Abstract
The essential product of the Duchenne muscular dystrophy (DMD) gene is dystrophin1, a rod-like protein2 that protects striated myocytes from contraction-induced injury3,4. Dystrophin-related protein (or utrophin) retains most of the structural and protein binding elements of dystrophin5. Importantly, normal thymic expression in DMD patients6 should protect utrophin by central immunologic tolerance. We designed a codon-optimized, synthetic transgene encoding a miniaturized utrophin (μUtro), deliverable by adeno-associated virus (AAV) vectors. Here, we show that μUtro is a highly functional, non-immunogenic substitute for dystrophin, preventing the most deleterious histological and physiological aspects of muscular dystrophy in small and large animal models. Following systemic administration of an AAV-μUtro to neonatal dystrophin-deficient mdx mice, histological and biochemical markers of myonecrosis and regeneration are completely suppressed throughout growth to adult weight. In the dystrophin-deficient golden retriever model, μUtro non-toxically prevented myonecrosis, even in the most powerful muscles. In a stringent test of immunogenicity, focal expression of μUtro in the deletional-null German shorthaired pointer model produced no evidence of cell-mediated immunity, in contrast to the robust T cell response against similarly constructed μDystrophin (μDystro). These findings support a model in which utrophin-derived therapies might be used to treat clinical dystrophin deficiency, with a favorable immunologic profile and preserved function in the face of extreme miniaturization.
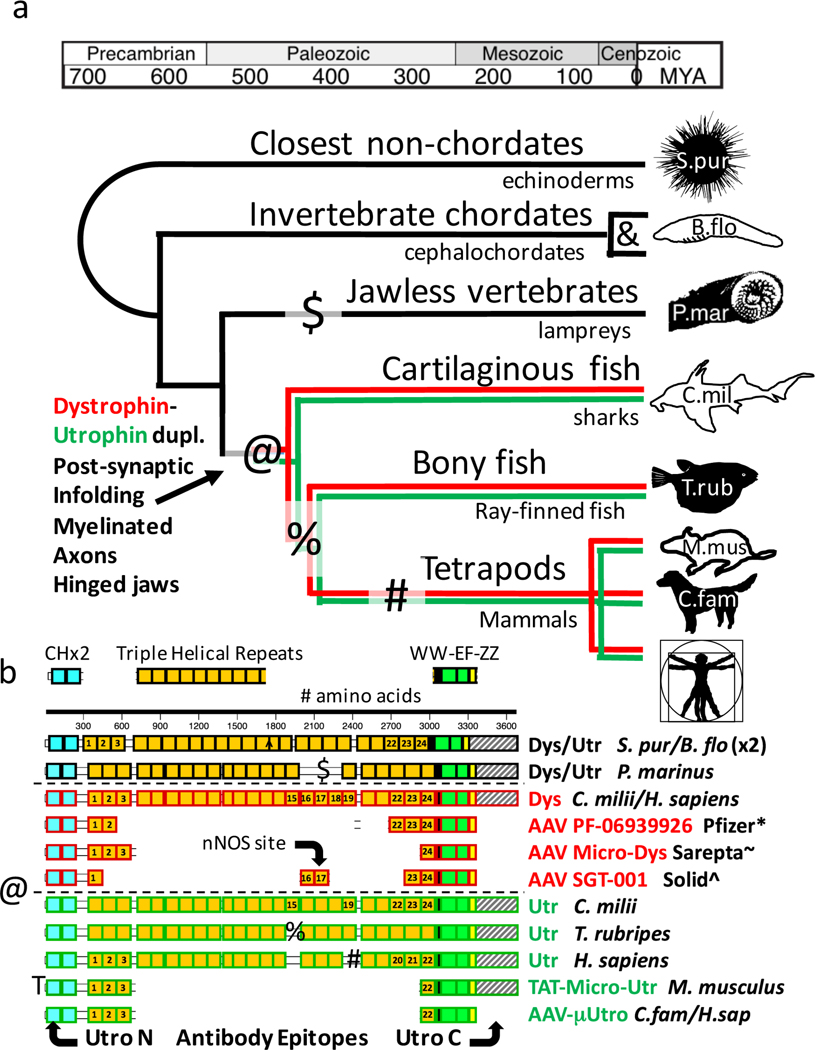
Although internally deleted vectors derived from human adenoviruses have been used to achieve somatic transfer of 12 kb cDNAs encoding full-length dystrophin, this approach has been abandoned because of the immunogenicity and limited biodistribution of the complex vector capsid7. Multiple vectors derived from human adeno-associated viruses (AAVs) have been shown to facilitate systemic gene transfer8–11, but their cloning capacity is limited to that of the wild-type virus (~5 kb), requiring removal by deletion of most of the dystrophin coding sequence, as in cases of Becker muscular dystrophy (BMD), which has a milder phenotype than Duchenne muscular dystrophy (DMD). An equally important second constraint on gene therapy for DMD is the deletional nature of the protein deficiency in most patients, with the potential for recombinant dystrophin as a ‘non-self’ protein to trigger autoimmune myositis12–14. Sponsors for clinical trials have recently announced holds for serious adverse events in response to regulatory concerns about vector-dose-dependent immunotoxicity15. We hypothesized that detailed analysis of the molecular evolution of dystrophin and its full-length paralogue utrophin might inform a synthetic biology approach to the challenge of maximizing functionality while minimizing immunogenicity. Our reconstruction of the remote history of dystrophin suggested that, at the protein’s inception, its rod domain contained 24 repeats of the ‘spectrin-like’ triple helical domain coopted from an N-terminal portion of another, much larger, strut-like cytoskeletal protein (C.D.G., manuscript in preparation). Crystal structures of triple helical repeats from dystrophin, utrophin and a closely related spectroplakin suggest that amino-acid side-chain interactions between adjacent repeats create an interlocking interface critical to the strength of the rod16,17. This principle may explain the phenotypes resulting from in-frame deletions and duplications in BMD patients and the rarity of deletions in chordate paralogues (for example, lamprey; Extended Data Fig. 1), as most disruptions of the native sequence of triple helical repeats have the potential to focally weaken the rod domain. To minimize the risk of creating a ‘weakest link’, we focused on deletions flanked on one side by the disordered domain classically labelled as ‘hinge 2’, and also deleted C-terminal sequences beyond the approximate end of the ZZ domain18,19. To take advantage of central immunological tolerance achieved through early developmental expression in the thymus6, we mapped these deletions in dystrophin onto the paralogous protein utrophin, which diverged from dystrophin early in vertebrate evolution (Extended Data Fig. 1). In light of these considerations, we synthesized transgenes based on wild-type utrophin mRNA sequences, and then improved expression by optimizing to the codon bias of mRNAs for the most abundant proteins of muscle: myosin and actin. Here we report on the results obtained in blinded pre-clinical studies using vectors based on AAV9 and the derived ancestral capsid ‘Anc80’20 to systemically deliver codon-optimized synthetic transgenes (μUtro, Canis familiaris and Homo sapiens) to all striated muscles (Extended Data Figs. 1b and 2a–e). Our transgenes are 22–27% divergent from those reported by Foster et al.21, Odom et al.22 and Sonnemann et al.23, reflecting differences in alignment, codon bias and primary protein structure, but most closely resemble the ‘delta R4–R23’ domain structures depicted as AAV-μDys (Sarepta-NCT02376816) and TAT-utrophin in Extended Data Fig. 1b. Like the lamprey dystrophin and two of the three transgenes selected for early clinical trials, our μUtro lacks a region corresponding to dystrophin R16–17 implicated in sarcolemmal nNOS localization (Extended Data Fig. 1b, AAV SGT-001 Solid-NCT03368742).
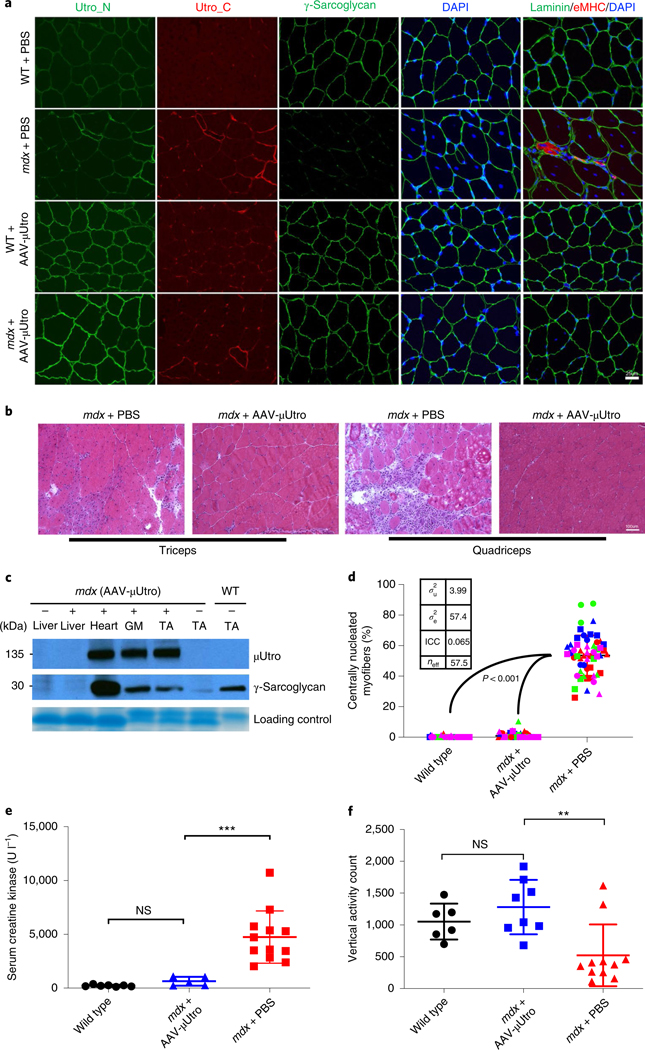
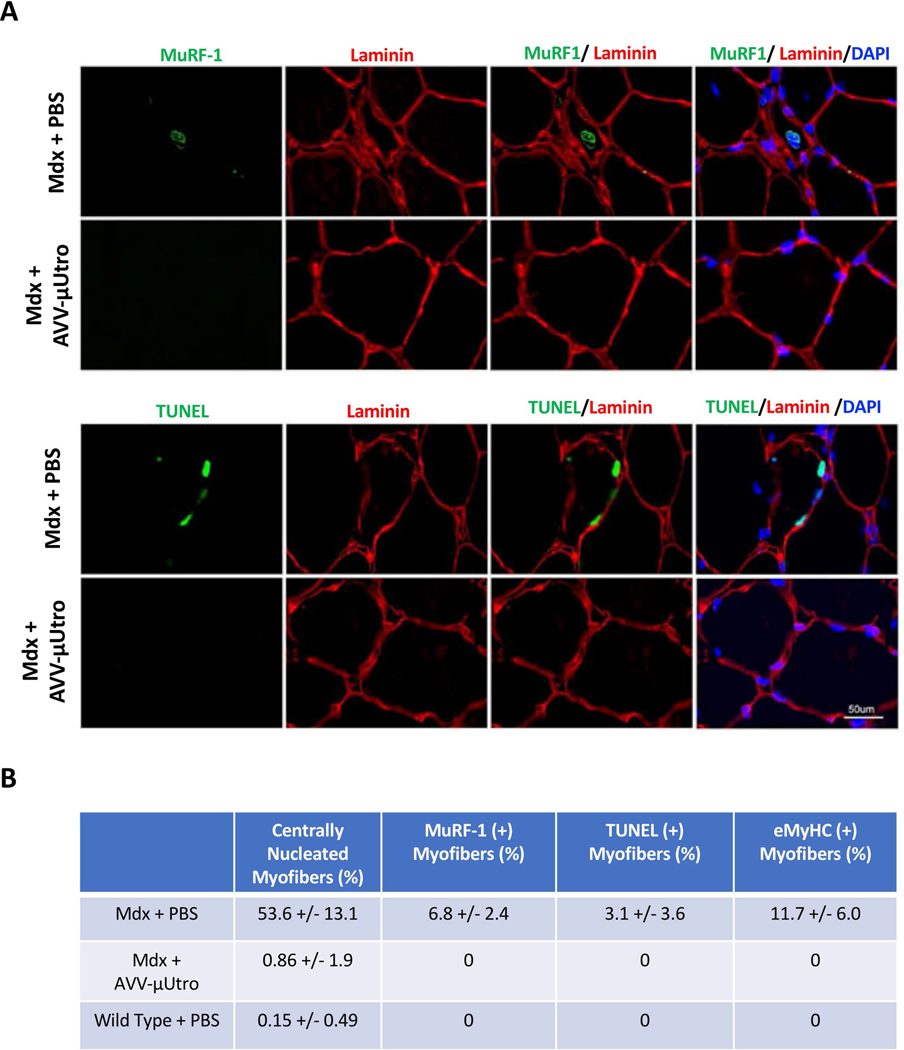
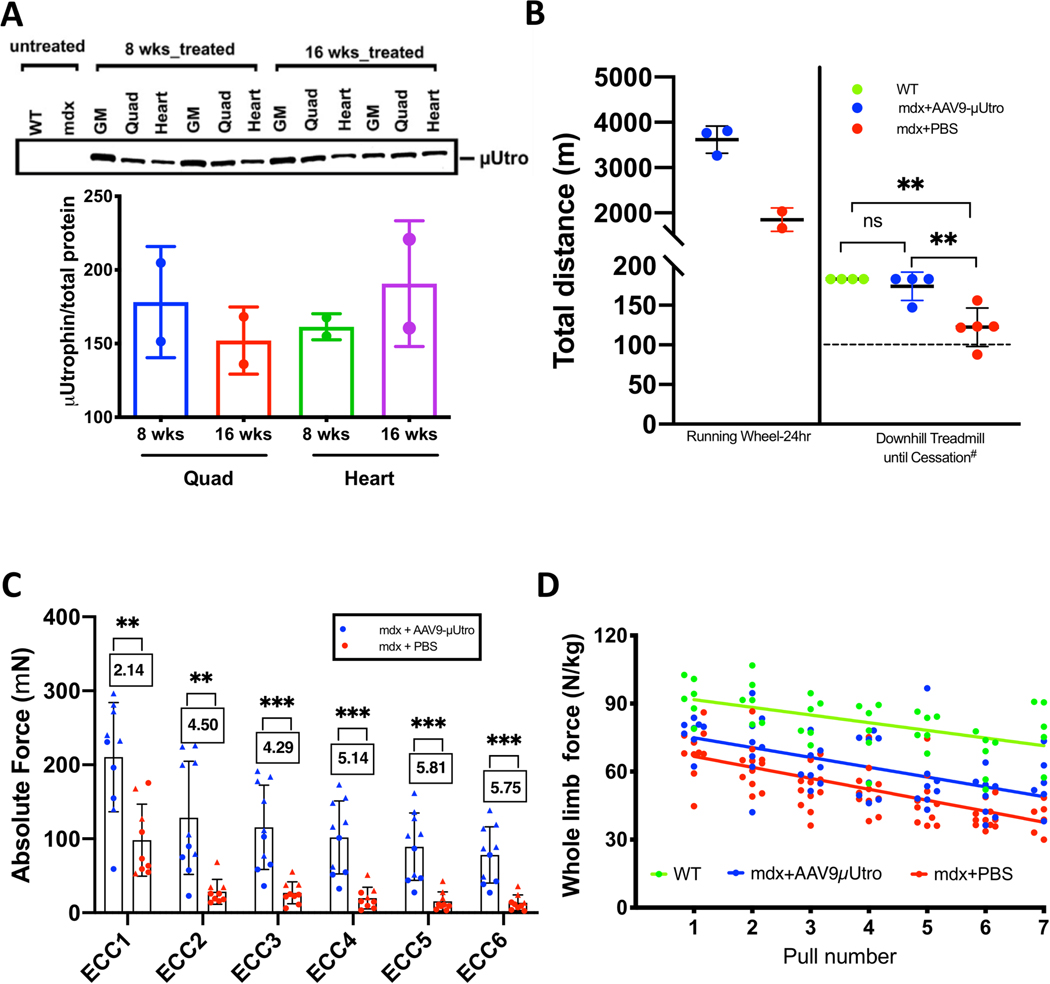
We initially performed intraperitoneal injections of doses up to 2.5 × 1012 vg (vector genomes) of AAV-μUtro into neonatal mdx mice weighing ~5 g and investigated the degree of myoprotection throughout muscle development. In these randomized, blinded studies, we observed equivalent global biodistribution to muscle with both AAV9 and Anc80, and both were well tolerated in mice without any signs of toxicity (Fig. 1a–c and Extended Data Figs. 2 and 3). At the 2.5 × 1012 vg per mouse dose, recombinant μUtrophin was expressed at a level sufficient for qualitatively complete suppression of all tested histological signs of muscular dystrophy, including myofiber centronucleation, embryonic myosin heavy chain (eMHC) expression, native utrophin upregulation, MuRF1 expression as a marker of protein degradation, myonuclear apoptosis (Fig. 1a–d and Extended Data Fig. 4a,b), ongoing myonecrosis and mononuclear cell infiltration. Of these signs, centronucleation is quantitatively the most sensitive indicator of myoprotection in mdx mice because it reflects previous cycles of regeneration. For the first time, we demonstrate normalization (or prevention) of centronucleation to a level biologically indistinguishable from wild type (Fig. 1d). This observed myoprotection was associated with sustained normalization of the dystrophin-associated glycoprotein complex (DGC) in the sarcolemma of cardiac and skeletal muscles (Fig. 1a). Western blot analysis further confirmed that expressed μUtro protein was sufficient to stabilize the DGC (Fig. 1c). Sustained expression of μUtro in skeletal and cardiac muscles was observed throughout a four-month period post vector delivery (the end point of study), indicating the durable level of myoprotection conferred by the single-dose treatment (Extended Data Fig. 5a). Serum creatine kinase (CK), a biomarker that reflects sarcolemmal permeability, was statistically indistinguishable from that of wild-type mice (Fig. 1e), suggestin g that codon optimization and recombinant protein overexpression early in development improved the response relative to administration of alternative transgenes via tail vein injection after the onset of myopathology10,22,24.
