Abstract
Introduction
Vascular endothelial growth factor A (VEGF-A) is a chemokine that induces proliferation and migration of vascular endothelial cells and is essential for both physiological and pathological angiogenesis. It is known for its high heritability (> 60%) and involvement in most common morbidities, which makes it a potentially interesting biomarker. Large GWAS studies have already assessed polymorphisms related to VEGF-A. However, no previous research has provided epigenome-wide insight in regulation of VEGF-A.
Methods
VEGF-A concentrations of healthy participants from the STANISLAS Family Study (n = 201) were comprehensively assessed for association with DNA methylation. Genome-wide DNA methylation profiles were determined in whole blood DNA using the 450K Infinium BeadChip Array (Illumina). VEGF-A concentration in PBMC extracts was detected using a high-sensitivity multiplex Cytokine Array (Randox Laboratories, UK).
Results
Epigenome-wide association analysis identified 41 methylation sites significantly associated with VEGF-A concentrations derived from PBMC extracts. Twenty CpG sites within 13 chromosomes reached Holm-Bonferroni significance. Significant values ranged from P = 1.08 × 10−7 to P = 5.64 × 10−15.
Conclusion
This study exposed twenty significant CpG sites linking DNA methylation to VEGF-A concentration. Methylation detected in promoter regions, such as TPX2 and HAS-1, could explain previously reported associations with the VEGFA gene. Methylation may also help in the understanding of the regulatory mechanisms of other genes located in the vicinity of detected CpG sites.
Keywords: EWAS, VEGF, Methylation, Epigenetics
Background
Recent developments and discoveries in epigenetics provided new insights into disease regulation, among which exploration of DNA methylation has become the most intriguing [1]. DNA methylation forms 5-methylcytosine on the CpG (cytosine-phosphate-guanine) site of a genome and normally results in silencing of the gene that is encoded in the sequence [2]. This particularity was researched in various epigenome-wide methylation studies (EWAS), which managed to relate individual CpGs with cardiovascular diseases [3], cancer [4] and other pathologies [5, 6]. In some cases, CpGs significantly associated with a certain disease are found on genes known to be involved with the aforementioned pathology or in promoter regions controlling gene expression [7]. In many cases, associations with chromosomal positions of methylated sites and disease are not obvious. Intergenic regions with CpG islands are thus systematically studied to elucidate the role of methylation in genomic regions distant from protein-coding regions [8].
Vascular endothelial growth factor A (VEGF-A) is a myogenic protein that induces angiogenesis, endothelial cell proliferation and plays an important role in the regulation of vasculogenesis [9]. VEGF-A is involved in the pathogenesis of cardiovascular disease [10], as well as other chronic diseases such as cancer [11], type 2 diabetes [12], osteoporosis, osteoarthritis [13] and chronic obstructive pulmonary disease (COPD) [14]. Anti-VEGF medications containing humanized antibody that blocks angiogenesis by inhibiting VEGF-A have already entered the market to treat a number of cancers, such as colon cancer, lung cancer, glioblastoma and renal cell carcinoma, as well as age-related macular degeneration [15–17].
The involvement of VEGF-A in various diseases makes it a universal biomarker with great potential for patient stratification in personalized medicine. The precise understanding of its biological and genetic regulation is required to fully appreciate its clinical potential. In previous years, a major effort has resulted in the discovery of several genetic variants with strong effects on growth factors, in particular VEGF-A concentration, using well-powered genome-wide association studies (GWAS). Ten genome-wide significant VEGF-A-associated SNPs [18, 19] that explained more than 50% of its individual variability have been identified. VEGF-A concentration is highly heritable reaching > 60% as demonstrated in the STANISLAS Family Study (SFS) [20]. Previous research has not yet investigated the role of epigenetics, such as DNA methylation on VEGF-A concentration. Therefore, epigenetic regulation could explain the missing heritability components [21].
Epigenetics is the study of gene transcription, regulation and expression that are not directly caused by the alteration of the genomic DNA sequence. DNA methylation occurs mostly on cytosine residues positioned in CpG islands (high density of CG dinucleotides) within a promoter region, transcription start site (TSS), first or second exons of a gene, in an enhancer region, or upstream from genes with CpG island shores (2 kb) or CpG shelves (2–4 kb) [2]. Previous studies have shown that epigenetics plays an important role in the regulation of promoter regions of VEGFA [22, 23] and VEGFR genes [24, 25], but no previous research studies have performed an EWAS of VEGF-A concentration to determine the methylation sites responsible for the regulation of VEGFA. As VEGF-A plays a distinct role in the development of several chronic diseases, the discovery of its epigenetic regulation mechanisms may contribute to a better understanding of these disorders and contribute in the research of new therapeutic possibilities.
To this end, we performed an EWAS on VEGF-A concentrations, measured from PBMC extracts in a healthy population, in order to identify possible epigenetic mechanisms involved in VEGF-A regulation before the pathological onset of chronic disease. We performed a large in silico analysis to detect possible repeating patterns of CpG chromosomal positions that could explain the role of each individual CpG site in VEGFA regulation.
Results
In this investigation, we set out to explore links between genome-wide DNA methylation and PBMC extract VEGF-A levels, in a population of 201 healthy individuals from the SFS. The characteristics of the studied population are presented in Table 1. Genome-wide methylation profiling of bisulfite-converted genomic DNA was performed by Illumina HumanMethylation450 bead array (Illumina Inc., San Diego, CA, USA).
Table 1.
