Abstract
In response to various stimuli, vascular smooth muscle cells (SMCs) can de-differentiate, proliferate and migrate in a process known as phenotypic modulation. However, the phenotype of modulated SMCs in vivo during atherosclerosis and the influence of this process on coronary artery disease (CAD) risk have not been clearly established. Using single cell RNA sequencing, we comprehensively characterized the transcriptomic phenotype of modulated SMCs in vivo in atherosclerotic lesions of both mouse and human arteries and found that these cells transform into unique fibroblast-like cells, termed “fibromyocytes”, rather than into a classical macrophage phenotype. SMC-specific knockout of TCF21, a causal CAD gene, markedly inhibited SMC phenotypic modulation in mice, leading to the presence of fewer fibromyocytes within lesions as well as within the protective fibrous cap of the lesions. Moreover, TCF21 expression was strongly associated with SMC phenotypic modulation in diseased human coronary arteries, and higher levels of TCF21 expression were associated with decreased CAD risk human CAD-relevant tissues. These results establish a protective role for both TCF21 and SMC phenotypic modulation in this disease.
INTRODUCTION
The most significant consequence of coronary artery disease (CAD) occurs when an “unstable” atherosclerotic lesion ruptures and triggers an occlusive thrombus, resulting in a myocardial infarction (MI). Compared to stable coronary lesions, these vulnerable plaques are characterized by a large necrotic lipid core and a thin overlying fibrous cap that is prone to rupture1,2. During atherosclerosis, smooth muscle cells (SMCs) from the vessel wall likely contribute to both the fibrous cap and to the underlying necrotic core3 via a process known as “phenotypic modulation”, in which SMCs de-differentiate, proliferate and migrate in response to atherogenic stimuli4,5. The current view is that phenotypically modulated SMCs can develop into one of two distinct phenotypes, depending on environmental cues, with very different potential consequences for plaque stability: i) pro-inflammatory, dysfunctional macrophage-like cells, characterized in vivo by the upregulation of the macrophage marker Lgals36, which may serve to destabilize the lesion, or ii) extracellular matrix-producing “synthetic” SMCs that may contribute to the protective fibrous cap, which would serve to prevent plaque rupture and myocardial infarction (MI)4,5. Despite the significant uncertainty regarding the phenotype of modulated SMCs, it has become increasingly clear that these cells are an important component of the developing plaque in animal models of atherosclerosis. A recent lineage tracing study of SMCs in the mouse aortic root revealed that phenotypically modulated SMCs contribute ~30% of all cells in the atherosclerotic plaque6. There is some evidence that SMC phenotypic modulation occurs in human atherosclerosis7, but the phenotype of these cells and their contribution to human disease remains to be elucidated.
TCF21, a basic helix-loop-helix transcription factor, is the causal gene at the coronary artery disease (CAD)-associated locus at 6q23.28-10. In murine cardiac development, Tcf21 is expressed in proepicardial cells that give rise to both cardiac fibroblasts and coronary artery smooth muscle cells (SMCs)11,12. In this context, Tcf21 is required for cardiac fibroblast development but is downregulated in cells that eventually become coronary artery SMCs13, suggesting that sustained Tcf21 expression shifts these precursor cells away from the SMC lineage. Similarly, in cultured human coronary artery SMCs (HCASMCs), TCF21 knockdown results in up regulation of SMC differentiation markers14. In adult mice, Tcf21 is primarily expressed in the adventitia surrounding the coronary arteries and the aortic root, and also sporadically in some cells in the medial layer of the aortic root14. During development of atherosclerotic disease in the aortic root of ApoE−/− mice, there is robust expression of Tcf21 in many cells within the lesion14. However, several fundamental questions remained: i) what cell type(s) express Tcf21 during lesion development; ii) how does Tcf21 affect the phenotype of these cells and iii) how does Tcf21 affect disease risk?
METHODS
Mouse strains
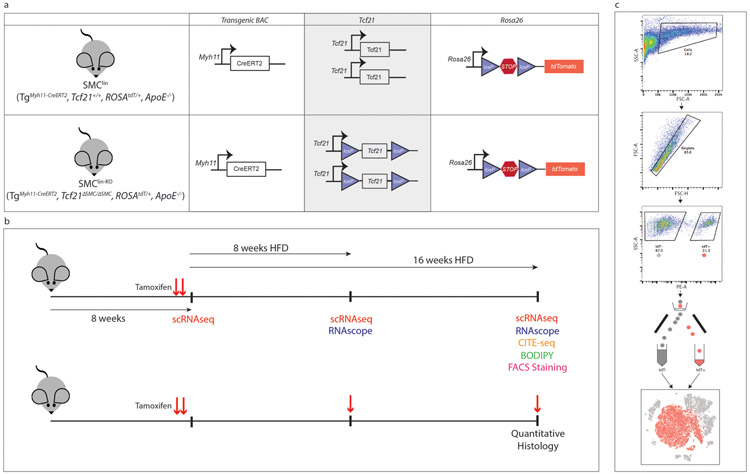
To enact SMC-specific lineage tracing and Tcf21 knockout, we used mice containing a well-characterized BAC transgene that expresses a tamoxifen-inducible Cre recombinase driven by the SMC-specific Myh11 promoter (TgMyh11-CreERT2, JAX# 019079)6,15,16. These mice were bred with a floxed tandem dimer tomato (tdT) fluorescent reporter line (B6.Cg-Gt(ROSA)26Sortm14(CAGtdTomato)Hze/J, JAX# 007914)17 to allow SMC-specific lineage tracing. A Tcf21flox allele was constructed by placing lox-P sites flanking the 5’-promoter region and first exon of the Tcf21 gene. All mice were bred onto the C56BL/6, ApoE−/− background. Final genotypes of SMC lineage-tracing (SMClin) mice were: TgMyh11-CreERT2, Tcf21+/+, ROSAtdT/+, ApoE−/−. Final genotypes of SMC lineage-tracing, Tcf21 knockout (SMClin-KO) mice were: TgMyh11-CreERT2, Tcf21ΔSMC/ΔSMC, ROSAtdT/+, ApoE−/−. As the Cre-expressing BAC was integrated into the Y chromosome, all lineage tracing mice in the study were male. The animal study protocol was approved by the Administrative Panel on Laboratory Animal Care (APLAC) at Stanford University.
Induction of lineage marker and Tcf21 knockout by Cre recombinase
For all scRNAseq, CITE-seq, RNAscope and immunohistochemistry experiments involving BODIPY, the tamoxifen gavage schedule was as follows: two doses of tamoxifen, at 0.2mg/gm bodyweight, were administered by oral gavage at 7 weeks of age, with each dose separated by 48 hours. Two doses were used to ensure complete activation of the Cre-ERT2. The recombination efficiency was estimated using the FACS-sorted scRNAseq data at the baseline time point - this revealed that 3538/3577 (98.9%) of cells classified as SMCs by scRNAseq in the baseline mouse were also FACS-positive for the tdT lineage marker. Approximately 48 hours after the second dose of tamoxifen, high fat diet (HFD) was started (Dyets #101511, 21% anhydrous milk fat, 19% casein, 0.15% cholesterol). For the quantitative immunohistochemistry and immunofluorescence (in situ) experiments, in addition to the two baseline doses of tamoxifen, a single additional dose of tamoxifen was administered by oral gavage after 8 weeks of HFD and then again at 16 weeks HFD, approximately 48 hours prior to sacrifice (Extended Data Fig. 1b). For these quantitative in situ experiments, we assessed the likelihood that the additional doses of tamoxifen at the 8 week and 16 week HFD timepoints could result in spurious recombination in lineage-negative cell types. We analyzed Myh11 expression in these lineage-negative cell types at 8 and 16 weeks of disease in the scRNAseq data and found that none of these cell types had upregulated Myh11 during disease and were therefore extremely unlikely to undergo recombination in response to the additional tamoxifen doses (data not shown).
Mouse aortic root/ascending aorta cell dissociation
Immediately after sacrifice, mice were perfused with PBS. The aortic root and ascending aorta were excised, up to the level of the brachiocephalic artery. Tissue was washed x3 in PBS, placed into an enzymatic dissociation cocktail (2 units/mL Liberase TM (Sigma #5401127001), 2 units/mL elastase (Worthington #LS002279 ) in HBSS), and minced. After incubation at 37C for 1 hour, the cell suspension was strained and then pelleted by centrifugation at 500xg for 5 minutes. The enzyme solution was then discarded and cells were resuspended in fresh HBSS. To increase biological replication, multiple mice were used to obtain single cell suspensions at each time point. For the SMClin genotype, three mice were used at baseline, and three mice were used at both 8 weeks and 16 weeks of disease. For the SMClin-KO genotype, one mouse was used at 8 weeks and three mice were used at 16 weeks.
Human coronary artery cell dissociation
Human coronary arteries used in this study were dissected from explanted hearts of transplant recipients, and were obtained from the Human Biorepository Tissue Research Bank under the Department of Cardiothoracic Surgery from consenting patients, with approval from the Stanford University Institutional Review Board. The basic clinical characteristics of the four patients included in this study are presented in Supplementary Table 5. The proximal to mid right coronary artery (RCA) was identified, excised, cleaned of peri-arterial fat, and then rinsed x3 in PBS. After excluding stented areas, atherosclerotic lesions were identified, ranging from mild, non-calcified plaques to more advanced lesions with areas of calcification. These areas were cut into approximately 50mg sections, and each section was placed into an enzymatic dissociation cocktail (10.4 units/mL Liberase TM, 8 units/mL elastase (Sigma #E7885) in 1mL HBSS), and minced. A total of approximately 120-240mg (~3-6 atherosclerotic sections) were used per patient. After incubation at 37C for 1 hour with periodic agitation, the cell suspension was pipetted up and down to break up any remaining tissue. The cell suspension was strained and then pelleted by centrifugation at 500xg for 5 minutes. The enzyme solution was then discarded and cells were resuspended in fresh HBSS.
FACS of mouse aortic root/ascending aorta cells
Cells were sorted on a BD Aria II instrument. An overview of the cell sorting process is illustrated in Extended Data Fig. 1c. Cells were gated on forward/side scatter parameters to exclude small debris and then gated on forward scatter height vs. forward scatter area to exclude obvious doublet events. Events passing these criteria were then sorted into one of two 1.5mL Eppendorf tubes based upon tdTomato fluorescence levels. tdTomato+ cells (considered to be of SMC lineage) and tdTomato− cells were then captured on separate but parallel runs of the same single-cell RNAseq workflow and datasets were later combined for all subsequent analyses.
FACS of human coronary artery cells
Cells were incubated with the calcein green viability reagent (Thermo Fisher #C34852) for 30 minutes at 4C prior to sorting, and a small portion of cells was left unstained as a negative control to determine gate placement. Cells were sorted on a Sony SH800s instrument. Cells were gated on forward/side scatter parameters to exclude small debris and then gated on forward scatter height vs. forward scatter area to exclude obvious doublet events. Calcein+ cells were then positively selected and sorted into a 1.5mL Eppendorf tube for further processing in the scRNAseq workflow.
Single cell capture and library preparation
All single cell capture and library preparation was performed at the Stanford Functional Genomics Facility (SFGF). Cells were loaded into a 10X Genomics microfluidics chip and encapsulated with barcoded oligo-dT-containing gel beads using the 10X Genomics Chromium controller according to the manufacturer’s instructions. Single-cell libraries were then constructed according to the manufacturer’s instructions. Libraries from individual samples were multiplexed into one lane prior to sequencing on an Illumina HiSeq4000 instrument.