Fig. 1 |. AAV-μutro treatment restores the DGC, prevents myofiber degeneration, normalizes serum CK level and improves muscle function in mdx mice.
a, Immunofluorescence staining of representative limb muscle (tibialis anterior) for epitopes shared by native and recombinant utrophin (Utro_N), for epitopes unique to native Utrophin (Utro_C), γ-sarcoglycan, eMHC and laminin. Scale bar, 25 μm. b, H&E of representative limb muscles. Scale bar, 100 μm. c, Western blot analysis detecting expression of recombinant μUtro and γ-sarcoglycan (see Source Data for full uncropped images). GM, gastrocnemius. Experiments from a–c were repeated independently at least two times with similar results. d, Percentage of centrally nucleated myofibers. For each group, n = 4 mice (distinct color), 3 muscles per mouse (distinct shape), >3 non-overlapping quantified fields/muscle. Wild type, 0.148 ± 0.483, mdx + AAV-μUtro 0.855 ± 1.837, mdx + PBS 53.564 ± 13.179; data presented as mean ± s.d. (the wide horizontal lines represent the group mean, and the associated error bars represent s.d.). Random-effect parameters (box) were calculated as defined in the Methods. ICC, intracluster correlation; σ2u, variance between clusters; σ2e, variance within clusters; neff, effective sample size. e, Quantification of serum CK levels in wild type (n = 7) 223.71 ± 85.69, mdx + AAV-μUtro (n = 5) 634.6 ± 413.15 and mdx + PBS mice (n = 12) 4,744.25 ± 2,430.92; data presented as mean ± s.d. Each symbol represents data from an individual animal, the wide horizontal line represents the group mean, and the associated error bars represent s.d. NS, not significant; ***P = 0.0008; statistical significance was assessed by one-way analysis of variance (ANOVA) with Tukey test multiple group comparison. f, Quantification of vertical activity 1 h post-grip strength in wild type (n = 6) 1,052 ± 282.09, mdx + AAV-μUtro (n = 8) 1,279 ± 428.64 and mdx + PBS mice (n = 11) 521.63 ± 485.85; data presented as mean ± s.d. Each symbol represents data from an individual animal, the wide horizontal line represents the group mean and associated error bars represent s.d. NS, not significant; **P = 0.0075; statistical significance was assessed by Kruskal–Wallis test with Dunn’s multiple group comparison.
To test whether AAV-μUtro confers functional improvement in mdx mice, we utilized an established hybrid assay that links force grip testing with a volitional component in which we noninvasively measure animals’ post-force grip vertical activity25. Our previous studies show that this hybrid test provides one of the most sensitive, clinically relevant parameters distinguishing mdx and wild-type mice, capturing behavior causally linked to the exaggerated fatigue response of dystrophin-deficient muscle26. This test demonstrated an objective, dramatic and statistically significant difference between untreated and AAV-μUtro treated mdx mice, while the latter group was indistinguishable from wild-type mice (Fig. 1f). Treated mdx mice not only showed increased voluntary wheel (eight weeks) and downhill treadmill running (16 weeks) distances relative to the untreated mice, but their ex vivo isolated extensor digitorum longus (EDL) muscles also displayed enhanced resistance to eccentric contraction-induced injury as well as enhanced muscle performance in vivo by force grip testing (Extended Data Fig. 5b–d). These findings suggest that early overexpression of μUtro is capable of full phenotypic amelioration in the absence of full-length dystrophin, despite the comparatively short length of the reverse-engineered protein’s rod-like linkage to the actin cytoskeleton and the lack of an R16–17 nNOS-binding motif.
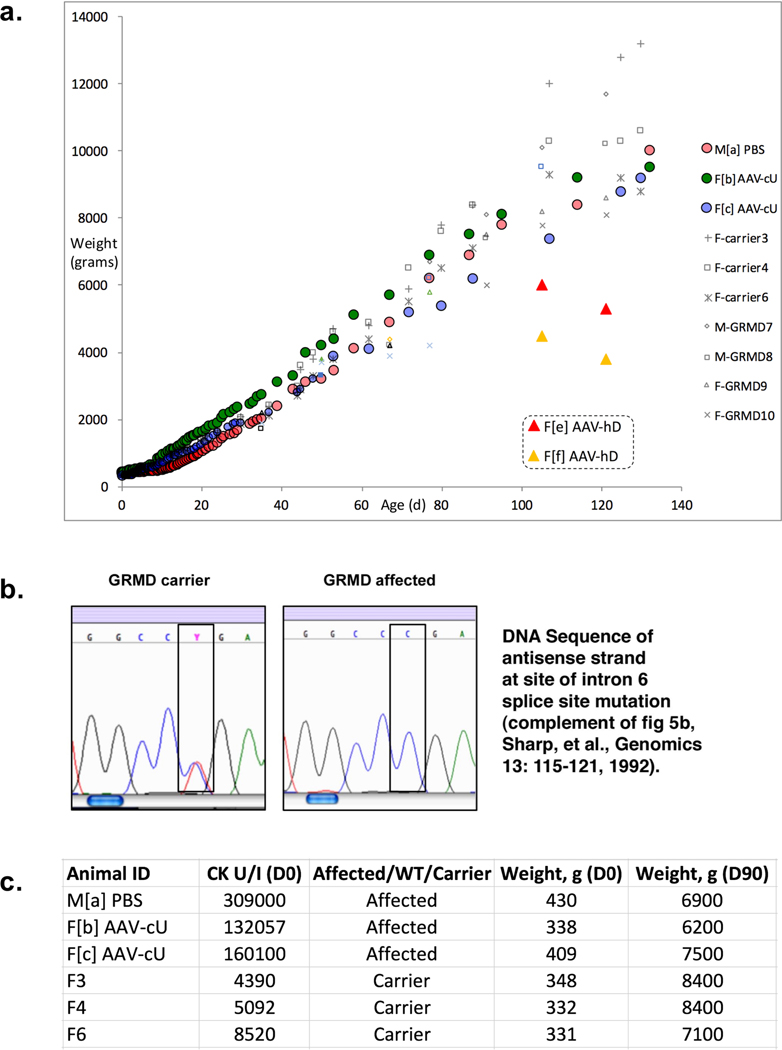
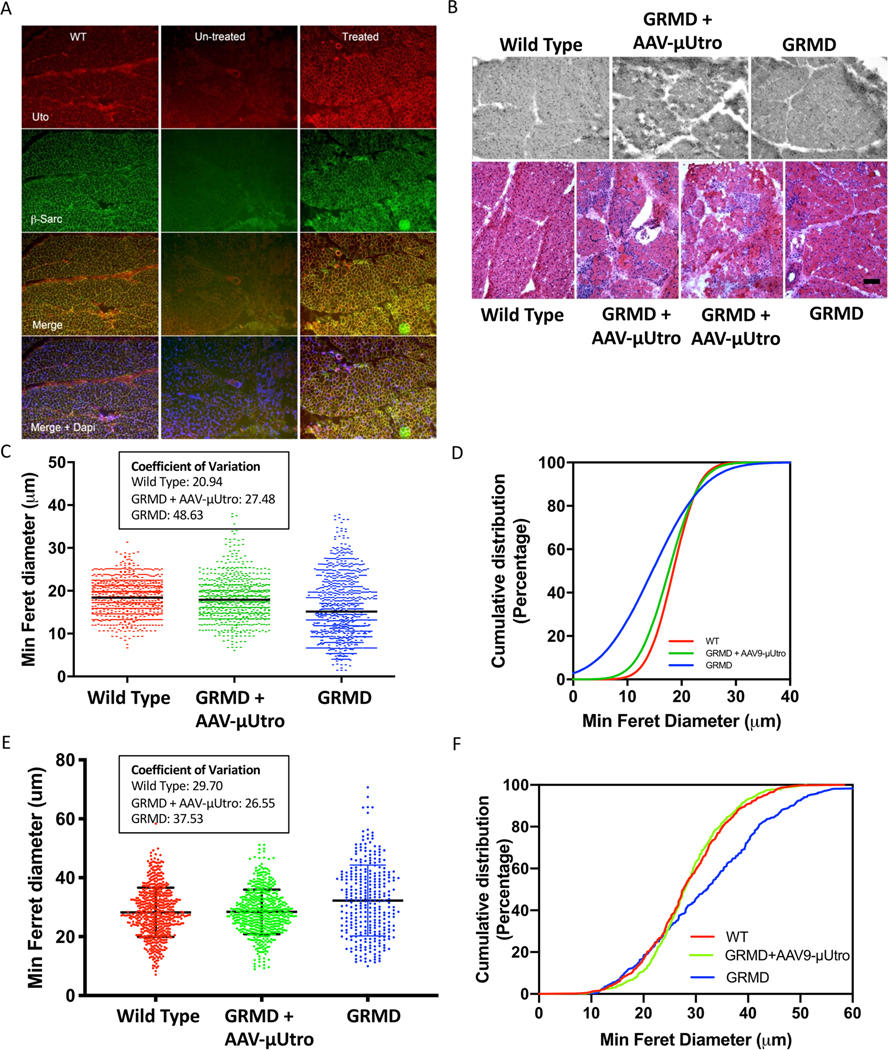
These results raised the hope of achieving full, rather than BMD-like partial reversal of the pathophysiology of DMD through systemic muscle transduction; however, it was not clear whether scale-dependent differences between small and large dystrophic animals would reveal limitations to this approach. The histological and immunological consequences of μUtro gene transfer were further investigated in a blinded study in which five golden retriever muscular dystrophy (GRMD) dogs, 4–7 days of age, were randomized to intravenous administration of AAV-μUtro at doses of 1 × 1013 and 3.2 × 1013 vg kg−1 at the time of injection, without immunosuppression. The treated dogs achieved a fourfold increase in weight, similar to that of carrier females (Extended Data Fig. 6a–c), in contrast to the previously reported weight decrement associated with immune myositis following systemic administration of xenogenic human dystrophin in the same GRMD model14. Six weeks post-injection we observed robust μUtro expression and stabilization of wild-type levels of sarcoglycan expression in the sarcolemma (Extended Data Fig. 7a). This sustained μUtro expression was associated with visibly reduced levels of myonecrosis, mononuclear infiltration (Extended Data Fig. 7b) and normalization of myofiber minimal Feret diameter (Extended Data Fig. 7c,d). At five and eight weeks post vector administration, canine interferon-γ ELISpot assays revealed no cell-mediated immunity against either the AAV capsid or the μUtro transgene product in our non-immunosuppressed treated GRMD dogs (Extended Data Fig. 8c). The major limitation of this proof-of-concept study stems from the 1,000-fold difference between the adult weight of mdx mice and GRMD dogs, 25 g and 25 kg, respectively, limiting our achievable AAV9 dose in the dog to 2.0 × 1012 vg kg−1 based on anticipated adult weight. At this dose, the dogs would inevitably ‘outgrow’ the vector, as did mdx mice treated with 2.15 × 1011 vg as 5 g neonates (Extended Data Fig. 3b). We therefore focused on early histological analysis to detect recombinant μUtro expression, myocyte protection and the immune response to systemic vector administration.
