we used glucose minimal medium. Next, in Harden et al., the pH under which the evolution took place was 4.6–4.8, while we evolved the culture at pH 5.5. Additionally, the incubation and stirring methods are different. In Harden et al., the culture was placed in 96-well plates with a culture volume of 200 µl per well and passaged daily at the stationary phase. The plates were shaken for 3 s every 15 minutes over the growth period. By contrast, our study had a culture volume of 15 ml in 35 ml tubes that were stirred continuously. The culture was passaged before reaching an OD600 of 0.3 to ensure selection at the exponential growth phase. Finally, our acid-adapted strains were evolved for 800 generations and 2.1×1012 CCDs; Harden et al. evolved the culture for a total 2000 generations, with no data available on CCDs. We typically use CCDs to describe the length of the evolution process, as it takes both the number of generations and population size into account.
We found five of the six acid-adapted strains to have mutations in rpoC, which functions as a subunit of RNA polymerase. The specific mutations include substitutions that change the encoded amino acids (AA2, AA3 and AA6) and deletions in the coding region that lead to shifts in the reading frame (AA4 and AA5). Based on the Pfam database, the substitutions occurred in the protein domains that contain the active site and the pore region that allows the entrance of nucleotides to the active site [43–45]. Also, the deletions that occurred at the end of the rpoC gene (base pairs 4106 and 4111 out of 4224) probably did not result in significant disruption in gene function. Harden et al. also observed missense mutations in subunits of RNA polymerase (rpoBCD) for all of the acid-adapted strains [24]. The authors of that study proposed several mechanisms to explain how mutations in the RNA polymerase complex might enhance fitness under acid stress. Here, however, we consider the mutations on rpoC to be associated with inducing faster growth rather than acid resistance. A comprehensive analysis of the 278 gene expression datasets of E. coli across diverse conditions has revealed that mutations in genes related to RNA polymerase typically lead to improved growth rate and reduced stress-related gene expression [46].
Other mutations found cover a range of cellular processes. However, none of the processes are directly related to the commonly known acid resistance mechanisms. Harden et al. identified mutations in genes related to the amino acid decarboxylase systems, and these mutations result in loss or downregulation of amino acid decarboxylase activities [24, 25]. The different mutations observed from the two studies are probably due to the different culture media in which the evolution took place. The culture medium used by Harden et al. was LBK medium, which is rich in amino acids. Activation of the amino acid decarboxylase systems requires the presence of amino acids in the medium [10–13]. According to Harden et al., amino acid decarboxylase systems protect E. coli from acid stress upon early exposure to the acidic environment, but incur fitness costs over the long term, where E. coli has developed other strategies to maintain the non-stress physiology. In our study, the culture medium was glucose minimal medium, in which the sole carbon source is glucose. Therefore, the amino acid decarboxylase systems were never activated under these conditions. Rather, we see mutations in genes that might be related to general cellular responses under stress conditions, such as transcription regulation (rho) and cellular physiology (csgD/csgB, yiaA).
The two acid-adapted strains with gene expression profiles available share several common general COG categories for upregulated genes under acid stress. However, besides sharing 14 DEGs in processes such as the TCA cycle, respiratory chain, ATP synthase and glutamate biosynthesis, the two strains have a number of DEGs with different cellular functions (Fig. 2). Both strains have SNPs in the rpoC gene, but in different protein domains according to the Pfam database mentioned earlier. It is interesting to find a large number of DEGs in strain AA2 with only point mutations in the rpoC gene, and relatively small number of DEGs in strain AA6 with several mutations in the rpoC gene and other genes. Specifically, the most up-regulated DEGs in strain AA2 (Table S4) cover processes including iron-related processes (exbD, feoA, ftnA, feoB), nitrate/nitrite metabolism (narG, narK, focA), carbohydrate transport metabolism (gatZ, gatA, pfkA, gpmM), and energy production and conversion (wrbA, frdA, frdB, sucB, sucC). The most down-regulated genes in AA2 cover a few processes such as carbohydrate transport metabolism (xylF, mglA, alsB) and cell-wall-related processes (yadN, phoE, yhiI). By contrast, the DEGs in strain AA6 (Table S5) are mainly associated with amino acid transport and metabolism, and energy production and conversion processes. The results here indicate that the specific rpoC point mutation in strain AA2 had a more significant and broader impact on gene expression change than the rpoC mutation in strain AA6. Such comparisons of the mutations and DEGs between the two strains show the different strategies used by E. coli under acid stress to adjust the level of gene transcripts. Similarly, a follow-up study on the work of Harden et al. also found different patterns of gene expression across four acid-adapted strains [25]. These two studies together demonstrate that regardless of the level of acid stress (pH 4.6–4.8 by Harden et al. and pH 5.5 in this study) and nutrient availability (LBK medium and glucose minimal medium), evolutionary pressure can drive E. coli to develop different strategies against acid stress.