CITE-seq
Cells were obtained from the atherosclerotic aortic root and ascending aorta of two SMClin mice on 16 weeks high fat diet as already described. After 1h of enzymatic dissociation, FBS (final concentration 10%) was added to the enzyme mixture, cells were centrifuged and supernatant was discarded. Cells were resuspended in 100uL cell staining buffer (2%BSA/0.01%Tween in PBS). A pool of TotalSeq antibodies was added at 1ug each (Cd16/Cd32 - BioLegend #101319, Cd11b - BioLegend #101265, Cd64 - BioLegend #139325, Cd86 - BioLegend #105047, F4/80 - BioLegend #123153) and cells were incubated for 30min at 4C, followed by washing x3 in staining buffer. Cells were isolated by FACS as previously described and captured on the 10X Chromium controller. Libraries were constructed according to the manufacturer’s protocol, with the following modifications according to the CITE-seq protocol (https://cite-seq.com/protocol): 1) during the cDNA amplification step, an additional primer (5’CCTTGGCACCCGAGAATT*C*C) was added to increase the yield of the Antibody Derived Tag (ADT) products; 2) during library preparation, ADT-derived and mRNA-derived cDNAs were separated by SPRI selection. The mRNA-derived cDNA fraction was used to construct 10X libraries according to the manufacturer’s instructions. The ADT-derived cDNA fraction was then purified using SPRI and then amplified using a 10x Genomics SI-PCR primer (5’AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGC*T*C) and a Illumina Small RNA RPI1 primer (5’CAAGCAGAAGACGGCATACGAGATCGTGATGTGACTGGAGTTCCTTGGCACCCGAGAATTC*C*A). PCR products were purified with SPRI and pooled with standard 10X libraries for sequencing on a HiSeq4000 instrument.
FACS analysis for macrophage markers
Cells were obtained from the atherosclerotic aortic root and ascending aorta of one SMClin mouse (16 weeks HFD) as already described. FBS (final concentration 10%) was added to the enzyme mixture, cells were centrifuged and supernatant discarded. Cells were resuspended in 100uL cell staining buffer (2%BSA/0.01%Tween in PBS). Cells were incubated with 1ug rat anti-CD16/32, BioLegend #101319) for 10min at 4C, washed two times and then incubated with 0.4ug BV421 goat anti-rat secondary (BD Biosciences # 565013) at 4C for 30min. Cells were then washed three times prior to FACS analysis.
Preparation of mouse aortic root sections
Immediately after sacrifice, mice were perfused with 0.4% PFA. The mouse aortic root and proximal ascending aorta, along with the base of the heart, was excised and immersed in 4% PFA at 4C for 12 hours (for IHC) to 24 hours (for RNAscope). After passing through a sucrose gradient, tissue was frozen in OCT to make blocks. Blocks were cut into 7um-thick sections for further analysis.
Immunohistochemistry
Slides were air-dried and OCT was removed with two washes in deionized (DI) water. Slides were immersed in 4% PFA for 2 minutes, followed by 4 washes in DI water. Slides were dried and sections were encircled with a liquid-blocking pen, followed by Peroxidazed (Biocare Medical # PX968) treatment for 5 minutes. Sections were washed x3 with DI water, and then incubated with Rodent Block M reagent (Biocare Medical # RBM961) for 30 minutes. Sections were washed x2 in TBS, then incubated overnight at 4C with an anti-SM22alpha rabbit polyclonal primary antibody (Abcam #ab14106, 1:300 dilution), an Lgals3 rat monoclonal antibody (Cedarlane Labs #CL8942AP, 1:100 dilution) or a CD68 rabbit polyclonal antibody (Abcam #ab125212, 1:300 dilution). Sections were washed for 5 minutes x2 with TBS and then incubated with the Rabbit-on-Rodent HRP Polymer (Biocare Medical # RMR622) or Rat Probe followed by Rat-on-Mouse HRP Polymer (Biocare Medical # RT517) for 30 minutes at room temperature (RT). Sections were washed x2 with TBS and then incubated with the Betazoid DAB chromogen reagents (Biocare Medical #BDB2004) for 4 minutes at RT. Sections were washed x2 in DI water and air-dried, followed by mounting with EcoMount medium (Biocare Medical #EM897L). The processed sections were visualized using Leica (model) under 5x and 10x (for lesion cap analysis) objective magnifications and images were obtained using Leica Application Suite X software. Sections obtained at equal distance measured from the superior margin of the aortic sinus were used for comparison. Areas of interest were quantified using ImageJ (NIH) software, and compared using a two-sided t-test. The lesion cap was defined as 30μm segment from the luminal surface as previously described6. Medial size was calculated by measuring the areas within the outer circumference and inner circumference of the medial layer. The difference between the two areas was used as the medial size/area. The Tagln immunostained images, which include the representative images shown in Figure 3I, were used to derive the medial area, as the Tagln staining delineated the medial layer clearly. Researchers were blinded to the genotype of the animals until completion of the analysis.
Cd68/BODIPY staining
Aortic root sections from SMClin mice immersed in water to remove OCT, then post-fixed in 4% PFA for 2 minutes. Sections were blocked with 1.5% goat serum in PBS for 30 minutes at RT, then incubated with anti-Cd68 (ab125212) at 1:400 overnight at 4C, washed, and then incubated with a goat anti-rabbit secondary antibody (Thermo Fisher #A21244, 1:500 dilution) for 30 minutes at RT. Directly after Cd68 immunostaining, BODIPY (Thermo Fisher # D3922, 1mg/mL DMSO stock), was diluted in PBS to 0.4ug/mL (1.53uM) and applied to sections for 30 minutes at RT, followed by washing x3 in PBS. Slides were mounted with Fluoroshield with DAPI (Sigma #46057).
RNAscope Assay
Slides were processed according to the manufacturer’s instructions, and all reagents were obtained from ACD Bio (Newark, CA). Slides were washed x1 in PBS, then immersed in 1X Target Retrieval reagent at 100C for 5 minutes. Slides were washed x2 in DI water, immersed in 100% ethanol, air-dried and sections were encircled with a liquid-blocking pen. Sections were incubated with Protease III reagent for 30 minutes at 40C, and then washed x2 with DI water. Sections were incubated with probes against mouse lumican (Lum), osteopontin (Spp1), human TNFRSF11B or a negative control probe for 2 hours at 40C. Multiplex fluorescence and colorimetric assays were performed per the manufacturer’s instructions.
Analysis of Single-Cell RNAseq Data
Fastq files from each experimental time point and mouse genotype were aligned to the reference genome individually using CellRanger Software (10X Genomics). Individual datasets were aggregated using the CellRanger aggr command without subsampling normalization. The aggregated dataset was then analyzed using the R package Seurat v.2.3.434,35. The dataset was trimmed of cells expressing fewer than 500 genes, and genes expressed in fewer than 5 cells. The number of genes, the number of unique molecular identifiers (UMIs) and the percentage of mitochondrial genes were examined to identify outliers. As an unusually high number of genes can result from a “doublet” event, in which two different cell types are captured together with the same barcoded bead, cells with > 3500 genes were discarded. Cells containing >7.5% mitochondrial genes were presumed to be of poor quality and were also discarded. The gene expression values then underwent library size normalization, in which raw gene counts from each cell were normalized relative to the total number of read counts present in that cell. The resulting expression values were then multiplied by 10,000 and log-transformed. Subsequent analyses were conducted using only the most highly-variable genes in the dataset. Principal component analysis was used for dimensionality reduction, followed by clustering in PCA space using a graph-based clustering approach34,35. t-SNE was then used for two-dimensional visualization of the resulting clusters. To estimate cell doublet rates, we used the baseline time point because the minimal time between tamoxifen gavage (tdTomato activation) and cell capture essentially excluded the possibility that trans-differentiation of SMCs to another cell type would affect the calculation. We determined the number FACS sorted tdTomato+ cells that had been assigned to cell clusters other than those known to express Myh11 at baseline (SMC1, SMC2, pericytes and a small number of phenotypically modulated SMCs). We then divided this number by the number of all tdTomato + cells. Out of 3707 tdTomato+ cells, 62 cells occurred in unexpected clusters, yielding a 62/3707 (1.7%) doublet rate. FASTQ files and matrices from single-cell RNAseq data that support the findings of this study have been deposited in the GEO database with primary accession code GSE131780.
Calculation of SMC Modulation and TCF21 Scores, and transcriptional ‘shift’
Top differentially-expressed genes distinguishing phenotypically modulated SMCs (fibromyocytes) from contractile SMCs (SMC1 and SMC2) at the 16 week high-fat diet time point were determined using the Seurat package. The SMC Modulation Score was then calculated using the top 20 genes that distinguished modulated SMCs and contractile SMCs as follows:
SMC Modulation Score = [1 + mean(Top20 upregulated modulated SMC genes)] / [1 + mean(Top20 upregulated contractile SMC genes)]. To construct the TCF21 score, we identified the top 20 TCF21-correlated and top 20 TCF21-anticorrelated genes across the “SMC” and “Fibromyocyte” clusters that also contained robust TCF21 ChIPseq peaks. The TCF21 score was then calculated as follows for each cell: TCF21 Score = [1 + mean(Top20 TCF21 correlated gene expression)] / [1 + mean(Top20 TCF21 anti-correlated gene expression)]. The transcriptional ‘shift’ of fibromyocytes towards or away from a given cell cluster ‘X’ in Fig. 2i was calculated as: (distance between the combined quiescent SMCs centroid and cluster ‘X’ centroid) - (distance between the fibromyocyte centroid and cluster ‘X’ centroid).
Human Coronary Artery Smooth Muscle Cell Culture
A total of 65 primary human coronary artery smooth muscle cell (HCASMC) lines were purchased from PromoCell (catalog #C-12511), Cell Applications (catalog # 350-05a), Lonza (catalog #CC-2583), Lifeline Cell Technology (catalog #FC-0031) and ATCC (catalog #PCS-100-021). Cells were cultured in smooth muscle growth medium (Lonza catalog #CC-3182) supplemented with hEGF, insulin, hFGF-b, and 5% FBS, according to the manufacturer’s instructions. All HCASMC lines were used at passages 4-8.
Pooled TCF21 Chromatin Immunoprecipitation Sequencing (ChIPseq)
Approximately 300,000 HCASMC cells from each cell line were cross linked in 1% formaldehyde for 10 min and then washed with PBS. Cell pellets were frozen at −80C. All cell pellets were thawed on ice, combined for a total of 19.5 million cross linked cells and resuspended in cold PBS. PBS was removed and replaced with hypotonic buffer (20mM Hepes pH 7.9, 10mM KCl, 1mM EDTA, pH 8, 10% glycerol) and cells incubated on ice for 6 min. Cells were dounce homogenized with 20 strokes on ice using a 7ml glass homogenizer. Nuclear lysates were sonicated using a Branson 250 Sonifier (power setting 4, constant duty for 12 rounds of 20 second pulses), resulting in chromatin fragments of 250-400 bp. Lysate was treated overnight at 4C with 5ug of anti-Tcf21 antibody (Sigma #HPA013189). Protein-DNA complexes were captured on Protein G agarose beads (Millipore Sigma #16-266) and eluted in 1% SDS TE buffer at 65C. After reverse cross linking, RNase A and proteinase K digestion, chromatin was purified using Qiagen PCR purification kit (Catalog #28106). ChIP DNA sequencing libraries were generated as previously described36 and sequenced on an Illumina HiSeq X instrument (150 bp paired-end reads). Fastq files were mapped to the Hg19 genome with the BWA-MEM aligner v.0.7.1237. ChIPseq peaks were then called with MACSv238 using default parameters. From this output, “robust” peaks were selected by specifying a minimum fold-enrichment of 10 and a minimum log10 q-value of 60. To determine which gene regions contained these robust TCF21 ChIPseq peaks, we expanded each NCBI Refseq-annotated gene region by 5kb in each direction and then determined the overlap of these gene regions with the TCF21 ChIPseq data.
TCF21 overexpression in HCASMCs
TCF21 cDNA was cloned into a 2nd generation lentiviral vector (pWPI, Addgene #12254) and packaged at the Stanford Gene Vector and Virus Core. HCASMCs were treated at 60% confluence with lentivirus at MOI of 5 for 24 hours. The virus was removed and cells were collected 48 hours later. Gene expression was assessed using TaqMan qPCR probes (Thermo Fisher) for TCF21 (Hs00162646_m1), Decorin (DCN, Hs00754870_s1), Lumican (LUM, Hs00929860_m1), and Matrix-Gla Protein (MGP, Hs00969490_m1) according to the manufacturer’s instructions on a ViiA7 Real-Time PCR system (Applied Biosystems, Foster City, CA). A total of 6 independent experiments were performed, each with 3 technical replicates. Fold-change values from control and overexpression conditions were compared with a two-sided Mann-Whitney U test in Prism version 8 (GraphPad).
