Abstract
Psychiatric disorders are highly heritable and associated with a wide variety of social adversity and physical health problems. Using genetic liability (rather than phenotypic measures of disease) as a proxy for psychiatric disease risk can be a useful alternative for research questions that would traditionally require large cohort studies with long-term follow up. Here we conducted a hypothesis-free phenome-wide association study in about 330,000 participants from the UK Biobank to examine associations of polygenic risk scores (PRS) for five psychiatric disorders (major depression (MDD), bipolar disorder (BP), schizophrenia (SCZ), attention-deficit/ hyperactivity disorder (ADHD) and autism spectrum disorder (ASD)) with 23,004 outcomes in UK Biobank, using the open-source PHESANT software package. There was evidence after multiple testing (p<2.55x10-06) for associations of PRSs with 294 outcomes, most of them attributed to associations of PRSMDD (n = 167) and PRSSCZ (n = 157) with mental health factors. Among others, we found strong evidence of association of higher PRSADHD with 1.1 months younger age at first sexual intercourse [95% confidence interval [CI]: -1.25,-0.92] and a history of physical maltreatment; PRSASD with 0.01% lower erythrocyte distribution width [95%CI: -0.013,-0.007]; PRSSCZ with 0.95 lower odds of playing computer games [95%CI:0.95,0.96]; PRSMDD with a 0.12 points higher neuroticism score [95%CI:0.111,0.135] and PRSBP with 1.03 higher odds of having a university degree [95%CI:1.02,1.03]. We were able to show that genetic liabilities for five major psychiatric disorders associate with long-term aspects of adult life, including socio-demographic factors, mental and physical health. This is evident even in individuals from the general population who do not necessarily present with a psychiatric disorder diagnosis.
Author summary
Psychiatric disorders are associated with a wide range of adverse health, social and economic problems. Our study investigated the association of genetic risk for five common psychiatric disorders with socio- demographics, lifestyle and health of about 330,000 participants in the UK Biobank using a systematic, hypothesis-free approach. We found that genetic risk for attention deficit/hyperactivity disorder (ADHD) and bipolar disorder were most strongly associated with lifestyle factors, such as time of first sexual intercourse and educational attainment. Genetic risks for autism spectrum disorder and schizophrenia were associated with altered blood cell counts and decreased risk of playing computer games, respectively. Increased genetic risk for depression was associated with other mental health outcomes such as neuroticism and irritability. In general, our results suggest that genetic risk for psychiatric disorders associates with a range of health and lifestyle traits that were measured in adulthood, in individuals from the general population who do not necessarily present with a psychiatric disorder diagnosis. However, it is important to note that these associations are not necessary causal but can also represent genetic correlation or be influenced by other factors, such as socio-economic factors and selection into the cohort. The findings should inform future research using causally informative designs.
Introduction
Family and twin research as well as large-scale genome-wide association studies (GWAS) have shown that psychiatric disorders are highly heritable [1] and that genetic risks for psychiatric disorders are associated with socio-economic factors, physical health outcomes as well as other psychiatric disorders [2–5]. Using genetic liability (rather than phenotypic measures of disease) as a proxy for psychiatric disease risk can be a useful alternative for research questions that would traditionally require long-term follow up and big datasets due to the low prevalence of some of the psychiatric disorders of interest in the population (e.g. adult-onset health consequences of child neurodevelopmental disorders). In addition, while high genetic risk for a psychiatric disorder is not always indicative of a diagnosis of psychiatric disease, it can index underlying subthreshold symptomatology that can still impact later adversities and quality of life [6]. Furthermore, psychiatric diagnosis may not always be available in any cohort, e.g. there are only very few self-reported diagnosis of Attention deficit/hyperactivity disorder (ADHD), autism spectrum disorder (ASD) or schizophrenia (SCZ) available in the UK Biobank sample, which would make direct comparisons between participants with and without a diagnosis impossible.
So far, studies have used hypothesis-driven approaches to investigate associations of genetic risk for psychiatric disorders with various psychiatric and health outcomes as well as lifestyle factors [7,8]. However, big data resources that are readily available, such as UK Biobank with about 500,000 participants, provide rich phenotypic information that can be used for hypothesis-free studies and offset the multiple testing burden. Phenome-wide association studies (pheWAS) are a type of hypothesis-free analysis where the association of a trait of interest is systematically tested with a potentially large number of phenotypes and can be hypothesis-generating by identifying an association when there is no prior reason to expect that an association may exist. As all available phenotypes are tested and the less ‘significant’ results published alongside those of greater ‘significance’, pheWAS can help to reduce biases associated with hypothesis-driven studies where researchers might only publish the most desirable or expected results.
In a Polygenic Risk Score (PRS) pheWAS (PRS-pheWAS) genetic risk is used as a proxy for lifelong liability for a disorder to explore associations of this genetic liability with a broad range of traits. Understanding these associations will be essential to inform prevention or early intervention strategies. However, conclusions about causality are limited due to the low predictive power and high pleiotropic effects of genetic risk scores for psychiatric conditions [8].
The aim of this study was to investigate the associations between genetic risk for five common psychiatric disorders–attention-deficit/ hyperactivity disorder (ADHD), autism spectrum disorder (ASD), schizophrenia (SCZ), major depression (MDD) and bipolar disorder (BP)—with a wide range of socio-demographic, lifestyle, physical and mental health outcomes in UK Biobank, using the systematic hypothesis-free PRS-pheWAS approach.
Results
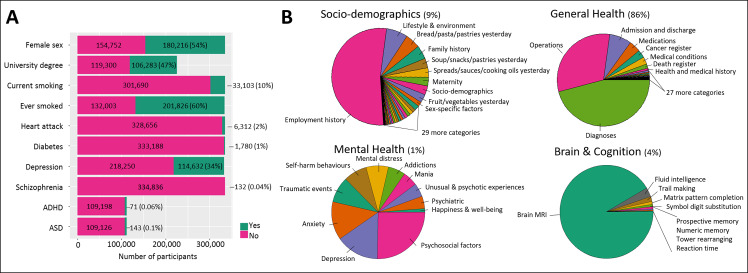
In total 334,976 participants of white British ancestry in UK Biobank were included in this study with an average age of 56 (standard deviation [SD] = 8) years. A descriptive overview of selected UK Biobank study sample characteristics is given in Fig 1A. The UK Biobank participants are known to be more educated and healthier than the average UK population which is reflected in the high percentage of people with a university degree (47%) and low prevalence of current smoking (10%) in the sample, which is comparable to the full UK Biobank release [9]. Furthermore, 34% of participants reported to have seen a general practitioner and 11% a psychiatrist for nerves, anxiety, tension or depression but there are few self-reported cases of schizophrenia (n = 132), ADHD (n = 71), ASD (n = 143) or bipolar disorder (n = 439). An overview of UK Biobank phenotype categories is given in Fig 1B.
Fig 1. Study overview.
(A) Descriptive overview of selected outcomes in UK Biobank. (B) Categories of UK Biobank with the size of pie chart sections indicating the number of included outcomes: socio-demographics (n = 2,057), general health (n = 19,740), mental health (n = 233), brain and cognition (n = 974).
The low number of self-reported ADHD, ASD, schizophrenia and bipolar disorder cases did not allow a direct test of predictive power for the respective PRS and we relied on the predictive accuracy reported in other studies [10–14]. The broad question whether participants have “Seen a psychiatrist for nerves, anxiety, tension or depression” was predicted by the PRSMDD (OR: 1.09 [95% confidence interval [CI]: 1.08,1.10] p = 5x10-52) and PRSSCZ (OR: 1.05 [95% CI:1.04,1.06] p = 2x10-17).
