Abstract
Mycobacterium avium subsp. paratuberculosis (MAP) is the causative agent of Johne’s disease (JD), a chronic infectious disease that causes intractable diarrhea in ruminants. To control the occurrence of JD in cattle, a national surveillance is conducted in Japan. Since 2013, real-time quantitative PCR has been used for definite diagnosis. In this study, we compared the amount of fecal MAP DNA with histopathological classification of ileocecal lesions. Multinomial logistic regression models enabled us to predict the probability of finding the histopathological classification from the amount of fecal MAP DNA. These results suggest that shedding level of MAP DNA could act as an indicator of JD progression.
Keywords: Mycobacterium avium subsp. paratuberculosis, pathological classification, quantitative real-time PCR
Mycobacterium avium subsp. paratuberculosis (MAP) is causative agent of Johne’s disease (JD), a chronic granulomatous inflammation of the distal small intestine and mesenteric lymph node in ruminants. After a long incubation period, this disease leads to emaciation, therapy-resistant diarrhea, and eventually death [5]. Based on the severity of the clinical signs, fecal shedding organisms, and the ease with which the disease may be detected using clinical examinations, bovine JD is divided into four stages: silent, subclinical, clinical, and advanced clinical disease [24]. As fecal shedding of MAP is intermittent in the subclinical stage [13], periodic survey is necessary for controlling JD.
In Japan, the first case of JD was reported in an imported dairy cow that died in 1927 [22]. As JD became endemic across the country after 1980s, national serological surveillance targeting MAP infection in cattle started in 1998 as a compulsory program enforced by law [26]. Since 2013, quantitative real-time PCR (qPCR) has been used for definite diagnosis and it enables the quantification of MAP shedding.
In the past two decades, various studies have been undertaken to identify robust indicators of JD progression. For example, González et al. compared histopathological findings and fecal culture results of JD [7]. Brady et al. investigated the relationships among the clinical signs, the extent and character of pathological changes, and the tissue distribution of MAP in infected cattle [2]. Furthermore, Kawaji et al. compared fecal MAP DNA amount and histopathological status [9], and de Silva et al. reported elevation in fecal MAP DNA was associated with progressing JD in sheep examination [6]. As the shedding level of MAP has been reported to correlate with histopathological findings, we investigated the possibility of using the amount of fecal MAP DNA to predict the histopathological classification of cattle using multinomial logistic regression models. Our principal objective was to determine the turning point of fecal MAP DNA amount in JD progression.
From October 2012 to July 2017, 153 cattle (102 Holstein Friesian cattle and 51 Japanese Black cattle) diagnosed with JD in Hokkaido prefecture were examined. No cattle showed profuse diarrhea except for three cattle. For definite diagnosis, Johne-spin (FASMAC, Atsugi, Japan) was used for DNA extraction from feces and the MAP IS900 gene was quantified by using Johne’s Gene Test Kit “KS” (Kyoritsu Seiyaku Co., Tokyo, Japan) and LightCycler® 480 Real-Time PCR System (Roche, Switzerland) according to manufacturer’s protocol. The kit protocol is based on the method described Kawaji et al. [10] with slight modification. In Japan, cattle detected over 1.00 × 10−3pg/well of MAP DNA are diagnosed as JD.