HCASMC genome and transcriptome sequencing
HCASMC genomic DNA was isolated using Qiagen DNeasy Blood & Tissue Kit (catalog # 69506). Libraries were prepared with Illumina’s TruSeq DNA PCR-Free Library Preparation Kit and sequenced on Illumina HiSeq X Ten System. RNA was extracted using Qiagen miRNeasy Mini Prep Kit (catalog # 74106). Libraries were made using Illumina TruSeq Stranded Total RNA Library Prep Kit (catalog # 20020597) and sequenced on HiSeq 2500 Platform. Whole-genome sequencing data were processed with the GATK best practices pipeline with hg19 as the reference genome. VCF records were phased with Beagle. Demultiplexed FASTQ files were mapped with STAR version 2.4.0i in 2-pass mode on hg19.
TCF21 expression quantitative loci (eQTL) analyses
We first examined all SNPs in the 6q23.2 locus that were associated with CAD at genome-wide significance (p<5x10−8, Fig. 6a). We queried individual eQTL relationships from the Gene-Tissue Expression (GTex) database and in our cohort of 52 HCASMC lines. To test for an additive effect of these cis CAD risk alleles on TCF21 expression in the HCASMC lines, we determined the relationship between the number of risk SNPs inherited and the TCF21 expression level in the HCASMC lines. Total read counts were determined using RNA-SeQC and transcript per million (TPM) values were then calculated for the TCF21 gene. To reduce noise, TCF21 TPM data from cell lines with identical haplotypes were averaged prior to analysis. We then examined a larger set of SNPs at the 6q23.2 locus associated with CAD at a p-value of <1e−5, extracted from CAD GWAS summary data (Nelson et al., www.cardiogramplusc4d.org). We also obtained cis-eQTL summary data from the STARNET database (dbGaP), derived from human aortic tissue, for these CAD-associated SNPs. We filtered for SNPs with absolute value of beta coefficients (log odds ratio (log(OR)) greater than 0.3 for both GWAS and eQTL. A linear mixed model was then used to compute a smooth local regression between the CAD GWAS beta and the eQTL log(OR). Pearson correlation coefficient r and p-value of significance were calculated using cor.test in R.
Statistical Methods
Differentially-expressed (DE) genes in the scRNAseq data were identified using a Wilcoxon rank sum test as implemented in the Seurat package v.2.3.4. In the scRNAseq data, the distribution of fibromyocytes and quiescent SMCs across the SMClin and SMClin-KO genotypes (n=3 mice in each genotype) was calculated using Pearson’s chi-squared test (chisq.test) in R. For the quantitative immunohistochemistry experiments, cohorts included 17 SMClin (WT) and 22 SMClin-KO (KO) mice. Due to occasional tissue block/section damage/folding, some animals had to be eliminated for some comparisons. In Fig. 3d, WT=16, KO=17. In Fig. 3e, WT=15, KO=18. In Fig. 3g, WT=16, KO=17. In Fig.3h, WT=14, KO=17. In Fig. 3j, WT=16, KO=22. In Fig. 3k, WT=15, KO=22. In Extended Data Fig. 3b, WT=15, KO=16. In Extended Data Fig. 3c, WT=16, KO=20. In Extended Data Fig. 3d, WT=16, KO=22. Comparisons in the mouse aortic root were made with a Student’s t-test (two-sided). For qPCR analysis comparing TCF21 over-expression versus control in HCASMCs, 6 independent experiments were performed, each with 3 technical replicates. Fold-change values (n=6 in each group) were analyzed using a two-sided Mann-Whitney U test in Prism version 8 (GraphPad). To test the relationship between the number of TCF21 risk SNPs and TCF21 expression in HCASMCs, and to test the relationship between the CAD GWAS beta value and the magnitude of the eQTL in the STARNET dataset, linear mixed models were used; p-value of significance was based on Pearson’s product moment correlation coefficient, and 95% confidence interval was based on Fisher’s Z-transform, as computed using cor.test in R. For HCASMCs, n=52 cell lines with 16 distinct haplotypes were analyzed. For STARNET data, n=36 SNPs were analyzed.
RESULTS
scRNAseq defines cellular composition of mouse atherosclerosis and reveals that Tcf21 expression is upregulated during SMC phenotypic modulation
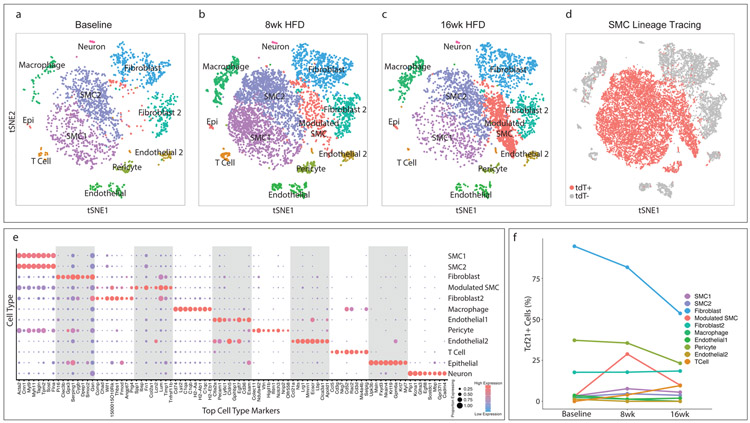
To lineage trace vascular SMCs, we used BAC transgenic mice expressing a tamoxifen-inducible Cre recombinase driven by the SMC-specific Myh11 promoter (TgMyh11-CreERT2)15,16, as well as a Cre-responsive reporter gene (tandem dimer Tomato, tdT) inserted at the ROSA26 locus (ROSAtdT/+)17 on the ApoE−/− background (SMClin mice, Extended Data Fig. 1a, top panel). When these mice were administered tamoxifen at 8 weeks of age, prior to high fat diet and disease onset, all SMCs (and any progeny resulting from subsequent proliferation) were permanently labeled with tdT fluorescence. To examine single-cell gene expression, we FACS sorted cells isolated from the aortic root and ascending aorta into two groups: tdT+ (SMC origin) and tdT− (all other cell types, Extended Data Fig. 1c) at baseline and after 8 and 16 weeks of high-fat diet (HFD, Extended Data Fig. 1b). We then performed scRNAseq on both groups of cells in parallel using the 10X Chromium platform and reagents. tdT+ and tdT− datasets for all time points were then merged for further analyses. The calculated cell doublet rate for these experiments was 1.7%, which is within the standard range for the 10X Chromium platform18, and the estimated recombination efficiency in SMCs was 98.9% (see methods). Figure 1 illustrates all major cell types identified by their gene expression profiles in the aortic root and ascending aorta at baseline prior to HFD (Fig. 1a), after eight weeks of HFD (Fig. 1b), and after sixteen weeks of HFD (Fig. 1c). Top cell type-specific genes are shown in Fig. 1e. Interestingly, the main SMC population was divided into two groups (SMC1 and SMC2), based on significant differences in gene expression patterns (Supplementary Tables 1-3).
Figure 1. Transcriptomic characterization of mouse aortic root atherosclerotic plaques and Tcf21 expression.
(a–c) t-Stochastic Neighbor Embedding (t-SNE) visualization of cell types present in the mouse aortic root at (a) baseline, n=3 mice, (b) after 8 weeks of high-fat diet (HFD), n = 3 mice and (c) 16 weeks of HFD, n= 3 mice , illustrating the appearance of a disease-specific cell type, the “modulated SMC” cluster. All cell cluster identities are indicated in a-c. (d) SMC lineage traced cells, identified by their expression of the tdT reporter gene via FACS, are labeled in red for all timepoints. tdT+, cells expressing tdT; tdt−, cells not expressing tdT. (e) The top 8 genes defining each type of cell cluster in (a–c) are listed. The size of each circle represents the fraction of cells in each cluster that express at least 1 detected transcript of each gene; the color scale indicates expression level (blue = low, red = high). (f) Percentage of cells of each cell type that contained detectable (non-zero) Tcf21 levels at baseline, 8 weeks and 16 weeks of disease. Epi = epithelial-like cell.
With the development of atherosclerosis, the most notable change was the appearance of a distinct group of cells that were juxtaposed in t-SNE space to the contractile SMC clusters (Fig. 1b-c, in red), increasing in prevalence from 8 weeks to 16 weeks of disease. Visualization of all cells that had been FACS sorted as tdT+ and were thus of SMC lineage revealed that the vast majority of cells in this disease-associated cell cluster were SMC-derived (Fig. 1d) and therefore represented phenotypically modulated SMCs. 11% of cells within the disease-associated cluster were lineage negative, suggesting an origin other than SMC. Interestingly, at the baseline timepoint a small proportion of SMCs (1.3%) were already classified as phenotypically modulated SMCs, consistent with recently-published data showing a population of Sca1+ SMCs in healthy mice19.
To characterize Tcf21 expression in the different vascular cell lineages, we measured the percentage of cells from each group that expressed detectable levels of Tcf21 (Fig. 1f). Fibroblasts had the highest percentage of Tcf21+ cells, but exhibited a decrease with disease progression. Other cell populations had only small changes in Tcf21+ cells during disease. An exception to this was found in the modulated SMC group - although derived from baseline SMCs that contained only 3.1% Tcf21+ cells, the modulated SMC cluster markedly upregulated Tcf21 (29% Tcf21+ cells) by eight weeks of disease. The percentage of Tcf21+ cells subsequently declined to 9.8% by 16 weeks of disease and did not return to the baseline levels seen in the contractile SMC clusters. Thus, the increase in Tcf21 expression within the lesion during disease was the result of a prominent upregulation of Tcf21 specifically in SMCs during phenotypic modulation.
