Abstract
Purpose:
Response to adjuvant chemotherapy (ACT) after tumor resection varies widely among non-small cell lung cancer (NSCLC) patients; therefore, it is of clinical importance to prospectively predict who will benefit from ACT before starting the treatment. The goal of this study is to validate a 12-gene ACT predictive signature developed from a previous study using a clinical grade assay.
Experimental Design:
We developed a clinical grade assay for formalin-fixed paraffin-embedded (FFPE) samples using the NanoString nCounter platform to measure the mRNA expression of the previously published 12-gene set. The predictive performance was validated in a cohort of 207 early stage resected NSCLC patients with matched propensity score of ACT.
Results:
The effects of ACT were significantly different in patients from the predicted ACT benefit-group and those in the predicted ACT non-benefit group (p=0.0056 for interaction between predicted risk group and ACT). Specifically, in the predicted ACT benefit group, the patients receiving ACT had significant RFS benefit (HR=0.34, p=0.016, ACT vs non-ACT), while in the predicted ACT non-benefit group, the patients receiving ACT actually had worse RFS (HR=1.86, p=0.14, ACT vs non-ACT) than those who did not receive ACT.
Conclusions:
This study validated that the 12-gene signature and the FFPE-based clinical assay predict that patients whose resected lung ADCs exhibit an ACT benefit gene expression pattern and who then receive ACT have significant survival advantage compared to patients whose tumors exhibit the benefit pattern but do not receive ACT.
Keywords: non-small cell lung cancer, predictive gene signatures, FFPF tissue samples
Introduction
Lung cancer is the leading cause of death in the United States and around the world, and non-small cell lung cancer (NSCLC) accounts for up to 85% of such deaths (1, 2) (3). Large clinical trials have demonstrated the survival benefits of adjuvant chemotherapy (ACT) and have established ACT as the standard of care for completely resected stage II-IIIA NSCLC, and shown the benefits of ACT for stage IB patients with tumors larger than 4 centimeters (4–9). However, the effect of ACT on prolonging survival is modest, while such treatment is also associated with serious adverse effects (4, 8, 10, 11). Therefore, the routine use of ACT is not justified for all patients with resected NSCLC (9), and it is critical to identify the subgroup of patients who will most likely benefit from ACT (12).
Recently, using a cohort of 442 Stage I-III NSCLC patients from the Lung Cancer Director Challenge Dataset (13), we discovered a 12-gene signature that predicts ACT response (14) in NSCLC patients. In addtion, this gene signature predicts patient prognosis in lung adenocarcinoma (ADC) patients, but not in squamous cell carcinoma (SCC) patients. This signature was derived and validated using microarray technology from fresh frozen tumor samples. The crucial next step in translating this discovery to clinical practice is to develop a clincal assay for this 12-gene signature in formalin-fixed paraffin-embedded (FFPE) samples. In this study, we developed a clinical grade assay using the NanoString nCounter GX platform to measure the expression of the 12-gene signature from FFPE samples. Both the prognostic and predictive performance of this clinical assay together with the original algorithm were validated in FFPE samples from early stage (defined as stage I and II) NSCLC patients, and we demonstrated that the benefit group predicted by this gene signature showed significant improvement in survival after ACT, while the predicted non-benefit group actually showed worse survival after ACT.
METHODS:
A list of 12 genes (ATP8A1, AURKA, C1orf116, COL4A3, DOCK9, HOPX, HSD17B6, IFT57, MBIP, NKX2–1, RRM2, TTC37) and the pre-defined risk prediction algorithm were extracted from our previous study (14). The NanoString nCounter platform was used to develop an assay that could measure the expression levels of these 12 genes from FFPE tissue samples. The assay was developed and optimized using a cohort of 30 NSCLC patients (with both FFPE and fresh frozen tissues), and the quality control measures were determined from this cohort. A cohort of 327 early stage (stage I and II) NSCLC patients was used for assay validation. The assay development and validation procedures are summarized in Figure 1 and also detailed in the following sections. The resulting Tables and Figures are summarized in Supplementary Table S1.
Figure 1.
