Abstract
Oculopharyngodistal myopathy (OPDM) is an adult-onset inherited neuromuscular disorder characterized by progressive ptosis, external ophthalmoplegia, and weakness of the masseter, facial, pharyngeal, and distal limb muscles. The myopathological features are presence of rimmed vacuoles (RVs) in the muscle fibers and myopathic changes of differing severity. Inheritance is variable, with either putative autosomal-dominant or autosomal-recessive pattern. Here, using a comprehensive strategy combining whole-genome sequencing (WGS), long-read whole-genome sequencing (LRS), linkage analysis, repeat-primed polymerase chain reaction (RP-PCR), and fluorescence amplicon length analysis polymerase chain reaction (AL-PCR), we identified an abnormal GGC repeat expansion in the 5′ UTR of GIPC1 in one out of four families and three sporadic case subjects from a Chinese OPDM cohort. Expanded GGC repeats were further confirmed as the cause of OPDM in an additional 2 out of 4 families and 6 out of 13 sporadic Chinese individuals with OPDM, as well as 7 out of 194 unrelated Japanese individuals with OPDM. Methylation, qRT-PCR, and western blot analysis indicated that GIPC1 mRNA levels were increased while protein levels were unaltered in OPDM-affected individuals. RNA sequencing indicated p53 signaling, vascular smooth muscle contraction, ubiquitin-mediated proteolysis, and ribosome pathways were involved in the pathogenic mechanisms of OPDM-affected individuals with GGC repeat expansion in GIPC1. This study provides further evidence that OPDM is associated with GGC repeat expansions in distinct genes and highly suggests that expanded GGC repeat units are essential in the pathogenesis of OPDM, regardless of the genes in which the expanded repeats are located.
Keywords: oculopharyngodistal myopathy, GIPC1, GGC repeat expansions, intranuclear inclusions, RNA-seq
Introduction
Oculopharyngodistal myopathy (OPDM [MIM: 164310]) is a rare adult-onset myopathy with putative autosomal-dominant or autosomal-recessive inheritance. The typical clinical manifestations are insidiously progressive ptosis, ophthalmoparesis, facial and masseter weakness, dysphagia, and muscle weakness of distal limbs. The myopathological features are myopathic changes of differing severity characterized by the presence of rimmed vacuoles (RVs) and filamentous intranuclear inclusions in the muscle fibers.1, 2, 3 Because the genetic causes and pathogenic mechanisms underlying OPDM have long remained elusive, OPDM-affected individuals have been diagnosed by clinical manifestations, histopathological findings, and genetic exclusion of similar conditions. Since OPDM was first described by Satoyoshi in 1977, about 200 OPDM-affected individuals from Turkey, Japan, the Netherlands, Italy, the United States, and China have been reported.1, 2, 3, 4, 5, 6, 7, 8, 9 It is important to identify the causative genes to diagnose and study the pathogenesis of OPDM. Our lab group and the lab group of Dr. Lochmuller have performed whole-exome/genome sequencing (WES/WGS) to explore the genetic causes of OPDM but have failed to identify likely pathogenic mutations.10,11
Despite the high similarity of clinical and pathological features in different individuals with OPDM, the inheritance modes, characterized by sporadic, putative autosomal-recessive, autosomal-dominant, or incomplete-penetrance patterns, have been unclear.10 Inspired by the striking similarity to the inheritance pattern of neuronal intranuclear inclusion disease (NIID [MIM: 603472]), as well as the intranuclear inclusions observed in both diseases, we hypothesize that OPDM may share a similar genetic basis with NIID—a disease caused by expanded GGC repeats in the 5′ untranslated region (UTR) of NOTCH2NLC (MIM: 618025).12, 13, 14 To explore the possibility of repeat expansions in OPDM, we performed long-read whole-genome sequencing (LRS) on the Oxford Nanopore platform. While our LRS data were being analyzed, a study reported that noncoding CGG repeat expansions in LRP12 (MIM: 618299) was identified as the disease-causing mutation in 22 out of 88 OPDM-affected individuals in a Japanese population.15 However, the genetic causes of 75% of OPDM cases remained unknown in that study.
In the present study, we aimed to explore the genetic etiology of OPDM in Chinese individuals. We performed LRS on a cohort of individuals with OPDM and identified trinucleotide repeat expansions in the 5′ UTR not only in LRP12 as it was described in Japanese individuals, but also in a different gene, GIPC1 (MIM: 605072), on chromosome 19 (chr19: 14,606,853–14,606,897). We also demonstrated that the GGC repeat expansions did not result in detectable changes of GIPC1 protein level in skeletal muscle of affected individuals. The observation of similar repeat expansions in two distinct genes strongly suggested that these repeat expansions caused OPDM possibly by shared pathogenesis, independently of their genome location and impact on the recipient gene.
Material and Methods
OPDM-Affected Individuals and Control Subjects
Eight families and 16 sporadic cases diagnosed with OPDM from mainland China were recruited from Peking University First Hospital. Families 1, 5, and 6 and sporadic individuals 1–3 had been reported previously.11 This diagnosis was supported both by clinical features including slowly progressive external ocular, laryngopharyngeal, facial, and distal limb muscle weakness and histopathological abnormities such as myopathic changes with formation of rimmed vacuoles (RVs). Genetic analysis indicated that no individuals had expanded GCN repeat of the polyadenylate binding protein nuclear 1 (PABPN1) gene (MIM: 602279)16 or expanded CTG repeat of the DM1 protein kinase (DMPK) gene (MIM: 605377).17 Meanwhile, all known genes associated with neuromuscular disorders were screened by targeted next-generation sequencing via Nextera kits (Illumina). No known gene mutation was found.11 The clinical data and laboratory findings were collected. Genomic DNA of peripheral blood leukocytes were obtained from 8 families, 16 sporadic cases, and 1,000 unaffected control subjects.
Besides Chinese OPDM cohorts, a cohort of Japanese individuals with OPDM diagnosed at National Center of Neurology and Psychiatry were also analyzed. Inclusion criteria are clinicopathologically suspected to have OPDM, but PABPN1 and LRP12 mutations were excluded.
This study was approved by the Local Ethics Committee of Peking University First Hospital in China. All participants gave informed consent.
Muscle Biopsy, Immunohistochemistry, Immunofluorescence, and Electron Microscopy
Open muscle biopsy was performed according to a standard procedure.11
For histological examination, serial frozen sections (8 μm) were stained by routine histochemistry, including hematoxylin & eosin (H&E), modified Gomori trichrome (mGT), Oil Red O, periodic acid Schiff, succinate dehydrogenase, nicotinamide adenine dinucleotide tetrazolium reductase (NADH-TR), cytochrome c oxidase, and muscle fiber ATPase at varying pH levels. For immunohistochemistry, frozen muscle sections were immunostained with specific anti-p62 (Abcam). All slides were visualized using a microscope.
For immunofluorescence, muscle sections were immunostained with specific anti-GIPC1 (ProteinTech Group) and a secondary anti-rabbit fluorescein isothiocyanate antibody, as well as anti-p62 (Abcam) and a secondary anti-murine tetramethylrhodamine antibody. Images were acquired at 60 × magnification by confocal microscopy (Nikon A1MP).
Electron microscopy was done as in previous studies.18,19 Skeletal muscle from OPDM-affected individuals was collected and fixed in 2.5% glutaraldehyde overnight at 4°C. Samples were sectioned on a Leica EM UC6/FC6 Ultramicrotome. After sections were transferred to copper grids, counter staining was performed with uranyl acetate and lead acetate before EM imaging.
