Abstract
The class of human genetic kidney diseases is extremely broad and heterogeneous. Accordingly, the range of associated disease phenotypes is highly variable. Many children and adults affected by inherited kidney disease will progress to ESKD at some point in life. Extensive research has been performed on various different disease models to investigate the underlying causes of genetic kidney disease and to identify disease mechanisms that are amenable to therapy. We review some of the research highlights that, by modeling inherited kidney disease, contributed to a better understanding of the underlying pathomechanisms, leading to the identification of novel genetic causes, new therapeutic targets, and to the development of new treatments. We also discuss how the implementation of more efficient genome-editing techniques and tissue-culture methods for kidney research is providing us with personalized models for a precision-medicine approach that takes into account the specificities of the patient and the underlying disease. We focus on the most common model systems used in kidney research and discuss how, according to their specific features, they can differentially contribute to biomedical research. Unfortunately, no definitive treatment exists for most inherited kidney disorders, warranting further exploitation of the existing disease models, as well as the implementation of novel, complex, human patient–specific models to deliver research breakthroughs.
Keywords: genetic renal disease, transgenic mouse, zebrafish, molecular genetics
Introduction
There are >150 human genetic kidney diseases that may affect kidney development or kidney tissue maintenance, leading to glomerular, tubular, and structural defects (1). The range of disease phenotypes associated with genetic diseases is huge. There are groups of kidney diseases where the phenotypes are largely kidney limited, such as autosomal-dominant tubulointerstitial kidney disease secondary to uromodulin (UMOD) mutations, where the molecular defect is restricted to the cells of the thick ascending limb of the loop of Henle (2). Glomerulopathies, such as inherited forms of nephrotic syndrome with podocyte-specific defects (3) and tubulopathies (4) such as Gitelman syndrome and nephrogenic diabetes insipidus, where encoded proteins are expressed in a specific tubular segment, would also be in this group. Other groups of inherited disorders are labeled as kidney disease but actually display phenotypes in multiple organs, such as autosomal dominant polycystic kidney disease (ADPKD) where cysts may be seen in organs aside from the kidney, as well as leading to berry aneurysms and valvular heart lesions (5,6). There are other examples of kidney genetic diseases with extra kidney manifestations, including Alport syndrome and related collagenopathies, which have prominent deafness and eye phenotypes. Inherited lysosomal storage disorders such as Fabry disease and disorders such as cystinosis, tuberous sclerosis, and von Hippel–Lindau syndrome have prominent kidney phenotypes but are really systemic diseases. Developmental causes of kidney disease such as mutations in transcription factors, for example HNF1β, have multisystem and highly variable phenotypes. Finally, there are ciliopathy disorders, such as nephronophthisis, which have dramatic kidney phenotypes but may be associated with multisystem phenotypes consistent with the underlying defect in primary cilia (see Glossary).
Glossary.
Centrosome/basal body: the centrosome is a cellular structure made of two centrioles (aggregates of microtubules arranged in a barrel-like shape). The centrosome is involved in cell division. When the cell is not dividing, a single centriole forms the basal body, which works as a template for the formation of the primary cilium.
Forward genetics: a genetic approach that, starting from the observation of a phenotype, identifies the associated genetic cause.
Primary cilium: an immotile, hairlike, microtubule-based cellular organelle found in many eukaryotic cells that protrudes from the surface of the cell, where it is thought to work as a cellular antenna for stimuli sensing. Primary cilia are made of nine doublets of microtubule filaments (9+0 configuration), as opposed to motile cilia that also contain a central doublet (9+2 configuration). A cell possesses only one primary cilium.
Reverse genetics: a genetic approach that identifies the function of a gene of interest by studying the phenotypic effects of alterations in the gene expression and/or sequence.
Site-directed mutagenesis: a method to generate changes in the DNA of a specific genomic sequence.
Transgene: an exogenous DNA sequence introduced in the genome of an organism.
Clustered regularly interspaced short palindromic repeat (CRISPR): works in combination with a CRISPR-associated endonuclease (Cas protein). Engineered CRISPR/Cas constitutes a genome-editing system that binds to a target DNA sequence complementary to a portion of a short synthetic RNA (guide RNA). Another portion of the guide RNA mediates the binding to the Cas protein that is responsible for cutting the DNA.
GAL4-UAS: this is based on the Saccharomyces cerevisiae transcription factor GAL4, which is put under the control of a promoter or enhancer and introduced into the genome of a Drosophila line. An effector line carries a target gene under the control of UAS, the GAL4 binding motif. When a GAL4-driver line is crossed with the effector line, the target gene will be expressed in the offspring.
RNA interference (RNAi): a conserved biologic process that acts on double-stranded RNA and results in its nucleolytic degradation. This process can be leveraged by researchers to achieve sequence-specific gene silencing by introducing into the organism short-interfering RNA (siRNA) that leads to the degradation of the targeted RNA.
Transcription activator-like effector nucleases (TALENs): a genome-editing technique that is based on a nuclease domain for DNA cleavage fused to a customizable DNA-binding domain.
Zinc-finger nucleases (ZFNs): these are based on customizable zinc-finger DNA-binding domains. A large number of engineered zinc-finger arrays are fused to a nonspecific nuclease domain to cleave a target DNA sequence. ZFNs function as dimers.
Human genetic diseases of the kidney may be grouped in terms of frequency. ADPKD remains the most common inherited cause of kidney disease leading to end stage kidney failure and research has often been championed in the field of cystic kidney disease and then subsequently pursued in other disease groups.
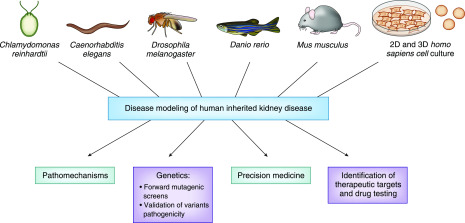
Extensive research has been conducted on disease models for inherited kidney disease to identify aberrant pathways and novel therapeutic targets (Figure 1). These models have also been used for drug screening and testing (Figure 1). Moreover, animal and cellular models represent a useful platform to validate the pathogenicity of novel mutations and variants of unknown significance, and to identify novel genetic causes of inherited kidney disease with forward and reverse genetic studies (Figure 1). Gene editing systems such as clustered regularly interspaced short palindromic repeat (CRISPR)/Cas now allow the precise introduction of the candidate mutation, allowing a fast and easy generation of humanized and patient-specific disease models, paving the way for precision-medicine approaches (Figure 1).
Figure 1.
Some of the most common disease models used in kidney research contribute to different aspects of our understanding of inherited kidney disease.
This review will discuss the features that characterize the different model systems most commonly used in kidney research—namely Chlamydomonas reinhardtii, Caenorhabditis elegans, Drosophila melanogaster (fruit fly), Danio rerio (zebrafish), Mus musculus (mouse), and mammalian cell cultures (Table 1)—and how they have differently contributed to the study of inherited kidney disorders in the context of the ever-evolving landscape of tools available for kidney genetics studies (Table 2). This information can prove useful in making the decision of which model should be used for future research, depending on the type of study and the features of each model (Figure 2). We also present some of the research highlights that, through the use of animal and cellular disease models, demonstrate how preclinical model systems have significantly contributed to the understanding of human inherited kidney diseases.
Table 1.
Summary of the specificities that distinguish C. reinhardtii, C. elegans, D. melanogaster, the zebrafish, the mouse, and two- and three-dimensional cell cultures as models in kidney research
| Model | Maintenance Costs | Availability of Tools for Gene Manipulation in Reverse Genetics | Suitability for High-Throughput Studies (Drug Screens and Forward Genetics) | Similarity to the Human Genome | Similarity to the Human Kidney |
|---|---|---|---|---|---|
| C. reinhardtii | Low | RNAi (gene silencing); CRISPR/Cas9 (gene editing) has been reported in Chlamydomonas (112,113), but with low editing efficiency | Yes. Characteristics such as small size, low maintenance cost, and genome haploidy during vegetative state make Chlamydomonas particularly suitable for high-throughput studies, such as mutagenesis and small molecule screens (114–116) | Chlamydomonas and humans share 706 protein families (117) and these are enriched in cilia and centrosome/basal body proteins (118) | The fact that the overall structure is well conserved between Chlamydomonas flagellum and human cilia makes it a good model to study kidney ciliopathies. Chlamydomonas flagellum has a 9+2 configuration, typical of motile cilia, as opposed to the 9+0 configuration of primary cilia in the kidney tubule |
| C. elegans | Low | RNAi; ZFNs (gene editing); TALENs (gene editing); CRISPR/Cas9 | Availability of strains with fluorescently tagged proteins for in vivo microscopy and easy analysis of cilia defects through functional tests and by verifying the ability to uptake lipophilic dye through the cilia make C. elegans a good model for high-throughput screens of ciliary phenotypes (119) | 38% of C. elegans protein-coding genes has a unique corresponding functional ortholog in human (120) | C. elegans has an excretory system in part functionally homologous to the human urinary tract but its relevance in kidney disease is mainly due to its role in cilia research. Ciliary structure is well conserved between C. elegans and human with many conserved ciliary and basal body proteins. Cilia configuration in C. elegans is of the 9+0 type, as in primary cilia of kidney epithelial cells |
| D. melanogaster (fruit fly) | Low | GAL4-UAS (transgenic tool for targeted gene expression); RNAi; CRISPR/Cas9 | Yes. Drosophila is an excellent model for large-scale studies due to its small size, low maintenance costs, rapid reproductive cycle, and the availability of transgenic lines for the simplified visualization of Drosophila excretory system | >75% of human disease genes have a Drosophila ortholog (30) | The fly excretory system clears unwanted substances and maintains water, salt, and pH homeostasis. As such, it is functionally homologous to the human excretory system. Malpighian tubules correspond to the kidney tubules and nephrocytes to the glomeruli |
| D. rerio (zebrafish) | Low | mRNA microinjection for overexpression studies; MO (gene silencing); ZFNs; TALENs, CRISPR/Cas9 | Yes. Small size and availability of transgenic lines to visualize the pronephros and cilia make it a good model for high-throughput screens in kidney research (41) | Approximately 70% of human genes have at least one functional homolog in zebrafish (39). Zebrafish has many duplicated genes due to a whole-genome duplication event in early teleost evolution (55) | The major difference between the excretory system of zebrafish and that of human is that zebrafish does not develop a metanephric kidney |
| M. musculus (mouse) | High | In contrast to other model systems, targeted mutagenesis in mouse embryonic stem cells exploiting homologous recombination had been possible, although not straightforward, well before the advent of CRISPR/Cas gene-editing system. CRISPR/Cas technologies have considerably simplified the generation of knockout and knockin murine models | Although it has been used in seminal forward genetic studies (121), due to high maintenance costs and relatively long reproductive cycle, the mouse is not often the model of choice for large screens | Mice and humans share approximately 70% of protein-coding genes (122) | The macroanatomy of the kidney is overall well conserved between mouse and human. Important differences in the regulation of master transcription factors in nephrogenesis account for differences such as the higher number of nephrons in human. A more detailed comparison can be found in (123) |
| Two-dimensional cell culture | Low | Cell cultures are extremely amenable to genetic manipulation. Among others, RNAi and CRISPR/Cas9 are probably the most commonly used tools to achieve knockdown and knockout/knockin, respectively, in two-dimensional cell cultures | Yes | Depends on the species of origin. It is possible to obtain patient-specific cells | N/A |
| Three-dimensional cell culture | For spheroid setting generally low, higher costs for kidney organoids due to employment of growth factors to direct differentiation to kidney lineages | mIMCD-3 spheroids were shown to be amenable to RNAi approaches (124). Due to low accessibility of the innermost core of kidney organoids, delivery of tools for transient gene knockdown or overexpression may prove difficult. Kidney organoids have been derived from iPSCs knockout lines for polycystic kidney disease genes generated with CRISPR/Cas9 (103) | Yes | Depends on the species of origin. It is possible to obtain patient-specific URECs to generate spheroids (80) or patient-specific iPSCs to generate kidney organoids (106) | Spheroids: N/A Kidney organoids: main limitations of the model that distinguish it from the human kidney are reduced size and vasculature, immaturity, lack/underrepresentation of certain cell types, and the absence of a branched collecting duct system (125) |
RNAi, RNA interference; CRISPR, clustered regularly interspaced short palindromic repeat; ZFNs, zinc finger nucleases; TALENs, transcription activator-like effector nucleases; MO, morpholino; N/A, not applicable; mIMCD-3, mouse inner medullary collecting duct cell line; iPSCs, induced pluripotent stem cells; URECs, urine-derived renal epithelial cells.
Table 2.
