Abstract
Background
During the last two decades, over 100 proteomics studies have identified a variety of potential biomarkers in CSF of Alzheimer’s (AD) patients. Although several reviews have proposed specific biomarkers, to date, the statistical relevance of these proteins has not been investigated and no peptidomic analyses have been generated on the basis of specific up- or down- regulation. Herein, we perform an analysis of all unbiased explorative proteomics studies of CSF biomarkers in AD to critically evaluate whether proteins and peptides identified in each study are consistent in distribution; direction change; and significance, which would strengthen their potential use in studies of AD pathology and progression.
Methods
We generated a database containing all CSF proteins whose levels are known to be significantly altered in human AD from 47 independent, validated, proteomics studies. Using this database, which contains 2022 AD and 2562 control human samples, we examined whether each protein is consistently present on the basis of reliable statistical studies; and if so, whether it is over- or under-represented in AD. Additionally, we performed a direct analysis of available mass spectrometric data of these proteins to generate an AD CSF peptide database with 3221 peptides for further analysis.
Results
Of the 162 proteins that were identified in 2 or more studies, we investigated their enrichment or depletion in AD CSF. This allowed us to identify 23 proteins which were increased and 50 proteins which were decreased in AD, some of which have never been revealed as consistent AD biomarkers (i.e. SPRC or MUC18). Regarding the analysis of the tryptic peptide database, we identified 87 peptides corresponding to 13 proteins as the most highly consistently altered peptides in AD. Analysis of tryptic peptide fingerprinting revealed specific peptides encoded by CH3L1, VGF, SCG2, PCSK1N, FBLN3 and APOC2 with the highest probability of detection in AD.
Conclusions
Our study reveals a panel of 27 proteins and 21 peptides highly altered in AD with consistent statistical significance; this panel constitutes a potent tool for the classification and diagnosis of AD.
Keywords: Alzheimer´s Disease, Biomarkers, Proteomic, Peptidomics
Background
Due to its high prevalence within the world population, Alzheimer´s disease (AD) is likely the most studied neurodegenerative disease [1]. A variety of studies have characterized AD pathology from a cognitive point of view [2] and/or using neuroimaging approaches [3]. While these approaches are necessary in order to distinguish the various hallmarks of AD progression, there is still a need to identify reliable biomarkers for the effective diagnosis and prognosis of AD.
Most studies aimed at the detection of AD biomarkers have been carried out on biological fluids such as blood or cerebrospinal fluid (CSF) (reviewed in [4–6]). The use of CSF represents the best approach to identify AD biomarkers (reviewed in [6]) since this fluid contacts the brain interstitial fluid directly and thus more accurately reflects biochemical changes related to central nervous system (CNS) processes. Indeed, studies of AD CSF have consistently demonstrated that amyloid-β (Aβ42), total tau (T-tau), and phosphorylated tau (P-tau) constitute CSF biomarkers relevant to AD diagnosis [6–8]. However, the heterogeneity of AD pathology calls for a deeper review of potential AD CSF biomarkers. Biomarker use may eventually help to predict disease progression, from asymptomatic stages to full-blown AD. Accordingly, over 100 different proteomic studies of potential CSF biomarkers in AD have been conducted during the past 15 years.
A number of reviews have been recently published which have examined these past studies and have proposed specific biomarkers for AD in CSF [9, 10]. However, only 3 proteomic reviews with controlled analyses directly comparing biomarkers in AD and control CSF have been conducted thus far. In 2016, Olsson et al. described which of the common biomarkers in both CSF and blood were the most altered in AD. They reported 5 CSF core biomarkers associated with AD [11]. Subsequently, Bastos et al. compiled data from 18 proteomic studies to obtain 309 proteins expressed differentially in CSF obtained from AD patients vs. controls [12]. The most recent CSF proteomic analysis, which appeared while we were finishing our study, included 29 studies, of which 25 specifically investigated differences between AD and controls, and reported that 478 proteins exhibited different levels in AD compared to controls [13]. This analysis includes most of the proteins that have been shown to differ between AD and controls. However, these data do not consider the statistical relevance of hits. Furthermore, none of the indicated reviews analyze all tryptic peptides on the basis of specific up- or down- regulation in AD. Therefore, the accuracy of the correlation of these potential protein biomarkers, and their specific tryptic peptides, with actual AD pathology remains unclear.
In the current review, by using various bioinformatics resources, we have generated a consistent data output which compiles protein changes presented in widely different formats in the various original independent studies. First, we searched the literature for unbiased CSF explorative proteomics studies that investigate AD and compiled data from 47 independent published studies, which considerably increases the number of studies collected so far, thereby expanding to 601 the total number of proteins identified in proteomic studies of CSF from AD patient samples. Additionally, we classified those studies as either descriptive or supported by quantitative data in order to exclusively extract those proteins whose levels were significantly altered in each of the studies. Thus, we have generated a panel of specific proteins that consistently appear in these studies to be over- or under-represented; and we detail their frequencies. Finally, as an important part of our study, we present a well-characterized and validated peptide analysis of all MS-data obtained from the proteins that show consistent changes (up- or down- regulation). A deep study of this novel AD peptide database, containing more than 3000 tryptic peptides, has led us to unveil a new panel consisting of the most commonly identified peptides in AD CSF, including the peptide direction change in each independent study; significance; and abundance in these clinical cohorts.
Methods
Data sources and search strategy
We searched MEDLINE (via PubMed) to December 2019. The keywords to perform the advanced search were “cerebrospinal fluid” and/or “CSF”, “proteomics”, “peptidomics” and “Alzheimer’s”. We also searched for additional terms such as “biomarkers” and for specific proteomic approaches (i.e. LC–MS/MS) (Fig. 1).
Fig. 1.
Methodological flow chart of the search strategy in the PubMed database. *Only those proteins that appeared consistently and statistically up- or down-regulated in AD in at least 2 studies were considered further
Inclusion criteria and construction of the database
To strengthen the study, we included only studies that identified more than one protein as an AD biomarker in CSF, removing 8 studies that focused on only one protein (see Fig. 1) or those that only provided qualitative information (n = 7). We also removed those studies that involved only Aβ and tau variants/isoforms (n = 13); studies focused exclusively on post-translational modifications (PTM) (n = 4); and others that, although containing the search parameters within the text, did not actually perform experimental proteomics research on CSF (for example, in plasma samples) (n = 27) (Fig. 1).
Neuropsychological criteria for the definition of diagnostic groups were heterogeneous (Table 1). Baseline characteristics to define healthy controls according to cognitive criteria were: CDR = 0, no neuropsychological deficits and/or not reaching mild cognitive impairment (MCI) criteria; score ≥ 23 on Montreal Cognitive Assessment; MMSE score of 30; and asymptomatic NCs, mutation non-carriers. Baseline criteria to define AD were the presence of self-reported cognitive complaints; cognitive criteria were CDR ≥ 1; score < 23 on the Montreal Cognitive Assessment; subjects who met criteria for probable AD dementia based on NIA-AA criteria; and familial Alzheimer disease (FAD) mutation carriers (MCs). In both control and AD groups, 23 out of the 47 studies confirmed an AD diagnosis using the CSF biomarkers Aβ42, T-tau and/or P-tau, and provided clinical evaluations that included the following: detailed informant-based history, assessment of medical records, medical history, family history, physical and neurologic examination, routine lab tests, brain CT or MRI, brain imaging neurological and cognitive examinations. Additional details are shown in Table 1. Thus, we have not included studies that contain the following: categories of pre-MCI individuals or subjective memory complaints (SMC); articles comparing Alzheimer’s disease proteins with Parkinson’s disease proteins; articles discussing protein differences between younger and older subjects; and articles that studied proteins before and after a specific treatment (n = 6).
Table 1.
Proteomic approaches, sample size and neuropsychological criteria for the groups used in the studies included
| No | Author and year | Proteomic approach | Controls samples | AD samples | Mean age (controls) | Mean age (AD) | Neuropsychological criteria | CSF biomarkers (control) | CSF biomarkers (AD) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Whelan CD et al. (2019) [86] | Olink™ ProSeek immunoassay | 565 | 176 | 72 | 74.6 | NINCDS-ADRDA criteria, MMSE score and CSF biomarkers |
Mean Aβ42 [pg/ml(SD)]: 752(253) Mean Aβ40 [pg/ml(SD)]: 5847(2042) Aβ42/40 ratio - log2 transformed (SD): 2.05(0.17) Mean total tau [pg/ml(SD)]: 292(89) Mean phospho-tau [pg/ml(SD)]: 37(13) |
Mean Aβ42 [pg/ml(SD)]: 305(132) Mean Aβ40 [pg/ml(SD)]: 5470(2179) Aβ42/40 ratio - log2 transformed (SD): 2.9(0.29) Mean total tau [pg/ml(SD)]: 649(221) Mean phospho-tau [pg/ml(SD)]: 123(47) |
| 2 | Khoonsari et al. (2019) [87] | Nano LC-MS/MS (7 T hybrid LTQ FT MS) | 45 | 76 | 88 | 72 | NINCDS-ADRDA criteria, DSM-IV criteria, MMSE and CSF biomarkers |
Aβ42 ng/l (Median[range]): 676(337–1343) p-tau ng/l (Median[range]) 414 (202–1121) t-tau ng/l (Median[range]): 63 (29-122) |
Aβ42 ng/l (Median[range]): 405(160–1160) p-tau ng/l (Median[range]) 82 (28–220) t-tau ng/l (Median[range]): 617 (160–1720) |
| 3 | Sathe et al. [88] | High-Resolution MS, TMT multiplexing technology and targeted PRM analysis | 5 | 5 | 71 | 71.6 | CDR, MOCA, CSF biomarkers and NIA-AA criteria | Mean Aβ42 [pg/ml(SD)]: 1185.86(389.29) | Mean Aβ42 [pg/ml(SD)]: 402.11(189.62) |
| 4 | Duits et al. (2018) [89] | PRM | 40 | 40 | 64.5 | 64.6 | MMSE, clinical evaluation, and CSF biomarkers |
Mean Aβ42 [pg/ml(SD)]: 1086(161) Mean total tau [pg/ml(SD)]: 228(64) Mean phospho-tau [pg/ml(SD)]: 40(9) |
Mean Aβ42 [pg/ml(SD)]: 640(91) Mean total tau [pg/ml(SD)]: 740(433) Mean phospho-tau [pg/ml(SD)]: 94(47) |
| 5 | Dayon et al. (2018) [90] | RP LC-MS/MS | 48 | 72 | 66 | 73.7 | CDR, APOEa genotype and CSF biomarkers |
Mean Aβ42 [pg/ml(SD)]: 957.4(194) Mean total tau [pg/ml(SD)]: 221.5(82.9) Mean phospho-tau [pg/ml(SD)]: 45.9(13.3) Mean phospho-tau (SD)/Aβ42: 0.049(0.015) |
Mean Aβ42 [pg/ml(SD)]: 774(281.5) Mean total tau [pg/ml(SD)]: 471.1(316.6) Mean phospho-tau [pg/ml(SD)]: 72.7(40.9) Mean phospho-tau (SD)/Aβ42: 0.114(0.097) |
| 6 | Brinkmalm et al. (2018) [14] | PRM-MS analysis and previously nanoflow LC-MS/MS | 15 | 10 | 64.9 | 62.7 | Clinical evaluation, computer tomography (CT) scan and CSF Aβ42 | Mean Aβ42 [pg/ml(SD)]: 877.2(206.3) | Aβ42 < 400 pg/ml;Mean Aβ42 [pg/ml(SD)]: 275.4(106.8) |
