Abstract
The DNA damage response of the multidrug-resistant pathogen Acinetobacter baumannii , which induces mutagenic UmuD′2C error-prone polymerases, differs from that of many bacteria. Acinetobacter species lack a LexA repressor, but induce gene transcription after DNA damage. One regulator, UmuDAb, binds to and represses the promoters of the multiple A. baumannii ATCC 17978 umuDC alleles and the divergently transcribed umuDAb and ddrR genes. ddrR is unique to the genus Acinetobacter and of unknown function. 5' RACE (rapid amplification of cDNA ends) PCR mapping of the umuDAb and ddrR transcriptional start sites revealed that their −35 promoter elements overlapped the UmuDAb binding site, suggesting that UmuDAb simultaneously repressed expression of both genes by blocking polymerase access. This coordinated control of ddrR and umuDAb suggested that ddrR might also regulate DNA damage-inducible gene transcription. RNA-sequencing experiments in 17 978 ddrR − cells showed that ddrR regulated approximately 25 % (n=39) of the mitomycin C-induced regulon, with umuDAb coregulating 17 of these ddrR-regulated genes. Eight genes (the umuDC polymerases, umuDAb and ddrR) were de-repressed in the absence of DNA damage, and nine genes were uninduced in the presence of DNA damage, in both ddrR and umuDAb mutant strains. These data suggest ddrR has multiple roles, both as a co-repressor and as a positive regulator of DNA damage-inducible gene transcription. Additionally, 57 genes were induced by mitomycin C in the ddrR mutant but not in wild-type cells. This regulon contained multiple genes for DNA replication, recombination and repair, transcriptional regulators, RND efflux, and transport. This study uncovered another regulator of the atypical DNA damage response of this genus, to help describe how this pathogen acquires drug resistance through its expression of the error-prone polymerases under DdrR and UmuDAb control.
Keywords: DNA damage, DdrR, UmuDAb, SOS response, LexA, repressor
Introduction
In many bacteria, DNA damage caused by UV light, mitomycin C (MMC) or antibiotics can induce a multitude of DNA damage response (SOS) genes that are under LexA control [1, 2]. LexA typically represses genes by binding to promoters of these genes [3] until it undergoes auto-cleavage by an activated RecA protein after DNA damage [4, 5]. Despite lacking a lexA gene, the Acinetobacter baumannii characteristics associated with DNA damage-induced mutagenesis are consistent with an SOS response: (i) repression of transcription in the absence of DNA damage [1–3], (ii) induction of RecA-facilitated repressor self-cleavage after DNA damage [6] caused by (iii) antibiotics (e.g. ciprofloxacin and tetracycline [7, 8]), UV light or MMC [9, 10], (iv) leading to the induction of antibiotic resistance (v) [7, 11, 12] in a process requiring protein synthesis.
One regulator of a minority of the DNA damage-induced genes in Acinetobacter species is UmuDAb. This homologue of the error-prone polymerase manager UmuD is encoded throughout the bacterial genus Acinetobacter [9], in addition to these species’ umuD genes encoded in operons with the umuC error-prone polymerase. UmuDAb shares with its UmuD homologues a catalytic C-terminal serine protease domain that enables its self-cleavage. Unlike umuD, however, the Acinetobacter umuDAb encodes an additional N-terminal domain that binds DNA [13] and is required for its repressor action [14]. These UmuDAb self-cleavage abilities [6], size [15] and function [14] resemble features of LexA. UmuDAb represses gene transcription until DNA damage (via antibiotics, radiation or chemical exposure [16]) triggers the induction of a subset of the typical SOS genes [17]. In A. baumannii ATCC strain 17 978 (hereafter abbreviated 17978), the UmuDAb-repressed regulon defined by both RNA-sequencing (RNA-seq) and microarray studies is notable for including the six error-prone Y-family polymerase umuDC genes (A1S_2008, 2015, 0636–637, 1173–1174) that contribute to SOS mutagenesis [10, 17]. The other members of the UmuDAb regulon include umuDAb itself (A1S_1389), and the ddrR gene (A1S_1388) transcribed divergently from it [13, 17].
We previously identified ddrR as a UmuDAb-regulated, DNA damage-inducible gene in Acinetobacter baylyi [15]. Its 246 bp ORF codes for 81 amino acids in A. baylyi ADP1 (78 amino acids in 17978 cells), and is preceded by an appropriately spaced ribosome binding site in both species. It is present in virtually all Acinetobacter species, including the pathogens A. baumannii and A. ursingii [9]. When present, it is always transcribed divergently from umuDAb; umuDAb is similarly always next to ddrR. ddrR shares a putative, ~282 bp promoter region with umuDAb in A. baumannii strains. The inclusion of ddrR, which is not encoded by microbes outside the genus Acinetobacter , in the UmuDAb regulon, as well as the conserved genomic organization of the umuDAb–ddrR gene pair within Acinetobacter species, suggested that it might play a role in the UmuDAb-mediated, atypical SOS response of this genus.
We hypothesized that inverted repeats in the umuDAb–ddrR intergenic region in A. baylyi (Fig. 1) served as a binding site for a regulatory protein [18]. Aranda et al. later identified a nearly identical palindromic sequence between umuDAb and ddrR in 17978 cells and demonstrated that UmuDAb binds to this DNA sequence in their promoters as well as to putative promoter regions upstream of the other umuDC homologues in 17 978 [13]. Mutations in the shared umuDAb–ddrR promoter region of A. baylyi significantly alter the transcription of both genes in a coordinated fashion [14]. Mutations in the most proximal (relative to umuDAb) region of the palindromic sequence abolish the repression typical of ddrR and umuDAb transcription under non-inducing conditions, and result in constitutive, high expression of both genes. However, mutations in the region most distal to umuDAb prevent the induction of transcription even under DNA-damaging conditions.
Fig. 1.
5' RACE (rapid amplification of cDNA ends) analyses reveal the relationship of the UmuDAb binding region to the ddrR-umuDAb promoters. 5' RACE experiments were conducted on RNA from MMC-treated 17978 and ADP1 cells. Upon mapping the first (+1) mRNA base of each gene, the −35 elements of each gene were observed to overlap either the defined UmuDAb binding site (for 17978) or an inverted repeat that, when mutated, abolished transcription of both genes in ADP1 cells [14]. The dotted box marks a UmuDAb binding site defined in 17978 through gel shift experiments [13], the dashed lines indicate the inverted repeats proposed to be regulatory binding sites in ADP1 [18], and the solid boxes represent −10 and −35 promoter consensus elements suggested by the +1 transcriptional start site(s). Distances between the +1 transcriptional start sites and the coding regions are shown in vertical boxes.
One need to decipher DNA damage regulatory mechanisms in Acinetobacter is that the opportunistic pathogen A. baumannii creates DNA damage-induced resistance to antibiotics that are used clinically [10]. As this species has been identified as a ‘serious threat’ to human health by the Centers for Disease Control [19] and other agencies given its multi-drug and pan-drug resistance, it is necessary to understand how its DNA damage response contributes to this problem. Furthermore, understanding how this genus induces ~150 genes after DNA damage [10, 13, 17], without encoding a lexA repressor gene [9], will advance our knowledge of novel transcriptional regulatory mechanisms.
We investigated the mechanism by which both ddrR and umuDAb were coordinately regulated by a promoter region that contained only one UmuDAb binding site. We further hypothesized that ddrR, like UmuDAb, was required for regulation of DNA damage-inducible genes. RNA-seq experiments in A. baumannii 17978 cells tested whether ddrR regulates any DNA damage-inducible genes, only UmuDAb-regulated genes or all of them. We found that in this species, all UmuDAb-repressed genes were corepressed by DdrR, and other UmuDAb-regulated genes required DdrR for their full induction after DNA damage. Finally, we saw that DdrR might participate in additional regulatory networks, as it regulated both DNA damage-inducible genes that do not require UmuDAb action and genes that were only induced in ddrR mutant cells.
Methods
Bacterial strains and growth conditions
All Acinetobacter strains described in Table 1 ( A. baylyi strains derived from ADP1 and A. baumannii strains derived from ATCC 17978) were grown at 37 °C in minimal media plus succinate [18] for RNA-seq transcriptome and real-time quantitative PCR (RT-qPCR) analyses. As described previously [17], for both RNA-seq transcriptome and RT-qPCR analyses, a 3 ml overnight culture, grown at 37 °C at 250 r.p.m., was diluted 1 : 25 into 5 ml of fresh medium and grown with shaking for 2 h, at which time the culture was split in two and 2 μg MMC ml−1 was added to one culture. This concentration of MMC was chosen to correspond to that used in previous studies, including RNA-seq studies, to facilitate direct comparison between results. [When measuring benA induction, sodium benzoate (3 mM) was added to the medium [18], instead of MMC.] Further incubation for 3 h allowed gene expression before isolation of total RNA. Kanamycin was added to LB medium at 30 μg ml−1, and gentamycin was added at 20 μg ml−1 for selection after transformation.