SMC phenotypic modulation in vivo during disease results in a specific fibroblast-like phenotype
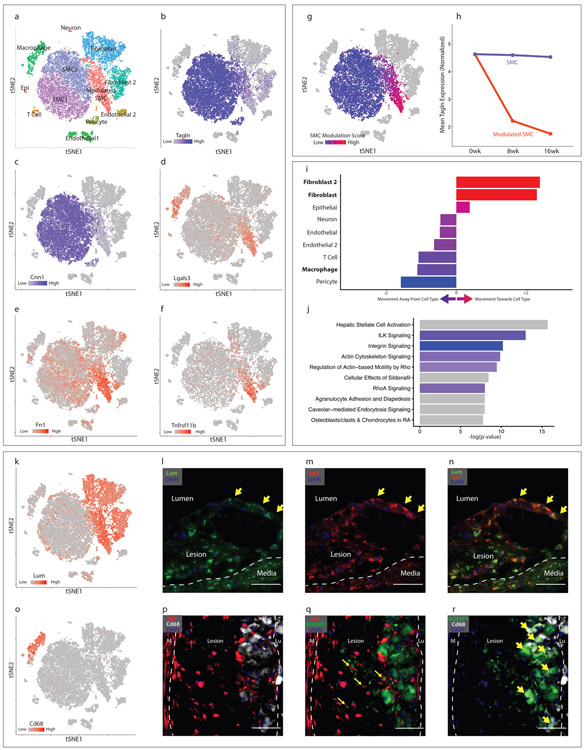
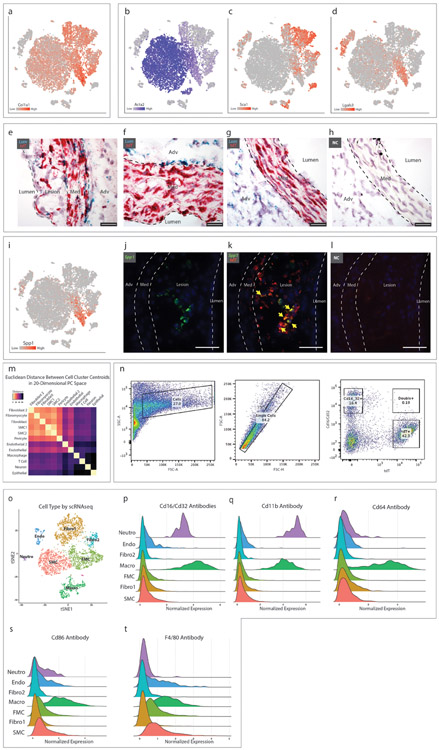
Data from SMClin mice at all time points are combined in Fig. 2a-g. In the phenotypically modulated SMC cluster, markers of SMC differentiation including transgelin (Tagln, Fig. 2b) and calponin (Cnn1, Fig. 2c) showed a gradient of decreasing expression from the parental SMC lineage, and a gradient of increased expression for Lgals3, a known marker of SMC phenotypic modulation (Fig. 2d), suggesting that these cells were undergoing SMC phenotypic modulation. There was marked upregulation of many other genes, including fibronectin 1 (FN1, Fig. 2e), osteoprotegerin (Tnfrsf11b, Fig. 2f) and collagen 1α1 (Col1α1, Extended Data Fig. 2a) in this cell group. In particular, during phenotypic modulation there was a striking upregulation of small leucine-rich proteoglycans (SLRPs) such as lumican (Lum, Fig. 2k), decorin (Dcn) and biglycan (Bgn), genes that are otherwise specific to the fibroblast and fibroblast 2 populations. We then used a gene expression score, derived from the top differentially-expressed genes distinguishing the modulated SMC cluster from the contractile SMC cluster (see Methods), to quantify the degree of SMC phenotypic modulation for each cell. Although the modulated SMC cluster was readily distinguished from contractile SMCs during clustering, we observed a progressive increase in the gene expression score in those cells more distant from the contractile SMC clusters (Fig. 2g). This transcriptional shift away from contractile SMCs also progressed over time (Fig. 2h). Phenotypically modulated SMCs have been previously noted to express markers associated with myofibroblasts (Acta2), mesenchymal stem cells (Sca1), and macrophages (Lgals3)6, and this information employed to suggest evidence of transdifferentiation into these cellular lineages. However, we found that the expression of these markers appears instead to reflect progression along a single path of phenotypic modulation, and not separation into distinct lineages (Extended Data Figs. 2b-d). To further investigate this possibility, we asked whether these phenotypically modulated SMCs were becoming more transcriptionally similar to other cell types within the lesion. We calculated the Euclidean distance between the centroids of all cell groups in 20-dimensional principal component (PC) space and determined, with contractile SMCs as a reference point, how the phenotypically modulated SMCs had shifted in relation to each cell cluster. This analysis revealed that during SMC phenotypic modulation, the transcriptional signature of these cells clearly shifts towards that of the fibroblast clusters (Fig. 2i). The relationship between all cell types is illustrated in Extended Data Fig. 2m. Molecular pathway analysis (IPA) performed on differentially-regulated genes between the phenotypically modulated SMCs and contractile SMCs also showed strong upregulation of genes expressed by fibroblasts (e.g., hepatic stellate cell activation) and down regulation of integrin and integrin-linked kinase signaling pathways (Fig. 2j). Specifically, downregulated were integrin-beta1 and integrin-alpha8, which comprise an integrin heterodimer restricted to SMCs20. Given the clear transition to a fibroblast-like phenotype, and their separation from other cell clusters, we termed the modulated SMCs “fibromyocytes” (FMCs), reflecting their origin from smooth muscle myocytes and their adoption of fibroblast phenotype.
Figure 2. Characterization of SMC phenotypic modulation in the mouse aortic root.
(a-g) t-SNE visualization of cell types present in the wild-type mouse aortic root from all timepoints combined (n=9 mice). (a) Cell types are indicated for each cluster. (b–f) t-SNE visualization at all timepoints combined, overlaid with expression of Tagln, Cnn1, Lgals3, Fn1 and Tnfrsf11b. Expression levels are indicated by scales in the lower left of each panel. n=9 mice. (g) A SMC modulation score was calculated for each cell based upon the expression of top differentially expressed genes between modulated SMC and contractile SMC clusters. Blue indicates more similarity to contractile SMC, red indicates more similarity to phenotypically modulated SMC. (h) Tagln expression in quiescent and modulated SMCs at 0, 8 and 16 weeks of high-fat diet (HFD). All expression levels are normalized for library size and log-transformed. (i) Transcriptional shift from contractile SMC to phenotypically modulated SMC phenotype at 16 weeks HFD, from the viewpoint of each non-SMC cell type within the lesion. Bars to the right of center indicate that, relative to contractile SMCs, modulated SMCs have shifted toward a given cell type. Bars to the left indicate that they have shifted away. Color represents the magnitude of shift (blue = farther away, red = closer towards). (j) Top enriched pathways for gene expression changes seen with SMC phenotypic modulation, as performed with Ingenuity Pathway Analysis (IPA). The top 200 differentially-expressed genes were analyzed with Fisher’s exact test (right-sided). Blue bars indicate negative Z-scores of predicted activation, and grey bars indicate that the pathway had not yet been annotated by IPA to yield an activity pattern. (k) t-SNE visualization at all timepoints combined, overlaid with expression of Lum. n=9 mice. (l-n) RNAscope staining in the mouse aortic root at 8 weeks of high-fat diet. Yellow arrows highlight cells at the fibrous cap expressing both Lum (green, in l) and tdT (red, in m), with merged Lum and tdT staining shown in (n). Dapi staining is shown in blue. Images in (l-n) are representative of 3 experiments, and scale bars represent 50μm. (o) t-SNE visualization at all timepoints combined, overlaid with expression of Cd68. n=9 mice. (p) tdT expression (red) and Cd68 immunostaining (white). Dapi staining is shown in blue. (q) tdT expression (red) and BODIPY lipid stain (green). Co-localization of tdT and BODIPY is highlighted with yellow arrows. (r) BODIPY lipid stain (green) and Cd68 immunostaining (white). Co-localization of BODIPY and Cd68 is highlighted with yellow arrows. Lu = lumen, M = media. Images in (p-r) are representative of 3 experiments, and scale bars represent 50μm.
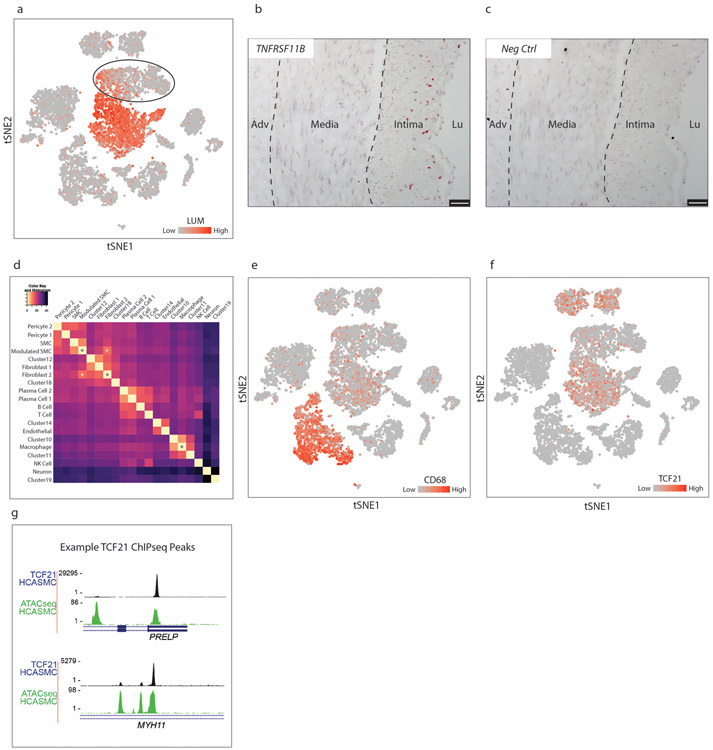
To localize these fibromyocytes within the plaque, we first searched for genes that were specific for the fibromyocyte cluster in the scRNAseq dataset. We found that the gene Lumican (Lum), when co-expressed with the tdT lineage marker, captured the majority of fibromyocytes with excellent (96%) specificity (Fig. 2k).
We performed RNAscope in situ hybridization for Lum and tdT genes in sections of the atherosclerotic mouse aortic root. This revealed that (Lum+/tdT+) cells were found throughout the lesion and also at the fibrous cap, (Fig. 2l-n), consistent with the notion that these cells migrate into the lesion during disease. There were also Lum+/tdT+ cells in the media underlying the lesion, but these cells were not present in the media of non-diseased areas of the vessel (Extended Data Fig. 2f-g), suggesting that medial SMCs begin to undergo phenotypic modulation prior to entering the plaque. We also performed RNAscope for osteopontin (Spp1), the expression of which is limited to more “extreme” modulated SMCs that also express multiple chondrocyte markers. These cells localized only to the lesion and not the media (Extended Data Fig. 2j-l), indicating that the Lum+/tdT+ cells in the media are likely at an earlier stage of phenotypic modulation.
A notable observation from these analyses is that at a transcriptional level, SMCs undergoing phenotypic modulation do not appear to be shifting towards a monocyte-derived macrophage phenotype within the plaque. Despite their shared expression of the macrophage marker Lgals3, fibromyocytes lack significant expression of virtually all other top markers that distinguish the macrophage cell cluster (Fig. 1e, 2o). Indeed, whole-transcriptome analyses revealed that, compared to contractile SMCs, fibromyocytes are actually more distant from macrophages, suggesting that these cells are becoming less similar to monocyte-derived macrophages in the mouse (Fig. 2i). We then sought to determine whether modulated SMCs express macrophage markers at the protein level using multiple techniques. We performed immunostaining for the macrophage marker Cd68 in mouse lesions, which did not identify significant Cd68 expression in tdT+ cells (Fig. 2p). We then incubated a single cell suspension from the atherosclerotic aortic root and ascending aorta of SMClin mice with antibodies against the macrophage markers Cd16 and Cd32 and performed flow cytometric analysis, which revealed that SMC-derived tdT+ cells do not express significant levels of Cd16 or Cd32 (Extended Data Fig. 2n). Finally, to integrate our transcriptional findings with protein-level data, we incubated a single cell suspension from the atherosclerotic aortic root and ascending aorta of two SMClin mice with a panel of six DNA-barcoded antibodies against commonly-used macrophage markers (Cd16, Cd32, Cd11b, Cd64, Cd86, and F4/80) prior to performing scRNAseq (CITE-seq21, Extended Data Fig. 2o-t). Antibody binding was then assessed by recovering the antibody-associated DNA barcodes in the cDNA library. This experiment confirmed that these macrophage markers are not upregulated in modulated SMCs compared with quiescent SMCs (Extended Data Fig. 2p-t), consistent with the transcriptomic data. We then assessed lipid uptake by SMC-derived cells within the lesion using the BODIPY neutral lipid stain. Consistent with previous reports22, we found that many modulated SMCs in the lesion do indeed contain lipid droplets (Fig. 2q). However, these were quantitatively and qualitatively distinct from the large macrophage-derived foam cells in the lesion (Fig. 2r). Taken together, these data suggest that although modulated SMCs in the plaque take up lipid, they do so without adopting a macrophage-like transcriptional phenotype.
