Abstract
Objectives:
To investigate how T-cell activation interacts with NSUN2 to influence HNSCC patient survival.
Materials and Methods:
The relationships between T-cell activation status (Activation, Intermediate, and Exhaustion), NSUN2 expression, and patient survival were evaluated using Kaplan-Meier survival curves and multivariate Cox regression models in a public dataset with 520 HNSCC patients. HPV status was determined based on a VirusScan analysis of RNA-seq data.
Results:
Among the patients with high NSUN2 expression, the Activation group exhibited longer survival than the Exhaustion group (trend P = 0.056). Adjusted hazards ratios (HRs) were 0.77 (95% CI: 0.49–1.19) for the Intermediate vs Exhaustion, and 0.61 (0.36 – 1.03) for Activation vs. Exhaustion. In contrast, there is a positive association between T-cell activation score and mortality in the patients with low NSUN2 expression (trend P = 0.016). The adjusted HRs were 1.97 (1.12–3.47) for the Intermediate vs Exhaustion, and 2.06 (1.16–3.68) for the Activation vs Exhaustion. In multivariate cox models with or without HPV status, the interaction between T-cell activation status and NSUN2 expression was statistically significant (P = 0.004 for with HPV status, and P = 0.002 for without, respectively). When not controlling for NSUN2 expression, there was no significant association between T-cell activation score and patient mortality (P = 0.84).
Conclusions:
An interaction between NSUN2 expression and T-cell activation status affects patient survival in HNSCC regardless of HPV status, suggesting that NSUN2 is a potential precision marker for immune-checkpoint blockade, and a potential therapeutic target.
Keywords: head and neck squamous carcinoma, NSUN2, prognosis, T-cell activation score
Introduction
Immune escape is a hallmark of human cancer including head and neck squamous-cell carcinoma (HNSCC). T-cell exhaustion is one of a plethora of mechanisms that can underlie the escape of human cancer from immune surveillance. Both cell-intrinsic and cell-extrinsic negative regulatory pathways play important roles in capitalizing on T-cell dysfunction. Cytotoxic T-lymphocyte antigen 4 (CTLA4) and programmed cell death-1 receptor (PD-1) are two extensively investigated negative regulators that dampen the function of effector T cells by engaging them as receptors of ligands [1]. Early proof-of-concept of these regulatory interactions led to approval of anti-CTLA4 (ipilimumab, tremelimumab) and anti-PD-1 (nivolumab, pembrolizumab) immunotherapy by the United States Food and Drug Administration (FDA) for the clinical management of several types of human cancer. The principle of immune checkpoint-based antibody immunotherapies is to reinvigorate effector CD8+ T-cells by releasing the ‘brake’ on the immune system’s proclivity to kill tumor cells. Burtness and colleagues recently reported that pembrolizumab alone or with chemotherapy significantly improved overall survival in a phase 3 study of recurrent and metastatic HNSCC with a relatively large sample size [2]. However, not all patients gain benefit from these therapies. A single-arm phase-II trial of nivolumab in 44 patients with recurrent and metastatic nasopharyngeal carcinoma provided an overall objective response rate (ORR) of 20.5%, and a 1-year overall survival rate of 59% (95% CI: 44.3–78.5%) [3]. Another phase-II trial of pembrolizumab included 27 patients with PD-L1-positive nasopharyngeal carcinoma, wherein the ORR was 25.9% (11.1–46.3%) over a median follow-up of 20 months [4]. An open-label phase-3 trial of nivolumab vs. standard chemotherapy (2:1) in 361 patients with recurrent HNSCC provided 2.4 months of overall survival benefit in the nivolumab group compared to standard therapy, and a response rate of 13.3% for nivolumab vs. 5.8% for standard therapy [5]. A phase-1b trial of pembrolizumab with 60 PD-L1-positive HNSCC patients provided 18% (8–32%) overall response [6]. Another phase 1b trial of pembrolizumab in 132 patients with recurrent and/or metastatic HNSCC produced 18% ORR (12–26%) [7]. A recent phase I/II single-arm trial of durvalumab (anti-PD-L1) and tremlimumab (anti-CTLA4) in combination is ongoing, and includes 35 patients with metastatic HNSCC, and no primary end-point results other than adverse events have been reached thus far [8]. These observations have motivated the search for other negative regulators of the immune response, such as LAG3, TIM3, and TIGIT, three regulators under active investigation [9, 10]. Numerous studies are investigating other cellular signals, such as the signals that affect the host response to immune-checkpoint blockade. For instance, CDK4/6 inhibitor treatment enhances antitumor efficacy of PD-1 immunotherapy, dramatically improving overall survival in animal models [11]. ADAR1 is an adenosine deaminase, which can bind and limit the sensing of endogenous double-stranded RNAs (dsRNAs) that leads to inflammation. Loss of ADAR1 can suppress PD-1 immunotherapy resistance by increasing tumor inflammation [12].
NOP2/Sun domain family member 2 protein (NSUN2), encoded in chromosome 10 by the NSUN2 gene, is a RNA-modification protein for m5C methylation in both cytoplasmic and mitochondrial tRNAs [13–15]. The methylation of tRNAs prevents tRNAs from endonucleolytic cleavage by angiogenin to generate tRNA-derived fragments (tRFs), consequently controlling the efficacy of protein translation [16–20], and leading to cytokine production and cellular metabolic changes in response to stress [21] . NSUN2-deficient cells exhibit reduced protein synthesis, dysregulation of cell cycle, abnormal cell differentiation and proliferation [17–20, 22]. NSUN2-knockout testes are completely absent of spermatids and sperm [23]. Over-expression of NSUN2 promotes the migration of neural cells toward the chemoattractant growth factor [24]. Both NSUN2 and IGF-II increase ovarian cancer mortality risk when expressed at high levels [25]. By interacting with ribosomal protein L6 (RPL6), NSUN2 drives the progression of gallbladder carcinoma [26]. Chen and colleagues recently demonstrated that NSUN2 promoted pathogenesis of bladder cancer via stabilizing mRNAs, and that overexpression of NSUN2 predicted poor prognosis of bladder cancer [27]. A previous study showed that high NSUN2 expression associated with poor prognosis in HNSCC [28]. In addition, NSUN2 has also been shown to modify mRNAs. High NSUN2 enhances CDK1 translation mediated by CDK1 mRNA methylation, consequently stimulating cell growth [29]. The stability of p16ink4 mRNA, a CDK inhibitor blocking the formation of cyclin D-CDK4/6 complex, increases due to NSUN2-mediated mRNA methylation [30, 31]. Moreover, RNA methylation-mediated switch in the secondary structure between double-stranded and single-stranded RNAs can affect immune response [12]. Given that immune cells sense tumors by utilizing pathogen and damage receptors, by which tumor-specific immunity is elicited, we hypothesized that NSUN2 expression level modifies the effect of T-cell activation on patients’ survival in HNSCC.