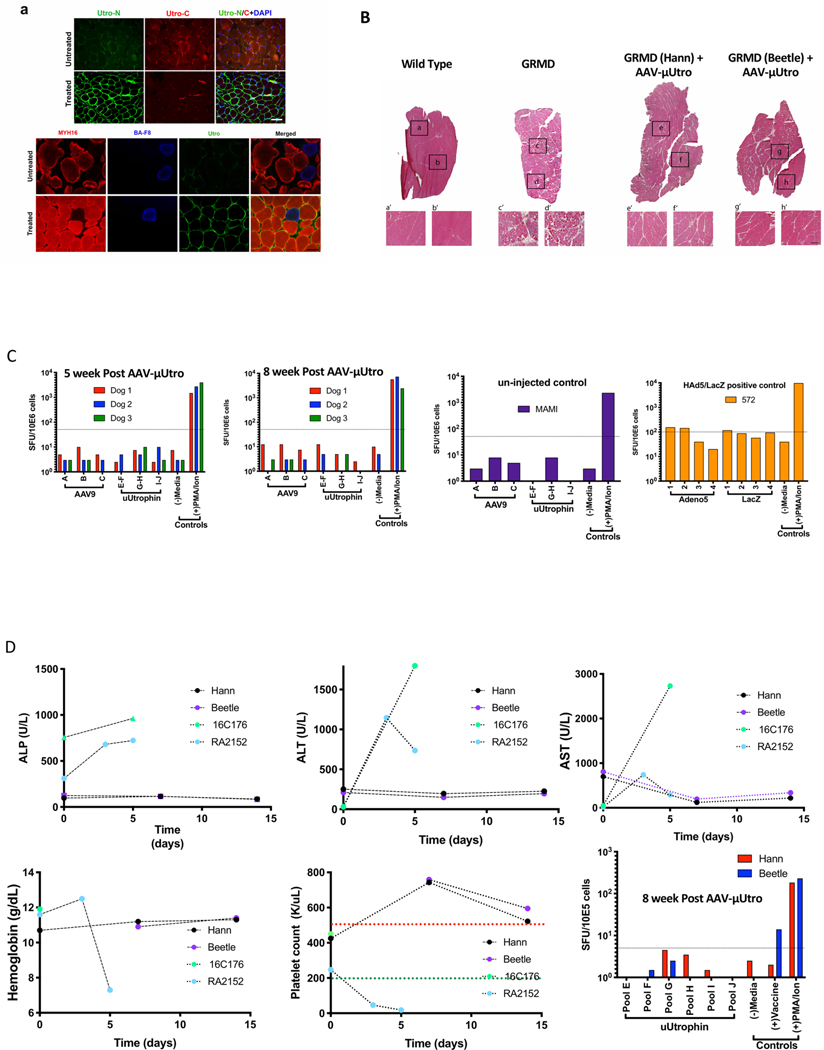
Our neonatal approach offers the possibility of early preventative treatment before the onset of irreversible muscle damage and also minimizes the risk of immune reaction against vector capsid antigens, as memory T cells recognizing the wild-type AAV develop through serial environmental exposure27,28. However, the majority of DMD patients are typically diagnosed after the age of two years, by which time massive muscle fiber degeneration, necrosis with mononuclear cell invasion and increased fiber size variability have already occurred29. To explore the feasibility of our approach in young boys with DMD, two juvenile GRMD dogs at 7.5 weeks of age (Hann and Beetle) were injected intravenously with AAV-μUtro at a dose as high as 1.25 × 1014 vg kg−1 at time of injection, during transient use of an anti-inflammatory dose of prednisone, 1 mg kg−1 daily30 (Fig. 2). Immunostaining of muscle biopsies, taken four weeks post-injection, showed homogeneous sarcolemmal expression of μUtro (Fig. 3a and Extended Data Fig. 8a), suppression of native utrophin (Fig. 3a), as well as rescue of the DGC (Fig. 3b). This was further confirmed by western blot (Fig. 3d).
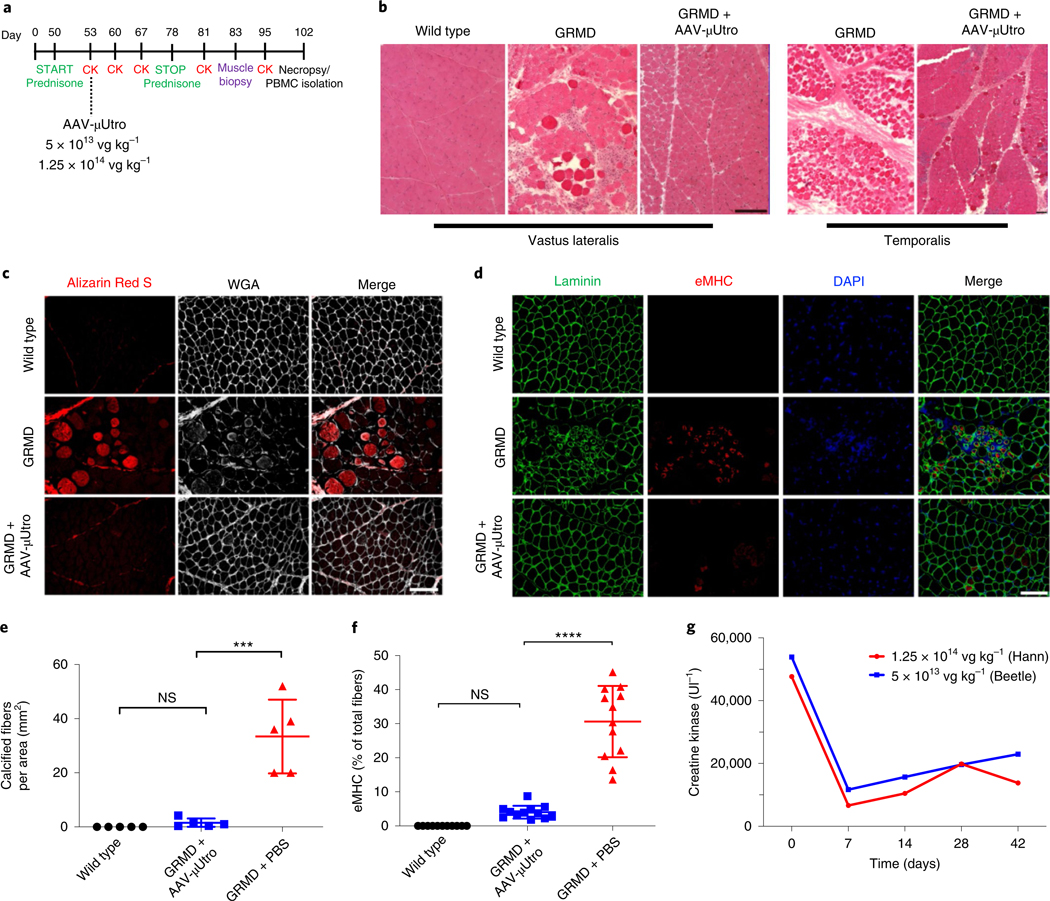
Fig. 2 |. Systemic delivery of AAV-μutro in GRMD dogs at seven weeks of age prevents myonecrosis and results in rapid reduction of serum CK levels.
a, Experimental timeline. b, Representative H&E of vastus lateralis and temporalis muscle. Experiments were repeated independently at least two times with similar results. c,e, Alizarin Red S and Wheat Germ Agglutinin (WGA) staining of tibialis anterior muscle (c; scale bar, 100 μm) with corresponding quantification of calcified fibers per area (e) for wild type (n = 5) 0 ± 0, GRMD + AAV-μUtro (n = 5) 1.496 × 10−6 ± 1.597 × 10−6 and GRMD + PBS (n = 5) 3.33 × 10−5 ± 1.363 × 10−5. Data are presented as mean ± s.d. Each symbol represents data from an individual animal, the wide horizontal line represents the group mean, and the associated error bars represent s.d. NS, not significant; ***P = 0.0009; statistical significance was assessed by the Kruskal–Wallis test with Dunn’s multiple group comparison. d,f, Immunofluorescence staining of tibialis anterior muscle for eMHC-positive fibers (d; scale bar, 100 μm), with corresponding quantification (f) for wild type (n = 5, ≥1 image field per muscle) 0 ± 0, GRMD + AAV-μUtro (n = 6, 2 image fields per muscle) 4.03 ± 1.87 and GRMD + PBS (n = 6, 2 image fields per muscle) 30.61 ± 10.45. Data are presented as mean ± s.d. Each symbol represents data from an individual animal, the wide horizontal line represents the group mean, and the associated error bars represent s.d. NS, not significant; ****P < 0.0001; statistical significance was assessed by one-way ANOVA with Tukey test multiple group comparison. g, Serum CK levels at various time points pre- and post-systemic infusion of AAV-μUtro at two doses: 1.25 × 1014 vg kg−1 (Hann) and 5 × 1013 vg kg−1 (Beetle).
