Abstract
Whole genome transcriptional profiling has become a standard genomic approach to investigate biological processes. RNA sequencing (RNAseq) in particular has witnessed myriad applications in genetics and various biomedical fields. RNAseq involves a relatively simple experimental protocol of RNA extraction and cDNA library preparation, and because of decreasing next-generation sequencing cost and lower computational burden for data processing, has obtained a central role in the modern biology. The recent application of RNAseq methodology to single cell transcriptional profiling has enabled the more precise characterization of cell lineage and cell state genetic profiles. The development of bioinformatic and statistical tools have provided for differential gene expression (DE) analysis, RNA isoform analysis, haplotype specific analysis of gene expression (allele specific expression - ASE), and analysis of expression quantitative trait loci (eQTL). We give an overview of these and recent developments in RNAseq methodology with emphasis on quality control, read mapping, feature counting, DE, ASE and eQTL analysis, and fusion transcript detection. We describe utilization of RNAseq as a diagnostic tool in Mendelian diseases, complex phenotypes and cancer and give an overview of long read RNAseq technology. Furthermore, we discuss in detail the recent revolution in single cell transcriptomics that is reshaping modern biology.
Keywords: transcriptomics, eQTL, long reads, RNA isoforms, single cell, gene expression, gene regulation, gene transcription, RNA sequencing
Subject Terms: Genetics, Gene Expression and Regulation
INTRODUCTION
Transcriptional profiling at a whole genome level was enabled by a number of technological advances as well as characterization of the genomic architecture in different species through sequencing of large numbers of expressed transcripts and whole genome sequencing. Initially, microarrays, or gene chips, of various designs were constructed by the immobilization of thousands of short DNA segments (probes) to a solid surface, and these arrays were queried by hybridization to fluorescently labeled cDNA “targets.” There were numerous technical and methodological challenges to this approach but a large number of microarray studies provided substantial breakthroughs in our understanding of biological processes. In the field of cardiovascular research, microarray studies profiled gene expression changes associated with vascular disease, cardiac failure, etc. Another approach to transcriptional profiling was the serial analysis of gene expression (SAGE) method, which introduced the concepts of sequencing and counting transcripts that would later be applied in RNA sequencing (RNAseq). SAGE was based on the generation of short cDNA sequences that were concatenated, sequenced, and the number of tags identified and summed for each RNA counted. Perhaps most importantly, these transcriptomic approaches provided for the development of innovative bioinformatic tools and spurred the development of gene ontology frameworks that continue to serve as a basis for evaluation of transcriptomic data and interpretation of differences in gene expression in the context of biological function.
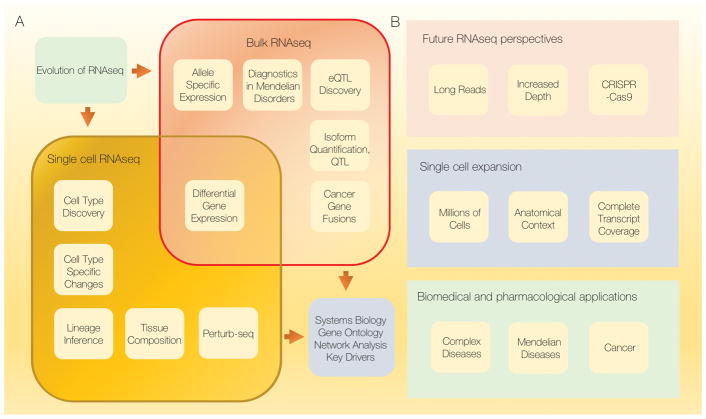
RNA sequencing, or RNAseq, is a type of whole genome transcriptome profiling based on the application of high throughput sequencing and it is generally used to quantify and characterize the entire RNA population present in a specific cell or tissue and under different biological contexts. RNAseq has a wide variety of applications in modern biological and biomedical research (Figure 1)1,2. RNAseq employs a straightforward experimental protocol for cellular RNA extraction and library preparation (Figure 2). The first two papers that used massively parallel sequencing of RNA molecules appeared in 20083,4. RNAseq has dramatically improved since its early years of development in terms of data quality and compared to microarrays offers a wider dynamic range of gene expression values, higher genomic coverage and less artefactual constraints such as cross-hybridization. RNAseq has been essential for systematizing non-translated RNA molecules into classes such as long non-coding RNA (lncRNA), microRNA (miRNA), and small nucleolar RNA (snRNA)5,6. RNAseq has found a specific application in cancer research, where it has been essential in detecting gene fusions7. Furthermore, recent development of single cell RNAseq methods has enabled accurate profiling of cellular diversity in tissues in their native in vivo environment using a limited number of reads from each cell8–10. RNAseq has made possible quantitation of RNA isoform species changing transcriptomics from a gene-centric paradigm to one focused on individual genetic isoforms. In this review, we present an overview of the main applications of RNAseq methodology on a cell population level, as well as recent advances in single cell transcriptomics and discuss potential future directions.
Figure 1. Evolution of RNA sequencing applications.
A) Current applications for bulk and single cell RNAseq methods. B) Ongoing adaptations and expansions of RNAseq methodology.
Figure 2. RNA sequencing at a cell population level.
A) Starting from heterogeneous cell population, mRNA is extracted using poly-A selection with biotin-labeled Poly-T probes for poly-A tail detection. Subsequently, mRNA is fragmented and fragments are primed with random hexamers to generate first strand cDNA fragments. Similarly using random primers second cDNA strand is generated, therefore producing double stranded DNA inserts. Next steps include end repair, adding the phosphoryl group to the 5′ end of fragments and blunt adding A nucleotide to the 3′end of fragments (A-tailing). In the next step adapters are added using their 3′ T overhang and ligated to both sides of the DNA insert. B) Next, two primers complementary to adapters add P5, Rd2 SP, and IndSP sequences to the fragment ends. To achieve multiplexing, such prepared DNA libraries are labeled with a unique identifier, or index, in the next step of library preparation together with adding P7 flow cell primer. Multiple samples are subsequently pooled into a single lane of a flow cell and jointly sequenced in one run. This allows time- and cost- effective sequencing of multiple RNA libraries from multiple samples (useful for large eQTL cohorts). RPKM- reads per kilobase per million mapped reads. Rd1 SP - Read one sequencing primer. Rd2 SP – Read two sequencing primer for paired-end sequencing applications. Ind - Index. P5, P7 - flow cell attachment sequence. IndSP –Index sequencing primer.
CELL POPULATION BASED RNASEQ
RNA library preparation, next gen sequencing, and read mapping
Preparing a high-quality sequencing library has a critical role to the outcome of RNAseq experiment (Figure 2). The decision of choosing the best library protocol depends on the primary objective of an experiment, i.e. whether the objective is to study mRNA, small RNA or the entire transcriptome. RNA purification generally falls into two categories, i.e. phenol-chloroform based (TRIZol) and the silica-gel based column methods (Qiagen), while mRNA enrichment is generally achieved either by poly(A)+ selection or by ribosomal RNA depletion. Ribosomal depletion allows for simultaneous assessment of polyadenylated and non-polyadenylated RNAs. However, ribosomal depletion methods might suffer from problems of high number of reads mapping to intronic regions. A comparative study showed that Poly(A)+ selection protocols produce the highest proportion of exonic reads regardless of the extraction method (exonic: 86% and 91%, intronic: 10% and 6%, TRIzol and Qiagen, respectively)11. Conversely, ribosomal depletion methods (RiboZero) produced between 15% to 35% intronic reads using Qiagen and TRIzol. Furthermore, the separation of nuclear and cytoplasmic fractions showed that the majority of intronic reads come from the nuclear fraction (where they can constitute up to 60%). In addition, the majority of the intronic reads were mapped in the same orientation as the underlying corresponding gene, which together indicates that their origin is not primarily anti-sense RNA, but rather unprocessed or partially processed RNAs species (hnRNA).
Detecting splicing events is directly dependent on the number of reads covering splice junctions. Library preparation can to some degree influence the number of splicing reads. It has been shown that the highest yield is produced with Qiagen/poly(A)+ RNA-seq purification and selection protocols (10.5% mapped reads), while TRIzol/poly(A)+ performed with similar yield (~9.2%)11. On the other hand, ribosomal depletion methods (RiboZero) performed less well (9.4% and 6.4%, Qiagen and TRIzol, respectively). Cytoplasmic and nuclear RNA isolates produced 10.6% and 2.9% of junction reads, indicating that mature RNA transcripts are being actively transported to the cytosol, while nuclear fraction contains more unspliced intermediates.
Modern sequencing machines and algorithms have dramatically improved the quality of sequencing in the last 10 years during the next generation sequencing expansion. Today, the various aspects of quality control (QC) are thus often overlooked when analyzing RNA sequencing data. Generic QC tools, such as FastQC modules, perform per base sequence quality control that informs on quality across the sequence, especially at the beginning and near the end of the read. In addition, FastQC modules report on per base/sequence GC content (detecting general or end-of-the-read GC/AT biases), library adapter content and per base N content (removal of poor quality base calls). FastQC also detects duplicated sequences (indicating PCR biases, particularly important in case RNAseq is used in allele specific expression analysis), overrepresented sequences (indicating either highly biologically significant sequences, like polyA stretches, or library contamination/duplication), and overrepresented k-mers (either biologically significant k-mers generally equally distributed across the read or sequencing errors appearing randomly throughout the read or increasing at the end of the read). For allele specific expression analysis12, it is recommended to remove duplicates using Picard MarkDuplicates or samtools rmdup modules. In addition, Picard contains the CollectRnaSeqMetrics module that produces RNA alignment metrics for a Sequence Alignment Map (SAM) file or its binary format (BAM). It estimates the 5′-3′ bias that results from errors of reverse transcriptase during library construction, and leads to the over-representation of ends of transcripts. Other tools that produce RNAseq specific QC metrics include RSeQC and RNA-SeQC13.
A significant issue with RNAseq is estimating the depth of sequencing that will profile the number of detected genes and transcripts to the point of saturation (Figure 3). In some cases, achieving the modest number of 30M reads is considered sufficient, even though it may be far from the saturation point. Saturation point may be estimated from mapped reads using tools such as Preseq. If the purpose of the experiment is to study differential expression on the gene level, such modest read numbers may be sufficient. However, gene transcripts can be expressed at very different levels, from a single copy to thousands and millions of copies per cell. Therefore, many low level transcripts require deeper sequencing of the transcriptome to be discovered. If the purpose is accurate quantification of known transcripts or to reveal novel transcripts, more extensive sequencing is needed. In experiments where sensitivity of detection is paramount, it is recommended to use a minimum sequencing depth of 100–200 M paired end reads with read length 75bp or longer. Technical variation in RNAseq can be determined by performing technical replicates, i.e., by repeatedly sequencing one cDNA library. Biological replicates, on the other hand, are performed by sequencing different cDNA libraries from repeated biological experiments. Preferably one should aim to design experimental setups so that both technical and biological replication is included. However, this may be cost-inefficient and may increase the computational burden in analysis. As biological replicates by default read out on technical variation, it is generally accepted that they obviate the need for technical variation in most experimental designs. On the other hand, technical variation could be estimated from the Poisson distribution. In an ideal RNAseq experiment counts of reads are multinomial and therefore follow the Poisson distribution. In that case, the variance is approximately equal to the mean, and the overall variance in counts that stems from the technical variation could be approximated using the abundance of transcripts. Therefore, it is possible to complete the statistical analysis without any technical replicates (as exemplified with count based tools, such as EdgeR and DESeq).
Figure 3. RNAseq downstream applications.
A) RNAseq application in genomic mechanistic studies. Discovery of mechanisms for how SNPs promote mRNA level changes (noncoding SNPs in promoters, enhancers and insulators and coding SNPs in coding enhancers). B) Allele-specific (AS) analysis involves discovery of differences in transcribed mRNA between parental alleles. Allele specific QTL discovery is possible in the context of SNPs that are not in the linkage disequilibrium with the AS SNP, while SNPs in LD with the AS SNP will have linked genotypes therefore making it impossible to assess the influence on AS trait. C) RNA isoform analysis with long reads may resolve mechanisms of distant exon pair co-association and mutual exclusion. D) The number of transcripts detected depends on the sequencing depth before reaching a stable plateau. E) Downstream experiments on model animals. F) eQTL discovery involves calculations of cis- and trans- eQTLs. In this example, variation regulates expression of gene G1 which encodes a transcription factor that regulates expression of gene G2. G) Comparison of various differential expression tools (see main text). FPR, false positive rate; TPR, true positive rate; logFC, log fold change; n, number of replicates.