Population characteristics
| Mean | SD | Median [interquartile range] | |
|---|---|---|---|
| Age (years) | 28.3 | 14.8 | 33.8 [13.25–42.08] |
| Sex (male %) | 50.2 | - | - |
| VEGF-A (pg/mL) | 59.3 | 75.5 | 43.4 [23.67–66.45] |
| BMI (kg/m2) | 21.6 | 4.0 | 21.3 [18.58–24.41] |
| Neutrophils (108/l) | 53.77 | 9.14 | 53.6 [47.5–60.6] |
| Lymphocytes (108/l) | 36.01 | 8.46 | 36.4 [29.7–41.2] |
| Monocytes (108/l) | 6.22 | 2.45 | 5.6 [4.6–7.4] |
| Eosinophils (108/l) | 2.84 | 1.98 | 2.2 [1.4–3.8] |
| Basophils (108/l) | 0.64 | 0.39 | 0.6 [0.4–0.9] |
SD standard deviation, VEGF-A vascular endothelial growth factor A, BMI body mass index. Neutrophils, lymphocytes, monocytes, eosinophils and basophils represent mean individual blood cell counts of studied population
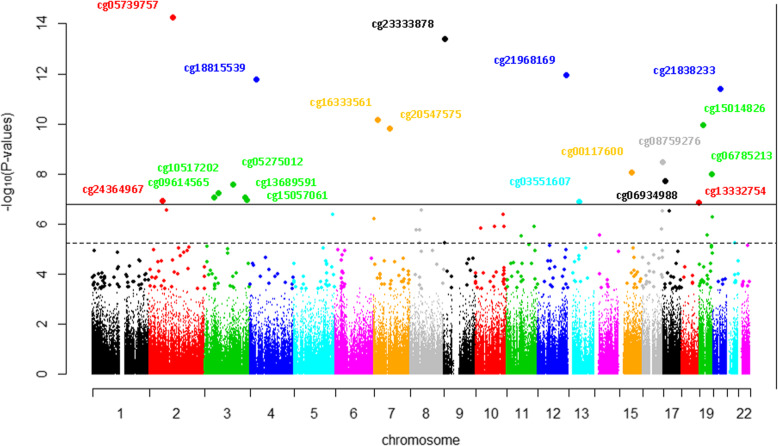
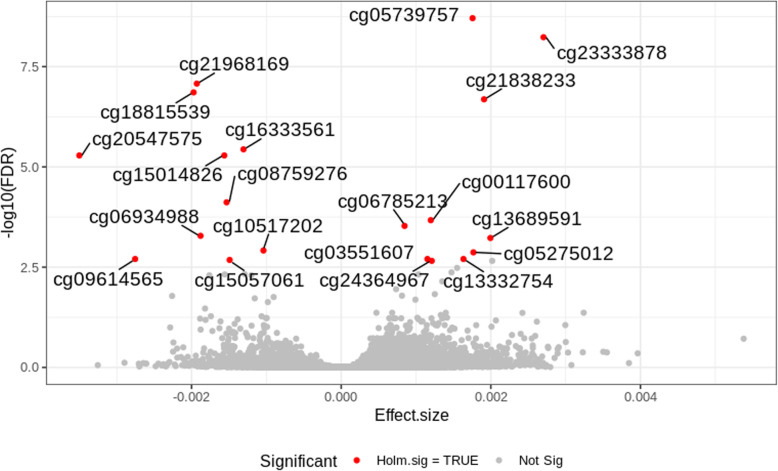
The results of our EWAS pointed out forty-one probes whose methylation was associated with VEGF-A concentration in cellular extracts (Sup. Table 1). Twenty probes were significant after Holm-Bonferroni adjustment (P < 1.6 × 10−7). The results for associations between DNA methylation and VEGF-A concentration are shown in Figs. 1 and 2. Manhattan plot shows that methylation is spread across different chromosomes. Chromosome 19 and chromosome 3 showed more significantly associated methylation sites than other chromosomes. The direction of all associations between DNA methylation and VEGF-A is presented with volcano plot.
Fig. 1.
Manhattan plot displaying adjusted P values of the association between methylation probes and VEGF-A concentration in cell extracts. The dotted line represents FDR value, and points above the full line indicate results that were significant after Holm-Bonferroni testing
Fig. 2.
Volcano plot showing the direction of all associations between DNA methylation and VEGF-A. CpG sites passing the multiple testing threshold are presented as red dots
Table 2 presents the list of twenty CpG sites that were significant after Holm-Bonferroni correction. Location and genes for CpG sites were retrieved from the annotation file of CpGassoc R package (CRAN). Significant P values have been detected; however, a small effect size was attributed to each CpG site. Six CpG sites did not have annotated genes. In silico analysis using the Ensembl browser was conducted to localize those CpG sites on the Human Genome (GRCh38.p10), in order to explore their genetic environment and define the nearest genes that could be impacted by CpG methylation. For each of the six CpG sites, the nearest upstream and downstream gene was identified. Results of the in silico analysis are presented in Table 3.
Table 2.
Novel associations between VEGFA levels and DNA methylation in a subset of the STANISLAS Family Study, after Holm-Bonferroni correction (P < 1.6 × 10−07). Data retrieved from the CpGassoc annotation file
| CpG Site | Chromosome | Gene | Location | Mean beta values | Effect size | P value |
|---|---|---|---|---|---|---|
| cg05739757 | 2q11.2 | RPL31 | TSS200 | 0.07 | 0.00176 | 5.64 × 10−15 |
| cg23333878 | 9p24.2 | GLIS3 | 5′UTR; 1stExon | 0.08 | 0.00270 | 3.86 × 10−14 |
| cg21838233 | 20q11.21 | TPX2 | 1stExon; 5′UTR | 0.04 | 0.00191 | 4.01 × 10−12 |
| cg18815539 | 4p15.2 | SEPSECS | 1stExon; 5′UTR | 0.04 | − 0.00197 | 1.70 × 10−12 |
| cg21968169 | 12q24.31 | LOC338799, SETD1B | TSS1500 | 0.09 | − 0.00189 | 1.11 × 10−12 |
| cg16333561 | 7q11.23 | ND | ND | 0.90 | − 0.00130 | 7.02 × 10−11 |
| cg20547575 | 7q11.22 | AUTS2 | Body | 0.03 | − 0.00349 | 1.52 × 10−10 |
| cg15014826 | 19p13.12 | ND | ND | 0.03 | − 0.00157 | 1.12 × 10−10 |
| cg00117600 | 15q21.3 | PIGB | 5′UTR; 1stExon | 0.04 | 0.00118 | 8.84 × 10−09 |
| cg08759276 | 16q24.1 | ND | ND | 0.78 | − 0.00153 | 3.31 × 10−09 |
| cg05275012 | 3p22.1 | ND | ND | 0.94 | 0.00176 | 8.81 × 10−08 |
| cg10517202 | 3q26.32 | ND | ND | 0.88 | − 0.00103 | 8.74 × 10−08 |
| cg09614565 | 3p14.3 | IL17RD | TSS200 | 0.02 | − 0.00283 | 5.69 × 10−08 |
| cg13689591 | 3q21.1–q21.2 | KALRN | Body | 0.88 | 0.00199 | 2.58 × 10−08 |
| cg06934988 | 17p13.1 | USP43 | TSS200 | 0.02 | − 0.00188 | 1.96 × 10−08 |
| cg06785213 | 19q13.4 | HAS1 | TSS200 | 0.20 | 0.00087 | 1.06 × 10−08 |
| cg13332754 | 18q22.3 | ND | ND | 0.95 | 0.00164 | 1.39 × 10−07 |
| cg03551607 | 13q14.2 | ESD | 5′UTR | 0.04 | 0.00115 | 1.34 × 10−07 |
| cg24364967 | 2p16.1 | CLHC1 | 5′UTR | 0.12 | 0.00122 | 1.22 × 10−07 |
| cg15057061 | 3q26.33 | SOX2OT | Body | 0.04 | − 0.00151 | 1.08 × 10−07 |
ND no data, Mean beta values mean value of the methylation occurring at the significant CpG sites (1 = methylated, 0 = non methylated), Effect size quantitative measure of the magnitude of the methylation effect on VEGF-A concentration
Table 3.