Whole-Genome Sequencing (WGS)
For short-read sequencing, the DNA sequencing library was constructed using a MGIEasy DNA Library Prep Kit following the manufacturer’s instructions (BGI, China) to generate DNA nanoballs (DNB). Each DNB library was sequenced on the MGISEQ-2000 instrument and 150 bp paired-end reads were generated. Burrows-Wheeler Alignment Maximal Exact Matches (BWA-MEM)20 was used for read mapping onto the hg38 human genome as a reference. Sequencing data passing quality control (QC) were subject to a computational pipeline for data processing and analysis, following the standard workflow.21 Calls with variant quality less than 20 were filtered out and 95% of the targeted bases were covered sufficiently to pass our thresholds for calling single-nucleotide polymorphisms (SNP) and small insertions or deletions (indels). GATK was used for calling single-nucleotide variants (SNVs)/indels, ANNOVAR was used for variant annotation, and we manually examined all potential disease variants in Integrative Genomics Viewer.
Long-Read Whole-Genome Sequencing (LRS)
DNA samples of affected individuals with OPDM and healthy individuals were sequenced using a PromethION sequencer (Oxford Nanopore Technologies). Library preparation was carried out using a 1D Genomic DNA ligation kit (SQKLSK109) according to the manufacturer’s protocol. For each individual, one PRO-002 (R9.4.1) flowcell was used. PromethION data base-calling was performed using guppy v.3.3.0 (Oxford Nanopore Technologies), and only pass reads (qscore ≥ 7) were used for subsequent analysis.
STR Detection and STR-Scoring Framework
Based on the repeatmasker file from UCSC Genome Browser,22 we prepared a gene-associated STR list spanning all GENCODE V19 genes. Genes upstream and downstream of the 10 kb region and the STR repeat unit ranged in length from 3 to 6 bp. The pass reads from PromethION were aligned to the reference genome hg19 using ngmlr v.0.2.7.23 For each repeat, the repeat count of each read that aligned with the STR locus was detected using repeatHMM24 without the peak calling step, which is not good with low coverage data.
As nanopore reads have highly biased errors, the nth highest repeat count was defined as the individual’s estimated repeat count (ERC), in which n is the largest integer not greater than the 20% sequencing depth in the STR locus. According to the above-mentioned description, we constructed ERC matrixes for the healthy individuals (Hm × n) and affected individuals (A m × s), where m, n, and s denote the number of STRs and the numbers of healthy and affected individuals, respectively. For example, Hi,j denotes the jth healthy individual’s ERC at ith STR locus for i = 1···m, j = 1···n.
To better screen the disease-associated STR, we designed a scoring framework, STR-Scoring, which can offset the impact of error bias and population common repeat expansion to prioritize the STR.
The scoring procedure was implemented as follows:
Repeat count change: for each repeat, we used cubic mean25 ci = to represent the STR repeat count in healthy individuals, and used di,j = max(Aij-ci, 0)/ci to represent the difference between each affected individual and all healthy individuals.
Population proportion: the proportion of healthy individuals with an ERC greater than the one affected individual was defined as the population proportion of the affected individual for every repeat, pi, j = sum(Hi· ≥ Ai,j)/n; for i = 1··· m, j = 1··· s. In addition, the number of affected individuals whose population proportion was less than 5% was normalized by the total number of affected individuals as a weighted score, qi = sum(pi, j < 0.05)/s for i = 1··· m, j = 1··· s.
Repeat location and pattern: if the repeat location (coding, UTR, promoter, exon of non-coding RNA, and intron) and pattern has been previously reported,25,26 fi = 2; otherwise, fi = 1. For the repeat pattern, the base order and reverse complement sequences were taken into account; for example, the pattern of CAG and its derived sequence AGC, GCA, CTG, GCT, TGC.
For each repeat, two methods were used to calculate the integrative scores. One is STR_Score = avg(di,j·(1-pi,j))qi·fi, and the other is STR_Score = avg(di,j·(1-pi,j))qi without weight coefficients of repeat location and pattern. Finally, based on the STR_Score, we ranked the repeats to screen the disease-associated locus.
Genome-wide Linkage Analysis
Genomic DNA was isolated from whole blood from 16 family members of family 1 (Figure 2B) and subjected to genome-wide linkage analysis using OmniZhongHua-8 v1.4 BeadChip (Illumina) containing 1,175,489 single-nucleotide polymorphism (SNP) markers as previously reported. 27,28 Genotype calling and quality control were processed with Illumina GenomeStudio Genotyping Module. SNPs with Mendelian errors or call rates lower than 100% were removed. Parametric linkage analysis was performed with Merlin v.1.1.2 to calculate the logarithm of the odds (LOD) scores. The inheritance model was considered as an incomplete penetrance autosomal dominant model with a rare disease allele frequency of 0.0001.
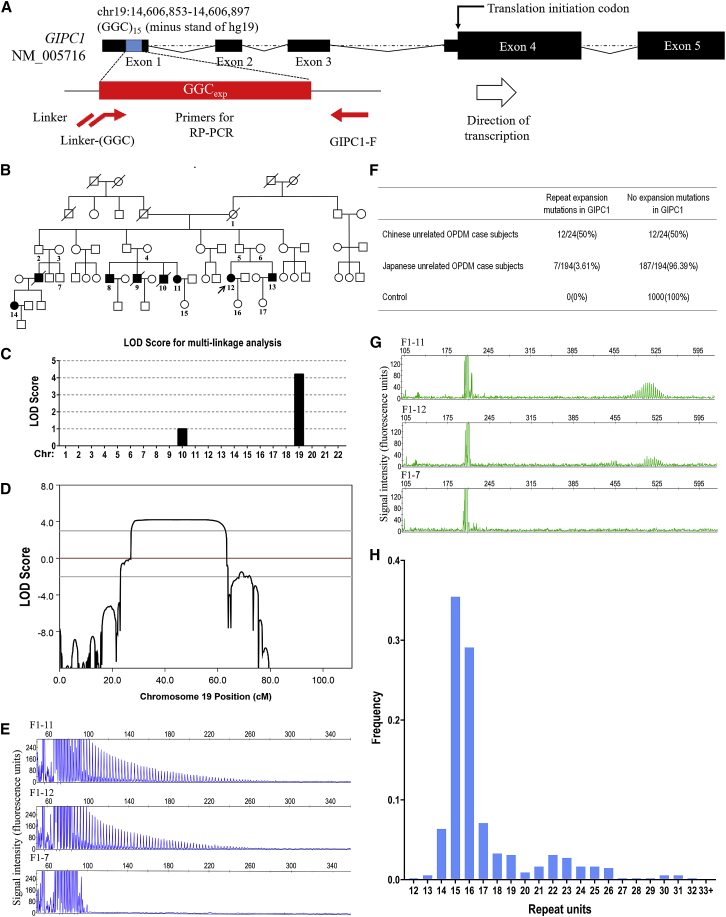
Figure 2.
Validation of GGC Repeat Expansions in GIPC1 and Variation of GGC Repeat Size among Normal Control Subjects
(A) Schematic representation of GIPC1 indicating the location of GGC repeat expansions. The primer set used for RP-PCR analysis is designed to detect expanded GGC repeats (red line and arrows).
(B) Pedigree chart of a family with OPDM (family 1). Squares and circles indicate males and females, respectively. A diagonal line through a symbol indicates a deceased individual. Affected individuals are indicated by filled symbols. The pedigree charts are simplified for confidentiality reasons. All the numbered individuals in family 1 were examined by RP-PCR and AL-PCR. As shown in the pedigree chart, seven individuals had repeat expansion variations, whereas ten unaffected individuals, including three married-in individuals, did not have repeat expansion mutations.
(C) Multipoint parametric linkage analysis demonstrated a linkage interval with maximum logarithm of the odds (LOD) scores of 4.21 in chromosome 19.
(D) Genetic linkage analysis indicated maximum LOD scores of 4.21 in chromosome 19, a 29.18-Mb region at 19p13.2–19q13.12 (chr19: 8,841,079–38,022,358).