Disorder specific effects
The PRS-pheWAS of each psychiatric disorder tested the association of the respective polygenic risk score, aggregated from independent, genome-wide significant SNPs, with 23,004 outcomes in UK Biobank, adjusted for age, sex and the first 10 genetic principal components. There was strong evidence after multiple testing correction based on the number of independent tests derived from spectral decomposition (p<2.55x10-6) for associations of either the ADHD, ASD, SCZ, MDD or BP PRS with 294 outcomes in 37 UK Biobank categories (Fig 2 and S1 Table) as described below. Of those, 290 outcomes also pass the more stringent Bonferroni threshold (2.17x10-6). Correlations among the PRS can be found in supplementary S2 Table. A detailed list of all PRS-pheWAS results generated by the open-source PHESANT software package can be found in S3 Table. Unless stated as a PHESANT result, estimates for continuous outcomes were generated by follow-up linear regressions to compute estimates on their original scale, as PHESANT automatically applied an inverse normal rank conversion to all continuous outcomes.
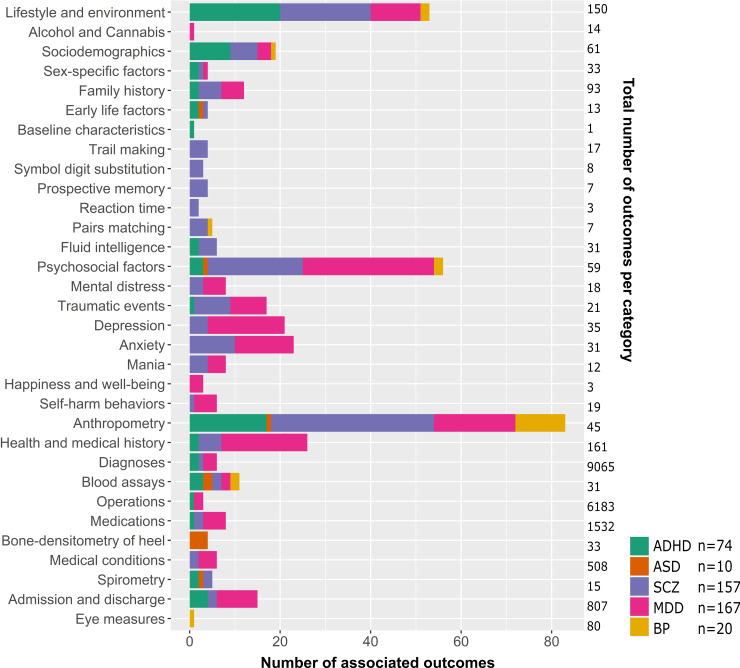
Fig 2. Overview of the distribution of disorder specific polygenic risk score (p<5x10-8) associated outcomes per category of the UK Biobank variables catalogue.
Shown are the number of associations with polygenic risk scores for attention deficit/hyperactivity disorder (ADHD), autism spectrum disorder (ASD), schizophrenia (SCZ), major depression (MDD) and bipolar disorder (BP).
Attention deficit/ hyperactivity disorder
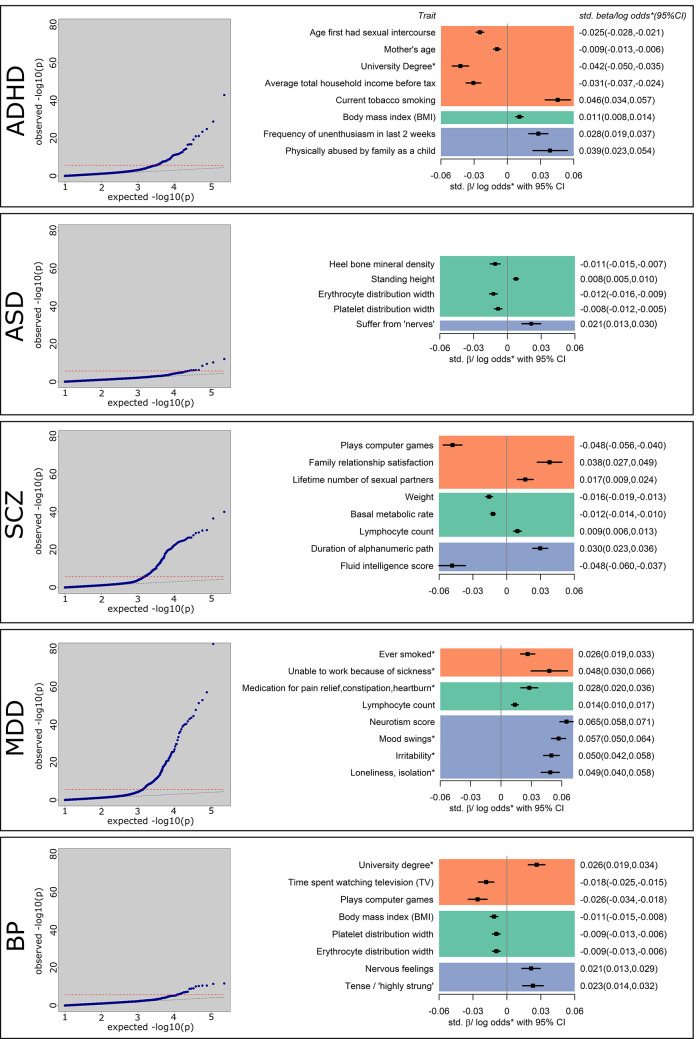
PRSADHD was strongly associated with 74 outcomes (Fig 3) including 36 socio-demographic factors, 32 general health and 6 mental health, brain and cognition outcomes. The strongest evidence of association with PRSADHD was seen for socio-demographic and lifestyle factors. 1 SD higher PRSADHD was associated with a 1.09 month younger age at first sexual intercourse [95% CI: -1.25,-0.92] (p = 2.0x10-16), and 0.96 lower odds of having a university degree [95% CI: 0.95, 0.97] (p = 1.7x10-29). In addition, higher PRSADHD was associated with younger age of their parents (-0.08 years [95%CI: -0.102,-0.050] p = 5.1x10-9; -0.10 years [95% CI: -0.136,-0.069] p = 1.9x10-9, for mother and father respectively), 0.97 lower odds of average household income [95%CI: 0.96,0.98] (p = 5.7x10-20), 1.05 higher odds of current smoking [95%CI: 1.03,1.06] (p = 5.7x10-15) and 1.04 higher odds of experiencing physical abuse as a child [95%CI: 1.02,1.06] (p = 4.4x10-6).
Fig 3. PRS-PheWAS results for attention deficit/hyperactivity disorder (ADHD), autism spectrum disorder (ASD), schizophrenia (SCZ), major depressive disorder (MDD) and bipolar disorder (BP).
Left hand panel: QQ plots of expected versus observed p-values for association of PRS with all outcomes in UK Biobank. Red line indicates the significance threshold derived by spectral decomposition (2.5x10-6). Right hand panel: selected results from different categories with p-values below the significance threshold and estimates generated by PHESANT. Results for continuous outcomes (std. β) are the standard deviation change of inverse-rank normal transformed outcome per 1 SD higher PRS.
Further, 1 SD increase in PRSADHD was associated with 15 physical health outcomes related to obesity, including 0.05 kg/m2 higher BMI [95%CI: 0.036,0.089] (p = 1.7x10-10), leg and arm fat mass, waist circumference and trunk fat mass. Furthermore, there was evidence for an association of PRSADHD with blood measures, such as 0.02 cells/L higher leukocyte count [95%CI: 0.011,0.025] (p = 2.5x10-7).