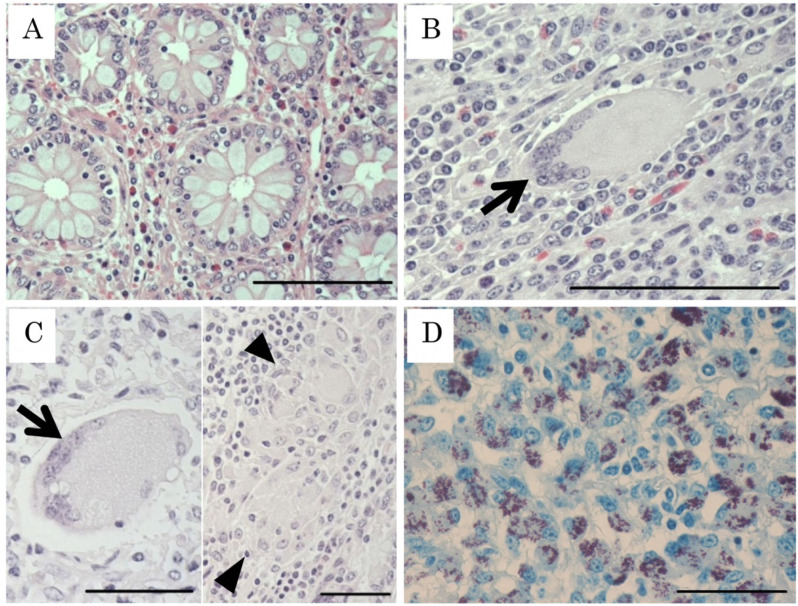
The diagnosed cattle were culled within 2 weeks and ileocecal samples were collected for histopathological analysis. Three cattle were less than 1.00 × 10−3pg/well of MAP DNA, however, farmers agreed with culling to prevent the spread of JD. Ileocecal valves were fixed with 10% neutral buffered formalin and embedded in paraffin. The lymph nodes were not treated because the intestinal tract is considered to be more directly related to MAP excretion than accessory lymph nodes. Therefore, only the ileocecal sample, which is one of the most important tissues for pathological diagnosis [7], was investigated in this study for the sake of simplicity. Sectioned specimens were stained with hematoxylin and eosin (HE), and by the Ziehl-Neelsen (ZN) method for acid-fast bacteria. In this study, we classified the JD lesion into four categories (Fig. 1), non-lesional (N) type, tuberculoid (T) type, lepromatous (L) type, and mixed (T/L) type. T type and L type were classified based on results of previous studies [3, 11]. Briefly, T type lesions were resistant to the MAP infection, including lymphocyte and macrophage infiltration in the apex of villus and the formation of Langhans giant cells. No or a few acid-fast bacilli are present in the cytoplasm of giant cells as observed by ZN staining. No epithelioid cells were observed. L type lesions were non-resistant to MAP infection. Epithelioid cells containing MAP infiltrated the mucosal walls of the ileum and accumulated predominantly in the lamina propria and submucosa, resulting in severe thickening of the mucosa. Few multinucleated giant cells were observed. T/L type lesions harbored combined characteristics of T and L type. Multinucleated giant cells, accumulation of epithelioid cells, and high abundance of MAP was observed. In N type lesions, the structure of intestinal villus was normal and no histopathological characteristic of JD was observed.
Fig. 1.
Histopathological classification of Johne’s disease lesions. (A) Non-lesional type (N type). Normal structure of crypta is observed. Hematoxylin Eosin (HE); Bar=100 µm. (B) Tuberculoid type (T type). Lymphocyte and macrophage infiltration in the apex of the villus and the formation of multinucleated giant cell (arrow) are observed. HE; Bar=100 µm. (C) Mixed type (T/L type). A multinucleated giant cell and accumulation of epithelioid cells (arrow head) are observed. HE; Bar=50 µm. (D) Lepromatous type (L type). Epithelioid cells containing Mycobacterium avium subsp. paratuberculosis (MAP) infiltrate in the ileal submucosa and accumulate predominantly between the lamina propria and muscular layer. Ziehl-Neelsen staining; Bar=50 µm.
