Abstract
Background:
Few studies have analyzed progressive demethylation in the path to cancer. This is of utmost importance, especially in populations such as African Americans, who display aggressive tumors at diagnosis, and for whom markers of early neoplastic transformation are needed. Here, we determined hypomethylated targets in the path to colorectal cancer (CRC) using Reduced Representation Bisulfite Sequencing (RRBS).
Methods:
DNA was extracted from fresh frozen tissues of patients with different colon lesions (normal, tubular adenoma, tubulovillous adenoma, and five cancers). RRBS was performed on these DNA extracts to identify hypomethylated gene targets. Alignment, mapping, and methylation analyses were performed. Pathways affected by the hypomethylated gene targets were determined using Ingenuity Pathway Analysis (IPA).
Results:
Pairwise analyses of samples led to the identification of the following novel hypomethylated genes: ELMO3 (Engulfment and cell motility 3), SLC6A2 (Solute carrier family 6 member 2), SYNM (Synemin), and HMX2 (Homeobox 2). The ELMO3 promoter was significantly hypomethylated at five CpG sites, SYNM at two CpG sites, SLC6A2 at one CpG site, and the HMX2 gene at one CpG site. IPA placed these genes within important carcinogenic pathways.
Conclusions:
This work provides insight into the role of hypomethylation in colon carcinogenesis in African Americans. The identified targets affected many important pathways, as demonstrated through IPA. These targets might serve as biomarkers for early diagnosis and potential targets for therapy.
Keywords: African Americans, colorectal cancer, DNA methylation, hypomethylation, RRBS
Introduction
Colorectal cancer (CRC) is the most common cancer in the United States,1 and is a major cause of cancer-related death.2 Different mechanisms and pathways associate with different clinical and pathological outcomes. African Americans (AAs) have a high CRC incidence,1,3,4 for which there are multiple reasons (environmental, socioeconomic, genetic, and epigenetic).1,5 Aberrant DNA methylation is an epigenetic factor that is involved in virtually every step of CRC neoplastic transformation.6 Within promotor regions, short stretches of CpG rich regions (CpG islands) are primary targets of methylation.7 In the path to cancer, there is a modest depletion of cytosine methylation but considerable aberrant methylation.8 This aberrant promoter methylation generally associates with lower gene expression or complete silencing of tumor suppressor and caretaker genes.8
Global hypomethylation has also been recognized as a cancer hallmark that associates with oncogenic transformation.3,9,10 Hypomethylation occurs commonly in highly and moderately repeated DNA sequences including heterochromatic DNA repeats, dispersed retrotransposons, and endogenous retroviral elements. Indeed, hypomethylation may directly affect karyotypic stability, and might initiate altered heterochromatic-euchromatic interactions favoring oncogenesis.11 Genome-wide DNA hypomethylation has been associated with genomic instability, which may confer poor prognosis.12
Long interspersed element (LINE-1) methylation levels have been used to measure global DNA methylation where activation of LINE-1 retrotransposons may lead to the transcription of adjacent genes, gene disruptions, chromosomal instability, or the generation of transcripts that regulate gene expression.13 Global methylation analysis revealed gradual hypomethylation of LINE sequences in the path to cancer, with less methylation in LINE sequences in cancer compared with adenoma samples, and in adenoma samples compared with normal colonic mucosa.3,13
Most CRCs develop from adenomatous polyps, while some develop through the serrated pathway.14 Aberrant methylation plays an important role in the malignant transformation of such polyps.15 However, the cumulative carcinogenic effect of parallel hypomethylation events in this process is still unknown.
In this research study, we investigated DNA hypomethylated targets in normal, adenoma, and cancer specimens from African Americans using Reduced Representation Bisulfite Sequencing (RRBS).
Materials and methods
Patients and tissue samples
The samples were obtained from AA patients who underwent colonoscopy or surgery for the removal of colonic lesions at Howard University Hospital (HUH) (Table 1). All samples were analyzed by an expert gastrointestinal pathologist of the HUH Department of Pathology and kept frozen until used for DNA extraction. The samples consisted of a blood sample, a normal colon tissue, a tubular adenoma, a tubullovillous adenoma and five cancers. Clinical, demographic, and pathology data were collected for all specimens (Table 1). Informed written consent was obtained from all participants. This research was approved by Howard University Institutional Review Board (IRB 06-MED-39).