Associations seen for brain and cognition include 0.04 points lower fluid intelligence score [95%CI: -0.051,-0.026] (p = 1.9x10-9).
Autism spectrum disorder
PRSASD was strongly associated with 10 outcomes (Fig 3), including 1 socio-demographic, 8 general health and 1 mental health outcome.
The strongest association of PRSASD was found for lower erythrocyte distribution width where 1 SD higher PRSASD associated with 0.01% lower erythrocyte distribution width [95% CI: -0.013, -0.007] (p = 6.3x10-10) and 0.98 lower odds of comparative body size at age 10 [95%CI:0.97,0.98] (p = 6.6x10-11). Furthermore, 1 SD higher PRSASD was associated with 0.001 g/cm2 lower heel bone mineral density (BMD) [95%CI:-0.002,-0.001] (p = 4.0x10-5).
The only mental health outcome that was associated with PRSASD was 1.02 higher odds of being a nervous person (“suffer from nerves”) [95%CI:1.01,1.03] (p = 7.9x10-7).
Schizophrenia
There was strong evidence of association for PRSSCZ with 157 outcomes (Fig 3), including 33 socio-demographic, 72 mental health and cognition and 52 general health outcomes.
The strongest evidence of an association with higher PRSSCZ was detected for time of completing an online cognitive function test (pairs matching) (231msec [95%CI: 190,273] p = 2.2x10-16), 1.06 higher odds of experiencing tense feelings[95%CI:1.05,1.07] (p = 3.2x10-37) and 0.95 lower odds of playing computer games [95%CI:0.95,0.96] (p = 7.6x10-30).
In addition, a 1 SD increased PRSSCZ was associated with 1.05 higher odds of consulting a psychiatrist for nerves, anxiety, tension or depression [95%CI:1.04,1.06] (p = 1.6x10-17).
Major depressive disorder
PRSMDD was associated with 167 outcomes (Fig 3), including 21 socio-demographic, 84 mental health and 62 general health outcomes.
Most of the associations (74%) were related to mental health, including an association of higher PRSMDD with higher odds of depression, anxiety, irritability, nervousness and mood swings. Strongest evidence of association with PRSMDD was found for 1.08 higher odds of “seen a doctor for nerves, anxiety, tension or depression” [95%CI:1.08,1.09] (p = 2.7x10-106), 0.12 points higher neuroticism score [95%CI:0.11,0.14] (p = 2.0x10-16) and 1.06 higher odds of having mood swings [95%CI:1.05,1.07] (p = 9.8x10-58).
Furthermore, there was strong evidence of 1 SD higher PRSMDD being associated with socio-demographic and lifestyle traits including 1.03 higher odds of ever smoking [95%CI:1.02,1.03] (p = 1.5x10-13) and 1.05 higher odds of cannabis use [95%CI:1.03,1.06] (p = 4.1x10-10).
Associated physical health measures included 1.03 higher odds of taking medication for pain relief, constipation or heartburn, e.g. paracetamol [95%CI:1.03,1.04] (p = 4.4x10-11) and 1.02 odds of more frequent feelings of pain, e.g. back pain [95%CI:1.02,1.03] (p = 1.1x10-9).
Bipolar disorder
PRSBP was associated with 20 outcomes (Fig 3), including 3 socio-demographic, 14 general health and 3 mental health outcomes.
Socio-demographic and lifestyle factors included associations of higher PRSBP with 1.03 higher odds of having a university degree [95%CI:1.02,1.03] (p = 2.4x10-12), 0.02 hours/day less time spent watching television [95%CI:-0.021,-0.010] (p = 2.8x10-8) and 0.97 lower odds of playing computer games [95%CI:0.97,0.98] (p = 1.1x10-9).
General health traits included 11 traits indicating an association of 1 SD higher PRSBP with 0.06kg/m2 lower BMI [95%CI:-0.07,-0.04] (p = 1.6x10-11) and 2 traits related to blood measures, such as 0.005% decreased platelet distribution width [95%CI:-0.006,-0.003] (p = 3.1x10-7).
Two traits related to mental health were nervous (OR:1.02 [95%CI:1.01,1.03] p = 2.3x10-7) and tense feelings (OR:1.02 [95%CI:1.01,1.03] p = 8.0x10-7).
Cross disorder considerations
The highest overlap of associated outcomes of the univariable PRS-pheWAS scans was seen between schizophrenia and depression (22 general health, 44 mental health and 9 sociodemographic outcomes in common), bipolar disorder (12 general health, 3 mental health and 2 sociodemographic outcomes in common) and ADHD (18 general health, 1 mental health, 2 brain and cognition and 4 sociodemographic outcomes in common). Large overlap was also seen between ADHD and MDD (22 general health, 3 mental health and 4 socio-economic outcomes) and BP (2 socio-economic and lifestyle and 7 general health outcomes) (Fig 4). However, the majority of the associations are directionally opposite for ADHD and BP. For example, higher PRSADHD showed evidence for associations with lower educational attainment and higher BMI, whereas higher PRSBP was associated with higher educational attainment and lower BMI.
Fig 4. Cross-disorder comparison.
Shown are standardized log odds (upper section in each panel) or standardized beta-values (lower section of each panel) of all outcomes associated with polygenic risk scores for either attention deficit/hyperactivity disorder (ADHD), autism spectrum disorder (ASD), schizophrenia (SCZ), major depressive disorder (MDD) or bipolar disorder (BP) at p<2.55x10-6 as indicated by stars (*). For outcomes categorized as multi-category, only one category is displayed. Only associations with anthropometric measures of the right side of the body are shown. Estimates were generated by PHESANT. Results for continuous outcomes (std. beta) are the standard deviation change of inverse-rank normal transformed outcome per 1 SD higher PRS.
Furthermore, all disorder PRSs showed some evidence for association with different blood cell counts, such as a decreased leukocyte count for PRSADHD and PRSMDD, or a decreased eosinophil count for PRSADHD and PRSSCZ.
There was very little overlap of highly associated outcomes between the neurodevelopmental domains (ADHD and ASD).
Sensitivity analysis
We repeated our tests of association for outcomes passing the spectral decomposition threshold, additionally adjusting for potential confounders (assessment centre, genotype chip and the first 40 principal components). These have not been included in the original analysis to reduce the possibility of them introducing collider bias. Estimates were highly consistent with our main results, as shown in S4 and S5 Tables.
Relaxing the p-value threshold for including SNPs in the PRS resulted in some changes in the results (S1 Fig). For ADHD, SCZ, MDD and BP the general trend was an inflation of p-values (S6 Table) and higher effect estimates with smaller confidence intervals. A different pattern was observed for autism spectrum disorder with inconsistent results for some of the outcomes, as described in detail in the supplementary S1 Text. Overall the strength of associations obtained for blood cell count traits across disorders varied between p-value thresholds, with weaker associations found for less stringent p-value thresholds.
When applying the more stringent Bonferroni correction for multiple testing we found that the number of strongly associated outcomes with the PRS reduced slightly to 71 outcomes for PRSADHD, 9 for PRSASD, 155 for PRSSCZ, 166 for PRSMDD and 19 for PRSBP. Outcomes that did not pass Bonferroni but phenoSPD were “Weight”, “Trunk fat mass” and “Forced expiratory volume in 1 second” for ADHD; “Forced vital capacity (FVC)” for ASD; “Word interpolation” and “Valsartan prescription” for SCZ; “Action taken following self-harm” for MDD and “Spherical power (left)” for BP.