Supplementary information retrieved by in silico analysis for methylation sites without annotated gene and chromosome position
| CpG site (strand) | cg16333561 (−) | cg15014826 (+) | cg08759276 (−) | cg05275012 (−) | cg10517202 (−) | cg13332754 (−) | |
|---|---|---|---|---|---|---|---|
| Chromosome | 7p21.2 | 19p13.12 | 16q24.1 | 3p22.1 | 3q26.32 | 18q22.3 | |
| Nearest genomic feature (product/strand) | Upstream | AC011287.1 (novel lincRNA/+) | ZSWIM4 (protein coding/+) | AC009108.4 (unknown/+) | AC122683.1 (lincRNA/−) | LINC00578 (lincRNA/+) | TSHZ1 (protein coding/+) |
| Downstream | AC005019.2 (novel lincRNA/−) | AC020916.1 (lincRNA/−) | AC009108.2 (lincRNA/−) | HMGN2P24 (pseudogene/−) | RN7SKP52 (misc RNA/−) | TSHZ1 (protein coding/+) | |
| Location | 7:13803079–13803128 | 19:13833585–13833634 | 16:86610656–86610705 | 3:40619738–40619787 | 3:177469455–177469504 | 18:75290150–75290199 | |
| Regulatory features (T cells, natural killer cells or B cells) | 1 enhancer and 2 CTCF binding sites | 8 promoters | 4 promoters and 1 CTCF binding site | 1 promoter, 1 enhancer and 3 CTCF binding sites | 1 promoter; 1 CTCF and 1 enhancer | 1 promoter; 2 enhancers and 1 CTCF binding site | |
| Nearest coding gene (strand) | Upstream | ARL4A (+) | ZSWIM4 (+) | FOXL1 (+) | ZNF621 (+) | TBL1XR1 (−) | TSHZ1, ZADH2 (+) |
| Downstream | ETV1 (−) | NANOS3 (+) | C16orf95 (−) | CTNNB1 (+) | KCNMB2 (+) | SMIM21 (−) |
Furthermore, we have analysed Holm-Bonferroni significant CpG sites using two different principles. Firstly, we have studied the genes encoded in the position of the CpG, focusing on their function, relation to diseases and association with VEGF-A (Sup. Table 2). Altogether, there were 28 genes retrieved from the annotation results file of the EWAS analysis and identified from in silico analysis, which were encoded in the proximity of 20 significant CpG sites. None of the genes was directly related to the VEGF-A protein. In order to reveal common genetic pathways, genomic environment of all CpG sites was studied in detail using Ensembl browser. Results are presented in the supplementary data (Sup. Figures 1 and 2). Secondly, we have studied the genetic environment of each CpG schematically, to detect possible common patterns related to the location of CpG on the genome (Sup. Figure 1 and 2).
The overall aim of such analysis was to explore publicly available databases in order to detect common biological pathways between VEGF-A and the concerned genes. Some of the identified genes were previously associated with VEGF-A (Sup. Table 2), but those associations were not explained genetically. DNA methylation could provide an answer to these associations. Furthermore, the patterns in which methylation occurs on the genome could provide us new information on methylation function and pave the way for novel hypotheses that could explain the function of CpGs in non-coding regions or locations without direct relation to a specific phenotype.
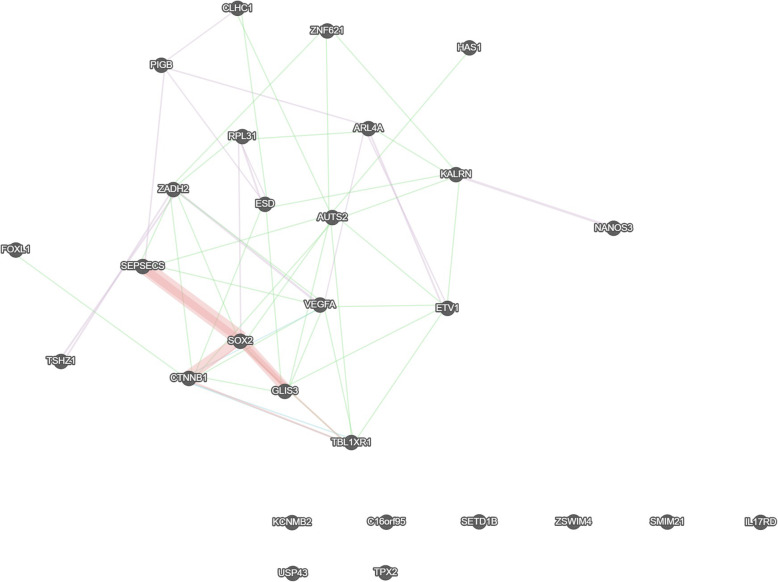
Interactions between annotated genes were further analysed with GeneMANIA app. GeneMANIA enables the construction of a composite gene-gene functional interaction network from a list of genes collected from many large, publicly available biological datasets [26]. A list of 28 genes has been input into GeneMANIA to research their possible relation to VEGFA: RPL31, GLIS3, TPX2, SEPSECS, LOC338799, SETD1B, AUTS2, PIGB, IL17RD, KALRN, USP43, HAS1, ESD, CLHC1, SOX2, ARL4A, ZSWIM4, FOXL1, ZNF621, TBL1XR1, TSHZ1, ZADH2, ETV1, NANOS3, C16orf95, CTNNB1, KCNMB2 and SMIM21. A gene network created as a result of this is presented in Fig. 3. Some of the input genes were not found by the bioinformatics tool and are thus not presented on the figure. Some genes, i.e. TPX, C16orf95, KCNMB2, ZSWIM4, SETD1B, SMIM21, IL17RD and USP43 were not related to any of genes input into GeneMania and are also not presented on the figure. Results revealed seven genes that were previously observed to have minor interactions with VEGF-A, namely ARL4A, ZADH2, SEPSECS, CTNNB1, TBL1XR1, GLIS3 and ETV1 [27]. ZADH2, SEPSECS, CTNNB1, TBL1XR1, GLIS3 and ETV1 had minor genetic interactions (presented with a green line in Fig. 3). ARL4A and ZADH2 had similar expression levels with VEGFA in gene expression studies (violate lines), as demonstrated in the analysis of the gene expression of the glioblastoma multiforme cancer cells [28] and study of steam cell populations [29], respectively. A blue line between CTNNB1 and VEGFA designates a common pathway, identified in pathway-based analysis of human functional protein network [30]. Most physical protein-protein interactions were detected within SEPSECS, SOX and AUTS2 genes (rose lines).