| 7 | Skillbäck et al. (2017) [45] | Quantification-driven proteomic | 40 | 40 | 64.5 | 64 | NIA and IWG-2 criteria, CSF biomarkers and MMSE |
Information extracted from graphs: Mean Aβ42 [pg/ml]: 950 Mean total tau [pg/ml]: 260 Mean phospho- tau [pg/ml]: 55 |
Information extracted from graphs: Mean Aβ42 [pg/ml]: 450 Mean total tau [pg/ml]: 600 Mean phospho- tau [pg/ml]: 85 |
| 8 | Wang et al. (2016) [46] | LC-MS/MS and lectin-enrichment chromatography | 4 | 4 | 74 | 75.7 | Neuropsychological tests | NI | |
| 9 | Paterson et al. (2016) [91] | Targeted proteomics: MRM-based triple quadrupole mass spectral assay | 36 | 46 | 58.5 | 62.9 | APOE# genotype and CSF biomarkers |
Mean Aβ42 [pg/ml(SD)]: 960(291) Mean total tau (range): 234.5(174.5–315.5) Mean phospho-tau [pg/ml(SD)]: 35.5(13.2) Mean phospho-tau (range)/Aβ42: 0.25(0.19-0.33) |
Mean Aβ42 [pg/ml(SD)]: 408(2168) Mean total tau (range): 947(760–1196-315.5) Mean phospho-tau [pg/ml(SD)]: 107.5(38.12) Mean phospho-tau (range)/Aβ42: 2.5(1.8-4.1) |
| 10 | Remnestal et al. (2016) [92] | Suspension bead array: HPA antibodies targeting the 280 proteins | 38 | 72 | 58 | 77.4 | NINCDS-ADRDA criteria and CSF biomarkers |
Mean Aβ42 [pg/ml(range)]: 706(559-1192) Mean total tau (range): 308(171-399) Mean phospho-tau (range): 47(29-60) |
Group AD1: Mean Aβ42 [pg/ml(range)]: 350(160-950) Mean total tau (range): 600(210–2430) Mean phospho-tau (range): 78(34–282)Group AD2: Mean Aβ42 [pg/ml(range)]: 453(260–639) Mean total tau (range): 834(190–3178) Mean phospho-tau (range): 86(59–179) |
| 11 | Khoonsari et al. (2016) [16] | RP nano LC- MS/MS andantibody suspension bead arrrays | 10 | 10 | 88.2 | 79.4 | Clinical evaluation, CSF biomarkers and MMSE | NI |
Mean Aβ42 [ng/l]: 420.1 Mean total tau [ng/l]: 652.1 Mean phospho-tau [ng/l]: 132.2 |
| 12 | Domenico et al. (2016) [93]** | 2-DE - WB - MS (2-DE gel/carbonyl immunoblot) | 6 | 6 | NI | NI | NINCDS-ADRDA criteria, neuropsychological test and APOE# genotype | NI | |
| 13 | Heywood et al. (2015) [94] | Label-free proteomic analyses (2-DE LC-MS) and Targeted proteomics: MRM-based triple quadrupole mass spectral assay | 15 | 16 | 61 | 68.1 | CSF biomarkers |
Mean Aβ42 [pg/ml(SD)]: 905.41(237) Mean total tau [pg/ml(SD)]:136.58(162) Mean phospho-tau [pg/ml(SD)]: 19.33(11.11) |
Mean Aβ42 [pg/ml(SD)]: 503.68(165.8) Mean total tau [pg/ml(SD)]:733.77(481.25) Mean phospho-tau [pg/ml(SD)]: 93.38(31.55) |
| 14 | Hendrickson et al. (2015) [20] | RP nano-HPLC coupled to an LTQ-FTMS hybrid mass spectrometer. SRM-MS | 30 | 30 | 69 | 70 | NINCDS-ADRDA criteria and APOE# genotype | NI | |
| 15 | Spellman et al. (2015) [95] | LC/MRM-MS | 85 | 66 | 75.6 | 75 | APOE# genotype, CSF biomarkers and MMSE |
Mean Aβ42 [SD]: 207.8(56.26) Mean phospho-tau (SD): 24.19(12.02) |
Mean Aβ42 [SD]: 141.12(37.39) Mean phospho-tau (SD): 41.95(20.6) |
| 16 | Hölttä et al. (2015) [15] | TMT, LC−ESI MS, LC−MALDI MS | 8 | 8 | 65 | 77 | DSM-IIIRand NINCDS-ADRDA criteria |
Mean Aβ42 [pg/ml(range)]: 700(545–817) Mean total tau (range): 405(330–523) |
Mean Aβ42 [pg/ml(range)]: 235(210–298) Mean total tau (range): 800(717–925) |
| 17 | Leung et al. (2015) [96] | MAP-RBM platform (XMAP Luminex platform) /ELISA | 325 | 344 | 71.7 | 74 | MMSE, CDR, NINDS-ADRDA criteria and CSF biomarkers | Aβ42 > 192 pg/ml | Aβ42 < 192 pg/ml |
| 18 | Khan et al. (2015) [97]** | XMAP Luminex platform | 88 | 65 | 75.8 | 74.6 | APOE# genotype, CSF biomarkers, and MMSE |
Mean Aβ42 [(SD)]: 205.7(57.2) Mean total tau (SD):69.2(27.9) Mean phospho-tau (SD): 24.9(13.2) |
Mean Aβ42 [(SD)]: 140.4(35.3) Mean total tau (SD):125.9(60.3) Mean phospho-tau (SD): 42.2(20.7) |
| 19 | Oláh et al. (2015) [98] | Master Antibody Microarray and WB | 25 | 25 | 74.5 | 72 | Clinial evaluation, NINCDS-ADRDA criteria, APOE# genotype and CSF biomarkers |
Mean Aβ42 [pg/ml(SD)]: 794(20) Mean total tau [pg/ml(SD)]: 341(171) Mean phospho-tau [pg/ml(SD)]: 23(2) |
Aβ42 < 500 pg/ml total tau >600 pg/ml phospho-tau>60 pg/ml |
| 20 | Alzate et al. (2014) [99] | MS and MS/MS and 2-DE multiplexed WB | 7 | 11 | AR: 62.8 to 81.5 | AR: 62.8 to 81.5 | APOE# genotype | NI | |
| 21 | Wildsmith et al. (2014)[100] | LC-MS/MS, Targeted LC-MRM and ELISA | 10 | 45 | 68.5 | 77.5 | MMSE and CSF biomarkers |
Information extracted from graphs: Mean Aβ42 [pg/ml]: 650 Mean total tau [pg/ml]: 60 Mean phospho- tau [pg/ml]: 25 |
Information extracted from graphs: Mean Aβ42 [pg/ml]: 400 Mean total tau [pg/ml]: 150Mean phospho- tau [pg/ml]: 60 |
| 22 | Chakrabarti et al. (2014) [101] | 2-DE, MALDI MS Analysis and co-immunoprecipitation | 11 | 11 | 67 | 71 | NINCDS-ADRDA criteria, DSM-IV criteria, MMSE, CDR and other tests | NI | |
| 23 | Choi et al. (2013) [49]** | Nano LC-MS/MS and nano LC-MRM/MS and ELISA | 3 | 3 | NI | NI | Confirmed by autopsy | NI | |
| 24 | Wijte et al. (2012) [102] | MALDI TOF/TOF MS | 20 | 20 | 77.7 | 75.95 | Postmortem neuropathological evaluation | NI | |
| 25 | Ringman et al. (2012) [103] | MS/MS, 2-DE LC/MS | 5 | 4 | 37.6 | 34.2 | CDR and FAD mutation carriers (MCs) and related noncarrieres (NCs)Π. APOE# genotype and CSF biomarkers |
Mean Aβ42 [pg/ml(SD)]: 618.4(100.1) Mean total tau [pg/ml(SD)]: 50.5(9.4) Mean phospho-tau [pg/ml(SD)]: 24.6(9.2) |
Mean Aβ42 [pg/ml(SD)]: 277.1(162.9) Mean total tau [pg/ml(SD)]: 140(59.9) Mean phospho-tau [pg/ml(SD)]: 67(29) |
| 26 | Manral et al. (2012) [104] | 2-DE and Nano LC-ESI-Q-TOF MS/MS system | 8 | 8 | 60.7 | 62.6 | NINCDS-ADRDA criteria, MMSE score and MRI | NI | |
| 27 | Vafadar-Isfahani et al. (2012) [105] | nLC-MALDI-TOF MS/MS and MALDI TOF/TOF MS | 23 | 33 | NI | NI | NINCDS-ADRDA criteria, DSM-IV and MMSE and clinical evaluation |
Information extracted from graphs: Mean Aβ42 [pg/ml]: 440 Mean Aβ40 [pg/ml]: 5000 Mean total tau [pg/ml]: 500 |
Information extracted from graphs: Mean Aβ42 [pg/ml]: 190 Mean Aβ40 [pg/ml]: 3900 Mean total tau [pg/ml]: 2000 |
| 28 | Jahn et al. (2011) [44] | CE-MS and MS/MS | 17 | 34 | 58.9 | 69 | NINCDS-ADRDA criteria, CSF biomarkers and MMSE score | NI |
Mean Aβ42 [pg/ml]: 300.91 Mean total tau [pg/ml]: 541.21 Mean phospho-tau [pg/ml]: 81.84 |
| 29 | Craig-Schapiro et al. (2011) [106]** | ELISA (targeted) | 242 | 28+63 | 71.6 | 76.8 | NINCDS-ADRDA criteria and CDR | NI | |
| 30 | Perrin et al. (2011) [50] | 2-DE DIGE / nano- LC-MS/MS | 198 | 29 | 71 | 76.5 | CDR, MMSE score and CSF biomarkers |
Mean Aβ42 [pg/ml(SD)]: 605(240) Mean total tau [pg/ml(SD)]: 304(161) Mean phospho-tau [pg/ml(SD)]: 55(25) |
Mean Aβ42 [pg/ml(SD)]: 351(118) Mean total tau [pg/ml(SD)]: 552(263) Mean phospho-tau [pg/ml(SD)]: 77(38) |
| 31 | Hu et al. (2010) [107]** | XMAP Luminex platform | 33 | 66 | NI | NI | MMSE, CSF biomarkers and confirmed by autopsy | NI | |
| 32 | Maarouf et al. (2009) [108] | 2-DE DIGE and MS | 43 | 47 | 84 | 79 | APOE# genotype, CERAD and the NIA-Reagan Institute | NI | |
| 33 | Yin et al. (2009) [109] | 1-DE and LC–MS/MS | 6 | 5 | 55.3 | 71 | NINCDS-ADRDA criteria | NI | |
| 34 | Zhang et al. (2008) [110] | Immunobead-based multiplex assays | 95 | 48 | 63 | 70 | Clinical evaluation | NI | |
| 35 | Jung et al. (2008) [66] | 2-DE, DE-MALDI-TOF MS | 30 | 27 | 73.2 | 77.7 | NINCDS-ADRDA criteria and CDR | NI | |
| 36 | Simonsen et al. (2007) [111] | MS | 32 | 85 | 68.3 | 69.15 | NINCDS-ADRDA criteria, MMSE score and clinical evaluation | NI | |
| 37 | Hu et al. (2007) [112]** | 2-DE DIGE, MS/MS and ELISA | 55 | 19 | NI | NI | CSF biomarkers, CDR and clinical evaluation |
Information extracted from graphs: Mean Aβ42 (range): 1(0–2) Mean total tau (range): 0.9(0–5)Mean phospho- tau (range): 0.8(0–5) |
Information extracted from graphs: Mean Aβ42 (range): 0.6(0–2) Mean total tau (range): 1.9(0–5)Mean phospho- tau (range): 1.1(0–5) |
| 38 | Korolainen et al. (2007) [113] | 2-DE and Nano LC-MS/MS | 8 | 11 | 64.7 | 73.54 | APOE# genotype, NINCDS-ADRDA criteria and MMSE score | NI | |
| 39 | Finehout et al. (2007) [114] | 2-DE and TOF/TOF MS | 12 | 44 | NI | NI | Postmortem confirmation | NI | |
| 40 | Simonsen et al. (2008) [115] | SELDI-TOF MS | 72 | 95 | 73.1 | 72.5 | NINCDS-ADRDA criteria, MMSE score and CSF biomarkers |
Group C1: Mean Aβ42 [pg/ml(SD)]: 876(209.2) Mean total tau (SD): 295.8 (153.6) Group C2: Mean Aβ42 [pg/ml(SD)]: 748.9(155.3) Mean total tau (SD): 332.9 (158.7) |
Group AD1: Mean Aβ42 [pg/ml(SD)]: 524.6(170.1) Mean total tau (SD): 739.1 (300.9) Group AD2: Mean Aβ42 [pg/ml(SD)]: 464(187.7) Mean total tau (SD): 657.7 (339.4) |
| 41 | Castaño et al. (2006) [116] | 2-DE and MALDI-TOF MS | 43 | 43 | 81 | 81 | Clinical diagnosis, Braak stages, and CERAD | NI | |
| 42 | Abdi et al. (2006) [52] | iTRAQ labeling and 2-DE LC, MS/MS and WB | 10 | 10 | 67 | 72 | MMSE, CDR, NINDS-ADRDA criteria and other test | NI | |
| 43 | Selle et al. (2005) [117] | MALDI-TOF-MS | 102 | 140 | 62.5 | 71 | ICD-10, NINCDS-ADRDA criteria, DSM-IV and MMSE | NI | |
| 44 | Zhang et al. (2005) [118]** | LC-MS ICAT | 20 | 32 | 70.5 | 71 | MMSE and CDR | NI | |
| 45 | Puchades et al. (2003) [68] | 2-DE and MALDI-TOF MS | 7 | 7 | 66 | 80 | NINCDS-ADRDA criteria and MMSE score | NI | |
| 46 | Carrette et al. (2003) [119] | SELDI-TOF MS using SAX2 Protein-chip arrays | 10 | 19 | 78 | 75 | NINCDS-ADRDA criteria | NI | |
| 47 | Davidsson et al. (2002) [120] | 2-DE, SYPRO Ruby staining and MS | 12 | 15 | 67.3 | 77.2 | NINCDS-ADRDA criteria and MMSE score | NI | |
| Total samples (Control) | Total samples (AD) | Average age (Control) | Average age (AD) | ||||||
| 2562 | 2022 | 70 | 73 | ||||||
ESI electrospray ionization, MRM multiple reaction monitoring, PRM parallel reaction monitoring, MS mass spectrometry, TMT tandem mass tag, RP-LC MS/MS reversed-phase LC-MS/MS, SRM reaction monitoring, CE-MS capillary electrophoresis mass spectrometry, SELDI-TOF-MS surface enhanced laser desorption/ionization time-of-flight mass spectrometry, WB Western Blot, AR age range, NI not information, CDR Clinical dementia rating scale, MOCA Montreal cognitive assessment, NINCDS-ADRDA National Institute of Neurological and Communicative Disorders and Stroke and the AD and Related Disorders Association, MMSE Mini-menta state exam, DSM Diagnostic and Statistical, FAD familial Alzheimer Disease, CERAD Consortium to Establish a Registry for Alzheimer’s Disease, ICD-10 10th revision of the International Statistical Classification of Diseases and Related Health Problems, IWG-2 International Working Group-2 criteria
#Studies based on APOE genotype
ΠStudy based on PSEN1 and APP mutations (this study was not considered to calculate the average age)
** Qualitative and not quantitative data
We examined both tables and information included in the main texts, as well as associated supplemental data, to extract information on protein and peptide sequences that were found to be altered in the CSF of AD samples. However, we included in our analysis only those articles that provided statistics for their results by showing significant differences between control and AD groups through numerous statistical methods such as the non-parametric Mann–Whitney U and Kruskal–Wallis tests, which were the most common statistical approaches among the different studies [14–16]. To solve the problem of heterogeneity in the annotation databases used in the various studies, we made all protein names consistent with the Uniprot database (https://www.uniprot.org) by filtering them through Ingenuity Pathway Analysis (IPA, http://www.ingenuity.com) and DAVID bioinformatics resources (http://david.abcc.ncifcrf.gov.), which enabled us to obtain the Entrez Gene ID of all proteins. To do this, it has been necessary to remove “hypothetical proteins”, “unidentified proteins”, “IgG light chain” “IgG heavy chain” and “predicted protein”. Variants of amyloid-β and tau, the focus of several reviews [10, 17] were not included. Of note, the FLJ00385 protein and all proteins depicted with Δ were not recognized by DAVID bioinformatics resources.