Mapping of RNAseq reads to the genome or transcriptome (in case the transcriptome assembly is available) represents a significant challenge, due to the non-contiguous structure of the transcriptome and relatively small sequencing read length. For example, the maximum read length of the majority of sequencing platforms (except those that are specialized in long read sequencing) is smaller than the average length of the exon. Current Illumina NovaSeq standard read length is 150bp for single end sequencing and 2×150bp for paired end sequencing, while exons possess the average length of 171 bp and the length of 20% of exons in the human genome is greater than 200bp14,15. Therefore, mapping tools need to be computationally sophisticated to decipher the origin of the read. This is difficult if a gene has several isoforms or gene paralogs that share significant homology as the mapping algorithm needs to be able to assign reads with sufficient certainty to the correct gene or gene isoform.
Various aligners have been developed to address the problem of accurate mapping of RNAseq reads to the genome. TopHat16, now superseded by HISAT2 for population-based mapping17, aligns reads to the genomes initially with a short read aligner Bowtie18, and subsequently identifies exon-intron junctions. Similar two-step strategy of initial read mapping that is used to discover splice junctions and drive subsequent final alignment has been applied in tools like MapSplice19 and GEM20. Unlike TopHat that is based on a short read mapper, STAR21 offers mapping of longer reads which is concurrent with the emerging third-generation sequencing technologies. STAR calls splice junctions in a single alignment protocol using the two-step sequential Maximal Mappable Prefix (MMP) concept (unlike e.g. Mummer22 that finds all possible Maximal Exact Matches), without any a priori database knowledge and without a preparatory contiguous read alignment, therefore dramatically reducing the mapping time. Second pass STAR mapping is a custom algorithm that makes a splice junction database from junctions found in the first mapping pass, filters out non-canonical junctions, generates a genome with junctions from the first pass and aligns reads across the previously detected junctions. Second step STAR mapping generally increases mapping percentage up to 2 percent. Reference free assemblers, such as Oases (based on preliminary assembly by Velvet)23,24 and Trinity25, produce transcripts in the absence of any genomic assembly. Mapping of RNA sequence reads is followed by the read counting step performed by read summarization tools such as, featureCounts26, SAMMate27 or htseq-count28.
Recently, mapping-free lightweight algorithms were developed, such as Sailfish29 and Kallisto, with an accompanying differential expression tool Sleuth30,31. Sailfish replaces the read mapping step and instead implements fragmenting reads into their constituent k-mers or ‘mini-reads’ and subsequently performs the ‘exact’ mapping of the k-mer. Salmon32, the successor to Sailfish software, avoids the need to build a k-mer size index, and therefore requires substantially less computational memory. By contrast, Kallisto introduced the idea of pseudo-alignments, i.e. determining transcripts which are compatible with the read, instead of determining where in each transcript the read aligns. This dramatically reduces the time and computing power of analysis, from hours to minutes, with up to 15 million processed reads per minute on a standard desktop computer, while preserving the accuracy of pseudo-aligning. Bootstrap analysis showed that the variance on a single subsample is equal to the variance calculated from various subsamples, emphasizing the high accuracy of this approach30. Kallisto therefore allows for efficient analysis and re-analysis of data against different transcriptomes, development of fast bootstrapping algorithms, and autonomy from high computing power and cloud services. Kallisto’s extension Sleuth utilizes Kallisto bootstraps to effectively distinguish between technical and biological sources of variance and subsequently estimate “true” biological variance31. In everyday practice, when fast and accurate transcript estimation is paramount, Kallisto or Salmon tools represent a balanced choice. However newer tools (such as pufferfish) that require much less computational memory and disk storage for hash table construction during the genome indexing step may become new standard in the future (current pseudo-alignment tools may use up to 150GB of RAM for an index of the human genome and up to 300GB for the collection of bacterial genomes, which vastly exceeds the 32–64GB memory size of moderate servers). Pufferfish proved faster and with lower memory and disk space requirements than kallisto in index construction, index loading and k-mer querying of the input reads.
Differential expression analysis strategies
Differential expression analysis tools have progressed significantly during the evolution of RNAseq. Since the first commonly accepted algorithm was developed, i.e. cuffdiff33, multiple tools have been developed and tested, such as DESeq, DESeq2, edgeR, limma, baySeq, NOISeq, PoissonSeq, SAMSeq, DEXSeq and DEGSeq. Choosing one of these that is appropriate for a particular experimental setup may represent a challenge. A recent study34, has shown that in case the number of biological replicates is low, i.e. generally less than 12, the true positive rate (TPR) is the highest with DEG-Seq, NOI-Seq, cuffdiff and edgeR, while for replicate numbers over 12, in which case the number of false positives (FPR) may significantly rise, DESeq was highlighted as the best performing tool in minimizing the number of false positives (Figure 3). When evaluated on the basis of fold-change (FC), for higher FC genes the number of replicates does not influence the true positive rate, while for lower and middle FC genes the number of replicates is crucial in maintaining TPR. Nevertheless, certain tools outperformed others for lower FC genes with smaller number of replicates, such as NOI-Seq, in maintaining high TPR, while tools like DESeq in case of lower FC and less replicates preserved lower rate of FPR. In addition, a previous comparative study35 recommended limma and DESeq in case biological replicates are less than 5 per condition, and found edgeR to be overly sensitive in this situation. In everyday experimental practice it is rare to have greater than 5 replicates, and DESeq represents a commonly used algorithm with balanced performance.
One of the drawbacks in RNAseq data analysis is the problem of over-dispersion36,37, i.e. the greater variance in data than what is approximated by a Poisson distribution, that itself represents an approximation of multinomial distribution of independently sampled reads across a population. EdgeR approximates a negative binomial distribution and a linear relationship between a mean and a variance, however this would be valid only for larger number of replicates per condition, while in practice these number of replicates are never achieved. DESeq, on the other hand, models mean-variance relationship for each gene by estimating a dispersion value, and then modeling a curve through these estimates and selecting a new dispersion value for each gene from the fitted line. DESeq2, conversely, seeks to remove mean-variance dependence by performing one of the two different transformations of the variance, regularized logarithm or variance stabilizing transformation.
Overall, the tools and strategies applied in RNAseq data analyses depend on the type of analysis in question, as three basic classifications emerge: differential gene expression (DGE), differential transcript usage (DTU) and differential transcript expression (DTE). DGE utilizes genes as a set of non-overlapping independent targets, to which counts could be unambiguously assigned. The problem appears when trying to apply count based method to transcript analysis, such as DTU or DTE, as transcripts of a single gene represent overlapping targets. It is an open question whether transcript-level counts could be analyzed in a manner of non-overlapping targets such as gene-level counts using count based edgeR/DESeq tools. Partitioning of the reads aligning to multiple transcripts will result in fractional estimated counts for transcripts, and whether their overall accuracy is perturbed with introduction of fractional estimated counts to the count based methods is still debated38. In addition, as the mean-variance relationship remains the same, transcript-level estimates when properly combined to the gene-level could be used in edgeR/DESeq, either summed up or by selecting the strongest isoform38.
DE genes can be classified into functional or structural categories, in order to assess the overall effect of the experimental treatment. The Gene Ontology (GO) Consortium defines classes to describe gene function via either molecular activities, compartmentalization or using larger pathways of gene products. Common ontology tools include DAVID, GOSTAT, REACTOME, ENRICHR or PANTHER. Ingenuity Pathway Analysis (IPA) clusters DE genes to define significantly affected signaling and metabolic pathways and it is showed to be informative in generating biochemical signaling cascades and simulating directional consequences of gene activity. Gene Set Enrichment Analysis (GSEA), instead of calculating thresholds, uses ranking information to correlate gene sets with the phenotypic class, thus determining whether a gene member shows a tendency to occur toward the top or bottom of the list.
RNAseq can define cellular and biochemical pathways that are perturbed in a disease state regardless of the underlying mutations, hence it could bridge different cardiovascular diseases (e.g., hypertrophic cardiomyopathy and dilated cardiomyopathy, fulminant myocarditis and chronic myocarditis) on a molecular level. Thus, RNAseq could aid in defining the possible disease pathways that could be targeted with therapy without knowing the exact mutation that disturbs the pathway.
A complete end-to-end RNAseq workflow consists of selection of individual tools for read aligning, expression modeling and differential expression and their incorporation into a functional computational workflow. The number of possible combinations increases exponentially the number of possible workflows. The choice of DE analysis carries the strongest impact, with more modest effects observed with different read aligners and expression modelers39.
Mapping quantitative expression trait loci with RNAseq
Expression quantitative trait loci (eQTLs) are genomic variants that modulate the expression of one or more genes. Such genes are usually proximal to the variant (cis-eQTLs), however they may also be distant genes (trans-eQTLs), whose expression is influenced through the regulation of a nearby transcription factor (Figure 3). Introduction of RNAseq in eQTL analysis represented a substantial improvement compared to the microarray approach. First, RNAseq carries information on the expression that originates from individual alleles, making it possible to study allele specific QTLs (i.e., if the variant influences allele specific expression of a gene in cis or in trans), in case variants do not fall into the same haploblock (Figure 3). Next, it generates an unparalleled body of information to study isoform expression, therefore making it possible to study the influence of a variant on particular RNA isoform expression (isoform percent spliced in – isoform psiQTL). Finally, it allows the study of exon eQTLs and exon splicing QTLs (also termed exon percent spliced in – exon psiQTL) that utilize only a subset of the information generated by RNAseq, focusing either on individual exons or individual exon-exon junctions, respectively. Mapping of eQTLs has now been accomplished by the Genotype-Tissue Expression (GTEx) consortium and made available through the GTEx portal. While such information provides fundamental insights into mechanisms of variation in gene expression in various biological contexts, it also serves as a critical type of information for the identification of causal variants and genes in regions of the human genome that have been associated with a specific trait or complex human disease. The vast majority of genetic variation that contributes to heritable disease risk are estimated to be a function of single nucleotide polymorphisms that serve as eQTLs to regulate causal gene function.
Allele-specific analysis for cis-effects, imprinting and nonsense-mediated decay
Allele specific expression (ASE) or allelic imbalance analysis refers to the analysis of expression variation between the two parental gene copies of a single gene in a diploid individual. ASE is usually distinguished by multiple heterozygous sites present in the coding sequence of the gene that are used to detect differences in allelic expression. ASE has become essential for studying various biological mechanisms, such as the effects of cis-regulatory variation on neighboring gene expression (Figure 3), genetic imprinting and nonsense-mediated decay (NMD), i.e., the induction of RNA degradation mechanism by variants that generate premature stop codons12. The limiting step in detecting ASE is the presence of heterozygous single nucleotide polymorphisms (SNP) in the exonic regions of genes. One of the advantages of ASE is that by using both alleles as a perfect pair of experimental and control variables, variants can be used even if their population frequencies are low (rare variants). In addition, given that inter-individual variation is not a factor as with eQTL analysis, the number of individuals that need to be included in the analysis is much smaller, making ASE a good tool for fine mapping GWAS causal variants and genes. However, it is important to emphasize that, as with eQTL variants, ASE variants do not necessarily confer causality, as due to linkage disequilibrium another variant in high LD might actually be causal or the allele specific effect may appear due to higher order epigenetic effects, such as imprinting.
Several tools have been developed to obtain allelic counts, and specially designed to address major pitfalls of the allelic analysis12,40,41. The major limitation to ASE data analysis is the reference allele bias42. This bias originates from the preferential mapping of the read that contains an allelic variant present in the reference genome compared to the read that contains the alternative variant. Mapping score will inevitably be higher for the read containing the reference allele, therefore it is expected that false positive ASE events are found in favor of the reference allele. Indeed, computational simulations with over 9.5 M common SNPs and indels (using simulated single-end 50 bp reads) revealed that 15.6% of variants will show reference bias and in some cases the complete loss of mapping to the alternate allele is observed43. The reference bias could be alleviated by constructing a diploid genome, either from parental genomes in case they are available or by incorporating personal variants into the reference genome and phasing heterozygous variants using 1000 Genomes Project reference panels, producing as a result two parental haplotypes. For example, in a recent study, using this approach to remove the reference bias, 63K total SNVs associated with ASE were detected using 382 individuals from the 1000 Genomes Project, with the reproducibility rate of 75%41. Another suggested solution, implemented in the software package WASP44, consists of flipping the detected allele in the read being sequenced and subsequently discarding the read in case the flipped read does not map to the same genomic location. Finally, ASEReadCounter12 has been developed and integrated into the GATK v.3.4 toolkit and it represents a post-mapping tool that utilizes already aligned RNAseq reads to the reference genome. After filtering reads based on mapping and quality control filters, ASEReadCounter finds bi-allelic heterozygous variants and counts the reference and alternative allele reads. Other sources of variation have been described and reviewed elsewhere. Nevertheless, for those genes where an informative exonic variant can be identified, ASE is a highly valuable tool for mapping causal variants and genes in the GWAS loci.