In order to reduce potential biases due to sample overlap with the MDD GWAS from Wray et al. [5], we computed a PRS based on the Wray et al. sample excluding UK Biobank and 23andMe participants (PRSMDDnoUKB). The corresponding pheWAS resulted in 51 outcomes passing the multiple testing threshold (S7 Table). Overall, effect estimates were attenuated for many previously identified outcomes and about one third of them did not pass the multiple testing threshold anymore. On the other hand, new associations especially with anthropometric traits could be observed.
Discussion
In this study, we conducted a PRS-pheWAS to examine the relationships between genetic liability for five major psychiatric disorders and 23,004 outcomes in about 330,000 UK Biobank participants.
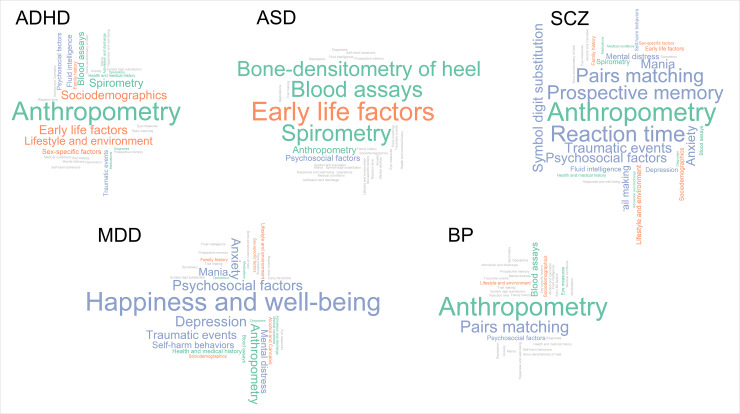
Our results build on a large body of literature supporting links between genetic risk for psychiatric disorders with a wide variety of outcomes including psychological well-being, lifestyle, socio-demographic factors and physical health [2,4,7,15,16]. Our findings also suggest that although psychiatric disorders show strong genetic overlap [7], genetic risk for distinct psychiatric disorders show differential associations with lifestyle, socio-demographic factors and physical health as highlighted in Fig 5. Genetic liability for ADHD and bipolar disorder showed the strongest associations with lifestyle and social environmental factors as well as physical health. On the other hand, genetic liability for major depression and schizophrenia was most strongly associated with psychological health and associations with lifestyle and socio-demographic factors were less robust.
Fig 5. Categories of highly associated outcomes with polygenic risk scores for attention deficit/hyperactivity disorder (ADHD), autism spectrum disorder (ASD), schizophrenia (SCZ), major depressive disorder (MDD) and bipolar disorder (BP).
Size of categories depends on the relative number of associated outcomes to the total number of outcomes within each category. Only categories with more than 1 variable are shown. Lifestyle and socio-demographic factors are shown in orange, physical health measures are shown in green and mental health, brain and cognition traits are shown in violet. Grey categories had zero hits for the corresponding disorder.
We were able to replicate previously reported associations between genetic liability for ADHD and lower educational attainment [17,18], higher prevalence of smoking [19], younger age at delivery [20] and higher body mass index [21]. While the previous findings for smoking and BMI were identified in young adults, our findings using an adult population-based sample with a mean age of 56 years, suggest that associations of childhood psychiatric disorder genetic liabilities with health and social outcomes persist into later adulthood. Associations of genetic liability for ADHD in childhood could represent effects of childhood ADHD or sub-threshold ADHD on long-term social and economic outcomes, or alternatively associations could be due to parental effects or horizontal pleiotropy (the same genetic variants affecting multiple traits). Hence, some of the observed associations in this study might also be more likely to act as a risk factor for the corresponding disorder, rather than being the consequence of it.
Interestingly ADHD genetic liability was also associated with a history of physical maltreatment. This result adds to findings from previous twin and adoption designs that have suggested that ADHD and ADHD genetic liability may have “evocative” effects on parent-child hostility [22–24].
Many of the associations of genetic liability for MDD with increased mood swings, irritability, feelings of loneliness and isolation are clinically known and have previously been reported [5]. Our results are also in line with a recent publication from the Brainstorm consortium investigating genetic correlations among psychiatric disorders with neurological and quantitative traits using LD score regression and GWAS summary statistics, reporting high genetic correlations between most psychiatric disorders and educational attainment and BMI [2,7]. However, we found little evidence for associations of genetic liability for ADHD and ASD with mental health outcomes, such as depressive symptoms, neuroticism or anxiety; and very few associations with cognitive or brain imaging outcomes, which might be because of the UK Biobank being a selected sample with lower rates of psychiatric disorders than the general population as discussed in the limitations section.
In addition to identifying previously reported associations, our PRS-pheWAS also revealed novel associations. We found a strong association of genetic liability for ASD with decreased heel bone mineral density, which furthers previous evidence from observational studies that children and adolescents with ASD have lower bone mineral density [25,26], higher frequency of bone fractures [27] and lower vitamin D levels [28,29], which is essential for bone metabolism. This might suggest that these observed associations may be due to pleiotropic effects of genetic variants associated with bone health.
In line with our results, other previous work in schizophrenia patients and their relatives identified an association between schizophrenia and longer performance duration on the Trail Making Test [30], which requires searching and connecting irregularly arranged targets (digits and letters) in ascending order and is widely used to test for executive function, cognitive ability and processing speed [31–36].
Altered blood cell counts were associated with genetic liability for all disorders. Many psychiatric disorders have been previously associated with allergic or inflammatory states [37–39], such as asthma [40] and atopic diseases [41,42] but it is unclear whether high inflammatory states are on the causal pathway to disorder manifestation or the result of comorbid and confounding behaviours associated with the disease, such as restricted diet, overweight, risky behaviours or medication. Our results support the possibility that altered blood cell counts could be a consequence of the disorder, but we cannot rule out contributions of horizontal pleiotropic effects that weaken or intensify the observed association when adding more SNPs into the PRS. Also, considering the inconsistent findings from the sensitivity analyses for blood count traits, results need further validation and should be treated with caution.
In addition to previous observations between psychiatric disorders and later outcomes that we were able to identify in our pheWAS, there are many reported and established associations that we did not observe in our study. For example, we did not observe an association between genetic risk for psychiatric disorders, apart from a negative association with PRSSCZ, and type 2 diabetes mellitus (T2D). Reasons for this lack of observations may include selection bias as discussed in the next paragraph, confounding of observational studies or complex relationships between disorders. Although there is evidence of genetic overlap between T2D and SCZ [43] the causal pathway is potentially complex and bidirectional, and needs to be investigated using a formal causal framework. However, being able to identify true causal relationships and shared biological pathways between psychiatric disorders and other health or socio-economic outcomes is essential in designing interventions. Hence, we encourage researchers to triangulate our findings using other causally informative studies.
Limitations
Patients with psychiatric disorders or high genetic liability for psychiatric disorders are known to be less likely to participate in studies in the first place and more likely to drop-out during an ongoing study [44]. Selection into a study as well as attrition can induce collider bias [45]. There is consensus that the UK Biobank sample is not representative of the UK population, with participants showing, for example, lower prevalence of current smoking and lower rates of mortality [9]. If both having a psychiatric disorder and a specific outcome (e.g. high socio-economic position) are associated with participation (the collider), this can induce an association between genetic risk for psychiatric disorders and the outcome (S2 Fig). Supplementary S8 Table provides an overview of the expected direction of the collider bias on the effect estimates for a simple model with positive or negative association with participation of the exposure or outcome variable, which can be used as a guide to expected direction of bias under some circumstances.