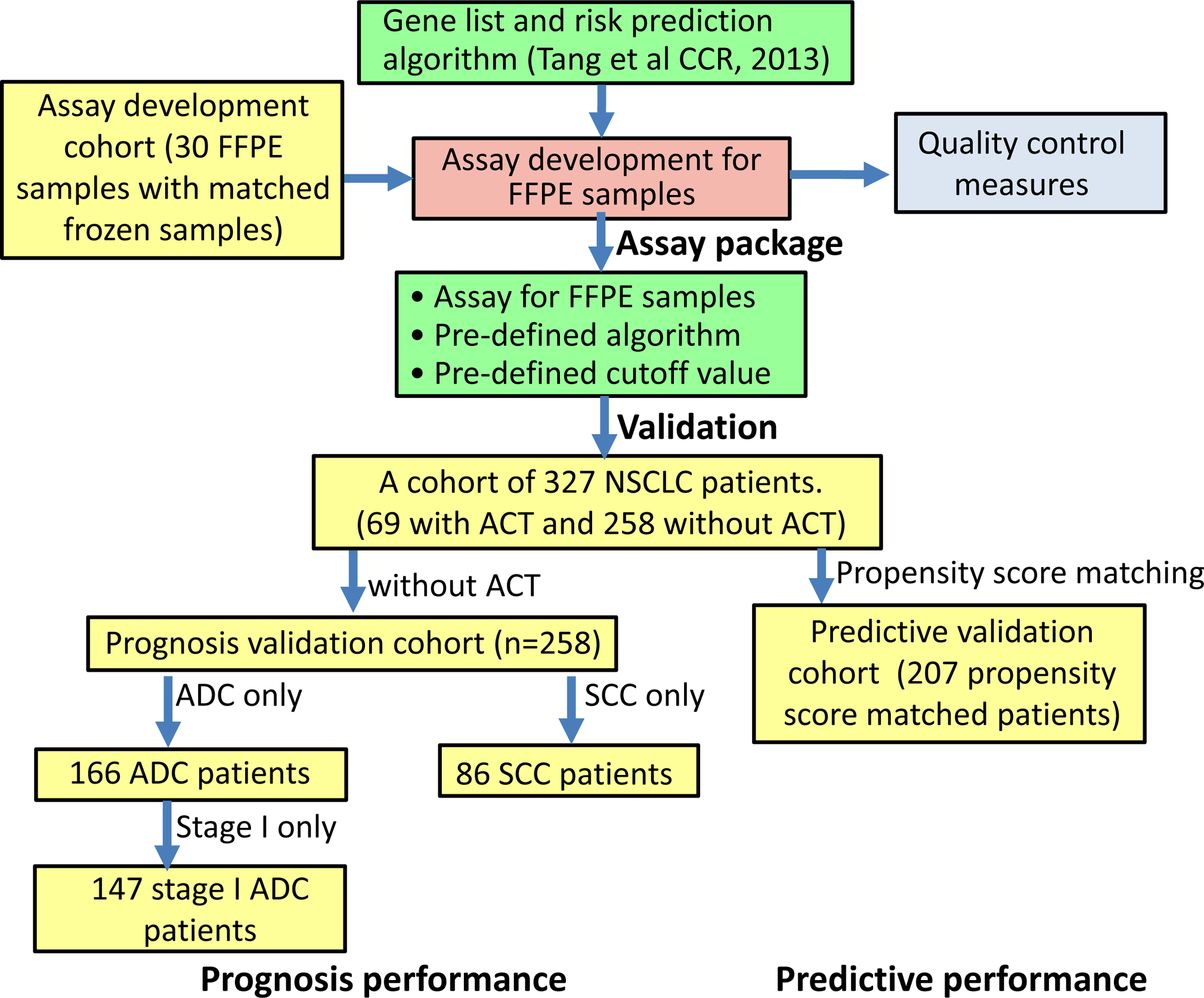
Flowchart for assay development and validation.
Patient Cohorts
The patients gave written consent; the study was approved by the ethics committee at the University of Texas, MD Anderson Cancer Center (IRB #LAB90–020 and # LAB03–0320); and the study was conducted in accordance with the Declaration of Helsinki.
Cohort for assay development (n=30):
A cohort of 30 NSCLC patients (with matched FFPE and fresh frozen tissues) was used for the assay development. The expression levels of the 12-gene set were measured in both FFPE and fresh frozen tissues using the Nanostring nCounter assay. The correlation between expression levels measured from FFPE tissue samples and those measured from corresponding fresh frozen samples was calculated for each individual gene to validate the measure accuracy of the assay for FFPE samples.
Cohort for validating prognostic performances (n=258):
The FFPE tissue samples of 327 early stage (stage I and II) NSCLC patients were obtained from the University of Texas Lung Cancer Specialized Program of Research Excellence (SPORE) Tissue Bank at the MD Anderson Cancer Center (MDACC, Houston, TX). This study was approved by the MD Anderson Institutional Review Board. Among these 327 early stage patients, 69 (21.1%) were treated with ACT, and the remaining 258 (78.9%) were not treated with ACT. None of the patients received neo-adjuvant chemotherapy. The 258 patients without ACT (166 ADC, 86 SCC and 6 others) were used as the validation cohort for prognostic performance of the 12-gene signature.
Cohort for validating predictive performances (n=207):
Since this is a retrospective cohort, in order to minimize the confounding factors, we used a propensity score matching technique to estimate the effect of ACT by accounting for the covariates (detailed in a later section). From the original 327 patients, we derived a cohort of 207 propensity score-matched patients, among which 69 patients (33.3%) were treated with ACT and 138 patients (66.7%) were not treated with ACT, to validate the predictive performance of the assay in FFPE samples.
Detailed information on the patients in the development cohort (n=30), the prognostic validation cohort (258 patients without ACT) and the predictive validation cohort (207 propensity score matched patients) are summarized in Table 1.
Table 1.