Summary of the strengths and limitations to guide the use of some common techniques used in kidney research
| Gene Expression Manipulation Technique | Effect | Strengths | Limitations | When It Should Be Used |
|---|---|---|---|---|
| siRNA transfection | RNAi (post-transcriptional gene silencing) | Inexpensive, rapid, and high-throughput | The transfection of synthetic siRNA usually leads to transient effects. Moderate risk of off-target effects. Can lead to variable and partial gene silencing | To perform a rapid experiment in organisms such as Chlamydomonas, C. elegans, or Drosophila, or in cell lines to assess the target gene function. It is good practice to use several siRNAs with different sequences that target the same gene to confirm the specificity of the observed effects |
| mRNA transfection/microinjection | Gene (over)expression | Inexpensive, rapid, and high throughput | The effects of mRNA transfection/microinjection strongly depend on the quantity of mRNA introduced into the organism. Careful interpretation of the results is warranted especially when the organism is forced to translate the exogenous mRNA at high, nonendogenous levels | Often used in rapid zebrafish and cell-based experiments to assess gene function or to perform rescue experiments (in combination with silencing of the same gene or of an interacting gene) |
| MO transfection/microinjection | Post-transcriptional gene silencing | High RNA affinity with superior ability to invade RNA secondary structure, resulting in high targeting predictability. Resistance to nucleases | The effects of transfection/microinjection of MO are transient. Moderate risk of off-target effects. Can lead to variable and partial gene silencing | Often used in rapid zebrafish and cell-based experiments. It is good practice to use several MOs with different sequences that target the same gene to confirm the specificity of the effects observed |
| ZFNs | Gene editing | Can target virtually any sequence in the genome | Engineering and design of ZFN is challenging and time consuming. Risk of off-target effects | When wanting to obtain a mutant organism or a stable cell line |
| TALENs | Gene editing | Design simplicity | Risk of off-target effects | When wanting to obtain a mutant organism or a stable cell line |
| CRISPR/Cas | Gene editing | Design simplicity and efficiency, which make this technique overall significantly less expensive and less time consuming than other gene editing systems | CRISPR/Cas system shares with other gene-editing systems the risk of off-target effects. Target sequence needs be adjacent to a specific protospacer adjacent motif | When wanting to obtain a mutant organism or a stable cell line. Can also be used for rapid gene silencing experiments without selection of the mutated founder organism or cell clone (e.g., to produce zebrafish crispants or mixed mutant cell populations). Due to its design simplicity and efficiency, it is to be considered the technique of choice to achieve gene editing in most cases. Whole-genome sequencing to assess specificity of gene editing in a mutant organism or stable cell line, albeit expensive, should be performed |
siRNA, short interfering RNA; RNAi, RNA interference; MO, morpholino; ZFNs, zinc finger nucleases; TALENs, transcription activator-like effector nucleases; CRISPR, clustered regularly interspaced short palindromic repeat.
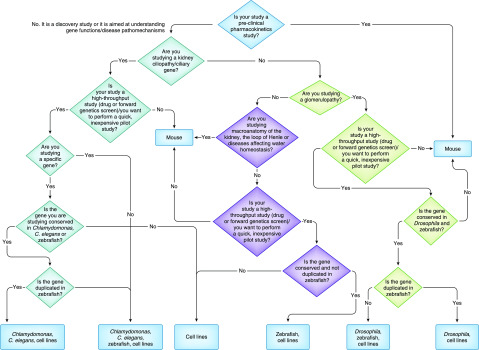
Figure 2.
Flow chart describing some of the considerations the researcher should make when deciding which model is the most appropriate for the study. Other considerations, such as the availability of animal facilities, ethics in place, and laboratory expertise are equally important in the decision process.
Seminal Studies in C. reinhardtii and C. elegans: the Golden Era of the Primary Cilium
Traditionally, until the early 2000s, nephrology research on animal models mainly involved the study of spontaneously arisen rodent models such as the cystic kidney disease cpk (7), bpk (8), and jck (9) mouse models and the cystic kidney PCK rat (10). These models had similarities to clinical features of human polycystic kidney disease (PKD) but, for several years, the underlying mutated gene in these animals remained unidentified, which hindered any further study on the associated pathomechanisms.
One of the greatest contributions to the understanding of pathomechanisms of kidney disease came from studies on two model organisms that do not possess a kidney, the flagellated unicellular green alga C. reinhardtii and the nematode C. elegans, which revealed for the first time a surprising but compelling link between defects in primary cilia and cystic kidney disease.
The first clues of a link between kidney cysts and cilia came from the study of two C. elegans genes required for male sensory behaviors, lov1 and pkd2, homologs of PKD1 and PKD2, the known genetic causes of ADPKD in humans. Barr and Starnberg (11) found that lov1 and pkd2 proteins were enriched in C. elegans sensory neuron cilia. Later studies then confirmed ciliary localization of the human ortholog proteins polycystin-1 and polycystin-2 in human and mouse kidney cells (12,13).
Confirmation of the link between cilia and cystic kidney disease came from a seminal study by Pazour et al. (14) who showed that ift88, the Chlamydomonas homolog of the gene mutated in the orpk (Tg737) mouse with PKD, encodes the intraflagellar transport subunit ift88, which is necessary for the assembly of Chlamydomonas flagella and mammalian cilia. Following this study, cloning of the mutated gene in the traditional spontaneous models of PKD confirmed that the disease-causing gene encoded a ciliary-associated protein also in the cpk mouse (15), in the bpk mouse (16,17), in the jck mouse (18), and in the PCK rat (19).
Identification of the genetic causes of several other monogenic cystic kidney disorders, together with the discovery that—in the majority of cases—the encoded proteins localized to the cilium, added a growing body of evidence confirming the association between primary cilia defects and cystic kidney disease (20).
Due to the fact that C. elegans is easy to maintain in culture and to genetically manipulate and that cilia are not required for its development or viability (21), it continues to be extensively used to investigate the ciliary functions of kidney disease genes such as BBS1 (22), RPGRIP1L (23), TMEM107 (24), and TTC21B (25). Discovery of the role of these genes in ciliary assembly and/or trafficking is essential to reveal ciliary-specific protein networks and the pathways involved in the pathogenicity of cystic kidney.
The Fly: an Insect Model for Human Glomerulopathies
D. melanogaster (fruit fly) has an excretory system responsible for clearance of substances and maintenance of water, salt, and pH homeostasis, which can therefore be considered functionally homologous to the human excretory system. The excretory system in Drosophila consists of the Malpighian tubules (26), analogous to kidney tubules and the nephrocytes, which correspond to some extent to the glomeruli. Nephrocytes are specialized cells that filtrate hemolymph (the Drosophila’s blood) and, through their endocytic activity, regulate its composition. There are two types of nephrocytes in the fly: the garland nephrocytes, along the esophagus, and the pericardial nephrocytes. Nephrocytes are surrounded by basement membrane and are characterized by cellular protrusions and folding of the plasma membrane, bridged by a diaphragm structure. This structure is very reminiscent of the mammalian glomerular filtration system (27). Drosophila orthologs of podocyte-specific proteins required for maintenance of the molecular sieve in the human glomeruli, such as nephrin and podocin, are expressed in the nephrocytes (28,29).
The conservation of several human disease genes in the fly (30) makes it an attractive model in biomedical research and, in the last decade, it became an established model in kidney research, particularly in the field of podocytopathies (27) and, more recently, of nephrolithiasis (kidney stones) (31,32).
Low maintenance costs and a fast reproductive cycle make Drosophila particularly suitable for large-scale screens. Chemical and insertional mutagenesis can be applied for forward genetic studies but the toolbox for targeted genetic manipulation is especially rich in Drosophila.
Largely used is the GAL4-UAS system, which allows spatially and temporally restricted expression of transgenes (33). The use of tissue-specific enhancers within this system allows specific targeting of nephrocytes. GAL4-UAS can be used to achieve silencing of the gene of interest, by combining it with RNA-interference techniques to knockdown the gene post-transcriptionally (34) or with the CRISPR/Cas9 gene-editing system for nephrocyte-specific gene knockout (35). Moreover, it is possible to perform rescue experiments by expressing the wild-type or mutant orthologous human protein in the corresponding Drosophila knockout line (36), which can be particularly useful to verify the pathogenicity of specific mutations.
Powerful functional studies to rapidly assess nephrocyte filtration and endocytosis are available and consist of monitoring the uptake by nephrocytes of tracers such as fluorescently labeled albumin or dextran ex vivo or in vivo.
The combination of all of these features makes the fly a very powerful model for gene-discovery studies (37) and to investigate the pathomechanism of inherited glomerulopathies (35).
Applying Drosophila models to the study of inherited forms of steroid-resistant nephrotic syndrome, Hermle et al. (35) were able to link the pathogenesis of COQ2-associated human nephropathy to the formation of mitochondrial reactive oxygen species, which may be particularly deleterious in podocytes. The same group used a Drosophila knockdown model to perform functional studies on GAPVD1, a novel genetic cause of monogenic steroid-resistant nephrotic syndrome (38).
A powerful in vivo system was recently established in Drosophila, which consists of a transgenic line characterized by the expression of a muscle-secreted fluorescent protein and easily detectable GFP-labeled nephrocytes, combined with a nephrocyte-specific driver for targeted gene knockdown (37). The availability of such a system can be extremely useful to screen for novel genetic causes of human glomerulopathies and makes Drosophila an important contributor in kidney research.
Zebrafish: Powerful Vertebrate Models of Kidney Disease Devoid of a Proper Kidney
A total of 70% of human genes have at least one zebrafish functional ortholog (39). Unlike human, zebrafish is devoid of a metanephric kidney and only a pronephros and subsequently a mesonephric kidney form during zebrafish development. Another major difference between human and zebrafish is the absence of a segment homologous to the mammalian loop of Henle in the zebrafish nephron. The zebrafish pronephros forms at around 24 hours post-fertilization and consists of a pair of segmented epithelial tubules with a fused glomerulus. The pronephros is fully functional at 48 hours post-fertilization (40).
Characteristics such as optical transparency, rapid organogenesis, small size, low maintenance costs, and high fecundity make zebrafish a useful model organism in genetic-medicine research and make it particularly suitable for large forward genetic and drug screens.
By screening mutagenized animals for phenotypes of interest, forward genetic screens allow for the discovery of previously unrecognized novel genetic causes of human disease. Zebrafish can be randomly mutagenized by exposing them to the mutagenic agent N-ethyl-N-nitrosourea (ENU; the mating scheme to generate mutant lines is exemplified in Figure 3) or gene function can be disrupted by using viruses to introduce exogenous DNA sequences in the zebrafish genome. Through a forward genetic approach, Sun et al. (41) identified ten genes associated with cystic pronephros phenotypes. Three of these genes were homologs of genes encoding Chlamydomonas intraflagellar transport components (ift57, ift81, and ift172), strengthening the connection between cilia defects and cystic kidney disease. Indeed, IFT81 and IFT172 are known to cause kidney phenotypes in human (42,43). Positional cloning to map the induced mutation traditionally constituted a bottleneck in the ENU mutagenesis pipeline, but improved annotation of the zebrafish genome and a reduction in sequencing costs now allow the use of whole-exome sequencing for rapid identification of new candidate genes that cause kidney phenotypes in zebrafish mutants (44).
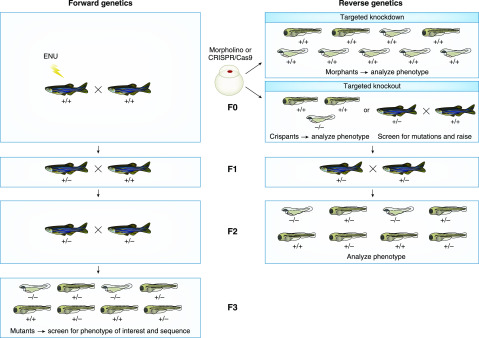
Figure 3.
Schematic of forward and reverse genetics in zebrafish. (Left) Forward genetics: the ENU-treated founder (F0) is a genetic mosaic with mutated germline. Crossing with wild-type fish generates an F1 progeny that is raised and outcrossed with wild-type fish. Half of the F2 progeny will be heterozygous for the ENU-induced mutation. Outcross with wild-type fish generates an F3 progeny that is 25% homozygous for the ENU-induced mutation. F3 zebrafish embryos or larvae are screened for phenotypes of interest and sequenced to identify the mutated gene. (Right) Reverse genetics: zebrafish embryos at the one- to four-cell stage can be injected with a morpholino directed against the gene of interest to induce post-transcriptional targeted gene knockdown. Phenotypes of morphant embryos or larvae can be analyzed to study gene function. Alternatively, targeted gene knockout can be obtained by injecting zebrafish embryos at the one- to two-cell stage with Cas9 (mRNA or protein) and guideRNA(s) directed against the gene of interest. Cas9 causes a double-strand break that is repaired through nonhomologous end joining, often resulting in indels that impair gene expression and/or function. Resulting crispants (F0) can be genetic mosaics. Phenotype and genotype of the F0 can be analyzed to study gene function. Alternatively, F0 crispants can be raised, genotyped, and the heterozygous founder is outcrossed with wild-type fish to generate an F1 progeny that will be 50% heterozygous for the Cas9-induced mutation. F1 fish are incrossed to generate an F2 progeny that will be 25% homozygous for the mutation. The phenotype of F2 embryos, larvae, or adults (if viable) is analyzed to study gene function. ENU, N-ethyl-N-nitrosourea.
