Abstract
Shigella spp., which are closely related to E. coli, can easily be maintained and stored in the laboratory. This unit includes protocols for routine growth conditions and media, for storage of the bacteria, and for monitoring the presence of the virulence plasmid.
Keywords: Shigella, Shigella flexneri
Introduction
Shigella species are members of the Enterobacteriaceae which cause bacillary dysentery, or shigellosis, in humans. Historically, the Shigella were classified as species distinct from Escherichia coli because of their pathogenicity and biochemical characteristics, but genome sequencing has shown that they are phylogenetically closely related (1). The Shigella species (Shigella boydii, Shigella dysenteriae, Shigella flexneri and Shigella sonnei) differ from most E. coli isolates in their inability to ferment lactose (Lac−), lack of motility, and lysine decarboxylase negative phenotype. Their distinct pathogenesis derives from proteins encoded by a large (~ 220 kb) plasmid (2) that is also found in pathogenic entero-invasive E. coli (EIEC) that produce similar symptoms (3–5). Specific chromosomal genes (6) and deletions (7), compared to E. coli K12, are also associated with Shigella pathogenicity.
Because the Shigella are members of the same family as E. coli, most of the culture media and techniques used for growth and maintenance of E. coli work well for the Shigella species. There are differences, however, and these will be detailed in this chapter. These specific protocols have been extensively used for S. flexneri, which is the best characterized of the Shigella species, and slight modifications may be needed for the other species.
If clinical isolates are being used, the antibiotic resistance profile should be determined before further use. Many strains show resistance to multiple antibiotics, including those that are commonly used for treatment of infections.
Caution: Shigella species are Risk Group 2 pathogens and all work should be done in a BSL-2 environment. Consult with your institutional biosafety committee before beginning any experiments with wild-type Shigella. Because Shigella are highly infectious, with an infectious dose of as few as 100 organisms, more stringent safety practices than used for other BSL-2 level organisms are recommended. In particular, S. dysenteriae may be subject to more stringent controls because it produces shiga toxin.
Strategic planning
Strain selection
For experiments involving genetic manipulations, it is highly recommended to use a strain for which the whole genome has been sequenced. S. flexneri contains large numbers of insertion sequences, remnants of phage genomes, and genomic islands, and these vary among isolates. Thus, while there may be little or no differences in coding sequences for most genes, the flanking sequences can vary. Examples of strains that are sequenced and have been used extensively in published studies are serotype 2a strains SF301 (8) and 2457 (9) and serotype 5a strain M90 (10).
Growth conditions
S. flexneri is a facultative anaerobe and can grow in the presence or absence of oxygen. It grows optimally at 37°C, in rich medium with aeration. It is acid tolerant, but the pH of laboratory media should be adjusted near neutral pH (6.8 −7.4) for best growth. S. flexneri can use a variety of carbon sources including amino acids and sugars; the most rapid growth is obtained with pyruvate or glucose as the carbon sources (11). Both complex and minimal, defined media support good growth of S. flexneri.
The Shigella virulence plasmid can be lost from strains upon repeated passage or growth at higher temperature (>35° C) (12, 13). For studies requiring maintenance of the virulence plasmid, Congo red agar (14, 15) is used to distinguish between wild type colonies and those that have lost or have deletions in the plasmid. The wild type strains absorb Congo red dye from the medium when grown at 37°C, producing red colonies (CR+), while those that have lost the plasmid or have deletions affecting virulence gene expression are white (CR−) (14, 16). The white colonies tend to be slightly larger and are less translucent than the red colonies. Note that some mutants, particular rough LPS mutants, will also bind the dye, even if they lack the virulence plasmid. These can be distinguished by their rough colony morphology.
Basic protocol 1: Growth of S. flexneri from frozen stocks or agar stabs
Materials
S. flexneri frozen stock
Wooden applicator stick, sterile
Inoculating loop, flame sterilized
Congo red agar plate
37°C incubator
-
1a.
For frozen stocks, scrape a small amount of frozen culture from the vial with the sterile applicator stick and transfer to the surface of a Congo red plate. Return the frozen stock to the −80°C freezer. Do not allow the stock to thaw, as repeated freeze thaw cycles reduce the viability of the stock.
-
1b.
For agar stabs, use a sterile inoculating needle or applicator stick and stab into the area of growth in the stab, then streak the needle or stick in a small area of the plate to transfer the bacteria.