Patient characteristics of the study cohorts. In this study, we developed and optimized the clinical assay using a cohort of 30 NSCLC patients. The prognostic performance of the assay was validated in the FFPE samples of a cohort with 258 NSCLC patients without ACT, and the predictive performance was validated in 207 propensity score-matched patients from the cohort of the 327 NSCLC patients. The patient characteristics of the three cohorts are summarized here:
| Assay development cohort (n=30) | Prognostic validation cohort (n=258) | Predictive validation cohort (n=207) | |
|---|---|---|---|
| Histology | |||
| ADC | 13 (43.3%) | 166 (64.3%) | 138 (66.7%) |
| SCC | 13 (43.3%) | 86 (33.3%) | 69 (33.3%) |
| Other | 4 (13.3%) | 6 (2.3%) | 0 (0.0%) |
| Stage | |||
| IA | 5 (16.7%) | 139 (53.9%) | 71 (34.3%) |
| IB | 6 (20.0%) | 80 (31.0%) | 50 (24.2%) |
| IIA | 6 (20.0%) | 23 (8.9%) | 46 (22.2%) |
| IIB | 8 (26.7%) | 16 (6.2%) | 40 (19.3%) |
| III | 4 (13.3%) | 0 (0.0%) | 0 (0.0%) |
| IV | 1 (3.3%) | 0 (0.0%) | 0 (0.0%) |
| Gender | |||
| Female | 14 (46.7%) | 129 (50.0%) | 97 (46.9%) |
| Male | 16 (53.3%) | 129 (50.0%) | 110 (53.1%) |
| Age (Year) | 68.77 | 68.08 | 63.85 |
| Follow-up time (Months) | 52.49 | 6.07 | 53.94 |
| Death | |||
| Yes | 23 (76.7%) | 193 (74.8%) | 151 (73.0%) |
| No | 7 (23.3%) | 65 (25.2%) | 56 (27.1%) |
| Adjuvant Therapy | |||
| Yes | 9 (30.0%) | 0 (0.0%) | 69 (33.3%) |
| No | 15 (50.0%) | 258 (100%) | 138 (66.7%) |
Propensity score matching
We used a propensity score matching technique (15, 16) to match the patients with and without ACT, in order to adjust for potential confounding factors in patient selection for ACT. The clinical variables, including histology, gender, age, smoking history and stage, were used in the propensity score matching, and the coefficient and p value of each variable in the logistic regression model for propensity score matching are summarized in Supplementary Table S2. In this study, we used a 1:2 ratio for ACT-treated and untreated patients, and the final matched patients include 69 ACT-treated (one patient was removed due to missing values in covariates) and 138 patients without ACT. The propensity score matching results are summarized in Supplementary Table S3. These 207 propensity score-matched patients were used in the following analyses to validate the predictive performance of the 12-gene assay. The propensity score matching was implemented using R package matchit (17).
RNA extraction
An unstained section was obtained from FFPE tissue blocks at 10μm thickness, attached to a non-coated glass slide and stored at −80°C freezer. The section was briefly baked at 65°C for 30 minutes and then de-paraffinized with CitriSolv (Fisher Scientific, #22-143-975). Tumor areas were macro-dissected and collected to a fresh Eppendorf micro-centrifuge tube containing lysis buffer. Total RNAs were extracted using AllPrep DNA/RNA FFPE extraction kit (QIAGEN, Cat#80234). RNA was quantified by NanoDrop 2000 and its quality was assessed by Agilent Bioanalyzer 1000 with Agilent RNA 6000 Nano Kit (Agilent, Cat# 5067–1511).
NanoString nCounter Gene Expression assay
NanoString nCounter technology uses unique color-coded molecular barcodes that can hybridize directly to target nucleic acid molecules, such as mRNA, to discover gene expression level with no need for amplification. The nCounter Gene Expression Assay can investigate multiple genes in a single reaction. Probe sequences were custom designed for target genes and manufactured by NanoString Technologies. This CodeSet includes seven housekeeping genes to correct for RNA input amount and/or quality differences. Housekeeping genes were selected from publicly available databases based on stability and detectable expression levels across the tissue type of interest. The manufacturer’s protocol was followed to perform an assay. A master mix of hybridization buffer and Reporter CodeSet was prepared at 10μl each, then 5 μl of total RNA sample and 5 μl of Capture ProbeSet were added to the reaction sequentially. After incubating at 65°C for hybridization for 20 to 22 hours, the reaction products were loaded to GEN2 Prep Station for washing and immobilizing signals to cartridge. The cartridge was imaged in Digital Analyzer at 555 fields of view (FOV), whose images would be interpreted to count for specified targets by nSolver software (NanoString Technologies).
Gene expression data preprocessing
We followed the procedure by Veldman-Jones et al (18) for gene expression data preprocessing. NAPPA R package (http://CRAN.R-project.org/package/NAPPA) from the CRAN (Comprehensive R Archive Network) was used to normalize nCounter data in three steps: (1) a Truncated Poisson correction was used to adjust the background signals in raw NanoString counts using internal negative controls. (2) The data were then normalized using internal positive controls. (3). A Sigmoid shrunken slope normalization was used to correct the data for input amount variation using the mean expression of housekeeping genes. If a raw count was below the average of the 8 internal negative control raw counts plus 2 SDs, the transcript was designated as not detected. Transcripts below the limit of detection (LOD) were not included in the data analysis. Finally, the data were log2 transformed.