Overall, we have studied the adaptive evolution of E. coli under acid stress, linking the improved phenotype to the underlying genotypes and levels of gene expression. The study provides a novel perspective on acid resistance mechanisms, as the commonly known acid resistance systems depend on rich medium or specific amino acids [47, 48]. In addition to the analysis of genetic mutations and DEGs, further analysis is needed to understand the change in regulatory actions using a recently developed approach [46]. Such analysis can be helpful in understanding the response to acid stress at the level of transcriptional regulation and in revealing potential drivers behind the global adjustment of cellular responses to acid stress.
Supplementary Data
Funding information
This work was supported by National Institute of General Medical Sciences of the National Institutes of Health Grant R01GM057089 and Novo Nordisk Foundation Grant NNF10CC1016517.
Acknowledgements
We would like to thank Laurence Yang for valuable discussions.
Author contributions
B.O.P. acquired the funding for this study. B.O.P., A.M.F. and B.D. conceptualized the research goals and aims. B.O.P., A.M.F., B.D. and C.A.O. designed the adaptive laboratory experiments. C.A.O., R.S., S.X., Y.G. and Y.H. performed the experiments, including adaptive laboratory evolution, whole genome sequencing and RNA-seq. B.D., A.V.S., X.F., P.V.P., K.C. and M.W. performed the data analysis. B.D., C.A.O., R.S. and B.O.P. wrote the manuscript. All authors read and approved the final manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: ALE, adaptive laboratory evolution; CCD, cumulative cell division; COG, cluster of orthologous group; DEG, differentially expressed gene; FDR, false discovery rate; RNA-seq, RNA sequencing; SRA, Sequence Read Archive.
Genomic sequencing data: NCBI SRA PRJNA546056.
RNA sequencing data: NCBI SRA PRJNA546062.
Two supplementary figures and five supplementary tables are available with the online version of this article.
Edited by: D. Grainger and T. Schneiders
References
- 1.Small P, Blankenhorn D, Welty D, Zinser E, Slonczewski JL. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J Bacteriol. 1994;176:1729–1737. doi: 10.1128/jb.176.6.1729-1737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin J, Lee IS, Frey J, Slonczewski JL, Foster JW. Comparative analysis of extreme acid survival in salmonella typhimurium, shigella flexneri, and Escherichia coli . J Bacteriol. 1995;177:4097–4104. doi: 10.1128/jb.177.14.4097-4104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin J, Smith MP, Chapin KC, Baik HS, Bennett GN, et al. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli . Appl Environ Microbiol. 1996;62:3094–3100. doi: 10.1128/aem.62.9.3094-3100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conner DE, Kotrola JS. Growth and survival of Escherichia coli O157:H7 under acidic conditions. Appl Environ Microbiol. 1995;61:382–385. doi: 10.1128/aem.61.1.382-385.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dlamini BC, Buys EM. Survival and growth of acid adapted Escherichia coli strains in broth at different ph levels. J Food Saf. 2009;29:484–497. doi: 10.1111/j.1745-4565.2009.00171.x. [DOI] [Google Scholar]
- 6.Vivijs B, Aertsen A, Michiels CW. Identification of genes required for growth of Escherichia coli MG1655 at moderately low pH. Front Microbiol. 2016;7:1672. doi: 10.3389/fmicb.2016.01672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans DF, Pye G, Bramley R, Clark AG, Dyson TJ, et al. Measurement of gastrointestinal pH profiles in normal ambulant human subjects. Gut. 1988;29:1035–1041. doi: 10.1136/gut.29.8.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ibekwe VC, Fadda HM, McConnell EL, Khela MK, Evans DF, et al. Interplay between intestinal pH, transit time and feed status on the in vivo performance of pH responsive ileo-colonic release systems. Pharm Res. 2008;25:1828–1835. doi: 10.1007/s11095-008-9580-9. [DOI] [PubMed] [Google Scholar]
- 9.Slonczewski JL, Fujisawa M, Dopson M, Krulwich TA. Cytoplasmic pH measurement and homeostasis in bacteria and archaea. Adv Microb Physiol. 2009;55:1–79.:317. doi: 10.1016/S0065-2911(09)05501-5. [DOI] [PubMed] [Google Scholar]