Fig. 3.
Network of genes related to CpG sites significantly associated with VEGF-A concentrations. Green lines present genetic associations, violet lines present co-expression, blue lines present common pathways and rose lines present physical interactions between connected genes
Significant results have been analysed using MethylGSA, a Bioconductor package to identify relevant physiological pathways. The analysis showed that CpG sites identified are involved in numerous molecular processes. The details are presented in the supplementary file (Sup. Figure 3).
Discussion
We have comprehensively explored the DNA methylome in a population of healthy individuals and have identified 41 methylation sites significantly associated with VEGF-A concentrations derived from PBMC extracts (Sup. Table 1). Significance levels after Bonferroni correction ranged from P = 1.08 × 10−7 (cg15057061) to P = 5.64 × 10−15 (cg05739757) for 20 CpG sites. Ten CpGs produced a positive effect size (range, 0.00087 to 0.0027), whereas the remaining ten methylation biomarkers possessed a negative effect size based on VEGF-A concentrations (range, 0.00349 to 0.00103). This study is the first epigenome-wide association study investigating the links between DNA methylation and VEGF-A concentration in a population of healthy individuals and the importance of its findings will be discussed below.
For all Holm-Bonferroni significant CpG sites, we looked for the nearest coding genes to understand the link between methylation of these genes and VEGF-A concentration. Fourteen genes were annotated in the result file obtained after performing EWAS (Table 2) using CpGassoc R package. To find the genomic features within the location of the rest six CpG sites, we have performed in silico analysis using Ensembl browser. None of the CpGs identified in this study was located within or near the VEGF gene family or its associated genes (Sup. Table 3). However, some CpGs identified in this study have been previously implicated in VEGF-related biological processes, such as cell proliferation, cell growth, angiogenesis and related diseases (Sup. Table 2). One such relation was found with TPX2 gene, significantly associated with cg21838233 (P = 4.01 × 10−12). The TPX2 gene is overexpressed in colon cancer, leading to vessel invasion and metastasis of colon cancer cells [31]. TPX2 gene silencing results in the inhibition of cell proliferation, and this effect has been linked to the down-regulation of the VEGFA expression [32]. Cg21838233 is located within the promoter region of TPX2, where methylation can play a crucial role in the control of gene expression [33]. The degree of methylation and location of the methylation site may directly affect the transcription and subsequent expression of the gene. Therefore, we could hypothesize that the expression of TPX2 is controlled by methylation of cg21838233, which in turn reflects further in the expression of VEGFA. However, such assumptions should be confirmed with other studies.
Another intriguing result was cg06785213 (P = 1.06 × 10−08) which was found in the vicinity of the HAS1 gene, 62 bp in 5′ upstream region. The HAS1 gene family encodes for hyaluronic acid (HA), which has an essential role in tissue development and homeostasis, and directs the initiation and progression of various pathological conditions, including angiogenesis [34]. Both proteins, HAS-1 and VEGF-A, have an important role in antigenic cascade [35, 36]. Thus, methylation in the vicinity of the HAS1 gene could play a distinct role in regulation of this process. Further research is required to confirm these hypotheses and elucidate new epigenetic pathways.
Five other genes in sup. Table 2 were related to angiogenic processes, namely ARL4A, ETV1, CTNNB1, TBL1XR1 and TSHZ1, located in the vicinity of CpGs detected in non-coding regions. In total, 6 out of 20 CpG sites from non-coding regions were significantly associated with VEGF-A (Table 3). Little is known whether such CpGs can have a real impact on genes in their proximity. However, it is known that it is not only the sequence in the immediate proximity of a region, such as promoter, that can influence gene activity [37]. DNA regions that were previously considered as “junk” DNA are now being considered as indispensable elements of regulation of gene expression [37]. Looking upstream and downstream of 6 annotated CpG sites, we have discovered that the most common genetic features in their immediate proximity were long non-coding RNAs (lncRNAs), which are emerging as regulators of gene expression in pathogenesis [38]. Cascade CpG-lincRNAs could take a part in regulation of coding genes (e.g. ARL4A, ETV1 or CTNNB1) and could thus impact on VEGF-A regulation. Certainly, all the above assumptions need to be verified. However, it is important that we consider all of the options that might, in the future, elucidate important regulation pathways. The small effect sizes of the significant CpG sites in this study showed that there was no mayor methylation site that would impact on VEGF-A concentration, but there is a sum of the small effect sizes that have a considerable epigenetic effect. Another interesting observation was the repartition of the effect sizes; half of them had positive values while other half had negative values, implying that VEGF-A regulation with methylation works in both directions, towards the increasing or decreasing of VEGF-A concentration (Fig. 4).
Fig. 4.