Table 1.
Strains used in this study
|
Strain name |
Genotype |
Characteristics |
Reference |
|---|---|---|---|
|
Acinetobacter baylyi strain ADP1 |
Wild type |
||
|
Acinetobacter baumannii ATCC 17978 |
Wild type |
ATCC [24] |
|
|
ACIAD2730 |
ADP1 −53_−1delddrR ΔddrR::tdk/kan |
Kmr |
[21] |
|
ACIAD0535 |
ADP1 ΔclpX::tdk/kan |
Kmr; source of the tdk–KanR cassette |
[21] |
|
JH1700 |
17978 ddrR:TnLK |
Kmr |
This study |
|
A. baylyi MSUcds2730 |
ADP1 ΔddrR::tdk/kan |
Kmr |
This study |
|
A. baylyi DR-Stop |
ADP1 ddrR 10C>T |
DdrR Q4X |
This study |
Construction of a 17978 ddrR::lacZkanR mutant
An A. baumannii ATCC 17978 ddrR::lacZ insertion mutant was constructed by allelic replacement of the wild type (WT) allele in a series of steps. First, a custom lacZ-kanR transposon was constructed and used in an in vitro transposition reaction, where it randomly transposed into ddrR contained in the plasmid p17UDDR. p17UDDR was constructed by PCR-amplifying a 2.8 kbp fragment of 17978 chromosomal DNA with primers Out17UDAbRev and Out17UDAbFor, and cloning the PCR product into pGEM-T Easy (Promega). p17UDDR encodes A1S_1389 (UmuDAb; genomic coordinates 1 641 271–1 631 882 in GenBank CP000521.1), A1S_ 1388 (DdrR; genomic coordinates 1 630 752–1 630 988), and hypothetical proteins A1S_1390 and A1S_3662. The transposon was constructed by ligating a 4.7 kb PstI fragment of pKOK6 [20], which contains a promoterless lacZ-kanamycin resistance gene cassette, into the PstI site of the EZ::TN pMOD-3<R6Kγori/MCS>Transposon Construction Vector (Epicentre) to produce plasmid pMOD3LK. This plasmid contained a 5.5 kb transposon that was named TnLK, and which was excised from pMOD3LK by PshAI digestion. Equimolar amounts of TnLK and the target plasmid p17UDDR were mixed with 1 U of EZ::TN transposase (Epicentre) and incubated at 37 °C for 2 h to allow in vitro transposition of TnLK into p17UDDR.
Second, the transformation of this transposition mixture into Escherichia coli DH5α yielded the plasmid pTn14, which contained TnLK inserted after the 46th base pair of ddrR, oriented with lacZ in the same orientation as ddrR (determined by DNA sequencing). Third, an 8.2 kb SpeI–NcoI fragment from pTn14 was subcloned into the SmaI site of the suicide vector pEX18Gm, which contains the sacB counter-selectable marker for recombination. Fourth, the resulting plasmid, pEXTn14, was electroporated into 17978 cells and selected for on gentamycin- and kanamycin-containing medium. Gentamycin- and kanamycin-resistant transformants were grown for 3 h in LB at 37 °C before plating on kanamycin- and 5 % (w/v) sucrose-containing LB agar for counterselection. PCR and DNA sequencing analyses of gentamycin-sensitive, kanamycin- and sucrose-resistant transformants confirmed that allelic replacement of the wild type ddrR allele with the insertion mutation had occurred and that the cells were not merodiploids. This strain was named JH1700.
A. baylyi mutant strain construction
A ΔddrR strain of ADP1 cells, MSUcds2730, was constructed in which the 246 bp ddrR ORF was replaced with the tdk/kan cassette that de Berardinis et al. previously used to make a library of single-gene deletion mutants of all genes in ADP1 [21]. In that library's ddrR mutant strain (ACIAD2730), 53 bp upstream of the ddrR ORF (that begins with a methionine codon and is preceded by a ribosome-binding site) was also deleted. This may have been a mis-annotation of the ORF, as the 53 bp are an ORF in ADP1 cells. (There is no similar ORF upstream of the A. baumannii ddrR.) Current annotations indicate a 246 bp ddrR ORF at base pairs 2 674 651–2 674 896 in the ADP1 genome GenBank file CR543861.1. To avoid possible interference with the promoter of ddrR, we deleted only the 246 bp ddrR ORF to form A. baylyi strain MSUcds2730.
Three PCR products were amplified from A. baylyi genomic DNA with NEB Long Amp Polymerase. PCR amplification with the primer pairs (i) ADP1drUDRevRC and 2730DStdk/KanFor, and (ii) CL-5 and 2730UStdk/KanRev was used to amplify the regions flanking the ddrR gene. The primer pair TDKKanFor and TDKKanRev used A. baylyi strain ACIAD0535 (ΔclpX::tdk/kan) as a source of the tdk–KanR cassette [21]. These three products were combined in equimolar amounts and used in splicing-overlap extension PCR amplification with primers CL-5 and ADP1drUDRevRC. The resulting 3.3 kb PCR product was transformed into ADP1 cells and selected for on LB-kanamycin plates. PCR and DNA sequencing confirmed the mutation.
The A. baylyi strain DR-Stop was constructed through two-piece splicing-overlap extension PCR to contain a nonsense (ochre) mutation of amino acid 4 of the ADP1 ddrR coding region. PCR amplification of ADP1 genomic DNA with Long Amp Polymerase and primer pairs (i) CL-4 and ADP1StopddrRFor, and (ii) To81Rev and ADP1StopddrRRev produced two products that were used in equimolar amounts in a third PCR with primer pair CL-4 and To81Rev. This 1.7 kb PCR product was transformed into A. baylyi strain ACIAD2729 (umuDAb::tdk-KanR) and plated on LB plates containing azithromycin at 200 μg ml−1 to counter-select for allelic replacement of the tdk-KanR cassette of umuDAb (ACIAD2729) with the mutated DNA PCR product. Azithromycin-resistant colonies were screened for kanamycin resistance, and kanamycin-sensitive colonies were tested with PCR and DNA sequencing to confirm the presence of the stop codon in ddrR. Strains, plasmids and primers are described in Tables 1–3, respectively.
Table 2.
Plasmids used in this study
|
Plasmid |
Description |
Source/reference |
|---|---|---|
|
pKOK6 |
Source of promoterless lacZ-Kmr gene cassette, with bidirectional transcriptional stop t sequence inserted between lacZ and Kmr; Kmr Ampr |
[20] |
|
pEX18Gm |
Counterselectable suicide vector containing sacB; Gmr |
[38] |
|
pGEM-T Easy |
Cloning vector; Ampr |
Promega |
|
EZ::TN pMOD-3<R6Kγori/MCS> |
Transposon construction vector; Ampr |
Epicentre |
|
p17UDDR |
2.8 kbp of 17978 chromosomal DNA (1 630 006–1 632 787) cloned into TA vector pGEM-T Easy; Ampr |
This study |
|
pMOD3LK |
pMOD-3<r6Kγori/MCS> vector containing lacZ-kanR cassette cloned into PshAI site to form transposon TnLK; Kmr Ampr |
This study |
|
pTn14 |
p17UDDR containing ddrR 46ins47TnLK; Kmr Ampr |
This study |
|
pEXTn14 |
pEX18Gm containing ~8 kbp SpeI–NcoI insert from pTn14 cloned into SmaI; Gmr Kmr |
This study |
Table 3.
