Abstract
Germline variation in PTEN results in variable clinical presentations, including benign and malignant neoplasia and neurodevelopmental disorders. Despite decades of research, it remains unclear how the PTEN genotype is related to clinical outcomes. In this study, we combined two recent deep mutational scanning (DMS) datasets probing the effects of single amino acid variation on enzyme activity and steady-state cellular abundance with a large, well-curated clinical cohort of PTEN-variant carriers. We sought to connect variant-specific molecular phenotypes to the clinical outcomes of individuals with PTEN variants. We found that DMS data partially explain quantitative clinical traits, including head circumference and Cleveland Clinic (CC) score, which is a semiquantitative surrogate of disease burden. We built logistic regression models that use DMS and CADD scores to separate clinical PTEN variation from gnomAD control-only variation with high accuracy. By using a survival-like analysis, we identified molecular phenotype groups with differential risk of early cancer onset as well as lifetime risk of cancer. Finally, we identified classes of DMS-defined variants with significantly different risk levels for classical hamartoma-related features (odds ratio [OR] range of 4.1–102.9). In stark contrast, the risk for developing autism or developmental delay does not significantly change across variant classes (OR range of 5.4–12.4). Together, these findings highlight the potential impact of combining DMS datasets with rich clinical data and provide new insights that might guide personalized clinical decisions for PTEN-variant carriers.
Keywords: PTEN, PTEN hamartoma tumor syndrome, PHTS, autism spectrum disorder, ASD, autism, cancer, deep mutational scanning, genotype-phenotype, multiplex assay for variant effect
Introduction
Germline variation of the tumor suppressor gene phosphatase and tensin homolog (PTEN [MIM: 601728]) manifests with variable and complex phenotypes, including macrocephaly (with increased occipital-frontal circumferences [OFC]), benign hamartomas affecting all three germ layers, malignant neoplasia across multiple tissues, and neurodevelopmental abnormalities, including autism spectrum disorder (ASD).1,2 This heterogeneity is reflected clinically with germline pathogenic PTEN variants found in variable subsets of defined syndromes, including Cowden syndrome and Bannayan-Riley-Ruvalcaba syndrome (CWS1 and BRRS [MIM: 158350]) as well as macrocephalic ASD (MAS [MIM: 605309]), among others.2, 3, 4, 5 Collectively, these syndromes have been termed PTEN hamartoma tumor syndrome (PHTS) when a germline PTEN variant is identified.1,2
The dramatic variability of these clinical presentations has sparked efforts to correlate PTEN variants with clinically relevant phenotypic classes. However, PTEN variants resist simple classification approaches based on secondary domain clustering or variant type. Recently, the mapping of a limited subset of germline PTEN variants onto the three-dimensional crystal structure failed to reveal a distinct pattern of distribution between ASD- or cancer-predisposition-associated variants.6 Classification efforts have also been impacted by sample sizes limited in terms of both functional data and PTEN variant cohorts.7,8 Additionally, PTEN has multiple functional roles in the cell apart from lipid phosphatase activity, which might also play a role in this phenotypic complexity.2,9, 10, 11
These challenges have prompted recent creative high-throughput methods to functionally measure the molecular phenotypes for thousands of nonsynonymous PTEN variants; these methods are collectively termed deep mutational scanning (DMS).8,12 We previously reported the effect of nearly all PTEN nonsynonymous variants on lipid phosphatase activity by utilizing a humanized yeast assay where lipid phosphatase activity was linked to cell survival (via the so-called fitness score).8 These data demonstrated that the solvent exposure of a wild-type (WT) residue is a critical determinant of mutational tolerance for lipid phosphatase fitness: solvent-exposed residues are much more tolerant to mutation. As expected, PTEN lipid phosphatase activity was generally intolerant to mutation in the catalytic pocket and phosphatase domain, though not without exception. Furthermore, in line with suggestions from prior, more limited functional studies,7 PTEN missense variants associated with ASD tended to retain partial lipid phosphatase activity.8
In a second independent study, the effect of ∼54% of all PTEN nonsynonymous variants on the steady-state cellular protein abundance (the so-called abundance score) was estimated via fluorescently tagged PTEN variants. It was observed that PTEN abundance is, in part, explained by the thermodynamic stability and cell-membrane interactions of a given variant. Although variant abundance inversely correlates with pathogenicity, notable exceptions are putative dominant-negative PTEN variants, which are highly stable but catalytically inactive.12
Although these two DMS studies have led to essential insights into the effect of PTEN variants on protein function, they were limited in their clinical analyses because both relied on previously published clinical reports and ClinVar database13 variants with varying degrees of validation and phenotypic description. In this study, to further uncover PTEN genotype-phenotype relationships and clarify individual risk for these diverse clinical presentations, we integrated these datasets with a large, prospectively accrued and comprehensively clinically characterized cohort of PTEN variant-positive individuals (the Cleveland Clinic [CC] cohort). These analyses demonstrate that molecular phenotypes associate with quantitative clinical traits. They also delineate differential lifetime cancer risk and indicate unexpected risk ratio relationships for neurodevelopmental and hamartoma-associated phenotypes.
Material and Methods
PTEN Variant Function Data and Imputation
We made use of two DMS datasets in this study.8,12 In brief, fitness scores were previously determined via the assessment of a PTEN variant’s ability to reverse toxicity by means of phosphatidylinositol (3,4,5)-triphosphate (PIP3) dephosphorylation in a humanized yeast system14 that expresses a hyperactive kinase.8 High-confidence fitness scores were previously generated for 86% of all variants, and a random-forest algorithm was used for imputation of fitness scores for the remaining unmeasured variants.8 Abundance scores were previously determined via the measurement of the steady-state abundance of PTEN variants with the VAMP-Seq assay in human cells.12 Abundance scores were generated for 54% of all variants.
By using a random-forest framework similar to what was used for the imputation of fitness scores, here, we imputed abundance scores for the remaining unmeasured variants (Figure S1). Modeling was implemented in Scikit-learn version 0.19.0 (“sklearn.ensemble.RandomForestRegressor,” “n_estimators” = 500, criterion = “mse,” “max_features” = 0.33, “random_state” = 0, “oob_score” = true). We determined feature importance by training random-forest models on the full dataset iteratively and randomly permuting a feature each time. The increase in error upon permutation of a feature is related to the importance of that feature.
Once we had calculated relative feature importance, we iteratively performed 10-fold cross-validation, i.e., we trained the model on 90% of data and tested on the remaining 10%. The starting model used the feature with the highest importance and the position average, i.e., the average score of all other substitution variants at that amino acid position and the n − 1 and n + 1 positions. If there were no measured variants at the n − 1, n, or n + 1 positions, we included the n − 2 and n + 2 positions. We then iteratively performed 10-fold cross-validation with models incorporating features in decreasing order of their importance until the Pearson correlations between predicted and observed scores plateaued. The final model was again assessed via 10-fold cross-validation (Figure S1; Table S1). Finally, we used the final model trained on all measured abundance scores to predict all unmeasured variants.
We classified the full set of missense protein variants (measured and imputed) as WT-like, hypomorphic, or truncation-like for fitness and abundance scores (Figure S2; Table S1). For fitness score, we considered variants WT-like if they were within the 2.5th or 97.5th percentile of synonymous WT fitness scores (Figures S2C and S2D). We considered variants truncation-like if their fitness scores were within the 2.5th or 97.5th percentile of nonsense variants at positions 1–350, excluding the regulatory tail because nonsense mutations in the tail are not damaging in the yeast assay. We considered variants hypomorphic for fitness score if they were between the WT-like and truncation-like bounds.