Methods
Data sources
This study includes 520 individuals with primary HNSCC, whose gene expression information and clinical data were retrieved from a TCGA dataset at cBioPortal (https://www.cbioportal.org). Among 518 patients with age information available, the average age was 60.9 years old (standard deviation 11.9, range 19–90). Among 519 patients whose sex was reported, 73.8% were men, and 26.2% were women. Patient ethnicity was Caucasian for 87.9% of reports (444 out of 505), with the remaining 12.1% reported as African American, Asian, and American Native. There were 117 non-smokers and 389 smokers. The majority of patients were diagnosed with the disease at advanced stage (282 at stage IV, 105 at stage III, 98 at stage II and 20 at stage I). Poor differentiation was observed in patient tumor tissues, at an incidence of 12.0% (62 out of 515) for grade I, 58.8% for grade II and 29% for grade III. Tumor sites (n = 519) included tongue (38.3%), pharynx (26.0%), and other (35.7%). Approximately half (52.6%, 271/515) had lymph invasion, and 35.2% (122/347) of patients had vascular invasion. Follow-up information was available for 517 patients, and the average overall survival was 21.5 months (range 0.07–210.8 months).
mRNA levels in Fragments Per Kilobase of transcript per Million mapped reads (FPKM), which were normalized by the upper quartile expectation maximization (RNA-seq V2 RSEM), were retrieved for NSUN2 as well as a panel of 14 genes that are associated with M2 macrophage and myeloid cells: NKG7, CCL4, CST7, PRF1, GZMA, GZMB, IFNG, CCL3, PDCD1 (aka PD-1), TIGIT, LAG3, TIM3, CTLA4 and CD274 (aka PD-L1) [32, 33]. Experimental data generation and processing were conducted as previously described [34]. No patients received neoadjuvant treatment.
No formal written consent is required for this type of study. The ethical standards of the institutional and/or national research committee and the 1964 Helsinki declaration and its later amendments or comparable ethical standards were followed in performing all procedures in this study involving human subjects. The study presented here complies with the current laws of USA.
RNA-seq-based HPV status determination
RNA sequencing–based HPV status determination was performed using VirusScan as described previously [35]. A cutoff of 100 HPV RNA viral transcript reads per hundred million (TPM) was specified as determinate of the HPV status for a tumor, with an HPV positive determination when there were ≥100 TPM, and HPV negative determination when there were <100 TPM. Using this cutoff, the concordance is 100% between RNA-seq-based HPV status and the HPV status determined by in situ hybridization performed on 248 tumors in the TCGA clinical data.
Gene-set enrichment analysis
Co-expression of genome-wide genes with NSUN2 was analyzed using Spearman correlation with multiple comparison correction. Gene set enrichment analysis was performed for the genes that had an adjusted P value < 0.001 [36].
Statistical analyses
A T-cell activation score was calculated for each subject as described previously [37], by taking the weighted average of log-transformed expression (FPKM +1) across the gene panel. The ‘overall survival’ was defined as the number of months from the initial diagnosis until death or the last follow-up. Spearman correlation was used to evaluate correlations. Survival analyses were performed using Kaplan-Meier survival curves and multivariate Cox proportional hazards, in which three groups—high (activation), intermediate, and low (exhaustion)—were assigned using the tertiles of the T-cell activation score. To investigate whether the NSUN2 expression modifies the effect of T-cell activation status on patients’ survival, we used the median of the NSUN2 expression level as the cutoff value to stratify the models. Patient age at diagnosis, disease stage, tumor grade, gender, and smoking history were included in the models to estimate adjusted hazards ratios (HRs) and their 95% confidence intervals (95% CIs). We also assessed the interaction between T-cell activation score and NSUN2 level across all patients by including their interactions in the Cox regression models. The proportional hazards assumption was examined. In all statistical analyses, a P value less than 0.05 was considered significant. Statistical analyses were performed using SAS version 9.4 (SAS Institute, Inc), and cgdsr and ReactomPA packages for R version 3.5.0 (https://www.R-project.org) were used for gene retrieval and pathway analysis.
Results
T-cell activation score and gene expression levels
Table 1 shows the distribution of T-cell activation score, and the expression of NSUN2. The averages were 0.21 with a standard deviation of 0.09 for T-cell activation score, 2221 FPKM for NSUN2. A significant negative correlation was found between NSUN2 expression and T cell activation score (P = 0.002). The Spearman correlation coefficient was −0.14 (95% CI: −0.22, −0.05).
Table 1.
Distribution of the expression level of NSUN2 and the T-cell activation score, and their spearman correlation
| Variable | n | Mean | SDa | Median | Range |
|---|---|---|---|---|---|
| NSUN2 | 520 | 2395 | 969 | 2221 | (540, 6577) |
| T cell activation score | 520 | 0.21 | 0.09 | 0.21 | (−0.01, 0.50) |
| Correlation coefficient (95% CIb) | −0.14 | (−0.22, −0.05) | |||
| P value | 0.002 | ||||
aSD: standard deviation
bCI: confidence interval
Table 2 and Figure 1 show the correlation and scatter plots of NSUN2 expression with CD163, CD33, and PDCD1 and myeloid cell marker CD45, respectively. There were significant negative correlations between NSUN2 expression and the expressions of M2-like macrophage markers CD163 (P = 0.0003), CD33 (P< 0.0001), PDCD1 (P < 0.0001), and CD45 (P < 0.0001; Table 2; Figure 1). The Spearman correlation coefficients were −0.16 (−0.24, −0.07) for CD163, −0.27 (−0.34, −0.18) for CD33, −031 (−0.47, −0.22) for PDCD1, and −0.25 (−0.32, −0.16) for CD45.
Table 2.
Spearman correlation between NSUN2 and markers of macrophage and myeloid cells
| Gene | n | Correlation coefficient | 95% CIa | P value |
|---|---|---|---|---|
| CD163 | 520 | −0.16 | (−0.24, −0.07) | 0.0003 |
| CD33 | 520 | −0.27 | (−0.34, −0.18) | <0.0001 |
| PDCD1 | 520 | −0.31 | (−0.47, −0.22) | <0.0001 |
| CD45 | 520 | −0.25 | (−0.32, −0.16) | <0.0001 |
CI: confidence interval
Figure 1.