Several other sources of errors exist in the ASE analysis. For example, errors could be introduced by overlapping mates in paired end RNAseq, since heterozygous variants could be counted twice for each read pair, which may introduce biases. In addition, removing low quality reads is necessary to prevent incorrect base calls at heterozygous sites. Furthermore, filtering out duplicate reads, originating from PCR amplification during library preparation, is necessary in case they are prevalent in the data set. The next type of error occurs when the homozygous variant is incorrectly called as heterozygous, which leads to preferential mapping to one allele leading to the false positive ASE signal. Another important bias is called the ambiguous mapping bias, and it occurs when reads containing one allele are preferentially mapped due to the random sequence homology of the reads with the alternative allele to another genomic location. Such reads will be mapped to multiple locations and removed due to mapping ambiguity leading to false positive ASE calls.
Isoform identification and differential isoform expression
As tracing down the precise transcript of origin for sequenced reads could be ambiguous, due to the existence of multi-transcript genes (alternative promoters and splicing), multi-gene families (gene paralogs) and pseudogenes, the precise attribution of reads to transcripts is currently impossible. For example, a read that spans an exon-exon junction common to all isoforms of a gene, could be equally attributed to any isoform. Due to this uncertainty, the Poisson distribution cannot approximate the read distribution, as the Poisson distribution requires non-overlapping genomic intervals, such as genes. Therefore, the count based methods, such as EdgeR that introduce Poisson modeling, only function on the level of genes as a whole, and would fail to model transcripts properly. In the analysis of genes, technical replicates become unnecessary as variance can be estimated from the mean in a Poisson distribution, therefore various methods focus on estimating biological variance by borrowing power across genes to obtain reliable variance estimates. On the other hand, transcript (isoform) analysis requires alternative statistical approaches to model read counts as they can only be estimated and not precisely determined45,46.
DEXSeq47 restricts the analysis to specific exons or exon-exon junctions, however this approach does not provide isoform information, and cannot detect biological phenomena such as isoform switches. However, while the direct transcript of origin of reads may be difficult or impossible to estimate, it is possible to infer a relative number of reads for each isoform, which requires a new statistical approach in differential expression analysis48. Recently, mapping free tools such as Sailfish and Kallisto, as discussed above, have introduced pseudo-alignments that produce a list of transcripts that are compatible with each read while avoiding mapping. These algorithms have brought substantial improvement in the number of (pseudo)aligned reads (86.5% Bowtie2; 90.8% kallisto), as well as in the accuracy in the estimated read count of each transcript, while showing similar performance on gene paralogs as other tools.
Splicing QTLs are estimated to be a major contributor to complex human traits, hence their accurate detection is paramount49,50. However, various technical challenges exist to define both individual exon-exon junctions and complete transcript isoforms using short-read data. Typically, single splice events are easier to study, such as skipping or inclusion of an exon, and exon extension or shortening from alternative 5′ or 3′ splice sites. In contrast to event-centric methods, transcript reconstruction methods commonly suffer from problems of ambiguous isoform calls, i.e. except for the simple scenarios, more than one combination of transcripts can explain the observed distribution of reads. Novel approaches including long read and Hybrid-Seq technologies may facilitate the completion of the full splicing catalogue, revealing complete transcripts. Recently, a tool called LeafCutter defined 42,716 clusters of alternatively excised introns and 5,774 sQTLs at 5% FDR using 372 lymphoblastoid cell line (LCL) RNA-seq samples from gEUVADIS51. LeafCutter defined from 10.8% to 19.3% unannotated alternatively spliced introns from the 14 GTEx tissues.
RNAseq in genetic diagnosis of Mendelian diseases, complex diseases and cancer
In addition to its main application in biomedical research, RNAseq could be effectively used as a complementary diagnostic tool for Mendelian diseases. Current genetic diagnostic tools, whole-exome sequencing (WES), and whole-genome sequencing or (WGS), lack the capacity for functional interpretation of genetic variants. In a recent study, transcriptome sequencing was performed in patients’ skeletal muscle cells in a cohort of 50 patients with rare and undiagnosed muscle disorders and compared to 184 GTEx control samples52. Only RNAseq was able to validate known mutations that alter splice sites and to discover novel splice-altering variants, giving a diagnosis rate of 35%. In addition, a novel de novo mutation in COL6A1 that was not identified in the 1000 Genomes Project was found and noted to introduce a splice-gain event that disrupted the triple helical domain of the encoded protein. This variant was highly recurrent as it was identified in 25% of unresolved patients in a separate cohort of muscular dystrophy patients in which previous genetic analyses were negative. Thus, RNAseq in relevant tissues provides for the identification of causal variants in rare diseases that were unable to be detected with current diagnostic methods, including WES and WGS.
In another study, RNAseq was combined with exome data from 1,812 cancer patients to identify in total ~900 somatic exonic variants that disrupt splicing, therefore implicating aberrant splicing as a mechanism for cancer pathogenicity53. In an allele-specific analysis, 163 somatic exonic variants were identified that promoted intron retention and exon skipping, respectively, providing robust evidence of causality. Allele-specific association, on the other hand, is not sensitive enough to detect splicing disruption that occurs at the intermediate level, where some transcripts will still show normal splicing. Therefore, authors applied ratio-based splicing analysis, where they calculated the ratio of abnormally to normally spliced RNAseq reads using the background distribution of ratios from ~2,500 RNAseq data sets obtained from normal tissues. This resulted in identification of 848 exonic SNVs linked to the five types of abnormal splicing (intron retention, exon skipping, intronic cryptic site activation, exonic cryptic site activation, combinatorial abnormal splicing), with exon skipping and intron retention as the most frequent events (503 and 338, respectively). Interestingly, variants causing intron retention were significantly enriched in tumor suppressor gene sets, including seven variants in TP53 and three in CDKN2A. In addition, a variant in SPSB3, a gene previously not associated with tumor suppression, promoted intron retention in two patients with kidney cancer, indicating the potential of RNAseq as a discovery tool for novel candidate genes in cancer.
The role of RNAseq in determining causal variants in complex diseases and phenotypes has been essential, especially in the context of defining eQTL variants and fine mapping of GWAS variants in order to define the true causal variant(s). In a recent study49, it was shown that splicing QTLs play a central role in the genetics of complex phenotypes, in some cases equal or even larger than that identified for expression QTLs. This study found 2,893 splicing QTLs in Yoruban lymphoblastoid cell lines. Although they exhibited no effect on gene expression levels, they were major genetic contributors to complex traits. For example, there was an enrichment of low p-value sQTLs in multiple sclerosis GWAS loci, and this predominance over eQTLs was robust with different detection cut-offs. A similar trend was observed for height GWAS loci, while eQTLs were dominant in rheumatoid arthritis GWAS loci. The study showed that QTLs identified in LCLs are informative for the estimation of contribution of different regulatory mechanisms to complex phenotypes, an effect that is probably the consequence of sharing of QTLs across tissues and ethnicities.
In coronary artery disease genomics54 (Figure 4A), RNAseq was used to determine eQTLs in ~600 coronary artery disease patients in the STARNET cohort55. RNA was isolated and sequenced from blood, internal mammary artery, atherosclerotic aortic root, subcutaneous fat, visceral abdominal fat, skeletal muscle, and liver with up to 30 million reads per sample. A total of 8 million eQTLs were discovered from the seven tissues, out of which ~4 million were unique SNP-gene pairs. Of the 19,926 genes that were expressed, 76.2% were defined as eGenes containing at least one cis-eQTL, indicating a large regulatory network across tissues. Meta-analysis of CAD GWAS has yielded 54 lead risk SNPs and 38 were cis-eQTLs identified in STARNET. This confirms the central role of gene expression profiling with RNAseq in fine-mapping of GWAS variants and inferring the causal variant.
Figure 4. RNAseq in complex diseases – coronary artery disease.
A) Schematic overview of coronary artery smooth muscle cells and progression to disease state in vivo, as well as modeling disease progression in vitro and RNAseq profiling. B) Fine mapping disease-associated risk SNPs from the GWAS catalog using various RNAseq applications. C) CAD GWAS candidate gene TCF21 overexpression in HCASMC leads to the perturbation of the transcriptome profile that recapitulates disease phenotype in vitro, as shown using gene ontologies. On the right, detection of immediate response genes after the treatment with PDGF that recapitulates similar disease cellular phenotype in vitro. D) Construction of the gene network using WGCNA modules or edge weighted force layout clustering that clustered genes from ~4000 public RNAseq datasets into smooth muscle- (TCF21, PDGFD, SMAD3, etc.) and lipid- (APOA1, APOB, etc.) related clusters. E) An excerpt of the network constructed using cis- and trans- eQTL connections defined with causal inference test in the STARNET data set. Multi tissue eQTL analysis enables the construction of the network of genes as shown with the example of TCF21 and PDGFD genes that are interconnected with the LIPA gene in the disease core network only when incorporating eQTLs from the liver with those from the diseased atherosclerotic aorta.
In the downstream analysis of complex disease variants, RNAseq has played a crucial role in providing evidence as to which variant or gene might be causal, by using a variety of approaches (DE genes, DE isoforms, isoform psiQTL, exon eQTL, exon psiQTL, aseQTL, etc.) (Figure 4B). In addition, downstream knock-out or overexpression studies of GWAS genes have proven beneficial. For instance, using RNAseq in cultured cells, coronary artery disease GWAS candidate gene TCF21 was confirmed to promote pro-migratory and synthetic phenotype of coronary artery smooth muscle cells recapitulating the disease phenotype in vitro56, similar to the treatment of pro-migratory factors such as PDGF (Figure 4C). RNAseq network analysis of CAD GWAS genes showed the existence of two distinct clusters of interconnected genes, one with smooth muscle cell- (TCF21, PDGFD, etc.) and the other one with lipid- related genes (APOA1, APOB, etc.)56 (Figure 4D). In the STARNET study55, the network constructed using cis- and trans- eQTL connections defined with multiple diseased tissues revealed a core network in HCASMC and liver tissues, consisting of TCF21, PDGFD, and LIPA genes (Figure 4E). RNAseq has also been beneficial in profiling differences in mouse disease models (e.g. high fat diet mouse model of CAD) and in knockout mice for the GWAS genes (Figure 3).
Long-read RNAseq as the future of transcriptome analysis
RNAseq using short-read sequencing carries a major limitation in the process of transcriptome reconstruction using fragmented cDNA molecules. As short-read sequencing can only capture the neighboring exon-exon junctions, how distant exon-exon pairs combine in a transcript, whether in a co-associated, mutually exclusive or independent manner (Figure 3), remains undetected. Therefore, studies have shifted towards long-read sequencing technologies (PacBio, Illumina MOLECULO, Oxford Nanopore Technologies, 10X Genomics) in order to capture full length mRNA molecules. By capturing the entire exon-intron structure of mRNA within a single read, even the rarest of isoforms become detectable, therefore revealing the full complexity of the transcriptome. RNAseq has been successfully conducted using both single-molecule long-read PacBio57,58 and synthetic long-read MOLECULO methodologies59. Using PacBio methodology ~25,600 distinct full-length isoforms were identified out of ~14,200 spliced GENCODE genes. PacBio provides high quality full length mRNA up to 1.4kb, while for longer genes the quality of 5′ sequence deteriorates, with their exon-intron structure still being well preserved. Illumina MOLECULO technology involves breaking DNA into 10kb fragments which are then marked with a unique barcode, sheared and sequenced on the standard platform and subsequently reconstructed using the barcode information into synthetic long reads that represent the original RNA molecules. Using MOLECULO, a total of 14.5% of all spliced reads in human brain had combinations of exons that are novel compared to the GENCODE transcript annotation, affecting ~13,800 human genes 59. Similar results were observed in the mouse brain, as 18% of reads showed novel combinations of splice sites affecting 8,600 genes. Oxford Nanopore MinION sequencing technology was successfully used in B1a cells on a transcriptome-wide single cell level to identify 1799 unannotated transcription start sites, 1435 unannotated end sites, 589 5′ splice sites and 536 3′ splice sites that did not match the GENCODE annotation60. The validity of the findings was confirmed with high degree of overlap with CAGE data derived from 5′ ends of transcripts.
