Abstract
The outbreak of viral pneumonia caused by the novel coronavirus SARS-CoV-2 that began in December 2019 caused high mortality. It has been suggested that the main protease (Mpro) of SARS-CoV-2 may be an important target to discover pharmaceutical compounds for the therapy of this life-threatening disease. Remdesivir, ritonavir and chloroquine have all been reported to play a role in suppressing SARS-CoV-2. Here, we applied a molecular docking method to study the binding stability of these drugs with SARS-CoV-2 Mpro. It appeared that the ligand–protein binding stability of the alliin and SARS-CoV-2 Mpro complex was better than others. The results suggested that alliin may serve as a good candidate as an inhibitor of SARS-CoV-2 Mpro. Therefore, the present research may provide some meaningful guidance for the prevention and treatment of SARS-CoV-2.
Keywords: : alliin, molecular docking, remdesivir, ritonavir, SARS-CoV-2 Mpro
METHOD SUMMARY
Molecular docking was used to assess the binding stability of various drugs with SARS-CoV-2 main protease (Mpro). PyMOL software was used to align the protein structures of SARS-CoV Mpro (ID: 5C5N) and SARS-CoV-2 Mpro (ID: 6LU7).
Since December 2019, many cases of viral pneumonia have been found in Wuhan City (Hubei Province, China), all of which were diagnosed with viral pneumonia and pulmonary infection. On 11 February 2020, the new coronavirus causing the pneumonia epidemic in Wuhan was officially named SARS-CoV-2 by the International Committee on Taxonomy of Viruses.
Viral pneumonia is inflammation of the lungs caused by a viral infection of the upper respiratory tract that spreads downward; it is an inhalation infection, transmitted via person-to-person droplets and contact. Viruses that cause viral pneumonia are common and include influenza viruses, parainfluenza virus, cytomegalovirus, adenovirus, rhinovirus and coronavirus. An outbreak of infectious atypical pneumonia in 2002–2003 was caused by the SARS virus SARS-CoV, a coronavirus [1,2].
It has been reported in the literature that the source of SARS-CoV-2 may be derived from bats. Moreover, homology analysis has shown that the similarity of the entire genome sequence of SARS-CoV-2 to SARS-CoV and the Middle East Respiratory Syndrome virus was 79.0 and 51.8%, respectively. The highest similarity was with the SARS-like coronavirus (SL-CoV ZC45 and ZXC21) carried by two chrysanthemum bats found in Zhoushan in 2005, ranging from 87.6 to 87.7%. The number of novel coronavirus infections is on the rise, which is a matter of great concern. Scientific research institutions and pharmaceutical companies around the world are committed to developing new vaccines and therapeutic drugs for SARS-CoV-2. There are two types of known drugs which are often discussed and may offer therapeutic benefit at present. One is ritonavir, an inhibitor of HIV protease; according to research, SARS and SARS-CoV-2 have a protease target similar to that of HIV. The other is remdesivir, a new nucleoside analog antiviral drug against Ebola. Chloroquine, a drug used for the treatment of malaria and rheumatic diseases, has also been proved to inhibit virus replication and has been shown to play a role in suppressing SARS-CoV-2 in cellular experiments [3].
The complex phytochemistry of garlic (Allium sativum) has often been studied. A large number of recent studies have identified the functional actions of garlic on cardiovascular disease [4] and cancer [5,6]. Furthermore, garlic has been demonstrated to possess immunomodulation, antiinflammatory [7], antimicrobial [8], antioxidant [9] and antiviral properties [10]. Alliin is the main active component of garlic and a wide range of biological activities of garlic are shown to originate from this compound.
SARS-CoV-2 main protease (Mpro), also known as chymotrypsin-like protease, is primarily responsible for cleaving polyproteins, while a papain-like protease also aids in the process. In an effort to better investigate inhibitors of SARS-CoV-2 Mpro, a molecular docking approach was used to examine the docking interaction between the three inhibitors (remdesivir, ritonavir and chloroquine) and SARS-CoV-2 Mpro. In addition, alliin was screened out to dock with SARS-CoV-2 Mpro, which may provide clues to the prevention and treatment of SARS-CoV-2.