Predict the risk score and assign risk groups
This is a prospective validation study, so the exact prediction model described in our previous study(14) was implemented. Specifically, pre-defined supervised principal component analysis was performed for the 12 genes. Using this predefined model, a risk score was assigned to each patient based on the patient’s gene expression level of the 12-gene signature.
Patients were separated into the high-risk (i.e. ACT benefit) or low-risk (i.e. ACT non-benefit) group based on the patient’s risk score. A pre-defined cutoff for risk score was needed to validate the prognostic and predictive performances of the developed assay. In this study, we determined the risk score cutoff based on the assay development cohort (n=30), and applied this pre-defined cutoff into the prognostic performance validation cohort (n=258) and the predictive performance validation cohort (n=207). The risk score was calculated by the expression level of the 12-gene signature in FFPE samples for each patient in the assay development cohort. The predicted risk scores of the 30 patients were plotted in Supplementary Figure S1(A), which indicates a bi-modal distribution of the predicted risk scores. In order to determine a data-driven threshold value of the high- and low-risk patient groups, a model-based clustering method (implemented by R package mclust) was used to fit the risk score distribution with a mixture model of two normal distributions, one corresponding to the high-risk group (red) and the other corresponding to the low-risk group (green). Based on the fitting of the mixture model, the probability of a specific patient being in the high-risk group could be calculated from the predicted risk score (derived from the expression of the 12-gene signature) of the patient. This functional relationship is shown in Supplementary Figure S1 (B).
Statistical analysis
Recurrence-free survival time (RFS) was calculated from the date of surgery until recurrence of lung cancer or death or the date of last follow-up contact. Survival curves were estimated using the Kaplan-Meier product-limit method (19). Differences in the survival outcomes between the predicted high- and low-risk groups were compared using a log-rank test. A multivariate Cox proportional-hazards model (20) was used to determine the association between the factors of interest (i.e. ACT, predicted risk-group and their interaction term) and the patient survival outcomes adjusted for other clinical variables, including age, gender, smoking status, histology and stage. Multivariate Cox proportional-hazards models were also used to determine the benefit of ACT adjusted for other clinical variables in the predicted high- and low-risk groups, respectively.
The time-dependent receiver-operation-characteristic (ROC) curves(21) at 24 months were calculated using R package survivalROC. The area under the curve (AUC) of each time-dependent ROC curve was calculated as a measure of prediction accuracy for patient prognosis. The ROC curve for the predictive analysis was calculated using R package tsm(22).
The concordance index (CI), defined by (23), was calculated using the R package survcomp.
RESULTS
mRNA expression measured from FFPE samples
The expression levels of the 12-gene set were measured from both FFPE and frozen tissue samples by nCounter gene expression assay in the development cohort (n=30) in order to compare the consistency of the measurements from the two types of tissue samples. The dynamic range included the average, maximum, and minimum count of each gene, and the Pearson correlation coefficients between the measurements in FFPE and fresh frozen tissue samples are summarized in Supplementary Table S5. The Pearson correlation coefficients for different genes ranged from 0.5 to 0.8, indicating that the measurements from FFPE samples were reliable and consistent with those from fresh frozen samples.
Prognostic performance of the 12-gene signature
The high- and low-risk groups defined by the 12-gene signature from FFPE tissue samples showed significant differences in recurrence-free survival (Figure 2C) for the 258 NSCLC patients without ACT treatment: the high-risk group had worse prognosis in recurrence-free survival (HR= 1.83 [1.19, 2.95], p = 0.0114). Prognoses for patients with different histology types ADC and SCC were also analyzed. The high-risk group had significantly worse RFS prognosis for ADC patients (Figure 2A, HR= 4.12 [2.07, 8.22], p= 1.25e-05), and stage I-only ADC patients (Figure 2B, HR= 3.91 [1.82, 8.38], p= 0.000161). The time-dependent ROCs were calculated in both the ADC patients (Supplementary Figure S2A, AUC=0.75) and the stage I ADC patients (Supplementary Figure S2B, AUC=0.74). Furthermore, the RFS differences were still significant in ADC patients (HR=4.20 [2.01, 8.79], p = 0.00014, Table 2) and in stage I ADC patients (HR=3.23 [1.26, 8.30], p=0.015, Supplementary Table S6) after adjusting for other clinical variables in the multivariate analysis. On the other hand, for SCC patients (n=90) the predicted high- and low-risk groups didn’t show any differences in recurrence-free survival (Figure 2D, HR= 1.16 [0.272, 4.92], p= 0.83). In summary, the prognostic value of the 12-gene signature is ADC-specific, which is consistent with the findings in our original study. Therefore, this study validated the prognostic performance of the 12-gene signature in FFPE tissue samples using a biomarker assay that can be developed into a CLIA-certified test for use in ADC patients, to move it further toward clinical application.
Figure 2.