PacBio systems, unlike Illumina’s sequencing of a clonal population of fragments, capture a single DNA molecule using polymerase immobilized to the bottom of the well and let the DNA strand pass through. PacBio systems can produce more than half of the reads with lengths greater than 20kb, and a maximum read length of over 60kb, depending on the stability of the polymerase. PacBio systems RSII and Sequel can achieve single pass accuracy of 86% with errors randomly distributed within each read. Therefore, sequencing with 20X and 40X coverage allows accuracy of 99.99% and 99.9999%, respectively, due to ‘consensus accuracy’ and rivals the accuracy of Sanger sequencing. Therefore, achieving the necessary fidelity of the long read systems is tightly correlated to the coverage of the sequenced transcript. Since a single run on the Sequel can produce 500K reads, multiple runs are still needed to achieve sufficient coverage for accurate sequencing of the complete human transcriptome that potentially contains more than 200K transcripts (GENCODE, version 27). This increases the cost burden of long range sequencing. The 10X Genomics emulsion-based system can partition large fragments up to ~100kb and amplify smaller fragments of DNA each with a barcode identifying the original large fragment61. Higher accuracy of 10X Genomics systems (usually coupled with Illumina sequencing) compared to PacBio is met with difficulties in reconstructing the transcripts, lack of the full coverage of a fragment, partially limited by the number of barcodes, and increased cost of the platform.
One of the emerging long-read technologies is Oxford Nanopore Technologies (ONT) with platforms like MinION and PromethION. Similar to PacBio, ONT brings longer read lengths (reaching up to 300kb) and real-time analysis, and in addition carries an advantage of portability of the device, ease of set up, and fast library preparations, making it a perfect choice for field applications. However, it suffers from higher error rates similar to PacBio’s single pass quality, which represents a drawback for applications in transcriptomics. ONT compared to all other technologies directly reads a sequence of the single-stranded DNA (ssDNA) by detecting the changes in electrical current as the native molecule is driven through the nanopore. MinION utilizes a DNA template and its complement connected via a hairpin adapter, hence, the template passes first followed by the adapter and the template, giving rise to ONT 1D and 2D reads (ONT 1D are template and complement reads, whereas the ONT 2D read represents a consensus of the two 1D reads and possesses higher accuracy). Comparative analysis of PacBio and ONT systems revealed that compared to PacBio error rates (14.20% for subreads and 1.72% for circular consensus sequence reads), ONT 1D reads carry higher error rates (20.19%), while ONT 2D is comparable to PacBio subread accuracy (13.40%)62. On the other hand, ONT produces higher yields per flow cell than PacBio, as a single nanopore may process multiple molecules, while the reusability is not an option in PacBio. Various Hybrid-Seq approaches (PacBio+Illumina, ONT+Illumina) show better performance compared to long reads in the transcriptome analyses, which may constitute the future of transcriptomics.
Non-coding RNA discovery
Various noncoding RNAs (ncRNAs) are produced by RNA polymerase II transcription from the mammalian genome (long non-coding RNAs or lncRNA, small regulatory RNAs such as miRNA, nonsense mediated decay transcripts, etc) with important contributions to gene expression and genome maintenance. In GENCODE version 27, out of 200,401 annotated human transcripts there are 27,908 long non-coding transcripts. As the typical lncRNA is biochemically identical to an mRNA, the general methods of identifying lncRNA involve RNA sequencing with either oligo(dT)- selected poly(A) RNAs or using libraries depleted of the rRNAs, and applying several filtering steps to remove artefactual transcripts and protein-coding genes, in order to annotate the remaining transcripts as lncRNAs63. On the other hand, in order to study specific classes of lncRNAs it is paramount to isolate pools that contain lncRNAs of interest. This could be achieved with immunoprecipitation-based approaches, such as RNA immunoprecipitation (RIP), in order to enrich for lncRNAs connected to particular proteins or protein complexes64. In order to eliminate the interactions of lncRNAs and proteins that appear through the DNA mediator, the isolates can be treated with enzymes such as RNase H (to degrade RNA in RNA-DNA complexes) and DNase I to digest DNA. The control library could be made with RNase A that fragments single stranded RNA molecules and RNase V1 that fragments RNA-RNA complexes. After elution of co-immunoprecipated molecules, RNA sequencing could identify lncRNAs that reside within the specific protein complexes. A similar method termed CLIP (cross-linking and immunoprecipitation), uses ultraviolet (UV) irradiation to cross-link RNAs to protein (but not protein to protein) and treatment with RNase to preserve only those RNA regions that are interacting with the protein. RNA-chromatin immunoprecipitation or RNA-ChIP, on the other hand identifies lncRNAs that bind chromatin.
GRO-Seq (Global Run-On sequencing) is a method to assay nascent RNA based on re-initiation of transcription by RNAPol II in vitro with labeled nucleotides65. GRO-Seq is highly efficient in detecting non-polyadenylated non-coding RNAs, such as enhancer RNAs (eRNAs). The number of eRNAs in the human genome was estimated to be from 40,000 to 65,000 using the cap analysis of gene expression (CAGE), which makes up a significant fraction of total transcriptome in the genome. However, eRNAs are still difficult to study due to their very labile nature, low abundance and high turnover rates. A more precise method named Precision nuclear Run-On and sequencing assay (PRO-seq) enables mapping transcriptionally-active Pol II at single base-pair resolution. Non-coding RNAs of low or transient expression such as eRNA could be more efficiently selected using CaptureSeq, which focuses on the RNAs of interest by capturing specific RNAs using pre-designed oligonucleotide probes.
SINGLE CELL RNASEQ
Developmental and disease mechanisms are most often cell type-specific processes. Therefore, bulk RNA sequencing of relevant tissues can be insensitive to cell type-specific changes in transcriptional activity. Even worse, these data can be misleading due to variation in the proportion of cell types present in different samples and confounding effects from other cell types that are active in the process being investigated. Although cell surface markers, and genetic lineage tracing in model systems, can be used to isolate specific populations of cells prior to sequencing, these methods depend on a small number of markers. The result is often imperfect with respect to sensitivity and specificity of selection, leading to isolation of only a subset of the cell type of interest or the inadvertent inclusion of multiple cell types. Recent advances have allowed RNA sequencing of hundreds to thousands of individual cells, overcoming many of these issues and revolutionizing our understanding of cell type-specific processes in vivo within the relevant tissue environment. The first transcriptomic analysis of a single cell was performed in 1988 by Rappolee and colleagues66 using gene-specific PCR, and the first whole-transcriptome amplification of a single cell was achieved in 1990 by Brady and Iscove67. Subsequent improvements in biochemical methods in conjunction with the breakthroughs in next generation sequencing have led to the current revolution in single cell RNAseq (scRNA-seq) which is providing unprecedented resolution to the study of complex developmental and human disease processes.
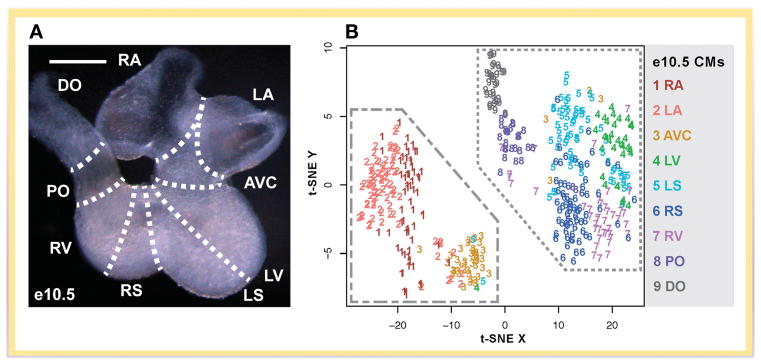
Thus far, single cell methodologies have had limited but demonstrative application to cardiovascular research. In 2016, two groups independently used scRNA-seq in anatomically defined areas of the developing mouse heart to create a single cell atlas of all major cardiac cell types during different stages of cardiac development68,69 (Fig 5A). From e8.5-e10.5 embryonic mouse hearts, Li et. al. used a random forest machine learning algorithm to predict the anatomical location of individual cardiomyocytes with >91% accuracy based upon their transcriptional profiles (Fig. 5B). This algorithm also showed good performance in predicting the localization of Isl-1 lineage-traced cells, which are known to specifically populate the right ventricle and outflow tract. DeLaughter et. al. characterized the transcriptomic profiles of anatomically separated cells at various time points from e9.5 to post-natal day 21. They identified the emergence of cardiac fibroblasts at the expected time point around e12.5, and catalogued the gene expression programs of cardiomyocytes (CMs) from development to maturity. They also used these developmental stage-specific transcriptional profiles to estimate the maturity of CMs derived from stem cells69. Both groups analyzed the effect of Nkx2-5 deficiency on CM gene expression and found defects in CM maturation. Further, Li et. al. found that Nkx2-5−/− CMs more closely resembled atrial rather than ventricular CMs68. Both papers illustrate the enormous potential of single-cell transcriptional profiling to identify cell type-specific defects in animal models of gene knockout. Likewise, the use of single-cell RNAseq in human tissue samples will allow investigation of cell type-specific defects in patients with pathogenic mutations, and reveal cell-type specific changes during various cardiovascular disease processes.
Figure 5. Cell atlas of the developing heart.
A) Embryonic day (e10.5) heart showing anatomical locations from which cells were isolated. B) t-SNE plot of cardiomyocytes from the e10.5 heart. Cardiomyocytes from different locations can be clearly distinguished by their transcriptional signatures.
Cell capture methods
All high-throughput methods for scRNA-seq rely on a suspension of viable single cells. While this is straightforward when dealing with cultured cells, obtaining a high-quality single cell suspension via enzymatic dissociation of a primary tissue is often challenging. Incomplete dissociation can bias the population towards cells that more readily detach from their microenvironment, whereas overly-aggressive dissociation can potentially alter the cells’ transcriptional profile and affect cell viability. Further, as the dissociation protocol can vary greatly depending on the organism and the tissue of interest, this must be carefully optimized for each scenario. Recent advances enabling RNA-seq of single nuclei (snRNA-seq)70 and incorporation of these methods into existing71,72 and novel73 high-throughput cell capture technologies are now allowing analysis of single-cell transcription in fresh frozen and possibly fixed tissue samples. Although the majority of reads arising from snRNA-seq are intronic, analysis of these unprocessed nuclear RNAs has been shown to correlate well with mature gene expression72–74.
The first scRNA-seq experiments relied on manual picking of small numbers of individual cells into separate tubes and/or wells75–77. Reverse transcription and amplification of cDNA was then carried out in microliter reaction volumes. The throughput for these plate-based methods was increased dramatically by sorting individual cells into 96- or 384-well plates followed by robotic automation of subsequent pipetting steps78,79. An alternative strategy was developed by the Quake group80, in which a suspension of single cells is loaded into a valve-based microfluidic chip. These microfluidic valves are then used to direct single cells into hydrodynamic capture sites and also to control the flow of reagents into individual nano-liter-sized cell reaction chambers. Cell lysis, reverse transcription, and PCR are automated on the chip. This confers a number of advantages, including simplifying the workflow and reducing reagent costs. In addition, the nano-liter reaction volume results in decreased amplification bias81 and increased sensitivity81,82 compared to the same chemistry conducted on a micro-liter scale. However, the capture of cells is dependent on cell size, requiring different microfluidics chips for cell of different sizes. In a biological sample containing cells of various sizes, this risks biasing cell capture to specific subsets of cells. Finally, although this method reduces reagent costs, the microfluidics chips are expensive. Other microfluidic approaches allow the culturing of single cells within capture sites, with resulting cell progeny being captured by downstream cell traps, and therefore allow tracking of transcriptional changes over multiple lineage generations83.
