Abstract
Kinesin-2 enables ciliary assembly and maintenance as an anterograde intraflagellar transport (IFT) motor. Molecular motor activity is driven by a heterotrimeric complex comprised of KIF3A and KIF3B or KIF3C plus one non-motor subunit, KIFAP3. Using exome sequencing, we identified heterozygous KIF3B variants in two unrelated families with hallmark ciliopathy phenotypes. In the first family, the proband presents with hepatic fibrosis, retinitis pigmentosa, and postaxial polydactyly; he harbors a de novo c.748G>C (p.Glu250Gln) variant affecting the kinesin motor domain encoded by KIF3B. The second family is a six-generation pedigree affected predominantly by retinitis pigmentosa. Affected individuals carry a heterozygous c.1568T>C (p.Leu523Pro) KIF3B variant segregating in an autosomal-dominant pattern. We observed a significant increase in primary cilia length in vitro in the context of either of the two mutations while variant KIF3B proteins retained stability indistinguishable from wild type. Furthermore, we tested the effects of KIF3B mutant mRNA expression in the developing zebrafish retina. In the presence of either missense variant, rhodopsin was sequestered to the photoreceptor rod inner segment layer with a concomitant increase in photoreceptor cilia length. Notably, impaired rhodopsin trafficking is also characteristic of recessive KIF3B models as exemplified by an early-onset, autosomal-recessive, progressive retinal degeneration in Bengal cats; we identified a c.1000G>A (p.Ala334Thr) KIF3B variant by genome-wide association study and whole-genome sequencing. Together, our genetic, cell-based, and in vivo modeling data delineate an autosomal-dominant syndromic retinal ciliopathy in humans and suggest that multiple KIF3B pathomechanisms can impair kinesin-driven ciliary transport in the photoreceptor.
Keywords: KIF3B, primary cilia, retinopathy, hepatic fibrosis, kinesin, whole-exome sequencing, zebrafish, feline genetics
Main Text
Kinesin family (KIF) genes encode a superfamily of microtubule-based molecular motors that transport intracellular cargo. Pathogenic variants in KIF-encoding loci have been associated with at least 14 distinct Mendelian disorders in humans, transmitted in both dominant or recessive inheritance fashions.1 One unifying feature of autosomal-dominant KIF-related disorders is the primary impact on the central nervous system as evidenced by phenotypes such as intellectual disability (KIF1A [MIM: 614255]), cortical dysplasia (KIF2A [MIM: 615411]), spastic paraplegia (KIF5A [MIM: 604187]), and microcephaly (KIF11 [MIM: 152950]). KIF genes mutated in autosomal-recessive disorders also impair neurological development and have been reported in X-linked intellectual disability (KIF4A [MIM: 300521]) or syndromic ciliopathies such as Acrocallosal syndrome (KIF7 [MIM: 200990]) and Meckel syndrome (KIF14 [MIM: 616258]).
Kinesin-2 subfamily motors, not currently associated with human disease phenotypes, have an established role in ciliogenesis. These macromolecular complexes ensure anterograde trafficking by ATP-dependent movement from the ciliary base toward the growing end of microtubules. In the context of kinesin-2 ablation, model organisms such as mouse or zebrafish display stunted or absent ciliary formation, leading to broad impairment of primary ciliary functions during vertebrate development.2
Protein synthesis does not occur in the cilium; the formation and maintenance of this organelle depends on intraflagellar transport (IFT) of protein cargo along the ciliary axoneme. KIF3A-KIF3B or KIF3A-KIF3C heterodimers and the non-motor kinesin-associated protein (KAP), encoded by KIFAP3, operate as the main effectors of kinesin-2 driven trafficking, together with homodimeric KIF17. Ciliopathies, defined as clinical entities caused by defects in ciliary structure or function, share common phenotypic features; these include obesity, hepatic fibrosis, retinitis pigmentosa (RP), cystic kidney disease, polydactyly, and intellectual disability.3 Here, we report affected individuals from a small nuclear family and an unrelated six-generation pedigree whose members harbor heterozygous nonsynonymous KIF3B variants and share partially overlapping ciliopathy clinical features.
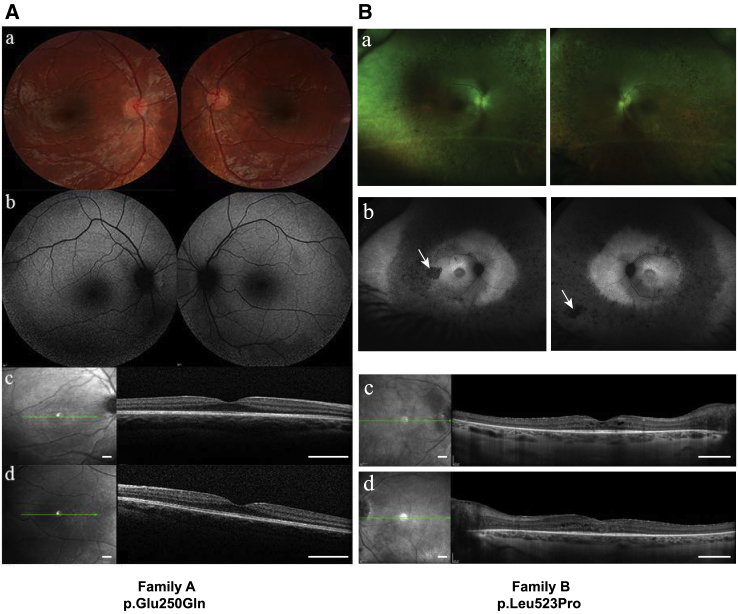
Individual 1 (family A, II-1; Table 1) is a male born from nonconsanguineous parents of northern European ancestry. At birth, postaxial hexadactyly of both hands and the right foot were observed. At 12 months, he presented with failure to thrive and hepatosplenomegaly. An abdominal ultrasound performed at 18 months showed a dysmorphic liver with dilatation of intrahepatic biliary ducts. A liver biopsy showed micronodular cirrhosis with modification of global hepatic architecture by annular fibrosis and persistence of peripheric biliary neoductules. Cardiac ultrasound showed a bicuspid aortic valve. At 24 months of age, he had esophageal varices and thrombocytopenia. At 4 years of age, because of suggestive clinical features of a ciliopathy, we performed an ophthalmological exam. The parents only reported their child to have difficulties moving at night. Fundus examination was normal (Figure 1Aa), but the fundus autofluorescence showed a modification of the peripheral autofluorescence and hyperautofluorescent perimacular points (Figure 1Ab). Electroretinograms showed alteration of the flicker and the scotopic responses (Figure S1). Spectral Domain Optical Coherence Tomography (SD-OCT) analysis showed a normal foveal profile (Figure 1Ac,d). At last ophthalmological examination at the age of 5 years, fundus examination showed retinal thinning with an increase in the visibility of the choroidal vascularization on the retinal periphery and on the temporal macula.
Table 1.
Clinical Summary of Individuals Harboring Nonsynonymous KIF3B Variants
| Center | Family A |
Family B |
|||||
|---|---|---|---|---|---|---|---|
| Nantes (France) | Houston (USA) | ||||||
| Individual identifier | 1 | 2 | 3 | 4 | 5 | 6 | |
| Affected individual | II-1 | IV-1 | IV-3 | V-4 | VI-2 | VI-1 | |
| Gender | male | male | male | female | female | male | |
| Age at last examination (years) | 5 | 50 | 55 | 52 | 23 | 21 | |
| Eye | retinitis pigmentosa | retinitis pigmentosa | retinitis pigmentosa | retinitis pigmentosa | retinitis pigmentosa | retinitis pigmentosa | |
| Eye: visual field area (OS;OD) [degrees diameter] | N/D | 10; 10 | 20; 22 | ND | 20 OU | constricted central vision | |
| Eye: age of first visual symptoms | diagnosed at 5 years | 6 years | diagnosed at 5 years | unknown | unknown | reports symptoms to be more severe than mother (V-4) or sister (VI-2) | |
| Liver | hepatic fibrosis | normal | normal | normal | normal | normal | |
| Skeletal malformations | postaxial hexadactyly of both hands and right foot | normal | normal | affected mother (IV-8) and affected brother (V-7) both reported to have postaxial hexadactyly | normal | normal | |
| Heart | bicuspid aortic valve | normal | normal | normal | normal | normal | |
| Kidney | normal | son (V-1) has kidney issues (dialysis) | normal | normal | normal | normal | |
Abbreviations are as follows: N/D, no data; OS, oculus sinister (left eye); OD, oculus dexter (right eye); OU, oculus uterque (both eyes).