A direct comparison of PHESANT estimates across the psychiatric disorders cannot be done without taking the differentially powered GWASs and derived PRS into account. This can affect the number and set of outcomes associated with each disorder, which only allows for a relative comparison among the PRS. We also cannot calculate the predictive power of the ADHD, ASD, schizophrenia and bipolar PRS in the UK Biobank sample due to very low number of self-reported diagnoses of these disorders. Further, the MDD GWAS used in the current study to calculate genetic risk scores included thirty thousand participants from UK Biobank (about 10% of the GWAS sample) which might have inflated our results for depression related items but is not expected to introduce bias in any other traits, such as blood counts. However, sensitivity analysis using a GWAS excluding the UK Biobank sample and hence much smaller sample size did confirm many of the found observations.
Although genetic risk scores were derived using variants associated at genome-wide significance level, they can still have horizontal pleiotropic effects on different disorders and traits. Hence, our reported associations cannot on their own inform about causality but should be followed up with other causally informative methods to assess the true direction of the causal effect as well as pleiotropy and heterogeneity between genetic variants. We therefore encourage triangulation of results using other study designs [46,47], such as two-sample MR, negative control or twin studies.
Conclusion
We were able to show that genetic liability for five common psychiatric disorders is associated with distinct domains of adult life, including socio-demographic factors, mental and physical health. This is evident even in individuals from the general population who do not necessarily present with a psychiatric disorder diagnosis, or for individuals who may have been diagnosed as a child but whose symptoms have decreased since. Our research has potential implications for both risk factors and consequences of mental health problems. Our findings indicate potential factors associated with genetic liability for psychiatric disorders, including some which have been identified before (e.g. irritability and depression) and also novel hypotheses (e.g. the association of genetic risk for ASD and reduced bone mineral density) that could be tested using different study designs. Finally the findings also support well-established research into the high long-term economic, societal and individual costs associated with mental health problems [48] and highlight that it is important for mental health scientists and clinicians to consider a broad range of lifestyle, socio-demographic and health risks beyond core diagnoses, in those at elevated familial/genetic risk as well as those with a psychiatric disorder.
Methods
Ethics statement
UK Biobank received ethical approval from the research ethics committee (reference 13/NW/0382). All participants provided informed consent to participate. This work was done under application number 16729 (using genetic data version 3 and phenotype dataset 21753).
Study population
Between 2006–2010 UK Biobank recruited 503,325 men and women in the England, Wales and Scotland at ages 40–69 years. The cohort contains a large dataset including physical measurements, blood/urine/saliva samples, health and lifestyle questionnaires as well as genotype (https://www.ukbiobank.ac.uk/).
For 463,010 participants genotyping was performed using the Affymetrix UK BiLEVE Axiom array or Affymetrix UK Biobank Axiom array. Participants with non-white British ancestry, defined as a self-reported non-white British ancestry in combination with a genetic principal component analysis (PCA) analysis conducted by UK Biobank (n = 54,757 non-white British or non-Caucasian) [49], and participants who had a kinship coefficient denoting a third-degree relatedness (based on Manichaikul et al. [50]; kinship coefficient < 0.0625, n = 73,277) were removed from an already quality checked dataset (excluding participants with withdrawn consent, sex mismatch or sex aneuploidy) [49,50], resulting in a dataset containing 334,976 participants (Fig 6).
Fig 6. Overview of study sample derivation.
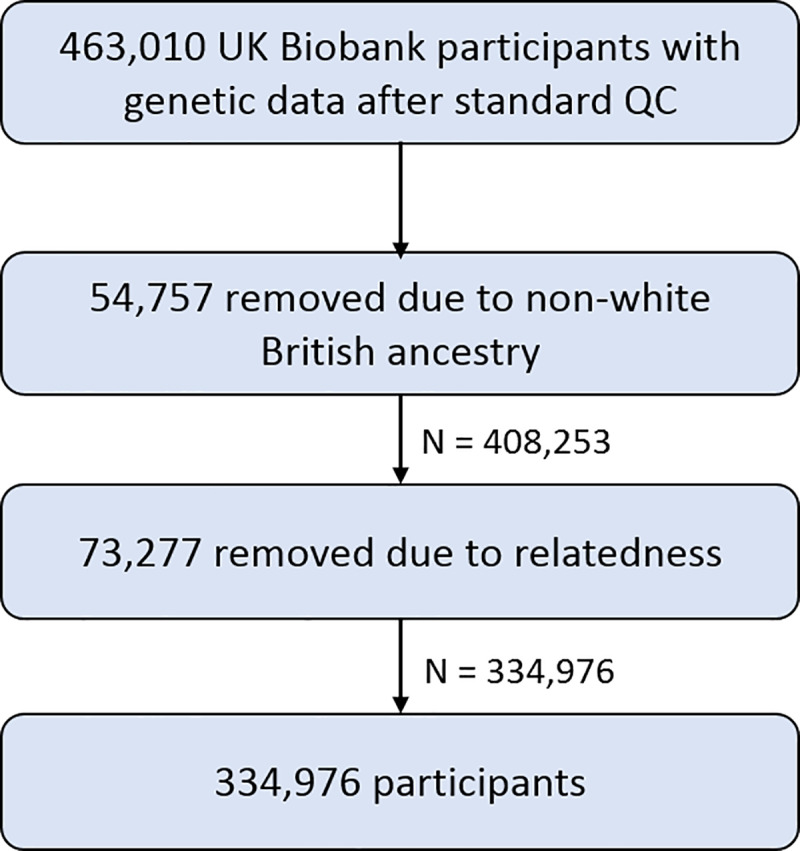
Participants with withdrawn consent, sex mismatch or sex aneuploidy were already removed from the dataset in standard QC steps. [49].
Polygenic risk scores
Genetic variants were identified from the most recent GWAS summary statistics listed in Table 1 with p<5x10-8 for ADHD, ASD, SCZ, MDD and BP. This stringent p-value cut-off was chosen to minimize bias introduced by horizontal pleiotropic effects of genetic variants. All summary statistics were subject to standard quality control including filtering for minor allele frequency (MAF>0.1) and imputation quality (INFO>0.8) and excluding the MHC region on chromosome 6 (26-33Mb) due to its complex linkage disequilibrium structure. Polygenic risk scores (PRS) were derived using independent risk alleles at p<5x10-8 in approximate linkage disequilibrium (R2<0.1 within 500kb distance) and computing a weighted, standardized mean score from these, as has been described previously [51,52].
Table 1. Details of GWAS used for calculating PRS.
| disorder | cases | controls | SNPs in PRS1 | Heritability | Source |
|---|---|---|---|---|---|
| ADHD | 20,183 | 35,191 | 10 | 5.5%2 | Demontis et al. (2019) [2] |
| ASD | 18,381 | 27,969 | 2 | 2.5%2 | Grove et al. (2017) [4] |
| Schizophrenia | 36,989 | 113,075 | 99 | 7%3 | Ripke et al. (2014) [3] |
| MDD | 135,458 | 344,901 | 44 | 1.9%3 | Wray et al. (2018) [5] |
| Bipolar disorder | 20,129 | 21,524 | 8 | 4%3 | Ruderfer et al. (2018) [53] |
1- PRS derived from genome-wide significant hits (p<5x10-8)
2- SNP heritability estimates (Nagelkerke’s R2) reported in the corresponding discovery sample
3-Percent of variance on the liability scale explained by PRS reported in corresponding discovery sample
ADHD–Attention deficit/hyperactivity disorder, MDD–Major depression, ASD–Autism spectrum disorder
Outcomes
UK Biobank provides a fully searchable data showcase (http://biobank.ctsu.ox.ac.uk/crystal/) which at the time of data download (March 2018) included 23,004 outcomes (see supplementary S2 Text), including lifestyle and environment, socio-demographic, early life factors, anthropometry, family history and depression outcomes.