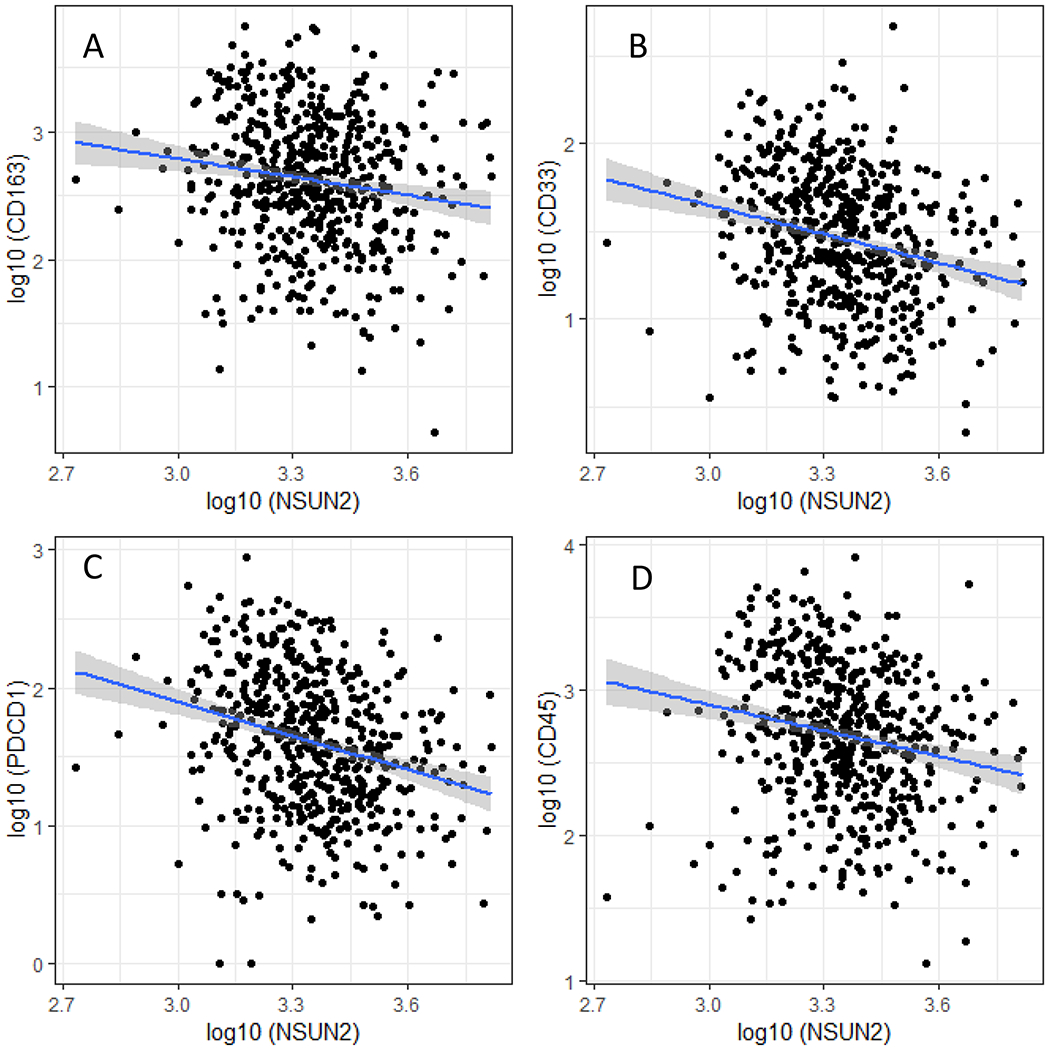
Scatter plot of NSUN2 expression with the markers for macrophage and myeloid cells. Expression (log10 FPKM) of NSUN2 is plotted against A) CD163, B) CD33, C) PDCD1 and D) CD45. The linear trend line is in blue, and the grey bound represents the 95% confidence interval.
Interaction between T-cell activation status and NSUN2 expression level in patient survival
In the survival analysis, we first tested the proportional-hazards assumption based on 1000 simulations for T-cell activation status, and use of the assumption was validated (P = 0.676 for all patients, P = 0.185 in the NSUN2 low expression group, and P = 0.922 in the NSUN2 high expression group).
We investigated the relationship between survival and the T-cell activation status (Activation, Intermediate, and Exhaustion) in the whole sample. No significant association was observed between T-cell activation status and overall survival in HNSCC (P = 0.97). The medians of overall survival were 65.8 months (32.2 – 89.3 months) for patients with a low T-cell activation score (Exhaustion), 49.4 months (34.5 – 71.2 months) for those with an intermediate T-cell activation score (Intermediate), and 54.9 months (95% CI: 37.3 – 68.4 months) for those with a high T-cell activation score (Activation; data not shown).
In the NSUN2 low expression group, patients in the Activation group showed inferior overall survival compared to those in the Exhaustion group (Figure 2A). The median of overall survival was 88.8 months (52.3–210.8 months) in the Exhaustion group, 49.4 months (35.5–156.4 months) in the Intermediate group, and 54.9 months (28.0–67.8 months) in the Activation group. Patients in the Activation group lived on average 34.9 months (almost 3 years) shorter than those in the Exhaustion group. The Activation group showed significantly increased risk of death (trend P = 0.012) compared to the Exhaustion group. The HRs of death were 1.67 (1.00–2.78) for Intermediate vs Exhaustion (p = 0.05), and 1.96 (1.16–3.29) for Activation vs Exhaustion (p = 0.012). In contrast, in the NSUN2 high expression group, patients with an Activation status showed better overall survival compared to those with an Exhaustion status (Figure 2B). The median of overall survival was 26.4 months (17.9–76.2 months) in the Exhaustion group, 45.8 months (27.5–84.4 months) in the Intermediate group, and 159.5 months (42.4–159.5 months) in the Activation group, respectively. On average, patients in the Activation group lived 133.1 months (over 11 yrs) longer than those in the Exhaustion group. The HRs of death were 0.75 (0.49–1.13) for Intermediate vs Exhaustion (p = 0.168), and 0.61 (0.38–0.99) for Activation vs Exhaustion (p = 0.044). The Activation group tended to have a lower risk in comparison to the Exhaustion group (trend P = 0.037). In the whole sample, a significant interaction was observed between the expression level of NSUN2 and the T-cell activation status in the interaction test (P = 0. 004).
Figure 2.