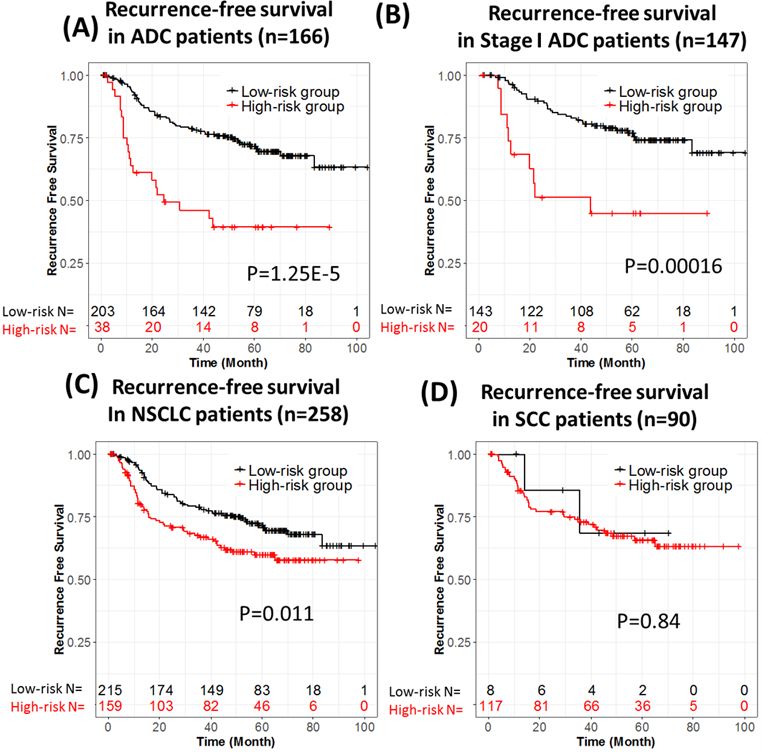
Prognostic performance of the 12-gene signature in ADC patients without ACT treatment. (A) Recurrence-free survival in ADC patients (n = 166). (B) Recurrence-free survival in stage I ADC patients (n = 147). (C) Recurrence-free survival in NSCLC patients (n = 258). (D) Recurrence-free survival in SCC patients (n = 90).
Table 2.
Multivariate analysis for validating the prognostic performance for recurrence-free survival of the 12-gene signature measured from FFPE samples after adjusting for other patient characteristics in the 166 ADC patients without ACT.
| HR | pv | |
|---|---|---|
| Smoking status (Yes vs. No) | 9.54 (1.3, 69.93) | 0.026 |
| Age (Year) | 1.00 (0.96, 1.04) | 0.88 |
| Gender (M vs. F) | 1.84 (0.99, 3.41) | 0.054 |
| Size (>=4cm vs. < 4cm) | 1.76 (0.62, 4.97) | 0.28 |
| Stage (II vs. I) | 2.30 (0.84, 6.30) | 0.11 |
| Group (High-risk vs. Low-risk) | 4.20 (2.01, 8.79) | 0.00014 |
Predictive performance of the 12-gene signature
The 207 early stage NSCLC patients (propensity score-matched for ACT treatment) in the validation cohort were placed by the assay into two groups: those predicted to benefit from ACT (ACT benefit) and those predicted not to benefit from ACT (non-benefit) group, using the same risk score and cutoff criteria. Figure 3 shows the recurrence-free survival curves for patients with and without ACT in the predicted ACT benefit and non-benefit groups, respectively. In the predicted ACT benefit group (high-risk group) (Figure 3A), the patients who received ACT had longer RFS time than those who did not receive ACT, while in the predicted ACT non-benefit group (low-risk group) (Figure 3B), patients who received ACT actually exhibited worse survival than those who did not receive ACT. Multivariate analyses for the efficacy of ACT treatment shows that ACT had significant RFS benefit (HR=0.34, p =0.016) in the predicted ACT benefit group (Figure 3C), while ACT was not associated with RFS benefit (HR=1.86, p=0.139) in the predicted ACT non-benefit group (Figure 3D) after adjusting for other clinical variables. To test the interaction between the predicted ACT benefit groups and ACT treatment effects, we performed multivariate analysis adjusting for clinical variables, including histology, smoking status, age, gender, tumor size, and stage, for all 207 patients (Table 3). This analysis indicated that, after adjusting for other clinical variables, ACT has a survival benefit for patients (HR=0.40, p = 0.020) and there is significant interaction (p=0.0056) between the effect of ACT and the predicted risk groups. We calculated the ROC curve for the predictive analysis (Supplementary Figure S3). The AUC for the predictive performance was 0.806 and the 95% confidence interval was (0.694, 0.865). This demonstrated that the effects of ACT are significantly different between the predicted ACT benefit group and predicted non-benefit group. This validated that the 12-gene signature could predict ACT survival benefit in early stage NSCLC patients and could be used to identify the patient subpopulation that would benefit from the treatment, thereby assisting the clinical decision for using ACT. Furthermore, the 12-gene signature could be used to identify patients in whom adjuvant chemotherapy might be harmful. Figure 3(B) shows that in the predicted ACT non-benefit group, the patients with ACT have significantly worse survival outcomes (pv=0.0154, log-rank test comparing the two survival curves).
Figure 3. Predictive performance of the 12-gene signature.