Figure 1.
Individuals Harboring Nonsynonymous KIF3B Variants Display Retinal Phenotypes
(A) Ophthalmological examination of individual 1 (family A) who harbors a de novo c.748G>C (p.Glu250Gln) KIF3B variant. Shown are (a) ophthalmological fundus imaging of the right and the left eyes; (b) fundus autofluorescence imaging of the right and the left eyes; (c) spectral domain optical tomography of the right fovea (scale: 1 mm); and (d) spectral domain optical tomography of the left fovea (scale: 1 mm). Refraction was performed under cyploplegia with chlorhydrate of cyclopentolate 0.5% (Alcon). Optical coherence tomography (OCT) analysis and the autofluorescence fundus imaging were conducted with an OCT spectal domain (Heidelberg Engineering, Spectralis HRA-OCT).
(B) Ophthalmological examination of individual VI-2 (family B) who harbors a c.1569T>C (p.Leu523Pro) KIF3B variant. White arrows indicate retinal pigments characteristic of retinitis pigmentosa. Shown are (a) ophthalmological fundus imaging of the right and the left eyes; (b) fundus autofluorescence imaging of the right and the left eyes; (c) spectral domain optical tomography of the right fovea (scale: 1 mm); and (d) spectral domain optical tomography of the left fovea (scale: 1 mm).
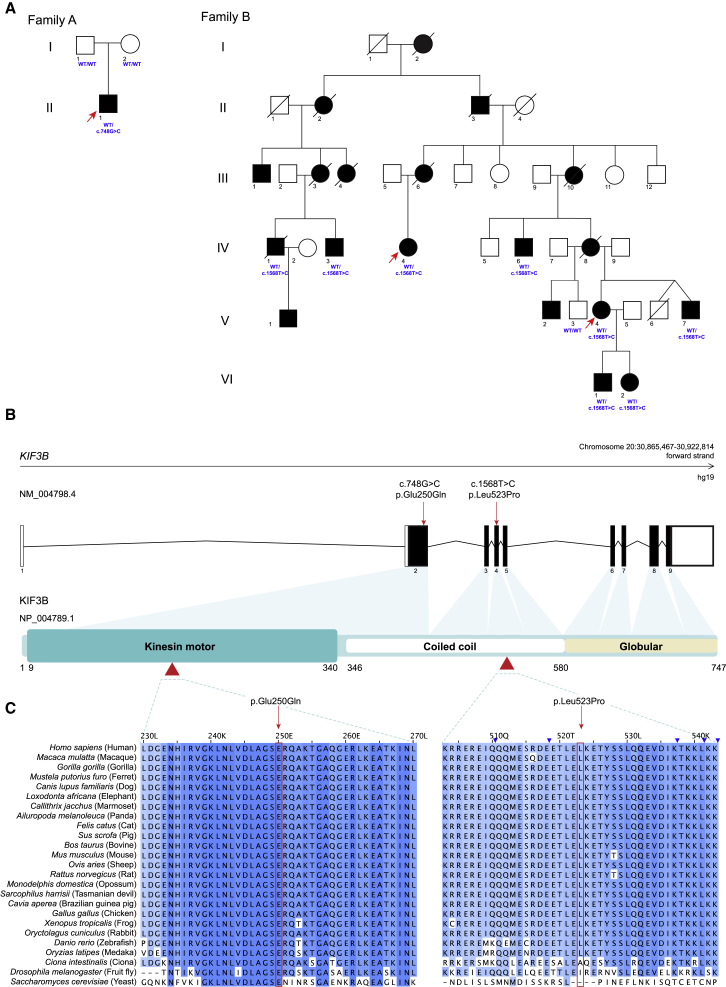
To investigate the genetic etiology of this individual’s clinical presentation, we consented the parent-child trio for all subsequent research procedures in accordance with ethical guidelines of Nantes University Hospital (Figure 2A). Array comparative genomic hybridization on the family A proband sample was negative, ruling out the possibility of large copy number variants. Next, we performed exome sequencing of the proband and achieved an average mean target coverage of 86× (Supplemental Methods). We retained variants with at least 9 reads and with a variant read frequency over 20% impacting exonic sequences or splice sites (±10 bp from the junction) and with an allele frequency <0.5% in 1000 Genomes, Genome Aggregation Database (gnomAD, 123,136 exomes and 15,496 whole-genome sequences; accessed on 11/10/2018), and in a local database of 952 exomes. We retained 280 rare variants (Table S1), including 13 that matched an X-linked or a recessive inheritance (homozygous, hemizygous, or at least 2 rare variants in the same gene) and 267 that matched a dominant inheritance (single heterozygous variant). We did not identify deleterious variants in any disease-associated gene, and we selected 88 variants of uncertain significance that were within an established human genetic disease-associated gene in either OMIM or Orphanet for further consideration (Table S1).
Figure 2.
Heterozygous Nonsynonymous Variants in KIF3B Segregate with Ciliopathy Phenotypes in Dominant Pedigrees
(A) Pedigrees of families A and B with segregation of KIF3B variants. Filled circles and squares represent affected female or male individuals, respectively; unfilled circles and squares represent unaffected female or male individuals; respectively. Deceased individuals are indicated by diagonal lines. Exome sequencing was performed for individuals marked with an arrow. Individual genotypes at the KIF3B locus are indicated in blue. All affected individuals in family B carried the KIF3B c.1568T>C (p.Leu523Pro) encoding variant (IV-1, IV-3, IV-4, IV-6, V-4, V-7, VI-1, VI-2) but the unaffected individual (V-3) was WT.
(B) Schematic of KIF3B gene organization and protein domains. Untranslated exons, white boxes; translated exons, black boxes; red arrows and arrowheads, KIF3B variants identified in humans. Kinesin motor, coiled coil, and globular domains (UniProtKB, PROSITE annotations, GenBank: NP_004789.1) are indicated.
(C) Conservation of 40 amino acid blocks impacted by nonsynonymous changes shown with a multiple sequence alignment (Clustal W v1.81) of 25 species sorted by pairwise identity. Red boxes, variant residues; blue shading of amino acids from dark to light represents most to least conserved, respectively. UniProtKB identifiers: Homo sapiens, O15066; Macaca mulatta, F6S877; Gorilla gorilla, G3RAF7; Mustela putorius furo, M3Z2F0; Canis lupus familiaris, E2QUS2; Loxodonta africana, G3T0G8; Callithrix jacchus, F7IBN6; Ailuropoda melanoleuca, G1M429; Felis catus, A0A2I2UKW2; Sus scrofa, F1S519; Bos taurus, F1N020; Mus musculus, Q61771; Ovis aries, W5NZV7; Rattus norvegicus, D3ZI07; Monodelphis domestica, F6RWN1; Sarcophilus harrisii, G3WA27; Cavia aperea, ENSCAPP00000010080 (ensembl, UniProtKB identifier not available); Gallus gallus, Q5F423; Xenopus tropicalis, F6R640; Oryctolagus cuniculus, G1U1D0; Danio rerio, F1QN54; Oryzias latipes, H2LAE9; Ciona intestinalis, F7B875; Drosophila melanogaster, P46867; Saccharomyces cerevisiae, P28742.