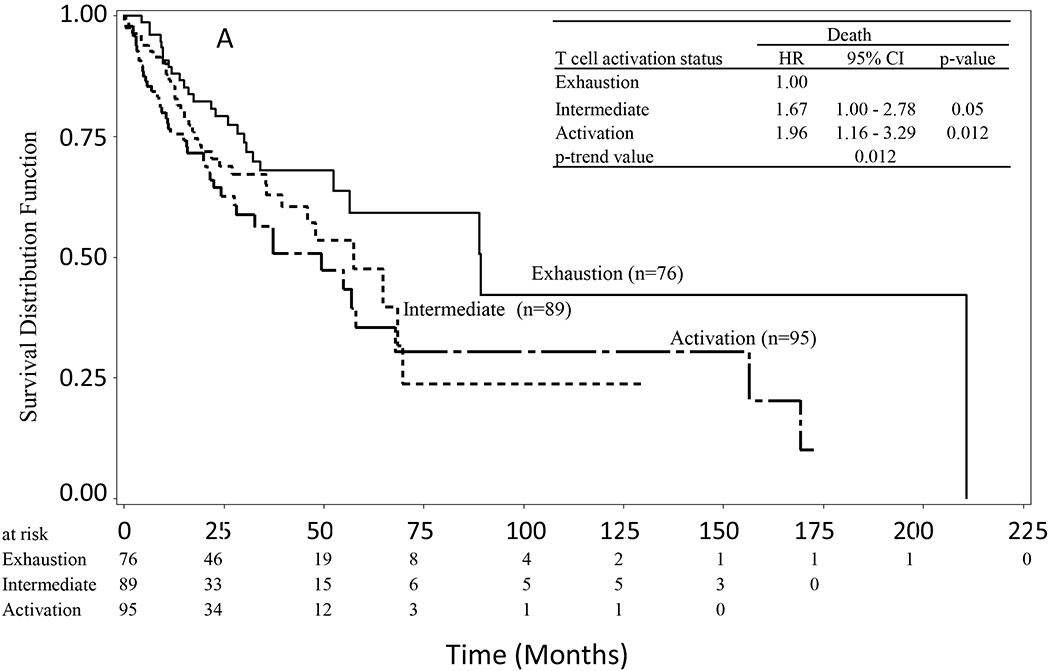
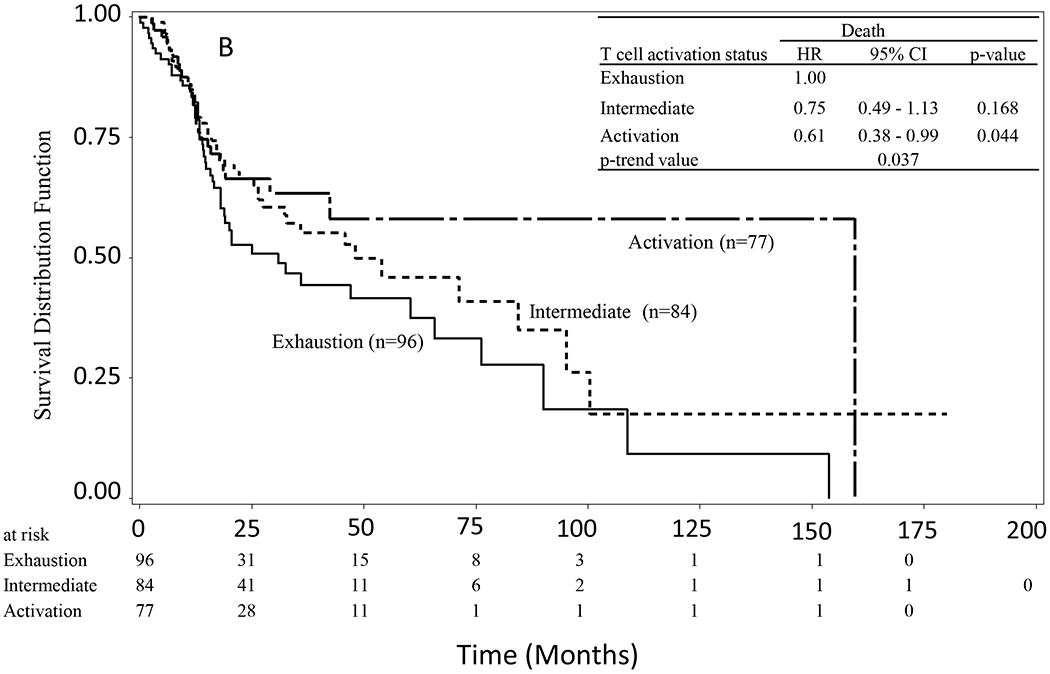
Kaplan-Meier survival curves of HNSCC stratified by T-cell activation status (score). A) In the subgroup with a low expression of NSUN2, patients in the Activation group had reduced overall survival compared to those in the Exhaustion group (P = 0.012). B) In the subgroup with a high expression of NSUN2, patients in the Activation group had longer overall survival than those in the Exhaustion group (P = 0.037).
We then performed multivariate Cox proportional-hazards models to adjust for potential confounding variables including the patient’s age at surgery, disease stage, tumor grade, gender, smoking status, and tumor site (“model 1”; Table 3). Similarly, there was no significant association between the T-cell activation status and death risk among all patients without accounting for the expression level of NSUN2 (Table 2). The adjusted HRs were 1.08 (0.77–1.50) for Intermediate vs Exhaustion (P = 0.664), and 1.03 (0.73–1.47) for Activation vs. Exhaustion (P = 0.852). However, when adjusting for covariates and including expression of NSUN2 in model 1, a significant interaction term is estimated between the expression level of NSUN2 and the T-cell activation status (P = 0.002). In the group with low NSUN2 expression, T-cell activation score was positively associated with increased death risk (trend P = 0.009). The adjusted HRs were 2.01 (1.15 – 3.52) for Intermediate vs Exhaustion (P = 0.015), and 2.16 (1.22–3.82) for Activation vs. Exhaustion (P = 0.008). In contrast, in the group with high expression of NSUN2, T-cell activation score was negatively associated with the disease risk (trend P = 0.028). The adjusted HRs were 0.74 (0.48–1.14) for Intermediate vs Exhaustion (P = 0.174), and 0.57 (0.34–0.96) for Activation vs Exhaustion (P = 0.033). Moreover, the interaction between the NSUN2 expression and T-cell activation status remained significant (P = 0.004) after addition of HPV status to the set of covariates (“model 2”). As with model 1, in the group with low NSUN2 expression, T-cell activation score was positively associated with an increased death risk (trend P = 0.016), whereas in the group with high NSUN2 expression, the Activation group trended toward a lower risk of death (P = 0.056, not significant). Among patients with low NSUN2 expression, the adjusted HRs were 1.97 (1.12–3.47) for Intermediate vs Exhaustion (p =0.019), and 2.06 (1.16–3.68) for Activation vs. Exhaustion (p = 0.014). Among patients with high NSUN2 expression, adjusted HRs were 0.77 (0.49–1.19) for Intermediate vs Exhaustion (p = 0.240), and 0.61 (0.36–1.03) for Activation vs Exhaustion (p = 0.063).