-
2.
Flame sterilize the inoculating loop and allow to cool or briefly touch the surface of the agar away from the inoculum to rapidly cool the wire. Spread the inoculum across one quadrant of the plate. Flame the loop, then streak from the first area onto a second quadrant of the plate, only streaking into the first quadrant, 1–2 times. Flame the loop, then repeat the process, on the third and then fourth quadrants of the plate, Fig. 1).
-
3.
Incubate the plate upside down at 37°C overnight. It is important to incubate at 37°C, since incubation at lower temperatures does not allow expression of genes required to detection of the CR+ phenotype (16). After overnight incubation, isolated colonies should be present in one of the quadrants. Well isolated, red colonies can be used to inoculate broth or plate cultures.
-
4.
Use parafilm to seal the cover onto the plate and store refrigerated (4°C). Colonies on refrigerated plates will remain viable for 7–10 days.
Figure 1:
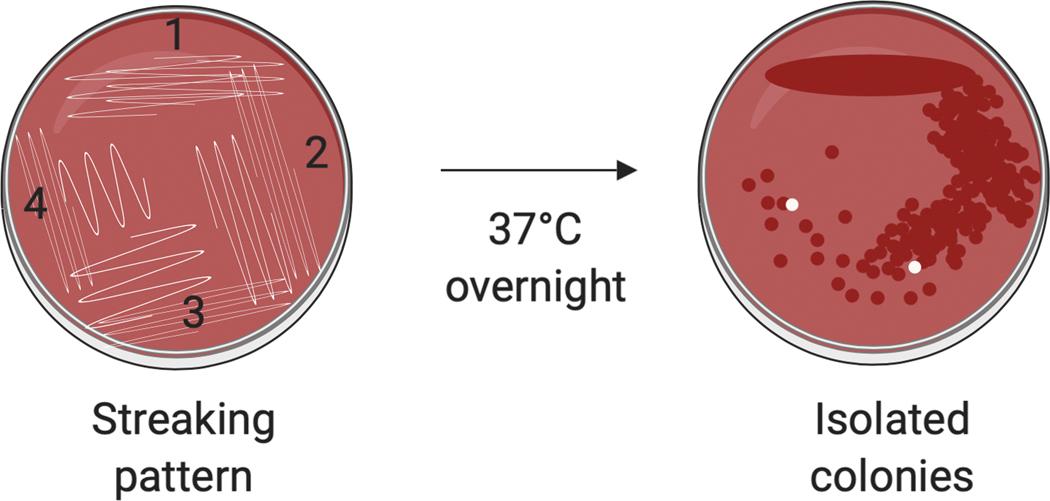
S. flexneri streaking for isolation on Congo red agar. As shown on the left, a small amount of the frozen stock or stab is spread on a small area of the plate (1). A sterile loop is used to streak the remaining quadrants of the plate (2, 3, and 4), as described in Basic Protocol 1. After overnight incubation at 37°C, wild‐type S. flexneri forms translucent colonies that bind the dye (Crb+) and appear red, as shown on the right. Cells that have lost or have deletions in the virulence plasmid produce larger, more opaque, white colonies (Crb−).
Basic protocol 2: Growth of S. flexneri in rich, liquid medium
Materials
TSB or LB medium, 3–5 ml in an 18 × 150 mm sterile, capped glass culture tube
Wooden applicator stick, sterile or flame-sterilized loop
Vortex mixer
37° rotary incubator or shaker
Pick a single, well-isolated, red colony from Congo red agar using a sterile applicator stick or loop.
Rapidly swish applicator in the broth. Remove applicator, cap the tube, and vortex.
Incubate with shaking until desired density is reached. Overnight incubation will yield a fully-grown culture (~ 2–4 X 109/ml). For Shigella spp. growing in rich media, A650 = 1.0 is approximately 8 X 108 cells/ml
For growing larger volumes of Shigella, begin with a 5 ml culture grown to mid-log phase (A650 = 0.3 – 0.7) and dilute 1:100 into a flask containing the growth medium. The flask should contain no more than 20% of its maximum volume to allow good aeration while shaking.