Table 1.
Demographical characteristics of the analyzed samples.
| Sample type | Gender | Age | Location |
|---|---|---|---|
| Normal blood | Male | 60 | na |
| Normal colon tissue | Male | 60 | Right |
| Tubular adenoma | Male | 59 | Left |
| Tubulovillous adenoma | Female | 76 | Right |
| Tumor | Male | 51 | Left |
| Tumor | Female | 69 | Right |
| Tumor | Female | 68 | Right |
| Tumor | Male | 55 | Left |
| Tumor | Female | 71 | Left |
na, not applicable.
DNA extraction
Genomic DNA was extracted from fresh frozen tissues. The samples were processed as previously described.16 Briefly, samples were homogenized in lysis buffer consisting of 100 mM Tris-HCl (pH 8.5), 5 mM EDTA, 0.2% SDS, 200 mM NaCl. Proteinase K was freshly added at a final concentration of 300 μg/ml. Samples were incubated overnight at 55°C to ensure that genomic DNA was completely dissociated from any DNA binding proteins. After digestion, genomic DNA was extracted using a Qiagen’s genomic DNA extraction kit, according to the manufacturer’s instructions (AllPrep DNA/RNA mini kit, Qiagen, Valencia, CA, USA). DNA quality and quantity were assessed using a NanoDrop spectrophotometer and 0.8% agarose gel electrophoresis.
EpiQuest library construction
EpiQuest libraries were prepared according to manufacturer’s protocol [Zymo Research (ZR), Irvine, CA, USA] and as previously described.16 Genomic DNA (200–500 ng) was digested with 60 units of TaqI and 30 units of MspI (NEB, Ipswich, MA, USA) sequentially. Size-selected TaqI-MspI fragments (40–120 bp and 120–350 bp) were filled-in and 3′-terminal-A extended, extracted with a ZR DNA Clean and Concentrator™-5 kit. Ligation to pre-annealed adapters containing 5′-methyl-cytosine was performed using the Illumina’s DNA preparation kit and protocol (Illumina, San Diego, CA, USA). Purified adaptor ligated fragments were bisulfite-treated using the EZ DNA Methylation-Direct™ Kit (ZR). Preparative-scale PCR was performed. DNA Clean and Concentrator™ -purified PCR products were subjected to a final size selection on a 4% NuSieve 3:1 agarose gel. SYBR-green-stained gel slices containing adaptor-ligated fragments of 130–210 bp or 210–460 bp in size was excised. Library material was recovered from the gel (Zymoclean™ Gel DNA Recovery Kit, ZR) and sequence on an Illumina HiSeq Genome Analyzer.
EpiQuest sequence alignments and data analysis
Sequence reads from bisulfite-treated EpiQuest libraries were identified using standard Illumina base-calling software and then analyzed using a Zymo Research proprietary analysis pipeline according to manufacture recommendations (Zymo research) and as previously described.16 Residual cytosines (Cs) in each read were first converted to thymines (Ts), with each such conversion noted for subsequent analysis. A reference sequence database was constructed from the 50-bp ends of each computationally predicted MspI-TaqI fragment in the 40–350 bp size range. All Cs in each fragment end then converted to Ts; converted reads were aligned to the converted reference by Bowtie. The number of mismatches in the induced alignment were then counted between the unconverted read and reference, ignoring cases in which a T in the unconverted read matched to a C in the unconverted reference. For a given read, the best alignment was kept if the second best alignment had two more mismatches; otherwise the read was discarded as nonunique. The methylation level of each sampled cytosine was estimated as the number of reads reporting a C, divided by the total number of reads reporting a C or T. Fisher’s exact test or t-test was performed for each CpG site that has at least five reads covered. Also, promoter, gene body, and CpG island annotations were added for each CpG. The software pipeline was implemented in Python.