Zebrafish has proven to be a powerful tool not only in forward genetic screens but also to study gene function and pathways associated with kidney disease (Table 3). Two decades ago, due to the absence of tools to efficiently generate knockout fish for the gene of interest, morpholino antisense oligonucleotides became a popular instrument to obtain rapid and cost effective transient gene knockdown in zebrafish embryos. Morpholinos can be injected in the zebrafish embryo at the one- to four-cell stage and, by interfering with transcript processing or translation, cause gene silencing without changing the genomic sequence (Figure 3). Some discrepancies between the phenotype of morphants and that of the corresponding mutants in several disease models, including kidney disease (41), raised concerns regarding the specificity of the effects induced by morpholinos in zebrafish. However, it has been demonstrated that phenomena such as genetic compensation could explain these discrepancies (45). Provided that control experiments are put in place to ensure the specificity of the effects observed (46), the use of morpholinos in zebrafish is still considered a valuable tool to study the function of genes involved in human inherited disorders, including kidney disease.
Table 3.
Examples of zebrafish and mouse models that have differentially contributed to our understanding of the pathomechanisms of human inherited kidney disease
| Human Kidney Disease | Zebrafish | Mouse |
|---|---|---|
| Glomerulopathies | ||
| Alport syndrome | The zebrafish mutant dragnet (alias col4a5) fails to recapitulate the human phenotype. Loss of col4a5 expression leads to defects in the laminar organization of the retinotectal projection, but no glomerular phenotype is observable (126) | Col4a3 knockout mouse model of autosomal Alport syndrome is characterized by progressive GN with microhematuria and proteinuria, consistent with the human disease, and focal multilaminated thickening and thinning of the glomerular basement membrane starting at 4 wk. End stage kidney failure happens at 14 wk with fibrotic glomeruli and collapsed capillaries (57). The finding that these mice display an upregulation of miR-21 and that inhibition of miR-21 slows kidney disease progression has identified a promising therapeutic target (58) |
| Nephrotic syndrome type 1 and 2 | The functions of the podocyte-specific proteins nephrin (nphs1) and podocin (nphs2) are highly conserved between the zebrafish pronephros and mammalian metanephros. Knockdown of nphs1 and nphs2 results in abnormal slit diaphragm and foot process architecture and in altered glomerular filtration (53) | Nphs1−/− and Nphs2−/− mice are born alive but die within 24 h and 5 wk, respectively, indicating a more severe phenotype when compared with human (127). Nphs2−/− mice develop diffuse mesangial sclerosis, indicating that, during glomerulogenesis, absence of podocin may lead to important alterations in the podocyte-endothelial-mesangial crosstalk (79) |
| Nephrotic syndrome type 4 | Knockdown of Wilms tumor 1a (wt1a) in zebrafish embryos leads to defects in podocyte development with effacement of foot processes, abnormalities in the slit diaphragm, and dysfunctional glomerular filtration, as assessed by injected dextran clearance rate (47) | Wt1 null mice are embryonic lethal, showing failure of kidney and gonad development. Metanephric mesenchyme cells are apoptotic and the ureteric bud does not stem from the Wolffian duct (128) |
| Tubulopathies/kidney ciliopathies | ||
| Autosomal dominant tubulointerstitial kidney disease–UMOD | Uromodulin (umod) morphants do not display kidney development defects, nor increased susceptibility to nephrotoxins, nor significant alterations in glomerular or tubule gene expression (129), perhaps consistently with a lack of a loop of Henle in the zebrafish | Umod null mice do not display anatomic or functional kidney alterations, but have altered levels of transporters in the loop of Henle (60) |
| Cystinosis | Mutant ctns zebrafish exhibit cystine accumulation, altered tubular reabsorption, and glomerular permeability, in line with the human phenotype (130) | Ctns−/− mice on the 129Sv × C57BL/6 genetic background, despite elevated kidney cystine levels, do not show an overt kidney phenotype (131), whereas Ctns−/− on the C57BL/6 background develop histologic kidney lesions but no clear glomerulopathy (78) |
| ADPKD | Zebrafish embryos microinjected with mRNA, encoding the C-terminal cytoplasmic portion of polycystin-1, form pronephric and liver cysts at 3 d post-fertilization, demonstrating the importance of pkd1 dosage to maintain pronephric and liver architecture in the zebrafish embryos (132). pkd2 was found mutated in an insertional mutagenic screen for zebrafish mutant embryos that displayed cystic pronephros phenotypes (41) | Pkd1 and Pkd2 null mice are embryonic lethal, whereas heterozygous mice are viable and progressively develop kidney cysts (62). Pkd2WS25 mice harbor an unstable allele. Localized absence of polycystin-2 in Pkd2WS25 cyst-lining epithelia implies a somatic loss of Pkd2 in kidney cysts (62) and argues in favor of a two-hit hypothesis for ADPKD pathogenicity; but the increased proliferation in kidney noncystic, polycystin-2–positive epithelia indicates that haploinsufficiency of Pkd2 is sufficient to cause cell proliferation in early polycystic kidney disease (64). The conditional Pkd1cond/cond mouse shows a variable onset of cystic kidney disease which depends on the time of genetic inactivation of Pkd1, indicating that the pathologic consequences of inactivation depend on the maturation status of the kidney (133) |
| Nephronophthisis type 3 | Knockdown of nphp3 in the zebrafish embryo causes hydrocephalus and body symmetry defects due to ciliary abnormalities at the Kupffer vesicle (134), a temporary developmental organ of left-right axis determination | The pcy mouse is an ortholog of NPHP3 and a slowly progressive model of cystic kidney disease, with kidney enlargement arising at 8 wk and thickened basement membrane (135). Pcy mice display a significant increase in kidney cAMP concentration and in the expression of V2R and AQP2, which is a gene positively regulated by cAMP (69) |
| Nephronophthisis type 6 | Knockdown of cep290 in zebrafish embryos results in retinal and brain defects as well as cyst formation in the pronephros, resembling the clinical features of patients with Joubert syndrome (136). cep290 morphant fish exhibit increased levels of γh2ax, a marker of DNA damage response signaling, suggesting a role for DNA damage or DNA damage response signaling in the pathophysiology of CEP290-associated kidney disease (137) | The Cep290−/− mice on a 129/Ola background are viable and present a mild kidney phenotype, with collecting duct small kidney cysts but no significant kidney enlargement, in line with a NPHP phenotype. Abnormalities in ciliary phenotype and in Hedgehog signaling are observable in cystic epithelia (80). Cep290−/− mice on a C57BL/6 background present severe hydrocephalus and are lethal (80,138), highlighting the possible contribution of genetic modifiers in the definition of the phenotype |
| Nephronophthisis type 9 | Knockdown of nek8 in zebrafish embryos leads to cyst formation and abnormal cardiac looping. Similar phenotypes are observed when zebrafish inversin (inv, orthologous of the genetic cause of NPHP2) is knocked down. Overexpression of nek8 through mRNA microinjection rescues these phenotypes in inv morphants, indicating that nek8 may act downstream of inv (139) | The jck mouse genetically is an orthologous model of NPHP9, with mutations in the Nek8 gene. Phenotypically it resembles ADPKD, with significantly enlarged kidneys at 4 wk. Kidneys present high cAMP levels and elevated PCNA staining in cyst-lining epithelia, revealing a proliferative phenotype and promising therapeutic targets (140–142) |
ADPKD, autosomal dominant polycystic kidney disease; NPHP, nephronophthisis; V2R, vasopressin V2 receptor; AQP2, aquaporin-2; PCNA, proliferating cell nuclear antigen.
By combining the injection of morpholinos with the injection of mRNA for the corresponding human wild-type or mutant ortholog, it is possible to perform rescue experiments to confirm the pathogenicity of novel mutations. For instance, this kind of experiment in zebrafish was performed to validate a novel missense mutation in WT1 (p.R458Q) as a cause of familial FSGS (47).
Engineered endonucleases such as zinc-finger nucleases, transcription activator-like effector nucleases, and the CRISPR/Cas9 gene-editing system have now emerged as tools for site-directed mutagenesis in zebrafish. In particular, the versatility and high efficiency of CRISPR/Cas9 make it the tool of choice in most laboratories for the rapid and cost-effective generation of knockout fish lines (mating scheme exemplified in Figure 3).
Several transgenic zebrafish lines that are useful in kidney research are available. A transgenic line Tg(wt1b:EGFP), where the sequence of the fluorescent protein GFP is downstream to the Wilms tumor 1b promoter, can facilitate the identification of the proximal part of the pronephros (48). Similarly, the transgenic line Tg(cldnb:Lyn-GFP) (49,50) can facilitate the visualization of the distal part of the pronephros and the cloaca. The transgenic line Tg(βact:ARL13b-GFP), where GFP is fused to the mouse ARL13B protein, can be useful in the study of kidney ciliopathies, because it allows for the visualization of the ciliary membrane (51). These transgenic lines can be used to study functions of the kidney gene of interest by microinjecting embryos with morpholinos or by crossing them with mutant lines. Moreover, they can be used in association with automated microscopy to analyze potential kidney effects of a library of compounds in therapeutic or toxicology drug screens (52).
Along with these powerful transgenic lines that allow monitoring of kidney phenotypes, powerful functional studies can be used to study kidney function, such as filtration assays using a fluorescent dextran tracer, where dextran is perfused into the vasculature and is used to monitor glomerular filtration (47,53,54).
Limitations of the model include the fact that the zebrafish has many duplicated genes due to a whole-genome duplication event in early teleost evolution (55), which can complicate the generation of orthologous models for human disease. A more specific issue for kidney research is represented by the fact that a true ortholog of AQP2 is lacking in the zebrafish, which makes it an unsuitable model for the study of the genetic diseases affecting water homeostasis (40,56).
Mouse Models of Inherited Kidney Disease: the Gold Standard
The availability of tools to induce site-directed mutagenesis in the mouse using embryonic stem cells has historically established it as the most-used in vivo vertebrate model organism for genetic-medicine research, well before the advent of CRISPR/Cas gene-editing systems. The fact that the mouse is a mammalian model and its small size contribute to its popularity as a model organism.
Several landmark mouse models have contributed substantially to the identification of pathomechanisms in kidney disease (Table 3), one such example is Cosgrove et al.’s (57) mouse model of Alport syndrome, characterized by progressive GN with microhematuria and proteinuria, consistent with the human disease. Studies on this mouse contributed to the elucidation of the molecular alterations that characterize the glomerular basement membrane in Alport syndrome (57). Recently, miR-21, a small noncoding RNA, was shown to be upregulated in this mouse and its inhibition significantly slowed kidney disease progression (58). Currently, anti–miR-21 (RG-012) is being evaluated in human studies for Alport syndrome in a phase 2 clinical trial (NCT02855268).
In regard to tubular disorders, there are many orthologous and nonorthologous models of tubulointerstitial and cystic kidney disease.
Mice with kidney-specific inactivation of HNF1β develop PKD and display downregulation of Umod, Pkhd1, and Pkd2 genes, showing that HNF1β is at the center of a transcriptional network in kidney disease (59).
Umod null mice do not display anatomic or functional kidney alterations in normal conditions but were shown to have altered levels of transporters in the loop of Henle (60). More recently, mouse experiments showed a significant expression of uromodulin in the early part of mouse distal connecting tubule, where it is critical for tubule function, structure, and plasticity (61).
Some very elegant orthologous mouse models of ADPKD have been studied to unravel the pathogenicity of this disease, contributing to the formulation of the “two-hit hypothesis” (62–64), which sees the formation of cysts as a result of a loss of heterozygosity in PKD genes in the tubular epithelium and to the investigation of the role of cilium in cyst formation (65,66).