Loss of Tcf21 in SMCs inhibits phenotypic modulation
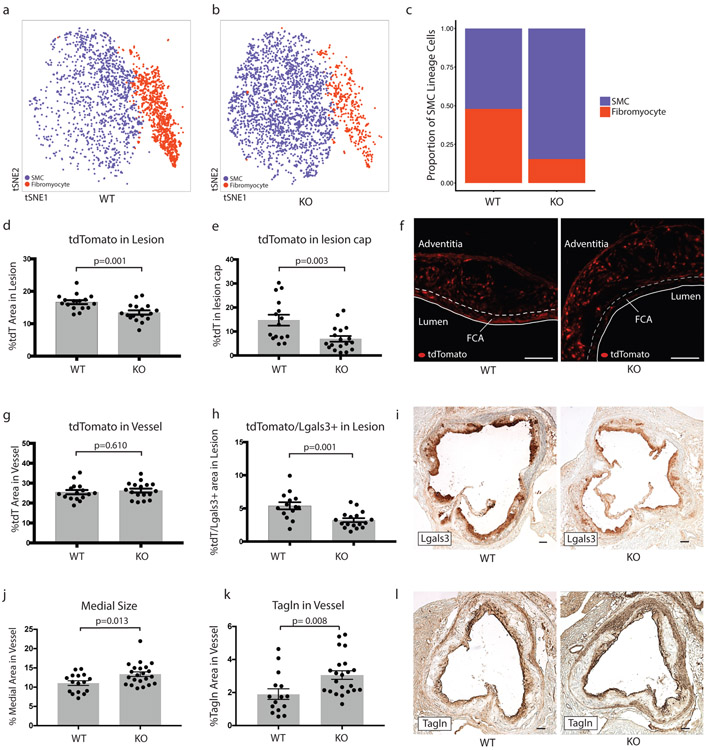
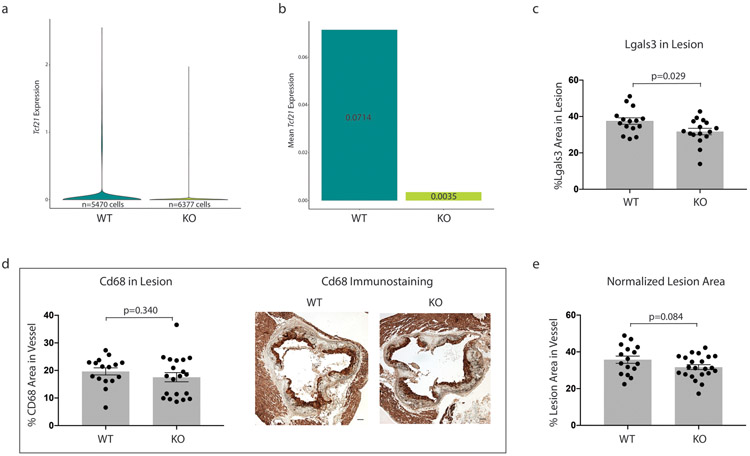
To determine the effect of Tcf21 on SMC phenotypic modulation, we performed scRNAseq in the aortic root and ascending aorta of SMC-specific conditional Tcf21 knockout mice (SMClin-KO). These mice were identical to the SMClin mice, except at the Tcf21 locus where both Tcf21 alleles were flanked with LoxP sites (Tcf21ΔSMC/ΔSMC, Extended Data Fig. 1a, bottom panel). Thus, when tamoxifen was administered to these SMClin-KO mice prior to HFD and disease onset, in addition to lineage marker activation the Tcf21 gene was permanently deleted in all SMCs and any progeny resulting from subsequent proliferation. As a control, we used SMClin mice. We first assessed the efficacy of Tcf21 deletion in the scRNAseq data and found that Tcf21 expression was reduced by 95% in SMC-derived cells from SMClin-KO mice relative to controls (Extended Data Fig. 3a). We measured the proportions of contractile SMCs and fibromyocytes in the scRNAseq data at the 16 week disease time point and found that, compared to SMCs from SMClin controls (Fig. 3a), SMCs from SMClin-KO mice exhibited a marked reduction in the ability to undergo phenotypic modulation (Fig. 3b). This reduction in SMC modulation was observed at both 8 weeks (8% in WT vs 1% in KO) and at 16 weeks of disease (Fig. 3c, 48% in WT vs 16% in KO, chi-square p = 2.2e−16). Plaque characteristics in the aortic root in a larger cohort of mice (n = 17 WT, 22 KO) supported the scRNAseq findings. At the 16 week disease time point, SMClin-KO mice exhibited a decrease in the proportion of lineage-traced tdT+ cells in the lesion relative to controls (Fig. 3d, p = 0.01), despite a similar tdT+ area within the whole vessel wall (Fig. 3g). Importantly, in the SMClin-KO mice, there was also a lower proportion of tdT+ SMC lineage-traced cells in the area of the fibrous cap (Figs. 3e-f, p = 0.003). The SMClin-KO mice also exhibited a reduction in the tdT+/Lgals3+ area within the lesion (Fig 3h, p = 0.001), more specifically showing fewer modulated SMCs within the lesion. The total Lgals3+ area within the lesion was also reduced (Fig. 3i, Extended Data Fig. 3b). In contrast, staining for the contractile SMC marker Tagln was increased (Fig. 3k-l, p = 0.008). The increase in Tagln area in the SMClin-KO group corresponded to an increased medial area in these mice (Fig. 3j, p = 0.01). As Lgals3 is also expressed in monocyte-derived macrophages, we stained for the macrophage-specific marker Cd68 to further exclude the possibility that changes in Lgals3 staining were caused by differences in macrophage content between the two groups. Indeed, there was no difference in Cd68 staining between SMClin and SMClin-KO mice (p = 0.34, Extended Data Fig. 3c). There was also no significant difference in lesion area between the two groups (Extended Data Fig. 3d). Together, these findings strongly suggest that loss of Tcf21 results in inhibition of SMC modulation and fewer fibromyocytes in the lesion and fibrous cap.
Figure 3. SMC-specific Tcf21 knockout markedly inhibits SMC phenotypic modulation in mice.
(a,b) Prevalence of contractile SMCs (blue) and fibromyocytes (red) at 16 weeks of disease in (a) SMClin (n=3 mice) and (b) SMClin-KO (n=3 mice). (c) Proportions of contractile (blue) and modulated (red) SMCs after 16 weeks of disease in SMClin and SMClin-KO mice (n=3 mice for each genotype, chi-square p = 2.2e−16). (d,e) Percentage of tdT-positive staining area in the lesion (d) and in the fibrous cap (e) defined as the area of the lesion within 30 μm of the luminal surface). (f) Representative images of tdT positive cells in SMClin and SMClin-KO mice. FCA = fibrous cap area. (g) Total tdT content of the vessel. (h) tdT+/Lgals3+ area in the lesion. (i) Representative images of Lgals3+ staining in the lesions of SMClin and SMClin-KO mice. Medial size (j) and Tagln content (k) in SMClin and SMClin-KO mice. Representative images of Tagln staining are shown in (l). All data in (d-l) were at 16 weeks of disease. Scale bars in (f,i and l) represent 100μm. Data in (d,e,g,h,j,k) were analyzed using a two-sided Student’s t-test. Error bars denote standard error.
Identification and characterization of modulated SMCs in human coronary arteries reveals a similar fibromyocyte phenotype
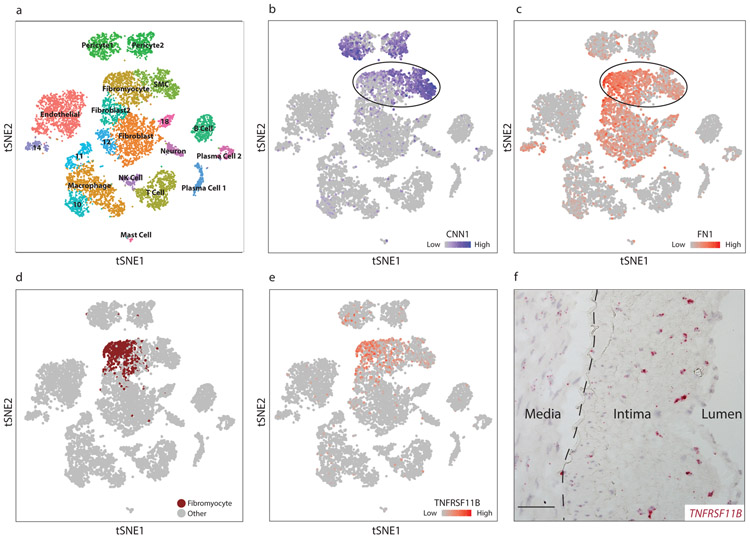
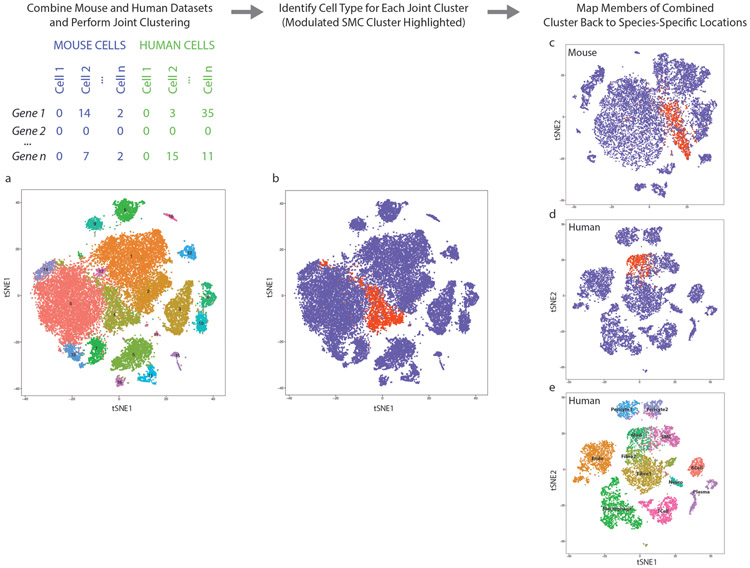
To determine whether our findings in the mouse could be observed in humans, we performed scRNAseq in dissociated cells from human atherosclerotic coronary arteries. Diseased segments within the right coronary artery of four cardiac transplant recipients (Supplementary Table 4) were dissociated and subjected to scRNAseq. Diseased segments ranged from non-calcified plaques to more advanced lesions with areas of calcification, and stented areas were excluded. Cell types were assigned to each cluster based upon the top defining genes in each cluster (Fig. 4a, Supplementary Table 5). Two contiguous clusters, labeled “SMC” and “Fibromyocyte” (Fig. 4a), were characterized by diminishing calponin (CNN1) expression (Fig. 4b), which appeared to parallel the gradual loss of CNN1 expression in murine fibromyocytes (Fig. 2c). The decrease in expression of CNN1 and other markers of SMC differentiation in these cell groups was accompanied by a corresponding increase in markers of SMC modulation, including fibronectin 1 (FN1, Fig. 4c) and lumican (LUM, Extended Data Fig. 4a), suggesting that these two clusters could represent SMCs undergoing phenotypic modulation to become fibromyocytes. We then sought to more definitively identify fibromyocytes in the human coronary artery based on whole-transcriptome similarity to bona fide lineage-traced fibromyocytes in the mouse. To this end, we combined the mouse and human datasets and, using the aligned canonical correlation analysis feature of the Seurat package, performed joint clustering on the combined mouse and human dataset (Extended Data Fig. 5). We found that this approach accurately clustered together known orthologous cell types from each species (Extended Data Fig. 5e). We then identified the human cells that had clustered with the bona fide, lineage-traced fibromyocytes in the mouse (Extended Data Fig. 5b, red cluster), and highlighted these cells within the context of the human single cell dataset (Fig. 4d, Extended Data Fig. 5d). We found that these cells mapped back to the same region undergoing loss of SMC markers and up regulation of fibromyocyte markers. We found that 86% of these cells mapped back to the “Fibromyocyte” cluster in Fig. 4a and 8% mapped to the immediately adjacent “Fibroblast 2” cluster, together accounting for 94% of all cells classified as fibromyocytes via joint clustering. The unbiased, whole-transcriptome similarity to mouse fibromyocytes and their independent compact clustering in the human dataset strongly suggest that these cells are indeed human fibromyocytes.
Figure 4. Identification of modulated SMCs in diseased human coronary arteries.
(a) t-SNE visualization of cell types isolated from the right coronary artery of four human patients, with assigned cell cluster identities indicated. (b-c) t-SNE visualization overlaid with expression of CNN1 (b) and FN1 (c). Expression levels are indicated by scales in the lower right of each panel. Data from n=4 patients. (d) In a joint mouse/human clustering analysis, a distinct population of human cells (brown) clustered together with lineage-traced fibromyocytes in the mouse. (e) t-SNE visualization overlaid with expression of TNFRSF11B. (f) RNAscope in-situ hybridization of TNFRSF11B in a human coronary artery. Image is representative of 4 experiments, and scale bar represents 50μm.
To localize these fibromyocytes in their anatomical context within the human coronary artery lesion, we first searched for markers that were highly specific for human fibromyocytes (Fig. 4d, brown) in the human scRNAseq dataset. We found that the gene TNFRSF11B, encoding osteoprotegerin (OPG), exhibited 97% specificity and 53% sensitivity for human fibromyocytes in the scRNAseq dataset (Fig. 4e). We then performed RNAscope in-situ hybridization to visualize the distribution of TNFRSF11B within the human lesions. This revealed strong TNFRSF11B staining primarily in the fibrous neointima (Fig. 4f, Extended Data Fig. 4b), with few cells strongly positive for TNFRSF11B in the media or the adventitia, consistent with the expected location of fibromyocytes. No staining was observed using a negative control probe (Extended Data Fig. 4c).