Bisulfite conversion and multiplex amplification
Samples were subjected to sodium bisulfite treatment using the EZ DNA Methylation-Direct™ Kit. Targeted amplification was done via Fluidigm 48, the 48 Access Array using Zymo Research’s targeted sequencing service protocol. Sample loading, harvesting, and pooling were performed according to manufacturer’s protocol (Fluidigm). Methylation profiling data from AA patients were compared with a normal candidate peripheral blood specimen as well as a normal colorectal tissue. A pairwise DNA methylation analysis was performed between colorectal neoplasia samples such as tubular adenoma, tubulovillous, and tumor versus normal tissue samples (blood and normal colon).
Barcoding/adapterization PCR
Amplicon pools for each sample was diluted 1:100 and then amplified using barcoded adaptor-linkers received from Fluidigm according to manufacturer’s protocols. Reactions were cleaned up using DNA Clean and Concentrator™-5, and the products were normalized by concentration and pooled. Sequencing, alignment, and data analysis libraries were denatured, diluted, and processed for sequencing on the Illumina MiSeq according to manufacturer’s protocols. The sequencing run was a 150-base paired-end run. Sequence reads were aligned and analyzed as described above.
Pathway analysis
Further analysis was performed using the Ingenuity Pathway Analysis (IPA) software. The IPA ‘Upstream Regulator Analysis’ predicts upstream regulators by combining the directional methylation changes from our RRBS targeted methylation-sequencing, and knowledge from prior experimental reports on causal effects between molecules such as TSG (tumor suppressor gene) and oncogenes, compiled in the IPA Knowledge Base. Upstream Regulator Analysis calculates a z-score based on the edge of dysregulation of all the downstream molecules and the uniformity of the existing evidence about the upstream–downstream relation, for every upstream regulator known to have a causal effect on at least four (activating and inhibiting up/down) dysregulated genes/transcripts. Z-scores of <0 and >0 respectively indicate a significant inhibition and activation state of the upstream regulator, regardless of the actual expression level of these molecules (http://pages.ingenuity.com/rs/ingenuity/images/0812%20upstream_regulator_analysis_whitepaper.pdf). The Network Generation Algorithm links molecules based on experimentally observed interactions, and orders based on their interconnectedness. In general, the more interactions with other network members, the more central a molecule will be in a network.
Results
RRBS data analysis revealed methylation target genes
RRBS sequencing data was analyzed in a pairwise manner to determine differentially hypermethylated and hypomethylated CpG sites (this study) with relevance to colorectal neoplastic transformation.16 Our results showed decreasing DNA methylation from normal to cancer, with an average methylation rate range from 0.22 (ELMO3) to 0.001 (SYNM) (Table 2). The RRBS analysis led to a list of genes that were hypomethylated in tubular adenoma, tubulovillous adenoma, and colorectal neoplasia verses normal samples (Table 2 and Figure 1). Novel hypomethylated genes are ELMO3, SYNM, SLC6A2, and HMX2. All hypomethylated genes (83 genes) were analyzed with regards to their relevance to CRC, cancer in general, other diseases, and known cancer pathways.
Table 2.
Novel identified hypomethylated genes in colon neoplasia progression.
| Gene | Transcript | Spearman correlation | p value (Spearman correlation) | Adjusted p value (false detection rate) | Normal | Adenoma tubular | Adenoma tubulovillous | Cancer |
|---|---|---|---|---|---|---|---|---|
| ELMO3 | NM_024712 | –0.8610 | 1.03E–012 | 1.43E–008 | 0.2220 | 0.1580 | 0.0000 | 0.0000 |
| SYNM | NM_145728; NM_015286 | –0.2899 | 6.30E–008 | 8.70E–004 | 0.0126 | 0.0138 | 0.0048 | 0.0008 |
| SLC6A2 | NM_001172504 | –0.2963 | 4.44E–007 | 0.0061 | 0.0566 | 0.0366 | 0.0743 | 0.0109 |
| HMX2 | NM_005519 | –0.2183 | 9.20E–007 | 0.0127 | 0.0087 | 0.0231 | 0.0771 | 0.0032 |
Figure 1.