The constellation of clinical phenotypes observed in individual 1 was suggestive of a ciliary disorder. We therefore overlaid the rare exome-sequencing variant set with 303 ciliary genes from the SYSCILIA Gold Standard (SCGSv.1).4 We identified five rare heterozygous nonsynonymous variants that intersected with the SCGSv.1 dataset (Table S1). Five variants were heterozygous alleles in genes already causally implicated in recessive ciliopathies. However, one variant in KIF3B (GenBank: NM_004798.4, c.748G>C [p.Glu250Gln]) was absent from gnomAD and predicted to be deleterious by SIFT,5 PolyPhen-2,6 and MutationTaster.7 Given the important role of KIF3B in anterograde IFT in cilia, and localization of this variant to the conserved kinesin motor domain (Figures 2B, 2C, S2, and S3), we tested familial segregation by Sanger sequencing and confirmed that the variant was de novo. We excluded false paternity and maternity by analysis of 11 microsatellite markers.
Through the data sharing platform GeneMatcher,8 we became aware of another rare KIF3B variant that had been identified as a possible cause of RP. This second variant (GenBank: NM_004798.4, c.1568T>C [p.Leu523Pro]) was found in two samples from a cohort of more than 200 families with autosomal-dominant RP ascertained in an ongoing study at the University of Texas Health Science Center in Houston.9 The two females were originally thought to be unrelated but the identification of rare variants during exome sequencing led to the eventual discovery that they were cousins (family B: IV-4 and V-4; Figure 2A; Table 1; Supplemental Methods). They are part of an extended Ashkenazi Jewish family that has RP segregating through at least six generations with more than 19 affected individuals reported (Figure 2A). There is no evidence of consanguinity in either branch of the family and male-to-male transmission of the disease excludes an X-linked mode of inheritance.
Affected members of family B exhibit classic symptoms of RP, with an average age of onset in the first decade. We performed detailed clinical assessment for five individuals (IV-1, IV-3, V-4, VI-2, and VI-1 [individuals 2–6]; Table 1; Figure 1B). Two members of the family have postaxial polydactyly (IV-8 and V-7), and one individual is reported to have kidney dysfunction (V-I; Table 1). Given their symptoms evocative of a ciliopathy, targeted genetic testing had been done previously on known genes for Bardet-Biedl syndrome (MIM: 209900) and dominant forms of inherited retinal degeneration. No likely pathogenic variants were identified, leading to additional testing by exome sequencing.
Bioinformatic analysis of exome data from family B was done using MendelScan as described,10 resulting in 28 rare variants shared between both individuals (Table S2). The majority of shared variants were present in multiple individuals in gnomAD, especially Ashkenazi Jewish (n = 23 variants with allele frequency > 0.05%; 50,040 individuals). The KIF3B variant was the only segregating variant absent in gnomAD. We performed segregation analysis of the four variants with the lowest overall incidence in gnomAD (KIF3B, PCNXL3, OR4D2, and TOB1) in an additional seven family members from the extended pedigree using bidirectional Sanger sequencing (six affected, one unaffected; Figure 2A). The KIF3B c.1568T>C (p.Leu523Pro) variant was the sole change that segregated with RP in the pedigree. To confirm that the KIF3B variant and the disease are not likely to be co-segregating by chance, we calculated a LOD score (logarithm of the odds) for the 14 informative meioses observed across the extended family. The LOD score of 4.2 (odds ratio 16,384:1) is strong statistical evidence supporting the KIF3B change as the cause of disease in the family. p.Leu523Pro is located in the coiled coil domain of KIF3B at a position conserved throughout vertebrates (Figures 2B, 2C, S2, and S3) is predicted to be pathogenic by SIFT,5 PolyPhen-2,6 and MutationTaster7 and is absent from all available databases, including gnomAD.
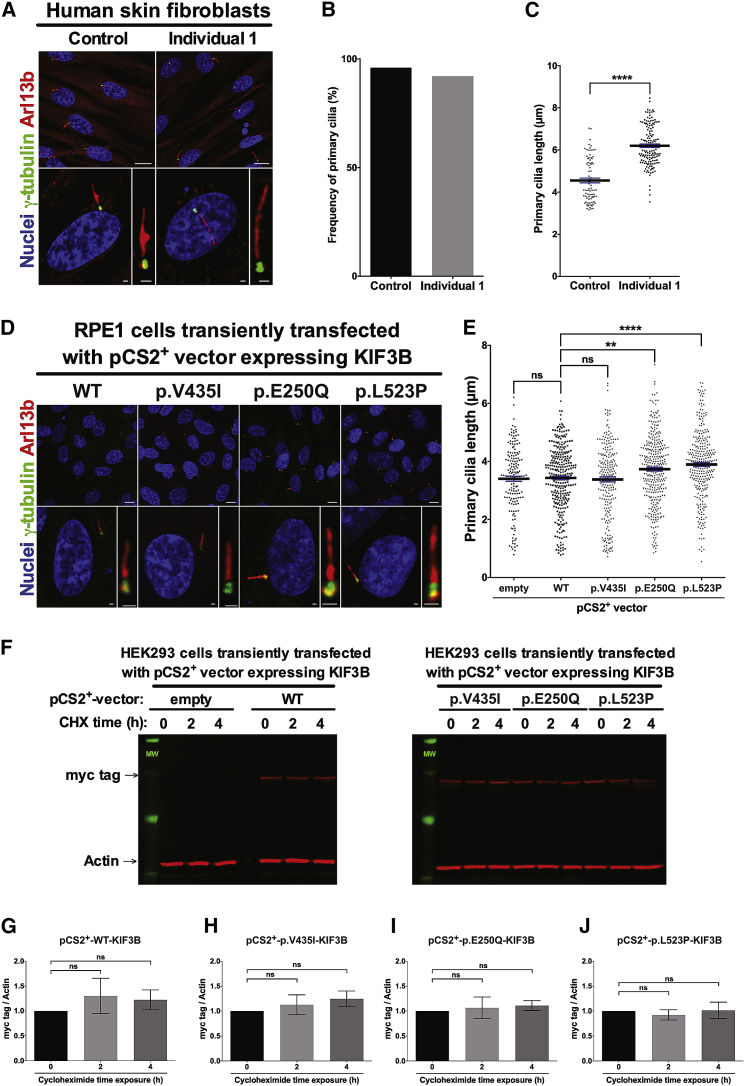
To characterize the cellular phenotypes induced by the dominantly inherited KIF3B variants identified in humans, we evaluated primary cilia length in skin fibroblasts isolated from individual 1 (family A; Supplemental Methods). We cultured fibroblasts under standard protocols of serum deprivation for 24 h to promote primary cilium elongation.11, 12, 13 We fixed case- and age-matched control cells and immunolabeled γ-tubulin and ARL13b as markers of the basal body and ciliary axoneme, respectively (Figure 3A; Table S3). We first noted that the proportion of ciliated cells from individual 1 was similar to those from the age-matched control (>90%; n = 66/72 and n = 147/153 fibroblasts/condition isolated from age-matched control and individual 1, respectively; Figure 3B). Next, we found that the primary cilia length was increased significantly in fibroblasts from individual 1 compared to age-matched control fibroblasts (36% average increase in case versus control cilia; p < 0.0001; Figure 3C).
Figure 3.