Materials & methods
Computational methods
Structures of SARS-CoV Mpro (ID: 5C5N) and SARS-CoV-2 Mpro (ID: 6LU7) were obtained from the Protein Data Bank. The protein structures were aligned using the PyMOL software (PyMOL Molecular graphics System, Version 1.4).
Molecular docking
Molecular docking is an efficient way to investigate the noncovalent binding of macromolecules or a macromolecule (receptor) and a small molecule (ligand) [11,12]. Molecular docking was performed using the molecular operating environment docking software (MOE 2015). We obtained the sdf structure formats of ligands from the PubChem database: remdesivir (CID: 121304016), ritonavir (CID: 392622), chloroquine (CID: 2719) and alliin (CID: 87310). Then the proteins were quick prepared. At the end of docking, the binding modes of four ligands to SARS-CoV-2 Mpro and SARS-CoV Mpro were observed to identify the critical residues.
Results & discussion
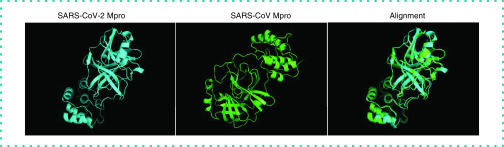
To understand the relationship between SARS-CoV-2 and SARS-CoV, we aligned the protein sequences of the two viruses. Figure 1 shows the coincidence of their 3D proteins; the root-mean-square deviation value between them is 0.585 Å, which shows that the two viruses’ Mpros are similar.
Figure 1. Protein alignment by PyMOL.
According to reports of the Wuhan Virus Research of the Chinese Academy of Sciences and the Institute of Toxicology and Pharmaceuticals of the Academy of Military Medical Sciences, remdesivir, ritonavir and chloroquine might play a role in suppressing SARS-CoV-2 at the cellular level. In this study, we used a molecular docking method to study the binding of these drugs with SARS-CoV-2 Mpro and compared the potential inhibitory effects of alliin on SARS-CoV-2 Mpro and SARS-CoV Mpro.
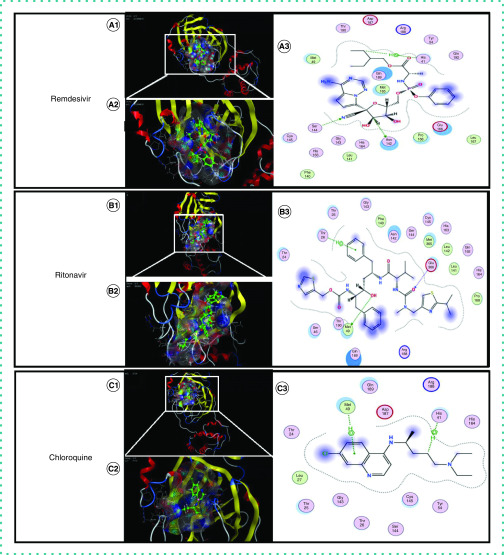
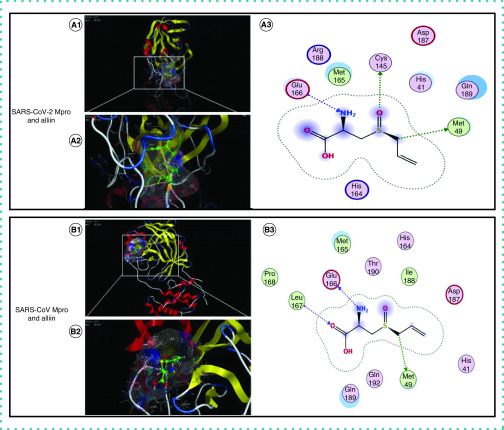
The molecular docking results revealed a high affinity between SARS-CoV-2 Mpro and the four ligands, as shown in Table 1; the molecular docking scores of SARS-CoV Mpro and alliin are also listed. As shown in Figure 2A, remdesivir was found to interact with SARS-CoV-2 Mpro at Asn-142, Ser-144 and His-41 with two H-bonds and one arene H-bond. Figure 2B shows that ritonavir was observed to interact with SARS-CoV-2 Mpro at Met-49, Glu-166 and Thr-26 with two H-bonds and one arene H-bond. As shown in Figure 2C, chloroquine was found to be interacting with SARS-CoV-2 Mpro at His-41 and Met-49 with two arene H-bonds. Finally, as shown in Figure 3, alliin was found to interact with SARS-CoV Mpro at Leu-167, Met-49 and Glu-166 with three H-bonds; for SARS-CoV-2 Mpro, the observed docking sites of alliin were Cys-145, Met-49 and Glu-166 with three H-bonds.