Location of hypomethylated CpG sites within the identified genes’ promoters: (a) ELMO3, (b) SYNM, (c) SLC6A2, (d) HMX2.
ELMO3 gene
The RRBS results for the ELMO3 (NM_024712) gene, located on chromosome 16, showed that the promoter was significantly hypomethylated at five CpG sites in adenoma.17 None of these CpG sites were methylated in tubulovillous adenoma and cancer samples compared with normal (Tables 2 and 3). The protein encoded by this gene is similar to a Caenorhabditis elegans protein that functions in phagocytosis of apoptotic cells and in cell migration. Other members of this small family of ELMO proteins have been shown to interact with the dedicator of cytokinesis 1 protein to promote phagocytosis and affect cell shape changes.
Table 3.
Number of hypomethylated CpG sites per identified gene.
| Genes | Normal | Tubular adenoma | Tubulovillous adenoma | Tumor |
|---|---|---|---|---|
| ELMO3 | 0 | 0 | 0 | 5 |
| SYNM | 0 | 0 | 0 | 2 |
| SLC6A2 | 0 | 0 | 0 | 1 |
| HMX2 | 0 | 0 | 0 | 1 |
SYNM gene
The promoter of the SYNM gene (NM_015286), located on chromosome 15, was significantly hypomethylated at two CpG sites in tumor samples. None of these CpG sites were found to be hypomethylated in normal tissues (Table 3). The protein encoded by this gene is an intermediate filament (IF) family member. IF proteins are cytoskeletal proteins that confer resistance to mechanical stress, and are encoded by a dispersed multigene family. This protein has been found to form a linkage between desmin, which is a subunit of the IF network, and the extracellular matrix, and provides an important structural support in muscle. Two alternatively spliced variants encoding different isoforms have been described for this gene.
SLC6A2 gene
For SLC6A2 gene (NM_001172501), located on chromosome 16, we found that one CpG site was significantly hypomethylated in tumor samples when compared with normal tissue (Table 3). This gene encodes a member of the sodium:neurotransmitter symporter family. This member is a multipass membrane protein responsible for the re-uptake of norepinephrine into presynaptic nerve terminals, and is a regulator of norepinephrine homeostasis. Mutations in this gene cause orthostatic intolerance, a syndrome characterized by lightheadedness, fatigue, altered mentation, and syncope. Alternatively, spliced transcript variants encoding different isoforms have been identified in this gene.
HMX2 gene
The promoter of the HMX2 gene (NM_005519), located on chromosome 10, was hypomethylated at one CpG site in tumor samples when compared with normal samples (Table 3). The protein encoded by this gene is a member of the NKL homeobox family of transcription factors. Members in this family play an important role in organ development during embryogenesis. A related mouse protein plays a role in patterning of inner ear structures. In humans, variations in a region containing this gene have been associated with inner ear malformations, vestibular dysfunction, and hearing loss.
Ingenuity pathway analysis
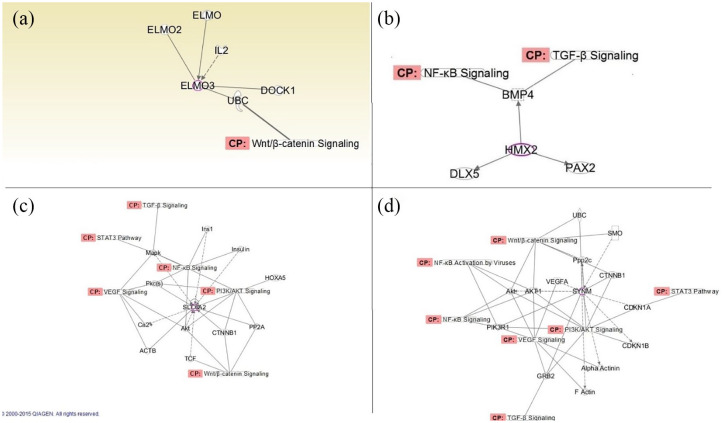
The hypomethylated markers were analyzed for their potential biological function(s) using IPA software. ELMO3 was mapped to the Wnt/β catenin pathway, HMX2 was mapped to the NF-kβ and TGF-β pathways, whereas SYNM and SLC6A2 were mapped to the Wnt/β, TGF-β, P13K/AKT, VEGF, NF-kβ, and STAT3 signaling pathways (Table 4 and Figure 2).