Importantly, the contribution of mouse models to the clarification of the pathomechanisms involved in kidney disease has also led to the identification of several promising therapeutic targets. In a few cases, compounds that were shown to modulate such targets and to ameliorate the kidney phenotype were then assessed for safety and efficacy in human studies. One major example of a drug now used in the treatment of inherited kidney disease as a result of successful translation of preclinical research findings in mouse is tolvaptan. Subsequent to the finding that kidney cAMP levels and expression of aquaporin-2 and vasopressin V2 receptor (V2R) were increased in Pkd2−/tm1Som (67), an orthologous model of ADPKD, and in other rodent models (68,69), the efficacy of V2R antagonists as modulators of the cystic phenotype was tested. Tolvaptan, a V2R antagonist, was shown to ameliorate kidney disease in PCK rats (70) and pcy mice (71). As a result of these preclinical studies, a landmark trial using tolvaptan in patients with ADPKD was published in 2012 and has significantly altered the treatment landscape for patients with this condition (72).
However, in terms of translatability of preclinical findings to clinical practice, tolvaptan represents the exception rather than the rule.
Concerns have been raised regarding the poor translational value of certain therapeutic conclusions drawn from preclinical models, including the mouse, in several research fields such as nephrology (73,74).
The use of nonorthologous mouse models and interspecies differences in gene expression and development may partially account for these discrepancies. Differences also exist in the end points considered when testing the efficacy of a treatment in mouse and human. Moreover, there is often a discrepancy in the stage of the disease when the treatment is administered, with mice often being treated at relatively early stages of the disease and patients often presenting with an established disease. Finally, in an attempt to minimize variance in animal experiments, inbred mouse strains are usually used in research. However, this genetic homogeneity hardly represents the genetic variability of the human population that is the prospective recipient of the treatment being tested (75).
Genetic background strongly influences the phenotype of inherited kidney disease in mouse; it has been shown to modulate the kidney phenotype in mouse models of Alport syndrome (76,77), cystinosis (78), nephrotic syndrome (79), and cystic kidney disease (80,81).
This should be taken into account when modeling kidney disease and can actually be used to gain information on how genetic modifiers contribute to kidney phenotype with important consequences on our understanding of kidney genetics and underlying disease mechanisms (76,81,82).
Mammalian Cellular Models: Growing Kidneys in a Dish
Monolayer cell cultures are attractive models for inherited kidney disease because they are cheap and easy to manipulate for the dissection of molecular pathways implicated in the disease and for the use in large high-throughput screens (83,84). There are several, well characterized cell lines commonly used in kidney research such as the mouse inner medullary collecting duct cell line (mIMCD-3) and the dog kidney cell line Madin–Darby canine kidney (MDCK).
Inherited kidney diseases are extremely heterogeneous and the phenotype can be strongly modulated by genetic background. Isolation and culture of primary cells from the patient can provide a useful human, patient-specific model of the disease that not only intrinsically possesses the primary genetic lesion without the need for gene editing but also conserves the genetic background information. Skin biopsy is an easy and minimally invasive way to obtain patient-specific fibroblasts that can be grown and studied in culture. Patient-specific fibroblasts have proven to be extremely useful to dissect the molecular pathomechanism of several inherited kidney diseases, such as nephronophthisis and cystic kidney disease in patients with Joubert syndrome (85,86). However, molecular pathways involved in kidney disease may not be active in nonkidney cells, highlighting the importance in studying certain mutations in the disease-relevant tissue context.
Urine-derived renal epithelial cells represent in this regard an extremely powerful, noninvasive source of disease-relevant, patient-specific primary material for kidney research. Kidney tubular epithelia are exposed to continuous passage of filtrate and living cells from these tubules are normally excreted daily within the urine. These cells can be isolated and specifically cultured to support proliferation of kidney epithelial cells (87).
It has recently been shown that urine-derived renal epithelial cells can be extremely powerful in vitro models to dissect pathomechanism of kidney diseases such as nephronophthisis (88) and Fabry disease (89). They can also be used to validate variants of unknown significance in patients with inherited kidney disease, as in the case of a synonymous variant in the nephronophthisis-associated gene NPHP3 that was shown to affect the gene splicing in a tissue-specific manner (90). Moreover, they can be used to identify novel therapeutic targets and test new therapies with a precision medicine approach (80,88,91).
In vitro two-dimensional (2D) cell models are therefore powerful models of kidney disease that are amenable to personalized and high-throughput studies (Figure 4).
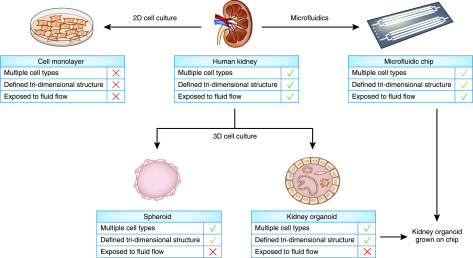
Figure 4.
The implementation of cell models for the study of inherited kidney disease is leading to complex human models that incorporate several features of the human kidney tissue. The human kidney is composed of multiple cell types (epithelial, interstitial, and endothelial cells) that are able to crosscommunicate and are arranged in a defined, hierarchical, tri-dimensional structure. The nephron epithelium is exposed to urine flow which may generate environmental cues potentially important for tissue homeostasis. The development of complex cell models holds the potential to reveal new pathophysiologic mechanisms of kidney disease.
However, the human kidney is characterized by a highly organized architecture and standard monolayer cultures fail to recapitulate cell-cell and cell–extracellular matrix interactions, polarity, and three-dimensional (3D) tissue structure of the kidney epithelium, which are particularly relevant in the pathogenicity of tubular disorders (92,93). To obtain more physiologically relevant 3D models to better mimic the phenotypes observed in vivo, many cell types traditionally used for 2D cell culture models of kidney disease have been adapted to grow in 3D spheroidal cellular aggregates. mIMDC-3, MDCK, and urine-derived renal epithelial cell lines can be grown in Matrigel to form luminated spheroids that mimic the polarized epithelial structure of kidney tubules and have been extensively used to study pathomechanisms of tubular disorders such as nephronophthisis (80,94–97) (Figure 4).
Another characteristic of the kidney tissue relevant when studying pathomechanisms of kidney disease is the coexistence within the kidney of multiple cell types. Transcriptomic analysis found evidences of crosstalk between cell types (98,99), which is essential to guide gene expression for correct response to stimuli and tissue homeostasis. Disturbance to this process can be extremely relevant for kidney disease (99) and the study of such mechanisms requires higher complexity systems.
Recent advances in stem cell research have led to new protocols that allow generation of kidney organoids from induced pluripotent stem cells (100–102). These organoids are characterized by unprecedented complexity due to the presence of segmented nephron and a rudimentary vasculature (Figure 4). These systems hold great potential in recapitulating kidney disease, as shown in a work focusing on kidney organoids with biallelic truncating mutations in PKD1 and PKD2, introduced with the CRISPR/Cas9 gene-editing system. Remarkably, fluid-filled cysts arose from tubular structures within PKD kidney organoids, albeit at low frequency in adhesion culture conditions (103). Interestingly, suspension culture of kidney organoids led to a tenfold increase in cystogenesis of PKD organoids, indicating that adherent cues and microenvironment may play a role in cyst formation (104).
Kidney organoids can be useful models not only for tubular disorders but also to study glomerulopathies. It has been shown that glomeruli sieved from kidney organoids display improved marker expression, polarity, and glomerular basement membrane matrisome when compared with podocyte 2D cultures; they are amenable to toxicity drug screens and can be used to model inherited glomerulopathies such as congenital nephrotic syndrome (105).
Despite these encouraging results, pilot studies to evaluate the ability of kidney organoids to mimic the phenotypic features of kidney disease have so far given variable results (103,104,106) and limitations such as reduced size and vasculature, immaturity (100,107), and lack/underrepresentation of certain cell types (107) may limit their utility as disease models. The introduction of environmental cues, such as exposure to flow stimuli, was shown to be essential for an enhanced vascularization and maturation of kidney organoids (108) (Figure 4). Indeed, growing kidney organoids under flow stimuli on millifluidic chips promotes the formation of vascular networks with perfusable lumens (108). These advances, together with efforts made using mouse embryonic stem cells to develop more anatomically realistic kidney organoids (102,109), can ultimately lead to the generation of functionally and structurally accurate human kidney models that have a urine drain.
Conclusions
Animal and cellular models are essential tools in the study of genetic kidney diseases. Such models have substantially expanded our knowledge of the genes and the pathways involved and have contributed to the identification of novel therapeutic targets.
So far, to draw conclusions on disease mechanisms to be ultimately applied to human, kidney and preclinical research in general have relied on the use of multiple disease models with complementary features that are summarized in this review.
Although the ease of manipulation of in vitro cellular models allows straightforward dissection of disease molecular mechanisms, the complexity of in vivo animal models supports the study of multiple cell types and tissue interactions in a pathophysiologic context. Mammalian cell cultures and zebrafish, due to their scalability, are particularly suitable for high-throughput studies such as forward genetics and drug screens. Such studies can then be taken forward in the translational pipeline, using the mouse for detailed study of pathomechanisms, as well as to investigate molecular mechanisms of action and pharmacokinetics of novel therapies. However, interspecies differences and the heterogeneity and the extremely low frequency of certain kidney disorders prompt for the implementation of more accurate and complex human patient–specific disease models.
Kidney organoids derived from patient-specific induced pluripotent stem cells—by combining in a single human model system several desirable features such as ease of manipulation, scalability (110,111), and a certain degree of complexity and suitability for precision medicine approaches (106)—hold great potential for the study of the pathomechanisms of kidney diseases. Improvements in morphologic and functional maturation, vascularization, and representation of ureteric epithelium within kidney organoids will be likely required to make them a fully competent tool for human disease modeling.
Several major questions in the nephrology field remain unanswered, such as the exact pathogenic mechanisms that lead to kidney fibrosis, kidney-cyst formation, and diabetic nephropathy. The hope is that the implementation of these powerful human models will in future significantly contribute to answer these questions while perhaps fueling new discoveries in the field of kidney regenerative medicine.
Disclosures
Dr. Sayer reports personal fees from Otsuka Pharmaceuticals Europe Ltd., during the conduct of the study. Dr. Molinari has nothing to disclose.
Funding
This work was supported by Kidney Research UK grant RP_006_20180227 and Northern Counties Kidney Research Fund grant 18/01.
Footnotes
Published online ahead of print. Publication date available at www.cjasn.org.