Ranking of the effect sizes of significant CpG sites in descending order
Methylation of CpGs, located on gene coding sections of DNA, has the potential to silence gene expression, which is especially important in disease development [39]. Abnormal patterns of DNA methylation have been observed in cancer, most commonly in CpG islands in gene promoter regions [40]. Schematic presentation of genetic regulatory elements in the vicinity of CpGs (Sup. Figure 1 and 2) demonstrated that most CpG sites significantly associated with VEGF-A concentrations were located within promoter regions, a regulatory region of DNA, where transcription is initiated. Normally, CpG islands within promoters are well characterized, but sometimes they are found in deserted areas [41]. However, there is evidence that some orphan CpG islands may initiate transcription and are likely to represent either uncharacterized promoters or promoters driving transcription of non-coding RNA [37]. CTCF binding sites present the second most common element highlighted. They enable CTCF zinc-finger transcription factor to bind and thus activate or repress the activity of various genes; moreover, they can act as enhancer-blocker [42]. Enhancers are the third regulatory elements found in the vicinity of CpGs. It enhances gene transcription by interactions with trans-acting factors, which allows specific control of gene activation, through chromatin looping of the intervening DNA [37].
We have noticed that regulatory elements are becoming less frequent with the distance from a CpG site. It means that identified CpG sites were located within regulatory vivid regions, indicating that CpGs could also be involved as an important element in regulation, without being located directly on the gene coding part.
All of the genes related to the 20 Holm-Bonferroni significant CpGs were also input into GeneMania to explore common genetic and physiological pathways. Seven genes were associated with VEGFA (ARL4A, ZADH2, SEPSECS, CTNNB1, TBL1XR1, GLIS3 and ETV1). For the genes highlighted in bold the relation with VEGF-A has been further confirmed with bibliographical research (Sup. Table 2).
A potential co-localisation of detected CpG sites and VEGF-A related genetic variants was also observed. Though 3 VEGF-A SNPs (i.e. rs10738760 and rs7043199, rs2639990) were localized at the same cytogenetic positions as 2 CpG sites (i.e. cg23333878 and cg13332754, respectively) considerable distances involved show that there is no co-localisation. The comparison of the epigenetic results with our previous GWAS [18, 19] shows that these new results shed additional light on the complexity of the mechanisms involving VEGF-A. The associations highlighted between VEGF-A and the CpG sites, as well as our previous VEGF GWAS [18, 19], all support the high heritability of VEGF-A, both at the genetic and epigenetic level.
Conclusion
We have found significant associations between DNA methylation and VEGF-A concentrations measured from the PBMCs cellular extracts. Significant CpG sites were located in vicinity of different coding genes, none of which was directly involved in VEGF-A regulation. Replication of these results in independent cohorts is important for their confirmation and could further provide new knowledge that could be used for the development of next-generation medications against VEGF-A-related diseases.
Methods
Populations
The SFS is a 10-year longitudinal survey with 3 visits at 5-year intervals, involving 1006 families from Vandoeuvre-lès-Nancy, France, first recruited between 1993 and 1995 [43, 44]. All subjects were of Caucasian origin, without the presence of chronic disorders, e.g. CVD or cancer, or previous personal history of such diseases. The study protocols were approved by the Comité Consultatif pour la Protection des Personnes dans la Recherche Biomédicale de Lorraine (Advisory Committee for the protection of people in biomedical research in Lorraine), and all subjects gave written informed consent for their participation in the study. All experiments were performed in accordance with relevant guidelines and regulations.
Data collection
Biological and clinical measurements were determined using appropriate, validated procedures. Blood samples were collected between 8 and 9 a.m. after overnight fasting. DNA was extracted by the Miller technique [45] and was stored at − 80 °C until further use. Body mass index (BMI) was calculated as weight (kg) divided by height2 (m2). All measurements were obtained by trained professionals.
Biological measurements
Isolation of PBMCs
Full blood from healthy donors was collected into sodium heparin tubes. Samples were homogenized with Hanks’ Balanced Salt Solution (SIGMA Aldrich, reference H6648) (VHanks = Vblood) and poured gently into a 15 mL tube with FicollTM paque (Sigma Aldrich, reference 17-1440-02) solution (VFicoll = VHanks + Vblood). The contents were centrifuged for 30 min at 300×g at room temperature.
High-density PBMC ring was retrieved and collected into a 15-mL tube, filled with Hanks Balanced Salt Solution and centrifuged for 10 min at 1000×g at room temperature (first washing). The supernatant was aspirated and 2 mL of Hanks Balanced Salt Solution was added. The tube was filled up to 15 mL with Hanks Balanced Salt Solution and centrifuged for a further 10 min at 1000×g at room temperature (second washing). The pellet was collected into an Eppendorf tube with 1 mL of Hanks Balanced Salt Solution. PBMCs populations were evaluated by microscopic observation after May-Grunwald-Giemsa staining and PBMCs concentration was normalized to 106 cells/mL in Hanks Buffer. After final centrifugation of 5 min at 1000×g at room temperature, the supernatant was aspirated and the pellet of PBMCs was processed immediately or stored at − 80 °C to maintain stability.
Total protein extraction
The lysis solution (lysate) was composed of 320 μL of cell lysis buffer (CelLyticTM-M, SIGMA Aldrich, reference C2978) and 1.6 μL of protease inhibitor (0.5%, Protease Inhibitor Cocktail, SIGMA Aldrich, reference P8215) for the samples with counted cells (> 106) and was added to the lymphocyte pellet. The mixture was stirred for 15 min at room temperature and centrifuged for 15 min at 12000×g and 4 °C. The supernatant was collected and was immediately used for further analysis or stored at − 80 °C to maintain stability.
VEGF-A measurement
PBMC extract concentrations of VEGF-A were estimated using the Randox high-sensitivity multiplex cytokine and growth factor array (Evidence Investigator Analyzer, Randox Laboratories Ltd., Crumlin, UK).
DNA methylation analysis
DNA methylation assay
DNA methylation patterns were investigated using a method, previously described in detail [46, 47]. Briefly, genome-wide methylation profiling of bisulfite-converted genomic DNA was performed by Illumina HumanMethylation450 bead array (Illumina Inc., San Diego, CA, USA). Illumina is using Infinium I and II arrays with probes for detection of methylated and unmethylated CpG sites. Methylation ratio, referred to as beta value by Ilumina’s software, is the proportion methylated/(methylated + unmethylated) for each CpG in the population of cells from which we extracted DNA.