We classified WT-like variants similarly for abundance score but with a slight adjustment to the distribution boundaries (Figures S2C and S2D). Because the abundance-score distribution tails were larger than the fitness-score distribution tails, we defined the bounds as the 5th percentile and the 95th percentile of synonymous WT distribution. We considered variants to be truncation-like for abundance score if they were within the range of the 5th percentile and the 95th percentile of nonsense variants at positions 30–300, in order to exclude experimental artifacts known to exist for variants near the protein termini as a result of the nature of the fusion protein used in the experiments.12
PTEN Population Variants from gnomAD
Data from the controls-only subset were downloaded from gnomAD v2.115 on January 10, 2019 (Table S2). For the cancer-incidence analysis and the clinical-outcomes odds ratio analyses, we included all controls-only gnomAD nonsynonymous variants (e.g., missense, nonsense, and indel frameshift). For the pathogenic versus benign analysis, we considered only gnomAD missense variants (i.e., we excluded frameshifting or truncation variants) with the exceptions of p.Arg173His (c.518G>A) and p.Lys289Glu (c.865A>G), which are classified as pathogenic or likely pathogenic in ClinVar. We also excluded p.Asp268Glu (c.804C>A), which occurs at a frequency greater than an order of magnitude above the frequency of most other variants.
Cleveland Clinic PTEN Cohort
This study was performed in accordance with the institutional review board 8458 protocol “Molecular Mechanisms Involved in Cancer Predisposition,” substudy PTEN, which has been approved by the Cleveland Clinic Institutional Review Board for Human Subjects’ Protection, and conducted with informed consent and in accordance with the World Medical Association Declaration of Helsinki. The CC cohort consists of 256 prospectively accrued individuals with germline PTEN nonsynonymous variants (145 missense and 111 nonsense variants; Tables 1, S3, and S4). The nonsense group includes nine individuals with insertions or deletions that result in an immediate termination codon. Genotype information concerning each individual’s germline PTEN variant as well as demographic and clinical data were also included. Collection and validation of clinical phenotypes were performed by experienced clinical personnel as detailed in a previous study.16 Demographic information includes age at the last clinical follow-up; sex; and age at diagnosis for various clinical phenotypes, including macrocephaly, neurodevelopmental pathologies (including ASD and developmental delay [DD]), and several different types of benign and malignant neoplasia. Adult individuals in the cohort were assigned a CC score, which is a derived sum of the weights of specific neurological, breast and gynecological, gastrointestinal, skin, endocrine, and genitourinary clinical features, assessed by clinical specialists. Both benign and malignant clinical features, including age of onset, are factored into the CC score. Moreover, the CC score is a validated, individualized estimate of the pretest probability of having a germline pathogenic PTEN variant. For example, a score of 15 indicates a 10% probability of one’s having a pathogenic variant. Given the methodology for calculating CC score, it also serves as a semiquantitative measure of the burden of disease: larger scores indicate an increasing disease burden and/or younger ages of onset. However, the scoring is only applicable to and validated for the adult population (individuals 18 years and older).16 OFC Z scores were calculated with published age-indexed tables.17
Table 1.
Cleveland Clinic Cohort of Individuals with Germline Nonsynonymous Variation in PTEN
| Phenotype |
Missense |
True truncations |
Total |
||||
|---|---|---|---|---|---|---|---|
| All n (%) | % Male | All n (%) | % Male | All n (%) | % Male | % Mis. | |
| All | 145 (100) | 40.7 | 111a (100) | 49.5 | 256 (100) | 44.5 | 56.6 |
| ASD/DD | 23 (15.9) | 78.3 | 6 (5.41) | 100.0 | 29 (11.3) | 82.8 | 79.3 |
| ASD/DD & PHTS | 32 (22.1) | 68.8 | 24 (21.6) | 75.0 | 56 (21.9) | 71.4 | 57.1 |
| PHTS | 90 (62.1) | 21.1 | 80 (72.1) | 38.8 | 170 (66.4) | 29.4 | 52.9 |
One individual did not have qualifying symptoms for any phenotype group.
Individuals were considered ASD/DD positive if they presented with ASD, DD, variable delay, or intellectual disability. Individuals were considered PHTS positive if they presented hamartomatous features, including any of the following: benign or malignant tumors, mucocutaneous lesions, arteriovenous malformation, lipomas, goiter, or uncommon skin lesions. Individuals with the common skin findings of skin tags, café-au-lait marks, or penile freckling in isolation, meaning without another hamartomatous feature, were not included in the PHTS group. Individuals who displayed both the neurodevelopmental and hamartomatous features were placed in the ASD/DD & PHTS grouping.
Logistic Regression Modeling for Pathogenic PTEN Variation
To test the accuracy of models with all combinations of features (e.g., fitness scores, abundance scores, and CADD scores), we determined the optimal regularization parameters (L1 versus L2 regularization and regularization strength) for each feature combination by using the GridSearchCV function within scikit learn. We assessed the performance of each model with 10-fold cross-validation, i.e., we iteratively trained models on 90% of the data and used that model to make predictions for the outstanding 10%. We repeated this procedure in order to make predictions for all variants. Once we determined that no multivariate model performed better than the univariate fitness score model, we retrained the fitness score model on the entire set of known pathogenic and benign variants. The optimal model used L1 regularization with strength of 1.0. We then used this model to predict the probability of pathogenicity for all single-amino-acid PTEN variants.
Cancer Incidence and Survival Analysis
For fitness- and abundance-score analyses, we classified all individuals from the CC cohort and gnomAD into WT-like missense, hypomorphic missense, truncation-like missense, or true truncation (i.e., nonsense or frameshifting) groups (Figure S2). For the combined molecular-score analysis, we used the hypomorphic cutoff to designate variants as deficient (minus) for fitness or abundance (i.e., −1.11 for fitness score and 0.71 for abundance score). We assumed the gnomAD individuals were cancer-free. The observation period for each subject was set from birth to age at the last clinical follow-up. For 26 of the 164 gnomAD individuals, we could unambiguously determine their age range by referring to data provided in the full gnomAD v2.1 variant call file. Because these data were provided in 5-year increments, we randomly selected a single year from this range for each individual. For the rest of the gnomAD control cohort, we obtained the distribution of ages from the gnomAD FAQ page. We randomly sampled an age range from the weighted distribution of age ranges and then randomly generated an age within that range. The imputed ages reflected the original age distribution in the gnomAD database well (goodness of fit chi-square = 0.30; degrees of freedom = 12, not significant). Differences in cancer incidence between the genotype groups were compared with the Kaplan-Meyer method and log-rank test. Analyses were performed for overall cancer incidence, and individuals were right-censored at the age at cancer diagnosis or age at the last clinical follow-up. Significant group differences were then examined via pairwise comparisons. In order to detect potential differences in early-onset cancer incidence, we further compared survival curves at age 35 years, and right-censoring occurred at age of early-onset (<35) cancer or otherwise at age 35.
Calculating Odds Ratios for Clinical Outcomes
Although all individuals with an identified germline pathogenic PTEN variant are clinically classified as belonging to the overarching classification of PHTS, we have developed clinical subgroupings with differing presentations in order to enable genotype-phenotype analyses in this study. All individuals with nonsense variants were treated as true truncations because all of these variants occurred upstream of both the final exon and the C-terminal tail. We used Statistical Package for the Social Sciences (SPSS) statistical software (version 25) from International Business Machines (IBM) to perform logistic regression modeling on ASD/DD or PHTS outcomes and survival analyses and used molecular phenotypes as exposures.