Data mining
We included in our analysis exclusively those studies which contained quantitative protein information, according to each study-specific cut-off point with a given statistical significance. The protein database, filtered as described above, was first analyzed to identify the unique proteins that change between AD and control samples. To analyze all proteins, we extracted related information based on different annotation aspects/terms which was inconsistent in the different studies. For example, Apolipoprotein J, APOJ, APO-J, CLUS_HUMAN, P10909, and TRPM-2 all are referred to Clusterin.
After generating a table containing all proteins that exhibit changes in AD (Additional file 1: Table S1), we calculated the number of articles that identify each specific protein and we further considered those proteins that appeared in 2 or more studies to generate a table that indicated whether they were found to increase or decrease in the context of AD.
An important part of our study consisted in the direct analysis of all MS-data to generate a database of all peptides corresponding to proteins with consistent changes (up- or down- regulation). This database includes all peptide sequences extracted from the proteins mentioned above, the observed peptide mass weight (Da), the significance difference statistics, the ratio of AD/Control or fold-change, and the direction change in each article. It should be noted that the significance of certain peptide changes had not been included in all studies, and so we independently calculated this parameter from raw data using paired t-tests. In addition, the peptide masses were calculated using the pI/Mw tool (https://web.expasy.org/compute_pi/). We considered only those peptides that appeared at least in two independent studies and analyzed whether their changes were supported by statistical analysis; and those observed in 3 or more independent studies were considered for discussion purposes as the most consistently observed in the context of AD.
Pathway analysis
Pathway, network and upstream regulator analysis were generated through the use of IPA (QIAGEN Inc., https://www.qiagenbio-informatics.com/products/ingenuity-pathway-analysis) with the proteins obtained in the current study, taking into consideration whether they increased or decreased in the context of AD. The threshold for the top canonical pathways was increased to ¡–Log (p value) 4.5, and only the most relevant network of proteins was considered. Upstream regulators were also investigated, not considering non-endogenous chemical drugs and toxicants. A bias correction of the z-score was performed and activation z-scores below 2 were not further considered in order to reduce artifactual results.
Risk of bias assessment
We have combined all of the statistical studies for comparative purposes, although the varying degrees of statistical significance intrinsic to the different proteomics and peptidomics approaches must be considered. Although in this study four independent investigators conducted literature searches to identify all possible protein data, we encountered difficulties in extracting the information within several articles, as the annotation datasets used differed between articles, and the statistical approaches were sometimes not clearly described. To allow the reader access to the original extracted information of proteins, we have included Additional file 1: Table S1.
As for the peptide database, it is important to note that only 17 articles reported actual peptide sequences; thus, this peptide analysis constitutes a different analysis than the one carried out with proteins.
Results
Our initial bibliographic screens support the idea that, among the different pathologies that affect the brain, AD is likely the most investigated from a proteomics point of view, with a total of 112 proteomics-related articles (Fig. 1). In order to generate a reliable database with only those studies that contain statistically significant proteomic data between AD and control samples- controls being exclusively healthy individuals (see Table 1 for information regarding neurophysiological criteria), we did not consider studies/data specific to MCI. After this screening, 47 articles were included in our analyses (Fig. 1). From these 47 articles, we generated a database that contains proteomic studies from a total of 2022 AD patients with a mean age of 73 years and a total of 2562 samples of healthy individuals with a mean age of 70 years (Table 1). Annotation databases used in the different articles to identify the proteins differed; therefore, a refinement of the database was needed to generate similar data outputs using IPA and DAVID bioinformatic resources. Information on the meaning of the abbreviations used for each protein and article (numbered from 1 to 47) is shown in Additional file 1: Table S1.
Analysis of the proteins that were found to change in AD vs control samples
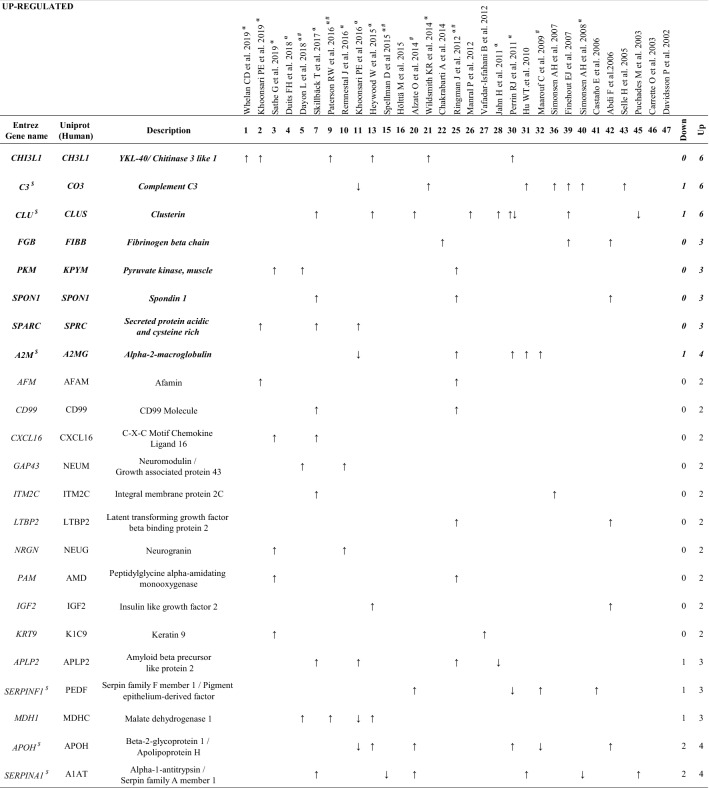
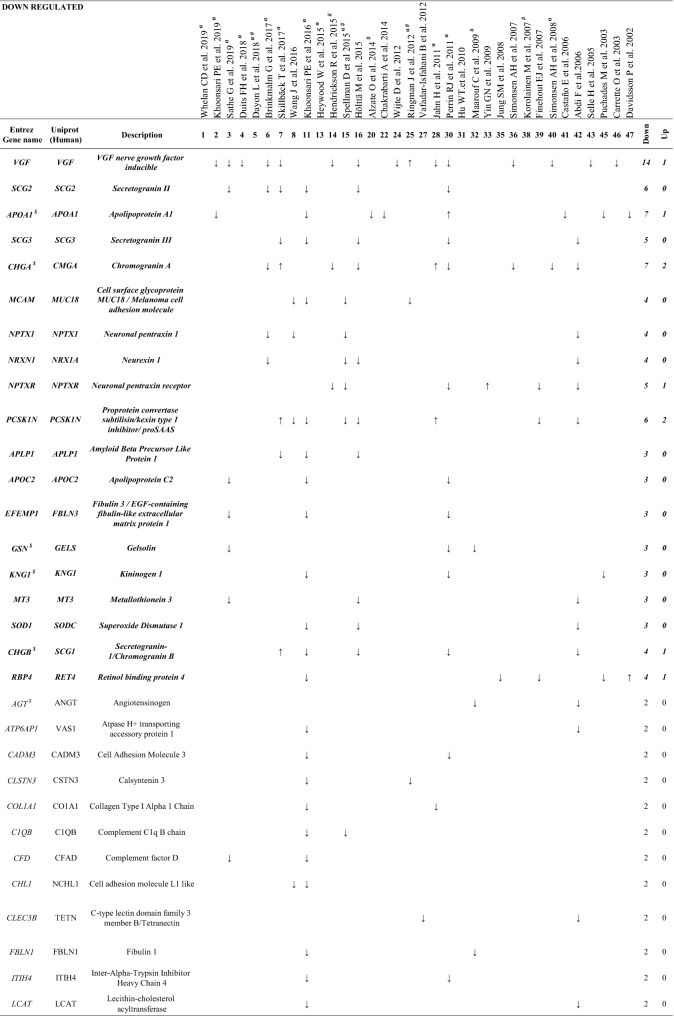
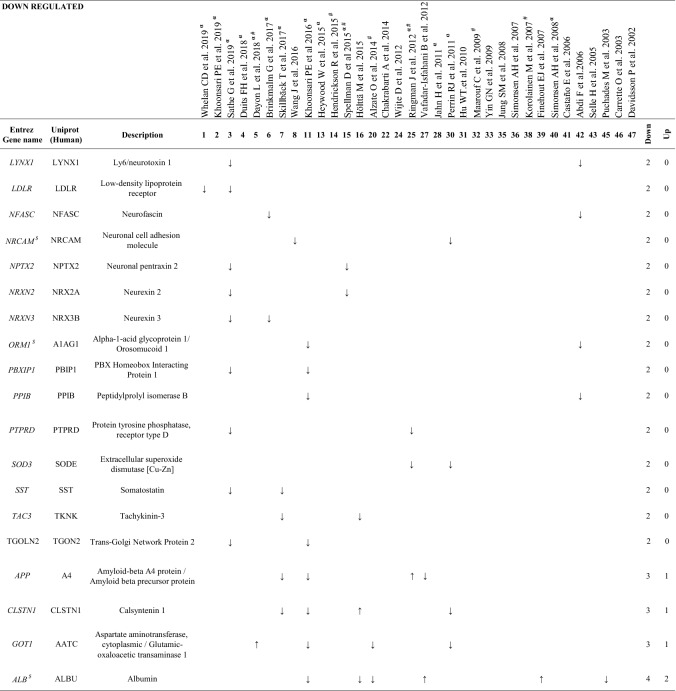
Additional file 2: Table S2 shows information related to the proteins identified as changing in AD, along with the number of articles that found changes in a particular protein. After matching the proteomic information between the different articles, a total of 601 unique proteins were identified (Additional file 2: Table S2). The proteins that appeared in at least 2 proteomic studies are shown in Table 2 (162 proteins). The most recurrent proteins were Apolipoprotein E (APOE), Nerve growth factor inducible (VGF) and Transthyretin (TTHY), being identified in 18, 15 and 14 independent proteomics studies, respectively (Table 2). In order to provide information on the faddabundance of each protein, the 50 most abundant proteins in the CSF of healthy individuals, according to published sources [18], are designated with $ in Tables 2, 3 and 4.
Table 2.
Proteins that show differences between AD and control groups across independent studies
| Number of articles | Gene (Protein) |
|---|---|
| 18 | APOE (APOE)$ |
| 15 | VGF (VGF) |
| 14 | TTR (TTHY)$ |
| 11 | CHGA (CMGA)$, CST3 (CYTC)$ |
| 10 | SERPINA1 (A1AT)$, APOA1 (APOA1)$ |
| 9 | C4 (CO4)$, CLU (CLUS)$, SPP1 (OSTP)$, PCSK1N (PCSK1N) |
| 8 | C3 (CO3)$ |
| 7 | PTGDS (PTGDS)$, B2M (B2MG), GC (VTDB)$ |
| 6 | ALB (ALBU)$, A2M (A2MG)$, APOH (APOH)$, CHI3L1 (CH3L1), NPTX1 (NPTX1), NPTXR (NPTXR), SCG2 (SCG2), RBP4 (RET4) |
| 5 | TF (TRFE)$, AGT (ANGT)$, FABP (FABPH), GSN (GELS)$, HP (HPT)$, SERPINA3 (AACT)$, CNDP1 (CNDP1)$, CP (CERU)$, CHGB (SCG1)$, ITM2B (ITM2B), SCG3 (SCG3) |
| 4 | A1BG (A1BG)$, FGB (FIBB), PLG (PLMN)$, NRXN1 (NRX1A), MDH1 (MDHC), APOD (APOD)$, CFB (CFAB)$, HPX (HEMO)$, IGFBP2 (IBP2), RETN (RETN), DAG1 (DAG1), APP (A4), FGA (FIBA), SPRC (SPARC), SPON1 (SPON1), GOT1 (AATC), APLP2 (APLP2), SCG5 (7B2), CLSTN1 (CSTN1), MCAM (MUC18), SERPINF1 (PEDF)$ |
| 3 | AMBP (AMBP), ITIH1 (ITIH1), LRG1 (A2GL), MT3 (MT3), NEGR1 (NEGR1), RNASE1 (RNASE1), SORT1 (SORT), APOC1 (APOC1), ALDOA (ALDOA), C2 (CO2), PKM (KPYM), AFM (AFAM), APOA2 (APOA2)$, CLEC3B (TETN), CFD (CFAD), HGF (HGF), PPY (PAHO), PSAP (SAP),VEGF (Q9UNS8), CNTN1 (CNTN1), NCAM1 (NCAM1), SPARCL1 (SPRL1)$, CXCL16 (CXL16), SPOCK1 (TICN1), VEGFA (VEGFA), EFEMP1 (FBLN3)$, BASP1 (BASP1), KNG1 (KNG1)$, NSG1 (NSG1), PRNP (PRIO), SERPINC1 (ANT3)$, SMOC1 (SMOC1), SOD1 (SODC), APLP1 (APLP1) |
| 2 | CTSD (CATD), ACTA2 (ACTA), ATP6AP1 (VAS1), AZGP1 (ZA2G)$, C1QB (C1QB), C5 (CO5), C6 (CO6), ENO2 (ENOG), ENPP2 (ENPP2)$, IGF2 (IGF2), KLK6 (KLK6)$, LCAT (LCAT), LTBP2 (LTBP2), LYNX1 (LYNX1), NFASC (NFASC), NRGN (NEUG), NRXN3 (NRX3B), PAM (AMD), PPIB (PPIB), PTPRD (PTPRD), PTPRN (PTPRN), SEZ6L (SE6L1), TAC1 (TKN1), GAP43 (NEUM), CHL1 (NCHL1)$, ORM1 (A1AG1)$, KRT9 (K1C9), IGFBP7 (IBP7), LDLR (LDLR), MMP10 (MMP10), TNFSF10 (TNF10), ADIPOQ1 (ADIPO), CPE (CBPE), CCL16 (CCL16), CD14 (CD14), CD40 (TNR5), C7 (CO7), C1R (C1R), DCD (DCD), HRG (HRG), IGFBP6 (IBP6), INS (A6XGL2), L1CAM (L1CAM), MMP2 (MMP2), MB (MYG), NBL1 (NBL1), NPDC1 (NPDC1), NRCAM (NRCAM)$, OGN (MIME), PRL (Q5THQ0), SERPINA7 (THBG), SERPINE1 (PAI1), CNTN2 (CNTN2), CD99 (CD99), ITM2C (ITM2C), PENK (PENK), SPOCK2 (TICN2), STT (SMS), TAC (TKNK), ITIH4 (ITIH4), SOD3 (SODE), VTN (VTNC), CADM3 (CADM3), CLSTN3 (CSTN3), COL1A1 (CO1A1), FBLN1 (FBLN1)$, IGSF8 (IGSF8), NCAN (NCAN), PBXIP1 (PBIP1), PGLYRP2 (PGRP2), TGOLN2 (TGON2) |
The number of studies which detected these changes is included. $ proteins which are abundant in CSF of healthy individuals (Schilde et al. 2018)
Table 3.