Basic protocol 3: Growth of S. flexneri in rich, defined medium
For experiments in which reproducibility and defined conditions are needed, the rich defined medium described by Neidhardt et al. (17) with slight modifications (EZ-RDM) (https://www.genome.wisc.edu/resources/protocols/ezmedium.htm) is used. This medium allows rapid growth and a high cell density, any component such as carbon or nitrogen source or micronutrients can be varied independently, background fluorescence is minimal, and results are reproducible. It is recommended for experiments such as RNA-Seq, gene expression analysis, and proteomics, or other situations where batch-to-batch variation in the medium and growth conditions must be minimized.
Materials
Shigella colonies on agar plate
EZ-RDM in capped tube (5 ml in 18 X 150 mm tube) or in flask at a volume of no more than 20% of the total flask volume.
Microfuge
Sterile 1.5 ml microfuge tubes
Because there can be an unpredictable lag time when inoculating Shigella colonies directly from a plate into defined or minimal media, the strain is first grown in LB, then diluted into the defined medium.
Inoculate 5 ml of LB with a single, isolated colony of Shigella as described in Basic Protocol 2. Incubate until the culture reaches late log phase (A650 = ~ 1.0) or overnight. For overnight culture, use a temperature of 30°C to reduce selection for mutants that have lost the virulence plasmid.
Centrifuge 1 ml of cells in a microcentrifuge for 5 min. and resuspend the pellet in EZ-RDM to a final A650 = 1.0. Dilute 1:100 – 1:200 into EZ-RDM and incubate at 37C with shaking until desired cell density is reached.
Some strains show a long lag when diluted from an overnight both culture into EZ-RDM. In this case, first do a relatively low dilution (1:20) by adding 250 μl of the broth culture to 5 ml EZ-RDM in an 18 X 150 mm capped tube, vortex, and incubate with shaking until late log phase and then centrifuge and dilute into EZ-RDM as above.
Basic protocol 4: Growth of S. flexneri in minimal media
Because Shigella are closely related to E. coli, many of the minimal media commonly used for E. coli are suitable for Shigella as well. Most Shigella isolates are auxotrophs and minimal media will require supplementation. The most common nutritional requirements are nicotinic acid, methione, and/or tryptophan (11).
Choice of minimal media: M9 or similar phosphate-buffered medium can be used for many purposes. However, for experiments requiring low phosphate, a tris-buffered medium such as T-medium, is preferred. This is particularly useful for studies of iron or other trace metals, since these can be found as contaminates of phosphates, or studies using 32P labeling.
Materials
Shigella colonies on agar plate
Minimal M9 medium or T-medium
Glucose or pyruvate (0.4% final concentration) as carbon source
Vitamin and amino acid supplements: Prepare stock at 100X the indicated concentration in water and filter sterilize. Add 10 ml per liter to minimal medium to give the following concentration.
Nicotinic acid 12.5 μg/ml final concentration
Methionine 45 μg/ml final concentration
Tryptophan 20 μg/ml final concentration
Inoculate 5 ml of LB with a single, isolated colony of Shigella as described in Basic Protocol 2. Incubate overnight.
Add 250 μl of the broth culture to 5 ml minimal medium in an 18 X 150 mm capped tube, vortex, and incubate with shaking until late log phase
Centrifuge 1 ml of cells in a microcentrifuge for 5 min. and resuspend the pellet in minimal medium to a final A650 = 1.0. Dilute 1:100 – 1:200 into minimal medium and incubate at 37° C with shaking until desired cell density is reached.
Troubleshooting:
Some strains of Shigella fail to grow in the minimal media. This is usually due to a nutritional requirement. While the addition of nicotinic acid, methionine, and tryptophan will support most strains, it may be necessary to add additional vitamins or amino acids. This can be tested by adding Supplement EZ described in in formula for EZ-Rich Defined Medium (EZ-RDM) below.
Basic protocol 5: Storage of S. flexneri in frozen stocks
Materials
LB agar plate
TSB-20% glycerol, sterile
5 ml pipette
Cryovial, Corning, 430658
Using a sterile loop, pick 2–3 well-isolated colonies from a Congo red plate and streak across the middle of the LB agar plate. Turn the plate, and without flaming the loop, streak across the initial streak covering the surface of the plate.
Incubate overnight at 37°C. The plate should have near-confluent growth.
Using a sterile pipette, pipette 3 ml of TSB-glycerol onto the surface of the plate and pipette the liquid up and down to resuspend the growth. Transfer 1 ml of the suspension into the cryovial.
Store frozen at −80°C. With minimal freeze-thawing, cultures are viable for >10 years.