Results
Distribution of Missense Variation across the Primary and Crystal Structures of PTEN
In order to examine PTEN genotype-phenotype relationships, we prospectively accrued a cohort of individuals with germline nonsynonymous variation in PTEN (Table 1). Previously, PHTS has been used as an umbrella term specifically for classically defined PTEN-related disorders (e.g., CWS1 and BRRS).18 Subsequently, as the phenotypic spectrum of pathogenic PTEN variants expanded, PHTS became a descriptor for all clinical presentations associated with germline PTEN variation.2 In order to explore potential differences between ASD/DD-related phenotypes and those associated with hamartoma and/or cancer phenotypes, we operationally grouped individuals with the classic hamartoma-related PTEN features as PHTS, whereas individuals with largely neurodevelopmental clinical features were designated as ASD/DD (Materials and Methods). Individuals with a combination of neurodevelopmental and hamartoma-related features were designated as a third group, ASD/DD & PHTS.
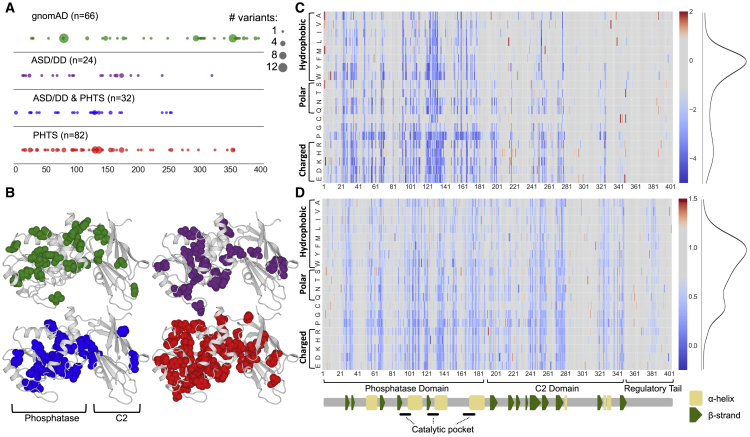
The cohort recapitulates the previously observed relative enrichment of ASD/DD phenotypes among those with missense as opposed to nonsense variation (16% versus 5%, respectively; Table 1).11,19 As a comparison, we also collated nonsynonymous variant data from control-only individuals in gnomAD, a database that aggregates sequencing studies.15 Because individuals with pediatric disorders are excluded from gnomAD, and because these individuals were specifically accrued as unaffected controls, they were assumed to be free of PTEN-related disorders. We categorized missense variants by associated clinical group and then mapped variants to the primary and crystal structures of PTEN; the functional domains are annotated, including the variants cataloged in gnomAD (Figures 1A and 1B). The clinical missense variants cluster most heavily in the dual-specificity phosphatase domain (residues 1–178) and are depleted in the C2 domain and tail (residues 179–403), reflecting the importance of the phosphatase domain to PTEN function (Figure 1A). Comparing all clinical variants with gnomAD variants demonstrates an enrichment of gnomAD variants in the C2 domain and tail, as compared to in the phosphatase domain (odds ratio = 5.19, 95% confidence interval (CI) = 2.5–10.8, p = 1.6 × 10−6, Fisher’s exact test; Figure 1B). In contrast, and consistent with similar studies,6 the distributions of variants were similar across clinical outcomes (p = 0.78 for ASD/DD versus PHTS, p = 0.71 for ASD/DD versus ASD/DD & PHTS, and p = 0.42 for PHTS versus ASD/DD & PHTS, Fisher’s exact test; Figure 1A). In three-dimensional space, gnomAD variants are significantly more solvent exposed (i.e., exposed at the surface of the protein) than the group of all clinical variants (medians of 87.4% versus 7.8%, respectively, p = 1.28 × 10−18, Mann-Whitney U test; Figure 1B). Variation at solvent-exposed positions is generally more tolerated than at positions that are not solvent exposed because these variants are less likely to disrupt protein structure.20
Figure 1.
Overview of the Datasets Used in this Study
(A) Diagram of PTEN primary protein structure with locations of PTEN missense variants found in the controls-only gnomAD population or associated with various clinical presentations in the CC cohort. The major protein domains and secondary-structure assignments are indicated. The size of the circle indicates the number of different amino acid variants at that position.
(B) Diagram of PTEN three-dimensional crystal structure with locations of PTEN missense variants found in the controls-only gnomAD population or associated with various clinical presentations in the CC cohort. The C-terminal tail was not solved in the crystal structure, and therefore, variants falling in this region are not shown. The color of the spheres indicates the same groups as shown in (A).
(C) Lipid-phosphatase fitness scores are displayed as a heatmap: blue corresponds to damaging variants (i.e., low lipid phosphatase fitness or low abundance), gray corresponds to WT-like variants, and red corresponds to putative fitness- or abundance-increasing variants (Materials and Methods). The distributions of all missense variants are shown as smoothed histograms on the right.
(D) Cellular abundance scores displayed as in (C).
Visualization and Imputation of Molecular Phenotypes of PTEN
We hypothesized that variant-level molecular-phenotype data might uncover new genotype-phenotype associations, on the basis of the reasoning that protein function data should correlate better with clinical outcome than with variant locations in primary or tertiary sequence space. We aggregated molecular phenotype information derived from recent DMS studies on the effect of thousands of variants on PTEN function, including inferred lipid-phosphatase activity (i.e., fitness score) and steady-state protein stability (i.e., abundance score).8,12 Previously, we demonstrated that using a random-forest-based machine-learning modeling approach allowed fitness scores of variants withheld from model training to be imputed with high accuracy. The model incorporated the position average effect of variants missing from a nearly complete DMS dataset (86% saturation) with biophysical, biochemical, and evolutionary data. Therefore, by using the imputations from this model, we previously constructed a comprehensive lipid-phosphatase functional map of fitness scores (Figure 1C).
We previously showed by downsampling the fitness dataset that a similar strategy could be used for less complete DMS datasets and still result in highly accurate predictions.8 We developed a similar modeling strategy for the protein abundance DMS dataset, which was at ∼54% saturation. Cross-validation showed the best-performing model could predict withheld abundance scores with an accuracy similar to that of biologic replicates (Pearson r = 0.75; Figure S1). Therefore, by using this approach, we imputed abundance scores for all missing missense variants (Figure 1D; Table S1). Combined, these complete datasets represent estimates of the effect of any given PTEN missense variant on the lipid phosphatase activity and steady-state abundance of PTEN. All analyses presented here used the combination of high-confidence-measured and imputed scores.
Fitness scores are modestly correlated with abundance scores (Pearson’s r = 0.43; Figure S2E), suggesting some information overlap but that each assay is also capturing (and failing to capture) unique variant effects on protein function. We used the distribution of programmed truncating (nonsense) and synonymous variants in these assays to define truncation-like, hypomorphic, and WT-like missense variant categories (Materials and Methods; Figures S2A–S2D). The missense variants in both datasets are bimodally distributed, and the majority of variants have WT-like scores (Figures 1C, 1D, and S2). We found that for both measures, variation in the phosphatase domain, i.e., truncation-like or hypomorphic variation, was generally more damaging than variation in the C2 domain or regulatory tail (fitness, 42% versus 12%; abundance, 50% versus 38%,; Figures 1C and 1D).