References
- 1.Devuyst O, Knoers NV, Remuzzi G, Schaefer F; Board of the Working Group for Inherited Kidney Diseases of the European Renal Association and European Dialysis and Transplant Association: Rare inherited kidney diseases: Challenges, opportunities, and perspectives. Lancet 383: 1844–1859, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Devuyst O, Olinger E, Weber S, Eckardt KU, Kmoch S, Rampoldi L, Bleyer AJ: Autosomal dominant tubulointerstitial kidney disease. Nat Rev Dis Primers 5: 60, 2019 [DOI] [PubMed] [Google Scholar]
- 3.Saleem MA: Molecular stratification of idiopathic nephrotic syndrome. Nat Rev Nephrol 15: 750–765, 2019 [DOI] [PubMed] [Google Scholar]
- 4.Sayer JA, Pearce SH: Diagnosis and clinical biochemistry of inherited tubulopathies. Ann Clin Biochem 38: 459–470, 2001 [DOI] [PubMed] [Google Scholar]
- 5.Harris PC, Torres VE: Polycystic kidney disease. Annu Rev Med 60: 321–337, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rossetti S, Harris PC: The genetics of vascular complications in autosomal dominant polycystic kidney disease (ADPKD). Curr Hypertens Rev 9: 37–43, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Russell E, McFarland E: Cystic kidneys, ck. Mouse News Lett 56: 40, 1977 [Google Scholar]
- 8.Nauta J, Ozawa Y, Sweeney WE Jr., Rutledge JC, Avner ED: Renal and biliary abnormalities in a new murine model of autosomal recessive polycystic kidney disease. Pediatr Nephrol 7: 163–172, 1993 [DOI] [PubMed] [Google Scholar]
- 9.Atala A, Freeman MR, Mandell J, Beier DR: Juvenile cystic kidneys (jck): A new mouse mutation which causes polycystic kidneys. Kidney Int 43: 1081–1085, 1993 [DOI] [PubMed] [Google Scholar]
- 10.Katsuyama M, Masuyama T, Komura I, Hibino T, Takahashi H: Characterization of a novel polycystic kidney rat model with accompanying polycystic liver. Exp Anim 49: 51–55, 2000 [DOI] [PubMed] [Google Scholar]
- 11.Barr MM, Sternberg PW: A polycystic kidney-disease gene homologue required for male mating behaviour in C. elegans. Nature 401: 386–389, 1999 [DOI] [PubMed] [Google Scholar]
- 12.Pazour GJ, San Agustin JT, Follit JA, Rosenbaum JL, Witman GB: Polycystin-2 localizes to kidney cilia and the ciliary level is elevated in orpk mice with polycystic kidney disease. Curr Biol 12: R378–R380, 2002 [DOI] [PubMed] [Google Scholar]
- 13.Yoder BK, Hou X, Guay-Woodford LM: The polycystic kidney disease proteins, polycystin-1, polycystin-2, polaris, and cystin, are co-localized in renal cilia. J Am Soc Nephrol 13: 2508–2516, 2002 [DOI] [PubMed] [Google Scholar]
- 14.Pazour GJ, Dickert BL, Vucica Y, Seeley ES, Rosenbaum JL, Witman GB, Cole DG: Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J Cell Biol 151: 709–718, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou X, Mrug M, Yoder BK, Lefkowitz EJ, Kremmidiotis G, D’Eustachio P, Beier DR, Guay-Woodford LM: Cystin, a novel cilia-associated protein, is disrupted in the cpk mouse model of polycystic kidney disease. J Clin Invest 109: 533–540, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cogswell C, Price SJ, Hou X, Guay-Woodford LM, Flaherty L, Bryda EC: Positional cloning of jcpk/bpk locus of the mouse. Mamm Genome 14: 242–249, 2003 [DOI] [PubMed] [Google Scholar]
- 17.Mohieldin AM, Haymour HS, Lo ST, AbouAlaiwi WA, Atkinson KF, Ward CJ, Gao M, Wessely O, Nauli SM: Protein composition and movements of membrane swellings associated with primary cilia. Cell Mol Life Sci 72: 2415–2429, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu S, Lu W, Obara T, Kuida S, Lehoczky J, Dewar K, Drummond IA, Beier DR: A defect in a novel Nek-family kinase causes cystic kidney disease in the mouse and in zebrafish. Development 129: 5839–5846, 2002 [DOI] [PubMed] [Google Scholar]
- 19.Ward CJ, Hogan MC, Rossetti S, Walker D, Sneddon T, Wang X, Kubly V, Cunningham JM, Bacallao R, Ishibashi M, Milliner DS, Torres VE, Harris PC: The gene mutated in autosomal recessive polycystic kidney disease encodes a large, receptor-like protein. Nat Genet 30: 259–269, 2002 [DOI] [PubMed] [Google Scholar]
- 20.Hildebrandt F, Otto E: Cilia and centrosomes: A unifying pathogenic concept for cystic kidney disease? Nat Rev Genet 6: 928–940, 2005 [DOI] [PubMed] [Google Scholar]
- 21.Ganner A, Neumann-Haefelin E: Genetic kidney diseases: Caenorhabditis elegans as model system. Cell Tissue Res 369: 105–118, 2017 [DOI] [PubMed] [Google Scholar]
- 22.Wei Q, Zhang Y, Li Y, Zhang Q, Ling K, Hu J: The BBSome controls IFT assembly and turnaround in cilia. Nat Cell Biol 14: 950–957, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen VL, Li C, Bowie RV, Clarke L, Mohan S, Blacque OE, Leroux MR: Formation of the transition zone by Mks5/Rpgrip1L establishes a ciliary zone of exclusion (CIZE) that compartmentalises ciliary signalling proteins and controls PIP2 ciliary abundance. EMBO J 34: 2537–2556, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lambacher NJ, Bruel AL, van Dam TJ, Szymańska K, Slaats GG, Kuhns S, McManus GJ, Kennedy JE, Gaff K, Wu KM, van der Lee R, Burglen L, Doummar D, Rivière JB, Faivre L, Attié-Bitach T, Saunier S, Curd A, Peckham M, Giles RH, Johnson CA, Huynen MA, Thauvin-Robinet C, Blacque OE: TMEM107 recruits ciliopathy proteins to subdomains of the ciliary transition zone and causes Joubert syndrome. Nat Cell Biol 18: 122–131, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niwa S: The nephronophthisis-related gene ift-139 is required for ciliogenesis in Caenorhabditis elegans. Sci Rep 6: 31544, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodan AR: The Drosophila Malpighian tubule as a model for mammalian tubule function. Curr Opin Nephrol Hypertens 28: 455–464, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Odenthal J, Brinkkoetter PT: Drosophila melanogaster and its nephrocytes: A versatile model for glomerular research. In: Methods in Cell Biology, Vol. 154, Amsterdam, The Netherlands, Elsevier, 2019, pp 217–240 [DOI] [PubMed] [Google Scholar]
- 28.Weavers H, Prieto-Sánchez S, Grawe F, Garcia-López A, Artero R, Wilsch-Bräuninger M, Ruiz-Gómez M, Skaer H, Denholm B: The insect nephrocyte is a podocyte-like cell with a filtration slit diaphragm. Nature 457: 322–326, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhuang S, Shao H, Guo F, Trimble R, Pearce E, Abmayr SM: Sns and Kirre, the Drosophila orthologs of Nephrin and Neph1, direct adhesion, fusion and formation of a slit diaphragm-like structure in insect nephrocytes. Development 136: 2335–2344, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reiter LT, Potocki L, Chien S, Gribskov M, Bier E: A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Res 11: 1114–1125, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chi T, Kim MS, Lang S, Bose N, Kahn A, Flechner L, Blaschko SD, Zee T, Muteliefu G, Bond N, Kolipinski M, Fakra SC, Mandel N, Miller J, Ramanathan A, Killilea DW, Brückner K, Kapahi P, Stoller ML: A Drosophila model identifies a critical role for zinc in mineralization for kidney stone disease. PLoS One 10: e0124150, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang H, Male M, Li Y, Wang N, Zhao C, Jin S, Hu J, Chen Z, Ye Z, Xu H: Efficacy of Hydroxy-L-proline (HYP) analogs in the treatment of primary hyperoxaluria in Drosophila Melanogaster. BMC Nephrol 19: 167, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brand AH, Perrimon N: Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415, 1993 [DOI] [PubMed] [Google Scholar]
- 34.Heigwer F, Port F, Boutros M: RNA interference (RNAi) screening in Drosophila. Genetics 208: 853–874, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hermle T, Braun DA, Helmstädter M, Huber TB, Hildebrandt F: Modeling monogenic human nephrotic syndrome in the Drosophila garland cell nephrocyte. J Am Soc Nephrol 28: 1521–1533, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu Y, Zhu JY, Richman A, Zhao Z, Zhang F, Ray PE, Han Z: A Drosophila model system to assess the function of human monogenic podocyte mutations that cause nephrotic syndrome. Hum Mol Genet 26: 768–780, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang F, Zhao Y, Han Z: An in vivo functional analysis system for renal gene discovery in Drosophila pericardial nephrocytes. J Am Soc Nephrol 24: 191–197, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hermle T, Schneider R, Schapiro D, Braun DA, van der Ven AT, Warejko JK, Daga A, Widmeier E, Nakayama M, Jobst-Schwan T, Majmundar AJ, Ashraf S, Rao J, Finn LS, Tasic V, Hernandez JD, Bagga A, Jalalah SM, El Desoky S, Kari JA, Laricchia KM, Lek M, Rehm HL, MacArthur DG, Mane S, Lifton RP, Shril S, Hildebrandt F: GAPVD1 and ANKFY1 mutations implicate RAB5 regulation in nephrotic syndrome. J Am Soc Nephrol 29: 2123–2138, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, Collins JE, Humphray S, McLaren K, Matthews L, McLaren S, Sealy I, Caccamo M, Churcher C, Scott C, Barrett JC, Koch R, Rauch GJ, White S, Chow W, Kilian B, Quintais LT, Guerra-Assunção JA, Zhou Y, Gu Y, Yen J, Vogel JH, Eyre T, Redmond S, Banerjee R, Chi J, Fu B, Langley E, Maguire SF, Laird GK, Lloyd D, Kenyon E, Donaldson S, Sehra H, Almeida-King J, Loveland J, Trevanion S, Jones M, Quail M, Willey D, Hunt A, Burton J, Sims S, McLay K, Plumb B, Davis J, Clee C, Oliver K, Clark R, Riddle C, Elliot D, Threadgold G, Harden G, Ware D, Begum S, Mortimore B, Kerry G, Heath P, Phillimore B, Tracey A, Corby N, Dunn M, Johnson C, Wood J, Clark S, Pelan S, Griffiths G, Smith M, Glithero R, Howden P, Barker N, Lloyd C, Stevens C, Harley J, Holt K, Panagiotidis G, Lovell J, Beasley H, Henderson C, Gordon D, Auger K, Wright D, Collins J, Raisen C, Dyer L, Leung K, Robertson L, Ambridge K, Leongamornlert D, McGuire S, Gilderthorp R, Griffiths C, Manthravadi D, Nichol S, Barker G, Whitehead S, Kay M, Brown J, Murnane C, Gray E, Humphries M, Sycamore N, Barker D, Saunders D, Wallis J, Babbage A, Hammond S, Mashreghi-Mohammadi M, Barr L, Martin S, Wray P, Ellington A, Matthews N, Ellwood M, Woodmansey R, Clark G, Cooper J, Tromans A, Grafham D, Skuce C, Pandian R, Andrews R, Harrison E, Kimberley A, Garnett J, Fosker N, Hall R, Garner P, Kelly D, Bird C, Palmer S, Gehring I, Berger A, Dooley CM, Ersan-Ürün Z, Eser C, Geiger H, Geisler M, Karotki L, Kirn A, Konantz J, Konantz M, Oberländer M, Rudolph-Geiger S, Teucke M, Lanz C, Raddatz G, Osoegawa K, Zhu B, Rapp A, Widaa S, Langford C, Yang F, Schuster SC, Carter NP, Harrow J, Ning Z, Herrero J, Searle SM, Enright A, Geisler R, Plasterk RH, Lee C, Westerfield M, de Jong PJ, Zon LI, Postlethwait JH, Nüsslein-Volhard C, Hubbard TJ, Roest Crollius H, Rogers J, Stemple DL: The zebrafish reference genome sequence and its relationship to the human genome [published correction appears in Nature 505: 248, 2014]. Nature 496: 498–503, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elmonem MA, Berlingerio SP, van den Heuvel LP, de Witte PA, Lowe M, Levtchenko EN: Genetic renal diseases: The emerging role of zebrafish models. Cells 7: 130, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun Z, Amsterdam A, Pazour GJ, Cole DG, Miller MS, Hopkins N: A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development 131: 4085–4093, 2004 [DOI] [PubMed] [Google Scholar]