Age, sex and the first 10 principal components derived from the genetic data were included as covariates in all regression models. Age was derived from the participants date of birth and the date of their first assessment centre visit. Sex was self-reported and validated using genetic data.
PHESANT PRS-pheWAS
PHESANT package (version 0.17) was used to test the association of each PRS with each outcome variable in Biobank. A detailed description of PHESANT’s automated rule-based method is given elsewhere [54,55]. In brief, decision rules are based on the variable field type and categorize each variable as one of four data types: continuous, ordered categorical, unordered categorical or binary. PHESANT then estimates the univariate association of the PRS (independent) with each outcome variable (dependent) in a regression model, respectively. Normality of continuous data is ensured by an inverse normal rank transformation prior to testing. To compute meaningful and better interpretable estimates, outcomes passing the multiple testing threshold that qualified as continuous outcomes were followed up on their original scale using a linear regression, excluding outliers and checking for normal distribution of residuals. All estimates correspond to 1 SD change of the PRS.
PHESANT assigns each UK Biobank outcome to one of 91 level 3 categories based on the 235 origin categories of the UK Biobank catalogue (a full list of categories is provided in S1 Table). Furthermore, three authors (BL, EW, ES) grouped these 91 categories into four prespecified higher level categories in order to aid result presentation: socio-demographics and lifestyle, brain and cognition, mental health and general health (Fig 1B).
To account for multiple testing (n = 23,004 tests) we used a previously derived threshold [55,56] based on an estimate of the number of independent phenotypes calculated using spectral decomposition (phenoSPD) (n = 19,645). The multiple testing adjusted significance threshold was p<2.55x10-6 (0.05/19,645). The amount of inflation of observed versus expected p-values is given as the ratio of the median chi-squared statistics for observed to expected median p-values, referred to as Lambda (λ). A conservative Bonferroni correction of multiple testing that assumes uncorrelated traits, would yield a similar p-value threshold of p<2.17x10-6 (0.05/23,004).
PHESANT sensitivity analysis
Analyses were re-run to assess residual confounding of assessment centre and genetic batch, including them as well as all 40 principal components as additional covariates for outcomes identified as strongly associated with either one of the disorders PRS. These covariates were not included in the first model because this could introduce collider bias if, for example, location of assessment centre is affected by both genetic predisposition and outcomes, as discussed in the limitations section.
Furthermore, PRS were derived using various p-value thresholds (p<0.01, p <0.1x10-3, p<1x10-4, p<1x10-5, p<1x10-6) with consequently increasing numbers of SNPs (S9 and S10 Tables) and the five PRS-pheWAS were re-run with the more relaxed PRS to capture a larger amount of explained variation in the disorders by accepting an increase in horizontal pleiotropic effects and adjunct noise. For MDD GWAS results were available for only 10,000 SNPs at these additional thresholds due to availability restrictions.
To assess biases due to a sample overlap between the Wray et al. MDD GWAS and UK Biobank, we computed a PRS based on the Wray et al. sample excluding the UK Biobank and 23andMe samples (43,204 cases and 95,680 controls, nSNPs in PRS = 3) as described in Howards et al. [57] and re-run the pheWAS based on this.
All analyses were performed in R version 3.2.4 ATLAS and R version 3.3.1, and the code is available at [https://github.com/MRCIEU/Psychiatric-disorder-pheWAS-UKBB]. Git tag v0.2 corresponds to the version presented here.
Supporting information
(TIFF)
(TIFF)
(TIFF)
(ADHD- attention defict/ hyperactivity disorder, ASD- autism spectrum disorder, SCZ- schizophrenia, MDD- major depressive disorder, BP- bipolar disorder)
(XLSX)
(ADHD- attention defict/ hyperactivity disorder, ASD- autism spectrum disorder, SCZ- schizophrenia, MDD- major depressive disorder, BP- bipolar disorder)
(XLSX)
Genetic risk scores were calculated as the weighted sum of all genome-wide significant risk alleles for each disorder. Estimates were generated by PHESANT. Results for continuous outcomes are the standard deviation change of inverse-rank normal transformed outcome per 1 SD higher PRS.
(XLSX)
Genetic risk scores were calculated as the weighted sum of all genome-wide significant risk alleles for each disorder. Estimates were generated by linear regression on the original variable scale per 1 SD higher PRS.
(XLSX)
Genetic risk scores were calculated as the weighted sum of all genome-wide significant risk alleles for each disorder.
(XLSX)
(XLSX)
Genetic risk score was calculated as the weighted sum of all genome-wide significant risk alleles for major depression using full GWAS summary statistics as reported in Wray et al. 2019 and GWAS summary statistics excluding the UK Biobank and 23andMe sample from the mentioned study (noUKB). Estimates were generated by PHESANT. Results for continuous outcomes are the standard deviation change of inverse-rank normal transformed outcome per 1SD higher PRS.
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
(DOCX)
Acknowledgments
This research was conducted using the UK Biobank resource under application number 16729. We acknowledge the members of the Psychiatric Genomics Consortium (PGC) and The Lundbeck Foundation Initiative for Integrative Psychiatric Research (iPSYCH) for the publicly available data used as the discovery samples in this article. Further we want to acknowledge Sean Harrison for providing the polygenic risk scores, Richard Anney for providing us the quality controlled GWAS summary statistics and Mick O’Donovan for his advice regarding the use of GWAS summary statistics.
Data Availability
All numeric data underlying the tables and figures is available in the supplementary material. The underlying data cannot be made public due to legal reasons but is available from UK Biobank upon request. Please refer to the guidelines at https://www.ukbiobank.ac.uk/ to apply for access or contact the access team via access@ukbiobank.ac.uk. All scripts to produce the results are available on GitHub (https://github.com/MRCIEU/Psychiatric-disorder-pheWAS-UKBB).