Proteins increased in AD CSF proteomic studies
Proteins with the same pattern of expression (toward the same direction) at least in 2 or 3 (bolditalic proteins) independent studies
$Proteins abundant in CSF of healthy individuals (Schilde et al. 2018)
αStudies that provide Aβ and Tau contents in CSF
#APOE genotype to diagnose AD
Table 4.
Proteins decreased in AD CSF proteomic studies
Proteins with the same pattern of expression (toward the same direction) at least in 2 or 3 (bolditalic proteins) independent studies
$Proteins abundant in CSF of healthy individuals (Schilde et al. 2018)
αStudies that provide Aβ and Tau contents in CSF
#APOE genotype to diagnose AD
We found that, of the proteins that were identified in more than one study, 63 proteins appeared both as increased and decreased in different proteomic studies and therefore were considered as inconsistent (Additional file 3: Table S3). On the other hand, we identified 23 proteins which were increased (Table 3), and 50 proteins decreased (Table 4) in AD samples in at least in 2 independent studies. Of these, a set of 27 proteins, indicated in bolditalic in Tables 3 and 4, represents the most consistent findings across the proteomic literature, being observed with the same pattern of expression at least in 3 independent studies. It is interesting to point out that although some of the studies included in our analysis used depletion kits prior to proteomics, from all possible proteins affected by the use of these kits, only 2, Albumin (ALB) and Fibrinogen beta chain (FIBB), were among the proteins detected in our study.
A pathway analysis of the subset of proteins exhibiting consistent changes (Tables 3 and 4) was performed using IPA, taking into account the significance of their increase or decrease. LRX/RXR activation appeared as the pathway with the most members of the study along with a significant negative z-score (Fig. 2a). Additionally, one major interacting network was identified, related to cell-to-cell signalling: cellular assembly and organization-nervous system development and function (Fig. 2b). Our IPA analysis revealed that the transcription factor NFE2L2/NRF2 (nuclear factor, erythroid 2-like 2; predicted to be inhibited) and the Serine/Threonine Kinase 11 (STK11 kinase; predicted to be activated) were the most consistent potential upstream regulators (both z-scores > 2; P value < 0.0001; Fig. 2c).
Fig. 2.
Pathways, network and upstream regulators analysis of the proteins increased or decreased in AD. a Canonical pathways were analyzed using IPA. The threshold for the top canonical pathways was set to –Log (p value) 4.5, and positive and negative z-scores are shown. b The most consistent protein network corresponds to cell-to-cell signalling-cellular assembly and organization-nervous system development and function. c Upstream regulators were investigated. γ, non-endogenous chemical drugs and toxicants were excluded. φ activation z-score was increased to 2. bias correction of the z-score is indicated. Green, proteins decreased in AD. Red, Proteins increased in AD
Analysis of the tryptic peptides of the proteins that change in AD vs control samples
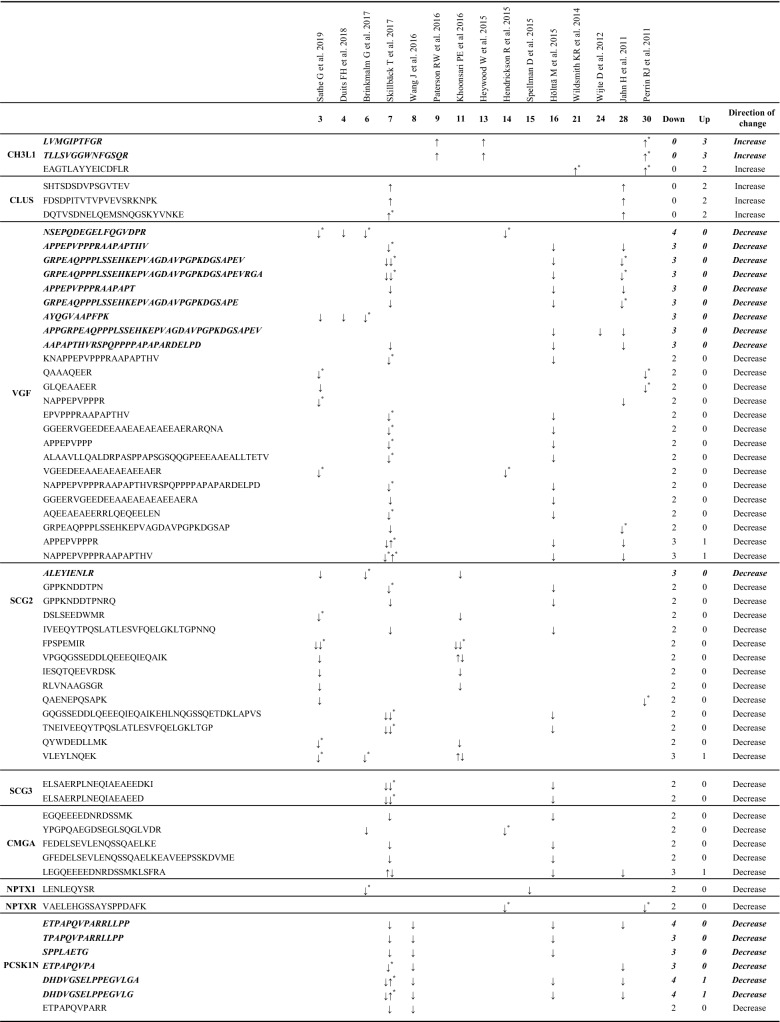
In order to identify changes at the peptide level, we performed a direct analysis of MS-data of the 27 proteins consistently recognized as altered in AD. Information regarding peptide masses of these proteins was available in 17 studies. With the information extracted from these articles we were able to generate a database with 3221 peptide sequences (Additional file 4: Table S4). Further analysis of this database revealed that 87 peptides [Table 5 and Additional file 6: Figure S1 (sequences in bolditalic and in red)], which correspond to a total of 13 proteins (2 proteins increased and 11 decreased in AD) maintained a direction change consistent with that observed by proteomics in relation to the AD pathology (Table 5). Other peptides from these proteins showed inconsistent distribution across the different studies (Additional file 5: Table S5). On the other hand, 21 peptides (Table 5 (bolditalic) and Additional file 6 Figure S1 (underlined)] were found in at least 3 independent studies. These peptides correspond to Chitinase 3 like 1 (CH3L1, 2 peptides), VGF (9 peptides), Secretogranin-2 (SCG2, 1 peptide), ProSAAS (PCSKN1, 6 peptides), EGF-containing fibulin-like extracellular matrix protein 1 (FBLN3, 1 peptide), and Apolipoprotein C2 (APOC2, 2 peptides). Interestingly, CH3L1 was the only protein with no inconsistency in all identified peptides, since all were found to increase in AD (Table 5).
Table 5.
Peptide sequences from the proteins altered in AD CSF proteomic studies
Peptide sequence with the same pattern of expression (toward the same direction) at least in 2 or 3 (bolditalic proteins) independent studies
*Peptide sequences identified which show changes between AD and control samples supported by statistical analysis
Discussion
In this comparative analysis we have compiled data from proteomic studies performed on human CSF samples obtained from AD and healthy individuals in order to construct a database of proteins and peptides that constitute the most reliable CSF biomarkers associated with an AD diagnosis. When comparing our hit proteins with the recently published CSF proteomics analysis performed by Wesenhagen et al., numerous differences are evident. An important distinction may be the use of different inclusion criteria in the two analyses. In order to generate a more accurate and reliable database, we exclusively took into consideration those proteins which were shown to be statistically significantly different between AD and control samples, according to each study-specific cut-off point. Therefore, our analysis relies exclusively on the basis of significant data, including hit distribution, direction change and significance, which could account for the discrepancies observed with Wesenhagen et al. We have also generated a novel database of peptides according to the proteomic findings that reveals the most consistently altered tryptic peptide biomarkers of AD within a given protein.
We initially identified APOE, TTHY, Osteopontin (OSTP) and Cystatin-C (CYTC) which are among the most abundant proteins in normal CSF [18] as the protein species whose levels change the most often in the context of AD. However, these 5 proteins appear both as increased and decreased in different proteomic studies. Although the number of studies included in Wesenhagen analysis was lower than ours, these findings are in agreement with their study [13], and prompts us to suggest that they cannot be considered as reliable CSF biomarkers of AD. While it is likely that heterogeneity in response direction may reflect irrelevant physiological or environmental factors, it is also interesting to speculate that this heterogeneity reflects unknown endophenotypes in AD. It is also possible that the changes in these 5 proteins provide an indication of the profound general protein dysregulation that occurs during AD progression. Additionally, our study reveals new information regarding proteins consistently altered in AD, including SPARC (SPRC), Kininogen-1 (KNG1) or Cell surface glycoprotein MUC18 (MUC18), whose role as potential CSF biomarkers for AD has not been investigated. These proteins are discussed below.
Among the most consistently down- and up-regulated AD proteomic biomarkers, our hit proteins with the highest occurrence are VGF and CH3L1, respectively. Both can be considered high quality CSF biomarkers, as they represent abundant proteins which consistently change in the same direction in 14 and 6 AD proteomics studies, respectively. VGF is a member of the granin family of proteins [19] previously proposed as a good marker for AD [10, 13], and a decrease in VGF-derived peptides in AD has been confirmed using other experimental approaches [14, 20, 21]. Interestingly, the 9 peptides that appear as reliable markers for the VGF down-regulation in AD correspond to both N- and C-terminal sequences, suggesting that the synthesis of the precursor protein itself is repressed in AD and not a specific derived peptide.
The levels of CH3L1, encoded by the CHI3L1 gene, were also found to be increased in all 6 proteomics studies in which it was identified, just as reported by Wesenhagen et al. [22]. We propose that CH3L1, which is expressed in the CNS by microglia and astrocytes, constitutes one of the most interesting potential biomarkers, mainly because its physiological role in brain remains speculative. Increased expression of CH3L1 is found in human brains from pathologically confirmed AD individuals, implicating CH3L1 in the neuroinflammatory response to Aβ deposition [23]. Moreover, 2 independent ELISA studies, conducted to evaluate the role of CH3L1 as a CSF biomarker, have confirmed the observation of increased levels of CH3L1 in AD patients [24, 25]. Given that CH3L1 has been proposed as a common neuroinflammatory biomarker of other neurodegenerative diseases, [26, 27], the fact that 2 peptides (see Table 5) were consistently changed in AD studies assumes increased relevance. Although the presence of these peptides may arise as a consequence of intrinsic properties which render them highly identifiable by mass spectrometry, further characterization of these 2 peptides as AD biomarkers among other neurodegenerative diseases deserves investigation.
Proteins consistently increased in AD CSF
In addition to CH3L1, other proteins were observed to increase in CSF samples from AD patients. Among those that stand out due to their consistency within the various studies (bolditalic proteins in Table 3) we found Complement C3 (CO3, 6 studies), Alpha-2-macroglobulin (A2MG, 4 studies), FIBB (3 studies), Pyruvate kinase (KPYM, 3 studies) and Spondin-1 (SPON1, 3 studies). This set of proteins has previously been reported to increase in AD CSF [13]. However, our analysis, which includes only significant data, permits the identification of proteins with consistently increased levels in AD CSF whose potential as reliable AD biomarkers was previously overlooked. For example, Clusterin (CLUS) was found to be increased in AD in 6 out of 7 studies. This chaperone protein is involved in lipid transport and metabolism and is produced and secreted predominantly by astrocytes within the CNS [28]. Various studies of CSF from AD patients, most using an ELISA approach, have proposed a role for CLUS as a potential AD biomarker [reviewed in [24]]; our study now provides additional support for this idea.