KIF3B p.Glu250Gln and p.Leu523Pro Variants Increase Primary Cilia Length
(A) Representative confocal images of primary human fibroblasts from the family A proband. Cells were methanol-fixed and immunostained with anti-ARL13b rabbit polyclonal antibody (red) and anti-γ-tubulin mouse IgG1 monoclonal antibody (green) and mounted with DAPI-Fluoromount G (blue) as markers of the primary cilium, basal body, and nuclei, respectively. Scale bars: 10 μm (top row) and 1 μm (bottom row). See Table S3 for details about antibodies used.
(B) Frequency of ciliated cells in primary skin fibroblasts from individual 1 (family A) and a matched control subject.
(C) Quantification of the primary cilia length measured on cells from a matched human control (4.55 ± 0.11 μm, n = 82) and individual 1 (family A) primary fibroblasts (6.20 ± 0.08 μm, n = 141).
For (B) and (C), six random images were assessed for each of control and affected; two independent replicates.
(D) Representative confocal images of hTERT-RPE1 cells transiently transfected with pCS2+-KIF3B-myc vectors and immunostained with anti-ARL13b (red) and anti-γ-tubulin (green) and mounted with DAPI-Fluoromount G as markers of the primary cilium, basal body, and nuclei, respectively. Scale bars: 10 μm (top row) and 1 μm (bottom row).
(E) Quantification of primary cilia length of hTERT-RPE1 cells transiently transfected with pCS2+-empty (3.40 ± 0.09 μm, n = 166), pCS2+-KIF3B-WT-myc (3.43 ± 0.06 μm, n = 302), pCS2+-KIF3B-p.V435I-myc (3.38 ± 0.09 μm, n = 233), pCS2+-KIF3B-p.E250Q-myc (3.73 ± 0.07 μm, n = 341), and pCS2+-KIF3B-p.L523P-myc (3.89 ± 0.07 μm, n = 303) vectors. Data are collected from six independent replicate experiments.
(F) Western blot analysis of myc tag (KIF3B protein levels) and actin in HEK293 cells transiently transfected with pCS2+-KIF3B-myc vectors and exposed 48 h post transfection to 50 μM cycloheximide for 0, 2, and 4 h.
(G–J) Quantitative analysis of immunoblotting results shown in (F); myc tag (KIF3B protein levels) was normalized to actin in transiently transfected HEK293 cells and data are shown relative to pCS2+-KIF3B-WT-myc vector. Performed in biological triplicates.
Data in all panels represent mean ± SEM. Statistical comparisons were performed with two-tailed unpaired t test (GraphPad PRISM software). ns, not significant, ∗∗p < 0.01; ∗∗∗∗p < 0.0001.
To validate the cilia length abnormalities observed in primary fibroblasts from individual 1, we evaluated the effect of KIF3B variants from both families in a ciliated context using hTERT-RPE1 cells, a commonly used immortalized retinal pigment epithelial cell line. We transfected C-terminally myc-tagged vectors encoding either WT or variant KIF3B (p.Glu250Gln; p.Leu523Pro or p.Val435Ile [negative control, rs41288638; 230/276,748 alleles in gnomAD]), fixed cells at 24 h post-transfection, performed immunostaining with γ-tubulin and ARL13B, and measured cilia length (Supplemental Methods; Tables S3 and S4). We observed no significant differences in primary cilia length between cells transfected with empty vector, KIF3B-WT-myc, or KIF3B-p.Val435Ile-myc constructs (Figures 3D and 3E). However, we detected a significant increase in primary cilia length for both case-associated variants compared to WT KIF3B (9% increase in p.Glu250Gln cells versus control, p < 0.01; and 13% increase in p.Leu523Pro cells versus control, p < 0.0001; Figures 3D and 3E). Notably, immunoblotting of myc-tag in whole-cell protein lysates isolated from transiently transfected HEK293 cells expressing KIF3B-myc variant constructs showed no significant differences in KIF3B protein levels compared to the KIF3B-WT-myc transfected condition (Supplemental Methods; Figure S4; Table S3).
To investigate further the stability of KIF3B protein harboring p.Glu250Gln or p.Leu523Pro, we monitored protein levels in a time course study after exposure to a protein synthesis inhibitor (Supplemental Methods; Table S3). We transiently transfected HEK293 cells with control and mutant KIF3B-myc constructs and treated cells with cycloheximide (50 μm for 2 and 4 h before protein harvest). Consistent with our initial results (Figure S4), we found no significant difference in KIF3B-myc protein levels across conditions exposed to cycloheximide for up to 4 h (Figures 3F–3J). These data support the notion that KIF3B protein harboring p.Glu250Gln or p.Leu523Pro do not undergo rapid degradation. Together, the cilia length and protein stability data indicate that primary cilia are markedly altered in the context of both KIF3B p.Glu250Gln and p.Leu523Pro variants but not for the KIF3B p.Val435Ile variant, supporting the specificity of this organellar defect.
To establish further a functional link between altered ciliary structure and overlapping human phenotypes observed in individuals with KIF3B variants, we considered prior knowledge from in vivo models of KIF3B dysfunction. Kif3b mouse models harboring a gene-trapped locus deleting a portion of exon 1 are embryonic lethal in homozygosity.14, 15, 16 A Cre-loxP conditional knockout mouse showed that loss of Kif3b results in a rapid photoreceptor degeneration associated with opsin mislocalization to rod inner segments, but Kif3b−/+ animals are viable and display no detectable retinal phenotypes.17 Additionally, kif3b−/− zebrafish models carrying a GenBank: NM_001100145.1; c.1105C>T (p.Gln369∗) encoding mutation display either absent or significantly reduced cilia length in multiple tissue types,18 contrary to our observations from KIF3B mutant primary fibroblasts with elongated cilia (Figures 3A and 3B).
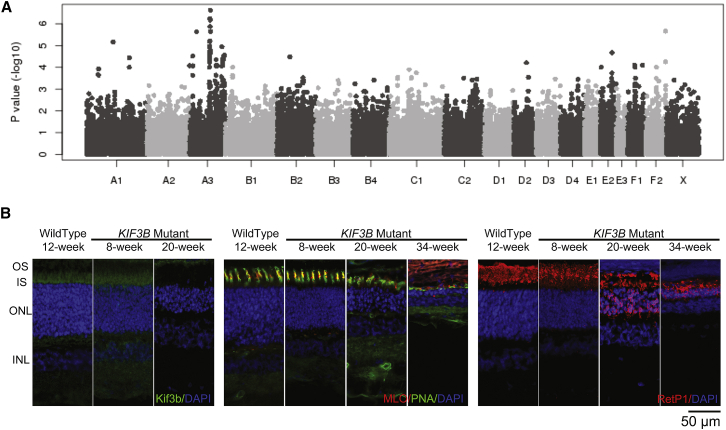
Moreover, as part of ongoing work to identify the molecular basis of clinically relevant traits in the domestic cat, we identified a nonsynonymous KIF3B change, ENSFCAT00000022266; c.1000G>A (p.Ala334Thr), as the likely cause of a recessive progressive retinal atrophy in the Bengal cat19 (Supplemental Note). We performed a genome-wide association study on 98 Bengal cats to localize an associated region on cat chromosome A3 (Supplemental Methods; Figure 4A; Table S5) and overlaid these data with whole-genome sequencing data from a trio of cats from the Bengal research colony (Table S6). The KIF3B p.Ala334Thr-encoding change was the sole variant that segregated with disease and is predicted to impair the kinesin motor domain (Figure S5). Importantly, we observed rhodopsin mislocalization to photoreceptor inner segments in KIF3B−/− mutant kittens as early as 8 weeks of age, consistent with structural and functional photoreceptor degeneration19 (Figure 4B; Supplemental Note; Supplemental Methods). Together, our in vitro data and in vivo observations from recessive KIF3B vertebrate models argue against a haploinsufficiency model for the affected humans in our study.
Figure 4.