Table 4.
Genes’ mapping to known molecular pathways.
| Pathways | Hypomethylated genes |
|---|---|
| Wnt/β catenin | ELMO3, SYNM, SLC6A2 |
| TGF-β | SYNM, SLC6A2, HMX2 |
| VEGF | SYNM, SLC6A2 |
| NF-k β | SYNM, SLC6A2, HMX2 |
| JAK/STAT3 | SYNM, SLC6A2 |
| P13k/AKT | SYNM, SLC6A2 |
Figure 2.
IPA of identified hypomethylated targets. (a) ELMO3, (b) SYNM, (c) SLC6A2, and (d) HMX2.
CP, conical pathway; IPA, ingenuity pathways analysis.
Pathway analysis of hypomethylated genes
Upstream and downstream regulator analysis identified MYOD, REB1, ZF5, and PAX5 as crosstalk regulators to SYNM, SLC6A2 and HMX2, respectively (Figure 1). Both endogenous and exogenous molecules are included in this analysis. All other regulator types can be found in Table 4, and Figure 2. Significantly dysregulated molecules were sorted by their function or involvement in pathophysiological processes.
Network of dysregulated molecules
The following functions are likely affected by the demethylation of the four identified genes (Figure 2):
Apoptosis response: ELMO3, SYNM, SLC6A2, and HMX2 are part of apoptosis response since they interact with the Wnt-β catenin, TGF-β, NF-k β, and VEGF pathways.
Proliferation response: SYNM, SLC6A2, and HMX2 are part of proliferation response since they interact with the TGF-β pathway.
Inflammatory response: SYNM, SLC6A2, and HMX2 are part of inflammatory response because they interact with the NF-k β pathway.
Discussion
Reduced representation bisulfite sequencing (RRBS) was used to establish genome-wide DNA methylation alterations. This technique’s single-base resolution has already been used on the same set of samples to establish hypermethylated targets in the path to cancer.16 Here, we re-examined the same data with an emphasis on hypomethylated gene targets and identified specific CpG sites in ELMO3, SYNM, SLC6A2, and HMX2 as primarily novel unmethylated targets in cancer specimens but not in other colonic lesions, these genes were also found to be implicated in many important pathways relevant to neoplastic transformation. In the TCGA database, three of these genes were neither hypo nor hypermethylated while HMX2 gene was reported as hypermethylated. It is worth noting that most specimens in the TCGA database are from Caucasian patients. As such, this might account for the discrepancy since all our samples are from AA patients
In mammals, the ELMO family consists of three proteins, ELMO1, ELMO2, and ELMO3. The ELMO1 protein has been shown to play important roles in cytoskeleton rearrangements during phagocytosis and cellular migration. However, while ELMO2 and ELMO3 are expected to possess similar functions as ELMO1, no transcriptional regulation information is currently available for ELMO3.18
ELMO3, as an oncogene, was found to be overexpressed in primary non-small cell lung cancer (NSCLC) in combination with its promoter hypomethylation; these cancer-specific events are associated with metastasis.19 ELMO3 was also found to play a role in the migration of epithelial cells from the crypt to the villus tip.20 IPA analysis mapped ELMO3 to Wnt/β catenin pathway, which plays a central role in colon carcinogenesis.