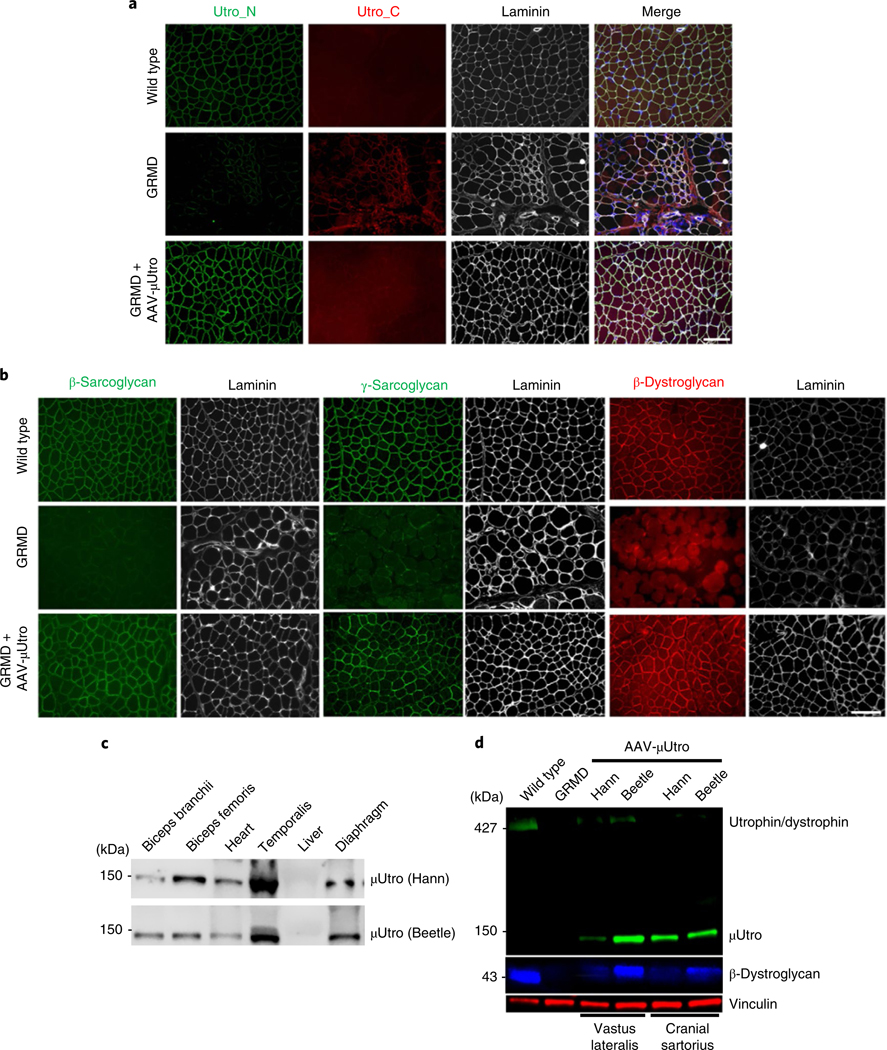
Fig. 3 |. Widespread expression of μutro rescues the DGC proteins in treated GRMD dogs after systemic delivery at the age of seven weeks.
a,b, Immunofluorescence staining of representative limb muscles against native and recombinant utrophin (Utro_N), native utrophin (Utro_C) and laminin (a) and β-sarcoglycan (green), γ-sarcoglycan (green) and β-dystroglycan (red) (b). Scale bars, 100 μm. c,d, Western blot analysis of μUtro (~135 kDa) of tissue collected at necropsy (c) and the expression of μUtro and β-dystroglycan in muscle biopsies of vastus lateralis and cranial sartorius (d). Vinculin serves as a loading control. (See Source Data for full uncropped images). Experiments in a–d were repeated independently at least two times with similar results. (See Source Data Fig. 3).
Histopathological characterization of limb muscle from treated GRMD dogs demonstrates near-complete suppression of ongoing muscle injury, as evidenced in untreated age-matched controls by a high rate of myonecrotic fibers, excessive calcium accumulation, clustered regenerating muscle fibers, abundant inflammatory cell infiltration and fat infiltration (Fig. 2b–f and Extended Data Fig. 8a,b). AAV-μUtro-treated dogs also showed nearly complete prevention of muscle degeneration and regeneration in masticatory muscles (Fig. 2b and Extended Data Fig. 8a), which are severely affected in untreated dogs because they express the uniquely powerful MYH16 myosin isoform31,32. Further western blot analysis at necropsy (3.5 months of age) showed persistent widespread expression of μUtro in skeletal and cardiac muscle (Fig. 3c). Consistent with our previous GRMD neonatal dog studies, interferon-γ ELISpot assays revealed no signal above background against μUtro (Extended Data Fig. 8c), Furthermore, no signs of severe acute toxicity were seen, in contrast to previous studies in GRMD dogs and non-human primates14,33,34 (Extended Data Fig. 8d). Importantly, an 80% drop in serum CK levels was measured one week post-infusion with AAV-μUtro in both dogs (Fig. 2g), a finding that is consistent with observed histological improvements (Extended Data Fig. 7e,f). To achieve durable myoprotection throughout muscle growth from infancy to skeletal maturity, dystrophic dogs and boys with DMD may require systemic administration of AAV vector at doses proportional to those required in mdx mouse pups to maintain robust, homogenous expression in the most severely affected muscle, the diaphragm35, for example 1 × 1015 vg kg−1 neonatal body weight (Extended Data Fig. 3b).
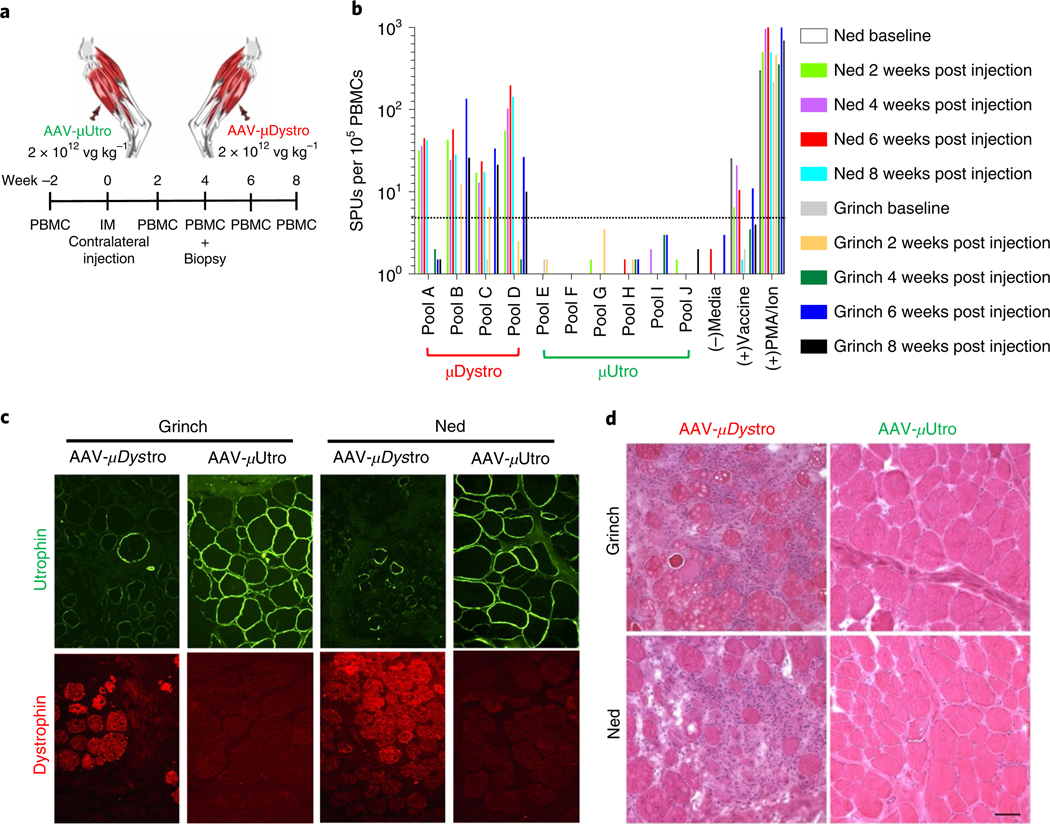
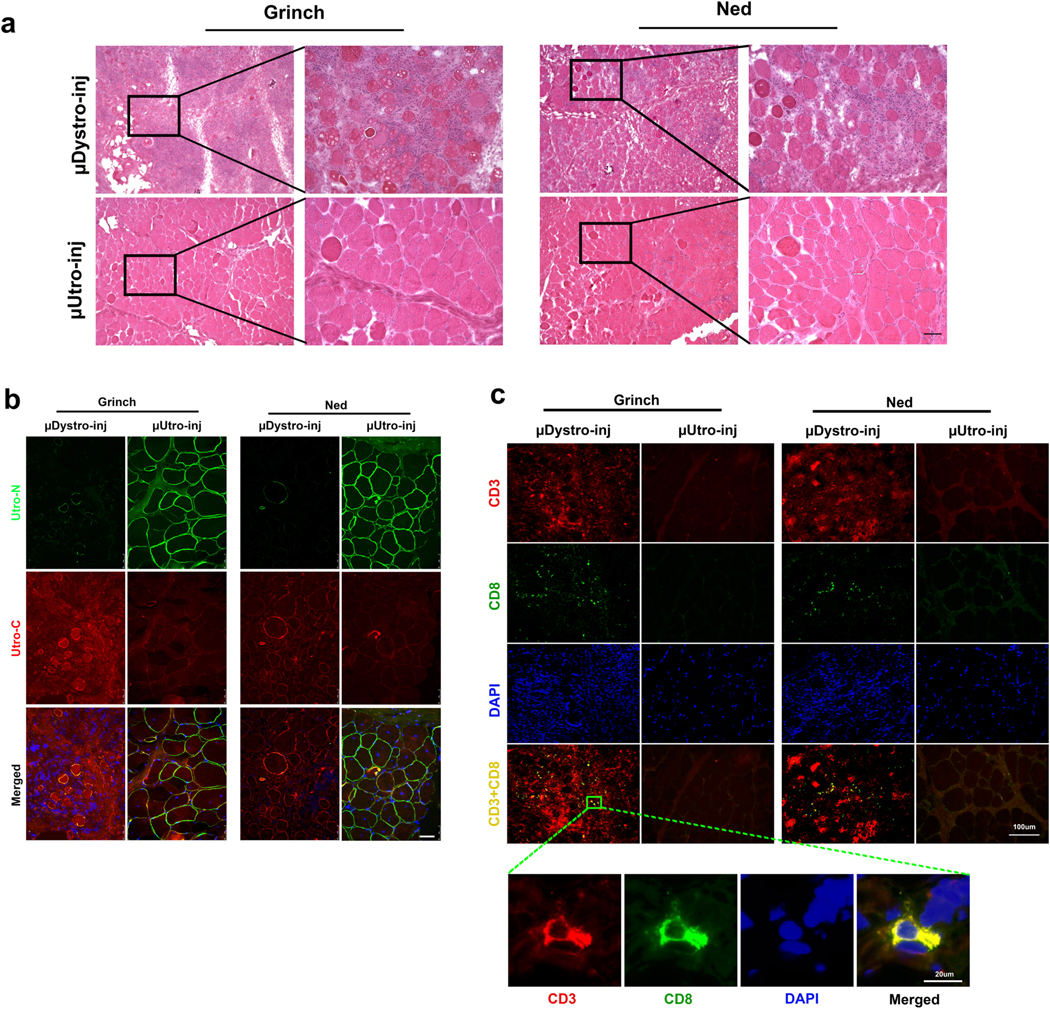
For the rigorous preclinical assessment of vector and/or transgene immunotoxicity, we took advantage of the unique German shorthaired pointer (GSHPMD) deletional-null canine model36,37. The GSHPMD model provides a superior platform for the study of central tolerance, because alternative splicing in the GRMD model allows detectable read-through of near-full-length dystrophin at potentially tolerizing levels, as might be expected to facilitate previously demonstrated therapeutic, long-term body-wide expression of AAV-encoded canine microdystrophins38–40. Adult GSHPMD dogs (Ned and Grinch) each received equivalent doses (2 × 1012 vg kg−1) of both AAV-μDystro and AAV-μUtro via intramuscular injection into contralateral tibialis compartments (Fig. 4a). Interferon-γ detection via ELISPOT revealed the presence of a strong systemic cell-mediated immune response against μDystro, as early as two weeks post-injection, but not to μUtro, despite expression from the constitutive CMV (cytomegalovirus) promoter, indicating the strength of central immunological tolerance (Fig. 4b). Immunostaining of muscle biopsies collected four weeks post-injection revealed persistent expression of μUtro, but only sparse amounts of μDystro (Fig. 4c). H&E analysis showed severe inflammation and mononuclear cell infiltration on the μDystro-injected side compared to their virtual absence on the μUtro-injected side (Fig. 4d and Extended Data Fig. 9). These findings indicate that the observed immune response is driven by μDystro and not the vector capsid, as equivalent doses of vector were injected to both limbs.
Fig. 4 |. Focal expression of μDystro, but not μutro, elicits a detectable peripheral and local immune response in the GSHP dystrophin deletional-null dog model.
a, Experimental timeline. Each dog (Grinch and Ned) received intra-muscular (IM) injections of both AAV-μDystro (right) and AAV-μUtro (left) at equivalent doses (1 × 1012 vg kg−1) into their tibialis anterior compartment. b, Peripheral blood mononuclear cells (PBMCs) collected pre- and two, four, six and eight weeks post-injection were cultured with synthetic peptides spanning the entire μDystro (pool A–D) and μUtro (pool E–J) peptide sequences, while vaccine peptides and phorbol myristate acetate (PMA)/ionomyocin served as positive controls. A positive result is interpreted as ≥5 spot forming units (SFU) per 1 × 105 PBMCs (dotted line). c, Immunofluorescence staining of muscle biopsies collected four weeks post-injection against utrophin (green) and dystrophin (red). Experiments were repeated independently at least two times with similar results. d, Representative H&E stain of muscle biopsies collected four weeks post-injection (see also Extended Data Fig. 9a). Experiments were repeated independently at least two times with similar results. (See Source Data Fig. 4).