Quality control
R package minfi (Bioconductor) was used to analyse and visualize Illumina Infinium methylation arrays [48]. The first step in microarray data preprocessing consisted of removing all probes that can generate artifactual data. Firstly, a detection P value was assigned to each probe. High detection P value normally corresponds to a probe with a low quality signal; therefore, probes with P > 0.05 were removed from all samples. Furthermore, probes missing in > 5% of the samples were excluded. To avoid spurious associations, probes containing locations on the genome where variation is already annotated in HumanMethlyation450 annotation file IlluminaHumanMethylation450kanno.ilmn12.hg19 (i.e. probes containing single-nucleotide polymorphism (SNP), sex chromosomes and a single-base extension (SBE) site) were excluded. Finally, probes containing cross-reactive and target polymorphic CpGs [49] were excluded, leaving 314 440 probes out of 484 777 for statistical analysis. In addition, one individual was excluded from our cohort after quality control checks of methylation array data (outlier of plotted median of the methylated against unmethylated intensity), leaving 200 individuals for the analysis.
Normalization
Second step in microarray data preprocessing was removing sources of variation, related to technical limitations—data normalization. Background correction, colour bias (dye bias) adjustment and Infinium I/II bias correction were carried out with Illumina background correction and SWAN [50] in the R package minfi.
Association study
CpGassoc (CRAN) was used to test for association between methylation at CpG sites across the genome and VEGF-A concentration in PBMC extracts [51]. As VEGF-A concentrations were not normally distributed in our population, a log-transformation has been applied to normalize the distribution. The random mixed-effects model included gender, age, BMI, family structure and individual blood cell counts (neutrophils, lymphocytes, monocytes, eosinophils and basophils) as covariates and chip array as random effect. In our model, cell counts were added as additional covariate terms to control for the confounding effects of variable leukocyte distribution for examination of the association between DNA methylation and VEGF-A concentration. Holm-Bonferroni correction for multiple testing was applied to the result.
In silico analysis
Ensembl browser [52] was used for localization of CpG sites on the Human Genome (GRCh38.p10), as well as for the establishment of regulatory features from their genomic environment. All annotated genes were investigated for interactions with VEGF-A gene using cytoscape app GeneMANIA [26]. MethylGSA R-package was used to relate significant genes or CpGs to known biological properties [53].
Supplementary information
Additional file 1: Sup. Figure 1. Genomic environment of six CpG sites. Dark green regions present a CpG site, numbers on left and right of the box indicate a location, within which CpG can be found. Nearest genomic features upstream (left) or downstream (right) are presented for each CpG. Distance (bp) between each CpG and genomic feature is indicated in light green regions. Turquoise squares present CTCF regions, red promoter region and yellow enhancers. Square brackets [ ] indicate that CpG is located within genomic feature. Seven different PBMC cell types were looked up (Sup. Table 1). Number 1-7/7 in each box of particular genomic region is indicating in what extent this genomic feature is presented in PBMCs. Diagrams on the top are presenting patterns of genomic features that can be found in the genomic environment of CpGs. We can see that in the immediate proximity of CpG enhancers are the most common and that with distance, genetic features become less common (regions of non-coding DNA). Sup. Figure 2. Genetic environment of fourteen CpG sites. Sup. Table 1. Forty-one significant CpG sites related to VEGF concentration derived from PBMCs extracts. Sup. Table 2. Summary table explaining the potential functionality and biological plausibility of each of the 20 significant CpGs and their nearby genes. Sup. Table 3. List of VEGF genes, VEGF receptor genes and VEGF-A-related genes. Genes in direct relation to VEGF-A were determined with STRING tool (http://version10.string-db.org/), the location was retrieved using Ensembl (www.ensembl.org/). Sup. Figure 3. Analysis of significant CpG sites. MethylGSA, a Bioconductor package was used to find relevant physiological pathways. Significant results are presented in the figure.
Acknowledgements
Not applicable.
Abbreviations
- BMI
Body mass index
- COPD
Chronic obstructive pulmonary disease
- DNA
Deoxyribonucleic acid
- EWAS
Epigenome-wide association studies
- GWAS
Genome-wide association studies
- PBMC
Peripheral blood mononuclear cell
- SBE
Single-base extension
- SD
Standard deviation
- SFS
STANISLAS Family Study
- SNP
Single-nucleotide polymorphism
- TSS
Transcription start site
- VEGF
Vascular endothelial growth factor
Authors’ contributions
VG participated in study design, performed data analysis and interpretation and drafted the manuscript. DRV participated in study design and performed data analysis and interpretation. SD participated in bioinformatics analysis and drafting of the manuscript. MGS and LD participated at analysis and drafting of the manuscript. EWAS analysis pipeline was designed by TX. HM, JL and PF contributed with biological measurements. CM prepared biological material and experiments. SVS was involved in study design and management. All authors participated in the interpretation of data and review of the manuscript. They all approved the final version of the manuscript.
Funding
Financial support was provided by the European Union within the frames of the Operational Programme FEDER-FSE Lorraine et Massif des Vosges 2014-2020, by Agence Nationale de la Recherche, programme d’Investissements d’avenir, grant number ANR-15RHU-0004. Additionally, this work was supported by the regional project CPER-ITM2P 2015-2020.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Study protocols were approved by the institutional ethics committee CCPPRB de Lorraine (Comité consultatif de protection des personnes dans la recherche biomédicale) and CNIL (Commission Nationale de l'Informatique et des Libertés). All subjects gave written informed consent for their participation in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13148-020-00874-w.