Primers used in this study
|
Primer |
Purpose |
Sequence |
|---|---|---|
|
Out17UDAbRev |
Amplify 17978 chromosomal DNA |
CGGTAGCGACTTATAATTTT |
|
Out17UDAbFor |
Amplify 17978 chromosomal DNA |
ACTCAGTGATAGATAATCGG |
|
ddrRRACE#2 |
ddrR 5′ RACE (ADP1) |
GATTACGCCAAGCTTGCATGTAGCTCTTGGGCATAACC |
|
ddrRRACE#3 |
ddrR 5′ RACE (ADP1) |
GATTACGCCAAGCTTCGTCATAATATGCTCGGCTTGTTCGG |
|
New5RaceumuDAb |
umuDAb 5′ RACE (ADP1) |
GATTACGCCAAGCTTCGCAATCACGATATCACCTGCTTTGGCCG |
|
5RACEudabADP1 |
umuDAb 5′ RACE (ADP1) |
GATTACGCCAAGCTTCTCGACATGCTCCTGTGCAGGTGATGG |
|
17 978umuDAbRACE |
umuDAb 5′ RACE (17978) |
GATTACGCCAAGCTTCCACACTTTAGGGGGCTGAAATTGGG |
|
17 978ddrRRACE |
ddrR 5′ RACE (17978) |
GATTACGCCAAGCTTGAGTGGGTAAGGGGATGTAAGCC |
|
CL-5 |
Construction of MSUcds2730 strain |
AGATCACGAGTTCTTGACC |
|
2730UStdk/KanRev |
TTTTTATGATTTGAATTGGAGGCTGGGTTTAAACTCCCTATCAGAAATT |
|
|
2730DStdk/KanFor |
CGATGAGTTTTTCTAAGCATGCGGAGCTGGATCTGGGTTTATTTTAGAGTAAC |
|
|
ADP1drUDRevRC |
CACAACAAATGACTGGACTT |
|
|
TDKKanFor |
Amplification of tdk-KanR cassette |
CCCAGCCTCCAATTCAAAT |
|
TDKKanRev |
CCAGCTCCGCATGCTTAG |
|
|
CL-4 |
Construction of ddrR nonsense mutation in DR-Stop strain |
CCTGCTTATGCAATGACAG |
|
To81Rev |
CTGAACGTATTTGATTGAGC |
|
|
ADP1StopddrRFor |
TTGCATCACGTTAATTCTTCATT |
|
|
ADP1StopddrRRev |
AATGAAGAATTAACGTGATGCAA |
RT-qPCR
Total RNA was purified from biological triplicates of 3 ml samples, processed with the Epicentre MasterPure RNA Purification kit and re-suspended at 200 ng µl−1. Further removal of contaminating DNA was performed using the Ambion DNA-free ‘rigorous’ DNase treatment. RT-qPCR was performed on an ABI 7300 Real-Time PCR system, as previously described [17], using primers described previously [14, 17] and in Table S1 (available in the online version of this article). Primers for A1S_1388 were designed after the transcriptional start of ddrR was determined; the reverse primer therefore comprised base pairs 19–41 of the ddrR ORF, located before the TnLK insertion between base pairs 46 and 47.
RNA-seq analyses
The Genomics Facility at the University of Louisville prepared RNA libraries with 1–2 µg of total 17978 RNA samples, using the Illumina TruSeq Stranded Total RNA LT Sample Prep Kit-Set B and the Ribo-Zero Gram-Negative Bacteria Kit for rRNA depletion and RNA fragmentation. Library validation (final fragment size: ~260 bp) was performed qualitatively on an Agilent Bioanalyzer using the Agilent DNA 1000 Kit. Sequencing library quantification was performed by a standard curve method of qPCR using the KAPA Library Quantitation Kit for Illumina Platforms DNA standards.
The samples were sequenced as 75 bp paired-end reads on the University of Louisville Center for Genetics and Molecular Medicine’s Illumina NextSeq 500 using the Nextseq500 Mid Output Kit (150 cycles), yielding an average of 18 million aligned paired-end reads per sample. Reads were mapped to GenBank reference sequences CP000521 (the 17 978 chromosome), CP000522 (plasmid pAB1) and CP000523 (plasmid pAB2) using TopHat2 alignment approaches. Differential expression was analysed with the Tuxedo suite using Cufflinks (v. 2.1.1) [22]. The transcript abundance (FPKM; fragments per kilobase of transcript per million fragments mapped reads) was calculated, and genes were considered differentially expressed after MMC treatment using CuffDiff analyses with a false discovery rate (FDR) cutoff value of q<0.01, and if they were induced (or repressed) more than two-fold. Sequence datasets were submitted to the NCBI Gene Expression Omnibus under accession number GSE104741. Locus tags with the prefix ‘A1S_’ refer to A. baumannii ATCC 17978 genes; the prefix ‘ACIAD’ indicates genes of A. baylyi strain ADP1.
5′ RACE
5′ RACE PCR was used to determine +1 sites for umuDAb and ddrR transcripts. mRNA was purified from MMC-treated ADP1 total RNA samples with Epicentre’s Terminator 5′-Phosphate-Dependent Exonuclease. Poly-A+ RNA from MMC-treated 17 978 total RNA samples was prepared with Takara PolyA polymerase before use in 5′ RACE reactions. These mRNA-enriched or poly-adenylated samples were used in the Clonetech SMARTer 5′/3′ RACE kit protocol to generate RACE-ready cDNA and perform 5′ RACE. Touchdown PCR cycling parameters based on primer melting temperatures were used for both species samples. Primers used to perform 5′ RACE reactions are listed in Table 3. All RACE PCR products were gel-purified and cloned into the Clontech linearized pRACE vector and sequenced (three clones for umuDAb in ADP1 and four plasmid clones for all other reactions).
Statistical analyses
GraphPad InStat software was used to conduct the ANOVA and t -test analyses as described in the text and legends for Figs 2–5.
Fig. 2.
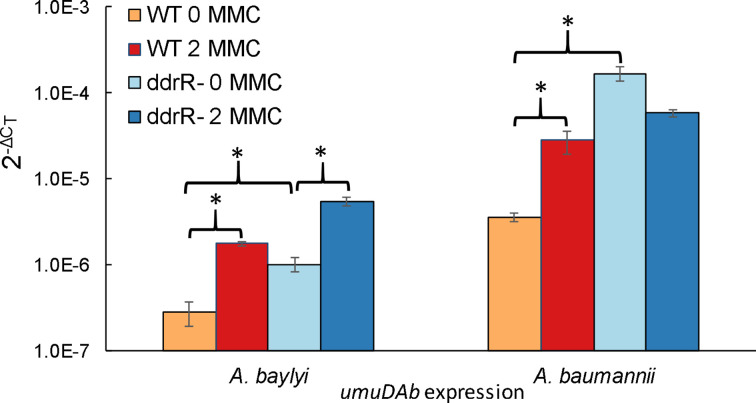
ddrR regulates expression of the UmuDAb repressor. RT-qPCR experiments measured umuDAb expression in wild type (WT) strains and ddrR mutant strains MSUcds2730 ( A. baylyi ) and JH1700 ( A. baumannii ), in the absence or presence of DNA damage (MMC, 2 μg ml−1). Mutation of ddrR resulted in the loss of the umuDAb repression that exists in WT cells in the absence of DNA damage. Additionally, expression of umuDAb after DNA damage was significantly induced in the ADP1 ddrR mutant MSUcds2730 (P<0.05 in a t -test). Asterisks indicate statistical significance in a Student’s t -test for P<0.05. The standard error of the mean from technical triplicates of biological triplicates is shown.
Fig. 3.
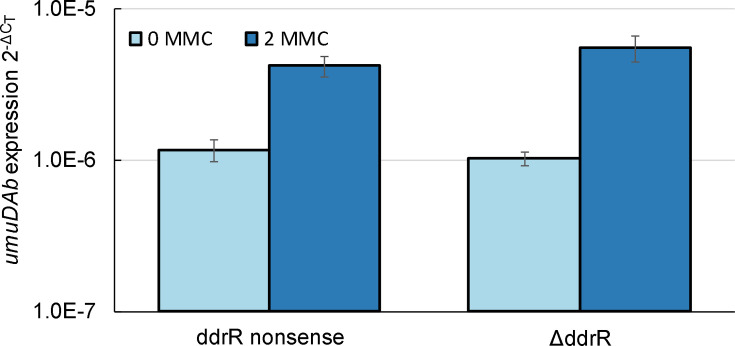
A DdrR protein probably exerts the regulatory actions of ddrR. RT-qPCR experiments measured the expression of umuDAb in two different ddrR mutants of ADP1 in the absence or presence of DNA damage (MMC, 2 μg ml−1). DR-Stop, a ddrR nonsense (stop codon) mutant, showed the same derepression of umuDAb in the absence of DNA damage as did the null ddrR mutant MSUcds2730. There was a significant induction in umuDAb expression (2−ΔCT) after DNA damage (P<0.01 in a one-tailed t -test) in each strain, but no significant difference between strains in their induction amount (P>0.05 in a two-tailed t -test comparing 2−ΔΔCT), or between expression in either the presence or the absence of MMC (P>0.05 in a two-tailed t -test.) The standard error of the mean from technical triplicates of biological triplicates is shown.
Fig. 4.