In summary, after leveraging a comparative phylogenomic approach to identify evolutionary constraints, we have reverse-engineered a highly therapeutic, 3.5 kb synthetic transgene for safe systemic delivery to muscles of murine and canine models for DMD. Our blinded studies reveal surprisingly complete myoprotection as long as the initial level of gene delivery is sufficient to accommodate subsequent muscle growth. Taken together, these findings may refocus the field towards the use of a functionally optimized, non-immunogenic utrophin-based gene therapy approach for DMD.
Methods
Bioinformatics and phylogenetic analysis.
Publicly available genomic DNA sequences for species listed in the figure legends and the main text were queried by multiple blast algorithms, in particular tBLASTn (refs. 41,42), to identify the coding sequences homologous to the full-length human dystrophin and utrophin. For most species, supporting evidence from mRNA sequences was available to define intron/exon boundaries. Where such evidence was lacking, FGENESH+ (Softberry) was used with organism-specific gene-finding parameters and a hidden Markov model plus similar protein-based gene prediction to identify putative coding sequences from assembled contigs. As an internal test of this approach, virtually all transcriptome-defined coding sequences were properly identified by the FGENESH+ program43. In the Aligned Sequence Guide (Supplementary Data), we list all the files generated by this latter approach, which are individually available as additional supplementary files. We recognize that these mRNA and protein sequence files are missing sporadic exons in indeterminate regions of publicly available genomic DNA. HMMER (http://hmmer.janelia.org/search/hmmscan) was used with E-value defined cutoffs to define protein coding domains matching hidden Markov models for calponin homology, spectrin-like repeat, WW, EF hand and ZZ domains. All deduced peptide sequence files were aligned by ClustalW using the default settings in MacVector version 13.5.1: Gonnet Series Matrix with parameters for pairwise alignment open gap penalty of 10, extend gap penalty of 0.1 and parameters for multiple alignment open gap series 10, extend gap penalty of 0.2 and delay divergence 30%. Phylogenetic reconstructions were generated with both full-length and truncated sequences using the neighborjoining tree building method with ties in trees resolved randomly and distances Poisson-corrected with gaps distributed proportionally or ignored to establish whether the choice impacted on tree topology. In all such cases, tree topology was insensitive to the management of gaps when the ‘best tree’ mode was selected. Alternative use of the bootstrap mode, with 10,000 replications, confirmed all nodes in the distance phylograms. Protein and DNA matrix analyses were based on the pam250 scoring matrix. The abbreviations used were as follows: purple sea urchin, Strongylocentrotus purpuratus, S.pur; amphioxus, Branchiostoma floridae, B.flo; elephant shark, Callorhinchus milii, C.mil; Chinese alligator, Alligator sinensis, A.sin; mouse, Mus musculus, M.mus; dog, Canis familiaris, C.fam; human, Homo sapiens, H.sap; Carolina anole, Anolis carolinensis, A.car; common chimpanzee, Pan troglodytes, P.tro; duck-billed platypus, Ornithorhynchus anatinus, O.ana.; Japanese pufferfish, Takifugu rubripes, T.rub; tropical clawed frog, Xenopus tropicalis, X.tro; dystrophin, D; utrophin, U.
μUtro transgene design and vector production.
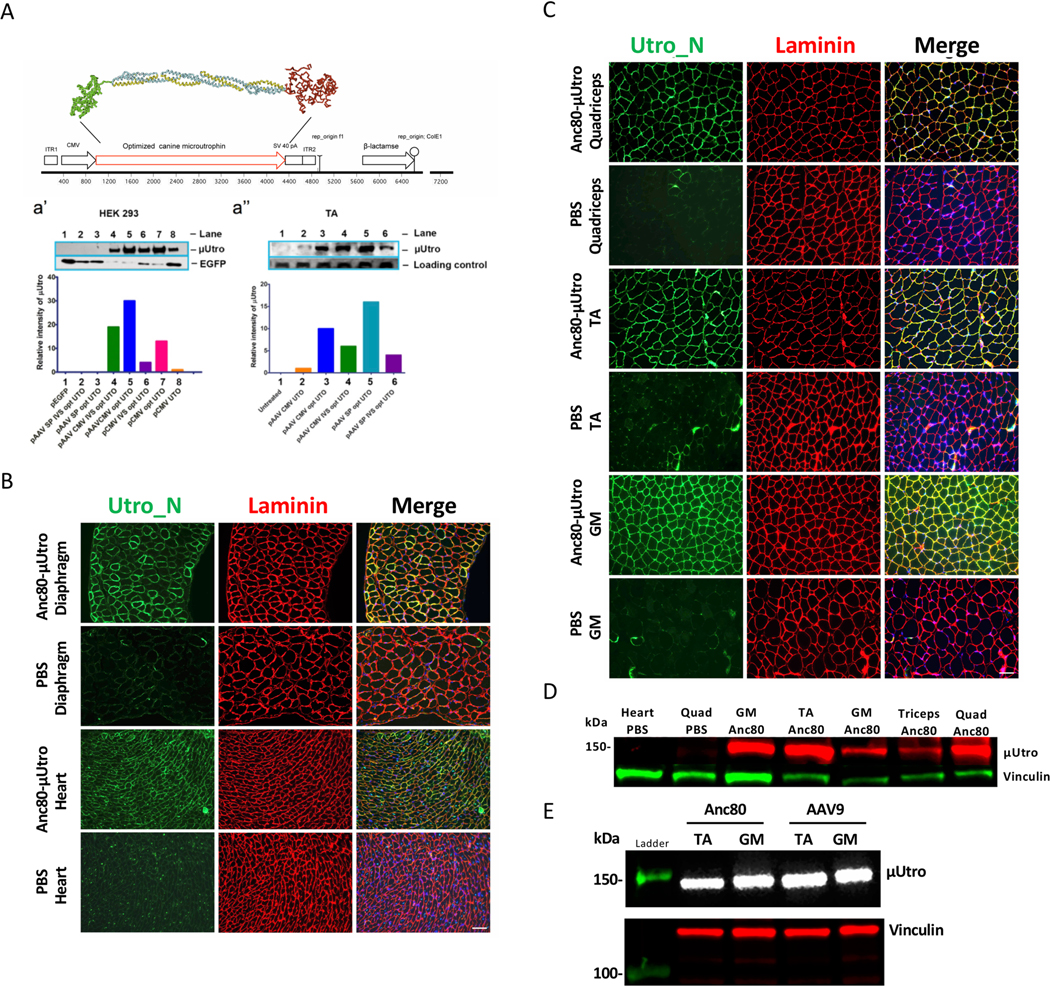
Based on phylogenomic analysis of sequence conservation and genotype–phenotype correlations among BMD/DMD patients, we modeled, in silico, the spectrum of AAV-encodable miniaturized utrophins that preserve the calponin homology domains, the first three and last three spectrin-like repeats, and the combination of the WW, EF hand and ZZ domains. To preserve amino-acid side-chain interactions between interhelical loops of adjacent spectrin-like repeats, we focused on only the subset of μUtro in which either the first or last three repeats were preserved intact. To minimize immunogenicity in dogs, we considered only those μUtro that could be created by the combination of a single internal deletion and a C-terminal truncation. Although the spectrin-like repeats are homologous to a consensus sequence, the divergence is such that no splice could be found between identical decapeptides in any of the mammalian utrophins. Thus, we used profile hidden Markov models, as implemented online at http://hmmer.janelia.org/search/hmmscan, to define and annotate spectrin-like triple helical repeat boundaries in the full-length murine, canine and human utrophin sequences (for example, 3,456 aa, XP_005615306). We used transgenes encoding proteins matching the canine and human proteins in neonatal mice, in which antigen-specific tolerance is easily induced by intraperitoneal injection of AAV44, but only the canine versions in neonatal and older dogs in which tolerance is anticipated to require earlier prenatal exposure to the isogenic native protein during immune ontogeny45. The μUtro transgenes were designed to contain the actin binding domain, triple helical repeats 1–3 and 22, a disordered, proline-rich region approximating that previously identified as ‘hinge’ 2, and the C-terminal WW, EF hand and ZZ domains, thus creating recombinant proteins designed to match the canine and human utrophin sequences with the exception of a single splice site at the deletion junction, thereby minimizing potential immunogenicity in dystrophin-deficient dogs, and ultimately humans, relative to previously reported transgenes8–11,22. The coding sequence chosen for use in our studies was selected as the highest-expressing candidate of a pool of cDNAs that were optimized and synthesized by competing biotech companies (GeneArt and DNA 2.0). Expression was determined by immunofluorescence staining and western blotting following electroporation of 50 μg DNA in tibialis anterior muscle of mdx mice. The synthetic coding sequence chosen for further use was found to drive expression ~30-fold higher in in vitro and in vivo assays than the wild-type canine cDNA sequence encoding a recombinant protein of identical primary structure. A notable difference between the best synthetic cDNA and the wild type is the level of codon bias, with only the optimized synthetic cDNA closely matching the extreme bias of the mammalian MHCs (for example, 154 CTG leucines, 0 TTA leucines). The synthetic canine μUtro cDNA was subcloned into an AAV2 expression vector cassette driven by either an 833 bp fragment of the CMV immediate early enhancer/promoter or a synthetic promoter spc5–12 (CMV and SP, respectively, in Extended Data Fig. 2a). AAV9 and Anc80 vectors were generated and purified by the University of Pennsylvania preclinical vector core using the triple transfection method in HEK 293 cells as previously described46. Vector preparations were assayed for quality, purity and endotoxin levels before pooling for injection of 2 × 1011 AAV-μUtro vg into the tibialis anterior muscles of mdx mice47. Although the measured level of expression in skeletal muscle following electroporation of plasmids was comparable between the CMV enhancer/promoter and SP cassettes, the yields of rAAV were consistently a factor of 10 lower for plasmids containing spc5–12 than CMV, so further studies were restricted to the CMV cassette. Unless otherwise noted for Anc80, AAV used to describe μUtro and μDystro vectors signifies AAV9.
Animals.
The Animal Care and Use Committee of the A&M University and the University of Pennsylvania approved all animal experiment protocols in mice and dogs.
Murine model vector administration.
Mouse strains C57BL/10SnJ and mdx were purchased from the Jackson Laboratory. This study involved 23 C57BL/10SnJ mice and 30 mdx mice, all injected at 9 ± 2 days of age. Before receiving intraperitoneal injection of AAV-μUtro or phosphate buffered saline (PBS), individual pups were toe-tattooed with the Aramis Micro tattoo kit (Ketchum Manufacturing) and randomly assigned to different dosage groups. Investigators were blinded during all injections and tissue harvesting. Based on this protocol, C57BL/10SnJ and mdx pups were injected with 50–250 μl of either PBS as negative control or AAV-μUtro diluted in PBS via a 32 gauge insulin syringe. Before injection, each mouse was weighed. After vector administration, all mice were returned to their litters and separated after weaning.
Murine model tissue procurement and storage.
At approximately eight weeks of age, mdx and C57BL/10SnJ mice underwent CO2 euthanasia in accordance with the institutional policy. The heart, tibialis anterior, gastrocnemius, quadriceps, triceps, abdominal, diaphragm, temporalis muscles and liver were harvested and further processed; others were stored but not utilized based on studies showing <100-fold lower off-target gene expression with AAV9 in mice48. Designated histological tissue samples were placed in OCT (Tissue-Tek) containing embedding molds (Richard-Allan Scientific) and rapidly frozen in liquid nitrogen-cooled isopentane. Additional designated biological tissue samples were placed in tissue containers and rapidly frozen in liquid nitrogen. All the specimens were stored at −80 °C. Cryosections of 5–7 μm thickness were cut on a cryostat (Microm HM550, Thermo Scientific) at −25 °C and mounted on glass slides (Superfrost Plus, Fisher Scientific).
Basic histology, H&E and Alizarin Red staining.
The 7-μm-thick cross-sections were air-dried for 15 min at room temperature. The slides were then stained with Harris’ hematoxylin dye for 2.5 min, rinsed in distilled water, dipped in 0.1% acetic acid for 15 s, followed by a repeat rinse in tap water for 4 min and counterstaining with 1% eosin for 1 min. As a final step, the slides were dehydrated in ethanol three times for 2 min each. Representative, non-overlapping high-powered fields were photographed for scoring. Alizarin Red staining was also carried out on 7-μmthick cross-sections. After 10 min fixation by 10% buffered formalin phosphate, the sections were washed three times for 5 min each with PBS and incubated with Alizarin Red dye for 15 min at room temperature. The slides were then washed in ethanol three times and mounted with Cytoseal 60 (Thermo Scientific).
Morphometric analysis.