Funding Statement
BL and LR are supported by the Wellcome Trust (grant ref: 204895/Z/16/Z) awarded to AT, GDS, ES and KT (https://wellcome.ac.uk/). BL, LACM, GDS, KT, EW and ES work in a unit that receives funding from the University of Bristol and the UK Medical Research Council (MC_UU_00011/1 and MC_UU_00011/3) (https://mrc.ukri.org/). LACM is funded by a University of Bristol Vice-Chancellor’s Fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Polderman TJC, Benyamin B, de Leeuw CA, Sullivan PF, van Bochoven A, et al. (2015) Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet 47: 702 10.1038/ng.3285 [DOI] [PubMed] [Google Scholar]
- 2.Demontis D, Walters RK, Martin J, Mattheisen M, Als TD, et al. (2019) Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet 51: 63–75. 10.1038/s41588-018-0269-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ripke S, Consortium SWGotPG (2014) Biological insights from 108 schizophrenia-associated genetic loci. Nature 511: 421–427. 10.1038/nature13595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grove J, Ripke S, Als TD, Mattheisen M, Walters RK, et al. (2019) Identification of common genetic risk variants for autism spectrum disorder. Nat Genet 51: 431–444. 10.1038/s41588-019-0344-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, et al. (2018) Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet 50: 668–681. 10.1038/s41588-018-0090-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thapar A, Cooper M, Rutter M (2017) Neurodevelopmental disorders. Lancet Psychiatry 4: 339–346. 10.1016/S2215-0366(16)30376-5 [DOI] [PubMed] [Google Scholar]
- 7.Anttila V, Bulik-Sullivan B, Finucane HK, Walters RK, Bras J, et al. (2018) Analysis of shared heritability in common disorders of the brain. Science 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wray NR, Lee SH, Mehta D, Vinkhuyzen AAE, Dudbridge F, et al. (2014) Research Review: Polygenic methods and their application to psychiatric traits. J Child Psychol Psychiatry 55: 1068–1087. 10.1111/jcpp.12295 [DOI] [PubMed] [Google Scholar]
- 9.Fry A, Littlejohns TJ, Sudlow C, Doherty N, Adamska L, et al. (2017) Comparison of Sociodemographic and Health-Related Characteristics of UK Biobank Participants With Those of the General Population. Am J Epidemiol 186: 1026–1034. 10.1093/aje/kwx246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin AR, Daly MJ, Robinson EB, Hyman SE, Neale BM (2019) Predicting Polygenic Risk of Psychiatric Disorders. Biol Psychiatry 86: 97–109. 10.1016/j.biopsych.2018.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dudbridge F (2013) Power and predictive accuracy of polygenic risk scores. PLoS Genet 9: e1003348 10.1371/journal.pgen.1003348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riglin L, Collishaw S, Thapar AK, Dalsgaard S, Langley K, et al. (2016) Association of Genetic Risk Variants With Attention-Deficit/Hyperactivity Disorder Trajectories in the General Population. JAMA Psychiatry 73: 1285–1292. 10.1001/jamapsychiatry.2016.2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.So H-C, Sham PC (2016) Exploring the predictive power of polygenic scores derived from genome-wide association studies: a study of 10 complex traits. Bioinformatics 33: 886–892. [DOI] [PubMed] [Google Scholar]
- 14.Jansen AG, Dieleman GC, Jansen PR, Verhulst FC, Posthuma D, et al. (2019) Psychiatric Polygenic Risk Scores as Predictor for Attention Deficit/Hyperactivity Disorder and Autism Spectrum Disorder in a Clinical Child and Adolescent Sample. Behav Genet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richardson TG, Harrison S, Hemani G, Davey Smith G (2019) An atlas of polygenic risk score associations to highlight putative causal relationships across the human phenome. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leppert B, Havdahl A, Riglin L, Jones HJ, Zheng J, et al. (2019) Association of Maternal Neurodevelopmental Risk Alleles With Early-Life Exposures. JAMA Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loe IM, Feldman HM (2007) Academic and Educational Outcomes of Children With ADHD. J Pediat Psychol 32: 643–654. [DOI] [PubMed] [Google Scholar]
- 18.Stergiakouli E, Martin J, Hamshere ML, Heron J, St Pourcain B, et al. (2017) Association between polygenic risk scores for attention-deficit hyperactivity disorder and educational and cognitive outcomes in the general population. Int J Epidemiol 46: 421–428. 10.1093/ije/dyw216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rhodes JD, Pelham WE, Gnagy EM, Shiffman S, Derefinko KJ, et al. (2016) Cigarette smoking and ADHD: An examination of prognostically relevant smoking behaviors among adolescents and young adults. Psychol Addict Behav 30: 588–600. 10.1037/adb0000188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flory K, Molina BS, Pelham WE, Jr., Gnagy E, Smith B (2006) Childhood ADHD predicts risky sexual behavior in young adulthood. J Clin Child Adolesc Psychol 35: 571–577. 10.1207/s15374424jccp3504_8 [DOI] [PubMed] [Google Scholar]
- 21.Hanc T, Cortese S (2018) Attention deficit/hyperactivity-disorder and obesity: A review and model of current hypotheses explaining their comorbidity. Neurosci Biobehav Rev 92: 16–28. 10.1016/j.neubiorev.2018.05.017 [DOI] [PubMed] [Google Scholar]
- 22.Schachar R, Taylor E, Wieselberg M, Thorley G, Rutter M (1987) Changes in Family Function and Relationships in Children Who Respond to Methylphenidate. J Am Acad Child Adolesc Psychiatry 26: 728–732. 10.1097/00004583-198709000-00019 [DOI] [PubMed] [Google Scholar]
- 23.Lifford KJ, Harold GT, Thapar A (2009) Parent-child hostility and child ADHD symptoms: a genetically sensitive and longitudinal analysis. J Child Psychol Psychiatry 50: 1468–1476. 10.1111/j.1469-7610.2009.02107.x [DOI] [PubMed] [Google Scholar]
- 24.Harold GT, Leve LD, Barrett D, Elam K, Neiderhiser JM, et al. (2013) Biological and rearing mother influences on child ADHD symptoms: revisiting the developmental interface between nature and nurture. J Child Psychol Psychiatry 54: 1038–1046. 10.1111/jcpp.12100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neumeyer AM, Cano Sokoloff N, McDonnell EI, Macklin EA, McDougle CJ, et al. (2018) Nutrition and Bone Density in Boys with Autism Spectrum Disorder. J Acad Nutr Diet 118: 865–877. 10.1016/j.jand.2017.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ekhlaspour L, Baskaran C, Campoverde KJ, Sokoloff NC, Neumeyer AM, et al. (2016) Bone Density in Adolescents and Young Adults with Autism Spectrum Disorders. J Autism Dev Disord 46: 3387–3391. 10.1007/s10803-016-2871-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neumeyer AM, O'Rourke JA, Massa A, Lee H, Lawson EA, et al. (2015) Brief report: bone fractures in children and adults with autism spectrum disorders. J Autism Dev Disord 45: 881–887. 10.1007/s10803-014-2228-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mostafa GA, Al-Ayadhi LY (2012) Reduced serum concentrations of 25-hydroxy vitamin D in children with autism: relation to autoimmunity. J Neuroinflammation 9: 201 10.1186/1742-2094-9-201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macova L, Bicikova M, Ostatnikova D, Hill M, Starka L (2017) Vitamin D, neurosteroids and autism. Physiol Res 66: S333–s340. 10.33549/physiolres.933721 [DOI] [PubMed] [Google Scholar]
- 30.Reitan RM (1958) Validity of the Trail Making Test as an indicator of organic brain damage. Perceptual and Motor Skills, 8, 271–276. [Google Scholar]
- 31.Aleman A, Hijman R, de Haan EH, Kahn RS (1999) Memory impairment in schizophrenia: a meta-analysis. Am J Psychiatry 156: 1358–1366. 10.1176/ajp.156.9.1358 [DOI] [PubMed] [Google Scholar]
- 32.Wolwer W, Gaebel W (2002) Impaired Trail-Making Test-B performance in patients with acute schizophrenia is related to inefficient sequencing of planning and acting. J Psychiatr Res 36: 407–416. 10.1016/s0022-3956(02)00050-x [DOI] [PubMed] [Google Scholar]