Finally, SPARC, also known as Osteonectin, was found to be consistently upregulated in CSF from AD patients (3 studies), which validates its novel utility as a reliable CSF biomarker. In brain, its expression is restricted to microglia and subcortical astrocytes, and a role for SPRC has been suggested in neuroinflammation [29, 30]. Specifically, high levels of SPRC have been shown in AD brain wherein it colocalizes to Aβ protein depots. It has been proposed that SPRC contributes to cerebral inflammation and subsequent tissue repair [31].
Proteins consistently reduced in AD CSF
Our analysis of proteins consistently reduced in AD identified 19 proteins representing the most consistent biomarkers (bolditalic proteins in Table 4). With our experimental approach, we identified 3 proteins previously reported by Wesenhagen et al., Neurexin 1A (NRX1A), APOC2 and FBLN3. In our study, NRX1A [and Neurexin-2 (NRX2A) and Neurexin-3-beta (NRX3B) to a lesser degree] appears consistently reduced in AD CSF across all of the 4 independent studies in which it was identified. These brain-specific proteins participate in synapse formation, plasticity and stability [32]. A recently published targeted proteomic study using Selected Reaction Monitoring, which monitors neurexin levels in control and preclinical AD patients, reinforces the data reported here by showing reduced levels of NRX2A and NRX3B in preclinical AD CSF [33]. Overall, the reduced levels of neurexins observed in CSF at the earliest preclinical AD stages support the idea of diminished synaptic density during AD progression. We also observed a downregulation of APOC2 in CSF of AD patients in 3 independent studies, again strengthening the notion that a change in lipid metabolism can be potentially related to cognitive status in AD. Supporting this idea, our study found Apolipoprotein A1 (APOA1) as consistently reduced in AD CSF (7 out of 8 studies). Considering that APOA1 is among the most abundant proteins within human CSF [18], the use of APOA1 as biomarker of AD progression is especially relevant to practical biomarker identification, as its reduction in the context of AD is easily measurable.
Finally, FBLN3 codified by the gene EFEMP1, is a glycoprotein associated with the extracellular matrix which is involved in cell proliferation and migration [34]. Although no direct evidence as to a specific role for FBLN3 in AD has been reported, FBLN3 has recently been described as an amyloidogenic protein [35]. FBLN3 was observed to be consistently downregulated in CSF from AD patients (3 studies). Interestingly, only one tryptic peptide (QTSPVSAMLVLVK) was observed to be down-regulated in all 3 studies, thus highlighting this peptide as the most relevant hit for the characterization of AD status.
Together with VGF, the members of the granin family Chromogranin-A (CMGA), Secretogranin-1 (SCG1), SCG2, Secretogranin-3 (SCG3) and PCSK1N were found to be consistently decreased in our study. CMGA has been the most frequently detected granin in proteomic CSF studies (7 studies), and this protein, together with SCG1 (found to be decreased in 4 out of 5 studies) represent the most abundant granins in human CSF. Immunoreactivity for CMGA, SCG1 and SCG2 has been observed in amyloid plaques of post-mortem brains from AD patients [36–39]. Taken together, the proteomic studies gathered in this review support the role of CMGA as a reliable CSF biomarker. It is especially relevant that the most consistent tryptic peptides of all proteins that decrease in AD correspond to granins (VGF, CMGA, SCG1, SCG2 SCG3, and PCSK1N). Their acidic isoelectric point and the presence of multiple dibasic cleavage sites could potentially favor their detection by MS-spectrometry, which might constitute an assay advantage, since a panel of these peptides could be designed as a direct tool for a fast spectrometric characterization of AD patients.
The granin family member PCSK1N, known as proSAAS, is a protein produced almost exclusively by neurons and endocrine cells, and was reduced in 6 out of the 8 AD proteomic studies where it was identified. Reduced PCSK1N levels in CSF may be related to increased brain retention of this protein within plaques and other aggregates, as previously observed [40–42]. In agreement with this idea, a recent transcriptomic study [43] found increased PCSK1N expression during AD progression. Interestingly, among all of the peptides included in our peptidomic study, the proSAAS peptide ETPAPQVPARRLLPP was the most consistent finding and corresponds to the C-terminal sequence known as BigLEN (LETPAPQVPARRLLPP). There is a consistent lack of the N-terminal Leucine on the retrieved peptides [15, 44–46] which suggests that this peptide corresponds to a biological fragment of proSAAS in CSF not resulting from tryptic cleavage, emphasizing the importance of this modified BigPEN peptide as a possible direct biomarker of AD in CSF.
Interestingly, both Neuronal pentraxin-1 (NPTX1) and Neuronal pentraxin receptor (NPTXR) showed consistent reduction (4 and 4 out of 5 studies, respectively). These proteins have been previously implicated in AD [47]. Although NPTX1 has been primarily considered as a plasma biomarker [48], the current analysis supports its role as a reliable CSF AD biomarker. As this protein likely constitutes a protein associated with both neuronal degradation and synaptic loss [49, 50], future work will be needed to determine the specificity of NPTX1 for AD versus other neurodegenerative diseases. The potential role of the NPTXR as a prognostic biomarker for AD has recently been studied by others [20, 51]. Begcevic et al., observed that CSF NPTXR levels decrease with the severity of AD [51], thereby supporting the significance of NPTXR as a valuable CSF biomarker for specific stages of AD progression. It is important to point out that from all proteomic studies included in our analysis, NPTX1 and NPTXR were identified together in 2 studies [52] which reinforces the importance of data-mining efforts to identify and consolidate reliable biomarkers of neurodegeneration.
MUC18, an adhesion molecule encoded by the MCAM-CD14 gene, was consistently reduced in AD CSF in 4 independent proteomic studies. We believe this hit is especially interesting since this protein has not been previously linked to the disease. MUC18 is expressed by a subpopulation of IL-17-secreting CD4 + and CD8 + human T cells (Th17 and Tc17 cells, respectively) [53–55]. Chronic neuroinflammation is a phenomenon commonly observed in AD [56], and various Th cell lineages, including Th1, Th17 and regulatory T cells, appear to play a complex role in AD-associated neurodegeneration [57–62]. In order to explain the significance of reduced MUC18 levels in AD CSF, we suggest that further study of the role of MUC18 in AD inflammation is important.
The Retinol-binding protein 4 (RBP4), found to be reduced in 4 out of 5 studies, can circulate as an adipokine, and is related to insulin metabolism and retinoic acid signalling, both AD-associated processes [63, 64]. Additionally, RBP4 binds to TTHY, a protein that is believed to modulate Aβ levels by transporting Aβ from brain to the periphery [65]. Studies in patients have shown reduced levels of RBP4 in AD [66–68]. Indeed, this decrease is associated with cognitive decline, suggesting that RBP4 might be a biomarker for AD progression [66–68]. Nevertheless, it has been recently shown that RBP4 levels are not altered in preclinical AD CSF samples [63], implying that RBP4 may not be a good biomarker at preclinical stages.
The Amyloid-like protein 1 (APLP1), a member of the APP family, was found to consistently decrease in 3 independent studies. This neuron-specific protein [69] is involved in the maintenance of dendritic spines and basal synaptic transmission [70]. APLP1 is a γ-secretase substrate [71], thereby, secreted APLP1 fragments might be of especial interest to investigate γ-secretase cleavage products in AD.
The actin-binding protein Gelsolin (GELS), which was consistently decreased in AD in 3 independent studies, has already been implicated in AD [72]. GELS specifically binds to Aβ, inhibits its aggregation and protects cells from Aβ-induced apoptosis [72]. Indeed, its role as a potential therapeutic strategy for AD treatment is currently being evaluated [73, 74].
The essential component of the coagulation pathway KNG1 [75], was consistently reduced in AD CSF in 3 independent proteomic studies. KNG1 is particularly attractive for further investigation since a direct relation between KNG1 and AD has not yet been reported. However, the only study which investigated KNG1 in AD (since KNG1 polymorphisms were shown to be associated with hypertension, and thereby, they hypothesized that could be cause for AD progression) showed that KNG1 polymorphisms are not correlated with the incidence of late onset AD in a 201 patient cohort [76].
Metallothionein-3 (MT3) was found to be consistently reduced in AD CSF (3 studies). This protein regulates CNS Cu2+ and Zn2+ transport and storage and inhibits the toxicity of these metals, thus representing a major component of metal homeostasis [77]; this protein thus may play an important role in AD progression.
Finally, Superoxide dismutase (SODC), which neutralizes superoxide oxygen radicals to hydrogen peroxide and molecular oxygen inside the cells [78], appeared to be consistently downregulated in CSF from AD patients (3 studies). Several studies have highlighted the role of SODC deficiency in the acceleration of Aβ oligomerization, neuronal inflammation, and memory impairment in AD [79, 80], thus establishing SODC as an important marker in the etiopathogenesis of this disease.
Given that we only included significant data in our study, the finding that the transcription factor NFE2L2/NRF2 was identified as the most important upstream regulator predicted to be repressed in AD patients is especially interesting. A reduction in this transcription factor could potentially result in several of the observed proteomics changes in AD [specifically underlying the reduced levels of Angiotensinogen (AGT), Complement factor D (CFAD), SCG1, Collagen alpha-1(I) chain (CO1A1), Peptidyl-prolyl cis–trans isomerase B (PPIB), SCG2, SODC and Extracellular superoxide dismutase (SODE) expression]. In this regard, it has been recently shown that NRF2 deficiency replicates the transcriptomic changes seen in Alzheimer’s patients and worsens APP and tau pathology [81]. Interestingly, we also identified STK11 [also known as liver kinase B1 (LKB1)], as an upstream regulator predicted to be activated in AD patients. STK11 has been described as a multifunctional master kinase which is involved in a variety of functions in the nervous system such as maintaining axon integrity, neural development, neural homeostasis, neuronal survival, and control of neurotransmitter release [82]. Indeed, its deletion leads to axon degeneration [83]. Additionally, dysregulation of STK11 has been shown to contribute to Aβ accumulation and tauopathy AD-associated [84, 85].
Conclusions
Data-mining of CSF proteomic studies from individuals suffering from AD retrieves and consolidates valuable information as to proteins and peptides clearly altered in AD, information that may be useful in the constitution of a screening panel to increase the accuracy of AD diagnosis. From a methodological perspective, there are still several challenges to solve, as it is unclear which proteins or specific peptides can readily be measured in patient samples; although ELISA would work for several candidates, the low abundance of other candidates (detected exclusively by mass spectrometry) renders their quantification directly in CSF more complex. Nevertheless, the evaluation of information regarding specific peptides has not been previously performed, and the data we present herein provides direct targets towards establishing the status of a given protein in AD. These data should support the development of a peptide array with verified biomarker candidates that could move into clinical practice, even within the next few years, to fulfill the need for early detection in order to better combat this widespread neurodegenerative disease. Names of the genes and proteins cited in the text are described in Tables 3, 4 and 5.
Supplementary information
Additional file 1: Table S1. Information regarding all the proteins extracted from the proteomic studies. The proteins identified in each article and their abbreviations are shown. Δ, proteins not recognized by DAVID bioinformatics resources, Π qualitative but no quantitative data.
Additional file 2: Table S2. Proteins identified with significant changes between AD and control samples. The number of studies that describe these changes are shown.
Additional file 3: Table S3. Proteins showing inconsistent changes in AD samples. $ proteins which are abundant in the CSF of healthy individuals [18]. Component CO4A and CO4B are indicated as A and B respectively.
Additional file 4: Table S4. Information regarding all peptides extracted from the proteomic studies. M.W. [Da]: Molecular weight obtained from each article. Theo. M.W. [Da]: theoretical molecular weight obtained from Compute pI/Mw tool (https://web.expasy.org/compute_pi/).
Additional file 5: Table S5. Peptide sequences showing inconsistent changes in CSF studies. Peptide sequences with different pattern of expression across independent studies or inconsistent direction change to that observed by proteomics in relation to AD pathology. *Peptide sequences with changes supported by statistical analysis between AD and control samples.
Additional file 6: Figure S1. Most frequently detected peptides within 13 protein sequences. Peptide sequences with the same pattern of expression in at least 2 (bold red) or 3 (bold red underlined) independent studies and maintain consistent direction change to that observed by proteomics in relation to the AD pathology.
Acknowledgements
Not applicable.