Three groups of mice, C57BL/Sn10J (wild type) and mdx randomized to either injection with PBS or AAV-μUtro, were studied by investigators blinded to specimen identification. Four to five randomly chosen areas from each tibialis anterior, gastrocnemius, quadriceps, triceps, temporalis and abdominal muscle were stained with H&E and screened with a light microscope for quantification of centrally nucleated myofibers. Areas at the myotendinous junctions were excluded from the measurements as they are rich in centrally nucleated fibers in both mdx and controls. In total, 11,649 fibers were evaluated.
Immunofluorescence staining procedures.
N-terminal and C-terminal utrophin double staining. Sections from all muscle specimens were processed for utrophin immunostaining by using both N-terminal polyclonal (against native and recombinant utrophin) as well as C-terminal monoclonal (against native utrophin) antibodies. After initial incubation for 20 min in a 1% solution of Triton X-100 (Roche Diagnostics) diluted in 0.01 M PBS (Roche Diagnostics), the specimens were rinsed three times in PBS for 5 min each (3 × 5 min). Sections were then incubated in 5% normal donkey serum for 15 min, followed by incubation with N-terminal utrophin antibody (N-19, sc-7460, goat polyclonal immunoglobulin-G (IgG), dilution 1:50) for 60 min at 37 °C. After a second cycle of 3 × 5 min PBS wash, the slides were incubated with 5% normal donkey serum for 15 min in room temperature. The prepared sections were then incubated in donkey anti-goat IgG-FITC (sc-2024, dilution 1:300) for 30 min at 37 °C. Following a third PBS wash for 3 × 5 min, the sections were first incubated with 10% normal goat serum (Invitrogen) for 15 min and then with the C-terminal utrophin antibody (MANCHO7, mouse monoclonal IgG2a, dilution 1:25) at 37 °C for 60 min. After a PBS wash for 3 × 5 min and incubation with 10% normal goat serum, the sections were incubated in goat anti-mouse IgG2a-Alexa Fluor594 (A-21140, Life Technologies, dilution 1:300) for 30 min at 37 °C. The sections were again washed in PBS for 3 × 5 min and mounted in Vectashield mounting medium (H-1000; Vector Laboratories) or mounting medium with DAPI (H-1500; Vector Laboratories. Photographs were taken with a Leica DM6000B microscope (Leica).
Double immunofluorescence staining for γ-sarcoglycan/laminin, MuRF1/laminin and eMHC/laminin, MYH16.
The staining procedures for these proteins followed the same protocol as described previously49. Rabbit anti-γ-sarcoglycan (NBP1–59744, Novus Biologicals) and MuRF1 (NBP1–31207, Novus Biologicals) polyclonal antibodies were used at a dilution of 1:50 in PBS with BSA. eMHC monoclonal antibody (F1.652; Developmental studies, Hybridoma Bank) was used at a dilution of 1:50–1:100 in PBS. Laminin chicken polyclonal antibody (ab14055–50, Abcam) at a dilution of 1:500–1:1,000 together with second goat anti-chicken IgY (TR) antibody (ab7116, Abcam, dilution 1:300) was applied to identify muscle fibers. MYH16 rabbit polyclonal antibody peptide sequence was generated using the human and canine sequence of MYH16’s ‘loop 2’ region. This was validated by demonstrating sensitive and completely specific binding to the dominant myosin isoform expressed in the canine temporalis. Peptide sequence: LLALLFKEEEAPAGS
TUNEL assay.
Sections were initially fixed in 10% buffered formalin phosphate (Fisher Scientific) for 20 min. In situ nick end labeling of fragmented DNA was then performed using a TACS2 TdT fluorescein apoptosis detection kit (Trevigen), as described in the manufacturer’s instructions.
Serum CK assay.
Blood serum was collected via venipuncture of the submandibular vein using a 5 mm animal lancet (Goldenrod Animal Lancet, Braintree Scientific). A total volume of 150 μl was collected in a heparinized blood collection tube (Terumo, cat. no. TMLH). The mice were carefully monitored for 30 min post blood withdrawal to observe for potential signs of distress. CK levels were determined by the Clinical Pathology Laboratory at the Matthew J. Ryan Veterinary Hospital of The University of Pennsylvania.
Ex vivo evaluation of EDL muscle contractile properties.
Ex vivo assessment was performed by the Muscle Physiology Assessment Core of The University of Pennsylvania. The physiological properties, including isometric twitch force, isometric tetanic force and force drop after eccentric contractions (ECCs), were quantified on freshly isolated EDL muscles from two-month-old mdx mice using an Aurora Mouse 1200A system equipped with Dynamic Muscle Control v.5.3 software50. All of these mice had undergone in vivo force grip testing 24 h prior to euthanasia and ex vivo testing. EDL muscles were maintained in continuously oxygenated Ringer’s solution (100 mM NaCl, 4.7 mM KCl, 3.4 mM CaCl2, 1.2 mM KH2PO4, 1.2 mM MgSO4, 25 mM HEPES and 5.5 mM d-glucose) at 24 °C. The twitch stimulation protocol applied was a single stimulus with a duration of 0.2 ms. For measuring tetanic maximal force generation, the same stimulus was repeated at a frequency of 120 Hz for 500 ms. Five minutes were allowed between two tetanic contractions to ensure muscle recovery. Muscle length was adjusted to obtain the maximal twitch response. This length was measured between the outermost visible tips of the myotendinous junctions and recorded as optimal length (L0). After testing the isometric properties of the EDL, a series of six ECCs (one every 5 min) were applied in cycles beginning with repeated 500 ms isometric contractions followed by stretching the muscle by 10% of L0 over a final 200 ms, while administering a maximal tetanic stimulation. The reported absolute force for each ECC corresponds to the peak force during the isometric phase of the ECC.
In vivo physiological testing in mdx mice.
To reduce the chances of bias and to ensure the consistency and robustness of the blinded experiments we used Institutional Animal Care and Use Committee-approved protocols as described in ref. 25. Voluntary running wheel distances were measured over 24 h. Baseline vertical activities were determined for 5 min, followed by 3 min of rest in the original cages. An axial force transducer was used to measure force. Downhill running distances were measured at +15° incline with treadmill speed increasing from 10 m min−1 (10 min) to 11 m min−1 (1 min) and then 12 m min−1 (6 min). Cessation was defined as either three consecutive shocks or the end of the protocol at 17 min.
Canine model.
Two groups of GRMD dog models were used for our experiment. The first group was bred in a colony at the A&M University and whelped at the University of Pennsylvania. This study involved five affected GRMD dogs and four age-matched littermates including one wild-type and three carrier females that served as a control group. All dystrophic dogs were identified by elevated serum CK levels and genotyped by PCR assay. All pups were randomly assigned to treatment groups, and the investigators were kept blinded during clinical and histological assessment. The pups were injected with AAV-μUtro at 6–10 days of age at a dose of 1.0 × 1013 vg kg−1 or 3.16 × 1013 vg kg−1 via an external jugular vein approach. Two pups were injected with low-dose AAV-μUtro, two were injected with the high dose, and the remaining one was injected with saline only. Each dog was weighed daily for the first six weeks and weekly thereafter.
The second canine group involved two GRMD dogs aged ~7.5 weeks. Three days before vector administration, the dogs were placed on an oral prednisone 1 mg kg−1 regimen for 25 days. AAV-μUtro was injected using the same approach at two different single doses of 1.25 × 1014 vg kg−1 (Hann) and 5.0 × 1013 vg kg−1 (Beetle), respectively. The dogs were randomly assigned to dose and the investigators were kept blinded during the clinical and histological assessments.
Deletional-null German shorthaired pointer (GSHPMD) dogs were bred and housed in Texas A&M University. Two seven-year-old affected GSHPMD dogs, Ned and Grinch, weighed 21 kg and 24.2 kg, respectively. Each dog received five intramuscular injections of AAV-μDystro (right) and AAV-μUtro (left), with a total equivalent dose of 1.0 × 1012 vg kg−1, into their tibialis anterior compartment. All five injection sites were tattooed, allowing us to pinpoint the injection sites for muscle biopsies four weeks post-injection. Peripheral blood was collected pre-, two, four, six and eight weeks post-injection in order to collect PBMCs.
Canine tissue procurement and storage.
The first group of GRMD dogs underwent needle biopsy of the cranial sartorius, vastus lateralis and triceps brachii muscles at approximately six weeks post vector injection. The specimens were stored with a set of blinding codes to prevent bias during analysis and interpretation. Biopsies were obtained through a spring-loaded 14-gauge needle trocar, thereby significantly minimizing the post-procedural pain in the animals. Muscle biopsies were then snap-frozen in liquid nitrogen-cooled isopentane, embedded in OCT medium (Sakuru) and stored at −80 °C. Blinding codes were broken after tissue analysis by individuals not associated with authorship of this study. One month after vector exposure the second group of injected dogs underwent open muscle biopsy of the same muscles. Seven weeks post vector administration these dogs were euthanized, and the harvested tissue was cryopreserved in the same way.
Canine histological analysis.
Transversely cut 7 μm serial sections were used for bright-field microscopy analysis and immunofluorescence staining to examine μUtro expression and sarcoglycan rescue. Muscle sections were stained with H&E for bright-field microscopy and mounted with Permount. For immunofluorescence staining, sections were blocked in 5% donkey serum in PBS for 45 min followed by incubation for 60 min at 37 °C using either a 1:350 dilution of polyclonal goat anti-utrophin antibody (N-19, sc-7460), a mouse monoclonal anti-dystrophin antibody (MANEX1011B(1C7), DSHB) or a 1:250 dilution of monoclonal β-sarcoglycan antibody (ab55683, Abcam). The sections were then rinsed three times in PBS and incubated for 45 min in Alexa488 donkey anti-goat secondary antibody or Alexa540 donkey anti-mouse secondary antibody at a dilution of 1:1,000. The slides were washed twice with PBS for 5 min followed by single wash of water and mounted with fade-resistant mounting medium containing DAPI (H-1500, Vector Labs). Images were captured at the same setting and processed in an identical way by setting the limits and gain throughout to avoid any disparity in immunofluorescence images using an Olympus B-65 fluorescent microscope. Minimal Feret’s diameter and the coefficients of variance were calculated according to TREAT-NMD protocol DMD_M.1.2.001 as updated on 1 January 2014.
Immunoblot analysis.
Immunoblot analysis was carried out by loading 20–40 μg per lane of whole cell or whole muscle lysate on 10% SDS–polyacrylamide gel. Protein was transferred to a polyvinylidene difluoride membrane. μUtro was detected by a goat polyclonal antibody against the N-terminal epitope at 1:500 dilution (N-19, sc-7460) and a secondary antibody, a donkey anti-goat antibody conjugated with horseradish peroxidase (Sigma-Aldrich) at 1:5,000 dilution. Protein detection and quantification was performed using the Odyssey infrared imaging system (LI-COR). Gamma-sarcolgycan was detected by a mouse monoclonal antibody (Vector Labs VP-G803) and a donkey, anti-mouse horseradish peroxidase conjugated secondary antibody (Santa Cruz Biotechnology).
Detection of neutralizing anti-AAV antibodies.
To assess the humoral immune response against AAV 9 capsid proteins, blood sera were collected at the day of birth, then at weeks 4 and 8 via peripheral vein. HEK 293 cells were seeded in a 48-well plate at a density of 105 cells per well in 200 μl DMEM containing 10% FBS. The cells were cultured for 3–4 h at 37 °C and allowed to adhere to the well. AAV9– green fluorescence protein (GFP) vector (1 × 108 particles) was incubated with mice sera at serial dilution with PBS for 2 h at 4 °C in a total volume of 25 μl. The mixture was then added to cells in a final volume of 200 μl that contained 4 × 106 particles of AAV9 and incubated for 24 or 48 h at 37 °C. Cells expressing GFP were counted under a fluorescent microscope. The neutralizing antibody titer was calculated using the highest dilution where the percentage of GFP-positive cells was 50% less than the control without sera.
Evaluation of cellular immune responses to AAV capsid, μUtro and μDystro derived peptides.
Cellular immune responses against AAV9 capsid, μDystro and μUtro antigens were evaluated with an interferon (IFN)-γ ELISpot assay51. The peptide library spanning AAV9 VP1 capsid consists of pools A–C (20 amino acids in length, overlapping by 10 residues), μDystro consists of pools A–D (15 amino acids in length, overlapping by 10 residues) and μUtro consists of pools E–J (15 amino acids in length, overlapping by 10 residues). A 96-well high protein binding Immobilon-P membrane was incubated overnight with canine IFN-γ monoclonal antibody. PBMCs were isolated on Ficoll hypaque gradients and cultured with synthetic peptides, PMA/ionomyocin or just medium at 37 °C, 5% CO2 for 36 h, followed by incubation with canine IFN-γ biotinylated monoclonal antibody for 1 h. Responses were considered positive when SFUs exceeded 50 per 106 PBMCs in duplicate wells.
Statistical analysis.