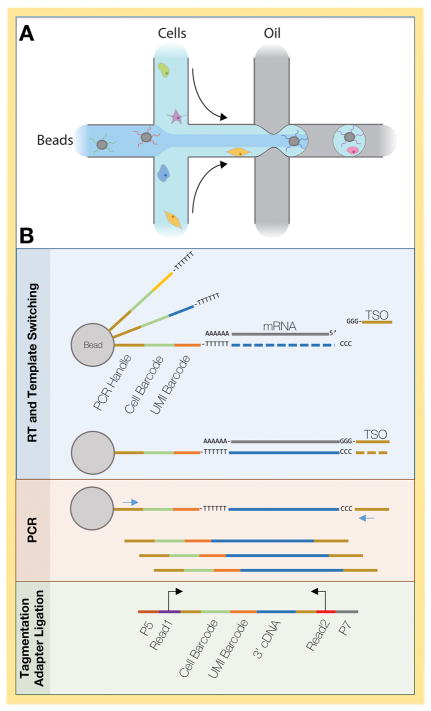
The application of droplet-based microfluidics to isolate single cells and capture their mRNA with cell- and molecule-specific primers now allows the routine processing of tens of thousands of cells in a single experiment. In these methods, two aqueous solutions, one containing a suspension of single cells and the other containing a lysis solution with oligo-dT primers affixed to a bead or hydrogel, are combined via separate microfluidics channels and then immediately “chopped” into individual droplets by orthogonal streams of an oil solution (Fig. 6A). Every primer on an individual bead or hydrogel contains the same cell barcode, but each primer also contains an additional unique barcode (unique molecular identifier, UMI, Fig. 6B). Thus, when a cell and primer-coated bead/hydrogel are encapsulated within the same droplet, the cell is lysed and its polyadenlyated RNA is captured by the oligo-dT primers. Reverse transcription then incorporates the cell and UMI barcodes from each primer into the resulting cDNA strands. In this way, all captured mRNA molecules from a given cell will contain the same cell-specific barcode, but each mRNA molecule will contain its own UMI barcode. The aqueous-oil emulsion is then broken, allowing library preparation in bulk solution. This technology was developed independently by the McCarroll and Kirschner labs in 2015, with the names Drop-seq and inDrop, respectively. These techniques require the purchase of specialized equipment, but the setup and consumable costs are modest, especially compared to commercially-available systems. The primary drawback of these techniques is that only the 3′ end of the mRNA is sequenced, making them unsuitable for studies of alternative splicing or allelic expression imbalance. While this method is typically limited to capturing tens of thousands of cells per experiment, a dataset of 1.3 million single cells from two embryonic mouse brains was recently accumulated by repeating cell capture many times75. However, this data set was generated by 10X Genomics, a commercial provider of the inDrop technology, and would currently be prohibitively expensive for the typical lab to generate.
Figure 6. Drop-seq microfluidics and chemistry.
A) Schematic of cell and bead partitioning into aqueous droplets, adapted from Mocosko, et. al.10. To avoid cell/bead doublets, cell and bead concentrations are adjusted so that only a small minority of droplets contain both a bead and a cell. B) Each bead-based oligo-dT primer contains the same cell barcode but different unique molecular identifier (UMI) barcodes. A template switching oligo (TSO) introduces a PCR handle, allowing cDNA amplification. Only the 3′ end of each cDNA molecule is incorporated during library construction. Barcodes and transcript sequences are associated via paired-end sequencing.
In 2017, two novel barcoding strategies were developed that have the potential to increase throughput to hundreds of thousands and even millions of single cells in a single experiment using widely-available laboratory tools73,84. These methods were developed independently by the Shendure and Seelig laboratories, and are named sci-RNA-seq and SPLiT-seq, respectively. A major advance integral to both techniques is the ability to perform reverse transcription and barcoding in situ in a suspension of fixed, permeabilized cells, essentially using each cell as its own reaction chamber. In this method, cells are seeded into 96- or 384-well plates, with each well containing a unique barcoded primer for reverse transcription (RT). After RT, cells are pooled together and split again into another 96-or 384-well plate with different well-specific barcodes. These techniques can be scaled up via multiple rounds of ‘split-pooled’ barcoding to allow the generation of hundreds of thousands to millions of uniquely barcoded single cell cDNA libraries in a single experiment, considerably reducing the cost of per-cell library preparation73,84. The authors used sci-RNA-seq to characterize the transcriptomes of 42,035 single cells from C. elegans at the L2 stage, effectively analyzing the single-cell transcriptional landscape of an entire organism. Likewise, SPLiT-seq was used to sequence the transcriptome of 109,069 single cells from an entire postnatal day 5 mouse brain, characterizing the transcriptome of an entire organ at single-cell resolution. Another attractive feature of these methods is that they use only readily available laboratory equipment and off-the-shelf reagents, which could help bring single-cell capability to the average laboratory.
In a vastly different approach, George Church and colleagues created a method to perform RNAseq, potentially from single cells, in situ in tissue sections85. Unlike the methods above, this approach preserves information regarding the cells’ location within the tissue environment. In this technique, cDNA produced by in situ RT is cross-linked and circularized, followed by rolling circle amplification and another cross-linking step. The amplified cDNA is then sequenced via the incorporation of fluorescently-labeled sequencing probes (SOLiD sequencing chemistry) using sophisticated microscopy and image processing techniques.
Single-cell chemistries
A number of different chemistries for generating amplified cDNA libraries have been developed over the past eight years. These chemistries differ most fundamentally in three key aspects: i) transcript coverage, ii) the method of cDNA amplification, and iii) barcoding strategy (Table 1). Most chemistries for single-cell RNAseq rely on oligo-dT based primers to capture cellular mRNA, except for SUPeR-seq87 and MATQ-seq88, which use random RT primers with a fixed anchor sequence and can therefore capture non-polyadenylated transcripts. Interestingly, one group found that long non-coding RNAs were also enriched with the use of single-nucleus RNAseq using an olido-dT based chemistry72.
Table 1.
| Cell Capture (rxn volume) |
Chemistry | High-Throughput Application |
Amplification | Barcoding Strategy | Transcript Coverage |
UMI | Spike-ins | Library Preparation |
Year |
|---|---|---|---|---|---|---|---|---|---|
| Plates(uL) or Valve-based Microfluidics (nL) | RT-PCR | Biomark Platform | PCR | N/A | Primer-specific | N/A | N/A | N/A | |
| STRT-seq75 | N/A | Single-primer PCR | Well-specific barcode added via template switching during RT | 5′ and 3′ | Yes | Yes | Pooled | 2011 | |
| CEL-seq86 | MARS-seq78 | in vitro transcription | Well-specific barcode incorporated into oligo(dT) primer | 3′ end | Yes | Yes | Pooled | 2014 | |
| CEL-seq276 | SORT-seq79 | in vitro transcription | Well- and UMI-specific barcodes incorporated into oligo(dT) primer | 3′ end | Yes | Yes | Pooled | 2016 | |
| Smart-seq277/SMARTer | Fluidigm C1 | PCR | Illumina indexes added during library preparation | Full-length | No | Yes | Individual | 2014 | |
| SUPeR-seq87 | N/A | PCR | Illumina indexes can be added during library preparation | Full-length | No | Yes | Individual | 2015 | |
| MATQ-seq88 | N/A | PCR | UMI added during second strand synthesis, Illumina indexes can be added during library preparation | Full-length | Yes | Yes | Individual | 2017 | |
| Droplet-based Microfluidics (nL) | Macosko10 | Drop-seq10 | PCR | Beads with cell- and UMI-barcoded oligo(dT) primers | 3′ end | Yes | Possible | Pooled | 2015 |
| CEL-seq86 | InDrop89 | in vitro transcription | Hydrogels with cell- and UMI-barcoded oligo(dT) primers | 3′ end | Yes | Possible | Pooled | 2015 | |
| Nano-wells (nL) | STRT-seq-2i90 | Wafergen Nanodispenser | PCR | UMI added during RT, cell indexes added during PCR and during tagmentation | 5′ end | Yes | Possible | Semi-pooled | 2017 |
| Pico-wells (pL) | Macosko10 | SeqWell91 | PCR | Beads with cell- and UMI-barcoded oligo(dT) primers | 3′ end | Yes | Possible | Pooled | 2017 |
| Plates(uL) | SPLiT-seq84 | SPLiT-seq84 | PCR | In situ barcoding at RT step via split-pooling | 3′ end | Yes | No | Pooled | 2017 |
| sci-RNA-seq73 | sci-RNA-seq73 | PCR | In situ barcoding during RT and PCR via split-pooling | 3′ end | Yes | No | Semi-pooled | 2017 |
While the majority of chemistries ultimately incorporate only the 3′ end of each mRNA into the final library, the STRT-seq75 and Smart-seq92 protocols and their derivatives use template switching during RT to incorporate a PCR “handle” after encountering the 5′ end of the mRNA molecule. In STRT-seq, this produces both 5′ and 3′ bias75, but in Smart-seq, long-range PCR is used to amplify the full-length cDNA molecule resulting in more even coverage across the transcript77,92, albeit still with a 3′ end bias. The chemistry used in Drop-seq also uses 5′ template switching during cDNA amplification, but selects for the 3′ end during library preparation. The SUPeR-seq87 and MATQ-seq88 chemistries also lead to greater coverage of the entire transcript and allow discovery of non-polyadenylated transcripts. The remaining chemistries produce libraries covering the 3′ end of the mRNA molecule, which can only be used to obtain gene counts. However, one advantage of limiting coverage to the 3′ end is that fewer sequencing reads are used on a given cDNA molecule.
Two primary methods of cDNA amplification (in vitro transcription and PCR) are used, each with distinct advantages and disadvantages. In vitro transcription (IVT, used in CEL-seq86 and CEL-seq276) is a linear amplification process, and results in fewer non-specific byproducts than does PCR amplification (used in STRT-seq75, Smart-seq92, Macosko10, STRT-seq-2i90, SPLiT-seq84 and sci-RNA-seq73). In addition, IVT is highly strand-specific, which allows accurate gene calling for areas in which different genes occupy the same location on sense and anti-sense strands. However, IVT is not able to achieve the same level of robust amplification inherent in an exponential PCR amplification reaction, and has a strong 3′ end bias. Despite the differences between these methods, a recent comparison between different single cell RNAseq chemistries showed little difference in sensitivity or accuracy of IVT-based methods compared to PCR-based methods, when performed using similar cell capture strategies82. Instead, these methods appeared to be more dependent on reaction volume, with the smaller nanoliter volumes conferring a significant benefit in sensitivity. For example, at a sequencing depth of 1 million reads per cell, CEL-seq2 has an estimated detection limit of 2 mRNA molecules per cell when performed at a nanoliter scale but 13 mRNA molecules when performed at a microliter scale82.
In general, most techniques incorporate cell and/or UMI barcoding during the RT step. With this strategy, each cell is exposed to a set of oligo-dT primers that contain the same cellular barcode. However, each primer within this set can also contain an additional unique 6–10 nucleotide barcode (unique molecular identifier, UMI). UMI barcodes are included to combat biases during PCR amplification; during analysis, all reads with the same UMI (i.e. PCR replicates) are collapsed into a single gene count. In most plate-based cell capture strategies, different cell/UMI barcodes are simply added to specific wells of each plate. In droplet-based microfluidic approaches, however, barcoded oligo-dT primers are affixed to a substrate (bead or hydrogel) to allow their segregation into droplets. Cellular barcodes can be added after cDNA generation (Smart-seq92/SMARTer), but this requires each cell’s cDNA to be processed separately. The application of unique cellular barcodes to each cell’s cDNA is the fundamental determinant of a method’s scalability - the random incorporation of any of a large number of possible barcodes (as in Drop-seq10 and inDrop89) or multiple rounds of randomly-incorporated barcodes (as in sci-RNA-seq73 and SPLiT-seq84) allow these techniques to surpass the scale of their predecessors by orders of magnitude.
The power to detect lowly-expressed genes is dependent on i) mRNA capture rate achieved with each cell capture/chemistry combination and ii) sequencing depth. The mRNA capture efficiency (i.e. the fraction of cellular mRNA that undergoes RT and barcoding) varies between ~10–50% for current techniques10,77,93,94. As noted above, smaller nanoliter reaction volumes have been shown to achieve greater sensitivity81,82, with valve-based microfluidics and their paired chemistries currently possessing the best sensitivity82. Sensitivity is also strongly dependent on the sequencing depth per cell82. However, sequencing to near-saturation (~1 million reads/cell)82 quickly becomes cost-limiting as the number of cells increases. Therefore, until the costs of sequencing decrease substantially, the higher-throughput methods will inevitably result in lower sequencing depth per cell and thus lower sensitivity.
Ultimately, the choice of cell capture method, chemistry, and depth of sequencing will be determined by the experimental question. Users wishing to survey the transcriptional landscape of a whole tissue or those interested in identifying rare cell types will choose higher-throughput techniques, whereas those who would like to study more subtle effects on gene expression in a readily-defined subtype of cells may choose lower-throughput methods at nanoliter scale with a near-saturation sequencing depth.
Single-cell RNAseq data analysis
Due to the large number of cells and genes captured by scRNA-seq techniques, the resulting datasets can confer great power to downstream analyses. However, it is important to recall that these data are derived from an extremely small amount of starting material (often <10pg RNA per cell) that has undergone many rounds of amplification in order to be detected. With the very low amounts of mRNA present in a single cell and relatively low mRNA capture efficiency, detection of transcripts in a given cell is inherently stochastic, particularly for genes with low expression levels. This leads to a high number of so-called ‘dropout’ events, in which a gene is expressed in a cell but is not detected, leading to a zero value. In addition, the large number of PCR cycles required to detect the initial cDNA molecules can also introduce significant biases based upon differential PCR efficiency for different transcripts and in different cells. Variation in library construction and sequencing depth between cells can also add to this technical noise. Layered on this is additional biological noise, which can come from differences in individual cell cycle state95 or the bursting kinetics of gene expression96. Thus, a number of novel analysis strategies have been developed to model these sources of noise in scRNA-seq data, bringing out clearer and more reliable signals from these data. Experimental methods to reduce technical noise are discussed first, followed by in silico normalization and noise modeling strategies.