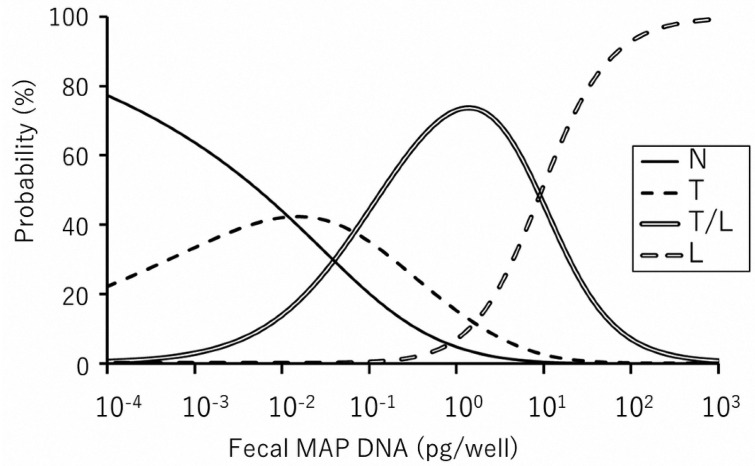
The examined 153 fecal and ileum samples were classified as shown in Table 1 (the results of individual cattle are shown in Supplementary Table). Among pathological classifications, the amount of MAP DNA ranged from 7.06 × 10−4 to 1.01 × 10−1 (median 6.83 × 10−3) pg/well in N type, 3.65 × 10−4 to 6.43 × 10−1 (median 1.98 × 10−2) pg/well in T type, 1.64 × 10−3 to 8.22 × 101 (median 3.60 × 10−1) pg/well in T/L type, and 2.72 × 10−0 to 1.99 × 103 (median 2.97 × 101) pg/well in L type. Our previous study [17] indicated that 1.0 pg DNA/well was equivalent to 6.3 × 103 CFU/g of feces by sedimentation culture method [18] using commercially available Herrold’s egg yolk medium. Multinomial logistic regression models predicting each disease classification associated with shedding level were developed using the vglm function in the VGAM package of the statistical software R [14, 23]. P-values less than 0.05 were considered statistically significant. Multinomial logistic regression models were generated to predict the disease classification using the amount of MAP DNA as a predictor variable. The results are shown in Fig. 2. This prediction model can be applied to predict the histopathological status using the amount of MAP DNA in fecal samples.
Table 1. The amounts of fecal Mycobacterium avium subsp. paratuberculosis (MAP) DNA and histopathological classification of 153 cattle (Holstein Friesian cattle and Japanese Black cattle).
| Fecal MAP DNA (pg/well) |
Histopathological classification |
Total | |||
|---|---|---|---|---|---|
| N type | T type | T/L type | L type | ||
| <10−2 | 24 (13, 11) | 17 (11, 6) | 6 (6,0) | 47 (30, 17) | |
| 10−2–10−1 | 17 (9, 8) | 18 (13, 5) | 4 (3, 1) | 39 (25, 14) | |
| 10−1–10° | 1 (1, 0) | 7 (5, 2) | 16 (15, 1) | 24 (21, 3) | |
| 10°–101 | 12 (10, 2) | 6 (4, 2) | 18 (14, 4) | ||
| 101–102 | 5 (3, 2) | 12 (6, 6) | 17 (9, 8) | ||
| ≥102 | 8 (3, 5) | 8 (3, 5) | |||
| Total | 42 (23, 19) | 42 (29, 13) | 43 (37, 6) | 26 (13, 13) | 153 (102, 51) |
N type: non-lesional type, T type: tuberculoid type, L type: lepromatous type, T/L type: mixed type.
Fig. 2.
Multivariate regression model comparing each disease classification and the amount of Mycobacterium avium subsp. paratuberculosis (MAP) DNA. The graph shows the probability of predicting histopathological lesion types at each amount of fecal MAP DNA. For example, in the presence of 1.0 × 10−3pg/well MAP DNA, the following can be predicted: 64% probability to find non-lesional type (N type), 33% probability to find tuberculoid type (T type), and less than 3% probability to find mixed type (T/L type) and lepromatous type (L type). Also, 1.0 × 102pg/well MAP DNA can lead to following prediction: 93% probability to find L type, 7% probability to find T/L type, and less than 1% probability to find N and T types.
In the past two decades, various methods to quantify MAP DNA have been developed [4] and several histopathological reports on JD progression have been published [2, 3, 7]. In agreement with previous reports, our results suggested that the severity of lesions is accompanied by a shift from N type to L type, and likewise, the fecal MAP amount is also increased during this type of shift. During MAP infection, MAP is taken up by macrophages and dendritic cells present in the subepithelial lamina propria. Interestingly, as apoptosis of infected macrophages is suppressed, MAP continues replication inside the macrophage during the natural lifespan (21–42 days) [8, 11]. The burst cells release MAP into the local environment after which the bacteria enter the intestinal lumen and are shed in the feces or remain localized and start a new cycle [11]. Both in macrophages as well as dendritic cells, infection with live MAP leads to an upregulation of the production of the suppressive cytokine, IL-10, and an arrest in mononuclear phagocyte maturation [11]. Rather than directly originating from lesions, it is most likely that early events of MAP-specific adaptive immunity are associated with uptake of MAP by pro-inflammatory dendritic cells or macrophages activated through unrelated events and migrating towards secondary lymphoid organs at the time of encountering MAP or MAP antigens [11]. The stage before activating adaptive immunity is histopathologically considered to be N type, and our study indicated median shedding level of MAP DNA to be 6.83 × 10−3pg/well (estimated 43 CFU/g of feces).