Fitness and Abundance Scores Explain Quantitative Clinical Traits
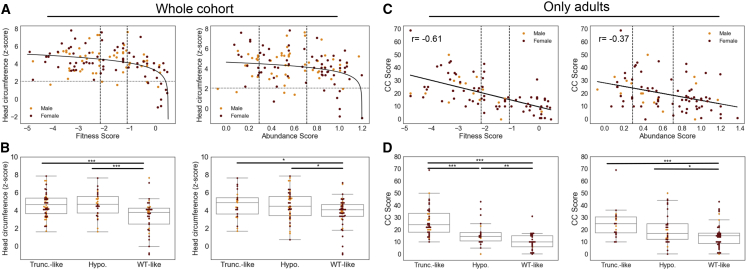
In an effort to link genotype to quantitative clinical phenotypes, we evaluated whether fitness and abundance scores of individuals’ PTEN missense variants could explain the degree of macrocephaly or phenotype burden. Burden was assessed by CC score, which takes into account neurological features as well as benign and malignant lesions of the body, for individuals over 18 years of age (Materials and Methods). Molecular phenotype scores were evaluated numerically as well as according to the defined functional categories (e.g., WT-like, hypomorphic, and truncation-like). We found a logarithmic relationship between fitness score and head size measured by OFC: Z scores plateaued around the hypomorphic cutoff (Figure 2A, left panel). Accordingly, we found a significant difference in OFC between the population of WT-like variants and truncation-like variants as well as between WT-like variants and hypomorphic variants (p = 4.3 × 10−5 and p = 6 × 10−4, respectively, Mann-Whitney U test), but we found no difference between hypomorphic variant and truncation-like variant fitness scores (Figure 2B, left panel). We also observed a logarithmic relationship between OFC and abundance score (Figure 2A, right panel). Treating abundance as a categorical variable revealed significant differences between WT-like and both truncation-like and hypomorphic variants (p = 0.02 and p = 0.01, respectively, Mann-Whitney U test). Similar to findings regarding the fitness score, there was no difference between the distribution of truncation-like and hypomorphic variants (Figure 2B, right panel).
Figure 2.
Relationships between Molecular-Phenotype Scores and Quantitative Clinical Traits for Missense Variants
(A) Occipital frontal circumference (OFC, Z score) plotted as a function of continuous fitness score or abundance score for all individuals with missense variants. Males are shown as orange, and females are shown as maroon. The vertical dashed lines indicate the hypomorphic and truncation-like cutoffs at −1.11 and −2.15, respectively. The horizontal dashed line indicates the threshold for macrocephaly (Z score = 2.054). The solid lines indicate logarithmic curves fit to the data; mean squared error = 2.06 and 2.30 for fitness and abundance scores, respectively.
(B) Boxplot of OFC Z scores for all individuals with missense variants with fitness or abundance scores in the WT-like, hypomorphic, or truncation-like ranges. Fitness scores are Cohen’s r = 0.35 for truncation-like variants versus WT-like variants and Cohen’s r = 0.40 for hypomorphic variants versus WT-like variants. Abundance scores are Cohen’s r = 0.24 for truncation-like variants versus WT-like variants and Cohen’s r = 0.22 for hypomorphic variants versus WT-like variants.
(C) CC score for adults with missense variants as a function of continuous fitness or abundance scores. Analyses are restricted to adults because CC score is not valid for individuals under 18 years of age. The vertical dashed lines indicate the hypomorphic and truncation-like cutoffs at −1.11 and −2.15, respectively. The solid lines indicate linear curves fit to the data.
(D) Boxplot of CC scores for adults with missense variants with fitness or abundance scores in the WT-like, hypomorphic, or truncation-like ranges. Fitness scores are Cohen’s r = 0.44 for truncation-like variants versus hypomorphic variants, Cohen’s r = 0.70 for truncation-like variants versus WT-like variants, and Cohen’s r = 0.34 for hypomorphic variants versus WT-like variants. Abundance scores are Cohen’s r = 0.42 for truncation-like variants versus WT-like variants and Cohen’s r = 0.20 for hypomorphic variants versus WT-like variants. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
In our analysis of phenotype burden, we found a significant linear relationship between missense variant fitness score and CC score (p = 3.7 × 10−10): fitness score explained 37% of the variation in CC score (Figure 2C, left panel). Similarly, by treating fitness score as a categorical variable, we found that more damaging groups of variants had distributions shifted toward higher (more severe) CC scores (Figure 2D, left panel). CC scores for truncation-like variants were significantly higher than those of hypomorphic variants (p = 2.5 × 10−4). Additionally, CC scores for hypomorphic variants were in turn significantly higher than those of WT-like variants (p = 9.2 × 10−3).
Alternatively, for abundance scores, although a significant linear relationship exists between CC score and abundance score (p = 3.2 × 10−4), it explains only 14% of the variation in CC score (Figure 2C, right panel). Likewise, when we treated abundance score as a categorical variable, we observed more trends that were more modest than those for fitness score. For hypomorphic variants, CC scores were nominally higher than for WT-like (p = 0.045) variants, but not significantly different from those for truncation-like variants (p = 0.08). CC scores for truncation-like variants were significantly higher than for WT-like variants (p = 6.7 × 10−4; Figure 2D, right panel). Combined, these results underscore the potential for molecular phenotypes to partially explain clinical outcomes.