Our results showed that ELMO3 promoter is hypomethylated in adenoma but not in normal samples, which might be explained by the involvement of this gene in intestinal epithelial cell migration. To our knowledge, this is the first time that ELMO3 has been cited as a novel hypomethylated gene in CRC. The present study suggests that DNA methylation of CpG sites located −133 to −118 bp upstream from the transcription start site of ELMO3 may affect the expression of this gene. CpG −133 to −118 are located in the MYOD binding site (Figure 1). MYOD is a transcription factor of the basic helix-loop-helix family and the myogenic factors subfamily. It is involved in muscle differentiation, induces fibroblasts to differentiate into myoblasts, and is essential for repair of damaged tissue. The impact of the hypomethylated ELMO3 gene has been reported in lung cancer, and is consistent with our results that this gene may play an important role in colorectal carcinogenesis.
The synemin (SYNM) gene encode proteins in the IF family. These are cytoskeletal proteins that confer resistance to mechanical stress, and provide important structural support in muscle.21 The silencing of synemin results in a significant decrease in tumor proliferation and increased tumor apoptosis,22 whereas its hypomethylation is associated with aggressive forms of breast cancer.23 Our results revealed that demethylation of SYNM promoter was significant in adenoma to tumor samples. This is the first implication of SYNM as a novel hypomethylated gene in CRC. The present study suggests that DNA methylation of CpG sites located −184 to −169 bp upstream from the transcription start site of SYNM may affect the expression of this gene. CpG −184 to −169 are located in the REB1 binding site. REB1 is frequently found in yeast promoters, and is capable of remodeling chromatin domains and enabling activation or repression by other transcription factors in diverse pathways.24 However, the most likely function of its binding site in the controlling region of SYNM is to prevent the formation of nucleosomes over the upstream activation sequence.25 SYNM was mapped to the Wnt/β, TGF-β, P13K/AKT, VEGF, NF-kβ, and STAT3 signaling pathways. The impact of the hypomethylated SYNM gene has been reported in breast cancer, and is in line with our findings regarding its important role in colorectal carcinogenesis.
The solute carrier family 6, member 2 gene, SLC6A2, is a norepinephrine transporter gene located on chromosome 16q12.2.26 This gene shows altered expression in cancer tissue, particularly head and neck cancer, which makes it a good candidate along with other genes to be used as a marker in determining molecular signatures in tumor tissues.27 The present study suggests that DNA methylation of CpG sites located −105 to –94 bp upstream from the transcription start site of SLC6A2 may affect the expression of this gene. CpG −105 to −94 are located in the binding site to which ZF5 can bind. ZF5 encodes a zinc finger protein, which contains five C2H2-type zinc fingers showing homology with the zinc finger of the Kruppel family, and binds to two sites in the C-MYC promoter.28 Our results showed that the SLC6A2 promoter is hypomethylated in tumor cells compared with normal samples. SLC6A2 was mapped to the Wnt/β, TGF-β, P13K/AKT, VEGF, NF-kβ, and STAT3 signaling pathways. To our knowledge. this is the first implication of SLC6A2 as a hypomethylated gene in AA with CRC. The impact of the hypomethylated SLC6A2 gene has been reported in head and neck cancer, and is consistent with our findings that this gene may be involved in colorectal carcinogenesis.
The H6 family homeobox 2 gene, HMX2, is a highly conserved homeobox transcription factor that functions as tumor suppressor gene.29 HMX2 showed significant hypomethylation in breast cancer patients in Denmark; the population with the highest incidence of breast cancer.30 Our results showed that the HMX2 promoter was hypomethylated in tumor compared with normal samples. This is the first time that HMX2 has been cited as a novel hypomethylated gene in AA with CRC. HMX2 was mapped to the NF-kβ and TGF-β pathways. The impact of the hypomethylated HMX2 gene has been reported in breast cancer, and is consistent with our results that this gene may play an important role in colorectal carcinogenesis. The present study suggests that DNA methylation of CpG sites located −97 to −70 bp upstream from the transcription start site of HMX2 may affect the expression of this gene. CpG −97 to −70 are located in Pax5 binding site. Pax5 is a critical regulator of B-cell maturation, where it controls the differentiation, function and identity of B lymphocytes, it has also been associated with human B-cell tumors.31
We are aware of the limitations of this study, such as the small sample size; this is due primarily to the high cost associated with RRBS methodology. The novel hypomethylated targets identified need to be further validated in a large cohort of AAs. Their specific hypomethylation status in this population needs to be validated in a cohort containing patients of other ethnic/racial groups. Once validated, these genes and associated pathways might serve as novel therapeutic targets.