A Recessive c.1000G>A (p.Ala334Thr) Mutation in KIF3B Is a Likely Cause for Progressive Retinal Atrophy in Bengal Cats
(A) Manhattan plot of GWAS for cat progressive retinal atrophy. After Bonferroni correction, several SNPs on cat chromosome A3 suggest a significant association with Bengal cat progressive retinal atrophy (Table S5). Cat chromosome A3 has genes homologous to human chromosome 20.
(B) Morphological and cellular changes as analyzed by immunohistochemistry. Left panel, anti-Kif3b antibody labels the region of the photoreceptor inner segment (IS) in wild-type cats. Similar labeling was detectable in sections from 8-week-old KIF3B homozygous mutants, but not in sections from 20-week-old mutant kittens. See Table S3 for details of antibodies used. Central panel, combination staining of the cone markers peanut agglutinin (PNA; cone sheath) with cone ML opsin (MLO). Cone morphology in KIF3B mutants at 8 weeks appears relatively normal but by 20 weeks there was distortion and stunting of outer segments with reduced ML opsin labeling. By 34 weeks of age, only short residual PNA labeling material remained with reduced ML opsin labeling. The outer nuclear layer (ONL) was progressively thinned. Right panel, labeling with a rhodopsin marker (RetP1). Rhodopsin is mislocalized in mutants to the inner segment as early as 8 weeks of age, then mislocalized to the outer nuclear layer cells bodies and synaptic terminals by 20 weeks. The rod inner and outer segments (OS) also showed some degeneration with disease progression.
To pursue the possibility of a dominant pathogenic effect for the p.Glu250Gln and p.Leu523Pro changes, we introduced mutant mRNAs into the developing zebrafish (Supplemental Methods; Table S4). The zebrafish genome encodes a single KIF3B ortholog (GRcZ11), for which the encoded protein (GenBank: NP_001093615.1) has 78% identity and 87% similarity to that of human (GenBank: NP_004789.1). Importantly, ablation of each of kif3b and its cognate kinesin-2 effectors encoded by kif3a and kif3c have been characterized previously in the zebrafish retina and perform similar trafficking functions to mammalian counterparts.18 We and others have shown that alteration of numerous genes involved in retinal degeneration can give rise to microphthalmia in zebrafish.20, 21, 22 As an initial test of the impact of either missense change on KIF3B function, we injected 100 pg WT or mutant KIF3B mRNA into embryos at the one-to-four cell stage and evaluated them live at 3 days post-fertilization (dpf) for external optic area in lateral bright field images using automated imaging and image analysis platforms (Supplemental Methods). We found no significant differences in eye size for clutches injected with p.Leu523Pro- or p.Val435Ile-encoding mRNA versus either WT or uninjected controls (Figures S6A and S6B). However, we observed marked lethality induced by p.Glu250Gln and assessed the effects of progressively decreasing amounts of mutant mRNA (50 pg, 25 pg, and 10 pg). The highest non-lethal doses resulted in a significant, dose-dependent, and reproducible reduction in eye area (19% reduction versus WT-injected at 50 pg; p < 0.0001; Figure S6C), an observation that persisted when normalized to larval body length to control for any possible developmental delay (Figure S6D). Co-injection of a 2:1 stoichiometric ratio of 100 pg WT with 50 pg p.Glu250Gln mRNA restored eye size to normal (not significantly different from equivalent doses of either WT or p.Glu250Gln alone; Figures S6A and S6B), supporting the specificity of p.Glu250Gln effect on eye morphology.
We hypothesized that the observed microphthalmia was caused by excessive cell death in the retina. In zebrafish and other vertebrates, the developing retina undergoes several waves of apoptosis that are necessary for the establishment of the retinal neuron networks. The first wave arises around 3 dpf and slows down at 5 dpf and the second wave takes place at 7 dpf.23 We evaluated apoptosis in retinal sections at 5 dpf by counting terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) positive cells (Supplemental Methods). We found no significant differences between larvae injected with either WT, p.Val435Ile-, or p.Leu523Pro-encoding mRNA compared to uninjected controls. However, we observed a modest but significant increase in apoptotic cells in larvae injected with p.Glu250Gln-encoding mRNA (50 pg; p < 0.05; Figures S7A and S7B), which is concordant with the microphthalmia phenotype observed in this condition (Figure S6).
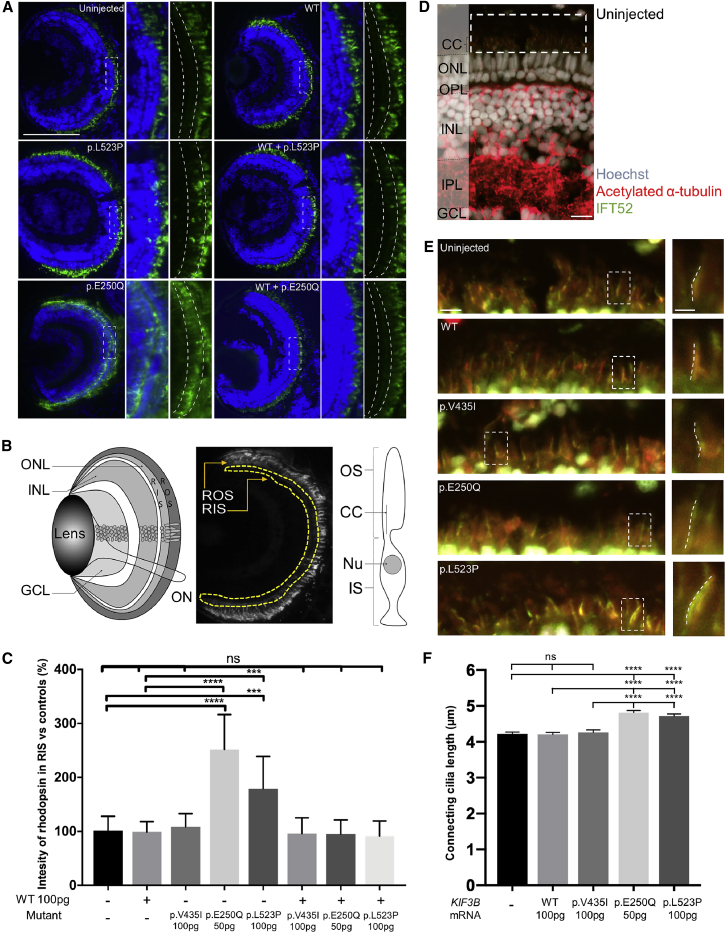
In the zebrafish retina, Kif3b plays a role in photoreceptor formation and function through the connecting cilium that is specific to low light intensity (scotopic) detection facilitated by rod cells.18 Further, kif3b mutants have documented defects in rhodopsin transport, consistent with other kinesin-2 mutants such as kif3a−/−.18 To determine whether zebrafish larvae with heterologous expression of mutant human KIF3B display defects in photoreceptor integrity, we evaluated rhodopsin localization in retinal sections at 5 dpf (Supplemental Methods; Table S3). We found no detectable differences between larvae injected with either WT or p.Val435Ile-encoding mRNA compared to uninjected controls (Figures 5A–5C). However, we observed a significant increase of rhodopsin in the rod inner segment for p.Leu523Pro (100 pg; p < 0.001), and to a greater extent, p.Glu250Gln-encoding (50 pg; p < 0.0001), versus WT mRNA (repeated with similar results, investigator masked to experimental condition; Figures 5A–5C). Consistent with the eye area studies, co-injection of 100 pg WT human KIF3B mRNA rescued the rhodopsin mislocalization defects of both variants such that they were indistinguishable from uninjected controls (Figures 5A and 5C)
Figure 5.