(E) Representative results of RP-PCR analysis showing GGC repeat expansions of individuals in family 1 (top and middle panels). In an unaffected individual from family 1, no GGC repeat expansions were detected (bottom panel). Experiments were conducted thrice with reproducible results.
(F) GGC repeat expansions in GIPC1 were observed in 12 out of the 24 Chinese OPDM-affected case subjects (8 familial and 16 sporadic case subjects). The repeat expansion mutations were also detected in 7 out of 194 Japanese individuals, but were not detected in any of the 1,000 control subjects.
(G) Representative results of AL-PCR analysis showing the numbers of expanded GGC repeats of individuals in family 1 (top and middle panels). In an unaffected individual from family 1, no GGC repeat expansions were detected (bottom panel).
(H) Frequency distribution of repeat units of GGC repeats of 550 control subjects in GIPC1, as revealed by fragment analysis.
Repeat-Primed Polymerase Chain Reaction (RP-PCR)
Genomic DNA from 8 families with affected individuals, 16 sporadic case subjects, and 1,000 unaffected control subjects were analyzed by repeat-primed PCR (RP-PCR). The PCR mix contained 0.25 U PrimeSTAR GXL DNA Polymerase, 1× PrimeSTAR GXL Buffer, 200 μM each dATP, dTTP, dCTP (Takara Bio, Shiga), and 7-deaza-dGTP (Sigma-Aldrich), 5% dimethyl sulfoxide (SigmaAldrich), 1M betaine (Sigma-Aldrich), 0.3 μM primer GIPC1-F and GIPC1-linker-R, 0.1 μM primer GIPC1-R, and 100 ng genomic DNA in a total reaction volume of 20 μL. The three primers were as follows: GIPC1-F: 5′-FAM-CAGACACATCCTTCTCGCAGAGGCCAC-3′; GIPC1-R: 5′-CAGGAAACAGCTATGACCGGCGGAGGCAGCGGCGGCGGC-3′; and GIPC1-linker-R: 5′-CAGGAAACAGCTATGACC-3′.
After incubation at 98°C for 10 min, the cycling conditions were followed by nine cycles of 98°C for 30 s, 66°C for 15 s with 1°C reduction per cycle and 72°C for 4 min, followed by 30 cycles of 98°C for 30 s, 58°C for 15 s and 72°C for 4 min, followed by a final elongation step of 72°C for 10 min. Electrophoresis was performed on a 3500xl Genetic analyzer (Thermo Fisher Scientific) and the data were analyzed using GeneMapper software (Thermo Fisher Scientific). A saw-tooth tail pattern in the electropherogram was considered to be the disease-associated repeat expansion.
Fluorescence Amplicon Length Analysis Polymerase Chain Reaction (AL-PCR)
The two primers were GIPC1-AL-F: 5′-VIC-CACATCCTTCTCGCAGAGGCCAC-3′ and GIPC1-AL-R: 5′-GAAGACGCGGATTGGCTGCGAGC-3′. The PCR mix and the thermal conditions were the same as in the RP-PCR protocol. Electrophoresis was performed on a 3500xl Genetic analyzer (Thermo Fisher Scientific) and the data were analyzed using GeneMapper software (Thermo Fisher Scientific). The length of the highest signal peak of expanded allele was used to calculate the repeat number.
AL-PCR was also performed for the fragment analysis of repeat sizes in controls with small modification. Alternatively, after incubation at 98°C for 10 min, the cycling conditions were followed by nine cycles of 98°C for 30 s, 66°C for 15 s with 1°C reduction per cycle, and 72°C for 30 s, followed by 30 cycles of 98°C for 30 s, 58°C for 15 s, and 72°C for 30 s, followed by a final elongation step of 72°C for 2 min.
Nanopore 5mC Methylation Modification Calling
We called 5mC methylation using the minimap229 and Nanopolish.30 The methylation level around the GGC repeats was compared between affected individuals and healthy individuals, and between expanded and non-expanded alleles using the Wilcoxon Rank Sum Test.
RNA-Seq and Bioinformatic Analysis
RNA sequencing was carried out in collaboration with Oebiotech. Total RNA was extracted using the mirVana miRNA Isolation Kit (Ambion) following the manufacturer’s protocol. RNA integrity was evaluated using the Agilent 2100 Bioanalyzer (Agilent Technologies). Samples with RNA Integrity Number (RIN) ≥ 7 were subjected to subsequent analysis. Libraries were constructed using TruSeq Stranded mRNA LTSample Prep Kit (Illumina) according to the manufacturer’s instructions. These libraries were sequenced on the Illumina sequencing platform (HiSeqTM 2500 or Illumina HiSeq X Ten) and 125 bp/150 bp paired-end reads were generated. Differentially expressed genes (DEGs) were identified using the DESeq R package31 functions estimateSizeFactors and nbinomTest. p value < 0.05 and foldchange > 2 or foldchange < 0.5 was set as the threshold for significantly differential expression. Hierarchical cluster analysis of DEGs was performed to explore gene expression patterns. GO enrichment and KEGG pathway enrichment analysis of DEGs were separately performed using R based on the hypergeometric distribution.32
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was isolated from skeletal muscle samples with TRizol reagent (Invitrogen). cDNA synthesis and qRT-PCR were performed as described.33 To detect GIPC1 mRNA levels in skeletal muscle from affected individuals with OPDM and control subjects, the following primers were used with GAPDH as a reference gene: GIPC1 forward: 5′-ACCAACGTCAAGGAGCTGTA-3′, GIPC1 reverse: 5′-CCACTTTGTGGGTGTTCAGG-3′; GAPDH forward: 5′-CTGGGCTACACTGAGCACC-3′, GAPDH reverse: 5′-AAGTGGTCGTTGAGGGCAATG-3′.
Western Blot Analysis
Skeletal muscle tissues from OPDM-affected individuals and control subjects were lysed with RIPA buffer (1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS [pH 7.4]) containing a cocktail of protease inhibitors (Roche). Lysates were analyzed by western blotting using the corresponding specific antibodies.
Statistical Analyses
Differences between two groups were analyzed using Student’s t test. The bar graphs with error bars represent mean ± standard deviation (SD).
Results
Identification of GGC Repeat Expansions in Individuals with OPDM
To determine the genetic cause of OPDM, we initially performed whole-genome sequencing (WGS) in OPDM-affected individuals, but we were unable to identify any likely causal variants. Then, we enrolled 5 individuals from 4 families with OPDM (Figure S1), 3 individuals with sporadic OPDM, and 100 healthy individuals and conducted long-read whole-genome sequencing (LRS) using ONT PromethION sequencing machines. The mean aligned coverage reached 18.6×. To better screen the disease-associated short tandem repeats (STR), we implemented a STR-Scoring method, which was a strategy to identify expanded repeats in the long reads based on comparisons between healthy and affected individuals (Figure 1). For the total 107,287 STRs that located in genic regions (including 10 Kb up-/downstream of genes), we obtained the Top10 candidates using the STR-Scoring method, of which the CCG/GGC repeats in the 5′ UTR of GIPC1 achieved a considerable higher score (4.45) compared to the other candidates (Figures 1 and S2, Table S1). To avoid potential drawbacks of the scoring framework which gave different weight factors to different genic regions and pattern, we re-evaluated the scores of those STRs without weight coefficients and the results confirmed that the CCG/GGC repeat in the 5′ UTR of the GIPC1 (Figure 2A) was still the most significant disease-causing locus with the highest score of 2.23 (Table S2).
Figure 1.
Detection of Expanded Repeats in the Long Reads Based on Comparisons between Healthy and Affected Individuals
The flow chart shows the schematic of the discovery pipeline for repeat expansion. Healthy individuals (green boxes) and affected individuals (red boxes) with OPDM were sequenced using ONT PromethION. The same pipeline is used to implement the alignment and STR detection, and then construct the ERC matrix. A method, STR-Scoring, was developed to prioritize the STR.