In conclusion, we have reported four new distinct CpG methylation targets in AAs with CRC. Our results reveal that most somatic cells are primed for global hypomethylation in regions of the genome that are typically hypomethylated in cancer cells. These markers were found to be involved in major carcinogenesis pathways and may be of a potential interest as CRC markers in AAs.
Acknowledgments
The authors would like to thank all patients who participated in this project.
Footnotes
Author contributions: HA, KW, and HB: Study design, data acquisition and analysis, and manuscript writing; AL, BS, and EL: Specimen identification, diagnosis, and data analysis; SV, AS, and SZ: Data acquisition and analysis.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and publication of this article: This project was supported (in part) by the National Institute on Minority Health and Health Disparities of the National Institutes of Health under Award Number G12MD007597.
Conflict of interest statement: The authors declare that there is no conflict of interest.
ORCID iD: Hassan Brim  https://orcid.org/0000-0002-3361-9243
https://orcid.org/0000-0002-3361-9243
Contributor Information
Hassan Ashktorab, Department of Pathology, and Medicine, Howard University, 2041 Georgia Avenue NW, Washington, DC 20059, USA.
Kareem Washington, Department of Genetics, Howard University Hospital, Washington, DC, USA.
Shatha Zarnogi, Department of Genetics, Howard University Hospital, Washington, DC, USA.
Afnan Shakoori, Department of Genetics, Howard University Hospital, Washington, DC, USA.
Sudhir Varma, Hithru Analytics, Laurel, MD, USA.
Edward Lee, Department of Pathology, Howard University Hospital, Washington, DC, USA.
Babak Shokrani, Department of Pathology, Howard University Hospital, Washington, DC, USA.
Adeyinka Laiyemo, Department of Medicine, Howard University Hospital, Washington, DC, USA.
Hassan Brim, Department of Pathology, and Medicine, Howard University, 2041 Georgia Avenue NW, Washington, DC 20059, USA.
References
- 1. Kumar K, Brim H, Giardiello F, et al. Distinct BRAF (V600E) and KRAS mutations in high microsatellite instability sporadic colorectal cancer in African Americans. Clin Cancer Res 2009; 15: 1155–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Al-Sohaily S, Biankin A, Leong R, et al. Molecular pathways in colorectal cancer. J Gastroenterol Hepatol 2012; 27: 1423–1431. [DOI] [PubMed] [Google Scholar]
- 3. Ashktorab H, Daremipouran M, Goel A, et al. DNA methylome profiling identifies novel methylated genes in African American patients with colorectal neoplasia. Epigenetics 2014; 9: 503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. National Cancer Institute. Surveillance, epidemiology, and end results program 2015, http://seer.cancer.gov/
- 5. Brim H, Ashktorab H. Genomics of colorectal cancer in African Americans. Next Gener Seq Appl 2016; 3: pii: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Esteller M, Corn PG, Baylin SB, et al. A gene hypermethylation profile of human cancer. Cancer Res 2001; 61: 3225–3229. [PubMed] [Google Scholar]
- 7. Nazemalhosseini Mojarad E, Kuppen PJ, Aghdaei HA, et al. The CpG island methylator phenotype (CIMP) in colorectal cancer. Gastroenterol Hepatol Bed Bench 2013; 6: 120–128. [PMC free article] [PubMed] [Google Scholar]
- 8. Markowitz SD, Bertagnolli MM. Molecular origins of cancer: molecular basis of colorectal cancer. N Engl J Med 2009; 361: 2449–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hon GC, Shen Y, McCleary DF, et al. Whole genome bisulfite sequencing reveals tissue-specific DNA methylation in a normal mouse. Cancer Res 2013; 73(Suppl. 8): abstract 1111. [Google Scholar]
- 10. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674. [DOI] [PubMed] [Google Scholar]
- 11. Das PM, Singal R. DNA methylation and cancer. J Clin Oncol 2004; 22: 4632–4642. [DOI] [PubMed] [Google Scholar]