References
- 1.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128(4):635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 3.Handy DE, Castro R, Loscalzo J. Epigenetic modifications: basic mechanisms and role in cardiovascular disease. Circulation. 2011;123(19):2145–2156. doi: 10.1161/CIRCULATIONAHA.110.956839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esteller M. Epigenetic gene silencing in cancer: the DNA hypermethylome. Hum Mol Genet. 2007;16 Spec No 1:R50-9. [DOI] [PubMed]
- 5.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28(10):1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 6.Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat Rev Genet. 2011;12(8):529–541. doi: 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9(6):465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 8.Medvedeva YA, Fridman MV, Oparina NJ, Malko DB, Ermakova EO, Kulakovskiy IV, et al. Intergenic, gene terminal, and intragenic CpG islands in the human genome. BMC Genomics. 2010;11(1):48. doi: 10.1186/1471-2164-11-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neufeld G, Cohen T, Gengrinovitch S, Poltorak Z. Vascular endothelial growth factor (VEGF) and its receptors. FASEB J. 1999;13(1):9–22. [PubMed] [Google Scholar]
- 10.Ferrara N, Gerber HP, LeCouter J. The biology of VEGF and its receptors. Nat Med. 2003;9(6):669–676. doi: 10.1038/nm0603-669. [DOI] [PubMed] [Google Scholar]
- 11.Carmeliet P. VEGF as a key mediator of angiogenesis in cancer. Oncology. 2005;69(Suppl. 3):4–10. doi: 10.1159/000088478. [DOI] [PubMed] [Google Scholar]
- 12.Awata T, Inoue K, Kurihara S, Ohkubo T, Watanabe M, Inukai K, et al. A common polymorphism in the 5′-untranslated region of the VEGF gene is associated with diabetic retinopathy in type 2 diabetes. Diabetes. 2002;51(5):1635–1639. doi: 10.2337/diabetes.51.5.1635. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Berendsen AD, Jia S, Lotinun S, Baron R, Ferrara N, et al. Intracellular VEGF regulates the balance between osteoblast and adipocyte differentiation. J Clin Invest. 2012;122(9):3101–3113. doi: 10.1172/JCI61209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding Y, Niu H, Li Y, He P, Li Q, Ouyang Y, et al. Polymorphisms in VEGF-A are associated with COPD risk in the Chinese population from Hainan province. J Genet. 2016;95(1):151–156. doi: 10.1007/s12041-016-0627-0. [DOI] [PubMed] [Google Scholar]
- 15.Hurwitz H, Fehrenbacher L, Novotny W, Cartwright T, Hainsworth J, Heim W, et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med. 2004;350(23):2335–2342. doi: 10.1056/NEJMoa032691. [DOI] [PubMed] [Google Scholar]
- 16.Sandler A, Gray R, Perry MC, Brahmer J, Schiller JH, Dowlati A, et al. Paclitaxel–carboplatin alone or with bevacizumab for non–small-cell lung cancer. N Engl J Med. 2006;355(24):2542–2550. doi: 10.1056/NEJMoa061884. [DOI] [PubMed] [Google Scholar]
- 17.Escudier B, Pluzanska A, Koralewski P, Ravaud A, Bracarda S, Szczylik C, et al. Bevacizumab plus interferon alfa-2a for treatment of metastatic renal cell carcinoma: a randomised, double-blind phase III trial. Lancet. 2007;370(9605):2103–2111. doi: 10.1016/S0140-6736(07)61904-7. [DOI] [PubMed] [Google Scholar]
- 18.Choi SH, Ruggiero D, Sorice R, Song C, Nutile T, Vernon Smith A, et al. Six novel loci sssociated with circulating VEGF levels identified by a meta-analysis of genome-wide association studies. PLoS Genet. 2016;12(2):e1005874. doi: 10.1371/journal.pgen.1005874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Debette S, Visvikis-Siest S, Chen MH, Ndiaye NC, Song C, Destefano A, et al. Identification of cis- and trans-acting genetic variants explaining up to half the variation in circulating vascular endothelial growth factor levels. Circ Res. 2011;109(5):554–563. doi: 10.1161/CIRCRESAHA.111.243790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berrahmoune H, Herbeth B, Lamont JV, Masson C, Fitzgerald PS, Visvikis-Siest S. Heritability for plasma VEGF concentration in the Stanislas family study. Ann Hum Genet. 2007;71(Pt 1):54–63. doi: 10.1111/j.1469-1809.2006.00298.x. [DOI] [PubMed] [Google Scholar]
- 21.Mayhew AJ, Meyre D. Assessing the heritability of complex traits in humans: methodological challenges and opportunities. Curr Genom. 2017;18(4):332–340. doi: 10.2174/1389202918666170307161450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groh A, Jahn K, Burkert A, Neyazi A, Schares L, Janke E, et al. Epigenetic regulation of the promotor region of vascular endothelial growth factor-A and nerve growth factor in opioid-maintained patients. Eur Addict Res. 2017;23(5):249–259. doi: 10.1159/000485030. [DOI] [PubMed] [Google Scholar]
- 23.Siddique AN, Nunna S, Rajavelu A, Zhang Y, Jurkowska RZ, Reinhardt R, et al. Targeted methylation and gene silencing of VEGF-A in human cells by using a designed Dnmt3a–Dnmt3L single-chain fusion protein with increased DNA methylation activity. J Mol Biol. 2013;425(3):479–491. doi: 10.1016/j.jmb.2012.11.038. [DOI] [PubMed] [Google Scholar]
- 24.Kim JY, Whang JH, Zhou W, Shin J, Noh SM, Song IS, et al. The expression of VEGF receptor genes is concurrently influenced by epigenetic gene silencing of the genes and VEGF activation. Epigenetics. 2009;4(5):313–321. doi: 10.4161/epi.4.5.9160. [DOI] [PubMed] [Google Scholar]
- 25.Kim J, Hwang J, Jeong H, Song H-J, Shin J, Hur G, et al. Promoter methylation status of VEGF receptor genes: a possible epigenetic biomarker to anticipate the efficacy of intracellular-acting VEGF-targeted drugs in cancer cells. Epigenetics. 2012;7(2):191–200. doi: 10.4161/epi.7.2.18973. [DOI] [PubMed] [Google Scholar]
- 26.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38(Web Server issue):W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin A, Wang RT, Ahn S, Park CC, Smith DJ. A genome-wide map of human genetic interactions inferred from radiation hybrid genotypes. Genome Res. 2010;20(8):1122–1132. doi: 10.1101/gr.104216.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Zhao W, Liu X, Guan G, Zhuang M. ARL3 is downregulated and acts as a prognostic biomarker in glioma. J Transl Med. 2019;17(1):210. doi: 10.1186/s12967-019-1914-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mallon BS, Chenoweth JG, Johnson KR, Hamilton RS, Tesar PJ, Yavatkar AS, et al. StemCellDB: the human pluripotent stem cell database at the National Institutes of Health. Stem Cell Res. 2013;10(1):57–66. doi: 10.1016/j.scr.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu G, Feng X, Stein L. A human functional protein interaction network and its application to cancer data analysis. Genome Biol. 2010;11(5):R53. doi: 10.1186/gb-2010-11-5-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei P, Zhang N, Xu Y, Li X, Shi D, Wang Y, et al. TPX2 is a novel prognostic marker for the growth and metastasis of colon cancer. J Transl Med. 2013;11:313. doi: 10.1186/1479-5876-11-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jian J, Huang Y, Liu L-Z, Li S, Deng F. TPX2 gene-silencing inhibits the proliferation and invasion of human colon cancer SW480 cells. Tumor. 2016;36(6):628-634.