Interestingly, using t-SNE visualization the “Fibromyocyte” cluster appeared to be continuous with the fibroblast 2 population. Calculating Euclidean distance in 20-dimensional PC space between cell cluster centroids in the human dataset confirmed that the “Fibromyocyte” cluster develops striking similarity to the fibroblast 2 cluster (Extended Data Fig. 4d). Both the “Fibromyocyte” and the “SMC” cluster remained significantly dissimilar to monocyte-derived macrophages (Extended Data Fig. 4d). Thus, consistent with the mouse data, SMCs undergoing phenotypic modulation in the human artery also appear to be acquiring a specific fibroblast-like “fibromyocyte” phenotype, characterized by strong upregulation of collagen, fibronectin 1, lumican and secreted proteoglycans. Of note, consistent with previous reports6,23, we did observe up regulation of the macrophage marker CD68 in human fibromyocytes relative to contractile SMCs (Extended Data Fig. 4e), but CD68 was also expressed in multiple non-macrophage cell types within the lesion, suggesting that it does not reflect the acquisition of macrophage-like properties.
TCF21 is associated with SMC phenotypic modulation in human coronary arteries
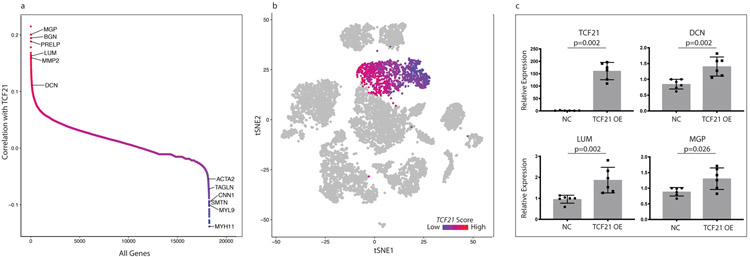
Given the marked effect of Tcf21 on SMC modulation in the mouse model of atherosclerosis, we sought to determine if TCF21 was also associated with SMC phenotypic modulation in human atherosclerotic coronary arteries. Across the “SMC” and “Fibromyocyte” clusters, we performed pairwise Spearman correlation between the expression of TCF21 and every other gene expressed in these cells. We found that TCF21 was highly anti-correlated with markers of differentiated SMCs, demonstrating that increased TCF21 expression in these cell clusters was associated with SMC de-differentiation (Fig. 5a). In addition, we found that TCF21 was highly correlated to many markers of fibromyocytes in both human and mouse. As Tcf21 was expressed at low levels (Extended Data Fig. 5f), we visualized behavior of the TCF21-associated gene program within the cell populations by creating a “TCF21 score” for each cell (see methods), which reflected the averaged expression of top TCF21-correlated genes across the “SMC” and “Fibromyocyte” clusters. Importantly, to establish a causal link with TCF21, all genes included in the score were required to also display robust TCF21 binding within the gene locus as assessed by TCF21 chromatin immunoprecipitation sequencing (ChIP-seq, example peaks shown in Extended Data Fig. 5g)24. This analysis clearly revealed a graded increase in this TCF21-associated gene expression program that correlated with SMC phenotypic modulation (Fig. 5b). Interestingly, this gradient extended into the fibroblast 2 cluster, and also into the main fibroblast cluster (data not shown), again supporting the notion that SMC phenotypic modulation, in part mediated by TCF21, could ultimately lead to a specific fibroblast-like cellular phenotype in humans.
Figure 5. TCF21 is associated with SMC modulation in human coronary arteries.
(a) Pairwise Pearson correlation of TCF21 with every other gene in cells across the “SMC” and “Fibromyocyte” clusters from Fig. 4a. Selected examples of genes regulated during SMC modulation are labeled. (b) t-SNE visualization of cell clusters of the human coronary samples (n=4 patients). The 20 most highly correlated and anti-correlated genes from Fig. 5a were used to calculate a TCF21-associated human cell gene expression score, which ranges from highly anti-correlated (blue) to highly correlated (red). (c) Expression of SMC modulation marker genes with TCF21 overexpression in HCASMCs. Results are from 6 independent experiments each with 3 technical replicates. Statistical significance was determined by comparing fold-change values using a two-sided Mann-Whitney U test. NC = empty vector negative control, OE = overexpression. Error bars denote standard deviation.
To further test a causative role for TCF21 in promoting a human fibromyocyte phenotype, we overexpressed TCF21 in human coronary artery smooth muscle cells (HCASMCs) and found that several key markers of SMC phenotypic modulation were upregulated in these cells by quantitative PCR (Fig. 5c). These findings, taken together with the correlation of a TCF21 -associated gene expression program with SMC phenotypic switching in vivo, suggest that TCF21 plays a causal role in SMC phenotypic modulation in human coronaries.
CAD risk alleles are associated with decreased TCF21 expression
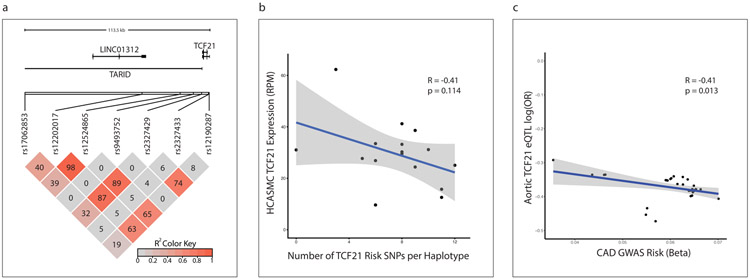
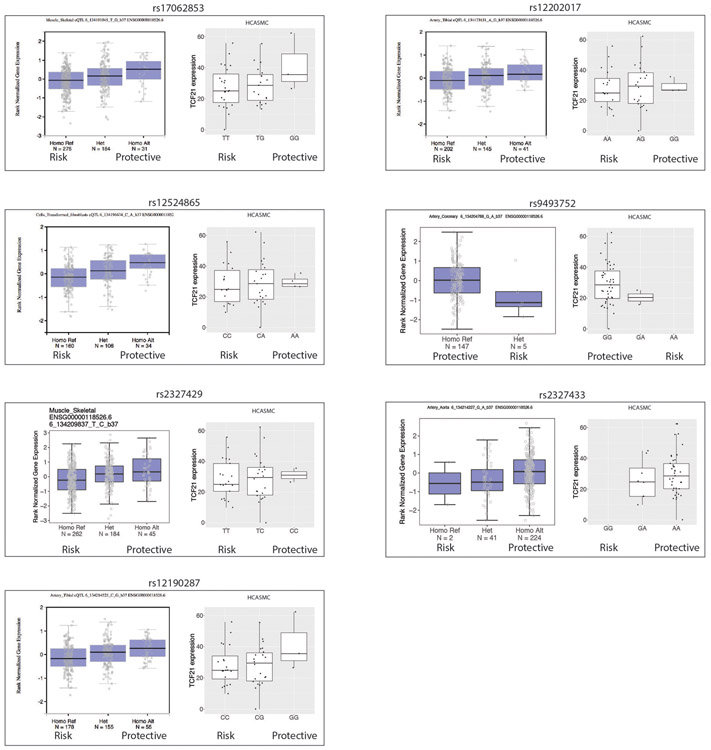
To further understand the impact of TCF21 expression and phenotypic modulation on human disease risk, we investigated the relationship between Genome Wide Association Study (GWAS) SNP genotypes at the 6q23.2 locus and TCF21 expression (cis-eQTL analysis). We first assessed seven SNPs associated with CAD at genome-wide significance. Figure 6a illustrates these SNPs and their linkage disequilibrium (LD) relationships. In a group of 52 HCASMC cell lines we found that, without exception, the CAD risk allele of each SNP was associated with decreased TCF21 expression (Extended Data Fig. 6). We also observed the same finding for these SNPs in highly CAD-relevant tissues in the Genotype-Tissue Expression (GTEx) database (Extended Data Fig. 6). In our HCASMC lines, these risk alleles showed an additive effect; haplotypes accumulating greater numbers of risk alleles were associated with progressively lower TCF21 expression (p = 0.114, R = −0.41, Fig. 6b). We then assessed a larger number of SNPs in the 6q23.2 locus that were associated with CAD risk at a false discovery rate of 1e−5. By performing eQTL analysis in aortic tissue from the STARNET database25 on this larger set of SNPs, we found that the magnitude of CAD risk imparted by each risk allele was also correlated with lower TCF21 expression from that allele (Fig. 6c, p = 0.013, R = −0.41). These eQTL findings from multiple independent SNPs in multiple CAD-relevant tissues strongly argue that TCF21 expression is protective against disease.
Figure 6. Reduced TCF21 expression is associated with increased coronary disease risk.
(a) Linkage disequilibrium relationships (LD, R2 measure) of all genome-wide significant CAD-associated SNPs at the 6q23.2 locus (bottom), relative to the position of TCF21 and long non-coding RNAs (LINC01312 and TARID) within the locus (top). The R2 color indicates the degree of LD between each pair of SNPs, and ranges from 0 (grey) to 1 (red). The corresponding R2 values are also shown in each box. (b) Relationship between the number of genome-wide significant CAD risk alleles in each haplotype (x-axis) and TCF21 expression (y-axis) in 52 primary human coronary artery smooth muscle cell (HCASMC) lines. (c) Correlation between the magnitude of CAD risk imparted by each risk allele (x-axis) with relative TCF21 expression from that allele (y-axis) in 36 CAD-associated SNPS at the 6q23.2 locus in aortic tissue from the STARNET database. p-value was calculated using Pearson’s moment correlation coefficient. Grey shaded areas indicate 95% confidence intervals are based on Fisher’s Z-transform.
DISCUSSION
The phenomenon of SMC phenotypic modulation has been studied primarily by exposing cultured SMCs to lipids and various growth factors26-29. These in vitro studies have consistently reported downregulation of SMC markers23,29, increased migration, proliferation, extracellular matrix secretion, upregulation of certain inflammatory cytokines30, macrophage markers23,29 and increased levels of phagocytic activity. However, SMC phenotypic modulation has been very difficult to study in vivo in mice and humans due to both reduced expression of canonical SMC markers and expression of some SMC markers by other cell types. In a recent landmark paper, Shankman, et. al. used smooth muscle cell lineage tracing to definitively identify phenotypically modulated cells of SMC origin in atherosclerotic lesions. However, assessment of modulated SMC phenotype with in situ studies was necessarily limited to small number of markers. Identification and characterization of modulated SMCs in human plaques has been even more challenging7,31 .
Based upon these studies, the current paradigm is that modulated SMCs can adopt either i) a pro-inflammatory macrophage-like phenotype characterized by Lgals3 expression4-6,23,29 that could result in plaque destabilization, ii) an extracellular matrix producing “synthetic” SMC phenotype4-6, which could contribute to the protective fibrous cap, or possibly iii) a mesenchymal stem cell-like population of unclear significance4-6. Because of this uncertainty regarding the phenotype of modulated SMCs within the lesion, it is also unclear whether the process of SMC phenotypic modulation leads to a more stable or less stable atherosclerotic plaque, and thus whether SMC modulation is protective or increases risk for CAD and MI.
In this study we found that, instead of assuming multiple distinct cell phenotypes, SMCs undergoing phenotypic modulation appear to exhibit a shift in gene expression along a continuous trajectory from a contractile SMC towards a fibroblast-like cell, which we term a “fibromyocyte”. This name was created to emphasize the opposite phenotypic trajectory of these cells compared to a fibroblast-derived “myofibroblast” that acquires properties of SMCs32. Although fibromyocytes display a decreasing gradient of SMC gene expression, they are a highly distinct population and cluster independently even at very low clustering resolutions. The transcriptional profile of SMC lineage-traced fibromyocytes in the mouse was employed to identify an orthologous human fibromyocyte population.
There has been much interest in the possibility that phenotypically modulated SMCs may adopt a detrimental macrophage-like phenotype during atherosclerosis, engulfing oxidized LDL and dying cells and eventually becoming plaque-destabilizing foam cells. SMCs subjected to cholesterol loading in culture accumulate intracellular lipid reminiscent of foam cells and display modest upregulation of the macrophage markers LGALS3 and CD6823 as well as a modest increase in phagocytic behavior23,29. In mice, Myh11 lineage-traced SMCs that migrate into the lesion express Lgals36. In human coronaries, CD68 expression was observed in cells within the lesion that also expressed a SMC-specific epigenetic mark6. Our data, combining SMC lineage tracing with scRNAseq measuring thousands of genes simultaneously, suggest that fibromyocytes in vivo do not acquire a macrophage-like transcriptional phenotype. We confirmed these findings using multiple methods at the protein level.