Table 3.
T-cell activation status and mortality from HNSCC, stratified by expression level of NSUN2
| Stratification | Death |
|||
|---|---|---|---|---|
| Variable | Variable | HRa | 95% CIb | P value |
| T-cell activation | ||||
| Exhaustion | 1.00 | |||
| Intermediate | 1.08 | 0.77–1.50 | 0.664 | |
| Activation | 1.03 | 0.73–1.47 | 0.852 | |
| P (for trend) | 0.84 | |||
| Model 1 | ||||
| Low NSUN2 | T-cell activation | |||
| Exhaustion | 1.00 | |||
| Intermediate | 2.01 | 1.15–3.52 | 0.015 | |
| Activation | 2.16 | 1.22–3.82 | 0.008 | |
| P (for trend) | 0.009 | |||
| High NSUN2 | T-cell activation | |||
| Exhaustion | 1.00 | |||
| Intermediate | 0.74 | 0.48–1.14 | 0.174 | |
| Activation | 0.57 | 0.34–0.96 | 0.033 | |
| P (for trend) | 0.028 | |||
| P value for the interaction between NSUN2 level and T-cell activation | 0.002 | |||
| Model 2 | ||||
| Low NSUN2 | T-cell activation | |||
| Exhaustion | 1.00 | |||
| Intermediate | 1.97 | 1.12–3.47 | 0.019 | |
| Activation | 2.06 | 1.16–3.68 | 0.014 | |
| P (for trend) | 0.016 | |||
| High NSUN2 | T-cell activation | |||
| Exhaustion | 1.00 | |||
| Intermediate | 0.77 | 0.49–1.19 | 0.240 | |
| Activation | 0.61 | 0.36–1.03 | 0.063 | |
| P (for trend) | 0.056 | |||
| P value for the interaction between NSUN2 level and T-cell activation | 0.004 | |||
aHR: adjusted hazard ratio, which was obtained from a multivariate Cox proportional hazards regression model with covariates of patient age at diagnosis (per 5 yrs), disease stage, tumor grade, gender (male vs female), smoking status (yes vs no) and primary tumor site (tongue, pharynx and other) in the model 1 (n =247 for low NSUN2 group and n =240 for high NSUN2). In model 2, HPV status (positive vs negative) was also included as a covariate (n = 237 for low NSUN2 group, and n =234 for high NSUN2).
bCI: confidence interval.
Pathway analysis for gene co-expressed with NSUN2
Among the 15 pathways that are enriched for genes coexpressed with NSUN2, the majority (11 pathways) are immune function- or inflammation-related pathways (such as interleukins, PD-1 signaling, TCR signaling, neutrophil degranulation, NF-kB and immunoregulatory interaction) (Figure 3) (adjusted p <0.001). Gene-set enrichment analysis showed that high expression of NSUN2 was associated with low expression of genes in pathways such as IFN-γ signaling (normalized enrichment score (NES) = - 2.94, false-discovery rate (FDR) =0.006), TCR signaling (NES = −2.80, FDR =0.006), innate immune system (NES = −3.13, FDR =0.006), adaptive immune system (NES = −3.75, FDR = 0.006), PD-1 signaling (NES = −3.75, FDR =0.006), Immunoregulatory interaction (NES = −4.29, FDR = 0.006), IL-2 signaling (NES = −2.67, FDR =0.006) , IL-3/IL-5 and GM-CSF signaling (NES = −2.37, FDR = 0.006). High expression of NSUN2 was associated with high expression of genes in pathways such as cell cycle checkpoint (NES = 2.60, FDR =0.006), epigenetic regulation of gene expression (NES = 2.49, FDR = 0.006), gene silencing by RNA (NES = 2.46, FDR = 0.006), mRNA splicing (NES = 2.81, FDR = 0.006), unfolded protein response (NES = 2.27, FDR = 0.006), and mitochondrial translation (NES = 2.27, FDR = 0.006) and tRNA modification (NES = 2.44, FDR =0.006) and processing (NES = 2.49, FDR = 0.006) (Supplementary Table S1).
Figure 3.
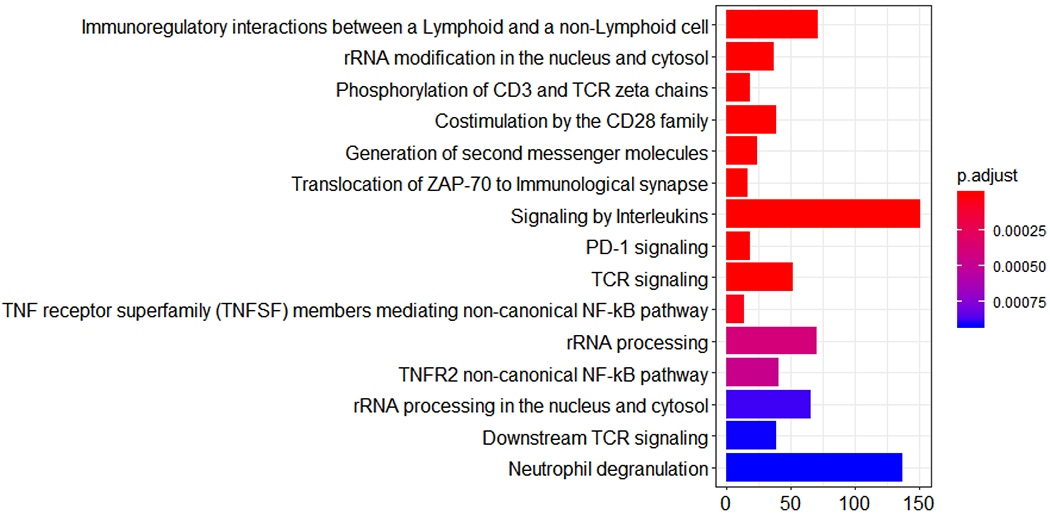
Bar plot of the pathways enriched for genes that are coexpressed with NSUN2. The size of the bar quantifies the number of genes in the pathway that are significantly coexpressed with NSUN2.