- 42.Perrault I, Halbritter J, Porath JD, Gérard X, Braun DA, Gee HY, Fathy HM, Saunier S, Cormier-Daire V, Thomas S, Attié-Bitach T, Boddaert N, Taschner M, Schueler M, Lorentzen E, Lifton RP, Lawson JA, Garfa-Traore M, Otto EA, Bastin P, Caillaud C, Kaplan J, Rozet JM, Hildebrandt F: IFT81, encoding an IFT-B core protein, as a very rare cause of a ciliopathy phenotype. J Med Genet 52: 657–665, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Halbritter J, Bizet AA, Schmidts M, Porath JD, Braun DA, Gee HY, McInerney-Leo AM, Krug P, Filhol E, Davis EE, Airik R, Czarnecki PG, Lehman AM, Trnka P, Nitschké P, Bole-Feysot C, Schueler M, Knebelmann B, Burtey S, Szabó AJ, Tory K, Leo PJ, Gardiner B, McKenzie FA, Zankl A, Brown MA, Hartley JL, Maher ER, Li C, Leroux MR, Scambler PJ, Zhan SH, Jones SJ, Kayserili H, Tuysuz B, Moorani KN, Constantinescu A, Krantz ID, Kaplan BS, Shah JV, Hurd TW, Doherty D, Katsanis N, Duncan EL, Otto EA, Beales PL, Mitchison HM, Saunier S, Hildebrandt F; UK10K Consortium: Defects in the IFT-B component IFT172 cause Jeune and Mainzer-Saldino syndromes in humans. Am J Hum Genet 93: 915–925, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ryan S, Willer J, Marjoram L, Bagwell J, Mankiewicz J, Leshchiner I, Goessling W, Bagnat M, Katsanis N: Rapid identification of kidney cyst mutations by whole exome sequencing in zebrafish. Development 140: 4445–4451, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.El-Brolosy MA, Kontarakis Z, Rossi A, Kuenne C, Günther S, Fukuda N, Kikhi K, Boezio GLM, Takacs CM, Lai SL, Fukuda R, Gerri C, Giraldez AJ, Stainier DYR: Genetic compensation triggered by mutant mRNA degradation. Nature 568: 193–197, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stainier DYR, Raz E, Lawson ND, Ekker SC, Burdine RD, Eisen JS, Ingham PW, Schulte-Merker S, Yelon D, Weinstein BM, Mullins MC, Wilson SW, Ramakrishnan L, Amacher SL, Neuhauss SCF, Meng A, Mochizuki N, Panula P, Moens CB: Guidelines for morpholino use in zebrafish. PLoS Genet 13: e1007000, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hall G, Gbadegesin RA, Lavin P, Wu G, Liu Y, Oh EC, Wang L, Spurney RF, Eckel J, Lindsey T, Homstad A, Malone AF, Phelan PJ, Shaw A, Howell DN, Conlon PJ, Katsanis N, Winn MP: A novel missense mutation of Wilms’ Tumor 1 causes autosomal dominant FSGS. J Am Soc Nephrol 26: 831–843, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perner B, Englert C, Bollig F: The Wilms tumor genes wt1a and wt1b control different steps during formation of the zebrafish pronephros. Dev Biol 309: 87–96, 2007 [DOI] [PubMed] [Google Scholar]
- 49.Haas P, Gilmour D: Chemokine signaling mediates self-organizing tissue migration in the zebrafish lateral line. Dev Cell 10: 673–680, 2006 [DOI] [PubMed] [Google Scholar]
- 50.Simms RJ, Hynes AM, Eley L, Inglis D, Chaudhry B, Dawe HR, Sayer JA: Modelling a ciliopathy: Ahi1 knockdown in model systems reveals an essential role in brain, retinal, and renal development. Cell Mol Life Sci 69: 993–1009, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Borovina A, Superina S, Voskas D, Ciruna B: Vangl2 directs the posterior tilting and asymmetric localization of motile primary cilia. Nat Cell Biol 12: 407–412, 2010 [DOI] [PubMed] [Google Scholar]
- 52.Westhoff JH, Giselbrecht S, Schmidts M, Schindler S, Beales PL, Tönshoff B, Liebel U, Gehrig J: Development of an automated imaging pipeline for the analysis of the zebrafish larval kidney. PLoS One 8: e82137, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kramer-Zucker AG, Wiessner S, Jensen AM, Drummond IA: Organization of the pronephric filtration apparatus in zebrafish requires Nephrin, Podocin and the FERM domain protein Mosaic eyes. Dev Biol 285: 316–329, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hinkes B, Wiggins RC, Gbadegesin R, Vlangos CN, Seelow D, Nürnberg G, Garg P, Verma R, Chaib H, Hoskins BE, Ashraf S, Becker C, Hennies HC, Goyal M, Wharram BL, Schachter AD, Mudumana S, Drummond I, Kerjaschki D, Waldherr R, Dietrich A, Ozaltin F, Bakkaloglu A, Cleper R, Basel-Vanagaite L, Pohl M, Griebel M, Tsygin AN, Soylu A, Müller D, Sorli CS, Bunney TD, Katan M, Liu J, Attanasio M, O’toole JF, Hasselbacher K, Mucha B, Otto EA, Airik R, Kispert A, Kelley GG, Smrcka AV, Gudermann T, Holzman LB, Nürnberg P, Hildebrandt F: Positional cloning uncovers mutations in PLCE1 responsible for a nephrotic syndrome variant that may be reversible. Nat Genet 38: 1397–1405, 2006 [DOI] [PubMed] [Google Scholar]
- 55.Glasauer SM, Neuhauss SC: Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol Genet Genomics 289: 1045–1060, 2014 [DOI] [PubMed] [Google Scholar]
- 56.Tingaud-Sequeira A, Calusinska M, Finn RN, Chauvigné F, Lozano J, Cerdà J: The zebrafish genome encodes the largest vertebrate repertoire of functional aquaporins with dual paralogy and substrate specificities similar to mammals. BMC Evol Biol 10: 38, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cosgrove D, Meehan DT, Grunkemeyer JA, Kornak JM, Sayers R, Hunter WJ, Samuelson GC: Collagen COL4A3 knockout: A mouse model for autosomal Alport syndrome. Genes Dev 10: 2981–2992, 1996 [DOI] [PubMed] [Google Scholar]
- 58.Gomez IG, MacKenna DA, Johnson BG, Kaimal V, Roach AM, Ren S, Nakagawa N, Xin C, Newitt R, Pandya S, Xia TH, Liu X, Borza DB, Grafals M, Shankland SJ, Himmelfarb J, Portilla D, Liu S, Chau BN, Duffield JS: Anti-microRNA-21 oligonucleotides prevent Alport nephropathy progression by stimulating metabolic pathways. J Clin Invest 125: 141–156, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gresh L, Fischer E, Reimann A, Tanguy M, Garbay S, Shao X, Hiesberger T, Fiette L, Igarashi P, Yaniv M, Pontoglio M: A transcriptional network in polycystic kidney disease. EMBO J 23: 1657–1668, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bachmann S, Mutig K, Bates J, Welker P, Geist B, Gross V, Luft FC, Alenina N, Bader M, Thiele BJ, Prasadan K, Raffi HS, Kumar S: Renal effects of Tamm-Horsfall protein (uromodulin) deficiency in mice. Am J Physiol Renal Physiol 288: F559–F567, 2005 [DOI] [PubMed] [Google Scholar]
- 61.Tokonami N, Takata T, Beyeler J, Ehrbar I, Yoshifuji A, Christensen EI, Loffing J, Devuyst O, Olinger EG: Uromodulin is expressed in the distal convoluted tubule, where it is critical for regulation of the sodium chloride cotransporter NCC. Kidney Int 94: 701–715, 2018 [DOI] [PubMed] [Google Scholar]
- 62.Wu G, D’Agati V, Cai Y, Markowitz G, Park JH, Reynolds DM, Maeda Y, Le TC, Hou H Jr., Kucherlapati R, Edelmann W, Somlo S: Somatic inactivation of Pkd2 results in polycystic kidney disease. Cell 93: 177–188, 1998 [DOI] [PubMed] [Google Scholar]
- 63.Pritchard L, Sloane-Stanley JA, Sharpe JA, Aspinwall R, Lu W, Buckle V, Strmecki L, Walker D, Ward CJ, Alpers CE, Zhou J, Wood WG, Harris PC: A human PKD1 transgene generates functional polycystin-1 in mice and is associated with a cystic phenotype. Hum Mol Genet 9: 2617–2627, 2000 [DOI] [PubMed] [Google Scholar]
- 64.Chang MY, Parker E, Ibrahim S, Shortland JR, Nahas ME, Haylor JL, Ong AC: Haploinsufficiency of Pkd2 is associated with increased tubular cell proliferation and interstitial fibrosis in two murine Pkd2 models. Nephrol Dial Transplant 21: 2078–2084, 2006 [DOI] [PubMed] [Google Scholar]
- 65.Ma M, Tian X, Igarashi P, Pazour GJ, Somlo S: Loss of cilia suppresses cyst growth in genetic models of autosomal dominant polycystic kidney disease. Nat Genet 45: 1004–1012, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walker RV, Keynton JL, Grimes DT, Sreekumar V, Williams DJ, Esapa C, Wu D, Knight MM, Norris DP: Ciliary exclusion of Polycystin-2 promotes kidney cystogenesis in an autosomal dominant polycystic kidney disease model. Nat Commun 10: 4072, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Torres VE, Wang X, Qian Q, Somlo S, Harris PC, Gattone VH 2nd: Effective treatment of an orthologous model of autosomal dominant polycystic kidney disease. Nat Med 10: 363–364, 2004 [DOI] [PubMed] [Google Scholar]
- 68.Gattone VH 2nd, Maser RL, Tian C, Rosenberg JM, Branden MG: Developmental expression of urine concentration-associated genes and their altered expression in murine infantile-type polycystic kidney disease. Dev Genet 24: 309–318, 1999 [DOI] [PubMed] [Google Scholar]
- 69.Gattone VH 2nd, Wang X, Harris PC, Torres VE: Inhibition of renal cystic disease development and progression by a vasopressin V2 receptor antagonist. Nat Med 9: 1323–1326, 2003 [DOI] [PubMed] [Google Scholar]
- 70.Wang X, Gattone V 2nd, Harris PC, Torres VE: Effectiveness of vasopressin V2 receptor antagonists OPC-31260 and OPC-41061 on polycystic kidney disease development in the PCK rat. J Am Soc Nephrol 16: 846–851, 2005 [DOI] [PubMed] [Google Scholar]
- 71.Aihara M, Fujiki H, Mizuguchi H, Hattori K, Ohmoto K, Ishikawa M, Nagano K, Yamamura Y: Tolvaptan delays the onset of end-stage renal disease in a polycystic kidney disease model by suppressing increases in kidney volume and renal injury. J Pharmacol Exp Ther 349: 258–267, 2014 [DOI] [PubMed] [Google Scholar]
- 72.Torres VE, Chapman AB, Devuyst O, Gansevoort RT, Grantham JJ, Higashihara E, Perrone RD, Krasa HB, Ouyang J, Czerwiec FS; TEMPO 3:4 Trial Investigators: Tolvaptan in patients with autosomal dominant polycystic kidney disease. N Engl J Med 367: 2407–2418, 2012. 23121377 [Google Scholar]
- 73.Mak IW, Evaniew N, Ghert M: Lost in translation: Animal models and clinical trials in cancer treatment. Am J Transl Res 6: 114–118, 2014 [PMC free article] [PubMed] [Google Scholar]
- 74.Molinari E, Sayer JA: Emerging treatments and personalised medicine for ciliopathies associated with cystic kidney disease. Expert Opin Orphan Drugs 5: 785–798, 2017 [Google Scholar]
- 75.Becker GJ, Hewitson TD: Animal models of chronic kidney disease: Useful but not perfect. Nephrol Dial Transplant 28: 2432–2438, 2013 [DOI] [PubMed] [Google Scholar]
- 76.Andrews KL, Mudd JL, Li C, Miner JH: Quantitative trait loci influence renal disease progression in a mouse model of Alport syndrome. Am J Pathol 160: 721–730, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Korstanje R, Caputo CR, Doty RA, Cook SA, Bronson RT, Davisson MT, Miner JH: A mouse Col4a4 mutation causing Alport glomerulosclerosis with abnormal collagen α3α4α5(IV) trimers. Kidney Int 85: 1461–1468, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nevo N, Chol M, Bailleux A, Kalatzis V, Morisset L, Devuyst O, Gubler MC, Antignac C: Renal phenotype of the cystinosis mouse model is dependent upon genetic background. Nephrol Dial Transplant 25: 1059–1066, 2010 [DOI] [PubMed] [Google Scholar]