- 33.Robertson KD, Jones PA. DNA methylation: past, present and future directions. Carcinogenesis. 2000;21(3):461–467. doi: 10.1093/carcin/21.3.461. [DOI] [PubMed] [Google Scholar]
- 34.Pardue EL, Ibrahim S, Ramamurthi A. Role of hyaluronan in angiogenesis and its utility to angiogenic tissue engineering. Organogenesis. 2008;4(4):203–214. doi: 10.4161/org.4.4.6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy JF, Lennon F, Steele C, Kelleher D, Fitzgerald D, Long AC. Engagement of CD44 modulates cyclooxygenase induction, VEGF generation, and proliferation in human vascular endothelial cells. FASEB J. 2005;19(3):446–448. doi: 10.1096/fj.03-1376fje. [DOI] [PubMed] [Google Scholar]
- 36.Rodgers LS, Lalani S, Hardy KM, Xiang X, Broka D, Antin PB, et al. Depolymerized hyaluronan induces vascular endothelial growth factor, a negative regulator of developmental epithelial-to-mesenchymal transformation. Circ Res. 2006;99(6):583–589. doi: 10.1161/01.RES.0000242561.95978.43. [DOI] [PubMed] [Google Scholar]
- 37.Barrett LW, Fletcher S, Wilton SD. Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell Mol Life Sci. 2012;69(21):3613–3634. doi: 10.1007/s00018-012-0990-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sas-Chen A, Aure MR, Leibovich L, Carvalho S, Enuka Y, Körner C, et al. EMBO Mol Med 2016;8(9):1052-1064. [DOI] [PMC free article] [PubMed]
- 39.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 40.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349(21):2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 41.Skinner MK, Guerrero-Bosagna C. Role of CpG deserts in the epigenetic transgenerational inheritance of differential DNA methylation regions. BMC Genomics. 2014;15(1):692. doi: 10.1186/1471-2164-15-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plasschaert RN, Vigneau S, Tempera I, Gupta R, Maksimoska J, Everett L, et al. CTCF binding site sequence differences are associated with unique regulatory and functional trends during embryonic stem cell differentiation. Nucleic Acids Res. 2014;42(2):774–789. doi: 10.1093/nar/gkt910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Visvikis-Siest S, Siest G. The STANISLAS Cohort: a 10-year follow-up of supposed healthy families. Gene-environment interactions, reference values and evaluation of biomarkers in prevention of cardiovascular diseases. Clin Chem Lab Med. 2008;46(6):733–747. doi: 10.1515/CCLM.2008.178. [DOI] [PubMed] [Google Scholar]
- 44.Siest G, Visvikis S, Herbeth B, Gueguen R, Vincent-Viry M, Sass C, et al. Objectives, design and recruitment of a familial and longitudinal cohort for studying gene-environment interactions in the field of cardiovascular risk: the Stanislas cohort. Clin Chem Lab Med. 1998;36(1):35–42. doi: 10.1515/CCLM.1998.007. [DOI] [PubMed] [Google Scholar]
- 45.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98(4):288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 47.Dedeurwaerder S, Defrance M, Bizet M, Calonne E, Bontempi G, Fuks F. A comprehensive overview of Infinium HumanMethylation450 data processing. Brief Bioinform. 2014;15(6):929–941. doi: 10.1093/bib/bbt054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Y-a C, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8(2):203–209. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maksimovic J, Gordon L, Oshlack A. SWAN: Subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biol. 2012;13(6):R44. doi: 10.1186/gb-2012-13-6-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barfield RT, Kilaru V, Smith AK, Conneely KN. CpGassoc: an R function for analysis of DNA methylation microarray data. Bioinformatics. 2012;28(9):1280–1281. doi: 10.1093/bioinformatics/bts124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aken BL, Ayling S, Barrell D, Clarke L, Curwen V, Fairley S, et al. The Ensembl gene annotation system. Database. 2016;2016. [DOI] [PMC free article] [PubMed]
- 53.Ren X, Kuan PF. methylGSA: a Bioconductor package and Shiny app for DNA methylation data length bias adjustment in gene set testing. Bioinformatics (Oxford, England) 2019;35(11):1958–1959. doi: 10.1093/bioinformatics/bty892. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Sup. Figure 1. Genomic environment of six CpG sites. Dark green regions present a CpG site, numbers on left and right of the box indicate a location, within which CpG can be found. Nearest genomic features upstream (left) or downstream (right) are presented for each CpG. Distance (bp) between each CpG and genomic feature is indicated in light green regions. Turquoise squares present CTCF regions, red promoter region and yellow enhancers. Square brackets [ ] indicate that CpG is located within genomic feature. Seven different PBMC cell types were looked up (Sup. Table 1). Number 1-7/7 in each box of particular genomic region is indicating in what extent this genomic feature is presented in PBMCs. Diagrams on the top are presenting patterns of genomic features that can be found in the genomic environment of CpGs. We can see that in the immediate proximity of CpG enhancers are the most common and that with distance, genetic features become less common (regions of non-coding DNA). Sup. Figure 2. Genetic environment of fourteen CpG sites. Sup. Table 1. Forty-one significant CpG sites related to VEGF concentration derived from PBMCs extracts. Sup. Table 2. Summary table explaining the potential functionality and biological plausibility of each of the 20 significant CpGs and their nearby genes. Sup. Table 3. List of VEGF genes, VEGF receptor genes and VEGF-A-related genes. Genes in direct relation to VEGF-A were determined with STRING tool (http://version10.string-db.org/), the location was retrieved using Ensembl (www.ensembl.org/). Sup. Figure 3. Analysis of significant CpG sites. MethylGSA, a Bioconductor package was used to find relevant physiological pathways. Significant results are presented in the figure.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.