When MAP is taken up and processed by activated macrophages, scar tissue, characterized by multinucleated giant cells, is formed. In early stage of JD, Th1-type associated cellular responses were predominant [25]. This stage is histopathologically considered to be T-type and the shedding level of MAP DNA is median 1.98 × 10−2pg /well (estimated 125 CFU/g of feces). Cell-mediated immunity was first detected 9 weeks after oral infection of goats with MAP while antibodies were detected at later on after infection (15 to 20 weeks) [20]. The progression to clinical disease is defined as a shift from potentially protective cell mediated immune response to non-protective antibody response [19, 21]. This stage is histopathologically considered as T/L type (fecal MAP DNA is median 3.60 × 10−1pg/well, estimated 2.3 × 103 CFU/g of feces), which, eventually, becomes L type (fecal MAP DNA is median 2.97 × 101pg/well, estimated 1.8 × 105 CFU/g of feces). The immunological switch has been attributed to infection load, T cell exhaustion, and several genetic triggers, such as hormonal changes during the periparturient period due to stress [11]. Recently, Sajiki et al. reported that prostaglandin E2 induction suppresses the Th1 immune responses in cattle with JD [15]. As prostaglandin E2 level is higher during late pregnancy [1], calving is considered to be a major risk for JD progression.
Magombedze et al. demonstrated an association between the ELISA value of a MAP-specific antibody and JD progression; however; compared with the amount of MAP shedding, the expression pattern of the Th2 response changed little [12]. Likewise, our results indicated a weak correlation between ELISA value and JD progression (Individual ELISA values are shown in Supplementary Table). Kawaji et al. identified an association between lesion progression and fecal MAP shedding in a study of sheep [9]. Although their histopathological classification was different from ours, the MAP DNA was found to be similar in the sheep. The difference between no lesion and primary lesion was markedly lower than that between the primary lesion and progressed lesion [9]. As described above, MAP-specific adaptive immunity is considered to occur via events that are unrelated to MAP [11]. This accidental immune activation may be the reason for the low peak in the probability calibration used for predicting the T type in Fig. 2. In a long-term study of cattle, Magombedze et al. defined a clinical stratum of cows that were shedding more than 100 CFU/g of feces and presented with weight loss and intermittent diarrhea; the highest level of fecal MAP shedding in their corresponding subclinical cattle group was 20 CFU/g [12]. Our previous study showed that the shedding level of MAP-infected cattle associated with a 90% probability of detection from environmental samples was 78 CFU/g of feces [17]. In this study, the likelihood of forming the T/L type, which is considered to represent a shift from potentially protective immune response to non-protective response, was increased from approximately 1.0 × 10−2pg/well, estimated 63 CFU/g of feces. This means that most of the cattle exhibiting the T/L type are considered to be an environmental contamination source. According the aforementioned research, cattle that shed more than 100 CFU/g may develop JD, and thus their early culling is critical for JD control. During JD progression, cattle that shed more than 1.0 pg/well DNA (estimated 6.3 × 103 CFU/g of feces) exhibit reduced productivity with respect to lactation yield and calving interval [16]. Our data show the likelihood of cattle forming L type lesions was increased from approximately 1.0 pg/well DNA. Therefore, cattle that form L type lesions, even if they exhibit no remarkable symptoms, tend to exhibit reduced productivity.
In conclusion, we revealed the association between the histopathological classification and the amount of fecal MAP DNA. Our results showed no significant differences between observations of Holstein Friesian and Japanese Black cattle. As MAP growth is associated with exhaustion of immune-cells and lesion formation, histopathological classification can be predicted from the amount of fecal MAP DNA. Our data suggest cattle shedding more than 1.0 × 10−2pg/well MAP DNA in feces may forming T/L type lesions and will progress JD in the future.