Basic protocol 6: Storage of S. flexneri in agar stabs
Materials
LB agar
Small, flat-bottomed, sterile glass vials with screw cap (e.g. 1 ml vials, VWR 470206–388)
Wooden applicator stick, sterile (Puritan 5.75 in wooden applicator stick) or flame-sterilized wire needle
Parafilm
Prepare LB agar and distribute into vials. Vials should be ½ - ¾ full. Let cool until agar is firm. Unused vials can be stored with the screw caps tight at room temperature or refrigerated for up to 1 year.
Using a sterile stick, touch a single, well-isolated colony and stab into the center of the agar to approximately 1/4” from the bottom of the agar. Loosely screw on the cap.
Incubate the inoculated vials at room temperature or at 37°C until a line of growth is visible along the site of the stab.
Tighten the screw cap and seal the top of the vial with Parafilm.
Store stabs at room temperature for up to 3 years
REAGENTS, MEDIA AND SOLUTIONS
Use deionized, distilled water or double-deionized water (e.g. Millipore Milli-Q system) for all reagents and media.
Congo red agar
30 g Trypticase soy broth (BD)
0.1 g Congo red dye
15 g agar
Add water to a final volume of 1000 ml and allow medium and dye to dissolve. Autoclave and cool to 55–65°C. The agar can be cooled and held in a 60°C water bath until ready to use and should be cool enough that the flask can be held without a hot pad.
Pour into sterile plastic petri dishes and allow to cool until the agar is completely firm.
Store the plates inverted in a plastic sleeve and refrigerated for up to 3 months.
Note: Use trypticase soy broth or tryptic soy broth plus agar, rather than purchasing trypticase soy agar or tryptic soy agar. The latter formulations lack glucose and phosphate buffer that are in the broth formulations. These are important for maximum adsorption of the Congo red dye.
LB (Lysogeny broth) medium
10 g tryptone
5 g yeast extract
10 g NaCl
Add water to 1000 ml
Adjust pH to 7.0 with NaOH. Distribute into clean bottles. Autoclave and store at room temperature. pour plates as described for Congo red agar.
LB agar
10 g tryptone
5 g yeast extract
10 g NaCl
15 g agar
Add water to 1000 ml
Adjust pH to 7.0 with NaOH. Autoclave and pour plates as described for Congo red agar.
M9 minimal medium
6 g Na2HPO4•7H2O
3 g KH2PO4
0.5 g NaCl
1.0 g NH4Cl
Add sterile water to 1000 ml
Autoclave, then add the following sterile, stock solutions:
20 ml glucose or other carbon source stock solution
1 ml 1 M MgSO4 stock solution
1 ml 100 mM CaCl2 stock solution
Amino acid and vitamin supplements as needed (see Protocol 4).
Stock solutions:
20% solution of the appropriate carbon source, glucose or sodium pyruvate, filter sterilize.
1 M MgSO4 Autoclave to sterilize.
100 mM CaCl2 Autoclave to sterilize.
T medium
This is a modification of the minimal medium described by Simon and Tessman (12).
5.8 g NaCl
3.7 g KCl
1.1 g NH4Cl
0.11 g CaCl2
0.272 g KH2PO4
12.1 g Tris(hydroxymethyl)amino methane
Add water to 1 liter. Adjust pH to 7.4 with 12N HCl.
Autoclave, then add the following sterile, stock solutions.
1 ml 1 M MgSO4 sterile stock solution as described for M9 minimal medium
1 ml 1 mM FeCl3 stock solution. The iron stock is filter sterilized and stored frozen at −20°C. If any precipitate is noted, discard, and make a fresh stock solution.
20 ml glucose or other carbon source stock solution
Amino acid and vitamin supplements as needed (see Protocol 4).
EZ-Rich Defined Medium (EZ-RDM)
This medium can be purchased from Technova as MOPS EZ Rich Defined Medium Kit, M2105. However, if any components are to be varied, the medium can be prepared as follows, omitting or varying any of the components as needed.
100ml 10X MOPS Mixture
10 ml 0.132 M K2HPO4
100 ml 10X ACGU
200 ml 5X Supplement EZ
580 ml Sterile H2O
10 ml Carbon source stock (100 X (20%) solution of the carbon source, filter sterilized)
10X MOPS mixture:
83.72 g MOPS
7.17 g Tricine
Add to 300 ml milliQ H2O in a 1 L beaker with a stir bar. Adjust pH to 7.4 with 10 N KOH.