- 10.Castanié-Cornet MP, Treffandier H, Francez-Charlot A, Gutierrez C, Cam K. The glutamate-dependent acid resistance system in Escherichia coli: essential and dual role of the His-Asp phosphorelay RcsCDB/AF. Microbiology. 2007;153:238–246. doi: 10.1099/mic.0.29278-0. [DOI] [PubMed] [Google Scholar]
- 11.Richard H, Foster JW. Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J Bacteriol. 2004;186:6032–6041. doi: 10.1128/JB.186.18.6032-6041.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diez-Gonzalez F, Karaibrahimoglu Y. Comparison of the glutamate-, arginine- and lysine-dependent acid resistance systems in Escherichia coli O157:H7. J Appl Microbiol. 2004;96:1237–1244. doi: 10.1111/j.1365-2672.2004.02251.x. [DOI] [PubMed] [Google Scholar]
- 13.Kashiwagi K, Suzuki T, Suzuki F, Furuchi T, Kobayashi H, et al. Coexistence of the genes for putrescine transport protein and ornithine decarboxylase at 16 min on Escherichia coli chromosome. J Biol Chem. 1991;266:20922–20927. [PubMed] [Google Scholar]
- 14.Hayes ET, Wilks JC, Sanfilippo P, Yohannes E, Tate DP, et al. Oxygen limitation modulates pH regulation of catabolism and hydrogenases, multidrug transporters, and envelope composition in Escherichia coli K-12. BMC Microbiol. 2006;6:89. doi: 10.1186/1471-2180-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maurer LM, Yohannes E, Bondurant SS, Radmacher M, Slonczewski JL. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J Bacteriol. 2005;187:304–319. doi: 10.1128/JB.187.1.304-319.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hong W, Wu YE, Fu X, Chang Z. Chaperone-dependent mechanisms for acid resistance in enteric bacteria. Trends Microbiol. 2012;20:328–335. doi: 10.1016/j.tim.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 17.Brown JL, Ross T, McMeekin TA, Nichols PD. Acid habituation of Escherichia coli and the potential role of cyclopropane fatty acids in low pH tolerance. Int J Food Microbiol. 1997;37:163–173. doi: 10.1016/S0168-1605(97)00068-8. [DOI] [PubMed] [Google Scholar]
- 18.Chang YY, Cronan JE. Membrane cyclopropane fatty acid content is a major factor in acid resistance of Escherichia coli . Mol Microbiol. 1999;33:249–259. doi: 10.1046/j.1365-2958.1999.01456.x. [DOI] [PubMed] [Google Scholar]
- 19.delaVega AL, Delcour AH. Cadaverine induces closing of E. coli porins. Embo J. 1995;14:6058–6065. doi: 10.1002/j.1460-2075.1995.tb00294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rowbury RJ, Goodson M, Wallace AD. The PhoE porin and transmission of the chemical stimulus for induction of acid resistance (acid habituation) in Escherichia coli . J Appl Bacteriol. 1992;72:233–243. doi: 10.1111/j.1365-2672.1992.tb01829.x. [DOI] [PubMed] [Google Scholar]
- 21.Kanjee U, Houry WA. Mechanisms of acid resistance in Escherichia coli . Annu Rev Microbiol. 2013;67:65–81. doi: 10.1146/annurev-micro-092412-155708. [DOI] [PubMed] [Google Scholar]
- 22.Dragosits M, Mattanovich D. Adaptive laboratory evolution - principles and applications for biotechnology. Microb Cell Fact. 2013;12:64. doi: 10.1186/1475-2859-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buermans HPJ, den Dunnen JT. Next generation sequencing technology: advances and applications. Biochim Biophys Acta. 1842;2014:1932–1941. doi: 10.1016/j.bbadis.2014.06.015. [DOI] [PubMed] [Google Scholar]
- 24.Harden MM, He A, Creamer K, Clark MW, Hamdallah I, et al. Acid-adapted strains of Escherichia coli K-12 obtained by experimental evolution. Appl Environ Microbiol. 2015;81:1932–1941. doi: 10.1128/AEM.03494-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He A, Penix SR, Basting PJ, Griffith JM, Creamer KE, et al. Acid evolution deletes amino-acid decarboxylases and reregulates catabolism of Escherichia coli K-12. Appl Environ Microbiol. 2017 doi: 10.1128/AEM.00442-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LaCroix RA, Sandberg TE, O'Brien EJ, Utrilla J, Ebrahim A, et al. Use of adaptive laboratory evolution to discover key mutations enabling rapid growth of Escherichia coli K-12 MG1655 on glucose minimal medium. Appl Environ Microbiol. 2015;81:17–30. doi: 10.1128/AEM.02246-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marotz C, Amir A, Humphrey G, Gaffney J, Gogul G, et al. Dna extraction for streamlined metagenomics of diverse environmental samples. Biotechniques. 2017;62:290–293. doi: 10.2144/000114559. [DOI] [PubMed] [Google Scholar]