Molecular-Phenotype Data Accurately Distinguish Likely Pathogenic from Benign Variation
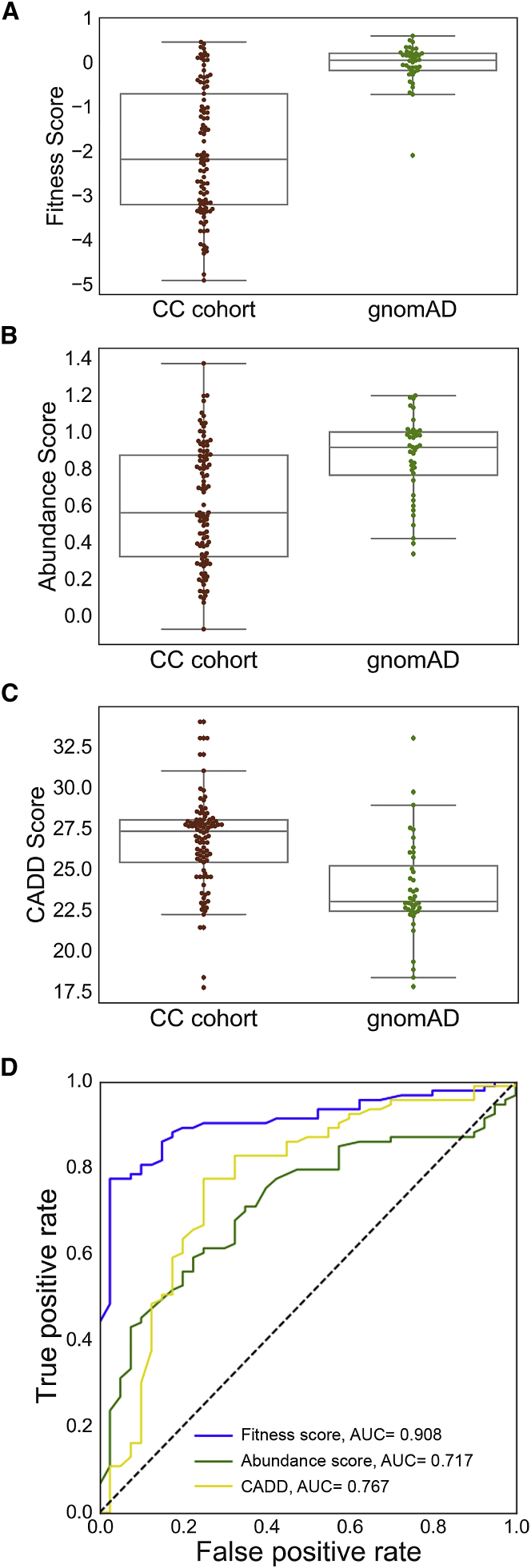
We previously showed that fitness scores discriminate ClinVar pathogenic or likely pathogenic PTEN variation from gnomAD putatively benign variation.8 Here, we examined whether molecular-phenotype data could identify pathogenic variation in this set of alleles and whether combining molecular-phenotype data could improve performance compared to univariate approaches. Thus, we contrasted the CC cohort of likely pathogenic PTEN variants with putatively benign population PTEN variants cataloged in the gnomAD control-only individuals (Materials and Methods; Figure 3; Table S5). We found that variants from the CC cohort were predicted to be significantly more damaging than variants from gnomAD by both fitness score (p = 6.5 × 10−13, Mann-Whitney U test) and abundance score (p = 7.6 × 10−6; Figures 3A and 3B). As a comparison, CADD scores21,22 are also more damaging for the CC cohort group than for the gnomAD group (p = 6.5 × 10−8; Figure 3C). Of these three predictors, fitness scores demonstrate the highest area under the receiver operating characteristic curve (AUC = 0.908, 10-fold cross-validation; Figure 3D). Although these predictors are correlated, the relationships are modest (Spearman’s rho = 0.52–0.59; Figure S3A), suggesting that multivariate models could yield improved performance. Therefore, we constructed logistic regression models by using various combinations of the molecular phenotype data and CADD scores to find the model that most accurately discriminates between the two groups (Figure S3B). We found that no multivariate model performed significantly better than the fitness-score univariate model (Figure S3B). Nevertheless, the substantial increase in predictive power of the fitness-score model over the CADD model highlights the power of empirical molecular-phenotype data to accurately predict pathogenicity.
Figure 3.
Molecular Phenotypes Discriminate Clinical from gnomAD Missense PTEN Variation
These analyses are done at the variant level so that recurrent variants do not bias the results. Two known pathogenic variants were removed from the gnomAD list (Materials and Methods).
(A)Boxplots comparing fitness scores of variants found in the CC cohort to the scores of variants found in gnomAD. Cohen’s r = 0.61.
(B) Boxplots comparing abundance scores of variants found in the CC cohort to the scores of variants found in gnomAD. Cohen’s r = 0.37.
(C) Boxplots comparing CADD scores of variants found in the CC cohort to the scores of variants found in gnomAD. Cohen’s r = 0.45.
(D) Receiver operator characteristic curves for univariate models. The feature weights are reported in Table S7.
Molecular Phenotypes Identify Subgroups with Distinct Cancer Susceptibility
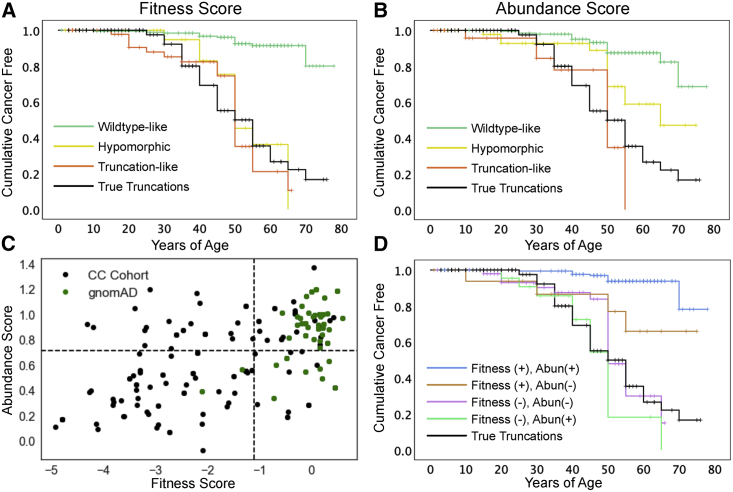
Although it is known that pathogenic PTEN variants dramatically increase an individual’s lifetime risk of developing specific cancers, we sought to understand whether molecular phenotypes could highlight functional classes of missense variants with differences in cancer susceptibility (Materials and Methods). As a comparison group, we included individuals with variants that are predicted to be truly truncating (e.g., nonsense and frameshifting). Survival functions were first compared between all classes of missense fitness or abundance scores and the true truncations, and were then compared with pairwise comparisons of survival functions when significant differences were found (Figures 4A and 4B). In this analysis, cancer-free status was considered as the survival criterion.
Figure 4.
Effects of Molecular Phenotype on Cumulative Cancer Incidence
(A) Survival-like analysis of individuals with different fitness-score classes of PTEN missense or true truncation variants. Here, “survival” is defined as being cancer free. The ticks represent right-censored individuals, i.e., the age at the last clinical follow-up. n = 204, WT-like variants; n = 35 hypomorphic variants; n = 68 truncation-like variants; and n = 114 true truncations.
(B) Survival-like analysis of individuals with different abundance-score classes of PTEN missense or true truncation variants. Here, “survival” is defined as being cancer free. The ticks represent right-censored individuals, i.e., the age at the last clinical follow-up. n = 210 WT-like variants; n = 65 hypomorphic variants; n = 32 truncation-like variants; and n = 114 true truncations.
(C) All CC cohort and gnomAD individuals plotted as a function of fitness and abundance scores. The dotted lines indicate WT-like thresholds (−1.11 and 0.71 for fitness and abundance scores, respectively).
(D) Survival-like analysis for individuals with PTEN missense variants falling in fitness-abundance quadrants or with true truncating variants. Here, “survival” is defined as being cancer free. The ticks represent right-censored individuals, i.e., the age at the last clinical follow-up. n = 29 fitness (−), abundance (+); n = 74 fitness (−), abundance (−); n = 23 fitness (+), abundance (−); n = 181 fitness (+), abundance (+); and n = 114 true truncations.
For fitness scores, survival functions were significantly different (p = 3.2 × 10−24, log rank; Figure 4A; Table S6). Pairwise comparisons showed that all of the reduced fitness-score categories' survival functions were similar to each other and significantly different from the WT-like survival function (Table S6). On the basis of the shape of the survival functions, we hypothesized that there might be a difference in early-onset risk. Therefore, we conducted a subanalysis with right-censoring at age 35, which again showed significant overall differences (p = 3.0 × 10−6, log rank). Pairwise comparisons showed that these differences were driven by truncation-like and true-truncation categories, each of which significantly deviated from WT-like variants (p = 1.0 × 10−5 and p = 2.4 × 10−7). The hypomorphic survival function appears visually to be intermediate between the groups. However, across this age range the hypomorphic function did not significantly differ from the WT-like function (p = 0.35) or either of the truncation-like or true-truncation categories (p = 0.155 and p = 0.122, respectively; Figure 4A).