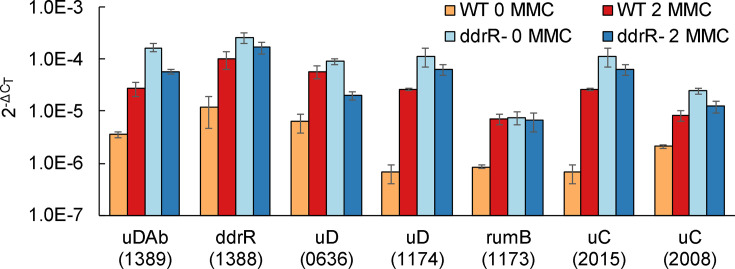
ddrR is required for repression of multiple error-prone polymerases in 17978. RT-qPCR experiments measured the expression of genes in 17978 WT and ddrR strain JH1700 in the absence or presence of DNA damage (MMC, 2 μg ml−1). Mutation of ddrR resulted in the derepression of all of these genes in the absence of DNA damage. Expression was measured in both un-induced and induced (MMC) conditions of WT or ddrR mutant cells. Genes are indicated by name or abbreviation (uDAb, umuDAb; uD, umuD; uC, umuC), and A1S gene locus number. Each gene was assayed in one RT-qPCR experiment (plate), with error bars indicating the standard error of the mean from technical triplicates of biological triplicates. For every gene, expression in WT cells was significantly (P<0.05 in a one-tailed t -test) increased after induction with MMC. Expression of every gene in the absence of DNA damage was significantly (P<0.05; in a one-tailed t -test) induced in ddrR mutant cells as compared to WT cells.
Fig. 5.
ddrR mutation does not affect the induction of genes that are not regulated by UmuDAb. RT-qPCR experiments measured the expression of genes (ACIAD0724 nrdA, ACIAD0445 gst or ACIAD1436 benA in ADP1; A1S_0408 gst or A1S_1215 benA in 17978) in un-induced (-I) and induced (‘+I’ for inclusion of inducing agent: MMC for nrdA and gst; or benzoate for benA) conditions for WT, ddrR and umuDAb strains of ADP1 and 17978. There was no significant difference in the induction level of any of these genes in the ddrR or umuDAb mutants (P>0.05; in a two-tailed t -test) after MMC or benzoate exposure (for benA gene expression only). nd , benA expression was not examined (not done) in umuDAb mutants of either 17978 or ADP1 by RT-qPCR, although previous experiments have shown that benzoate-mediated induction of benA is unaffected by the umuDAb mutation [18]. Each gene was assayed in one RT-qPCR experiment (plate), with error bars indicating the standard error of the mean from technical triplicates of biological triplicates.
Results
umuDAb, as a umuD and lexA homologue [18], is a canonical SOS gene and hence its regulation by, and induction after, DNA damage was expected. However, the ddrR gene, which is highly induced after DNA damage [18], is of unknown function and does not encode a protein with homology to any other proteins. It is found only among members of the genus Acinetobacter [9], where every umuDAb allele and ddrR allele (which range in size from 231 to 252 bp) are co-located and oriented divergently from each other.
ddrR and umuDAb promoters overlap with the UmuDAb binding site
We sought to determine how the one UmuDAb binding site suggested to exist in the ddrR–umuDAb intergenic region could accomplish the regulation of both umuDAb and ddrR expression. To map the relationship between the UmuDAb binding site and these genes’ putative promoters, we identified the transcriptional start sites of ddrR and umuDAb in A. baylyi strain ADP1 (ACIAD2730 and ACIAD2729) and A. baumannii ATCC 17978 (A1S_1388 and A1S_1389). We hypothesized that ddrR and umuDAb might have overlapping promoters that could allow one UmuDAb binding event to regulate both genes’ expression coordinately.
5′ RACE experiments for ddrR indicated a +1 site 41 bp upstream of ddrR in ADP1, and 180 and 182 bp upstream of ddrR in 17 978 (Fig. 1). 5′ RACE determined the +1 site for umuDAb transcription to be 30–31 bp upstream of umuDAb in ADP1 and 30 bp upstream of umuDAb in 17 978. These +1 sites for ddrR and umuDAb predict adjacent −35 promoter consensus elements for umuDAb and ddrR in both species.
These data, combined with previous data showing a loss of transcription when these bases have been mutated [14], suggest that UmuDAb simultaneously represses both genes by binding to DNA and blocking RNA polymerase access to both of the umuDAb and ddrR promoters. This coregulation of both genes suggested that the gene products, once expressed, might be used in the same pathway or process.
ddrR encodes a protein that regulates gene expression after DNA damage
The coregulation of ddrR and umuDAb transcription by UmuDAb suggested that a ddrR mutant might, like a umuDAb mutant, also cause dysregulation or derepression of DNA damage-induced genes in Acinetobacter . This hypothesis was tested in RT-qPCR experiments performed on RNA harvested from ADP1 WT and MSUcds2730 (ΔddrR::tdk/kan), and 17978 WT and JH1700 (ddrR:TnLK) cells grown in inducing (2 μg MMC ml−1) and control conditions. Expression of umuDAb in these ddrR mutants and in the extended deletion mutant strain ACIAD2730 (data not shown; P<0.05) was significantly derepressed under control conditions, relative to the WT strains (P<0.05) (Fig. 2). In each species, umuDAb expression was significantly (P<0.05) higher in the ddrR mutant cells than in WT cells. In ADP1 but not 17978, umuDAb was further significantly induced (5.4-fold) in the ΔddrR cells after MMC treatment (Fig. 2), even after the loss of repression in the absence of MMC treatment.
It is not known whether ddrR, uniquely found in the genus Acinetobacter , encodes a protein or whether it exerts its action as an RNA transcript. We mutated codon 4 of the ADP1 ddrR to form a stop codon and noted that the induction and expression of umuDAb (in both the absence and the presence of MMC) was the same in this ADP1 ddrR nonsense mutant strain (DR-Stop) as in the null ddrR mutant MSUcds2730 (Fig. 3). These results suggest that the changes in transcription seen in MSUcds2730 were due to the absence of a DdrR protein. Similar to umuDAb expression being equally induced in MSUcds2730 and DR-Stop, ddrR expression in DR-Stop also showed 2.11-fold induction (P<0.05 in a one-tailed t -test; data not shown) after MMC treatment.
We next tested whether ddrR regulation of umuDAb transcription was simply due to umuDAb’s proximity next to ddrR, or if DdrR also repressed itself and other genes in the UmuDAb regulon: the umuC error-prone polymerases and umuD polymerase managers umuDC A1S_0636–0637; umuDrumB A1S_1174–1173; umuC A1S_2008; and umuC A1S_2015 [13, 17]. RT-qPCR experiments in JH1700 showed significantly increased expression of umuDAb and its regulon in the absence of DNA damage (Fig. 4), as did umuDAb mutants of 17 978 on umuDC expression [17]. ddrR also repressed its own expression, as evidenced by the similar increase in its expression in JH1700 cells in the absence of DNA damage (Fig. 4).
Further RT-qPCR experiments in both species showed that ddrR did not, however, regulate the expression of all DNA damage-induced genes. Several DNA damage-induced genes that are not regulated by UmuDAb [17], such as the glutathione S-transferase gene gst (A1S_0408 and ACIAD0445) and the nucleotide reductase gene nrdA (ACIAD0724), were not regulated by ddrR either (Fig. 5). Additionally, benzoate-induced benA genes (ACIAD1436 and A1S_1215), whose expression is unaffected by the umuDAb mutation, were similarly unaffected by ddrR mutation in JH1700 (Fig. 5).
The ddrR transcriptome shows coregulation of DNA-damage inducible genes with UmuDAb
To test whether ddrR also repressed (or otherwise regulated) additional genes, we conducted RNA-seq experiments on JH1700 cells that were either untreated or treated with MMC. Genes that were induced in this ddrR mutant more than two-fold after MMC treatment and had an FDR of less than 0.01 were considered to be differentially expressed (n=182). Roughly two-thirds of these (n=113) had been identified previously as MMC-inducible in WT cells, so their differential expression in JH1700 suggested that DdrR was not required for their induction. (None were derepressed in the absence of MMC treatment and further induced in the presence of MMC.) Most (95 %) of these DdrR-independent genes were located in the three cryptic prophages designated CP5, CP9 and CP14 [23] that contain ~90 % of the genes induced in the WT 17978 cells after MMC treatment [17].
Approximately 25 % (n=39 genes) of the established MMC-induced regulon of WT 17978 cells [13, 17] was regulated by ddrR (i.e. the genes were not MMC-induced in JH1700 cells; Table 4). UmuDAb also regulated the expression of 17 of these 39 ddrR-regulated genes (Fig. 6), which were flagged for further study (Table 4). Within the ddrR- and umuDAb-regulon was a group of DNA damage-inducible genes that are repressed before DNA damage and are induced after repression is lifted. These include the umuC error-prone polymerases and umuD polymerase managers, as well as umuDAb and ddrR, all of which are repressed by UmuDAb before MMC treatment [13, 17]. These eight genes were the only DNA damage-inducible genes whose expression was derepressed in JH1700 cells in the absence of DNA damage. RT-qPCR experiments validated the derepression seen in the RNA-seq data of each of these genes (Fig. 4).