The repeat counts of the STR locus in one out of four families and three sporadic case subjects (F1-11, F1-12, S2, S3, S4) were more than 100, while repeat counts were between 13 and 32 in the healthy individuals (Figures 1 and S3). In addition, the population proportion of this repeat was zero in all GIPC1-affected individuals with OPDM (F1-11, F1-12, S2, S3, S4), which indicated that no healthy individual has a repeat count higher than that in the GIPC1-affected individuals with OPDM. Thus, these results suggested that GGC repeat expansions in the 5′ UTR of GIPC1 were associated with OPDM. In contrast, only one individual (F2-4) in this cohort had expanded GGC repeat units in LRP12 based on LRS.
To validate our LRS data, linkage analysis was performed in the six-generation Chinese OPDM-affected family (family 1) (Figure 2B). The results identified a 29.18-Mb candidate region at 19p13.2–19q13.12 (chr19: 8,841,079–38,022,358) with a maximum logarithm of odds (LOD) score of 4.21 (Figures 2C and 2D). Therefore, we confirmed that GIPC1 (chr19: 14,606,853–14,606,897) with GGC repeat expansion was located in this candidate region.
RP-PCR and AL-PCR Analysis of Repeat Expansions in GIPC1
RP-PCR analysis with primers targeting the GGC repeat expansions located in the 5′ UTR of GIPC1 was conducted to verify the segregation of GIPC1 variant in family 1 (Figures 2A and 2B). The GIPC1 repeat expansions were demonstrated by a saw-tooth pattern in seven affected family members (F1-8, F1-9, F1-10, F1-11, F1-12, F1-13, F1-14) but not in ten unaffected family members, indicating a family co-segregation in 17 individuals from family 1 (Figures 2B, 2E, and S4). Subsequently, GGC repeat expansions were further examined in the index individuals of 7 additional families (Figure S1), all 16 sporadic Chinese individuals, 194 unrelated Japanese individuals (both clinically and pathologically diagnosed and known gene excluded), as well as 1,000 unaffected Chinese control subjects. In total, 3 out of 8 families, 9 out of 16 sporadic Chinese individuals, and seven out of 194 unrelated Japanese individuals showed repeat expansions in the 5′ UTR of GIPC1, while none of the 1,000 unaffected Chinese control subjects showed repeat expansions (Figures 2F, S5, and S6).
We then determined the GGC repeat size using AL-PCR and found that all the affected individuals carried expanded GGC repeats, with the numbers of GGC repeat units ranging from 88 to 164 in our cohort and 73 to 161 in the Japanese cohort (Figures 2G and S4–S6). It should be noted that both RP-PCR and AL-PCR had failed to detect GGC repeat expansion in GIPC1 in an unaffected individual (F1-5), who had a GGC repeat expansion with more than 500 times in one allele identified by LRS (Figure S3), suggesting LRS was more reliable on detecting extremely long repeat expansion in human genome. The GGC repeat sizes in the GIPC1 gene ranged from 12 to 32 in 550 unaffected control subjects determined by fragment analysis (Figure 2H). Together, these results strongly indicated that GGC repeat expansions in the 5′ UTR of GIPC1 were associated with OPDM.
Clinical Characteristics of GIPC1-Affected Chinese Individuals with OPDM
Three familial probands and nine sporadic case subjects with OPDM associated with GGC repeat expansions in GIPC1 were clinically evaluated in this study (see Supplemental Note). Details of clinical features of GIPC1-affected OPDM familial probands and sporadic subjects are summarized in Table 1. The mean age of onset was 27.0 ± 7.2 years (ranging between 14 and 38 years). Disease duration ranged from 1 to 24 years. Initial symptoms included muscle weakness of distal limb in ten individuals, ptosis in one individual, and weakness in closing eyes in one individual. The full OPDM phenotype developed slowly in all individuals with disease progression, manifesting ptosis, ophthalmoparesis, facial and bulbar muscle weakness, and distal limb muscles predominantly affected.
Table 1.
The Clinical Features and GGC Repeat Size of 12 Chinese OPDM-Affected Individuals with GIPC1 Mutation
| Patient No. | Sex | Age of Onset | Disease Duration (year) | Initial Symptom |
Main Clinical Features |
EMG Pattern | Serum Creatine Kinasea(IU/L) | Rimmed Vacuoles on Muscle Pathology | GGC Repeat Size | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ptosis | External Ophthalmoplegia | Bulbar Muscle Weakness | Facial Muscle Weakness | Predominant Distal Limb Muscle Weakness | |||||||||
| F1-12 | F | 14 | 24 | weakness in closing eyes | + | + | + | + | + | MC | 849 | + | 117 |
| F7-1 | F | 31 | 5 | weakness in bilateral legs | + | + | + | + | + | MC | 277 | + | 115 |
| F8-1 | F | 38 | 7 | weakness in bilateral legs | + | - | + | + | + | MC | 358 | + | 108 |
| S1 | F | 30 | 19 | weakness in climbing up stairs | + | + | + | + | + | MC | 174 | + | 136 |
| S2 | M | 20 | 8 | weakness in the right leg | + | + | + | + | + | MC | 1,090 | + | 117 |
| S3 | F | 28 | 9 | bilateral ptosis | + | + | + | + | + | MC | 581 | + | 108 |
| S4 | F | 22 | 7 | weakness in climbing up stairs | + | + | + | + | + | MC | 790 | + | 120 |
| S5 | M | 33 | 8 | weakness in bilateral legs | + | + | + | + | + | MC | 2,524 | + | 124 |
| S6 | F | 24 | 11 | weakness in bilateral legs | + | + | + | + | + | MC & NC | 154 | + | 129 |
| S7 | M | 26 | 1 | weakness in bilateral legs | + | + | + | + | + | NA | >1,000 | + | 97 |
| S8 | M | 21 | 3 | weakness in bilateral legs | + | + | + | + | + | MC | 455 | + | 106 |
| S9 | M | 37 | 12 | weakness of four limbs | + | + | + | + | + | N/A | 345 | + | 103 |
Abbreviations: M, male; F, female; MC, myopathic change; NC, neurogenic change; N/A, not available.
Normal limits: 70–170 IU/L.
Laboratory data are shown in Table 1. Eleven case subjects (91.7%) of OPDM showed mildly to moderately elevated serum creatine kinase (CK) levels. Electromyography (EMG) revealed myogenic changes in ten individuals (83.3%) and neurogenic changes in one individual (8.3%). Muscle MRI examinations were done in seven GIPC1-affected individuals with OPDM. Muscle imaging showed mild to severe fatty replacement, and the distal muscles were much more involved than the proximal muscles (Figures 3A, 3B, and S7).
Figure 3.
Muscle MRI and Myopathological Changes of the GIPC1-Affected Individuals with OPDM
(A and B) Muscle MRI of individual S3 showed fatty infiltration of lower limb muscles, with the distal muscles (B, calf level) more severely affected than the proximal muscles (A, thigh level).
(C–F) Hematoxylin & eosin (H&E) (C and D) and modified Gomori trichrome (mGT) (E and F) staining of muscle sections from individuals S6 and S7, showing dystrophic change with variation in fiber size and endomysial fibrosis, and fibers with rimmed vacuoles (marked by arrow).
(G and H) P62 staining of muscle section from individual S6, inclusion bodies shown in both rimmed vacuole (RV) (marked by arrow) and nuclei (marked by arrowhead and higher magnification).
(I) Electron microscopy of muscle tissue from individual S7. RV filled with myeloid body and higher magnification showed cytoplasmic tubulofilamentous inclusion bodies (marked by arrow).