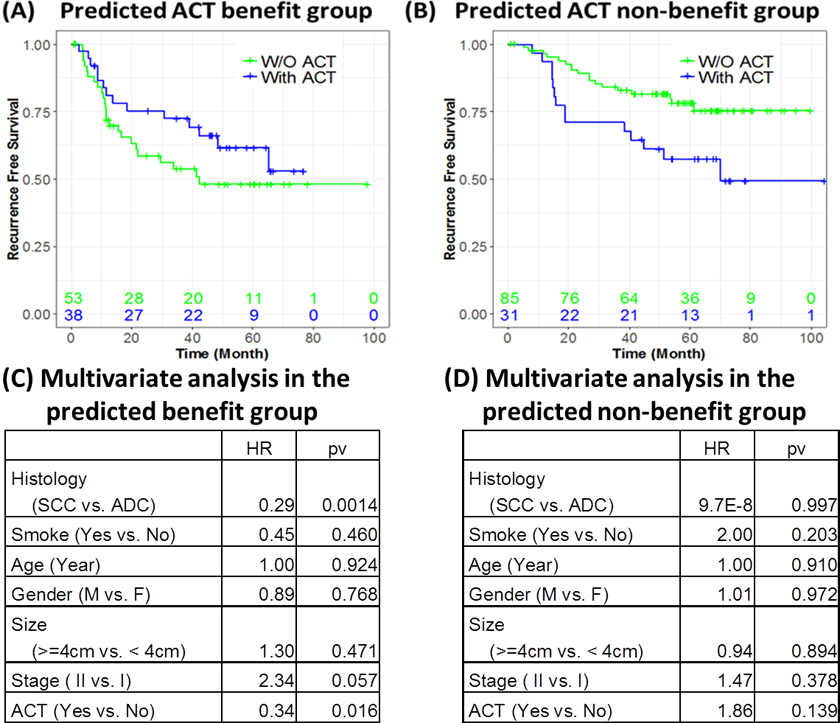
The effects of ACT on recurrence-free survival (RFS) of patients in benefit and non-benefit groups. The RFS time compared between ACT-treated patients vs. non-ACT treated patients in the predicted benefit group (A), and non-benefit group (B). Multivariate analyses of ACT efficacy adjusted for other clinical variables in the predicted ACT benefit group (C) and predicted ACT non-benefit group (D), respectively. In the ACT benefit group (A & C) predicted by the 12-gene signature, the patients with ACT (blue line) had better prognosis than those without ACT treatment. In the predicted ACT non-benefit group (B & D), the patients with ACT (blue line) had worse prognosis than those without ACT treatment (pv=0.0154, log rank test comparing the two survival curves). Furthermore, the multivariate analysis in the predicted ACT benefit group shows the significant benefit of ACT (HR=0.34, pv=0.016) after adjusting for patient clinical variables. In the ACT non-benefit group, the patients with ACT (blue line) had worse prognosis than those without ACT treatment. This indicates that this group of patients did not benefit from ACT treatment. The multivariate analysis in the predicted non-ACT benefit group (D) shows that ACT treatment is associated with worse prognosis (HR=1.86, pv=0.139) after adjusting for other clinical variables.
Table 3.
Multivariate analysis to test the interaction between the ACT treatment effect and the risk groups predicted by the 12-gene signature measured from FFPE samples for recurrence-free survival among the 207 propensity score-matched NSCLC patients.
| HR | pv | |
|---|---|---|
| Histology (SCC vs. ADC) | 0.29 (0.14, 0.58) | 0.00051 |
| Smoking status (Yes vs. No) | 1.54 (0.59, 4.03) | 0.37 |
| Age (Year) | 1.00 (0.97, 1.04) | 0.88 |
| Gender (M vs. F) | 0.96 (0.57, 1.61) | 0.87 |
| Size (>=4cm vs. < 4cm) | 1.15 (0.66, 1.99) | 0.62 |
| Stage (II vs. I) | 1.73 (0.95, 3.15) | 0.075 |
| ACT (Yes vs. No) | 0.40 (0.18, 0.87) | 0.020 |
| Group (Benefit vs. Non-benefit) | 0.14 (0.07, 0.31) | 1.1E–6 |
| Interaction (ACT * Group) | 4.11 (1.51, 11.18) | 0.0056 |
DISCUSSION
Response to standard chemotherapy in lung cancer varies widely among patients. Therefore, it is of substantial clinical importance to be able to predict who will benefit from ACT before starting treatment. Multiple studies have been conducted to identify clinical factors that are associated with chemotherapy response in NSCLC (9, 24–27). Cancer and Leukemia Group B (CALGB) recently demonstrated that there is no significant survival benefit of ACT in stage IB NSCLC patients based on a randomized trial (p-value 0.12), while a statistically-significant survival advantage was observed for stage IB patients with tumors ≥ 4 cm(9). The Lung Adjuvant Cisplatin Evaluation (LACE) study showed that the chemotherapy effect was higher in patients with better performance status, and there was no interaction between the chemotherapy effect and gender, age, histology, type of surgery, planned radiotherapy, or planned total dose of cisplatin (27). Currently, a patient’s TNM stage is the main clinical variable that provides prognostic information to suggest which patients need ACT. However, the TNM information (or the specific tumor histopathologic subtype) does not predict which patients within a TNM-stage category will derive survival benefit from ACT. Therefore, identifying and validating molecular markers with clinical assays to predict ACT response is important.