Supplementary
REFERENCES
- 1.Berisha B., Schams D., Rodler D., Sinowatz F., Pfaffl M. W.2018. Changes in the expression of prostaglandin family members in bovine corpus luteum during the estrous cycle and pregnancy. Mol. Reprod. Dev. 85: 622–634. doi: 10.1002/mrd.22999 [DOI] [PubMed] [Google Scholar]
- 2.Brady C., O’Grady D., O’Meara F., Egan J., Bassett H.2008. Relationships between clinical signs, pathological changes and tissue distribution of Mycobacterium avium subspecies paratuberculosis in 21 cows from herds affected by Johne’s disease. Vet. Rec. 162: 147–152. doi: 10.1136/vr.162.5.147 [DOI] [PubMed] [Google Scholar]
- 3.Buergelt C. D., Hall C., McEntee K., Duncan J. R.1978. Pathological evaluation of paratuberculosis in naturally infected cattle. Vet. Pathol. 15: 196–207. doi: 10.1177/030098587801500206 [DOI] [PubMed] [Google Scholar]
- 4.Chaubey K. K., Gupta R. D., Gupta S., Singh S. V., Bhatia A. K., Jayaraman S., Kumar N., Goel A., Rathore A. S., Sahzad, Sohal J. S., Stephen B. J., Singh M., Goyal M., Dhama K., Derakhshandeh A.2016. Trends and advances in the diagnosis and control of paratuberculosis in domestic livestock. Vet. Q. 36: 203–227. doi: 10.1080/01652176.2016.1196508 [DOI] [PubMed] [Google Scholar]
- 5.Chiodini R. J., Van Kruiningen H. J., Merkal R. S.1984. Ruminant paratuberculosis (Johne’s disease): the current status and future prospects. Cornell Vet. 74: 218–262. [PubMed] [Google Scholar]
- 6.de Silva K., Begg D. J., Plain K. M., Purdie A. C., Kawaji S., Dhand N. K., Whittington R. J.2013. Can early host responses to mycobacterial infection predict eventual disease outcomes? Prev. Vet. Med. 112: 203–212. doi: 10.1016/j.prevetmed.2013.08.006 [DOI] [PubMed] [Google Scholar]
- 7.González J., Geijo M. V., García-Pariente C., Verna A., Corpa J. M., Reyes L. E., Ferreras M. C., Juste R. A., García Marín J. F., Pérez V.2005. Histopathological classification of lesions associated with natural paratuberculosis infection in cattle. J. Comp. Pathol. 133: 184–196. doi: 10.1016/j.jcpa.2005.04.007 [DOI] [PubMed] [Google Scholar]
- 8.Kabara E., Coussens P. M.2012. Infection of primary bovine macrophages with Mycobacterium avium subspecies paratuberculosis suppresses host cell apoptosis. Front. Microbiol. 3: 215. doi: 10.3389/fmicb.2012.00215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawaji S., Begg D. J., Plain K. M., Whittington R. J.2011. A longitudinal study to evaluate the diagnostic potential of a direct faecal quantitative PCR test for Johne’s disease in sheep. Vet. Microbiol. 148: 35–44. doi: 10.1016/j.vetmic.2010.07.022 [DOI] [PubMed] [Google Scholar]
- 10.Kawaji S., Taylor D. L., Mori Y., Whittington R. J.2007. Detection of Mycobacterium avium subsp. paratuberculosis in ovine faeces by direct quantitative PCR has similar or greater sensitivity compared to radiometric culture. Vet. Microbiol. 125: 36–48. doi: 10.1016/j.vetmic.2007.05.002 [DOI] [PubMed] [Google Scholar]
- 11.Koets A. P., Eda S., Sreevatsan S.2015. The within host dynamics of Mycobacterium avium ssp. paratuberculosis infection in cattle: where time and place matter. Vet. Res. (Faisalabad) 46: 61. doi: 10.1186/s13567-015-0185-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magombedze G., Shiri T., Eda S., Stabel J. R.2017. Inferring biomarkers for Mycobacterium avium subsp. paratuberculosis infection and disease progression in cattle using experimental data. Sci. Rep. 7: 44765. doi: 10.1038/srep44765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mortier R. A. R., Barkema H. W., Orsel K., Wolf R., De Buck J.2014. Shedding patterns of dairy calves experimentally infected with Mycobacterium avium subspecies paratuberculosis. Vet. Res. (Faisalabad) 45: 71. doi: 10.1186/s13567-014-0071-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.R Core Team. 2016. A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. [Google Scholar]