(J) Electron microscopy of muscle tissue from individual F1-12. Intranuclear inclusions contained filamentous aggregates in the nuclei of the muscle fiber (marked by arrowhead and shown at higher magnification).
All 12 GIPC1-affected individuals with OPDM underwent skeletal muscle biopsies. All individuals showed a myopathic pattern to various degrees characterized by fiber size variation, endomysial fibrosis, and rimmed vacuoles (RVs) on H&E and modified GT staining (Figures 3C–3F). RV is a kind of autophagy-related vacuole and is immunoreactive to anti-p62 antibody.34 To investigate p62-positive inclusions in skeletal muscle sections from individuals with OPDM, we performed immunohistochemistry (IHC) with anti-p62 antibody. The results showed that p62-positive inclusions were present not only in RVs (marked by arrows in Figure 3G) but also in nuclei (marked by arrowheads in Figures 3G and 3H). Electron microscopy revealed that RVs were filled with myeloid bodies and tubulofilamentous inclusions (Figure 3I) and intranuclear inclusions containing filamentous aggregates (Figure 3J).
Methylation Status of Expanded GGC Repeats and Expression of GIPC1 in Skeletal Muscle from OPDM
We examined the 5-methylcytosine (5mC) modification in the region (chr19: 14,606,353–14,607,397) including the GGC repeat and the adjacent CpG island, using the ONT PromethION sequencing data. The methylation level around the GGC repeats was low and no significant differences in methylation between affected individuals and healthy individuals were detected (p = 0.334) (Figures 4A and 4B). Moreover, 5mC modifications in both expanded and non-expanded alleles were examined, and no significant differences in methylation were detected (p = 0.509) while the methylation levels were significantly increased in the Alu sequences35 in all affected individuals as positive controls (Figures 4C, 4D, and S8). Protein abundance was also unaltered in the skeletal muscle samples from OPDM-affected individuals with GGC repeat expansions in GIPC1 compared with age-matched control subjects, confirmed by western blot using the antibody to GIPC1 protein that was previously validated (Figures 4E and 4F).36 However, qRT-PCR results showed that GIPC1 mRNA levels were significantly increased in skeletal muscle samples from GIPC1-affected individuals with OPDM compared with unaffected age-matched control subjects (Figure S9). These data suggested that the GGC repeat expansions in the 5′ UTR might affect the transcriptional but not translational level of GIPC1.
Figure 4.
Methylation and Expression at GIPC1 Locus
(A) Methylation status across the expanded GGC repeat region in GIPC1 from whole blood DNA was determined using LRS data from five affected individuals (F1-11, F1-12, S2, S3, S4) and 100 healthy individuals; no significant difference in methylation was detected between OPDM-affected individuals and control subjects.
(B) Quantification of methylation level between OPDM-affected individuals and control subjects.
(C) Methylation status between expanded and non-expanded alleles of GIPC1 from whole blood DNA was determined using LRS data from five affected individuals (F1-11, F1-12, S2, S3, S4), and no significant difference was detected between expanded and non-expanded alleles.
(D) Quantification of methylation level between expanded and non-expanded alleles.
All data were analyzed using Wilcoxon rank sum test in (A) and (C), or Student’s t test in (B) and (D).
(E) Western blot analysis of GIPC1 protein levels in five OPDM-affected individuals with expanded GGC repeats in GIPC1 and four age-matched control subjects. GAPDH was used as a loading control.
(F) Quantification of relative GIPC1 protein level in each group; no significant difference was observed between OPDM-affected individuals with expanded GGC repeats and controls. Data were analyzed using Student’s t test; ns, not significant.
GIPC1 Distribution and RNA-Seq Profiling in Skeletal Muscle from OPDM
To investigate the distribution of GIPC1 protein in muscle fibers derived from OPDM-affected individuals with GGC repeat expansions in the 5′ UTR of GIPC1, immunofluorescence was carried out in two GIPC1-affected individuals (S6, S7) and an age-matched control. The results showed that GIPC1 protein was frequently distributed in the cytoplasm in skeletal muscle from both control and GIPC1-affected individuals with OPDM (Figure 5A). GIPC1 protein was partially co-localized with p62 in the RVs (marked by arrows in Figure 5A), as well as in the intranuclear inclusions (marked by arrowheads in Figure 5A), in the skeletal muscle fibers derived from GIPC1-affected individuals with OPDM. These results suggested that loss of function of GIPC1 might affect the pathogenesis of OPDM.
Figure 5.
GIPC1 Distribution in Skeletal Muscle and RNA-Seq Profiling
(A) Immunofluorescence staining reveals the distribution of GIPC1 in muscle fibers. GIPC1 partially co-localized with: p62-positive inclusions in rimmed vacuole marked with arrows; and p62-positive intranuclear inclusions marked with arrowheads.
(B) Heatmap showing hierarchical clustering of the differentially expressed mRNAs, with annotated genes in the p53 signaling pathway, vascular smooth muscle contraction, ubiquitin-mediated proteolysis, and ribosome.
To explore the molecules and pathways involved in the mechanisms of OPDM, the skeletal muscle biopsy samples showing myopathic changes with formation of RVs from three OPDM-affected subjects with GGC repeat expansions in GIPC1 (F7-1, S5, S7) and five age-matched control subjects (C1-C5) were used in RNA-sequencing. The genome-wide mRNA expression profiles in muscle biopsies were detected, and dysregulated genes between the two groups are listed in Table S3. As shown in the volcano plot and heatmap, 791 genes were differentially expressed, with 184 downregulated and 607 upregulated (Figure S10). The significant Gene Ontology (GO) items of the whole differential mRNAs indicated that translation initiation, actin nucleation, and sarcomere organization were relevant to OPDM (Figure S11). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses revealed that differentially expressed genes most significantly corresponded to components of vascular smooth muscle contraction, ubiquitin mediated proteolysis, and ribosome that were downregulated, while components of the p53 signaling pathway were upregulated (Figures S12A and S12B), shown in the annotated heatmap (Figure 5B).
Discussion
Oculopharyngodistal myopathy (OPDM) is an adult-onset hereditary muscle disease, characterized by progressive ptosis, external ophthalmoplegia, facial muscle weakness, distal limb muscle weakness and atrophy, and bulbar involvements, resulting in dysphagia and dysarthria, with autosomal-dominant or autosomal-recessive inheritance.2,4,6 Muscle biopsy and histochemistry examination show myopathic changes of different severity with formation of rimmed vacuoles (RVs).7 The genetic mutations causing OPDM are not yet fully understood, although a recent study indicated that about one fourth of OPDM-affected individuals carried CGG repeat expansions in LRP12 in a cohort of Japanese individuals.15 Here, using a comprehensive strategy combining LRS, linkage analysis, RP-PCR, and AL-PCR, we identified abnormal GGC repeat expansions in the 5′ UTR of GIPC1 (chr19: 14,606,853-14,606,897) as a genetic cause of OPDM, representing about 50% of incidence in a cohort of 24 familial and sporadic OPDM-affected Chinese individuals. However, 3.61% of OPDM-affected Japanese individuals carried the GGC repeat expansions in GIPC1, suggesting distinct genetic causes of OPDM in different populations or different ethnic groups.
The identification of causative repeat expansions has usually been accomplished by classic genetic linkage analysis.37 In this study, we developed an STR-Scoring method based on long-read whole-genome sequencing (LRS) to directly detect repeat expansions from LRS data and discovered the causative mutations through the scoring procedure as indicated in Figure 1. Importantly, the results of STR-Scoring were validated by linkage analysis and RP-PCR. Since our STR-Scoring procedure does not require classic linkage analysis, it can be applied to families with limited members available, as well as sporadic individuals without family histories. This method has proven to be promising for the discovery of repeat expansions in the short tandem repeats (STR) with 3–6 bp repeats in the untranslated region of the human genome that could be pathogenic. The search for repeat expansions is expected to further expand our knowledge about the genetic architecture of a wide range of human diseases.