Variant classes defined by abundance scores also had significantly different survival functions (p = 6.2 × 10−18, log rank; Figure 4B; Table S6). In contrast to the fitness scores, pairwise comparisons revealed a stepwise relationship for abundance scores: hypomorphic missense variants conferred greater lifetime hazard than WT-like missense variants (p = 1.3 × 10−4) and truncation-like missense variants conferred greater hazard than the hypomorphic abundance class (p = 0.024; Figure 4B). True truncations might confer greater hazard than hypomorphic missense variants, but this comparison was not significant (p = 0.07). Right-censoring at age 35 identified significantly different survival functions for the abundance-defined variant classes as well (p = 1.1 × 10−5). As for the fitness-score analysis, these differences were driven by truncation-like and true-truncation functions that were significantly different from WT-like functions between birth and age of 35 (p = 2.5 × 10−5 and p = 1.0 × 10−6, respectively). The hypomorphic survival function was visually intermediate between WT-like function (p = 0.06) and the truncation-group functions (p = 0.15 and p = 0.20), but none of the comparisons were significantly different.
We next leveraged the two-dimensional molecular-phenotype data to separate missense variants into four categories on the basis of deficiencies in fitness score, abundance score, or both. For this analysis, we combined hypomorphic and truncation-like scoring variants as the negative group for PTEN function for each score in order to keep adequate group sizes. A larger proportion of gnomAD individuals, compared to the CC cohort, have variants in the fitness-positive, abundance-positive quadrant (Figure 4C). The survival functions for these combined molecular-phenotype-defined groups were significantly different (p = 6.3 × 10−24, log rank). Pairwise comparisons showed that variants retaining WT-like fitness (+) and abundance (+) have the lowest overall hazard and have a survival function that is significantly different from that of all other groups (Table S6). The remaining three classes are deficient for fitness, abundance, or both scores and are not significantly different from each other or the true truncations (Figure 4D). These data provide high-resolution comparisons of cancer risk for different variant classes, and these comparisons can be further clarified by larger sample sizes.
Molecular Phenotypes Identify Distinct Risk Profiles for ASD/DD and PHTS Subgroupings
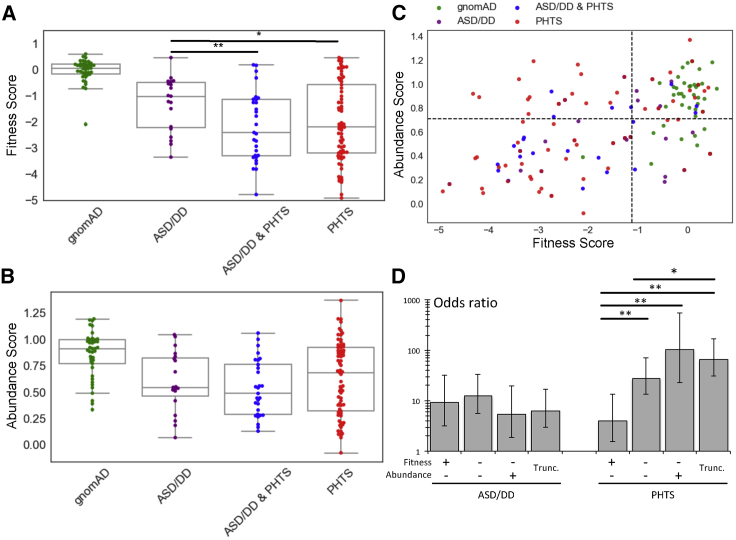
Understanding the molecular differences between the variants that associate with ASD/DD versus PHTS (especially cancer occurrence) outcomes is critical for understanding PTEN pathobiology, and such understanding ultimately guides clinical management. Consistent with our and others’ previous findings,7,8 fitness scores of individuals in the ASD/DD group are less damaging than those of the PHTS-positive groups (p = 5.5 × 10−3 and p = 0.011 for ASD/DD versus ASD/DD & PHTS and for ASD/DD versus PHTS, respectively, Mann-Whitney U test; Figure 5A). However, abundance scores do not differ between the clinical phenotype groups (Figure 5B).
Figure 5.
Association of Molecular Phenotypes with Specific Clinical Outcomes
(A) Boxplots showing fitness scores of PTEN variants occurring in individuals in gnomAD or different clinical categories. Cohen’s r = 0.36 for ASD/DD versus ASD/DD & PHTS and Cohen’s r = 0.25 for ASD/DD versus PHTS.
(B) Boxplots showing abundance scores of PTEN variants occurring in individuals in gnomAD or different clinical categories.
(C) PTEN variants plotted according to fitness and abundance scores, colored by clinical group. The dashed lines indicate hypomorphic cutoffs (−1.11 for fitness scores and 0.71 for abundance scores).
(D) ORs for developing ASD/DD or PHTS symptoms for different variant classes. ORs represent the comparison of that class with variants in the fitness-positive, abundance-positive quadrant [top right in (C)]. Error bars represent 95% CIs. ORs and 95% CIs are reported in Table S8. ∗p < 0.05, ∗∗p < 0.01.
Next, we tested whether the severity of variant molecular phenotype, as assessed by fitness or abundance score, affected the odds of developing ASD/DD or PHTS symptoms (regardless of the presence or absence of the other qualifying symptoms). We included all members of the CC cohort as well as gnomAD individuals (Materials and Methods). By using a logistic-regression model, we calculated ORs for ASD/DD and PHTS as a function of fitness or abundance scores. WT-like variants were used as the reference group. For both molecular phenotypes, more severe missense variants do not significantly increase the odds of an individual developing ASD/DD (OR ranges = 3.9–6.1 and 4.2–7.8 for fitness and abundance scores, respectively). The odds for true truncation variants are marginally decreased in comparison to the odds for the missense variant classes, though this trend is not significant (Figure S4B). In contrast, for fitness and abundance scores, the odds of an individual developing qualifying symptoms for a PHTS classification increase as mutation severity increases in a stepwise manner; stronger differences in risk were observed for the abundance score (OR ranges = 20.4–51.3 and 5.1–28.7 for fitness and abundance scores, respectively; Figure S4B).
We next tested whether two-dimensional molecular-phenotype data would provide additional insights into the risk for developing ASD/DD or PHTS symptoms. Whereas variants from the control individuals from gnomAD clearly cluster in the fitness-positive, abundance-positive quadrant, the affected individuals populate the other three quadrants (Figure 5C). Interestingly, compared to the PHTS-positive categories, the ASD/DD category has a larger fraction of individuals in the fitness-positive, abundance-positive quadrant (30% versus 10% and 20% for ASD/DD versus ASD/DD & PHTS and PHTS, respectively) and a smaller fraction in the putatively dominant-negative, fitness-compromised, abundance-positive quadrant (5% versus 21% and 25% for ASD/DD versus ASD/DD & PHTS and PHTS, respectively; Figure S4A).
Using a logistic-regression approach, we generated ORs for the combined two-dimensional molecular phenotypes. We again observed no major differences in the odds for developing ASD/DD in any missense groups or the true truncation category (OR range = 5.4–12.4). In contrast, the odds for developing PHTS are highly dependent on the variant grouping (OR range = 4.1–102.9). Missense variants that maintain lipid-phosphatase activity but are low abundance show the lowest odds for an individual’s developing qualifying symptoms for a PHTS classification (OR = 4.1, 95% CI = 1.5–10.7; Figure 5D). Missense variants that were fitness and abundance negative showed a significantly different intermediate risk (OR = 27.6, 95% CI = 13.5–56.5). Variants that have WT-like abundance but abrogated lipid-phosphatase activity (putative dominant-negative variants) have the highest odds of an individual’s developing qualifying symptoms for a PHTS classification (OR = 102.9, 95% CI = 22.8–464.0), though the odds are not significantly different from those of variants in the fitness-negative, abundance-negative, or true-truncation categories (Figure 5D).