Discussion
In this study, we investigated the association of T-cell activation score with patient survival and the interaction between NSUN2 expression and the T-cell activation status in HNSCC patient survival. In our analysis of 520 patients with primary HNSCC, the relationship between T-cell activation score and patient survival was in the opposite direction for tumors with high expression of NSUN2 as for tumors with low expression of NSUN2. We found that in the subgroup with a low expression of NSUN2, patients with a high T-cell activation score lived 34.9 months shorter on average than those with a low T-cell activation score, whereas in the subgroup with high expression of NSUN2, patients with a high T-cell activation score lived 133.1 months longer on average than those with a low T-cell activation score. There was no significant difference in median survival between patients with high and low T-cell activation scores when not stratifying by NSUN2 expression level. Interaction analysis with a multivariate Cox model suggested a statistically significant interaction between expression of NSUN2 and T-cell activation score in HNSCC. When we included HPV status in the model—which reduced the sample size to 500 patients—the relationship remained statistically significant. This consistency implies that the NSUN2 expression level modifies the effect of T-cell activation on patient survival in HNSCC regardless of HPV status.
T-cell exhaustion is a hallmark of human cancer—including HNSCC—and is characterized by high expression of deactivating immune checkpoint proteins such as PD-1/PD-L1, CTLA4, LAG3, TIM3 and TIGIT. Reinvigoration of effector CD8+ T cells by immune-checkpoint blockades such as anti-PD-1/anti-PD-L1 and anti-CTLA4 improves patient survival in several human solid cancers, including HNSCC [3–5]. However, not all patients respond well to the treatment. Unexpectedly, even some patients treated with immunotherapy paradoxically showed hyper-progression of tumor growth [38, 39]. Lo Russo and colleagues reported that hyper-progression occurred in about 25% patients (39 out of 152) with non-small cell lung cancer (NSCLC) who received PD-1/PD-L1 blockade [38]. They found enriched tumor-associated macrophages (TAM) with M2-like CD163+CD33+PD-L1+ phenotype in the hyper-progression patients. Patient-derived xenograft (PDX) mouse models reproduced the phenotype and the clustering of TAMs. Saada-Bouzid and colleagues retrospectively evaluated the response of patients with recurrent and/or metastatic HNSCC to anti-PD-1/PD-L1 immunotherapy, and found 29% patients with hyper-progression, which was positively associated with a shorter progression-free survival [40]. By performing next-generation sequencing analysis on 155 patients with a stage-IV cancer who received immunotherapy, Kato and colleagues demonstrated that those with hyper-progression showed MDM2 family amplification or EGFR aberrations or DNMT3A alterations [41]. A potential mechanism was hypothesized: that anti-PD-1/PD-L1 induced IFN-γ-activated JAK-STAT signaling, consequently increasing interferon regulator factor (IRF)-8 expression that further resulted in expression of MDM2 [41–43]. MDM2 overexpression inhibits the p53 tumor suppressor [41]. Champiat and colleagues retrospectively evaluated 218 consecutive patients enrolled and treated in phase-I clinical trials of anti-PD-1/PD-L1 therapy, and found that 9% (12 of 131) patients had hyperprogression after the immunotherapy treatment, and that hyper-progression was associated with an elevated death risk [44]. Ferrara and colleagues reported that 13.8% (56 of 406) of patients treated with immunotherapy and 5.1% (3 of 59) of patients treated with single-agent chemotherapy had hyper-progression in NSCLC, and that these hyper-progressive patients had poor prognosis [45]. However, the potential molecular mechanisms underlying the hyper-progression are unclear and have yet to be characterized. The reactivation of CD8+ T-cells by PD-1 blockade results in elevated IFN-γ, which has been shown to activate tumor immunosuppressive myeloid cells, and increased inhibitory metabolites (such as indoleamine 2,3 –dioxygenase) controlling Treg differentiation, or leading to aberrant inflammation and a microenvironment that favors immune escape and tumor growth [46–49].
NSUN2, as a tRNA methyltransferase, modifies both tRNAs and mRNAs by adding m5C methylation to RNAs. NSUN2 is essential factor for maintenance of self-renewal and differentiation in stem cells [24, 50, 51]. CD8+ T-cells of sufficient stemness provide the capacity within the immune system to respond the stimuli of tumor neoantigens, and keep CD8+ T cells at the enough ‘ammunition’ even with chronic antigen exposure. It has been reported that NSUN2 promotes the translation of intercellular adhesion molecule 1 (ICAM-1) by methylating ICAM-1 mRNA [52], which can further suppress tumor metastasis by inhibiting M2-macrophage polarization. In this study, we found a statistically significant negative association between expression of NSUN2 and the expression of markers of M2 macrophages, including CD163, CD33, PDCD1 (aka PD-1), and myeloid cell marker CD45. This observation suggests that a low NSUN2 expression is accompanied with a high level of M2 macrophage and myeloid cells. We found that, conditioning on low expression of NSUN2, patients with a high T-cell activation score exhibited inferior overall survival than those with a low T-cell activation score, suggesting that a phenomenon similar to hyper-progression could be occurring at high immune response. We do not know what molecular mechanisms underlie this phenomenon—it may be complicated, but it warrants exploration. Indeed, in the context of tumors, the immune system may have dual functions of both eradicating ‘foreign antigens’, and contributing to tumor growth through direct or indirect mechanisms such as DNA damage and angiogenesis/tumor tissue remodeling induced by inflammation and free radicals [53–55]. Recently, research utilizing animal models demonstrated that RNA-editing induced RNA structure switching from single-strand RNA (ssRNA) to dsRNA could increase tumor inflammation and enhance the efficacy of PD-1 blockade [12]. Gene-set enrichment analysis for NSUN2 co-expressed genes suggests that low expression of NSUN2 could lead to excessive inflammation and immune response (negatively associated with IFN-γ signaling), and could synergize with high T-cell activation score to result in increased mortality.
Conclusions
This study is the first one to demonstrate the interaction between T-cell activation score and expression of NSUN2 affects patient survival in HNSCC. T-cell activation score was negatively associated with NSUN2 expression, but was not associated with patient survival overall. However, in the subgroup of patients with low NSUN2 expression, high T-cell activation score significantly increased the risk of mortality, whereas an opposite association was observed in the subgroup of patients with high expression of NSUN2, where a (non-significant) reduction in the risk of mortality manifested for those with high T-cell activation score. These findings suggest that immune checkpoint inhibitors could benefit patients with high NSUN2 expression by reinvigorating effector T cells, but may be detrimental to patients who have a low NSUN2 expression. NSUN2 expression is a potential marker for precision immunotherapy in HNSCC, and the outcome of immunotherapy might be able to be improved by targeting NSUN2. Studies with larger sample sizes are warranted to further examine how NSUN2 expression level affects the patients’ response to immune checkpoint blockade.