Table 4.
DNA damage-inducible genes regulated by DdrR
|
Regulation |
Gene locus* |
Gene name/function |
Location† |
COG(s)‡ |
|---|---|---|---|---|
|
Repressed by DdrR and UmuDAb |
||||
|
|
A1S_1388 |
ddrR |
||
|
|
A1S_1389 |
umuDAb |
KT |
|
|
|
A1S_0636 A1S_0637 |
umuD |
pAB3 |
KT |
|
|
umuC |
pAB3 |
L |
|
|
|
A1S_1173 A1S_1174 |
rumB |
CP5 |
L |
|
|
umuD |
CP5 |
KT |
|
|
|
A1S_2008 |
umuC |
L |
|
|
|
A1S_2015 |
umuC |
CP9 |
L |
|
Activated by DdrR and UmuDAb |
||||
|
|
A1S_0278 |
trnT; Trp tRNA |
||
|
|
A1S_0421 |
infA; translation factor IF-1 |
J |
|
|
|
A1S_1144 |
repressor; S24 family peptidase |
CP5 |
K, KT |
|
|
A1S_3622 |
hypothetical |
CP5 |
N, T, M |
|
|
A1S_2014 |
SOS response-associated peptidase |
CP9 |
S |
|
|
A1S_2037 A1S_3774 A1S_3775 |
esvI; transcriptional regulator/repressor |
CP9 |
KT |
|
|
hypothetical |
CP9 |
||
|
|
hypothetical |
CP9 |
||
|
|
A1S_3704 |
holin |
CP14 |
|
|
Regulated by DdrR§ |
||||
|
|
A1S_1147 |
site-specific DNA methylase-like |
CP5 |
L |
|
|
A1S_3611 A1S_3612 A1S_3613 A1S_1148 A1S_1149 A1S_1151 |
hypothetical |
CP5 |
|
|
|
HNH endonuclease |
CP5 |
V |
|
|
|
hypothetical |
CP5 |
||
|
|
hypothetical |
CP5 |
S |
|
|
|
hypothetical |
CP5 |
||
|
|
hypothetical |
CP5 |
S |
|
|
|
A1S_3603 |
hypothetical |
CP5 |
|
|
|
A1S_3604 |
hypothetical |
CP5 |
|
|
|
A1S_3608 |
hypothetical |
CP5 |
|
|
|
A1S_3615 |
hypothetical |
CP5 |
L |
|
|
A1S_3621 |
hypothetical |
CP5 |
GEPR |
|
|
A1S_2031 |
phage protein |
CP9 |
S |
|
|
A1S_3755 |
holin |
CP9 |
GEPR |
|
|
A1S_3772 |
hypothetical |
CP9 |
|
|
|
A1S_3778 |
hypothetical |
CP9 |
|
|
|
A1S_3779 |
hypothetical |
CP9 |
|
|
|
A1S_3693 |
hypothetical |
CP14 |
|
|
|
A1S_3695 |
hypothetical |
CP14 |
|
|
|
A1S_3696 |
hypothetical |
CP14 |
|
|
|
A1S_3699 |
hypothetical |
CP14 |
|
|
|
A1S_3705 |
hypothetical |
CP14 |
*Gene loci appearing together in the same box indicate that these loci reside in the same operon.
†Where no location is given, a chromosomal location outside of a prophage is indicated.
‡COG, Cluster of Orthologous Groups; a method of functional annotation of genes based on orthology in complete microbial genomes [39].
§These genes were induced >2× after DNA damage, but their FDR values did not fall below 0.01 [or 0.05, with the exception of A1S_1149 (q=0.013) and A1S_3699, (q=0.016)], so they were not differentially expressed in the ddrR mutant and were thus considered to be regulated by DdrR.
Fig. 6.
Proportion of the DNA damage-inducible genes that are regulated by ddrR and umuDAb. The relationship between 17978 genes induced after MMC in WT cells and ddrR- and umuDAb-dependent genes induced is shown in an area proportional Venn diagram constructed using the BxToolBox at bioinforx.com.
Another group of nine DNA damage-inducible genes was co-regulated not through repression, but by being activated by both DdrR and UmuDAb. Expression of some of these genes did not increase in JH1700 cells after DNA damage. These included infA (translation initiation factor IF-1, A1S_0421) and three CP9 genes: esvI (putative phage repressor A1S_2037 [24]), a putative SOS response-associated peptidase (A1S_2014 [25]) and a hypothetical ORF (A1S_3774). The expression of the other five genes in this group increased more than two-fold after DNA damage, in both the ddrR and the umuDAb mutants (exception: A1S_0278, which was induced 1.8-fold in the umuDAb mutant), but were not considered differentially expressed. These showed, as a trend, lower expression after DNA damage than in WT cells. These included a putative CP5 phage repressor (A1S_1144), the tRNA gene trnT (A1S_0278) and three hypothetical ORFs located in the cryptic prophages (Table 4).
DdrR regulates some DNA-damage inducible genes without UmuDAb cooperation
Our analysis also identified 22 DNA damage-inducible genes whose induction was regulated by DdrR but not UmuAb (Table 4). These were located in the three cryptic prophages CP5, CP9 and CP14 and encoded mostly hypothetical phage proteins (Table 4). They were dependent upon RecA for their induction, like 99 % of the DNA damage-inducible genes in 17 978 [17]. These genes were not differentially expressed after DNA damage in JH1700 cells, although they were all induced more than two-fold (median 6.5-fold induced) and displayed lower expression in the absence of MMC treatment than in WT cells (P<0.05 in a t -test).
As a test of whether the lower expression in JH1700 was merely a characteristic of this strain, we evaluated the expression of 20 randomly chosen genes that were not DNA damage-inducible (Table S2). There was no significant difference (P>0.2 in a non-parametric repeated-measures ANOVA using a Friedman test) between expression of these genes in the WT and JH1700 strains in either the control or the MMC-treated condition.
DdrR also regulates genes that are not DNA damage-induced in WT cells
A fourth set of ddrR-regulated genes was differentially expressed and induced in JH1700 but had not been induced in WT cells (n=69). Further analysis comparing these ddrR RNA-seq reads to FPKM reads from a previous RNA-seq study [17] and also to a microarray study [13] indicated that 12 of these genes were induced by MMC in WT cells (five genes were induced in both studies). This reduced the number of ddrR-dependent, inducible genes to 57 (Table 5). These were largely chromosomally encoded, except for two genes on pAB3, and outside of cryptic prophages (except for three CP14 genes). This regulon contained multiple genes related to DNA replication, recombination and repair: a helicase, recX, ruvA, ruvB, parE and others (see Table 5 for gene identities). Multiple genes were identified as transcriptional regulators: a phage repressor A1S_1582 in CP14, as well as four AraC family, an AsnC family and an AcrR/TetR family regulators. General resistance–nodulation–cell division (RND) efflux (A1S_0008, 3146), RND transporters (adeFGH; A1S_2304–2306) and other transport functions (A1S_0023, 0030, 2068–2070, 2977) were also commonly represented.
Table 5.