- 15.Sajiki Y., Konnai S., Okagawa T., Nishimori A., Maekawa N., Goto S., Ikebuchi R., Nagata R., Kawaji S., Kagawa Y., Yamada S., Kato Y., Nakajima C., Suzuki Y., Murata S., Mori Y., Ohashi K.2018. Prostaglandin E2 induction suppresses the Th1 immune responses in cattle with Johne’s disease. Infect. Immun. 86: e00910–e00917. doi: 10.1128/IAI.00910-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakakibara S., Kanno H., Tachibana S.2016. Evaluation of the production losses in daily and beef herd with paratuberculosis infection. Jpn. J. Anim. Hyg. 42: 173–180. [Google Scholar]
- 17.Sakakibara S., Kanno H., Tachibana S.2017. The detection of the Mycobacterium avium subspecies paratuberculosis in environmental samples as a screening test to determine a herd’s paratuberculosis status. Nippon Juishikai Zasshi 70: 511–515. [Google Scholar]
- 18.Stabel J. R.1997. An improved method for cultivation of Mycobacterium paratuberculosis from bovine fecal samples and comparison to three other methods. J. Vet. Diagn. Invest. 9: 375–380. doi: 10.1177/104063879700900406 [DOI] [PubMed] [Google Scholar]
- 19.Stabel J. R.2000. Transitions in immune responses to Mycobacterium paratuberculosis. Vet. Microbiol. 77: 465–473. doi: 10.1016/S0378-1135(00)00331-X [DOI] [PubMed] [Google Scholar]
- 20.Storset A. K., Hasvold H. J., Valheim M., Brun-Hansen H., Berntsen G., Whist S. K., Djønne B., Press C. M., Holstad G., Larsen H. J. S.2001. Subclinical paratuberculosis in goats following experimental infection. An immunological and microbiological study. Vet. Immunol. Immunopathol. 80: 271–287. doi: 10.1016/S0165-2427(01)00294-X [DOI] [PubMed] [Google Scholar]
- 21.Sweeney R. W., Jones D. E., Habecker P., Scott P.1998. Interferon-gamma and interleukin 4 gene expression in cows infected with Mycobacterium paratuberculosis. Am. J. Vet. Res. 59: 842–847. [PubMed] [Google Scholar]
- 22.Takehara K.1930. Occurrence of bovine paratuberculosis. Chuo Juikai Zasshi 43: 311–320 [in Japanese]. [Google Scholar]
- 23.Thomas W. Y., Wild C. J.1996. Vector generalized additive models. J. R. Stat. Soc. B 58: 481–493. [Google Scholar]
- 24.Whitlock R. H., Buergelt C.1996. Preclinical and clinical manifestations of paratuberculosis (including pathology). Vet. Clin. North Am. Food Anim. Pract. 12: 345–356. doi: 10.1016/S0749-0720(15)30410-2 [DOI] [PubMed] [Google Scholar]
- 25.Wu C. W., Glasner J., Collins M., Naser S., Talaat A. M.2006. Whole-genome plasticity among Mycobacterium avium subspecies: insights from comparative genomic hybridizations. J. Bacteriol. 188: 711–723. doi: 10.1128/JB.188.2.711-723.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamamoto T., Murai K., Hayama Y., Kobayashi S., Nagata R., Kawaji S., Osaki M., Sakakibara S. I., Tsutsui T.2018. Evaluation of fecal shedding and antibody response in dairy cattle infected with paratuberculosis using national surveillance data in Japan. Prev. Vet. Med. 149: 38–46. doi: 10.1016/j.prevetmed.2017.10.009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.