Retinal Phenotypes in Zebrafish Larvae with Heterologous Mutant KIF3B Expression
(A) Representative images of optic sections obtained from 5 dpf larvae and immunostained with anti-rhodopsin mouse monoclonal antibody (green) and Hoechst stain (blue). Dashed boxes indicate insets (right; with and without Hoescht). Dashed curved lines in insets demarcate rod inner segment (RIS). Scale bar: 50 μm, with equivalent scaling for each condition.
(B) Schematic of the zebrafish larval eye (left). GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer; ON, optic nerve. Schematic to indicate measurement of rhodopsin intensity in RIS of optic sections obtained from 5 dpf larvae (center). Area of interest is within yellow dashed lines; ROS, rod outer segment. Schematic of a rod photoreceptor (right). IS, inner segment; OS, outer segment.
(C) Quantification of rhodopsin intensity of RIS in optic sections (A and B) normalized to controls in 5 dpf larvae. n = 10 larvae per condition, repeated twice with similar results. Error bars indicate standard deviation (SD) of the mean.
(D) Representative merged image of an optic section obtained from a 5 dpf uninjected larva that was immunostained with anti-acetylated α-tubulin mouse monoclonal antibody (red, ciliary axoneme), anti-IFT52 rabbit polyclonal antibody (green, anterograde IFT), and Hoechst staining (gray; nuclei). CC, connecting cilium; OPL, outer plexiform layer; IPL, inner plexiform layer. Scale bar, 10 μm. Note that in addition to ciliary microtubules, anti-acetylated α-tubulin stains axon tracts throughout the retina.
(E) Left, representative magnified views with enhanced contrast to enable visualization of connecting cilia (equivalent across images; see dashed white box in D), scale bar: 5 μm, with equivalent scaling for each condition; right, insets show a magnified view of a representative connecting cilium (see dashed white box at left); dashed lines indicate ciliary length measurement, scale bar: 2 μm, with equivalent scaling for each condition.
(F) Quantification of photoreceptor connecting cilia length in 5 dpf retinas. n = 10–12 larvae per condition, n = 174–246 cilia per retina, repeated twice with similar results. Error bars indicate standard error of the mean (SEM).
Statistical comparisons were performed with a non-parametric one-way ANOVA followed by Tukey’s multiple comparison (GraphPad PRISM software; v.7.0c). ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001; ns, not significant; WT, wild type; p.Glu250Gln and p.Leu523Pro are variants identified in cases; p.Val435Ile is a negative control (rs41288638; 230/276,748 alleles in gnomAD). See Table S3 for antibodies used.
We next wondered whether the observed rhodopsin mislocalization was coincident with altered cilia length in zebrafish with ectopic expression of KIF3B mutant mRNAs. To paint photoreceptor-connecting cilia, we performed antibody staining of transverse cryosections from 5 dpf larvae with two ciliary markers, acetylated α-tubulin and IFT52, and measured length of structures with double labeling (Supplemental Methods; Figure 5D; Table S3). We found no significant differences in connecting cilium length between larvae injected with either WT or p.Val435Ile encoding mRNA compared to uninjected controls (Figures 5E and 5F). However, we observed a significant elongation of connecting cilia for p.Leu523Pro-encoding (100 pg; p < 0.0001) and p.Glu250Gln-encoding (50 pg; p < 0.0001) versus WT mRNA (repeated with similar results; Figures 5E and 5F). We detected no differences in length measurements when acetylated α-tubulin and IFT52 signal was assessed individually, arguing against sequestration of IFT-B components as the molecular mechanism underpinning the long cilia phenotype (Figure S8). In aggregate, these data reinforce the relevance of KIF3B function to photoreceptor integrity and opsin trafficking in rod photoreceptors and support the pathogenicity of p.Glu250Gln and p.Leu523Pro.
The molecular motor is essential for kinesin complexes to move along microtubules in an ATP-dependent manner24 and multiple dominant pathogenic amino acid substitutions in kinesin genes have been identified in this domain. The Glu250 position is an ultra-conserved residue in 41/42 human kinesin motor domain protein sequences, except KIF26B (Figure S2) and vertebrate KIF3B orthologs (Figure 2C). This residue was substituted by a lysine in KIF1A (GenBank: NM_001244008.1; c.757G>A [p.Glu253Lys]) in two individuals with neuroanatomical phenotypes described by Lee et al.24 Notably, individuals carrying this variant exhibited a severe phenotype, consistent with the more acute presentation of individual 1 (family A) in our study. Five case subjects with substitutions of the equivalent residue in KIF5C by a Lys or Val were also reported.25 Rice et al. observed that when the glutamic acid was substituted by alanine (p.Glu236Ala), the ATPase activity of the kinesin was reduced at least 1,000-fold.26 Hence, this variant might act in a dominant manner by altering the ability of the KIF3A/B heterodimer to move along the ciliary microtubules, disorganizing ciliogenesis and thus causing the augmented primary cilia length of the fibroblasts of individual 1 (Figure 3).
Kittens homozygous for the KIF3B mutation showed a rapid loss of photoreceptors. KIF3B immunolabeling showed that initially the mutant protein was detectable in the region of inner and outer segments; however, with progressive degeneration of both layers and loss of photoreceptor nuclei Kif3b immunoreactivity was no longer detectable (Figure 4B). This loss of immunoreactivity was accompanied by pronounced mislocalization of rhodopsin. Abnormal trafficking of opsin to the outer segment with retention in the inner segment and cell body has been reported in a number of retinal degeneration conditions including RP in humans and animal models.27, 28, 29, 30, 31, 32, 33 The KIF3B p.Ala334Thr variant in the Bengal cat might not totally ablate protein function resulting in a recessive retinal phenotype, in contrast to previously reported zebrafish and mouse recessive KIF3B mutants that are embryonic lethal.14, 15, 16,18,34 Further studies are required to investigate impact of p.Ala334Thr on cilia length and KIF3B protein stability in the feline retina.
The ciliopathies are known for exhibiting variable expressivity of endophenotypes,35 and this phenomenon is exemplified by the two pedigrees in our study and the Bengal cat. Inter- and intra-familial variability within this disease spectrum has been attributed to allelism at the same locus,36 second-site modification in trans,37 and for dominant disorders, variable expression of the wild-type allele.38 Two different KIF3B protein domains are impacted by each of the deleterious variants characterized by this study (kinesin motor, family A versus coiled coil domain, family B), leading us to speculate that allele effect could be sufficient to drive phenotype differences between the two families. Our in vivo assessment of rhodopsin trafficking in the zebrafish retina supports this posit (Figure 5). However, we cannot exclude the possibility that the family A proband has an exacerbated syndromic phenotype due to genetic interaction with additional variation in other loci that interact either directly or indirectly with KIF3B. We recognize that a limitation of this study is the fact that only coding regions were queried for variation; whole-genome sequencing will be required to obtain a complete list of contributory sites that determine phenotype in both families.
In conclusion, our study adds to the repertoire of ciliary genes that result in human pathology and characterize at clinical and molecular levels a ciliopathy hallmarked by RP, polydactyly, and liver fibrosis likely caused by an alteration of kinesin-2-driven transport. Most ciliopathies, especially those caused by IFT motor or raft proteins, are autosomal-recessive disorders.39, 40, 41 However, we add another exception to this paradigm by reporting humans who harbor dominant de novo or inherited heterozygous variants, placing them in the class of dominant ciliopathies inclusive of conditions such as polycystic kidney disease (MIM: 173900) and orofacial digital syndrome (MIM: 311200).
Declaration of Interests
N.K. is a paid consultant for and holds significant stock of Rescindo Therapeutics, Inc.