- 79.Roselli S, Heidet L, Sich M, Henger A, Kretzler M, Gubler MC, Antignac C: Early glomerular filtration defect and severe renal disease in podocin-deficient mice. Mol Cell Biol 24: 550–560, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hynes AM, Giles RH, Srivastava S, Eley L, Whitehead J, Danilenko M, Raman S, Slaats GG, Colville JG, Ajzenberg H, Kroes HY, Thelwall PE, Simmons NL, Miles CG, Sayer JA: Murine Joubert syndrome reveals Hedgehog signaling defects as a potential therapeutic target for nephronophthisis. Proc Natl Acad Sci U S A 111: 9893–9898, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Woo DD, Nguyen DK, Khatibi N, Olsen P: Genetic identification of two major modifier loci of polycystic kidney disease progression in pcy mice. J Clin Invest 100: 1934–1940, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ramsbottom S, Miles C, Sayer J: Murine Cep290 phenotypes are modified by genetic backgrounds and provide an impetus for investigating disease modifier alleles. F1000 Res 4: 590, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wheway G, Schmidts M, Mans DA, Szymanska K, Nguyen TT, Racher H, Phelps IG, Toedt G, Kennedy J, Wunderlich KA, Sorusch N, Abdelhamed ZA, Natarajan S, Herridge W, van Reeuwijk J, Horn N, Boldt K, Parry DA, Letteboer SJF, Roosing S, Adams M, Bell SM, Bond J, Higgins J, Morrison EE, Tomlinson DC, Slaats GG, van Dam TJP, Huang L, Kessler K, Giessl A, Logan CV, Boyle EA, Shendure J, Anazi S, Aldahmesh M, Al Hazzaa S, Hegele RA, Ober C, Frosk P, Mhanni AA, Chodirker BN, Chudley AE, Lamont R, Bernier FP, Beaulieu CL, Gordon P, Pon R, Donahue C, Barkovich AJ, Wolf L, Toomes C, Thiel CT, Boycott KM, McKibbin M, Inglehearn CF, Stewart F, Omran H, Huynen MA, Sergouniotis PI, Alkuraya FS, Parboosingh JS, Innes AM, Willoughby CE, Giles RH, Webster AR, Ueffing M, Blacque O, Gleeson JG, Wolfrum U, Beales PL, Gibson T, Doherty D, Mitchison HM, Roepman R, Johnson CA; UK10K Consortium;University of Washington Center for Mendelian Genomics: An siRNA-based functional genomics screen for the identification of regulators of ciliogenesis and ciliopathy genes. Nat Cell Biol 17: 1074–1087, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim YJ, Kim S, Jung Y, Jung E, Kwon HJ, Kim J: Eupatilin rescues ciliary transition zone defects to ameliorate ciliopathy-related phenotypes. J Clin Invest 128: 3642–3648, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shimada H, Lu Q, Insinna-Kettenhofen C, Nagashima K, English MA, Semler EM, Mahgerefteh J, Cideciyan AV, Li T, Brooks BP, Gunay-Aygun M, Jacobson SG, Cogliati T, Westlake CJ, Swaroop A: In Vitro modeling using ciliopathy-patient-derived cells reveals distinct cilia dysfunctions caused by CEP290 mutations. Cell Reports 20: 384–396, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Alkanderi S, Molinari E, Shaheen R, Elmaghloob Y, Stephen LA, Sammut V, Ramsbottom SA, Srivastava S, Cairns G, Edwards N, Rice SJ, Ewida N, Alhashem A, White K, Miles CG, Steel DH, Alkuraya FS, Ismail S, Sayer JA: ARL3 mutations cause Joubert syndrome by disrupting ciliary protein composition. Am J Hum Genet 103: 612–620, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhou T, Benda C, Duzinger S, Huang Y, Li X, Li Y, Guo X, Cao G, Chen S, Hao L, Chan YC, Ng KM, Ho JC, Wieser M, Wu J, Redl H, Tse HF, Grillari J, Grillari-Voglauer R, Pei D, Esteban MA: Generation of induced pluripotent stem cells from urine. J Am Soc Nephrol 22: 1221–1228, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Srivastava S, Ramsbottom SA, Molinari E, Alkanderi S, Filby A, White K, Henry C, Saunier S, Miles CG, Sayer JA: A human patient-derived cellular model of Joubert syndrome reveals ciliary defects which can be rescued with targeted therapies. Hum Mol Genet 26: 4657–4667, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Slaats GG, Braun F, Hoehne M, Frech LE, Blomberg L, Benzing T, Schermer B, Rinschen MM, Kurschat CE: Urine-derived cells: A promising diagnostic tool in Fabry disease patients. Sci Rep 8: 11042, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Molinari E, Decker E, Mabillard H, Tellez J, Srivastava S, Raman S, Wood K, Kempf C, Alkanderi S, Ramsbottom SA, Miles CG, Johnson CA, Hildebrandt F, Bergmann C, Sayer JA: Human urine-derived renal epithelial cells provide insights into kidney-specific alternate splicing variants. Eur J Hum Genet 26: 1791–1796, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ramsbottom SA, Molinari E, Srivastava S, Silberman F, Henry C, Alkanderi S, Devlin LA, White K, Steel DH, Saunier S, Miles CG, Sayer JA: Targeted exon skipping of a CEP290 mutation rescues Joubert syndrome phenotypes in vitro and in a murine model. Proc Natl Acad Sci U S A 115: 12489–12494, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wilson PD: Apico-basal polarity in polycystic kidney disease epithelia. Biochim Biophys Acta 1812: 1239–1248, 2011 [DOI] [PubMed] [Google Scholar]
- 93.Schlüter MA, Margolis B: Apicobasal polarity in the kidney. Exp Cell Res 318: 1033–1039, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ghosh AK, Hurd T, Hildebrandt F: 3D spheroid defects in NPHP knockdown cells are rescued by the somatostatin receptor agonist octreotide. Am J Physiol Renal Physiol 303: F1225–F1229, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Otto EA, Hurd TW, Airik R, Chaki M, Zhou W, Stoetzel C, Patil SB, Levy S, Ghosh AK, Murga-Zamalloa CA, van Reeuwijk J, Letteboer SJ, Sang L, Giles RH, Liu Q, Coene KL, Estrada-Cuzcano A, Collin RW, McLaughlin HM, Held S, Kasanuki JM, Ramaswami G, Conte J, Lopez I, Washburn J, Macdonald J, Hu J, Yamashita Y, Maher ER, Guay-Woodford LM, Neumann HP, Obermüller N, Koenekoop RK, Bergmann C, Bei X, Lewis RA, Katsanis N, Lopes V, Williams DS, Lyons RH, Dang CV, Brito DA, Dias MB, Zhang X, Cavalcoli JD, Nürnberg G, Nürnberg P, Pierce EA, Jackson PK, Antignac C, Saunier S, Roepman R, Dollfus H, Khanna H, Hildebrandt F: Candidate exome capture identifies mutation of SDCCAG8 as the cause of a retinal-renal ciliopathy. Nat Genet 42: 840–850, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chaki M, Airik R, Ghosh AK, Giles RH, Chen R, Slaats GG, Wang H, Hurd TW, Zhou W, Cluckey A, Gee HY, Ramaswami G, Hong CJ, Hamilton BA, Cervenka I, Ganji RS, Bryja V, Arts HH, van Reeuwijk J, Oud MM, Letteboer SJ, Roepman R, Husson H, Ibraghimov-Beskrovnaya O, Yasunaga T, Walz G, Eley L, Sayer JA, Schermer B, Liebau MC, Benzing T, Le Corre S, Drummond I, Janssen S, Allen SJ, Natarajan S, O’Toole JF, Attanasio M, Saunier S, Antignac C, Koenekoop RK, Ren H, Lopez I, Nayir A, Stoetzel C, Dollfus H, Massoudi R, Gleeson JG, Andreoli SP, Doherty DG, Lindstrad A, Golzio C, Katsanis N, Pape L, Abboud EB, Al-Rajhi AA, Lewis RA, Omran H, Lee EY, Wang S, Sekiguchi JM, Saunders R, Johnson CA, Garner E, Vanselow K, Andersen JS, Shlomai J, Nurnberg G, Nurnberg P, Levy S, Smogorzewska A, Otto EA, Hildebrandt F: Exome capture reveals ZNF423 and CEP164 mutations, linking renal ciliopathies to DNA damage response signaling. Cell 150: 533–548, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Choi HJC, Lin JR, Vannier JB, Slaats GG, Kile AC, Paulsen RD, Manning DK, Beier DR, Giles RH, Boulton SJ, Cimprich KA: NEK8 links the ATR-regulated replication stress response and S phase CDK activity to renal ciliopathies. Mol Cell 51: 423–439, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen L, Lee JW, Chou CL, Nair AV, Battistone MA, Păunescu TG, Merkulova M, Breton S, Verlander JW, Wall SM, Brown D, Burg MB, Knepper MA: Transcriptomes of major renal collecting duct cell types in mouse identified by single-cell RNA-seq. Proc Natl Acad Sci U S A 114: E9989–E9998, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Park J, Shrestha R, Qiu C, Kondo A, Huang S, Werth M, Li M, Barasch J, Suszták K: Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science 360: 758–763, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Takasato M, Er PX, Chiu HS, Maier B, Baillie GJ, Ferguson C, Parton RG, Wolvetang EJ, Roost MS, Chuva de Sousa Lopes SM, Little MH: Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526: 564–568, 2015 [DOI] [PubMed] [Google Scholar]
- 101.Morizane R, Lam AQ, Freedman BS, Kishi S, Valerius MT, Bonventre JV: Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat Biotechnol 33: 1193–1200, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Taguchi A, Nishinakamura R: Higher-order kidney organogenesis from pluripotent stem cells. Cell Stem Cell 21: 730–746.e6, 2017 [DOI] [PubMed] [Google Scholar]
- 103.Freedman BS, Brooks CR, Lam AQ, Fu H, Morizane R, Agrawal V, Saad AF, Li MK, Hughes MR, Werff RV, Peters DT, Lu J, Baccei A, Siedlecki AM, Valerius MT, Musunuru K, McNagny KM, Steinman TI, Zhou J, Lerou PH, Bonventre JV: . Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat Commun 6: 8715, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cruz NM, Song X, Czerniecki SM, Gulieva RE, Churchill AJ, Kim YK, Winston K, Tran LM, Diaz MA, Fu H, Finn LS, Pei Y, Himmelfarb J, Freedman BS: Organoid cystogenesis reveals a critical role of microenvironment in human polycystic kidney disease. Nat Mater 16: 1112–1119, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hale LJ, Howden SE, Phipson B, Lonsdale A, Er PX, Ghobrial I, Hosawi S, Wilson S, Lawlor KT, Khan S, Oshlack A, Quinlan C, Lennon R, Little MH: 3D organoid-derived human glomeruli for personalised podocyte disease modelling and drug screening. Nat Commun 9: 5167, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Forbes TA, Howden SE, Lawlor K, Phipson B, Maksimovic J, Hale L, Wilson S, Quinlan C, Ho G, Holman K, Bennetts B, Crawford J, Trnka P, Oshlack A, Patel C, Mallett A, Simons C, Little MH: Patient-iPSC-Derived kidney organoids show functional validation of a ciliopathic renal phenotype and reveal underlying pathogenetic mechanisms. Am J Hum Genet 102: 816–831, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wu H, Uchimura K, Donnelly EL, Kirita Y, Morris SA, Humphreys BD: Comparative analysis and refinement of human PSC-derived kidney organoid differentiation with single-cell transcriptomics. Cell Stem Cell 23: 869–881.e8, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Homan KA, Gupta N, Kroll KT, Kolesky DB, Skylar-Scott M, Miyoshi T, Mau D, Valerius MT, Ferrante T, Bonventre JV, Lewis JA, Morizane R: Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nat Methods 16: 255–262, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mills CG, Lawrence ML, Munro DAD, Elhendawi M, Mullins JJ, Davies JA: Asymmetric BMP4 signalling improves the realism of kidney organoids. Sci Rep 7: 14824, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Czerniecki SM, Cruz NM, Harder JL, Menon R, Annis J, Otto EA, Gulieva RE, Islas LV, Kim YK, Tran LM, Martins TJ, Pippin JW, Fu H, Kretzler M, Shankland SJ, Himmelfarb J, Moon RT, Paragas N, Freedman BS: High-throughput screening enhances kidney organoid differentiation from human pluripotent stem cells and enables Automated multidimensional phenotyping. Cell Stem Cell 22: 929–940.e4, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kumar SV, Er PX, Lawlor KT, Motazedian A, Scurr M, Ghobrial I, Combes AN, Zappia L, Oshlack A, Stanley EG, Little MH: Kidney micro-organoids in suspension culture as a scalable source of human pluripotent stem cell-derived kidney cells. Development 146: dev172361, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shin SE, Lim JM, Koh HG, Kim EK, Kang NK, Jeon S, Kwon S, Shin WS, Lee B, Hwangbo K, Kim J, Ye SH, Yun JY, Seo H, Oh HM, Kim KJ, Kim JS, Jeong WJ, Chang YK, Jeong BR: CRISPR/Cas9-induced knockout and knock-in mutations in Chlamydomonas reinhardtii. Sci Rep 6: 27810, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Baek K, Kim DH, Jeong J, Sim SJ, Melis A, Kim JS, Jin E, Bae S: DNA-free two-gene knockout in Chlamydomonas reinhardtii via CRISPR-Cas9 ribonucleoproteins. Sci Rep 6: 30620, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li X, Zhang R, Patena W, Gang SS, Blum SR, Ivanova N, Yue R, Robertson JM, Lefebvre PA, Fitz-Gibbon ST, Grossman AR, Jonikas MC: An indexed, mapped mutant library enables reverse genetics studies of biological processes in Chlamydomonas reinhardtii. Plant Cell 28: 367–387, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cheng X, Liu G, Ke W, Zhao L, Lv B, Ma X, Xu N, Xia X, Deng X, Zheng C, Huang K: Building a multipurpose insertional mutant library for forward and reverse genetics in Chlamydomonas. Plant Methods 13: 36, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jack B, Avasthi P: Chemical screening for flagella-associated phenotypes in Chlamydomonas reinhardtii. In: Plant Chemical Genomics, edited by Fauser F, Jonikas M, Vol. 1795, New York, NY, Springer New York, 2018, pp 203–221 [DOI] [PubMed] [Google Scholar]