- 12. Ogino S, Nosho K, Kirkner GJ, et al. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst 2008; 100: 1734–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ogino S, Lochhead P, Chan AT, et al. Molecular pathological epidemiology of epigenetics: emerging integrative science to analyze environment, host, and disease. Mod Pathol 2013; 26: 465–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Colussi D, Brandi G, Bazzoli F, et al. Molecular pathways involved in colorectal cancer: implications for disease behavior and prevention. Int J Mol Sci 2013; 14: 16365–16385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schuebel KE, Chen W, Cope L, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet 2007; 3: 1709–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ashktorab H, Shakoori A, Zarnogi S, et al. Reduced representation bisulfite sequencing determination of distinctive DNA hypermethylated genes in the progression to colon cancer in African Americans. Gastroenterol Res Pract 2016; 2016: 2102674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pan C, Zhang Y, Meng Q, et al. Down regulation of the expression of ELMO3 by COX2 inhibitor suppresses tumor growth and metastasis in non-small-cell lung cancer. Front Oncol 2019; 9: 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. East MP, Bowzard JB, Dacks JB, et al. ELMO domains, evolutionary and functional characterization of a novel GTPase-activating protein (GAP) domain for Arf protein family GTPases. J Biol Chem 2012; 287: 39538–39553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Søes S, Daugaard IL, Sørensen BS, et al. Hypomethylation and increased expression of the putative oncogene ELMO3 are associated with lung cancer development and metastases formation. Oncoscience 2014; 1: 367–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kristensen LS, Soes S, Hansen LL. ELMO3: a direct driver of cancer metastasis? Cell Cycle 2014; 13: 2483–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pan Y, Jing R, Pitre A, et al. Intermediate filament protein synemin contributes to the migratory properties of astrocytoma cells by influencing the dynamics of the actin cytoskeleton. FASEB J 2008; 22: 3196–3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kapoor S. Synemin: an evolving role in tumor growth and progression. J Cachexia Sarcopenia Muscle 2014; 5: 347–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Noetzel E, Rose M, Sevinc E, et al. Intermediate filament dynamics and breast cancer: aberrant promoter methylation of the Synemin gene is associated with early tumor relapse. Oncogene 2010; 29: 4814–4825. [DOI] [PubMed] [Google Scholar]
- 24. Badis G, Chan ET, van Bakel H, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell 2008; 32: 878–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Scott EW, Baker HV. Concerted action of the transcriptional activators REB1, RAP1, and GCR1 in the high-level expression of the glycolytic gene TPI. Mol Cell Biol 1993; 13: 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ono K, Iwanaga Y, Mannami T, et al. Epidemiological evidence of an association between SLC6A2 gene polymorphism and hypertension. Hypertens Res 2003; 26: 685–689. [DOI] [PubMed] [Google Scholar]
- 27. Kuriakose MA, Suresh A. Diagnostic tests for predicting prognosis, recurrence, resistance or sensitivity to therapy and metastatic status in cancer. Patent number 20140342946, USA, 2014. [Google Scholar]
- 28. Numoto M, Yokoro K, Koshi J. ZF5, which is a Kruppel-type transcriptional repressor, requires the zinc finger domain for self-association. Biochem Biophys Res Commun 1999; 256: 573–578. [DOI] [PubMed] [Google Scholar]
- 29. Feng Y, Xu Q. Pivotal role of hmx2 and hmx3 in zebrafish inner ear and lateral line development. Dev Biol 2010; 339: 507–518. [DOI] [PubMed] [Google Scholar]
- 30. Wojdacz TK, Windeløv JA, Thestrup BB, et al. Identification and characterization of locus-specific methylation patterns within novel loci undergoing hypermethylation during breast cancer pathogenesis. Breast Cancer Res 2014; 16: R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McManus S, Ebert A, Salvagiotto G, et al. The transcription factor Pax5 regulates its target genes by recruiting chromatin-modifying proteins in committed B cells. EMBO J 2011; 30: 2388–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]