Abbreviations
- AD
Alzheimer´s disease
- ADP
Adenosine diphosphate
- AR
Age range
- Aβ42
Amyloid-beta 1-42
- BBB
Blood-brain barrier
- CDR
Clinical dementia rating scale
- CE-MS
Capillary electrophoresis mass spectrometry
- CERAD
Consortium to Establish a Registry for Alzheimer’s Disease
- CNS
Central nervous system
- CSF
Cerebrospinal fluid
- CT
Computed tomography
- DAVID
Database for annotation, visualization and integrated discovery
- DA
Daltons
- DNA
Deoxyribonucleic acid
- DSM
Diagnostic and statistical
- ELISA
Enzyme-linked immunosorbent assay
- ESI
Electrospray ionization
- FAD
Familial alzheimer Disease
- IAPP
Islet amyloid polypeptide
- ICD-10
10th revision of the International Statistical Classification of Diseases and Related Health Problems
- ID
Identifier digital
- IWG-2
International Working Group-2 criteria
- IPA
Ingenuity pathway analysis
- LC–MS/MS
Liquid chromatography-tandem mass spectrometry
- MAP-RBM
Multi-analyte profiling technology platform- rules based medicine
- MCI
Mild cognitive impairment
- MCs
Mutation carriers
- MEDLINE
Medical literature analysis and retrieval system online
- MMSE
Mini-menta state exam
- MOCA
Montreal cognitive assessment
- MRI
Magnetic resonance imaging
- MRM
Multiple reaction monitoring
- mRNA
Messenger ribonucleic acid
- MS
Mass spectrometry
- NA
Not Applicable
- NCs
Mutation non-carriers
- NI
Not information
- NIA-AA
National Institute on Aging and Alzheimer’s Association
- NINCDS-ADRDA
National Institute of Neurological and Communicative Disorders and Stroke and the AD and Related Disorders Association
- PRM
Parallel reaction monitoring
- PSEN1
Presenilin 1
- p-tau
Phosphorylated tau
- RNA
Ribonucleic acid
- ROS
Reactive oxygen species
- RP-LC MS/MS
Reversed-phase LC–MS/MS
- SELDI-TOF-MS
Surface enhanced laser desorption/ionization time-of-flight mass spectrometry
- SMC
Subjective memory complaints
- TMT
Tandem mass tag
- T-tau
Total tau
- WB
Western blot
Authors’ contributions
All authors have made substantial contributions to the conception, drafting and editing of this work and have approved the final manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by a Grant from MINECO (SAF2016-79311-R) to Mario Durán-Prado. Yoana Rabanal-Ruiz is financed by FEDER (2018/D/LD/MC/8).
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Juan R. Peinado and Yoana Rabanal-Ruiz shared senior authorship
Contributor Information
Juan R. Peinado, Email: JuanRamon.Peinado@uclm.es
Yoana Rabanal-Ruiz, Email: Yoana.Rabanal@uclm.es.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12014-020-09276-9.
References
- 1.Scheltens P, Blennow K, Breteler MM, de Strooper B, Frisoni GB, Salloway S, et al. Alzheimer’s disease. Lancet. 2016;388(10043):505–517. doi: 10.1016/S0140-6736(15)01124-1. [DOI] [PubMed] [Google Scholar]
- 2.Levy B, Tsoy E, Gable S. Developing cognitive markers of alzheimer’s disease for primary care: implications for behavioral and global prevention. J Alzheimer’s Dis. 2016;54(4):1259–1272. doi: 10.3233/JAD-160309. [DOI] [PubMed] [Google Scholar]
- 3.Ledig C, Schuh A, Guerrero R, Heckemann RA, Rueckert D. Structural brain imaging in Alzheimer’s disease and mild cognitive impairment: biomarker analysis and shared morphometry database. Sci Rep. 2018;8(1):11258. doi: 10.1038/s41598-018-29295-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cellu Proteomics. 2002;1(11):845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 5.Baird AL, Westwood S, Lovestone S. Blood-based proteomic biomarkers of alzheimer’s disease pathology. Front Neurol. 2015;6:236. doi: 10.3389/fneur.2015.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blennow K, Zetterberg H. Biomarkers for Alzheimer’s disease: current status and prospects for the future. J Intern Med. 2018;284(6):643–663. doi: 10.1111/joim.12816. [DOI] [PubMed] [Google Scholar]
- 7.Blennow K. Cerebrospinal fluid protein biomarkers for Alzheimer’s disease. NeuroRx J Am Soc Exp NeuroTher. 2004;1(2):213–225. doi: 10.1602/neurorx.1.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenmann H. CSF biomarkers for amyloid and tau pathology in Alzheimer’s disease. J Mol Neurosci. 2012;47(1):1–14. doi: 10.1007/s12031-011-9665-5. [DOI] [PubMed] [Google Scholar]
- 9.Bjerke M, Engelborghs S. Cerebrospinal fluid biomarkers for early and differential alzheimer’s disease diagnosis. J Alzheimer’s Dis. 2018;62(3):1199–1209. doi: 10.3233/JAD-170680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Portelius E, Brinkmalm G, Pannee J, Zetterberg H, Blennow K, Dahlen R, et al. Proteomic studies of cerebrospinal fluid biomarkers of Alzheimer’s disease: an update. Exp Rev Proteomics. 2017;14(11):1007–1020. doi: 10.1080/14789450.2017.1384697. [DOI] [PubMed] [Google Scholar]
- 11.Olsson B, Lautner R, Andreasson U, Ohrfelt A, Portelius E, Bjerke M, et al. CSF and blood biomarkers for the diagnosis of Alzheimer’s disease: a systematic review and meta-analysis. Lancet Neurol. 2016;15(7):673–684. doi: 10.1016/S1474-4422(16)00070-3. [DOI] [PubMed] [Google Scholar]
- 12.Bastos P, Ferreira R, Manadas B, Moreira PI, Vitorino R. Insights into the human brain proteome: disclosing the biological meaning of protein networks in cerebrospinal fluid. Crit Rev Clin Lab Sci. 2017;54(3):185–204. doi: 10.1080/10408363.2017.1299682. [DOI] [PubMed] [Google Scholar]
- 13.Wesenhagen KEJ, Teunissen CE, Visser PJ, Tijms BM. Cerebrospinal fluid proteomics and biological heterogeneity in Alzheimer’s disease: a literature review. Crit Rev Clin Lab Sci. 2019;57:1–13. doi: 10.1080/10408363.2019.1670613. [DOI] [PubMed] [Google Scholar]
- 14.Brinkmalm G, Sjodin S, Simonsen AH, Hasselbalch SG, Zetterberg H, Brinkmalm A, et al. A parallel reaction monitoring mass spectrometric method for analysis of potential csf biomarkers for alzheimer’s disease. Proteomics Clin Appl. 2018;12(1):1700131. doi: 10.1002/prca.201700131. [DOI] [PubMed] [Google Scholar]
- 15.Hölttä M, Minthon L, Hansson O, Holmen-Larsson J, Pike I, Ward M, et al. An integrated workflow for multiplex CSF proteomics and peptidomics-identification of candidate cerebrospinal fluid biomarkers of Alzheimer’s disease. J Proteome Res. 2015;14(2):654–663. doi: 10.1021/pr501076j. [DOI] [PubMed] [Google Scholar]
- 16.Khoonsari PE, Haggmark A, Lonnberg M, Mikus M, Kilander L, Lannfelt L, et al. Analysis of the cerebrospinal fluid proteome in alzheimer’s disease. PLoS ONE. 2016;11(3):e0150672. doi: 10.1371/journal.pone.0150672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galozzi S, Marcus K, Barkovits K. Amyloid-beta as a biomarker for Alzheimer’s disease: quantification methods in body fluids. Exp Rev Proteomics. 2015;12(4):343–354. doi: 10.1586/14789450.2015.1065183. [DOI] [PubMed] [Google Scholar]
- 18.Schilde LM, Kosters S, Steinbach S, Schork K, Eisenacher M, Galozzi S, et al. Protein variability in cerebrospinal fluid and its possible implications for neurological protein biomarker research. PLoS ONE. 2018;13(11):e0206478. doi: 10.1371/journal.pone.0206478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartolomucci A, Possenti R, Mahata SK, Fischer-Colbrie R, Loh YP, Salton SR. The extended granin family: structure, function, and biomedical implications. Endocr Rev. 2011;32(6):755–797. doi: 10.1210/er.2010-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hendrickson RC, Lee AY, Song Q, Liaw A, Wiener M, Paweletz CP, et al. High resolution discovery proteomics reveals candidate disease progression markers of alzheimer’s disease in human cerebrospinal fluid. PLoS ONE. 2015;10(8):e0135365. doi: 10.1371/journal.pone.0135365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Llano DA, Devanarayan P, Devanarayan V. VGF in cerebrospinal fluid combined with conventional biomarkers enhances prediction of conversion from MCI to AD. Alzheimer Dis Assoc Disord. 2019;33(4):307–314. doi: 10.1097/WAD.0000000000000328. [DOI] [PubMed] [Google Scholar]
- 22.Eipper BA, Stoffers DA, Mains RE. The biosynthesis of neuropeptides: peptide alpha-amidation. Annu Rev Neurosci. 1992;15:57–85. doi: 10.1146/annurev.ne.15.030192.000421. [DOI] [PubMed] [Google Scholar]
- 23.Bonneh-Barkay D, Wang G, Starkey A, Hamilton RL, Wiley CA. In vivo CHI3L1 (YKL-40) expression in astrocytes in acute and chronic neurological diseases. J Neuroinflamm. 2010;7:34. doi: 10.1186/1742-2094-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dhiman K, Blennow K, Zetterberg H, Martins RN, Gupta VB. Cerebrospinal fluid biomarkers for understanding multiple aspects of Alzheimer’s disease pathogenesis. Cell Mol Life Sci. 2019;76(10):1833–1863. doi: 10.1007/s00018-019-03040-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosen C, Andersson CH, Andreasson U, Molinuevo JL, Bjerke M, Rami L, et al. Increased levels of chitotriosidase and YKL-40 in cerebrospinal fluid from patients with alzheimer’s disease. Demen Geriatr Cogn Disord Extra. 2014;4(2):297–304. doi: 10.1159/000362164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baldacci F, Lista S, Palermo G, Giorgi FS, Vergallo A, Hampel H. The neuroinflammatory biomarker YKL-40 for neurodegenerative diseases: advances in development. Exp Rev Proteomics. 2019;16(7):593–600. doi: 10.1080/14789450.2019.1628643. [DOI] [PubMed] [Google Scholar]
- 27.Llorens F, Thune K, Tahir W, Kanata E, Diaz-Lucena D, Xanthopoulos K, et al. YKL-40 in the brain and cerebrospinal fluid of neurodegenerative dementias. Mol Neurodegener. 2017;12(1):83. doi: 10.1186/s13024-017-0226-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyles JK, Pitas RE, Wilson E, Mahley RW, Taylor JM. Apolipoprotein E associated with astrocytic glia of the central nervous system and with nonmyelinating glia of the peripheral nervous system. J Clin Investig. 1985;76(4):1501–1513. doi: 10.1172/JCI112130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lloyd-Burton S, Roskams AJ. SPARC-like 1 (SC1) is a diversely expressed and developmentally regulated matricellular protein that does not compensate for the absence of SPARC in the CNS. J Comp Neurol. 2012;520(12):2575–2590. doi: 10.1002/cne.23029. [DOI] [PubMed] [Google Scholar]
- 30.Lloyd-Burton SM, York EM, Anwar MA, Vincent AJ, Roskams AJ. SPARC regulates microgliosis and functional recovery following cortical ischemia. J Neurosci. 2013;33(10):4468–4481. doi: 10.1523/JNEUROSCI.3585-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strunz M, Jarrell JT, Cohen DS, Rosin ER, Vanderburg CR, Huang X. Modulation of SPARC/Hevin proteins in alzheimer’s disease brain injury. J Alzheimer’s Dis. 2019;68(2):695–710. doi: 10.3233/JAD-181032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sindi IA, Tannenberg RK, Dodd PR. Role for the neurexin-neuroligin complex in Alzheimer’s disease. Neurobiol Aging. 2014;35(4):746–756. doi: 10.1016/j.neurobiolaging.2013.09.032. [DOI] [PubMed] [Google Scholar]
- 33.Lleo A, Nunez-Llaves R, Alcolea D, Chiva C, Balateu-Panos D, Colom-Cadena M, et al. Changes in synaptic proteins precede neurodegeneration markers in preclinical alzheimer’s disease cerebrospinal fluid. Mol Cell Proteomics. 2019;18(3):546–560. doi: 10.1074/mcp.RA118.001290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Marmorstein LY. Focus on molecules: fibulin-3 (EFEMP1) Exp Eye Res. 2010;90(3):374–375. doi: 10.1016/j.exer.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tasaki M, Ueda M, Hoshii Y, Mizukami M, Matsumoto S, Nakamura M, et al. A novel age-related venous amyloidosis derived from EGF-containing fibulin-like extracellular matrix protein 1. J Pathol. 2019;247(4):444–455. doi: 10.1002/path.5203. [DOI] [PubMed] [Google Scholar]
- 36.Kaufmann WA, Barnas U, Humpel C, Nowakowski K, DeCol C, Gurka P, et al. Synaptic loss reflected by secretoneurin-like immunoreactivity in the human hippocampus in Alzheimer’s disease. Eur J Neurosci. 1998;10(3):1084–1094. doi: 10.1046/j.1460-9568.1998.00121.x. [DOI] [PubMed] [Google Scholar]
- 37.Lechner T, Adlassnig C, Humpel C, Kaufmann WA, Maier H, Reinstadler-Kramer K, et al. Chromogranin peptides in Alzheimer’s disease. Exp Gerontol. 2004;39(1):101–113. doi: 10.1016/j.exger.2003.09.018. [DOI] [PubMed] [Google Scholar]
- 38.Marksteiner J, Kaufmann WA, Gurka P, Humpel C. Synaptic proteins in Alzheimer’s disease. J Mol Neurosci. 2002;18(1–2):53–63. doi: 10.1385/JMN:18:1-2:53. [DOI] [PubMed] [Google Scholar]