Table 1. . Molecular docking results of SARS-CoV-2 main protease and severe acute respiratory syndrome coronavirus main protease.
| Receptor | Ligand | Score | RMSD |
|---|---|---|---|
| SARS-CoV-2 Mpro | remdesivir | -9.4339 | 3.1145 |
| SARS-CoV-2 Mpro | ritonavir | -8.9094 | 1.8379 |
| SARS-CoV-2 Mpro | chloroquine | -7.0456 | 1.3582 |
| SARS-CoV-2 Mpro | alliin | -12.6107 | 1.5180 |
| SARS-CoV Mpro | alliin | -4.9641 | 1.6326 |
Mpro: Main protease; RMSD: Root-mean-square deviation; SARS-CoV: Severe acute respiratory syndrome coronavirus; SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2.
Figure 2. Interactions of known drugs with hydrophobic pockets of SARS-CoV-2 main protease.
(A) 1 and 2 show interactions of remdesivir with hydrophobic pockets of SARS-CoV-2 Mpro; 3 shows molecular interactions between remdesivir and SARS-CoV-2 Mpro. (B) 1 and 2 show interactions of ritonavir with hydrophobic pockets of SARS-CoV-2 Mpro; 3 shows molecular interactions between ritonavir and SARS-CoV-2 Mpro. (C) 1 and 2 show interactions of chloroquine with hydrophobic pockets of SARS-CoV-2 Mpro; 3 shows molecular interactions between chloroquine and SARS-CoV-2 Mpro.
Mpro: Main protease; SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2.
Figure 3. Interactions of alliin with hydrophobic pockets of SARS-CoV-2 main protease and severe acute respiratory syndrome coronavirus main protease.
(A) 1 and 2 show interactions of alliin with hydrophobic pockets of SARS-CoV-2 Mpro; 3 shows molecular interactions between alliin and SARS-CoV-2 Mpro. (B) 1 and 2 show interactions of alliin with hydrophobic pockets of SARS-CoV Mpro; 3 shows molecular interactions between alliin and SARS-CoV Mpro.
Mpro: Main protease; SARS-CoV: Severe acute respiratory syndrome coronavirus; SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2.
It has been reported that inhibiting the main protease would help to restrict viral maturation, thereby reducing the SARS-CoV-2 infection in humans [13]. The active site of SARS-CoV-2 Mpro is located in the gap between domains I (residues 8–101) and II (residues 102–184) and has a Cys–His catalytic dyad (Cys145 and His41) [14,15]. A recent article indicated that montelukast, a potential drug acting on SARS-CoV-2 Mpro, was well fitted into the active pocket of Mpro, in which hydrophobic amino acids (Thr24, Leu27, His41, Phe140, Cys145, His163, Met165, Pro168 and His172) compose a relatively hydrophobic environment to contain the compound and stabilize its conformation [16]. In another study, Qamar et al. revealed that SARS-CoV-2 has a Cys–His catalytic dyad (Cys-145 and His-41) consistent with the SARS chymotrypsin-like protease (Cys-145 and His-41) [17]. The authors indicated that 5,7,3′,4′-tetrahydroxy-2′-(3,3-dimethylallyl) isoflavone, a potential candidate for treatment of SARS-CoV-2, formed strong hydrogen bonds with the catalytic dyad residues (Cys-145 and His-41) and had significant interactions with the receptor-binding residues Thr24, Thr25, Thr26, Cys44, Thr45, Ser46, Met49, Asn142, Gly143, His164, Glu166 and Gln189 [18]. Consistent with this, in our study alliin was well fitted into the active pocket of SARS-CoV-2 Mpro, containing many amino acids, such as His41, His164, Met165, Asp187, Arg188 and Gln189, which formed significant interactions with the compound and stabilized its conformation. In addition, the docking scores indicated the strength of the ligand–protein complexes, with lower values indicating greater stability. It can therefore be inferred that the binding stability of the alliin and SARS-CoV-2 Mpro ligand–protein complex was better than that of the others tested.