Informative group sizes for AAV vector injections in mdx mice were estimated based on one of the most sensitive and widely used histological assays available: the proportion of centrally nucleated muscle fibers from mice necropsied at eight weeks of age. To maximize the statistical power for the number of animals used, we adopted the approach described in ref. 52, to accommodate dependency among the multiple high-powered fields within a mouse. Mixed effects models accounting for clustering within mouse, using an exchangeable correlation structure, were used to compare the three groups defined by genotype and treatment. Estimation of the intracluster correlation from these models indicated a low value (<10%), suggesting a relatively high effective sample size, despite the necessarily small numbers of mice per group (at least four). Other random-effects parameters calculated in this analysis include the variance between clusters σ2u, the variance within clusters σ2e and the effective sample size neff.
To characterize the distribution of minimum Feret diameter in wild-type, treated and untreated dystrophic dogs, dots representing individual measurements from representative high-powered fields were plotted. Due to the small number of dogs, this analysis is entirely descriptive.
All analyses except the proportion of centrally nucleated muscle fibers are presented as mean ± s.d. Statistical analysis was processed using Prism 7 software (GraphPad). The statistical significance as P values is indicated in the figures: *P < 0.05; **P < 0.001; ***P < 0.0001; NS, not significant.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1 |. Evolution of dystrophin and utrophin and the framework for therapeutic vector design.
a, Reconstructed timeline of the dystrophin/utrophin divergence illustrating the timing of two independent duplications and three subsequent lineage-specific deletions (&, @, $, %, #). The timing of the dystrophin/utrophin duplication correlates with the emergence of hinged jaws, oligodendrocytes and membrane in-folding at the neuromuscular junctional site where utrophin is most highly expressed. MYA, million years ago. b, Delineation of domain conservation relative to recombinant proteins compatible with the size limit for AAV transgenes, including dystrophin-based constructs in early-stage clinical investigation and others described in the text. ClinicalTrials.gov reference numbers: *NCT03362502, ~NCT02376816 and ^NCT03368742. Symbols (@, $, %, #) indicate the positions of naturally occurring lineage-specific deletions; localization of epitopes recognized by anti-utrophin antibodies Utro N and C are depicted at the extreme lower edge of the figure.
Extended Data Fig. 2 |. Optimization of canine μutro transgene expression.
a, Schematic representation of μUtro based on CnN3D renderings of calponin homology domain (green), four spectrin-like repeats (yellow/blue) and combined WW-EF hand-ZZ domain (red). Disordered domains (‘hinges’) 1, 2 and 4 not depicted. a′, Western blot analysis and quantification of μUtro expression in HEK 293 cells using eight distinct plasmid vectors. a′, Western blot analysis and quantification of μUtro expression one week post intramuscular injections in the tibialis anterior (TA) followed by electroporation of six distinct μUtro containing vectors, as labeled (UTO, μ-Utro cDNA; opt, optimized; SP, synthetic promoter C5–1253; loading control, α-actin; CMV, cytomegalovirus promoter). b,c, Immunofluorescence staining against utrophin N terminus (Utro_N) and laminin in mdx mice injected with either Anc80-μUtro or PBS. Muscles collected include diaphragm, heart (b), quadriceps (quad), TA and gastrocnemius (GM) (c). d, Western blot analysis of μUtro expression in tissue samples, with vinculin serving as a loading control. e, Western blot analysis comparing μUtro expression in the TA and GM after systemic deliver using an AAV9 or Anc80 vector. Vinculin serving as a loading control. (See Source Data for full uncropped gel images.) All experiments were repeated independently at least two or more times with similar results. (See Source Data Extended Data Fig. 2).
Extended Data Fig. 3 |. Neonatal systemic administration of AAV-μutro in mdx mice shows histopathological amelioration in limb and diaphragm muscle eight weeks post injection.
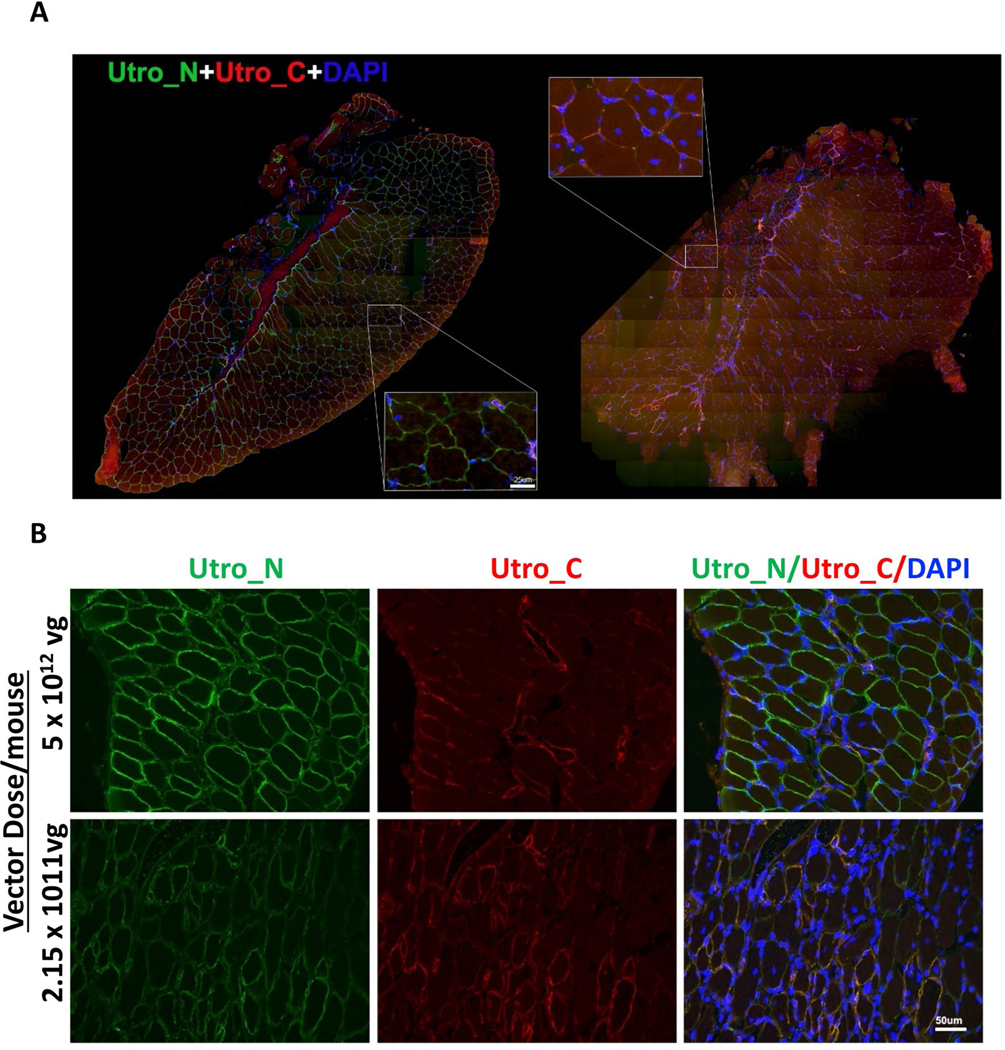
a, Double immunofluorescence staining against an epitope shared by native and recombinant utrophin (Utro_N) and an epitope unique to native utrophin (Utro_C) of the entire tibialis anterior muscle. Scale bar, 100 μm. b, Representative immunofluorescence staining of sectioned diaphragm35 at two distinct vector doses per mouse. Scale bar, 50 μm. All experiments were repeated independently at least two times with similar results.
Extended Data Fig. 4 |. Neonatal systemic administration of AAV-μutro eliminates active proteolysis, apoptosis and centrally nucleated myofibers in eight-week-old mdx limb muscle.
a, Representative immunofluorescence staining with MuRF1 (top panel) and TUNEL (bottom panel). Scale bar, 50 μm. b, Table summarizing mean and s.d. of centrally nucleated myofibers (Fig. 1d), MuRF1, TUNEL and embryonic positive myofibers (for example, see Fig. 1a) in mdx + PBS, mdx + AAV-μUtro and wild type + PBS. Data are presented as mean ± s.d.
Extended Data Fig. 5 |. Systemic delivery of AAV-μutro in neonatal mdx mice provides sustained expression of μutro and results in in vivo and ex vivo functional improvement.
a, Quantification of μUtro expression via western blot, at eight (n = 2 mice) and 16 weeks (n = 2 mice) of age, in GM, quad (quadriceps) and heart muscle. (See Source Data for full uncropped gel images.) b, Quantification of total distance covered on a running wheel over a 24 h time period by mdx + AAV-μUtro (n = 3) 3,614.11 ± 300.08 and mdx + PBS (n = 2) 1,848.96 ± 259.15. Distance was covered via downhill treadmill until cessation (# criteria for constituting cessation in the Methods). The dashed line represents distance covered during acclimatization prior to a ramped speed increase. Wild type (n = 4) 183 ± 0, mdx + AAV-μUtro (n = 4) 174.05 ± 17.9 and mdx + PBS (n = 5) 122.28 ± 24.12. Data are presented as mean ± s.d. Each symbol represents data from an individual animal and the wide horizontal line represents the group mean with associated error bars representing s.d. NS, not significant; **P < 0.01; statistical significance was assessed by one-way ANOVA with Tukey test multiple group comparison. c, Absolute force after seven cycles of eccentric contractions (ECCs) of extensor digitorum longus (EDL) muscle in mdx + AAV-μUtro mice (n = 10) and mdx + PBS mice (n = 9). Boxed values represent the fold difference of absolute force for mdx + AAV-μUtro (ECC1 210.37 ± 73.86, ECC2 128.36 ± 76.6, ECC3 115.54 ± 57.08, ECC4 101.83 ± 49.34, ECC5 89.28 ± 45.41, ECC6 78.21 ± 38.08) and mdx + PBS (ECC1 98.19 ± 48.78, ECC2 28.46 ± 16.81, ECC3 26.93 ± 14.91, ECC4 19.8 ± 14.74, ECC5 15 ± 12.8, ECC6 13.58, s.d. = 10.57). Data are presented as mean ± s.d. Each symbol represents data from an individual animal and the wide horizontal line represents the group mean with associated error bars representing s.d. **P < 0.01, ***P < 0.001; statistical significance was assessed by two-tailed unpaired t-test. d, Seven serial whole limb force measurements of wild type (n = 6), mdx + AAV-μUtro (n = 8) and mdx + PBS (n = 11). (See Source Data Extended Data Fig. 5).
Extended Data Fig. 6 |. Normal growth of GRMD dogs randomized to AAV-μutro as evidence against immune-mediated myositis.
a, Individual weights of dogs randomized to the highest doses (3.16 × 1013 vg kg−1) of AAV-cU (AAV-μUtro) without immunosuppression (F[b] AAV-cU, F[c] AAV-cU), as well as relevant controls including littermates randomized to PBS and other littermate carrier females (F-carrier3, F-carrier4, F-carrier6) and non-littermate GRMD males (M-GRMD7, M-GRMD8) and females (F-GRMD9, F-GRMD10). Also included for comparison are relevant weights of previously reported GRMD females receiving human μDystro (F[e] AAV-hD, F[f] AAV-hD) showing rapid weight loss immediately prior to euthanasia and necropsy showing signs of systemic myositis14. b, Representative results of PCR assays for GRMD genotype. c, Graphical depiction of Animal ID#, creatine kinase (CK) levels at birth, GRMD status by PCR, and weight at two time points for littermates of AAV-μUtro injected dogs.
Extended Data Fig. 7 |. Homogeneously expressed recombinant μutro normalizes β-sarcoglycan, reverses myopathology and normalizes fiber size in GRMD dogs after systemic gene delivery.
a, Immunofluorescence staining detecting the utrophin N terminus (Utro N, red) and β-sarcoglycan (green) of vastus lateralis muscle from wild type (WT), treated (AAV-μUtro) or untreated (PBS). The experiment was repeated independently at least two times with similar results. b, H&E staining of vastus lateralis muscle biopsies. Scale bar, 100 μm c, Distribution of minimum Feret diameter of myofibers in vastus lateralis muscle biopsies pooled from age-matched wild type (n = 920) 19 ± 4.01, GRMD littermates randomized to AAV-μUtro (n = 758) 17.63 ± 4.9, and PBS (n = 1,014) 14.49 ± 7.24. Data are presented as mean ± s.d. Coefficients of variation for all groups are reported in the box. d, Cumulative distribution (percentage) plot of minimum Feret diameter from c. e, Distribution of minimum Feret diameter of myofibers in vastus lateralis muscle biopsies obtained at necropsy, wild type (n = 569) 28.26 ± 8.39, AAV-μUtro (n = 542) 28.4 ± 7.54 and PBS (n = 317) 32.26 ± 12.01. Data are presented as mean ± s.d. Coefficients of variation for all groups are reported in the box. f, Cumulative distribution (percentage) plot of minimum Feret diameter from e. Coefficients of variation for all groups are reported in the box. (See Source Data Extended Data Fig. 7).