Discussion
Despite two decades of effort, we still lack a clear understanding of how the PTEN genotype affects specific clinical phenotypes. Recent advances in DNA synthesis and sequencing technologies allow for a new experimental paradigm in which the effects of thousands of variations on protein function can be empirically measured in parallel. Two such experiments recently explored the effects of PTEN variation on lipid phosphatase activity (fitness score) and steady-state cellular abundance (abundance score).8,12 Using imputation, we generated estimated functional scores for all possible PTEN missense variants. In order to understand how molecular-phenotype data relates to clinical outcomes, we integrated these data with clinical information from the CC cohort of PTEN variant-positive individuals. These analyses have validated the clinical utility of comprehensive multi-dimensional functional scores and have uncovered unexpected insights into the PTEN genotype-phenotype map.
Our analyses demonstrate that molecular-phenotype scores are correlated with quantitative clinical traits. Fitness and abundance scores showed a logarithmic relationship with the most penetrant PTEN phenotype, macrocephaly (∼95% of PTEN patients).23 In previous work, we designed an algorithm to determine an individual’s a priori risk for having a germline pathogenic PTEN variant (CC score). CC score is also a surrogate measure of an adult’s phenotypic burden and takes age of onset into account.16 CC scores and functional scores have a linear relationship wherein more severe phenotypic burden associates with worse functional scores. We then demonstrated that molecular-phenotype data can be used for modeling and thus predicting likely pathogenic variants with higher accuracy than can CADD, a completely in silico approach.
Although broadly predicting pathogenicity has value in a clinical setting and can help resolve PTEN variants of uncertain significance (VUSs), we were also interested in exploring whether these molecular phenotypes could provide additional insights into the diverse clinical outcomes associated with germline PTEN disruption. Our analyses showed that molecular phenotypes can define subgroups of individuals with common or unique age-related cancer hazards. Although putative true truncating variants, such as nonsense variants, showed high lifetime cancer risk, highly damaging missense variants as defined by the molecular phenotypes appear to be at least as impactful. Moreover, our data from single molecular phenotypes show truncation-like and true truncation survival functions separate from WT-like functions over an early-onset age range. Hypomorphic functions are potentially intermediate over this early-onset range but not yet significantly different from the WT-like functions. Combining molecular phenotype scores provides further granularity for these cancer risks.
A growing number of studies have provided important insight into the question of whether genotypes drive diverse phenotypic outcomes for carriers of germline PTEN variants. However, these studies have generally been limited by small sample sizes. For instance, Spinelli et al. investigated the lipid-phosphatase activity and protein stability of seven ASD-associated and five PHTS-associated PTEN missense variants by using virally infected U87MG cells.7 They found ASD-associated variants retained partial phosphatase function but exhibited dramatically decreased stability, whereas PHTS-associated variants lost phosphatase function but exhibited relatively better stability.7 These findings form the basis for Leslie and Longy’s hypothesis in which ASD/DD results from hypomorphic PTEN variants but traditional PHTS (i.e., hamatomatous and malignant growth) results from more damaging variants.11 Our previous work using fitness scores (i.e., inferred lipid-phosphatase activity) and ASD/DD- or PHTS-associated variants from the literature lent support to this hypothesis.8
Here, by using the largest set of clinically annotated variants examined to date, we strengthen these previous findings by showing that ASD/DD-associated PTEN variants, on average, retain hypomorphic lipid-phosphatase activity, whereas those associated with either ASD/DD and PHTS or PHTS alone are more damaging. Moreover, the fraction of missense variants and the distribution of variants according to fitness and abundance scores are more similar between the two PHTS-associated groups, suggesting that they are in fact molecularly similar. We made the surprising discovery, however, that risk for developing ASD/DD is not dramatically altered across different variant loss-of-function categories, whereas the risk for PHTS can increase by an order of magnitude. Thus, it appears that although all individuals with pathogenic PTEN variants are at a substantial risk for developing ASD/DD, the risk, and thus the subsequent penetrance, of PHTS symptoms (i.e., hamartomatous and malignant growth) is significantly greater for true truncations and truncation-like missense variants. These differential risk profiles would then explain the lower fraction of true truncations in cohorts recruited primarily on the basis of an ASD/DD diagnosis.11,19
The biologic basis of these differential risk profiles remains unclear. The retention of any lipid-phosphatase activity of the variant allele, coupled with the second functional PTEN allele, might be sufficient to prevent the formation of hamartomas in some cases. There are numerous PTEN functions that are not described by the molecular phenotypes included in this study. Lipid phosphatase-independent functions might also modulate risk. For example, recent studies have shown a potential relationship between altered PTEN subcellular localization and clinical outcomes; in these studies, PTEN variants showing aberrant nuclear depletion were associated with ASD/DD.24, 25, 26, 27 Ideally, a comprehensive analysis would include the effect of variation on PTEN’s protein-phosphatase activity, subcellular localization, nuclear function, and protein-protein interaction. New high-throughput assays might make such datasets available in the near future.
Given that the majority of ASD/DD diagnoses are of children or young adults, an important open question is “What will their lifetime risk for neoplasia truly be?” Longitudinal tracking to definitively assess neoplasia risk in this cohort will improve the allocation of clinical resources and guide the delivery of precision care. Our current data suggest that certain subsets of individuals with PTEN-associated ASD/DD are more likely to have higher cancer risk than are other subsets. Either longitudinal follow-up with these individuals or new prospective recruitment efforts will be needed for researchers to answer this question definitively.
Declaration of Interests
C.E. is an external strategic advisor to N-of-One and is the pro bono chief medical officer of Family Care Path and Covariance Diagnostics. All other authors declare no competing interests.
Acknowledgments
We thank A.C. Adey, D.M. Fowler, K.A. Matreyek, J. Zonana, G. Mandel, P.J. Stork, K.M. Wright, I.N. Smith, and M. Seyfi for helpful discussions. We thank Martha Atherton and the Atherton Foundation for their support of the NARSAD awards. This work was supported, in part, by a NARSAD Young Investigator Grant from the Brain and Behavior Research Foundation through the NARSAD-Atherton Foundation Young Investigator Award (22935 to B.J.O.), a Sloan Research Fellowship in Neurosciences (Alfred P. Sloan Foundation; FG-2015-65608 to B.J.O.), the Ambrose Monell Foundation (to C.E.), the Zacconi Program of PTEN Research Excellence (to C.E.), and internal funds (C.E. and B.J.O.). T.L.M received support from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (F31HD095571). T.L.M. is an ARCS scholar (Achievement Rewards for College Scientists Foundation, Inc., Oregon Chapter), and B.J.O. is a Klingenstein-Simons Fellow (Esther A. and Joseph Klingenstein Fund, Simons Foundation). C.E. is the Sondra J. and Stephen R. Hardis Endowed Chair of Cancer Genomic Medicine at the Cleveland Clinic and is an American Cancer Society (ACS) clinical research professor.
Published: May 21, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.04.014.
Contributor Information
Charis Eng, Email: engc@ccf.org.
Brian J. O’Roak, Email: oroak@ohsu.edu.