Genes induced after DNA damage in ddrR mutant but not induced in WT
|
Gene locus* |
Name/function |
COG |
|---|---|---|
|
A1S_0005† |
put.‡ cytochrome b precursor |
C |
|
A1S_0008 |
put. RND type efflux pump |
P |
|
A1S_0023 |
put. malic acid transport protein |
P |
|
A1S_0030 |
alkanesulfonate transport protein |
P |
|
A1S_0094 |
lrp regulon transcriptional regulator (AsnC family) |
K |
|
A1S_0170 |
put. outer membrane copper receptor (OprC family) |
P |
|
A1S_0276† A1S_0277† |
trnY |
|
|
trnG |
|
|
|
A1S_0310 |
excinuclease ABC subunit C (esvL; ethanol-stimulated virulence protein [24]) |
L |
|
A1S_0422† |
put. transcriptional regulator (AraC family) |
K |
|
A1S_0564 |
put. translation initiation inhibitor (yjgF family) |
J |
|
A1S_0663 |
put. DNA helicase on plasmid pAB3 |
L |
|
A1S_0666 |
TrbL/VirB6 plasmid conjugal transfer protein on plasmid pAB3 |
U |
|
A1S_1124 |
transcriptional regulator (AraC family) |
K |
|
A1S_1377† |
transcriptional regulator (AcrR family) |
K |
|
A1S_1383 A1S_3661 |
surface Ag |
|
|
hypothetical |
|
|
|
A1S_1582† |
put. bacteriophage repressor C2 of prophage CP14 |
K, KT |
|
A1S_1614† |
hypothetical |
S |
|
A1S_1746 |
put. transcriptional regulator |
K |
|
A1S_1761 |
acetyltransferase |
KR |
|
A1S_1762† |
hypothetical |
K, E |
|
A1S_1963 |
recX regulatory protein |
R |
|
A1S_2068 |
put. benzoate membrane transport |
Q |
|
A1S_2069 A1S_2070† |
put. Mg2+ transporter transmembrane |
S |
|
mgtA; P-type ATPase Mg2+ ATPase transporter |
P |
|
|
A1S_2148 A1S_2149† A1S_2150 |
put. acetyl-CoA synthetase/AMP-(fatty) acid ligase |
I |
|
put. acyl CoA dehydrogenase oxidoreductase |
I |
|
|
oxidoreductase short-chain dehydrogenase/reductase family |
IQR |
|
|
A1S_2304 A1S_2305 A1S_2306 |
adeF RND family drug transporter |
M |
|
adeG cation/multidrug efflux pump |
V |
|
|
adeH put. RND family drug transporter |
MU |
|
|
A1S_2586 A1S_2587 A1S_2588† |
dGTP triphosphohydrolase |
F |
|
ruvA Holliday junction helicase subunit A |
L |
|
|
ruvB Holliday junction helicase subunit B |
L |
|
|
A1S_2963† |
purK; phosphoribosylaminoimidazole carboxylase ATPase subunit |
F |
|
A1S_2970 |
put. glutathione-like synthetase |
E |
|
A1S_2977 |
cation diffusion facilitator family transporter |
P |
|
A1S_3139† |
put. signal peptide |
|
|
A1S_3146† |
mdfA; Multidrug efflux transport protein |
GEPR |
|
A1S_3326 |
put. membrane protein |
S |
|
A1S_3359† |
parE; topoisomerase IV subunit B |
L |
|
A1S_3361 |
hypothetical |
R, R |
|
A1S_3385† |
put. membrane protein |
|
|
A1S_3428 A1S_3429 |
put. glucose dehydrogenase precursor |
G |
|
hypothetical |
S |
|
|
A1S_3472 |
plasmid replication (pAB2) |
L |
|
A1S_3485 |
hypothetical |
S |
|
A1S_3563 |
hypothetical; between nrdA and nrdB |
|
|
A1S_3574† |
peptide between 30S ribosomal proteins rpsG (S7) and rpsL (S12) |
|
|
A1S_3662 |
general stress protein |
R |
|
A1S_3690† |
hypothetical (CP14) |
|
|
A1S_3692† |
hypothetical (CP14) |
|
|
A1S_3709 |
hypothetical |
|
|
A1S_3865 |
hypothetical |
|
|
A1S_3877† |
hypothetical peptide |
|
*Gene loci appearing together in the same box indicate that these loci reside in the same operon.
†These genes were also induced in the umuDAb mutant but not in WT 17978 cells [17].
‡“put.” = Putative function.
Finally, 28 genes were repressed in JH1700, 14 of which were previously observed to be repressed in WT cells [17] (Table 6). In all but two genes out of these 28, the expression pattern (of being repressed or not repressed) was the same in the umuDAb mutant as in WT cells. None of these genes were located in the cryptic prophages.
Table 6.
A. baumannii genes repressed in the ddrR mutant
|
Gene locus |
Name/function |
Log2-fold change in ddrR mutant |
Repressed in wild type*? |
|---|---|---|---|
|
A1S_0292 |
put. outer membrane protein W |
−1.69 |
yes |
|
A1S_0391 |
50S ribosomal protein L31 type B |
−2.70 |
|
|
A1S_0548 |
put. transcriptional regulator (TetR family) |
−1.50 |
yes |
|
A1S_0549 |
hypothetical |
−1.42 |
yes |
|
A1S_0891 |
hemerythrin-like metal-binding protein |
−1.71 |
yes |
|
A1S_1216 A1S_1217 A1S_1218 |
LysR regulator |
−1.49 |
|
|
heavy metal translocating P-type ATPase |
−2.60 |
|
|
|
hmrR, copper-responsive HTH regulator (MerR family) |
−1.99 |
|
|
|
A1S_1319 |
hypothetical |
−1.59 |
* |
|
A1S_1467 |
put. glutamate symport transmembrane protein |
−1.54 |
|
|
A1S_1476 |
hypothetical |
−1.30 |
|
|
A1S_1734 |
hypothetical |
−1.31 |
|
|
A1S_1811 |
ankyrin-related protein |
−1.26 |
|
|
A1S_1924 A1S_1926 |
cydA, cytochrome d terminal oxidase |
−2.00 |
yes |
|
put. membrane protein |
−2.12 |
yes |
|
|
A1S_1927 |
put. acetyltransferase |
−1.24 |
* |
|
A1S_2098 |
put. alcohol dehydrogenase |
−1.77 |
|
|
A1S_2102 |
aldehyde dehydrogenase 1 |
−1.31 |
|
|
A1S_2210 |
hypothetical |
−1.69 |
|
|
A1S_2317 |
putative lipoprotein precursor (RlpA-like) |
−2.03 |
yes |
|
A1S_2936 |
copper resistance protein A precursor |
−2.11 |
|
|
A1S_3402 A1S_3403 A1S_3404 |
rocF hydrolase |
−1.69 |
yes |
|
hutI imidazolonepropionase |
−1.65 |
yes |
|
|
proline transport protein (APC family) |
−1.39 |
yes |
|
|
A1S_3627 |
hypothetical |
−2.15 |
yes |
|
A1S_3748 |
hypothetical |
−1.24 |
yes |
|
A1S_3794 |
hypothetical |
−1.68 |
yes |
|
A1S_3858 |
hypothetical |
−1.73 |
yes |
*Response for UmuDAb regulatory status is the same as for WT cells, except for where an asterisk symbol appears, denoting that the gene is also repressed in umuDAb mutant cells.
Discussion
Defining the source of ddrR–umuDAb transcriptional coregulation
Based on the overlap of the ADP1 inverted repeats (IRs) [18] with the ddrR and umuDAb −35 promoter consensus elements, and the similar overlap of these −35 elements with the 17 978 UmuDAb binding site [13], we propose that UmuDAb binding to DNA concurrently represses the transcription of both of these genes by blocking RNA polymerase access. In a previous study, A. baylyi strain JH100 showed no induction of ddrR and umuDAb expression under DNA damaging conditions [14]. JH100 possesses mutations in what this study defined as the ddrR −35 consensus element (changing it from TTGACG to GAGACG), and the umuDAb −35 consensus element (from TTGAAT to CAACGT). These mutations in the −35 elements are the most likely explanation for this loss of gene expression in this strain. The newly identified ddrR −35 element includes bases of the IRs identified in our previous study [18], which are identical in both species (Fig. 1), although it does not overlap the UmuDAb binding site proposed for 17978 cells [13]. These results also suggest that the required UmuDAb binding region may be larger than previously suggested [13]. It is interesting that the σ70 −35 promoter consensus elements are not located in the two separate IRs that compose the UmuDAb binding site, which contains TTGAA(A/T) inverted repeats. Rather, both of the −35 elements overlap the same IR, which is farthest from umuDAb. This arrangement allows the tightly coordinated expression of DdrR and UmuDAb. Our observations are consistent with the fact that other repressors (e.g. LexA) do not always bind to the same area within a promoter. Sometimes the repressor binding site overlaps with the −35 or −10 element, while other promoters contain the binding site between these two elements, downstream of the promoter elements, or even in the ORF itself [26]. The UmuDAb binding site was similarly placed upstream of umuDAb in both species but a variable distance upstream of ddrR as there is a longer intergenic region present in 17 978 relative to ADP1.
DdrR coregulates multiple genes with UmuDAb
Repression of six error-prone umuDC homologues, umuDAb and ddrR required both DdrR and UmuDAb. The ddrR mutation caused umuDAb overexpression but also yielded more expression of rumB, umuD, umuC, umuDAb and ddrR, which are repressed by UmuDAb. These observations suggest that DdrR might act as a corepressor to aid in UmuDAb repressive DNA binding. The UmuDAb N-terminal domain, which may possess a helix-turn-helix (HTH) structure like the DNA damage response repressor LexA, is required to repress DNA damage-induced genes [14]. In this model, the lack of DdrR might prevent UmuDAb repression from these genes’ promoters in a manner consistent with our observations. This action of ddrR suggests that DdrR is a corepressor, with UmuDAb, of a specific set of error-prone polymerases in Acinetobacter . The DdrR protein did not repress all DNA damage-inducible genes, but only corepressed the UmuDAb-repressed regulon. These results represent the first example of a LexA-like repressor using a corepressor to control host chromosomal genes (namely, A1S_2008, 1388 and 1389), although LexA often regulates horizontally acquired genetic elements such as plasmid or prophages via corepressors that may respond to nutritional or environmental factors [27–30].