Bring total volume to 440 ml
Add 10 ml of a freshly made iron solution (0.028 g FeSO4•7H2O dissolved in 10 ml H2O)
Add the following in the order shown:
50 ml NH4Cl stock
10 ml K2SO4 stock
0.25 ml CaCl2•2H2O stock
2.1 ml MgCl2 stock
100 ml NaCl stock
0.2 ml Micronutrient stock
387 ml sterile milliQ H2O
Filter sterilize with a 1 L capacity 0.2 micron filter. Aliquot into sterile plastic bottles and freeze at −20° C.
Stock solutions for 10X MOPS mixture:
Make each solution separately, store at room temperature.
NH4Cl stock
50.82 g NH4Cl add H2O to 500 ml
K2SO4 stock
4.8 g K2SO4 add H2O to 100 ml
CaCl2•2H2O stock
0.294 g CaCl2•2H2O add H2O to 100 ml
MgCl2 stock
50.75 g MgCl2 add H2O to 100 ml
NaCl stock
292.2 g NaCl add H2O to 1000 ml
Micronutrient stock
Add the following to 40 ml milliQ H2O
0.009 g (NH4)6Mo7O24•4H2O
0.062 g H3BO3
0.018 g CoCl2
0.006 g CuSO4
0.040 g MnCl2
0.007 g ZnSO4
Bring total volume to 50 ml. Store at room temperature.
g 23.0 g K2HPO4
MilliQ H2O to 1000 ml
Autoclave and store at room temperature.
ACUG stock (10X)
0.270 g adenine
0.222 g cytosine
0.224 g uracil
0.302 g guanine
Dissolve together in 1 L 0.015 M KOH. Filter sterilize and store at −20° C.
Supplement EZ (5X)
5 ml alanine stock
65 ml arginine stock
40 ml asparagine stock
5 ml aspartic acid stock
50 ml cysteine stock
5 ml glutamic acid stock
25 ml glutamine stock
5 ml glycine stock
5 ml histidine stock
10 ml isoleucine stock
80 ml leucine stock
5 ml lysine stock
5 ml methionine stock
40 ml phenylalanine stock
5 ml proline stock
125 ml serine stock
5 ml threonine stock
10 ml tryptophan stock
100 ml tyrosine stock
10 ml valine stock
50 ml VA Vitamin Solution
350 ml H2O
Mix and filter sterilize with 1 L capacity 0.2 micron filter. Aliquot into sterile plastic bottles and store frozen at −20° C.
Supplement stocks
Prepare each stock separately in the volume of water shown and store frozen at −20° C.
1.78 g alanine (free) 25 ml
8.44 g arginine (HCl) 100 ml
0.66 g asparagine (free) 100 ml
1.71 g aspartic acid (K salt) 25 ml
0.09 g cysteine (HCl H2O) 50 ml
2.78 g glutamic acid (K salt) 25 ml
1.8 g glutamine (free) 100 ml
1.5 g glycine (free) 25 ml
1.05 g histidine (HCl H2O) 25 ml
0.65 g isoleucine (free) 25 ml
0.66 g leucine (free) 100 ml
1.83 g lysine 25 ml
0.75 g methionine (free) 25 ml
0.83 g phenylalanine (free) 100 ml
1.15 g proline (free) 25 ml
4.2 g serine (free) 100 ml
1.19 g threonine (free) 25 ml
0.26 g tryptophan (free) 25 ml
0.045 g tyrosine (free) 25 ml 0.01 M KOH
0.88 g valine (free) 25 ml
VA Vitamin Solution (100X)
Prepare each solution separately, then combine
0.169 g thiamine HCl dissolved in 25 ml H2O
0.238 g calcium pantothenate dissolved in 25 ml H2O
0.069 g p-aminobenzoic acid dissolved in 25 ml 0.02 M KOH
0.069 g p-hydroxybenzoic acid dissolved in 25 ml 0.02 M KOH
0.077 g 2,3-dihydroxybenzoic dissolved in 25 ml 0.02 M KOH
Add 375 ml milliQ H2O
Mix and store at −20°C.