- 33.Heinrichs RW, Zakzanis KK (1998) Neurocognitive deficit in schizophrenia: a quantitative review of the evidence. Neuropsychol 12: 426–445. [DOI] [PubMed] [Google Scholar]
- 34.Zalla T, Joyce C, Szoke A, Schurhoff F, Pillon B, et al. (2004) Executive dysfunctions as potential markers of familial vulnerability to bipolar disorder and schizophrenia. Psychiatry Res 121: 207–217. 10.1016/s0165-1781(03)00252-x [DOI] [PubMed] [Google Scholar]
- 35.Sitskoorn MM, Aleman A, Ebisch SJ, Appels MC, Kahn RS (2004) Cognitive deficits in relatives of patients with schizophrenia: a meta-analysis. Schizophr Res 71: 285–295. 10.1016/j.schres.2004.03.007 [DOI] [PubMed] [Google Scholar]
- 36.Perianez JA, Rios-Lago M, Rodriguez-Sanchez JM, Adrover-Roig D, Sanchez-Cubillo I, et al. (2007) Trail Making Test in traumatic brain injury, schizophrenia, and normal ageing: sample comparisons and normative data. Arch Clin Neuropsychol 22: 433–447. 10.1016/j.acn.2007.01.022 [DOI] [PubMed] [Google Scholar]
- 37.Zhou RY, Wang JJ, Sun JC, You Y, Ying JN, et al. (2017) Attention deficit hyperactivity disorder may be a highly inflammation and immune-associated disease (Review). Mol Med Rep 16: 5071–5077. 10.3892/mmr.2017.7228 [DOI] [PubMed] [Google Scholar]
- 38.Galecki P, Talarowska M (2018) Inflammatory theory of depression. Psychiatr Pol 52: 437–447. 10.12740/PP/76863 [DOI] [PubMed] [Google Scholar]
- 39.Savitz J, Harrison NA (2018) Interoception and Inflammation in Psychiatric Disorders. Biol Psychiatry Cogn Neurosci Neuroimaging 3: 514–524. 10.1016/j.bpsc.2017.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tonacci A, Billeci L, Ruta L, Tartarisco G, Pioggia G, et al. (2017) A systematic review of the association between allergic asthma and autism. Minerva Pediatr 69: 538–550. 10.23736/S0026-4946.16.04623-5 [DOI] [PubMed] [Google Scholar]
- 41.Billeci L, Tonacci A, Tartarisco G, Ruta L, Pioggia G, et al. (2015) Association Between Atopic Dermatitis and Autism Spectrum Disorders: A Systematic Review. Am J Clin Dermatol 16: 371–388. 10.1007/s40257-015-0145-5 [DOI] [PubMed] [Google Scholar]
- 42.Schans JV, Cicek R, de Vries TW, Hak E, Hoekstra PJ (2017) Association of atopic diseases and attention-deficit/hyperactivity disorder: A systematic review and meta-analyses. Neurosci Biobehav Rev 74: 139–148. 10.1016/j.neubiorev.2017.01.011 [DOI] [PubMed] [Google Scholar]
- 43.Hackinger S, Prins B (2018) Evidence for genetic contribution to the increased risk of type 2 diabetes in schizophrenia. 8: 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taylor AE, Jones HJ, Sallis H, Euesden J, Stergiakouli E, et al. (2018) Exploring the association of genetic factors with participation in the Avon Longitudinal Study of Parents and Children. Int J Epidemiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Munafò MR, Tilling K, Taylor AE, Evans DM, Davey Smith G (2018) Collider scope: when selection bias can substantially influence observed associations. Int J Epidemiol 47: 226–235. 10.1093/ije/dyx206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lawlor DA, Tilling K, Davey Smith G (2016) Triangulation in aetiological epidemiology. Int J Epidemiol 45: 1866–1886. 10.1093/ije/dyw314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davey Smith G, Ebrahim S (2001) Epidemiology—is it time to call it a day? Int J Epidemiol 30: 1–11. 10.1093/ije/30.1.1 [DOI] [PubMed] [Google Scholar]
- 48.World Health Organisation WHO (2003) Investing in mental health. Geneva.
- 49.Mitchell R, Elsworth B, Mitchell R, Raistrick C, Paternoster L, et al. (2019) MRC IEU UK Biobank GWAS pipeline version 2. 10.5523/bris.pnoat5528cxo5520u5552p5526ynfaekeigi. [DOI]
- 50.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, et al. (2010) Robust relationship inference in genome-wide association studies. Bioinformatics 26: 2867–2873. 10.1093/bioinformatics/btq559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martin J, Taylor MJ, Rydell M, Riglin L, Eyre O, et al. (2018) Sex-specific manifestation of genetic risk for attention deficit hyperactivity disorder in the general population. J Child Psychol Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Richardson TG, Harrison S, Hemani G, Davey Smith G (2019) An atlas of polygenic risk score associations to highlight putative causal relationships across the human phenome. eLife 8: e43657 10.7554/eLife.43657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ruderfer DM, Ripke S, McQuillin A, Boocock J, Stahl EA, et al. (2018) Genomic Dissection of Bipolar Disorder and Schizophrenia, Including 28 Subphenotypes. Cell 173: 1705–1715.e1716. 10.1016/j.cell.2018.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Millard LAC, Davies NM, Gaunt TR, Davey Smith G, Tilling K (2017) Software Application Profile: PHESANT: a tool for performing automated phenome scans in UK Biobank. Int J Epidemiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Millard LAC, Davies NM, Tilling K, Gaunt TR, Davey Smith G (2019) Searching for the causal effects of body mass index in over 300 000 participants in UK Biobank, using Mendelian randomization. PLoS Genet 15: e1007951 10.1371/journal.pgen.1007951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng J, Richardson TG, Millard LAC, Hemani G, Elsworth BL, et al. (2018) PhenoSpD: an integrated toolkit for phenotypic correlation estimation and multiple testing correction using GWAS summary statistics. Gigascience 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Howard DM, Adams MJ, Clarke T-K, Hafferty JD, Gibson J, et al. (2019) Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci 22: 343–352. 10.1038/s41593-018-0326-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIFF)
(TIFF)
(TIFF)
(ADHD- attention defict/ hyperactivity disorder, ASD- autism spectrum disorder, SCZ- schizophrenia, MDD- major depressive disorder, BP- bipolar disorder)
(XLSX)
(ADHD- attention defict/ hyperactivity disorder, ASD- autism spectrum disorder, SCZ- schizophrenia, MDD- major depressive disorder, BP- bipolar disorder)
(XLSX)
Genetic risk scores were calculated as the weighted sum of all genome-wide significant risk alleles for each disorder. Estimates were generated by PHESANT. Results for continuous outcomes are the standard deviation change of inverse-rank normal transformed outcome per 1 SD higher PRS.
(XLSX)
Genetic risk scores were calculated as the weighted sum of all genome-wide significant risk alleles for each disorder. Estimates were generated by linear regression on the original variable scale per 1 SD higher PRS.
(XLSX)
Genetic risk scores were calculated as the weighted sum of all genome-wide significant risk alleles for each disorder.
(XLSX)
(XLSX)
Genetic risk score was calculated as the weighted sum of all genome-wide significant risk alleles for major depression using full GWAS summary statistics as reported in Wray et al. 2019 and GWAS summary statistics excluding the UK Biobank and 23andMe sample from the mentioned study (noUKB). Estimates were generated by PHESANT. Results for continuous outcomes are the standard deviation change of inverse-rank normal transformed outcome per 1SD higher PRS.
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
(DOCX)
Data Availability Statement
All numeric data underlying the tables and figures is available in the supplementary material. The underlying data cannot be made public due to legal reasons but is available from UK Biobank upon request. Please refer to the guidelines at https://www.ukbiobank.ac.uk/ to apply for access or contact the access team via access@ukbiobank.ac.uk. All scripts to produce the results are available on GitHub (https://github.com/MRCIEU/Psychiatric-disorder-pheWAS-UKBB).