The strength of the connection between DdrR and UmuDAb regulatory action was seen not just in ddrR corepressing UmuDAb-repressed genes, but also in ddrR coregulating genes that require UmuDAb for their increased expression after DNA damage. UmuDAb has a role in repressing transcription of specific genes before DNA damage [12, 14, 17], where UmuDAb self-cleavage at a conserved C-terminal site relieves this repression and induces gene expression [6]. However, some evidence suggests additional possible roles in the DNA damage response system of Acinetobacter , specifically in allowing or causing induction of genes after DNA damage [13]. In this study, besides the DdrR–UmuDAb corepressed genes, nine genes were coregulated by DdrR and UmuDAb, showing no significant increase in target gene expression after DNA damage in both the ddrR and umuDAb mutants. This suggests an additional coregulatory process involving both genes’ products where DdrR may act together with UmuDAb in this role as well as its actions as a repressor.
In the coregulated class of nine DNA damage-inducible proteins, it is striking that three are either phage repressors [A1S_1144 and A1S_2017 (esvI), an ethanol-stimulated virulence factor identified in a Caenorhabditis elegans screen of 17 978 [24]] or putatively associated with DNA damage sensing (A1S_2014). A1S_2014 was identified through comparative genetics as a member of a new SOS response-associated peptidase (SRAP) family [25], and is transcribed upstream of the umuC homologue A1S_2015, but was regulated differently from it in this study. These similarities suggest that besides UmuDAb, DdrR might work with additional repressors or regulatory proteins in a broader DNA stress response. Additionally, three of these nine genes appear to be, based on analyses of conserved gene pairs with OperonDB [31], encoded as part of the same CP9 operon: esvI and two hypothetical proteins A1S_3774 and 3775. DdrR and UmuDAb also coregulate the gene encoding initiation factor IF-1 (A1S_0421), which is responsible for translational initiation [32]. Mutation in either ddrR or umuDAb might result in an inability to induce the amounts of IF-1 needed to produce sufficient DNA damage response proteins to respond to DNA damaging conditions.
Compared to the ddrR- and umuDAb-coregulated genes, it is possible that the functions of the genes regulated only by umuDAb require less tight repression and earlier expression than the mutagenic, error-prone polymerases corepressed by ddrR, which are typically expressed late in the SOS response [33]. Alternatively, it may be that ddrR responds to an additional nutritional signal that is not related to UmuDAb-regulated genes that carry out different functions. This would be consistent with corepressor actions seen previously [27, 28, 34].
Regulation by DdrR can occur without UmuDAb involvement
This study showed that some (n=22) DdrR-regulated genes are not codependent on UmuDAb. In total, 99 % of the 17 978 DNA damage-inducible transcriptome [12, 17], and all but one of the 28 UmuDAb-regulated genes, are dependent upon RecA for their regulation (the notable exceptions being recA itself, and the hypothetical phage gene A1S_2020).
Finally, one-third (n=20) of the 57 genes whose expression was differentially induced in JH1700 cells but not in WT cells were located in seven potential operons or gene clusters of two or more adjacently transcribed genes. These operons are predicted by the OperonDB database [31] and/or MicrobesOnline predictions [35], which further supports the validity of this classification of ddrR-dependent genes. These gene clusters included a triphosphohydrolase and Holliday junction helicase genes ruvAB A1S_2586–2588, Mg2+ transport genes A1S_2069–2070, fatty acid ligase and oxidoreductases A1S_2148–50, RND efflux transporters A1S_2304–2306, and multiple genes of unknown function. The A1S_2304–2306 operon is significant because these are known virulence genes in A. baumannii that when overexpressed are associated with biofilm formation and drug resistance [36] and induced by non-DNA damage stresses such as NaCl [37]. Using FPKM comparisons to an earlier study [17], we observed that 20 of these 57 genes had also been induced in a umuDAb mutant but not in the WT cells. These did not constitute particular clusters of genes, however, with typically one, or none, of three genes in a cluster showing regulation by UmuDAb. This induction was recA-dependent in nearly all of these 57 genes except A1S_1383 and A1S_3661, which were identified as co-located in an operon, and the hypothetical A1S_3877. This RecA-dependence allows for the possibility that DdrR may be working through a RecA-sensitive DNA damage-sensing component, or is itself RecA-sensitive.
In these experiments, we examined the role of the ddrR gene in two different Acinetobacter species. Both the model soil microbe A. baylyi ADP1 and the opportunistic pathogen A. baumannii strain 17978 show corepression of the regulatory pair ddrR–umuDAb in the absence of DNA damage. However, the A. baylyi ddrR mutant strains, which expressed umuDAb in the uninduced condition similar to that of WT cells after DNA damage (Fig. 3), still further induced expression of these genes from that higher expression level. 17978 cells showed no additional induction in any UmuDAb-DdrR corepressed gene. As ADP1 does not possess any umuDC operons or unassociated umuC gene targets of DdrR–UmuDAb repression, we could not directly test whether this observation extended to other genes in ADP1, as we could for 17 978. One speculation is that a DdrR–UmuDAb–promoter physical interaction is stronger in A. baumannii , perhaps due to selection for tighter control of its several umuDC error-prone polymerases. DdrR and UmuDAb are 60 and 79% identical between the two species, respectively, allowing for this possibility.
The coordinate DdrR–UmuDAb repression of 17 target genes in A. baumannii , and of ddrR–umuDAb in both species, is facilitated by the joint auto-regulation of DdrR and UmuDAb. In our RNA-seq analyses, we identified multiple regulons of DdrR-regulated genes, including a regulon corepressed in conjunction with UmuDAb and a regulon coregulated with UmuDAb, which lacked induction when either protein was absent. In addition, DdrR regulated a group of 22 genes that do not depend on UmuDAb for their increased expression after DNA damage, as well as a group of 57 genes that were only differentially regulated (and induced) when ddrR was mutated. The unusual and atypical mechanisms and genes that this pathogen uses to control and cause its mutagenic responses to DNA damage highlight the importance of deciphering the response system used by the genus Acinetobacter .
Supplementary Data
Funding information
NIH R15GM085722 to J.H. supported this work. Part of this work was performed with the assistance of the University of Louisville Genomics Facility, which is supported by NIH NIGMS P20GM103436 (KY IDeA Networks of Biomedical Research Excellence), NIH P30GM106396 to the University of Louisville J. G. Brown Cancer Center Phase III CoBRE, the J. G. Brown Foundation, and user fees.
Author contribution
M. P. developed the methodology, M. P., A. G., J. H. performed the experiments; A. G. and J. H. validated the experimental results; J. H. conducted the statistical analyses; J. H. acquired the financial support and strain resources; M. P. and J. H. prepared the visuals; J. H. wrote the manuscript; M. P., A. G., J. H. reviewed and edited the manuscript; all the authors have read the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: FDR, false discovery rate; FPKM, fragments per kilobase of transcript per million fragments mapped reads; IR, inverted repeat; MMC, mitomycin C; RACE, rapid amplification of cDNA ends; RNA-seq, RNA sequencing; RND, resistance–nodulation–cell division; RT-qPCR, real-time quantitative PCR.
Two supplementary tables are available with the online version of this article.