Spike-ins, UMIs and batch effects
The addition of a known amount of RNA spike-ins to each cell’s lysis environment (usually 1–5% of expected endogenous mRNA molecules86) can help control for the effect of between-cell variability in lysis, RT, PCR, library preparation and sequencing depth. This is most commonly performed using a synthetic mixture of 92 unique poly-adenylated RNAs developed by the External RNA Controls Consortium (ERCC)97. Because the transcripts in the ERCC mix span a concentration range of 106, they can also be used to determine the relative levels of endogenous transcripts. As these spike-in RNAs are relatively short (250–2000nt), have short (20nt) poly-A tails and are not capped at the 5′ end, they can lead to under-estimates of transcript abundance by as much as an order of magnitude82 and therefore cannot be used for absolute quantification. The optimal amount of spike-in needs to be determined beforehand for each cell population under examination to ensure that they are detected but do not overwhelm the endogenous mRNA signal.
The incorporation of unique molecular identifiers (UMIs) is another widely-used method to reduce technical noise from biases in PCR amplification and sequencing depth. Compared to simply counting the number of reads mapping to a given gene, which includes many PCR replicates, collapsing all reads containing a given UMI (and thus derived from the same cDNA molecule) into a single read count can reduce technical noise by approximately two-fold93. UMI counting can in theory also be used for quantification, but this requires each transcript be counted at least once and thus requires each cell be sequenced to saturation98. Importantly, UMIs can be used together with exogenous RNA spike-ins to account for between-cell variability in the experimental protocol. Because the same number of spike-in molecules are added to each cell’s reaction environment, and each captured spike-in molecule is tagged with a UMI, it is possible to create a relative scaling factor for each cell based upon the cell’s average spike in UMI count. This scaling factor can help normalize for technical variation in most steps in the experimental workflow, including relative RT efficiency. Many of these considerations are also nicely reviewed by Grun, et. al99 and Stegle, et. al100.
Batch effects can be confounding when two different experimental conditions are processed completely separately, leading to uncertainty regarding whether differences observed between the two conditions are the result of an important biological property or simply result from the differences in sample processing. To avoid confounding batch effects in scRNA-seq experiments, the goal is to merge the different experimental conditions at the earliest possible stage in sample processing. In the most ideal situation, different experimental conditions would be identifiable on the transcriptional level (i.e. a strongly-expressed transgene present in one sample but not the other) such that these samples could be pooled prior to cell capture. Alternatively, using plate-based technologies, cells from different experimental conditions could be sorted into known subsets of wells, allowing the two groups to be identified later. However, as these ideal conditions are most often not met, cell capture, lysis, RT, and library preparation occur separately for different experimental conditions. In these cases, it is common practice for libraries to be pooled together and sequenced on the same sequencing lane(s) to avoid complete confounding. If feasible, inclusion of multiple biological replicates can help minimize the effects of confounding.
Normalization and cell modeling to reduce technical noise
Because of between-cell differences in total RNA content, cell lysis, capture efficiency, PCR efficiency and sequence depth, data should be normalized prior to analysis. Normalization techniques from bulk RNA-seq analysis can be used to account for these effects, such as library size normalization, upper quartile normalization or normalization to a reference ‘cell’ comprised of averaged gene values101. However, the low amount of starting material and significant amplification required in scRNA-seq experiments lead to sparse and potentially biased datasets that can pose challenges for these normalization methods98. Thus, normalization strategies have been designed specifically for scRNA-seq analyses. One approach, scran102, pools groups of cells together within the broader population to perform normalization, and then deconvolves the pooled scaling factors to estimate cell-specific scaling factors. SCNorm103 uses quantile regression to account for the effect of sequencing depth on transcript expression, and then uses this information to estimate gene-specific scaling factors. Because of the 3′ end bias in scRNA-seq technologies, adjusting for transcript length is problematic and should be avoided.
Once the data have been normalized, the next step is to identify highly-variable genes (HVGs). This is critical for two reasons: i) HVGs, by their nature, are the most likely genes that will distinguish different cell types from one another within the population, and ii) reduction in the number of genes tested generally increases the power of downstream analyses104. However, identifying true HVGs in scRNA-seq datasets can be confounded by significant technical noise. For instance, the stochastic nature of transcript ‘capture’ or sampling in scRNA-seq becomes increasingly evident as the mean expression level of a gene decreases, leading to disproportionate (technical) variance for genes with low to moderate expression levels. As a result, variance in a gene’s expression is highly dependent on expression level (Fig. 7B). Therefore, to determine whether a given gene is a true HVG, its variance is often compared to the observed variance of other genes at a similar level of expression (Fig. 7B, pink dots)104,105. Alternatively, the PAGODA package models a gene’s expression across cells as a mixture of two probabilistic processes: i) successful amplification and ii) drop-out106. This model is then applied to each individual cell to determine the probability that a given gene is expressed at a given level (including the probability of a zero value representing a drop-out event). The significance of a gene’s variance is then evaluated against the expected variance at similar expression levels107. Additional methods for modeling technical108 and unwanted biological95,108 variation such as cell cycle state are also available.
Figure 7. General workflow for scRNA-seq analysis.
A) Starting with a cell-gene readcount matrix, data are normalized and low-quality cells are excluded. B) Highly-variable genes (HVGs, pink dots) are identified after accounting for the relationship between expression level and variance (squared coefficient of variation, CV2). Figure obtained with permission from Brennecke, et. al105. C) HVGs are used in a dimensionality reduction method such as principal component analysis (PCA), and cells are evaluated based upon their principal component (PC) dimensions. D) Cells are assigned to clusters in PC space or in gene expression space. E) Downstream analyses include the identification of cell-specific markers (left), cell type-specific transcriptional changes in response to experimental manipulations (middle), and the inference of possible lineage relationships (right).
Detection of Cell Types
A major goal of scRNA-seq analysis is the identification and characterization of distinct cell sub-types within a heterogenous cell population. Once HVGs are identified, these genes are used for unsupervised dimensionality reduction and clustering to identify distinct cell types. Dimensionality reduction methods used in scRNA-seq analyses include principal component analysis (PCA, Fig. 7C)104,109, independent component analysis (ICA)110, t-distributed stochastic neighbor embedding (t-SNE, Fig. 7D)104,111, and gene co-expression analysis112. A cellular distance matrix computed either in gene expression or in dimension-reduced space can then be used to perform unsupervised clustering, which is a general machine learning problem that groups similar cells together in clusters while also distinguishing distinct clusters from one another. Common approaches used in scRNA-seq packages include hierarchical clustering107, K-nearest neighbor graph-based clustering104,113, and k-means clustering109,114,115. The results of these clustering methods can be visualized directly via dendogram or heatmap, or can be subjected to further non-linear dimensionality reduction and visualization, such as with t-SNE (Figs. 5B and 7D)104. Alternatively, other approaches perform clustering directly on a cellular distance matrix without prior dimensionality reduction113–116.
Differential Expression Analyses
A common goal in RNA-seq experiments is to determine differential expression between two samples that have been subjected to different experimental perturbations. In bulk RNA-seq experiments of complex tissues, differential expression analyses can be confounded by differences in cell composition between samples. For example, in a mouse model of atherosclerosis, aortic samples taken from different mice may contain significantly different amounts of disease. Thus, gene expression changes between samples may result more from differential disease burden than the experimental perturbation. This can be partially corrected by increasing the number of biological replicates in each experimental condition, but still remains a significant confounding factor in bulk RNA-seq experiments.
A key advantage of scRNA-seq, especially in complex tissues, is the ability to restrict differential expression analyses to specific cell types within the broader population. This allows identification of the specific cell type(s) affected by the experimental perturbation. And, because signals are not averaged across multiple cell types, this type of analysis is i) potentially more sensitive for detecting cell type-specific gene expression changes and ii) much more resistant to confounding by differences in tissue composition. Evaluation of differential expression is a key component in most available single cell analysis packages.
Lineage inference using pseudotemporal ordering
An important goal in understanding many developmental and disease processes is to determine how cells adapt or differentiate over time at a transcriptional level. This requires the ability to link cells in different stages of differentiation together to infer lineage relationships. Traditionally, this has also required the assessment of cells at multiple time points, which can be laborious and expensive to carry out, especially at a single cell level. To address these challenges, several groups have developed algorithms to infer lineage relationships between single cells using a large number of cells at a single time point78,117–119. These methods are based upon the concept that, at the time the cell population is captured, there are likely to be cells in various stages along a lineage path towards differentiation and/or cell adaptation. Two of these approaches, Monocle117 and Waterfall118, create an inferred lineage path through dimension-reduced space using a minimum spanning tree approach. Another approach uses a diffusion map for dimensionality reduction and then computes a distance measure (diffusion pseudotime) between each pair of cells on the map to allow inference of lineage relationships120. Bacher, et. al. also nicely review these techniques in greater depth121. For the frequent scenarios in which traditional fluorescence-based lineage tracing systems are not available, these techniques have significant potential to reveal important novel relationships between cell types.
SUMMARY
In this review, we have summarized the major applications of RNAseq, currently the most robust and generally useful method of transcriptional profiling. RNA sequencing has emerged as one of the essential methodologies in contemporary biological and medical sciences. The relative simplicity of experimental RNA extraction and facilitated library preparation have made possible scaling up to hundreds or even thousands of samples. The future of RNAseq development includes migration towards single cell experiments, which parallels an analog to digital transition in deconstructing the average signal in gene expression of a cell population. On the other hand, compared to bulk RNA sequencing, single cell sequencing comes with several challenges including the expense of commercial library preparation kits and sequencing, as well as an elevated computational burden for analysis. However, this trend may be only temporary and it is reasonable to expect a decrease in per-cell library costs and more efficient computing with newer methods. In addition, with increasing read length, RNAseq is expected to fully sequence complete transcripts in the future and to decipher isoform content without the need of reconstruction using shorter read lengths. Once the sequencing depth increases substantially and reaches saturation levels, it should be expected to migrate isoform analysis to the single cell level, without the need of imputation of missing values. By detailing the transcriptome in each cell at the deepest level, it would be possible to fully unravel the complexity of cell populations and tissues, resolving even the most subtle of cell state changes. In a relatively short period of scientific and technological improvement, RNAseq profiling has provided a blueprint of cellular identity. Further improvement of this technology will make it possible to catalogue the complete transcript collection for each individual cell in a tissue or organism.
The future of next generation sequencing technologies will inherently bring the improvement in read length and accuracy and decrease in cost of long read technologies such as PacBio and Oxford Nanopore Technologies, which will ultimately eliminate the need for assembling genomes and transcriptomes using short reads or hybrid technologies. For the moment, short read technologies remain more accessible to wider scientific community due to their lower cost, which will in the near future generate many novel advances in algorithmic approaches in genome/transcriptome reconstruction. If cardiovascular research closely follows these advances, this field will inevitably benefit in terms of defining various molecular mechanisms responsible for the genetic modulation of disease.
Acknowledgments
SOURCES OF FUNDING
NIH F32HL129670 (R.C.W)
NIH R01HL109512, R01HL134817, R33HL120757, R01DK107437 and R01HL139478 (T.Q.)