Conclusion
The lack of effective anti-SARS-CoV-2 agents is a current problem and there is urgent demand to discover anti-SARS-CoV-2 inhibitors to combat this deadly disease. SARS-CoV-2 Mpro is an important target for the design of therapeutically useful drugs. In the present study, alliin was screened out to dock with SARS-CoV-2 Mpro and the mechanisms of alliin and known inhibitors were investigated. The results suggested that alliin may be a good candidate for the prevention and treatment of SARS-CoV-2.
Future perspective
Our results are based on in silico analysis. We have not conducted further in vivo and in vitro antiviral experiments yet, because we want to share the results with other researchers as quickly as possible. In the next study, we will be close to in vivo and in vitro evaluations and will prepare for clinical trial applications.
Author contributions
T Li designed and revised the work. B Cheng analyzed the data and wrote the manuscript. Both authors approved the final version of the manuscript.
Acknowledgments
The authors are grateful for the assistance of Jilin Agricultural Science and Technology University.
Financial & competing interests disclosure
This study was supported in part by Fund project of Jilin Agricultural Science and Technology University (no. 20190277) and in part by Fund project of Jilin Agricultural Science and Technology University (no. [2019]XZ003). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest.
No writing assistance was utilized in the production of this manuscript.
Open access
This work is licensed under the Attribution-NonCommercial-NoDerivatives 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
References
- 1.Jo S, Kim S, Shin DH. et al. Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzym. Inhib. Med. Chem. 35(1), 145–151 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang L, Bao BB, Song GQ. et al. Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: chemical synthesis, biological evaluation, molecular docking and 3D-QSAR study. Eur. J. Med. Chem. 137(1), 450–461 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu HZ. Drug treatment options for the 2019-new coronavirus (2019-nCoV). BioSci. Trends 14(1), 69–71 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Banerjee SK, Maulik SK. Effect of garlic on cardiovascular disorders: a review. Nutr. J. 1(4), 1–14 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Puccinelli MT, Stan SD. Dietary bioactive diallyl trisulfide in cancer prevention and treatment. Int. J. Mol. Sci. 18(8), 1645–1646 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Li S, Meng X. et al. Dietary natural products for prevention and treatment of breast cancer. Nutrients 9(7), 728–730 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Georgia SF, Catherine K. The immunomodulation and anti-inflammatory effects of garlic organosulfur compounds in cancer chemoprevention. Anti-Cancer Agents Med. Chem. 14(2), 233–240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morozova EA, Revtovich SV, Anufrieva NV. et al. Alliin is a suicide substrate of Citrobacter freundii methionine γ-lyase: structural bases of inactivation of the enzyme. Acta Crystallogr. Sect. D-Struct. Biol. 70(11), 3034–3042 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Banerjee SK, Mukherjee PK, Maulik SK. Garlic as an antioxidant: the good, the bad and the ugly. Phytother. Res. 17(2), 97–106 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Ankri S, Mirelman D. Antimicrobial properties of allicin from garlic. Microbes Infect. 1(2), 125–129 (1999). [DOI] [PubMed] [Google Scholar]
- 11.Fu Y, Chen Z, Sun J. Random drift particle swarm optimisation algorithm for highly flexible protein-ligand docking. J. Theor. Biol. 457, 180–189 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Bazgier V, Berka K, Otyepka M. et al. Exponential repulsion improves structural predictability of molecular docking. J. Comput. Chem. 37(28), 2485–2494 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Navneet B, Sowmya K, Gopalakrishnan B. et al. De novo design of new chemical entities (NCEs) for SARS-CoV-2 using artificial intelligence (2020) (Epub ahead of print). https://chemrxiv.org/articles/De_Novo_Design_of_New_Chemical_Entities_NCEs_for_SARS-CoV-2_Using_Artificial_Intelligence/11998347/1
- 14.Jin Z, Du X, Xu Y. et al. Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature (2019) (Epub ahead of print). [DOI] [PubMed] [Google Scholar]
- 15.Yang HT, Xie WQ, Xue XY. et al. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 3(10), 1742–1752 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu C, Liu Y, Yang Y. et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. (2020) (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang HT, Yang MJ, Ding Y. et al. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl Acad. Sci. USA 100(23), 13190–13195 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qamar MT, Alqahtani SM, Alamri MA. et al. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J. Pharm. Sci. (2020) (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]