Edited by: D. Grainger and D. Lee
References
- 1.Friedberg EC. DNA Repair and Mutagenesis. Washington, DC: ASM Press; 1995. [Google Scholar]
- 2.Battista JR, Donnelly CE, Ohta T, Walker GC. The SOS response and induced mutagenesis. Prog Clin Biol Res. 1990;340A:169–178. [PubMed] [Google Scholar]
- 3.Mount DW, Low KB, Edmiston SJ. Dominant mutations (Lex) in Escherichia coli K-12 which affect radiation sensitivity and frequency of ultraviolet lght-induced mutations. J Bacteriol. 1972;112:886–893. doi: 10.1128/jb.112.2.886-893.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Little JW, Edmiston SH, Pacelli LZ, Mount DW. Cleavage of the Escherichia coli lexA protein by the recA protease. Proc Natl Acad Sci USA. 1980;77:3225–3229. doi: 10.1073/pnas.77.6.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Little JW, Mount DW, Yanisch-Perron CR. Purified lexA protein is a repressor of the recA and lexA genes. Proc Natl Acad Sci USA. 1981;78:4199–4203. doi: 10.1073/pnas.78.7.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hare JM, Adhikari S, Lambert KV, Hare AE, Grice AN. The Acinetobacter regulatory UmuDAb protein cleaves in response to DNA damage with chimeric LexA/UmuD characteristics. FEMS Microbiol Lett. 2012;334:57–65. doi: 10.1111/j.1574-6968.2012.02618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jara LM, Cortés P, Bou G, Barbé J, Aranda J. Differential roles of antimicrobials in the acquisition of drug resistance through activation of the SOS response in Acinetobacter baumannii . Antimicrob Agents Chemother. 2015;59:4318–4320. doi: 10.1128/AAC.04918-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Macguire AE, Ching MC, Diamond BH, Kazakov A, Novichkov P, et al. Activation of phenotypic subpopulations in response to ciprofloxacin treatment in Acinetobacter baumannii . Mol Microbiol. 2014;92:138–152. doi: 10.1111/mmi.12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hare JM, Bradley JA, Lin CL, Elam TJ. Diverse responses to UV light exposure in Acinetobacter include the capacity for DNA damage-induced mutagenesis in the opportunistic pathogens Acinetobacter baumannii and Acinetobacter ursingii . Microbiology. 2012;158:601–611. doi: 10.1099/mic.0.054668-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Norton MD, Spilkia AJ, Godoy VG. Antibiotic resistance acquired through a DNA damage-inducible response in Acinetobacter baumannii . J Bacteriol. 2013;195:1335–1345. doi: 10.1128/JB.02176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aranda J, López M, Leiva E, Magán A, Adler B, et al. Role of Acinetobacter baumannii UmuD homologs in antibiotic resistance acquired through DNA damage-induced mutagenesis. Antimicrob Agents Chemother. 2014;58:1771–1773. doi: 10.1128/AAC.02346-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Norton MD, Spilkia AJ, Godoy VG. Antibiotic resistance acquired through a DNA damage-inducible response in Acinetobacter baumannii . J Bacteriol. 2013;195:1335–1345. doi: 10.1128/JB.02176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aranda J, Poza M, Shingu-Vázquez M, Cortés P, Boyce JD, et al. Identification of a DNA-damage-inducible regulon in Acinetobacter baumannii . J Bacteriol. 2013;195:5577–5582. doi: 10.1128/JB.00853-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Witkowski TA, Grice AN, Stinnett DB, Wells WK, Peterson MA, et al. UmuDAb: an error-prone polymerase accessory homolog whose N-terminal domain is required for repression of DNA damage inducible gene expression in Acinetobacter baylyi . PLoS One. 2016;11:e0152013. doi: 10.1371/journal.pone.0152013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hare JM, Perkins SN, Gregg-Jolly LA, Constitutively Expressed A. A constitutively expressed, truncated umuDC operon regulates the recA-dependent DNA damage induction of a gene in Acinetobacter baylyi strain ADP1. Appl Environ Microbiol. 2006;72:4036–4043. doi: 10.1128/AEM.02774-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelley WL. Lex marks the spot: the virulent side of SOS and a closer look at the LexA regulon. Mol Microbiol. 2006;62:1228–1238. doi: 10.1111/j.1365-2958.2006.05444.x. [DOI] [PubMed] [Google Scholar]
- 17.Hare JM, Ferrell JC, Witkowski TA, Grice AN. Prophage induction and differential RecA and UmuDAb transcriptome regulation in the DNA damage responses of Acinetobacter baumannii and Acinetobacter baylyi . PLoS One. 2014;9:e93861. doi: 10.1371/journal.pone.0093861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hare JM, Perkins SN, Gregg-Jolly LA. A constitutively expressed, truncated umuDC operon regulates the recA-dependent DNA damage induction of a gene in Acinetobacter baylyi strain ADP1. Appl Env Microbiol. 2006;72:4036–4043. doi: 10.1128/AEM.02774-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Centers for Disease Control and Prevention Antibiotic Resistance Threats in the United States. Atlanta, GA: CDC; 2013. pp. 1–114. [Google Scholar]
- 20.Kokotek W, Lotz W. Construction of a lacZ-kanamycin-resistance cassette, useful for site-directed mutagenesis and as a promoter probe. Gene. 1989;84:467–471. doi: 10.1016/0378-1119(89)90522-2. [DOI] [PubMed] [Google Scholar]
- 21.de Berardinis V, Vallenet D, Castelli V, Besnard M, Pinet A, et al. A complete collection of single-gene deletion mutants of Acinetobacter baylyi ADP1. Mol Syst Biol. 2008;4:174. doi: 10.1038/msb.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di Nocera PP, Rocco F, Giannouli M, Triassi M, Zarrilli R. Genome organization of epidemic Acinetobacter baumannii strains. BMC Microbiol. 2011;11:224. doi: 10.1186/1471-2180-11-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith MG, Gianoulis TA, Pukatzki S, Mekalanos JJ, Ornston LN, et al. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 2007;21:601–614. doi: 10.1101/gad.1510307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aravind L, Anand S, Iyer LM. Novel autoproteolytic and DNA-damage sensing components in the bacterial SOS response and oxidized methylcytosine-induced eukaryotic DNA demethylation systems. Biol Direct. 2013;8:20. doi: 10.1186/1745-6150-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schnarr M, Oertel-Buchheit P, Kazmaier M, Granger-Schnarr M. Dna binding properties of the LexA repressor. Biochimie. 1991;73:423–431. doi: 10.1016/0300-9084(91)90109-E. [DOI] [PubMed] [Google Scholar]
- 27.Kamenšek S, Browning DF, Podlesek Z, Busby SJW, Žgur-Bertok D, et al. Silencing of DNase colicin E8 gene expression by a complex nucleoprotein assembly ensures timely colicin induction. PLoS Genet. 2015;11:e1005354. doi: 10.1371/journal.pgen.1005354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Butala M, Sonjak S, Kamenšek S, Hodošček M, Browning DF, et al. Double locking of an Escherichia coli promoter by two repressors prevents premature colicin expression and cell lysis. Mol Microbiol. 2012;86:129–139. doi: 10.1111/j.1365-2958.2012.08179.x. [DOI] [PubMed] [Google Scholar]
- 29.Fornelos N, Butala M, Hodnik V, Anderluh G, Bamford JK, et al. Bacteriophage GIL01 gp7 interacts with host LexA repressor to enhance DNA binding and inhibit RecA-mediated auto-cleavage. Nucleic Acids Res. 2015;43:7315–7329. doi: 10.1093/nar/gkv634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quinones M, Kimsey HH, Waldor MK. Lexa cleavage is required for CTX prophage induction. Mol Cell. 2005;17:291–300. doi: 10.1016/j.molcel.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 31.Pertea M, Ayanbule K, Smedinghoff M, Salzberg SL. OperonDB: a comprehensive database of predicted operons in microbial genomes. Nucleic Acids Res. 2009;37:D479–D482. doi: 10.1093/nar/gkn784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laursen BS, Sørensen HP, Mortensen KK, Sperling-Petersen HU. Initiation of protein synthesis in bacteria. Microbiol Mol Biol Rev. 2005;69:101–123. doi: 10.1128/MMBR.69.1.101-123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli . Genetics. 2001;158:41. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nedialkova LP, Denzler R, Koeppel MB, Diehl M, Ring D, et al. Inflammation fuels colicin Ib-dependent competition of Salmonella serovar typhimurium and E. coli in enterobacterial blooms. PLoS Pathog. 2014;10:e1003844. doi: 10.1371/journal.ppat.1003844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price MN, Huang KH, Alm EJ, Arkin AP. A novel method for accurate operon predictions in all sequenced prokaryotes. Nucleic Acids Res. 2005;33:880–892. doi: 10.1093/nar/gki232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He X, Lu F, Yuan F, Jiang D, Zhao P, et al. Biofilm formation caused by clinical Acinetobacter baumannii isolates is associated with overexpression of the AdeFGH efflux pump. Antimicrob Agents Chemother. 2015;59:4817–4825. doi: 10.1128/AAC.00877-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hood MI, Jacobs AC, Sayood K, Dunman PM, Skaar EP. Acinetobacter baumannii increases tolerance to antibiotics in response to monovalent cations. Antimicrob Agents Chemother. 2010;54:1029. doi: 10.1128/AAC.00963-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/S0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 39.Galperin MY, Makarova KS, Wolf YI, Koonin EV. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2015;43:D261–D269. doi: 10.1093/nar/gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.