NON-STANDARD ABBREVIATIONS AND ACRONYMS
- ASE
allele specific expression
- CAGE
cap analysis of gene expression
- CEL-seq
cell expression by linear amplification and sequencing
- CLIP
cross-linking and immunoprecipitation
- CMs
cardiomyocytes
- DE
differential expression
- DGE
differential gene expression
- DTE
differential transcript expression
- DTU
differential transcript usage
- eQTL
expression quantitative trait loci
- ERCC
External RNA Controls Consortium
- eRNAs
enhancer RNAs
- exon psiQTL
exon percent spliced in QTL
- FC
fold-change
- FPR
false positive rate
- GRO-Seq
Global Run-On sequencing
- GSEA
Gene Set Enrichment Analysis
- GTEx
Genotype-Tissue Expression
- HVG
highly-variable gene
- ICA
independent component analysis
- LCL
lymphoblastoid cell line
- lncRNA
long non-coding RNA
- MATQ-seq
multiple annealing and dC-tailing-based quantitative single-cell RNA-seq
- miRNA
microRNA
- MMP
Maximal Mappable Prefix
- ncRNAs
noncoding RNAs
- NMD
nonsense-mediated decay
- nt
nucleotides
- ONT
Oxford Nanopore Technologies
- PAGODA
pathway and gene overdispersion analysis
- PCA
principal component analysis
- PRO-seq
Precision nuclear Run-On and sequencing assay
- QC
quality control
- RIP
RNA immunoprecipitation
- RT
reverse transcription
- SAGE
serial analysis of gene expression
- sci-RNA-seq
single cell combinatorial indexing RNA sequencing
- scRNA-seq
single cell RNA sequencing
- SMARTer
switching mechanism at 5′ end of RNA transcript
- SNP
single nucleotide polymorphisms
- snRNA
small nucleolar RNA
- snRNA-seq
single nucleus RNA sequencing
- SPLiT-seq
split pool ligation-based transcriptome sequencing
- STRT-seq
single cell tagged reverse transcription sequencing
- SUPeR-seq
single-cell universal poly(A)-independent RNA sequencing
- TPR
true positive rate
- t-SNE
t-distributed stochastic neighbor embedding
- UMI
unique molecular identifier
- WES
whole-exome sequencing
- WGS
whole-genome sequencing
Footnotes
DISCLOSURES
None
REFERENCES CITED
- 1.Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, Szcześniak MW, Gaffney DJ, Elo LL, Zhang X, Mortazavi A. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016;17:13. doi: 10.1186/s13059-016-0881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bähler J. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- 5.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun L, Zhang Z, Bailey TL, Perkins AC, Tallack MR, Xu Z, Liu H. Prediction of novel long non-coding RNAs based on RNA-Seq data of mouse Klf1 knockout study. BMC Bioinformatics. 2012;13:331. doi: 10.1186/1471-2105-13-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar S, Vo AD, Qin F, Li H. Comparative assessment of methods for the fusion transcripts detection from RNA-Seq data. Sci Rep. 2016;6:21597. doi: 10.1038/srep21597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, Teichmann SA. The Technology and Biology of Single-Cell RNA Sequencing. Mol Cell. 2015;58:610–620. doi: 10.1016/j.molcel.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 9.Gawad C, Koh W, Quake SR. Single-cell genome sequencing: current state of the science. Nat Rev Genet. 2016;17:175–188. doi: 10.1038/nrg.2015.16. [DOI] [PubMed] [Google Scholar]
- 10.Macosko EZ, Basu A, Satija R, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sultan M, Amstislavskiy V, Risch T, Schuette M, Dökel S, Ralser M, Balzereit D, Lehrach H, Yaspo M-L. Influence of RNA extraction methods and library selection schemes on RNA-seq data. BMC Genomics. 2014;15:675. doi: 10.1186/1471-2164-15-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castel SE, Levy-Moonshine A, Mohammadi P, Banks E, Lappalainen T. Tools and best practices for data processing in allelic expression analysis. Genome Biol. 2015;16:195. doi: 10.1186/s13059-015-0762-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeLuca DS, Levin JZ, Sivachenko A, Fennell T, Nazaire M-D, Williams C, Reich M, Winckler W, Getz G. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinforma Oxf Engl. 2012;28:1530–1532. doi: 10.1093/bioinformatics/bts196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gudlaugsdottir S, Boswell DR, Wood GR, Ma J. Exon size distribution and the origin of introns. Genetica. 2007;131:299–306. doi: 10.1007/s10709-007-9139-4. [DOI] [PubMed] [Google Scholar]
- 15.Sakharkar MK, Chow VTK, Kangueane P. Distributions of exons and introns in the human genome. In Silico Biol. 2004;4:387–393. [PubMed] [Google Scholar]
- 16.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langmead B. Aligning short sequencing reads with Bowtie. Curr Protoc Bioinforma Ed Board Andreas Baxevanis Al. 2010 doi: 10.1002/0471250953.bi1107s32. CHAPTER:Unit-11.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, He X, Mieczkowski P, Grimm SA, Perou CM, MacLeod JN, Chiang DY, Prins JF, Liu J. MapSplice: Accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010;38:e178–e178. doi: 10.1093/nar/gkq622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marco-Sola S, Sammeth M, Guigó R, Ribeca P. The GEM mapper: fast, accurate and versatile alignment by filtration. Nat Methods. 2012;9:1185–1188. doi: 10.1038/nmeth.2221. [DOI] [PubMed] [Google Scholar]
- 21.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schulz MH, Zerbino DR, Vingron M, Birney E. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics. 2012;28:1086–1092. doi: 10.1093/bioinformatics/bts094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grabherr MG, Haas BJ, Yassour M, et al. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general-purpose program for assigning sequence reads to genomic features. Bioinformatics. 2013:btt656. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 27.Xu G, Deng N, Zhao Z, Judeh T, Flemington E, Zhu D. SAMMate: a GUI tool for processing short read alignments in SAM/BAM format. Source Code Biol Med. 2011;6:2. doi: 10.1186/1751-0473-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anders S, Pyl PT, Huber W. HTSeq – A Python framework to work with high-throughput sequencing data. Bioinformatics. 2014:btu638. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patro R, Mount SM, Kingsford C. Sailfish enables alignment-free isoform quantification from RNA-seq reads using lightweight algorithms. Nat Biotechnol. 2014;32:462–464. doi: 10.1038/nbt.2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 31.Pimentel H, Bray NL, Puente S, Melsted P, Pachter L. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods. 2017;14:687–690. doi: 10.1038/nmeth.4324. [DOI] [PubMed] [Google Scholar]
- 32.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schurch NJ, Schofield P, Gierliński M, Cole C, Sherstnev A, Singh V, Wrobel N, Gharbi K, Simpson GG, Owen-Hughes T, Blaxter M, Barton GJ. How many biological replicates are needed in an RNA-seq experiment and which differential expression tool should you use? RNA. 2016;22:839–851. doi: 10.1261/rna.053959.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seyednasrollah F, Laiho A, Elo LL. Comparison of software packages for detecting differential expression in RNA-seq studies. Brief Bioinform. 2015;16:59–70. doi: 10.1093/bib/bbt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fang Z, Martin J, Wang Z. Statistical methods for identifying differentially expressed genes in RNA-Seq experiments. Cell Biosci. 2012;2:26. doi: 10.1186/2045-3701-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pachter L. Models for transcript quantification from RNA-Seq. ArXiv11043889 Q-Bio Stat [Internet] 2011 Available from: http://arxiv.org/abs/1104.3889.
- 38.Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research. 2015;4:1521. doi: 10.12688/f1000research.7563.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams CR, Baccarella A, Parrish JZ, Kim CC. Empirical assessment of analysis workflows for differential expression analysis of human samples using RNA-Seq. BMC Bioinformatics. 2017;18:38. doi: 10.1186/s12859-016-1457-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Romanel A, Lago S, Prandi D, Sboner A, Demichelis F. ASEQ: fast allele-specific studies from next-generation sequencing data. BMC Med Genomics. 2015;8:9. doi: 10.1186/s12920-015-0084-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen J, Rozowsky J, Galeev TR, Harmanci A, Kitchen R, Bedford J, Abyzov A, Kong Y, Regan L, Gerstein M. A uniform survey of allele-specific binding and expression over 1000-Genomes-Project individuals. Nat Commun. 2016;7:ncomms11101. doi: 10.1038/ncomms11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Degner JF, Marioni JC, Pai AA, Pickrell JK, Nkadori E, Gilad Y, Pritchard JK. Effect of read-mapping biases on detecting allele-specific expression from RNA-sequencing data. Bioinforma Oxf Engl. 2009;25:3207–3212. doi: 10.1093/bioinformatics/btp579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Panousis NI, Gutierrez-Arcelus M, Dermitzakis ET, Lappalainen T. Allelic mapping bias in RNA-sequencing is not a major confounder in eQTL studies. Genome Biol. 2014;15:467. doi: 10.1186/s13059-014-0467-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van de Geijn B, McVicker G, Gilad Y, Pritchard JK. WASP: allele-specific software for robust molecular quantitative trait locus discovery. Nat Methods. 2015;12:1061–1063. doi: 10.1038/nmeth.3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang H, Wong WH. Statistical inferences for isoform expression in RNA-Seq. Bioinforma Oxf Engl. 2009;25:1026–1032. doi: 10.1093/bioinformatics/btp113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BMG, Haag JD, Gould MN, Stewart RM, Kendziorski C. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinforma Oxf Engl. 2013;29:1035–1043. doi: 10.1093/bioinformatics/btt087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012;22:2008–2017. doi: 10.1101/gr.133744.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li YI, van de Geijn B, Raj A, Knowles DA, Petti AA, Golan D, Gilad Y, Pritchard JK. RNA splicing is a primary link between genetic variation and disease. Science. 2016;352:600–604. doi: 10.1126/science.aad9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goldstein LD, Cao Y, Pau G, Lawrence M, Wu TD, Seshagiri S, Gentleman R. Prediction and Quantification of Splice Events from RNA-Seq Data. PLOS ONE. 2016;11:e0156132. doi: 10.1371/journal.pone.0156132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li YI, Knowles DA, Humphrey J, Barbeira AN, Dickinson SP, Im HK, Pritchard JK. Annotation-free quantification of RNA splicing using LeafCutter. Nat Genet. 2018;50:151–158. doi: 10.1038/s41588-017-0004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cummings BB, Marshall JL, Tukiainen T, et al. Improving genetic diagnosis in Mendelian disease with transcriptome sequencing. Sci Transl Med. 2017;9:eaal5209. doi: 10.1126/scitranslmed.aal5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jung H, Lee D, Lee J, Park D, Kim YJ, Park W-Y, Hong D, Park PJ, Lee E. Intron retention is a widespread mechanism of tumor-suppressor inactivation. Nat Genet. 2015;47:1242–1248. doi: 10.1038/ng.3414. [DOI] [PubMed] [Google Scholar]
- 54.Pjanic M, Miller CL, Wirka R, Kim JB, DiRenzo DM, Quertermous T. Genetics and Genomics of Coronary Artery Disease. Curr Cardiol Rep. 2016;18:102. doi: 10.1007/s11886-016-0777-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Franzén O, Ermel R, Cohain A, et al. Cardiometabolic risk loci share downstream cis- and trans-gene regulation across tissues and diseases. Science. 2016;353:827–830. doi: 10.1126/science.aad6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim JB, Pjanic M, Nguyen T, Miller CL, Iyer D, Liu B, Wang T, Sazonova O, Carcamo-Orive I, Matic LP, Maegdefessel L, Hedin U, Quertermous T. TCF21 and the environmental sensor aryl-hydrocarbon receptor cooperate to activate a pro-inflammatory gene expression program in coronary artery smooth muscle cells. PLoS Genet. 2017;13:e1006750. doi: 10.1371/journal.pgen.1006750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharon D, Tilgner H, Grubert F, Snyder M. A single-molecule long-read survey of the human transcriptome. Nat Biotechnol. 2013;31:1009–1014. doi: 10.1038/nbt.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tilgner H, Grubert F, Sharon D, Snyder MP. Defining a personal, allele-specific, and single-molecule long-read transcriptome. Proc Natl Acad Sci. 2014;111:9869–9874. doi: 10.1073/pnas.1400447111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tilgner H, Jahanbani F, Blauwkamp T, Moshrefi A, Jaeger E, Chen F, Harel I, Bustamante CD, Rasmussen M, Snyder MP. Comprehensive transcriptome analysis using synthetic long-read sequencing reveals molecular co-association of distant splicing events. Nat Biotechnol. 2015;33:736–742. doi: 10.1038/nbt.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Byrne A, Beaudin AE, Olsen HE, Jain M, Cole C, Palmer T, DuBois RM, Forsberg EC, Akeson M, Vollmers C. Nanopore long-read RNAseq reveals widespread transcriptional variation among the surface receptors of individual B cells. Nat Commun. 2017;8:16027. doi: 10.1038/ncomms16027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17:333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weirather JL, de Cesare M, Wang Y, Piazza P, Sebastiano V, Wang X-J, Buck D, Au KF. Comprehensive comparison of Pacific Biosciences and Oxford Nanopore Technologies and their applications to transcriptome analysis. F1000Research. 2017;6:100. doi: 10.12688/f1000research.10571.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ulitsky I. Evolution to the rescue: using comparative genomics to understand long non-coding RNAs. Nat Rev Genet. 2016;17:601–614. doi: 10.1038/nrg.2016.85. [DOI] [PubMed] [Google Scholar]