Although a large number of cancer biomarkers have been reported, few have been translated into real clinical tools. The major bottleneck in translating biomarker discovery to improved patient outcomes is the availability of accurate clinical tests (assays) that will allow treatments to be optimized and tailored to an individual’s needs. In this study, we developed and tested a clinical-grade assay measuring mRNA expression level from FFPE tumor samples, together with a pre-defined risk stratification algorithm for patient response to ACT after tumor resection. The gene signature measured by the clinical-grade assay demonstrated promising results in predicting adjuvant chemotherapy response of NSCLC patients.
Thus far, the dominant effort to improve the clinical outcomes of lung cancer patients with ACT has been put toward testing the benefits of adding targeted therapies; for example, the National Cancer Institute of Canada JBR.19 trial added gefitinib to ACT for resected NSCLC patients, and the Eastern Cooperative Oncology Group E1505 trial added bevacizumab. Although molecular targeted therapies are promising, only a small proportion of early-stage NSCLC patients have mutations that are currently targetable for these therapies. Standard ACTs are currently the primary treatment choice for lung cancer patients. Therefore, being able to identify a sub-group of patients who would benefit from ACT would be an effective way to improve the clinical outcomes of lung cancer patients.
This is a retrospective study with propensity score matching to minimize the effect of potential confounders for treatment effects and biomarker effects. Prospective clinical trials are needed to further validate the predictive signature for ACT. It would also be interesting to test whether this 12-gene signature could predict chemotherapy response in late stage lung cancer patients, and other types of cancer. Finally, multiple measurements of the same specimen on different days will be helpful for technical validation of the performance of the clinical-grade assay for future clinical application.
Supplementary Material
STATEMENT OF TRANSLATIONAL RELEVANCE.
Response to standard chemotherapy in lung cancer varies widely among patients. Therefore, it is of substantial clinical importance to be able to predict who will benefit from adjuvant chemotherapy (ACT) before starting treatment. Although a large number of cancer biomarkers have been reported, few have been translated into real clinical tools. The major bottleneck in translating biomarker discovery to improved patient outcomes is the availability of accurate clinical tests (assays) that will allow treatments to be optimized and tailored to an individual’s needs. In this study, we developed and validated a clinical-grade assay to measure mRNA expression levels from formalin-fixed paraffin-embedded (FFPE) tumor samples, together with validating a pre-defined risk stratification algorithm for patient response to ACT after tumor resection. We validated that the previously published 12-gene signature measured from FFPE tumor samples provided prognostic information in resected lung adenocarcinoma (ADC) patients who do not receive ACT. Importantly, this gene signature and the FFPE-based clinical assay predict that patients whose resected lung ADCs exhibit an ACT benefit gene expression pattern and who then receive ACT have a significant survival advantage, compared to patients whose tumors exhibit the benefit pattern but do not receive ACT.
Funding:
This work was supported by the National Institutes of Health [5R01CA152301, P50CA70907, 5P30CA142543, 1R01GM115473 and 1R01CA172211], the Cancer Prevention and Research Institute of Texas [RP120732] and the National Key Research and Development Plan of China [2016YEE0103400] and National Natural Science Foundation of China [81572288].
Footnotes
Conflict of Interest: None.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, et al. Cancer statistics, 2008. CA Cancer J Clin 2008; 58:71–96. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin 2007; 57:43–66. [DOI] [PubMed] [Google Scholar]
- 3.Tsuboi M, Ohira T, Saji H, Miyajima K, Kajiwara N, Uchida O, et al. The present status of postoperative adjuvant chemotherapy for completely resected non-small cell lung cancer. Ann Thorac Cardiovasc Surg 2007; 13:73–7. [PubMed] [Google Scholar]
- 4.Strauss GM, Herndon J, Maddaus MA, Johnstone DW, Johnson EA, Watson DM, et al. Randomized clinical trial of adjuvant chemotherapy with paclitaxel and carboplatin following resection in stage ib non-small cell lung cancer (nsclc): Report of cancer and leukemia group b (calgb) protocol 9633. J Clin Oncol (Meeting Abstracts) 2004; 22:7019–. [Google Scholar]
- 5.Douillard JY, Rosell R, De Lena M, Carpagnano F, Ramlau R, Gonzales-Larriba JL, et al. Adjuvant vinorelbine plus cisplatin versus observation in patients with completely resected stage ib-iiia non-small-cell lung cancer (adjuvant navelbine international trialist association [anita]): A randomised controlled trial. Lancet Oncol 2006; 7:719–27. [DOI] [PubMed] [Google Scholar]