Web Resources
CADD, https://cadd.gs.washington.edu/
ClinVar Database, https://www.clinicalgenome.org/data-sharing/clinvar/
Consurf, https://consurfdb.tau.ac.il/
gnomAD, https://gnomad.broadinstitute.org/downloads/
OMIM, https://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
Provean, http://provean.jcvi.org/
Supplemental Data
References
- 1.Yehia L., Eng C. 65 years of the double helix: One gene, many endocrine and metabolic syndromes: PTEN-opathies and precision medicine. Endocr. Relat. Cancer. 2018;25:T121–T140. doi: 10.1530/ERC-18-0162. [DOI] [PubMed] [Google Scholar]
- 2.Yehia L., Ngeow J., Eng C. PTEN-opathies: from biological insights to evidence-based precision medicine. J. Clin. Invest. 2019;129:452–464. doi: 10.1172/JCI121277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liaw D., Marsh D.J., Li J., Dahia P.L., Wang S.I., Zheng Z., Bose S., Call K.M., Tsou H.C., Peacocke M. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat. Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 4.Marsh D.J., Dahia P.L.M., Zheng Z., Liaw D., Parsons R., Gorlin R.J., Eng C. Germline mutations in PTEN are present in Bannayan-Zonana syndrome. Nat. Genet. 1997;16:333–334. doi: 10.1038/ng0897-333. [DOI] [PubMed] [Google Scholar]
- 5.Butler M.G., Dasouki M.J., Zhou X.P., Talebizadeh Z., Brown M., Takahashi T.N., Miles J.H., Wang C.H., Stratton R., Pilarski R., Eng C. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J. Med. Genet. 2005;42:318–321. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith I.N., Thacker S., Jaini R., Eng C. Dynamics and structural stability effects of germline PTEN mutations associated with cancer versus autism phenotypes. J. Biomol. Struct. Dyn. 2019;37:1766–1782. doi: 10.1080/07391102.2018.1465854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spinelli L., Black F.M., Berg J.N., Eickholt B.J., Leslie N.R. Functionally distinct groups of inherited PTEN mutations in autism and tumour syndromes. J. Med. Genet. 2015;52:128–134. doi: 10.1136/jmedgenet-2014-102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mighell T.L., Evans-Dutson S., O’Roak B.J. A saturation mutagenesis approach to understanding PTEN lipid phosphatase activity and genotype-phenotype relationships. Am. J. Hum. Genet. 2018;102:943–955. doi: 10.1016/j.ajhg.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orloff M.S., Eng C. Genetic and phenotypic heterogeneity in the PTEN hamartoma tumour syndrome. Oncogene. 2008;27:5387–5397. doi: 10.1038/onc.2008.237. [DOI] [PubMed] [Google Scholar]
- 10.Mester J., Eng C. When overgrowth bumps into cancer: the PTEN-opathies. Am. J. Med. Genet. C. Semin. Med. Genet. 2013;163C:114–121. doi: 10.1002/ajmg.c.31364. [DOI] [PubMed] [Google Scholar]
- 11.Leslie N.R., Longy M. Inherited PTEN mutations and the prediction of phenotype. Semin. Cell Dev. Biol. 2016;52:30–38. doi: 10.1016/j.semcdb.2016.01.030. [DOI] [PubMed] [Google Scholar]
- 12.Matreyek K.A., Starita L.M., Stephany J.J., Martin B., Chiasson M.A., Gray V.E., Kircher M., Khechaduri A., Dines J.N., Hause R.J. Multiplex assessment of protein variant abundance by massively parallel sequencing. Nat. Genet. 2018;50:874–882. doi: 10.1038/s41588-018-0122-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landrum M.J., Lee J.M., Riley G.R., Jang W., Rubinstein W.S., Church D.M., Maglott D.R. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodríguez-Escudero I., Roelants F.M., Thorner J., Nombela C., Molina M., Cid V.J. Reconstitution of the mammalian PI3K/PTEN/Akt pathway in yeast. Biochem. J. 2005;390:613–623. doi: 10.1042/BJ20050574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv. 2019 doi: 10.1101/531210. [DOI] [Google Scholar]
- 16.Tan M.-H., Mester J., Peterson C., Yang Y., Chen J.-L., Rybicki L.A., Milas K., Pederson H., Remzi B., Orloff M.S., Eng C. A clinical scoring system for selection of patients for PTEN mutation testing is proposed on the basis of a prospective study of 3042 probands. Am. J. Hum. Genet. 2011;88:42–56. doi: 10.1016/j.ajhg.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roche A.F., Mukherjee D., Guo S.M., Moore W.M. Head circumference reference data: birth to 18 years. Pediatrics. 1987;79:706–712. [PubMed] [Google Scholar]
- 18.Marsh D.J., Kum J.B., Lunetta K.L., Bennett M.J., Gorlin R.J., Ahmed S.F., Bodurtha J., Crowe C., Curtis M.A., Dasouki M. PTEN mutation spectrum and genotype-phenotype correlations in Bannayan-Riley-Ruvalcaba syndrome suggest a single entity with Cowden syndrome. Hum. Mol. Genet. 1999;8:1461–1472. doi: 10.1093/hmg/8.8.1461. [DOI] [PubMed] [Google Scholar]
- 19.Tan M.-H., Mester J.L., Ngeow J., Rybicki L.A., Orloff M.S., Eng C. Lifetime cancer risks in individuals with germline PTEN mutations. Clin. Cancer Res. 2012;18:400–407. doi: 10.1158/1078-0432.CCR-11-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saunders C.T., Baker D. Evaluation of structural and evolutionary contributions to deleterious mutation prediction. J. Mol. Biol. 2002;322:891–901. doi: 10.1016/s0022-2836(02)00813-6. [DOI] [PubMed] [Google Scholar]
- 21.Rentzsch P., Witten D., Cooper G.M., Shendure J., Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47(D1):D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mester J.L., Tilot A.K., Rybicki L.A., Frazier T.W., 2nd, Eng C. Analysis of prevalence and degree of macrocephaly in patients with germline PTEN mutations and of brain weight in Pten knock-in murine model. Eur. J. Hum. Genet. 2011;19:763–768. doi: 10.1038/ejhg.2011.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frazier T.W., Embacher R., Tilot A.K., Koenig K., Mester J., Eng C. Molecular and phenotypic abnormalities in individuals with germline heterozygous PTEN mutations and autism. Mol. Psychiatry. 2015;20:1132–1138. doi: 10.1038/mp.2014.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tilot A.K., Bebek G., Niazi F., Altemus J.B., Romigh T., Frazier T.W., Eng C. Neural transcriptome of constitutional Pten dysfunction in mice and its relevance to human idiopathic autism spectrum disorder. Mol. Psychiatry. 2016;21:118–125. doi: 10.1038/mp.2015.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tilot A.K., Gaugler M.K., Yu Q., Romigh T., Yu W., Miller R.H., Frazier T.W., 2nd, Eng C. Germline disruption of Pten localization causes enhanced sex-dependent social motivation and increased glial production. Hum. Mol. Genet. 2014;23:3212–3227. doi: 10.1093/hmg/ddu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fricano-Kugler C.J., Getz S.A., Williams M.R., Zurawel A.A., DeSpenza T., Jr., Frazel P.W., Li M., O’Malley A.J., Moen E.L., Luikart B.W. Nuclear excluded autism-associated phosphatase and tensin homolog mutations dysregulate neuronal growth. Biol. Psychiatry. 2018;84:265–277. doi: 10.1016/j.biopsych.2017.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.