- 64.Lee C, Kikyo N. Strategies to identify long noncoding RNAs involved in gene regulation. Cell Biosci. 2012;2:37. doi: 10.1186/2045-3701-2-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li W, Notani D, Rosenfeld MG. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet. 2016;17:207–223. doi: 10.1038/nrg.2016.4. [DOI] [PubMed] [Google Scholar]
- 66.Rappolee DA, Wang A, Mark D, Werb Z. Novel method for studying mRNA phenotypes in single or small numbers of cells. J Cell Biochem. 1989;39:1–11. doi: 10.1002/jcb.240390102. [DOI] [PubMed] [Google Scholar]
- 67.Brady G, Barbara M, Iscove N. Representative in vitro cDNA amplification from individual hemopoietic cells and colonies. Methods Mol Cell Biol. 1990:17–25. [Google Scholar]
- 68.Li G, Xu A, Sim S, Priest JR, Tian X, Khan T, Quertermous T, Zhou B, Tsao PS, Quake SR, Wu SM. Transcriptomic Profiling Maps Anatomically Patterned Subpopulations among Single Embryonic Cardiac Cells. Dev Cell. 2016;39:491–507. doi: 10.1016/j.devcel.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.DeLaughter DM, Bick AG, Wakimoto H, McKean D, Gorham JM, Kathiriya IS, Hinson JT, Homsy J, Gray J, Pu W, Bruneau BG, Seidman JG, Seidman CE. Single-Cell Resolution of Temporal Gene Expression during Heart Development. Dev Cell. 2016;39:480–490. doi: 10.1016/j.devcel.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grindberg RV, Yee-Greenbaum JL, McConnell MJ, et al. RNA-sequencing from single nuclei. Proc Natl Acad Sci U S A. 2013;110:19802–19807. doi: 10.1073/pnas.1319700110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lake BB, Ai R, Kaeser GE, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352:1586–1590. doi: 10.1126/science.aaf1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zeng W, Jiang S, Kong X, El-Ali N, Ball AR, Ma CI-H, Hashimoto N, Yokomori K, Mortazavi A. Single-nucleus RNA-seq of differentiating human myoblasts reveals the extent of fate heterogeneity. Nucleic Acids Res. 2016;44:e158. doi: 10.1093/nar/gkw739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cao J, Packer JS, Ramani V, Cusanovich DA, Huynh C, Daza R, Qiu X, Lee C, Furlan SN, Steemers FJ, Adey A, Waterston RH, Trapnell C, Shendure J. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science. 2017;357:661–667. doi: 10.1126/science.aam8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gaidatzis D, Burger L, Florescu M, Stadler MB. Analysis of intronic and exonic reads in RNA-seq data characterizes transcriptional and post-transcriptional regulation. Nat Biotechnol. 2015;33:722–729. doi: 10.1038/nbt.3269. [DOI] [PubMed] [Google Scholar]
- 75.Islam S, Kjällquist U, Moliner A, Zajac P, Fan J-B, Lönnerberg P, Linnarsson S. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 2011;21:1160–1167. doi: 10.1101/gr.110882.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hashimshony T, Senderovich N, Avital G, Klochendler A, de Leeuw Y, Anavy L, Gennert D, Li S, Livak KJ, Rozenblatt-Rosen O, Dor Y, Regev A, Yanai I. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 2016;17:77. doi: 10.1186/s13059-016-0938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Picelli S, Björklund ÅK, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 78.Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A, Amit I. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Muraro MJ, Dharmadhikari G, Grün D, Groen N, Dielen T, Jansen E, van Gurp L, Engelse MA, Carlotti F, de Koning EJP, van Oudenaarden A. A Single-Cell Transcriptome Atlas of the Human Pancreas. Cell Syst. 2016;3:385–394. e3. doi: 10.1016/j.cels.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Treutlein B, Brownfield DG, Wu AR, Neff NF, Mantalas GL, Espinoza FH, Desai TJ, Krasnow MA, Quake SR. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509:371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu AR, Neff NF, Kalisky T, Dalerba P, Treutlein B, Rothenberg ME, Mburu FM, Mantalas GL, Sim S, Clarke MF, Quake SR. Quantitative assessment of single-cell RNA-sequencing methods. Nat Methods. 2014;11:41–46. doi: 10.1038/nmeth.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Svensson V, Natarajan KN, Ly L-H, Miragaia RJ, Labalette C, Macaulay IC, Cvejic A, Teichmann SA. Power analysis of single-cell RNA-sequencing experiments. Nat Methods. 2017;14:381–387. doi: 10.1038/nmeth.4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kimmerling RJ, Lee Szeto G, Li JW, Genshaft AS, Kazer SW, Payer KR, de Riba Borrajo J, Blainey PC, Irvine DJ, Shalek AK, Manalis SR. A microfluidic platform enabling single-cell RNA-seq of multigenerational lineages. Nat Commun. 2016;7:10220. doi: 10.1038/ncomms10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rosenberg AB, Roco C, Muscat RA, Kuchina A, Mukherjee S, Chen W, Peeler DJ, Yao Z, Tasic B, Sellers DL, Pun SH, Seelig G. Scaling single cell transcriptomics through split pool barcoding. bioRxiv. 2017:105163. [Google Scholar]
- 85.Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Ferrante TC, Terry R, Turczyk BM, Yang JL, Lee HS, Aach J, Zhang K, Church GM. Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat Protoc. 2015;10:442–458. doi: 10.1038/nprot.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hashimshony T, Wagner F, Sher N, Yanai I. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2012;2:666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 87.Fan X, Zhang X, Wu X, Guo H, Hu Y, Tang F, Huang Y. Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol. 2015;16:148. doi: 10.1186/s13059-015-0706-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sheng K, Cao W, Niu Y, Deng Q, Zong C. Effective detection of variation in single-cell transcriptomes using MATQ-seq. Nat Methods. 2017;14:267–270. doi: 10.1038/nmeth.4145. [DOI] [PubMed] [Google Scholar]
- 89.Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hochgerner H, Lönnerberg P, Hodge R, et al. STRT-seq-2i: dual-index 5′ single cell and nucleus RNA-seq on an addressable microwell array. Sci Rep. 2017;7:16327. doi: 10.1038/s41598-017-16546-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gierahn TM, Wadsworth MH, Hughes TK, Bryson BD, Butler A, Satija R, Fortune S, Love JC, Shalek AK. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat Methods. 2017;14:395–398. doi: 10.1038/nmeth.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ramsköld D, Luo S, Wang Y-C, Li R, Deng Q, Faridani OR, Daniels GA, Khrebtukova I, Loring JF, Laurent LC, Schroth GP, Sandberg R. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Grün D, Kester L, van Oudenaarden A. Validation of noise models for single-cell transcriptomics. Nat Methods. 2014;11:637–640. doi: 10.1038/nmeth.2930. [DOI] [PubMed] [Google Scholar]
- 94.Islam S, Zeisel A, Joost S, La Manno G, Zajac P, Kasper M, Lönnerberg P, Linnarsson S. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat Methods. 2014;11:163–166. doi: 10.1038/nmeth.2772. [DOI] [PubMed] [Google Scholar]
- 95.Buettner F, Natarajan KN, Casale FP, Proserpio V, Scialdone A, Theis FJ, Teichmann SA, Marioni JC, Stegle O. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat Biotechnol. 2015;33:155–160. doi: 10.1038/nbt.3102. [DOI] [PubMed] [Google Scholar]
- 96.Suter DM, Molina N, Gatfield D, Schneider K, Schibler U, Naef F. Mammalian genes are transcribed with widely different bursting kinetics. Science. 2011;332:472–474. doi: 10.1126/science.1198817. [DOI] [PubMed] [Google Scholar]
- 97.Baker SC, Bauer SR, Beyer RP, et al. The External RNA Controls Consortium: a progress report. Nat Methods. 2005;2:731–734. doi: 10.1038/nmeth1005-731. [DOI] [PubMed] [Google Scholar]
- 98.Vallejos CA, Risso D, Scialdone A, Dudoit S, Marioni JC. Normalizing single-cell RNA sequencing data: challenges and opportunities. Nat Methods. 2017;14:565–571. doi: 10.1038/nmeth.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Grün D, van Oudenaarden A. Design and Analysis of Single-Cell Sequencing Experiments. Cell. 2015;163:799–810. doi: 10.1016/j.cell.2015.10.039. [DOI] [PubMed] [Google Scholar]
- 100.Stegle O, Teichmann SA, Marioni JC. Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet. 2015;16:133–145. doi: 10.1038/nrg3833. [DOI] [PubMed] [Google Scholar]
- 101.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lun ATL, Bach K, Marioni JC. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 2016;17:75. doi: 10.1186/s13059-016-0947-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bacher R, Chu L-F, Leng N, Gasch AP, Thomson JA, Stewart RM, Newton M, Kendziorski C. SCnorm: robust normalization of single-cell RNA-seq data. Nat Methods. 2017;14:584–586. doi: 10.1038/nmeth.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Brennecke P, Anders S, Kim JK, Kołodziejczyk AA, Zhang X, Proserpio V, Baying B, Benes V, Teichmann SA, Marioni JC, Heisler MG. Accounting for technical noise in single-cell RNA-seq experiments. Nat Methods. 2013;10:1093–1095. doi: 10.1038/nmeth.2645. [DOI] [PubMed] [Google Scholar]
- 106.Kharchenko PV, Silberstein L, Scadden DT. Bayesian approach to single-cell differential expression analysis. Nat Methods. 2014;11:740–742. doi: 10.1038/nmeth.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fan J, Salathia N, Liu R, Kaeser GE, Yung YC, Herman JL, Kaper F, Fan J-B, Zhang K, Chun J, Kharchenko PV. Characterizing transcriptional heterogeneity through pathway and gene set overdispersion analysis. Nat Methods. 2016;13:241–244. doi: 10.1038/nmeth.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, Linsley PS, Gottardo R. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16:278. doi: 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kiselev VY, Kirschner K, Schaub MT, Andrews T, Yiu A, Chandra T, Natarajan KN, Reik W, Barahona M, Green AR, Hemberg M. SC3: consensus clustering of single-cell RNA-seq data. Nat Methods. 2017;14:483–486. doi: 10.1038/nmeth.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Adamson B, Norman TM, Jost M, et al. A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell. 2016;167:1867–1882. e21. doi: 10.1016/j.cell.2016.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.van der Maaten L, Hinton G. Visualizing Data using t-SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 112.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xu C, Su Z. Identification of cell types from single-cell transcriptomes using a novel clustering method. Bioinforma Oxf Engl. 2015;31:1974–1980. doi: 10.1093/bioinformatics/btv088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Marco E, Karp RL, Guo G, Robson P, Hart AH, Trippa L, Yuan G-C. Bifurcation analysis of single-cell gene expression data reveals epigenetic landscape. Proc Natl Acad Sci U S A. 2014;111:E5643–5650. doi: 10.1073/pnas.1408993111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Grün D, Lyubimova A, Kester L, Wiebrands K, Basak O, Sasaki N, Clevers H, van Oudenaarden A. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 2015;525:251–255. doi: 10.1038/nature14966. [DOI] [PubMed] [Google Scholar]
- 116.Olsson A, Venkatasubramanian M, Chaudhri VK, Aronow BJ, Salomonis N, Singh H, Grimes HL. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature. 2016;537:698–702. doi: 10.1038/nature19348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shin J, Berg DA, Zhu Y, Shin JY, Song J, Bonaguidi MA, Enikolopov G, Nauen DW, Christian KM, Ming G, Song H. Single-Cell RNA-Seq with Waterfall Reveals Molecular Cascades underlying Adult Neurogenesis. Cell Stem Cell. 2015;17:360–372. doi: 10.1016/j.stem.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zeng C, Mulas F, Sui Y, Guan T, Miller N, Tan Y, Liu F, Jin W, Carrano AC, Huising MO, Shirihai OS, Yeo GW, Sander M. Pseudotemporal Ordering of Single Cells Reveals Metabolic Control of Postnatal β Cell Proliferation. Cell Metab. 2017;25:1160–1175. e11. doi: 10.1016/j.cmet.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Haghverdi L, Büttner M, Wolf FA, Buettner F, Theis FJ. Diffusion pseudotime robustly reconstructs lineage branching. Nat Methods. 2016;13:845–848. doi: 10.1038/nmeth.3971. [DOI] [PubMed] [Google Scholar]
- 121.Bacher R, Kendziorski C. Design and computational analysis of single-cell RNA-sequencing experiments. Genome Biol. 2016;17:63. doi: 10.1186/s13059-016-0927-y. [DOI] [PMC free article] [PubMed] [Google Scholar]