The inheritance is variable in different STR expansion-related diseases. For example, Baratela-Scott syndrome (BSS [MIM: 300881]) has autosomal-recessive inheritance38 while LRP12-affected OPDM has autosomal-dominant inheritance15 and oculopharyngeal muscular dystrophy (OPMD [MIM: 164300]) is either an autosomal-dominant or autosomal-recessive pattern determining its phenotypic variation.16 Despite OPDM exhibiting variable inheritance patterns in different studies, all the affected individuals with GGC repeat expansions in GIPC1 were heterozygous in the present study. This indicated the inherited mode associated with GIPC1 was in an autosomal-dominant pattern. Whether there is genetically definite recessive OPDM awaits further investigation.
Diseases caused by STR usually show genetic instability and anticipation with variable severity and incomplete penetrance, such as fragile X-associated tremor/ataxia syndrome (FXTAS [MIM: 300623]) with CGG repeats in FMR1 (MIM: 309550).39 Among the GIPC1-affected individuals with OPDM in our study, genetic anticipation was not observed in family 1, and a correlation between the repeat size and the onset age was not found with available clinical data. Therefore, anticipation was not clearly seen in GIPC1-affected individuals with OPDM. Surprisingly, our LRS data indicated that an asymptomatic individual (F1-5) carried more than 500 GGC repeats in the 5′ UTR of GIPC1 while his offspring were both symptomatic and carriers with 117 repeats (F1-12) and 113 repeats (F1-13), respectively. This highly suggested an autosomal-dominant inheritance with incomplete penetrance in this GIPC1-affected OPDM family, although the exploration of the underlying mechanism is very challenging. Since GGC repeat sizes in all GIPC1-affected OPDM individuals ranged from 73 to 164 in this study, it is possible that the GGC repeat sizes in the 5′ UTR of GIPC1 might have a certain range to cause OPDM. For example, a range of 55 to 200 CGG repeats in the 5′ UTR of FMR1 causes FXTAS while more than 200 CGG repeats leads to fragile X syndrome (FXS [MIM: 300624]) with different pathogenic mechanisms.40 We speculated that more than 500 GGC repeats in the 5′ UTR of GIPC1 might not to be causative for OPDM, which needs to be further investigated in more OPDM-affected families. In addition, different forms of trinucleotide repeats might also have different contributions to the pathogenic mechanism, because we observed not only GGC repeats, but also different repeat units, such as GCA from our LRS data (Figure S2). Unfortunately, we were unable to compare the correlation between different repeat interruptions and the severity of each affected individual in this cohort, considering the high error rate of LRS in single nucleotide reading. Thus, more accurate methods are required for further analyses.
GGC repeat expansions in the 5′ UTR frequently result in excessive methylation and decreased gene expression, such as FXS or BSS.38,41 However, our results showed that the methylation around the GGC repeats was not significantly different between GIPC1-affected individuals with OPDM and unaffected individuals. Additionally, the protein levels of GIPC1 were unaltered in GIPC1-affected individuals with OPDM, though the GIPC1 mRNA levels were increased. It was reported that FMR1 mRNA was elevated in FXTAS, suggesting that FXTAS might represent a toxic RNA gain-of-function effect.40,42 Thus, it is likely that elevated GIPC1 mRNA levels may result in RNA toxicity with the transcribed expanded GGC repeats.
GIPC1, GIPC PDZ domain containing family member 1, was first found to interact specifically with the C terminus of RGS-GAIP to regulate vesicular trafficking.43 A recent study indicated that a GIPC1 mutant displayed angiogenesis defects in zebrafish model, suggesting the role of GIPC1 in vascular development.36 We found that GIPC1 protein partially co-localized with p62 in the aggregates both in RVs and nucleus, suggesting that loss-of-function of GIPC1 might affect the pathogenesis of OPDM. RNA-seq data indicated that genes in the p53 signaling pathway were upregulated, while genes in vascular smooth muscle contraction were downregulated. It is reported that downregulated GIPC1 resulted in increased p53 expression,44 suggesting that p53 signaling pathway activation in GIPC1-affected individuals with OPDM might result from the loss-of-function of GIPC1. Due to GIPC1 functioning in vascular development,36 it is possible that downregulation of components in vascular smooth muscle contraction could be relevant to loss of function of GIPC1. Together, GIPC1 protein partially localized in the aggregates might affect the pathogenesis of OPDM.
Whole-genome mRNA expression profiling in GIPC1-affected individuals with OPDM indicated downregulation of components in ubiquitin-mediated proteolysis and ribosome, which could be a pathogenic mechanism in this disease. It is possible that inhibition of the ubiquitin-mediated proteolysis pathway results in protein aggregates in RVs and the nucleus. Interestingly, a number of components of the ribosome were downregulated in GIPC1-affected individuals with OPDM, which was also reported in individuals with frontotemporal dementia (FTD [MIM: 600274]) and amyotrophic lateral sclerosis (ALS [MIM: 612069]) with the C9orf72 (MIM: 614260) mutation.45 The (GGGGCC)n repeats in C9ORF72 are translated into aggregating dipeptide-repeat proteins in FTD/ALS,46 which could reduce ribosome levels.45 It is possible that GGC repeat expansions in the 5′ UTR of GIPC1 could be translated into toxic peptides, like the aggregating dipeptide-repeat proteins from the C9ORF72 mutation, which could similarly reduce ribosome levels.
Identification of the GGC repeat expansions as a genetic cause of OPDM will help to elucidate the molecular pathogenesis in this disease. The diseases FXTAS, NIID, OPMD, and OPDM are caused by repeat expansions and have some overlaps in clinical presentations. It is possible that abnormal translations of expanded GGC repeats are a common mechanism in these diseases, regardless of the genes in which the expanded repeats are located. Although there are two distinct genes, GIPC1 and LRP12, that contribute to the pathogenesis of OPDM, the transcribed expanded GGC repeats RNA or translated toxic peptides via repeat-associated non-ATG (RAN) translation47, 48, 49, 50 could be the common pathogenic mechanisms for OPDM with GGC repeat expansions either in GIPC1 or LRP12. However, the hypothesis still needs to be investigated in cellular and animal models.
In conclusion, our study identified the expansion of GGC repeats in GIPC1 as the most frequent genetic cause in Chinese OPDM-affected individuals, and indicated molecules and pathways involved in the mechanisms of OPDM. Our findings will benefit the clinical diagnosis and the understanding of the molecular pathogenesis of OPDM in the future.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We appreciated the cooperation of the individuals and their families. We are grateful to Dr. Nicolas Charlet-Berguerand for helpful suggestions. We thank Dr. Wanjin Chen for the help on linkage analysis. The work was supported by the National Natural Science Foundation of China (No. 81571219 to Z.W.), Double thousand talents program of Jiangxi province (to D.H.), Intramural Research Grant (29-4 to I.N.; 30-9 to A.I.) for Neurological and Psychiatric Disorders of NCNP, and AMED under grant number JP19ek0109285h0003 and Joint Usage and Joint Research Programs, the Institute of Advanced Medical Sciences, Tokushima University (2019, A9 to A.I.).
Published: May 14, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.04.011.
Contributor Information
Daojun Hong, Email: hongdaojun@hotmail.com.
Zhaoxia Wang, Email: drwangzx@163.com.