Acknowledgments
We are grateful to the families who participated in this study. We thank Kevin Adams, Westley Heydeck, and Julien Philippe for technical support and advice. We thank Brian Perkins for the IFT52 antibody, and his assistance with zebrafish antibody staining protocols. We also thank Stefan Scholz and Elisabet Teixido from UFZ as well as Tobias Kießling from “Scientific Software Solutions” (www.tks3.com) for use and assistance with FishInspector software. This work was supported by US National Institutes of Health grants HD042601 (N.K.), GM121317 (N.K.), DK072301 (N.K. and E.E.D.), and EY007142 (S.P.D.), and by grants from the Foundation Fighting Blindness USA (S.P.D.) and the William Stamps Farish Fund (S.P.D.). N.K. is a Distinguished Valerie and George D. Kennedy Professor. This project was funded in part previously by the National Center for Research Resources R24 RR016094 and the Office of Research Infrastructure Programs OD R24OD010928, the Phyllis and George Miller Feline Health Fund, Center for Companion Animal Health, School of Veterinary Medicine, University of California – Davis (2007-38-FM, 2008-08-F), the Winn Feline Foundation (MT07-012, W12-022), and Cat Health Network (D12FE-509) and the University of Missouri, College of Veterinary Medicine Gilbreath McLorn Endowment (L.A.L.). We appreciate the dedicated assistance of Bengal cat breeders for their participation in the research. S.M.P.-J. is funded by the Myers-Dunlap Endowment at Michigan State University. This work was supported by grants from the National Eye Institute (R01EY012910 to E.A.P., R01EY026904 to K.B. and E.A.P., and P30EY014104 for MEEI core support) and the Foundation Fighting Blindness (EGI-GE-1218-0753-UCSD to K.B. and E.A.P.).
Published: May 7, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.04.005.
Contributor Information
Erica E. Davis, Email: eridavis@luriechildrens.org.
Bertrand Isidor, Email: bertrand.isidor@chu-nantes.fr.
99 Lives Consortium:
Reuben M. Buckley, Danielle Aberdein, Paulo C. Alves, Gregory S. Barsh, Rebecca R. Bellone, Tomas F. Bergström, Adam R. Boyko, Jeffrey A. Brockman, Margret L. Casal, Marta G. Castelhano, Ottmar Distl, Nicholas H. Dodman, N. Matthew Ellinwood, Jonathan E. Fogle, Oliver P. Forman, Dorian J. Garrick, Edward I. Ginns, Jens Häggström, Robert J. Harvey, Daisuke Hasegawa, Bianca Haase, Christopher R. Helps, Isabel Hernandez, Marjo K. Hytönen, Maria Kaukonen, Christopher B. Kaelin, Tomoki Kosho, Emilie Leclerc, Teri L. Lear, Tosso Leeb, Ronald H.L. Li, Hannes Lohi, Maria Longeri, Mark A. Magnuson, Richard Malik, Shrinivas P. Mane, John S. Munday, William J. Murphy, Niels C. Pedersen, Max F. Rothschild, Clare Rusbridge, Beth Shapiro, Joshua A. Stern, William F. Swanson, Karen A. Terio, Rory J. Todhunter, Wesley C. Warren, Elizabeth A. Wilcox, Julia H. Wildschutte, and Yoshihiko Yu
Web Resources
99 Lives Cat Genome Sequencing Initiative, http://felinegenetics.missouri.edu/99lives
1000 Genomes, http://www.internationalgenome.org/
ClustalW, http://www.clustal.org/
Ensembl Genome Browser, http://www.ensembl.org/index.html
ExAC Browser, http://exac.broadinstitute.org/
Fiji, https://fiji.sc/
GeneMatcher, https://genematcher.org/
gnomAD Browser, https://gnomad.broadinstitute.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, https://www.omim.org/
QuickChange, http://www.genomics.agilent.com/primerDesignProgram.jsp
UCSC Genome Browser, http://genome.ucsc.edu
UCSF Chimera software, https://www.cgl.ucsf.edu/chimera/
UniProt, http://www.uniprot.org/
Supplemental data
References
- 1.Reilly M.L., Stokman M.F., Magry V., Jeanpierre C., Alves M., Paydar M., Hellinga J., Delous M., Pouly D., Failler M. Loss of function mutations in KIF14 cause severe microcephaly and kidney development defects in humans and zebrafish. Hum. Mol. Genet. 2019;28:778–795. doi: 10.1093/hmg/ddy381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goetz S.C., Anderson K.V. The primary cilium: a signalling centre during vertebrate development. Nat. Rev. Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Badano J.L., Mitsuma N., Beales P.L., Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- 4.van Dam T.J., Wheway G., Slaats G.G., Huynen M.A., Giles R.H., SYSCILIA Study Group The SYSCILIA gold standard (SCGSv1) of known ciliary components and its applications within a systems biology consortium. Cilia. 2013;2:7. doi: 10.1186/2046-2530-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 6.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwarz J.M., Rödelsperger C., Schuelke M., Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 8.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sullivan L.S., Bowne S.J., Birch D.G., Hughbanks-Wheaton D., Heckenlively J.R., Lewis R.A., Garcia C.A., Ruiz R.S., Blanton S.H., Northrup H. Prevalence of disease-causing mutations in families with autosomal dominant retinitis pigmentosa: a screen of known genes in 200 families. Invest. Ophthalmol. Vis. Sci. 2006;47:3052–3064. doi: 10.1167/iovs.05-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koboldt D.C., Larson D.E., Sullivan L.S., Bowne S.J., Steinberg K.M., Churchill J.D., Buhr A.C., Nutter N., Pierce E.A., Blanton S.H. Exome-based mapping and variant prioritization for inherited Mendelian disorders. Am. J. Hum. Genet. 2014;94:373–384. doi: 10.1016/j.ajhg.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pampliega O., Orhon I., Patel B., Sridhar S., Díaz-Carretero A., Beau I., Codogno P., Satir B.H., Satir P., Cuervo A.M. Functional interaction between autophagy and ciliogenesis. Nature. 2013;502:194–200. doi: 10.1038/nature12639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stoetzel C., Bär S., De Craene J.-O., Scheidecker S., Etard C., Chicher J., Reck J.R., Perrault I., Geoffroy V., Chennen K. A mutation in VPS15 (PIK3R4) causes a ciliopathy and affects IFT20 release from the cis-Golgi. Nat. Commun. 2016;7:13586. doi: 10.1038/ncomms13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rozycki M., Lodyga M., Lam J., Miranda M.Z., Fátyol K., Speight P., Kapus A. The fate of the primary cilium during myofibroblast transition. Mol. Biol. Cell. 2014;25:643–657. doi: 10.1091/mbc.E13-07-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nonaka S., Tanaka Y., Okada Y., Takeda S., Harada A., Kanai Y., Kido M., Hirokawa N. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 1998;95:829–837. doi: 10.1016/s0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- 15.Takeda S., Yonekawa Y., Tanaka Y., Okada Y., Nonaka S., Hirokawa N. Left-right asymmetry and kinesin superfamily protein KIF3A: new insights in determination of laterality and mesoderm induction by kif3A-/- mice analysis. J. Cell Biol. 1999;145:825–836. doi: 10.1083/jcb.145.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alsabban A.H., Morikawa M., Tanaka Y., Takei Y., Hirokawa N. Kinesin Kif3b mutation reduces NMDAR subunit NR2A trafficking and causes schizophrenia-like phenotypes in mice. EMBO J. 2020;39:e101090. doi: 10.15252/embj.2018101090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jimeno D., Lillo C., Roberts E.A., Goldstein L.S.B., Williams D.S. Kinesin-2 and photoreceptor cell death: requirement of motor subunits. Exp. Eye Res. 2006;82:351–353. doi: 10.1016/j.exer.