- 39.Willis M, Leitner I, Jellinger KA, Marksteiner J. Chromogranin peptides in brain diseases. J Neural Transm. 2011;118(5):727–735. doi: 10.1007/s00702-011-0648-z. [DOI] [PubMed] [Google Scholar]
- 40.Hoshino A, Helwig M, Rezaei S, Berridge C, Eriksen JL, Lindberg I. A novel function for proSAAS as an amyloid anti-aggregant in Alzheimer’s disease. J Neurochem. 2014;128(3):419–430. doi: 10.1111/jnc.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peinado JR, Sami F, Rajpurohit N, Lindberg I. Blockade of islet amyloid polypeptide fibrillation and cytotoxicity by the secretory chaperones 7B2 and proSAAS. FEBS Lett. 2013;587(21):3406–3411. doi: 10.1016/j.febslet.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wada M, Ren CH, Koyama S, Arawaka S, Kawakatsu S, Kimura H, et al. A human granin-like neuroendocrine peptide precursor (proSAAS) immunoreactivity in tau inclusions of Alzheimer’s disease and parkinsonism-dementia complex on Guam. Neurosci Lett. 2004;356(1):49–52. doi: 10.1016/j.neulet.2003.11.028. [DOI] [PubMed] [Google Scholar]
- 43.Mathys H, Davila-Velderrain J, Peng Z, Gao F, Mohammadi S, Young JZ, et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature. 2019;570(7761):332–337. doi: 10.1038/s41586-019-1195-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jahn H, Wittke S, Zurbig P, Raedler TJ, Arlt S, Kellmann M, et al. Peptide fingerprinting of Alzheimer’s disease in cerebrospinal fluid: identification and prospective evaluation of new synaptic biomarkers. PLoS ONE. 2011;6(10):e26540. doi: 10.1371/journal.pone.0026540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skillbäck T, Mattsson N, Hansson K, Mirgorodskaya E, Dahlén R, van der Flier W, et al. A novel quantification-driven proteomic strategy identifies an endogenous peptide of pleiotrophin as a new biomarker of Alzheimer’s disease. Sci Rep. 2017;7(1):13333. doi: 10.1038/s41598-017-13831-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J, Cunningham R, Zetterberg H, Asthana S, Carlsson C, Okonkwo O, et al. Label-free quantitative comparison of cerebrospinal fluid glycoproteins and endogenous peptides in subjects with Alzheimer’s disease, mild cognitive impairment, and healthy individuals. Proteomics Clin Appl. 2016;10(12):1225–1241. doi: 10.1002/prca.201600009. [DOI] [PubMed] [Google Scholar]
- 47.Swanson A, Wolf T, Sitzmann A, Willette AA. Neuroinflammation in Alzheimer’s disease: pleiotropic roles for cytokines and neuronal pentraxins. Behav Brain Res. 2018;347:49–56. doi: 10.1016/j.bbr.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma QL, Teng E, Zuo X, Jones M, Teter B, Zhao EY, et al. Neuronal pentraxin 1: a synaptic-derived plasma biomarker in Alzheimer’s disease. Neurobiol Dis. 2018;114:120–128. doi: 10.1016/j.nbd.2018.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choi YS, Hou S, Choe LH, Lee KH. Targeted human cerebrospinal fluid proteomics for the validation of multiple Alzheimer’s disease biomarker candidates. J Chromatogr B Anal Technol Biomed Life Sci. 2013;930:129–135. doi: 10.1016/j.jchromb.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perrin RJ, Craig-Schapiro R, Malone JP, Shah AR, Gilmore P, Davis AE, et al. Identification and validation of novel cerebrospinal fluid biomarkers for staging early Alzheimer’s disease. PLoS ONE. 2011;6(1):e16032. doi: 10.1371/journal.pone.0016032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Begcevic I, Tsolaki M, Brinc D, Brown M, Martinez-Morillo E, Lazarou I, et al. Neuronal pentraxin receptor-1 is a new cerebrospinal fluid biomarker of Alzheimer’s disease progression. F1000Research. 2018;7:1012. doi: 10.12688/f1000research.15095.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abdi F, Quinn JF, Jankovic J, McIntosh M, Leverenz JB, Peskind E, et al. Detection of biomarkers with a multiplex quantitative proteomic platform in cerebrospinal fluid of patients with neurodegenerative disorders. J Alzheimer’s Dis. 2006;9(3):293–348. doi: 10.3233/jad-2006-9309. [DOI] [PubMed] [Google Scholar]
- 53.Dagur PK, Biancotto A, Stansky E, Sen HN, Nussenblatt RB, McCoy JP. Secretion of interleukin-17 by CD8 + T cells expressing CD146 (MCAM) Clin Immunol. 2014;152(1–2):36–47. doi: 10.1016/j.clim.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dagur PK, Biancotto A, Wei L, Sen HN, Yao M, Strober W, et al. MCAM-expressing CD4(+) T cells in peripheral blood secrete IL-17A and are significantly elevated in inflammatory autoimmune diseases. J Autoimmun. 2011;37(4):319–327. doi: 10.1016/j.jaut.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Larochelle C, Lecuyer MA, Alvarez JI, Charabati M, Saint-Laurent O, Ghannam S, et al. Melanoma cell adhesion molecule-positive CD8 T lymphocytes mediate central nervous system inflammation. Ann Neurol. 2015;78(1):39–53. doi: 10.1002/ana.24415. [DOI] [PubMed] [Google Scholar]
- 56.Sommer A, Winner B, Prots I. The Trojan horse–neuroinflammatory impact of T cells in neurodegenerative diseases. Mol Neurodegener. 2017;12(1):78. doi: 10.1186/s13024-017-0222-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oberstein TJ, Taha L, Spitzer P, Hellstern J, Herrmann M, Kornhuber J, et al. Imbalance of circulating T(h)17 and regulatory T cells in alzheimer’s disease: a case control study. Front Immunol. 2018;9:1213. doi: 10.3389/fimmu.2018.01213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saresella M, Calabrese E, Marventano I, Piancone F, Gatti A, Alberoni M, et al. Increased activity of Th-17 and Th-9 lymphocytes and a skewing of the post-thymic differentiation pathway are seen in Alzheimer’s disease. Brain Behav Immun. 2011;25(3):539–547. doi: 10.1016/j.bbi.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 59.Zhang J, Ke KF, Liu Z, Qiu YH, Peng YP. Th17 cell-mediated neuroinflammation is involved in neurodegeneration of abeta1-42-induced Alzheimer’s disease model rats. PLoS ONE. 2013;8(10):e75786. doi: 10.1371/journal.pone.0075786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dansokho C, Ait Ahmed D, Aid S, Toly-Ndour C, Chaigneau T, Calle V, et al. Regulatory T cells delay disease progression in Alzheimer-like pathology. Brain Journal Neurol. 2016;139(Pt 4):1237–1251. doi: 10.1093/brain/awv408. [DOI] [PubMed] [Google Scholar]
- 61.Larbi A, Pawelec G, Witkowski JM, Schipper HM, Derhovanessian E, Goldeck D, et al. Dramatic shifts in circulating CD4 but not CD8 T cell subsets in mild Alzheimer’s disease. J Alzheimer’s Dis. 2009;17(1):91–103. doi: 10.3233/JAD-2009-1015. [DOI] [PubMed] [Google Scholar]
- 62.Saresella M, Calabrese E, Marventano I, Piancone F, Gatti A, Calvo MG, et al. PD1 negative and PD1 positive CD4 + T regulatory cells in mild cognitive impairment and Alzheimer’s disease. J Alzheimer’s Dis. 2010;21(3):927–938. doi: 10.3233/JAD-2010-091696. [DOI] [PubMed] [Google Scholar]
- 63.Ishii M, Iadecola C. Metabolic and non-cognitive manifestations of alzheimer’s disease: the hypothalamus as both culprit and target of pathology. Cell Metab. 2015;22(5):761–776. doi: 10.1016/j.cmet.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sodhi RK, Singh N. Retinoids as potential targets for Alzheimer’s disease. Pharmacol Biochem Behav. 2014;120:117–123. doi: 10.1016/j.pbb.2014.02.016. [DOI] [PubMed] [Google Scholar]
- 65.Buxbaum JN, Reixach N. Transthyretin: the servant of many masters. Cell Mol Life Sci. 2009;66(19):3095–3101. doi: 10.1007/s00018-009-0109-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jung SM, Lee K, Lee JW, Namkoong H, Kim HK, Kim S, et al. Both plasma retinol-binding protein and haptoglobin precursor allele 1 in CSF: candidate biomarkers for the progression of normal to mild cognitive impairment to Alzheimer’s disease. Neurosci Lett. 2008;436(2):153–157. doi: 10.1016/j.neulet.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 67.Maury CP, Teppo AM. Immunodetection of protein composition in cerebral amyloid extracts in Alzheimer’s disease: enrichment of retinol-binding protein. J Neurol Sci. 1987;80(2–3):221–228. doi: 10.1016/0022-510x(87)90156-0. [DOI] [PubMed] [Google Scholar]
- 68.Puchades M, Hansson SF, Nilsson CL, Andreasen N, Blennow K, Davidsson P. Proteomic studies of potential cerebrospinal fluid protein markers for Alzheimer’s disease. Brain Res Mol Brain Res. 2003;118(1–2):140–146. doi: 10.1016/j.molbrainres.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 69.Kim TW, Wu K, Xu JL, McAuliffe G, Tanzi RE, Wasco W, et al. Selective localization of amyloid precursor-like protein 1 in the cerebral cortex postsynaptic density. Brain Res Mol Brain Res. 1995;32(1):36–44. doi: 10.1016/0169-328x(95)00328-p. [DOI] [PubMed] [Google Scholar]
- 70.Schauenburg L, Liebsch F, Eravci M, Mayer MC, Weise C, Multhaup G. APLP1 is endoproteolytically cleaved by γ-secretase without previous ectodomain shedding. Scientific Reports. 2018;8(1):1916. doi: 10.1038/s41598-018-19530-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sjödin S, Andersson KKA, Mercken M, Zetterberg H, Borghys H, Blennow K, et al. APLP1 as a cerebrospinal fluid biomarker for γ-secretase modulator treatment. Alzheimer’s Res Ther. 2015;7(1):77. doi: 10.1186/s13195-015-0160-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chauhan V, Ji L, Chauhan A. Anti-amyloidogenic, anti-oxidant and anti-apoptotic role of gelsolin in Alzheimer’s disease. Biogerontology. 2008;9(6):381–389. doi: 10.1007/s10522-008-9169-z. [DOI] [PubMed] [Google Scholar]
- 73.Carro E. Gelsolin as therapeutic target in Alzheimer’s disease. Exp Opinion Ther Targets. 2010;14(6):585–592. doi: 10.1517/14728222.2010.488222. [DOI] [PubMed] [Google Scholar]
- 74.Ji L, Zhao X, Hua Z. Potential therapeutic implications of gelsolin in Alzheimer’s disease. J Alzheimer’s Dis. 2015;44(1):13–25. doi: 10.3233/JAD-141548. [DOI] [PubMed] [Google Scholar]
- 75.Brunel H, Massanet R, Martinez-Perez A, Ziyatdinov A, Martin-Fernandez L, Souto JC, et al. The central role of KNG1 gene as a genetic determinant of coagulation pathway-related traits: exploring metaphenotypes. PLoS ONE. 2016;11(12):e0167187. doi: 10.1371/journal.pone.0167187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Deng Y, Hou D, Tian M, Li W, Feng X, Yu Z. Relationship between the gene polymorphisms of kallikrein-kinin system and Alzheimer’s disease in a Hunan Han Chinese population. Int J Clin Exp Pathol. 2015;8(12):15550–15562. [PMC free article] [PubMed] [Google Scholar]
- 77.Vašák M, Meloni G. Mammalian metallothionein-3: new functional and structural insights. Int J Mol Sci. 2017;18(6):1117. doi: 10.3390/ijms18061117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Estacio SG, Leal SS, Cristovao JS, Faisca PF, Gomes CM. Calcium binding to gatekeeper residues flanking aggregation-prone segments underlies non-fibrillar amyloid traits in superoxide dismutase 1 (SOD1) Biochem Biophys Acta. 2015;1854(2):118–126. doi: 10.1016/j.bbapap.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 79.McLimans KE, Clark BE, Plagman A, Pappas C, Klinedinst B, Anatharam V, et al. Is cerebrospinal fluid superoxide dismutase 1 a biomarker of Tau but not amyloid-induced neurodegeneration in alzheimer’s disease? Antioxid Redox Signal. 2019;31(8):572–578. doi: 10.1089/ars.2019.7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Murakami K, Murata N, Noda Y, Tahara S, Kaneko T, Kinoshita N, et al. SOD1 (copper/zinc superoxide dismutase) deficiency drives amyloid beta protein oligomerization and memory loss in mouse model of Alzheimer disease. J Biol Chem. 2011;286(52):44557–44568. doi: 10.1074/jbc.M111.279208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rojo AI, Pajares M, Rada P, Nunez A, Nevado-Holgado AJ, Killik R, et al. NRF2 deficiency replicates transcriptomic changes in Alzheimer’s patients and worsens APP and TAU pathology. Redox Biol. 2017;13:444–451. doi: 10.1016/j.redox.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kuwako KI, Okano H. Versatile roles of LKB1 kinase signaling in neural development and homeostasis. Front Mol Neurosci. 2018;11:354. doi: 10.3389/fnmol.2018.00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sun G, Reynolds R, Leclerc I, Rutter GA. RIP2-mediated LKB1 deletion causes axon degeneration in the spinal cordand hind-limb paralysis. Dis Model Mech. 2011;4(2):193–202. doi: 10.1242/dmm.006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kodamullil AT, Younesi E, Naz M, Bagewadi S, Hofmann-Apitius M. Computable cause-and-effect models of healthy and Alzheimer’s disease states and their mechanistic differential analysis. Alzheimer’s Demen. 