- 6.Kato H, Ichinose Y, Ohta M, Hata E, Tsubota N, Tada H, et al. A randomized trial of adjuvant chemotherapy with uracil-tegafur for adenocarcinoma of the lung. N Engl J Med 2004; 350:1713–21. [DOI] [PubMed] [Google Scholar]
- 7.The International Adjuvant Lung Cancer Trial Collaborative Group. Cisplatin-based adjuvant chemotherapy in patients with completely resected non-small-cell lung cancer. N Engl J Med 2004; 350:351–60. [DOI] [PubMed] [Google Scholar]
- 8.Winton T, Livingston R, Johnson D, Rigas J, Johnston M, Butts C, et al. Vinorelbine plus cisplatin vs. Observation in resected non-small-cell lung cancer. N Engl J Med 2005; 352:2589–97. [DOI] [PubMed] [Google Scholar]
- 9.Strauss GM, Herndon JE II, Maddaus MA, Johnstone DW, Johnson EA, Harpole DH, et al. Adjuvant paclitaxel plus carboplatin compared with observation in stage ib non-small-cell lung cancer: Calgb 9633 with the cancer and leukemia group b, radiation therapy oncology group, and north central cancer treatment group study groups. J Clin Oncol 2008; 26:5043–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Douillard JY. [adjuvant chemotherapy for non-small cell lung cancer--which agents for which patients?]. Rev Mal Respir 2005; 22:8S118–23. [PubMed] [Google Scholar]
- 11.Olaussen KA, Mountzios G, Soria JC. Ercc1 as a risk stratifier in platinum-based chemotherapy for nonsmall-cell lung cancer. Curr Opin Pulm Med 2007; 13:284–9. [DOI] [PubMed] [Google Scholar]
- 12.Olaussen KA, Dunant A, Fouret P, Brambilla E, Andre F, Haddad V, et al. DNA repair by ercc1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med 2006; 355:983–91. [DOI] [PubMed] [Google Scholar]
- 13.Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, et al. Gene expression-based survival prediction in lung adenocarcinoma: A multi-site, blinded validation study. Nat Med 2008; 14:822–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang H, Xiao G, Behrens C, Schiller J, Allen J, Chow CW, et al. A 12-gene set predicts survival benefits from adjuvant chemotherapy in non-small cell lung cancer patients. Clin Cancer Res 2013; 19:1577–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.ROSENBAUM PR RUBIN DB. The central role of the propensity score in observational studies for causal effects. Biometrika 1983; 70:41–55. [Google Scholar]
- 16.Rosenbaum PR, Rubin DB. Constructing a control group using multivariate matched sampling methods that incorporate the propensity score. The American Statistician 1985; 39:33–8. [Google Scholar]
- 17.Ho D, Imai K, King G, Stuart E. Matching as nonparametric preprocessing for reducing model dependence in parametric causal inference. Political Analysis 2007; 15:199–236. [Google Scholar]
- 18.Veldman-Jones MH, Brant R, Rooney C, Geh C, Emery H, Harbron CG, et al. Evaluating robustness and sensitivity of the nanostring technologies ncounter platform to enable multiplexed gene expression analysis of clinical samples. Cancer Research 2015; 75:2587–93. [DOI] [PubMed] [Google Scholar]
- 19.Kaplan ELM P Nonparametric estimation from incomplete observations. Journal of the American Statistical Association 1958; 53:457–81. [Google Scholar]
- 20.Collett D Modelling survival data in medical research Chapman & Hall/CRC 2003. [Google Scholar]
- 21.Heagerty PJ, Lumley T, Pepe MS. Time-dependent roc curves for censored survival data and a diagnostic marker. Biometrics 2000; 56:337–44. [DOI] [PubMed] [Google Scholar]
- 22.Y H, PB G, H J. Assessing treatment‐selection markers using a potential outcomes framework. Biometrics 2012; 68:687–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schroder MS, Culhane AC, Quackenbush J, Haibe-Kains B. Survcomp: An r/bioconductor package for performance assessment and comparison of survival models. Bioinformatics 2011; 27:3206–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berghmans T, Paesmans M, Meert AP, Mascaux C, Lothaire P, Lafitte JJ, et al. Survival improvement in resectable non-small cell lung cancer with (neo)adjuvant chemotherapy: Results of a meta-analysis of the literature. Lung Cancer 2005; 49:13–23. [DOI] [PubMed] [Google Scholar]
- 25.Hotta K, Matsuo K, Ueoka H, Kiura K, Tabata M, Tanimoto M. Role of adjuvant chemotherapy in patients with resected non-small-cell lung cancer: Reappraisal with a meta-analysis of randomized controlled trials. J Clin Oncol 2004; 22:3860–7. [DOI] [PubMed] [Google Scholar]
- 26.Sedrakyan A, Van Der Meulen J, O’Byrne K, Prendiville J, Hill J, Treasure T. Postoperative chemotherapy for non-small cell lung cancer: A systematic review and meta-analysis. J Thorac Cardiovasc Surg 2004; 128:414–9. [DOI] [PubMed] [Google Scholar]
- 27.Pignon JP, Tribodet H, Scagliotti GV, Douillard JY, Shepherd FA, Stephens RJ, et al. Lung adjuvant cisplatin evaluation: A pooled analysis by the lace collaborative group. J Clin Oncol 2008; 26:3552–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