Web Resources
ANNOVAR, http://www.openbioinformatics.org/annovar/annovar_region.html
GENCODE V19, https://www.gencodegenes.org/human/release_19.html
Genomestudio (v.2011.1), https://support.illumina.com/array/array_software/genomestudio/downloads.html/
Integrative Genomics Viewer, https://software.broadinstitute.org/software/igv/
MERLIN (v.1.1.2), http://csg.sph.umich.edu/abecasis/Merlin/download/
OMIM, https://www.omim.org/
RepeatHMM, https://github.com/WGLab/RepeatHMM
UCSC Genome Browser, https://genome.ucsc.edu
Supplemental Data
References
- 1.Durmus H., Laval S.H., Deymeer F., Parman Y., Kiyan E., Gokyigiti M., Ertekin C., Ercan I., Solakoglu S., Karcagi V. Oculopharyngodistal myopathy is a distinct entity: clinical and genetic features of 47 patients. Neurology. 2011;76:227–235. doi: 10.1212/WNL.0b013e318207b043. [DOI] [PubMed] [Google Scholar]
- 2.Satoyoshi E., Kinoshita M. Oculopharyngodistal myopathy. Arch. Neurol. 1977;34:89–92. doi: 10.1001/archneur.1977.00500140043007. [DOI] [PubMed] [Google Scholar]
- 3.Lu H., Luan X., Yuan Y., Dong M., Sun W., Yan C. The clinical and myopathological features of oculopharyngodistal myopathy in a Chinese family. Neuropathology. 2008;28:599–603. doi: 10.1111/j.1440-1789.2008.00924.x. [DOI] [PubMed] [Google Scholar]
- 4.Minami N., Ikezoe K., Kuroda H., Nakabayashi H., Satoyoshi E., Nonaka I. Oculopharyngodistal myopathy is genetically heterogeneous and most cases are distinct from oculopharyngeal muscular dystrophy. Neuromuscul. Disord. 2001;11:699–702. doi: 10.1016/s0960-8966(01)00227-9. [DOI] [PubMed] [Google Scholar]
- 5.Mignarri A., Carluccio M.A., Malandrini A., Sicurelli F., Galli L., Mazzei M.A., Federico A., Orrico A., Dotti M.T. The first Italian patient with oculopharyngodistal myopathy: case report and considerations on differential diagnosis. Neuromuscul. Disord. 2012;22:759–762. doi: 10.1016/j.nmd.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 6.Uyama E., Uchino M., Chateau D., Tomé F.M.S. Autosomal recessive oculopharyngodistal myopathy in light of distal myopathy with rimmed vacuoles and oculopharyngeal muscular dystrophy. Neuromuscul. Disord. 1998;8:119–125. doi: 10.1016/s0960-8966(98)00002-9. [DOI] [PubMed] [Google Scholar]
- 7.van der Sluijs B.M., ter Laak H.J., Scheffer H., van der Maarel S.M., van Engelen B.G.M. Autosomal recessive oculopharyngodistal myopathy: a distinct phenotypical, histological, and genetic entity. J. Neurol. Neurosurg. Psychiatry. 2004;75:1499–1501. doi: 10.1136/jnnp.2003.025072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amato A.A., Jackson C.E., Ridings L.W., Barohn R.J. Childhood-onset oculopharyngodistal myopathy with chronic intestinal pseudo-obstruction. Muscle Nerve. 1995;18:842–847. doi: 10.1002/mus.880180807. [DOI] [PubMed] [Google Scholar]
- 9.Lu X.H., Pu C.Q., Huang X.S., Liu J.X., Mao Y.L. Clinical, pathological and molecular study of two Chinese families with oculopharyngodistal myopathy. Chin. J. Neurol. 2012;45:557–560. [Google Scholar]
- 10.Wagner M., Laval S., Mueller J., Hacer D., Oflazer P., Lochmuller H. P88 Using whole exome sequencing to identify the mutation causing oculopharyngodistal myopathy. Neuromuscul. Disord. 2012;22:S31–S32. [Google Scholar]
- 11.Zhao J., Liu J., Xiao J., Du J., Que C., Shi X., Liang W., Sun W., Zhang W., Lv H. Clinical and muscle imaging findings in 14 mainland chinese patients with oculopharyngodistal myopathy. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0128629. e0128629–e0128629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sone J., Mitsuhashi S., Fujita A., Mizuguchi T., Hamanaka K., Mori K., Koike H., Hashiguchi A., Takashima H., Sugiyama H. Long-read sequencing identifies GGC repeat expansions in NOTCH2NLC associated with neuronal intranuclear inclusion disease. Nat. Genet. 2019;51:1215–1221. doi: 10.1038/s41588-019-0459-y. [DOI] [PubMed] [Google Scholar]
- 13.Deng J., Gu M., Miao Y., Yao S., Zhu M., Fang P., Yu X., Li P., Su Y., Huang J. Long-read sequencing identified repeat expansions in the 5'UTR of the NOTCH2NLC gene from Chinese patients with neuronal intranuclear inclusion disease. J. Med. Genet. 2019;56:758–764. doi: 10.1136/jmedgenet-2019-106268. [DOI] [PubMed] [Google Scholar]
- 14.Tian Y., Wang J.-L., Huang W., Zeng S., Jiao B., Liu Z., Chen Z., Li Y., Wang Y., Min H.-X. Expansion of Human-Specific GGC Repeat in Neuronal Intranuclear Inclusion Disease-Related Disorders. Am. J. Hum. Genet. 2019;105:166–176. doi: 10.1016/j.ajhg.2019.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishiura H., Shibata S., Yoshimura J., Suzuki Y., Qu W., Doi K., Almansour M.A., Kikuchi J.K., Taira M., Mitsui J. Noncoding CGG repeat expansions in neuronal intranuclear inclusion disease, oculopharyngodistal myopathy and an overlapping disease. Nat. Genet. 2019;51:1222–1232. doi: 10.1038/s41588-019-0458-z. [DOI] [PubMed] [Google Scholar]
- 16.Brais B., Bouchard J.-P., Xie Y.-G., Rochefort D.L., Chrétien N., Tomé F.M.S., Lafrenière R.G., Rommens J.M., Uyama E., Nohira O. Short GCG expansions in the PABP2 gene cause oculopharyngeal muscular dystrophy. Nat. Genet. 1998;18:164–167. doi: 10.1038/ng0298-164. [DOI] [PubMed] [Google Scholar]
- 17.Brook J.D., McCurrach M.E., Harley H.G., Buckler A.J., Church D., Aburatani H., Hunter K., Stanton V.P., Thirion J.-P., Hudson T. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 18.Deng J., Wang P., Chen X., Cheng H., Liu J., Fushimi K., Zhu L., Wu J.Y. FUS interacts with ATP synthase beta subunit and induces mitochondrial unfolded protein response in cellular and animal models. Proc. Natl. Acad. Sci. USA. 2018;115:E9678–E9686. doi: 10.1073/pnas.1806655115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng J., Yang M., Chen Y., Chen X., Liu J., Sun S., Cheng H., Li Y., Bigio E.H., Mesulam M. FUS Interacts with HSP60 to Promote Mitochondrial Damage. PLoS Genet. 2015;11:e1005357. doi: 10.1371/journal.pgen.1005357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandes F., Freitas A.T. slaMEM: efficient retrieval of maximal exact matches using a sampled LCP array. Bioinformatics. 2014;30:464–471. doi: 10.1093/bioinformatics/btt706. [DOI] [PubMed] [Google Scholar]