CRITICAL PARAMETERS:
Because the virulence plasmid can be lost from Shigella isolates, care should be taken to ensure that the plasmid is present for any studies involving pathogenicity. Strains should be streaked onto Congo red agar before use, and red colonies picked for further growth. Overnight broth cultures should be grown at 30°C to reduce selection for cells that have lost the plasmid.
ACKNOWLEDGEMENT:
This work was supported by grant NIAID AI16935 from the National Institutes for Health.
References
- 1.Devanga Ragupathi NK, Muthuirulandi Sethuvel DP, Inbanathan FY, Veeraraghavan B. 2017. Accurate differentiation of Escherichia coli and Shigella serogroups: challenges and strategies. New Microbes New Infect 21:58–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sansonetti PJ, Kopecko DJ, Formal SB. 1982. Involvement of a plasmid in the invasive ability of Shigella flexneri. Infect Immun 35:852–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hazen TH, Leonard SR, Lampel KA, Lacher DW, Maurelli AT, Rasko DA. 2016. Investigating the Relatedness of Enteroinvasive Escherichia coli to other E. coli and Shigella Isolates by Using Comparative Genomics. Infect Immun 84:2362–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sansonetti PJ, d’Hauteville H, Formal SB, Toucas M. 1982. Plasmid-mediated invasiveness of “Shigella-like” Escherichia coli. Ann Microbiol (Paris) 133:351–355. [PubMed] [Google Scholar]
- 5.Sansonetti PJ, d’Hauteville H, Ecobichon C, Pourcel C. 1983. Molecular comparison of virulence plasmids in Shigella and enteroinvasive Escherichia coli. Ann Microbiol (Paris) 134A:295–318. [PubMed] [Google Scholar]
- 6.Hale TL. 1991. Genetic basis of virulence in Shigella species. Microbiol Rev 55:206–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maurelli AT, Fernández RE, Bloch CA, Rode CK, Fasano A. 1998. “Black holes” and bacterial pathogenicity: A large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc Natl Acad Sci 95:3943–3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin Q, Yuan Z, Xu J, Wang Y, Shen Y, Lu W, Wang J, Liu H, Yang J, Yang F, Zhang X, Zhang J, Yang G, Wu H, Qu D, Dong J, Sun L, Xue Y, Zhao A, Gao Y, Zhu J, Kan B, Ding K, Chen S, Cheng H, Yao Z, He B, Chen R, Ma D, Qiang B, Wen Y, Hou Y, Yu J. 2002. Genome sequence of Shigella flexneri 2a: insights into pathogenicity through comparison with genomes of Escherichia coli K12 and O157. Nucleic Acids Res 30:4432–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei J, Goldberg MB, Burland V, Venkatesan MM, Deng W, Fournier G, Mayhew GF, Plunkett G, Rose DJ, Darling A, Mau B, Perna NT, Payne SM, Runyen-Janecky LJ, Zhou S, Schwartz DC, Blattner FR. 2003. Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect Immun 71:2775–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Onodera NT, Ryu J, Durbic T, Nislow C, Archibald JM, Rohde JR. 2012. Genome sequence of Shigella flexneri serotype 5a strain M90T Sm. J Bacteriol 194:3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waligora EA, Fisher CR, Hanovice NJ, Rodou A, Wyckoff EE, Payne SM. 2014. Role of intracellular carbon metabolism pathways in Shigella flexneri virulence. Infect Immun 82:2746–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saeed A, Johansson D, Sandström G, Abd H. 2012. Temperature Depended Role of Shigella flexneri Invasion Plasmid on the Interaction with Acanthamoeba castellanii. Int J Microbiol 2012:917031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schuch R, Maurelli AT. 1997. Virulence plasmid instability in Shigella flexneri 2a is induced by virulence gene expression. Infect Immun 65:3686–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Payne SM, Finkelstein RA. 1977. Detection and differentiation of iron-responsive avirulent mutants on Congo red agar. Infect Immun 18:94–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maurelli AT, Blackmon B, Curtiss R. 1984. Loss of pigmentation in Shigella flexneri 2a is correlated with loss of virulence and virulence-associated plasmid. Infect Immun 43:397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maurelli AT, Blackmon B, Curtiss R. 1984. Temperature-dependent expression of virulence genes in Shigella species. Infect Immun 43:195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neidhardt FC, Bloch PL, Smith DF. 1974. Culture medium for enterobacteria. J Bacteriol 119:736–47. [DOI] [PMC free article] [PubMed] [Google Scholar]