Interestingly, we found a small number of modulated SMCs at the baseline timepoint, which is consistent with the recent finding of a rare Sca1+ SMC population in healthy mice19. While the authors of this recent study did not demonstrate that these Sca1+ SMCs are an exclusive contributor to lesional SMC-derived cells, many modulated SMCs within the lesion do express Sca1 (Extended Data Fig. 2c). The oligoclonal nature of SMC contribution to atherosclerosis has already been established by multiple groups using multi-colored lineage tracing methods19,22,33. When interpreted in the context of our findings, these studies suggest that a small number SMCs undergo phenotypic modulation and expand to create the fibromyocyte population. However, it is not currently possible to determine whether some or all fibromyocytes arise from a distinct population of modulated SMCs that exist homeostatically within the healthy vessel wall.
SMC-specific Tcf21 knockout revealed that Tcf21 promotes phenotypic modulation in vivo. It is interesting that during phenotypic modulation SMCs up regulate Tcf21 and transform into a fibroblast-like phenotype, and that loss of Tcf21 inhibits this phenotypic transition. In embryogenesis, Tcf21 regulates fundamental cell fate decisions in the developing epicardium, serving as a fate determining factor for the divergence of coronary vascular smooth muscle cell and cardiac fibroblast lineages. In this setting, Tcf21 is downregulated in cells that are fated to become differentiated coronary SMC and expression remains in the interstitial and adventitial fibroblast lineage. Strikingly, Tcf21−/− mice show a near-complete absence of cardiac fibroblasts13, indicating that Tcf21 expression is required for fibroblast development. Thus, during atherosclerosis, Tcf21 appears to be recapitulating its developmental role by directing cell fate decisions away from SMCs and towards a fibroblast gene expression program.
This study is the first to show a gene that is causally associated with CAD at genome-wide significance can fundamentally alter the process of SMC phenotypic modulation in vivo. Given our findings that Tcf21 expression promoted SMC phenotypic modulation in vivo, we had a unique opportunity to assess whether TCF21 and SMC phenotypic modulation were protective or deleterious during CAD. We identified multiple, independent lines of evidence suggesting that TCF21 expression is causally associated with reduced risk of CAD. Taken together, these data suggest that both TCF21 expression and SMC phenotypic modulation are beneficial during the disease process, although likely in a time and context-dependent manner. Based on the finding that loss of Tcf21 results in fewer fibromyocytes in the lesion and at the protective fibrous cap, it is quite plausible that Tcf21 exerts its protective effect by promoting the infiltration of fibromyocytes into the lesion and the fibrous cap.
Extended Data
Extended Data Figure 1. Design of mouse experiments.
(a) Alleles present in SMClin and SMClin-KO mice. KO (knockout) refers to Tcf21, lin = lineage tracing, Tg = transgene, ΔSMC = SMC cell-specific KO (b) Mice were maintained on chow diet from birth until 7 weeks of age, then underwent gavage and high-fat diet (HFD) treatment. For single-cell RNAseq (scRNAseq), RNAscope, CITE-seq, histology involving BODIPY and FACS staining experiments (upper timeline), mice were gavaged only at 7 weeks of age, prior to onset of HFD, as denoted by red arrows. For scRNAseq experiments, mice were sacrificed at baseline (72 hours after initial tamoxifen gavage), or after 8 weeks or 16 weeks of HFD. For RNAscope experiments, mice were sacrificed after either 8 weeks or 16 weeks of HFD. For the CITE-seq experiment, mice were sacrificed after 16 weeks of HFD. For BODIPY studies, mice were sacrificed after 16 week of HFD. For the FACS staining experiment two mice, one after 12 weeks HFD and another after 15 weeks HFD were used. For quantitative histology experiments (lower timeline), mice were gavaged at 7 weeks of age, after 8 weeks of HFD and after 16 weeks of HFD (48 hours prior to sacrifice) as denoted by red arrows. For these quantitative histology experiments, all mice were sacrificed after 16 weeks of HFD. (c) Fluorescence activated cell sorting (FACS) workflow for isolating single cells from the mouse aortic root.
Extended Data Figure 2. SMC phenotypic modulation in the mouse aortic root.
(a-d) t-SNE visualization of cell types present in the wild-type mouse aortic root from all timepoints overlaid with expression of Col1a1, Acta2, Sca1 and Lgals3. n=9 mice. (e-f) RNAscope staining for lumican (Lum, green) and tdT (red) in (e) a plaque after 8 weeks HFD, (f) the non-diseased media of a mouse on 16 weeks HFD and (g) in a baseline healthy aorta. (h) RNAscope negative control. Images in (e-h) are representative from 2 experiments and scale bars indicate 25μm. (i) t-SNE visualization of cell types present in the wild-type mouse aortic root from all timepoints overlaid with osteopontin (Spp1) expression. n=9 mice. (j-k) RNAscope co-localization of Spp1 (green) and tdT (red) in a plaque after 16 weeks HFD. Yellow arrows indicate co-localization of Spp1 and tdT. (l) RNAscope negative control. Images from (j-l) are representative of 4 experiments, and scale bars indicate 50μm. (m) Heatmap representation of the Euclidean distance between cell cluster centroids in 20-dimensional principal component space with smallest distances in yellow and largest distances in black. Data are after 16 weeks of HFD. (n) Staining of a single cell suspension from the atherosclerotic mouse aortic root and ascending aorta with antibodies against the macrophage markers Cd16 and Cd32, and analysis of co-expression with the tdT SMC lineage marker. Data are from one experiment and n=2 mice (after 12 and 15 weeks HFD). (o-t) Single cells from the atherosclerotic mouse aortic root and ascending aorta at 16 weeks HFD were incubated with DNA-barcoded antibodies against the macrophage markers Cd16, Cd32, Cd11b, Cd64, Cd86 and F4/80 prior to undergoing scRNAseq (CITE-seq), yielding simultaneous transcriptomic and antibody binding data within each individual cell. (o) Cell type assignments were determined with scRNAseq as described previously. (p-t) Quantitative antibody binding within each cell type. Results are from one experiment and n=2 mice.
Extended Data Figure 3. Additional characteristics of SMClin vs SMClin-KO mice.
(a-b) Tcf21 expression in SMC lineage-labeled cells from SMClin (WT) and SMClin-KO (KO) mice from all timepoints combined. n=13 mice. (a) Tcf21 expression for all WT cells (left, min=0, max=2.55, mean=0.071) and all KO cells (right, min=0, max=1.97, mean=0.004). (b) Mean Tcf21 expression visualized for all SMC lineage-labeled WT and KO cells. (c) Total Lgals3+ area in the lesion is reduced in SMClin-KO mice. (d) Cd68 immunohistochemistry quantification (left) and representative images (right). Scale bars represent 100μm. (e) Lesion area, normalized to the total vessel area. Data from (c-e) are after 16 weeks HFD, and analyzed using a two-sided student’s t-test. Error bars indicate standard error.
Extended Data Figure 4. Human phenotypically modulated SMCs.
(a) t-SNE visualization of celltypes in the right coronary artery of 4 patients, overlaid with LUM expression. Expression levels are indicated by scales in the lower right. (b) TNFRSF11B RNAscope staining in a human coronary artery section. Hybridization events are seen as red dots. (c) Negative control RNAscope probe shows no staining. Images in (b-c) are representative of 4 experiments, and scale bars represent 50μm. (d) Heatmap representation of the Euclidean distance between cell cluster centroids in 20-dimensional principal component space, with smallest distances in yellow and largest distances in black. Relationship between “Fibromyocyte” and “Fibroblast 2” clusters is highlighted with white asterisks. The “Fibromyocyte”, “SMC” and the main “Macrophage” clusters are denoted by black asterisks. (e-f) t-SNE visualization of celltypes in the right coronary artery of 4 patients overlaid with (e) CD68 expression and (f) TCF21 expression. (g) UCSC Genome Browser shots of representative TCF21 ChIPseq peaks within the PRELP and MYH11 genes, which are highly correlated and anti-correlated, respectively, with TCF21 and the fibromyocyte phenotype. Images are from one ChIPseq experiment.
Extended Data Figure 5. Joint clustering approach identifies human phenotypically modulated SMCs.
(a) Joint clustering of mouse and human datasets using canonical correlation analysis (CCA) as per the Seurat package. (b) The shared mouse/human cluster containing bona fide SMC lineage-traced, phenotypically modulated SMCs (fibromyocytes) from the mouse is highlighted in red. (c) Mouse cells in the shared mouse/human fibromyocyte cluster in (b) are highlighted in the independently-clustered mouse dataset, confirming their location within the known fibromyocyte cell cluster. (d) Human cells in the shared mouse/human fibromyocyte cluster in (b) are highlighted in the independently-clustered human dataset, illustrating their location predominantly in the “Fibromyocyte” cluster (also shown in brown in Fig. 4d). (e) All joint mouse/human clusters in (a) were mapped back to the human dataset. Agreement is identified in cell type assignment between the joint clustering approach and the independently-clustered human dataset.
Extended Data Figure 6. Association of genome-wide significant CAD risk SNPs at the 6q23.2 locus with TCF21 expression.
Seven SNPs in the 6q23.2 locus were associated with CAD at genome-wide significance. The association between risk and protective genotypes and TCF21 expression for each of these SNPs was determined using the gene-tissue expression database (GTEx) in CAD-relevant tissues and a cohort of 52 HCASMC lines. Number of independent tissue samples included for each SNP is indicated in the GTEx data (‘N’), and n=52 cell lines for the HCASMC data. In each box plot, the middle line represents the median, box represents the 1st to 3rd quartile range, and whiskers represent 1.5 times the interquertile range.
Supplementary Material
Supplementary Table 1. Mouse cell cluster markers - all timepoints. The top 100 gene markers distinguishing each cluster (reference cluster) from the remaining clusters in the mouse scRNAseq dataset. Data were analyzed from all timepoints combined in wild-type mice (n=9 mice). Cluster names are noted at left. p_val = p-value. log_FC = average log2 fold-change. pct.1 = percentage of cells in the reference cluster that express at least 1 transcript of the gene. pct.2 = percentage of cells in all other clusters that express at least 1 transcript of the gene. p_val_adj = Bonferroni-adjusted p-value corrected for comparison with all genes in the dataset.
Supplementary Table 2. Mouse cell cluster markers - 8 weeks high-fat diet. The top 100 gene markers distinguishing each cluster (reference cluster) from the remaining clusters in the mouse scRNAseq dataset. Data were analyzed at the 8 week timepoint in wild-type mice (n=3 mice). Cluster names are noted at left. p_val = p-value. log_FC = average log2 fold-change. pct.1 = percentage of cells in the reference cluster that express at least 1 transcript of the gene. pct.2 = percentage of cells in all other clusters that express at least 1 transcript of the gene. p_val_adj = Bonferroni-adjusted p-value corrected for comparison with all genes in the dataset.
Supplementary Table 3. Mouse cell cluster markers - 16 weeks high-fat diet. The top 100 gene markers distinguishing each cluster (reference cluster) from the remaining clusters in the mouse scRNAseq dataset. Data were analyzed at the 16 week timeopint in wild-type mice (n=3 mice). Cluster names are noted at left. p_val = p-value. log_FC = average log2 fold-change. pct.1 = percentage of cells in the reference cluster that express at least 1 transcript of the gene. pct.2 = percentage of cells in all other clusters that express at least 1 transcript of the gene. p_val_adj = Bonferroni-adjusted p-value corrected for comparison with all genes in the dataset.
Supplementary Table 4. Human cell cluster markers. The top 100 gene markers distinguishing each cluster (reference cluster) from the remaining clusters in the human scRNAseq dataset (n=4 patients). Cluster names are noted at left. p_val = p-value. log_FC = average log2 fold-change. pct.1 = percentage of cells in the reference cluster that express at least 1 transcript of the gene. pct.2 = percentage of cells in all other clusters that express at least 1 transcript of the gene. p_val_adj = Bonferroni-adjusted p-value corrected for comparison with all genes in the dataset.