- 28.Deatherage DE, Barrick JE. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol. 2014;1151:165–188. doi: 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen S, Huang T, Zhou Y, Han Y, Xu M, et al. AfterQC: automatic filtering, trimming, error removing and quality control for fastq data. BMC Bioinformatics. 2017;18:80. doi: 10.1186/s12859-017-1469-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, et al. Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wood EJ. Data for biochemical research. Biochemistry and Molecular Biology Education. (3rd ed) 1987;15:97. doi: 10.1016/0307-4412(87)90110-5. p. [DOI] [Google Scholar]
- 34.Helmann JD. Rna polymerase: a nexus of gene regulation. Methods. 2009;47:1–5. doi: 10.1016/j.ymeth.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klein-Marcuschamer D, Santos CNS, Yu H, Stephanopoulos G. Mutagenesis of the bacterial RNA polymerase alpha subunit for improvement of complex phenotypes. Appl Environ Microbiol. 2009;75:2705–2711. doi: 10.1128/AEM.01888-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sandberg TE, Pedersen M, LaCroix RA, Ebrahim A, Bonde M, et al. Evolution of Escherichia coli to 42 °C and subsequent genetic engineering reveals adaptive mechanisms and novel mutations. Mol Biol Evol. 2014;31:2647–2662. doi: 10.1093/molbev/msu209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sandberg TE, Lloyd CJ, Palsson BO, Feist AM. Laboratory evolution to alternating substrate environments yields distinct phenotypic and genetic adaptive strategies. Appl Environ Microbiol. 2017;83 doi: 10.1128/AEM.00410-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCloskey D, Xu S, Sandberg TE, Brunk E, Hefner Y, et al. Evolution of gene knockout strains of E. coli reveal regulatory architectures governed by metabolism. Nat Commun. 2018;9:3796. doi: 10.1038/s41467-018-06219-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conrad TM, Frazier M, Joyce AR, Cho BK, Knight EM, et al. Rna polymerase mutants found through adaptive evolution reprogram Escherichia coli for optimal growth in minimal media. Proc Natl Acad Sci USA. 2010;107:20500–20505. doi: 10.1073/pnas.0911253107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng KK, Lee BS, Masuda T, Ito T, Ikeda K, et al. Global metabolic network reorganization by adaptive mutations allows fast growth of Escherichia coli on glycerol. Nat Commun. 2014;5:3233. doi: 10.1038/ncomms4233. [DOI] [PubMed] [Google Scholar]
- 41.Wytock TP, Fiebig A, Willett JW, Herrou J, Fergin A, et al. Experimental evolution of diverse Escherichia coli metabolic mutants identifies genetic loci for convergent adaptation of growth rate. PLoS Genet. 2018;14:e1007284. doi: 10.1371/journal.pgen.1007284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Severinov K, Mustaev A, Kukarin A, Muzzin O, Bass I, et al. Structural modules of the large subunits of RNA polymerase. Introducing archaebacterial and chloroplast split sites in the beta and beta' subunits of Escherichia coli RNA polymerase. J Biol Chem. 1996;271:27969–27974. doi: 10.1074/jbc.271.44.27969. [DOI] [PubMed] [Google Scholar]
- 45.Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 46.Sastry AV, Gao Y, Szubin R, Hefner Y, Xu S, et al. The Escherichia coli transcriptome consists of independently regulated modules. bioRxiv. doi: 10.1101/620799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Foster JW. Escherichia coli acid resistance: tales of an amateur acidophile. Nat Rev Microbiol. 2004;2:898–907. doi: 10.1038/nrmicro1021. [DOI] [PubMed] [Google Scholar]
- 48.Lund P, Tramonti A, De Biase D. Coping with low pH: molecular strategies in neutralophilic bacteria. FEMS Microbiol Rev. 2014;38:1091–1125. doi: 10.1111/1574-6976.12076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genomic sequence data have been deposited in the NCBI Sequence Read Archive (SRA) under accession number PRJNA546056. The RNA-seq data have been deposited in the NCBI SRA under accession number PRJNA546062.