- 117.Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, Marshall WF, Qu LH, Nelson DR, Sanderfoot AA, Spalding MH, Kapitonov VV, Ren Q, Ferris P, Lindquist E, Shapiro H, Lucas SM, Grimwood J, Schmutz J, Cardol P, Cerutti H, Chanfreau G, Chen CL, Cognat V, Croft MT, Dent R, Dutcher S, Fernández E, Fukuzawa H, González-Ballester D, González-Halphen D, Hallmann A, Hanikenne M, Hippler M, Inwood W, Jabbari K, Kalanon M, Kuras R, Lefebvre PA, Lemaire SD, Lobanov AV, Lohr M, Manuell A, Meier I, Mets L, Mittag M, Mittelmeier T, Moroney JV, Moseley J, Napoli C, Nedelcu AM, Niyogi K, Novoselov SV, Paulsen IT, Pazour G, Purton S, Ral JP, Riaño-Pachón DM, Riekhof W, Rymarquis L, Schroda M, Stern D, Umen J, Willows R, Wilson N, Zimmer SL, Allmer J, Balk J, Bisova K, Chen CJ, Elias M, Gendler K, Hauser C, Lamb MR, Ledford H, Long JC, Minagawa J, Page MD, Pan J, Pootakham W, Roje S, Rose A, Stahlberg E, Terauchi AM, Yang P, Ball S, Bowler C, Dieckmann CL, Gladyshev VN, Green P, Jorgensen R, Mayfield S, Mueller-Roeber B, Rajamani S, Sayre RT, Brokstein P, Dubchak I, Goodstein D, Hornick L, Huang YW, Jhaveri J, Luo Y, Martínez D, Ngau WC, Otillar B, Poliakov A, Porter A, Szajkowski L, Werner G, Zhou K, Grigoriev IV, Rokhsar DS, Grossman AR: The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, Li H, Blacque OE, Li L, Leitch CC, Lewis RA, Green JS, Parfrey PS, Leroux MR, Davidson WS, Beales PL, Guay-Woodford LM, Yoder BK, Stormo GD, Katsanis N, Dutcher SK: Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell 117: 541–552, 2004 [DOI] [PubMed] [Google Scholar]
- 119.Masyukova SV, Landis DE, Henke SJ, Williams CL, Pieczynski JN, Roszczynialski KN, Covington JE, Malarkey EB, Yoder BK: A screen for modifiers of cilia phenotypes reveals novel MKS alleles and uncovers a specific genetic interaction between osm-3 and nphp-4. PLoS Genet 12: e1005841, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shaye DD, Greenwald I: OrthoList: A compendium of C. elegans genes with human orthologs [published correction appears in PLoS One 9(1), 2014]. PLoS One 6: e20085, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV: Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature 426: 83–87, 2003 [DOI] [PubMed] [Google Scholar]
- 122.Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, Shen Y, Pervouchine DD, Djebali S, Thurman RE, Kaul R, Rynes E, Kirilusha A, Marinov GK, Williams BA, Trout D, Amrhein H, Fisher-Aylor K, Antoshechkin I, DeSalvo G, See LH, Fastuca M, Drenkow J, Zaleski C, Dobin A, Prieto P, Lagarde J, Bussotti G, Tanzer A, Denas O, Li K, Bender MA, Zhang M, Byron R, Groudine MT, McCleary D, Pham L, Ye Z, Kuan S, Edsall L, Wu YC, Rasmussen MD, Bansal MS, Kellis M, Keller CA, Morrissey CS, Mishra T, Jain D, Dogan N, Harris RS, Cayting P, Kawli T, Boyle AP, Euskirchen G, Kundaje A, Lin S, Lin Y, Jansen C, Malladi VS, Cline MS, Erickson DT, Kirkup VM, Learned K, Sloan CA, Rosenbloom KR, Lacerda de Sousa B, Beal K, Pignatelli M, Flicek P, Lian J, Kahveci T, Lee D, Kent WJ, Ramalho Santos M, Herrero J, Notredame C, Johnson A, Vong S, Lee K, Bates D, Neri F, Diegel M, Canfield T, Sabo PJ, Wilken MS, Reh TA, Giste E, Shafer A, Kutyavin T, Haugen E, Dunn D, Reynolds AP, Neph S, Humbert R, Hansen RS, De Bruijn M, Selleri L, Rudensky A, Josefowicz S, Samstein R, Eichler EE, Orkin SH, Levasseur D, Papayannopoulou T, Chang KH, Skoultchi A, Gosh S, Disteche C, Treuting P, Wang Y, Weiss MJ, Blobel GA, Cao X, Zhong S, Wang T, Good PJ, Lowdon RF, Adams LB, Zhou XQ, Pazin MJ, Feingold EA, Wold B, Taylor J, Mortazavi A, Weissman SM, Stamatoyannopoulos JA, Snyder MP, Guigo R, Gingeras TR, Gilbert DM, Hardison RC, Beer MA, Ren B; Mouse ENCODE Consortium: A comparative encyclopedia of DNA elements in the mouse genome. Nature 515: 355–364, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lindström NO, McMahon JA, Guo J, Tran T, Guo Q, Rutledge E, Parvez RK, Saribekyan G, Schuler RE, Liao C, Kim AD, Abdelhalim A, Ruffins SW, Thornton ME, Baskin L, Grubbs B, Kesselman C, McMahon AP: Conserved and divergent features of human and mouse kidney organogenesis. J Am Soc Nephrol 29: 785–805, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Giles RH, Ajzenberg H, Jackson PK: 3D spheroid model of mIMCD3 cells for studying ciliopathies and renal epithelial disorders. Nat Protoc 9: 2725–2731, 2014 [DOI] [PubMed] [Google Scholar]
- 125.Little MH, Combes AN: Kidney organoids: Accurate models or fortunate accidents. Genes Dev 33: 1319–1345, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Xiao T, Baier H: Lamina-specific axonal projections in the zebrafish tectum require the type IV collagen Dragnet. Nat Neurosci 10: 1529–1537, 2007 [DOI] [PubMed] [Google Scholar]
- 127.Putaala H, Soininen R, Kilpeläinen P, Wartiovaara J, Tryggvason K: The murine nephrin gene is specifically expressed in kidney, brain and pancreas: Inactivation of the gene leads to massive proteinuria and neonatal death. Hum Mol Genet 10: 1–8, 2001 [DOI] [PubMed] [Google Scholar]
- 128.Kreidberg JA, Sariola H, Loring JM, Maeda M, Pelletier J, Housman D, Jaenisch R: WT-1 is required for early kidney development. Cell 74: 679–691, 1993 [DOI] [PubMed] [Google Scholar]
- 129.Gorski M, Tin A, Garnaas M, McMahon GM, Chu AY, Tayo BO, Pattaro C, Teumer A, Chasman DI, Chalmers J, Hamet P, Tremblay J, Woodward M, Aspelund T, Eiriksdottir G, Gudnason V, Harris TB, Launer LJ, Smith AV, Mitchell BD, O’Connell JR, Shuldiner AR, Coresh J, Li M, Freudenberger P, Hofer E, Schmidt H, Schmidt R, Holliday EG, Mitchell P, Wang JJ, de Boer IH, Li G, Siscovick DS, Kutalik Z, Corre T, Vollenweider P, Waeber G, Gupta J, Kanetsky PA, Hwang SJ, Olden M, Yang Q, de Andrade M, Atkinson EJ, Kardia SL, Turner ST, Stafford JM, Ding J, Liu Y, Barlassina C, Cusi D, Salvi E, Staessen JA, Ridker PM, Grallert H, Meisinger C, Müller-Nurasyid M, Krämer BK, Kramer H, Rosas SE, Nolte IM, Penninx BW, Snieder H, Fabiola Del Greco M, Franke A, Nöthlings U, Lieb W, Bakker SJ, Gansevoort RT, van der Harst P, Dehghan A, Franco OH, Hofman A, Rivadeneira F, Sedaghat S, Uitterlinden AG, Coassin S, Haun M, Kollerits B, Kronenberg F, Paulweber B, Aumann N, Endlich K, Pietzner M, Völker U, Rettig R, Chouraki V, Helmer C, Lambert JC, Metzger M, Stengel B, Lehtimäki T, Lyytikäinen LP, Raitakari O, Johnson A, Parsa A, Bochud M, Heid IM, Goessling W, Köttgen A, Kao WH, Fox CS, Böger CA: Genome-wide association study of kidney function decline in individuals of European descent. Kidney Int 87: 1017–1029, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Elmonem MA, Khalil R, Khodaparast L, Khodaparast L, Arcolino FO, Morgan J, Pastore A, Tylzanowski P, Ny A, Lowe M, de Witte PA, Baelde HJ, van den Heuvel LP, Levtchenko E: Cystinosis (ctns) zebrafish mutant shows pronephric glomerular and tubular dysfunction. Sci Rep 7: 42583, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cherqui S, Sevin C, Hamard G, Kalatzis V, Sich M, Pequignot MO, Gogat K, Abitbol M, Broyer M, Gubler MC, Antignac C: Intralysosomal cystine accumulation in mice lacking cystinosin, the protein defective in cystinosis. Mol Cell Biol 22: 7622–7632, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Low SH, Vasanth S, Larson CH, Mukherjee S, Sharma N, Kinter MT, Kane ME, Obara T, Weimbs T: Polycystin-1, STAT6, and P100 function in a pathway that transduces ciliary mechanosensation and is activated in polycystic kidney disease. Dev Cell 10: 57–69, 2006 [DOI] [PubMed] [Google Scholar]
- 133.Piontek K, Menezes LF, Garcia-Gonzalez MA, Huso DL, Germino GG: A critical developmental switch defines the kinetics of kidney cyst formation after loss of Pkd1. Nat Med 13: 1490–1495, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhou W, Dai J, Attanasio M, Hildebrandt F: Nephrocystin-3 is required for ciliary function in zebrafish embryos. Am J Physiol Renal Physiol 299: F55–F62, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Takahashi H, Calvet JP, Dittemore-Hoover D, Yoshida K, Grantham JJ, Gattone VH 2nd: A hereditary model of slowly progressive polycystic kidney disease in the mouse. J Am Soc Nephrol 1: 980–989, 1991 [DOI] [PubMed] [Google Scholar]
- 136.Sayer JA, Otto EA, O’Toole JF, Nurnberg G, Kennedy MA, Becker C, Hennies HC, Helou J, Attanasio M, Fausett BV, Utsch B, Khanna H, Liu Y, Drummond I, Kawakami I, Kusakabe T, Tsuda M, Ma L, Lee H, Larson RG, Allen SJ, Wilkinson CJ, Nigg EA, Shou C, Lillo C, Williams DS, Hoppe B, Kemper MJ, Neuhaus T, Parisi MA, Glass IA, Petry M, Kispert A, Gloy J, Ganner A, Walz G, Zhu X, Goldman D, Nurnberg P, Swaroop A, Leroux MR, Hildebrandt F: The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat Genet 38: 674–681, 2006 [DOI] [PubMed] [Google Scholar]
- 137.Slaats GG, Saldivar JC, Bacal J, Zeman MK, Kile AC, Hynes AM, Srivastava S, Nazmutdinova J, den Ouden K, Zagers MS, Foletto V, Verhaar MC, Miles C, Sayer JA, Cimprich KA, Giles RH: DNA replication stress underlies renal phenotypes in CEP290-associated Joubert syndrome. J Clin Invest 125: 3657–3666, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rachel RA, Yamamoto EA, Dewanjee MK, May-Simera HL, Sergeev YV, Hackett AN, Pohida K, Munasinghe J, Gotoh N, Wickstead B, Fariss RN, Dong L, Li T, Swaroop A: CEP290 alleles in mice disrupt tissue-specific cilia biogenesis and recapitulate features of syndromic ciliopathies. Hum Mol Genet 24: 3775–3791, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Fukui H, Shiba D, Asakawa K, Kawakami K, Yokoyama T: The ciliary protein Nek8/Nphp9 acts downstream of Inv/Nphp2 during pronephros morphogenesis and left-right establishment in zebrafish. FEBS Lett 586: 2273–2279, 2012 [DOI] [PubMed] [Google Scholar]
- 140.Smith LA, Bukanov NO, Husson H, Russo RJ, Barry TC, Taylor AL, Beier DR, Ibraghimov-Beskrovnaya O: Development of polycystic kidney disease in juvenile cystic kidney mice: Insights into pathogenesis, ciliary abnormalities, and common features with human disease. J Am Soc Nephrol 17: 2821–2831, 2006 [DOI] [PubMed] [Google Scholar]
- 141.Bukanov NO, Smith LA, Klinger KW, Ledbetter SR, Ibraghimov-Beskrovnaya O: Long-lasting arrest of murine polycystic kidney disease with CDK inhibitor roscovitine. Nature 444: 949–952, 2006 [DOI] [PubMed] [Google Scholar]
- 142.Husson H, Moreno S, Smith LA, Smith MM, Russo RJ, Pitstick R, Sergeev M, Ledbetter SR, Bukanov NO, Lane M, Zhang K, Billot K, Carlson G, Shah J, Meijer L, Beier DR, Ibraghimov-Beskrovnaya O: Reduction of ciliary length through pharmacologic or genetic inhibition of CDK5 attenuates polycystic kidney disease in a model of nephronophthisis. Hum Mol Genet 25: 2245–2255, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]