Extended Data Fig. 8 |. μutro expression provides histological improvement without any signs of toxicity in injected GRMD dogs.
a, Representative images of temporalis muscle from age-matched AVV-μUtro (treated) and PBS (untreated) GRMD dogs stained against an N-terminal epitope shared by native and recombinant utrophin (Utro N) and a C-terminal epitope unique to native utrophin (Utro C) (top) and MYH16, BA-F8 (slow twitch myofiber) and Utro N (bottom). Scale bar, 20 μm. Experiment was repeated independently at least two or more times with similar results. b, H&E stain. Top: A whole view of vastus lateralis muscle obtained from a muscle biopsy four weeks post-injection. Botom: A higher magnification of the corresponding boxed region. The experiment was repeated independently at least two times with similar results. c, γ-Interferon production was quantified by counting the spot forming units (SFUs) per million peripheral blood mononuclear cells (PBMCs). A response above the dotted line is considered positive and above the background signal. Peptide libraries used represent the full sequence of AAV9 capsid (A–C), μUtro (E–J), Adenovirus5 (1–4) and LacZ (1–4). d, Measurement of alkaline phosphatase (ALP), alanine aminotransferase (ALT), aspartate aminotransferase (AST), hemoglobin, platelets and γ-interferon before and after AAV-μUtro injection in two juvenile GRMD dogs (Hann/Beetle). Non-human primate (16C176 and RA2152) data are incorporated to illustrate the absence of disseminated intravascular coagulation (DIC)33,34. (See Source Data Extended Data Fig. 7).
Extended Data Fig. 9 |. μDystro, but not μutro, elicits a local cell mediated immune response after an intramuscular injection in the GSHPMD dogs.
Muscle biopsies were obtained four weeks post-intramuscular injection of μDystro and μUtro. a, Representative H&E stain of muscle biopsies. Right panel for each dog shows higher magnification of the corresponding boxed region. Scale bar, 50 μm. b, Immunofluorescence staining of muscle biopsies against Utro_N and Utro_C. Scale bar, 50 μm. c, Immunofluorescence staining of muscle biopsies against CD3 and CD8; the bottom merged panel offers a validation of both antibodies. All experiments were repeated independently at least two times with similar results.
Supplementary Material
Acknowledgements
This work was supported by grants from the NIH (H.S., R01NS042874, U01NS052476 and S10RR028027; L.M., T32AR053461; C.D.G., T32HL007954; J.K., U24NS059696; A.M., F.B. and M.P., T32AR053461; Y.S. and histology, P30AR050950), the Muscular Dystrophy Association (H.S. and J.K.) and the families of O. Soot and M. Haider, to whom this work is dedicated. Additional services were supported by P30DK047757 and the NHLBI Gene Therapy Resource Center as well as the University of Pennsylvania Vector and Immunology Cores. We thank L. Vandenberghe for providing helper plasmid (AAP2) for Anc80 production, A. Stout and J. Zhao as well as the UPenn CDB microscopy core for their technical assistance and resource access, and V. Arruda, J. Bennett, J. Johnston, R. Calcedo, F. Wright and J. Plotkin for support and expertise. Dedicated to the memory of Mohammed Haider and Beth Stedman.
Footnotes
Data availability
All data generated or analyzed during this study are included in this published article or in the Supplementary Information files.
Competing interests
H.H.S. is an inventor on several patents assigned to the University of Pennsylvania and subject to the institutional patent policies, which may include royalty distribution in the event of licensure. None of the patents are currently licensed, their potential valuation may go up or down with the publication of this manuscript, all are in the public domain, some have recently expired.
Additional information
Extended data is available for this paper at https://doi.org/10.1038/s41591-019-0594-0.
Supplementary information is available for this paper at https://doi.org/10.1038/s41591-019-0594-0.
Peer review information Brett Benedetti and Kate Gao were the primary editors on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, statements of code and data availability and associated accession codes are available at https://doi.org/10.1038/s41591-019-0594-0.
References
- 1.Hoffman EP, Brown RH Jr. & Kunkel LM Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell 51, 919–928 (1987). [DOI] [PubMed] [Google Scholar]
- 2.Koenig M, Monaco AP & Kunkel LM The complete sequence of dystrophin predicts a rod-shaped cytoskeletal protein. Cell 53, 219–226 (1988). [DOI] [PubMed] [Google Scholar]
- 3.Petrof BJ, Shrager JB, Stedman HH, Kelly AM & Sweeney HL Dystrophin protects the sarcolemma from stresses developed during muscle contraction. Proc. Natl Acad. Sci. USA 90, 3710–3714 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ibraghimov-Beskrovnaya O. et al. Primary structure of dystrophin-associated glycoproteins linking dystrophin to the extracellular matrix. Nature 355, 696–702 (1992). [DOI] [PubMed] [Google Scholar]
- 5.Tinsley JM et al. Primary structure of dystrophin-related protein. Nature 360, 591–593 (1992). [DOI] [PubMed] [Google Scholar]
- 6.Mesnard-Rouiller L, Bismuth J, Wakkach A, Poea-Guyon S. & Berrih-Aknin S. Thymic myoid cells express high levels of muscle genes. J. Neuroimmunol 148, 97–105 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Clemens PR et al. In vivo muscle gene transfer of full-length dystrophin with an adenoviral vector that lacks all viral genes. Gene Ther. 3, 965–972 (1996). [PubMed] [Google Scholar]
- 8.Wang B, Li J. & Xiao X. Adeno-associated virus vector carrying human minidystrophin genes effectively ameliorates muscular dystrophy in mdx mouse model. Proc. Natl Acad. Sci. USA 97, 13714–13719 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harper SQ et al. Modular flexibility of dystrophin: implications for gene therapy of Duchenne muscular dystrophy. Nat. Med 8, 253–261 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Gregorevic P. et al. Systemic delivery of genes to striated muscles using adeno-associated viral vectors. Nat. Med 10, 828–834 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gregorevic P. et al. rAAV6-microdystrophin preserves muscle function and extends lifespan in severely dystrophic mice. Nat. Med 12, 787–789 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore MJ & Flotte TR Autoimmunity in a genetic disease—a cautionary tale. N. Engl. J. Med 363, 1473–1475 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Mendell JR et al. Dystrophin immunity in Duchenne’s muscular dystrophy. N. Engl. J. Med 363, 1429–1437 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kornegay JN et al. Widespread muscle expression of an AAV9 human mini-dystrophin vector after intravenous injection in neonatal dystrophin-deficient dogs. Mol. Ther 18, 1501–1508 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duan D. & Systemic AAV Micro-dystrophin gene therapy for Duchenne muscular dystrophy. Mol. Ther 26, 2337–2356 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muthu M, Richardson KA & Sutherland-Smith AJ The crystal structures of dystrophin and utrophin spectrin repeats: implications for domain boundaries. PLoS One 7, e40066 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ortega E. et al. The structure of the plakin domain of plectin reveals an extended rod-like shape. J. Biol. Chem 291, 18643–18662 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishikawa-Sakurai M, Yoshida M, Imamura M, Davies KE & Ozawa E. ZZ domain is essentially required for the physiological binding of dystrophin and utrophin to beta-dystroglycan. Hum. Mol. Genet 13, 693–702 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Hnia K. et al. ZZ domain of dystrophin and utrophin: topology and mapping of a beta-dystroglycan interaction site. Biochem. J 401, 667–677 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zinn E. et al. In silico reconstruction of the viral evolutionary lineage yields a potent gene therapy vector. Cell Rep. 12, 1056–1068 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foster H. et al. Codon and mRNA sequence optimization of microdystrophin transgenes improves expression and physiological outcome in dystrophic mdx mice following AAV2/8 gene transfer. Mol. Ther 16, 1825–1832 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Odom GL, Gregorevic P, Allen JM, Finn E. & Chamberlain JS Microutrophin delivery through rAAV6 increases lifespan and improves muscle function in dystrophic dystrophin/utrophin-deficient mice. Mol. Ther 16, 1539–1545 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sonnemann KJ et al. Functional substitution by TAT-utrophin in dystrophin-deficient mice. PLoS Med. 6, e1000083 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kennedy TL et al. Micro-utrophin improves cardiac and skeletal muscle function of severely affected D2/mdx mice. Mol. Ther. Methods Clin. Dev 11, 92–105 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song Y. et al. Suite of clinically relevant functional assays to address therapeutic efficacy and disease mechanism in the dystrophic mdx mouse. J. Appl. Physiol 122, 593–602 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobayashi YM et al. Sarcolemma-localized nNOS is required to maintain activity after mild exercise. Nature 456, 511–515 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nichols T. et al. Translational data from adeno-associated virus-mediated gene therapy of hemophilia B in dogs. Hum. Gene Ther. Clin. Dev. 26, 5–14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calcedo R. et al. Adeno-associated virus antibody profiles in newborns, children and adolescents. Clin. Vaccine Immunol 18, 1586–1588 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yiu EM & Kornberg AJ Duchenne muscular dystrophy. J. Paediatr. Child Health 51, 759–764 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Liu JM et al. Effects of prednisone in canine muscular dystrophy.Muscle Nerve 30, 767–773 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Stedman HH et al. Myosin gene mutation correlates with anatomical changes in the human lineage. Nature 428, 415–418 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Toniolo L. et al. Masticatory myosin unveiled: first determination of contractile parameters of muscle fibers from carnivore jaw muscles. Am. J. Physiol. Cell Physiol 295, C1535–C1542 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Hinderer C. et al. Severe toxicity in nonhuman primates and piglets following high-dose intravenous administration of an adeno-associated virus vector expressing human SMN. Hum. Gene Ther 29, 285–298 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hordeaux J. et al. The neurotropic properties of AAV-PHP.B are limited to C57BL/6J mice. Mol. Ther 26, 664–668 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stedman HH et al. The mdx mouse diaphragm reproduces the degenerative changes of Duchenne muscular dystrophy. Nature 352, 536–539 (1991). [DOI] [PubMed] [Google Scholar]
- 36.Schatzberg SJ et al. Molecular analysis of a spontaneous dystrophin ‘knockout’ dog. Neuromuscul. Disord 9, 289–295 (1999). [DOI] [PubMed] [Google Scholar]
- 37.VanBelzen DJ, Malik AS, Henthorn PS, Kornegay JN & Stedman HH Mechanism of deletion removing all dystrophin exons in a canine model for DMD implicates concerted evolution of X chromosome pseudogenes. Mol. Ther. Methods Clin. Dev 4, 62–71 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schatzberg SJ et al. Alternative dystrophin gene transcripts in golden retriever muscular dystrophy. Muscle Nerve 21, 991–998 (1998). [DOI] [PubMed] [Google Scholar]
- 39.Yue Y. et al. Safe and bodywide muscle transduction in young adult Duchenne muscular dystrophy dogs with adeno-associated virus. Hum. Mol. Genet 24, 5880–5890 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le Guiner C. et al. Long-term microdystrophin gene therapy is effective in a canine model of Duchenne muscular dystrophy. Nat. Commun 8, 16105 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altschul SF, Gish W, Miller W, Myers EW & Lipman DJ Basic local alignment search tool. J. Mol. Biol 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 42.Altschul SF et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Solovyev V, Kosarev P, Seledsov I. & Vorobyev D. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 7, S10.11–S10.12 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi Y, Falahati R, Zhang J, Flebbe-Rehwaldt L. & Gaensler KM Role of antigen-specific regulatory CD4+ CD25+ T cells in tolerance induction after neonatal IP administration of AAV-hF.IX. Gene Ther. 20, 987–996 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davey MG et al. Induction of immune tolerance to foreign protein via adeno-associated viral vector gene transfer in mid-gestation fetal sheep. PLoS One 12, e0171132 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vandenberghe LH et al. Efficient serotype-dependent release of functional vector into the culture medium during adeno-associated virus manufacturing. Hum. Gene Ther 21, 1251–1257 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lock M, Alvira MR & Wilson JM Analysis of particle content of recombinant adeno-associated virus serotype 8 vectors by ion-exchange chromatography. Hum. Gene Ther. Methods 23, 56–64 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zincarelli C, Soltys S, Rengo G. & Rabinowitz JE Analysis of AAV serotypes 1–9 mediated gene expression and tropism in mice after systemic injection. Mol. Ther 16, 1073–1080 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Song Y, Forsgren S, Yu J, Lorentzon R. & Stal PS Effects on contralateral muscles after unilateral electrical muscle stimulation and exercise. PloS One 7, e52230 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mishra MK, Loro E, Sengupta K, Wilton SD & Khurana TS Functional improvement of dystrophic muscle by repression of utrophin: let-7c interaction. PLoS One 12, e0182676 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mingozzi F. et al. AAV-1-mediated gene transfer to skeletal muscle in humans results in dose-dependent activation of capsid-specific T cells. Blood 114, 2077–2086 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aarts E, Verhage M, Veenvliet JV, Dolan CV & van der Sluis S. A solution to dependency: using multilevel analysis to accommodate nested data. Nat. Neurosci 17, 491–496 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Li X, Eastman EM, Schwartz RJ & Draghia-Akli R. Synthetic muscle promoters: activities exceeding naturally occurring regulatory sequences. Nat. Biotechnol 17, 241–245 (1999). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.