Supplementary Table 5. Clinical characteristics of patients in the study. Basic clinical characteristics of each patient from which samples were obtained for the study. Patient samples (proximal-to-mid right coronary artery) were used for scRNAseq as described in the methods section.
ACKNOWLEDGEMENTS
The authors are grateful to Dr. Gary Owens for providing TgMyh11-Cre, ApoE−/− animals and experimental protocols. The authors are also grateful to Dr. Libette Roman, DVM, for her assistance in developing the human coronary digestion protocol. This work was supported by NIH grant F32HL129670 and AHA grant 18CDA34110206 (RW), NIH grants K08HL133375 (JBK), R00HL125912 (CLM), R01HL109512 (TQ), R01HL134817 (TQ), R33HL120757 (TQ), R01DK107437 (TQ), R01HL139478 (TQ), and 1R01HL141371 (JCW). The sequencing data was generated on an Illumina HiSeq 4000 that was purchased with funds from NIH under award number S10OD018220. Cell sorting was performed on an instrument in the Shared FACS Facility obtained using NIH S10 Shared Instrument Grant S10RR025518-01.
Footnotes
DECLARATION OF COMPETING INTERESTS
The authors claim no conflicts of interest relating to this manuscript.
DATA AVAILABILITY
High throughput sequencing data (FASTQ) files for all scRNA-seq, CITE-seq and ChIP-seq, as well as cell-gene count matrices for all scRNAseq and CITE-seq experiments, have been deposited at Gene Expression Omnibus (GEO) with SuperSeries reference number GSE131780. These data were used to generate images in Figs. 1-5 and Extended Data Figs. 2-5. FASTQ files and processed data are also available from the corresponding author upon request.
REFERENCES
- 1.Davies MJ, Richardson PD, Woolf N, Katz DR & Mann J Risk of thrombosis in human atherosclerotic plaques: role of extracellular lipid, macrophage, and smooth muscle cell content. British heart journal 69, 377–381 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Libby P & Aikawa M Stabilization of atherosclerotic plaques: new mechanisms and clinical targets. Nature medicine 8, 1257–1262, doi: 10.1038/nm1102-1257 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Ross R & Glomset JA Atherosclerosis and the arterial smooth muscle cell: Proliferation of smooth muscle is a key event in the genesis of the lesions of atherosclerosis. Science (New York, N.Y.) 180, 1332–1339 (1973). [DOI] [PubMed] [Google Scholar]
- 4.Bennett MR, Sinha S & Owens GK Vascular Smooth Muscle Cells in Atherosclerosis. Circulation research 118, 692–702, doi: 10.1161/circresaha.115.306361 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gomez D & Owens GK Smooth muscle cell phenotypic switching in atherosclerosis. Cardiovascular research 95, 156–164, doi: 10.1093/cvr/cvs115 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shankman LS et al. KLF4-dependent phenotypic modulation of smooth muscle cells has a key role in atherosclerotic plaque pathogenesis. Nature medicine 21, 628–637, doi: 10.1038/nm.3866 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gomez D, Shankman LS, Nguyen AT & Owens GK Detection of histone modifications at specific gene loci in single cells in histological sections. Nature methods 10, 171–177, doi: 10.1038/nmeth.2332 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schunkert H et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nature genetics 43, 333–338, doi: 10.1038/ng.784 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller CL et al. Disease-related growth factor and embryonic signaling pathways modulate an enhancer of TCF21 expression at the 6q23.2 coronary heart disease locus. PLoS genetics 9, e1003652, doi: 10.1371/journal.pgen.1003652 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller CL et al. Coronary heart disease-associated variation in TCF21 disrupts a miR-224 binding site and miRNA-mediated regulation. PLoS genetics 10, e1004263, doi: 10.1371/journal.pgen.1004263 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dettman RW, Denetclaw W Jr., Ordahl CP & Bristow J Common epicardial origin of coronary vascular smooth muscle, perivascular fibroblasts, and intermyocardial fibroblasts in the avian heart. Developmental biology 193, 169–181, doi: 10.1006/dbio.1997.8801 (1998). [DOI] [PubMed] [Google Scholar]
- 12.Winter EM & Gittenberger-de Groot AC Epicardium-derived cells in cardiogenesis and cardiac regeneration. Cellular and molecular life sciences : CMLS 64, 692–703, doi: 10.1007/s00018-007-6522-3 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Acharya A et al. The bHLH transcription factor Tcf21 is required for lineage-specific EMT of cardiac fibroblast progenitors. Development (Cambridge, England) 139, 2139–2149, doi: 10.1242/dev.079970 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nurnberg ST et al. Coronary Artery Disease Associated Transcription Factor TCF21 Regulates Smooth Muscle Precursor Cells That Contribute to the Fibrous Cap. PLoS genetics 11, e1005155, doi: 10.1371/journal.pgen.1005155 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herring BP, Hoggatt AM, Burlak C & Offermanns S Previously differentiated medial vascular smooth muscle cells contribute to neointima formation following vascular injury. Vascular cell 6, 21, doi: 10.1186/2045-824x-6-21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wirth A et al. G12-G13-LARG-mediated signaling in vascular smooth muscle is required for salt-induced hypertension. Nature medicine 14, 64–68, doi: 10.1038/nm1666 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Madisen L et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature neuroscience 13, 133–140, doi: 10.1038/nn.2467 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng GX et al. Massively parallel digital transcriptional profiling of single cells. Nature communications 8, 14049, doi: 10.1038/ncomms14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobnikar L et al. Disease-relevant transcriptional signatures identified in individual smooth muscle cells from healthy mouse vessels. Nature communications 9, 4567, doi: 10.1038/s41467-018-06891-x (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitchen CM, Cowan SL, Long X & Miano JM Expression and promoter analysis of a highly restricted integrin alpha gene in vascular smooth muscle. Gene 513, 82–89, doi: 10.1016/j.gene.2012.10.073 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stoeckius M et al. Simultaneous epitope and transcriptome measurement in single cells. Nature methods 14, 865–868, doi: 10.1038/nmeth.4380 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobsen K et al. Diverse cellular architecture of atherosclerotic plaque derives from clonal expansion of a few medial SMCs. JCI insight 2, doi: 10.1172/jci.insight.95890 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vengrenyuk Y et al. Cholesterol loading reprograms the microRNA-143/145-myocardin axis to convert aortic smooth muscle cells to a dysfunctional macrophage-like phenotype. Arteriosclerosis, thrombosis, and vascular biology 35, 535–546, doi: 10.1161/atvbaha.114.304029 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sazonova O et al. Characterization of TCF21 Downstream Target Regions Identifies a Transcriptional Network Linking Multiple Independent Coronary Artery Disease Loci. PLoS genetics 11, e1005202, doi: 10.1371/journal.pgen.1005202 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franzen O et al. Cardiometabolic risk loci share downstream cis- and trans-gene regulation across tissues and diseases. Science (New York, N.Y.) 353, 827–830, doi: 10.1126/science.aad6970 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clement N et al. Notch3 and IL-1beta exert opposing effects on a vascular smooth muscle cell inflammatory pathway in which NF-kappaB drives crosstalk. Journal of cell science 120, 3352–3361, doi: 10.1242/jcs.007872 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Pidkovka NA et al. Oxidized phospholipids induce phenotypic switching of vascular smooth muscle cells in vivo and in vitro. Circulation research 101, 792–801, doi: 10.1161/circresaha.107.152736 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Dandre F & Owens GK Platelet-derived growth factor-BB and Ets-1 transcription factor negatively regulate transcription of multiple smooth muscle cell differentiation marker genes. American journal of physiology. Heart and circulatory physiology 286, H2042–2051, doi: 10.1152/ajpheart.00625.2003 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Rong JX, Shapiro M, Trogan E & Fisher EA Transdifferentiation of mouse aortic smooth muscle cells to a macrophage-like state after cholesterol loading. Proceedings of the National Academy of Sciences of the United States of America 100, 13531–13536, doi: 10.1073/pnas.1735526100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rong JX, Berman JW, Taubman MB & Fisher EA Lysophosphatidylcholine stimulates monocyte chemoattractant protein-1 gene expression in rat aortic smooth muscle cells. Arteriosclerosis, thrombosis, and vascular biology 22, 1617–1623 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Allahverdian S, Chehroudi AC, McManus BM, Abraham T & Francis GA Contribution of intimal smooth muscle cells to cholesterol accumulation and macrophage-like cells in human atherosclerosis. Circulation 129, 1551–1559, doi: 10.1161/circulationaha.113.005015 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Kanisicak O et al. Genetic lineage tracing defines myofibroblast origin and function in the injured heart. Nature communications 7, 12260, doi: 10.1038/ncomms12260 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chappell J et al. Extensive Proliferation of a Subset of Differentiated, yet Plastic, Medial Vascular Smooth Muscle Cells Contributes to Neointimal Formation in Mouse Injury and Atherosclerosis Models. Circulation research 119, 1313–1323, doi: 10.1161/circresaha.116.309799 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature biotechnology 36, 411–420, doi: 10.1038/nbt.4096 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Satija R, Farrell JA, Gennert D, Schier AF & Regev A Spatial reconstruction of single-cell gene expression data. Nature biotechnology 33, 495–502, doi: 10.1038/nbt.3192 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kasowski M et al. Variation in transcription factor binding among humans. Science (New York, N.Y.) 328, 232–235, doi: 10.1126/science.1183621 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H & Durbin R Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England) 25, 1754–1760, doi: 10.1093/bioinformatics/btp324 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y et al. Model-based analysis of ChIP-Seq (MACS). Genome biology 9, R137, doi: 10.1186/gb-2008-9-9-r137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Mouse cell cluster markers - all timepoints. The top 100 gene markers distinguishing each cluster (reference cluster) from the remaining clusters in the mouse scRNAseq dataset. Data were analyzed from all timepoints combined in wild-type mice (n=9 mice). Cluster names are noted at left. p_val = p-value. log_FC = average log2 fold-change. pct.1 = percentage of cells in the reference cluster that express at least 1 transcript of the gene. pct.2 = percentage of cells in all other clusters that express at least 1 transcript of the gene. p_val_adj = Bonferroni-adjusted p-value corrected for comparison with all genes in the dataset.
Supplementary Table 2. Mouse cell cluster markers - 8 weeks high-fat diet. The top 100 gene markers distinguishing each cluster (reference cluster) from the remaining clusters in the mouse scRNAseq dataset. Data were analyzed at the 8 week timepoint in wild-type mice (n=3 mice). Cluster names are noted at left. p_val = p-value. log_FC = average log2 fold-change. pct.1 = percentage of cells in the reference cluster that express at least 1 transcript of the gene. pct.2 = percentage of cells in all other clusters that express at least 1 transcript of the gene. p_val_adj = Bonferroni-adjusted p-value corrected for comparison with all genes in the dataset.
Supplementary Table 3. Mouse cell cluster markers - 16 weeks high-fat diet. The top 100 gene markers distinguishing each cluster (reference cluster) from the remaining clusters in the mouse scRNAseq dataset. Data were analyzed at the 16 week timeopint in wild-type mice (n=3 mice). Cluster names are noted at left. p_val = p-value. log_FC = average log2 fold-change. pct.1 = percentage of cells in the reference cluster that express at least 1 transcript of the gene. pct.2 = percentage of cells in all other clusters that express at least 1 transcript of the gene. p_val_adj = Bonferroni-adjusted p-value corrected for comparison with all genes in the dataset.
Supplementary Table 4. Human cell cluster markers. The top 100 gene markers distinguishing each cluster (reference cluster) from the remaining clusters in the human scRNAseq dataset (n=4 patients). Cluster names are noted at left. p_val = p-value. log_FC = average log2 fold-change. pct.1 = percentage of cells in the reference cluster that express at least 1 transcript of the gene. pct.2 = percentage of cells in all other clusters that express at least 1 transcript of the gene. p_val_adj = Bonferroni-adjusted p-value corrected for comparison with all genes in the dataset.
Supplementary Table 5. Clinical characteristics of patients in the study. Basic clinical characteristics of each patient from which samples were obtained for the study. Patient samples (proximal-to-mid right coronary artery) were used for scRNAseq as described in the methods section.