Supplementary Material
Supplementary Table 1. Gene-set enrichment (GSEA) analysis results
Acknowledgement:
We thank Dr. Barbara Burtness at Yale School of Medicine for her critical reading and discussion in writing manuscript.
Funding: Vincent L Cannataro was supported by Yale Cancer Biology Postdoctoral Training grant (NIH T32 CA193200).
Availability of data and material: The data that support the findings of this study are available from head and neck squamous carcinoma (TCGA, Provisional) at cBioPortal for Cancer Genomics (https://www.cbioportal.org).
Footnotes
Conflict of interest: None declared.
Declarations:
Ethics approval and consent to participate: no formal consent is required for this type of study. The ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards were followed in performing all procedures in this study involving human subjects. The study presented here complies with the current laws of the United States of America.
Consent for publication: not applicable.
References:
- [1].Chen L, Han X. Anti-PD-1/PD-L1 therapy of human cancer: past, present, and future. J Clin Invest. 2015;125:3384–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Burtness B, Harrington KJ, Greil R, Soulieres D, Tahara M, Castro G, et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOT-048): a randomized, open-lable, phase 3 study. Lancet. 2019. [DOI] [PubMed] [Google Scholar]
- [3].Ma BBY, Lim WT, Goh BC, Hui EP, Lo KW, Pettinger A, et al. Antitumor Activity of Nivolumab in Recurrent and Metastatic Nasopharyngeal Carcinoma: An International, Multicenter Study of the Mayo Clinic Phase 2 Consortium (NCI-9742). J Clin Oncol. 2018;36:1412–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hsu C, Lee SH, Ejadi S, Even C, Cohen RB, Le Tourneau C, et al. Safety and Antitumor Activity of Pembrolizumab in Patients With Programmed Death-Ligand 1-Positive Nasopharyngeal Carcinoma: Results of the KEYNOTE-028 Study. J Clin Oncol. 2017;35:4050–6. [DOI] [PubMed] [Google Scholar]
- [5].Ferris RL, Blumenschein G Jr., Fayette J, Guigay J, Colevas AD, Licitra L, et al. Nivolumab for Recurrent Squamous-Cell Carcinoma of the Head and Neck. N Engl J Med. 2016;375:1856–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Seiwert TY, Burtness B, Mehra R, Weiss J, Berger R, Eder JP, et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase 1b trial. Lancet Oncol. 2016;17:956–65. [DOI] [PubMed] [Google Scholar]
- [7].Chow LQM, Haddad R, Gupta S, Mahipal A, Mehra R, Tahara M, et al. Antitumor Activity of Pembrolizumab in Biomarker-Unselected Patients With Recurrent and/or Metastatic Head and Neck Squamous Cell Carcinoma: Results From the Phase Ib KEYNOTE-012 Expansion Cohort. J Clin Oncol. 2016;34:3838–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bahig H, Aubin F, Stagg J, Gologan O, Ballivy O, Bissada E, et al. Phase I/II trial of Durvalumab plus Tremelimumab and stereotactic body radiotherapy for metastatic head and neck carcinoma. BMC Cancer. 2019;19:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Fourcade J, Sun Z, Pagliano O, Guillaume P, Luescher IF, Sander C, et al. CD8(+) T cells specific for tumor antigens can be rendered dysfunctional by the tumor microenvironment through upregulation of the inhibitory receptors BTLA and PD-1. Cancer Res. 2012;72:887–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wherry EJ. T cell exhaustion. Nat Immunol. 2011;12:492–9. [DOI] [PubMed] [Google Scholar]
- [11].Zhang J, Bu X, Wang H, Zhu Y, Geng Y, Nihira NT, et al. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature. 2018;553:91–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ishizuka JJ, Manguso RT, Cheruiyot CK, Bi K, Panda A, Iracheta-Vellve A, et al. Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature. 2019;565:43–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Brzezicha B, Schmidt M, Makalowska I, Jarmolowski A, Pienkowska J, Szweykowska-Kulinska Z. Identification of human tRNA:m5C methyltransferase catalysing intron-dependent m5C formation in the first position of the anticodon of the pre-tRNA Leu (CAA). Nucleic Acids Res. 2006;34:6034–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Shinoda S, Kitagawa S, Nakagawa S, Wei FY, Tomizawa K, Araki K, et al. Mammalian NSUN2 introduces 5-methylcytidines into mitochondrial tRNAs. Nucleic Acids Res. 2019;47:8734–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Van Haute L, Lee SY, McCann BJ, Powell CA, Bansal D, Vasiliauskaite L, et al. NSUN2 introduces 5-methylcytosines in mammalian mitochondrial tRNAs. Nucleic Acids Res. 2019;47:8720–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014;33:2020–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43:613–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sobala A, Hutvagner G. Small RNAs derived from the 5’ end of tRNA can inhibit protein translation in human cells. RNA Biol. 2013;10:553–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol Cell. 2010;40:228–37. [DOI] [PubMed] [Google Scholar]
- [20].Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, et al. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol. 2012;19:900–5. [DOI] [PubMed] [Google Scholar]
- [21].Liu S, Chen Y, Ren Y, Zhou J, Ren J, Lee I, et al. A tRNA-derived RNA Fragment Plays an Important Role in the Mechanism of Arsenite -induced Cellular Responses. Sci Rep. 2018;8:16838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Popis MC, Blanco S, Frye M. Posttranscriptional methylation of transfer and ribosomal RNA in stress response pathways, cell differentiation, and cancer. Curr Opin Oncol. 2016;28:65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hussain S, Tuorto F, Menon S, Blanco S, Cox C, Flores JV, et al. The mouse cytosine-5 RNA methyltransferase NSun2 is a component of the chromatoid body and required for testis differentiation. Mol Cell Biol. 2013;33:1561–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Flores JV, Cordero-Espinoza L, Oeztuerk-Winder F, Andersson-Rolf A, Selmi T, Blanco S, et al. Cytosine-5 RNA Methylation Regulates Neural Stem Cell Differentiation and Motility. Stem Cell Reports. 2017;8:112–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yang JC, Risch E, Zhang M, Huang C, Huang H, Lu L. Association of tRNA methyltransferase NSUN2/IGF-II molecular signature with ovarian cancer survival. Future Oncol. 2017;13:1981–90. [DOI] [PubMed] [Google Scholar]