2015;11(11):1329–1339. doi: 10.1016/j.jalz.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 85.Wang JW, Imai Y, Lu B. Activation of PAR-1 kinase and stimulation of tau phosphorylation by diverse signals require the tumor suppressor protein LKB1. J Neurosci. 2007;27(3):574–581. doi: 10.1523/JNEUROSCI.5094-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Whelan CD, Mattsson N, Nagle MW, Vijayaraghavan S, Hyde C, Janelidze S, et al. Multiplex proteomics identifies novel CSF and plasma biomarkers of early Alzheimer's disease. Acta Neuropathol Commun. 2019;7(1):169. doi: 10.1186/s40478-019-0795-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Khoonsari PE, Shevchenko G, Herman S, Remnestal J, Giedraitis V, Brundin R, et al. Improved Differential Diagnosis of Alzheimer's Disease by Integrating ELISA and Mass Spectrometry-Based Cerebrospinal Fluid Biomarkers. J Alzheimers Dis. 2019;67(2):639–651. doi: 10.3233/JAD-180855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sathe G, Na CH, Renuse S, Madugundu AK, Albert M, Moghekar A, et al. Quantitative Proteomic Profiling of Cerebrospinal Fluid to Identify Candidate Biomarkers for Alzheimer's Disease. Proteomics Clin Appl. 2019;13(4):e1800105. doi: 10.1002/prca.201800105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Duits FH, Brinkmalm G, Teunissen CE, Brinkmalm A, Scheltens P, Van der Flier WM, et al. Synaptic proteins in CSF as potential novel biomarkers for prognosis in prodromal Alzheimer's disease. Alzheimers Res Ther. 2018;10(1):5. doi: 10.1186/s13195-017-0335-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dayon L, Nunez Galindo A, Wojcik J, Cominetti O, Corthesy J, Oikonomidi A, et al. Alzheimer disease pathology and the cerebrospinal fluid proteome. Alzheimers Res Ther. 2018;10(1):66. doi: 10.1186/s13195-018-0397-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Paterson RW, Heywood WE, Heslegrave AJ, Magdalinou NK, Andreasson U, Sirka E, et al. A targeted proteomic multiplex CSF assay identifies increased malate dehydrogenase and other neurodegenerative biomarkers in individuals with Alzheimer's disease pathology. Transl Psychiatry. 2016;6(11):e952. doi: 10.1038/tp.2016.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Remnestal J, Just D, Mitsios N, Fredolini C, Mulder J, Schwenk JM, et al. CSF profiling of the human brain enriched proteome reveals associations of neuromodulin and neurogranin to Alzheimer's disease. Proteomics Clin Appl. 2016;10(12):1242–1253. doi: 10.1002/prca.201500150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Di Domenico F, Pupo G, Giraldo E, Badia MC, Monllor P, Lloret A, et al. Oxidative signature of cerebrospinal fluid from mild cognitive impairment and Alzheimer disease patients. Free Radic Biol Med. 2016;91:1–9. doi: 10.1016/j.freeradbiomed.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 94.Heywood WE, Galimberti D, Bliss E, Sirka E, Paterson RW, Magdalinou NK, et al. Identification of novel CSF biomarkers for neurodegeneration and their validation by a high-throughput multiplexed targeted proteomic assay. Mol Neurodegener. 2015;10:64. doi: 10.1186/s13024-015-0059-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Spellman DS, Wildsmith KR, Honigberg LA, Tuefferd M, Baker D, Raghavan N, et al. Development and evaluation of a multiplexed mass spectrometry based assay for measuring candidate peptide biomarkers in Alzheimer's Disease Neuroimaging Initiative (ADNI) CSF. Proteomics Clin Appl. 2015;9(7–8):715–731. doi: 10.1002/prca.201400178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Leung YY, Toledo JB, Nefedov A, Polikar R, Raghavan N, Xie SX, et al. Identifying amyloid pathology-related cerebrospinal fluid biomarkers for Alzheimer's disease in a multicohort study. Alzheimers Dement (Amst). 2015;1(3):339–348. doi: 10.1016/j.dadm.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Khan W, Aguilar C, Kiddle SJ, Doyle O, Thambisetty M, Muehlboeck S, et al. A Subset of Cerebrospinal Fluid Proteins from a Multi-Analyte Panel Associated with Brain Atrophy, Disease Classification and Prediction in Alzheimer's Disease. PloS one. 2015;10(8):e0134368. doi: 10.1371/journal.pone.0134368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Oláh Z, Kalman J, Toth ME, Zvara A, Santha M, Ivitz E, et al. Proteomic analysis of cerebrospinal fluid in Alzheimer's disease: wanted dead or alive. Journal of Alzheimer's disease : JAD. 2015;44(4):1303–1312. doi: 10.3233/JAD-140141. [DOI] [PubMed] [Google Scholar]
- 99.Alzate O, Osorio C, DeKroon RM, Corcimaru A, Gunawardena HP. Differentially charged isoforms of apolipoprotein E from human blood are potential biomarkers of Alzheimer's disease. Alzheimers Res Ther. 2014;6(4):43. doi: 10.1186/alzrt273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wildsmith KR, Schauer SP, Smith AM, Arnott D, Zhu Y, Haznedar J, et al. Identification of longitudinally dynamic biomarkers in Alzheimer's disease cerebrospinal fluid by targeted proteomics. Mol Neurodegener. 2014;9:22. doi: 10.1186/1750-1326-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chakrabarti A, Chatterjee A, Sengupta MB, Chattopadhyay P, Mukhopadhyay D. Altered levels of amyloid precursor protein intracellular domain-interacting proteins in Alzheimer disease. Alzheimer Dis Assoc Disord. 2014;28(3):283–290. doi: 10.1097/WAD.0000000000000011. [DOI] [PubMed] [Google Scholar]
- 102.Wijte D, McDonnell LA, Balog CI, Bossers K, Deelder AM, Swaab DF, et al. A novel peptidomics approach to detect markers of Alzheimer's disease in cerebrospinal fluid. Methods. 2012;56(4):500–507. doi: 10.1016/j.ymeth.2012.03.018. [DOI] [PubMed] [Google Scholar]
- 103.Ringman JM, Schulman H, Becker C, Jones T, Bai Y, Immermann F, et al. Proteomic changes in cerebrospinal fluid of presymptomatic and affected persons carrying familial Alzheimer disease mutations. Arch Neurol. 2012;69(1):96–104. doi: 10.1001/archneurol.2011.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Manral P, Sharma P, Hariprasad G. Chandralekha, Tripathi M, Srinivasan A. Can apolipoproteins and complement factors be biomarkers of Alzheimer's disease? Curr Alzheimer Res. 2012;9(8):935–943. doi: 10.2174/156720512803251110. [DOI] [PubMed] [Google Scholar]
- 105.Vafadar-Isfahani B, Ball G, Coveney C, Lemetre C, Boocock D, Minthon L, et al. Identification of SPARC-like 1 protein as part of a biomarker panel for Alzheimer's disease in cerebrospinal fluid. J Alzheimers Dis. 2012;28(3):625–636. doi: 10.3233/JAD-2011-111505. [DOI] [PubMed] [Google Scholar]
- 106.Craig-Schapiro R, Kuhn M, Xiong C, Pickering EH, Liu J, Misko TP, et al. Multiplexed immunoassay panel identifies novel CSF biomarkers for Alzheimer's disease diagnosis and prognosis. PloS one. 2011;6(4):e18850. doi: 10.1371/journal.pone.0018850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hu WT, Chen-Plotkin A, Arnold SE, Grossman M, Clark CM, Shaw LM, et al. Novel CSF biomarkers for Alzheimer's disease and mild cognitive impairment. Acta neuropathologica. 2010;119(6):669–678. doi: 10.1007/s00401-010-0667-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Maarouf CL, Andacht TM, Kokjohn TA, Castano EM, Sue LI, Beach TG, et al. Proteomic analysis of Alzheimer's disease cerebrospinal fluid from neuropathologically diagnosed subjects. Curr Alzheimer Res. 2009;6(4):399–406. doi: 10.2174/156720509788929318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yin GN, Lee HW, Cho J-Y, Suk K. Neuronal pentraxin receptor in cerebrospinal fluid as a potential biomarker for neurodegenerative diseases. Brain Res. 2009;1265:158–170. doi: 10.1016/j.brainres.2009.01.058. [DOI] [PubMed] [Google Scholar]
- 110.Zhang J, Sokal I, Peskind ER, Quinn JF, Jankovic J, Kenney C, et al. CSF multianalyte profile distinguishes Alzheimer and Parkinson diseases. Am J Clin Pathol. 2008;129(4):526–529. doi: 10.1309/W01Y0B808EMEH12L. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Simonsen AH, McGuire J, Hansson O, Zetterberg H, Podust VN, Davies HA, et al. Novel panel of cerebrospinal fluid biomarkers for the prediction of progression to Alzheimer dementia in patients with mild cognitive impairment. Arch Neurol. 2007;64(3):366–70. doi: 10.1001/archneur.64.3.366. [DOI] [PubMed] [Google Scholar]
- 112.Hu Y, Hosseini A, Kauwe JS, Gross J, Cairns NJ, Goate AM, et al. Identification and validation of novel CSF biomarkers for early stages of Alzheimer's disease. Proteomics Clin Appl. 2007;1(11):1373–1384. doi: 10.1002/prca.200600999. [DOI] [PubMed] [Google Scholar]
- 113.Korolainen MA, Nyman TA, Nyyssonen P, Hartikainen ES, Pirttila T. Multiplexed proteomic analysis of oxidation and concentrations of cerebrospinal fluid proteins in Alzheimer disease. Clin Chem. 2007;53(4):657–665. doi: 10.1373/clinchem.2006.078014. [DOI] [PubMed] [Google Scholar]
- 114.Finehout EJ, Franck Z, Choe LH, Relkin N, Lee KH. Cerebrospinal fluid proteomic biomarkers for Alzheimer's disease. Ann Neurol. 2007;61(2):120–129. doi: 10.1002/ana.21038. [DOI] [PubMed] [Google Scholar]
- 115.Simonsen AH, McGuire J, Podust VN, Davies H, Minthon L, Skoog I, et al. Identification of a novel panel of cerebrospinal fluid biomarkers for Alzheimer's disease. Neurobiol Aging. 2008;29(7):961–968. doi: 10.1016/j.neurobiolaging.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 116.Castaño EM, Roher AE, Esh CL, Kokjohn TA, Beach T. Comparative proteomics of cerebrospinal fluid in neuropathologically-confirmed Alzheimer's disease and non-demented elderly subjects. Neurol Res. 2006;28(2):155–163. doi: 10.1179/016164106X98035. [DOI] [PubMed] [Google Scholar]
- 117.Selle H, Lamerz J, Buerger K, Dessauer A, Hager K, Hampel H, et al. Identification of novel biomarker candidates by differential peptidomics analysis of cerebrospinal fluid in Alzheimer's disease. Comb Chem High Throughput Screen. 2005;8(8):801–806. doi: 10.2174/138620705774962391. [DOI] [PubMed] [Google Scholar]
- 118.Zhang J, Goodlett DR, Quinn JF, Peskind E, Kaye JA, Zhou Y, et al. Quantitative proteomics of cerebrospinal fluid from patients with Alzheimer disease. J Alzheimers Dis. 2005;7(2):125–33. doi: 10.3233/jad-2005-7205. [DOI] [PubMed] [Google Scholar]
- 119.Carrette O, Demalte I, Scherl A, Yalkinoglu O, Corthals G, Burkhard P, et al. A panel of cerebrospinal fluid potential biomarkers for the diagnosis of Alzheimer's disease. Proteomics. 2003;3(8):1486–1494. doi: 10.1002/pmic.200300470. [DOI] [PubMed] [Google Scholar]
- 120.Davidsson P, Westman-Brinkmalm A, Nilsson CL, Lindbjer M, Paulson L, Andreasen N, et al. Proteome analysis of cerebrospinal fluid proteins in Alzheimer patients. Neuroreport. 2002;13(5):611–615. doi: 10.1097/00001756-200204160-00015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Information regarding all the proteins extracted from the proteomic studies. The proteins identified in each article and their abbreviations are shown. Δ, proteins not recognized by DAVID bioinformatics resources, Π qualitative but no quantitative data.
Additional file 2: Table S2. Proteins identified with significant changes between AD and control samples. The number of studies that describe these changes are shown.
Additional file 3: Table S3. Proteins showing inconsistent changes in AD samples. $ proteins which are abundant in the CSF of healthy individuals [18]. Component CO4A and CO4B are indicated as A and B respectively.
Additional file 4: Table S4. Information regarding all peptides extracted from the proteomic studies. M.W. [Da]: Molecular weight obtained from each article. Theo. M.W. [Da]: theoretical molecular weight obtained from Compute pI/Mw tool (https://web.expasy.org/compute_pi/).
Additional file 5: Table S5. Peptide sequences showing inconsistent changes in CSF studies. Peptide sequences with different pattern of expression across independent studies or inconsistent direction change to that observed by proteomics in relation to AD pathology. *Peptide sequences with changes supported by statistical analysis between AD and control samples.
Additional file 6: Figure S1. Most frequently detected peptides within 13 protein sequences. Peptide sequences with the same pattern of expression in at least 2 (bold red) or 3 (bold red underlined) independent studies and maintain consistent direction change to that observed by proteomics in relation to the AD pathology.
Data Availability Statement
Not applicable.