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 18.Zhao C., Omori Y., Brodowska K., Kovach P., Malicki J. Kinesin-2 family in vertebrate ciliogenesis. Proc. Natl. Acad. Sci. USA. 2012;109:2388–2393. doi: 10.1073/pnas.1116035109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ofri R., Reilly C.M., Maggs D.J., Fitzgerald P.G., Shilo-Benjamini Y., Good K.L., Grahn R.A., Splawski D.D., Lyons L.A. Characterization of an Early-Onset, Autosomal Recessive, Progressive Retinal Degeneration in Bengal Cats. Invest. Ophthalmol. Vis. Sci. 2015;56:5299–5308. doi: 10.1167/iovs.15-16585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y.P., Bosch D.G.M., Siemiatkowska A.M., Rendtorff N.D., Boonstra F.N., Möller C., Tranebjærg L., Katsanis N., Cremers F.P.M. Putative digenic inheritance of heterozygous RP1L1 and C2orf71 null mutations in syndromic retinal dystrophy. Ophthalmic Genet. 2017;38:127–132. doi: 10.3109/13816810.2016.1151898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li L., Nakaya N., Chavali V.R.M., Ma Z., Jiao X., Sieving P.A., Riazuddin S., Tomarev S.I., Ayyagari R., Riazuddin S.A., Hejtmancik J.F. A mutation in ZNF513, a putative regulator of photoreceptor development, causes autosomal-recessive retinitis pigmentosa. Am. J. Hum. Genet. 2010;87:400–409. doi: 10.1016/j.ajhg.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shu X., Zeng Z., Gautier P., Lennon A., Gakovic M., Cheetham M.E., Patton E.E., Wright A.F. Knockdown of the zebrafish ortholog of the retinitis pigmentosa 2 (RP2) gene results in retinal degeneration. Invest. Ophthalmol. Vis. Sci. 2011;52:2960–2966. doi: 10.1167/iovs.10-6800. [DOI] [PubMed] [Google Scholar]
- 23.Biehlmaier O., Neuhauss S.C., Kohler K. Onset and time course of apoptosis in the developing zebrafish retina. Cell Tissue Res. 2001;306:199–207. doi: 10.1007/s004410100447. [DOI] [PubMed] [Google Scholar]
- 24.Lee J.-R., Srour M., Kim D., Hamdan F.F., Lim S.-H., Brunel-Guitton C., Décarie J.-C., Rossignol E., Mitchell G.A., Schreiber A. De novo mutations in the motor domain of KIF1A cause cognitive impairment, spastic paraparesis, axonal neuropathy, and cerebellar atrophy. Hum. Mutat. 2015;36:69–78. doi: 10.1002/humu.22709. [DOI] [PubMed] [Google Scholar]
- 25.Michels S., Foss K., Park K., Golden-Grant K., Saneto R., Lopez J., Mirzaa G.M. Mutations of KIF5C cause a neurodevelopmental disorder of infantile-onset epilepsy, absent language, and distinctive malformations of cortical development. Am. J. Med. Genet. A. 2017;173:3127–3131. doi: 10.1002/ajmg.a.38496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rice S., Lin A.W., Safer D., Hart C.L., Naber N., Carragher B.O., Cain S.M., Pechatnikova E., Wilson-Kubalek E.M., Whittaker M. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- 27.Adamian M., Pawlyk B.S., Hong D.-H., Berson E.L. Rod and cone opsin mislocalization in an autopsy eye from a carrier of X-linked retinitis pigmentosa with a Gly436Asp mutation in the RPGR gene. Am. J. Ophthalmol. 2006;142:515–518. doi: 10.1016/j.ajo.2006.03.061. [DOI] [PubMed] [Google Scholar]
- 28.Concepcion F., Chen J. Q344ter mutation causes mislocalization of rhodopsin molecules that are catalytically active: a mouse model of Q344ter-induced retinal degeneration. PLoS ONE. 2010;5:e10904. doi: 10.1371/journal.pone.0010904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao J., Cheon K., Nusinowitz S., Liu Q., Bei D., Atkins K., Azimi A., Daiger S.P., Farber D.B., Heckenlively J.R. Progressive photoreceptor degeneration, outer segment dysplasia, and rhodopsin mislocalization in mice with targeted disruption of the retinitis pigmentosa-1 (Rp1) gene. Proc. Natl. Acad. Sci. USA. 2002;99:5698–5703. doi: 10.1073/pnas.042122399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishimura D.Y., Fath M., Mullins R.F., Searby C., Andrews M., Davis R., Andorf J.L., Mykytyn K., Swiderski R.E., Yang B. Bbs2-null mice have neurosensory deficits, a defect in social dominance, and retinopathy associated with mislocalization of rhodopsin. Proc. Natl. Acad. Sci. USA. 2004;101:16588–16593. doi: 10.1073/pnas.0405496101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mowat F.M., Gornik K.R., Dinculescu A., Boye S.L., Hauswirth W.W., Petersen-Jones S.M., Bartoe J.T. Tyrosine capsid-mutant AAV vectors for gene delivery to the canine retina from a subretinal or intravitreal approach. Gene Ther. 2014;21:96–105. doi: 10.1038/gt.2013.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beltran W.A., Cideciyan A.V., Lewin A.S., Iwabe S., Khanna H., Sumaroka A., Chiodo V.A., Fajardo D.S., Román A.J., Deng W.-T. Gene therapy rescues photoreceptor blindness in dogs and paves the way for treating human X-linked retinitis pigmentosa. Proc. Natl. Acad. Sci. USA. 2012;109:2132–2137. doi: 10.1073/pnas.1118847109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hollingsworth T.J., Gross A.K. Defective trafficking of rhodopsin and its role in retinal degenerations. Int. Rev. Cell Mol. Biol. 2012;293:1–44. doi: 10.1016/B978-0-12-394304-0.00006-3. [DOI] [PubMed] [Google Scholar]
- 34.Feng D., Chen Z., Yang K., Miao S., Xu B., Kang Y., Xie H., Zhao C. The cytoplasmic tail of rhodopsin triggers rapid rod degeneration in kinesin-2 mutants. J. Biol. Chem. 2017;292:17375–17386. doi: 10.1074/jbc.M117.784017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davis E.E., Katsanis N. The ciliopathies: a transitional model into systems biology of human genetic disease. Curr. Opin. Genet. Dev. 2012;22:290–303. doi: 10.1016/j.gde.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Braun D.A., Hildebrandt F. Ciliopathies. Cold Spring Harb. Perspect. Biol. 2017;9 doi: 10.1101/cshperspect.a028191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kousi M., Katsanis N. Genetic modifiers and oligogenic inheritance. Cold Spring Harb. Perspect. Med. 2015;5:5. doi: 10.1101/cshperspect.a017145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rivolta C., McGee T.L., Rio Frio T., Jensen R.V., Berson E.L., Dryja T.P. Variation in retinitis pigmentosa-11 (PRPF31 or RP11) gene expression between symptomatic and asymptomatic patients with dominant RP11 mutations. Hum. Mutat. 2006;27:644–653. doi: 10.1002/humu.20325. [DOI] [PubMed] [Google Scholar]
- 39.Afzelius B.A. A human syndrome caused by immotile cilia. Science. 1976;193:317–319. doi: 10.1126/science.1084576. [DOI] [PubMed] [Google Scholar]
- 40.Huynh Cong E., Bizet A.A., Boyer O., Woerner S., Gribouval O., Filhol E., Arrondel C., Thomas S., Silbermann F., Canaud G. A homozygous missense mutation in the ciliary gene TTC21B causes familial FSGS. J. Am. Soc. Nephrol. 2014;25:2435–2443. doi: 10.1681/ASN.2013101126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bujakowska K.M., Zhang Q., Siemiatkowska A.M., Liu Q., Place E., Falk M.J., Consugar M., Lancelot M.-E., Antonio A., Lonjou C. Mutations in IFT172 cause isolated retinal degeneration and Bardet-Biedl syndrome. Hum. Mol. Genet. 2015;24:230–242. doi: 10.1093/hmg/ddu441. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.