- 21.Tsai E.A., Shakbatyan R., Evans J., Rossetti P., Graham C., Sharma H., Lin C.-F., Lebo M.S. Bioinformatics Workflow for Clinical Whole Genome Sequencing at Partners HealthCare Personalized Medicine. J. Pers. Med. 2016;6:12. doi: 10.3390/jpm6010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haeussler M., Zweig A.S., Tyner C., Speir M.L., Rosenbloom K.R., Raney B.J., Lee C.M., Lee B.T., Hinrichs A.S., Gonzalez J.N. The UCSC Genome Browser database: 2019 update. Nucleic Acids Res. 2019;47(D1):D853–D858. doi: 10.1093/nar/gky1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sedlazeck F.J., Rescheneder P., Smolka M., Fang H., Nattestad M., von Haeseler A., Schatz M.C. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods. 2018;15:461–468. doi: 10.1038/s41592-018-0001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Q., Zhang P., Wang D., Gu W., Wang K. Interrogating the “unsequenceable” genomic trinucleotide repeat disorders by long-read sequencing. Genome Med. 2017;9:65. doi: 10.1186/s13073-017-0456-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitsuhashi S., Frith M.C., Mizuguchi T., Miyatake S., Toyota T., Adachi H., Oma Y., Kino Y., Mitsuhashi H., Matsumoto N. Tandem-genotypes: robust detection of tandem repeat expansions from long DNA reads. Genome Biol. 2019;20:58. doi: 10.1186/s13059-019-1667-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang H., Kirkness E.F., Lippert C., Biggs W.H., Fabani M., Guzman E., Ramakrishnan S., Lavrenko V., Kakaradov B., Hou C. Profiling of Short-Tandem-Repeat Disease Alleles in 12,632 Human Whole Genomes. Am. J. Hum. Genet. 2017;101:700–715. doi: 10.1016/j.ajhg.2017.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li M., Mei L., He C., Chen H., Cai X., Liu Y., Tian R., Tian Q., Song J., Jiang L. Extrusion pump ABCC1 was first linked with nonsyndromic hearing loss in humans by stepwise genetic analysis. Genet. Med. 2019;21:2744–2754. doi: 10.1038/s41436-019-0594-y. [DOI] [PubMed] [Google Scholar]
- 28.Chourabi M., Liew M.S., Lim S., H’mida-Ben Brahim D., Boussofara L., Dai L., Wong P.M., Foo J.N., Sriha B., Robinson K.S. ENPP1 Mutation Causes Recessive Cole Disease by Altering Melanogenesis. J. Invest. Dermatol. 2018;138:291–300. doi: 10.1016/j.jid.2017.08.045. [DOI] [PubMed] [Google Scholar]
- 29.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simpson J.T., Workman R.E., Zuzarte P.C., David M., Dursi L.J., Timp W. Detecting DNA cytosine methylation using nanopore sequencing. Nat. Methods. 2017;14:407–410. doi: 10.1038/nmeth.4184. [DOI] [PubMed] [Google Scholar]
- 31.Anders S., Huber W. Differential expression of RNA-Seq data at the gene level – the DESeq package. 2012. http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.359.7464&rep=rep1&type=pdf
- 32.Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., Katayama T., Kawashima S., Okuda S., Tokimatsu T., Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fiorese C.J., Schulz A.M., Lin Y.F., Rosin N., Pellegrino M.W., Haynes C.M. The Transcription Factor ATF5 Mediates a Mammalian Mitochondrial UPR. Curr. Biol. 2016;26:2037–2043. doi: 10.1016/j.cub.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vittonatto E., Boschi S., CHIADò-Piat L., Ponzalino V., Bortolani S., Brusa C., Rainero I., Ricci F., Vercelli L., Mongini T. Differential diagnosis of vacuolar muscle biopsies: use of p62, LC3 and LAMP2 immunohistochemistry. Acta Myol. 2017;36:191–198. [PMC free article] [PubMed] [Google Scholar]
- 35.Kochanek S., Renz D., Doerfler W. DNA methylation in the Alu sequences of diploid and haploid primary human cells. EMBO J. 1993;12:1141–1151. doi: 10.1002/j.1460-2075.1993.tb05755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carretero-Ortega J., Chhangawala Z., Hunt S., Narvaez C., Menéndez-González J., Gay C.M., Zygmunt T., Li X., Torres-Vázquez J. GIPC proteins negatively modulate Plexind1 signaling during vascular development. eLife. 2019;8:e30454. doi: 10.7554/eLife.30454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pulst S.M. Genetic linkage analysis. Arch. Neurol. 1999;56:667–672. doi: 10.1001/archneur.56.6.667. [DOI] [PubMed] [Google Scholar]
- 38.LaCroix A.J., Stabley D., Sahraoui R., Adam M.P., Mehaffey M., Kernan K., Myers C.T., Fagerstrom C., Anadiotis G., Akkari Y.M., University of Washington Center for Mendelian Genomics GGC Repeat Expansion and Exon 1 Methylation of XYLT1 Is a Common Pathogenic Variant in Baratela-Scott Syndrome. Am. J. Hum. Genet. 2019;104:35–44. doi: 10.1016/j.ajhg.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hagerman P. Fragile X-associated tremor/ataxia syndrome (FXTAS): pathology and mechanisms. Acta Neuropathol. 2013;126:1–19. doi: 10.1007/s00401-013-1138-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Glineburg M.R., Todd P.K., Charlet-Berguerand N., Sellier C. Repeat-associated non-AUG (RAN) translation and other molecular mechanisms in Fragile X Tremor Ataxia Syndrome. Brain Res. 2018;1693(Pt A):43–54. doi: 10.1016/j.brainres.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kong H.E., Zhao J., Xu S., Jin P., Jin Y. Fragile X-Associated Tremor/Ataxia Syndrome: From Molecular Pathogenesis to Development of Therapeutics. Front. Cell. Neurosci. 2017;11:128. doi: 10.3389/fncel.2017.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oostra B.A., Willemsen R. FMR1: a gene with three faces. Biochim. Biophys. Acta. 2009;1790:467–477. doi: 10.1016/j.bbagen.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Vries L., Lou X., Zhao G., Zheng B., Farquhar M.G. GIPC, a PDZ domain containing protein, interacts specifically with the C terminus of RGS-GAIP. Proc. Natl. Acad. Sci. USA. 1998;95:12340–12345. doi: 10.1073/pnas.95.21.12340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang L., Lau J.S., Patra C.R., Cao Y., Bhattacharya S., Dutta S., Nandy D., Wang E., Rupasinghe C.N., Vohra P. RGS-GAIP–Interacting Protein Controls Breast Cancer Progression . Mol. Cell Res. 2010;8:1591–1600. doi: 10.1158/1541-7786.MCR-10-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hartmann H., Hornburg D., Czuppa M., Bader J., Michaelsen M., Farny D., Arzberger T., Mann M., Meissner F., Edbauer D. Proteomics and C9orf72 neuropathology identify ribosomes as poly-GR/PR interactors driving toxicity. Life Sci Alliance. 2018;1:e201800070. doi: 10.26508/lsa.201800070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mori K., Weng S.-M., Arzberger T., May S., Rentzsch K., Kremmer E., Schmid B., Kretzschmar H.A., Cruts M., Van Broeckhoven C. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 47.Zu T., Gibbens B., Doty N.S., Gomes-Pereira M., Huguet A., Stone M.D., Margolis J., Peterson M., Markowski T.W., Ingram M.A.C. Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl. Acad. Sci. USA. 2011;108:260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pinto B.S., Saxena T., Oliveira R., Méndez-Gómez H.R., Cleary J.D., Denes L.T., McConnell O., Arboleda J., Xia G., Swanson M.S., Wang E.T. Impeding Transcription of Expanded Microsatellite Repeats by Deactivated Cas9. Mol. Cell. 2017;68:479–490.e5. doi: 10.1016/j.molcel.2017.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mizielinska S., Grönke S., Niccoli T., Ridler C.E., Clayton E.L., Devoy A., Moens T., Norona F.E., Woollacott I.O.C., Pietrzyk J. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science. 2014;345:1192–1194. doi: 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sellier C., Buijsen R.A.M., He F., Natla S., Jung L., Tropel P., Gaucherot A., Jacobs H., Meziane H., Vincent A. Translation of Expanded CGG Repeats into FMRpolyG Is Pathogenic and May Contribute to Fragile X Tremor Ataxia Syndrome. Neuron. 2017;93:331–347. doi: 10.1016/j.neuron.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.