- [26].Gao Y, Wang Z, Zhu Y, Zhu Q, Yang Y, Jin Y, et al. NOP2/Sun RNA methyltransferase 2 promotes tumor progression via its interacting partner RPL6 in gallbladder carcinoma. Cancer Sci. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chen X, Li A, Sun BF, Yang Y, Han YN, Yuan X, et al. 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat Cell Biol. 2019;21:978–90. [DOI] [PubMed] [Google Scholar]
- [28].Lu L, Zhu G, Zeng H, Xu Q, Holzmann K. High tRNA Transferase NSUN2 Gene Expression is Associated with Poor Prognosis in Head and Neck Squamous Carcinoma. Cancer investigation. 2018;36:246–53. [DOI] [PubMed] [Google Scholar]
- [29].Xing J, Yi J, Cai X, Tang H, Liu Z, Zhang X, et al. NSun2 Promotes Cell Growth via Elevating Cyclin-Dependent Kinase 1 Translation. Mol Cell Biol. 2015;35:4043–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhang X, Liu Z, Yi J, Tang H, Xing J, Yu M, et al. The tRNA methyltransferase NSun2 stabilizes p16INK(4) mRNA by methylating the 3’-untranslated region of p16. Nat Commun. 2012;3:712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sharpless NE. INK4a/ARF: a multifunctional tumor suppressor locus. Mutat Res. 2005;576:22–38. [DOI] [PubMed] [Google Scholar]
- [32].Cancer Genome Atlas N Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Cannataro VL, Gaffney SG, Sasaki T, Issaeva N, Grewal NKS, Grandis JR, et al. APOBEC-induced mutations and their cancer effect size in head and neck squamous cell carcinoma. Oncogene. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Yu G, He QY. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol Biosyst. 2016;12:477–9. [DOI] [PubMed] [Google Scholar]
- [37].Lu L, Bai Y, Wang Z. Elevated T cell activation score is associated with improved survival of breast cancer. Breast Cancer Res Treat. 2017;164:689–96. [DOI] [PubMed] [Google Scholar]
- [38].Lo Russo G, Moro M, Sommariva M, Cancila V, Boeri M, Centonze G, et al. Antibody-Fc/FcR Interaction on Macrophages as a Mechanism for Hyperprogressive Disease in Non-small Cell Lung Cancer Subsequent to PD-1/PD-L1 Blockade. Clinical cancer research : an official journal of the American Association for Cancer Research. 2019;25:989–99. [DOI] [PubMed] [Google Scholar]
- [39].Knorr DA, Ravetch JV. Immunotherapy and Hyperprogression: Unwanted Outcomes, Unclear Mechanism. Clinical cancer research : an official journal of the American Association for Cancer Research. 2019;25:904–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Saada-Bouzid E, Defaucheux C, Karabajakian A, Coloma VP, Servois V, Paoletti X, et al. Hyperprogression during anti-PD-1/PD-L1 therapy in patients with recurrent and/or metastatic head and neck squamous cell carcinoma. Annals of oncology : official journal of the European Society for Medical Oncology. 2017;28:1605–11. [DOI] [PubMed] [Google Scholar]
- [41].Kato S, Goodman A, Walavalkar V, Barkauskas DA, Sharabi A, Kurzrock R. Hyperprogressors after Immunotherapy: Analysis of Genomic Alterations Associated with Accelerated Growth Rate. Clinical cancer research : an official journal of the American Association for Cancer Research. 2017;23:4242–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zhou JX, Lee CH, Qi CF, Wang H, Naghashfar Z, Abbasi S, et al. IFN regulatory factor 8 regulates MDM2 in germinal center B cells. J Immunol. 2009;183:3188–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Waight JD, Netherby C, Hensen ML, Miller A, Hu Q, Liu S, et al. Myeloid-derived suppressor cell development is regulated by a STAT/IRF-8 axis. J Clin Invest. 2013;123:4464–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Champiat S, Dercle L, Ammari S, Massard C, Hollebecque A, Postel-Vinay S, et al. Hyperprogressive Disease Is a New Pattern of Progression in Cancer Patients Treated by Anti-PD-1/PD-L1. Clinical cancer research : an official journal of the American Association for Cancer Research. 2017;23:1920–8. [DOI] [PubMed] [Google Scholar]
- [45].Ferrara R, Mezquita L, Texier M, Lahmar J, Audigier-Valette C, Tessonnier L, et al. Hyperprogressive Disease in Patients With Advanced Non-Small Cell Lung Cancer Treated With PD-1/PD-L1 Inhibitors or With Single-Agent Chemotherapy. JAMA Oncol. 2018;4:1543–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Akbay EA, Koyama S, Liu Y, Dries R, Bufe LE, Silkes M, et al. Interleukin-17A Promotes Lung Tumor Progression through Neutrophil Attraction to Tumor Sites and Mediating Resistance to PD-1 Blockade. J Thorac Oncol. 2017;12:1268–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Baban B, Chandler PR, Sharma MD, Pihkala J, Koni PA, Munn DH, et al. IDO activates regulatory T cells and blocks their conversion into Th17-like T cells. J Immunol. 2009;183:2475–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Spranger S, Spaapen RM, Zha Y, Williams J, Meng Y, Ha TT, et al. Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci Transl Med. 2013;5:200ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Huang B, Pan PY, Li Q, Sato AI, Levy DE, Bromberg J, et al. Gr-1+CD115+ immature myeloid suppressor cells mediate the development of tumor-induced T regulatory cells and T-cell anergy in tumor-bearing host. Cancer Res. 2006;66:1123–31. [DOI] [PubMed] [Google Scholar]
- [50].Frye M, Harada BT, Behm M, He C. RNA modifications modulate gene expression during development. Science. 2018;361:1346–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Blanco S, Kurowski A, Nichols J, Watt FM, Benitah SA, Frye M. The RNA-methyltransferase Misu (NSun2) poises epidermal stem cells to differentiate. PLoS genetics. 2011;7:e1002403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Luo Y, Feng J, Xu Q, Wang W, Wang X. NSun2 Deficiency Protects Endothelium From Inflammation via mRNA Methylation of ICAM-1. Circ Res. 2016;118:944–56. [DOI] [PubMed] [Google Scholar]
- [53].Guo X, Zhai L, Xue R, Shi J, Zeng Q, Gao C. Mast Cell Tryptase Contributes to Pancreatic Cancer Growth through Promoting Angiogenesis via Activation of Angiopoietin-1. Int J Mol Sci. 2016;17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Pollard JW. Tumour-educated macrophages promote tumour progression and metastasis. Nat Rev Cancer. 2004;4:71–8. [DOI] [PubMed] [Google Scholar]
- [55].DeNardo DG, Andreu P, Coussens LM. Interactions between lymphocytes and myeloid cells regulate pro- versus anti-tumor immunity. Cancer Metastasis Rev. 2010;29:309–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Gene-set enrichment (GSEA) analysis results


